Abstract
Magnetic resonance (MR) imaging can provide a longitudinal view of neurological disease through repeated imaging of patients at successive stages of impairment. Until recently, the difficulty of manual delineation has limited volumetric analyses of MR datasets to a few select regions and a small number of subjects. Increased throughput offered by faster imaging methods, automated segmentation and deformation-based morphometry have recently been applied to overcome this limitation with mouse models of neurological conditions. We use automated analyses to produce an unbiased view of volumetric changes in a transgenic mouse model for Alzheimer’s disease (AD) at two points in the progression of disease: immediately prior to and shortly after the onset of amyloid formation. In addition to the cortex and hippocampus, where atrophy has been well documented in AD patients, we identify volumetric losses in the pons and substantia nigra where neurodegeneration has not been carefully examined. We find that deficits in cortical volume precede amyloid formation in this mouse model, similar to pre-symptomatic atrophy seen in patients with familial AD. Unexpectedly, volumetric losses identified by MR outside of the forebrain predict locations of future amyloid formation, such as the inferior colliculus and spinal nuclei, which develop pathology at very late stages of disease. Our work provides proof-of-principle that MR microscopy can expand our view of AD by offering a complete and unbiased examination of volumetric changes that guide us in revisiting the canonical neuropathology.
Introduction
Alzheimer’s disease (AD) has been intensely studied because of its high prevalence, debilitating progression, and final outcome. Despite dramatic advancements in cognitive assessment, biomarker analysis, and brain imaging, definitive diagnosis of AD is still based on histological confirmation of pathological features, including neurofibrilary tangles, amyloid plaques, and cortical degeneration. Non-invasive tools such based on magnetic resonance imaging (MRI) are currently being developed to gain a longitudinal view of the disease from pre-symptomatic stages to severe degeneration. MRI has been most widely used in AD research to map the onset and advance of neurodegeneration, with the goal of identifying particular brain regions that can predict the likelihood of progressing from mild cognitive impairment (MCI) to dementia (Mosconi et al., 2007), (Busatto et al., 2008), (Thompson et al., 2007), (Ries et al., 2008), (O’Brien et al., 1997), (Petrella et al., 2003), (Leow et al., 2009; Scahill and Fox, 2007). Such morphometric studies have traditionally relied on manual segmentation of pre-selected regions-of-interest (ROI), such as the hippocampus and cortex, which are known to become severely degenerated in AD (Van Broeck et al., 2008). This approach takes advantage of the non-invasive nature of MRI to provide longitudinal information on when degeneration begins and how quickly it progresses. However, it neglects another strength of MRI, which is its ability to provide information on the entire brain from the same data set used to study one ROI. More recent studies have adopted the use of automated tools for structural comparison without a priori focus on particular brain regions (Thompson et al., 2007), (Leow et al., 2009) but large variation in the human brain, in particular the cortical surface, make their use particularly challenging (Busatto et al., 2008).
By comparison, studies done on mice can be thought as more amenable to automated analysis. Their brains are structurally much simpler than humans, and lack the individual variations that make analyses of human MR images considerably more challenging. Moreover, there are a wide variety of transgenic mice available as models of neurodegenerative disease: the Jackson Laboratory now distributes over 90 mouse strains designed to recapitulate various aspects of AD (http://jaxmice.jax.org/list/ra1593.html). In the current study we applied two automated, non-biased methods to screen for volume loss throughout the brain of a newly developed, controllable transgenic model for Alzheimer’s disease. Our studies focused on two stages early in disease, just prior to and just following the onset of amyloid plaque formation, for which we obtained high-resolution T1, T2, and T2* images from active-contrast enhanced fixed tissue. Automated segmentation of the T1 data sets, using a high-resolution mouse brain atlas as a template, produced volume measurements for 33 anatomic structures (Sharief et al., 2008); while local structural differences were assessed by deformation-based morphometry (DBM) based on the vector fields required to warp each brain image to a reference template. While conducting the volumetric studies we discovered that, at least in fixed tissue, high-resolution T2 echoes and T2* GRASS images are capable of detecting very early-stage amyloid deposits without the need for plaque-specific contrast agents. Our results demonstrate the power of unbiased analysis techniques combined with whole-brain high-resolution MRI data to reveal unexpected areas of neurodegeneration and predict unrecognized sites of amyloid deposition in parts of the brain that often go unexplored in Alzheimer’s disease.
Methods
Animals
We studied a mouse model of Alzheimer’s disease in which a double mutant form of the amyloid precursor protein (APP, encoding the Swedish and Indiana familial mutations, APPswe/ind) (Jankowsky et al., 2005) was controlled by the tetracycline-responsive transactivator (TTA, (Gossen and Bujard, 1992)). The system is designed to allow temporal control over when APPswe/ind is expressed: under normal conditions, TTA is active and APPswe/ind is transcribed, however, in the presence of tetracycline or its more stable analog doxycycline, TTA is inactivated and transgenic APP transcription is blocked. We took advantage of this system to generate animals in which the disease was initiated after the brain had fully matured. Animals were reared on doxycycline (F4845, 200 mg/kg doxycycline in Purina 5001 rodent chow, BioServe, Frenchtown, NJ) to suppress APP overexpression for the first 6 weeks of life (P1–P42) then the drug was removed to initiate transgenic APP expression. Mice for this study were generated from intercrossing the TTA (CaMKII-tTA; (Mayford et al., 1996)) and APPswe/ind (tetO-APPswe/ind102) transgenic lines to produce double transgenic APP/TTA and single transgenic TTA offspring. Both double and single transgenic animals expressed the tetracycline-responsive transactivator TTA, but only the double transgenic animals were capable of making APPswe/ind and developing amyloid pathology. Both lines used in this work had been backcrossed to C57BL6/J for more than 10 generations, making all animals for this study congenic on the B6 background. Backcrossed offspring of Line B CaMKIIα-TTA and Line 102 tetO-APPswe/ind have been deposited with Jackson Laboratories for distribution to non-profit institutions under strain numbers 7004 and 7051.
Two time points were chosen for analysis: 7 weeks after APP induction, just prior to the onset of amyloid formation, and 4 months after APP induction, when a moderate amyloid burden had accumulated. Six age-matched APP/TTA and six TTA mice (4 males and 2 females) were imaged at each time point.
Tissue Preparation
All experiments were conducted in accordance with NIH guidelines, using protocols approved by the Duke University Institutional Animal Care and Use Committee. The mice were anesthetized with 100 mg/kg pentobarbital (i.p.) and then transcardially perfusion fixed, first with 0.9% saline plus gadoteridol - ProHance (Bracco Diagnostics, Princeton, NJ) (10:1, v:v), followed by a mixture of 10% formalin and ProHance (10:1, v:v), (Johnson et al., 2002a). The heads were stored overnight in formalin, and then trimmed to remove the lower jaw and muscle. The brains were scanned within the cranial vault to a avoid distortions or damage to the tissue during excision from the skull. Imaging of each specimen was begun 48 hours after fixation to ensure reproducible contrast across samples.
Image Acquisition
Fixed specimens were imaged using a 9.4 T (400 MHz) vertical bore Oxford magnet with a GE EXCITE console (Epic 11.0) and a 14-mm diameter solenoid RF coil. We used a 3D spin warp sequence with the readout gradient applied along the long axis of the brain (anterior-posterior). Three imaging protocols were applied to each sample. A T1 weighted sequence was acquired with the following parameters: echo time (TE) 5.1 ms, repetition time (TR) 50 ms, 62.5 kHz bandwidth, field of view (FOV) of 11×11×22 mm. A T2 weighted sequence was acquired with a Carr Purcell Meiboom Gill sequence using the same FOV and bandwidth, with TR of 400 ms and TE of 7 ms. Post-processing included a Fourier transform along the echo time line to produce data heavily dependent on T2 differences (Sharief and Johnson, 2006). A T2* weighed imaging sequence was acquired with a 3D GRASS sequence, same FOV, bandwidth 31.25 kHz, TR 50 ms, TE 6.5 ms, alpha 60°. Asymmetric sampling of k-space with dynamic adjustment of receiver gain and partial zero filling of k-space were used to achieve an image matrix size of 1024×512×512, resulting in an isotropic 21.5 μm resolution, in 2 hours 7 minutes for the T1, and T2* data set. A matrix of 512×512×256 with isotropic resolution of 43 μm was generated for the T2 weighted data, with total acquisition time of 4 h 20 minutes.
Brain Segmentation and Volumetry
The mouse brains were segmented using a reference brain atlas at 43 μm resolution with 33 labeled regions (Badea et al., 2007). To match the atlas resolution, the T1 MRM datasets were down-sampled to 43 μm. The datasets were skull-stripped applying a sequence of morphological operations in ImageJ (http://rsb.info.nih.gov/ij/). The algorithm (Badea et al., 2007) starts with smoothing, followed by thresholding and erosion with a spherical structuring element. Region growing starts from a seed point located in the center of the volume. Voxels with values between fixed thresholds and connected to the initial region are added to the brain mask. Dilation is then applied for the same number of times (6) as erosion. This process removed most of the skull surrounding the brain; remaining non-brain areas were removed manually. Following skull-stripping, the voxel values were intensity normalized and all brain images were brought into a common space using a rigid transform. The atlas brain was then registered to each test brain using an affine transform, followed by a non-rigid transform. All registration steps are performed using a three step multi-resolution scheme (Rueckert et al., 1999). The transform required to map the atlas onto each brain was then applied to the atlas labels, generating the segmentation of each test brain into 33 structures.
Absolute volumes for individual structures were calculated by multiplying the number of voxels within the structure by the voxel volume. Composite volume structures were calculated as follows: ‘brain’ as the sum of all individual structures; ‘thalamus’ as the sum of ventral thalamic nuclei, laterodorsal thalamic nuclei, geniculate bodies and the remaining unlabeled regions of thalamus; ‘medulla and midbrain’ as the sum of medulla, periaqueductal gray, substantia nigra, interpeduncular nucleus, cochlear nucleus, mesencephalic reticular nucleus (plus red nucleus), anterior pretectal nucleus, and trigeminal tract plus the remaining unlabeled regions of midbrain; ‘brainstem’ as the sum of medulla, midbrain, and pons.
Comparisons between segmented regions were assessed in MATLAB (The Mathworks, Natick, MA) using one-way analysis of variance (ANOVA) with volume as the dependent variable and genotype (or age) as the independent variable.
Mapping local volume changes with DBM
We tested for volumetric changes occurring within anatomical structures using the local deformation fields computed during the registration of individual brains to the C57BL6/J template described above (Chung et al., 2001). The determinants of the local Jacobians (J) characterize the change in volume at each voxel required to register the atlas template to the sample brain. Jacobian determinants smaller than 1 represent local tissue loss, while values greater than 1 represent local tissue growth. Smoothing of the images prior to analysis was due to downsampling required to match the 21.5 μm T1W images to the 43 μm resolution of the atlas. The Jacobian maps were smoothed with a Gaussian kernel (FWHM ~ 182 μm). Statistical analyses on the Jacobian determinants were done using SurfStat (Worsley KJ et al., 2009). We have compared changes in local volumes in DBL transgenic mice versus single transgenic TTA controls, and between two time points (7 weeks and 4 months of APP overexpression), using a linear mixed effect model to account for the fixed effects of age and genotype, and the interaction between age and genotype. The t statistic on the determinants of the local Jacobians was estimated, in the case of equal samples and variances, as where σ12 is the pooled standard deviation: . The results of Student t-tests were considered significant at p < 0.05.
Data are shown without false discovery rate correction.
The volume data are available for public download from the Duke CIVM web site at: http://www.civm.duhs.duke.edu/neuro200901/.
Cortical Layer Thickness
Cortical thickness through the somatosensory cortex was measured manually using the Measure function in ImageJ. All brains were reoriented to symmetrically display the right and left hemispheres and were leveled to a flat ventral surface. Two coronal T2*W slices at the level of the anterior commissure (corresponding to a coronal slice located 0.14 mm from bregma in the (Franklin and Paxinos, 2007) atlas, and approximately 1.72 mm lateral from the middsagittal plane were selected for analysis from each animal. This location corresponds approximately to the border between the hindlimb and forelimb areas of the primary somatosensory cortex. We measured the distance between the corpus callosum and the cortical surface along a line drawn perpendicular to the corpus callosum. The measurement was repeated bilaterally at two sites within the somatosensory cortex of each slice so that the average of 8 measures was used to represent cortical thickness for each animal. All measurements were performed by the same rater (AB), with ± 5% reproducibility in the measured values.
Amyloid Histology
Following MR imaging, fixed brains were cryoprotected in 30% sucrose, frozen on dry ice, and sectioned at 35 μm. Floating sections were stored in 25% glycerol/25% ethylene glycol/0.1M phosphate buffer at −20 ° C until use.
Campbell-Switzer silver stain
A detailed protocol for this stain can be found online at the NeuroScience Associates website http://www.nsalabs.com/documents/publications/campbell-switzer_protocol.htm
Results
Relative Benefits of Different Imaging Strategies
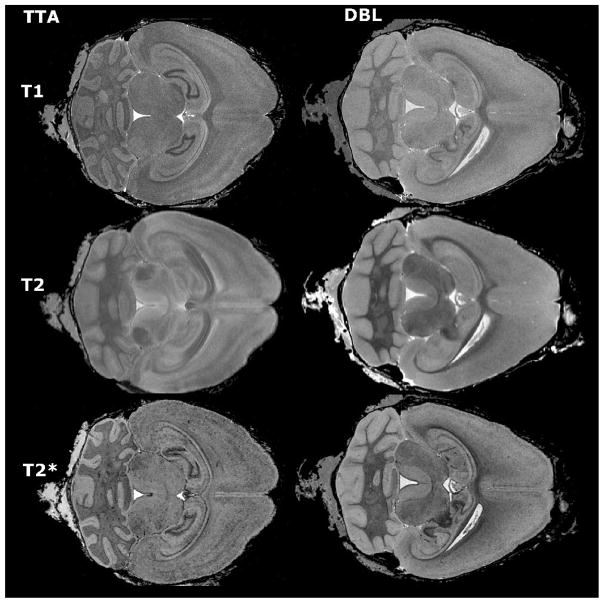
We explored three strategies for imaging amyloid pathology and measuring structural degeneration in a mouse model of Alzheimer’s disease. All three protocols took advantage of active staining during tissue fixation, which allows high resolution imaging with increased contrast and shortened scan times due to the presence of a T1-shortening contrast agent (Johnson et al., 2002b). Of the T1, T2, and T2* data obtained, we found that the T1-weighted spin echo sequence produced the best overall anatomical contrast and was chosen for our registration-based segmentation. Detection of amyloid plaques was most robust in high-resolution T2*-weighted images (21.5 μm GRASS) and in the lower-resolution (43 μm) T2 images (Figure 1) where contrast may be enhanced by iron contained in the amyloid plaques. In addition to the T2 images obtained via MEFIC processing of the multi echo CPMG sequence, individual echoes, in particular the second echo, provided excellent contrast for the identification of amyloid deposits. Larger plaques could be seen in T2 images, while smaller plaques were visible in T2* images. Additional sources of contrast in these GRASS images made it necessary to browse through several contiguous slices to ensure that hypointense spots were not small blood vessels. Thus each imaging sequence provided complementary data: T1 yielded good structural resolution, T2 good amyloid resolution, and T2* good sensitivity for smaller deposits (Figure 2a).
Figure 1.
Different imaging spectrums offer complementary information about brain structure. Multispectral images were acquired for both APP/TTA double transgenic mice (DBL) and TTA single transgenic controls (TTA). T1-weighted images provided the best structural definition and were used for segmentation (top row), T2-weighted images generated the best signal to noise ratio for detecting large amyloid plaques (middle row), while T2* (GRASS) images offered the best resolution for smaller deposits (bottom row).
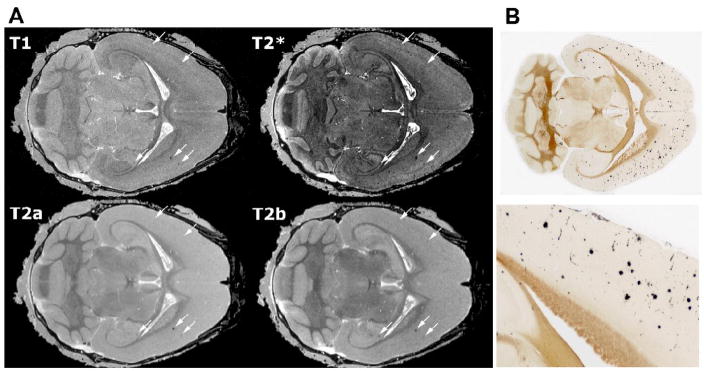
Figure 2.
Amyloid deposits are readily apparent in T2- and T2* weighted (GRASS) images. A. The largest plaques can be seen with all 3 sequences (arrows), although the best sensitivity is provided by T2*-weighted (GRASS) images and the best specificity by the MEFIC-processed T2-weighted images. All images shown here were taken from a DBL transgenic mouse 4 mo after APP induction; the top row displays high-resolution (21.5 μm) T1- and T2*-weighted (GRASS) images and the bottom row shows the first two echoes from a multiecho MEFIC processed T2-weighted image (a: 7 ms TE, b: 14 ms TE). While plaques can be seen in all images, the second echo (T2b) and the T2*-weighted images are better at discriminating smaller plaques. B. H istological staining confirms that DBL transgenic mice have a mild amyloid burden throughout the cortex and hippocampus by 4 mo of age. The lower panel shows the boxed region of the upper section at a higher magnification. Histological sections shown in B and MR slices shown in A are taken from different mice so no alignment of deposits with hypointensities is shown.
Amyloid detected by MRM was confirmed by standard histology in several animals from the older (4 mo) cohort (Figure 2b). As predicted from earlier studies, DBL mice that had overexpressed APP for 4 months carried a mild amyloid burden across the forebrain. In contrast, no amyloid or other histological abnormalities were found in the age-matched single transgenic controls.
Volumetric differences – automated segmentation
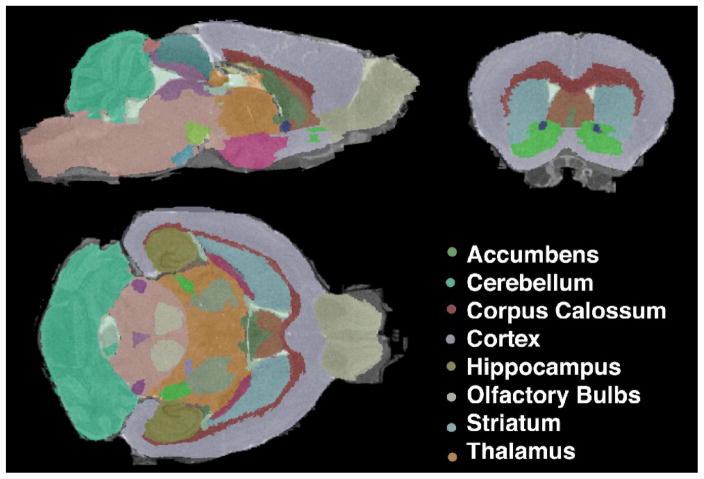
A regional brain template derived from adult C57BL/6J mice was used for segmenting the T1 data sets from DBL and age-matched TTA mice into 33 major structures (Figure 3). The atlas based segmentation relies on automated registration which aligned well strong anatomical features such as the hippocampal layers and the anterior commissure; although the presence of randomly positioned amyloid lesions altered the contrast of images from the older DBL mice, it did not appear to affect the quality of atlas registration.
Figure 3.
A high resolution C57BL/6 mouse brain atlas was used to segment T1-weighted images from DBL and TTA transgenic mice. Illustrated here is the segmentation of a DBL transgenic mouse following 4 months of APP overexpression.
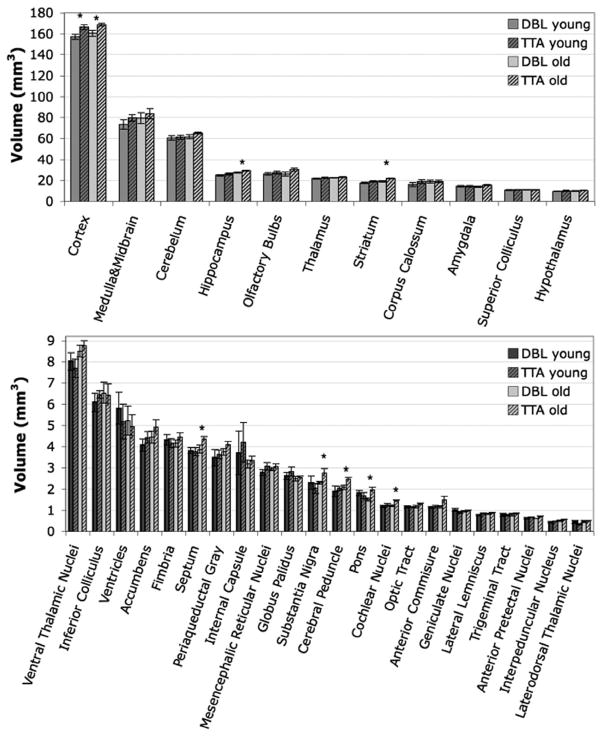
Segmented regions ranged in volume from a minimum of 0.2–0.6 mm 3 for the interpeduncular and laterodorsal thalamic nuclei, to a maximum of ~149–175 mm3 for the cortex (Figure 4).
Figure 4.
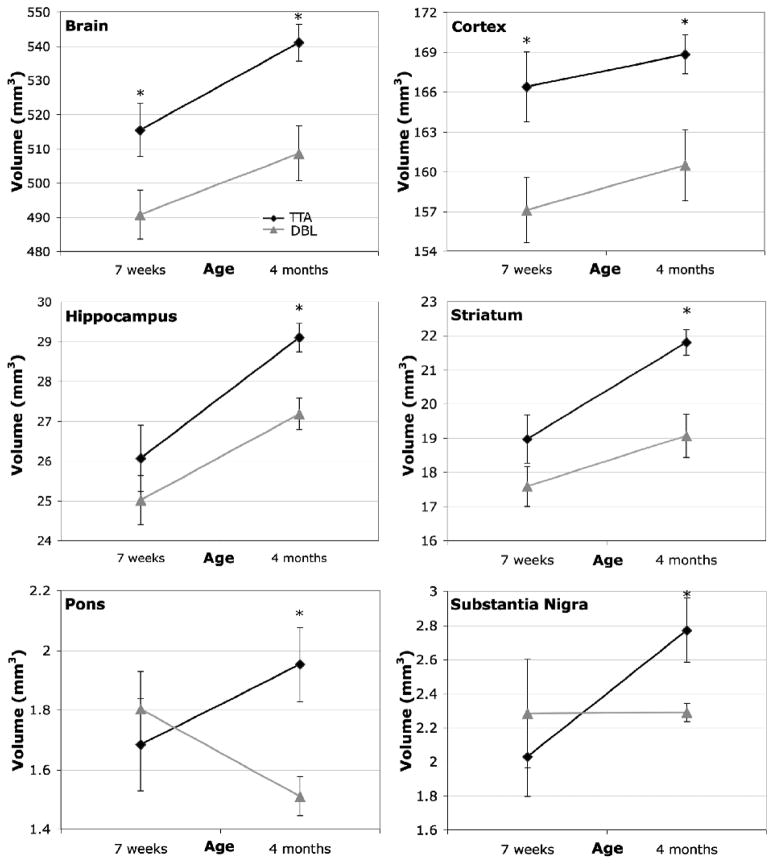
Mean volumes (± SEM) for the 33 analyzed structures analyzed by atlas-based segmentation. Each genotype (DBL and TTA) was assessed at two ages (7 weeks and 4 months after inducing APP overexpression). Structures showing significant volume differences are indicated with asterisks and are expanded in Figure 5.
The most overt change in brain volume between the Alzheimer’s DBL mice and their TTA siblings was a decrease of ~5–6% overall brain volume. Importantly, this reduction was apparent prior to the formation of amyloid, and persisted after its appearance in the older animals (young DBL (490.83 ± 17.49 mm3) vs. young TTA (515.44 ± 19.02 mm3); p<0.04, ci=[−48.11, −1.11]; old DBL (508.62±19.80 mm3) vs. old TTA (541.08±13.19 mm3); p<0.007, ci=[−54.10, −10.83]; Figure 5). The change in overall brain size was accompanied by a 6% reduction in cortical volume (young DBL (157.10 ± 6.99 mm3) vs. young TTA (166.39 ± 6.41 mm3); p<0.03, ci=[−17.28, −1.3]; old DBL (160.48±6.56 mm3) vs. old TTA (168.82± 3.52 mm3); p<0.02, ci=[−15.11, −1.58]).
Figure 5.
Eight of the 33 structures analyzed differed in volume between genotypes at one or both ages examined. Six of the eight are highlighted here. Significant changes were observed in overall brain volume (p<0.04, ci=[−48.11, −1.11]) and cortex (p<0.03, ci=[−17.28, −1.3]) prior to the onset of amyloid in the DBL mice (7 weeks). Volume deficits in these two regions persisted in the older amyloid-positive animals (overall brain (p<0.007, ci=[−54.10, −10.83]) and cortical volume (p<0.02, ci=[−15.11, −1.58])), with additional volume losses developing in the hippocampus (p<0.005, ci=[−3.11, −0.73]), striatum (p<0.004, ci = [−4.38, −1.09]), pons (p< 0.01, ci=[−0.76, −0.13]), and substantia nigra (p<0.03, ci=[−0.91,−0.05]), cochlear nucleus (p< 0.008, ci=[−0.36, −0.07]; not shown) and cerebral peduncle (p<0.03, ci=[−0.67, −0.06]; not shown).
Regional volume changes became more widespread with age. While the overall brain and cortex were smaller in the young DBL mice, several additional structures became significantly diminished once amyloid pathology ensued, including the striatum (−5% in DBL (160.48 ± 6.56 mm3) vs. TTA (168.82 ± 3.52 mm3); p<0.004, ci=[=4.38, −1.09]), hippocampus (−6.6% in DBL (27.18±0.96 mm3) vs. TTA (29.1±0.88 mm3); p<0.005, ci=[−3.11, −0.73]), pons (−22% in DBL (1.51 ± 0.16 mm3) vs. TTA (1.95 ± 0.30 mm3); p< 0.01, ci=[−0.76, −0.13]), substantia nigra (−17% in DBL (2.29±0.03 mm3) vs. TTA (2.77±0.53 mm3); p<0.03, ci=[−0.91, −0.05]), cerebral peduncle (−15% in DBL (2.08±0.25 mm3) vs. TTA (2.44 ± 0.22 mm3); p<0.03, ci=[−0.67, −0.06]), and cochlear nucleus (−15% in DBL (1.22 ± 0.11 mm3) vs. TTA (1.44 ± 0.12 mm3); p< 0.008, ci=[−0.36, −0.07]). The trend noted in most of these structures was a slower rate of growth in the DBL compared to TTA control mice; that is, many of these areas grew in size between the two ages we imaged, but grew less in the DBL than in the TTA animals. Surprisingly, the pons, an area not usually associated with AD, was the only area to show a dramatic loss of volume with age in the DBL transgenic mice. A number of other structures showed reductions that approached significance (p<0.1) in the 4 mo DBL animals compared to the TTA controls, including the amygdala, cerebellum, olfactory bulbs, septum and anterior commissure. These findings indicate that changes in cortical volume precede amyloid formation, but that many additional structures, including regions well beyond the temporal lobe, are diminished once deposits appear.
Deformation Based Morphometry
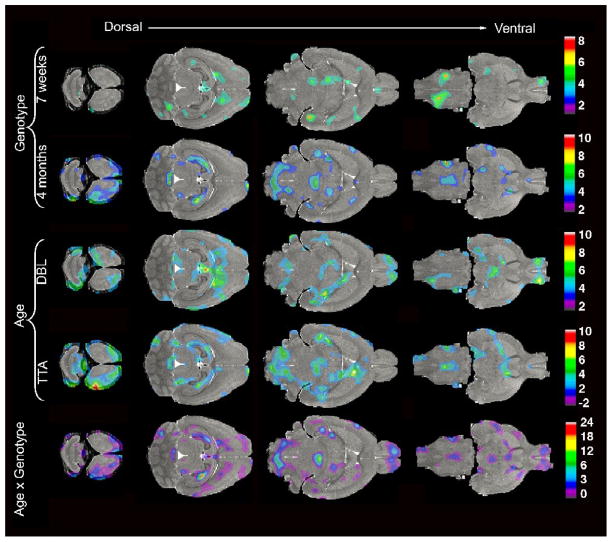
To assess local, rather than regional, changes in the brains of our AD mice we analyzed the deformation fields required to map individual brains into the space of the reference brain (Figure 6). Visual inspection of voxelwise t maps, thresholded at p<0.05, revealed significant differences between genotypes in multiple areas of the cortex, indicative of cortical thinning, in both young and old animals. The hippocampus was also affected in the young DBL mice and became more pronounced with age. The bilateral involvement of the dorsal hippocampus is one of the most dramatic features of the Jacobian analysis in older animals. Patchy areas of significant change in the young DBLs were also noted in the hindbrain reticular nuclei, ventral thalamus, septum, and olfactory bulbs. Several of these areas expanded with age, involving additional thalamic (i.e., red nucleus) and hindbrain nuclei (i.e. spinal nuclei and solitary tract), and structures around the anterior commissure (i.e. nucleus accumbens and septum).
Figure 6.
Statistical analyses of the Jacobian fields (t maps thresholded at levels corresponding to p < 0.05) reveal local structural changes in the DBL transgenic mice. Anatomical differences between genotypes are apparent 7 weeks after APP induction in areas of the cortex, olfactory bulbs and hippocampus (top row). In the older age group (4 months) between-genotype comparisons identify changes in parts of the dorsal hippocampus, anterior-frontal cortex, olfactory bulbs, cerebellum, brainstem, and inferior colliculus (second row). Within-genotype comparisons of the two age groups highlight the normal growth of many regions throughout the brains of TTA control mice (fourth row). Age-related changes in DBL mice are more difficult to discern, likely due to the opposing effects of growth and degeneration (third row). The interaction between age and genotype is significant in the dorsal cortex, hippocampus, inferior colliculus, medial septum, and regions of the brainstem (bottom row).
Perhaps counter-intuitively, within-genotype effects of age were more pronounced in the control animals than in the AD mice, spanning widespread areas of the cortical sheet, hippocampus, cerebellum, hindbrain, and thalamus. Segmental analysis shown in Figures 4 and 5 indicate that all of these areas undergo substantial growth with age in the TTA mice. In contrast, very few areas of significant change were noted across age for the DBL mice, where the interplay between growth and degeneration appears to limit volumetric differences.
Analysis of the interaction between age and genotype allowed us to examine differences in the rate at which local structures expanded (or contracted) in DBL and TTA mice. As expected, many of the same areas that were prominent on the independent genotype and age comparisons persisted as significant in the analysis of their interaction. Again, the dorsal hippocampus displayed the most dramatic changes, both in extent and significance. Other regions familiar from single-component analyses included scattered areas of the cortical sheet (including parts of the frontal, insular, temporal, and entorhinal cortices), nuclei of the hindbrain (spinal and reticular nuclei and solitary tract), multiple areas of the ventral thalamus, and structures along the anterior commissure (nucleus accumbens, caudate putamen, and septal nuclei).
Cortical thinning
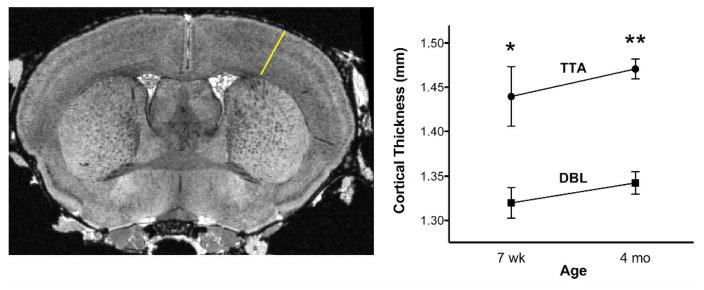
Cortical thinning has been observed in distributed association areas of patients with mild AD (Dickerson and Sperling, 2008) (Dickerson et al., 2009) but it is unclear whether subtle differences in cortical thickness overlap spatially or temporally with the formation of amyloid plaques. We tested our AD mice for this feature of the human disease by manually measuring cortical thickness from the T2*-weighted images. The Jacobian maps indicated strong, bilateral losses in the somatosensory cortex which guided our manual measurements of cortical thickness. We found that the primary somatosensory (SI) cortex was thinner in DBL transgenic mice than in the TTA controls (Figure 7). This difference was significant after Bonferroni (4 groups, 6 independent comparisons) correction (F=12.45, p=7.98*10 −5), and appeared early in the disease prior to the formation of amyloid lesions (7 wk: 1.31 ± 0.04 mm in DBL versus 1.43 ± 0.08 mm in TTA controls); p < 0.01, ci = [−0.21, −0.03], consistent with observations of cortical thinning in pre-symptomatic patients (Dickerson and Sperling, 2008). As in human AD (McEvoy et al., 2009), differences in cortical thickness persist as the disease progresses (4 mo: 1.34 ± 0.03 mm in DBL versus 1.47 ± 0.03 mm in the TTA controls; p < 1.94*10−5, ci=[−0.21,−0.04]).
Figure 7.
Cortical thinning appears prior to amyloid formation. Measurements through the somatosensory cortex were made from the same rostral-caudal plane for each animal (white line in left panel); graph displays mean ± SEM for each age and genotype (7 wk: p < 0.01, ci = [−0.21, −0.03]; 4 mo: p < 1.94*10−5, ci=[−0.21,−0.04]).
Mid- and Hindbrain Volumetric Changes Predict Later Appearance of Amyloid
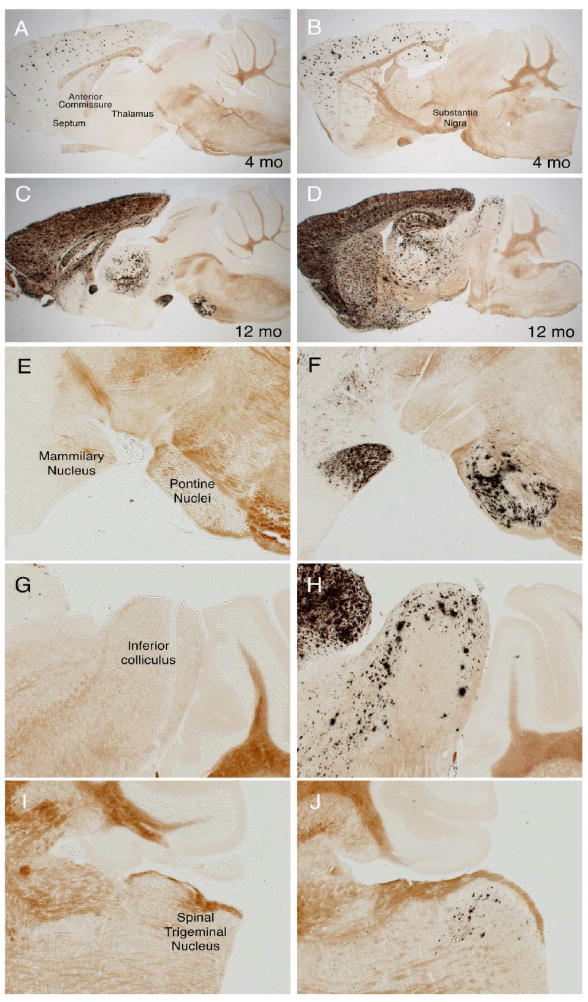
While many sites of volumetric differences identified from the MR analyses were in structures commonly associated with AD such as the cortex, septum, striatum, and hippocampus (Busatto et al., 2008; Scahill and Fox, 2007), (Thompson et al., 2007), (Ries et al., 2008), several other sites we found were located in unexpected regions of the mid- and hindbrain. This led us to re-examine the course of amyloid pathology in our APP/TTA transgenic mice with a renewed focus on the pons, substantia nigra, cochlear nucleus, cerebral peduncle (identified from the segmental analysis), the inferior colliculus, hindbrain (dorsal raphe and red nucleus), and brainstem (solitary tract, spinal nuclei, and pontine reticular nuclei) identified by deformation-based morphometry. At the disease stage used for MR imaging (following 7 weeks or 4 months of transgenic APP overexpression), none of these areas showed any sign of amyloid pathology. But when examined at a considerably later stage of disease, several of these areas had become decorated with amyloid. For this analysis, we used mice taken from the same transgenic line (APP102 × TTA), but which had been harvested (without MR imaging) following up to 12 months of APP overexpression. Amyloid pathology in the forebrain, where the APP transgene is active, becomes severe during this period (Figure 8). Careful examination of the oldest animals, between 9 and 12 months of age, revealed the beginnings of amyloid deposition in many areas of the mid- and hindbrain identified by MR as having volumetric changes 5–8 months earlier. Substantial aggregation was found in the inferior colliculus and pontine nucleus, while a smattering of diffuse amyloid was found in the cerebral peduncle and substantia nigra. Amazingly, aggregates were seen as far down the brainstem as the spinal trigeminal nucleus situated caudal to the cerebellum. None of these areas is thought to express the CaMKIIα promoter used to control APP expression in this model, suggesting that the early volumetric loss as well as the later plaque formation are due to afferent connections from neurons in the forebrain.
Figure 8.
MR analyses predict unexpected sites of future amyloid formation in mid- and hindbrain structures. A, C, E, G and I. Volumetric MR analyses detect significant loss or diminished growth in multiple structures outside of the neocortex following 4 months of APP overexpression, but at this stage of disease amyloid pathology appears only in the forebrain. B. D, F, H, and J. Much later in the disease (after 12 months of APP overexpression) several regions identified by MR ultimately develop amyloid pathology (arrows in B and D), suggesting that structural alterations occur long before amyloid formation. Because overexpression of transgenic APP is limited to the forebrain, damage to these mid- and hindbrain structures must arise through different connections to rostral structures. All panels show double transgenic APP102/TTA sections stained with Campbell-Switzer silver method for amyloid plaques; animals pictured here were not used for MR imaging. A–D show low magnification images from two different sagittal planes, E–F show higher magnification images of miodbrain structures in A and B; G–J show higher magnification images of mid- and hindbrain structures in C and D.
Volume Losses Are Brain-Specific
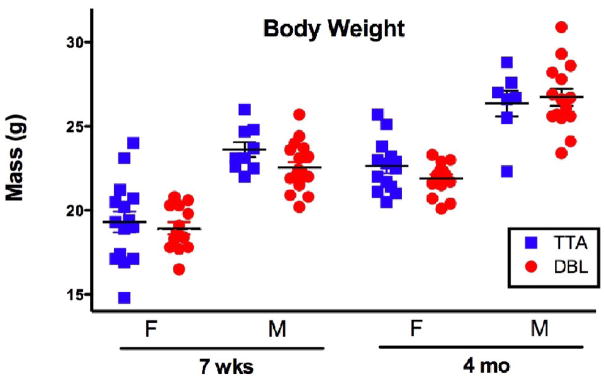
Importantly, the changes we observed in brain volume were not caused by a general decrease in body mass. Often, APP transgenic animals with high levels of Aβ production are visibly smaller than their non-transgenic siblings. This is likely caused by effects of transgene expression during development which are unrelated to the adult-onset disease process. We avoided this potential confound by using the TTA system to withhold transgene expression until the mice reached sexual maturity at 6 weeks of age. Body weights measured in similarly reared congenic mice confirm that the DBL and TTA animals were identical in mass at each of the ages we tested (Figure 9). Weight measurements from the animals used for MR imaging is consistent with these findings. Thus, the changes in regional brain volume we report here were due to specific effects on brain morphology in the adult animal rather than general effects on body mass in the maturing pups.
Figure 9.
Body weight varies with gender and age but not with genotype. Comparison of male (M) and female (F) DBL and TTA transgenic animals at each time point used for imaging indicates no effect of genotype on overall body weight. Each data point represents a single animal; lines show mean ± SEM. Animals used for weight analysis were distinct from those used for imaging but were derived from the same congenic colony and reared under the same postnatal doxycycline regimen. No significant differences in mean weight were found between genotypes matched for gender and age (n=15, 13, 9, 18, 14, 14, 7, 15, per group as shown on graph, Student’s t-test). One comparison (M, 7 wk) approached significance (p=0.0653, ci −2.218 to 0.07365), however, this difference was not apparent 9 weeks later (p=0.6849, ci −1.53 to 2.282).
Discussion
Structural atrophy in the human brain predicts conversion from MCI to AD (for a review see (Ries et al., 2008), (Mosconi et al., 2007)), with atrophy rates accelerating as dementia progresses (Buckner et al., 2005). Until recently our view of structural changes during the progression of AD has been fairly myopic – the majority of published MRI studies having focused on a limited number of structures known to degenerate in the disease, largely because the manual segmentation required to isolate regions for analysis is tedious and time-consuming. However, MRI data sets contain far more information than we currently extract. The development of automated segmentation and deformation-based mophometry methods would allow us to tap the full potential of MRI as a non-invasive, longitudinal imaging tool. Whole-brain automated analyses would provide an unbiased view of neurodegeneration and the possibility of identifying unrecognized temporal and spatial patterns of atrophy. Here we demonstrate the potential of two automated methods for the analysis of high resolution, active staining, contrast-enhanced MRI to identify wide-ranging volumetric changes in a mouse model of AD. We found that both methods detected volume loss prior to plaque formation in regions of the brain commonly measured in MRI studies of AD. More importantly, both methods also detected volume differences in brain structures well outside the areas thought to be affected in AD. In several cases, mid- and hindbrain structures identified by MR were found to develop amyloid pathology much later in the disease – the MR analyses predicted later histological abnormalities that would not have been recognized otherwise. High-resolution MR analysis has thus helped us locate degeneration and pathology in remote, but interconnected, areas through the use of automated, non-biased analyses. Our findings provide a rationale for expanding the use of automated structural analysis in human AD at very early stages, where we might find similarly unsuspected changes that broaden our view of the disease.
Given the interest in advancing the capability of MR for use in longitudinal studies of neurodegenerative disease, other groups have also used MR to view amyloid burden (Lee et al., 2004) (Zhang et al., 2004) (Wengenack et al., 2008) and assess volumetric changes (Lau et al., 2008) in the brains of APP transgenic mice. Several past studies have used manually segmented MRI to identify pre-amyloid decreases in dentate gyrus (Redwine et al., 2003), cerebrum (Van Broeck et al., 2008), and total brain volume (Delatour et al., 2006), (Van Broeck et al., 2008) of young transgenic mice. Because of the labor required to manually segment MR volumes, these studies examined only one or two (or at most five) structures, each of which had been previously implicated in AD. Our work extends this region-of-interest approach to an unbiased look at the entire brain, where we have discovered that cortical volume is significantly diminished early in disease, and with it, confirmed the decrease in total brain volume. We identified significant differences in cortical sheet thickness, cortical volume, and overall brain volume prior to the onset of amyloid plaques, suggesting that the pathogenesis of AD begins before histological lesions are apparent. These structural changes likely have functional consequences, as recent studies have shown that cognitive impairments in several mouse models can begin prior to the appearance of overt pathology (Moechars et al., 1999), (Westerman et al., 2002), (Kelly et al., 2003)
Three recent studies have taken a more expansive look at the whole brain using automated techniques for MR analysis of APP transgenic mice. Similar to the work we present here, (Maheswaran et al., 2009) compare automated atlas-based segmentation to deformation-based morphometry in an APP x PS1 model of AD, but start imaging after the onset of amyloid pathology. Suprisingly, they report that parts of the hippocampus, deep layers of cortex, and thalamus are all bigger, with some structures growing faster, in the double transgenic mice than in their WT controls. Our results are more consistent with those of (Lau et al., 2008) who apply both a deformation-based approach and limited manual segmentation to longitudinal in vivo MR images. Lau et al. study a separate APP × PS1 line, with two time points prior to and two after the onset of amyloid. They examine a wide range of structures throughout the brain, and like us, find that most regions continue to grow in the DBL transgenic mice, albeit at a lower rate than in their WT siblings, with significant differences in the interaction between age and genotype detected for specific areas of the cortex, hippocampus, thalamus, striatum, and septum. In both of these studies, as well as in our own work, there is generally good correlation between the two techniques. The advantage of DBM over atlas-based segmentation is the ability to examine local changes that might be too small to impart a significant effect on the overall structure. In our study, for example, DBM identified significant changes in hippocampal volume of young DBL mice relative to controls that were not yet apparent in the automated segmentation. Similarly, DBM was able to detect changes in particular nuclei of the pons that were indistinguishable in the segmental analysis. We presented the uncorrected statistics, which are easier to interpret. An FDR correction at 0.001 level identified as significant a small number of sparse voxels, some localized in the dorsal hippocampal region, a region well-known to be affected in AD. The FDR correction proved to give an overly conservative view of the data, however we were able to use the uncorrected statistics to discover novel pathology in an independent cohort – pathology that wouldn’t have been discovered had we relied on corrected values. The generality of segmentation analysis is complemented by the local view of DBM, and conversely the complex interpretation of small changes in DBM benefits from the simple outcomes of segmentation.
Consistent with the general agreement between DBM and segmentation, both methods identified the most surprising results to come out of our automated, whole-brain volumetric analyses. While we had expected degeneration in the neocortex based on the spatial overlap with pathology, we had not anticipated significant volume differences in the pons, inferior colliculus, substantia nigra, cerebral peduncle, and spinal nuclei. The presence of atrophy in these structures may be due to the artificial pattern of Aβ production in the brains our mice; however, transgene expression under the CaMKIIα promoter is limited primarily to the forebrain and is absent from the mid- and hindbrain structures we’ve identified (Mayford et al., 1996; Ochiishi et al., 1998) (Kamata et al., 2006). Although the transgene is not thought to be expressed in these regions, each connects to structures where the transgene is active. As a result of the MR findings, we revisited a timed series of amyloid stained-sections from this line to find diffuse amyloid appearing much later in disease in several (although not all) of the structures identified by MR as showing volumetric deficits 5–8 months earlier. This outcome highlights the strength of non-biased, automated MR volumetric analyses for their unique ability to assess wide-ranging changes and make predictions that inform our understanding of disease pathogenesis.
It is worth noting that not all areas of the brain were affected equally in our mice. Even though the transgene is widely expressed throughout the forebrain, the volume losses we detected were fairly restricted. Two structures from our segemental analysis that stand out for their relative preservation are the amygdala and olfactory bulbs. These areas develop amyloid pathology at approximately the same rate as the cortex and hippocampus, yet they show no significant difference in volume at either of the ages we examined. The DBM analysis suggests a somewhat more complex picture, in which local losses occurred but were not extensive enough to affect the size of the structure as a whole. Conversely, our segmental analysis identified volume loss in the cortex substantial enough to involve the entire structure, while morphometry mapped these changes to more limited sub-regions. The somatosensory cortex suffered the brunt of the damage, but other areas such as the motor, cingulate, and visual cortices were conspicuously spared. It is unclear why some regions were susceptible and others relatively resistant, but the specificity revealed through these MR analyses offers a comparison that future studies might exploit to understand how the human disease primarily affects areas involved in learning and memory while leaving other structures untouched.
Although our study has been performed in an artificial mouse model, several of our findings relate surprisingly well to what is known about the course of neurodegeneration in human AD. The losses we noted in cortex, hippocampus, and whole brain volumes are found consistently in both manually segmented and voxel-based morphological studies of human AD (reviewed in (Mosconi et al., 2007), (Busatto et al., 2008), (Thompson et al., 2007), (Ries et al., 2008)). More impressive than the overlap of atrophy in these regions commonly associated with AD is the overlap with regions not often considered in the disease. As in our AD mice, the pons can be dramatically affected in AD patients, where there are several reports of degeneration or cytoskeletal pathology in the locus ceruleus (Haglund et al., 2006), (Strong et al., 1991), (Busch et al., 1997), (Lyness et al., 2003), (Zarow et al., 2003), (German et al., 2005), (O’Neil et al., 2007), pontine parabrachial nuclei, subpeduncular nucleus, tegmentopontine reticular nucleus, reticular formation, and pontine nucleus (Rub et al., 2001b), (Rub et al., 2001a), (Iseki et al., 1989). Also reported, but poorly studied, is amyloid pathology in the inferior and superior colliculi of AD patients (Iseki et al., 1989), (Leuba and Saini, 1995), (Parvizi et al., 2001). In contrast, involvement of the substantia nigra in AD has become so well recognized that its presence is subclassified as dementia with Lewy-bodies or Parkinson’s disease with dementia.
The relationship of pre-amyloid cortical atrophy in our mice to the human disease is somewhat more elusive: by the time cases of sporadic AD present with symptoms, many have already developed amyloid pathology (Kemppainen et al., 2007), (Forsberg et al., 2008). The pre-plaque stage at which our mice were imaged can only be studied in humans with autosomal dominant forms of inherited AD. Familial cases of AD are rare, but a handful of studies have examined these patients for signs of brain atrophy and loss of connectivity prior to the onset of symptoms. Longitudinal assessment of brain volume by MRI revealed significant atrophy in the entorhinal cortex and hippocampus several years before the appearance of memory impairments (Fox et al., 2001; Fox et al., 1996), (Schott et al., 2003). The pre-amyloid cortical atrophy we describe in our transgenic mice is also seen in the rare cases of human AD where it can be examined with certainty of future prognosis. Thus, despite the artificial overexpression of a synthetic APP mutation, our transgenic mice reproduce many features of the human disease and serve as a rational model for examining pre- and post-amyloid volumetric changes.
The automated methods developed for analyzing mouse brain MRI data have allowed us to identify structural changes in parts of the brain we never would have considered a priori given the commonly established wisdom about degeneration in AD. These techniques show the true potential of MRI for structural studies: by extracting information from the entire data set, we’ve generated a temporal and spatial map of where atrophy begins and progresses during these early stages of disease. Future studies can now focus more closely on some of these unexpected sites of degeneration to explore what forebrain connections make them vulnerable, and whether the same regions are at risk in other mouse models of AD. More importantly, we can begin to understand what role these areas play in the progression of symptoms characteristic of the disease. For now, the tools available for automated analyses of longitudinal mouse brain MRI data provide a window into AD that the human disease cannot yet afford. But with major neuroimaging initiatives already underway for AD, we may soon have the opportunity to see the brain as broadly as we have here to confirm in human patients the novel sites of atrophy we’ve identified from the mouse models.
Acknowledgments
We thank Dr. Robert Switzer III of NeuroScience Associates for sharing the Campbell-Switzer silver technique with us and providing stained sections for our late-stage histopathology analysis. We are also grateful to Sidali Benazouz and Beth Olsen for help maintaining the mouse colony. This work was supported by grants NIH/NCRR grants U24 RR021760 (GAJ) and P41-RR005959 (GAJ), NIH/NIA grant K01-AG026144 (JLJ), Alzheimer’s Association grant NIRG 062582 (JLJ), and NIH Director’s New Innovator Award DP2 OD001734 (JLJ).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Badea A, Ali-Sharief AA, Johnson GA. Morphometric analysis of the C57BL/6J mouse brain. Neuroimage. 2007;37:683–693. doi: 10.1016/j.neuroimage.2007.05.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckner RL, Snyder AZ, Shannon BJ, LaRossa G, Sachs R, Fotenos AF, Sheline YI, Klunk WE, Mathis CA, Morris JC, Mintun MA. Molecular, structural, and functional characterization of Alzheimer’s disease: evidence for a relationship between default activity, amyloid, and memory. J Neurosci. 2005;25:7709–7717. doi: 10.1523/JNEUROSCI.2177-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busatto GF, Diniz BS, Zanetti MV. Voxel-based morphometry in Alzheimer’s disease. Expert Rev Neurother. 2008;8:1691–1702. doi: 10.1586/14737175.8.11.1691. [DOI] [PubMed] [Google Scholar]
- Busch C, Bohl J, Ohm TG. Spatial, temporal and numeric analysis of Alzheimer changes in the nucleus coeruleus. Neurobiol Aging. 1997;18:401–406. doi: 10.1016/s0197-4580(97)00035-3. [DOI] [PubMed] [Google Scholar]
- Chung MK, Worsley KJ, Paus T, Cherif C, Collins DL, Giedd JN, Rapoport JL, Evans AC. A unified statistical approach to deformation-based morphometry. Neuroimage. 2001;14:595–606. doi: 10.1006/nimg.2001.0862. [DOI] [PubMed] [Google Scholar]
- Delatour B, Guegan M, Volk A, Dhenain M. In vivo MRI and histological evaluation of brain atrophy in APP/PS1 transgenic mice. Neurobiol Aging. 2006;27:835–847. doi: 10.1016/j.neurobiolaging.2005.04.011. [DOI] [PubMed] [Google Scholar]
- Dickerson BC, Bakkour A, Salat DH, Feczko E, Pacheco J, Greve DN, Grodstein F, Wright CI, Blacker D, Rosas HD, Sperling RA, Atri A, Growdon JH, Hyman BT, Morris JC, Fischl B, Buckner RL. The cortical signature of Alzheimer’s disease: regionally specific cortical thinning relates to symptom severity in very mild to mild AD dementia and is detectable in asymptomatic amyloid-positive individuals. Cereb Cortex. 2009;19:497–510. doi: 10.1093/cercor/bhn113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickerson BC, Sperling RA. Functional abnormalities of the medial temporal lobe memory system in mild cognitive impairment and Alzheimer’s disease: insights from functional MRI studies. Neuropsychologia. 2008;46:1624–1635. doi: 10.1016/j.neuropsychologia.2007.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forsberg A, Engler H, Almkvist O, Blomquist G, Hagman G, Wall A, Ringheim A, Langstrom B, Nordberg A. PET imaging of amyloid deposition in patients with mild cognitive impairment. Neurobiol Aging. 2008;29:1456–1465. doi: 10.1016/j.neurobiolaging.2007.03.029. [DOI] [PubMed] [Google Scholar]
- Fox NC, Crum WR, Scahill RI, Stevens JM, Janssen JC, Rossor MN. Imaging of onset and progression of Alzheimer’s disease with voxel-compression mapping of serial magnetic resonance images. Lancet. 2001;358:201–205. doi: 10.1016/S0140-6736(01)05408-3. [DOI] [PubMed] [Google Scholar]
- Fox NC, Warrington EK, Stevens JM, Rossor MN. Atrophy of the hippocampal formation in early familial Alzheimer’s disease. A longitudinal MRI study of at-risk members of a family with an amyloid precursor protein 717Val-Gly mutation. Ann N Y Acad Sci. 1996;777:226–232. doi: 10.1111/j.1749-6632.1996.tb34423.x. [DOI] [PubMed] [Google Scholar]
- Franklin KB, Paxinos G. The Mouse Brain in Stereotaxic Coordinates. 3. Academic Press; San Diego: 2007. [Google Scholar]
- German DC, Nelson O, Liang F, Liang CL, Games D. The PDAPP mouse model of Alzheimer’s disease: locus coeruleus neuronal shrinkage. J Comp Neurol. 2005;492:469–476. doi: 10.1002/cne.20744. [DOI] [PubMed] [Google Scholar]
- Gossen M, Bujard H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc Natl Acad Sci U S A. 1992;89:5547–5551. doi: 10.1073/pnas.89.12.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haglund M, Sjobeck M, Englund E. Locus ceruleus degeneration is ubiquitous in Alzheimer’s disease: possible implications for diagnosis and treatment. Neuropathology. 2006;26:528–532. doi: 10.1111/j.1440-1789.2006.00725.x. [DOI] [PubMed] [Google Scholar]
- Iseki E, Matsushita M, Kosaka K, Kondo H, Ishii T, Amano N. Distribution and morphology of brain stem plaques in Alzheimer’s disease. Acta Neuropathol. 1989;78:131–136. doi: 10.1007/BF00688200. [DOI] [PubMed] [Google Scholar]
- Jankowsky JL, Slunt HH, Gonzales V, Savonenko AV, Wen JC, Jenkins NA, Copeland NG, Younkin LH, Lester HA, Younkin SG, Borchelt DR. Persistent amyloidosis following suppression of Abeta production in a transgenic model of Alzheimer disease. PLoS Med. 2005;2:e355. doi: 10.1371/journal.pmed.0020355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GA, Cofer GP, Gewalt SL, Hedlund LW. Morphologic phenotyping with magnetic resonance microscopy: the visible mouse. Radiology. 2002a;222:789–793. doi: 10.1148/radiol.2223010531. [DOI] [PubMed] [Google Scholar]
- Johnson GA, Cofer GP, Gewalt SL, Hedlund LW. Morphologic phenotyping with MR microscopy: the visible mouse. Radiology. 2002b;222:789–793. doi: 10.1148/radiol.2223010531. [DOI] [PubMed] [Google Scholar]
- Kamata A, Takeuchi Y, Fukunaga K. Identification of the isoforms of Ca2+/calmodulin-dependent protein kinase II and expression of brain-derived neurotrophic factor mRNAs in the substantia nigra. J Neurochem. 2006;96:195–203. doi: 10.1111/j.1471-4159.2005.03531.x. [DOI] [PubMed] [Google Scholar]
- Kelly PH, Bondolfi L, Hunziker D, Schlecht HP, Carver K, Maguire E, Abramowski D, Wiederhold KH, Sturchler-Pierrat C, Jucker M, Bergmann R, Staufenbiel M, Sommer B. Progressive age-related impairment of cognitive behavior in APP23 transgenic mice. Neurobiol Aging. 2003;24:365–378. doi: 10.1016/s0197-4580(02)00098-2. [DOI] [PubMed] [Google Scholar]
- Kemppainen NM, Aalto S, Wilson IA, Nagren K, Helin S, Bruck A, Oikonen V, Kailajarvi M, Scheinin M, Viitanen M, Parkkola R, Rinne JO. PET amyloid ligand [11C]PIB uptake is increased in mild cognitive impairment. Neurology. 2007;68:1603–1606. doi: 10.1212/01.wnl.0000260969.94695.56. [DOI] [PubMed] [Google Scholar]
- Lau JC, Lerch JP, Sled JG, Henkelman RM, Evans AC, Bedell BJ. Longitudinal neuroanatomical changes determined by deformation-based morphometry in a mouse model of Alzheimer’s disease. Neuroimage. 2008;42:19–27. doi: 10.1016/j.neuroimage.2008.04.252. [DOI] [PubMed] [Google Scholar]
- Lee SP, Falangola MF, Nixon RA, Duff K, Helpern JA. Visualization of beta-amyloid plaques in a transgenic mouse model of Alzheimer’s disease using MR microscopy without contrast reagents. Magn Reson Med. 2004;52:538–544. doi: 10.1002/mrm.20196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow AD, Yanovsky I, Parikshak N, Hua X, Lee S, Toga AW, Jack CR, Jr, Bernstein MA, Britson PJ, Gunter JL, Ward CP, Borowski B, Shaw LM, Trojanowski JQ, Fleisher AS, Harvey D, Kornak J, Schuff N, Alexander GE, Weiner MW, Thompson PM. Alzheimer’s Disease Neuroimaging Initiative: A one-year follow up study using Tensor-Based Morphometry correlating degenerative rates, biomarkers and cognition. Neuroimage. 2009 doi: 10.1016/j.neuroimage.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leuba G, Saini K. Pathology of subcortical visual centres in relation to cortical degeneration in Alzheimer’s disease. Neuropathol Appl Neurobiol. 1995;21:410–422. doi: 10.1111/j.1365-2990.1995.tb01078.x. [DOI] [PubMed] [Google Scholar]
- Lyness SA, Zarow C, Chui HC. Neuron loss in key cholinergic and aminergic nuclei in Alzheimer disease: a meta-analysis. Neurobiol Aging. 2003;24:1–23. doi: 10.1016/s0197-4580(02)00057-x. [DOI] [PubMed] [Google Scholar]
- Maheswaran S, Barjat H, Bate ST, Aljabar P, Hill DL, Tilling L, Upton N, James MF, Hajnal JV, Rueckert D. Analysis of serial magnetic resonance images of mouse brains using image registration. Neuroimage. 2009;44:692–700. doi: 10.1016/j.neuroimage.2008.10.016. [DOI] [PubMed] [Google Scholar]
- Mayford M, Bach ME, Huang YY, Wang L, Hawkins RD, Kandel ER. Control of memory formation through regulated expression of a CaMKII transgene. Science. 1996;274:1678–1683. doi: 10.1126/science.274.5293.1678. [DOI] [PubMed] [Google Scholar]
- McEvoy LK, Fennema-Notestine C, Roddey JC, Hagler DJ, Jr, Holland D, Karow DS, Pung CJ, Brewer JB, Dale AM. Alzheimer disease: quantitative structural neuroimaging for detection and prediction of clinical and structural changes in mild cognitive impairment. Radiology. 2009;251:195–205. doi: 10.1148/radiol.2511080924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moechars D, Dewachter I, Lorent K, Reverse D, Baekelandt V, Naidu A, Tesseur I, Spittaels K, Haute CV, Checler F, Godaux E, Cordell B, Van Leuven F. Early phenotypic changes in transgenic mice that overexpress different mutants of amyloid precursor protein in brain. J Biol Chem. 1999;274:6483–6492. doi: 10.1074/jbc.274.10.6483. [DOI] [PubMed] [Google Scholar]
- Mosconi L, Brys M, Glodzik-Sobanska L, De Santi S, Rusinek H, de Leon MJ. Early detection of Alzheimer’s disease using neuroimaging. Exp Gerontol. 2007;42:129–138. doi: 10.1016/j.exger.2006.05.016. [DOI] [PubMed] [Google Scholar]
- O’Brien JT, Desmond P, Ames D, Schweitzer I, Chiu E, Tress B. Temporal lobe magnetic resonance imaging can differentiate Alzheimer’s disease from normal ageing, depression, vascular dementia and other causes of cognitive impairment. Psychol Med. 1997;27:1267–1275. doi: 10.1017/s0033291797005606. [DOI] [PubMed] [Google Scholar]
- O’Neil JN, Mouton PR, Tizabi Y, Ottinger MA, Lei DL, Ingram DK, Manaye KF. Catecholaminergic neuronal loss in locus coeruleus of aged female dtg APP/PS1 mice. J Chem Neuroanat. 2007;34:102–107. doi: 10.1016/j.jchemneu.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochiishi T, Yamauchi T, Terashima T. Regional differences between the immunohistochemical distribution of Ca2+/calmodulin-dependent protein kinase II alpha and beta isoforms in the brainstem of the rat. Brain Res. 1998;790:129–140. doi: 10.1016/s0006-8993(98)00058-4. [DOI] [PubMed] [Google Scholar]
- Parvizi J, Van Hoesen GW, Damasio A. The selective vulnerability of brainstem nuclei to Alzheimer’s disease. Ann Neurol. 2001;49:53–66. doi: 10.1002/1531-8249(200101)49:1<53::aid-ana30>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- Petrella JR, Coleman RE, Doraiswamy PM. Neuroimaging and early diagnosis of Alzheimer disease: a look to the future. Radiology. 2003;226:315–336. doi: 10.1148/radiol.2262011600. [DOI] [PubMed] [Google Scholar]
- Redwine JM, Kosofsky B, Jacobs RE, Games D, Reilly JF, Morrison JH, Young WG, Bloom FE. Dentate gyrus volume is reduced before onset of plaque formation in PDAPP mice: a magnetic resonance microscopy and stereologic analysis. Proc Natl Acad Sci U S A. 2003;100:1381–1386. doi: 10.1073/pnas.242746599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ries ML, Carlsson CM, Rowley HA, Sager MA, Gleason CE, Asthana S, Johnson SC. Magnetic resonance imaging characterization of brain structure and function in mild cognitive impairment: a review. J Am Geriatr Soc. 2008;56:920–934. doi: 10.1111/j.1532-5415.2008.01684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rub U, Del Tredici K, Schultz C, Thal DR, Braak E, Braak H. The autonomic higher order processing nuclei of the lower brain stem are among the early targets of the Alzheimer’s disease-related cytoskeletal pathology. Acta Neuropathol. 2001a;101:555–564. doi: 10.1007/s004010000320. [DOI] [PubMed] [Google Scholar]
- Rub U, Schultz C, Del Tredici K, Braak H. Early involvement of the tegmentopontine reticular nucleus during the evolution of Alzheimer’s disease-related cytoskeletal pathology. Brain Res. 2001b;908:107–112. doi: 10.1016/s0006-8993(01)02598-7. [DOI] [PubMed] [Google Scholar]
- Rueckert D, Sonoda LI, Hayes C, Hill DL, Leach MO, Hawkes DJ. Nonrigid registration using free-form deformations: application to breast MR images. IEEE Trans Med Imaging. 1999;18:712–721. doi: 10.1109/42.796284. [DOI] [PubMed] [Google Scholar]
- Scahill RI, Fox NC. Longitudinal imaging in dementia. Br J Radiol. 2007;80(Spec No 2):S92–98. doi: 10.1259/bjr/78981552. [DOI] [PubMed] [Google Scholar]
- Schott JM, Fox NC, Frost C, Scahill RI, Janssen JC, Chan D, Jenkins R, Rossor MN. Assessing the onset of structural change in familial Alzheimer’s disease. Ann Neurol. 2003;53:181–188. doi: 10.1002/ana.10424. [DOI] [PubMed] [Google Scholar]
- Sharief AA, Badea A, Dale AM, Johnson GA. Automated segmentation of the actively stained mouse brain using multi-spectral MR microscopy. Neuroimage. 2008;39:136–145. doi: 10.1016/j.neuroimage.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharief AA, Johnson GA. Enhanced T2 contrast for MR histology of the mouse brain. Magn Reson Med. 2006;56:717–725. doi: 10.1002/mrm.21026. [DOI] [PubMed] [Google Scholar]
- Strong R, Huang JS, Huang SS, Chung HD, Hale C, Burke WJ. Degeneration of the cholinergic innervation of the locus ceruleus in Alzheimer’s disease. Brain Res. 1991;542:23–28. doi: 10.1016/0006-8993(91)90992-5. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Hayashi KM, Dutton RA, Chiang MC, Leow AD, Sowell ER, De Zubicaray G, Becker JT, Lopez OL, Aizenstein HJ, Toga AW. Tracking Alzheimer’s disease. Ann N Y Acad Sci. 2007;1097:183–214. doi: 10.1196/annals.1379.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Broeck B, Vanhoutte G, Pirici D, Van Dam D, Wils H, Cuijt I, Vennekens K, Zabielski M, Michalik A, Theuns J, De Deyn PP, Van der Linden A, Van Broeckhoven C, Kumar-Singh S. Intraneuronal amyloid beta and reduced brain volume in a novel APP T714I mouse model for Alzheimer’s disease. Neurobiol Aging. 2008;29:241–252. doi: 10.1016/j.neurobiolaging.2006.10.016. [DOI] [PubMed] [Google Scholar]
- Wengenack TM, Jack CR, Jr, Garwood M, Poduslo JF. MR microimaging of amyloid plaques in Alzheimer’s disease transgenic mice. Eur J Nucl Med Mol Imaging. 2008;35(Suppl 1):S82–88. doi: 10.1007/s00259-007-0706-9. [DOI] [PubMed] [Google Scholar]
- Westerman MA, Cooper-Blacketer D, Mariash A, Kotilinek L, Kawarabayashi T, Younkin LH, Carlson GA, Younkin SG, Ashe KH. The relationship between Abeta and memory in the Tg2576 mouse model of Alzheimer’s disease. J Neurosci. 2002;22:1858–1867. doi: 10.1523/JNEUROSCI.22-05-01858.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worsley KJ, JETCMKCEDBBOLMBACE SurfStat: A Matlab toolbox for the statistical analysis of univariate and multivariate surface and volumetric data using linear mixed effects models and random field theory. 15th Annual Meeting of the Organization for Human Brain Mapping; San Francisco, CA. 2009. [Google Scholar]
- Zarow C, Lyness SA, Mortimer JA, Chui HC. Neuronal loss is greater in the locus coeruleus than nucleus basalis and substantia nigra in Alzheimer and Parkinson diseases. Arch Neurol. 2003;60:337–341. doi: 10.1001/archneur.60.3.337. [DOI] [PubMed] [Google Scholar]
- Zhang J, Yarowsky P, Gordon MN, Di Carlo G, Munireddy S, van Zijl PC, Mori S. Detection of amyloid plaques in mouse models of Alzheimer’s disease by magnetic resonance imaging. Magn Reson Med. 2004;51:452–457. doi: 10.1002/mrm.10730. [DOI] [PubMed] [Google Scholar]