Abstract
During infection, hepatitis C virus (HCV) NS4B protein remodels host membranes to form HCV replication complexes (RC) which appear as foci under fluorescence microscopy (FM). To understand the role of Rab proteins in forming NS4B foci, cells expressing the HCV replicon were examined biochemically and via FM. First, we show that an isolated NS4B-bound subcellular fraction is competent for HCV RNA synthesis. Further, this fraction is differentially enriched in Rab1, 2, 5, 6 & 7. However, when examined via FM, NS4B foci appear to be selectively associated with Rab5 & Rab7 proteins. Additionally, dominant negative (DN) Rab6 expression impairs Rab5 recruitment into NS4B foci. Further, silencing of Rab5 or Rab7 resulted in a significant decrease in HCV genome replication. Finally, expression of DN Rab5 or Rab7 led to a reticular NS4B subcellular distribution, suggesting that endocytic proteins Rab5 and Rab7, but not Rab11, may facilitate NS4B foci formation.
INTRODUCTION
As a member of the Flaviviridae family (Miller and Purcell, 1990; Simmonds et al., 2005) of viruses, hepatitis C virus (HCV) is a positive-strand RNA virus responsible for chronic liver disease and hepatocellular carcinoma. The 9.6 kb virus genome consists of a 5’-untranslated region (5’ UTR), a long open reading frame (ORF) encoding a polyprotein of approximately 3010 amino acids, and a 3’ UTR. Both the 5’ and 3’ UTRs play a role in HCV genome replication (Moradpour, Penin, and Rice, 2007). In addition, the ORF encodes at least three structural proteins (core, E1 and E2), the highly hydrophobic p7 peptide, and six nonstructural (NS) proteins (NS2, N3, NS4A, NS4B, NS5A & NS5B). The NS proteins including NS3, NS4A, NS4B, NS5A and NS5B are known to be sufficient for HCV genome replication in vitro (Blight, Kolykhalov, and Rice, 2000; Lohmann et al., 1999). Specifically, the N terminus of NS3 has serine protease activity responsible for cleavage of the precursor NS proteins NS3-4A, NS4A-B, NS4B-5A and NS5A-B, whereas its C terminal helicase activity is required for unwinding HCV RNA (Kim et al., 1995; Tackett et al., 2001). NS3 activity requires its co-factor, NS4A, which assists NS3 in binding to host membranes (Wolk et al., 2000). NS5A may have multiple functions including inhibition of the interferon response, HCV RNA binding and putative genome encapsidation (Appel et al., 2008; Guo, Bichko, and Seeger, 2001; Huang et al., 2005). NS5B is the HCV RNA-dependent RNA polymerase (RdRp) (Moradpour, Penin, and Rice, 2007) responsible for HCV RNA synthesis. However, efficient HCV genome replication requires the active participation of both viral and host factors (Bartenschlager and Sparacio, 2007; Moradpour, Penin, and Rice, 2007; Stone et al., 2007).
Many positive-strand RNA viruses such as HCV replicate their genome in association with cytosolic membranes. Some of these viruses remodel internal membranes. For HCV, a novel membrane structure, termed the membranous web (Egger et al., 2002), has been observed in cells expressing mature NS4B protein (Egger et al., 2002; Konan et al., 2003), the precursor NS4A-B (Konan et al., 2003), the subgenomic replicon (Gosert et al., 2003), or in HCV-infected cells (Rouille et al., 2006). In fluorescence microscopy (FM), the web appears as foci, and disruption of these foci has been associated with an impairment of HCV genome replication (Aligo et al., 2009; Elazar et al., 2004; Lundin et al., 2003; Stone et al., 2007; Targett-Adams, Boulant, and McLauchlan, 2008). NS4B foci contain the HCV replication complex (RC), which is composed of the replicase proteins (NS3, NS4A, NS4B, NS5A and NS5B), host factors and viral RNA. The determinants of NS4B-induced foci formation are the subject of intense investigation. Our current understanding suggests that the NS4B N-terminal and C-terminal domains play an important role in the formation of these structures (Aligo et al., 2009). However, it is anticipated that several host factors contribute to the formation of NS4B foci. For example, we have reported that the early endosome (EE) protein Rab5 is associated with NS4B foci, binds to NS4B and is required for HCV genome replication, perhaps through the formation of NS4B foci (Stone et al., 2007). Additionally, a recent report indicates that Rab5 is required for cell culture JFH1 genome replication (Berger et al., 2009) further strengthening the idea of a role for Rab5 in HCV replication. Finally, we have shown that the NS4B C-terminal domain might play a role in Rab5 recruitment into NS4B foci (Aligo et al., 2009).
Rab proteins (Rabs) belong to the Ras superfamily of small GTPases, which include more than 50 members. The Rabs are involved in regulating vesicle budding, transport and fusion with target membranes (Cai, Reinisch, and Ferro-Novick, 2007; Grosshans, Ortiz, and Novick, 2006; Markgraf, Peplowska, and Ungermann, 2007) while cycling between active (GTP-bound), and inactive (GDP-bound) forms (Bucci et al., 1995; Hoffenberg et al., 1995). To hydrolyze GTP, Rabs require the activity of their cognate GTPase-activating proteins (GAPs), whereas the guanine nucleotide exchange factors (GEFs) catalyze Rab GTP binding. Thus, constitutively active Rabs are GTPase-defective, whereas dominant negative (DN) Rabs are locked in the GDP conformation and sequester the GEFs needed for Rab activation (Li and Stahl, 1993; Prada-Delgado et al., 2005). Rabs are associated with specific host membranes. For instance, Rab5 is bound to the EE where it regulates endocytosis and EE fusion (Grosshans, Ortiz, and Novick, 2006). Rab7 is found in the late endosome (LE) and facilitates transport from LE to lysosomes (LYS) (Gutierrez et al., 2004; Jager et al., 2004). Rab6 is mostly associated with EE recycling to the Golgi as well as intra-Golgi traffic (Mallard et al., 2002; Martinez et al., 1994). Finally Rab1 and 2 are mostly found in the Golgi and regulate traffic between ER and the Golgi apparatus (Grosshans, Ortiz, and Novick, 2006; Tisdale et al., 1992). DN Rabs have been used as tools to understand the membrane trafficking pathways hijacked by viruses or bacteria to replicate in their respective hosts. Using this approach, Rab5 was found to play a role in HCV and foot-and-mouth disease virus entry into the cell (Johns et al., 2009; Meertens, Bertaux, and Dragic, 2006), whereas Rab1 is involved in Legionella pneumophila replication in vacuoles (Derre and Isberg, 2004).
In this report, we describe two approaches used to better understand the contribution of Rabs in the formation of the active HCV RC. Since NS4B foci are derived from the ER compartment and yet contain Rab5, an EE regulating protein, our hypothesis stipulates that multiple Rabs might be involved in the recruitment of Rab5 into NS4B foci. We found that endocytic Rabs (Rab5 & 7) are strongly associated with HCV NS4B foci, whereas secretory Rabs (Rab1, 2 and 6) are less so. Further, some DN Rabs were found to disrupt NS4B foci and/or its association with Rab5. Finally, silencing of Rab2, 5 and 7 resulted in a significant decrease in HCV RNA synthesis. The significance of these Rabs in HCV replication will be discussed.
RESULTS
The isolated NS4B-bound fraction is competent for HCV RNA-dependent RNA polymerase activity
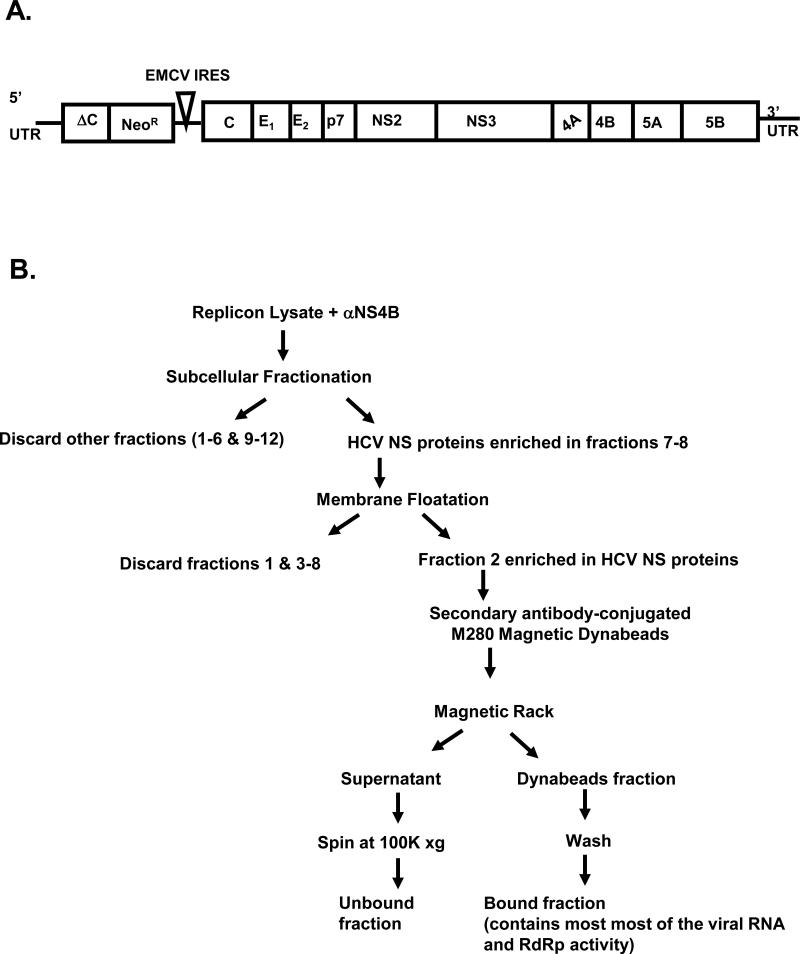
We previously reported the isolation of a HCV RC fraction containing the EE proteins Rab5 and EEA1, and found that Rab5 is required for HCV genome replication (Stone et al., 2007). Further, Berger et al. (Berger et al., 2009) recently confirmed these findings when they showed that Rab5 and EEA1 are needed for Con1 replicon or cell culture JFH1 replication. To determine whether other cellular Rabs play a role in HCV genome replication, we have slightly modified the isolation procedure to include a membrane floatation assay such that membrane-bound NS4B protein is enriched in the final isolation step. Fig. 1B outlines these steps including subcellular fractionation to identify fractions enriched in HCV replicase proteins, membrane floatation (Elazar et al., 2004) to concentrate membrane-bound replicase proteins, and immunoisolation to separate NS4B-associated proteins from others. In our previous report, we used cells expressing Con1 subgenomic replicon, with a C-terminal GFP fusion to NS5A (NS5AGFP replicon) (Moradpour et al., 2004) as a source of the replicase complex (Stone et al., 2007). However, since NS4B expression levels are relatively high in full-length replicon cells (C5B replicon; Fig. 1A) (Ikeda et al., 2002), these cells were used for the current study. Further, the C5B replicon allows for study of the HCV RC in the context of all of the viral proteins.
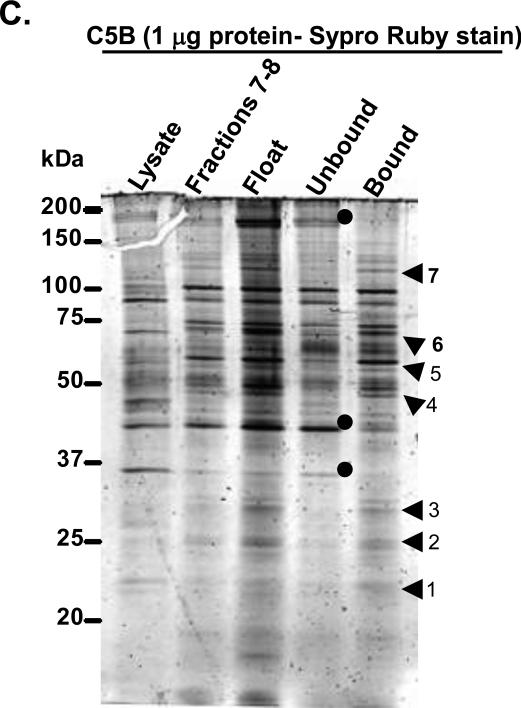
FIG. 1.
A. Schematic representation of the full-length replicon construct (Ikeda et al., 2002) used in this study. B. Outline of the three-step procedure used for isolation of a membrane fraction enriched in HCV replicase proteins. The three steps include, i) subcellular fractionation of cell lysate containing NS4B antibody, ii) membrane flotation of two fractions enriched in replicase proteins, and iii) use of secondary antibody-coated magnetic Dynabeads to isolate a fraction containing HCV proteins. C. One microgram of each extract was separated on 10% SDS-PAGE, followed by Sypro Ruby staining. The protein bands preferentially enriched in the bound fraction are indicated by arrowheads, whereas those in the unbound fraction are denoted by circles. Numbers 1-7 refer to the bands in Table 1, which were examined by mass spectrometry analysis (supplemental material).
Following the outline in Fig. 1B, we used rabbit polyclonal NS4B antibody and magnetic beads conjugated to secondary anti rabbit antibody to obtain two subcellular fractions including the supernatant (Unbound) and the Dynabeads-associated proteins (Bound). To examine the protein profiles in the isolated fractions, one microgram of protein from unbound or bound fraction was separated by 10% SDS-PAGE and stained with Sypro Ruby. As shown in Fig. 1C, stronger bands appeared in the floatation lane (Float) in comparison to Lysate lane, suggesting protein enrichment in the Float lane. Further, a few protein bands (arrowheads) were exclusively enriched in the bound fraction, whereas others (dots) could only be observed in the unbound fraction. The bands from both fractions could be found in the float lane, the source of the immunoisolated proteins. These data indicate the presence of both a shared and distinct population of proteins in the bound and unbound fractions. Similar to our previous report (Stone et al., 2007), we detected HCV NS3, 4B, 5A and 5B proteins in both the bound and unbound fractions by immunoblotting (data not shown), indicating that an antibody specific to NS4B protein can be used successfully to immunoisolate HCV replicase proteins.
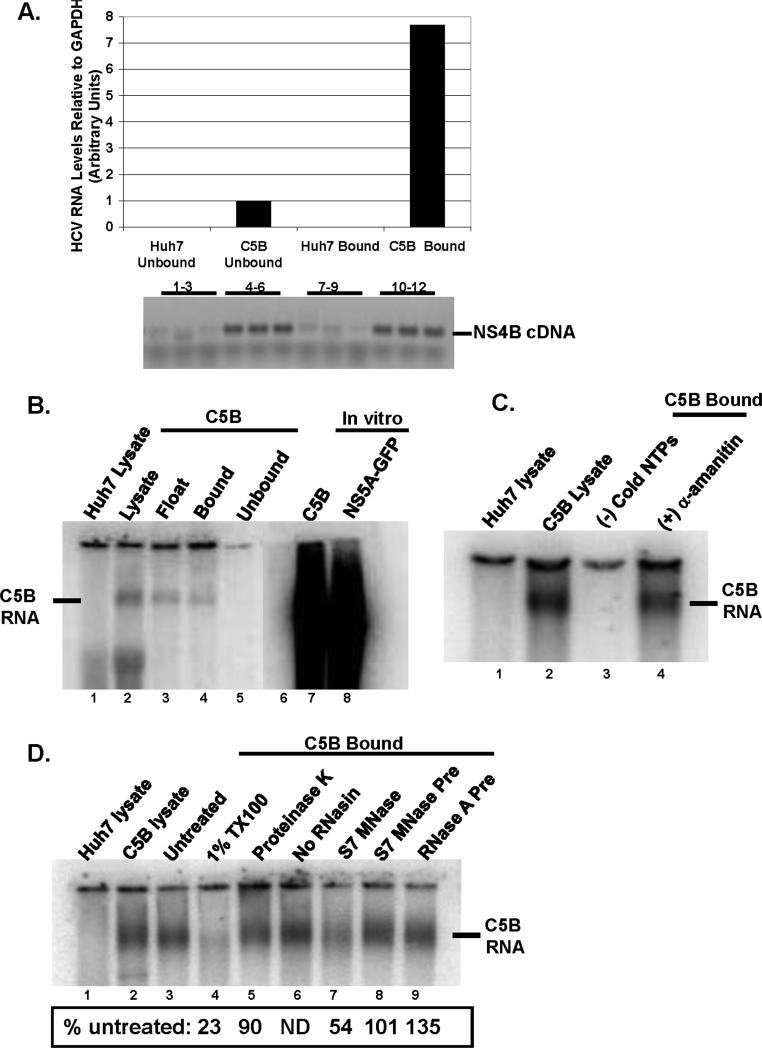
Crude membrane fractions showing HCV RNA-dependent RNA polymerase (RdRp) activity have been reported previously (Aizaki et al., 2004; El-Hage and Luo, 2003; Hardy et al., 2003; Lai et al., 2003; Quinkert, Bartenschlager, and Lohmann, 2005). However, it was not clear whether the isolated fractions were competent for in vitro HCV RNA synthesis. To test for such activity, we first examined the relative amount of HCV RNA present in both bound and unbound fractions by quantitative real time PCR analysis. The relative amount of HCV RNA was approximately seven times higher in the bound fraction than in the unbound (Fig. 2A) indicating that HCV RNA is mostly associated with the bound fraction. To determine whether the bound fraction was competent for HCV RNA synthesis, it was added to a reaction mixture containing actinomycin D, [α32P] CTP and cold NTPs, but no exogenous viral RNA template. The reaction was incubated at 37°C for 1h after which the RNA was isolated, separated on a formaldehyde-denaturing gel, dried and visualized with a phosphorImager. As seen in Fig. 2B, an RNA band similar in size to the in vitro transcribed full-length replicon RNA was synthesized in the lysate sample, the float fraction and the bound fraction; no such band was found with parental Huh7 lysate. To ensure that the RNA product in Fig. 2B was synthesized by the HCV RdRp and not host DNA-dependent RNA polymerases, the in vitro RdRp assay was performed in the presence of both actinomycin D and α-amanitin (an inhibitor of eukaryotic RNA polymerase II). As shown in Fig. 2C, the RdRp is resistant to both actinomycin D and α-amanitin. Further, RNA synthesis was not observed in the absence of exogenous NTPs. These findings suggest that a terminal transferase (Hardy et al., 2003; Ranjith-Kumar et al., 2001) was not adding [α32P] CTP at the end of endogenous HCV genome.
FIG.2.
A. RNA was extracted from bound and unbound fractions from Huh7 and C5B cells. The relative amount of RNA was determined using real-time PCR and was normalized to GAPDH levels. cDNA products were also run on a 1% agarose gel: lanes 1-3 (Huh7 unbound) and 7-9 (Huh7 bound) show no product whereas lanes 4-6 (C5B unbound) and 10-12 (C5B bound) display HCV-specific cDNA. B. The samples from different steps in the isolation procedure were tested for in-vitro RdRp activity in the absence of exogenous HCV RNA template. In-vitro transcribed and radiolabeled C5B (lane 7) and NS5AGFP (lane 8) replicon RNAs were included as markers. Notice the presence of a product in C5B lysate (lane 2) and C5B bound (lane 4). No such product is present in Huh7 lysate (lane 1). C. The bound fractions were tested for RdRp activity in the absence of cold NTPs (lane 3) or in the presence of an inhibitor of host RNA polymerase II, α-amanitin (lane 4). Notice that no RNA was detectable when RdRp assay was performed in the absence of cold NTPs (lane 3), but α-amanitin coupled with actinomycin D had little effect on RdRp activity (compare lane 4 to lane 1). D. The samples were tested for in-vitro RdRp activity in the presence of 1% TX100 (lane 4), 0.5 U/ml S7 MNase (lane 7), 1mg/ml proteinase K (lane 5) or preincubated with 0.5 U/ml RNases (lanes 8 and 9). In lane 6, RdRp activity was tested in the absence of RNase inhibitor, RNasin. The reaction mixtures were processed as in FIG. 2 B & C. Panels A through D are representative results from at least two independent experiments. No RNAsin (Fig. 4D, lane 6) was done only once.
Reported features of the HCV replicase complex in crude membrane fractions include its sensitivity to detergent treatment, whereas RNases and proteases have no effect on its activity (Ali, Tardif, and Siddiqui, 2002; Miyanari et al., 2003; Quinkert, Bartenschlager, and Lohmann, 2005). To determine whether the bound fraction has similar characteristics, RdRp activity was tested in the presence of detergent (TX100), protease (proteinase K) or nucleases [RNase A and micrococcal nuclease (S7 MNase)]. As shown in Fig. 2D, HCV RNA accumulation was decreased roughly 4-fold in the presence of TX100 (lane 4). However, when compared to untreated sample (lane 3), HCV RNA levels were not greatly affected by proteinase K treatment (lane 5) or preincubation of the bound fraction with S7 MNase or RNase A (lanes 8 & 9). These results are consistent with the interpretation that the replicase complex in the bound fraction is membrane-enclosed or that the RdRp activity is sensitive to detergent treatment. Incubation of the in vitro reaction with S7 MNase (without inactivation by EGTA; lane 7) resulted in a decrease in viral RNA levels, indicating perhaps that a fraction of the newly synthesized RNA was exposed to nuclease degradation. However, there was no decrease in the levels of nascent RNA when the RdRp assay was performed in the absence of RNase inhibitor, RNasin (lane 6). These results suggest that the bound fraction contains negligible nuclease activity.
The bound fraction contains both secretory and endocytic Rabs
Two monomeric GTPases normally associated with endocytosis and cargo recycling (Rab5 and 4) were previously detected in the bound fraction (Stone et al., 2007). To determine whether other Rabs were associated with HCV replicase proteins, selected bands in the bound fraction (indicated by arrowheads in Fig. 1C) were subjected to mass spectrometry. As shown in Table 1 (supplemental data), several reported HCV-associated proteins as well as newly identified host factors were found in the bound fraction. Of interest to us were the Rab proteins including Rab2A, 6A, 11A and 11B. Rab2 is required for retrograde traffic from the Golgi complex to the ER (Tisdale, 1999; Tisdale and Balch, 1996; Tisdale et al., 1992). By contrast, Rab11 and Rab6 are associated with recycling endosomes (RE) and the Golgi apparatus. Specifically, Rab11 is involved in recycling cargo to the plasma membrane (Junutula et al., 2004; Ullrich et al., 1996; Wilcke et al., 2000), whereas Rab6 plays a role in retrograde traffic from the LE to the trans-Golgi or within the Golgi complex (Mallard et al., 2002; Martinez et al., 1994).
To confirm the presence of these Rabs in the bound fraction, fifty micrograms of protein were separated on 10% SDS-PAGE and examined by immunoblotting with Rab-specific antibodies. As shown in Fig. 3A, Rab 1A, 2, 4A, 5 (A&B), 6 and 7 were differentially enriched in the bound fraction in comparison to the crude lysate. The most enriched Rabs (over 2-fold) were Rab 2 and 7, followed by Rab 1A and 6 (1.5- to 2-fold) (Fig. 3B). Surprisingly, the least enriched Rabs were the EE Rabs (Rab4A and 5) previously found to colocalize with NS4B protein by FM. These results indicate selective enrichment of Rabs in the bound fraction. We were unable to detect Rab11 with Rab11-specific antibodies (data not shown).
FIG. 3.
A. Fifty micrograms of cell extracts were separated on 10% SDS-PAGE and subjected to immunoblotting using antibodies specific to Rab1A, 2, 4A, 5A, 5B, 6 and 7 proteins. B. Quantitation of the band volume of each Rab protein in the C5B bound fraction relative to C5B lysate. Notice the preferential enrichment of Rab2, 6 and 7 in the C5B bound fraction. Results are the average from two independent experiments.
Rab7 colocalizes with NS4B protein in C5B cells
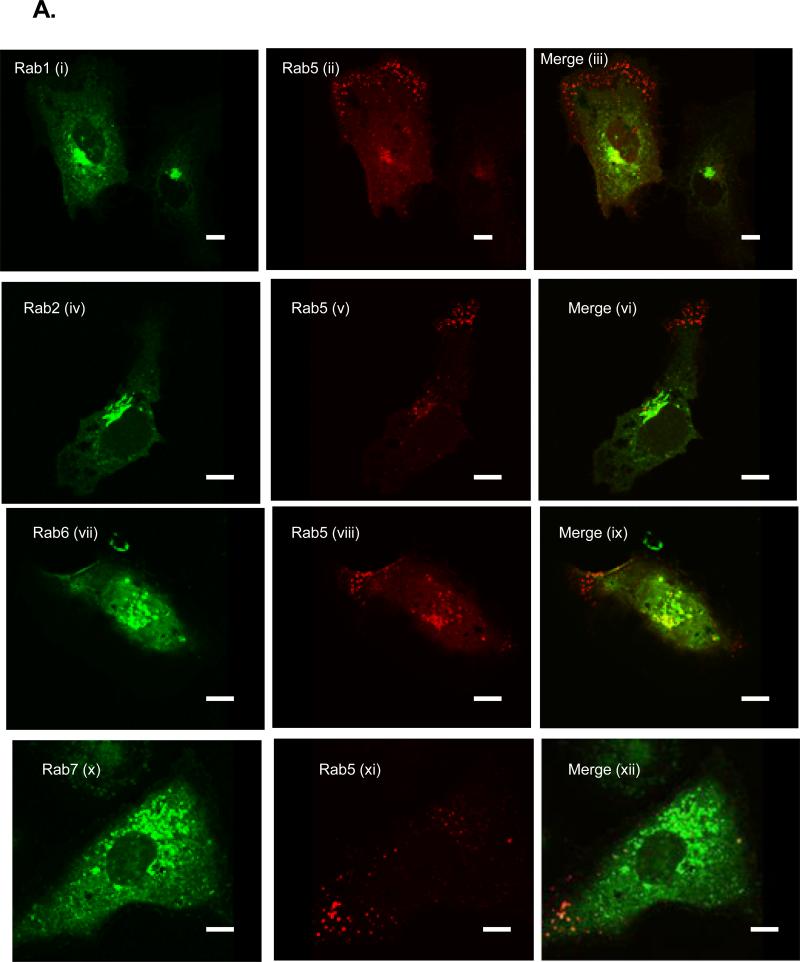
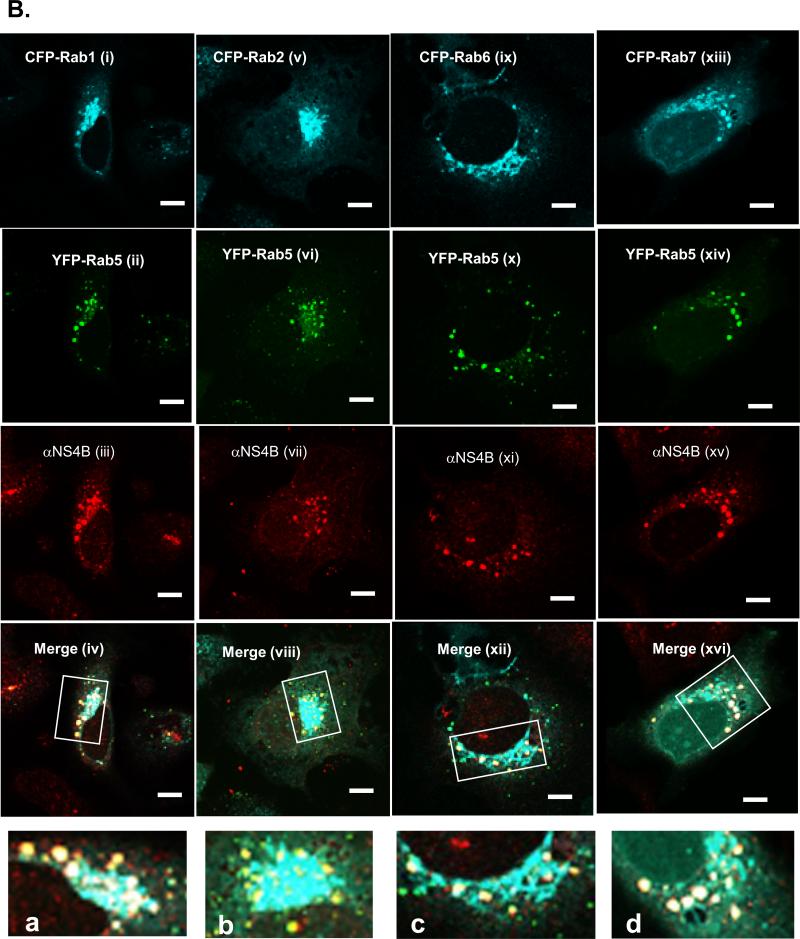
The differential association of the Rabs with the bound fraction could be explained by, 1) the nonspecific recruitment of a large population of ER- and LE/LYS-derived membranes into the bound fraction or, 2) the preferential recruitment of specific Rabs into that fraction. To test these hypotheses, we used FM to examine the subcellular localization of the Rabs (Fig. 3) both in parental (Huh7) and C5B replicon cells. In our previous report, we showed co-localization of endogenous Rab5 with NS4B protein in C5B cells (Stone et al., 2007). However, this approach was not used in this study because of the poor staining for most of the Rab proteins in Huh7 or C5B cells. Instead, for consistency, we have overexpressed all the Rabs with an N-terminal CFP, GFP or YFP tag. As seen in Fig. 4A (i-v), expression of these proteins in Huh7 cells resulted in the Golgi-like fluorescence for Rab1, 2 and 6, whereas the endocytic Rabs (Rab5 and 7) displayed the characteristic foci and perinuclear pattern associated with these proteins. Next, each Rab protein was expressed in C5B cells and examined by confocal microscopy to assess putative co-localization with NS4B protein. As displayed in Fig. 4 (B & C), expression of Rab1 (i-iii; a), 2 (iv-vi; b) or 6 (x-xii; c) resulted in minimal colocalization of each protein with NS4B [Pearson coefficient (PC) is less than 0.4]. However, Rab5 (vii-ix; c) and Rab7 (xiii-xv; d) foci significantly merged with NS4B (PC at least 0.5). These data suggest that EE and LE/LYS Rabs are selectively associated with NS4B foci.
FIG. 4.
A. Huh7 cells were transfected with DNA constructs expressing various Rab proteins with an N-terminal CFP (Rab1, 2, 6 &7) or GFP (Rab5) fusion. At 24 h post-transfection, the cells were processed and pictures were taken with a confocal microscope (i-v). Also shown are controls including CFP-expressing Huh7 (vi) as well as Huh7 (vii) and C5B (viii) cells stained for NS4B expression. Notice the diffuse and nuclear fluorescence of CFP alone and the absence of NS4B staining in Huh7 cells. B. C5B cells were transfected with DNA constructs expressing CFP-Rab1 (i-iv; a), CFP-Rab2 (iv-vi; b), GFP-Rab5 (vii-ix; c), CFP-Rab6 (x-xii; d) and CFP-Rab7 (xiii-xv; e). At 24 h post-transfection, the cells were stained with NS4B-specific antibody and samples were examined via confocal microscopy. Magnified areas are indicated by rectangles. Note that the CFP-expressing Rab proteins were pseudo-colored green. C. Quantitation results are from at least five Rab- and NS4B-expressing cells. Pearson colocalization results are significant when the coefficient (PC) is at least 0.5. Notice the moderate to strong merging of Rab5 or 7 with NS4B foci. There is weak colocalization of NS4B with Rab1, 2 and 6. Bars = 10 μm.
Rab5 and 7 proteins colocalize in C5B cells
The findings that Rab5 or 7 merged with NS4B foci, when expressed singly, led us to hypothesize that these Rabs would co-localize in C5B cells. To test this prediction, we first examined the colocalization of these Rabs in Huh7 cells. Since YFP-Rab5 was pseudo-colored red and the other Rabs (CFP-Rab1, 2, 6 & 7) were green, the merging of Rab5 with these Rabs would result in a yellow color. As seen in Fig. 5A, co-expression of Rab5 with Rab1 (i-iii), 2 (iv-vi), 6 (vii-ix) or 7 (x-xii) resulted in negligible merging of their fluorescence.
FIG.5.
A. Huh7 cells were co-transfected with DNA constructs expressing YFP-Rab5 and CFP-Rab1 (i-iii), 2 (iv-vi), 6 (vii-ix) or 7 (x-xii) proteins. At 24 h post-transfection, the cells were processed and pictures were taken with a confocal microscope. Note that the CFP-expressing Rab proteins were pseudo-colored green whereas Rab5 is red. Also notice the lack of a significant merging of Rab5 fluorescence with the other Rab proteins. B. C5B cells were transfected with DNA constructs expressing YFP-Rab5 and CFP-Rab1 (i-iv; a), 2 (v-viii; b), 6 (ix-xii; c) or 7 (xiii-xvi; d) proteins. At 24 h post-transfection, the cells were stained with NS4B-specific antibody and samples were examined as in (B). CFP-fused Rabs are blue, Rab5 is green and NS4B is red. Merging of the three colors results in white fluorescence. Magnified areas are indicated by rectangles. C. Quantitation results are from at least five cells co-expressing Rab proteins and NS4B. Notice the colocalization of Rab5 and 7 with NS4B in C5B cells (xiii-xvi; d) as displayed by the Pearson colocalization results in (C) and the white foci in (B). There is weak merging of Rab5 & 6 (ix-xii; c) or Rab5 & 1/2 (i-viii; a & b) with NS4B foci. Bars = 10 μm.
To examine the putative colocalization of CFP-fused Rab1, 2, 6 or 7 with YFP-fused Rab5 in C5B cells, the respective Rabs were co-expressed, followed by immunostaining with NS4B-specific antibody and Alexa Fluor 594-conjugated secondary antibody. Since YFP-Rab5 (pseudo-colored green) and NS4B foci merge, cells expressing Rab5 and NS4B were expected to display yellow color. Further, any association of the CFP-fused Rabs (1, 2, 6 or 7) with YFP-Rab5 and NS4B was predicted to merge as white fluorescence. As seen in Fig. 5 (B & C), co-expression of Rab1, 2 or 6 with Rab5 resulted in a weaker merging of these Rabs in C5B cells [(i-xii; a-c); PC is 0.5 or less]. In contrast, a stronger merging was observed between Rab7 and Rab5 when they were co-expressed in C5B cells (xiii-xvi; d) as indicated by the PC above 0.5. These findings suggest that Rabs are selectively recruited into NS4B foci, with Rab 5 and 7 being the most prevalent.
DN Rab6 protein partially disrupts Rab5 association with NS4B foci
To understand the selective recruitment of endocytic Rabs (5 & 7) into NS4B foci, we postulated that ER-derived NS4B-bound secretory vesicles are transported to the plasma membrane (PM) and retrieved back into the cytosol via the endocytic pathway, thus allowing the incorporation of Rab 5 and 7 into NS4B foci. If correct, we predicted that expression of DN Rab 1 or 2 (Derre and Isberg, 2004; Dupre et al., 2006; Tisdale et al., 1992), which partially inhibits ER-to-Golgi traffic, could disrupt NS4B association with Rab5 protein. Alternatively, if Rab5 is recruited into NS4B foci while NS4B-bound vesicles are formed, we predict that DN Rab1 or 2 would have no effect on Rab5 association with NS4B foci. As seen in fig. 6, the DN forms of CFP-fused Rab1 (i), 2 (v) and 6 (ix & xiii) exhibited a more diffuse subcellular distribution with some nuclear fluorescence (v, ix & xiii) in comparison to their Wt counterparts (Fig. 4 & 5). Further, co-expression of DN Rab1 (i-iv; a) or 2 (v-viii; b) with Wt Rab5 had no appreciable effect on the merging of NS4B and Rab5 foci. In contrast, when DN Rab6 was co-expressed with Wt Rab5 in C5B cells, Rab5 association with NS4B protein was partially disrupted as indicated by the red NS4B staining in C5B cells (Fig. 6, ix-xvi; c and d). These data suggest that ER-to-Golgi secretory traffic may not be required for Rab5 recruitment into NS4B foci. Instead, LE to Golgi traffic may be involved in Rab5 incorporation into NS4B foci. However, given that some of the NS4B foci were preformed before the expression of DN Rab1 and 2 proteins, we cannot completely rule out a role for the secretory pathway in Rab5 recruitment into NS4B foci. Finally, since merging of YFP-Rab5 and NS4B foci results in yellow fluorescence, the white foci shown in (a), (b), (c) & (d) are probably the result of the diffuse subcellular distribution of the CFP-fused DN Rab proteins.
FIG. 6.
C5B cells were transfected with DNA constructs expressing Wt Rab5 and DN Rab1 (i-iv; a), 2 (v-viii; b) or 6 (ix-xvi; c and d). At 24 h post-transfection, the cells were stained with NS4B-specific antibody and samples were examined by confocal microscopy. Magnified areas are indicated by rectangles. Notice the significant merging of Wt Rab5 and NS4B foci in cells expressing DN Rab1 (a) or 2 (b). There is less overlap of Wt Rab5 & NS4B in cells expressing DN Rab6 (c and d), as exhibited by the red NS4B fluorescence. Bars = 10 μm.
Some DN Rabs disrupt NS4B foci
The findings that Rab5 and 7 were recruited into NS4B foci led us to test whether these Rabs might be needed to form NS4B foci. This is also in light of the recent reports showing that both Rab5 and 7 are required for HCV genome replication and that DN Rab5 expression disrupts NS4B foci in replicon cells (Berger et al., 2009; Stone et al., 2007; Tai et al., 2009). We also tested Rab6 since DN Rab6 expression disrupted NS4B co-localization with Rab5 (Fig. 6). As displayed in Fig. 7A, expression of Wt Rab5 (i-iii; a), 6 (vii-ix; c) or 7 (xiii-xv; e) resulted in the formation of the typical Wt NS4B foci. In contrast, expression of DN Rab5 (iv-vi; b) and 7 (xvi-xviii; f) proteins led to a more reticular NS4B subcellular distribution (compare b to a and f to e). A minor disruption of NS4B foci was observed following DN Rab6 expression (x-xii) (compare d to c). These results suggest that Rab5, 7, and to a lesser extent Rab6, are involved in the formation of NS4B foci which contain active HCV RC. Since partial inhibition of the endocytic pathway by DN Rab Rab5 and 7 resulted in a disruption of NS4B foci, we tested whether inhibition of the endocytic recycling pathway by DN Rab11 would have a similar effect on NS4B foci. We also examined the role of other secretory Rabs (Rab1 and Rab2) on NS4B foci formation. As shown the in Fig. 7B, expression of DN Rab11 (iv-vi; b), 1 (x-xii; d) or 2 (xvi-xviii; f) had no significant effect on the formation of NS4B foci (compare b to a, d to c and f to e). These data indicate that secretory Rabs (1 and 2) and the recycling endosome Rab11 have a negligible effect on the formation of NS4B foci.
FIG. 7.
A. C5B cells were transfected with DNA constructs expressing Wt and DN Rab5, (i-vi; a & b), 6 (vii-xii; c & d) or 7 (xiii-xviii; e & f). At 24 h post-transfection, the cells were stained with NS4B-specific antibody and samples were examined via confocal microscopy. B. C5B cells were transfected with DNA constructs expressing Wt and DN Rab11, (i-vi; a & b), 1 (vii-xii; c & d) or 2 (xiii-xviii; e & f). Magnified areas are indicated by rectangles and Rabs are pseudo-colored green. Notice that expression of DN Rab5 (b), Rab7 (f), and to a lesser extent Rab6 (d), results in a disruption of NS4B foci. DN Rab11 (b), Rab1 (d), or Rab2 (f) does not siginificantly disrupt NS4B foci. Bars = 10 μm.
Silencing of Rab2, 5 or 7 results in a significant decrease in HCV RNA synthesis
We and others have shown that Rab5 is required for HCV genome replication (Berger et al., 2009; Stone et al., 2007; Tai et al., 2009). Further, a recent report suggests a similar role for Rab7 (Tai et al., 2009). To confirm these findings and address the role of secretory Rabs in HCV replication, siRNAs specific to Rab2, 5A and 7 were used to silence the expression of the cognate proteins in replicon cells. As shown in Fig. 8B, expression of each siRNA resulted in a significant decrease in the cognate Rab levels in the replicon cells. Additionally, silencing of Rab2, 5A or 7 did not affect the expression of the other Rabs (Fig. 8C). However, knockdown of Rab2, 5A or 7 resulted in 50-60% decrease in HCV RNA levels in comparison to siCONTROL-transfected cells (Fig. 8A). These results suggest that Rab2, 5 and 7 play a role HCV genome replication.
FIG. 8.
Effect of Rab2, 5 or 7 silencing on HCV genome replication. A. Replicon cells were transfected with siCONTROL, Rab2, 5 or 7 siRNA. At 48 h posttransfection, total cellular RNAs were isolated and used to determine the level of HCV RNA by quantitative real-time PCR. Results were normalized to GAPDH. Notice that silencing of Rab2, 5 or 7 results in 50-60% decrease in HCV RNA synthesis. B. Immunoblot analyses were performed using antibodies specific to Rab2, 5A or 7 protein. Notice that each siRNA resulted in a significant decrease of the cognate Rab protein. C. Immunoblot analyses were performed using antibodies specific to Rab2, 5A, 7 or GAPDH. Notice that silencing of Rab2 did not affect the expression of the other Rabs. Conversely, silencing of Rab5A or 7 did not affect the expression of Rab2.
DISCUSSION
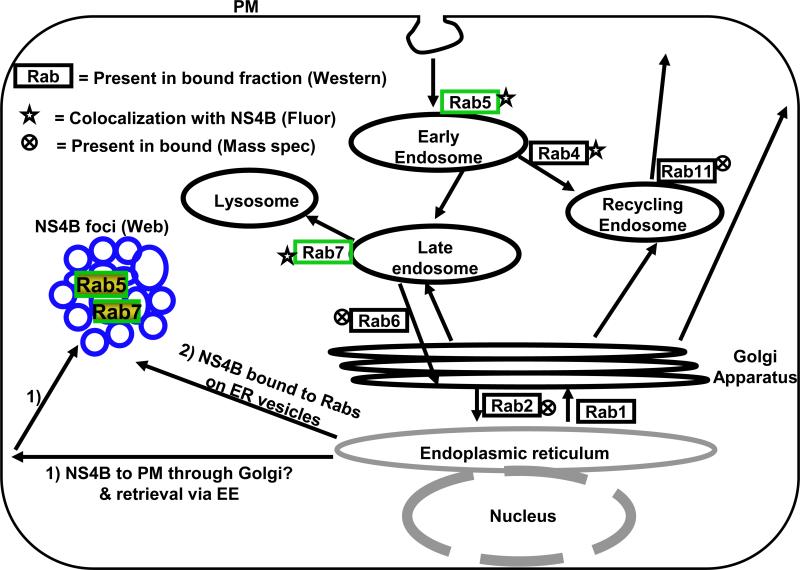
The goal for this study was to identify cellular Rabs implicated in the formation of NS4B foci, which contain the HCV RC. First, using immunoblotting and proteomic analysis, we show that an isolated replication-competent subcellular fraction contains several Rabs including Rab1, 2, 4, 5, 6, 7, and 11. However, examination of C5B replicon cells via FM indicates that only two endocytic Rabs, Rab5 and 7, are strongly associated with NS4B foci. These results suggest that, 1) some of the Rabs identified in the subcellular fraction were nonspecifically isolated with the HCV RC, 2) we isolated some inactive complexes containing NS4B protein or, 3) these Rabs are associated with viral structural proteins, which might be in the bound fraction. Secondly, we show that Rab5 and 7 co-localize in NS4B foci, suggesting that, 1) NS4B foci may contain membrane transport and fusion proteins from both the EE and LE/LYS compartments or, 2) NS4B interacts with both Rab5 and Rab7 proteins. Third, we found that Rabs involved in the secretory pathway may not be required for Rab5 recruitment into NS4B foci, suggesting that Rab5 association with NS4B foci may occur directly on the ER membranes or through an alternate route. We show that partial inhibition of EE membrane fusion, LE to LYS membrane fusion, or LE to Golgi membrane fusion, results in a disruption of NS4B foci. Additionally, silencing of Rab2, 5A or 7 results in a significant decrease in HCV RNA synthesis. These data are consistent with the interpretation that Rab5, 6 & 7 play a role in the formation of HCV RC and may explain in part why silencing of a single Rab protein does not result in severe impairment in HCV genome replication. Interestingly, one endocytic Rab protein, Rab11, was not associated with NS4B. These results suggest that signals in Rab5 and Rab7 or their cognate effectors might contribute to the incorporation of these Rabs into NS4B foci. We chose to examine the role of Rabs in HCV replication for several reasons. First, Rabs play an essential role in membrane transport, docking and fusion in the cell (Cai, Reinisch, and Ferro-Novick, 2007; Grosshans, Ortiz, and Novick, 2006), suggesting that viruses might exploit them to facilitate different processes in their life cycle. Second, Rabs are associated with specific intracellular membranes, suggesting that these proteins could be used as tools to identify the intracellular membrane origin of NS4B foci. The findings that Rab5 and 7 are required for HCV genome replication (Berger et al., 2009; Stone et al., 2007; Tai et al., 2009) may be explained in part by the fact that Rab5 and 7 may play a role in the formation of HCV RC represented by NS4B foci. However, Rab5 and 7 are not normally associated with the ER compartment, the site where both NS4B synthesis and initiation of NS4B foci may take place. Thus, our current model (Fig. 9) states that, 1) NS4B-bound vesicles are transported to the PM and retrieved into the cytosol via the endocytic pathway where Rab5 and 7 are recruited into NS4B foci. Alternatively, 2) Rab5 and 7 are recruited into NS4B foci while NS4B is on ER-derived membrane vesicles. Our current data are consistent with the second hypothesis since putative inhibition of ER-to-Golgi traffic by DN Rab1 or 2 did not seem to affect NS4B foci or their association with Rab5. However, we cannot rule out the involvement of the secretory Rabs since Rab6 (required for LE to Golgi or intra-Golgi traffic) appears to play some role in the formation of NS4B foci and Rab2 siRNA also impairs HCV genome replication. In fact, it is plausible that either hypothesis is correct since it has been reported that there are two types of HCV RCs, one which is smaller, motile and presumably associated with microtubules while the other is larger and static (Lai et al., 2008; Wolk et al., 2008). Partial inhibition of Rab5 and 7 trafficking to their cognate membranes could explain in part the impairment of HCV genome replication by Rab2 siRNA. Additionally, since NS4B foci were examined in replicon cells, it is conceivable that the preformed foci are less sensitive to the DN Rabs that inhibit the secretory pathway. Thus, studies are underway to test these Rabs in the context of JFH1 infection. Finally, a Rab-GAP TBC domain has been reported to bind NS5A and mediate HCV replication (Sklan et al., 2007). Identification of the Rab regulated by this Rab-GAP could shed more light on Rab proteins involved in HCV replication.
FIG. 9.
Model for Rab recruitment into NS4B foci. In model 1, NS4B-bound vesicles are transported to the PM and retrieved into the cytosol via the endocytic pathway where Rab5 and 7 are recruited into NS4B foci. In model 2, Rab5 and 7 are recruited into NS4B foci while NS4B is on ER membranes. The Rabs identified in this study by mass spectrometry (Mass Spec) analysis, FM (Fluor), or western blot are shown. Rab4 has been previously reported to colocalize with NS4B foci (Stone et al., 2007).
When observed via transmission electron microcopy, NS4B foci appear as a cluster of vesicles, each approximately 200 nm in size (Aligo et al., 2009). Such vesicles are much larger than the average COPII coat vesicle, about 87 nm in diameter (Miller et al., 2002; Shimoni et al., 2000). Therefore our data may suggest that Rab5 and 7 are recruited by NS4B to facilitate the fusion of ER-derived vesicles to form NS4B foci. Consistent with this interpretation, we have previously shown that Rab5 interacts with NS4B protein (Stone et al., 2007), but it is not known whether NS4B binds to Rab7 as well. Our data are also consistent with the notion that NS4B-derived vesicles may have several origins, including EE vesicles with a diameter of 100-250 nm (Cataldo et al., 1997). In addition to Rab5, we have previously found Rab4 and EEA1 to be associated with NS4B foci (Stone et al., 2007), and a recent report also indicates that EEA1 is required for HCV replication (Berger et al., 2009), further implicating the EE compartment in the formation of functional HCV RC.
In summary, we have identified endocytic Rabs as important players in the formation of NS4B foci. Our data and others suggest that NS4B-derived membrane vesicles are formed by a mechanism more complex than only a simple remodeling of the ER membranes.
MATERIALS AND METHODS
Cells
Cells expressing full-length replicon, C5B, have been described by Ikeda et al. (Ikeda et al., 2002). Parental (Huh7) and replicon cells (C5B) were grown as monolayers in advanced Dulbecco's modified Eagle's medium (DMEM) supplemented with 1.5% fetal bovine serum (FBS), non-essential amino acids (NEAA), 100 units/ml penicillin and 100 mg/ml of streptomycin at 37 °C in a 5% CO2 incubator. For replicon cells, the media also contained 0.3-0.5 mg/ml G418 (Geneticin). Huh7 and C5B cells were generously provided by Stanley Lemon (University of Texas Medical Branch, Galveston, TX). Described reagents were purchased from Invitrogen (Rockville, MD).
Antibodies
Rabbit polyclonal antibody to HCV NS4B was obtained from Covance (Denver, PA). Rabbit polyclonal antibodies to HCV NS3, NS5A and NS5B were kindly provided by Craig Cameron (Penn State, University Park, PA). Antibodies to Rab 1A, 2, 4A, 5A & B, 6 (Santa Cruz Biotechnology, Santa Cruz, CA), and Rab7 (Cell Signaling, Danvers, MA) were used in these studies. Horseradish peroxidase-conjugated secondary antibodies (used in chemiluminescence) were obtained from Vector laboratories (Burlingame, CA). Alexa Fluor 594-conjugated secondary antibodies (used in immunofluorescence) were from Invotrogen.
Plasmids
CFP-fused Wt Rab constructs (1, 2, 6, 7 and 11), their dominant negative versions (Rab1A-S25N, Rab2-S20N, Rab6A-T27N & Rab7-T22N), and GFP-fused Rab5A-S34N were provided by Won Do Heo (Korea Advanced Institute of Science and Technology, Korea). Wt GFP-Rab5 was kindly provided by Brian Knoll, Baylor College of Medicine (Houston, TX). To construct the YFP-Rab5 vector, pEYFP-C1 (Clontech, Mountain View, CA) was digested with NheI and NotI. The purified YFP fragment was subcloned into NheI- and NotI-cleaved GFP-Rab5 vector, thus replacing GFP.
DNA Transfection
For each experiment, Huh7 or C5B replicon cells were trypsinized and grown overnight in 6-well plates to obtain 60% confluent monolayer cells. Prior to transfection, the cells were washed with phosphate-buffered saline (PBS) and fed with 2 ml of complete medium. Cells were transfected according to the TransIT-LT1 protocol from Mirus (Madison, WI). The DNA mixture was added to each dish and incubated at 37°C for 24 or 48 h. With this procedure, DNA transfection efficiency was usually 70-90%.
Immunoisolation of a subcellular fraction containing HCV replicase proteins
The immunoisolation was done according to Stone et al. (Stone et al., 2007) with a slight modification. Briefly, 4-6 × 100 mm dishes (1 × 106 cells/dish), each of parental (Huh7) and replicon cells (C5B), were grown for 48h. The cells were trypsinized, washed, and resuspended in phosphate-buffered saline (PBS) solution containing 0.25M sucrose (PBS/sucrose) and protease inhibitors (1mM PMSF and 1 tablet of Complete, Mini, EDTA-free; Roche, Nutley, NJ). The cells were then lysed with 7 passages in a ball-bearing homogenizer (Balch and Rothman, 1985) to ensure approximately 90% lysis. Cell lysates were spun at 2500 xg/10 min at 4°C to pellet cellular debris and nuclei. The resulting supernatant will be referred to as the crude lysate or lysate throughout this paper. Purified NS4B antibody (1:200 dilution; Covance, Denver, PA) was added to equal amount (4 mg) of protein from either parental or replicon cells, and the 1.6 ml mixture (adjusted with PBS/sucrose and 3% final BSA) was incubated overnight at 4°C with constant rotation. The resulting homogenate was overlaid on a discontinuous iodixanol (Sigma-Aldrich, St. Louis, MO) gradient (4%, 8%, 12%, 20%, 30%, and 35%) in PBS/sucrose and the gradient was spun at 120,000 xg/1h 45min at 4°C in an Ti 80 rotor. A total of 12 fractions (630 l each) were collected from top to bottom. Typically, fractions 7 and 8 are endoplasmic reticulum (ER)-enriched, as determined by Calnexin-positive bands; in the replicon lysates, these fractions also contain viral replicase proteins including NS3, NS4B, NS5A and NS5B.
For membrane floatation assay, fractions 7 and 8 were combined and mixed with 60% iodixanol. A discontinuous iodixanol gradient (5%, 25% and 30%) (Elazar et al., 2004) was layered on the top of the combined fractions and the samples were spun at 120,000 xg for 4h 25min at 4°C in a Ti80 Rotor. A total of 8 fractions (867 μl each) were collected from top to bottom. Typically, the floatation gradient fraction 2 contained most of the HCV replicase proteins.
To immunoisolate a subcellular fraction containing NS4B and associated host factors, secondary anti rabbit antibody-coated magnetic Dynabeads M-280 (4.3 × 107/ml; Invitrogen, Carlsbad, CA) were added to the floated membrane fraction 2 with 3% final BSA. After overnight incubation at 4°C with constant rotation, the mixture was placed in a magnetic rack (Invitrogen) for 2-5 min. The resulting supernatant was collected and labeled as “Unbound”. NS4B positive membrane-bound beads were labeled as “Bound”. The bound fraction was washed four times to get rid off loosely bound proteins or membranes, whereas the unbound supernatant microsomes were spun twice at 100,000 xg, each for 1 h at 4°C. Bound and unbound fractions were resuspended in 4x sample buffer (240 mM Tris pH 6.8, 4% SDS, 40% glycerol, 4% β-mercaptoethanol, 0.01% bromophenol blue) for immunoblot. For HCV RNA-dependent RNA polymerase activity, Bound and unbound fractions were resuspended in diethylpyrocarbonate (DEPC)-treated PBS.
Sypro Ruby staining of replicase fraction
To examine the protein profile in the different fractions, an equal amount of protein (1 μg) was separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The gel was fixed with 50% methanol/ 7% acetic acid and stained overnight with 75% Sypro Ruby Protein Gel Stain (Invitrogen, Carlsbad, California). After two washes in 10% methanol/7% acetic acid, the protein bands were visualized on a phosphorImager (Typhoon 8600, Molecular Dynamics, Sunnyvale, CA).
Protein identification by capillary LC-ESI-MS/MS
Protein bands mostly enriched in the bound fraction, but not the unbound fraction, were excised, divided into pieces no larger than 1 mm and subjected to in-gel digestion overnight with sequencing grade trypsin (Promega Corp., Madison, WI). Samples were analyzed by a capillary liquid chromatography–tandem mass spectrometry system with a nanospray ionization source (capLC-nanoESI-MS/MS) that consisted of a Surveyor Micro AS autosampler, a Surveyor MS Pump, and an LTQ linear ion trap mass spectrometer (Thermo Electron Corporation, Waltham, MA). The digests were desalted on-line and eluted by using a 30-min linear acetonitrile gradient (2% to 40 % acetonitrile in 0.1% formic acid) at a flow rate of ~0.3 μL/min obtained by pre-column splitting of a flow of 200 μL/min. The analytical column was a PicoFrit column (New Objective) packed in-house with 5 μm wide pore C18 particles (Supelco Co, Bellefonte, PA). From a survey scan performed over m/z 550 -1500, tandem mass spectrometry (e.g. collision induced dissociation or CID) was triggered for precursor ions above an intensity threshold determined based on the baseline. Raw tandem mass spectra were processed and searched against the protein sequence database using a locally licensed and maintained Mascot 2.2 search engine to identify proteins. Mass tolerance was 3 amu and 2 amu for precursor and product ions, respectively. Up to 2 missed cleavages were allowed for digestion by trypsin and methionine oxidation was considered as a variable modification. At least two unique peptides with a Mascot peptide score higher than 40 were required to confidently identity a protein.
Quantitative Real-time PCR
Total cellular RNA was prepared from bound and unbound samples by using the TRIZOL method (Invitrogen, Carlsbad, CA), followed by precipitation with isopropanol and resuspension in DEPC-treated water. First strand cDNA was synthesized from the DNA-free RNA using random primers and the High Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). Triplicate samples of cDNA were mixed with a Taqman probe and a set of forward and reverse primers specific for either HCV NS4B (1b, strain N) or GAPDH and the mixture was subjected to real-time quantitative PCR using the ABI 7300 Sequence Detection System (Applied Biosystems, Foster City, CA).
In-vitro HCV RNA synthesis
Typically, 25 μl of crude lysate, bound or unbound fraction was resuspended in a 50 μl reaction containing 1 mM ATP, GTP and UTP (CTP at 40 μM), 1 mCi of (α32P) CTP per ml, 1 × buffer (50 mM Tris-Cl pH 7.8, 50 mM KCl, 5 mM mgCl2, 10 mM DTT, 5 mM creatine phosphate and 25 μg/ml creatine kinase), 10 μg/ml actinomycin D and 800 U/ml RNasin. The reaction was incubated at 37°C for 1h, with gentle mixing (every 30 min), followed by RNA extraction using the TRIZOL method (Invitrogen, Carlsbad, CA). The RNA was precipitated with isopropanol, washed in ethanol, dried and resuspended in DEPC-treated water. The RNA was separated on 1% formaldehyde denaturing gel. After drying, the gel was exposed to a phosphor screen and the RNA was visualized with a phosphorImager.
Immunoblot of HCV and host proteins
To visualize proteins from the bound and unbound fractions, 50 μg of protein were resuspended in 4x sample buffer, while 50 μg of crude lysates were used as controls. The samples were separated on 10% SDS-PAGE, followed by transfer onto an Immobilon-P membrane (PVDF; Millipore, Billerica, MA). Following binding with the respective primary antibody and HRP-conjugated secondary antibody, proteins were visualized by enhanced chemiluminescence detection method (ECL western blotting substrate, Pierce, Rockford, Il).
Indirect immunofluorescence
Cells on coverslips were washed in PBS and fixe d in 4% formaldehyde in PBS for 15 min and permeabilized with 0.05% Triton X-100 for 5 min at room temperature (RT). The cells were washed three times with PBS and incubated in 3% bovine serum albumin (BSA) for 30 min. Primary antibodies were diluted in 3% BSA and incubated with the cells for 1 h at room temperature. After three washes, Alexa fluor 594 (1:1000)-conjugated secondary antibody (Molecular Probes, Eugene, OR) was added to the cells for 1 h at RT. After three more washes in PBS, the cells were stained with DAPI/PBS for 10 min at room temperature, followed by three additional washes in PBS. The cells were mounted on glass slides in Vectashield (Vector Laboratories, Inc., Burlingame, CA). The samples were then examined with a confocal laser scanning microscope (Olympus Fluoroview FV1000). Image analysis was performed using Olympus Fluoroview version 1.7c software. Pearson's co-efficient (PC) for colocalization is calculated on a pixel by pixel basis; it is from a scale of −1 to 1 and is based on two variables (x and y) and the linear relationship between these variables. Numbers below 0.5 suggest little to no colocalization, whereas those above 0.5 indicate moderate to strong association. All the images were taken using similar conditions. Co-localization of green (FITC) and red (cy3) signals produces yellow, whereas co-localization of blue, green and red results in white. All images were saved as TIFF files, imported to and processed in Adobe Photoshop.
siRNA Transfection
The day before transfection, C5B replicon cells were trypsinized and grown overnight in 6-well plates to yield a roughly 60% confluent monolayer. Prior to transfection, the cells were washed with phosphate-buffered saline (PBS) and fed with 1 ml of complete medium. Cells were transfected with 50nM siRNA according to the TransIT-TKO protocol from Mirus (Madison, WI). siGENOME SMARTpool siRNA was obtained from Dharmacon (Lafayette, CO), and was resuspended as a 20 uM stock in 1x siRNA buffer (Dharmacon). 50 nM of siCONTROL nontargeting siRNA pool was used as a control. Transfected cells were incubated for 48 h in complete media, and processed for RT-PCR analysis or western blotting.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Stanley Lemon and Craig Cameron for reagents, Craig Cameron and Joseph Reese for suggestions and critical reading of the manuscript.
This work was supported by K22 CA129241 from the National Institute of Health.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Aizaki H, Lee KJ, Sung VM, Ishiko H, Lai MM. Characterization of the hepatitis C virus RNA replication complex associated with lipid rafts. Virology. 2004;324(2):450–61. doi: 10.1016/j.virol.2004.03.034. [DOI] [PubMed] [Google Scholar]
- Ali N, Tardif KD, Siddiqui A. Cell-free replication of the hepatitis C virus subgenomic replicon. J Virol. 2002;76(23):12001–7. doi: 10.1128/JVI.76.23.12001-12007.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aligo J, Jia S, Manna D, Konan KV. Formation and function of hepatitis C virus replication complexes require residues in the carboxy-terminal domain of NS4B protein. Virology. 2009;393(1):68–83. doi: 10.1016/j.virol.2009.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Appel N, Zayas M, Miller S, Krijnse-Locker J, Schaller T, Friebe P, Kallis S, Engel U, Bartenschlager R. Essential role of domain III of nonstructural protein 5A for hepatitis C virus infectious particle assembly. PLoS Pathog. 2008;4(3):e1000035. doi: 10.1371/journal.ppat.1000035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balch WE, Rothman JE. Characterization of protein transport between successive compartments of the Golgi apparatus: asymmetric properties of donor and acceptor activities in a cell-free system. Arch Biochem Biophys. 1985;240(1):413–25. doi: 10.1016/0003-9861(85)90046-3. [DOI] [PubMed] [Google Scholar]
- Bartenschlager R, Sparacio S. Hepatitis C virus molecular clones and their replication capacity in vivo and in cell culture. Virus Res. 2007 doi: 10.1016/j.virusres.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Berger KL, Cooper JD, Heaton NS, Yoon R, Oakland TE, Jordan TX, Mateu G, Grakoui A, Randall G. Roles for endocytic trafficking and phosphatidylinositol 4-kinase III alpha in hepatitis C virus replication. Proc Natl Acad Sci U S A. 2009;106(18):7577–82. doi: 10.1073/pnas.0902693106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blight KJ, Kolykhalov AA, Rice CM. Efficient initiation of HCV RNA replication in cell culture. Science. 2000;290(5498):1972–4. doi: 10.1126/science.290.5498.1972. [DOI] [PubMed] [Google Scholar]
- Bucci C, Lutcke A, Steele-Mortimer O, Olkkonen VM, Dupree P, Chiariello M, Bruni CB, Simons K, Zerial M. Co-operative regulation of endocytosis by three Rab5 isoforms. FEBS Lett. 1995;366(1):65–71. doi: 10.1016/0014-5793(95)00477-q. [DOI] [PubMed] [Google Scholar]
- Cai H, Reinisch K, Ferro-Novick S. Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell. 2007;12(5):671–82. doi: 10.1016/j.devcel.2007.04.005. [DOI] [PubMed] [Google Scholar]
- Cataldo AM, Barnett JL, Pieroni C, Nixon RA. Increased neuronal endocytosis and protease delivery to early endosomes in sporadic Alzheimer's disease: neuropathologic evidence for a mechanism of increased beta-amyloidogenesis. J Neurosci. 1997;17(16):6142–51. doi: 10.1523/JNEUROSCI.17-16-06142.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derre I, Isberg RR. Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect Immun. 2004;72(5):3048–53. doi: 10.1128/IAI.72.5.3048-3053.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupre DJ, Robitaille M, Ethier N, Villeneuve LR, Mamarbachi AM, Hebert TE. Seven transmembrane receptor core signaling complexes are assembled prior to plasma membrane trafficking. J Biol Chem. 2006;281(45):34561–73. doi: 10.1074/jbc.M605012200. [DOI] [PubMed] [Google Scholar]
- Egger D, Wolk B, Gosert R, Bianchi L, Blum HE, Moradpour D, Bienz K. Expression of hepatitis C virus proteins induces distinct membrane alterations including a candidate viral replication complex. J Virol. 2002;76(12):5974–84. doi: 10.1128/JVI.76.12.5974-5984.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hage N, Luo G. Replication of hepatitis C virus RNA occurs in a membrane-bound replication complex containing nonstructural viral proteins and RNA. J Gen Virol. 2003;84(Pt 10):2761–9. doi: 10.1099/vir.0.19305-0. [DOI] [PubMed] [Google Scholar]
- Elazar M, Liu P, Rice CM, Glenn JS. An N-terminal amphipathic helix in hepatitis C virus (HCV) NS4B mediates membrane association, correct localization of replication complex proteins, and HCV RNA replication. J Virol. 2004;78(20):11393–400. doi: 10.1128/JVI.78.20.11393-11400.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R, Egger D, Lohmann V, Bartenschlager R, Blum HE, Bienz K, Moradpour D. Identification of the hepatitis C virus RNA replication complex in Huh-7 cells harboring subgenomic replicons. J Virol. 2003;77(9):5487–92. doi: 10.1128/JVI.77.9.5487-5492.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosshans BL, Ortiz D, Novick P. Rabs and their effectors: achieving specificity in membrane traffic. Proc Natl Acad Sci U S A. 2006;103(32):11821–7. doi: 10.1073/pnas.0601617103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo JT, Bichko VV, Seeger C. Effect of alpha interferon on the hepatitis C virus replicon. J Virol. 2001;75(18):8516–23. doi: 10.1128/JVI.75.18.8516-8523.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez MG, Munafo DB, Beron W, Colombo MI. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J Cell Sci. 2004;117(Pt 13):2687–97. doi: 10.1242/jcs.01114. [DOI] [PubMed] [Google Scholar]
- Hardy RW, Marcotrigiano J, Blight KJ, Majors JE, Rice CM. Hepatitis C virus RNA synthesis in a cell-free system isolated from replicon-containing hepatoma cells. J Virol. 2003;77(3):2029–37. doi: 10.1128/JVI.77.3.2029-2037.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffenberg S, Sanford JC, Liu S, Daniel DS, Tuvin M, Knoll BJ, Wessling-Resnick M, Dickey BF. Biochemical and functional characterization of a recombinant GTPase, Rab5, and two of its mutants. J Biol Chem. 1995;270(10):5048–56. doi: 10.1074/jbc.270.10.5048. [DOI] [PubMed] [Google Scholar]
- Huang L, Hwang J, Sharma SD, Hargittai MR, Chen Y, Arnold JJ, Raney KD, Cameron CE. Hepatitis C virus nonstructural protein 5A (NS5A) is an RNA-binding protein. J Biol Chem. 2005;280(43):36417–28. doi: 10.1074/jbc.M508175200. [DOI] [PubMed] [Google Scholar]
- Ikeda M, Yi M, Li K, Lemon SM. Selectable subgenomic and genome-length dicistronic RNAs derived from an infectious molecular clone of the HCV-N strain of hepatitis C virus replicate efficiently in cultured Huh7 cells. J Virol. 2002;76(6):2997–3006. doi: 10.1128/JVI.76.6.2997-3006.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jager S, Bucci C, Tanida I, Ueno T, Kominami E, Saftig P, Eskelinen EL. Role for Rab7 in maturation of late autophagic vacuoles. J Cell Sci. 2004;117(Pt 20):4837–48. doi: 10.1242/jcs.01370. [DOI] [PubMed] [Google Scholar]
- Johns HL, Berryman S, Monaghan P, Belsham GJ, Jackson T. A dominant-negative mutant of rab5 inhibits infection of cells by foot-and-mouth disease virus: implications for virus entry. J Virol. 2009;83(12):6247–56. doi: 10.1128/JVI.02460-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Junutula JR, Schonteich E, Wilson GM, Peden AA, Scheller RH, Prekeris R. Molecular characterization of Rab11 interactions with members of the family of Rab11-interacting proteins. J Biol Chem. 2004;279(32):33430–7. doi: 10.1074/jbc.M404633200. [DOI] [PubMed] [Google Scholar]
- Kim DW, Gwack Y, Han JH, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215(1):160–6. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- Konan KV, Giddings TH, Jr., Ikeda M, Li K, Lemon SM, Kirkegaard K. Nonstructural protein precursor NS4A/B from hepatitis C virus alters function and ultrastructure of host secretory apparatus. J Virol. 2003;77(14):7843–55. doi: 10.1128/JVI.77.14.7843-7855.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai CK, Jeng KS, Machida K, Lai MM. Association of hepatitis C virus replication complexes with microtubules and actin filaments is dependent on the interaction of NS3 and NS5A. J Virol. 2008;82(17):8838–48. doi: 10.1128/JVI.00398-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai VC, Dempsey S, Lau JY, Hong Z, Zhong W. In vitro RNA replication directed by replicase complexes isolated from the subgenomic replicon cells of hepatitis C virus. J Virol. 2003;77(3):2295–300. doi: 10.1128/JVI.77.3.2295-2300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Stahl PD. Structure-function relationship of the small GTPase rab5. J Biol Chem. 1993;268(32):24475–80. [PubMed] [Google Scholar]
- Lohmann V, Korner F, Koch J, Herian U, Theilmann L, Bartenschlager R. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science. 1999;285(5424):110–3. doi: 10.1126/science.285.5424.110. [DOI] [PubMed] [Google Scholar]
- Lundin M, Monne M, Widell A, Von Heijne G, Persson MA. Topology of the membrane-associated hepatitis C virus protein NS4B. J Virol. 2003;77(9):5428–38. doi: 10.1128/JVI.77.9.5428-5438.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallard F, Tang BL, Galli T, Tenza D, Saint-Pol A, Yue X, Antony C, Hong W, Goud B, Johannes L. Early/recycling endosomes-to-TGN transport involves two SNARE complexes and a Rab6 isoform. J Cell Biol. 2002;156(4):653–64. doi: 10.1083/jcb.200110081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markgraf DF, Peplowska K, Ungermann C. Rab cascades and tethering factors in the endomembrane system. FEBS Lett. 2007;581(11):2125–30. doi: 10.1016/j.febslet.2007.01.090. [DOI] [PubMed] [Google Scholar]
- Martinez O, Schmidt A, Salamero J, Hoflack B, Roa M, Goud B. The small GTP-binding protein rab6 functions in intra-Golgi transport. J Cell Biol. 1994;127(6 Pt 1):1575–88. doi: 10.1083/jcb.127.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meertens L, Bertaux C, Dragic T. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J Virol. 2006;80(23):11571–8. doi: 10.1128/JVI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller E, Antonny B, Hamamoto S, Schekman R. Cargo selection into COPII vesicles is driven by the Sec24p subunit. EMBO J. 2002;21(22):6105–13. doi: 10.1093/emboj/cdf605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller RH, Purcell RH. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci U S A. 1990;87(6):2057–61. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyanari Y, Hijikata M, Yamaji M, Hosaka M, Takahashi H, Shimotohno K. Hepatitis C virus non-structural proteins in the probable membranous compartment function in viral genome replication. J Biol Chem. 2003;278(50):50301–8. doi: 10.1074/jbc.M305684200. [DOI] [PubMed] [Google Scholar]
- Moradpour D, Evans MJ, Gosert R, Yuan Z, Blum HE, Goff SP, Lindenbach BD, Rice CM. Insertion of green fluorescent protein into nonstructural protein 5A allows direct visualization of functional hepatitis C virus replication complexes. J Virol. 2004;78(14):7400–9. doi: 10.1128/JVI.78.14.7400-7409.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D, Penin F, Rice CM. Replication of hepatitis C virus. Nat Rev Microbiol. 2007;5(6):453–63. doi: 10.1038/nrmicro1645. [DOI] [PubMed] [Google Scholar]
- Prada-Delgado A, Carrasco-Marin E, Pena-Macarro C, Del Cerro-Vadillo E, Fresno-Escudero M, Leyva-Cobian F, Alvarez-Dominguez C. Inhibition of Rab5a exchange activity is a key step for Listeria monocytogenes survival. Traffic. 2005;6(3):252–65. doi: 10.1111/j.1600-0854.2005.00265.x. [DOI] [PubMed] [Google Scholar]
- Quinkert D, Bartenschlager R, Lohmann V. Quantitative analysis of the hepatitis C virus replication complex. J Virol. 2005;79(21):13594–605. doi: 10.1128/JVI.79.21.13594-13605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ranjith-Kumar CT, Gajewski J, Gutshall L, Maley D, Sarisky RT, Kao CC. Terminal nucleotidyl transferase activity of recombinant Flaviviridae RNA-dependent RNA polymerases: implication for viral RNA synthesis. J Virol. 2001;75(18):8615–23. doi: 10.1128/JVI.75.18.8615-8623.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouille Y, Helle F, Delgrange D, Roingeard P, Voisset C, Blanchard E, Belouzard S, McKeating J, Patel AH, Maertens G, Wakita T, Wychowski C, Dubuisson J. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J Virol. 2006;80(6):2832–41. doi: 10.1128/JVI.80.6.2832-2841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimoni Y, Kurihara T, Ravazzola M, Amherdt M, Orci L, Schekman R. Lst1p and Sec24p cooperate in sorting of the plasma membrane ATPase into COPII vesicles in Saccharomyces cerevisiae. J Cell Biol. 2000;151(5):973–84. doi: 10.1083/jcb.151.5.973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmonds P, Bukh J, Combet C, Deleage G, Enomoto N, Feinstone S, Halfon P, Inchauspe G, Kuiken C, Maertens G, Mizokami M, Murphy DG, Okamoto H, Pawlotsky JM, Penin F, Sablon E, Shin IT, Stuyver LJ, Thiel HJ, Viazov S, Weiner AJ, Widell A. Consensus proposals for a unified system of nomenclature of hepatitis C virus genotypes. Hepatology. 2005;42(4):962–73. doi: 10.1002/hep.20819. [DOI] [PubMed] [Google Scholar]
- Sklan EH, Serrano RL, Einav S, Pfeffer SR, Lambright DG, Glenn JS. TBC1D20 is a Rab1 GTPase-activating protein that mediates hepatitis C virus replication. J Biol Chem. 2007;282(50):36354–61. doi: 10.1074/jbc.M705221200. [DOI] [PubMed] [Google Scholar]
- Stone M, Jia S, Heo WD, Meyer T, Konan KV. Participation of rab5, an early endosome protein, in hepatitis C virus RNA replication machinery. J Virol. 2007;81(9):4551–63. doi: 10.1128/JVI.01366-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tackett AJ, Wei L, Cameron CE, Raney KD. Unwinding of nucleic acids by HCV NS3 helicase is sensitive to the structure of the duplex. Nucleic Acids Res. 2001;29(2):565–72. doi: 10.1093/nar/29.2.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai AW, Benita Y, Peng LF, Kim SS, Sakamoto N, Xavier RJ, Chung RT. A functional genomic screen identifies cellular cofactors of hepatitis C virus replication. Cell Host Microbe. 2009;5(3):298–307. doi: 10.1016/j.chom.2009.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targett-Adams P, Boulant S, McLauchlan J. Visualization of double-stranded RNA in cells supporting hepatitis C virus RNA replication. J Virol. 2008;82(5):2182–95. doi: 10.1128/JVI.01565-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale EJ. A Rab2 mutant with impaired GTPase activity stimulates vesicle formation from pre-Golgi intermediates. Mol Biol Cell. 1999;10(6):1837–49. doi: 10.1091/mbc.10.6.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale EJ, Balch WE. Rab2 is essential for the maturation of pre-Golgi intermediates. J Biol Chem. 1996;271(46):29372–9. doi: 10.1074/jbc.271.46.29372. [DOI] [PubMed] [Google Scholar]
- Tisdale EJ, Bourne JR, Khosravi-Far R, Der CJ, Balch WE. GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J Cell Biol. 1992;119(4):749–61. doi: 10.1083/jcb.119.4.749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich O, Reinsch S, Urbe S, Zerial M, Parton RG. Rab11 regulates recycling through the pericentriolar recycling endosome. J Cell Biol. 1996;135(4):913–24. doi: 10.1083/jcb.135.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcke M, Johannes L, Galli T, Mayau V, Goud B, Salamero J. Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-golgi network. J Cell Biol. 2000;151(6):1207–20. doi: 10.1083/jcb.151.6.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk B, Buchele B, Moradpour D, Rice CM. A dynamic view of hepatitis C virus replication complexes. J Virol. 2008;82(21):10519–31. doi: 10.1128/JVI.00640-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolk B, Sansonno D, Krausslich HG, Dammacco F, Rice CM, Blum HE, Moradpour D. Subcellular localization, stability, and trans-cleavage competence of the hepatitis C virus NS3-NS4A complex expressed in tetracycline- regulated cell lines. J Virol. 2000;74(5):2293–304. doi: 10.1128/jvi.74.5.2293-2304.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.