Abstract
The development of safe and efficient polymer carriers for DNA vaccine delivery requires mechanistic understanding of structure-function relationship of the polymer carriers and their interaction with antigen-presenting cells. Here we have synthesized a series of diblock copolymers with well-defined chain-length using atom transfer radical polymerization and characterized the influence of polycation chain-length on the physico-chemical properties of the polymer/DNA complexes as well as the interaction with dendritic cells. The copolymers consist of a hydrophilic poly(ethylene glycol) block and a cationic poly(aminoethyl methacrylate) (PAEM) block. The average degree of polymerization (DP) of the PAEM block was varied among 19, 39, and 75, with nearly uniform distribution. With increasing PAEM chain-length, polyplexes formed by the diblock copolymers and plasmid DNA had smaller average particle size and showed higher stability against electrostatic destabilization by salt and heparin. The polymers were not toxic to mouse dendritic cells (DCs) and only displayed chain-length-dependent toxicity at a high concentration (1 mg/mL). In vitro gene transfection efficiency and polyplex uptake in DCs were also found to correlate with chain-length of the PAEM block with the longer polymer chain favoring transfection and cellular uptake. The polyplexes induced a modest up-regulation of surface markers for DC maturation that was not significantly dependent on PAEM chain-length. Finally, the polyplex prepared from the longest PAEM block (DP of 75) achieved an average of 20% enhancement over non-condensed anionic dextran in terms of uptake by DCs in the draining lymph nodes 24 hours after subcutaneous injection into mice. Insights gained from studying such structurally well-defined polymer carriers and their interaction with dendritic cells may contribute to improved design of practically useful DNA vaccine delivery systems.
Keywords: Block copolymers, DNA vaccine, Dendritic cells, Gene Delivery
1. Introduction
The major component of a DNA vaccine is a plasmid DNA that encodes for protein antigens of interest. Antigen presenting cells (APCs) transfected with the DNA vaccine express the encoded antigen endogenously, process and present the antigen through the Major Histocompatibility Complex (MHC) molecules, resulting in the generation of antigen-specific humoral and cellular immune responses in vivo. DNA vaccination is a simple, scalable method for inducing immune responses without the many limitations of conventional vaccines based on attenuated pathogens and subunit vaccines [1]. In recent years DNA vaccines have shown considerable promise in defending against a wide range of bacterial and viral pathogens [2] and have been evaluated as immunotherapeutics for cancer [3], where the proper mobilization of both arms of the adaptive immunity is desired.
Dendritic cells (DCs) are the most important APCs and play a central role in regulating innate and adaptive immunity. Similar to macrophages, DCs are involved in acquiring antigens at the peripheral tissues and presenting antigens to naïve B and T lymphocytes in the spleen and draining lymph nodes. More importantly, DCs serve as a “master switch” of the immune system – between generating potent immunity to reject foreign antigen and inducing tolerance to self-antigen, the decision largely depends on the “context” (such as the maturation state of DCs), within which the antigen is presented [4]. Therefore, it is logical to focus on the ability to modulate the behavior of DCs when it comes to certain practical applications and clinical outcomes – for example, immune activation is required for effective vaccination and immunotherapy, whereas immune tolerance is desired for improving biocompatibility of biomaterial implants [5,6].
The success of DNA vaccination hinges on the ability of targeting DNA vaccines to DCs. There are two ways of accomplishing this: the ex vivo and the in vivo methods. For the so called ex vivo DC therapy, immature DCs from a patient are isolated, transfected with DNA encoding disease-associated antigens, activated with adjuvants, and introduced back to the same patient [7]. This method is being evaluated in clinical trials but is clearly a laborious and costly process. On the other hand, targeting DNA vaccines directly to DCs in vivo is more ideal, yet targeting and transfecting DCs efficiently, especially using nonviral vectors, have been quite challenging, given the complex nature of the anatomy and physiology of the immune system [5,8].
A large variety of nonviral polymers has been developed for gene delivery in general [9,10] and some of them have been evaluated for DNA vaccine delivery [11]. Microspheres of biodegradable polymers of various chemical compositions have been used to encapsulate DNA vaccines and shown to generate robust immune responses in vivo, presumably due to the sustained supply of antigens and passive targeting of micrometer-size particles to DCs [12]. A number of nano-scale molecular complexes of polymer/DNA vaccine have also been evaluated, using synthetic polycations such as polyethylenimine (PEI) [13] and polylysine [14], naturally derived polycations such as chitosan [15], as well as biodegradable cationic polyesters [16] and block copolymer surfactants [17]. Despite much effort in the past, it remains unclear about the design principles of polymer carrier specific for targeting DNA vaccine to DCs, although it is apparent that one must overcome a multitude of cellular and subcellular barriers generic to gene delivery to any cell type [18], in addition to meeting unique challenges of modulating antigen presentation and the maturation state of DCs. Moreover, the lack of control over molecular structure and properties of the polymer carriers makes it difficult to study mechanistically and systematically the interaction between polymer/DNA vaccine formulations and DCs in vitro and in vivo.
In this study we focus on a diblock copolymer system that may potentially serve as a molecular platform for the elucidation of polymer-DC interaction in the context of DNA vaccine delivery. The diblock copolymer, polyethylene glycol-block-poly(aminoethyl methacrylate) (PEG-b-PAEM), shown in Fig. 1, consists of a simple cationic block with primary amines for DNA binding and a PEG block for steric stabilization of polymer/DNA complexes. PEG-b-PAEM copolymers can be synthesized using a controlled polymerization technique, atom transfer radical polymerization (ATRP), resulting in polymer blocks of defined length that have been previously shown to influence the complexation with heparin [21] and oligonucleotides [31]. Here we report results of physico-chemical characterization of PEG-b-PAEM/DNA plasmid complexes, their interactions with DCs in vitro (including uptake, cytotoxicity, gene transfection, and phenotypic maturation), and targeted uptake by DCs in vivo, aiming to illustrate how synthetic polymers with well-defined molecular features such as polycation chain-length may have a profound influence on the delivery of cargos to DCs.
Fig. 1.
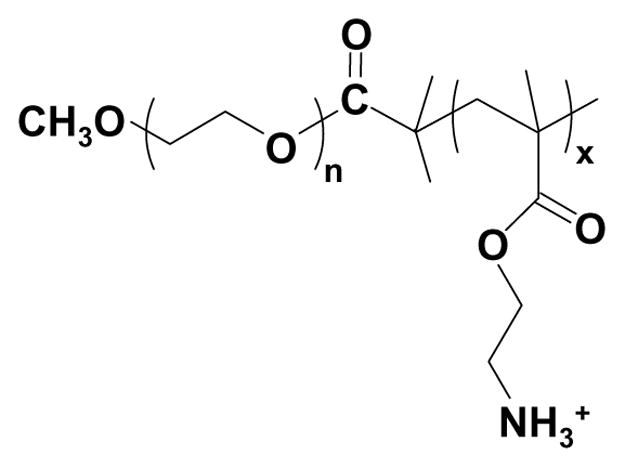
Chemical structure of the PEG-b-PAEM diblock copolymer.
2. Materials and methods
2.1. Chemicals
Monomethoxy-PEG (average Mn of 2,000) and PEI (branched, 25 kDa) were purchased from Polysciences. PEG was used after vacuum drying at 80 °C for 2 h. Toluene (Aldrich) was dried by refluxing over sodium and distilled. The PEG macro-initiator for ATRP and monomer (N(tert-butoxycarbonyl)aminoethyl methacrylate) (tBAM), was synthesized as described elsewhere [19,20]. Copper (I) chloride (CuCl) and 2,2′-dipyridyl (bPy) were purchased from Sigma. Other chemicals and solvents were purchased from Sigma and used without further purification.
2.2 Synthesis of diblock copolymers
Diblock copolymers with Boc-protected side-chains (PEG-b-PtBAM) were synthesized via ATRP using a process modified from [21]. A glass two-neck flask was charged with PEG macro-initiator, tBAM, CuCl, bPy, and the system was degassed three times. Dried degassed toluene was added, and the mixture was heated at 80 °C for 6 h. The reaction solution was then added dropwise to stirred hexane to obtain the crude product, which was further purified by dissolving in tetrahydrofuran (THF) and passing through a basic aluminum oxide column to remove the Cu complex. The resulting product was precipitated again in hexane, filtered, and dried in vacuum at room temperature. Three different initiator-to-monomer feed ratios were used in order to obtain PtBMA blocks with varying chain-length (Table 1). To synthesize PEG-b-PAEM, 5 g of PEG-b-PtBAM was dissolved in 5 mL of trifluoroacetic acid (TFA) and stirred for 1 h at room temperature. TFA was then removed by evaporation, and the oil residue was rinsed three times with diethyl ether. The resultant precipitate was collected by filtration, washed twice by diethyl ether, and dried overnight in vacuum.
Table 1.
Condition of polymerization and characterization of diblock copolymers.
| Polymer | Initiator:Monomer (feed molar ratio) | Yield (%) | Mna (×104) | Mwa (×104) | PDIa |
|---|---|---|---|---|---|
| PEG-b-PtBAM19b | 1:30 | 83 | 0.66 | 0.76 | 1.15 |
| PEG-b-PtBAM39b | 1:50 | 80 | 1.10 | 1.25 | 1.14 |
| PEG-b-PtBAM75b | 1:100 | 85 | 1.92 | 2.09 | 1.09 |
| PEG-b-PAEM19 | - | 97 | 0.68 | 0.79 | 1.16 |
| PEG-b-PAEM39 | - | 98 | 1.39 | 1.57 | 1.13 |
| PEG-b-PAEM75 | - | 96 | 2.18 | 2.40 | 1.10 |
Average molecular weight and polydispersity were determined by GPC using PEG standards;
Average degree of polymerization was determined by NMR analysis.
2.3 Characterization of diblock copolymers
The 1H-NMR spectra of the polymers were acquired on a Varian Unity spectrometer (300 MHz) using CDCl3 (for PEG-b-PtBAM) and D2O (for PEG-b-PAEM) as solvents. Chemical shifts were recorded in ppm and referenced against tetramethylsilane (TMS) and D2O, respectively. The molecular weight distribution and polydispersity of PEG-b-PtBAM were determined by gel permeation chromatography (GPC) analysis relative to polystyrene calibration (Waters 590 HPLC pump, a Waters 2410 differential refractometer, and a series of Waters Styragel columns (HR1, HR2, and HR3) using THF as eluent at a flow rate of 1.0 mL/min at 35 °C. The molecular weight and polydispersity of PEG-b-PBAM were determined by aqueous GPC analysis relative to PEG standard calibration (Waters 1525 binary HPLC pump, a Waters 2414 differential refractometer, and a Waters YMC HPLC column) using a sodium acetate/acetic acid buffer (pH 4.0) as eluent at a flow rate of 1.0 mL/min at 35 °C. All sample solutions were filtered through a 0.45 μm filter before analysis.
2.4 Preparation of polyplexes and gel retardation assays
Polymer/DNA complexes (polyplexes) of N:P ratios ranging from 1:8 to 16:1 were prepared by adding 25 μL of polymer solution in 20 mM HEPES (pH 7.4) to 25 μL of DNA plasmid solution (0.2 μg/μL in 20 mM HEPES, pH 7.4), vortexed for 10 seconds, and the dispersions were incubated for 30 min at room temperature. To ascertain DNA binding, the polyplexes were analyzed by gel electrophoresis on a 1.0% agarose gel containing 0.5 μg/mL ethidium bromide. To determine the strength of DNA binding by polymers with varying cationic chain-length, polyplexes at N:P ratio of 8:1 were incubated with increasing concentrations of heparin (0.1~1.5 IU per μg of DNA) for 10 min at room temperature and analyzed by agarose gel electrophoresis.
2.5 Dynamic light scattering (DLS)
The average hydrodynamic diameter and polydispersity index of polyplexes in HEPES buffer (20 mM) at 25 °C were determined using a ZetaPlus Particle Analyzer (Brookhaven Instruments Corporation, Holtsville, NY; 27 mW laser; 658 nm incident beam, 90° scattering angle). Polyplexes with N:P ratios ranging from 1:4 to 16:1 were prepared as described above and diluted 20 times to a final volume of 2 mL in HEPES buffer before measurement. To determine colloidal stability of polyplexes against physiological medium, polyplexes of N:P ratio of 8:1 were incubated in 20 mM HEPES buffer (pH 7.4) containing 150 mM NaCl at 37 °C. The change in average hydrodynamic diameter of the polyplexes was monitored continuously for up to 2 to 4 h.
2.6 Transmission electron microscopy (TEM)
The morphology and size of polyplexes were observed using a JEOL JEM-1210 transmission electron microscope. Samples for TEM were prepared by dropping the polyplex solution onto carbon-coated EM grids followed by negative staining of phosphotungstic acid (1.0%, pH 4~5).
2.7 Cytotoxicity assay
The cytotoxicity of polymers was evaluated by an MTT (3-(4,5-dimethyl-thiazol-2-yl)-2,5-diphenyl tetrazolium bromide) assay [22]. Murine DC 2.4 cells (ATCC) were seeded into 96-well plates at 6000 cells/well and cultured with polymers of various concentrations for 24 h in DC2.4 media (DMEM low glucose, 10% FBS, 10 mM HEPES, 100 U/mL penicillin/streptomycin) at 5% CO2 and 37 °C. MTT in PBS (5 mg/mL, 20 μL) was added to each well reaching a final concentration of 0.5 mg/mL. After 4 hours unreacted MTT was removed by aspiration. The formazan crystals were dissolved in 100 μL DMSO and the absorbance was measured at 570 nm using a Bio-Tek Synergy HT plate reader. Cell viability was calculated by [Absorbance of cells exposed to polymers]/[Absorbance of cells cultured without polymers] in percentage.
2.8 Uptake by DCs in vitro
Polymers were complexed with dextran labeled with Oregon Green 488 (anionic, 70 kDa, Invitrogen) at a mass ratio of 1:1 in 20 mM HEPES and incubated for 15 min at room temperature. The polyplexes were then added to DC 2.4 cells in a 6-well plate in 2 mL of serum-free media with each well containing 2 μg of dextran. Cells were incubated for 4 h at 37 °C and washed three times with 1 mL of FACS buffer (PBS, 1% bovine serum albumin, 0.02% sodium azide). Cells were then removed from plates using 0.25% trypsin-EDTA, washed in PBS and resuspended in FACS buffer. The level of dextran fluorescence of cells was quantified by flow cytometry using a FACSCalibur flow cytometer (Becton Dickson). Data analysis was done using the software, Flowjo.
2.9 DC transfection in vitro
To prepare polyplexes for DC transfection, polymers were mixed with plasmid DNA containing a CMV promoter and a luciferase reporter gene (Elim Biopharmaceuticals) at N:P ratios of 8:1. DC 2.4 cells were seeded in 6-well plates 200,000 cells per well and cultured for 24 h. Polyplexes were added at 4 μg DNA per well in 2 mL of serum-free media, with or without 30 mM hydroxychloroquine (Sigma), and were incubated for 4 h. The cells were then washed and cultured in fresh media with 10% serum for 20 h. Luciferase expression was determined using the Bright Glo Luciferase Assay kit (Promega) following manufacturer’s instruction. Protein content of tranfected cells was determined using a BCA protein assay kit (Pierce). The level of luciferase expression was calculated as relative light units (RLU) per mg of total cell protein.
2.10 DC maturation in vitro
Murine bone-marrow-derived dendritic cells (BMDCs) were derived as described in [23]. Four to six-week-old C57BL/6 male mice were purchased from Jackson labs (Bar Harbor, ME) and housed and used in accordance with guidelines from the University of Minnesota Institutional Animal Care and Use Committee. To harvest bone marrow progenitor cells, mice were sacrificed and the femur and tibia were removed, cleaned, and soaked in 70% ethanol for 1 min. The bones were then washed twice with PBS and transferred into RPMI medium (Gibco) supplemented with 2% FCS (RP-2). The bones were cut at both ends and marrow was flushed out with RP-2. The cell suspension was filtered through a cell strainer (Gibco) and centrifuged at 1000 rpm for 5 min. The cells were resuspended in 10 mLs of ACK lysis buffer (Lonza) for 5 min at 37°C to lyse red blood cells, then washed with excess volume of RP-2, centrifuged and resuspended in RPMI medium supplemented with 10% FCS, 100 U/ml penicillin/streptomycin, 2 mM L-glutamine, 1 mM sodium pyruvate and 50 μM 2-mercaptoethanol (RP-10 complete medium). A 3 vol% of conditioned medium containing GM-CSF secreted by J558L cells (courtesy of Dr. Kristin Hogquist, Center for Immunology, University of Minnesota) was added to stimulate cell differentiation into DCs. The cells were maintained for 6 days at 37 °C in a humidified atmosphere containing 5% CO2. On days 2 and 4 the media was replaced with fresh RP-10 complete medium supplemented with GM-CSF. On day 6, the clusters of developing DCs were harvested and plated at 1×106 cells/well in 6-well plates. An aliquot of cells from every batch of preparation was stained with APC-anti-mouse CD11c antibody to determine the percentage of CD11c+ DCs. Typical purity of CD11c+ DCs in these cultures was approximately 80~95%.
To examine the effect on DC maturation state, polyplexes with N:P ratio of 8:1 were added to BMDCs 6-well plates at 4 μg DNA per well and cultured for 24 h. The cells were washed and stained with FITC- or PE-labeled anti-mouse-CD80, CD86, and CD40 antibodies (Biolegend, San Diego, CA) and analyzed by flow cytometry. Untreated cells were of the immature phenotype (iDC, CD80lowCD86lowCD40low). Mature DCs were generated by stimulating with 2 μg/mL of lipopolysaccharide (LPS, from E. coli strain 026:B6, Sigma). To exclude the possibility of endotoxin contamination, all cell culture media, buffers, polymer stock solutions, and DNA plasmid stock solution were tested using a LAL endotoxin detection kit following manufacturer’s instruction.
2.11 Uptake by DCs in vivo
Polymer/dextran complexes were prepared at a weight ratio of 1:1 in 20 mM HEPES as described in section 2.8. Complexes containing 143 μg of dextran were injected in a total volume of 50 μL subcutaneously at the tail base of C57/BL6 mice. For comparison, some mice were injected with 50 μL of 1% solution of 100-nm fluorescent polystyrene particles (“Fluospheres”, Invitrogen) in 20 mM HEPES buffer. After 24 hours mice were sacrificed and the inguinal and popliteal lymph nodes were removed. Lymph nodes were digested in complete RPMI media containing 400 U/mL of collagenase at 37°C for 30 min, followed by addition of 10 mM EDTA and mixing for 3 min. The mixture was then diluted with 10 mL of HEPES buffered saline solution (HBSS, Gibco) containing 5 mM EDTA and 2% serum and passed through a cell strainer. The resulting single-cell suspension was washed with PBS containing 1% bovine serum albumin and resuspended in FACS buffer. CD11c+ DCs were stained as described above. The fluorescence intensity from the dextran and polystyrene nanoparticles taken up by CD11c+ DCs was quantified by flow cytometry.
3. Results and discussion
3.1 Synthesis and characterization of PEG-b-PAEM copolymers
Engineering nonviral carriers for DNA vaccine delivery is highly important in developing safe and clinically useful immunotherapeutics against diseases such as cancer and infectious diseases. The motivation of the current study is to explore a simple synthetic polymer system with well-defined molecular structure and properties that can be further tailored for efficient delivery of cargos to antigen-presenting dendritic cells. The design of the PEG-b-PAEM diblock copolymer embodies two essential principles for achieving efficient gene transfer in vitro and in vivo: 1) a cationic segment for condensing DNA and 2) a PEG block for steric stabilization of the polyplex. As shown in Fig. 1, the PAEM block has a flexible backbone of polymethacrylate with side-chains terminated in primary amines. Polymethacrylate derivatives such as PAEM can be synthesized by controlled radical polymerization (such as ATRP) that offers a convenient and effective means to generate polymers with well-defined chain-length, composition, and molecular architecture [21, 24, 31]. Therefore, PEG-b-PAEM may serve as a model system, which not only allows one to probe mechanistically the structure-function relationship in the context of DNA vaccine delivery to DCs, but with further modification, may also emerge as practically useful DNA vaccine formulations.
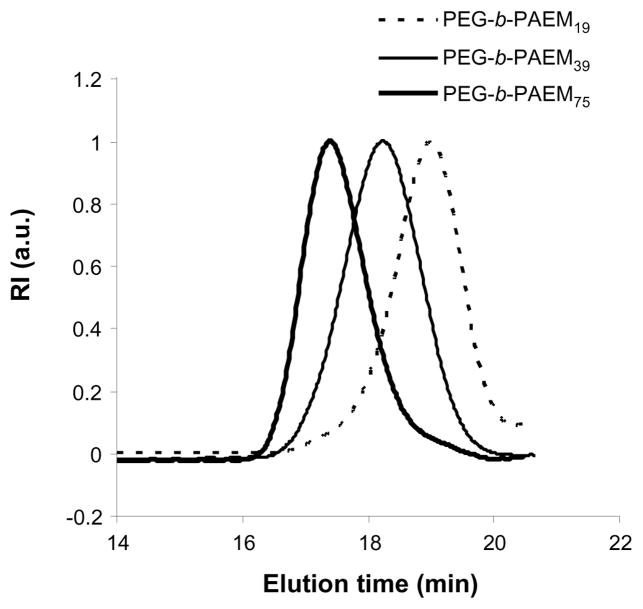
Here we have synthesized PEG-b-PAEM diblock copolymers using ATRP via a previously reported pathway [21]. In the first step, the ATRP of Boc-protected aminoethyl methacrylate monomer (tBMA) was initiated by a mono-functional PEG (2000 Da) macro-initiator using Cu (I)-dipyridyl complex as catalyst. The molar ratio of the initiator and monomer was varied to achieve PtBMA blocks with different length (Table 1). Three PEG-b-PtBMA polymers were synthesized with over 80% yield and characterized by GPC and NMR. Based on GPC analysis, the number average molecular weight (Mn) of the three polymers was calculated to be 0.66, 1.10, and 1.92×104 with the polydispersity index (PDI) of 1.15, 1.14, and 1.09, respectively (Table 1). The narrow molecular weigh distribution of the polymers indicates that the ATRP reaction was successful. Proton NMR analysis (not shown) confirms the chemical structure of all three PEG-b-PtBMA polymers, and the average degree of polymerization (DP) of the PtBMA blocks was calculated to be 19, 39, and 75. In the second step, the Boc-protecting group was removed by acid treatment, yielding the final products of PEG-b-PAEM. Proton NMR analysis showed complete disappearance of the diagnostic methyl proton signal of the Boc group at δ 1.46 ppm, confirming that all Boc groups have been removed. The polymers were easily soluble in water and methanol at room temperature. The aqueous GPC chromatograms of the three PEG-b-PAEM polymers are shown in Fig. 2. All three polymers had single peaks of molecular weight distribution and small PDI values of 1.16, 1.13, and 1.10. Although the single-step ATRP of aminoethyl methacrylate was recently reported [25], the two-step method used here offers excellent control of the polymerization, resulting in block copolymers of well-defined chain-length.
Fig. 2.
Aqueous GPC chromatograms of PEG-b-PAEM diblock copolymers.
3.2 DNA binding and condensation
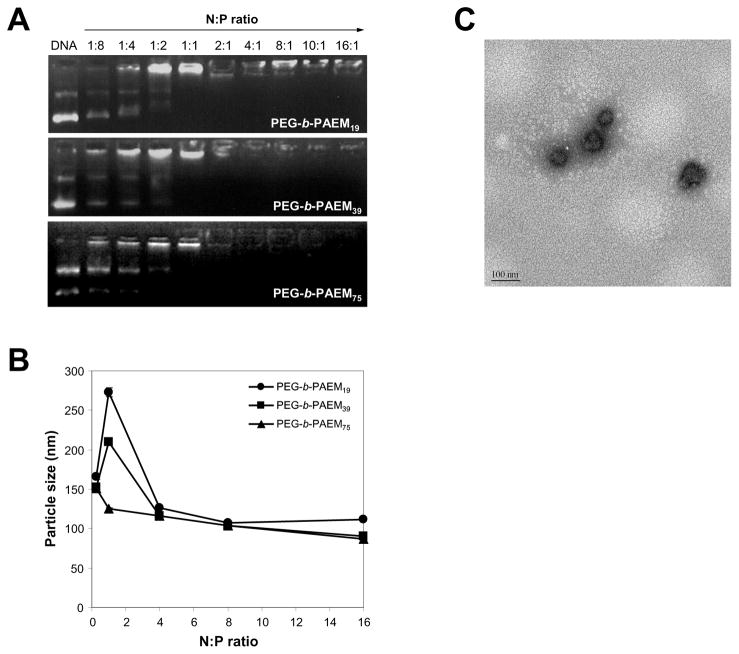
Gel retardation assay shows that PEG-b-PAEM block copolymers were able to bind to and completely neutralize the negative charges of a DNA plasmid at the N:P ratio of 1:1 and beyond, regardless of the chain-length of the PAEM block (Fig. 3A). This is expected since the positive charge density is the same among all three polymers. DLS reveals that the average particle size of polyplexes in aqueous buffer spanned a range of 100 to 300 nm with much dependence on the N:P ratio (Fig. 3B). The size distribution of each type of polyplex was also quite narrow with PDI ranging from 0.05 to 0.2. The distinct dependence of particle size on polycation chain-length was most prominently seen at N:P ratio of 1:1. Under such neutral charge condition, the particle size of the polyplexes (ca. 120, 210, 275 nm) increased with decreasing chain-length of the PAEM block (average DP of 19, 39, 75, respectively) (Fig. 3B), suggesting higher DNA condensing capacity of longer chains of polycations. At N:P ratio of 4:1 and beyond, average particle size stabilizes around 100 nm for all three polyplexes (Fig. 3B). Discrete, compact polyplexes formed at N:P ratio of 8:1 (Fig. 3C) were selected for further characterization to understand the influence of polycation chain-length on polyplex stability. Taken together, these results illustrate the ability of using block copolymers with defined chain-length to control the formation of defined and narrowly dispersed polyplexes, which is highly important in light of the notion that DC-specific targeting and internalization can be much affected by the size of the particulates [26,27].
Fig. 3.
Characterization of plasmid DNA binding and condensation by PEG-b-PAEM. (A) Gel retardation assay of polyplexes prepared at various N:P ratios. (B) DLS determination of the average particle size of polyplexes in aqueous buffer (20 mM HEPES, pH 7.4) as a function of N:P ratio. (C) A typical TEM image of polyplexes (PEG-b-PAEM75/DNA, N:P ratio of 8:1) showing discrete, condensed nanoparticles of ~100 nm in size. Scale bar: 100 nm.
The structure of PAEM is reminiscent of that of polylysine. Much is known about DNA condensation by polylysine [28] and PEG-polylysine block copolymers with narrow molecular weight distribution [29]. The polymethacrylate backbone of PAEM is conceivably more flexible than the polyamide backbone of polylysine, and PAEM also has a higher positive charge density than polylysine. Both of these are features favoring the condensation of DNA into compact, small nano-structures. A direct comparison between PAEM and polylysine in terms of DNA binding and gene transfection properties may be an interesting topic for future investigation. While polymers containing secondary and tertiary amines such as poly(dimethyl aminoethyl methacrylate) (PDAEMA) and a variety of other cationic moieties have been postulated to facilitate subcellular trafficking and DNA release [30], primary amines of PAEM maintain the maximum capacity of condensing DNA at physiological pH and can easily incorporate other types of amines through chemical transformation or copolymerization [21, 31].
3.3 Stability of polyplexes
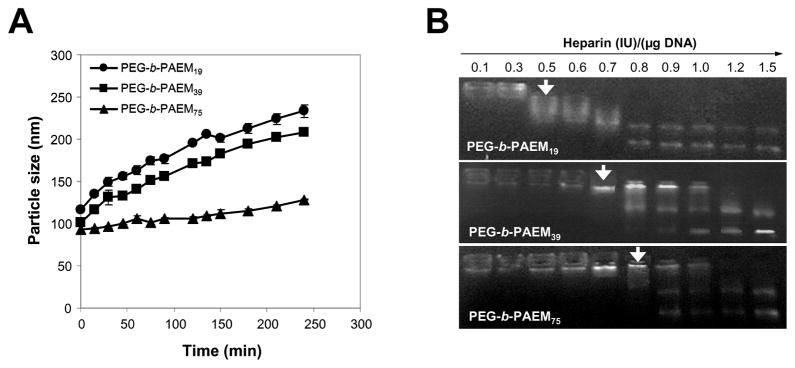
The stability of polyplexes at N:P ratio of 8:1 was studied at physiological ionic strength (150 mM NaCl, pH 7.4) and in the presence of increasing amount of heparin. First, the average size dependence of polyplexes on incubation time was determined by DLS measurements in 150 mM NaCl. As the presence of salt in the buffer screens and weakens the electrostatic interaction between the polymer and DNA, it is expected that the polyplexes would become unstable and aggregate over time. This was indeed the case for polyplexes formed by PEG-b-PAEM19 and PEG-b-PAEM39, almost doubling their size over a period of 4 h (Fig. 4A). On the contrary, polyplexes formed by PEG-b-PAEM75 only enlarged slightly over the same period of time. Second, polyplexes were analyzed by agarose gel electrophoresis after incubation with increasing amount of heparin, a polyanion that can compete with DNA for the binding to the polycation. As shown in Fig. 4B, the threshold concentration of heparin at which polyplex disruption occurred was 0.5 IU for polyplexes of PEG-b-PAEM19, and 0.7~0.8 IU for PEG-b-PAEM39 and PEG-b-PAEM75. Both of these experiments demonstrated that precisely controlled chain-length of cationic block copolymers has a profound impact on the colloidal stability of polyplexes.
Fig. 4.
Stability of polyplexes of plasmid DNA and PEG-b-PAEM at N:P ratio of 8:1 in 20 mM HEPES, pH 7.4. (A) Time-dependent aggregation of polyplexes in 150 mM NaCl. (B) Dissociation of DNA from polymer induced by increasing amount of heparin. Arrows point to the lowest heparin concentration at which disruption of polyplex was observed.
3.4 Cytotoxicity, in vitro transfection of DCs, and cellular uptake
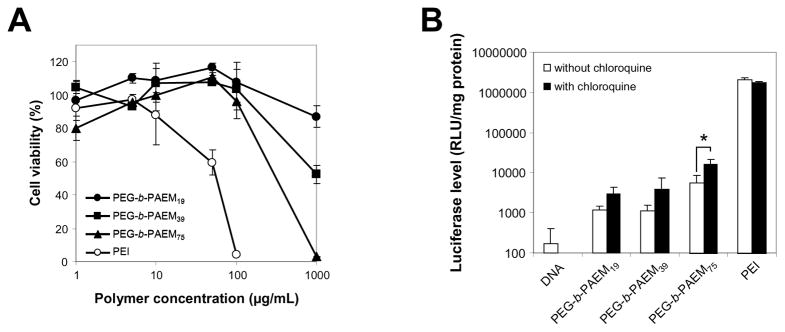
A murine dendritic cell line, DC 2.4, was used for the evaluation of toxicity and transfection efficiency of the polymers. All three polymers were not toxic to DC 2.4 cells at concentrations as high as 0.1 mg/mL (Fig. 5A). In contrast, branched PEI (25 kDa) was quite toxic, killing most DCs at 0.1 mg/mL (Fig. 5A). The low cytotoxicity of PEG-b-PAEM was expected and may be largely due to the presence of PEG. Nonetheless, at a rather high concentration of 1 mg/mL, toxicity was observed for all three polyplexes, and the level of toxicity correlated well with the chain-length of the PAEM block (Fig. 5A).
Fig. 5.
Cytotoxicity of the PEG-b-PAEM polymers and transfection efficiency of polyplexes (N:P 8:1 at a DNA concentration of 20 μg/mL) in DC 2.4 cells. (A) Cytotoxicity determined using an MTT assay. Cells treated with culture media only were taken as the 100% viability. (B) Transfection efficiency using luciferase as reporter in the absence or presence of 30 mM hydroxychloroquine. * P<0.05, unpaired t test.
Transfection efficiency of DC 2.4 cells was determined using luciferase as the reporter gene. Luciferase expression in cells transfected with the PEG-b-PAEM polyplexes was 10 to 100-times higher than the naked plasmid but 100-times lower than branched PEI polyplexes (Fig. 5B). Among the three polyplexes, the longest PAEM block appeared to result in the highest transfection whereas the other two shorter blocks were approximately the same (Fig. 5B). The observation that chain-length of polycations is related to transfection efficiency has been noted in numerous studies on polymers of diverse chemical structure [9,10], such as PEI [32,33], polylysine [34], PDAEMA [35,36], chitosan [37], poly(β-amino esters) [38], just to name a few. Because polymers with high polydispersity were often used, the correlation between polycation chain-length and transfection could only be probed reliably between average molecular weight values far enough apart from one another. On the other hand, narrowly-disperse polymers with precisely controlled chain-length, such as the PEG-b-PAEM, are more suitable for uncovering finer details of the chain-length dependence of nonviral gene delivery.
Endosomal escape is an important step of gene transfer that affects transfection efficiency. As the side-chains of PEG-b-PAEM are exclusively primary amines, there is a lack of buffering capacity at acidic endosomal pH that is required for the “proton sponge” mechanism to work [39]. We show here that the addition of 30 mM hydroxychloroquine, an endosomalytic compound, improved transfection slightly, yet only the improvement on the PEG-b-PAEM75 polyplexes was significant statistically (*P<0.05) (Fig. 5B). Chloroquine concentration higher than 30 mM was prohibited due to severe toxicity to the DCs. Further tuning of the length and side-chain structure of the polycationic block may be required to improve the gene transfection efficiency of PEG-b-PAEM75 in DCs.
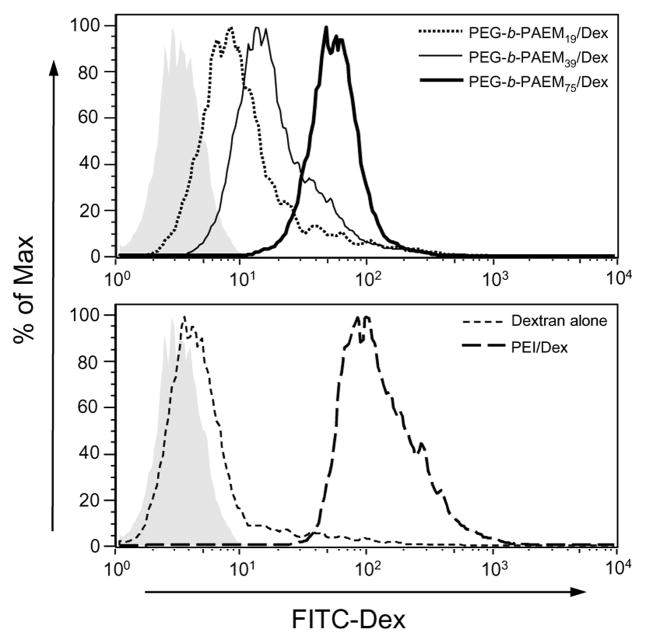
Another factor that affects directly the efficiency of transfection is cellular uptake of the polyplexes. To assess the polycation chain-length dependence on cellular uptake, polyplexes were prepared by condensing anionic fluorescently-labeled dextran (70 kDa), incubated with DCs, and the amount of uptake of dextran was quantified by flow cytometry. There is a clear correlation between the amount of uptake and the chain-length of the PAEM block, with the longest PEG-b-PAEM75 polyplexes showing the highest uptake followed by the shorter PEG-b-PAEM39 and PEG-b-PAEM19 (Fig. 6, upper panel). Longer PAEM chains are capable of stronger electrostatic binding to polyanions, as demonstrated in Fig. 4, thereby resulting in more stable polyplexes and higher uptake by DCs through phagocytosis. Moreover, the uptake of PEG-b-PAEM/Dex was less than PEI/Dex, presumably due to neutral PEGylated particle surface, but more importantly, it was much higher than Dex alone (Fig. 6, lower panel). The latter finding is encouraging, because it suggests that PEG-b-PAEM could be a useful material for ex vivo DC therapy where enhanced loading of DCs with DNA vaccine or protein antigen without causing cytotoxicity is highly desirable [40]. For in vivo DNA vaccine applications a PEGylated polyamidoamine has been shown recently to be instrumental in generating immune responses in animals against a DNA-encoded antigen delivered via tattooing, whereas the non-PEGylated polyamidoamine was complete ineffective [41]. Therefore, despite their modest in vitro gene transfection efficiency, the PEG-b-PAEM polymers may potentially be advantageous for delivering DNA vaccines to DCs in vivo.
Fig. 6.
In vitro uptake of polyplexes of PEG-b-PAEM and anionic fluorescently-labeled dextran (Dex) in DC 2.4 cells. Gray shaded area: untreated cells.
3.5 DC maturation
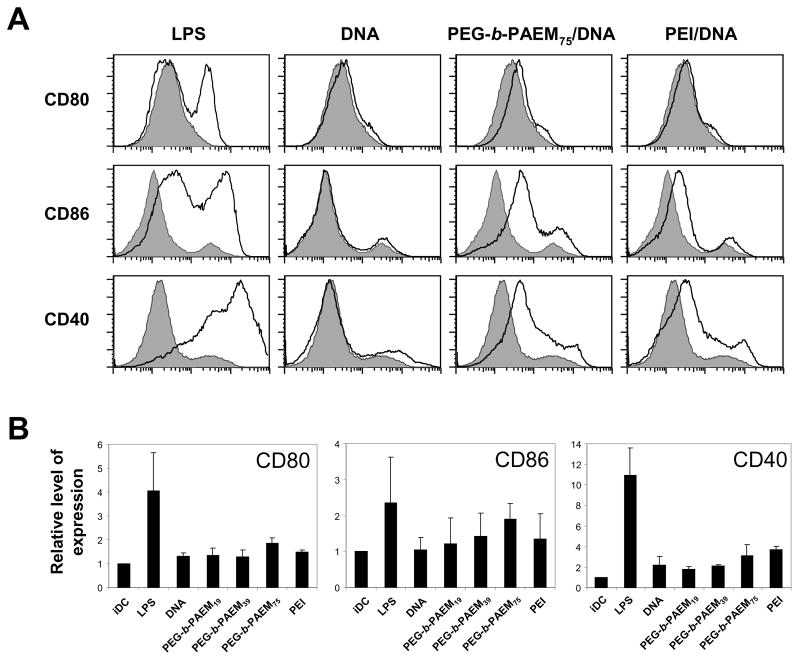
The maturation state of DCs is an important parameter to consider in the context of vaccination and implantable controlled drug delivery. Adjuvants are routinely used in vaccine formulations with one major objective being the phenotypic maturation of antigen-presenting cells [42]. In contrast, achieving perfect biocompatibility of implantable biomaterials demands that DCs remain in the immature state so that the immune system is not activated [42]. Several studies have reported the maturation or the lack of maturation of mouse or human DCs after in vitro culture with various polymeric biomaterials such as biodegradable polyesters and polycations [26,43], with somewhat conflicting conclusions. Here we examined the maturation state of mouse BMDCs after 24-h-culture with polymer/DNA polyplexes in vitro. As shown in Fig. 7, treatment with endotoxin (LPS) up-regulated the expression of CD80, CD86, and CD40 in BMDCs. Naked DNA plasmid did not have any effect on any of the markers, presumably due to low internalization by DCs. Exposure to the PEG-b-PAEM/DNA polyplexes induced a modest up-regulation of the maturation markers, but the correlation between maturation and chain-length of PAEM was not pronounced (Fig. 7). The same was observed for DCs treated with PEI/DNA polyplexes (Fig. 7), suggesting that the surface character of polyplexes does not seem to have much impact on DC maturation. The implication of these findings is two-fold. First, for practical use in DNA vaccine delivery, the PEG-b-PAEM copolymers must be modified to include modalities that trigger DC maturation. Second, the PEG-b-PAEM copolymers may have excellent biocompatibility and could be used safely for controlled delivery in vivo without much concern of immune activation.
Fig. 7.
Expression of co-stimulatory surface markers (CD80, CD86, CD40) in BMDCs treated with polyplexes. Cells treated with LPS were the positive control. Polyplexes were prepared from various polymers and DNA plasmid at N:P ratio of 8:1 and added to cells at a DNA concentration of 20 μg/mL. (A) A representative set of flow cytometry histograms. Gray shaded area: untreated cells. (B) Normalized expression level of maturation markers compiled from three independent experiments.
3.6 In vivo uptake in DCs
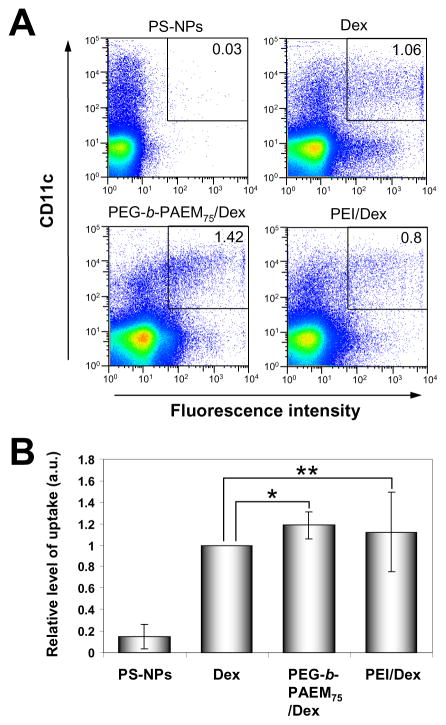
Of the three PEG-b-PAEM copolymers, we selected PEG-b-PAEM75 with the longest PAEM block for further evaluation of in vivo delivery to DCs. To track the uptake of polyplexes, fluorescent Dex was used. Freshly prepared polyplexes were injected subcutaneously in mice. After 24 hours, the mice were sacrificed and the draining lymph nodes were analyzed to determine what fraction of DCs contained fluorescent Dex. Polymer-condensed Dex and non-condensed Dex all drained efficiently into the lymph nodes and were taken up by DCs, whereas 100-nm polystyrene nanoparticles were poorly taken up (Fig. 8A). Importantly, Dex condensed by PEG-b-PAEM75 showed consistently higher uptake by DCs than free Dex (P<0.03) and the average enhancement of uptake was ~20% (Fig. 8B). PEI/Dex, on the other hand, improved DC uptake on average, but failed to achieve statistical significance (P>0.05). These preliminary in vivo results strongly suggest that PEG-b-PAEM copolymers with well-defined structure may be practically useful materials for enhancing the delivery of anionic cargos such as DNA vaccine to dendritic cells.
Fig. 8.
In vivo uptake of polyplexes by DCs in the draining lymph nodes 24 h after s.c. injection in mice. Polyplexes were prepared from PEG-b-PAEM75 or PEI with anionic fluorescently-labeled dextran (Dex). For comparison, Dex alone and 100-nm fluorescent polystyrene nanoparticles (PS-NPs) were also injected. (A) Flow cytometry analysis of a representative experiment showing the percentages of CD11+ DCs in the draining lymph nodes that have taken up fluorescent Dex or PS-NPs. (B) Normalized level of uptake of fluorescent Dex or PS-NPs. Each group consists of four mice. * P<0.03, ** P>0.05, paired t test.
4. Conclusion
We have demonstrated that a simple cationic diblock copolymer, PEG-b-PAEM, with well-defined chemical composition and nearly uniform distribution of chain-length, may serve as a model system for probing the interaction between synthetic DNA vaccine carriers and dendritic cells. We showed that chain-length of the cationic PAEM block had a profound impact on the physico-chemical properties of the polyplexes, including average particle size and stability, as well as biological properties such as cytotoxicity, gene transfection, cellular uptake, and dendritic cell maturation. With further improvement, these diblock copolymers may be practically useful carriers for delivering DNA-based vaccines and therapeutics to dendritic cells.
Supplementary Material
Acknowledgments
The GM-CSF secreting cell line (J558L) was kindly provided by Dr. Kristin Hogquist, Center for Immunology, University of Minnesota. The authors thank Dr. Robert Tranquillo for the use of fluorescence plate-reader. Emily Krogstad thanks the University of Minnesota UROP Program for partial support. This work is partially funded by the NIH (Grants R21CA121832, R01CA129189), the Wallace H. Coulter Foundation (Early Career Translational Research Award), and the US Department of Defense.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liu MA. DNA vaccines: a review. J Internal Med. 2003;253:402–410. doi: 10.1046/j.1365-2796.2003.01140.x. [DOI] [PubMed] [Google Scholar]
- 2.Laddy DJ, Weiner DB. From plasmids to protection: a review of DNA vaccines against infectious diseases. Int Rev Immunol. 2006;25:99–123. doi: 10.1080/08830180600785827. [DOI] [PubMed] [Google Scholar]
- 3.Rice J, Ottensmeier CH, Stevenson FK. DNA vaccines: precision tools for activating effective immunity against cancer. Nature Rev Cancer. 2008;8:108–120. doi: 10.1038/nrc2326. [DOI] [PubMed] [Google Scholar]
- 4.Mellman I, Steinman RM. Dendritic cells: specialized and regulated antigen processing machines. Cell. 2001;106:255–258. doi: 10.1016/s0092-8674(01)00449-4. [DOI] [PubMed] [Google Scholar]
- 5.Reddy ST, Swartz MA, Hubbell JA. Targeting dendritic cells with biomaterials: developing the next generation of vaccines. Trends Immunol. 2006;27:573–579. doi: 10.1016/j.it.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Chan G, Mooney DJ. New materials for tissue engineering: towards greater control over the biological response. Trends Biotechnol. 2008;26:382–392. doi: 10.1016/j.tibtech.2008.03.011. [DOI] [PubMed] [Google Scholar]
- 7.Gilboa E. DC-based cancer vaccines. J Clin Invest. 2007;117:1195–1203. doi: 10.1172/JCI31205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tacken PJ, de Vries IJ, Torensma R, Figdor CG. Dendritic-cell immunotherapy: from ex vivo loading to in vivo targeting. Nature Rev Immunol. 2007;7:790–802. doi: 10.1038/nri2173. [DOI] [PubMed] [Google Scholar]
- 9.Putnam D. Polymers for gene delivery across length scales. Nature Mater. 2006;5:439–451. doi: 10.1038/nmat1645. [DOI] [PubMed] [Google Scholar]
- 10.Luten J, van Nostrum CF, De Smedt SC, Hennink WE. Biodegradable polymers as non-viral carriers for plasmid DNA delivery. J Controlled Release. 2008;126:97–110. doi: 10.1016/j.jconrel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen DN, Green JJ, Chan JM, Langer R, Anderson DG. Polymeric materials for gene delivery and DNA vaccination. Adv Mater. 2008;20:1–21. doi: 10.1002/adma.200801478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.O’Hagan DT, Singh M, Ulmer JB. Microparticles for the delivery of DNA vaccines. Immunol Rev. 2004;199:191–200. doi: 10.1111/j.0105-2896.2004.00153.x. [DOI] [PubMed] [Google Scholar]
- 13.Lisziewicz J, Trocio J, Whitman L, Varga G, Xu J, Bakare N, Erbacher P, Fox C, Woodward R, Markham P, Arya S, Behr JP, Lori F. DermaVir: a novel topical vaccine for HIV/AIDS. J Invest Dermatol. 2005;124:160–169. doi: 10.1111/j.0022-202X.2004.23535.x. [DOI] [PubMed] [Google Scholar]
- 14.Locher CP, Putnam D, Langer R, Witt SA, Ashlock BM, Levy JA. Enhancement of a human immunodeficiency virus env DNA vaccine using a novel polycationic nanoparticle formulation. Immunol Lett. 2003;90:67–70. doi: 10.1016/j.imlet.2003.02.001. [DOI] [PubMed] [Google Scholar]
- 15.Roy K, Mao HQ, Huang SK, Leong KW. Oral gene delivery with chitosan--DNA nanoparticles generates immunologic protection in a murine model of peanut allergy. Nature Med. 1999;5:387–391. doi: 10.1038/7385. [DOI] [PubMed] [Google Scholar]
- 16.Greenland JR, Liu H, Berry D, Anderson DG, Kim WK, Irvine DJ, Langer R, Letvin NL. β-amino ester polymers facilitate in vivo DNA transfection and adjuvant plasmid DNA immunization. Mol Ther. 2005;12:164–170. doi: 10.1016/j.ymthe.2005.01.021. [DOI] [PubMed] [Google Scholar]
- 17.Vilalta A, Mahajan RK, Hartikka J, Rusalov D, Martin T, Bozoukova V, Leamy V, Hall K, Lalor P, Rolland A, Kaslow DC. I. Poloxamer-formulated plasmid DNA-based human cytomegalovirus vaccine: evaluation of plasmid DNA biodistribution/persistence and integration. Human Gene Ther. 2005;16:1143–1150. doi: 10.1089/hum.2005.16.1143. [DOI] [PubMed] [Google Scholar]
- 18.Rettig GR, Rice KG. Non-viral gene delivery: from the needle to the nucleus. Expert Opin Biol Ther. 2007;7:799–808. doi: 10.1517/14712598.7.6.799. [DOI] [PubMed] [Google Scholar]
- 19.Kuroda K, DeGrado WF. Amphiphilic polymethacrylate derivatives as antimicrobial agents. J Am Chem Soc. 2005;127:4128–4129. doi: 10.1021/ja044205+. [DOI] [PubMed] [Google Scholar]
- 20.Jankova K, Chen X, Kops J, Batsberg W. Synthesis of amphiphilic PS-b-PEG-b-PS by atom transfer radical polymerization. Macromolecules. 1998;31:538–541. [Google Scholar]
- 21.Dufresne MH, Leroux JC. Study of the micellization behavior of different order amino block copolymers with heparin. Pharm Res. 2004;21:160–169. doi: 10.1023/b:pham.0000012164.60867.c6. [DOI] [PubMed] [Google Scholar]
- 22.Mosmann T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 23.Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S, Muramatsu S, Steinman RM. Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony-stimulating factor. J Exp Med. 1992;176:1693–1702. doi: 10.1084/jem.176.6.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Matyjaszewski K, Xia JH. Atom transfer radical polymerization. Chem Rev. 2001;101:2921–2990. doi: 10.1021/cr940534g. [DOI] [PubMed] [Google Scholar]
- 25.He L, Read ES, Armes SP, Adams DJ. Direct synthesis of controlled-structure primary amine-based methacrylic polymers by living radical polymerization. Macromolecules. 2007;40:4429–4438. [Google Scholar]
- 26.Jilek S, Merkle HP, Walter E. DNA-loaded biodegradable microparticles as vaccine delivery systems and their interaction with dendritic cells. Adv Drug Delivery Rev. 2005;57:377–390. doi: 10.1016/j.addr.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 27.Manolova V, Flace A, Bauer M, Schwarz K, Saudan P, Bachmann MF. Nanoparticles target distinct dendritic cell populations according to their size. Eur J Immunol. 2008;38:1404–1413. doi: 10.1002/eji.200737984. [DOI] [PubMed] [Google Scholar]
- 28.Mann A, Richa R, Ganguli M. DNA condensation by poly-L-lysine at the single molecule level: role of DNA concentration and polymer length. J Controlled Release. 2008;125:252–262. doi: 10.1016/j.jconrel.2007.10.019. [DOI] [PubMed] [Google Scholar]
- 29.Itaka K, Yamauchi K, Harada A, Nakamura K, Kawaguchi H, Kataoka K. Polyion complex micelles from plasmid DNA and poly(ethylene glycol)-poly(L-lysine) block copolymer as serum-tolerable polyplex system: physicochemical properties of micelles relevant to gene transfection efficiency. Biomaterials. 2003;24:4495–4506. doi: 10.1016/s0142-9612(03)00347-8. [DOI] [PubMed] [Google Scholar]
- 30.Dubruel P, Schacht E. Vinyl polymers as non-viral gene delivery carriers: current status and prospects. Macromol Biosci. 2006;6:789–810. doi: 10.1002/mabi.200600110. [DOI] [PubMed] [Google Scholar]
- 31.Dufresne MH, Elsahahy M, Leroux JC. Characterization of polyion complex micelles designed to address the challenges of oligonucleotide delivery. Pharm Res. 2008;25:2083–2093. doi: 10.1007/s11095-008-9591-6. [DOI] [PubMed] [Google Scholar]
- 32.Godbey WT, Wu KK, Mikos AG. Size matters: molecular weight affects the efficiency of poly(ethylenimine) as a gene delivery vehicle. J Biomed Mater Res. 1999;45:268–275. doi: 10.1002/(sici)1097-4636(19990605)45:3<268::aid-jbm15>3.0.co;2-q. [DOI] [PubMed] [Google Scholar]
- 33.Kunath K, von Harpe A, Fisher D, Petersen H, Bickel U, Voigt K, Kissel T. Low-molecular-weight polyethylenimine as a non-viral vector for DNA delivery: comparison of physicochemical properties, transfection efficiency and in vivo distribution with high-molecular-weight polyethylenimine. J Controlled Release. 2003;89:113–125. doi: 10.1016/s0168-3659(03)00076-2. [DOI] [PubMed] [Google Scholar]
- 34.Wolfert MA, Seymour LW. Atomic force microscopic analysis of the influence of the molecular weight of poly(L)lysine on the size of polyelectrolyte complexes formed with DNA. Gene Ther. 1996;3:269–273. [PubMed] [Google Scholar]
- 35.van de Wetering P, Cherng JY, Talsma H, Hennink WE. Relation between transfection efficiency and cytotoxicity of poly(2-(dimethylamino)ethyl methacrylate)/plasmid complexes. J Controlled Release. 1997;49:59–69. [Google Scholar]
- 36.Layman JM, Ramirez SM, Green MD, Long TE. Influence of polycation molecular weight on poly(2-dimethylaminoethyl methacrylate)-mediated DNA delivery in vitro. Biomacromolecules. 2009;10:1244–1252. doi: 10.1021/bm9000124. [DOI] [PubMed] [Google Scholar]
- 37.Huang M, Fong CW, Khor E, Lim LY. Transfection efficiency of chitosan vectors: effect of polymer molecular weight and degree of deacetylation. J Controlled Release. 2005;106:391–406. doi: 10.1016/j.jconrel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 38.Akinc A, Anderson DG, Lynn DM, Langer R. Synthesis of poly(β-amino ester)s optimized for highly effective gene delivery. Bioconjugate Chem. 2003;14:979–988. doi: 10.1021/bc034067y. [DOI] [PubMed] [Google Scholar]
- 39.Cho YW, Kim JD, Park K. Polycation gene delivery systems: escape from endosomes to cytosol. J Pharm Pharmacol. 2003;55:721–734. doi: 10.1211/002235703765951311. [DOI] [PubMed] [Google Scholar]
- 40.Steinman RM, Banchereau J. Taking dendritic cells into medicine. Nature. 2007;449:419–426. doi: 10.1038/nature06175. [DOI] [PubMed] [Google Scholar]
- 41.van den Berg JH, Oosterhuis K, Hennink WE, Storm G, van der Aa LJ, Engbersen JF, Haanen JB, Beijnen JH, Schumacher TN, Nuijen B. Shielding the cationic charge of nanoparticle-formulated dermal DNA vaccines is essential for antigen expression and immunogenicity. J Controlled Release. 2009 Sep 12; doi: 10.1016/j.jconrel.2009.09.005. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 42.Apostolopoulos V. Methods of delivery to antigen-presenting cells: development of new and improved vaccines. Mol Pharm. 2007;4:1–3. doi: 10.1021/mp060102h. [DOI] [PubMed] [Google Scholar]
- 43.Babensee JE. Interaction of dendritic cells with biomaterials. Sem Immunol. 2008;10:101–108. doi: 10.1016/j.smim.2007.10.013. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.