Abstract
How organ size is controlled in mammals is not currently understood. In Drosophila the Hippo signaling pathway functions to suppress growth in imaginal discs and has been suggested to control organ size. To investigate the role of hippo signaling in regulation of mammalian organ size we have generated conditional alleles of Sav1, mst1, and mst2, orthologs of Drosophila Salvador and hippo, respectively. Specific deletion of both mst1 and mst2 in hepatocytes results in significantly enlarged livers due to excessive proliferation. By the age of 5–6 months, mst1/2 conditional mutant livers have multiple foci of liver tumors, indicating that the combined activities of mst1 and mst2 act as redundant tumor suppressors in hepatocytes. Similar findings were obtained with liver-specific deletion of Sav1, a second core Hippo signaling component that facilitates activation of mst1 and mst2. Tumors from sav1 mutants exhibited varied morphology, suggesting a mixed-lineage origin of tumor-initiating cells. Transcriptional profiling of liver tissues from both mst1/2 and sav1 conditional mutants revealed a network of Hippo signaling regulated genes with specific enrichment for genes involved in immune and inflammatory responses. Histological and immunological characterization of mst1/2 double mutant liver tissues revealed abundant accumulation of adult facultative stem cells termed oval cells in periductal regions. Because oval cells induction is commonly associated with liver injury and tumor formation, it is likely that these cells contribute to the enlarged livers and hepatomas that we observe in sav1 and mst1/2 mutants. Taken together, our results demonstrate that the Hippo signaling pathway is a critical regulator of mammalian liver growth and a potent suppressor of liver tumor formation.
Keywords: hepatocellular carcinoma, oval cell response, Hippo signaling
The Hippo signaling pathway has recently been identified in Drosophila as an essential regulator of cell proliferation and apoptosis during development (1, 2). Key components of the hippo pathway include two kinases, hippo and warts that function in a cascade to phosphorylate the transcriptional activator protein yorkie, resulting in retention of yorkie in the cytoplasm. When Hippo signaling is attenuated, yorkie phosphorylation is reduced or absent, leading to its nuclear localization, binding to the sequence-specific DNA-binding protein scalloped and regulation of target genes. In Drosophila, transcriptional targets of hippo signaling include cyclinE and diap1 that promote proliferation and survival, respectively. Hence, Hippo signaling coordinately regulates organ growth by affecting rates of cell division and apoptosis.
In mammals, each component of the core Drosophila Hippo signaling cascade has at least one conserved ortholog and biochemical studies indicate that they function in a similar manner to affect nuclear versus cytoplasmic localization of the mammalian yorkie orthologs yap and taz (3, 4). Furthermore, studies in cell culture and in vivo suggest that nuclear localization of yap and taz drives cell proliferation and survival, consistent with a conserved role for Hippo signaling in regulating organ size in mammals.
Despite conservation of biochemical function and the ability of nuclear yap to drive cell proliferation and survival, little is known about the requirements for mammalian Hippo signaling during normal development and in regulating organ size. Previous loss-of-function studies have been hampered by early lethality of mammalian core Hippo signaling pathway mutants or by potential redundancy between individual orthologs (5 –9). Hence, whether Hippo signaling is normally required to regulate mammalian organ size, to repress proliferation and promote apoptosis, and to suppress tumor formation is not known.
Here we employ a conditional mutagenesis strategy in mice to address the function of core Hippo signaling pathway components in regulation of organ size and in repressing tumor formation. Specifically, we have used conditional alleles of mammalian orthologs of hippo, the serine-threonine kinases mst1 and mst2, to address requirements for hippo signaling in the postnatal liver, a tissue previously shown to be responsive to elevated levels of nuclear yap. Our results indicate that in the liver the combined activities of mst1 and mst2 are required to repress proliferation of mature hepatocytes, to prevent activation of facultative adult liver stem cells (oval cells), and to inhibit tumor formation. Additionally, we show using a conditional allele of sav1, that an adaptor that potentiates mst1 and mst2 activity is likewise required to suppress growth in the mature liver and to prevent tumor formation. Hence, taken together, our results define previously undescribed essential functions for mammalian hippo signaling in regulation of organ size, cell proliferation and survival, and tumor suppression.
Results and Discussion
Hippo Signaling Is Required to Suppress Liver Growth In Vivo.
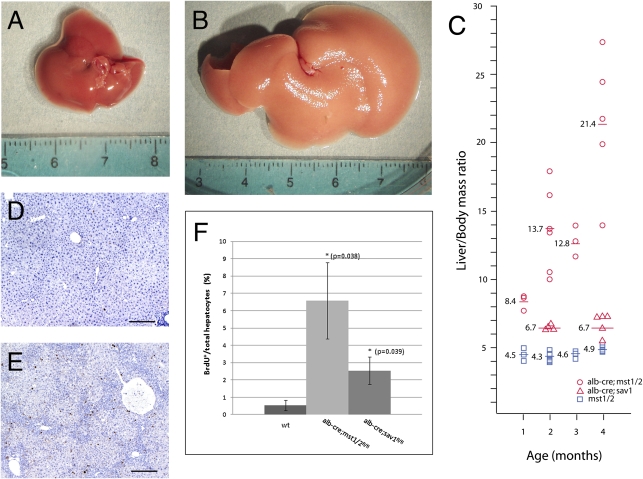
To investigate the role of mammalian hippo signaling in vivo we generated mice that selectively inactivate the hippo serine–threonine kinase orthologs mst1 and mst2 in hepatocytes, using albumin-cre (10). Combined liver-specific removal of mst1 and mst2 (hereafter referred to as mst1/2 double mutants or mst1/2 mutants) resulted in progressive hepatomegaly with a 2-fold increase in liver mass relative to total body mass at 1 month of age and a 3-fold increase by 3 months of age (Fig. 1 A–C, Fig. S1). Similar results were obtained for sav1 conditional mutants (hereafter referred to as sav1 mutants), with a modest, but significant 10% increase in liver size by 4 months of age (Fig. 1C). Deletion of mst1 or mst2 alone did not affect liver size, indicating redundancy for these two closely related kinases in regulating the proliferation and growth of hepatocytes. The increased liver mass in mst1/2 double mutants and sav1 mutants was associated with an increased proliferation index (Fig. 1 D–F). By 3 months of age the majority of liver hepatocytes in wild-type mice are not undergoing cell cycling as evidenced by the small numbers of cells that incorporate BrdU after a 24-h pulse. In contrast, mst1/2 double mutants exhibit marked elevated levels of hepatocyte BrdU incorporation, indicating the combined activities of mst1 and mst2 repress cell cycle entry of hepatocytes and maintain their quiescence in the mature liver. Increased BrdU incorporation was also observed in Salvador mutant livers, although to a lesser extent (Fig. 1F). Modest increases in cell death were observed relative to wild type in both mst1/2 double and sav1 mutant hepatocytes (Fig. S2). These findings suggest that enhanced hepatocyte proliferation drives excessive growth upon reduction of Hippo signaling in the mammalian liver.
Fig. 1.
Mst1 and mst2 are required to restrict liver growth and to maintain hepatocyte quiescence. In contrast to wild type (A), albumin-cre;mst1/2 double mutants (B) have significantly enlarged livers at 2 months of age. (C) A plot of liver/body as a function of age shows increased liver size of mst1/2 mutants at 1 month. Continued growth is evident at 2 and 3 months of age. Modest increases in albumin-cre;sav1 mutant liver sizes are seen at 2 and 4 months of age. Few wild-type hepatocytes incorporate BrdU at 2 months of age (D), although albumin-cre;mst1/2 mutants show abundant BrdU incorporation (E). (F) Quantification of BrdU labeling. Albumin -cre;mst1/2 double mutants and albumin-cre;sav1 mutants exhibit 12-fold and 5-fold increases in BrdU incorporation respectively.
Reduced Phosphorylation of lats1/2 and yap in mst1/2 Double Mutant Hepatocytes.
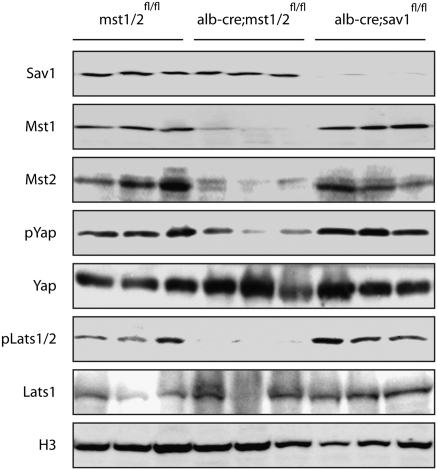
The Hippo signaling pathway as defined in Drosophila and in mammalian tissue culture is a kinase cascade with mst1/2 functioning to phosphorylate lats1/2, which in turn phosphorylates yap, resulting in its cytoplasmic retention by 14-3-3 proteins (3, 4). To determine whether the Hippo kinase cascase operates in vivo in mammalian hepatocytes, we examined the phosphorylation status of lats1/2 and yap in mst1/2 and sav1 double mutants (Fig. 2). Western analysis demonstrates significant reduction of mst1 and mst2 protein levels in mst1/2 double mutants and of sav1 in sav1 mutants, indicating efficient deletion of floxed alleles in hepatocytes using albumin-cre. Relative to wild type, yap and lats1/2 phosphorylation is significantly reduced in mst1/2 double mutants whereas the levels of unphosphorylated yap and lats2 are relatively unaffected. In contrast, levels of phospho-yap or phospho-lats1/2 are not affected in sav1 mutants, suggesting either that sav1 is not required in hepatocytes for mst1/2 kinase activity or that additional relevant substrates are affected by sav1 deletion. Alternatively, mst1/2 kinase activity may depend on sav1 in a context-dependent manner and is not required at the time points analyzed. Nevertheless, reduced phosphorylation of yap and lats1/2 in mst1/2 double mutants is consistent with an in vivo requirement for mst1/2 kinase activity in activation of the Hippo signaling pathway in hepatocytes.
Fig. 2.
Western analysis of albumin-cre;sav1 and albumin-cre;mst1/2 double mutant tissues. Cells enriched for hepatocytes show loss of sav1 protein in albumin-cre;sav1 mutants and reduced amounts of mst1 and mst2 proteins in albumin-cre;mst1/2 mutants. Each lane represents proteins extracted from independent mutant livers. Phosphorylation of Yap and Lats is reduced in albumin-cre;mst1/2 double mutant hepatocytes but not in albumin-cre;sav1 mutant cells. Histone H3 (H3) was used as a loading control.
Hippo Signaling Is a Potent Tumor Suppressor in the Mammalian Liver.
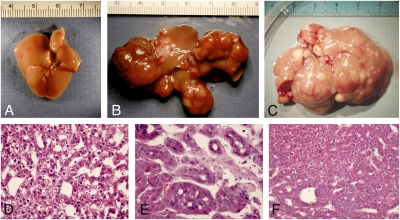
To determine whether Hippo signaling functions as a tumor suppressor pathway in the mammalian liver, we examined sav1 and mst1/2 double conditional mutant mice for signs of liver tumorigenesis. At 13–14 months of age all albumin-cre;sav1 floxed mice (n = 7) examined had developed large liver tumors (Fig. 3B) with histological characteristics of both hepatocellular carcinoma and cholangiocarcinoma (Fig. 3 D and E). No control mice of similar age had liver tumors (n = 8). The tumors formed at multiple foci, indicating that multiple independent initiating events had likely occurred in Salvador mutant mice. We also observed tumors in sav1 mice with independent cre drivers, including MMTV-cre (11) (n = 4) and caggs-creER(T2) (12) (n = 5). Both MMTV-cre and caggs-creER(T2) are reported to have widespread constitutive activity, including low levels in the liver. Like the albumin-cre;sav1 floxed mice, all animals examined had liver tumors by 14 months of age, indicating that the tumor phenotype is highly penetrant and independent of genetic background. We did not observe tumors in other tissues of either MMTV-cre or caggs-creER(T2);sav1 mutant mice, suggesting a degree of specificity for sav1 tumor suppressive activity. In the case of mst1/2 double mutants (n = 3), livers were highly enlarged and had multiple focal tumors visible on gross examination at 5–6 months of age (Fig. 3C). Histological examination revealed a disorganized parenchyma with the majority of the liver exhibiting a dysplastic partially transformed phenotype. The histopathology of tumor nodules was consistent with a diagnosis of hepatocellular carcinoma (Fig. 3F). To determine the age of onset of liver tumors in mst1/2 double mutants, we killed animals at earlier time points. At 1 and 2 months of age no tumors were observed (n = 9). By 3 months of age one or two small tumors (<5 mm) could be detected in two of three mice examined. By 4 months of age all mice (n = 5) had one or more tumors. Hence, the majority of mst1/2 mutants are likely to develop liver tumors by 4 months of age. Taken together, our results demonstrate a potent tumor suppressor role for Hippo signaling in the mammalian liver.
Fig. 3.
Hepatoma formation in Hippo pathway mutants. Wild-type livers display a normal appearance, devoid of tumor foci (A). In contrast, conditional deletion of sav1 in hepatocytes results in large, multifocal tumors (B). Likewise, albumin-cre;mst1/2 mutant livers are significantly enlarged relative to wild type and display multiple focal tumor nodules (C). Histological examination (D and E) reveals both well and poorly differentiated hepatocellular carcinoma (D and F) and cholangiocarcinoma (E).
Transcriptional Profiling of sav1 and mst1/2 Mutant Tissues Reveals Common Hippo Pathway Targets.
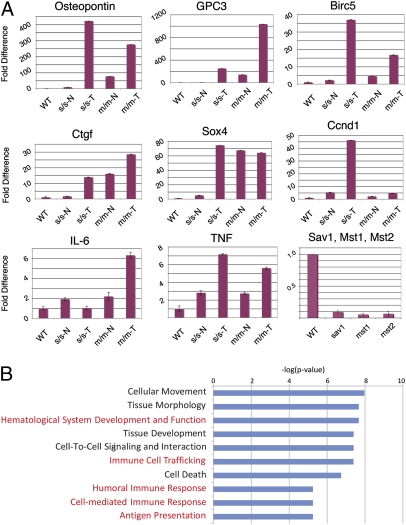
To gain insight into transcriptional regulation by Hippo signaling in the mammalian liver we performed microarray analysis of tissues from sav1 and mst1/2 mutants, including both normal and tumor tissues. Comparison of the expression profiles of 18 sav1, 7 mst1/2 mutant, and 8 matched wild-type controls revealed 2,899 features that displayed a significant difference between wild-type and mutant samples (Fig. S3A). To validate and extend the microarray data set, we determined the relative transcript levels for six genes that were previously shown to be up-regulated by liver-specific overexpression of a constitutively active mutant yap protein (13) by quantitative RT-PCR. All transcripts were up-regulated in mutant versus wild-type tissues (Fig. 4A). For example, osteopontin was up-regulated 9-fold in sav1 mutant tissue relative to wild type and >400-fold in tumor tissue. In mst1/2 mutants we found a 77-fold increase in osteopontin transcripts in mutant tissue at 2 months of age and a 275-fold increase by 5 months of age. Similar observations were found for glypican-3 (gpc3), sox4, connective tissue growth factor (ctgf), birc5 (survivin), and cyclin D1 (ccnd1). Additionally, quantative RT-PCR analysis confirms reduction of sav1, mst1, and mst2 transcripts (Fig. 4A) in hepatocytes isolated from mutant hepatocytes. Hence, the transcriptional profile of both sav1 and mst1/2 double mutants is consistent with activation of targets of the downstream transcriptional activator yap.
Fig. 4.
Transcriptional analysis of sav1 and mst1/2 mutant liver tissue. (A) Quantitative RT-PCR analysis of gpc3, osteopontin, ctgf, sox4, survivin, cyclin D1, IL-6, and TNF-α validates and extends the microarray data indicating differential expression of these transcripts. Reduction of sav1, mst1, and mst2 transcript levels is readily apparent in hepatocyte-enriched cell fractions from albumin-cre;sav1 and albumin-cre;mst1/2 mutants. (B) Gene set enrichment analysis reveals overrepresentation of transcripts involved in immune response in Hippo pathway mutant tissues.
Transcripts Affected by Reduced Hippo Signaling in Hepatocytes Are Involved in Cell Movement and Immune Response.
Gene set enrichment analysis of differentially regulated transcripts in either sav1 or mst1/2 mutant tissues revealed an unexpected overrepresentation of genes involved in cell movement and immune response (Fig. 4B). This finding is consistent with either an inflammatory response or an oval cell response, both of which occur following liver injury in situations where hepatocyte proliferation is attenuated (14 –16). To explore this possibility further at the transcriptional level, we determined the relative expression of interleukin-6 (IL-6) and tumor necrosis factor alpha (TNF-α, Fig. 4A), both of which have been implicated in oval and inflammatory responses in the liver (17 –20). For both IL-6 and TNF-α we observe modest, but significant increases in transcript level consistent with a generalized inflammatory and/or oval cell response (Fig. 3B).
Hippo Signaling Is Required to Repress Adult Liver Stem Cell Activation.
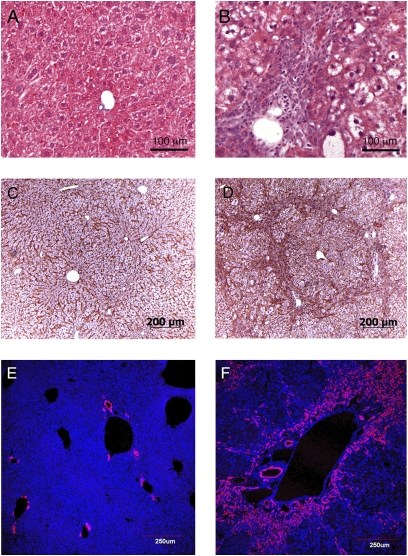
To determine whether an inflammatory and/or oval cell response occurs upon reduction of Hippo signaling in the mammalian liver we initially compared the histological appearance of livers from wild-type mice with that of livers from mice that contained a combined mst1 and mst2 deletion (Fig. 5). At 1 month after birth, the normal liver is composed of regularly spaced hepatocytes that line the liver sinusoids. In contrast, mst1/2 double mutants have an irregular arrangement of hepatocytes that obscures the normal radiating pattern of liver sinusoids (Fig. 5 A–D). A second prominent feature of mst1/2 double mutant tissues is the presence of hepatocytes that exhibit a clear cell phenotype (Fig. 5B). Abundant hepatocytes with clear cytoplasm might suggest altered glycogen and/or lipid metabolism. However, Oil red O staining revealed no difference between wild-type and mst1/2 double mutant livers, indicating that accumulation of long chain fatty acids is unlikely to account for the abnormal appearance of mst1/2 mutant hepatocytes. Periodic acid Schiff staining was similar between wild-type and mutant mice, suggesting that glycogen metabolism is not severely altered in mst1/2 mutant hepatocytes. A third feature of mst1/2 double mutants is the presence of large numbers of small periductal cells that are visible at 2 months of age (Fig. 5B). These cells appear to be highly invasive and by 2 months have infiltrated extensively into the liver parenchyma and by 3 months surround the hepatic plates (Fig. 5D). Taken together, these histological findings are consistent with chronic liver damage. However, we did not detect elevated serum levels of alanine aminotransferase (ALT) and aspartate aminotransferase (AST) at 2 months of age in mst1/2 double mutants, indicating that although there are histological features indicative of liver damage, overall liver function is not significantly affected at this stage.
Fig. 5.
Histology and oval cell response in albumin-cre;mst1/2 double mutants are consistent with chronic liver injury. The normal liver (A) is composed of regularly spaced plates of hepatocytes surrounding the portal vein. In contrast, albuin-cre;mst1/2 mutants display a disorganized arrangements of hepatocytes, many of which have an abnormal clear cytoplasm with H&E stain. Many small periductal cells are visible that are not present in wild-type liver sections. Overall hepatocyte disorganization and infiltrating periductal cells are prominently visualized with cell membrane β-catenin staining of wild type (C) versus mutant (D). The A6 antibody, a marker for oval and ductal cells, stains only ductal cells in wild-type (E) tissues, but stains both ductal cells and periductal cells in albumin-cre;mst1/2 double mutants (F).
The histological appearance of small proliferating periductal cells is consistent with several possibilities: an inflammatory response, a fibrotic response, an oval cell response, or a combination of these responses. Trichrome and alpha-smooth muscle actin immunostaining were negative in this cell population, indicating that the infiltrating periductal cells are not mounting a fibrotic response at 1–3 months of age. F4/80 antibody staining was moderately elevated in mutant tissues, suggesting that macrophage accumulation may contribute to periductal infiltration (Fig. S4). In contrast, abundant and intense staining was obtained for antigens that are expressed in oval cells (Fig. 5 E and F and Fig. S5). Oval cells are not normally present in healthy liver, but are induced in response to injury when hepatocyte proliferation is blocked (15, 16). They are facultative stem cells that can contribute to both the hepatocyte and biliary lineages. At present no single antibody can unambiguously define murine oval cells, likely due to their heterogeneity. However, most murine oval cells are positive for ductal markers such as M1C1-1C3 and many stain with the OC2-1D11 and A6 antibodies (21, 22). Using this panel of antibodies, we consistently observe intense staining of periductal cells in mst1/2 mutant livers, indicating that mst1/2 signaling is required to repress oval cell induction and proliferation. Similar findings were found for A6 staining of sav12 mutant tissue (Fig. S5). Hence, we conclude that Hippo signaling represses oval cell activation in the murine liver.
Regulation of Liver Growth and Tumor Suppressor Activity of the Mammalian Hippo Signaling Pathway.
Deletion of the core Hippo signaling components sav1 and combined deletion of mst1 and mst2 result in enhanced liver size and in hepatocellular carcinoma. Hence our results demonstrate a previously undescribed essential requirement for Hippo signaling in maintaining hepatocytes in a quiescent state, in modulating liver size, and in suppressing tumor formation. Additionally, we provide evidence that Hippo signaling is required to repress facultative stem cell activation in the mature liver. These results are significant because although previous studies have indicated that overexpression of an activated form of yap is sufficient to induce liver growth and tumor formation (13, 23), whether hippo signaling is normally required to repress liver growth and tumor formation has not been previously addressed.
In principle, Hippo signaling might modulate liver size by acting to repress growth when the organ achieves a mature size. Alternatively, Hippo signaling may act constitutively to repress hepatocyte proliferation. In the mouse, the liver grows significantly during the postnatal period along with the growth of the animal (24) although the liver/body weight ratio remains relatively constant (Fig. 1C). Our results indicate that Hippo signaling is required both to restrict liver growth during the proliferative postnatal phase and to maintain hepatocyte quiescence in the adult. Hence, Hippo signaling is likely active at all stages and is required to repress liver growth not only when the mature liver size is achieved, but also during growth in the perinatal period.
Whereas Hippo signaling is clearly a critical regulator of liver size, the detailed molecular mechanism of how Hippo signaling regulates this process is not clear, nor is it known how attenuation of Hippo signaling leads to tumor formation. Enhanced hepatocyte proliferation is likely central to increased liver size in both mst1/2 and sav1 liver conditional mutant mice. Most likely this effect is mediated by a decreased phosphorylation and enhanced nuclear localization of yap as overexpression of a constitutive form of YAP that cannot be phosphorylated by lats kinases results in reversible hepatomegaly (13, 23). Several genes that promote proliferation are up-regulated in sav1 and mst1/2 mutants as well as in yap-overexpressing livers, consistent with the proposal that both sav1 and mst1/2 act through phosphorylation of yap. Whether sav1 and/or mst1/2 have effects independent of YAP phosphorylation is not known at this time and will require further experimentation. Inhibition of apoptosis is unlikely to contribute significantly to enhanced liver size in hippo pathway mutants as levels of apoptosis in wild-type and Hippo pathway mutant liver tissues are generally very low. Our transcriptional profiling studies and those of Dong et al. (13) reveal several shared transcripts that are overexpressed when Hippo signaling is reduced that may be relevant to liver disease progression in Hippo pathway mutants. Glypican-3, osteopontin, and sox4 are abundantly expressed in sav1 and mst1/2 mutant livers as well as following overexpression of yap. As glypican-3, osteopontin, and sox4 expression is commonly elevated in human cancers (25 –27), including hepatocellular carcinoma, their up-regulation may contribute significantly to tumor formation and/or progression in both sav1 and mst1/2 mouse mutants.
The histological appearance and presence of an abundant oval cell response in mst1/2 double mutant liver tissue are suggestive of a response to chronic liver damage (16). This observation is of particular interest for several reasons. First, an abundant oval cell response is likely to contribute to enhanced liver tumor formation in mice with attenuated hippo signaling activity. Oval cells are bipotental hepatic progenitor cells that are induced following carcinogen exposure in experimental models of hepatocellular carcinoma and are thought to function as cancer stem cells in these models (28, 29). That we observe tumors of both biliary and hepatocyte lineages in sav1 mutant mice is consistent with an important contribution of oval cell activation in tumor formation. Second, an oval cell response is typically initiated only when hepatocytes are unable to respond to injury through compensatory proliferation, suggesting that proliferating hepatocytes actively repress oval cell activation (15). The signal or signals that mediate this inhibition are not known. In the case of mst1/2 double mutants, an abundant oval cell response is apparent despite elevated hepatocyte proliferation, suggesting that Hippo signaling directly or indirectly suppresses a hepatocyte-derived signal that functions to repress oval cell activation. Alternatively or additionally, Hippo signaling might be required in oval cells to repress their proliferation and expansion. At present we cannot unambiguously distinguish between these possibilities as albumin-cre marks both oval cell and hepatocyte lineages (30, 31).
Taken together, our results define a critical role for Hippo signaling in modulating liver size and in suppressing tumor formation in mice. In addition, we provide evidence that Hippo signaling regulates progenitor cell expansion in the liver by repressing oval cell activation. The combined effects of Hippo signaling on repressing hepatocyte and oval cell proliferation likely contribute significantly to liver tumor formation in mice that have attenuated Hippo signaling activity and may help explain our observations that sav1 mutant mice selectively develop liver tumors with several independent and widely expressed cre drivers. Our findings are also relevant to human liver cancer in that a majority of human hepatocellular carcinomas have elevated levels of nuclear YAP localization (32), indicative of attenuated Hippo signaling in these tumors. It is likely that abnormal YAP localization in these tumors reflects attenuation of the activities of key core components of hippo signaling. The definition of Hippo signaling as a potent suppressor of liver growth and tumorigenesis will allow further definition of pathways that operate together with Hippo signaling to drive tissue growth during development and regeneration as well as to discover collaborating pathways that lead to tumor formation.
Methods
Generation and Breeding of sav1 fl/fl, mst1 fl/fl, and mst2 fl/fl Mice.
Targeting constructs were prepared by subcloning of PCR-amplified DNA from BAC clones (Children’s Hospital Oakland Research Institute, BacPac Resources) containing sav1, mst1, and mst2. The strategy for construction of the targeting vectors is detailed in Fig. S1. Vectors were electroporated into C57BL/6;129SvEvTac hybrid RJ2.2 cells (available on request) and targeted clones were identified by Southern hybridization with external probes. Chimeras were generated by aggregation with CD1 morulae as described (33). Germline transmission was achieved by mating high percentage chimeric males to CD1 females and assaying pups for coat color and presence of the targeted allele. FRT-flanked neo cassettes were removed in vivo by crossing to Flpe deleter mice (34). Mice were subsequently bred to albumin-cre (10), MMTV-cre (11), and Caggs-creER(T2) (12) mice followed by backcrossing to homozygous floxed animals. All mice were housed in a conventional facility with a 12-h light/dark schedule and access to food and water ad libidum. All procedures were approved by the University of Texas, M. D. Anderson Cancer Center Animal Care and Use Committee.
Histology, BrdU labeling, and Immunostaining.
Liver tissues were fixed in 4% paraformaldehyde overnight at 4°C and processed for paraffin embedding. Paraffin sections from each sample were cut at 5 μm and stained with H&E. For labeling of cells undergoing DNA synthesis in vivo, 0.01 mL/g body weight of a 3 mg/mL solution of BrdU (Sigma, B9285) in PBS was injected i.p. 24 h before euthanizing the mice. BrdU staining was carried out using the BrdU In Situ Detection Kit (BD Pharmingen, 550803). For quantification of BrdU-positive hepatocytes, five different areas in each sample were photographed and counted. The result was statistically analyzed by Student’s t test. Immunofluorescent staining was performed as previously described (21). Briefly, freshly isolated liver tissues were embedded in optimal cutting temperature (OCT) embedding medium without fixation and samples were frozen in an isopentane/liquid nitrogen slurry. Sections were cut at 6 μm and fixed in cold acetone (−20°C) for 10 min before applying the primary antibodies. The A6 antibody was obtained from Valentina Factor (National Institutes of Health) and the OC2-1D11 and MIC1-1c3 antibodies were provided by Craig Dorrell (Oregon Health Sciences Center). Visualization of primary antibody staining was achieved with Cy3-conjugated goat anti-rat (Jackson Immunoresearch, 112-165-167) and imaged with confocal microscopy. F4-80 (BD Biosciences) was used for macrophage stainings. β-Catenin (Santa Cruz Biotechnology, sc-7199) staining was performed on paraffin sections using peroxidase-conjugated anti-rabbit secondary antibodies (ImmPRESS Anti-Rabbit Ig Kit; Vector Laboratories, MP-7401), followed by 3,3′-diaminobenzidine (DAB) (Vector Laboratories, SK-4105) to visualize the peroxidase activity. Sections were then counterstained and Permount sealed for photography.
Hepatocyte Isolation and Western Analysis.
A parenchymal cell fraction enriched in hepatocytes was prepared using a standard two-step perfusion method (35). These cells were lysed and extacts prepared and processed according to published procedures (36). Briefly, Western blots were run on a 10% acrylamide gel. The Western blots were blocked in 5% milk in TBST and antibodies were incubated in 5% BSA in tris-buffered saline with tween-20 (TBST). Antibodies used were Sav1 (1:500, Cell Signaling), Mst1 (1:500, Cell Signaling), Mst2 (1:500, Cell Signaling), Phospho-YAP (Ser127) (1:500, Cell Signaling), YAP (1:500, Cell Signaling), Phospho-LATS1 (Ser909) (1:500, Cell Signaling), LATS1 (C66B5) (1:500, Cell Signaling), and Histone H3 (1:1000, Cell Signaling).
Microarray and Quantitative RT-PCR Analysis.
Liver samples were dissected into ≈30-mg cubes and stored in RNAlater stabilization reagent (Qiagen) until further processing. Total RNA was isolated from mouse tissues by using an RNeasy total RNA isolation Kit (Qiagen) according to manufacturer’s protocol. Biotin-labeled cRNA samples for hybridization were prepared by using an Illumina Total Prep RNA Amplification Kit (Ambion). Five hundred nanograms of total RNA was used for the synthesis of cDNA, followed by amplification and biotin labeling. A total of 1.5 μg of biotinylated cRNAs was hybridized to the Illumina mouse-6 BeadChip v.2 microarray. Signals were developed by Amersham fluorolink streptavidin-Cy3 (GE Healthcare Bio-Sciences). Gene expression data were collected by using an Illumina bead Array Reader confocal scanner (BeadStation 500GXDW). All statistical analysis was performed using BRB Arraytools Version 3.6 (37) and under the R language environment. The microarray data were normalized using the quantile normalization method in the Linear Models for Microarray Data package (38). The random variance t test was applied to identify the genes significantly different between two groups compared. In the analysis to identify enriched gene sets, Fisher’s exact test was applied to genes whose expression was significantly altered (P < 0.001) by using gene sets defined in the Ingenuity Pathway Analysis database. A heat map of expression was generated by Treeview (39). Complete datasets containing features differentially expressed in wild-type versus Hippo pathway mutant tissues are contained in Tables S1–S3. Quantitative RT-PCR analysis was carried out using TaqMan gene expression assays (Applied Biosystems) according to the manufacturer’s instructions.
Supplementary Material
Acknowledgments
We thank Richard Behringer for technical support and valuable suggestions, Jenny Deng for assistance in ES cell culture, Jan Parker-Thornburg and Jennifer Alana for morula aggregations, Dae-Sik Lim and Yingzi Yang for sharing results before publication, and Guijin Lu for genotyping. This work was supported by National Institutes of Health Grants HD060579 and HD052785 (to R.L.J.) and by a support grant to the M. D. Anderson Cancer Center from the National Cancer Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0911427107/DCSupplemental.
References
- 1.Edgar BA. From cell structure to transcription: Hippo forges a new path. Cell. 2006;124:267–273. doi: 10.1016/j.cell.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 2.Harvey K, Tapon N. The Salvador-Warts-Hippo pathway - an emerging tumour-suppressor network. Nat Rev Cancer. 2007;7:182–191. doi: 10.1038/nrc2070. [DOI] [PubMed] [Google Scholar]
- 3.Zeng Q, Hong W. The emerging role of the hippo pathway in cell contact inhibition, organ size control, and cancer development in mammals. Cancer Cell. 2008;13:188–192. doi: 10.1016/j.ccr.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 4.Zhao B, Lei QY, Guan KL. The Hippo-YAP pathway: New connections between regulation of organ size and cancer. Curr Opin Cell Biol. 2008;20:638–646. doi: 10.1016/j.ceb.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.St John MA, et al. Mice deficient of Lats1 develop soft-tissue sarcomas, ovarian tumours and pituitary dysfunction. Nat Genet. 1999;21:182–186. doi: 10.1038/5965. [DOI] [PubMed] [Google Scholar]
- 6.McPherson JP, et al. Lats2/Kpm is required for embryonic development, proliferation control and genomic integrity. EMBO J. 2004;23:3677–3688. doi: 10.1038/sj.emboj.7600371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yabuta N, et al. Lats2 is an essential mitotic regulator required for the coordination of cell division. J Biol Chem. 2007;282:19259–19271. doi: 10.1074/jbc.M608562200. [DOI] [PubMed] [Google Scholar]
- 8.Lee JH, et al. A crucial role of WW45 in developing epithelial tissues in the mouse EMBO J . 2008;27:1231–1242. doi: 10.1038/emboj.2008.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anguera MC, Liu M, Avruch J, Lee JT. Characterization of two Mst1-deficient mouse models. Dev Dyn. 2008;237:3424–3434. doi: 10.1002/dvdy.21764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Postic C, Magnuson MA. DNA excision in liver by an albumin-Cre transgene occurs progressively with age. Genesis. 2000;26:149–150. doi: 10.1002/(sici)1526-968x(200002)26:2<149::aid-gene16>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 11.Wagner KU, et al. Spatial and temporal expression of the Cre gene under the control of the MMTV-LTR in different lines of transgenic mice. Transgenic Res. 2001;10:545–553. doi: 10.1023/a:1013063514007. [DOI] [PubMed] [Google Scholar]
- 12.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 13.Dong J, et al. Elucidation of a universal size-control mechanism in Drosophila and mammals. Cell. 2007;130:1120–1133. doi: 10.1016/j.cell.2007.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tacke F, Luedde T, Trautwein C. Inflammatory pathways in liver homeostasis and liver injury. Clin Rev Allergy Immunol. 2009;36:4–12. doi: 10.1007/s12016-008-8091-0. [DOI] [PubMed] [Google Scholar]
- 15.Duncan AW, Dorrell C, Grompe M. Stem cells and liver regeneration. Gastroenterology. 2009;137:466–481. doi: 10.1053/j.gastro.2009.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bird TG, Lorenzini S, Forbes SJ. Activation of stem cells in hepatic diseases. Cell Tissue Res. 2008;331:283–300. doi: 10.1007/s00441-007-0542-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Matthews VB, Klinken E, Yeoh GC. Direct effects of interleukin-6 on liver progenitor oval cells in culture. Wound Repair Regen. 2004;12:650–656. doi: 10.1111/j.1067-1927.2004.12605.x. [DOI] [PubMed] [Google Scholar]
- 18.Knight B, et al. Impaired preneoplastic changes and liver tumor formation in tumor necrosis factor receptor type 1 knockout mice. J Exp Med. 2000;192:1809–1818. doi: 10.1084/jem.192.12.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Streetz KL, et al. Interleukin 6/gp130-dependent pathways are protective during chronic liver diseases. Hepatology. 2003;38:218–229. doi: 10.1053/jhep.2003.50268. [DOI] [PubMed] [Google Scholar]
- 20.Streetz K, et al. Tumor necrosis factor alpha in the pathogenesis of human and murine fulminant hepatic failure. Gastroenterology. 2000;119:446–460. doi: 10.1053/gast.2000.9364. [DOI] [PubMed] [Google Scholar]
- 21.Dorrell C, et al. Surface markers for the murine oval cell response. Hepatology. 2008;48:1282–1291. doi: 10.1002/hep.22468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Faktor VM, et al. [Common antigens of oval cells and cholangiocytes in the mouse. Their detection by using monoclonal antibodies] Ontogenez. 1990;21:625–632. [PubMed] [Google Scholar]
- 23.Camargo FD, et al. YAP1 increases organ size and expands undifferentiated progenitor cells. Curr Biol. 2007;17:2054–2060. doi: 10.1016/j.cub.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 24.Gall GAE, Kyle WH. Growth of the laboratory mouse. Theor Appl Genet. 1968;38:304–308. doi: 10.1007/BF01297571. [DOI] [PubMed] [Google Scholar]
- 25.Kandil DH, Cooper K. Glypican-3: A novel diagnostic marker for hepatocellular carcinoma and more. Adv Anat Pathol. 2009;16:125–129. doi: 10.1097/PAP.0b013e3181992455. [DOI] [PubMed] [Google Scholar]
- 26.Bellahcène A, Castronovo V, Ogbureke KU, Fisher LW, Fedarko NS. Small integrin-binding ligand N-linked glycoproteins (SIBLINGs): Multifunctional proteins in cancer. Nat Rev Cancer. 2008;8:212–226. doi: 10.1038/nrc2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Penzo-Mendez AI. Critical roles for SoxC transcription factors in development and cancer. Int J Biochem Cell Biol. 2009 doi: 10.1016/j.biocel.2009.07.018. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knight B, Matthews VB, Olynyk JK, Yeoh GC. Jekyll and Hyde: Evolving perspectives on the function and potential of the adult liver progenitor (oval) cell. BioEssays. 2005;27:1192–1202. doi: 10.1002/bies.20311. [DOI] [PubMed] [Google Scholar]
- 29.Sell S, Leffert HL. Liver cancer stem cells. J Clin Oncol. 2008;26:2800–2805. doi: 10.1200/JCO.2007.15.5945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Matthews VB, Rose-John S, Yeoh GC. Genetic manipulations utilizing albumin and alpha-fetoprotein promoter/enhancers affect both hepatocytes and oval cells. Hepatology. 2004;40:759. doi: 10.1002/hep.20396. author reply 760. [DOI] [PubMed] [Google Scholar]
- 31.Yechoor V, et al. Neurogenin3 is sufficient for transdetermination of hepatic progenitor cells into neo-islets in vivo but not transdifferentiation of hepatocytes. Dev Cell. 2009;16:358–373. doi: 10.1016/j.devcel.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu MZ, et al. Yes-associated protein is an independent prognostic marker in hepatocellular carcinoma. Cancer. 2009;115:4576–4585. doi: 10.1002/cncr.24495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eakin GS, Hadjantonakis AK. Production of chimeras by aggregation of embryonic stem cells with diploid or tetraploid mouse embryos. Nat Protoc. 2006;1:1145–1153. doi: 10.1038/nprot.2006.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rodríguez CI, et al. High-efficiency deleter mice show that FLPe is an alternative to Cre-loxP. Nat Genet. 2000;25:139–140. doi: 10.1038/75973. [DOI] [PubMed] [Google Scholar]
- 35.Seglen PO. Preparation of isolated rat liver cells. Methods Cell Biol. 1976;13:29–83. doi: 10.1016/s0091-679x(08)61797-5. [DOI] [PubMed] [Google Scholar]
- 36.Bossuyt W, et al. Atonal homolog 1 is a tumor suppressor gene. PLoS Biol. 2009;7:e39. doi: 10.1371/journal.pbio.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Simon R, et al. Analysis of gene expression data using BRB-Array tools. Cancer Inform. 2007;3:11–17. [PMC free article] [PubMed] [Google Scholar]
- 38.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 39.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.