Abstract
Natural protein crystals (polyhedra) armour certain viruses, allowing them to survive for years under hostile conditions. We have determined the structure of polyhedra of the baculovirus Autographa californica multiple nucleopolyhedrovirus (AcMNPV), revealing a highly symmetrical covalently cross-braced robust lattice, the subunits of which possess a flexible adaptor enabling this supra-molecular assembly to specifically entrap massive baculoviruses. Inter-subunit chemical switches modulate the controlled release of virus particles in the unusual high pH environment of the target insect's gut. Surprisingly, the polyhedrin subunits are more similar to picornavirus coat proteins than to the polyhedrin of cytoplasmic polyhedrosis virus (CPV). It is, therefore, remarkable that both AcMNPV and CPV polyhedra possess identical crystal lattices and crystal symmetry. This crystalline arrangement must be particularly well suited to the functional requirements of the polyhedra and has been either preserved or re-selected during evolution. The use of flexible adaptors to generate a powerful system for packaging irregular particles is characteristic of the AcMNPV polyhedrin and may provide a vehicle to sequester a wide range of objects such as biological nano-particles.
Keywords: baculovirus, microcrystals, polyhedra, protein structure
Introduction
The virions of a number of insect viruses from disparate virus families (including baculoviruses, cytoplasmic polyhedrosis viruses (CPVs) and entomopoxviruses) are specifically occluded within robust crystalline particles (Rohrmann, 1986). Typically 1–3 μm in size, these tend to have a characteristic shape, and the well-ordered lattices are built mainly from a single viral protein (Di et al, 1991; Anduleit et al, 2005). The occlusion bodies act as protective packages allowing infectious virions to survive for long periods in harsh environments, providing a delivery system between hosts by oral-faecal routes and resisting solubilization until exposed to the alkaline insect midgut (Rohrmann, 1986).
Baculoviruses are a family of large DNA viruses, which replicate and assemble within the host nucleus. They are divided into two genera, alpha- and beta-baculoviruses, referred to as alphaBV and betaBV, respectively (Jehle et al, 2006). During an infection, the cylinder-shaped (30–50 nm diameter, 200–300 nm long) dsDNA-containing nucleocapsids can be embedded while within the nucleus of the insect cell, in occlusion bodies, named polyhedra for alphaBVs (Rohrmann, 2008). AlphaBV polyhedra are usually 0.15–3 μm in size (Ackermann and Smirnoff, 1983) and contain virus nucleocapsids surrounded by an envelope derived from the nuclear membrane into which are incorporated at least five different virus-encoded proteins, some of which presumably interact with polyhedrin (the polyhedra protein) to orchestrate occlusion. Some baculovirus strains pack a single virion within this envelope, whereas others (the so-called multiple nuclear polyhedrosis viruses) pack a bunch of ∼5–15. Overall, the total number of virions in a polyhedra ranges from one to several hundred (Ackermann and Smirnoff, 1983), scattered apparently randomly within the polyhedra (Scharnhorst et al, 1977). Finally, the entire alphaBV polyhedra is wrapped in a sheath of carbohydrate and virus-encoded proteins (Gross et al, 1994). The ∼29 kDa polyhedrin is one of the most conserved proteins of the virus (Supplementary Figure 1). Naturally occurring amino-acid substitutions in polyhedrin, many of them are single point mutations, produce a variety of phenotypic changes ranging from large, cuboid polyhedra, which occlude no or few virions (Carstens et al, 1986; Katsuma et al, 1999; Lin et al, 2000), to overall changes in polyhedra shape (Cheng et al, 1998). A nuclear localization signal (NLS), K32RKK35, directs the polyhedrin to the nucleus (Jarvis et al, 1991). The structure of the polyhedrin and its organization within the polyhedra has remained obscure, although analysis of partially dissolved baculovirus polyhedra suggested the presence of dodecameric or disulphide-linked octameric molecules of polyhedrin (Rohrmann, 1977; Scharnhorst and Weaver, 1980).
In comparison with the rod-shaped baculoviruses, CPV is 75 nm in diameter and the dsRNA genome is contained in an icosahedral protein capsid, which is occluded directly into the non-enveloped polyhedra (Mertens et al, 2005). Similar to baculoviruses, the shape of CPV polyhedra can be altered by point mutations of polyhedrin (Ikeda et al, 1998). The crystal structures of both virus-containing and recombinant empty Bombyx mori CPV type 1 polyhedra have been determined (Coulibaly et al, 2007), revealing the polyhedra to be beautifully assembled from rigid 28.6 kDa polyhedrin building blocks, which assemble with the aid of ribonucleoside triphosphates (NTPs) to form a lattice that contains virtually no solvent. The polyhedrin is formed from a β-barrel core with helical extensions and was dissimilar to any other known protein structure. The polyhedrin lattice possesses cubic symmetry, so that its strength will be maximally isotropic and it matches, as closely as is possible in a crystal lattice, the icosahedral symmetry of the CPV particle.
Remarkably, X-ray powder diffraction analysis of baculovirus and CPV polyhedra (Fujiwara et al, 1984; Di et al, 1991; Anduleit et al, 2005) indicated that, despite no obvious amino-acid sequence similarity, they possess crystal lattices, which are essentially indistinguishable in terms of size (a=10 nm) and symmetry (body centred lattice with 23 symmetry), suggesting that these parameters, which presumably underpin the biological properties of the polyhedra, are strongly conserved. To establish whether this arises, as has been predicted (Anduleit et al, 2005; Coulibaly et al, 2007), from similarities in the 3D structures of the proteins that build the lattice, we have determined such structures for wild-type and mutant Autographa californica multiple nucleopolyhedrovirus (AcMNPV) polyhedra and find, surprisingly, that while the space group is identical to CPV polyhedrin (I23), AcMNPV polyhedrin in fact resembles more closely the canonical capsid protein structure of the picornavirus lineage of viruses (Bamford et al, 2005). Although not providing conclusive evidence for the evolutionary relationships between these viral polyhedra, the AcMNPV polyhedrin structure explains various functional aspects, including pH-dependent disassembly and adaptation to the nuclear life style of baculoviruses.
Results
Structure determination
We have determined, from in vivo grown crystals, the structures of both virus-containing wild-type and a virus-empty mutant (G25D) polyhedra from AcMNPV, the type species of alphaBVs. The crystals were derived from infected insect cells (see Materials and methods), with the wild-type crystals being usually <5 μm in maximum linear dimension while many G25D crystals attained 5–10 μm. Despite their still tiny size, it was possible to determine the high-resolution (1.84 Å) structure for the larger mutant polyhedra by X-ray analysis using a tuneable micro-focus synchrotron beam (we used conventional seleno-methionine (Se-Met) labelling to solve the phase problem), Table 1. Owing to the small size of the crystals and their low-solvent content (nominally 21%), structure determination was challenging; however, the final structure is reliable (Rwork=0.164, Rfree=0.217 from 36.3 to 1.84 Å resolution, Table 1). Although the majority of the structure is precisely determined, the electron density is poorly defined for residues 3–7 (for which only the backbone could be traced) and 142–147, whereas residues 1–2, 32–48 and 174–186 could not be positioned at all. The final 1.8 Å resolution structure contains 88 water molecules, with no ions, buffer molecules or other small molecules visible. The structure of wild-type AcMNPV polyhedra was then determined at 3.0 Å resolution by molecular replacement (Table 1). The structures of mutant and wild-type polyhedra are very similar and because of its higher quality, we usually describe the mutant structure below; however, the wild-type structure is a little better packed (as described below) such that some of the disordered residues are clearly visible and this structure helped in the final interpretation of these regions.
Table 1.
Data collection, phasing and refinement statistics
| G25D nativea | G25D Se-Meta | Wild-type nativea | |
|---|---|---|---|
| Data collection | |||
| Space group | I23 | ||
| Cell dimensions | |||
| a=b=c (Å) | 102.58 | 102.60 | 101.56 |
| Resolution (Å) | 50.0–1.84 (1.91–1.84) | 40.0–3.0 (3.11–3.00) | 50.0–3.0 (3.11–3.00) |
| Unique reflections | 15809 (1576) | 3721 (366) | 3329 (326) |
| Rmerge | 0.210 (0.000b) | 0.248 (0.629) | 0.383 (0.000b) |
| I/σI | 12.1 (1.0) | 23.4 (7.9) | 4.4 (1.4) |
| Completeness (%) | 100 (100) | 100 (100) | 99.6 (100) |
| Redundancy | 14.7 (10.9) | 24.3 (24.3) | 6.8 (6.4) |
| Refinement | |||
| Resolution (Å) | 36.3–1.84 | 41.5–3.0 | |
| Number of reflections | 15762 | 3325 | |
| Rwork/Rfree | 0.164/0.217 | 0.205/0.238 | |
| Number of atoms | |||
| Protein | 1752 | 1723 | |
| Water | 88 | ||
| B-factors | |||
| Protein | 34.1 | 28.8 | |
| Water | 34.5 | ||
| r.m.s. deviations | |||
| Bond lengths (Å) | 0.007 | 0.007 | |
| Bond angles (deg) | 1.04 | 0.94 | |
| Values in parentheses are for the highest resolution shell. | |||
| aSeventeen G25D native crystals, 31 G25D Se-Met crystals and seven wild-type native crystals were merged, respectively. | |||
| bR⩾1. | |||
Subunit structure
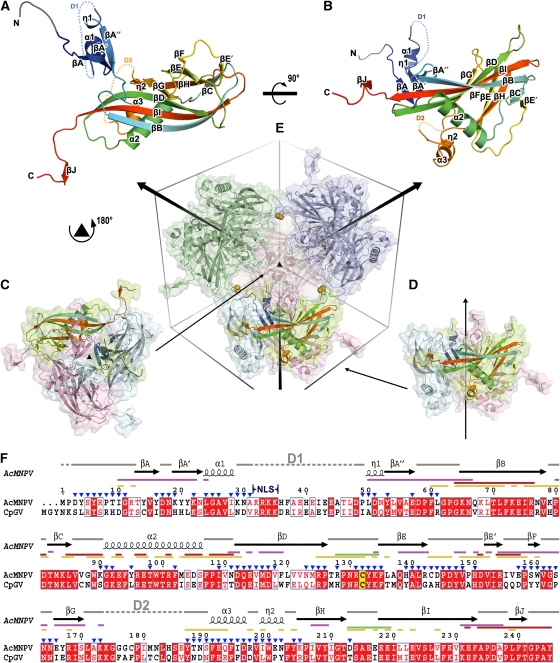
AcMNPV polyhedra belong to space group I23 with one polyhedrin subunit in the crystal asymmetric unit, so that 24 copies of the 28.6 kDa (245 amino acids) subunit are tightly packed in the unit cell. The polyhedrin subunit can be broken down into three parts: N-terminal head, a β-barrel body (comprising the majority of the subunit) and C-terminal tail (Figure 1A and B). The β-barrel body is reminiscent of that of the capsid proteins of picorna-like viruses and we retain that strand nomenclature here. Four β-strands (BIDG) form the core of an extended first sheet with strands CHEF forming the second sheet, which contains fewer, shorter strands (Supplementary Figure 2A). Three helices (α2, α3 and η2) cap the exposed side of the sheets, enclosing the hydrophobic core and forming a groove between α2 and the BIDG sheet. Extending from strand βI is a C-terminal hook comprising 13 highly conserved residues (residues 233–245, Figure 1F; Supplementary Figure 1). The head domain (residues 1–31 and 49–63) consists of a hook-shaped loop, three anti-parallel β-strands (A, A′, A′′), an α-helix and a 310 helix. As residues 32–48 were not seen in the electron density map, there is an ambiguity in connecting the N-terminal residues 1–31 to the rest of the subunit. In principle, 12 different links are feasible; however, for clarity, only one of these is shown in Figure 1A and B (we justify this choice below).
Figure 1.
Overview of structure. (A, B) The structure of AcMNPV polyhedrin, in two orthogonal views, shown as cartoon representations coloured from blue at the N-terminus to red at the C-terminus. Residues 3–7, which were built as poly-ala are drawn in grey. Disordered segments D1 (residues 32–48) and D2 (residues 174–186) are shown as dots. (C, D) Cartoon and surface representations of polyhedrin trimers shown in two orientations aligned to (E). Subunits are coloured by chain with one in each trimer coloured as in (A). (E) Dodecameric unit of four trimers linked by disulphide bonds (represented as orange spheres). The unit cell and three-fold axes are drawn to facilitate observation. (F) Amino-acid sequence conservation between the type species of the alphaBVs (AcMNPV) and betaBVs (CpGV). Conserved residues drawn in red boxes, similar residues in red type. The secondary structure assignment for AcMNPV is drawn above. Conserved cysteine is highlighted in yellow. Interactions involved in various stages of oligomization are mapped as lines coloured thus; purple for trimer, green for dodecamer, red for interactions through C-terminal and yellow for other interactions in the unit cell. The position of the nuclear localization signal (NLS) is indicated and residues exposed to the cavity are marked by blue triangles. The sequences were aligned with ClustalW (Chenna et al, 2003) and visualized using ESPript (Gouet et al, 1999).
Mapping functional mutations
A number of naturally occurring mutations have been described for baculovirus polyhedrin, some of the best characterized are described in Table 2. A significant number fall into one of the two classes: (i) mutations, which disrupt the formation of polyhedra and (ii) mutations, which result in the formation of larger polyhedra, sometimes in the cytoplasm. In general, mutations of class (i) tend to occur within the core β-barrel of the polyhedrin, in which they presumably significantly disrupt the molecule (Supplementary Figure 3); an example is the valine to phenylalanine mutation at residue 118 in which it would be difficult to accommodate the bulk of the aromatic side chain. In contrast, mutations of class (ii) tend to cluster in or near the disordered regions of the structure (Supplementary Figure 3). We have obtained structures for the wild-type virus and for one class (ii) mutant, G25D. The mutant polyhedra are larger than the wild-type and are found, empty of viruses, in the nucleus of the infected cells. Structurally, the change from glycine to aspartic acid is quite modest, and yet there are significant differences between the two types of polyhedra—in the wild–type, the smaller glycine amino acid provides space for a nearby side chain (Asp51), resulting in a better local packing and a slight (∼1 Å) reduction in the size of the unit cell. Although this change is small, it is completely reproducible and the changes observed are likely to be representative of the situation in vivo.
Table 2.
List of AcMNPV and BmNPV polyhedra mutant viruses adapted from Ribeiro et al (2009)
| Polyhedrin mutation | Position in structure | Virus occlusion | Occlusion morphology | Reference | |
|---|---|---|---|---|---|
| Class i | |||||
| M29/AcMNPV | L85P | Within βC (core) | No | Dispersed protein mass | Carstens (1987) |
| vSynlitx1B12P/AcMNPV | V118F | Within βD (core) | No | Dispersed protein mass | Ribeiro et al (2009) |
| #136/BmNPV | L141F | Within βE (core) | No | Dispersed protein mass | Katsuma et al (1999) |
| Class ii | |||||
| AcMNPV-Tkmt513/AcMNPV | G25D | Within α1, close to D1 | No | Large and cuboid | Lin et al (2000) |
| M5/AcMNPV | P59L | Within loop between βA′′ and B | Yes/no | Large and cuboid | Carstens et al (1986) |
| #211/BmNPV | E144L | Within loop between βE and E′ | Yes | Irregularly shaped or small polyhedra | Katsuma et al (1999) |
| #128/BmNPV | L171P | Close to D2 | No | As wild-type occlusion | Katsuma et al (1999) |
| Others | |||||
| #220/BmNPV | D58N I222T | Within loop between βA′′ and B Within βI | Yes | Cuboid | Katsuma et al (1999) |
| #126/BmNPV | C178Y | Within D2 | No | Dispersed protein mass | Katsuma et al (1999) |
| M934/AcMNPV | L183F | Within D2 | No | Dispersed protein mass | Carstens et al (1992) |
A structural similarity to viral coat proteins, and perhaps to CPV polyhedrin
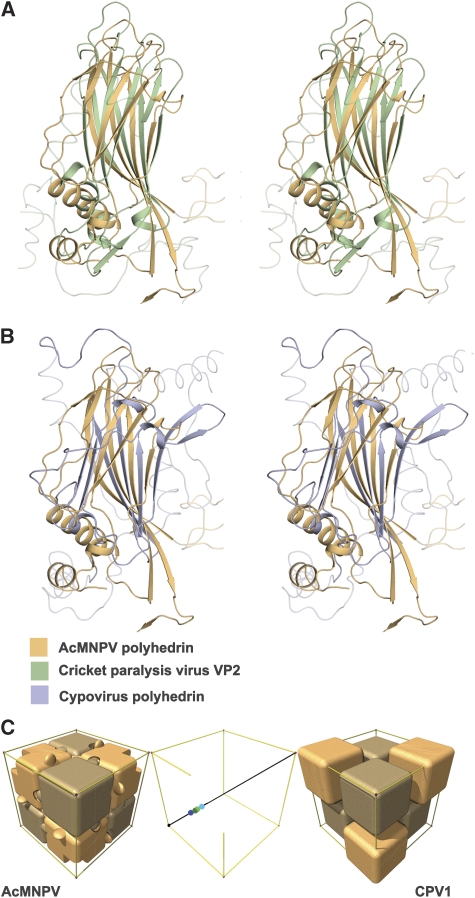
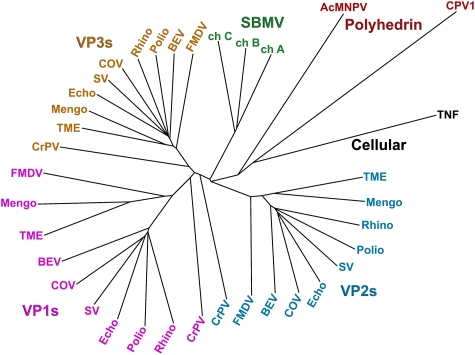
Automated structural comparisons against the Protein Data Bank (Dali, Holm et al, 2008; SSM, Krissinel and Henrick, 2004) reveal that the AcMNPV polyhedrin has a jelly-roll fold similar to that observed in the capsid proteins of picorna-like viruses (Figure 2A; Supplementary Table I). Indeed, the most similar structures tend to be picornavirus VP3 major coat proteins and the ‘insect picornavirus' cricket paralysis virus coat proteins. More detailed comparison with the program SHP (Stuart et al, 1979) reveals that the closest similarity is with VP2 of cricket paralysis virus, 119 equivalent Cαs can be superposed with r.m.s. deviation 3.2 Å. Although there are many structural differences, there are also points of similarity beyond the basic sheet topology, thus the BIDG sheet is the more extended in both the picornavirus capsid proteins and AcMNPV polyhedrin, and the topological positions of some of the helices match. This similarity is of a level that has earlier been used to infer divergence from a common ancestor, and this is graphically shown in Figure 3. A somewhat lower level of similarity is shown with CPV polyhedrin (Figure 2B), 86 Cαs superpose with r.m.s. deviation 3.5 Å. The Dali Z-score for this comparison is correspondingly low at 1.5 (Supplementary Table I); however, the program does not find any structures with significantly greater similarity to CPV polyhedrin and detailed SHP analysis confirms that cellular jelly-roll proteins such as tumour necrosis factor (TNF) are no more similar (Figure 3).
Figure 2.
Structural superpositions. Stereo views showing the alignment of (A) AcMNPV polyhedrin (orange) with cricket paralysis virus VP2 (green) (Tate et al, 1999) and (B) AcMNPV polyhedrin (orange) with CPV1 polyhedrin (purple). The program SHP (Stuart et al, 1979) was used to perform the superpositions. (C) Schematic representation of the packing of both AcMNPV and CPV1 polyhedrin trimers (shown as blocks) within their unit cells. The trimeric units are positioned rather differently on the three-fold body diagonal—this is represented in the central panel in which the centres of the trimers of differing polarity are reflected into the (0,0,0) to (½, ½, ½) portion. The AcMNPV trimer centres (shown in shades of green) lie close to (¼, ¼, ¼), whereas the CPV1 trimers (centres shown in shades of blue) are offset by about 6 Å. This accounts for the differences seen in the outer panels.
Figure 3.
Phylogenetic tree showing the relationship between the polyhedrin proteins of AcMNPV and CPV1, against picorna-like virus capsid proteins. Superpositions were performed using SHP (Stuart et al, 1979) and the phylogenetic tree calculated with PHYLIP (Felsenstein, 1989). The following abbreviations are used: AcMNPV, Autographa californica baculovirus; BEV, bovine enterovirus; ch, chain; COV, coxsackievirus A3; CPV1, cytoplasmic polyhedrosis virus type 1; CrPV, cricket paralysis virus; Echo, echovirus; FMDV, foot-and-mouth disease virus; Mengo, mengo virus; Polio, poliovirus; Rhino, rhinovirus; SBMV, southern bean mosaic virus; SV, swine vesicular disease virus; TME, theiler murine encephalomyelitis virus; TNF, tumour necrosis factor. Note that CPV1 polyhedrin is closest to AcMNPV polyhedrin, which is, however, closer to the picornavirus capsid structures than CPV1.
Subunit organization in the crystal lattice
The AcMNPV subunits form tightly packed trimers, which resemble a cube with one quadrant removed—this region houses the disordered portions of the molecule, as we discuss below (Figures 1C, D and 4). The jelly-roll β-barrels of each subunit are packed orthogonal to each other (reminiscent to that seen in the non-viral TNF trimer; Jones et al, 1989), and define the basic size of the crystal building block. The extent of the interface in the trimer depends on the assignment of the linkage of the N-terminal head, which as mentioned above is ambiguous, as 17 residues are missing and these occupy a region close to the crystallographic origin in which four three-fold and three two-fold symmetry axes converge, so that 12 different links are in principle feasible (it is conceivable, but unlikely that within a crystal the N-terminal residues are not always connected in the same way). Nevertheless, given the positioning of the symmetry axes, the distances involved and the nature of the subunit interactions described below, we consider it most likely that the N-terminal portion is attached to one of the subunits within a trimer (Supplementary Figure 4). The N-terminal head contributes to the clamping sheet fundamental to the trimer assembly, part of a robust trimer interface comprising a mixture of hydrophobic interactions, salt bridges and hydrogen bonds, which buries between 20 and 30% (∼3000–4000 Å2) of the surface area of each polyhedrin subunit (Figure 1F; Supplementary Figure 5). At the base of the trimer, the N-termini come together to form a short stalk. Helices α1 and η1 form the base and strands βA, A′ and A′′ the upper region (these N-terminal strands clamp onto and expand the BIDG sheet from adjacent subunits; Supplementary Figure 2B). The C-terminal hooks protrude at right angles from three edges of the trimer cube.
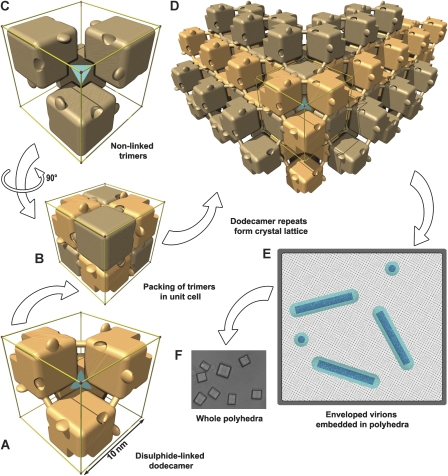
Figure 4.
Schematic representations of polyhedra organization. Polyhedrin trimers are depicted as simplified cubic blocks, with the C-terminal hooks and pockets present. To clarify interpretation, the edges of the unit cell are shown in gold and a cyan tetrahedron symbolizes the cell centre. Within a unit cell, disulphide-linked trimers with one polarity are coloured light beech (A) and those with the opposite polarity are coloured light brown (C). (B) All eight trimers in the unit cell. The disulphide bond connecting adjoining trimers is shown as a dowel. (D) The crystal lattice is built up from repeats of the dodecameric unit. (E) Sketch of a cross-section through a polyhedron. The lattice spacing of the unit cells is illustrated as a dot pattern into which are embedded nucleocapsids (dark blue) surrounded by an envelope (cyan). (F) Light microscopy image of G25D mutant AcMNPV polyhedra.
Eight of the cube-like trimers are arranged neatly to fill the crystal unit cell (Figure 4). These eight trimers in fact form two nested sets of four, one arrayed at the corners of the cell and the other at the centre, the only difference between these two sets being in the polarity of the trimer with respect to the three-fold axes (four trimers point inwards towards the centre of the unit cell, whereas the other four point outwards towards the corners). The regularity of the packing seen in Figure 4 arises because the trimers are situated almost exactly mid-way along the body diagonal between the centre and the corner of the crystal cell (Figures 1E and 2C). This arrangement leaves the C-terminal hooks jutting outwards and brings the absent quadrants of the trimers together around the centre and corner points, to form cavities (volume 110 000 Å3), which harbour the disordered portions of polyhedrin (Figure 4C; Supplementary Figure 6).
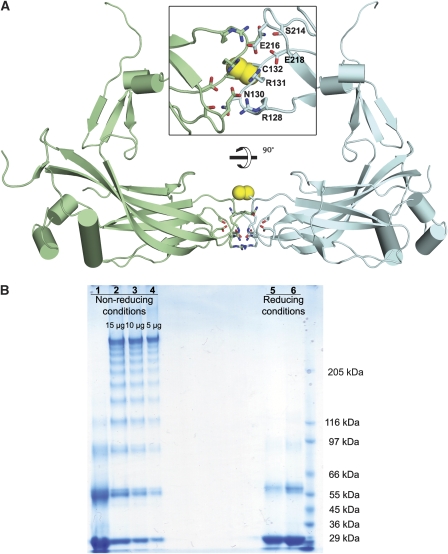
Assuming that the N-terminal heads exchange within trimers, the closest interactions between trimers of the same polarity come at the centre of each face of the unit cell in which the βD-βE and βH-βI loops of subunits from neighbouring trimers come together. The interaction here is not extensive (800 Å2) and would not normally be sufficient to achieve a strong interaction; however, four salt bridges between highly conserved glutamate, arginine and asparagine side chains and a covalent disulphide lock between two symmetry-related copies of Cys132 tether this extended dimer (Figure 5A). Crystallographic and biochemical analyses show that in wild-type polyhedra, the disulphide is fully formed, whereas in the mutant polyhedra, it is partly reduced (Figure 5B). As Cys132 is the only cysteine fully conserved across both the alpha- and beta-baculoviruses (Supplementary Figure 1), it seems likely that this disulphide stabilization is biologically relevant. These interactions link four trimers of the same polarity in a tetrahedral arrangement to form a dodecameric cage (Figures 1E and 4A). Dodecamers, and smaller oligomers, can be observed by non-reducing SDS–PAGE with shorter heating times (Figure 5B) and chemical cross-linking (Scharnhorst and Weaver, 1980). The formation of the disulphide bonds between trimers presumably occurs relatively late, after disruption of the reducing environment of the nucleus. The assembly of the crystal is completed by clipping dodecamers together (Figure 4D). In this lattice, the disulphide bonds provide diagonal cross-braces, whereas stability in the direction of the unit cell edges is provided by a very extensive interface (∼10 000 Å2), which includes protruding C-terminal hooks, which engage adjacent dodecamers through a series of conserved residues (Figure 1F; Supplementary Figure 1).
Figure 5.
Inter-subunit disulphide bond. (A) Cartoon diagram of two disulphide-bonded polyhedrin related by a two-fold rotation forming an extended dimer. Structures are coloured by chains and the helices are shown in cylinders. Residues involved in intermolecular salt bridges are drawn as sticks and disulphides are shown as yellow spheres in two orthogonal views. (B) SDS–PAGE of AcMNPV polyhedra under reducing and non-reducing conditions when dissolved in bicarbonate buffer, pH 10.5. Polyhedra were incubated at 80°C for 2 h to inactivate the protease associated with polyhedra before exposure to alkaline buffer. Samples were electrophoresed on 4–8% gradient SDS gels with 1 min (lanes 2–4 and 6) and 5 min (lanes 1 and 5) heating at 100°C.
Comparison of the lattice arrangement between baculovirus and CPV polyhedrins
Although the protein folds and the interactions, which stabilize the crystal lattice, differ between AcMNPV and CPV (AcMNPV uses disulphide bonds and CPV uses NTPs), there are some broad similarities in the lattice architecture. Not only the size of the unit cell (∼102–103 Å), but also the space groups are identical (I23) and both are assembled from similarly sized trimeric building blocks arrayed on the body diagonals of the cubic lattice. However, the CPV trimers are translated about 6 Å along this diagonal (Figure 2C) and the subunit jelly rolls rotated by ∼137° compared with AcMNPV.
Virus occlusion
Despite forming an extremely robust lattice, the AcMNPV polyhedra nonetheless harbour pools of flexibility at the crystallographic origin and symmetry-related points (Supplementary Figure 6). These contain the region between residues 32 and 48, which shows the lowest amino-acid sequence conservation across the baculoviruses (Figure 1F; Supplementary Figure 1), suggesting that it may confer specificity for the packaging of a particular virus strain. Baculoviruses, unlike CPVs, do not possess icosahedral symmetry, so there needs to be physical flexibility in the areas, which engage proteins on the surface of the virion envelope, as there is an inevitable symmetry mismatch with the irregular packets of baculoviruses (Figure 4E). This hypothesis is supported by the observation that certain mutations in this region interfere with the packaging of virus particles and by the structural differences we observe between the non-occluding G25D mutant and wild-type virus, in which the more flexible wild-type glycine might allow the protein to sample enough conformational space to efficiently attach to the virus. At present, we cannot precisely map the interacting residues, and the identities of the interacting proteins on the surface of the membrane-coated virions remain unknown (Rohrmann, 2008). The remarkable ability to form a crystalline lattice around pleiomorphic bundles of virions, each of which excludes some 20 000 polyhedrin subunits, probably comes at a cost in terms of lattice strain, as mutants, which do not occlude viruses, often generate crystals larger than the typical few hundred unit cell-wide virus-containing polyhedra.
Regulation of polyhedra assembly
We would expect that if polyhedrin subunits accumulate in any cellular compartment, they will assemble into polyhedra, unless this is suppressed by a specific mechanism. AcMNPV polyhedrin is made in the cytoplasm and transported to the nucleus. As polyhedra do not build up in the cytoplasm unless there is a deficiency in nuclear transport (e.g. a loss of the NLS; Jarvis et al, 1991), it seems plausible that premature cytoplasmic assembly is prevented by sequestration of polyhedrin by importin, through NLS interactions (Figure 1F). The primary regulation of polyhedra assembly in the nucleus is then presumably by late expression of the polyhedrin gene (Rohrmann, 2008). In contrast, CPV virus particles and polyhedra assemble in the cytoplasm and mRNA production from virus cores is probably roughly equal for all proteins (although a host repression system may lead to differential protein expression, as polyhedrin is reported to accumulate a little later than some other viral proteins; Payne and Mertens, 1983). We suggest that the specific incorporation of a number of NTPs at key points in the CPV crystal lattice (Coulibaly et al, 2007) may provide a further mechanism for the regulation of polyhedra assembly in the CPVs, namely the NTP pool concentration. Thus, depletion of the NTP pool during the early phase of infection may retard assembly until complete CPV particles are available for occlusion. This strategy would not be appropriate for nuclear viruses such as AcMNPV, as NTP pool levels are much lower in the nucleus (Plagemann, 1972).
Dissolution through pH-dependent switches
Although mature polyhedra are extremely robust, withstanding pH 2 and years of exposure in the environment, they readily dissolve in the carbonate-buffered alkaline pH environment of the insect midgut to release the virus particles (dissolution in carbonate begins at ∼pH 10). It seems that there are similarities and differences in the mechanism of this dissolution between AcMNPV and CPV polyhedra. Presumably, an early stage in the disassembly of AcMNPV polyhedra is baculovirus specific—the breakage of the –S–S– bonds, which is accelerated at high pH, especially with carbonate (Donovan and White, 1971). The second stage in the dissolution, which has common features between CPVs and NPVs, is the disassembly of the non-covalent interfaces between and within trimers. It seems that tyrosine side chains (pKa 10.1) act as pH-dependent switches to direct this process. The fraction of tyrosine residues in AcMNPV (6%) is roughly double that in a typical protein (Brooks and Fresco, 2002), and these are clustered at the trimer interface (Supplementary Figure 7). A similar situation pertains in CPV (Coulibaly et al, 2007).
Discussion
We have found, unexpectedly, that the baculovirus polyhedrin is strikingly similar in structure to a picornavirus coat protein, at a level suggestive of homology. It is indeed feasible that these two molecules share a common ancestor, as there is a precedent for lateral gene transfer between baculoviruses and RNA viruses (e.g. gp64 of baculoviruses and gp75 of thogotovirus; Kadlec et al, 2008) and the insect picornavirus cricket paralysis virus resembles a primitive picornavirus (no cellular proteins show such a strong similarity to the baculovirus poyhedrin, Figure 3). This suggests that either the picornavirus-like capsids or the baculovirus polyhedrin might have originated through co-infection of an insect. The latter seems the more attractive hypothesis, as the picornavirus superfamily includes, for instance, plant viruses, which are likely more ancient (Figure 3), but are less similar to AcMNPV polyhedrin. Furthermore, although polyhedra are not required for viral transmission, the acquisition of a self-assembling fragment of a picornavirus capsid protein could confer a significant selective advantage on an ancestral baculovirus.
The relatively strong similarity of AcMNPV polyhedrin with a capsid jelly roll contrasts with the lower level of similarity between the AcMNPV and CPV polyhedrins. Until now it had been assumed that the essentially identical lattice parameter displayed by the two polyhedra indicated that they would possess closely related structures, despite having no detectable amino-acid sequence similarity (Anduleit et al, 2005; Coulibaly et al, 2007). We have now found that this identity extends to the crystallographic space group, but is not reflected in the 3D structure of the polyhedra, which are markedly different. This remarkable finding could have fundamentally different explanations: convergence to a common lattice driven by functional requirements or divergence from a common ancestor with the crystal lattice being strictly conserved. Both explanations imply that the lattice possesses particular geometric features that render it exquisitely fit for purpose. As the inter-subunit interactions are not conserved between the two polyhedra, this geometry presumably reflects either (i) inherent mechanical properties, (ii) features that facilitate viral inclusion or (iii) features that engage in some unsuspected way with complementary cellular geometry. The I23 lattice will provide isometric strength—each stabilizing interaction providing strength in 12 regularly spaced directions, although other space groups might be expected to provide similar advantages (e.g. P23, I432, etc). As to the identity of cell length, if the subunit is to be a robust single domain structure, then it is likely to have a size of no more than ∼40 kDa (whereas some very small structures might be hard pressed to accomplish the dual functions of lattice formation and viral inclusion). These restraints do not convincingly explain a similarity in cell edge at the level of ∼1%. As there are radical differences between the protein-only icosahedral CPV and the complex, pleiomorphic packages of AcMNPV, it is difficult to see that viral inclusion would be a principal driver to crystal identity—although high point group symmetry, such as 23 (possessed by space group I23), allows virus engagement modules to project in appropriate directions into voids in the lattice, facilitating occlusion. Finally, geometric matching with some cellular component is conceivable, but as yet there is no evidence of the involvement of cellular factors modulating polyhedra formation (with the exception of NTPs for CPV, as discussed above). In summary, although there are obvious reasons why this particular lattice is well suited to its function, it seems likely that a whole array of alternative solutions might serve equally well, indeed different solutions to the virus packaging problem do exist in nature (e.g. the entomopoxviruses polyhedrin is far larger; Marlow et al, 1998). This weakens the argument for the inevitable convergence to a unique lattice. So is it possible that despite the marked drift in tertiary structure, the commonality in the crystal lattice betrays the common origins of these proteins? CPV does possess some similarities with the AcMNPV polyhedrin (Figures 2B,C and 3), at least as much as to any other known protein structure. Although this alone is not a compelling argument for homology, there is also similarity in the trimeric assembly and broad organization within the unit cell. Thus, it is conceivable that polyhedrin could have been transferred to CPV through recombination and fixed by the selective advantage of fortuitous occlusion of CPV particles. Whatever the evolutionary relationship of CPV with AcMNPV, the strong conservation of cell symmetry and size between very different strains of CPV (with little similarity in amino-acid sequence; Anduleit et al, 2005) shows that these crystal parameters are a strongly conserved phenotype.
The discovery, by structural analysis, of serial homologues might in the future clarify the picture of polyhedra evolution, but for now the modular structure of AcMNPV polyhedrin provides an ideal framework for nano-engineering. The disordered region from residues 32–48 containing the NLS and probably responsible for virus occlusion is variable in sequence between different strains of baculovirus (Figure 1F) and as it does not seem to be required for polyhedra assembly, it can probably be radically modified. Thus, by swapping in other specific attachment modules, one could design robust containers that might be used to either deliver specific, and potentially complex, payloads, or to sequester, in a chemically inert framework, targets to be removed.
Materials and methods
Expression and purification of polyhedra
Sf9 cells (Invitrogen) were grown in Sf-900 II serum-free medium (SFM, Gibco-BRL) at 27°C. The polyhedrin gene from AcMNPV was sub-cloned into the transfer vector pBacPAK9 (Clontech) and site-directed mutagenesis was used to generate a G25D mutant, which produces larger polyhedra (Lin et al, 2000). The recombinant virus was produced by co-transfection of linearized baculovirus DNA and the transfer vector after a standard protocol using Cellfectin (Invitrogen) (Zhao et al, 2003). Wild-type and mutant AcMNPV polyhedra were expressed and purified as described earlier (Anduleit et al, 2005).
To grow seleno-methionine-labelled polyhedra, Sf9 cells in SFM were seeded at a density of 1 × 106/ml and infected with a multiplicity of infection of 1. Twenty hours post-infection, the media was changed to SF-900 II SFM methionine- and cysteine-free medium supplemented with heat-inactivated dialysed FBS (Gibco-BRL) to 10% (v/v) and 0.15 g/l cysteine. After 4 h of growth at 27°C, Se-Met was added to the medium to achieve the final concentration of 250 mg/l and cells were grown further for 72 h before harvesting.
Data collection
Polyhedra crystals were suspended in a solution of 10 mM Hepes pH 7.5 and 50% ethylene glycol spread on MicroMesh mounts (MiTeGen, Ithaca, USA) and flash frozen in a stream of nitrogen gas at 100 K. X-ray datasets were collected at 100 K at beamline I24 (Diamond) on a Rayonix MX300 detector with the beam focused to 8 × 8 μm2 at λ=0.9778 Å. Data were processed with HKL2000/Denzo/Scalepack (Otwinowski and Minor, 1997) in space group I23. Seventeen native crystals and 31 seleno-methionine-substituted crystals of G25D mutant as well as seven wild-type crystals were merged to give three data sets with adequate redundancy (Table 1).
Structure determination and refinement
The structure of the G25D mutant was solved using single isomorphous replacement with anomalous scattering (SIRAS) and the wild-type was solved from the mutant structure by molecular replacement. Heavy atom sites were found by SHELX (Sheldrick, 2008) and refined at 3 Å by SHARP (de La Fortelle and Bricogne, 1997). SHARP figures of merit were (centric/acentric) 0.30/0.31 (over all data) and 0.10/0.11 (3.1–3.0 Å). Phasing power (isomorphous/anomalous) was 0.797/0.490. Solvent flattened electron density maps from SOLOMON (Abrahams and Leslie, 1996) and DM (Cowtan, 1994) (3 Å) were used for the initial model building using Coot (Emsley and Cowtan, 2004) (solvent content 30%, figures of merit 0.76 (30–6 Å) to 0.60 (3.2–3.0 Å)). Iterative model building and refinement were performed by Coot and BUSTER (Blanc et al, 2004), indeed of a number of programs-only BUSTER allowed progress and model completion. Final refinement used PHENIX (Adams et al, 2002) at a resolution of 1.84 Å. The final model contains residues 3–31, 49–173 and 187–245 and 88 water molecules. The electron density was very weak for residues 3–7 and 142–147 and the former was simply built as a poly-ala backbone from a low-resolution map. The final model was validated by MolProbity (Davis et al, 2007). A total of 97.6% of residues lie within favoured regions of the Ramachandran plot and the remainder (with one outlier Y147) lie in additionally allowed regions. The wild-type structure was solved by rigid body refinement using CNS (Brunger et al, 1998) and further refined by autoBUSTER to 3 Å resolution. The final models of the well-defined part are essentially identical, although residues 142–147 showed some differences.
Molecule interfaces and oligomerization were analysed by the PISA server at the European Bioinformatics Institute (Krissinel and Henrick, 2007). Illustrations were generated by Pymol (DeLano Scientific).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Table I
Supplementary Information References
Review Process File
Acknowledgments
We thank Drs Jingshan Ren, Robert Esnouf and Stephen Graham for patient and expert advice and assistance and the staff of Diamond Light Source for the support of beamline I24. The work was supported by the Medical Research Council UK and SPINE2-COMPLEXES LSHG-CT-2006-031220. Diamond is supported by the UK government through the STFC and by the Wellcome Trust. XJ performed most experiments and contributed to all other aspects of the work. GS and DIS were mainly involved in experimental design and writing the paper, all authors contributed to data collection. Coordinates and structure factors have been deposited, with accession codes 2wux and 2wuy (G25D mutant and wild-type, respectively).
Footnotes
The authors declare that they have no conflict of interest.
References
- Abrahams JP, Leslie AG (1996) Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr D Biol Crystallogr 52(Pt 1): 30–42 [DOI] [PubMed] [Google Scholar]
- Ackermann HW, Smirnoff WA (1983) A morphological investigation of 23 baculoviruses. J Invert Pathol 41: 269–280 [Google Scholar]
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58(Pt 11): 1948–1954 [DOI] [PubMed] [Google Scholar]
- Anduleit K, Sutton G, Diprose JM, Mertens PP, Grimes JM, Stuart DI (2005) Crystal lattice as biological phenotype for insect viruses. Protein Sci 14: 2741–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bamford DH, Grimes JM, Stuart DI (2005) What does structure tell us about virus evolution? Curr Opin Struct Biol 15: 655–663 [DOI] [PubMed] [Google Scholar]
- Blanc E, Roversi P, Vonrhein C, Flensburg C, Lea SM, Bricogne G (2004) Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr 60(Pt 12 Pt 1): 2210–2221 [DOI] [PubMed] [Google Scholar]
- Brooks DJ, Fresco JR (2002) Increased frequency of cysteine, tyrosine, and phenylalanine residues since the last universal ancestor. Mol Cell Proteomic 1: 125–131 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Carstens EB (1987) Identification and nucleotide sequence of the regions of Autographa californica nuclear polyhedrosis virus genome carrying insertion elements derived from Spodoptera frugiperda. Virology 161: 8–17 [DOI] [PubMed] [Google Scholar]
- Carstens EB, Krebs A, Gallerneault CE (1986) Identification of an amino acid essential to the normal assembly of Autographa californica nuclear polyhedrosis virus polyhedra. J Virol 58: 684–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carstens EB, Williams GV, Faulkner P, Partington S (1992) Analysis of polyhedra morphology mutants of Autographa californica nuclear polyhedrosis virus: molecular and ultrastructural features. J Gen Virol 73(Pt 6): 1471–1479 [DOI] [PubMed] [Google Scholar]
- Cheng XW, Carner GR, Fescemyer HW (1998) Polyhedrin sequence determines the tetrahedral shape of occlusion bodies in Thysanoplusia orichalcea single-nucleocapsid nucleopolyhedrovirus. J Gen Virol 79(Pt 10): 2549–2556 [DOI] [PubMed] [Google Scholar]
- Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG, Thompson JD (2003) Multiple sequence alignment with the Clustal series of programs. Nucleic Acids Res 31: 3497–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulibaly F, Chiu E, Ikeda K, Gutmann S, Haebel PW, Schulze-Briese C, Mori H, Metcalf P (2007) The molecular organization of cypovirus polyhedra. Nature 446: 97–101 [DOI] [PubMed] [Google Scholar]
- Cowtan K (1994) ‘dm': An automated procedure for phase improvement by density modification. Joint CCP4 and ESF-EACBM Newsl Protein Crystallogr 31: 34–38 [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB III, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35 (Web Server issue): W375–W383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de La Fortelle E, Bricogne G (1997) SHARP: a maximum-likelihood heavy-atom parameter refinement program for the MIR and MAD methods. In Methods in Enzymology, Carter CW, Sweet RM (eds), pp 472–494. New York: Academic Press [DOI] [PubMed] [Google Scholar]
- Di X, Sun YK, McCrae MA, Rossmann MG (1991) X-ray powder pattern analysis of cytoplasmic polyhedrosis virus inclusion bodies. Virology 180: 153–158 [DOI] [PubMed] [Google Scholar]
- Donovan JW, White TM (1971) Alkaline hydrolysis of the disulfide bonds of ovomucoid and of low molecular weight aliphatic and aromatic disulfides. Biochemistry 10: 32–38 [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60 (Pt 12 Pt 1): 2126–2132 [DOI] [PubMed] [Google Scholar]
- Felsenstein J (1989) PHYLIP—phylogeny inference package (version 3.2). Cladistics 5: 164–166 [Google Scholar]
- Fujiwara T, Yukibuchi E, Tanaka Y, Yamamoto Y, Tomita Ki, Hukuhara T (1984) X-ray diffraction studies of polyhedral inclusion bodies of nuclear and cyto-plasmic polyhedrosis viruses. Appl Entomol Zool Jpn 19: 402–403 [Google Scholar]
- Gouet P, Courcelle E, Stuart DI, Metoz F (1999) ESPript: analysis of multiple sequence alignments in PostScript. Bioinformatics 15: 305–308 [DOI] [PubMed] [Google Scholar]
- Gross CH, Russell RL, Rohrmann GF (1994) Orgyia pseudotsugata baculovirus p10 and polyhedron envelope protein genes: analysis of their relative expression levels and role in polyhedron structure. J Gen Virol 75(Pt 5): 1115–1123 [DOI] [PubMed] [Google Scholar]
- Holm L, Kaariainen S, Rosenstrom P, Schenkel A (2008) Searching protein structure databases with DaliLite v.3. Bioinformatics 24: 2780–2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda K, Nakazawa H, Alain R, Belloncik S, Mori H (1998) Characterizations of natural and induced polyhedrin gene mutants of Bombyx mori cytoplasmic polyhedrosis viruses. Arch Virol 143: 241–248 [DOI] [PubMed] [Google Scholar]
- Jarvis DL, Bohlmeyer DA, Garcia A Jr (1991) Requirements for nuclear localization and supramolecular assembly of a baculovirus polyhedrin protein. Virology 185: 795–810 [DOI] [PubMed] [Google Scholar]
- Jehle JA, Blissard GW, Bonning BC, Cory JS, Herniou EA, Rohrmann GF, Theilmann DA, Thiem SM, Vlak JM (2006) On the classification and nomenclature of baculoviruses: a proposal for revision. Arch Virol 151: 1257–1266 [DOI] [PubMed] [Google Scholar]
- Jones EY, Stuart DI, Walker NP (1989) Structure of tumour necrosis factor. Nature 338: 225–228 [DOI] [PubMed] [Google Scholar]
- Kadlec J, Loureiro S, Abrescia NG, Stuart DI, Jones IM (2008) The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines. Nat Struct Mol Biol 15: 1024–1030 [DOI] [PubMed] [Google Scholar]
- Katsuma S, Noguchi Y, Shimada T, Nagata M, Kobayashi M, Maeda S (1999) Molecular characterization of baculovirus Bombyx mori nucleopolyhedrovirus polyhedron mutants. Arch Virol 144: 1275–1285 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2004) Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr D Biol Crystallogr 60: 2256–2268 [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372: 774–797 [DOI] [PubMed] [Google Scholar]
- Lin G, Zhong J, Wang X (2000) Abnormal formation of polyhedra resulting from a single mutation in the polyhedrin gene of autographa californica multicapsid nucleopolyhedrovirus. J Inverteb Pathol 76: 13–19 [DOI] [PubMed] [Google Scholar]
- Marlow SA, Wilson LE, Lawrie AM, Wilkinson N, King LA (1998) Assembly of Amsacta moorei entomopoxvirus spheroidin into spheroids following synthesis in insect cells using a baculovirus vector. J Gen Virol 79(Pt 3): 623–628 [DOI] [PubMed] [Google Scholar]
- Mertens PPC, Attoui H, Duncan R, Dermody TS (2005) In Virus Taxonomy: Eighth Report of the International Committee on Taxonomy of Viruses, Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds), pp 447–454. London: Elsevier/Academic Press [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. In Methods in Enzymology. Macromolecular Crystallography, Part A, Carter CWJ, Sweet RM (eds), pp 307–326. San Diego, CA: Academic Press [DOI] [PubMed] [Google Scholar]
- Payne CC, Mertens PP (1983) Cytoplasmic polyhedrosis viruses. In The Reoviridae, Joklik WK (ed), pp 425–504. New York: Plenum Publishing Corp. [Google Scholar]
- Plagemann PG (1972) Nucleotide pools in Novikoff rat hepatoma cells growing in suspension culture. 3. Effects of nucleosides in medium on levels of nucleotides in separate nucleotide pools for nuclear and cytoplasmic RNA synthesis. J Cell Biol 52: 131–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro BM, Generino AP, Acacio CN, Kalapothakis E, Bao SN (2009) Characterization of a new Autographa californica multiple nucleopolyhedrovirus (AcMNPV) polyhedra mutant. Virus Res 140: 1–7 [DOI] [PubMed] [Google Scholar]
- Rohrmann GF (1977) Characterization of N-polyhedrin of two baculovirus strains pathogenic for Orgyia pseudotsugata. Biochemistry 16: 1631–1634 [DOI] [PubMed] [Google Scholar]
- Rohrmann GF (1986) Polyhedrin structure. J Gen Virol 67 (Pt 8): 1499–1513 [DOI] [PubMed] [Google Scholar]
- Rohrmann GF (2008) Baculovirus molecular biology http://www.ncbi.nlm.nih.gov/bookshelf/br.fcgi?book=bacvir
- Scharnhorst DW, Saving KL, Vuturo SB, Cooke PH, Weaver RF (1977) Structural studies on the polyhedral inclusion bodies, virions, and DNA of the nuclear polyhedrosis virus of the cotton bollworm Heliothis zea. J Virol 21: 292–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scharnhorst DW, Weaver RF (1980) Structural analysis of the matrix protein from the nuclear polyhedrosis virus of Heliothis zea. Virology 102: 468–472 [DOI] [PubMed] [Google Scholar]
- Sheldrick GM (2008) A short history of SHELX. Acta Crystallogr A 64 (Pt 1): 112–122 [DOI] [PubMed] [Google Scholar]
- Stuart DI, Levine M, Muirhead H, Stammers DK (1979) Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 Å. J Mol Biol 134: 109–142 [DOI] [PubMed] [Google Scholar]
- Tate J, Liljas L, Scotti P, Christian P, Lin T, Johnson JE (1999) The crystal structure of cricket paralysis virus: the first view of a new virus family. Nat Struct Biol 6: 765–774 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Chapman DA, Jones IM (2003) Improving baculovirus recombination. Nucleic Acids Res 31: E6–E6 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Figure 4
Supplementary Figure 5
Supplementary Figure 6
Supplementary Figure 7
Supplementary Table I
Supplementary Information References
Review Process File