Abstract
Genetic screens in the yeast Saccharomyces cerevisiae have identified many proteins involved in the secretory pathway, most of which have orthologues in higher eukaryotes. To investigate whether there are additional proteins that are required for secretion in metazoans but are absent from yeast, we used genome-wide RNA interference (RNAi) to look for genes required for secretion of recombinant luciferase from Drosophila S2 cells. This identified two novel components of the secretory pathway that are conserved from humans to plants. Gryzun is distantly related to, but distinct from, the Trs130 subunit of the TRAPP complex but is absent from S. cerevisiae. RNAi of human Gryzun (C4orf41) blocks Golgi exit. Kish is a small membrane protein with a previously uncharacterised orthologue in yeast. The screen also identified Drosophila orthologues of almost 60% of the yeast genes essential for secretion. Given this coverage, the small number of novel components suggests that contrary to previous indications the number of essential core components of the secretory pathway is not much greater in metazoans than in yeasts.
Keywords: Golgi, Gryzun, Kish, TMEM167A, TMEM167B
Introduction
The secretory pathway is a defining feature of eukaryotic cells, and allows the synthesis of membrane and secreted proteins in the endoplasmic reticulum (ER) to be separated from the constraints of the cell surface (Bonifacino and Glick, 2004). In the ER proteins are folded, assembled, and covalently modified with features such as N-linked glycans. The pathway that carries these newly made proteins to their destination organelle starts with transport to the Golgi apparatus where further post-translation modification occurs, and from which escaped ER residents are returned back (Jackson, 2009). At the trans side of the Golgi, proteins are sorted into carriers for transport to the plasma membrane or other destination organelles. Over the past 20 years many components of the machinery that mediates this traffic of proteins and lipids have been identified. Some of these studies have been based on the in vitro reconstitution of trafficking events from mammalian cells, in particular transport within the Golgi (Balch et al, 1984), but many have used genetic screens in the budding yeast Saccharomyces cerevisiae. Secretion is necessary for yeast growth, but conditional alleles of essential secretory (SEC) genes can be isolated by virtue of their accumulating internal membranes at the non-permissive temperature (Novick et al, 1980). Combining the analysis of these SEC genes with data from biochemical studies of transport events has revealed how the individual components act together in specific parts of the secretory pathway. As a result the formation of the COPII-coated vesicles that mediate ER to Golgi transport, the retrograde traffic back to the ER from the Golgi by COPI-coated vesicles, and the fusion of membranes by SNAREs, is increasing well understood at a molecular level (Lee et al, 2004; Cai et al, 2007; Wickner and Schekman, 2008).
Most of the components of the secretory pathway identified in yeast have orthologues in higher eukaryotes that have been found to have similar roles. However it is at present unclear if the secretory pathway in higher eukaryotes also requires additional components. Unlike S. cerevisiae, the metazoan Golgi is arranged in a stack and is adjacent to ER exit sites (Ivan et al, 2008; Glick and Nakano, 2009). In addition, the compartments of the secretory pathway are substantially larger in metazoans than in yeast and hence might require additional components for their organisation and structural integrity. Finally, in metazoans the motility of vesicles and organelles is mostly based on microtubules whereas in budding yeast it appears entirely actin based (Vale, 2003; Fagarasanu and Rachubinski, 2007). To search for new components of the secretory pathway in metazoans, we used the ease of genome-wide RNA interference (RNAi) screening in Drosophila cultured cells. Genes can be efficiently knocked down in these cells by treatment with dsRNAs. Moreover, Drosophila has not undergone the whole-genome duplications that occurred in the vertebrate lineage of metazoans, and so genes tend not to have potentially redundant paralogues. Our screen for genes required for efficient secretion identified several novel proteins, at least two of which are required for secretory pathway function in both flies and human cells. One has been previously identified in Drosophila for its potential role in learning and memory. The other has a yeast orthologue that was missed in previous studies probably because of its small size.
Results
A genome-wide RNAi screen for proteins required for secretion
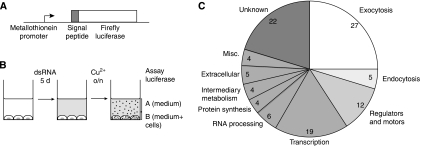
To establish a secretion assay that is compatible with high-throughput screening, we generated a Drosophila S2 cell line that stably expresses a form of firefly luciferase preceded by a signal peptide. This secreted reporter is under the control of a metallothionein promoter, allowing expression to be induced by the addition of copper sulphate to the growth medium (Figure 1A). This cell line was plated in 384-well plates and transfected in duplicate with >24 000 dsRNAs covering the whole Drosophila genome. After 5 days of dsRNA treatment, expression of the reporter was induced with copper sulphate, the medium was harvested and assayed for secreted luciferase activity (Figure 1B). To correct for cell number, we also determined luciferase activity in the cells. We used the ratio of secreted luciferase versus the total amount of luciferase in each well to normalise the data. As the genes involved in secretion are not expected to be clustered in a particular plate, we used the variation within each plate for statistical analysis, and assigned a z-score, that is the number of standard deviations by which the result from one well differs from the mean value for the whole plate. Genes for which both duplicates scored below a z-score threshold of −2 (i.e. secretion was reduced by more than two standard deviations below the mean) were considered as ‘primary hits'. We also set aside the next highest set (i.e. both z-scores being at least 1.5 standard deviations below the mean) and considered them as being ‘secondary hits' and possibly significant.
Figure 1.
A genome-wide RNAi screen for proteins required for secretion. (A) Schematic diagram of the reporter construct used to generate the stable S2 cell line used for the screen. The metallothionein promoter and the BiP signal peptide are from Drosophila. (B) Summary of screening protocol. Secretory pathway function was quantified by determining the proportion of total luciferase activity in the well that was in the medium fraction (i.e. A/A+B). (C) Distribution of the functions of the 108 hits from the screen (for more details see Supplementary Table 1).
Hits from the screen are enriched in known components of membrane traffic
The screen identified 108 primary hits (Supplementary Table I). Further examination revealed that 32 (30%) encode known components of membrane traffic, the large majority of which are involved in exocytosis (27 of 32, i.e. 25% of the total hits; Figure 1C). Of the remaining 76, 19 are known to have a role in transcription, including 7 of the 13 TATA binding protein-associated factors, and a further 23 are known components of other cellular processes. Some of these proteins could be required for sustained expression of the secreted luciferase reporter or may reflect indirect effects or false positives. The screen also recovered 22 genes with no known function (of which 11 have human orthologues), and 12 encoding proteins such as motor proteins, kinases, or G-protein regulators, functions that could conceivably impact on the secretory pathway. These 34 genes were selected for re-screening. Among the 198 secondary hits (at least 1.5 standard deviations below the mean), 21 (11%) are known components of membrane traffic, of which 12 are involved in exocytosis (i.e. only 6% of the total hits), indicating a higher level of background than for the primary hits. Of the remaining 177 genes, 13 encode proteins that are of unknown function but have a detectable human orthologue. These 13 were also included in those selected for re-screening to reduce the chance that a novel component of the secretory pathway was missed.
Re-screening of candidate novel components of secretion
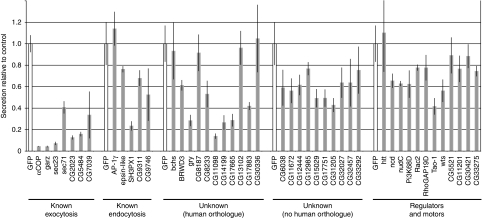
To validate the putative components of membrane traffic identified in the primary screen, we used the same assay as before but with a different set of dsRNAs designed to avoid off-target effects (Kulkarni et al, 2006; Ma et al, 2006). As positive controls, we included several primary hits that affect known components of the secretory pathway. Also included were the five genes from the screen that encode known components of endocytic traffic. The re-screen was performed in triplicate in a 96-well plate format, and the results are shown in Figure 2.
Figure 2.
Re-screen of potential hits from the genome-wide screen. Secretion of luciferase from S2 cells treated with dsRNA corresponding to the indicated genes. Secretion was quantified as in Figure 1 (i.e. the proportion of total activity present in the well that was in the medium), and normalised with respect to the GFP control included with each set (means of triplicates, error bars indicating standard deviation). Of the ‘known exocytosis' hits CG2023, CG5484, and CG7039 are the Drosophila orthologues of Sec20, Yif1, and ARFRP1 respectively. In total 13 of the hits for known components of exocytosis were re-screened with different amplicons and all were positive again, indicating the robustness of the assay system.
Knocking down the known components of the exocytic pathway led to a substantial reduction in luciferase secretion as expected. dsRNA against the novel genes produced a range of effects with several causing a reduction of secretion comparable to that seen with known secretory pathway components. Inhibition of the genes encoding proteins presumed to act in other processes, but which could impact on the secretory pathway, gave weaker effects on secretion, with the strongest inhibition seen after knockdown of the putative protein kinase Tao-1. Given that the positive controls showed a reduction in luciferase secretion of 60–90%, we decided to focus on all those genes whose knockdown showed a reduction of at least 50% and which had a detectable orthologue in humans. Thus, six novel genes were taken forward for further study, these being Gryzun, CG8233, CG11098, CG14199, CG17665, and CG17883. We also re-examined the Tao-1 kinase and SH3PX1 as the latter was the only ‘endocytic' gene that had a substantial effect on secretion upon re-screen. Below we briefly describe the results of our preliminary analysis of the six novel genes as well as of Tao-1 and SH3PX1.
Localisation of the eight candidate proteins
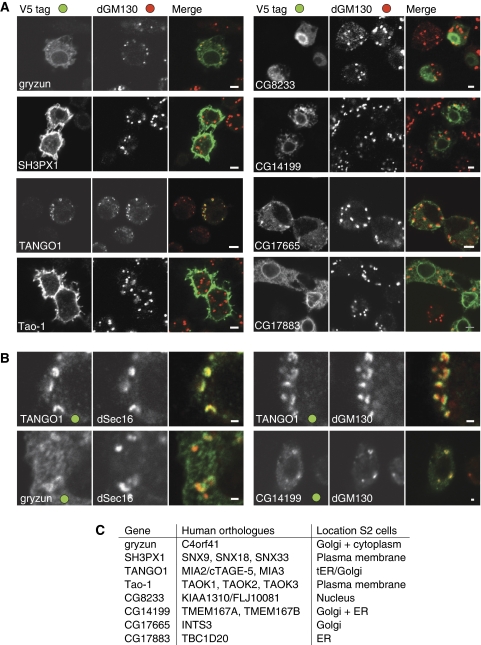
To characterise the site of action of the proteins encoded by the eight selected genes, we first expressed the genes in S2 cells with an epitope tag at the C terminus, and determined the intracellular localisation of the tagged protein by immunofluorescence. The results are shown in Figure 3A. In the rest of the paper we report a more detailed investigation of two, that is Gryzun and CG14199, but first we briefly discuss the results for the other six proteins that we did not investigate further as they did not seem likely to be involved directly in secretion, or in two cases were identified as components of the secretory pathway in reports from others (CG11098 and CG17883).
Figure 3.
Localisation of tagged forms of the proteins encoded by selected genes identified in the screen. (A, B) Confocal micrographs of S2 cells transfected with plasmids expressing the indicated genes tagged at the C terminus with the V5 epitope. Cells were labelled for the V5 tag and the Golgi marker dGM130, or the ER exit site component dSec16. Scale bars 5 μm (A) or 1 μm (B). (C) Summary of the localisation data in (A, B) for the eight selected genes along with the gene names for their human orthologues.
SH3PX1. This is a soluble protein that contains SH3, PX, and BAR domains. Of its three mammalian orthologues, SNX9 has been shown to interact with dynamin and to be involved in clathrin-dependent endocytosis (Soulet et al, 2005). By contrast, SNX18 and SNX33 have been suggested to exert their effect in vesicle budding from endosomes (Haberg et al, 2008; Heiseke et al, 2008). Tagged SH3PX1 is predominantly localised to the cell surface. RNAi against SH3PX1 causes cells to enlarge, although they still contained a single nucleus (data not shown).
Tao-1. Tao-1 is a predicted protein kinase of the Ste20 family. It has three paralogues in humans, Tao1, Tao2, and Tao3, which are MAP kinase kinase kinases (Raman et al, 2007). The tagged Drosophila protein was found localised at the plasma membrane. As with SH3PX1, RNAi leads to the appearance of enlarged mononucleate cells with an apparently normal Golgi. It is conceivable that RNAi of Tao-1 alters the phosphorylation of a specific set of proteins that includes components of the secretory pathway.
CG11098/TANGO1. This protein is predicted to have a leader peptide and a single transmembrane domain. It has an SH3-related domain following the leader peptide and is predicted to have a long coiled-coil domain in its cytosolic tail. It is conserved in some metazoans, and has at least two mammalian orthologues (MIA3 and MIA2/cTAGE-5) (Stoll et al, 2001; Stoll and Bosserhoff, 2008). There are two further relatives in mammals that appear to encode just the leader peptide and the N-terminal SH3 domain. For one of these, melanoma inhibitory activity/cartilage-derived retinoic-acid-sensitive protein (MIA/CD-RAP), the N-terminal SH3 domain was isolated as a secreted product of melanomas, and reported to affect growth and adhesion (Lougheed et al, 2001; Stoll et al, 2001). CG11098 was identified in a similar RNAi screen for Drosophila genes required for secretion in S2 cells, and was named ‘transport and Golgi organisation 1' (TANGO1) (Bard et al, 2006). The C-terminally tagged form is found close to the Golgi apparatus, but recently the human TANGO1 orthologue MIA3 has been reported to be in ER exit sites and proposed to be involved in sorting of cargo into COPII vesicles (Saito et al, 2009). In S2 cells ER exit sites are next to the Golgi and indeed Drosophila TANGO1 colocalises with the exit site marker dSec16, but is displaced from the cis-Golgi marker dGM130 (Figure 3B). Given the recent studies of TANGO1 and its orthologues by others, we did not pursue it further.
CG8233. This is a soluble protein that is present only in metazoans. It appears to be nuclear, and as such it seems unlikely to be directly involved in membrane traffic. Indeed it has been reported to be part of a complex of unknown function that contains the MOF lysine acetyltransferase subunit that is also present in the dosage compensation complex that regulates chromatin structure in males (Mendjan et al, 2006). It is conceivable that RNAi of CG8233 alters the expression of a specific set of genes that includes those that encode components of the secretory pathway. Recently CG8233 was also identified in an RNAi screen for defects in centriole duplication and also found to be nuclear (Rcd1; Dobbelaere et al, 2008).
CG17665. A soluble protein whose mammalian orthologue is the Int3 subunit of the ‘integrator complex', which binds to RNA polymerase II. This complex appears to be involved in the processing of small nuclear RNAs. It contains at least 12 subunits, one of which (Int11) is related to RNA-processing enzymes (Baillat et al, 2005; Egloff et al, 2007). However, the functions of the other subunits are not known, and Int3 is not related to any known protein. The subunits of the complex are conserved from mammals to plants, but are absent from fungi. We find that, surprisingly, tagged Int3 is predominantly localised to the Golgi apparatus. Golgi localisation may reflect a Drosophila-specific adaptation as epitope-tagged mammalian Int3 appeared nuclear with no detectable Golgi staining (data not shown).
CG17883. This protein is predicted to have a C-terminal transmembrane domain and a TBC domain that, in other proteins, has been found to have GAP activity on Rab family members (Pan et al, 2006). It has a single human orthologue, TBC1D20, which was recently reported to have GAP activity on the small G-protein Rab1 and to be required for secretory pathway function (Haas et al, 2007; Sklan et al, 2007). We found that the tagged protein was localised to the ER, consistent with the distribution reported for mammalian cells. Therefore our results provide confirmation that CG17883 is likely to be a genuine component of the secretory pathway.
Of the above six proteins, four seem likely to affect secretion indirectly, perhaps by participating in endocytosis or transcription, and only two appear to be strong candidates to be directly involved in the secretory pathway (TANGO1 and CG17883/TBC1D20). However, these two proteins have been investigated recently by others in flies or mammals. Therefore we confirmed that the RNAi knockdowns had worked as expected (Supplementary Figure 1), and then concentrated in detail on the two remaining proteins from the eight selected (Gryzun and CG14199). We describe below what is known about them and provide further evidence that they are important for protein secretion in Drosophila and other species.
Gryzun and CG14199 are potential novel components of the secretory pathway
Of the eight candidate proteins that we localised, Gryzun and CG14199 appear good candidates for a role in the secretory pathway, but have not been previously characterised.
Gryzun (CG17569). Predicted to be a large soluble protein, it is conserved from mammals to plants, but absent from some yeasts including S. cerevisiae. It was named Gryzun after one of Pavlov's dogs as it is encoded by one of 60 genes identified in a P-element insertion screen for Drosophila mutants that cause defects in long-term memory (Dubnau et al, 2003). The P-element insertion site was not precisely mapped but appears to lie upstream of the gene and is thus unlikely to cause a null allele. Our PSI-BLAST searches reveal that Gryzun is distantly related to TRS130/TRAPPC10 (Supplementary Figure 2). This protein is a subunit of the TRAPP complex that is involved in vesicle transport within the Golgi (Sacher et al, 1998, 2008). The C-terminally tagged form of Gryzun shows mostly cytoplasmic staining in S2 cells, but some accumulation on the Golgi was also visible (Figure 3A). The Golgi in S2 cells is localised next to ER exit sites, but the two can be resolved by light microscopy (Ivan et al, 2008). This is illustrated by comparison of TANGO1 with dSec16 (ER exit sites) and dGM130 (cis-Golgi). TANGO1 has been recently shown to be in ER exit sites (Saito et al, 2009), and is coincident with Sec16 but slightly displaced from GM130 (Figure 3B). Gryzun is less clearly resolved due to the cytosolic pool but the Golgi-associated material appears clearly displaced from dSec16 (Figure 3B), and so it is unlikely to be in concentrated in ER exit sites.
CG14199. This is a small protein of only 72 residues that is predicted to contain two transmembrane domains. It is exceptionally well conserved in evolution with a single orthologue present in most species from protozoa to humans (TMEM167A), with vertebrates also having a second paralogue (TMEM167B /C1orf119). The tagged Drosophila protein accumulates in the ER and Golgi (Figure 3A), with both nuclear envelope staining but also some colocalisation with GM130, with the latter being clearer at lower expression levels (Figure 3B). This, combined with data described below, strongly suggests a role in secretory pathway and we have named this gene kish (ksh) (transliterated from the Hungarian for small).
Gryzun and Kish are required for secretion in Drosophila tissues
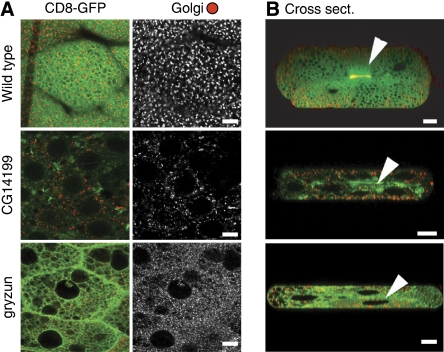
To assess the function of the two selected proteins in intact tissues, we turned to transgenic RNAi in Drosophila. To monitor secretion, we created a transgene expressing CD8-GFP, a transmembrane protein often used as a marker of the plasma membrane (Lee and Luo, 1999; Xu et al, 2002). In this background, RNAi constructs targeted against Gryzun or Kish were expressed in the larval salivary gland, an organ specialised in secretion. Figure 4 shows that as previously reported wild-type CD8-GFP accumulates on the apical surface of the secretory cells that line the duct of the salivary gland (Xu et al, 2002). In contrast, the RNAi construct against Gryzun caused a reduction in apical expression of CD8-GFP and also appeared to disrupt the normal appearance of the Golgi. Note also that the glue granules (which form in the Golgi) are larger and more heterogeneous than in the wild type, and that some CD8-GFP now appears on the basal surface of the cells, phenotypes consistent with a perturbation of Golgi function. Expression of the RNAi construct against Kish resulted in markedly reduced CD8-GFP expression and loss of surface accumulation, indicating severe perturbation of the secretory pathway. Consistent with this disruption in secretory function, expression of either RNAi construct in wing discs caused lethality in pupae. In addition, a recent genome-wide gene disruption project using the Minos transposable element has generated an insertion in the gryzun coding region at residue 447 of the 1338 total (Mi[ET1]gryMB06920), and we found that flies homozygous for this allele die at the end of embryogenesis.
Figure 4.
Effect of RNAi against CG14199/kish and gryzun on CD8-GFP expression in the Drosophila salivary gland. (A, B) Confocal micrographs of larval L3 salivary glands expressing CD8-GFP and stained for the Golgi marker dGolgin-245. The flies either express no dsRNA or express a dsRNA against the indicated gene in the salivary gland using the Gal4 system. In the absence of dsRNA, CD8-GFP accumulates as expected on the apical plasma membrane that lines the secretory duct (arrowhead in panel B). Nuclei are the very large black holes visible in the cross sections and some of the horizontal sections. For CG14199 and gryzun z-stacks of several salivary glands were analysed for each, with the same phenotypes observed in all cases, including loss of the concentration of the CD8-GFP reporter on the apical membrane. The images are taken at the same settings so that GFP levels can be compared directly. Scale bars=10 μm.
Human orthologues of Gryzun and Kish are required for normal secretion
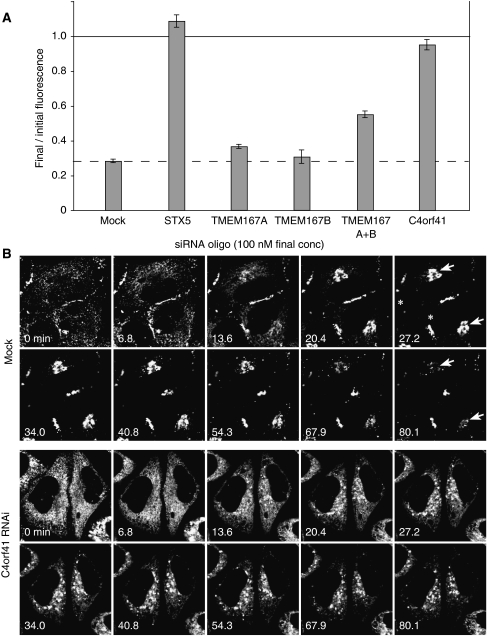
The human genome contains a single orthologue of Gryzun encoded by the gene C4orf41, and two paralogues of Kish (TMEM167A and TMEM167B). To examine their role in secretion, we used a cultured cell line expressing a GFP-labelled fusion protein (GFP-FM4-hGH) that has been engineered to aggregate reversibly in the ER so that it can be released into the secretory pathway by the addition of a small molecule (Rivera et al, 2000). Secretion can then be followed by FACS analysis of the amount of GFP fusion remaining associated with the cell. Figure 5A shows that the loss of GFP-FM4-hGH from the cells is blocked by treating cells with siRNAs directed against C4orf41, to a degree close to that seen with the positive control (siRNA against syntaxin-5). siRNAs directed against either TMEM167A or TMEM167B individually did not show a significant effect, but combining the two caused a partial, yet reproducible, reduction in secretion (Figure 5A).
Figure 5.
C4orf41 depletion blocks constitutive secretion from mammalian cells. (A) siRNAs against C4orf41, TMEM167A, TMEM167B, and syntaxin 5 (STX5) were transfected into a HeLaM-derived cell line (C1) that expresses a regulated secretory reporter. At 4 days after RNAi treatment, secretion of the GFP-FM4-hGH reporter was induced by adding the aggregation preventing small molecule AP21998. The fluorescence remaining in the cells was determined at the start and after 80 min using flow cytometry. The result is expressed as the proportion of material that remains in the cell (error bars show fractional deviation of triplicate data points, and the experiment was repeated twice). To confirm that the C4orf41 secretion phenotype was genuine and not an off-target effect, the C4orf41 siRNA pool was deconvoluted and three out of the four oligonucleotides found to give a substantial block in secretion. (B) Confocal time-lapse imaging of the release of the GFP-FM4-hGH reporter from C1 cells that had been mock-treated, or with siRNAs for C4orf41. Release was initiated by the addition of AP21998 after 1 min, with frames shown for the indicated time points (minutes). There is a low level of basal secretion that results in GFP accumulation on the surface at cell contact sites (asterisks at 27.2 min). In mock-treated cells the material released from the ER enters the Golgi (arrowheads at 27.2 min) and then leaves. In C4orf41 siRNA-treated cells the reduction in basal secretion causes an increase in ER labelling at the start of the experiment. Nonetheless, a substantial fraction of the material leaves the ER and accumulates in scattered structures in the cytoplasm. The full time series are in Supplementary Movies 1 and 2.
The effects of the siRNAs against C4orf41 were examined further using time-lapse imaging to follow the movement of GFP-FM4-hGH after release from the ER. In mock-treated cells the GFP reporter moves from the ER to the Golgi from which it exits in small carriers. In cells treated with C4orf41 siRNAs, a substantial amount of the GFP fusion still exited the ER but then accumulated in scattered structures from which it did not appear to leave, indicating that secretion had been blocked after exit from the ER (Figure 5B; Supplementary Movies 1 and 2).
C4orf41, the human orthologue of Gryzun is required for traffic through the Golgi
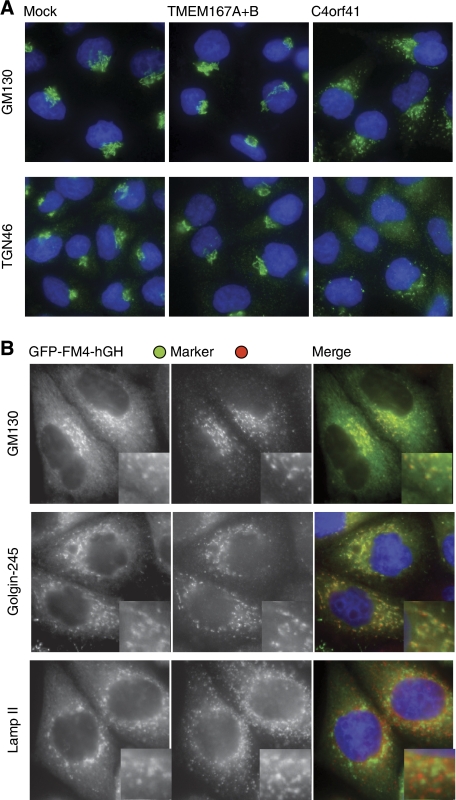
The post-ER block in secretion in cells treated with siRNAs against C4orf41 suggests a defect in Golgi function. To investigate this possibility in more detail, we initially examined the distribution of Golgi markers. Upon C4orf41 siRNA treatment the cis-Golgi marker GM130 becomes scattered (Figure 6A). In addition, the TGN recycling proteins TGN46 becomes severely disrupted, or apparently absent, a phenotype seen previously with other treatments seen that disrupt the Golgi (Levine and Munro, 1998). We did not detect substantial defects in cells treated with the siRNAs against both Kish paralogues, consistent with the milder phenotype on secretion. The scattered structures in which the GFP-labelled cargo accumulates after C4orf41 siRNA treatment colocalised with the Golgi markers GM130 and golgin-245 (Figure 6B). In contrast the ER and lysosomes appeared unperturbed (Figure 6B; Supplementary Movie 2; data not shown). Taken together these results indicate that C4orf41 is required for maintaining the Golgi's integrity and competence to allow secreted proteins to pass through to the cell surface.
Figure 6.
Depletion of C4orf41 fragments the Golgi ribbon and prevents secretory cargo moving beyond the Golgi fragments. (A) Wide-field micrographs of HeLaM cells treated with the indicated siRNAs and stained for the Golgi markers GM130 or TGN46, and with DAPI to visualise the nucleus (blue). Quantitation of mock-, TMEM167-, and C4orf41-treated cells revealed perturbation of GM130 in 0, 0, and 93% respectively (n=70), and of TGN46 in 0, 5, and 95% respectively (n=110). (B) Wide-field micrographs of C1 cells treated with siRNA against C4orf41 and then fixed 80 min after induction of the release of GFP-FM4-hGH from the ER as in Figure 4. The cells were labelled with the indicated markers for the cis-Golgi (GM130), the trans-Golgi (golgin-245), and lysosomes (Lamp II). To confirm the Golgi fragmentation phenotype, we performed each staining experiment with two different oligonucleotides against C4orf41, and representative results are shown.
Ksh1, the yeast orthologue of Kish, has a role in the secretory pathway
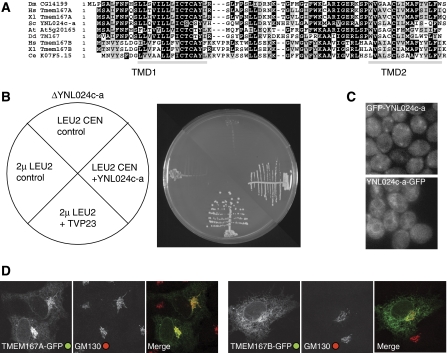
The above experiments suggest that both the Drosophila and the human orthologues of Kish have a role in secretion, but the siRNA experiments in human cells showed only a partial block. This may reflect the need to knock down two proteins making it harder to get close to a null phenotype, but it precluded identifying the site of action. To seek more evidence that this protein is involved in the secretory pathway, we examined the yeast S. cerevisiae. As noted above S. cerevisiae and some other budding yeasts have lost Gryzun, but they do have an orthologue of Kish that is encoded by the previously uncharacterised open reading frame (ORF) YNL024c-a. This yeast protein has only 72 residues and is 56% identical to the Drosophila protein, and we will refer to it as Ksh1 (Figure 7A). It was not included in the initial annotation of the yeast genome as it was below the minimum ORF size threshold of 100 residues. This small size may have also reduced the chance of detection in genetic screens. It has been reported to be essential for growth of yeast (Kastenmayer, 2006), an observation that we have confirmed (Figure 7B).
Figure 7.
Analysis of the yeast and mammalian orthologues of CG14199/Kish. (A) An alignment of CG14199/Kish with orthologues from the indicated species (Hs, Homo sapiens; Xl, Xenopus laevis; Sc, S. cerevisiae; At, Arabidopsis thaliana; Dd, Dictyostelium discoideum; Ce, Caenorhabditis elegans). Shading indicates residues that are identical (black) or conserved (grey) in five or more of the nine sequences, and with grey bars being predicted transmembrane domains. Vertebrates have two paralogues of Kish called TMEM167A and TMEM167B. (B) Growth of S. cerevisiae with a deletion in YNL024c-a (KSH1), the orthologue of CG14199. Strains were created that lacked the genomic copy of YNL024c-a but were kept alive by the YNL024c-a gene on a URA3 CEN ‘covering' plasmid, and also contained the indicated CEN (pRS415) or 2 μ LEU2 plasmids. The strains were streaked on 5-fluororotic acid (5-FOA) to eject the URA3 covering plasmid and then incubated for 3 days at 25°C before photography. The LEU2 plasmids containing YNL024c-a and TVP23 expressed from their own promoters support growth in the absence of genomic YNL024c-a, but this was not seen with the control plasmids (an empty CEN plasmid, or a 2μ plasmid expressing an irrelevant gene (LRC6)). (C) Wide-field micrographs of wild-type strain BY4741 with GFP integrated at the N or C terminus of the endogenous YNL024c-a gene by PCR-based homologous recombination. For the N-terminal fusion expression was promoted by a constitutive form of the PHO5 promoter. (D) Confocal micrographs of COS cells expressing the human orthologues of Kish with GFP attached to the C terminus, and labelled for the Golgi marker GM130.
Despite extensive efforts, we were unable to generate a reliable conditional allele of KSH1. Therefore, to obtain an indication of its function, we looked for genes that could suppress a KSH1 deletion when overexpressed. Thus we screened a 2μ-based overexpression genomic library for the ability to rescue growth following the loss of a plasmid bearing the sole copy in the strain of the KSH1 gene. From 10 000 transformants four plasmids containing KSH1 were recovered. In addition, we also recovered a plasmid encoding Tvp23, a small polytopic membrane protein of the Golgi apparatus (Inadome et al, 2007). Examination of glycoproteins expressed in the rescued strain revealed defects in processing (data not shown), suggesting Golgi malfunction. The function of Tvp23 is unknown, and although not essential for growth, it is conserved from protozoa to humans. Recently Tvp23 was identified in another yeast multicopy suppressor screen as being able to suppress a temperature-sensitive allele of the SNARE protein Vti1 suggesting a role for Tvp23 in endosome to Golgi traffic (Stein et al, 2009). Vti1 functions in both the Golgi with Sed5 and endosomes with Pep12 (Lupashin et al, 1997; von Mollard et al, 1997), but Tvp23 only suppresses a Golgi-specific allele of Vti1 and not an endosome-specific allele indicating that excess Tvp23 is able to suppress a defect in early Golgi function (Stein et al, 2009). Attempts to localise the Ksh1 protein were hampered by our inability to generate a tagged form that could rescue the lethality of the null strain (tags at both the N and C termini were tested). Notwithstanding this caveat, an N-terminally tagged form of Ksh1 gave a punctate pattern characteristic of the Golgi (Figure 7C). A similar pattern was seen for the tagged forms of the fly and human orthologues (Figures 3 and 7D). Taken together, these results from flies, human cells, and yeast strongly suggest a role for Kish/Kis/TMEM167 in the early part of the secretory pathway.
Discussion
The genome-wide screen reported here has identified two previously uncharacterised proteins that are involved in secretion. These two proteins are highly conserved in evolution suggesting that they may be core components of the secretory pathway. The small size of Ksh1 probably explains why it has not emerged from previous studies in yeast, especially as the small ORFs, which were not included in the initial genome annotation, have also been omitted from most of the subsequent high-throughput screens. Gryzun shows a distant sequence similarity to the Trs130 subunit of the TRAPP complex and has orthologues in plants and protozoa. Human Gryzun is more closely related to plant Gryzun than it is to human Trs130, indicating that the two proteins diverged before the last common ancestor of eukaryotes. Interestingly, Gryzun orthologues are also present in most filamentous fungi and budding yeasts, but the gene appears to have been lost from budding yeasts of the Saccharomyces complex. The orthologue in zebrafish is one of several genes identified in a screen for mutant embryos that have enlarged livers, and thus named ‘foie gras' (Sadler et al, 2005). The foie gras mutant is a recessive embryonic lethal and also shows defects in the formation of other tissues, although the underlying cause was not resolved. The role of Trs130 itself is somewhat unclear as the only undisputed function of TRAPP is as an exchange factor for Ypt1, the yeast orthologue of Rab1, and this activity requires only four of the core subunits of TRAPP (Bet3, Bet5, Trs23, and Trs31) and not Trs130 (Sacher et al, 1998, 2008). Interestingly, when the human TRAPP complex was purified using TAP-tagged human Bet3 it was noted to co-precipitate with several proteins that were not orthologues of known TRAPP subunits, and one of these is the human orthologue of Gryzun (‘hypothetical protein AL136752.1'; Gavin et al, 2002). Thus, it seems possible that Gryzun is an alternative/additional TRAPP subunit, although our attempts to use immunoprecipitation to detect an interaction between tagged forms of Gryzun and Bet3 co-expressed in Drosophila cultured cells were not successful, indicating that further studies may require analysis of the native complex (unpublished observations).
The evolutionary pressure for compaction of fungal genomes means that S. cerevisiae will inevitably lack some features of the secretory pathway that are present in higher eukaryotes. However, the results of our screen suggest that there may not be a large number of core essential components of the metazoan secretory pathway that are absent from S. cerevisiae. Our screen identified the Drosophila orthologues of many of the known essential components of the yeast secretory pathway. Some components such as COG or exocyst subunits are essential in yeast but do not appear to be essential for secretion in Drosophila or S2 cells (Farkas et al, 2003; Stuart et al, 2007). If these are set aside, our screen identified Drosophila orthologues of 44% of the known essential components of the yeast secretory pathway (Supplementary Figure 3). Despite this substantial coverage, only four novel components were found: Gryzun, TANGO1, Kish, and dTBC1D20. Of these, only the latter two have orthologues in S. cerevisiae. If the secondary hits are also included in the analysis, then the coverage of yeast genes increases to 59%. However, when the 13 secondary hits of unknown function that had human orthologues were re-tested in the re-screen, none had a detectable role in secretion (Supplementary Table 2). Clearly our screen may well have missed some of the essential components of the secretory pathway, but given the level of coverage the number missed is unlikely to be large.
A previous RNAi screen in S2 cells identified 110 genes putatively required for secretion. Among these, only 26 were known trafficking components whereas most of the remainder were novel (Bard et al, 2006). Fourteen of these novel genes were characterised further and named TANGO1-14. Only two of them, TANGO1 and TANGO3, were recovered in our screen. RNAi against TANGO1 had a significant effect on secretion even after re-screening, and therefore appears to be a real hit. In our study, RNAi against TANGO3, which has no known function, did not have strong effect upon re-screening. It is a membrane protein that has many close relatives in Drosophilidae, but no clear orthologues outside insects (TreeFam family TF327319; Li et al, 2006). It is of course conceivable that the other TANGOs have more subtle roles in the secretory pathway that our assay did not detect. However, our screen was validated after the potential for off-target effects was realised (Kulkarni et al, 2006; Ma et al, 2006). We were therefore able to re-screen each of our hits with distinct amplicons that were specifically designed to minimise off-target problems. Indeed TANGO8, a 58 residue ORF that lacks detectable orthologues even in other Drosophila species, contains a CAN trinucleotide repeat of the type now known to be responsible for off-target effects. In addition, our screen benefited from an inbuilt correction for variation in cell number. This was achieved by normalising the amount of secreted luciferase to that present in the cell fraction. Normalisation is likely to be crucial for any functional readout sensitive to cell number and hence cell viability and growth. Therefore, it is probably unwise to assume that all of the TANGO proteins, and the other approximately 70 mostly uncharacterised genes identified in the previous screen, are directly involved in the secretory pathway. Indeed, recent studies have revealed that TANGO7 is a translation initiation factor (Luke-Glaser et al, 2007), and TANGO11 is a mitochondrial protein required for constitutive and induced fission of mitochondria and peroxisomes (Gandre-Babbe and van der Bliek, 2008). Moreover, mice lacking both mammalian orthologues of TANGO13 are viable and show no apparent defect in protein secretion (Westmuckett et al, 2008).
Our data suggest that the secretory pathway of metazoans may not involve many more essential components than those that have been identified in S. cerevisiae. Of course it seems certain that metazoans have evolved additional components and regulators that are required for the formation of complex structures such as ER exit sites, the Golgi ribbon, and connections to microtubules. Such features are likely to optimise secretion in the context of survival and reproductive fitness of the whole organism. Likewise, we expect that components involved in specialist or polarised secretory events that are not recapitulated in S2 cells remain to be found. Nonetheless, we may be nearing the identification of all of the core components of the eukaryotic secretory pathway and this should allow a system-level approach and modelling of the fundamental mechanisms of the pathway and thus provide a framework for understanding its regulation and cell-type-specific specialisation.
Materials and methods
Genome-wide RNAi screen
To construct a reporter for secretion, the firefly luciferase gene was cloned between the XbaI and EcoRI sites of plasmid pMT/BiP/V5-His (Invitrogen). The resulting plasmid was stably transfected in S2 cells by co-transfection and selection with hygromycin. The genome-wide screen was performed at the Drosophila RNAi Screening Center (DRSC, Harvard Medical School). In brief, 20 μl of cells (approximately 15–18 000 cells) in serum-free Schneider's medium were seeded in 384-well plates (white solid bottom) containing 0.1 μg (5 μl) dsRNA and incubated for 1 h. Schneider's medium (40 μl) containing 16.25% fetal calf serum (FCS) was then added and plates were incubated for 5 days at 25°C. Plates were then centrifuged and 40 μl supernatant was aspirated using a 384-well CyBi-Well vario workstation (CyBio). Medium was replaced by 20 μl complete Schneider's medium containing CuSO4 (100 μM final) and incubated for an additional day. Subsequently 20 μl of the conditioned medium was transferred into a new plate and an additional 5 μl from the plate containing the cells was aspirated to leave the cells and 20 μl of medium. After adding an equal amount (20 μl) of firefly luciferase substrate in lysis buffer (Dual-Glo; Promega) to both the medium and the cells-plus-medium fractions, light emission was assayed using a plate reader (Analyst GT; Molecular Devices). The secondary screen was performed in 96-well plates using a similar protocol, with cell number, dsRNA concentration, and medium volume adjusted accordingly. dsRNA was produced from independent re-screen amplicons provided by the DRSC (RNAeasy; Qiagen). For RT–PCR validation, total RNA was purified using SV Total RNA Isolation System (Promega), and cDNA was produced from 1 μg of RNA per sample, using random primers and ImProm-II Reverse Transcription System (Promega). Each PCR was then performed with 1 μl cDNA.
Fly stocks
UAS-RNAi lines against CG14199 (ID 40884) and gryzun (ID 41657) were obtained from the Vienna Drosophila RNAi Center. To investigate salivary glands, we crossed these with a line containing AB1-Gal4 (salivary gland driver) and the tubulin-CD8-GFP transgene, which is expressed throughout the larva. To construct the latter, we obtained a Bluescript-derived plasmid encoding mouse CD8 fused to EGFP from Lee and Luo (1999). The CD8-GFP fragment was excised with XhoI (blunted) and XbaI and ligated into the NotI (blunted) and XbaI sites of a variant of CaSpeR4 carrying the tubulin1α promoter and an SV40 polyadenylation site (Lee and Luo, 1999). The stock carrying the Minos insertion in Gryzun (w[1118]; Mi{ET1}gry[MB06920]/TM6C, Sb[1]) was obtained from the Bloomington Drosophila Stock Center.
Pharmacologically regulated constitutive secretion assay
The vector (pC4S1-FM4-FCS-hGH) encoding the pharmacologically regulated reporter construct was obtained from Ariad Pharmaceuticals, Cambridge, MA (Rivera et al, 2000). The reporter construct was modified so that EGFP was cloned in-frame between the signal sequence and the first FM4 aggregation domain. The insert encoding the reporter construct was then moved into the retroviral expression vector pQXCIP (Clontech). A stable cell line (C1) expressing the reporter construct was generated by virally transducing HeLaM cells as previously described (Peden et al, 2004). The properties of this cell line will be described in detail elsewhere (Gordon and Peden, unpublished). To initiate secretion C1 cells were incubated with 1 μM AP21998 (Ariad Pharmaceuticals, Cambridge, MA) at 37°C in 5% CO2 for 80 min. Secretion was halted by placing the C1 cells on ice. The cells were then detached by incubation with trypsin-EDTA (PAA Laboratories) at 4°C for two hours. The trypsin was quenched using DME supplemented with FCS and the samples were then analysed for EGFP fluorescence using a FACSCalibur flow cytometer (BD Biosciences). Dead cells were excluded using 7-aminoactinomycin-D. The mean fluorescence (FL1) for each sample was then calculated using the software FlowJo (Tree Star Inc.). Live cell microscopy of C1 cells was at 37°C on an incubated Zeiss LSM510 confocal using a Plan-Apochromat × 63/1.4 oil DIC lens.
siRNA transfection
C1 or HeLaM cells were transfected with OnTargetPlus SMARTpool siRNA oligonucleotides (100 nM final concentration; Dharmacon) using Oligofectamine (Invitrogen) as previously described (Motley et al, 2003). Secretion analysis or microscopy was performed 96 h after the initial transfection. To measure the amount of EGFP secreted, each set of siRNA oligonucleotides was transfected in duplicate. The first sample was used for the initial time point (0 min) and the second sample was used for the final time point (80 min). The mean fluorescence of each sample was measured and the proportion of GFP remaining in the cells was calculated. Single siRNAs for gryzun (e.g. CATTTGATGTTGCGGTTAA and CATTTGATGTTGCGGTTAA) gave the same time-lapse and immunofluorescence phenotypes as the OnTargetPlus SMARTpool.
Immunofluorescence
Drosophila S2 cells (DMel; Invitrogen) were treated with dsRNA as described previously (Bettencourt-Dias et al, 2005), or transfected using Cellfectin (Invitrogen) with plasmids expressing cDNAs tagged at the C terminus with the V5 epitope tag. The plasmids were generated by cloning cDNAs (Geneservice) into pMT/V5 (Invitrogen), and expression was induced by overnight incubation with 100 μM CuSO4 or for 45 min with 500 μM CuSO4 followed by a 2 h chase. Cells were fixed with 4% paraformaldehyde, permeabilised with 1% Triton X-100, and processed for immunofluorescence as described previously (Sinka et al, 2008). Primary antibodies against the V5 tag (Invitrogen), dGM130 (Abcam), dSec16 (Ivan et al, 2008), and dGolgin-245 (Sinka et al, 2008) were detected with Alexa-labelled secondary antibodies (Invitrogen), and imaged using a confocal microscope (LSM 510; Carl Zeiss).
C1 and HeLaM cells were fixed, permeabilised, and labelled as previously described (Peden et al, 2004). Mouse anti-GM130, anti-golgin-245 (BD Biosciences), and rabbit anti-TGN46 (kind gift from Dr Matthew Seaman, University of Cambridge) were detected by fluorescence microscopy with a Zeiss Axioplan microscope equipped with a × 63 oil objective, a MicroMAX CCD camera (Princeton Instruments), and a SEDAT quad pass filter set (Chroma Technology).
Supplementary Material
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Table 1
Supplementary Table 2
Supplementary Movie 1
Supplementary Movie 2
Review Process File
Acknowledgments
We thank Bernard Mathey-Prevot, Norbert Perrimon, and members of the DRSC for important advice during the genome-wide screen; Ram Dasgupta for helpful comments in the initial stages; Maria Gagliardi for assistance in generating dsRNA; Catherine Rabouille for reagents; and Katja Röper for comments on the paper. AAP was supported by a career development award from the Medical Research Council.
Footnotes
The authors declare that they have no conflict of interest.
References
- Baillat D, Hakimi MA, Naar AM, Shilatifard A, Cooch N, Shiekhattar R (2005) Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123: 265–276 [DOI] [PubMed] [Google Scholar]
- Balch WE, Dunphy WG, Braell WA, Rothman JE (1984) Reconstitution of the transport of protein between successive compartments of the Golgi measured by the coupled incorporation of N-acetylglucosamine. Cell 39: 405–416 [DOI] [PubMed] [Google Scholar]
- Bard F, Casano L, Mallabiabarrena A, Wallace E, Saito K, Kitayama H, Guizzunti G, Hu Y, Wendler F, Dasgupta R, Perrimon N, Malhotra V (2006) Functional genomics reveals genes involved in protein secretion and Golgi organization. Nature 439: 604–607 [DOI] [PubMed] [Google Scholar]
- Bettencourt-Dias M, Sinka R, Frenz L, Glover DM (2005) RNAi in Drosophila cell cultures. In Gene Silencing by RNA Interference Sohail M (ed). pp 147–166. Boca Raton: CRC Press [Google Scholar]
- Bonifacino JS, Glick BS (2004) The mechanisms of vesicle budding and fusion. Cell 116: 153–166 [DOI] [PubMed] [Google Scholar]
- Cai H, Reinisch K, Ferro-Novick S (2007) Coats, tethers, Rabs, and SNAREs work together to mediate the intracellular destination of a transport vesicle. Dev Cell 12: 671–682 [DOI] [PubMed] [Google Scholar]
- Dobbelaere J, Josué F, Suijkerbuijk S, Baum B, Tapon N, Raff J (2008) A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. Plos Biol 6: e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubnau J, Chiang AS, Grady L, Barditch J, Gossweiler S, McNeil J, Smith P, Buldoc F, Scott R, Certa U, Broger C, Tully T (2003) The staufen/pumilio pathway is involved in Drosophila long-term memory. Curr Biol 13: 286–296 [DOI] [PubMed] [Google Scholar]
- Egloff S, O'Reilly D, Chapman RD, Taylor A, Tanzhaus K, Pitts L, Eick D, Murphy S (2007) Serine-7 of the RNA polymerase II CTD is specifically required for snRNA gene expression. Science 318: 1777–1779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagarasanu A, Rachubinski RA (2007) Orchestrating organelle inheritance in Saccharomyces cerevisiae. Curr Opin Microbiol 10: 528–538 [DOI] [PubMed] [Google Scholar]
- Farkas RM, Giansanti MG, Gatti M, Fuller MT (2003) The Drosophila Cog5 homologue is required for cytokinesis, cell elongation, and assembly of specialized Golgi architecture during spermatogenesis. Mol Biol Cell 14: 190–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM (2008) The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell 19: 2402–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavin AC, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Höfert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T et al. (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415: 141–147 [DOI] [PubMed] [Google Scholar]
- Glick BS, Nakano A (2009) Membrane traffic within the Golgi apparatus. Annu Rev Cell Dev Biol 25: 113–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas AK, Yoshimura S, Stephens DJ, Preisinger C, Fuchs E, Barr FA (2007) Analysis of GTPase-activating proteins: Rab1 and Rab43 are key Rabs required to maintain a functional Golgi complex in human cells. J Cell Sci 120: 2997–3010 [DOI] [PubMed] [Google Scholar]
- Haberg K, Lundmark R, Carlsson SR (2008) SNX18 is an SNX9 paralog that acts as a membrane tubulator in AP-1-positive endosomal trafficking. J Cell Sci 121: 1495–1505 [DOI] [PubMed] [Google Scholar]
- Heiseke A, Schobel S, Lichtenthaler SF, Vorberg I, Groschup MH, Kretzschmar H, Schatzl HM, Nunziante M (2008) The novel sorting nexin SNX33 interferes with cellular PrP formation by modulation of PrP shedding. Traffic 9: 1116–1129 [DOI] [PubMed] [Google Scholar]
- Inadome H, Noda Y, Kamimura Y, Adachi H, Yoda K (2007) Tvp38, Tvp23, Tvp18 and Tvp15: novel membrane proteins in the Tlg2-containing Golgi/endosome compartments of Saccharomyces cerevisiae. Exp Cell Res 313: 688–697 [DOI] [PubMed] [Google Scholar]
- Ivan V, de Voer G, Xanthakis D, Spoorendonk KM, Kondylis V, Rabouille C (2008) Drosophila Sec16 mediates the biogenesis of tER sites upstream of Sar1 through an arginine-rich motif. Mol Biol Cell 19: 4352–4365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CL (2009) Mechanisms of transport through the Golgi complex. J Cell Sci 122: 443–452 [DOI] [PubMed] [Google Scholar]
- Kastenmayer J (2006) Functional genomics of genes with small open reading frames (sORFs) in S. cerevisiae. Genome Res 16: 365–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni MM, Booker M, Silver SJ, Friedman A, Hong P, Perrimon N, Mathey-Prevot B (2006) Evidence of off-target effects associated with long dsRNAs in Drosophila melanogaster cell-based assays. Nat Methods 3: 833–838 [DOI] [PubMed] [Google Scholar]
- Lee MC, Miller EA, Goldberg J, Orci L, Schekman R (2004) Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol 20: 87–123 [DOI] [PubMed] [Google Scholar]
- Lee T, Luo L (1999) Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22: 451–461 [DOI] [PubMed] [Google Scholar]
- Levine TP, Munro S (1998) The pleckstrin homology domain of oxysterol-binding protein recognises a determinant specific to Golgi membranes. Curr Biol 8: 729–739 [DOI] [PubMed] [Google Scholar]
- Li H, Coghlan A, Ruan J, Coin LJ, Heriche JK, Osmotherly L, Li R, Liu T, Zhang Z, Bolund L, Wong GK, Zheng W, Dehal P, Wang J, Durbin R (2006) TreeFam: a curated database of phylogenetic trees of animal gene families. Nucleic Acids Res 34: D572–D580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lougheed JC, Holton JM, Alber T, Bazan JF, Handel TM (2001) Structure of melanoma inhibitory activity protein, a member of a recently identified family of secreted proteins. Proc Natl Acad Sci USA 98: 5515–5520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luke-Glaser S, Roy M, Larsen B, Le Bihan T, Metalnikov P, Tyers M, Peter M, Pintard L (2007) CIF-1, a shared subunit of the COP9/signalosome and eukaryotic initiation factor 3 complexes, regulates MEL-26 levels in the Caenorhabditis elegans embryo. Mol Cell Biol 27: 4526–4540 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupashin VV, Pokrovskaya ID, McNew JA, Waters MG (1997) Characterization of a novel yeast SNARE protein implicated in Golgi retrograde traffic. Mol Biol Cell 8: 2659–2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y, Creanga A, Lum L, Beachy PA (2006) Prevalence of off-target effects in Drosophila RNA interference screens. Nature 443: 359–363 [DOI] [PubMed] [Google Scholar]
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, Wilm M, Stunnenberg HG, Saumweber H, Akhtar A (2006) Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell 21: 811–823 [DOI] [PubMed] [Google Scholar]
- Motley A, Bright NA, Seaman MN, Robinson MS (2003) Clathrin-mediated endocytosis in AP-2-depleted cells. J Cell Biol 162: 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Field C, Schekman R (1980) Identification of 23 complementation groups required for post-translational events in the yeast secretory pathway. Cell 21: 205–215 [DOI] [PubMed] [Google Scholar]
- Pan X, Eathiraj S, Munson M, Lambright DG (2006) TBC-domain GAPs for Rab GTPases accelerate GTP hydrolysis by a dual-finger mechanism. Nature 442: 303–306 [DOI] [PubMed] [Google Scholar]
- Peden AA, Oorschot V, Hesser BA, Austin CD, Scheller RH, Klumperman J (2004) Localization of the AP-3 adaptor complex defines a novel endosomal exit site for lysosomal membrane proteins. J Cell Biol 164: 1065–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman M, Earnest S, Zhang K, Zhao Y, Cobb MH (2007) TAO kinases mediate activation of p38 in response to DNA damage. EMBO J 26: 2005–2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera VM, Wang X, Wardwell S, Courage NL, Volchuk A, Keenan T, Holt DA, Gilman M, Orci L, Cerasoli F, Rothman JE, Clackson T (2000) Regulation of protein secretion through controlled aggregation in the endoplasmic reticulum. Science 287: 826–830 [DOI] [PubMed] [Google Scholar]
- Sacher M, Jiang Y, Barrowman J, Scarpa A, Burston J, Zhang L, Schieltz D, Yates JR, Abeliovich H, Ferro-Novick S (1998) TRAPP, a highly conserved novel complex on the cis-Golgi that mediates vesicle docking and fusion. EMBO J 17: 2494–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sacher M, Kim YG, Lavie A, Oh BH, Segev N (2008) The TRAPP complex: insights into its architecture and function. Traffic 9: 2032–2042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler KC, Amsterdam A, Soroka C, Boyer J, Hopkins N (2005) A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development 132: 3561–3572 [DOI] [PubMed] [Google Scholar]
- Saito K, Chen M, Bard F, Chen S, Zhou H, Woodley D, Polischuk R, Schekman R, Malhotra V (2009) TANGO1 facilitates cargo loading at endoplasmic reticulum exit sites. Cell 136: 891–902 [DOI] [PubMed] [Google Scholar]
- Sinka R, Gillingham AK, Kondylis V, Munro S (2008) Golgi coiled-coil proteins contain multiple binding sites for Rab family G proteins. J Cell Biol 183: 607–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklan EH, Serrano RL, Einav S, Pfeffer SR, Lambright DG, Glenn JS (2007) TBC1D20 is a Rab1 GTPase-activating protein that mediates hepatitis C virus replication. J Biol Chem 282: 36354–36361 [DOI] [PubMed] [Google Scholar]
- Soulet F, Yarar D, Leonard M, Schmid SL (2005) SNX9 regulates dynamin assembly and is required for efficient clathrin-mediated endocytosis. Mol Biol Cell 16: 2058–2067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein IS, Gottfried A, Zimmermann J, Fischer von Mollard G (2009) TVP23 interacts genetically with the yeast SNARE VTI1 and functions in retrograde transport from the early endosome to the late Golgi. Biochem J 419: 229–236 [DOI] [PubMed] [Google Scholar]
- Stoll R, Bosserhoff A (2008) Extracellular SH3 domain containing proteins—features of a new protein family. Curr Protein Pept Sci 9: 221–226 [DOI] [PubMed] [Google Scholar]
- Stoll R, Renner C, Zweckstetter M, Brüggert M, Ambrosius D, Palme S, Engh RA, Golob M, Breibach I, Buettner R, Voelter W, Holak TA, Bosserhoff AK (2001) The extracellular human melanoma inhibitory activity (MIA) protein adopts an SH3 domain-like fold. EMBO J 20: 340–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart LM, Boulais J, Charriere GM, Hennessy EJ, Brunet S, Jutras I, Goyette G, Rondeau C, Letarte S, Huang H, Ye P, Morales F, Kocks C, Bader JS, Desjardins M, Ezekowitz RA (2007) A systems biology analysis of the Drosophila phagosome. Nature 445: 95–101 [DOI] [PubMed] [Google Scholar]
- Vale RD (2003) The molecular motor toolbox for intracellular transport. Cell 112: 467–480 [DOI] [PubMed] [Google Scholar]
- von Mollard GF, Nothwehr SF, Stevens TH (1997) The yeast v-SNARE Vti1p mediates two vesicle transport pathways through interactions with the t-SNAREs Sed5p and Pep12p. J Cell Biol 137: 1511–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westmuckett AD, Hoffhines AJ, Borghei A, Moore KL (2008) Early postnatal pulmonary failure and primary hypothyroidism in mice with combined TPST-1 and TPST-2 deficiency. Gen Comp Endocrinol 156: 145–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner W, Schekman R (2008) Membrane fusion. Nat Struct Mol Biol 15: 658–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H, Brill JA, Hsien J, McBride R, Boulianne GL, Trimble WS (2002) Syntaxin 5 is required for cytokinesis and spermatid differentiation in Drosophila. Dev Biol 251: 294–306 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1
Supplementary Figure 2
Supplementary Figure 3
Supplementary Table 1
Supplementary Table 2
Supplementary Movie 1
Supplementary Movie 2
Review Process File