Abstract
The aim of this paper is to develop an intrinsic regression model for the analysis of positive-definite matrices as responses in a Riemannian manifold and their association with a set of covariates, such as age and gender, in a Euclidean space. The primary motivation and application of the proposed methodology is in medical imaging. Because the set of positive-definite matrices do not form a vector space, directly applying classical multivariate regression may be inadequate in establishing the relationship between positive-definite matrices and covariates of interest, such as age and gender, in real applications. Our intrinsic regression model, which is a semiparametric model, uses a link function to map from the Euclidean space of covariates to the Riemannian manifold of positive-definite matrices. We develop an estimation procedure to calculate parameter estimates and establish their limiting distributions. We develop score statistics to test linear hypotheses on unknown parameters and develop a test procedure based on a resampling method to simultaneously assess the statistical significance of linear hypotheses across a large region of interest. Simulation studies are used to demonstrate the methodology and examine the finite sample performance of the test procedure for controlling the family-wise error rate. We apply our methods to the detection of statistical significance of diagnostic effects on the integrity of white matter in a diffusion tensor study of human immunodeficiency virus. Supplemental materials for this article are available online.
Keywords: Diffusion tensor, Intrinsic regression, Positive-definite matrix, Riemannian manifold, Score statistic
1. INTRODUCTION
Symmetric positive-definite matrices (SPD) arise in many applications including diffusion tensor imaging (DTI), computational anatomy, and statistics. For instance, DTI, which tracks the effective diffusion of water molecules in the human brain in vivo, contains a 3 × 3 positive-definite matrix D, called a diffusion tensor, at each voxel (three-dimensional pixel) of an imaging space. Because water molecules tend to diffuse along the pathways of white matter fibers, tracking their diffusion with DTI allows investigators to map the microstructure and organization of those pathways. Specifically, the three eigenvalue-eigenvector pairs {(λi, vi) : i = 1, 2, 3} of D (λ1 ≥ λ2 ≥ λ3) can quantify the degree and direction, respectively, of diffusivity and the principal directions (v1) of the diffusion tensors (DTs) have been widely used to reconstruct the white matter fiber tracts (Basser, Mattiello, and LeBihan 1994a, 1994b; Zhu et al. 2007). Statistical analysis of DTI measures (e.g., eigenvalues and eigenvectors), diffusion tensors, and fiber tracts can reveal disruptions in structural organization and the neuronanatomical connectivity of the brains of persons with neurological and neuropsychiatric illnesses, and substance use disorders (Lim and Helpern 2002; Focke et al. 2008; Vernooij et al. 2008).
In computational anatomy, a symmetric positive-definite deformation tensor S = (JJT)1/2 at each location of the image can capture the directional information of shape change decoded in the Jacobian matrices J (Grenander and Miller 1998; Lepore et al. 2008). The directional shape change information decoded in deformation tensors has been demonstrated to be more discriminative than the determinant of J in several structural MRI based studies of HIV/AIDS (Lepore et al. 2008). More generally, deformation tensors can be used to detect subtle changes of brain structures across groups and along time. An appropriate statistical analysis of deformation tensors is important for understanding normal brain development, the neural bases of neuropsychiatric disorders, and the joint effects of environmental and genetic factors on brain structure and function. A fundamental issue in many applications is modeling covariance matrices for multivariate data, longitudinal data, spatial data, and time series data (Chiu, Leonard, and Tsui 1996; Pourahmadi 1999, 2000; Anderson 2003).
Despite the extensive use of SPD matrices, a formal statistical framework for using a set of covariates in a Euclidean space to predict SPD matrices as responses has not yet been developed. One may ignore the fact that SPD matrices are in a nonlinear space and then directly apply classical multivariate regression to studying the association between SPD matrices and covariates. However, this method is not adequate in many applications. As an illustration, we consider the “mean” of SPD matrices. In Euclidean space, the mean of SPD matrices is just the empirical average of SPD matrices, which can lead to a swelling effect in imaging processing (Chefdhotel et al. 2004). In contrast, if one regards SPD matrices as points in Riemannian manifold and calculates their Frechét mean (Frechét 1948; Karcher 1977), then the swelling effect completely disappears (Pennec, Fillard, and Ayache 2006). Moreover, even when SPD matrices are regarded as random elements in Riemannian manifold, the existing statistical methods can only compare differences between the means of the two (or multiple) groups of SPD matrices, which is equivalent to modeling SPD matrices with a single categorical variable (Bhattacharya and Patrangenaru 2003, 2005; Schwartzman 2006; Whitcher et al. 2007). For instance, Schwartzman (2006) has developed several parametric models for SPD matrices and derived distributions of several test statistics for testing differences in two groups of SPD matrices. However, the use of parametric models to characterize SPD matrices in real applications requires further research. Recently, Kim and Richards (2008) have studied the problem of deconvolution density estimation on the space of SPD matrices. Although much effort has been devoted to appropriately modeling covariance matrices in longitudinal data and time series settings (Pourahmadi 1999, 2000), these covariance matrices are not response variables as is the case in medical imaging.
To the best of our knowledge, this is the very first paper that develops a semiparametric regression model with SPD matrices as responses in a Remannian manifold and covariates in Euclidean space. Compared with the existing literature, we extend the two-group model studied by Schwartzman (2006) to allow for general covariates and generalize the normal distributional modeling in Schwartzman (2006) by assuming only that certain appropriately defined residuals have mean zero. Our regression model is semiparametric, and thus it avoids specifying parametric distributions for random SPD matrices. We propose an inference procedure to estimate the regression coefficients in this semiparametric model. We establish asymptotic properties, including consistency and asymptotic normality, of the estimates of the regression coefficients. We develop score statistics to test linear hypotheses on unknown parameters. Finally, we develop a test procedure based on a resampling method to simultaneously assess the statistical significance of linear hypotheses across a large region of interest.
The rest of this paper is organized as follows. We will first review some basic results about the Riemannian geometrical structure for the space of SPD matrices. We will next present the semiparametric regression model and estimation methods for estimating the regression coefficients. Then we will develop score statistics to carry out hypothesis testing and develop a test procedure for correcting for multiple comparisons. Simulation studies will assess the empirical performance of the estimation algorithms and test statistics. Finally, we will analyze a neuroimaging dataset to illustrate an application of these methods, before offering some concluding remarks.
2. THEORY
In this section, we formally introduce a semiparametric regression model for a SPD matrix response, develop an estimation algorithm for estimating regression coefficients, and develop a score statistic for testing linear hypotheses of the regression coefficients. We then present a test procedure based on a resampling method for correcting multiple statistical comparisons.
2.1 Model Formulation
We develop a formal regression model for SPD matrices. Suppose we observe a m × m SPD matrix Si and a k × 1 covariate vector xi, such as diagnostic status and age, for i = 1,…, n. Let Sym+(m) be the set of m × m SPD matrices, β be a p × 1 vector of regression coefficients in Rp and Σ(·,·) be a map from Rk × Rp to Sym+(m). The regression model is obtained by modeling the “conditional mean” of Si given xi, denoted by Σi(β) = Σ(xi, β) ∈ Sym+(m). Note that we just borrow the term “conditional mean” from Euclidean space, but we will formalize the notion of a “conditional mean” explicitly below.
Following Pourahmadi (2000), we consider a general specification of Σ(x, β) using the Cholesky decomposition of Σ(x, β). For the ith observation, through a lower triangular matrix Ci(β) = C(xi, β) = (cjk(xi, β)) (Pourahmadi 2000), the Cholesky decomposition of Σ(xi, β) is given by
| (1) |
The specification in (1) has been widely used due to computational simplicity (Pourahmadi 1999, 2000). Now we need to specify the explicit forms of cjk(xi, β) for all j ≥ k in order to determine all entries in Σi(β). These functions cjk(xi, β) in Ci(β) can be regarded as link functions in generalized linear models (McCullagh and Nelder 1989). For instance, for m = 2, we may choose
| (2) |
where and β(j) for j = 1, 2, 3 are subvectors of β. To ensure the uniqueness of the Cholesky decomposition, we may impose additional constraints that cjj(xi, β) > 0 for all j = 1, … ,m. For instance, we may assume that Ci(β) takes the form
| (3) |
Compared with (3), the link function in (2) leads to a simpler computation. However, there is an ambiguous sign problem associated with (2), since Σi(β) = Ci(β)PPTCi(β)T, where P = diag(±1, …,±1). In practice, we can standardize all covariates and impose additional sign constraints on the intercept terms of β(1) and β(3) to remove this ambiguity. The link functions in (2) and (3) can be easily generalized to m ≥ 3. When xi = 0, Σ(0, β) = C(0, β)C(0, β)T represents an intercept matrix, which is similar to the intercept in a linear model.
We may consider two direct reparametrizations of Σi(β). The first is a matrix-logarithmic model. If we consider the spectral decomposition of Σ ∈ Sym+(m) given by OVOT, where O is a m × m orthonormal matrix and V is a m × m diagonal matrix with positive entries, then the matrix logarithm of Σ is log(Σ) = log(OVOT) = O log(V)OT, where log(V) ∈ diag(m) with diagonal elements equal the logarithm of the diagonal entries of V. Let vecs(U) represent (U11, …, U1m, U22, …, U2m, …, Umm)T for any m × m symmetric matrix U = (Uij) and let l(x, β) be a map from Rk × Rp to Rm(m+1)/2. The matrix-logarithmic model assumes that the matrix logarithm of Σi(β) can be written as
| (4) |
A graphical illustration is given in Figure 1. For instance, let B be an m(m + 1)/2 × (k + 1) matrix. If we set l(xi, β) = Bzi and β contains all unknown parameters in B, then we obtain the matrix-logarithmic model for the covariance matrix in Chiu, Leonard, and Tsui (1996). For m = 1, we have . Thus, exp(β1, 1) can be represented as an intercept when xi = 0, whereas β1, (2) characterizes the “slope” vector.
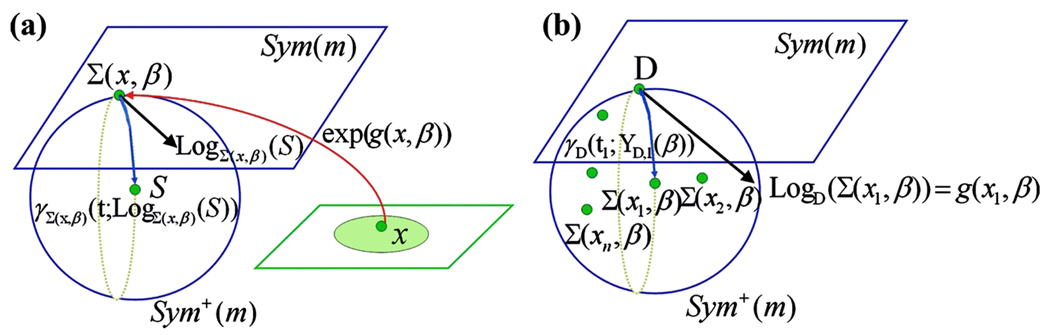
Figure 1.
Graphical illustration of two link functions in the (a) matrix-logarithmic and (b) geodesic models. In (a), the link function for the matrix-logarithmic model equals Σ(x, β) = exp(g(x, β)) or log(Σ(x, β)) = g(x, β). In (b), the link function for the geodesic model equals LogD(Σ(x, β)) = g(x, β), where D is the intercept matrix.
The second is a “geodesic” model for Σ(xi, β). Intuitively, a geodesic on the manifold Sym+(m) is a “straight line” on Sym+(m). Let GL(m) be the set of m × m matrices of nonzero determinant. For any C ∈ GL(m), we define the matrix exponential transformation of C, denoted by exp(C), as , where C0 = Im is the m × m identity matrix and Ck is the ordinary matrix multiplication of C k times. We consider an intercept matrix D ∈ Sym+(m) and any square root of D, denoted by B, such that B ∈ GL(m) and Σ(0, β) = D = BBT, when xi = 0. Then, for a given l(xi, β), we consider a “directional” matrix YD(xi, β) = YD,i(β) in the space of m × m symmetric matrices, denoted by Sym(m), such that vecs(YD,i(β)) = l(xi, β) and l(0, β) = 0. The geodesic model assumes that
| (5) |
for i = 1, …, n. As shown in proposition 2.2.7 of Schwartzman (2006), Σ(xi, β) is closely related to the geodesic passing through D = BBT in the direction of YD,i(β) and the specification of Σ(xi, β) in (5) is unique. We will formalize the notions of “geodesic” and “direction” in Section 2.2. A graphical illustration is given in Figure 1(b). Because the specifications (4) and (5) involve the matrix exponential transformation, they can be much more computationally difficult than specifications (2) and (3).
We introduce a definition of “residual” to ensure that Σi(β) is the proper “conditional mean” of Si given xi. For instance, in the classical linear model, the response is the sum of a regression function and a residual term. Then, the regression function is the conditional mean of the response only when the conditional mean of the residual equals zero. Given two points Si and Σi(β) on the manifold Sym+(m), we need to define the residual or “difference” between Si and Σi(β). At Σi(β), we have a tangent space of the manifold Sym+(m), denoted by TΣi(β) Sym+(m), which is a Euclidean space representing a first-order approximation of the manifold Sym+(m) near Σi(β). Then, we calculate the projection of Si onto TΣi(β) Sym+(m), denoted by LogΣi(β)(Si), which is given by
| (6) |
Thus, LogΣi(β)(Si) on TΣi(β) Sym+(m) can be regarded as the difference between Si and Σi(β) for i = 1, …, n. Since LogΣi(β)(Si) for different i are in different tangent spaces (or Euclidean spaces), we must translate them back to the same tangent space, such as the tangent space at the identity matrix Im, denoted by TIm Sym+(m) (Schwartzman 2006). This translation can be done using the group action of GL(m) introduced in Section 2.2. Specifically, to translate LogΣi(β)(Si) ∈ TΣi(β) Sym+(m) into TIm Sym+(m), we follow Schwartzman (2006) to define the residual of Si with respect to Σi(β) as
| (7) |
for i = 1, …, n. The εi(β) are uniquely defined, because the Cholesky decomposition is unique and LogΣi(β)(Si) is independent of the choice of the square root of Σi(β). Moreover, because all εi(β) are in the same tangent space TIm Sym+(m) and TIm Sym+(m) is a Euclidean space, we can apply classical multivariate analysis techniques in Euclidean space to the analysis of εΣi(β)(Si) (Anderson 2003).
The intrinsic regression model for SPD matrices is then defined by
| (8) |
for i = 1, …, n, where the expectation is taken with respect to the conditional distribution of Si given xi. Model (8) does not assume any parametric distribution for Si given xi, and thus it allows for a large class of distributions. In contrast, let Ω be an m × m matrix in Sym+(m), Schwartzman (2006, p. 42) considered a symmetric matrix variate normal distribution of εi (Gupta and Nagar 2000), whose density is given by
| (9) |
Then, using Si = Ci(β) exp(εi)Ci(β)T, the density of Si, denoted by p(Si; β), is
| (10) |
where J(Ci(β)−1SiCi(β)−T) is the Jacobian of the transformation εi = Ci(β)−1SiCi(β)−T. Because the true distribution of εi may deviate from the Gaussian distribution in (9), it can be very restrictive to assume a parametric distribution, such as (10), for Si. In addition, our model (8) does not assume homogeneous variance across all i. This is also desirable for applications, such as the analysis of imaging data, including diffusion tensor data, because between-subject and between-voxel variability in the imaging measures (e.g., DT) can be substantial.
2.2 Geometrical Structure of Sym+(m)
We summarize some basic results from Schwartzman (2006) about the geometrical structure of Sym+(m) as a Riemannian manifold (Do Carmo 1992; Lang 1999). The space Sym+(m) is a submanifold of the Euclidean space Sym(m). Geometrically, the spaces Sym+(m) and Sym(m) are differentiable manifolds of m(m + 1)/2 dimensions and they are homeomorphically related by the matrix exponential transformation and logarithm.
We first introduce the tangent vector and tangent space at any D ∈ Sym+(m). For a small scalar δ > 0, let C(t) be a differentiable map from (−δ, δ) to Sym+(m) such that it passes through C(0) = D. The tangent vector at D is defined as the derivative of the smooth curve C(t) with respect to t at t = 0. The set of all tangent vectors at D forms the tangent space of Sym+(m) at D, denoted by TD Sym+(m). As shown in proposition 2.2.3 of Schwartzman (2006), TD Sym+(m) is identified with a copy of Sym(m). For instance, when D = Im, we consider the curve C(t) = exp(tY) ∈ Sym+(m) satisfying C(0) = Im for any Y ∈ Sym(m) and t ∈ (−δ, δ). Then, the derivative of C(t) at t = 0 is just Y and thus TIm Sym+(m) = Sym(m). Second, we introduce an inner product of any two tangent vectors in the same tangent space, which varies smoothly along the manifold. We consider the scaled Frobenius inner product of any two tangent vectors YD and ZD in TD Sym+(m), which is defined by
| (11) |
Let B ∈ GL(m) be any square root of D ∈ Sym+(m) such that D = BBT and let γD(t; YD) be the geodesic on Sym+(m) passing through D in the direction of the tangent vector YD ∈ TD Sym+(m). As shown in proposition 2.2.7 of Schwartzman (2006), γD(t; YD) is uniquely given by
| (12) |
Given the geodesic γD(t; YD), the Riemannian exponential map ExpD(YD), which maps the tangent vector YD ∈ TD Sym+(m) to a point X ∈ Sym+(m), is uniquely defined as
| (13) |
When D = Im, ExpD reduces to the matrix exponential exp(·). Recall that in the geodesic model, we assume that Σ(0, β) = D = BBT and Σ(xi, β) = γD(1; YD,i(β)), in which vecs(YD,i(β)) = l(xi, β) and l(0, β) = 0.
The Riemannian logarithmic map at D, denoted by LogD(·), maps X ∈ Sym+(m) onto the tangent vector YD in TD Sym+(m). Specifically, LogD : Sym+(m) → TD Sym+(m) at D = BBT ∈ Sym+(m) is uniquely defined as
| (14) |
The Riemannian exponential and logarithmic maps satisfy
| (15) |
Recall that in the definition of “conditional residual,” we have two points D = Σ(xi, β) and X = Si on Sym+(m) and then we define LogΣ(xi, β)(Si) on TΣ(xi, β) Sym+(m). Because LogΣ(xi, β) (Si) is independent of the square root of Σ(xi, β), we can choose the Cholesky decomposition of Σ(xi, β) = Ci(β)Ci(β)T in the definition of LogΣ(xi, β) (Si).
A nice property of Sym+(m) is that a group action of GL(m) on Sym+(m) can relate any two (X, D) ∈ Sym+(m) and the tangent spaces at X and D, respectively. Specifically, the group action of GL(m) on Sym+(m) consists of all transformations ϕB(X) = BXBT for any B ∈ GL(m). For any (X, D) ∈ Sym+(m), there exists B ∈ GL(m) such that BXBT = D. The group action of GL(m) on Sym+(m) induces a group action between TX Sym+(m) and TϕB(X) Sym+(m). Explicitly, if Y ∈ TX Sym+(m), then ϕB(Y) = BYBT ∈ TϕB(X) Sym+(m). Particularly, we have applied the group action to translate LogΣi(β) (Si) ∈ TΣi(β) Sym+(m) to Ci(β)−1 LogΣi(β) (Si) × Ci(β)−T ∈ TCi(β)−1 Σi(β)Ci(β)−T Sym+(m) = TIm Sym+(m) for i = 1, …, n.
We consider the geodesic distance between any two points on Sym+(m). Let B ∈ GL(m) be any square root of D, the geodesic distance between D and X in Sym+(m) is uniquely given by
| (16) |
The geodesic distance has many nice properties. For instance, the geodesic distance is a proper metric satisfying positive definiteness, symmetry, and the triangle inequality. Specifically, d(D, X) = d(X, D). The geodesic distance is also invariant under group actions, that is, d(D, X) = d(BDBT , BXBT) for any B ∈ GL(m).
2.3 Estimation
We calculate an intrinsic least squares estimator (ILSE) of the parameter vector β, denoted by β̂, by minimizing the total residual sum of squares:
| (17) |
Thus, β̂ solves the estimating equations given by
| (18) |
where ∂ denotes partial differentiation with respect to a parameter vector, such as β. The ILSE is closely related to the intrinsic mean ΣIM of S1, …, Sn ∈ Sym+(m), which is defined as
| (19) |
where d(·, ·) has been defined in equation (16). In this case, both Σi and Ci(β) are independent of i and the covariates. Let β = (c11, c21, c22, …, cm1, …, cmm) be the entries of the lower triangular portion of C(β) = (cjk) such that Σ = C(β)C(β)T. It follows from (16) that , which leads to Σ̂IM = C(β̂ )C(β̂)T.
We now develop an annealing optimization algorithm for obtaining β̂. Our annealing algorithm consists of five steps. In Step 1, we first calculate the Cholesky decompositions of all Si, denoted by , and then we fit a multivariate linear regression model with the lower part of C̃i as the response and the corresponding part of Ci(β) as the mean function, which yield the initial value β(0). Then, starting from β(0), we use the downhill simplex method to search for another initial value β(1) (Nelder and Mead 1965). In Step 2, starting from β(k), we use the Gibbs sampler coupled with the Metropolis–Hasting algorithm to iteratively draw N0 (say, 200) dependent observations {β(k,l) : l = 1, … ,N0} from exp(−Gn(β)/τk). Among the {β(k,l) : l = 1, …, N0}, we find the optimal β̃(k) = argminβ(k,l) {Gn(β)}. In Step 3, we use the downhill simplex method with β̃(k) as the starting point to search for β(k+1). In Step 4, we iterate between Steps 2 and 3 for k = 1, …, M0 (say, M0 = 4) and increase τk at each iteration. We choose τ1 to achieve an acceptance rate of the Metropolis–Hasting algorithm within the Gibbs sampler around 0.25 and then we slightly reduce the acceptance rate to around 0.15 as k increases. We save the optimal βB, which minimizes Gn(β), among all β(k,l) and β(k) during the iterations. In Step 5, starting from βB, we use the Newton–Raphson algorithm to calculate β̂ as described below. Compared with other optimization methods, such as the simulated annealing algorithm, we can efficiently find β̂ due to several nice features. According to our experience, β(1) is usually close to β̂, and we always use the downhill simplex method to search for a new optimal estimator after drawing a small number of Monte Carlo samples. Thus, we avoid using large values of N0 and M0.
Let ∂βGn(β(t)) and , respectively, denote the first and second-order partial derivatives of Gn(β) with respect to β evaluated at β(t). The Newton–Raphson algorithm yields
| (20) |
In addition, we choose 0 < ρ = 1/2k0 ≤ 1 for some k0 ≥ 0 such that Gn(β(t+1)) ≤ Gn(β(t)). The Newton–Raphson algorithm stops when the absolute difference between consecutive β(t)’s is smaller than a predefined small number, say 10−4. At the final iteration, we set β̂ = β(t). In addition, because may not be positive definite, we approximate in order to stabilize the Newton–Raphson algorithm. Details regarding ∂βGn(β(t)), and and its approximation are given in lemma 1 of the supplemental document.
2.4 Asymptotic Properties
We first introduce some notation to describe the limiting behavior of the ILSE for SPDs. Let β* be the true value of β such that (8) holds at β*. Let Ci* denote Ci(β*) and ; ℬ denotes the parameter space for β; ‖ · ‖ denotes the Euclidean norm of a vector or a matrix; a⊗2 = aaT for any vector a; and →L denotes convergence in distribution.
We establish consistency and asymptotically normality of β̂. We obtain the following theorems, whose detailed assumptions and proofs can be found in the supplemental document.
Theorem 1
(a) For model (8), if assumptions (C1), (C2), and (C3) in the supplemental document are true, then β̂ converges to β* in probability.
(b) For model (8), under assumptions (C1)–(C4), we have
| (21) |
as n → ∞.
Theorem 1 has several important applications. Theorem 1(a) establishes weak convergence of β̂. According to Theorem 1(b), the covariance matrix of β̂ under model (8) can be consistently estimated by
| (22) |
Moreover, we can use Theorem 1(b) to construct confidence cones of β and its functions. Since Theorem 1 only establishes the asymptotic properties of β̂ when the sample size is large, these properties may be inadequate to characterize the finite sample behavior of β̂ for relatively small sample sizes. In the case of small and moderate sample sizes, we may have to resort to higher-order approximations, such as saddlepoint approximations and bootstrap methods (Davison and Hinkley 1997; Butler 2007).
2.5 Testing Linear Hypotheses
Our choice of hypotheses to test is motivated by scientific questions, which involve a comparison of SPDs across diagnostic groups or detecting change in SPDs across time (Schwartzman 2006; Whitcher et al. 2007; Lepore et al. 2008). These questions usually can be formulated as testing linear hypotheses of β as follows:
| (23) |
where R is an r × p matrix of full row rank and b0 is an r × 1 specified vector. We test the null hypothesis H0 : Rβ = b0 using a score test statistic Wn defined by
| (24) |
where μ = Rβ, , in which β̃ denotes the estimate of β under H0 and the explicit expressions of Ûi, μ (β̃) and Ln, μ are given in the supplemental document (Rotnitzky and Jewell 1990).
Theorem 2
For model (8), if assumptions (C1)–(C4) in the supplemental document are true, then the statistic Wn is asymptotically distributed as χ2(r), a chi-square distribution with r degrees of freedom, under the null hypothesis H0.
An asymptotically valid test can be obtained by comparing sample values of the test statistic with the critical value of the right-hand tail of the χ2(r) distribution at a prespecified significance level α. That is, we reject H0 if , and do not reject H0 otherwise, where is the upper α-percentile of the χ2(r) distribution.
2.6 Multiple Hypotheses
In imaging applications, we need to test the hypotheses H0 against H1 across multiple brain regions or across the many voxels of the imaging volume. Thus, the next step entails using statistical methods (e.g., random field theory, false discovery rates, permutation methods) to adjust p-values for these multiple statistical tests. To test H0 in all voxels of the region under study, we consider the maximum of the score test statistics given by
| (25) |
where Wn(d) denotes the score test statistic at voxel d and 𝒟 denotes the brain region. The maximum statistic Wn, 𝒟 plays a crucial role in controlling the family-wise error rate.
We propose to use a test procedure that is based on a resampling method to approximate the null distribution of Wn,𝒟 (Kosorok 2003; Lin 2005). The test procedure is implemented as follows:
Step 1. We calculate the score test statistic Wn(d) at each voxel d, and then we compute Wn,𝒟 = maxd∈𝒟 Wn(d).
Step 2. Generate n realizations of ε denoted by , where ε equals ±1 with equal probability.
Step 3. Calculate Wn(d)(k) = Ln, μ(d)(k)T [Îμμ(d)]−1 × Ln, μ(d)(k), and then compute , where .
Step 4. Repeat Steps 2–3 K0 times and calculate . Finally, the p-value of Wn,𝒟 is approximated by . We reject the null hypothesis H0 : Rβ = b0 across all voxels of the region when pμ,𝒟 is smaller than a prespecified value α, say 0.05.
Step 5. Calculate the adjusted p-value of Wn(d) at each voxel d of the region according to .
There are several advantages of using the resampling method in the above test procedure. The above procedure needs to compute Ûi,μ(β̃, d) and Ĩμμ only once. It also avoids repeated analyses of permuted datasets in the permutation methods, because fitting the regression models for SPD matrices across all voxels of an imaging volume can take up to more than 20 hours for each permuted dataset. In contrast, the proposed resampling method takes less than 5 minutes for K0 = 1,000.
3. SIMULATION STUDIES
We conducted three sets of Monte Carlo simulations. All computations for these simulation studies were done in C++ on an IBM ThinkCentre M50 workstation. The first set of simulations was to evaluate the accuracy of the parameter estimates and their associated variance estimates for the proposed intrinsic regression model. We set m = 3 and generated the simulated data as follows. We considered the Cholesky decomposition (1) of Σ(xi, β) with , in which zi = (1, xi1)T was a 2 × 1 vector of covariates of interest, for i = 1, …, n. We generated xi1 independently from a Gaussian generator with zero mean and unit variance. Thus, is a 12 × 1 vector. Then, we simulated εi from a N(0, Ω) distribution and calculated Ci(β*) exp(εi)Ci(β*)T, in which we set for k = 1, …, 6. We chose two different Ω’s as follows: Ω1 = 0.6I3 and , where 13 = (1, 1, 1)T. For each Ω, we set n = 20 and 80 and then simulated 500 data sets for each case. For each simulated dataset, we applied the annealing optimization algorithm with N0 = 200 and M0 = 4, which took an average CPU time of about 30 seconds to obtain β̂ and Cov(β̂) in (22).
Based on 500 parameter estimates, we calculated the bias, the mean of the estimated standard error estimates (SE), the standard deviation of the estimated standard error estimates (SD–SE) and the root-mean-square error (RMS) (Table 1). All relative efficiencies (the ratio of the mean of the standard deviation estimates to the root mean-square error) are close to 1.0, indicating that matrix (22) is an accurate estimate of diag(Cov(β̂)). As expected, the root mean-square error decreases as the sample size increases. Moreover, comparing the results from Ω1 and Ω2, we note that increasing the correlation in Ω only slightly decreases the bias and SD of β̂.
Table 1.
Bias (×10−2), RMS (×10−2), SE (×10−2), SD–SE (×10−2), and RS of all 12 parameters under Ω1 and Ω2. BIAS denotes the bias of the mean of the ILSE estimates; RMS denotes the root-mean-square error; SE denotes the mean of the standard deviation estimates; SD–SE denotes the standard deviation of the standard deviation estimates; RS denotes the ratio of RMS over SD. Two different sample sizes {20, 80} and 500 simulated datasets were used for each case
| n = 20 | n = 80 | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| BIAS | RMS | SE | SD–SE | RS | BIAS | RMS | SE | SD–SE | RS | |
| Ω1 | ||||||||||
| β1 | 2.60 | 6.10 | 6.73 | 2.20 | 1.10 | 0.58 | 3.59 | 3.37 | 0.68 | 0.94 |
| β2 | 1.78 | 6.10 | 6.61 | 1.72 | 1.08 | 0.06 | 3.90 | 3.61 | 0.55 | 0.92 |
| β3 | 1.88 | 7.06 | 6.96 | 1.32 | 0.98 | 0.69 | 3.91 | 3.56 | 0.35 | 0.91 |
| β4 | 1.15 | 6.86 | 6.89 | 1.30 | 1.01 | 0.35 | 3.83 | 3.51 | 0.33 | 0.92 |
| β5 | 3.83 | 15.34 | 17.24 | 4.22 | 1.12 | 1.08 | 8.35 | 8.58 | 1.20 | 1.02 |
| β6 | 2.83 | 15.07 | 16.97 | 3.76 | 1.12 | 0.54 | 8.45 | 8.46 | 0.95 | 1.01 |
| β7 | 1.43 | 8.75 | 8.07 | 1.42 | 0.92 | −0.37 | 4.19 | 4.10 | 0.39 | 0.98 |
| β8 | 0.48 | 8.32 | 7.98 | 1.40 | 0.96 | −0.44 | 4.12 | 4.06 | 0.38 | 0.98 |
| β9 | 5.14 | 29.38 | 32.06 | 7.57 | 1.09 | 1.6 | 14.84 | 16.07 | 2.23 | 1.08 |
| β10 | 3.97 | 28.88 | 31.59 | 7.14 | 1.09 | 1.05 | 14.84 | 15.85 | 1.77 | 1.07 |
| β11 | 3.62 | 20.32 | 19.91 | 3.87 | 0.98 | 1.00 | 10.63 | 10.15 | 0.85 | 0.96 |
| β12 | 2.69 | 20.11 | 19.68 | 3.83 | 0.98 | 0.53 | 10.48 | 10.03 | 0.84 | 0.96 |
| Ω2 | ||||||||||
| β1 | 2.76 | 6.87 | 6.73 | 2.21 | 0.97 | 0.59 | 4.00 | 3.7 | 0.57 | 0.93 |
| β2 | 1.81 | 6.72 | 6.61 | 1.63 | 0.98 | 0.46 | 3.97 | 3.7 | 0.53 | 0.93 |
| β3 | 1.96 | 7.74 | 7.23 | 1.27 | 0.93 | 0.23 | 3.72 | 3.54 | 0.36 | 0.95 |
| β4 | 1.24 | 7.43 | 7.15 | 1.26 | 0.96 | 0.03 | 3.67 | 3.5 | 0.36 | 0.95 |
| β5 | 3.08 | 11.63 | 12.61 | 2.85 | 1.08 | 1.02 | 6.87 | 6.33 | 0.73 | 0.92 |
| β6 | 1.87 | 11.78 | 12.41 | 2.31 | 1.05 | 0.86 | 6.9 | 6.3 | 0.7 | 0.91 |
| β7 | 1.56 | 8.44 | 8.26 | 1.46 | 0.98 | 0.11 | 4.5 | 4.2 | 0.34 | 0.93 |
| β8 | 0.62 | 8.09 | 8.17 | 1.44 | 1.01 | 0.04 | 4.5 | 4.1 | 0.34 | 0.91 |
| β9 | 2.75 | 18.90 | 19.75 | 4.14 | 1.04 | 1.06 | 10.74 | 9.93 | 1.09 | 0.93 |
| β10 | 1.33 | 18.84 | 19.46 | 3.68 | 1.03 | 0.86 | 10.79 | 9.82 | 0.93 | 0.91 |
| β11 | 3.87 | 16.68 | 15.49 | 2.54 | 0.93 | −0.58 | 7.45 | 7.44 | 0.66 | 0.99 |
| β12 | 2.88 | 16.46 | 15.31 | 2.51 | 0.94 | −0.91 | 7.22 | 7.37 | 0.65 | 1.02 |
The second set of simulations examined the finite sample performance of the score statistic Wn. We used the same setup as the first set of simulations except that we set β = 0, and then varied the second component of βj = (βj,1, βj,2)′ for j = 1, …, 6. We were interested in testing whether the effect of the covariate xi1 is significant. Letting β·,2 = (β1,2, … , β6,2)T, we tested the following hypotheses:
To assess the type I and II error rates for Wn, we chose four different values for β·,2: 0 × 16, 0.2 × 16, 0.4 × 16, and 0.6 × 16, and chose Ω1 and Ω2. For each Ω, we set n = 20, 40, and 80 and then simulated 1,000 datasets for each case.
The score statistic Wn performs reasonably well for relatively small sample sizes (Table 2). The type I error rates were not excessive even for both the 5% and 1% significance levels at n = 40. Increasing the sample size can increase the power of rejecting the null hypothesis. Comparing the results from Ω1 and Ω2, we note that increasing the correlation in Ω only slightly influences the finite sample performance of Wn.
Table 2.
Comparisons of the rejection rates for score test statistics under Ω1 and Ω2. Three differing sample sizes {20, 40, 80} and 1,000 simulated datasets were used for each case and two significance levels, 5% and 1%, were considered. The null and alternative hypotheses were, respectively, given by H0 : β·,2 = 0 and H1 : β·,2 ≠ 0. Two methods including the resampling method (RE) and χ2 distribution [χ2(6)] were used to calculate the rejection rates
| n = 20 | n = 40 | n = 80 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 5% | 1% | 5% | 1% | 5% | 1% | |||||||
| β·,2 | RE | χ2(6) | RE | χ2(6) | RE | χ2(6) | RE | χ2(6) | RE | χ2(6) | RE | χ2(6) |
| Ω1 | ||||||||||||
| 0 × 16 | 0.143 | 0.031 | 0.037 | 0 | 0.067 | 0.043 | 0.026 | 0.007 | 0.067 | 0.037 | 0.017 | 0.003 |
| 0.2 × 16 | 0.513 | 0.177 | 0.253 | 0.011 | 0.957 | 0.883 | 0.796 | 0.461 | 1 | 1 | 0.991 | 0.971 |
| 0.4 × 16 | 0.597 | 0.213 | 0.293 | 0.022 | 0.993 | 0.951 | 0.832 | 0.481 | 1 | 1 | 1 | 1 |
| 0.6 × 16 | 0.773 | 0.442 | 0.520 | 0.042 | 1 | 1 | 1 | 0.983 | 1 | 1 | 1 | 1 |
| Ω2 | ||||||||||||
| 0 × 16 | 0.126 | 0.023 | 0.037 | 0 | 0.063 | 0.037 | 0.017 | 0.003 | 0.061 | 0.033 | 0.013 | 0.003 |
| 0.2 × 16 | 0.581 | 0.221 | 0.302 | 0.010 | 0.977 | 0.953 | 0.851 | 0.491 | 1 | 1 | 0.991 | 0.991 |
| 0.4 × 16 | 0.602 | 0.227 | 0.321 | 0.032 | 0.991 | 0.981 | 0.871 | 0.562 | 1 | 1 | 1 | 1 |
| 0.6 × 16 | 0.903 | 0.51 | 0.611 | 0.051 | 1 | 1 | 1 | 0.981 | 1 | 1 | 1 | 1 |
In the third set of simulations, we examined the finite sample performance of Wn,𝒟. We generated 3 × 3 SPDs at all 2,500 pixels of a 50 by 50 image slice for each of all n subjects. We used the same setup as the first set of simulations with an exception. We only consider Ω1, the variance of εi(d), at each pixel and for any two pixels d and d′, the correlation of εi,k(d) and εi,k(d′) equals ρ‖d−d′‖, where ρ ∈ [0, 1]. We tested the null hypothesis H0 : β·,2(d) = 0 at all pixels on the slice. We set n = 40 and 80. We first assumed β·,2(d) = 0 at all pixels on the slice to assess the family-wise error rate. To assess the power, we selected a region-of-interest (ROI) with 40 pixels on the reference slice and set β·,2(d) at 0.3 × 16, 0.6 × 16, and 0.9 × 16 for any point d in the ROI. For each simulated dataset, we applied the annealing optimization algorithm with N0 = 200 and M0 = 5 to the simulated data at each pixel and the test procedure to all 2,500 pixels, which took an average CPU time of about 8 hours.
We used the family-wise error rate as the type I error rate and estimated it based on 100 replications with significance level α = 5%. We also calculated the average of the probabilities of rejecting each of the 40 pixels in the ROI as an estimate of the average power using 100 replications and a significance level of α = 5%.
For the test statistic Wn,𝒟, our test procedure worked reasonably well for relatively small sample sizes (Table 3). The family-wise error rates for our test procedure were not particularly accurate for n = 40 when the correlation is zero; in contrast, they improved at n = 40 and ρ = 0.5. Thus, the sample size could somewhat influence the finite sample performance of our test procedure, particularly when the sample sizes are small.
Table 3.
Comparisons of the family-wise error rates and average powers for the test procedure under two different correlations ρ = 0.0 and 0.5. We considered two different sample sizes {40, 80} and 100 simulated datasets for each case at the 5% significance level. In all voxels, the null and alternative hypotheses were, respectively, given by H0 : β·,2(d) = 06 and H1 : β·,2 (d) ≠ 06. We considered four different β·,2(d) {0.0 × 16, 0.3 × 16, 0.6 × 16, 0.9 × 16} for all voxels within the region of interest, whereas we set β·,2(d) = 06 for all voxels outside the region of interest. FWR denotes the family wise error rate and Apower denotes the average rejection rate for voxels inside the region of interest
| n = 40 | n = 80 | |||||||
|---|---|---|---|---|---|---|---|---|
| ρ = 0.0 | ρ = 0.5 | ρ = 0.0 | ρ = 0.5 | |||||
| β·,2(d) | FWR | Apower | FWR | Apower | FWR | Apower | FWR | Apower |
| 0.0 × 16 | 0.12 | 0.00 | 0.06 | 0.00 | 0.08 | 0.00 | 0.07 | 0.00 |
| 0.3 × 16 | 0.18 | 0.10 | 0.12 | 0.10 | 0.06 | 0.56 | 0.06 | 0.57 |
| 0.6 × 16 | 0.14 | 0.67 | 0.06 | 0.68 | 0.02 | 1.00 | 0.03 | 1.00 |
| 0.9 × 16 | 0.12 | 0.83 | 0.10 | 0.85 | 0.08 | 1.00 | 0.06 | 1.00 |
4. HIV IMAGING DATA
We assess the integrity of white matter in human immunodeficiency virus (HIV). White matter is one of the three main solid components of the central nervous system (CNS) and is composed of bundles of myelinated nerve cell processes (or axons), which connect various gray matter areas (the locations of nerve cell bodies) of the brain to each other, and carry nerve impulses between neurons. The white matter is important for passing messages between different areas of gray matter within the CNS. After initial HIV infection, the virus is detectable in the CNS before antibodies are detectable in the blood and HIV could be cultured from brain tissue as early as 15 days (Davis et al. 1992; Rausch and Davis 2001). Since DTI can detect the subtle disruption of white matter structural integrity by assessing the degree to which fiber tracts within the white matter have lost their directional organization (Basser, Mattiello, and LeBihan 1994a, 1994b; Lim and Helpern 2002; Focke et al. 2008; Vernooij et al. 2008), DTI may be an important tool for detecting the early CNS HIV involvement.
We considered 47 subjects, in which 29 were HIV+ subjects (21 males and 8 females) and 18 were healthy (9 males and 9 females) controls from a cross-sectional study. The ages of the HIV+ subjects ranged from 30 to 52 years (mean: 40.0, SD: 5.6 years) and those of healthy controls ranged from 27 to 54 years (mean: 41.2, SD: 7.4 years). For each subject, both diffusion-weighted images and T1 weighted images were acquired. Diffusion gradients with a b-value of 1,000 s/mm2 were applied in six noncollinear directions, (1, 0, 1), (−1, 0, 1), (0, 1, 1), (0, 1, −1), (1, 1, 0), and (−1, 1, 0). A b = 0 reference scan was also obtained for diffusion tensor matrix calculations. Forty-six contiguous slices with a slice thickness of 2 mm covered a field of view (FOV) of 256 × 256 mm2 with an isotropic voxel size of 2 × 2 × 2 mm3. Eighteen acquisitions were used to improve the signal-to-noise ratio (SNR) in the images. High resolution T1 weighted (T1W) images were acquired using a 3D MP-RAGE sequence. Then, a weighted least squares estimation method was used to construct the diffusion tensors (Basser, Mattiello, and LeBihan 1994b; Zhu et al. 2007).
All images were visually inspected before analysis to ensure no bulk motion. A two-step image registration approach was utilized to spatially normalize the DTI parameters. The first step used a B-spline model based on a bidirectional elastic registration method to align the T1 weighted images of all subjects including both normal controls and HIV+ subjects to the T1 weighted images (template) of an arbitrarily chosen 41-year-old female healthy subject. In order to minimize biases induced by the choice of the template, the symmetry between the image pair to be coregistered was ensured through enforcing the consistency between the forward (e.g., from subject X to the template) and backward (from the template to subject X) transformations. The second step was to align each subject’s DTI images to his/her own anatomical T1 weighted images with a 12-parameter affine registration tool in FSL 3.2 (Analysis Group, FMRIB, Oxford, U.K.), so that DTI results can be spatially normalized through the spatial transformation obtained from high resolution T1 elastic registration. The tensor reorientation matrix at each voxel was derived as a rotation matrix approximating the transformation matrix (from T1 registration) through a singular value decomposition (SVD) (Alexander et al. 2001; Xu et al. 2003).
To control for the effects of covariates (diagnosis, age, and gender), we considered model (2) for Ci(β) for diffusion tensors at each voxel. The zi = (1, x1i, x2i, x3i)T is a 4 × 1 vector, in which x1i is Age/10, x2i is gender, and x3i denotes the diagnosis (1 HIV+ and 0 Healthy control). Moreover, we limited the statistical analysis within the major white matter regions which contains 17,444 voxels with mean fractional anisotropy (FA) value in normal volunteers greater than 0.4. We applied the annealing optimization algorithm with N0 = 200 and M0 = 4, which took an average CPU time of approximately 80 hours to carry out the statistical analysis.
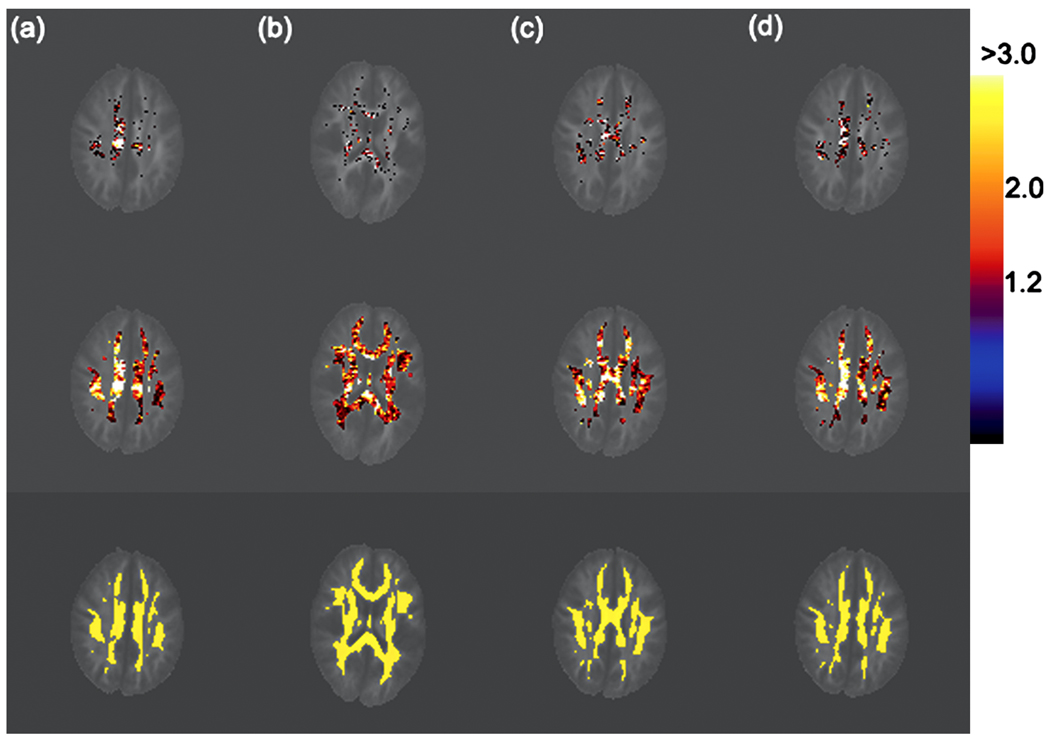
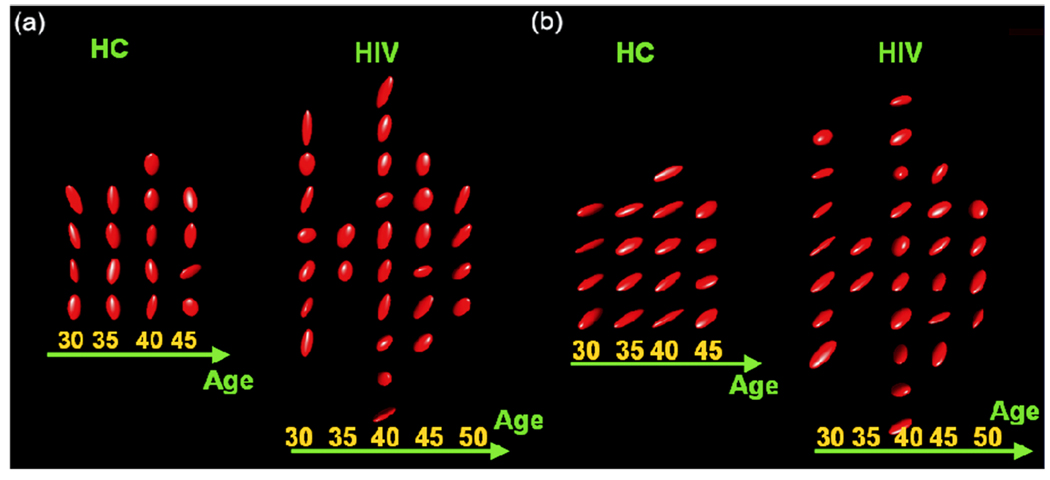
We detected the statistical significance of diagnosis (or age) on the integrity of white matter at all voxels with FA value greater than 0.4. Here, R is a 6 × 24 matrix and b0 = (0, 0, 0, 0, 0, 0)T for the hypotheses on either diagnosis or age. The uncorrected p-values based on the score statistics were color coded at each voxel in the selected regions of the reference brain (Figure 2). To correct for multiple comparisons, we applied our test procedure to calculate the adjusted p-value pD(d) at each voxel in the selected regions of the reference brain (Figure 2). Color-coded maps of p-values using either the uncorrected p(d) alone or the corrected pD(d) indicated several large-scale diagnosis and age effects for white matter integrity. The test procedure for correcting for multiple hypotheses, however, captured far fewer points of differences in the selected regions of the reference brain (Figure 2). As previously reported in studies of HIV (Kure et al. 1990; Pomara et al. 2001), the patients with HIV+ exhibited the subtle disruption of white matter structural integrity in the internal capsule and inferior longitudinal fasciculus. Our studies based on diffusion tensors thus seem to confirm these earlier results obtained with FA values (Pomara et al. 2001). We also picked two voxels and presented their diffusion tensors using an ellipsoid representation (Figure 3). We observed significant differences between HIV+ subjects and healthy controls (Figure 3a) and observed different age trends between HIV+ subjects and healthy controls (Figure 3b).
Figure 2.
Significance testing of diagnosis effect: color-coded maps of raw and adjusted p-values in four selected ROIs of the reference brain. The color scale reflects the magnitude of the values of −log10(P), with black to blue representing smaller values (0–1) and red to white representing larger values (1.88–3). Row 1: adjusted −log10(P) values of the score statistics based on our test procedure for the correction of multiple comparisons. Row 2: raw −log10(P) values of the score statistics based on a χ2 distribution. Row 3: selected ROIs with FA values greater than 0.4. After correcting for multiple comparisons, statistically significant diagnosis effects remain in the superior internal capsule area.
Figure 3.
Ellipsoid representation of diffusion tensors from two voxels to illustrate significant effects of diagnosis and age. Panel (a): testing diagnosis effect. Panel (b): testing age effect.
5. DISCUSSION
We have developed a general statistical framework for an intrinsic regression model of positive-definite matrices as responses in a Riemannian manifold and their association with a set of covariates, such as age and gender, in Euclidean space. The intrinsic regression model is based on the first moment of imaging measures and therefore it avoids any parametric assumptions regarding SPD matrices. We have proposed several link functions to map covariates in Euclidean space to positive-definite matrices in the Riemannian manifold Sym+(m). We have developed an annealing optimization algorithm to search for the ILSE of β. The test procedure based on the resampling method not only accounts for multiple comparisons across the entire region of interest under investigation, but it also asymptotically preserves the dependence structure among the test statistics. Our simulation studies have demonstrated that the methodology developed here provides relatively accurate control of the family-wise error rate.
We also note several limitations of our procedures. There is a computational issue with the estimation procedure for computing β̂. Since Gn(β) is generally not concave and can have many local minima, the annealing optimization algorithm proposed here cannot guarantee to locating the global minimum β̂ of Gn(β) for arbitrary link function. More advanced optimization algorithms are needed for more complex link functions. Our test procedure based on Wn,𝒟 and the resampling method performs reasonably well for relatively larger sample sizes, say 100. However, further improvement on controlling the family-wise error rates is needed. We may either use other computationally extensive methods, such as permutation method, in order to achieve better control of the family-wise error rates, or use other multiple comparison procedures, such as the false discovery rate.
Many issues still merit further research. One major issue is to develop diagnostic measures for assessing the influence of individual observations in semiparametric regression for SPD matrices. Another major issue is to construct goodness-of-fit statistics for testing possible misspecification in (7). Moreover, it is of interest to develop nonparametric regression methods for SPD matrices (Fan and Gijbels 1996). We will study these issues in our future work.
Supplementary Material
Acknowledgments
This work was supported in part by NSF grants SES-06-43663 and BCS-08-26844 and NIH grants UL1-RR025747-01, R01MH086633, and R21 AG033387 to Dr. Zhu, NIH grants GM 70335 and CA 74015 to Dr. Ibrahim and NIH grants R01NS055754 to Dr. Lin. We thank the Editor, an associate editor, and two referees for helpful suggestions, which have improved substantially the present form of this article.
Footnotes
SUPPLEMENTAL MATERIALS
Supplemental materials include assumptions, proofs of Theorems 1 and 2, and a figure for significance testing of age effects.
REFERENCES
- Alexander DC, Pierpaoli C, Basser PJ, Gee JC. Spatial Transformations of Diffusion Tensor Magnetic Resonance Images. IEEE Transactions on Medical Imaging. 2001;20:1131–1139. doi: 10.1109/42.963816. [DOI] [PubMed] [Google Scholar]
- Anderson TW. An Introduction to Multivariate Statistical Analysis (3rd ed.). Wiley Series in Probability and Statistics. Hoboken, NJ: Wiley; 2003. [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D. MR Diffusion Tensor Spectroscopy and Imaging. Biophysical Journal. 1994a;66:259–267. doi: 10.1016/S0006-3495(94)80775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D. Estimation of the Effective Self-Diffusion Tensor From the NMR Spin Echo. Journal of Magnetic Resonance, Ser. B. 1994b;103:247–254. doi: 10.1006/jmrb.1994.1037. [DOI] [PubMed] [Google Scholar]
- Bhattacharya R, Patrangenaru V. Large Sample Theory of Intrinsic and Extrinsic Sample Means on Manifolds-I. The Annals of Statistics. 2003;31:1–29. [Google Scholar]
- Bhattacharya R, Patrangenaru V. Large Sample Theory of Intrinsic and Extrinsic Sample Means on Manifolds-II. The Annals of Statistics. 2005;33:1225–1259. [Google Scholar]
- Butler RW. Saddlepoint Approximations With Applications. New York: Cambridge University Press; 2007. [Google Scholar]
- Chefd’hotel C, Tschumperlé D, Deriche R, Faugeras O. Regularizing Flows for Constrained Matrix-Valued Images. Journal of Mathematical Imaging and Vision. 2004;20:147–162. [Google Scholar]
- Chiu TYM, Leonard T, Tsui KW. The Matrix-Logarithmic Covariance Model. Journal of the American Statistical Association. 1996;91:198–210. [Google Scholar]
- Davis LE, Hjelle BL, Miller VE, Palmer DL, Llewellyn AL, Merlin TL, Young SA, Mills RG, Wachsman W, Wiley CA. Early Viral Brain Invasion in Iatrogenic Human Immunodeficiency Virus Infection. Neurology. 1992;42:1736–1739. doi: 10.1212/wnl.42.9.1736. [DOI] [PubMed] [Google Scholar]
- Davison AC, Hinkley DV. Bootstrap Methods and Their Application. Cambridge: Cambridge University Press; 1997. [Google Scholar]
- Do Carmo MP. Riemannian Geometry. Boston: Birkhäuser, Basel; 1992. [Google Scholar]
- Fan J, Gijbels I. Local Polynomial Modeling and Its Applications. London: Chapman & Hall; 1996. [Google Scholar]
- Focke NK, Yogarajah M, Bonelli SB, Bartlett PA, Symms MR, Duncan JS. Voxel-Based Diffusion Tensor Imaging in Patients With Mesial Temporal Lobe Epilepsy and Hippocampal Sclerosis. NeuroImage. 2008;40:728–737. doi: 10.1016/j.neuroimage.2007.12.031. [DOI] [PubMed] [Google Scholar]
- Fréchet M. Les éléments Aléatoires De Nature Quelconque Dans Un Espace Distancé. Annales de l’Institut Henri Poincaré. 1948;10:215–230. [Google Scholar]
- Grenander U, Miller MI. Computational Anatomy: An Emerging Discipline. Quarterly of Applied Mathematics. 1998;56:617–694. [Google Scholar]
- Gupta AK, Nagar DK. Matrix Variate Distributions. Boca Raton, FL: Chapman & Hall/CRC; 2000. [Google Scholar]
- Karcher H. Riemannian Center of Mass and Mollifier Smoothing. Communications on Pure and Applied Mathematics. 1977;30:509–541. [Google Scholar]
- Kim PT, Richards DS. The Pennsylvania State University, Dept. of Statistics; “Deconvolution Density Estimation on Spaces of Positive Definite Symmetric Matrices,” technical report. 2008
- Kosorok MR. Bootstraps of Sums of Independent but Not Identically Distributed Stochastic Processes. Journal of Multivariate Analysis. 2003;84:299–318. [Google Scholar]
- Kure K, Lyman WD, Weidenheim KM, Dickson DW. Cellular Localization of an HIV-1 Antigen in Subacute AIDS Encephalitis Using an Improved Double-Labeling Immunohistochemical Method. American Journal of Pathology. 1990;136:1085–1092. [PMC free article] [PubMed] [Google Scholar]
- Lang S. Fundamentals of Differential Geometry. New York: Springer-Verlag; 1999. [Google Scholar]
- Lepore N, Brun CA, Chou Y, Chiang M, Dutton RA, Hayashi KM, Luders E, Lopez OL, Aizenstein HJ, Toga AW, Becker JT, Thompson PM. Generalized Tensor-Based Morphometry of HIV/AIDS Using Multivariate Statistics on Deformation Tensors. IEEE Transactions in Medical Imaging. 2008;27:129–141. doi: 10.1109/TMI.2007.906091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim KO, Helpern JA. Neuropsychiatric Applications of DTI—a Review. NMR in Biomedicine. 2002;15:587–593. doi: 10.1002/nbm.789. [DOI] [PubMed] [Google Scholar]
- Lin DY. An Efficient Monte Carlo Approach to Assessing Statistical Significance in Genomic Studies. Bioinformatics. 2005;21:781–787. doi: 10.1093/bioinformatics/bti053. [DOI] [PubMed] [Google Scholar]
- McCullagh P, Nelder JA. Generalized Linear Models. 2nd ed. London: Chapman & Hall; 1989. [Google Scholar]
- Nelder JA, Mead R. A Simplex Method for Function Minimization. Computer Journal. 1965;7:308–313. [Google Scholar]
- Pennec X, Fillard P, Ayache N. A Riemannian Framework for Tensor Computing. International Journal of Computer Vision. 2006;66:41–66. [Google Scholar]
- Pomara N, Crandalla DT, Choia SJ, Johnson G, Lim KO. White Matter Abnormalities in HIV-1 Infection: A Diffusion Tensor Imaging Study. Psychiatry Research: Neuroimaging. 2001;106:15–24. doi: 10.1016/s0925-4927(00)00082-2. [DOI] [PubMed] [Google Scholar]
- Pourahmadi M. Joint Mean–Covariance Models With Applications to Longitudinal Data, I: Unconstrained Parametrization. Biometrika. 1999;86:677–690. [Google Scholar]
- Pourahmadi M. Maximum Likelihood Estimation of Generalized Linear Models for Multivariate Normal Covariance Matrix. Biometrika. 2000;87:425–435. [Google Scholar]
- Rausch DM, Davis MR. HIV in the CNS: Pathogenic Relationships to Systemic HIV Disease and Other CNS Diseases. Journal of Neurovirology. 2001;7:85–96. doi: 10.1080/13550280152058744. [DOI] [PubMed] [Google Scholar]
- Rotnitzky A, Jewell NP. Hypothesis Testing of Regression Parameters in Semiparametric Generalized Linear Models for Cluster Data. Biometrika. 1990;77:485–489. [Google Scholar]
- Schwartzman A. Ph.D. thesis. Stanford University; 2006. Random Ellipsoids and False Discovery Rates: Statistics for Diffusion Tensor Imaging Data. [Google Scholar]
- Vernooij MW, de Groot M, van der Lugt A, Ikram MA, Krestin GP, Hofman A, Niessen WJ, Breteler MMB. White Matter Atrophy and Lesion Formation Explain the Loss of Structural Integrity of White Matter in Aging. NeuroImage. 2008;43:470–477. doi: 10.1016/j.neuroimage.2008.07.052. [DOI] [PubMed] [Google Scholar]
- Whitcher B, Wisco JJ, Hadjikhani N, Tuch DS. Statistical Group Comparison of Diffusion Tensors via Multivariate Hypothesis Testing. Magnetic Resonance in Medicine. 2007;57:1065–1074. doi: 10.1002/mrm.21229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Mori S, Shen D, van Zijl P, Davatzikos C. Spatial Normalization of Diffusion Tensor Fields. Magnetic Resonance in Medicine. 2003;50:175–182. doi: 10.1002/mrm.10489. [DOI] [PubMed] [Google Scholar]
- Zhu HT, Zhang HP, Ibrahim JG, Peterson BG. Statistical Analysis of Diffusion Tensors in Diffusion-Weighted Magnetic Resonance Image Data. Journal of the American Statistical Association. 2007;102:1085–1102. (with discussion) [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.