SYNOPSIS
In collagen, strands of sequence XaaYaaGly form a triple helical structure. The Yaa residue is often (2S,4R)-4-hydroxyproline (Hyp). The inductive effect of the hydroxyl group of Hyp residues greatly increases collagen stability. Here, electron withdrawal by the hydroxyl group in Hyp and its 4S diastereomer (hyp) is increased by the addition of an acetyl group or trifluoroacetyl group. The crystalline structure of AcHyp(C(O)CH3)OMe and Achyp(C(O)CH3)OMe are similar to those of AcHypOMe and AcProOMe, respectively. The O-acylation of AcHypOMe and AchypOMe increases the 13C chemical shift of its Cγ atom: AcHyp(C(O)CF3)OMe ≅ Achyp(C(O)CF3)OMe > AcHyp(C(O)CH3)OMe ≅ Achyp(C(O)CH3)OMe > AcHypOMe ≅ AchypOMe. This increased inductive effect is not apparent in the thermodynamics or kinetics of amide bond isomerization. Despite apparently unfavorable steric interactions, (ProHypGly)10 that is O-acylated with 10 acetyl groups forms a triple helix that has intermediate stability: (ProHypGly)10 > (ProHyp(C(O)CH3)Gly)10 ≫ (ProProGly)10. Thus, the benefit to collagen stability endowed by the hydroxyl group of Hyp residues is largely retained by an acetoxyl group.
INTRODUCTION
Collagen is the most prevalent component of the extracellular matrix in animals.1,2 Collagen has a characteristic tertiary structure consisting of three left-handed polyproline II-type helices wound around a common axis to form a triple helix with a shallow right-handed superhelical pitch. The close packing of the polypeptide chains in the triple helix requires that every third residue be glycine. Each polypeptide chain in fibrillar collagen contains about 300 repeats of the sequence XaaYaaGly, in which Xaa is often (2S)-proline (Pro) and Yaa is often (2S,4R)-4-hydroxyproline (Hyp). The Hyp residues arise from a post-translational modification of Pro residues by the enzyme prolyl 4-hydroxylase. The thermal stability of collagen has a positive correlation with its Hyp content.3,4
The effects of Hyp stereochemistry and sequence on collagen stability have been explored with peptide mimics.5,6 In seminal work, Prockop and coworkers showed that [(ProHypGly)10]3 is more stable than [(ProProGly)10]3, whereas (HypProGly)10 does not exhibit triple helix formation.7,8 The diastereomer (2S,4S)-4-hydroxyproline (hyp) in the Xaa or Yaa position precludes triple helix formation.9 More recent studies have shown that Hyp can enhance triple helical stability in the Xaa position, but not if the Yaa residue is Pro.10,11
Previous reports from our laboratory have shown that replacing Hyp in (ProHypGly)n (n = 7 or 10) with (2S,4R)-4-fluoroproline (Flp) (but not its diastereomer (2S,4S)-4-fluoroproline (flp)) greatly enhances the stability of its triple helices (Table I).12–14 The increase in stability derives from the enhanced electron-withdrawing ability of the fluoro group. That inductive effect constrains the pucker of the pyrrolidine ring, and thereby preorganizes the peptide backbone dihedral angles of the individual strands into conformations that are favorable for triple helix formation.15,16
Table I.
Effect of Hyp and Flp on the conformational stability of collagen triple helices
No known reagent can be used to replace the hydroxyl group of Hyp residues with a fluoro group in aqueous solution.17 The electron-withdrawing ability of a hydroxyl group can, however, be increased by its chemical modification. Here, we synthesize and characterize O-acyl derivatives of AcHypOMe and AchypOMe. We also determine the effects of O-acetylation on the conformational stability of a [(ProHypGly)10]3 triple helix. We find that the influence of the hydroxyl group remains largely intact after its acylation.
RESULTS AND DISCUSSION
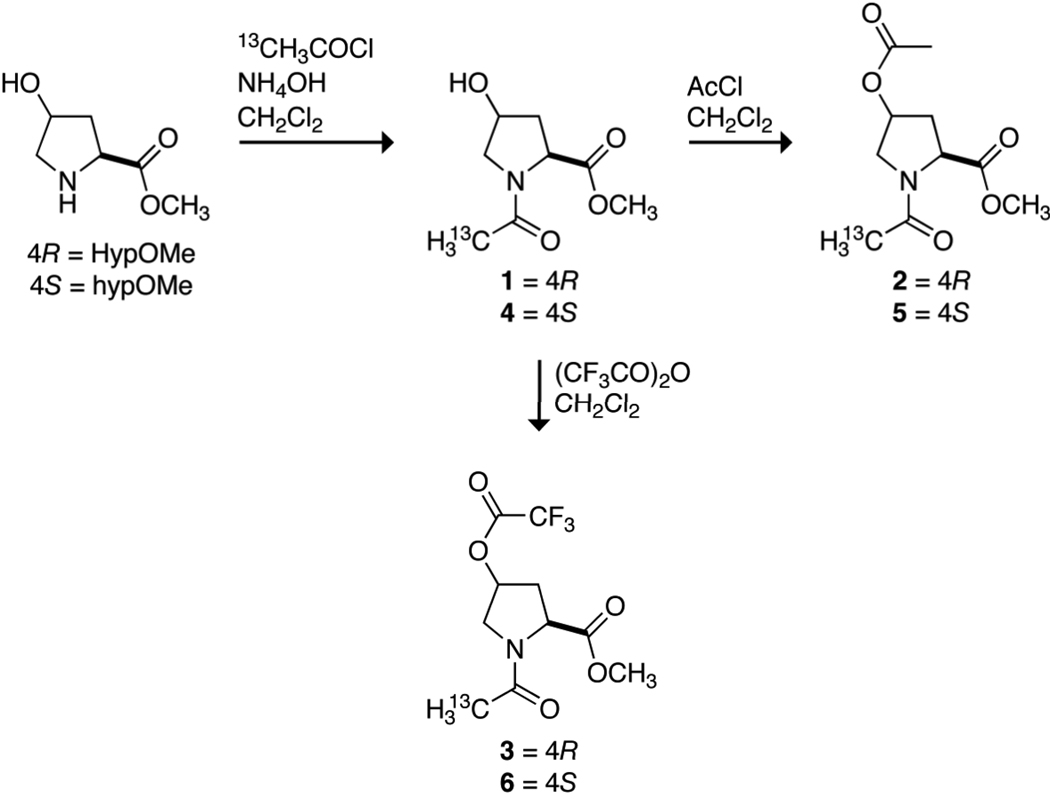
Acyl groups could be installed into natural collagen by the O-acylation of the hydroxyl groups of Hyp residues. Hydroxyl, acetoxyl, and trifluoroacetoxyl groups confer an especially wide range of electron-withdrawing ability. These functional groups have conjugate acid pKa values of 15.6, 4.76, and 0.23, respectively,18 and F values (which report on inductive effects) of 0.33, 0.42, and 0.58, respectively.19 To reveal the consequences of O-acylation of 4-hydroxyproline residues, compounds 1–6 were synthesized by the routes shown in Scheme 1. In 1–6, hydroxyl, acetoxyl, and trifluoroacetoxyl groups are installed in each configuration at C1γ of AcProOMe.
Scheme 1.
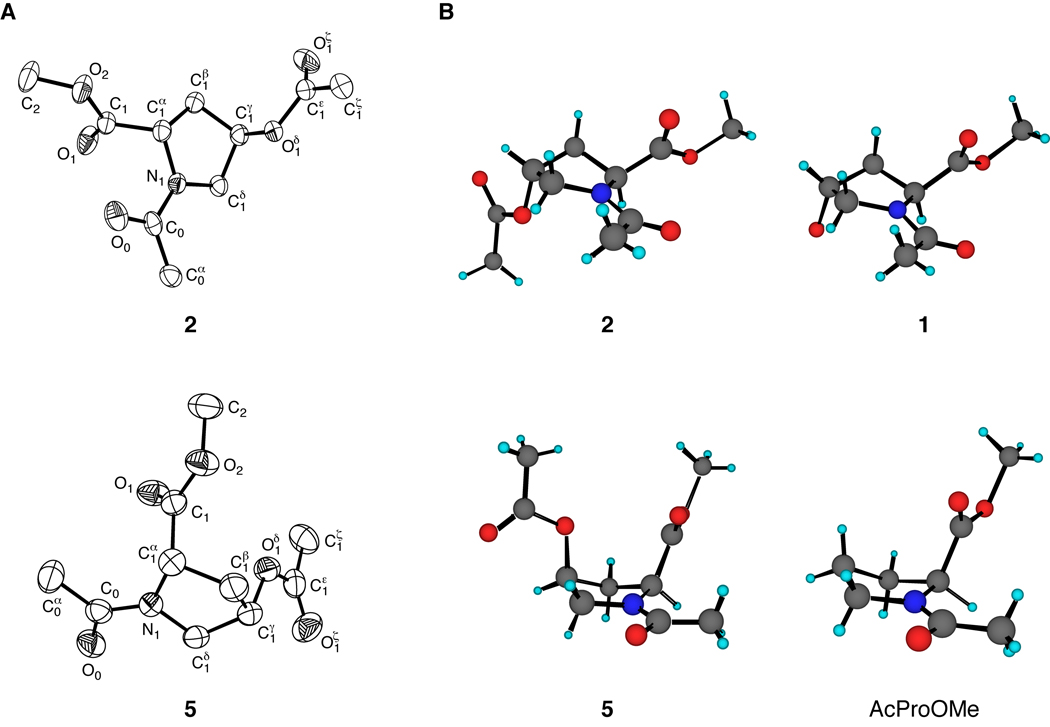
The structures of crystalline 2 and 5 were determined by X-ray diffraction analysis. These structures are shown in Figure 1, along with those of crystalline 1 and AcProOMe.20 Compounds 1 and 2 crystallized with their pyrrolidine rings in the Cγ-exo conformation and a trans amide bond, whereas compound 5 and AcProOMe crystallized in the Cγ-endo conformation and a cis amide bond. The ring puckers of 2 and 5 are fully consistent with the manifestation of a “gauche effect”—the tendency of molecules to adopt the conformation that has the maximum number of gauche interactions between adjacent polar bonds (here, C1δ–N1 and C1γ–O1δ).21,15,16 The ϕ and ψ angles of 2 and 5 are similar to those for proline residues with Cγ-exo and Cγ-endo puckers, respectively (Table II).22 For example, the backbone dihedral angles for 2 are intermediate between those of 1 and AcFlpOMe (7), and are favorable for triple helix formation.23,24
FIGURE 1.
(A) ORTEP diagrams of crystalline 2 (50% probability ellipsoids) and 5 (40% probability ellipsoids). (B) Comparison of pyrrolidine ring conformations of crystalline 2 and 1 (top),20 and 5 and AcProOMe (bottom).20
Table II.
Backbone dihedral angles of crystalline AcProOMe, 1, 2, 5, and 7 derived from X-ray diffraction analysis a
| Compound | Ring pucker | ϕ | ψ | ω |
|---|---|---|---|---|
| AcProOMea | Cγ-endo | −78.9 | 176.7 | −3.1 |
| 5 | Cγ-endo | −73.9 | –170.9 | −3.4 |
| 1b | Cγ-exo | −62.0 | 156.4 | −180.0 |
| 2 | Cγ-exo | −58.2 | 142.0 | −179.4 |
| 7b | Cγ-exo | −55.4 | 140.6 | −177.0 |
ϕ, Ci−1−Ni−Ciα−Ci; ψ (herein), Ni−Ciα−Ci−Oi+1; ω, Oi−1−Ci−1−Ni−Ciα
Data are from ref 20.
13C NMR chemical shifts can report on electron withdrawal by pendant function groups.25 The relative electron-withdrawing ability of the hydroxyl, acetoxyl, and trifluoroacetoxyl groups in 1–6 were assessed by comparing the 13C NMR chemical shifts of their Cγ atoms with those of AcProOMe, AcFlpOMe (7), and AcflpOMe (8). These 13C NMR chemical shifts (Table III) indicate that electron withdrawal increases in the order: hydroxyl (1, 4) < acetoxyl (2, 5) < trifluoroacetoxyl (3, 6), in accord with the conjugate acid pKa and F values of these groups.
Table III.
13Cγ chemical shift (δ) of AcProOMe and compounds 1–8 a
| Compound | δ | Compound | δ |
|---|---|---|---|
| AcProOMe | 21.9 | ||
| 1 | 70.4 | 4 | 70.7 |
| 2 | 73.7 | 5 | 73.7 |
| 3 | 78.5 | 6 | 78.6 |
| 7 | 93.3 | 8 | 93.5 |
Values were obtained at 25°C in 1,4-dioxane-d8.
The cis-to-trans equilibrium constant (KZ/E) for prolyl peptide bonds is close to unity. For example, the Gly–Pro bond in the pentapeptide AcAlaGlyProAlaLysNH2 has a value of KZ/E = 6.3 at 23°C and pH 6.26 Yet, all of the peptide bonds in a collagen triple helix are in the trans conformation. Accordingly, substitutions that favor the trans isomer can enhance collagen stability.21,12,13
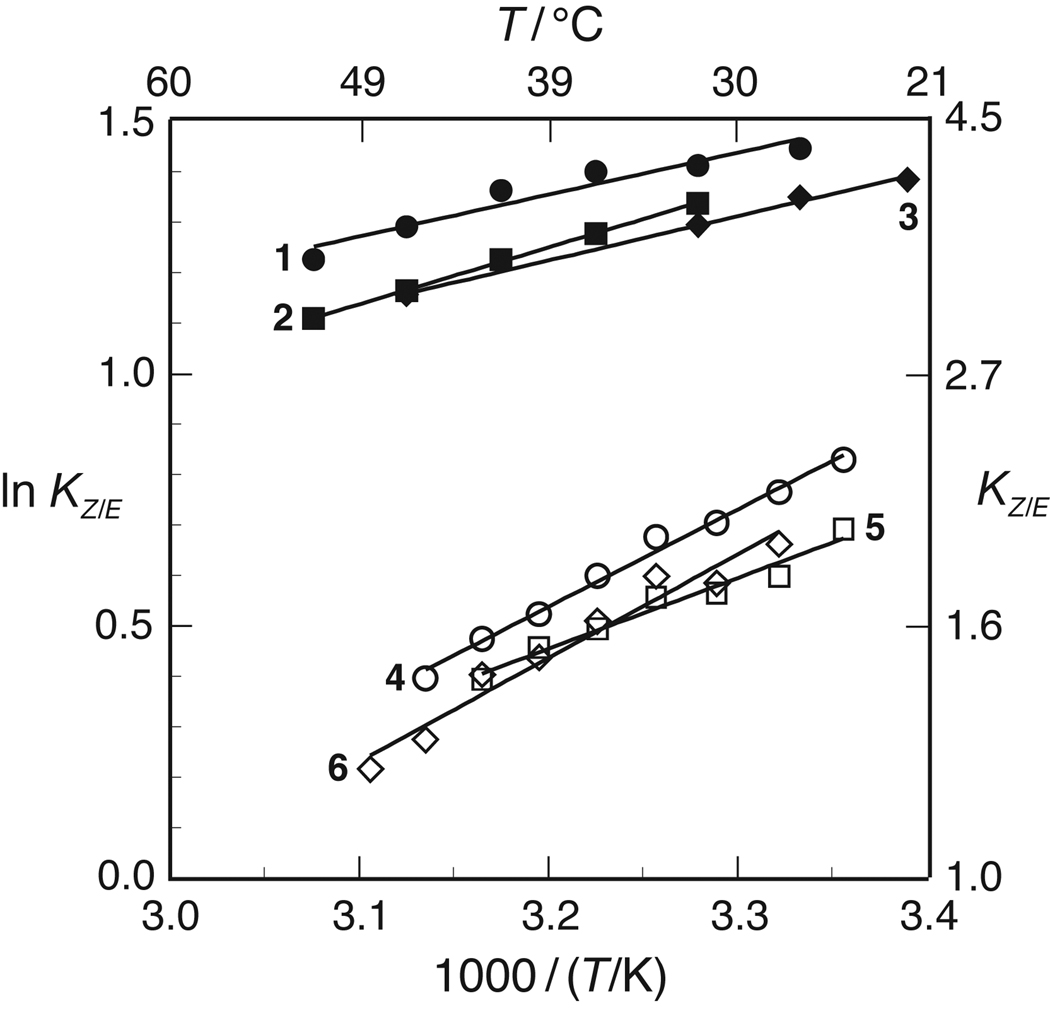
The value of KZ/E for 1–6 was assessed by NMR spectroscopy.27–30,21 At 37°C, the KZ/E values within a series (4R versus 4S) are within error of each other (Table IV). The similarity of these values within each series suggests that any enhancement of preorganization due to increased electron withdrawal at the 4-position is either undetectable or counteracted by other factors. The value of KZ/E is greater for the 4R series than the 4S series, which is consistent with previous work.31,14,32,16
Table IV.
Thermodynamic parameters for amide bond isomerization in compounds 1–6 a
| Compound | ΔH° (kcal/mol) | ΔS° [cal/(mol·deg)] | KZ/E (37°C) |
|---|---|---|---|
| 1 | −1.65 ± 0.12 | −2.59 ± 0.38 | 4.0 ± 0.4 |
| 2 | −2.24 ± 0.02 | −4.68 ± 0.07 | 3.3 ± 0.3 |
| 3 | −1.75 ± 0.04 | −3.15 ± 0.13 | 3.5 ± 0.4 |
| 4 | −3.82 ± 0.08 | −11.19 ± 0.25 | 1.8 ± 0.2 |
| 5 | −2.78 ± 0.12 | −8.01 ± 0.39 | 1.6 ± 0.4 |
| 6 | −4.08 ± 0.18 | −12.18 ± 0.56 | 1.6 ± 0.6 |
Values (± S.E.) are derived from the data in Figure 2.
The effect of temperature on the values of KZ/E for 1–6 was also assessed by NMR spectroscopy. The resulting van’t Hoff plots are shown in Figure 2. Values for ΔH° and ΔS° (± S.E.) were calculated from linear least-squares fits of these data to eq 1:
| (1) |
These values are listed in Table IV. This analysis assumes that the enthalpic and entropic differences between the cis and trans isomers are independent of temperature, that is, ΔCp° = 0 for the isomerization reaction. The linearity of the data in Figure 2 indicate that this assumption is likely to be valid.
FIGURE 2.
van’t Hoff plot for the cis-to-trans amide bond isomerization of compounds 1–6 in 1,4-dioxane.
In all conditions studied herein, KZ/E > 1: the trans isomer of 1–6 is always more stable than the cis isomer (Figure 2). The values of both ΔH° and ΔS° for compounds 4–6 are more negative than those for 1–3, indicating that isomerization is more enthalpically favorable but entropically unfavorable in the 4S series than the 4R series (Table III).
Electron withdrawal at Cγ in 1–6 should increase the bond order in the amide C–O bond and decrease the bond order in the amide C–N bond. Indeed, this effect of electron withdrawal was manifested in amide bond isomerization being faster in AcFlpOMe than AcProOMe.21 An analogous difference was not apparent, however, in the rate of cis–trans isomerization of 1–3 and 4–6 (see Supporting Information). Apparently, the differential electron withdrawal by hydroxyl, acetoxyl, and trifluoroacetoxyl groups is too small to yield a measurable difference in the rate of amide bond isomerization of 1–3 and 4–6.
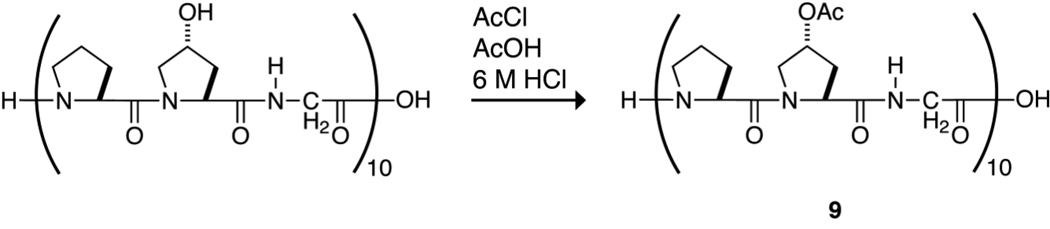
The physicochemical properties of 2 suggest that O-acetylation of (ProHypGly)10 would not preclude its forming a triple helix and could enhance the stability of that helix. This hypothesis was tested by reacting the hydroxyl groups of (ProHypGly)10 with acetyl chloride in 6 M HCl, as shown in Scheme 2. The reaction was done under acidic conditions to preclude acetylation at the N-terminus of the peptide.33 Mass spectrometry indicated that the acetylation proceeded to completion and without byproducts to produce peptide 9 (Figure 3). Attempts to acylate (ProHypGly)10 with trifluoroacetyl groups were unsuccessful because the harsher conditions required for selective O-trifluoroacetylation led to decomposition of the peptide. Moreover, trifluoroacetoxyl groups are much more labile than acetoxyl groups in water, which would make obtaining Tm data of a homogeneous O-trifluoroacetylated peptide problematic.
Scheme 2.
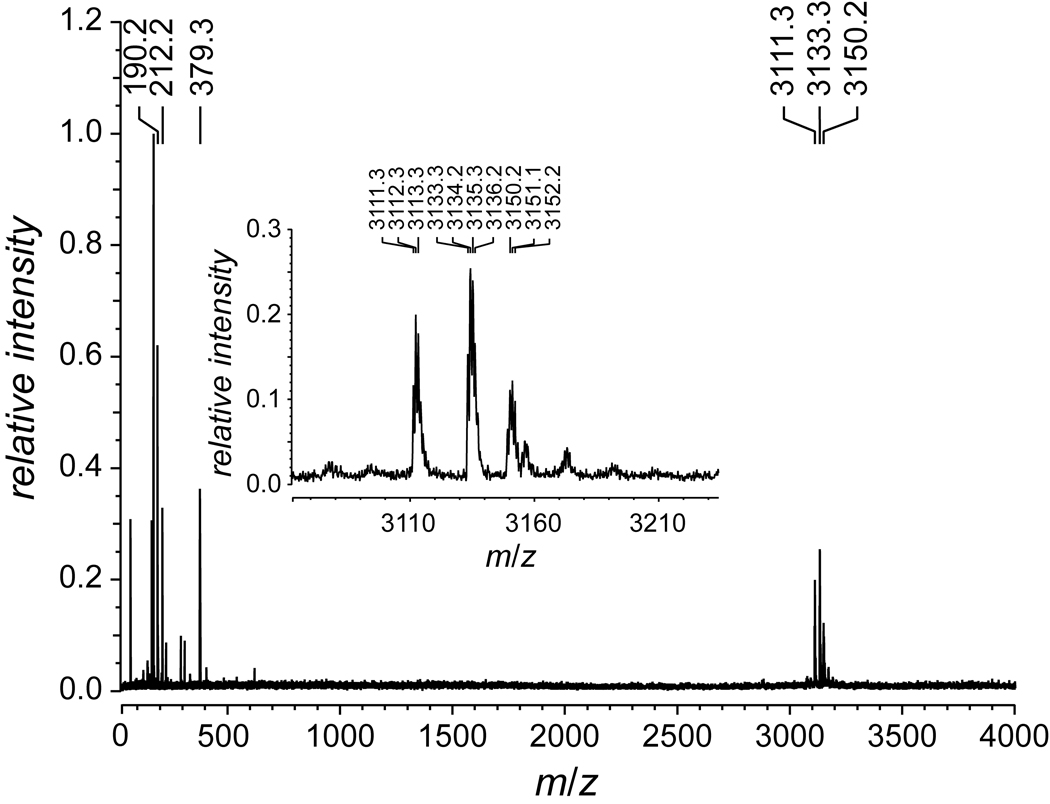
FIGURE 3.
MALDI–TOF mass spectrum of peptide 9 to give m/z 3111.3 (MH+ 3110.4).
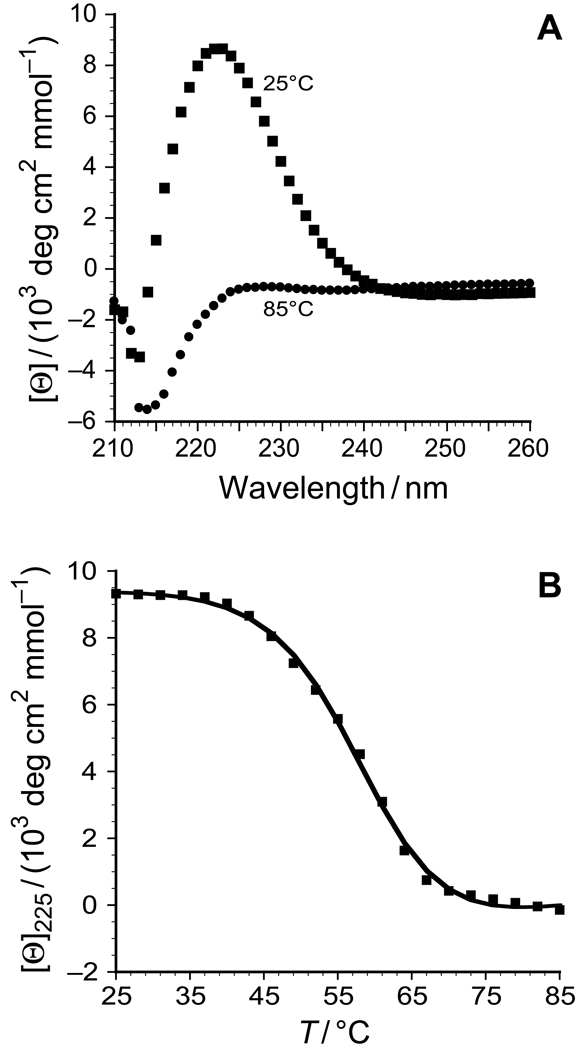
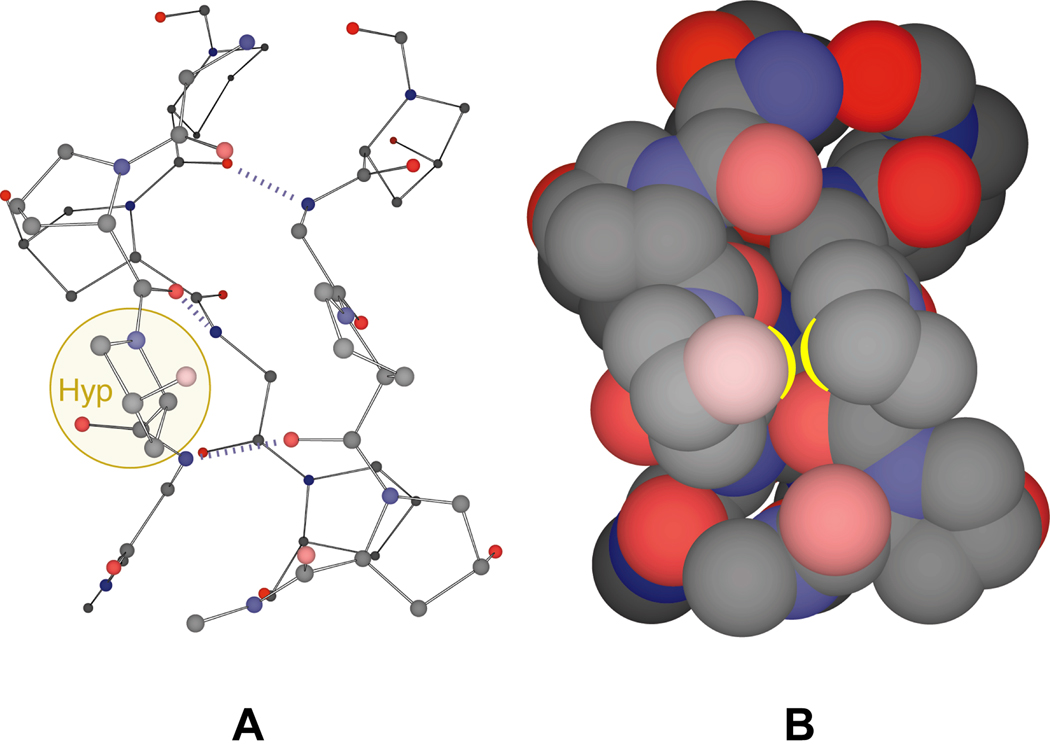
After incubation of the peptide 9 in 50 mM acetic acid at 4°C for 24 h, CD spectra at 25°C indicated that triple helix formation had occurred (Figure 4A). In this solution, the value of Tm of the triple helix (which is the temperature at the midpoint of the thermal transition from native to denatured states) was 57.5°C (Figure 4B), compared to 69°C for [(ProHypGly)10]3.12,13 What is the origin of this decrease in conformational stability? Examination of the structure of the collagen triple helix reveals that addition of an acetyl group to the pseudo-axial hydroxyl group of Hyp could cause significant steric clashes between that acetyl group and a proline residue in a neighboring strand (Figure 5).
FIGURE 4.
(A) Circular dichroism spectra of peptide 9 at 25°C (squares) and 85°C (circles). (B) Thermal denaturation curve for triple helical peptide 9 in 50 mM ascetic acid. The squares represent the data points, and the line represents the curve-fit of the data to give Tm = 57.5°C.
FIGURE 5.
Segment of a (ProHypGly)n triple helix. (A) Ball-and-stick model with a Hyp residue highlighted. (B) Space-filling model with an interstrand Hyp)(Pro interaction highlighted. The N-terminus of the triple helical segment is at the top of the figure. Carbon is gray, nitrogen is blue, and oxygen is red. Atomic coordinates are from PDB entry 1CAG.38
In an earlier report, O-acetylation was found to decrease greatly the stability of a (ProHypGly)10 triple helix (ΔTm = −33°C).34 In this earlier report, neither the integrity of the peptide after acetylation nor the extent of its acetylation were apparent. Regardless, the authors used their data to argue that the large decrease in stability was due to the lesser ability of acetoxyl groups (compared to hydroxyl groups) to form water-mediated hydrogen bonds between strands. In marked contrast, we find a small decrease in the stability of [(ProHyp(C(O)CH3)Gly)10]3 compared to [(ProHypGly)10]3 (ΔTm = −11°C). This lower stability could arise from the steric demands of its 30 acetyl groups (Figure 5) as well as the slight decrease in the value of KZ/E upon O-acetylation (Table III). Despite the loss of its hydroxyl groups, (ProHyp(C(O)CH3)Gly)10 forms a more stable triple helix than does (ProProGly)10 (ΔTm = 17°C). Other evidence also argues against the importance of bridging water molecules in mediating collagen stability.12,6
CONCLUSIONS
Electron withdrawal by the hydroxyl group is known to have dramatic effects on the physicochemical properties of Hyp and hyp residues, and thereby on the conformational stability of triple helical collagen. We synthesized derivatives of AcHypOMe and AchypOMe containing O-acetoxyl and O-trifluoroacetoxyl groups, which are more electron-withdrawing than hydroxyl groups. These changes had little impact on the three-dimensional structure of these residues or the energetics of their cis–trans amide bond isomerization. Chemical modification of the collagen mimic (ProHypGly)10 by O-acetylation leads to a modest decrease in the stability of its triple helix. Apparently, O-acylation of Hyp residues (in contrast to replacing Hyp with Flp residues) creates a steric conflict in the triple helix.
EXPERIMENTAL SECTION
General
Reagents were obtained from Aldrich Chemical (Milwaukee, WI) or Fisher Scientific (Hanover Park, IL) and used without further purification. Amino acids and their derivatives were obtained from Fisher Scientific, Bachem Bioscience (King of Prussia, PA), or Novabiochem (San Diego, CA). Dichloromethane was drawn from a Baker Cycletainer. Thin-layer chromatography was performed by using aluminum-backed plates coated with silica gel containing F254 phosphor and visualized by UV illumination or staining with I2, p-anisaldehyde stain, or phosphomolybdic acid stain. NMR spectra were obtained with Bruker AC-300 and Varian UNITY-500 spectrometers.
N-(2-13C-Acetyl)-(2S,4R)-4-hydroxyproline methyl ester (1)
(2S,4R)-4-Hydroxyproline methyl ester hydrochloride (1.821 g, 10.0 mmol) was suspended in CH2Cl2 (30 mL), and this suspension was cooled to 0°C. 2-13C-Acetyl chloride (0.43 mL, 6.33 mmol) was added in one portion. A mixture of concentrated NH4OH (1.0 mL) and H2O (10 mL) was then added dropwise. The reaction mixture was stirred at 0°C for 3 h. The layers were separated and then concentrated under reduced pressure. The organic layer contained the product, which was purified by flash chromatography, eluting with MeOH (5–10% v/v) in CH2Cl2 to yield 1 as a colorless oil (273 mg, 14.5%). 1H NMR (300 MHz, CDCl3, two rotamers): δ 4.59–4.50 (m, 2H), 4.21 (d, J = 3.9 Hz, 1H), 3.78, 3.73 (two s, 3H), 3.77–3.71 (m, 1H), 3.55–3.50 (m, 1H), 2.50–2.25 (m, 1H), 2.08, 1.96 (two d, J = 129.0, 129.0 Hz, 3H), 2.21–2.00 (m, 1H). 13C NMR (75 MHz, CDCl3, two rotamers): δ 172.8, 170.1 (d, J = 51.7 Hz), 69.7, 68.1, 58.6, 57.4, 55.8, 54.3, 52.6, 39.5, 37.7, 22.1 (enriched), 21.5 (enriched). ESI–MS: m/z 211.0849 ([M + Na]+); 211.0854 ([M + Na]+, calcd).
N-(2-13C-Acetyl)-(2S,4R)-4-acetoxyproline methyl ester (2)
Compound 1 (102 mg, 0.54 mmol) was dissolved in dry CH2Cl2 (1.0 mL). Acetyl chloride (0.10 mL, 1.41 mmol) was added to the resulting solution in two portions. The reaction mixture was stirred for 4 h, and then concentrated under reduced pressure to give 2 as a colorless oil in quantitative yield. 1H NMR (300 MHz, CDCl3, two rotamers): δ 5.41–5.26 (m, 1H), 4.56–4.52 (m, 1H), 3.95–3.60 (m, 2H), 3.80, 3.75 (two s, 3H), 2.65–2.15 (m, 2H), 2.09, 2.00 (two d, J = 128.1, 128.1 Hz, 3H), 2.08 (s, 3H). 13C NMR (75 MHz, CDCl3, major rotamer): δ 172.1, 170.1, 168.3 (d, J = 51.6 Hz), 72.5, 57.1, 53.0, 52.2, 34.8, 22.1 (enriched), 21.6 (minor rotamer, enriched), 20.8. ESI–MS: m/z 253.0875 ([M + Na]+); 253.0881 ([M + Na]+, calcd).
N-(2-13C-Acetyl)-(2S,4R)-4-trifluoroacetoxyproline methyl ester (3)
Compound 1 (102 mg, 0.54 mmol) was dissolved in dry CH2Cl2 (1.0 mL). Trifluoroacetic anhydride (0.09 mL, 0.64 mmol) was added to the resulting solution in one portion. The reaction mixture was stirred for 2 h, and then concentrated under reduced pressure to give 3 as a colorless oil in quantitative yield. 1H NMR (300 MHz, CDCl3, two rotamers): δ 5.62–5.50 (m, 1H), 4.65–4.57 (m, 1H), 4.11–4.03 (m, 1H), 3.82, 3.77 (two s, 3H), 3.81–3.79 (m, 1H), 2.73–2.33 (m, 2H), 2.15, 2.06 (two d, J = 128.7, 128.4 Hz, 3H). 13C NMR (75 MHz, CDCl3, two rotamers): δ 171.4, 171.2, 170.4 (d, J = 51.8 Hz), 158.5 (app. d, J = 41.5 Hz), 156.7 (app. d, J = 43.1 Hz), 114.1 (q, J = 283.8 Hz), 76.6, 75.2, 58.1, 57.1, 53.0, 52.5, 51.2, 36.4, 34.4, 21.7 (enriched), 20.8 (enriched). 19F NMR (282 MHz, CDCl3) δ −77.6, −79.2. ESI–MS: m/z 307.0609 ([M + Na]+); 307.0599 ([M + Na]+, calcd).
N-(2-13C-Acetyl)-(2S,4S)-4-hydroxyproline methyl ester (4)
(2S,4S)-4-Hydroxyproline methyl ester hydrochloride (0.334 g, 1.84 mmol) was dissolved in DMF (4 mL), and the resulting solution was cooled to 0°C. Triethylamine (0.8 mL, 5.74 mmol) was added, leading to the formation of a white solid. 2-13C-Acetyl chloride (0.43 mL, 6.33 mmol) was then added, and the reaction mixture was stirred for 3 h while being allowed to warm to room temperature. The reaction mixture was concentrated under reduced pressure. The residue was suspended in EtOAc, and this suspension was filtered. The filtrate was concentrated to a yellow oil, and the product was purified by flash chromatography, eluting with MeOH (5% v/v) in CH2Cl2 to yield 4 as a colorless oil (0.235 g, 68%). 1H NMR (300 MHz, CDCl3, two rotamers): δ 4.52 (dd, J = 9.6, 2.4 Hz, 1H), 4.49–4.42 (m, 1H), 4.04 (d, J = 8.1 Hz, 1H), 3.79, 3.77 (two s, 3H), 3.73–3.57 (m, 2H), 2.40–2.29 (m, 1H), 2.15–1.08 (m, 1H), 2.10, 2.02 (two d, J = 129.0, 129.0 Hz, 3H). 13C NMR (75 MHz, CDCl3, two rotamers): δ 174.2, 170.3 (d, J = 51.2 Hz), 70.7, 68.6, 58.8, 57.3, 56.4, 55.1, 52.5, 39.3, 37.1, 22.1 (enriched), 21.9 (enriched). ESI–HRMS: m/z 211.0772 ([M + Na]+); 211.0776 ([M + Na]+, calcd).
N-(2-13C-Acetyl)-(2S,4S)-4-(2-13C)acetoxyproline methyl ester (5)
Compound 4 (132 mg, 0.700 mmol) was dissolved in dry CH2Cl2 (1.5 mL). Acetyl chloride (75 µL, 1.055 mmol) was added to the resulting solution in one portion. The reaction mixture was stirred at room temperature for 18 h, and then concentrated under reduced pressure. The resulting oil was azeotroped twice with toluene and placed under high vacuum overnight to give a colorless oil in quantitative yield. 1H NMR (300 MHz, CDCl3, two rotamers) δ 5.32-5.25 (m, 1H), 4.75-4.45 (two m, 1H), 3.91–3.64 (m, 2H), 3.79, 3.74 (two s, 3H), 2.58–1.95 (m, 2H), 2.10, 2.05 (two d, J = 128.4 Hz, 3H), 2.03, 1.98 (two s, 3H). 13C NMR (75 MHz, CDCl3, major rotamer): δ 171.3, 170.1, 169.3 (d, J = 50.5 Hz), 72.7, 56.8, 53.0, 52.2, 34.7, 22.1 (enriched), 21.9 (minor rotamer, enriched), 20.7. ESI–MS: m/z 231.1069 ([M + H]+); 231.1062 ([M + H]+, calcd.)
N-(2-13C-Acetyl)-(2S,4S)-4-trifluoroacetoxyproline methyl ester (6)
Compound 4 (30 mg, 0.16 mmol) was dissolved in CH2Cl2 (1.0 mL). Trifluoroacetic anhydride (22.5 µL, 0.16 mmol) was added to the resulting solution in one portion. The reaction mixture was stirred for 2 h, at which point thin-layer chromatography indicated that starting material was still present. Another portion of trifluoroacetic anhydride (22.5 µL, 0.16 mmol) was added. The reaction mixture was stirred an additional 2 h, concentrated under reduced pressure, and placed under vacuum overnight to give 6 as a colorless oil in quantitative yield. 1H NMR (300 MHz, CDCl3, two rotamers): δ 5.57–5.52 (m, 1H), 4.43 (dd, J = 8.1, 2.7 Hz, 0.5 H), 4.62 (d, J = 8.4 Hz, 0.5H), 4.03–3.82 (m, 2H), 3.78, 3.72 (two s, 3H), 2.76–2.48 (m, 2H), 2.17, 2.13 (two d, J = 129.0, 129.0 Hz, 3H). 13C NMR (75 MHz, CDCl3, two rotamers): δ 171.9 (d, J = 50.1 Hz), 159.3–156.4(m), 114.5 (q, J = 284.0), 77.4, 59.1, 57.5, 53.3, 53.1, 52.9, 52.5, 36.9, 35.1, 22.1 (enriched), 22.0 (enriched). 19F NMR (282 MHz, CDCl3) δ −74.0, −74.1. ESI–MS: m/z 285.0768 ([M + H]+); 285.0779 ([M + H]+, calcd).
(ProHyp(C(O)CH3)Gly)10 (9)
(Pro-Hyp-Gly)10 (Peptides International, Louisville, KY; 3.9 mg, 1.3 µmol) was dissolved in 6 M HCl (5.6 µL) and glacial acetic acid (11.2 µL), and the resulting solution was cooled to 0°C. Acetyl chloride (30 µL, 0.42 mmol) was added, and the resulting mixture was agitated gently for approximately 25 min. The product was precipitated from diethyl ether and collected by centrifugation. The resulting solid was washed several times with small portions of diethyl ether and then dried under vacuum to yield (ProHyp(C(O)CH3)Gly)10 (9) as a white solid (3.0 mg, 75%). MS (MALDI) m/z 3111.3 (MH+ 3110.4).
X-Ray Crystallography
The crystals of 2 and 5 used for X-ray structure determination grew slowly from the oils obtained after their purification by chromatography. The structure determinations were performed as described previously.35,36 The experimental procedures and tables of atomic coordinates, bond lengths, bond angles, and torsion angles are provided in the Supporting Information.
Measurement of KZ/E
Values of KZ/E for 1–6 were determined by NMR spectroscopy using a Bruker NMR spectrometer operating at 300.00 MHz for 1H NMR (1, 3, and 6) or at 74.43 MHz for 13C NMR using a 5-mm broadband probe (1, 2, 4, and 5). Identical results were obtained for 1 using either 1H or 13C NMR spectroscopy. NMR samples of 1–6 were prepared at concentrations of 10 mM in 1,4-dioxane-d8 (99 atom %). Doubling or halving the concentration of 1 and 3 in the sample did not alter the values, indicating the absence of effects on KZ/E from intermolecular interactions. Experiments were performed using a temperature range of 22–52°C. The temperature settings of the spectrometer were calibrated to within 1°C by reference to a 100% ethylene glycol standard.
Measurement of Tm
The structure and stability of a triple helix of peptide 9 was assessed by CD spectroscopy using an Aviv 202 SF instrument equipped with an automated temperature controller. A 0.2 mM solution of peptide 9 in 50 mM HOAc was incubated at 4°C for 24 h. Aliquots of 300 µL were placed in 0.1-cm pathlength quartz cuvettes that had been equilibrated at 5°C. Wavelength scans were performed from 200–260 nm at 25 and 85°C, with a slit width of 1 nm and an averaging time of 3 s. Thermal denaturation experiments were performed by raising the temperature from 5 to 85°C in 3-°C steps, equilibrating for 5 min at each temperature, and monitoring at 225 nm with a 20-s averaging time. Values of Tm, which is the temperature at the midpoint of the thermal transition, were determined in triplicate by fitting the data to a two-state model using the software package NLREG v4.0 (Philip Sherrod).
Supplementary Material
Acknowledgments
We are grateful to F.W. Kotch for assistance. This work was supported by grant AR44276 (NIH to R.T.R.). C.L.J. was supported by Chemistry–Biology Interface Training Grant GM08506 (NIH).
REFERENCES
- 1.Ramachandran GN, Reddi AH, editors. Biochemistry of Collagen. New York: Plenum Press; 1976. [Google Scholar]
- 2.Nimni ME, editor. Collagen 1–4. Boca Raton, FL: CRC Press; 1988. [Google Scholar]
- 3.Burjanadze TV. Biopolymers. 1979;18:931–938. doi: 10.1002/bip.1979.360180413. [DOI] [PubMed] [Google Scholar]
- 4.Burjanadze TV. Biopolymers. 2000;53:523–528. doi: 10.1002/(SICI)1097-0282(200005)53:6<523::AID-BIP8>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- 5.Fields GB, Prockop DJ. Biopolymers. 1996;40:345–357. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C345::AID-BIP1%3E3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 6.Jenkins CL, Raines RT. Nat Prod Rep. 2002;19:49–59. doi: 10.1039/a903001h. [DOI] [PubMed] [Google Scholar]
- 7.Berg RA, Prockop DJ. Biochem Biophys Res Comm. 1973;52:115–120. doi: 10.1016/0006-291x(73)90961-3. [DOI] [PubMed] [Google Scholar]
- 8.Inouye K, Kobayashi Y, Kyogoku Y, Kishida Y, Sakakibara S, Prockop DJ. Arch Biochem Biophys. 1982;219:198–203. doi: 10.1016/0003-9861(82)90149-7. [DOI] [PubMed] [Google Scholar]
- 9.Inouye K, Sakakibara S, Prockop DJ. Biochim Biophys Acta. 1976;420:133–141. doi: 10.1016/0005-2795(76)90352-4. [DOI] [PubMed] [Google Scholar]
- 10.Bann JG, Bächinger HP. J Biol Chem. 2000;275:24466–24469. doi: 10.1074/jbc.M003336200. [DOI] [PubMed] [Google Scholar]
- 11.Mizuno K, Hayashi T, Bachinger HP. J Biol Chem. 2003;278:32373–32379. doi: 10.1074/jbc.M304741200. [DOI] [PubMed] [Google Scholar]
- 12.Holmgren SK, Taylor KM, Bretscher LE, Raines RT. Nature. 1998;392:666–667. doi: 10.1038/33573. [DOI] [PubMed] [Google Scholar]
- 13.Holmgren SK, Bretscher LE, Taylor KM, Raines RT. Chem Biol. 1999;6:63–70. doi: 10.1016/S1074-5521(99)80003-9. [DOI] [PubMed] [Google Scholar]
- 14.Bretscher LE, Jenkins CL, Taylor KM, DeRider ML, Raines RT. J Am Chem Soc. 2001;123:777–778. doi: 10.1021/ja005542v. [DOI] [PubMed] [Google Scholar]
- 15.Improta R, Benzi C, Barone V. J Am Chem Soc. 2001;123:12568–12577. doi: 10.1021/ja010599i. [DOI] [PubMed] [Google Scholar]
- 16.DeRider ML, Wilkens SJ, Waddell MJ, Bretscher LE, Weinhold F, Raines RT, Markley JL. J Am Chem Soc. 2002;124:2497–2505. doi: 10.1021/ja0166904. [DOI] [PubMed] [Google Scholar]
- 17.Ibrahim H, Togni A. Chem Commun. 2004:1147–1155. [Google Scholar]
- 18.Dippy JFJ, Hughes SRC, Rozanski A. J Chem Soc. 1959:2492–2498. [Google Scholar]
- 19.Hansch C, Leo A, Taft RW. Chem Rev. 1991;91:165–195. [Google Scholar]
- 20.Panasik N, Jr, Eberhardt ES, Edison AS, Powell DR, Raines RT. Int J Pept Protein Res. 1994;44:262–269. doi: 10.1111/j.1399-3011.1994.tb00169.x. [DOI] [PubMed] [Google Scholar]
- 21.Eberhardt ES, Panasik N, Jr, Raines RT. J Am Chem Soc. 1996;118:12261–12266. doi: 10.1021/ja9623119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vitagliano L, Berisio R, Mastrangelo A, Mazzarella L, Zagari A. Protein Sci. 2001;10:2627–2632. doi: 10.1110/ps.ps.26601a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berisio R, Vitagliano L, Mazzarella L, Zagari A. Biopolymers. 2001;56:8–13. doi: 10.1002/1097-0282(2000)56:1<8::AID-BIP1037>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 24.Berisio R, Vitagliano L, Mazzarella L, Zagari A. Protein Sci. 2002;11:262–270. doi: 10.1110/ps.32602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Friebolin H. Basic One- and Two-Dimensional NMR Spectroscopy. 3rd Ed. New York: VCH; 1998. [Google Scholar]
- 26.Reimer U, Scherer G, Drewello M, Kruber S, Schutkowski M, Fischer G. J Mol Biol. 1998;279:449–460. doi: 10.1006/jmbi.1998.1770. [DOI] [PubMed] [Google Scholar]
- 27.Forsén S, Hoffman RA. J Chem Phys. 1963;39:2892–2901. [Google Scholar]
- 28.Grathwohl C, Wüthrich K. Biopolymers. 1981;20:2623–2633. [Google Scholar]
- 29.Led JJ, Gesmar H. J Mag Res. 1982;49:444–463. [Google Scholar]
- 30.Eberhardt ES, Loh SN, Raines RT. Tetrahedron Lett. 1993;34:3055–3056. doi: 10.1016/S0040-4039(00)93377-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bretscher LE, Taylor KM, Raines RT. In: Fields GB, Tam JP, Barany G, editors. Peptides for the New Millennium: Proceedings of the Sixteenth American Peptide Symposium; Dordrecht, The Netherlands: Kluwer Academic; 2000. pp. 355–356. [Google Scholar]
- 32.Renner C, Alefelder S, Bae JH, Budisa N, Huber R, Moroder L. Angew Chem Int Ed Engl. 2001;40:923–925. [PubMed] [Google Scholar]
- 33.Wilchek M, Patchornik A. J Org Chem. 1964;29:1629–1630. [Google Scholar]
- 34.Weber RW, Nitschmann H. Helv Chim Acta. 1978;61:701–708. [Google Scholar]
- 35.Jenkins CL, Bretscher LE, Guzei IA, Raines RT. J Am Chem Soc. 2003;125:6422–6427. doi: 10.1021/ja034015j. [DOI] [PubMed] [Google Scholar]
- 36.Jenkins CL, Lin G, Duo J, Rapolu D, Guzei IA, Raines RT, Krow GR. J Org Chem. 2004;69 doi: 10.1021/jo049242y. In Press. [DOI] [PubMed] [Google Scholar]
- 37.Shaw BR, Schurr JM. Biopolymers. 1975;14:1951–1985. [Google Scholar]
- 38.Bella J, Eaton M, Brodsky B, Berman HM. Science. 1994;266:75–81. doi: 10.1126/science.7695699. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.