Abstract
Background
Tungsten carbide (WC) and tungsten carbide cobalt (WC-Co) nanoparticles are of occupational health relevance because of the increasing usage in hard metal industries. Earlier studies showed an enhanced toxic potential for WC-Co compared to WC or cobalt ions alone. Therefore, we investigated the impact of these particles, compared to cobalt ions applied as CoCl2, on the global gene expression level in human keratinocytes (HaCaT) in vitro.
Results
WC nanoparticles exerted very little effects on the transcriptomic level after 3 hours and 3 days of exposure. In contrast, WC-Co nanoparticles caused significant transcriptional changes that were similar to those provoked by CoCl2. However, CoCl2 exerted even more pronounced changes in the transcription patterns. Gene set enrichment analyses revealed that the differentially expressed genes were related to hypoxia response, carbohydrate metabolism, endocrine pathways, and targets of several transcription factors. The role of the transcription factor HIF1 (hypoxia inducible factor 1) is particularly highlighted and aspects of downstream events as well as the role of other transcription factors related to cobalt toxicity are considered.
Conclusions
This study provides extensive data useful for the understanding of nanoparticle and cobalt toxicity. It shows that WC nanoparticles caused low transcriptional responses while WC-Co nanoparticles are able to exert responses similar to that of free cobalt ions, particularly the induction of hypoxia-like effects via interactions with HIF1α in human keratinocytes. However, the enhanced toxicity of WC-Co particles compared to CoCl2 could not be explained by differences in gene transcription.
Background
Engineered nanomaterials are used in large amounts in several industries and an increasing demand, including new types of particles, is anticipated in the future [1]. Their physico-chemical properties, i.e. the small size and the high surface to volume ratio are one of the most interesting characteristics, which is useful for many applications in medicine, chemistry, material sciences and physics. However, these physico-chemical characteristics may be associated with undesired health effects not known for, or different from, the bulk materials. Hence, the field of nanotoxicology is emerging to assess possible hazards of nanomaterials. Several reviews have summarised the potential cellular mechanisms of nanoparticles toxicity such as increase in the production of reactive oxygen species (ROS) and induction of inflammatory responses [2-4]. The cellular responses appear to be dependent on the physical and chemical properties of the particles, such as particle size, dissolution behaviour, surface reactivity and binding ability [2,5,6].
So far, the majority of in vivo and in vitro studies in nanotoxicology have focussed on endpoints such as vitality, production of reactive oxygen species, immunological parameters or cell death. However, the elucidation of the mode of action and identification of subacute effects with potential implications for chronic toxicity are difficult to obtain from these studies. Therefore, modern toxicogenomic approaches established already in pharmacology and toxicology [7-10] could be used to unravel the toxicodynamics of nanomaterials. First studies on the effects of nano- or ultrafine particles on global gene expression patterns revealed compound-specific but no general responses due to the exposure to particles [11-14]. Hence, the chemical composition of the particles seems to play a major role for transcriptional responses. Griffitt and colleagues [11] showed that metal ions (silver and copper) caused similar expression patterns as nanoparticles of the same materials in zebrafish, but the numbers of affected genes were always higher after exposure to the particles. In an in vitro study by Waters et al. [12] it was found that changes in cell viability provoked by silica exhibited a higher correlation with particle surface area than either particle mass or number in macrophages. The majority of biological processes represented by the differentially expressed genes were nearly identical, irrespective of particle diameter.
A toxicogenomic approach has been used in this study to analyse the mode of action of tungsten carbide (WC) and tungsten carbide cobalt (WC-Co) nanoparticles. These nanoparticles are intended to be increasingly used in hard metal industries for the production of wear resistant and hard tools. The major advantage of using WC and WC-Co nano-scaled instead of micro-scaled particles is the increased hardness of resulting composite materials and therefore a prolonged wear lifespan of tools and other products [15]. Cobalt serves as binding agent improving the sintering of hard metals from WC nanoparticles. Therefore, the use of WC-Co particles is favoured in hard metal industries. Potential health implications may be of concern for workers involved in the manufacturing process. Previous studies using μm-sized particles have indicated a lack of toxicity for WC particles but a hazardous potential for cobalt metal particles in vivo and in vitro [16-18]. A mixture of these μm-scaled powders (WC-Co) exhibited an enhanced toxicity if compared to cobalt metal powder alone [16-20]. Our previous research also showed toxicity enhancing effects for nano-sized WC-Co compared to WC or CoCl2 [21,22]. The increased toxicity was proposed to result from specific interactions of WC and cobalt. Since the International Agency for Research on Cancer (IARC) has classified cobalt as 'possibly carcinogenic' and tungsten carbide cobalt as 'probably carcinogenic' to humans [23] research on the elucidation of the mode of action of nano-sized particles of these materials is of high relevance for occupational health. Ionic cobalt (Co2+) is known to exert hypoxia like responses via stabilising the α subunit of the hypoxia inducible transcription factor (HIF1) [24-26]. Ubiquitously expressed HIF1α is degraded via oxygen-dependent prolyl-4-hydroxylation under normoxic conditions [27]. These degradation processes are blocked by cobalt binding or oxygen deficiency (hypoxia) which results in enriched HIF1α levels in the cells. HIF1 is a transcription factor that mediates response to hypoxia by regulating the transcription of genes encoding proteins that play key roles in angiogenesis, glucose and energy metabolism, cell survival and proliferation, iron metabolism, and vascular functions [28,29]. Comparative gene expression studies showed HIF1-mediated responses to be similar for hypoxia and CoCl2 exposure [8,30].
Whether metallic cobalt in nanoparticles, particularly in combination with tungsten, provokes specific toxic effects deviating from or exceeding those observed for dissolved ionic cobalt is not finally clarified yet. Lison and co-workers [31] described the formation of reactive oxygen species (ROS) after a rapid dissolution of cobalt ions out of WC-Co micrometer sized particles in a cell free system, a phenomenon that could not be found with metal cobalt particles or a combination of WC particles with CoCl2. Furthermore, another study that evaluated the role of ROS in the interactive toxicity of carbide-cobalt mixtures found no evidence that production of ROS contributed to the toxicity of WC-Co in macrophages [32]. Lombaert and co-workers [33] investigated gene expression in macrophages exposed to micrometer sized WC-Co particles (a mixture of cobalt metal with a median particle size (d50) of 4 μm and WC particles d50 < 1 μm). They identified differential expression of genes involved in apoptosis regulation, stress response, glucose metabolism, cell signalling, immune response and other pathways. The effects were discussed to be at least partially provoked by dissolved cobalt ions.
In a previous study [21] we have investigated the impact of WC and WC-Co nanoparticles on the vitality of various mammalian cells (lung, skin, colon and oligodendrocyte cell lines; primary neural cell and astroglial cultures). Significant cytotoxic effects were observed for nano-sized WC-Co (33 μg/ml). About 15% of WC and 76% of Co were found to be dissolved after 1 week of storage of the stock solution. Interestingly, WC-Co particles (consisting of 30 μg/ml WC and 3 μg/ml cobalt) showed a higher toxicity than equivalent concentrations of CoCl2 (3 μg/ml) indicating that leaching of cobalt alone may not explain the toxic effects. It was also demonstrated that the tungsten based nanoparticles could enter various cell types [21,22]. Based on this study we selected the human skin cell line HaCaT to investigate the effects of WC and WC-Co nanoparticles and cobalt ions on gene expression patterns. Our major goal was to elucidate (1) whether the observed effects indicate specific mode of actions of WC-Co nanoparticles and/or (2) whether the effects can primarily be explained by dissolved Co.
Results
We compared the mode of action of WC and WC-Co nanoparticles and dissolved CoCl2 in HaCaT cells by recording changes in transcription profiles by microarray analysis. HaCaT cells were exposed to the lowest concentration of WC-Co causing a reduction of cell vitality (33 μg/ml; [21]) and corresponding concentrations of WC and CoCl2. RNA isolated after 3 h and 3 d of exposure from 5 independent biological replicates per treatment was analysed using a commercial human whole genome microarray. Various analyses routines were performed to identify differentially expressed genes, treatment clusters and affected biological pathways.
Identification of differentially expressed genes
SAM analysis of normalised microarray fluorescence intensities for all treatments revealed 1956 significantly differentially expressed genes with about 1146 showing an induction or repression of more than 2fold. The highest number of genes with a significantly altered expression above 2fold was observed after 3 d of exposure (Table 1). Among the different treatments, exposure to CoCl2 provoked the strongest changes in gene expression (373 and 826 genes for 3 h and 3 d of exposure, respectively) followed by WC-Co (37 and 248, respectively) and WC nanoparticles (28 and 49 respectively). Comparison of the genes affected by the different treatments revealed a considerable overlap of transcription profiles. The highest commonalities were observed between the gene expression patterns of CoCl2 and WC-Co after 3 d of exposure (184 genes differentially expressed in both treatments), followed by the exposure to CoCl2 at 3 h and 3 d (134 genes) and WC/WC-Co at 3 d (31 genes). A list of the genes with the strongest differential expression (>5fold) can be found in Table 2, the complete set of genes is available in the Additional file 1.
Table 1.
Number of significant repressed or induced genes > 2fold per treatment*.
| Treatment | up | down | total | Treatment | Overlapping genes between treatments | ||||
|---|---|---|---|---|---|---|---|---|---|
| WC3h | 26 | 2 | 28 | WC3h | WC3h | ||||
| WCCo3h | 13 | 24 | 37 | WCCo3h | 8 | WCCo3h | |||
| CoCl3h | 242 | 131 | 373 | CoCl3h | 8 | 17 | CoCl3h | ||
| WC3d | 18 | 31 | 49 | WC3d | 3 | 2 | 11 | WC3d | |
| WCCo3d | 141 | 107 | 248 | WCCo3d | 2 | 8 | 29 | 31 | WCCo3d |
| CoCl3d | 541 | 285 | 826 | CoCl3d | 8 | 15 | 134 | 19 | 184 |
* identified with SAM; the pair wise comparison indicates genes that were differentially expressed in both of the considered treatments
Table 2.
Genes with most prominent changes in expression levels#.
| WC 3 h | WCCo 3 h | CoCl2 3 h | WC 3 d | WCCo 3 d | CoCl2 3 d | Gene Name | Accession | Description |
|---|---|---|---|---|---|---|---|---|
| up | up | up | up | up | PPFIA4 | NM_015053 | protein tyrosine phosphatase, receptor type, f polypeptide (PTPRF), interacting protein (liprin), alpha 4 (PPFIA4), mRNA [NM_015053] | |
| up | up | up | up | BNIP3 | NM_004052 | BCL2/adenovirus E1B 19 kDa interacting protein 3 (BNIP3), nuclear gene encoding mitochondrial protein, mRNA [NM_004052] | ||
| up | up | up | up | PDK1 | NM_002610 | pyruvate dehydrogenase kinase, isozyme 1 (PDK1), nuclear gene encoding mitochondrial protein, mRNA [NM_002610] | ||
| up | up | up | CA9 | NM_001216 | carbonic anhydrase IX (CA9), mRNA [NM_001216] | |||
| up | up | up | EGLN3 | NM_022073 | egl nine homolog 3 (C. elegans) (EGLN3), mRNA [NM_022073] | |||
| up | up | up | LOXL2 | NM_002318 | lysyl oxidase-like 2 (LOXL2), mRNA [NM_002318] | |||
| up | up | up | THC2369600 | THC2369600 | ALU6_HUMAN (P39193) Alu subfamily SP sequence contamination warning entry, partial (12%) [THC2369600] | |||
| up | up | PLOD2 | NM_182943 | procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (PLOD2), transcript variant 1, mRNA [NM_182943] | ||||
| up | up | PPP1R3C | NM_005398 | protein phosphatase 1, regulatory (inhibitor) subunit 3C (PPP1R3C), mRNA [NM_005398] | ||||
| up | up | CST6 | NM_001323 | cystatin E/M (CST6), mRNA [NM_001323] | ||||
| up | up | FN1 | NM_212482 | fibronectin 1 (FN1), transcript variant 1, mRNA [NM_212482] | ||||
| up | up | THC2316753 | THC2316753 | Q91TG6 (Q91TG6) T130, partial (7%) [THC2316753] | ||||
| up | up | VLDLR | NM_003383 | very low density lipoprotein receptor (VLDLR), transcript variant 1, mRNA [NM_003383] | ||||
| up | up | AML1a | ENST00000358356 | mRNA for AML1a protein, complete cds. [D43967] | ||||
| up | up | HK2 | NM_000189 | hexokinase 2 (HK2), mRNA [NM_000189] | ||||
| up | up | PTGS2 | NM_000963 | prostaglandin-endoperoxide synthase 2 (prostaglandin G/H synthase and cyclooxygenase) (PTGS2), mRNA [NM_000963] | ||||
| up | up | SLC2A14 | BC060766 | solute carrier family 2 (facilitated glucose transporter), member 14, mRNA (cDNA clone MGC:71510 IMAGE:5297510), complete cds. [BC060766] | ||||
| up | up | GPI | NM_000175 | glucose phosphate isomerase (GPI), mRNA [NM_000175] | ||||
| up | up | ENO2 | NM_001975 | enolase 2 (gamma, neuronal) (ENO2), mRNA [NM_001975] | ||||
| up | up | CDH2 | NM_001792 | cadherin 2, type 1, N-cadherin (neuronal) (CDH2), mRNA [NM_001792] | ||||
| up | up | AK021874 | ENST00000366930 | cDNA FLJ11812 fis, clone HEMBA1006364. [AK021874] | ||||
| up | up | up | up | ASB2 | NM_016150 | ankyrin repeat and SOCS box-containing 2 (ASB2), mRNA [NM_016150] | ||
| up | up | up | up | ANGPTL4 | NM_139314 | angiopoietin-like 4 (ANGPTL4), transcript variant 1, mRNA [NM_139314] | ||
| up | up | up | LOC653068 | XM_925841 | PREDICTED: similar to TBP-associated factor 9L (LOC653068), mRNA [XM_925841] | |||
| up | up | TCF19 | BC033086 | transcription factor 19 (SC1), mRNA (cDNA clone MGC:45652 IMAGE:3160434), complete cds. [BC033086] | ||||
| up | up | AIPL1 | NM_014336 | aryl hydrocarbon receptor interacting protein-like 1 (AIPL1), transcript variant 1, mRNA [NM_014336] | ||||
| up | up | LOC200726 | XM_117266 | PREDICTED: hypothetical LOC200726 (LOC200726), mRNA [XM_117266] | ||||
| up | up | LUZPP1 | AJ312775 | mRNA for leucine zipper protein 3 (LUZP3 gene). [AJ312775] | ||||
| up | C9orf65 | NM_138818 | chromosome 9 open reading frame 65 (C9orf65), mRNA [NM_138818] | |||||
| up | THC2411387 | THC2411387 | ALU8_HUMAN (P39195) Alu subfamily SX sequence contamination warning entry, partial (9%) [THC2411387] | |||||
| up | SOX6 | NM_033326 | SRY (sex determining region Y)-box 6 (SOX6), transcript variant 2, mRNA [NM_033326] | |||||
| up | SULT2A1 | NM_003167 | sulfotransferase family, cytosolic, 2A, dehydroepiandrosterone (DHEA)-preferring, member 1 (SULT2A1), mRNA [NM_003167] | |||||
| up | C1orf67 | BC042869 | cDNA clone IMAGE:5270407. [BC042869] | |||||
| up | CRB1 | NM_201253 | crumbs homolog 1 (Drosophila) (CRB1), mRNA [NM_201253] | |||||
| up | up | EDN2 | NM_001956 | endothelin 2 (EDN2), mRNA [NM_001956] | ||||
| up | CBLN4 | NM_080617 | cerebellin 4 precursor (CBLN4), mRNA [NM_080617] | |||||
| up | G65686 | ENST00000332107 | A117 Human STS cDNA, sequence tagged site. [G65686] | |||||
| up | XAGE2 | NM_130777 | X antigen family, member 2 (XAGE2), mRNA [NM_130777] | |||||
| up | PELO | AF118075 | PRO1770 mRNA, complete cds. [AF118075] | |||||
| up | SAA3P | AY209188 | truncated serum amyloid A3 precursor (SAA3) mRNA, complete cds. [AY209188] | |||||
| up | ITIH5 | NM_030569 | inter-alpha (globulin) inhibitor H5 (ITIH5), transcript variant 1, mRNA [NM_030569] | |||||
| up | MCHR1 | NM_005297 | melanin-concentrating hormone receptor 1 (MCHR1), mRNA [NM_005297] | |||||
| up | MGAT4A | NM_012214 | mannosyl (alpha-1,3-)-glycoprotein beta-1,4-N-acetylglucosaminyltransferase, isozyme A (MGAT4A), mRNA [NM_012214] | |||||
| up | GPR65 | NM_003608 | G protein-coupled receptor 65 (GPR65), mRNA [NM_003608] | |||||
| up | DFNB31 | AK056190 | cDNA FLJ31628 fis, clone NT2RI2003344, weakly similar to PRESYNAPTIC PROTEIN SAP97. [AK056190] | |||||
| up | LRP8 | NM_033300 | low density lipoprotein receptor-related protein 8, apolipoprotein e receptor (LRP8), transcript variant 2, mRNA [NM_033300] | |||||
| dn | up | up | up | SORCS3 | NM_014978 | sortilin-related VPS10 domain containing receptor 3 (SORCS3), mRNA [NM_014978] | ||
| dn | up | HLA-DPA1 | NM_033554 | major histocompatibility complex, class II, DP alpha 1 (HLA-DPA1), mRNA [NM_033554] | ||||
| up | dn | dn | AKR1C1 | BC040210 | aldo-keto reductase family 1, member C1, mRNA (cDNA clone MGC:42600 IMAGE:4825338), complete cds. [BC040210] | |||
| up | dn | up | WDR64 | NM_144625 | WD repeat domain 64 (WDR64), mRNA [NM_144625] | |||
| dn | up | ESCO2 | NM_001017420 | establishment of cohesion 1 homolog 2 (S. cerevisiae) (ESCO2), mRNA [NM_001017420] | ||||
| dn | up | CACNG5 | NM_145811 | calcium channel, voltage-dependent, gamma subunit 5 (CACNG5), transcript variant 1, mRNA [NM_145811] | ||||
| dn | up | ANPEP | NM_001150 | alanyl (membrane) aminopeptidase (aminopeptidase N, aminopeptidase M, microsomal aminopeptidase, CD13, p150) (ANPEP), mRNA [NM_001150] | ||||
| dn | up | GPX7 | NM_015696 | glutathione peroxidase 7 (GPX7), mRNA [NM_015696] | ||||
| dn | dn | up | TTLL7 | NM_024686 | tubulin tyrosine ligase-like family, member 7 (TTLL7), mRNA [NM_024686] | |||
| dn | dn | up | THC2371963 | THC2371963 | AIP1_HUMAN (Q86UL8) Atrophin-1 interacting protein 1 (Atrophin-1 interacting protein A) (MAGI-2), partial (3%) [THC2371963] | |||
| up | dn | dn | GDF15 | NM_004864 | growth differentiation factor 15 (GDF15), mRNA [NM_004864] | |||
| dn | dn | ALDH1A1 | NM_000689 | aldehyde dehydrogenase 1 family, member A1 (ALDH1A1), mRNA [NM_000689] | ||||
| dn | dn | DCN | NM_001920 | decorin (DCN), transcript variant A1, mRNA [NM_001920] | ||||
| dn | dn | SOX2 | NM_003106 | SRY (sex determining region Y)-box 2 (SOX2), mRNA [NM_003106] | ||||
| dn | dn | TAF9B | NM_015975 | TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31 kDa (TAF9B), mRNA [NM_015975] | ||||
| dn | dn | THC2302184 | THC2302184 | GAL2_HUMAN (Q01415) N-acetylgalactosamine kinase (GalNAc kinase) (Galactokinase 2), partial (21%) [THC2302184] | ||||
| dn | dn | PTPRZ1 | NM_002851 | protein tyrosine phosphatase, receptor-type, Z polypeptide 1 (PTPRZ1), mRNA [NM_002851] | ||||
| dn | dn | DMRT2 | NM_006557 | doublesex and mab-3 related transcription factor 2 (DMRT2), transcript variant 1, mRNA [NM_006557] | ||||
| dn | dn | CLEC3A | NM_005752 | C-type lectin domain family 3, member A (CLEC3A), mRNA [NM_005752] | ||||
| dn | dn | ATP10B | AB018258 | mRNA for KIAA0715 protein, partial cds. [AB018258] | ||||
| dn | dn | PROS1 | NM_000313 | protein S (alpha) (PROS1), mRNA [NM_000313] | ||||
| dn | dn | RAXLX | NM_001008494 | RAX-like homeobox (RAXLX), mRNA [NM_001008494] | ||||
| dn | dn | OLFM4 | NM_006418 | olfactomedin 4 (OLFM4), mRNA [NM_006418] | ||||
| dn | dn | DSG4 | NM_177986 | desmoglein 4 (DSG4), mRNA [NM_177986] | ||||
| dn | dn | dn | IQGAP2 | NM_006633 | IQ motif containing GTPase activating protein 2 (IQGAP2), mRNA [NM_006633] | |||
| dn | dn | dn | MAL | NM_002371 | mal, T-cell differentiation protein (MAL), transcript variant a, mRNA [NM_002371] | |||
| dn | dn | dn | KRT1 | NM_006121 | keratin 1 (epidermolytic hyperkeratosis) (KRT1), mRNA [NM_006121] | |||
| dn | dn | dn | MMP1 | NM_002421 | matrix metallopeptidase 1 (interstitial collagenase) (MMP1), mRNA [NM_002421] | |||
| dn | dn | dn | LAMP3 | NM_014398 | lysosomal-associated membrane protein 3 (LAMP3), mRNA [NM_014398] | |||
| dn | dn | dn | HLA-DMB | NM_002118 | major histocompatibility complex, class II, DM beta (HLA-DMB), mRNA [NM_002118] | |||
| dn | dn | dn | HERC5 | NM_016323 | hect domain and RLD 5 (HERC5), mRNA [NM_016323] | |||
| dn | BTN3A3 | NM_006994 | butyrophilin, subfamily 3, member A3 (BTN3A3), transcript variant 1, mRNA [NM_006994] | |||||
| dn | C6orf130 | NM_145063 | chromosome 6 open reading frame 130 (C6orf130), mRNA [NM_145063] | |||||
| dn | CRY2 | NM_021117 | cryptochrome 2 (photolyase-like) (CRY2), mRNA [NM_021117] | |||||
| dn | TMEM140 | NM_018295 | transmembrane protein 140 (TMEM140), mRNA [NM_018295] | |||||
| dn | ZNF438 | NM_182755 | zinc finger protein 438 (ZNF438), mRNA [NM_182755] | |||||
| dn | DNMT3L | NM_013369 | DNA (cytosine-5-)-methyltransferase 3-like (DNMT3L), transcript vari ant 1, mRNA [NM_013369] | |||||
| dn | CTAGE3 | AF338231 | CTAGE-3 protein mRNA, complete cds. [AF338231] | |||||
| dn | MOSPD2 | NM_152581 | motile sperm domain containing 2 (MOSPD2), mRNA [NM_152581] | |||||
| dn | OR4N4 | NM_001005241 | olfactory receptor, family 4, subfamily N, member 4 (OR4N4), mRNA [NM_001005241] | |||||
| dn | SPATA7 | NM_018418 | spermatogenesis associated 7 (SPATA7), transcript variant 1, mRNA [NM_018418] | |||||
| dn | TMC1 | NM_138691 | transmembrane channel-like 1 (TMC1), mRNA [NM_138691] | |||||
# listed genes were statistically significant different and exhibited at least a 5fold induction or repression in one of the treatments (with respect to controls and based on normalised fluorescence intensity ratios in the microarray analysis); type of gene regulation is indicated as "up" for induction (> 2fold) and "dn" for repression (> 2fold); fields are empty when induction or repression was < 2fold; full table is provided as Additional File 1
Confirmation of microarray data
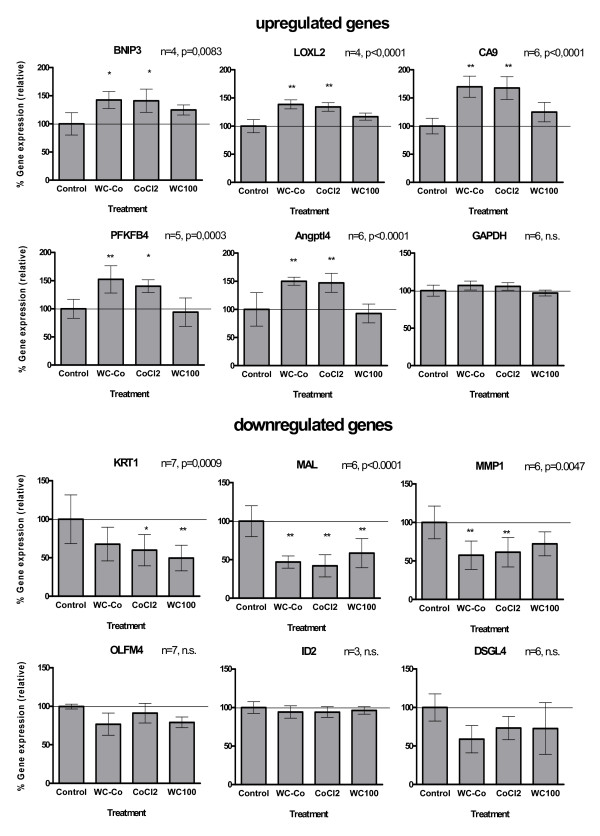
In order to verify the microarray results, RT-PCR analysis was conducted using the same set of samples used for the microarrays as well as RNA samples from independent exposure experiments. Twelve genes with significantly differential expression and a minimum of 2fold up- or downregulation were selected for RT-PCR confirmation. However, care was taken that genes with weak (close to 2fold differential expression) and strong changes (up to 23fold) in expression were included (Figure 1). For eight of the selected genes the significant changes of expression could be confirmed (BNIP3, LOXL2, ANGPTL4, CA9, PFKFB4, KRT1, MAL, MMP1). Trends (induction or repression) were conserved between microarray and RT-PCR data for each treatment. The remaining genes (GAPDH, ID2, OLFM4, DSG4) exhibited a high variability and could not be confirmed as statistically significant from controls by RT-PCR.
Figure 1.
Validation of microarray data by RT-PCR. Relative gene expression of arbitrarily selected genes in HaCaT cells after 3 d of exposure to 30 μg/ml WC and 33 μg/ml WC-Co nanoparticles and 3 μg/ml CoCl2 was analysed by semiquantiative RT-PCR. Selected genes represent genes with significant changes (2 to 23fold) of expression levels in microarrays. Gene expression values were converted to percent of the mean of controls and are presented as mean + standard deviation (SD). Statistical differences were analysed with one-way ANOVA followed by Dunnett's post test (treatment vs. control). Values of p < 0.05 were considered statistically significant; *p < 0.05, **p < 0.01.
PCA and cluster analyses
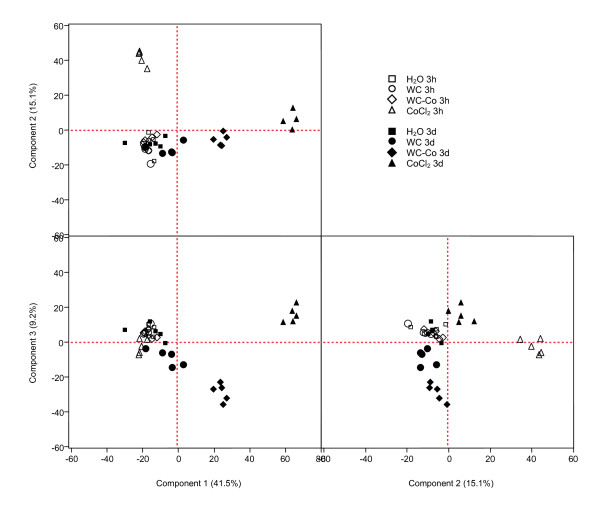
Two methods of descriptive statistics - PCA (principal component analysis) and HCA (hierarchical cluster analysis) - were applied to identify commonalities or differences between treatments based on the patterns of significantly differentially expressed genes. By PCA analysis about 65% of the variability in different treatments was represented by the first 3 components. Four clearly separated clusters, i.e. cells treated with CoCl2 for 3 h, the same treatment for 3 d, cells treated with WC-Co for 3 d and cells treated with WC for 3 d were identified (Figure 2). All other treatments, including the controls, were not separated and formed a large cluster with apparently weak gene expression changes if compared to controls. WC-treated cells were less clearly separated from controls. This was indicated by the observation that a clear distinct cluster was only demonstrated for PC axis 2 and 3.
Figure 2.
Principle component analyses. PCA of differentially expressed genes in HaCaT cells exposed for 3 h and 3 d to 30 μg/ml WC, 33 μg/ml WC-Co nanoparticles and 3 μg/ml CoCl2. Each symbol represents a biological replicate.
Similar results were obtained by HCA, which identified 3 treatment clusters. Both CoCl2 treatments and the 3 d exposures of WC-Co formed a distinct cluster. All other treatments (controls, WC3d, WC3h, WC-Co3h) were grouped in one cluster (Additional file 2). Gene clustering revealed two clusters with strongly induced genes. Genes of these clusters (i.e. LOXL2, BNIP3, CA9, PDK1, ASB2, EGLN3, ANKRD37, PNCK) are coding for proteins with diverse functions, but some of them are known to be direct targets of the transcription factor HIF1α (see below). For the remaining clusters it was not possible to identify predominating groups of gene ontology. Therefore, gene clusters were not analysed in detail but two types of pathway analysis were used to identify signalling pathways and biological functions associated with the differentially expressed genes.
Gene set enrichment analysis (GSEA) and identification of affected signalling pathways
Pathway analysis was performed by two approaches, with (1) GSEA software [34,35] using the complete set of gene expression data from the microarray experiments, and (2) the DAVID database [36] using the list of more then 2fold differentially expressed genes previously identified with SAM.
GSEA identified communalities with existing gene sets (enrichment) primarily for induced genes. The highest number of affected gene sets was found for the 3 d WC-Co treatment. The majority of pathways associated with down-regulated gene sets were found after 3 h of exposure with WC-Co. An overview of pathway related gene sets with the highest commonality (based on statistical significance analysis) to the observed patterns of differentially expressed genes is given in Table 3.
Table 3.
Gene sets with strongest overlaps with observed differentially expressed genes.*
| up-regulated gene sets | description | WC 3 h | WCCo 3 h | CoCl2 3 h | WC 3 d | WCCo 3 d | CoCl2 3 d |
|---|---|---|---|---|---|---|---|
| hypoxia related gene sets | |||||||
| HIF1_TARGETS | Hif-1 (hypoxia-inducible factor 1) transcriptional targets | 0.17 | 0.01 | ||||
| HIFPATHWAY | BIOCARTA: Under normal conditions, HIF-1 is degraded; under hypoxic conditions, it activates transcription of genes controlled by hypoxic response elements (HREs) | 0.02 | 0.20 | ||||
| HYPOXIA_FIBRO_UP | Up-regulated by hypoxia in normal fibroblasts from both young and old donors | 0.20 | 0.65 | 0.05 | |||
| HYPOXIA_NORMAL_UP | Up-regulated by hypoxia in normal, RPTEC renal cells | 0.50 | 0.02 | ||||
| HYPOXIA_REG_UP | Up-regulated by hypoxia in renal cells, and down-regulated with reoxygenation | 0.05 | 0.01 | 0.03 | |||
| HYPOXIA_REVIEW | Genes known to be induced by hypoxia | 0.22 | 0.75 | 0.00 | |||
| MANALO_HYPOXIA_UP | Genes up-regulated in human pulmonary endothelial cells under hypoxic conditions or after exposure to AdCA5, an adenovirus carrying constitutively active hypoxia-inducible factor 1 (HIF-1alpha) | 0.24 | 0.01 | 0.21 | |||
| MENSE_HYPOXIA_UP | List of Hypoxia-induced genes found in both Astrocytes and HeLa Cell | 0.00 | 0.02 | 0.13 | |||
| RESPONSE_TO_HYPOXIA | GO:0001666. Change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating lowered oxygen tension | 0.12 | 0.08 | ||||
| carbohydrate metabolism | |||||||
| POLYSACCHARIDE_METABOLIC_PROCESS | GO:0005976. Chemical reactions and pathways involving polysaccharides, a polymer of more than 10 monosaccharide residues joined by glycosidic linkages | 0.06 | |||||
| FRUCTOSE_AND_MANNOSE_METABOLISM | Genes involved in fructose and mannose metabolism | 0.21 | 0.02 | ||||
| HSA00010_GLYCOLYSIS_AND_ GLUCONEOGENESIS | KEGG: Genes involved in glycolysis and gluconeogenesis | 0.00 | 0.14 | ||||
| GLYCOGEN_METABOLISM | Genes involved in glycogen metabolism | 0.04 | |||||
| GALACTOSE_METABOLISM | Genes involved in galactose metabolism | 0.04 | |||||
| PENTOSE_PHOSPHATE_PATHWAY | Genes involved in pentose phosphate pathway | 0.61 | 0.14 | ||||
| STARCH_AND_SUCROSE_ METABOLISM | Genes involved in starch and sucrose metabolism | 0.01 | 0.50 | ||||
| endocrine metabolism | |||||||
| GN_CAMP_GRANULOSA_UP | Up-regulated in human granulosa cells by the gonadotropins LH and FSH, as well as by cAMP- stimulator forskolin | 0.01 | 0.17 | ||||
| LH_GRANULOSA_UP | Up-regulated in human granulosa cells stimulated with luteinizing hormone (LH) | 0.01 | |||||
| FSH_GRANULOSA_UP | Up-regulated in human granulosa cells stimulated with follicle stimulation hormone (FSH) | 0.01 | |||||
| BREAST_CANCER_ESTROGEN_SIGNALING | Genes preferentially expressed in breast cancers, especially those involved in estrogen-receptor- dependent signal transduction | 0.05 | |||||
| PROSTAGLANDIN_SYNTHESIS_ REGULATION | WIKIPATHWAYS: Prostaglandin Synthesis and Regulation | 0.09 | |||||
| HSA04150_MTOR_SIGNALING_ PATHWAY | KEGG: Genes involved in mTOR signalling pathway | 0.19 | |||||
| cell adhesion, structure, cytoskeleton | |||||||
| HSA04510_FOCAL_ADHESION | KEGG: Genes involved in focal adhesion | 0.07 | 0.46 | ||||
| ACTIN_CYTOSKELETON | GO:0015629. Part of the cytoskeleton (the internal framework of a cell) composed of actin and associated proteins | 0.04 | |||||
| ACTIN_BINDING | GO:0003779. Interacting selectively with monomeric or multimeric forms of actin, including actin Filaments | 0.06 | |||||
| CYTOSKELETON_DEPENDENT_ INTRACELLULAR_TRANSPORT | GO:0030705. The directed movement of substances along cytoskeletal elements such as microfilaments or microtubules within a cell | 0.07 | |||||
| ANATOMICAL_STRUCTURE_FORMATION | GO:0048646. Process pertaining to the initial formation of an anatomical structure from unspecified parts | 0.04 | |||||
| VASCULATURE_DEVELOPMENT | GO:0001944. Process whose specific outcome is the progression of the vasculature over time, from its formation to the mature structure | 0.04 | |||||
| ANGIOGENESIS | GO:0001525. Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels | 0.05 | |||||
| HSA04512_ECM_ RECEPTOR_INTERACTION | KEGG: Genes involved in ECM-receptor interaction | 0.06 | 0.29 | ||||
| miscellaneous | |||||||
| G13_SIGNALING_PATHWAY | G13 signaling pathway | 0.10 | 0.51 | ||||
| NUCLEOTIDE_BIOSYNTHETIC_PROCESS | GO:0009165. Chemical reactions and pathways resulting in the formation of nucleotides | 0.09 | |||||
| CARBON_CARBON_LYASE_ACTIVITY | GO:0016830. Catalysis of the cleavage of C-C bonds by other means than by hydrolysis or oxidation, or conversely adding a group to a double bond | 0.10 | |||||
| ALKPATHWAY | Activin receptor-like kinase 3 (ALK3) is required during gestation for cardiac muscle development | 0.29 | |||||
| CARDIACEGFPATHWAY | BIOCARTA: Cardiac hypertrophy, a response to high blood pressure, is stimulated by GPCR ligands such as angiotensin II that activate the EGF pathway | 0.21 | 0.01 | ||||
| WNT_SIGNALING | Wnt signaling genes | 0.01 | |||||
| HSA05211_RENAL_CELL_CARCINOMA | Genes involved in renal cell carcinoma | 0.07 | |||||
| NKTPATHWAY | BIOCARTA: T cell differentiation into Th1 and Th2 cells occurs by differential chemokine receptor expression, which mediates tissue localization and immune response | 0.29 | |||||
| BIOGENIC_AMINE_SYNTHESIS | WIKIPATHWAYS: Genes involved in synthesis of biogenic amines | 0.23 | |||||
| HSA00591_LINOLEIC_ACID_METABOLISM | Genes involved in linoleic acid metabolism | 0.22 | |||||
| HSA00361_GAMMA_HEXACHLOROCYCLOHEXANE_DEGRADATION | KEGG: Genes involved in gamma-hexachlorocyclohexane degradation | 0.17 | |||||
| down-regulated gene sets | description | WC 3 h | WCCo 3 h | CoCl2 3 h | WC 3 d | WCCo 3 d | CoCl2 3 d |
| RNA metabolism and processing | |||||||
| MRNA_METABOLIC_PROCESS | GO:0016071. Chemical reactions and pathways involving mRNA | 0.23 | |||||
| RIBONUCLEOPROTEIN_COMPLEX_ | GO:0022613. The cellular process by which a complex containing RNA and proteins, is synthesized, | 0.25 | |||||
| BIOGENESIS_AND_ASSEMBLY | aggregates, and bonds together | ||||||
| RNA_PROCESSING | GO:0006396. Any process involved in the conversion of one or more primary RNA transcripts into one or more mature RNA molecules | 0.20 | |||||
| RNA_SPLICING__VIA_TRANSESTERIFICATION_ REACTIONS | GO:0000375. Splicing of RNA via a series of two transesterification reactions | 0.20 | |||||
| SEQUENCE_SPECIFIC_ DNA_BINDING | GO:0043565. Interacting selectively with DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding | 0.33 | |||||
| TRNA_METABOLIC_PROCESS | GO:0006399. Chemical reactions and pathways involving tRNA | 0.27 | |||||
| nucleus and the nuclear membrane related gene sets | |||||||
| PORE_COMPLEX | GO:0046930. Any small opening in a membrane that allows the passage of gases and/or liquids. | 0.28 | |||||
| NUCLEAR_PORE | GO:0005643. Any of the numerous similar discrete openings in the nuclear envelope of a eukaryotic cell, where the inner and outer nuclear membranes are joined | 0.19 | |||||
| NUCLEAR_LUMEN | GO:0031981. The volume enclosed by the nuclear inner membrane | 0.31 | |||||
| NUCLEAR_MEMBRANE | GO:0031965. Either of the lipid bilayers that surround the nucleus and form the nuclear envelope; excludes the intermembrane space | 0.31 | |||||
| enzyme and receptor activity | |||||||
| UBIQUITIN_PROTEIN_ LIGASE_ACTIVITY | GO:0004842. Catalysis of the reaction: ATP + ubiquitin + protein lysine = AMP + diphosphate + protein N-ubiquityllysine | 0.32 | |||||
| SMALL_PROTEIN_CONJUGATING_ ENZYME_ACTIVITY | GO:0008639. Catalysis of the covalent attachment of small proteins, such as ubiquitin or ubiquitin-like proteins, to lysine residues on a target protein. This function may be performed alone or in conjunction with an E3, ubiquitin-like protein ligase | 0.21 | |||||
| CASPASEPATHWAY | BIOCARTA: Caspases are cysteine proteases active in apoptosis | 0.30 | |||||
* identified by GSEA (gene set enrichment analysis), using the databases MSigDB C2 and C5; number of entries is limited to gene sets with a false discovery rate (FDR) < 0.35 in at least one of the treatments and FDR < 0.1 for the WC-Co 3 d treatment; only gene sets obviously related to biochemical pathways or cellular organelles were selected; full table provided as Additional File 3
Gene sets related to the hypoxia pathway as well as carbohydrate metabolism were induced by WC-Co and CoCl2 after 3 d. A significant association with the induction of the hypoxia gene sets was also observed after 3 h of exposure with WC-Co. As indicated by the enrichment of genes for the transcription factor HIF1α (hypoxia inducible factor 1 alpha), regulation via HIF1α may play a major role in provoking the observed changes in hypoxia and carbohydrate metabolism genes. Furthermore, GSEA detected an enrichment of genes related to RNA metabolism and processing as well as genes coding for proteins of the nucleus and the nuclear membrane. These gene sets referred mainly to genes down-regulated after 3 h of exposure to WC-Co nanoparticles. Some of the genes with strong differential repression (> 5fold; e.g. MAL, KRT1, GDF15, MMP1; identified by SAM) were not found to be included in these pathways.
DAVID revealed similar results as GSEA. However, small gene sets, for instance a down-regulation of metallothioneins in the 3 h CoCl2 exposure, were additionally identified by DAVID. Furthermore, genes coding for several proteins containing a functional prolyl-4-hydroxylase alpha subunit were highlighted as up-regulated for the 3 d of exposure with CoCl2 and WC-Co.
The complete results of the GSEA and DAVID analyses are provided in the Additional file 3.
Discussion
The increasing use of nanoparticles may also lead to an increased human exposure and adverse health effects. Occupational exposure is one of the most relevant exposure routes. In order to estimate the potential human health impacts of nanoparticles a precise knowledge on their mechanism of action is indispensable. This knowledge allows, for instance, clarifying whether effects are specifically associated with or enhanced by the nano-sized dimensions or whether the same type of effects as known for corresponding bulk material or dissolved compounds occurs.
In the present paper we focussed on WC and WC-Co nanoparticles which are used in hard metal industries. Dermal uptake, inhalation or accidental oral uptake present possible routes for occupational exposure for these particles. Our previous study has demonstrated their incorporation into various types of cells. Toxicity was low but enhanced for WC-Co compared to pure WC particles [21,22]. A transcriptome analysis of human macrophages exposed to μm-sized WC-Co revealed differential expression of genes known to be affected by cobalt as well [33], providing first evidence that dissolved cobalt seems to play a role in WC-Co toxicity. However, no direct comparison of transcription patterns provoked by nano-sized WC-Co, WC and dissolved cobalt is available so far. In order to model human skin exposure, the human keratinocyte cell line (HaCaT) was selected as experimental model to perform microarray analyses. A number of statistical methods and database analysis tools were used to compare the data sets and perform a detailed pathway analysis.
Transcriptional changes in WC exposed cells
Identification of significantly altered genes revealed only little changes for the exposure of HaCaT cells to WC. Similar observations were made for WC in larger particle size in vitro and in vivo [16,19]. The weak transcriptomic response may be explained by the physicochemical characteristics, since WC nanoparticles were shown to be chemically inert [21]. The genes detected as differentially expressed with WC were mostly also affected by WC-Co and CoCl2 (e.g. EGLN3, CA9, BNIP3, LOXL2, PDK1, KRT1, MMP1). This might be due to traces of cobalt and other metals in WC nanoparticle preparations that have been reported at low concentrations of about 5 × 10-4 μM (described by Bastian et al., 2009 [21]). Some of the genes, however, showed a reciprocal differential expression pattern. For example, while induced by CoCl2, a repression was detected for WC and WC-Co nanoparticles, for e.g. TTLL7, KIT, CHST6, NODAL, WDR64, DES, HS6ST3, DLX2, GPR158. In order to identify potential effects associated with the dimensions of nanoparticles but not related to the chemical compound, we compared our expression data set with 503 genes that were found to be affected by exposure to amorphous silica nanoparticles [12]. In this study, transcriptomic profile of macrophages exposed to amorphous silica particles in two different sizes and different concentrations were recorded. Similar to WC, amorphous silica is known to be chemically inert. Only 29 out of 503 of the silica-sensitive genes were also found to be differentially expressed in our study. Since these genes showed an altered expression with CoCl2 rather than with WC particles, they may reflect a general unspecific stress response.
Transcriptional changes in WC-Co and CoCl2 exposed cells
Compared to the effects with WC nanoparticles, more genes were affected by the WC-Co nanoparticles. Most of them were altered by CoCl2 as well. We found strong overlaps of the expression data of WC-Co and CoCl2 treated cells, whereas the highest number of genes differentially expressed was found with CoCl2. As demonstrated by GSEA analysis, the differentially expressed genes involved in the transcriptional response to WC-Co and CoCl2 could be associated to various biological functions or signalling pathways which are discussed in detail in the following paragraphs.
Whereas most of the affected genes were induced, a number of genes repressed by WC, WC-Co and CoCl2 exposure have been found as well (e.g. MAL, OLFM4, KRT1, CLCA2, MMP1, IQGAP2). For most of these genes the mechanisms of transcriptional regulation are not known and special pathways related to this group of genes could not be identified.
The role of HIF1 for differential gene expression in WC-Co and CoCl2 treated cells
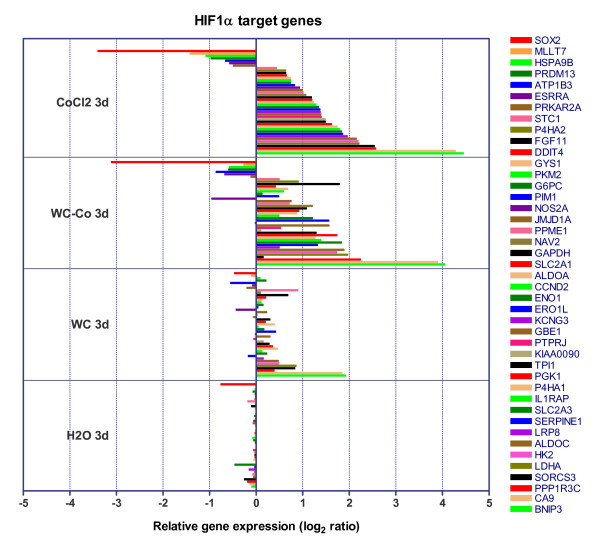
Comparison of the pattern of significant genes and gene set enrichment analyses demonstrated similar responses and signalling pathways for cells exposed to WC-Co and CoCl2, e.g. genes involved in the metabolism of glycolysis and gluconeogenesis, cell adhesion and the response to hypoxia. Under hypoxic conditions, the α subunit of hypoxia inducible factor 1 (HIF1α) accumulates and induces transcription of diverse target genes. HIF1α is a transcription factor that is ubiquitously expressed but rapidly degraded under normoxic conditions. Cobalt ions are known to stabilise HIF1α under normoxic conditions and therefore exert hypoxia-like cellular responses [24-26,37]. Several genes sorted into gene sets related to hypoxia and other pathways e.g. glycolysis and gluconeogenesis, are primary targets of the transcription factor HIF1. To analyse whether such HIF1 target genes were affected by our treatments we generated a list of HIF1target genes (list contained two gene sets from the GSEA C3 TFT database and the "HIF1_Targets" gene set (C2) that was generated after Semenza (2001) [28]). When the expression of HIF1 primary targets is compared, WC-Co nanoparticles provoke almost the same pattern of induction or repression as CoCl2 (Figure 3).
Figure 3.
Expression of HIF1α target genes. HIF1α target genes and their expression levels after 3 d of exposure of HaCaT cells to 30 μg/ml WC and 33 μg/ml WC-Co nanoparticles and 3 μg/ml CoCl2. Bars indicate the mean microarray expression levels of 5 biological replicates. This figure represents all affected HIF target genes identified as significantly differentially expressed by SAM without a fold change threshold.
HIF1 as an initial factor for downstream regulation
The list of affected HIF1 targets contained transcription factors that could be involved in the regulation of secondary HIF1 targets. One example is SOX2. SOX2 contains a putative HIF1 promotor binding site and was found to be strongly repressed after 3 d of exposure with WC-Co and CoCl2. SOX2 is known to play a key role in stem cell generation and pluripotency [38-40]. Greber et al. (2007) [41] studied the transcription profile of embryonic stem cells and embryonic carcinoma cells following a knock down of SOX2. In HaCaT cells exposed to CoCl2, 97 genes with differential expression (26 repressed, 71 induced genes) showed a similar expression pattern if compared to the SOX2 knock down. None of these genes is known as direct HIF1 targets or exhibit HIF1 binding sites and quite a few of them were mentioned in the context of hypoxia earlier. These genes might be regulated by the SOX2 transcription factor as potential secondary HIF1 targets.
Endothelin 2 (EDN2) is another example for a gene with a putative promoter HIF1 binding site that could be involved in the differential expression of genes in the CoCl2 treatment. In agreement with the HIF1 promotor binding site, Na et al. (2008) [42] reported the induction of EDN2 after 3 and 6 hours of hypoxic treatment in granulosa cells. Similarly, EDN2 was found to be induced in HaCaT cells exposed to WC-Co and CoCl2 after 3 h. An induction of other genes of the endothelin complex (EDNRB, EDNRA, ECE2) was detected with the same treatments after 3 d. The induction of collagen mRNA levels and the repression of the matrix metallopeptidase 1 (MMP1) by the endothelin complex was described by Shi-Wen and colleagues [43]. Indeed, an induction of the collagen gene COL5A1 and the repression of the endothelin downstream target MMP1 were detected in HaCaT cells exposed to WC-Co and CoCl2 for 3 d.
In addition to the sets of genes regulated by HIF1, GSEA identified sets of genes that are regulated by other transcription factors. However, similar to HIF1, transcripts of the genes encoding the transcription factors themselves were not found to be differentially expressed. The list (see Additional file 3) contained transcription factors known to be HIF1α interaction partners - e.g. ARNT (aryl hydrocarbon nuclear translocator, [44]) - or HIF1 supporting factors - e.g. (AP1, [45]; Smad3/4; [46]), but also a number of the enriched gene sets not known to be related to HIF1 or targets of HIF1 (such as BACH2, NEF2, ALX4, PAX3).
By comparing HIF1 target genes with known hypoxia responsive genes it becomes obvious that only part of the hypoxia related genes are known to be direct or indirect targets of HIF1. Nevertheless, the fact that most of the genes and pathways affected with WC-Co and CoCl2 were also observed in toxicogenomic studies investigating hypoxia [8,47,48] led us to conclude that the stabilisation of HIF1α via cobalt is an initial step and most of the reactions that are not directly related to HIF1α might reflect downstream events.
Cobalt ions as co-factor substitute
In CoCl2 treated cells the YY1 transcription factor was identified as a potential master regulating factor with GSEA. YY1 is a ubiquitous transcription factor with fundamental biological functions. Its role in cancer biology is also intensely discussed [49]. An interaction of YY1 with cobalt was not yet described but might be conceivable, because YY1 contains four zinc finger domains. The substitution of zinc ions and other divalent metal ions by Co2+ is often discussed to play a role in transcription factor domains, DNA repair mechanisms and calcium metabolism [50-52]. The transcriptional changes of YY1 target genes after the CoCl2 exposure indicate an induction or enrichment of the YY1 protein but it remains unclear, whether the substitution of zinc ions by Co2+ is responsible for that.
In HaCaT cells, a significant depletion of intracellular Zn2+ and Mg2+ after CoCl2 exposure was described by Ortega and colleagues [53]. A substitution of magnesia ions by Co2+ may result in the interruption of ATPases and the energy balance of the cell [54]. It is proposed that ion substitution plays a role in uptake mechanisms of cobalt ions into cells, which evidently happens via cation-dependent ionic pumps [50,53]. Although we found gene sets related to metal ion or cation ion binding proteins to be affected, we did not detect an enrichment of gene sets connected to the described effects resulting from ion substitution.
Differential expression of protein kinases and phosphatases
It was noticed that in WC-Co and CoCl2 exposures several kinases and phosphatases exhibited a differential expression. Kinases are a major group of proteins involved in endocytosis [55,56]. In a knock-down study of Pelkmans and colleagues (2005) [56] 209 kinases with known important functions in several pathways were identified to be involved in endocytosis. Interestingly, most of the pathways they could link with endocytotic processes were also affected in our study (e.g. mTOR, Wnt, integrin/adhesion, RTKs/RSTKs, GPCR).
Protein tyrosine phosphatases (PTPs) and kinases as well as enzymes of the phosphoinositol-3-kinase family (PI3Ks) seem to play a special role in the response to cobalt. The latter have been described as possible upstream regulators of HIF1α [57-60] and have functions in some of the pathways found to be induced (e.g. Insulin-, IGF1-, PPARα-pathway; based on GSEA analysis). PTPs are known to be signalling molecules that regulate a variety of cellular processes including cell growth, differentiation, mitotic cycle, and oncogenic transformation. Some genes coding for PTPs were not only affected by the cobalt containing treatments but also by WC (PPFIA4, PTPRT, PTPRZ1). Since kinases and phosphatases are also involved in the cellular response to various kinds of environmental stress, their altered expression may be related to a cobalt-induced and/or a particle uptake related stress response.
Oxidative stress and transcriptional response
The production of reactive oxygen species (ROS) and the subsequent induction of oxidative stress are discussed as major modes of action of nanoparticles [2,61,62] and was also described to be involved in the cellular response to cobalt ions [54,63,64]. Nevertheless, genes or gene clusters that are related to oxidative stress responses could not be found within our data set of differentially expressed genes. This was confirmed by a lack of ROS production in HaCaT cells for any of the treatments (manuscript in preparation).
Cobalt ions and WC-Co toxicity
Cobalt is an essential trace element for humans, but becomes toxic at high concentrations. In a previous study, we analysed acute toxicity by measuring cell viability of HaCaT cells after same exposure conditions as performed in this study [21]. Altered proliferation or morphological changes of the cells were not observed. Toxicity of CoCl2 was indicated by decreasing cell viability at concentration of 100 μM (corresponds to 6 μg/ml) and above. Lower concentrations of CoCl2 have not been observed to cause acute toxicological reactions in several cells in vitro, including in HaCaT cells [21,54,65]. Intense transcriptional changes were observed in this study at concentrations slightly below those causing in vitro toxicity. The differentially expressed genes may serve as indicators for potential long term effects and may also be useful for investigations of molecular mechanisms.
WC-Co nanoparticles exhibited an increased toxicity in previous studies performed in different types of cell lines (human and fish) when compared to WC particles and CoCl2 [21,22]. Viability of HaCaT cells was slightly (15%) decreased after 3 days of exposure. This enhanced toxicity was discussed as either a result of increased cellular cobalt uptake associated with the uptake of WC particles - the so called "Trojan horse" hypothesis [61] - or a result of unknown combinatory effects of WC particles and cobalt. The "trojan horse" theory is supported by studies showing increased toxicity of nanoparticles with leaching ions compared to the ions alone [11,66-68]. However, analysing the global transcriptional response of HaCaT cells to WC-Co nanoparticles and equivalent WC and cobalt treatments, no evidence for either of these theories could be provided. The number of transcriptional changes was more pronounced in CoCl2 exposed cells, but particularly the regulation of genes resulting from cobalt dependent stabilisation of HIF1α was similar for both, WC-Co and CoCl2. The patterns of transcriptional regulation clearly indicate that the majority of the effects were associated with cobalt ions and did not indicate a special type of interaction between WC and cobalt. However, the enhanced toxicity of WC-Co with respect to CoCl2 appears to be mediated via unknown non-transcriptionally regulated pathways.
Conclusion
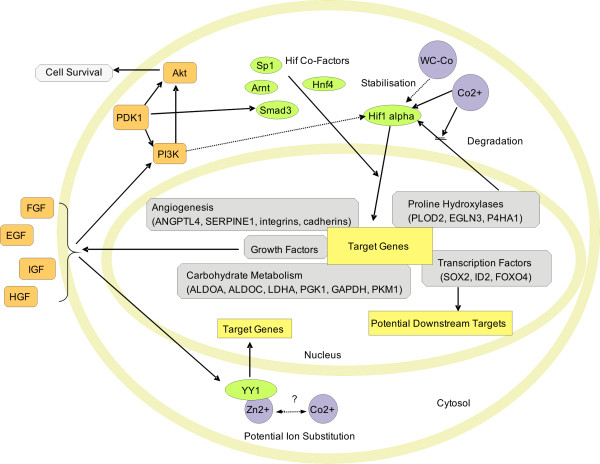
Analysis of gene expression patterns in the human keratinocyte cell line HaCaT demonstrated that the transcriptional response to WC-Co nanoparticles is mainly caused by cobalt ions leaching from the particles. While WC nanoparticles alone do only show very weak effects in expression patterns, WC-Co and CoCl2 exhibited significant transcriptional changes in genes involved in carbohydrate metabolism, hypoxia response, endocrine pathways, cell adhesion and others. The cobalt-sensitive transcription factor HIF1 plays an important role in the regulation of genes involved in these pathways, showing that WC-Co nanoparticles exert hypoxia-like responses similar to CoCl2. The subacute response to CoCl2 was analysed and discussed with respect to downstream events of HIF1 and involvement of other transcription factors (e.g. SOX2, YY1) in cobalt toxicity. A simplified scheme of potential major pathways resulting from cobalt reactions within the cell is provided in Figure 4.
Figure 4.
Scheme of affected cellular pathways. Illustration of the major cellular signalling pathways that were indicated by analyses of the transcriptional responses to WC-Co nanoparticles and cobalt chloride. Arrows indicate known (full lines) or potential (dashed lines) interactions. (Complex Proteins = orange, Transcription Factors = green).
However, while detailed analyses of transcriptional regulations clearly indicate that leached cobalt is likely to be the major trigger for gene regulation in cells exposed to WC-Co, the changes in transcription patterns do not explain the enhanced toxicity of WC-Co if compared to equivalent concentrations of WC or CoCl2. This enhanced toxicity is suggested to be mediated by unknown combinatory effects of WC and cobalt not reflected primarily at the transcriptional response level. However, support or rejection of this hypothesis requires further investigations.
Methods
Preparation and characterisation of particle suspensions and cobalt chloride solutions
Particles and exposure conditions used in this study were identical to those in a previous study which also describes details of the particle preparation, characterisation and behaviour in cell culture media [21]. Briefly, particle suspensions with a concentration of 100 μg/ml were prepared from tungsten carbide (WC) and tungsten carbide cobalt (WC-Co; 10 mass % cobalt content) powders as described in detail by Bastian et al. [21]. Particles exhibited a Brunauer-Emmett-Teller (BET) specific surface area of 6.9 m2/g (WC) and 6.6 m2/g (WC-Co). A mean particle size 56 and 62 nm was calculated from BET values (dBET) for WC and WC-Co, respectively. By dynamic light scattering a mean particle size of 145 ± 5 nm for both WC and WC-Co was estimated (calculated according to DIN ISO 13321 [69]). Similar ranges of particle size distribution and morphology were measured for both types of particles. Particle size was shown to be stable in cell culture media supplemented with FBS (see also [21]). One week after the preparation of the suspension about 6% of tungsten from WC and 15% of tungsten and 76% of cobalt from WC-Co were found to be dissolved [21].
Exposure of cells to nanoparticles was performed using stock suspensions of 300 μg/ml WC (in water) and 330 μg/ml WC-Co (consisting of 300 μg/ml WC and 30 μg/ml cobalt, in 0.03% sodium polyphosphate, see Bastian et al. [21]). Particle suspensions were sterilised by autoclaving and treated for 10 min with ultrasound (Merck Eurolab, Darmstadt, Germany) to disperse aggregates before exposure of cells. Previous investigations did not reveal any changes in particle characteristics by autoclaving and re-dispersing [21].
A cobalt chloride (Fluka/Sigma-Aldrich, Seelze, Germany) stock solution of 10 mM was prepared in distilled water, sterilised by autoclaving and diluted with cell culture grade water (PAA Laboratories, Pasching, Austria). All suspensions and solutions were kept at 4°C.
HaCaT cell culture and exposure of cells
The permanent human keratinocyte cell line, HaCaT (purchased from CLS - Cell Lines Service, Eppelheim, Germany) [70], was maintained in RPMI medium ('Roswell Park Memorial Institute' medium; Biochrom, Karlsruhe, Germany) supplemented with 5% (v/v) FBS and 1% (v/v) penicillin/streptomycin. Cells were cultured in monolayers at 37°C in a humidified, 5% (v/v) CO2-atmosphere and sub-cultured twice a week in 75 cm2 flasks (Techno Plastic Products AG, Trasadingen, Switzerland); passages 30 to 40 were used for experiments. For sub-culturing, cells were washed three times with Versene (Invitrogen/Gibco, Berlin, Germany) and detached by trypsin (0.25% (v/v) in phosphate-buffered saline (Biowest, Renningen, Germany).
Cells were counted using a haemocytometer and seeded at densities of 2 × 105 cells/ml for 3 d of exposure or 5 × 105 cells/ml for 3 h of exposure, respectively in a final volume of 10 ml per 75 cm2 flasks. In order to synchronise proliferation prior to exposure with nanoparticles, cells were allowed to grow for 24 h in RPMI with 5% FBS and subsequently for 24 h in RPMI without FBS for synchronisation [71]. Subsequently, cells were exposed to 30 μg/ml WC, 33 μg/ml WC-Co (cobalt content was 3 μg/ml), or 3 μg/ml cobalt chloride by mixing RPMI containing 5% FBS with 10 fold concentrated stock solutions. Exposure was performed in the dark with 5 independent replicates (performed at different days using different cell passage numbers).
Controls were performed with the water used for the preparation of particle suspensions. The WC-Co suspension also contained polyphosphate (0.003% v/v). However, polyphosphate was not included in controls since the final polyphosphate concentration did not exceed the normal sodium phosphate concentration in cell culture media. Furthermore, no evidences for any effect of low polyphosphate concentrations on cell vitality and function was observed in a previous study [21]. As also shown previously [21], nanoparticles did not aggregate during the exposure period if exposure was performed in FBS supplemented cell culture medium.
RNA extraction
Total RNA was extracted from 75 cm2 cell culture flasks with 1 ml Trizol reagent (Invitrogen, Karlsruhe, Germany) according to the manufacturer's instructions. RNA samples were additionally purified using the RNeasy Kit (Qiagen, Hilden, Germany), RNA qualities and quantities were determined with the Experion detection system (Biorad, Munich, Germany).
Microarray experiments
The effect of the different treatments on transcription profiles of HaCaT cells was compared by microarray analysis (whole genome human 44K array, Agilent Technologies, Böblingen, Germany). Therefore, microarray hybridisations were performed for each treatment (control, WC 30 μg/ml, WC-Co 33 μg/ml, CoCl2 3 μg/ml; 3 h and 3 d exposure each) with 5 independent biological replicates. All hybridisations were performed against a common reference RNA [72] consisting of a mixture of equal amounts of RNA from all treatments. Synthesis of cDNA, cRNA and cRNA-labeling was performed with the Agilent Low RNA Input Linear Amplification Kit according to the manufacturer's instructions. cRNA was labelled with Cy3 (controls and treatments) and Cy5 (common reference). Cy3 and Cy5 labelled cRNA were combined and hybridised to the microarray slides in the DNA Microarrays Hybridisation Oven (Agilent Technologies). Slides were scanned with the Agilent DNA Microarray Scanner (Agilent Technologies). Hybridisation and scanning were performed according to standard protocols of the manufacturer.
Microarray data analysis
Dye-normalised fluorescent intensities of individual microarray spots were extracted using the Agilent Feature Extraction software 9.5. Data were further normalised by dividing the Cy3/Cy5 ratio of each treatment by the mean Cy3/Cy5 ratio of the controls. Data were then analysed using the TMEV software version 4.3 (http://www.tm4.org/) [73]. Genes with significantly altered expression patterns were identified by a modified t-statistic (SAM = significance analysis of microarrays) [74]. Multiple comparison of the complete data set was performed using the lowest possible false discovery rate that allows identification of significantly differentially expressed gene (FDR < 0.03%). Further descriptive analysis by hierarchical clustering (TMEV 4.3) and principal component analysis (PCA, JMP 8.0, SAS institute; http://www.jmp.com) was restricted to the statistically significant genes. Fold changes (FC) of expression levels were calculated using the mean values of each treatment and the mean of the respective controls. A complete list of FC-values of all significantly differentially expressed genes is included in the supplementary information section of this paper (Additional file 1). The microarray data have been submitted to the Gene Expression Omnibus (GEO) database (series no. GSE16727, http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE16727).
Gene set enrichment and pathway analysis
In order to identify biological pathways and functions associated with the changes in gene expression patterns, transcription profiles were analysed by Gene Set Enrichment Analysis (GSEA) [34,35](http://www.broad.mit.edu/gsea/). GSEA is based on ranking of the genes according to their statistical significance and comparison of the patterns to sets of predefined genes. These predefined gene sets are provided by the Molecular Signatures Database (MSigDB) and include five different types of databases (C1 to C5). For our analyses we used the databases C2 (gene sets collected from various sources such as online pathway databases, publications in PubMed including microarray studies, and knowledge of domain experts), C3 (transcription factor targets, i.e. genes that share a transcription factor binding site defined in the TRANSFAC database version 7.4, http://www.gene-regulation.com/) and C5 (gene sets of the Gene Ontology (GO) database, http://www.geneontology.org). Further details are explained on the MSigDB homepage http://www.broad.mit.edu/gsea/msigdb/index.jsp. Since GSEA does not allow the analysis of multiple datasets, analysis was performed pair wise comparing each treatment with the control.
Furthermore, pathway analysis was performed by means of the Database for Annotation, Visualisation and Integrated Discovery (DAVID) [36] (http://david.abcc.ncifcrf.gov/)using the list of differentially expressed genes identified by SAM (see above).
RT-PCR
cDNA was synthesised from RNA using the RevAid™ First Strand cDNA Synthesis Kit (MBI Fermentas, St. Leon-Rot, Germany) according to the manufacturer's instructions. Primers were designed using the computer program Primer3 [75] or Beacon Designer 7 (Premier Biosoft, Palo Alto, USA; (http://www.PremierBiosoft.com) and purchased from Invitrogen. Primer sequences are listed in Table 4.
Table 4.
Sequences of primers used for the validation of microarray data by RT-PCR
| Gene Name | GenBank Accession | Forward Primer Sequence | Reverse Primer Sequence |
|---|---|---|---|
| RPL41 | NM_021104 | AAGATGAGGCAGAGGTCCAA | TCCAGAATGTCACAGGTCCA |
| LOXL2 | NM_002318 | AGCTTCTGCTTGGAGGACACA | TGAAGGAACCACCTATGTGGCA |
| ANGPTL4 | NM_139314 | GTCCTCGCACCTGGAACCC | CTTCGGGCAGGCTTGGCCAC |
| PFKFB4 | NM_004567 | TCCCCACGGGAATTGACAC | GAGAGTTGGGCAGTTGGTCAT |
| BNIP3 | NM_004052 | ACACCACAAGATACCAACAGG | TCTTCATGACGCTCGTGTTCCTC |
| GAPDH | NM_002046 | AGGCTGAGAACGGGAAGC | AGAGGGGGCAGAGATGATG |
| CA9 | NM_001216 | AACCAGACAGTGATGCTGAGT | TGGCATAATGAGCAGGACAGGA |
| MAL | NM_002371 | AAACATTGCTGCCGTGGTGTTC | AGGTTAGACACAGCAAGCTCCCA |
| OLFM4 | NM_006418 | ATTGGGTGGCGCCATTGAATA | TGGTGTTCATAGTACGGGTGGCA |
| ID2 | NM_002166 | GACCCGATGAGCCTGCTATAC | AATAGTGGGATGCGAGTCCAG |
| DSG4 | NM_177986 | TGAAGATGAAGGTCGACCAGC | GGGTTGCACACATGGATCAGCAT |
| KRT1 | NM_006121 | AGAATGCCCTCAAGGATGCCA | TTCTCCGGTAAGGCTGGGACAAA |
| MMP1 | NM_002421 | AAGAGGCTGGGAAGCCATCAC | TCAGTGAGGACAAACTGAGCCA |
Target genes and the reference gene RPL41 [76] were amplified from 1 μl of cDNA using 1 unit of Taq Polymerase (Promega, Mannheim, Germany), 50 mM TRIS-HCl (pH 9.0, Serva, Heidelberg, Germany), 1.5 mM MgCl2 (Sigma, Steinheim, Germany), 15 mM (NH4)2SO4 (Sigma), 0.1% (v/v) Triton-X 100 (Merck, Darmstadt, Germany), 0.2 mM dNTPs (MBI Fermentas) and 0.6 μM of each primer in a 25 μl reaction volume. The number of cycles was adjusted to obtain amplified DNA during the exponential phase of the reaction. Annealing was performed at 55°C. PCR-fragments were analysed by agarose gel electrophoresis (1.5% w/v agarose) and ethidium bromide staining (0.005% w/v). mRNA abundance was evaluated by either visual comparison of band intensity or densitometric analysis using the image analysis software ImageJ (Version 1.33u, available at http://rsb.info.nih.gov/ij/). Relative gene expression levels were calculated by normalisation of band intensities to the reference gene. These relative gene expression values were converted to percent of the average control values. Statistical differences were analysed after confirmation of normal distribution (Kolmogorov-Smirnov test) with one-way ANOVA followed by Dunnett's post test using GraphPad Prism 4.0 software (GraphPad Software, San Diego California USA, http://www.graphpad.com). Values of p < 0.05 were considered statistically significant.
Abbreviations
BET: Brunauer-Emmett-Teller specific surface area; DLS: dynamic light scattering; FBS: foetal bovine serum; FC: fold change; FDR: false discovery rate; GO: gene ontology; GSEA: gene set enrichment analyses; HCA: hierarchical cluster analyses; IARC: International Agency for Research on Cancer; LOEC: lowest observed effect level; MSigDB: Molecular Signature Database; NOEC: no observed effect concentration; PCA: principal component analyses; PI3Ks: phosphoinositol-3-kinases; ROS: reactive oxygen species; RPMI: Roswell Park Memorial Institute medium; RT-PCR: reverse transcriptase polymerase chain reaction; SAM: significance analyses of microarrays; TFT: transcription factor targets; v: volume; w: weight; WC: tungsten carbide; WC-Co: tungsten carbide cobalt; xPCS: mean particle size
Authors' contributions
WB carried out the experiments, the computational analyses and manuscript preparation. DK assisted with the cell culture and exposures. KS and SS conceived and supervised the study, participated in the design of the study and in manuscript preparation. SS helped with the data analyses. All authors have read and approved the manuscript.
Supplementary Material
Table contains all significantly altered expressed genes and fold changes for all treatments
Heat map of differentially expressed genes and hierarchical clustering of all replicates
Table contains all gene sets and pathways identified as enriched by GSEA and DAVID
Contributor Information
Wibke Busch, Email: wibke.busch@ufz.de.
Dana Kühnel, Email: dana.kuehnel@ufz.de.
Kristin Schirmer, Email: kristin.schirmer@eawag.ch.
Stefan Scholz, Email: stefan.scholz@ufz.de.
Acknowledgements
This research was supported by a PhD - scholarship to WB by the Max Buchner Forschungsstiftung and a grant of the German Federal Ministry for Education and Research (BMBF) to the project INOS (Identification and Evaluation of Health and Environmental Effects of Technical Nanoscaled Particles; grant # 03X0013C).
We would like to thank Dr. Hans Mollenkopf from the Max Planck Institute for Infection Biology Berlin (MPI-IB) and Julia Jarells from the Max Planck Institute of Cell Biology and Genetics Dresden (MPI-CBG) for providing facilities and advise for microarray scanning and data extraction. Dr. Andreas Beyer from the Biotechnology Centre of the University of Dresden is acknowledged for support of data analyses.
References
- The Royal Society and the Royal Academy of Engineering. Nanoscience and Nanotechnologies: Opportunities and Uncertainties. London: Royal Society Publications; 2004. pp. 1–23. [Google Scholar]
- Nel A, Xia T, Madler L, Li N. Toxic potential of materials at the nanolevel. Science. 2006;311(5761):622–627. doi: 10.1126/science.1114397. [DOI] [PubMed] [Google Scholar]
- Medina C, Santos-Martinez MJ, Radomski A, Corrigan OI, Radomski MW. Nanoparticles: pharmacological and toxicological significance. Br J Pharmacol. 2007;150(5):552–558. doi: 10.1038/sj.bjp.0707130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Xia T, Nel AE. The role of oxidative stress in ambient particulate matter-induced lung diseases and its implications in the toxicity of engineered nanoparticles. Free Radic Biol Med. 2008;44(9):1689–1699. doi: 10.1016/j.freeradbiomed.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warheit DB. How meaningful are the results of nanotoxicity studies in the absence of adequate material characterization? Toxicol Sci. 2008;101(2):183–185. doi: 10.1093/toxsci/kfm279. [DOI] [PubMed] [Google Scholar]
- Oberdorster G, Oberdorster E, Oberdorster J. Nanotoxicology: an emerging discipline evolving from studies of ultrafine particles. Environ Health Perspect. 2005;113(7):823–839. doi: 10.1289/ehp.7339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waring JF, Gum R, Morfitt D, Jolly RA, Ciurlionis R, Heindel M, Gallenberg L, Buratto B, Ulrich RG. Identifying toxic mechanisms using DNA microarrays: evidence that an experimental inhibitor of cell adhesion molecule expression signals through the aryl hydrocarbon nuclear receptor. Toxicology. 2002;181-182:537–550. doi: 10.1016/S0300-483X(02)00477-8. [DOI] [PubMed] [Google Scholar]
- Vengellur A, Phillips JM, Hogenesch JB, LaPres JJ. Gene expression profiling of hypoxia signaling in human hepatocellular carcinoma cells. Physiol Genomics. 2005;22(3):308–318. doi: 10.1152/physiolgenomics.00045.2004. [DOI] [PubMed] [Google Scholar]
- Andrew AS, Warren AJ, Barchowsky A, Temple KA, Klei L, Soucy NV, O'Hara KA, Hamilton JW. Genomic and proteomic profiling of responses to toxic metals in human lung cells. Environ Health Perspect. 2003;111(6):825–835. doi: 10.1289/ehp.111-1241504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stierum R, Heijne W, Kienhuis A, van Ommen B, Groten J. Toxicogenomics concepts and applications to study hepatic effects of food additives and chemicals. Toxicol Appl Pharmacol. 2005;207(2 Suppl):179–188. doi: 10.1016/j.taap.2005.01.050. [DOI] [PubMed] [Google Scholar]
- Griffitt RJ, Hyndman K, Denslow ND, Barber DS. Comparison of molecular and histological changes in zebrafish gills exposed to metallic nanoparticles. Toxicol Sci. 2009;107(2):404–415. doi: 10.1093/toxsci/kfn256. [DOI] [PubMed] [Google Scholar]
- Waters KM, Masiello LM, Zangar RC, Tarasevich BJ, Karin NJ, Quesenberry RD, Bandyopadhyay S, Teeguarden JG, Pounds JG, Thrall BD. Macrophage responses to silica nanoparticles are highly conserved across particle sizes. Toxicol Sci. 2009;107(2):553–569. doi: 10.1093/toxsci/kfn250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottipolu RR, Wallenborn JG, Karoly ED, Schladweiler MC, Ledbetter AD, Krantz T, Linak WP, Nyska A, Johnson JA, Thomas R. et al. One-month diesel exhaust inhalation produces hypertensive gene expression pattern in healthy rats. Environ Health Perspect. 2009;117(1):38–46. doi: 10.1289/ehp.11647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson EM, Williams A, Yauk CL, Vincent R. Toxicogenomic analysis of susceptibility to inhaled urban particulate matter in mice with chronic lung inflammation. Part Fibre Toxicol. 2009;6:6. doi: 10.1186/1743-8977-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter V, Von Ruthendorf M. On hardness and toughness of ultrafine and nanocrystalline hard materials. International Journal of Refractory Metals & Hard Materials. 1999;17(1-3):141–152. doi: 10.1016/S0263-4368(99)00003-7. [DOI] [Google Scholar]
- Lison D, Lauwerys R. Study of the mechanism responsible for the elective toxicity of tungsten carbide-cobalt powder toward macrophages. Toxicol Lett. 1992;60(2):203–210. doi: 10.1016/0378-4274(92)90275-O. [DOI] [PubMed] [Google Scholar]
- Lasfargues G, Lison D, Maldague P, Lauwerys R. Comparative study of the acute lung toxicity of pure cobalt powder and cobalt-tungsten carbide mixture in rat. Toxicol Appl Pharmacol. 1992;112(1):41–50. doi: 10.1016/0041-008X(92)90277-Y. [DOI] [PubMed] [Google Scholar]
- Lasfargues G, Lardot C, Delos M, Lauwerys R, Lison D. The delayed lung responses to single and repeated intratracheal administration of pure cobalt and hard metal powder in the rat. Environ Res. 1995;69(2):108–121. doi: 10.1006/enrs.1995.1032. [DOI] [PubMed] [Google Scholar]
- Lison D, Lauwerys R. In vitro cytotoxic effects of cobalt-containing dusts on mouse peritoneal and rat alveolar macrophages. Environ Res. 1990;52:187–198. doi: 10.1016/S0013-9351(05)80253-1. [DOI] [PubMed] [Google Scholar]
- Roesems G, Hoet PH, Dinsdale D, Demedts M, Nemery B. In vitro cytotoxicity of various forms of cobalt for rat alveolar macrophages and type II pneumocytes. Toxicol Appl Pharmacol. 2000;162(1):2–9. doi: 10.1006/taap.1999.8824. [DOI] [PubMed] [Google Scholar]
- Bastian S, Busch W, Kühnel D, Springer A, Meissner T, Holke R, Scholz S, Iwe M, Pompe W, Gelinsky M. et al. Toxicity of tungsten carbide and cobalt-doped tungsten carbide nanoparticles in mammalian cells in vitro. Environ Health Perspect. 2009;117(4):530–536. doi: 10.1289/ehp.0800121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühnel D, Busch W, Meissner T, Springer A, Potthoff A, Richter V, Gelinsky M, Scholz S, Schirmer K. Agglomeration of tungsten carbide nanoparticles in exposure medium does not prevent uptake and toxicity toward a rainbow trout gill cell line. Aquat Toxicol. 2009;93(2-3):91–99. doi: 10.1016/j.aquatox.2009.04.003. [DOI] [PubMed] [Google Scholar]
- IARC. Cobalt in hard-metals and cobalt sulfate, gallium arsenide, indium phosphide and vanadium pentoxide. IARC Monographs on the Evaluation of Carcinogenic Risks to Humans. 2006. p. 86. [PMC free article] [PubMed]
- Yuan Y, Hilliard G, Ferguson T, Millhorn DE. Cobalt inhibits the interaction between hypoxia-inducible factor-alpha and von Hippel-Lindau protein by direct binding to hypoxia-inducible factor-alpha. J Biol Chem. 2003;278(18):15911–15916. doi: 10.1074/jbc.M300463200. [DOI] [PubMed] [Google Scholar]
- Vengellur A, LaPres JJ. The role of hypoxia inducible factor 1alpha in cobalt chloride induced cell death in mouse embryonic fibroblasts. Toxicol Sci. 2004;82(2):638–646. doi: 10.1093/toxsci/kfh278. [DOI] [PubMed] [Google Scholar]
- Kaczmarek M, Cachau RE, Topol IA, Kasprzak KS, Ghio A, Salnikow K. Metal ions-stimulated iron oxidation in hydroxylases facilitates stabilization of HIF-1 alpha protein. Toxicol Sci. 2009;107(2):394–403. doi: 10.1093/toxsci/kfn251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruick RK, McKnight SL. A conserved family of prolyl-4-hydroxylases that modify HIF. Science. 2001;294(5545):1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Hypoxia-inducible factor 1: oxygen homeostasis and disease pathophysiology. Trends Mol Med. 2001;7(8):345–350. doi: 10.1016/S1471-4914(01)02090-1. [DOI] [PubMed] [Google Scholar]
- Wenger RH. Cellular adaptation to hypoxia: O2-sensing protein hydroxylases, hypoxia-inducible transcription factors, and O2-regulated gene expression. FASEB J. 2002;16(10):1151–1162. doi: 10.1096/fj.01-0944rev. [DOI] [PubMed] [Google Scholar]
- Olbryt M, Jarzab M, Jazowiecka-Rakus J, Simek K, Szala S, Sochanik A. Gene expression profile of B 16(F10) murine melanoma cells exposed to hypoxic conditions in vitro. Gene Expr. 2006;13(3):191–203. doi: 10.3727/000000006783991818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lison D, Carbonnelle P, Mollo L, Lauwerys R, Fubini B. Physicochemical mechanism of the interaction between cobalt metal and carbide particles to generate toxic activated oxygen species. Chem Res Toxicol. 1995;8(4):600–606. doi: 10.1021/tx00046a015. [DOI] [PubMed] [Google Scholar]
- Lison D, Lauwerys R. Evaluation of the role of reactive oxygen species in the interactive toxicity of carbide-cobalt mixtures on macrophages in culture. Arch Toxicol. 1993;67(5):347–351. doi: 10.1007/BF01973706. [DOI] [PubMed] [Google Scholar]
- Lombaert N, Lison D, Van Hummelen P, Kirsch-Volders M. In vitro expression of hard metal dust (WC-Co)--responsive genes in human peripheral blood mononucleated cells. Toxicol Appl Pharmacol. 2008;227(2):299–312. doi: 10.1016/j.taap.2007.11.002. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E. et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Moroz E, Carlin S, Dyomina K, Burke S, Thaler HT, Blasberg R, Serganova I. Real-time imaging of HIF-1alpha stabilization and degradation. PLoS One. 2009;4(4):e5077. doi: 10.1371/journal.pone.0005077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JB, Zaehres H, Wu G, Gentile L, Ko K, Sebastiano V, Arauzo-Bravo MJ, Ruau D, Han DW, Zenke M. et al. Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature. 2008;454(7204):646–650. doi: 10.1038/nature07061. [DOI] [PubMed] [Google Scholar]
- Loh YH, Ng JH, Ng HH. Molecular framework underlying pluripotency. Cell Cycle. 2008;7(7):885–891. doi: 10.4161/cc.7.7.5636. [DOI] [PubMed] [Google Scholar]
- Pei D. Regulation of pluripotency and reprogramming by transcription factors. J Biol Chem. 2009;284(6):3365–3369. doi: 10.1074/jbc.R800063200. [DOI] [PubMed] [Google Scholar]
- Greber B, Lehrach H, Adjaye J. Silencing of core transcription factors in human EC cells highlights the importance of autocrine FGF signaling for self-renewal. BMC Dev Biol. 2007;7:46. doi: 10.1186/1471-213X-7-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na G, Bridges PJ, Koo Y, Ko C. Role of hypoxia in the regulation of periovulatory EDN2 expression in the mouse. Can J Physiol Pharmacol. 2008;86(6):310–319. doi: 10.1139/Y08-025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi-Wen X, Denton CP, Dashwood MR, Holmes AM, Bou-Gharios G, Pearson JD, Black CM, Abraham DJ. Fibroblast matrix gene expression and connective tissue remodeling: role of endothelin-1. J Invest Dermatol. 2001;116(3):417–425. doi: 10.1046/j.1523-1747.2001.01256.x. [DOI] [PubMed] [Google Scholar]
- Jiang BH, Rue E, Wang GL, Roe R, Semenza GL. Dimerization, DNA binding, and transactivation properties of hypoxia-inducible factor 1. J Biol Chem. 1996;271(30):17771–17778. doi: 10.1074/jbc.271.30.17771. [DOI] [PubMed] [Google Scholar]
- Yamashita K, Discher DJ, Hu J, Bishopric NH, Webster KA. Molecular regulation of the endothelin-1 gene by hypoxia. Contributions of hypoxia-inducible factor-1, activator protein-1, GATA-2, AND p300/CBP. J Biol Chem. 2001;276(16):12645–12653. doi: 10.1074/jbc.M011344200. [DOI] [PubMed] [Google Scholar]
- Sanchez-Elsner T, Botella LM, Velasco B, Langa C, Bernabeu C. Endoglin expression is regulated by transcriptional cooperation between the hypoxia and transforming growth factor-beta pathways. J Biol Chem. 2002;277(46):43799–43808. doi: 10.1074/jbc.M207160200. [DOI] [PubMed] [Google Scholar]
- Mense SM, Sengupta A, Zhou M, Lan C, Bentsman G, Volsky DJ, Zhang L. Gene expression profiling reveals the profound upregulation of hypoxia-responsive genes in primary human astrocytes. Physiol Genomics. 2006;25(3):435–449. doi: 10.1152/physiolgenomics.00315.2005. [DOI] [PubMed] [Google Scholar]
- Manalo DJ, Rowan A, Lavoie T, Natarajan L, Kelly BD, Ye SQ, Garcia JG, Semenza GL. Transcriptional regulation of vascular endothelial cell responses to hypoxia by HIF-1. Blood. 2005;105(2):659–669. doi: 10.1182/blood-2004-07-2958. [DOI] [PubMed] [Google Scholar]
- Gordon S, Akopyan G, Garban H, Bonavida B. Transcription factor YY1: structure, function, and therapeutic implications in cancer biology. Oncogene. 2006;25(8):1125–1142. doi: 10.1038/sj.onc.1209080. [DOI] [PubMed] [Google Scholar]
- Kasten U, Hartwig A, Beyersmann D. Mechanisms of cobalt(II) uptake into V79 Chinese hamster cells. Arch Toxicol. 1992;66(8):592–597. doi: 10.1007/BF01973391. [DOI] [PubMed] [Google Scholar]
- Kopera E, Schwerdtle T, Hartwig A, Bal W. Co(II) and Cd(II) substitute for Zn(II) in the zinc finger derived from the DNA repair protein XPA, demonstrating a variety of potential mechanisms of toxicity. Chem Res Toxicol. 2004;17(11):1452–1458. doi: 10.1021/tx049842s. [DOI] [PubMed] [Google Scholar]
- Beyersmann D, Hartwig A. Carcinogenic metal compounds: recent insight into molecular and cellular mechanisms. Arch Toxicol. 2008;82(8):493–512. doi: 10.1007/s00204-008-0313-y. [DOI] [PubMed] [Google Scholar]
- Ortega R, Bresson C, Fraysse A, Sandre C, Deves G, Gombert C, Tabarant M, Bleuet P, Seznec H, Simionovici A. et al. Cobalt distribution in keratinocyte cells indicates nuclear and perinuclear accumulation and interaction with magnesium and zinc homeostasis. Toxicol Lett. 2009;188(1):26–32. doi: 10.1016/j.toxlet.2009.02.024. [DOI] [PubMed] [Google Scholar]
- Karovic O, Tonazzini I, Rebola N, Edstrom E, Lovdahl C, Fredholm BB, Dare E. Toxic effects of cobalt in primary cultures of mouse astrocytes. Similarities with hypoxia and role of HIF-1alpha. Biochem Pharmacol. 2007;73(5):694–708. doi: 10.1016/j.bcp.2006.11.008. [DOI] [PubMed] [Google Scholar]
- Doherty GJ, McMahon HT. Mechanisms of endocytosis. Annu Rev Biochem. 2009;78:857–902. doi: 10.1146/annurev.biochem.78.081307.110540. [DOI] [PubMed] [Google Scholar]
- Pelkmans L, Fava E, Grabner H, Hannus M, Habermann B, Krausz E, Zerial M. Genome-wide analysis of human kinases in clathrin- and caveolae/raft-mediated endocytosis. Nature. 2005;436(7047):78–86. doi: 10.1038/nature03571. [DOI] [PubMed] [Google Scholar]
- Jin HO, An S, Lee HC, Woo SH, Seo SK, Choe TB, Yoo DH, Lee SB, Um HD, Lee SJ. et al. Hypoxic condition- and high cell density-induced expression of Redd1 is regulated by activation of hypoxia-inducible factor-1alpha and Sp1 through the phosphatidylinositol 3-kinase/Akt signaling pathway. Cell Signal. 2007;19(7):1393–1403. doi: 10.1016/j.cellsig.2006.12.014. [DOI] [PubMed] [Google Scholar]
- Tang TT, Lasky LA. The forkhead transcription factor FOXO4 induces the down-regulation of hypoxia-inducible factor 1 alpha by a von Hippel-Lindau protein-independent mechanism. J Biol Chem. 2003;278(32):30125–30135. doi: 10.1074/jbc.M302042200. [DOI] [PubMed] [Google Scholar]
- Wang X, Li C, Chen Y, Hao Y, Zhou W, Chen C, Yu Z. Hypoxia enhances CXCR4 expression favoring microglia migration via HIF-1[alpha] activation. Biochemical and Biophysical Research Communications. 2008;371(2):283–288. doi: 10.1016/j.bbrc.2008.04.055. [DOI] [PubMed] [Google Scholar]
- Mottet D, Dumont V, Deccache Y, Demazy C, Ninane N, Raes M, Michiels C. Regulation of Hypoxia-inducible Factor-1{alpha} Protein Level during Hypoxic Conditions by the Phosphatidylinositol 3-Kinase/Akt/Glycogen Synthase Kinase 3{beta} Pathway in HepG2 Cells. J Biol Chem. 2003;278(33):31277–31285. doi: 10.1074/jbc.M300763200. [DOI] [PubMed] [Google Scholar]
- Limbach LK, Wick P, Manser P, Grass RN, Bruinink A, Stark WJ. Exposure of engineered nanoparticles to human lung epithelial cells: influence of chemical composition and catalytic activity on oxidative stress. Environ Sci Technol. 2007;41(11):4158–4163. doi: 10.1021/es062629t. [DOI] [PubMed] [Google Scholar]
- Schins RP, Knaapen AM. Genotoxicity of poorly soluble particles. Inhal Toxicol. 2007;19(Suppl 1):189–198. doi: 10.1080/08958370701496202. [DOI] [PubMed] [Google Scholar]
- Zou W, Yan M, Xu W, Huo H, Sun L, Zheng Z, Liu X. Cobalt chloride induces PC12 cells apoptosis through reactive oxygen species and accompanied by AP-1 activation. J Neurosci Res. 2001;64(6):646–653. doi: 10.1002/jnr.1118. [DOI] [PubMed] [Google Scholar]
- Chandel NS, Maltepe E, Goldwasser E, Mathieu CE, Simon MC, Schumacker PT. Mitochondrial reactive oxygen species trigger hypoxia-induced transcription. Proc Natl Acad Sci USA. 1998;95(20):11715–11720. doi: 10.1073/pnas.95.20.11715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ermolli M, Menne C, Pozzi G, Serra MA, Clerici LA. Nickel, cobalt and chromium-induced cytotoxicity and intracellular accumulation in human hacat keratinocytes. Toxicology. 2001;159(1-2):23–31. doi: 10.1016/S0300-483X(00)00373-5. [DOI] [PubMed] [Google Scholar]
- Li H, Zhang J, Wang T, Luo W, Zhou Q, Jiang G. Elemental selenium particles at nano-size (Nano-Se) are more toxic to Medaka (Oryzias latipes) as a consequence of hyper-accumulation of selenium: a comparison with sodium selenite. Aquat Toxicol. 2008;89(4):251–256. doi: 10.1016/j.aquatox.2008.07.008. [DOI] [PubMed] [Google Scholar]
- Navarro E, Piccapietra F, Wagner B, Marconi F, Kaegi R, Odzak N, Sigg L, Behra R. Toxicity of silver nanoparticles to Chlamydomonas reinhardtii. Environ Sci Technol. 2008;42(23):8959–8964. doi: 10.1021/es801785m. [DOI] [PubMed] [Google Scholar]
- Xia T, Kovochich M, Liong M, Madler L, Gilbert B, Shi H, Yeh JI, Zink JI, Nel AE. Comparison of the mechanism of toxicity of zinc oxide and cerium oxide nanoparticles based on dissolution and oxidative stress properties. ACS Nano. 2008;2(10):2121–2134. doi: 10.1021/nn800511k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DIN ISO 13321. Partikelgrößenanalyse - Photonenkorrelationsspektroskopie. DIN ISO 13321 2004-10.
- Boukamp P, Petrussevska RT, Breitkreutz D, Hornung J, Markham A, Fusenig NE. Normal keratinization in a spontaneously immortalized aneuploid human keratinocyte cell line. J Cell Biol. 1988;106(3):761–771. doi: 10.1083/jcb.106.3.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khammanit R, Chantakru S, Kitiyanant Y, Saikhun J. Effect of serum starvation and chemical inhibitors on cell cycle synchronization of canine dermal fibroblasts. Theriogenology. 2008;70(1):27–34. doi: 10.1016/j.theriogenology.2008.02.015. [DOI] [PubMed] [Google Scholar]
- Sterrenburg E, Turk R, Boer JM, van Ommen GB, den Dunnen JT. A common reference for cDNA microarray hybridizations. Nucleic Acids Res. 2002;30(21):e116. doi: 10.1093/nar/gnf115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed AI, Sharov V, White J, Li J, Liang W, Bhagabati N, Braisted J, Klapa M, Currier T, Thiagarajan M. et al. TM4: a free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- Tusher VG, Tibshirani R, Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci USA. 2001;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Waxman S, Wurmbach E. De-regulation of common housekeeping genes in hepatocellular carcinoma. BMC Genomics. 2007;8:243. doi: 10.1186/1471-2164-8-243. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table contains all significantly altered expressed genes and fold changes for all treatments
Heat map of differentially expressed genes and hierarchical clustering of all replicates
Table contains all gene sets and pathways identified as enriched by GSEA and DAVID