Abstract
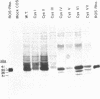
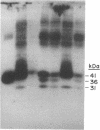
To investigate the role of different cysteine residues in bovine rhodopsin, a series of mutants were prepared in which the cysteine residues were systematically replaced by serines. The mutant genes were expressed in monkey kidney cells (COS-1) and the mutant opsins were evaluated for their levels of expression, glycosylation patterns, and ability to form the chromophore characteristic of rhodopsin and to activate transducin. Substitution of the three cytoplasmic cysteines (Cys-316, Cys-322, and Cys-323) and the four membrane-embedded cysteines (Cys-140, Cys-167, Cys-222, and Cys-264) produced proteins with wild-type phenotype. Also, single substitutions of Cys-185 gave rise to a wild-type phenotype. In contrast, substitution of the three intradiscal cysteines (Cys-110, Cys-185, and Cys-187) or single substitution of Cys-110 or Cys-187 gave proteins that were expressed at reduced levels, glycosylated abnormally, and unable to bind 11-cis-retinal. Thus, of the 10 cysteines in bovine rhodopsin, only intradiscal Cys-110 and Cys-187 are essential for the correct tertiary structure of the protein.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Al-Saleh S., Gore M., Akhtar M. On the disulphide bonds of rhodopsins. Biochem J. 1987 Aug 15;246(1):131–137. doi: 10.1042/bj2460131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Applebury M. L., Hargrave P. A. Molecular biology of the visual pigments. Vision Res. 1986;26(12):1881–1895. doi: 10.1016/0042-6989(86)90115-x. [DOI] [PubMed] [Google Scholar]
- Barclay P. L., Findlay J. B. Labelling of the cytoplasmic domains of ovine rhodopsin with hydrophilic chemical probes. Biochem J. 1984 May 15;220(1):75–84. doi: 10.1042/bj2200075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonner T. I., Buckley N. J., Young A. C., Brann M. R. Identification of a family of muscarinic acetylcholine receptor genes. Science. 1987 Jul 31;237(4814):527–532. doi: 10.1126/science.3037705. [DOI] [PubMed] [Google Scholar]
- Burnette W. N. "Western blotting": electrophoretic transfer of proteins from sodium dodecyl sulfate--polyacrylamide gels to unmodified nitrocellulose and radiographic detection with antibody and radioiodinated protein A. Anal Biochem. 1981 Apr;112(2):195–203. doi: 10.1016/0003-2697(81)90281-5. [DOI] [PubMed] [Google Scholar]
- Davison M. D., Findlay J. B. Modification of ovine opsin with the photosensitive hydrophobic probe 1-azido-4-[125I]iodobenzene. Labelling of the chromophore-attachment domain. Biochem J. 1986 Mar 1;234(2):413–420. doi: 10.1042/bj2340413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Grip W. J., Daemen F. J. Sulfhydryl chemistry of rhodopsin. Methods Enzymol. 1982;81:223–236. doi: 10.1016/s0076-6879(82)81035-5. [DOI] [PubMed] [Google Scholar]
- Dixon R. A., Kobilka B. K., Strader D. J., Benovic J. L., Dohlman H. G., Frielle T., Bolanowski M. A., Bennett C. D., Rands E., Diehl R. E. Cloning of the gene and cDNA for mammalian beta-adrenergic receptor and homology with rhodopsin. Nature. 1986 May 1;321(6065):75–79. doi: 10.1038/321075a0. [DOI] [PubMed] [Google Scholar]
- Dixon R. A., Sigal I. S., Candelore M. R., Register R. B., Scattergood W., Rands E., Strader C. D. Structural features required for ligand binding to the beta-adrenergic receptor. EMBO J. 1987 Nov;6(11):3269–3275. doi: 10.1002/j.1460-2075.1987.tb02645.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon R. A., Sigal I. S., Rands E., Register R. B., Candelore M. R., Blake A. D., Strader C. D. Ligand binding to the beta-adrenergic receptor involves its rhodopsin-like core. Nature. 1987 Mar 5;326(6108):73–77. doi: 10.1038/326073a0. [DOI] [PubMed] [Google Scholar]
- Ferretti L., Karnik S. S., Khorana H. G., Nassal M., Oprian D. D. Total synthesis of a gene for bovine rhodopsin. Proc Natl Acad Sci U S A. 1986 Feb;83(3):599–603. doi: 10.1073/pnas.83.3.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Findlay J. B., Pappin D. J. The opsin family of proteins. Biochem J. 1986 Sep 15;238(3):625–642. doi: 10.1042/bj2380625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke R. R., Sakmar T. P., Oprian D. D., Khorana H. G. A single amino acid substitution in rhodopsin (lysine 248----leucine) prevents activation of transducin. J Biol Chem. 1988 Feb 15;263(5):2119–2122. [PubMed] [Google Scholar]
- Fung B. K., Hurley J. B., Stryer L. Flow of information in the light-triggered cyclic nucleotide cascade of vision. Proc Natl Acad Sci U S A. 1981 Jan;78(1):152–156. doi: 10.1073/pnas.78.1.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagen D. C., McCaffrey G., Sprague G. F., Jr Evidence the yeast STE3 gene encodes a receptor for the peptide pheromone a factor: gene sequence and implications for the structure of the presumed receptor. Proc Natl Acad Sci U S A. 1986 Mar;83(5):1418–1422. doi: 10.1073/pnas.83.5.1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargrave P. A., McDowell J. H., Curtis D. R., Wang J. K., Juszczak E., Fong S. L., Rao J. K., Argos P. The structure of bovine rhodopsin. Biophys Struct Mech. 1983;9(4):235–244. doi: 10.1007/BF00535659. [DOI] [PubMed] [Google Scholar]
- Hong K., Hubbell W. L. Lipid requirements for Rhodopsin regenerability. Biochemistry. 1973 Oct 23;12(22):4517–4523. doi: 10.1021/bi00746a033. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lefkowitz R. J., Caron M. G. Adrenergic receptors. Models for the study of receptors coupled to guanine nucleotide regulatory proteins. J Biol Chem. 1988 Apr 15;263(11):4993–4996. [PubMed] [Google Scholar]
- Lo K. M., Jones S. S., Hackett N. R., Khorana H. G. Specific amino acid substitutions in bacterioopsin: Replacement of a restriction fragment in the structural gene by synthetic DNA fragments containing altered codons. Proc Natl Acad Sci U S A. 1984 Apr;81(8):2285–2289. doi: 10.1073/pnas.81.8.2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Nathans J., Hogness D. S. Isolation, sequence analysis, and intron-exon arrangement of the gene encoding bovine rhodopsin. Cell. 1983 Oct;34(3):807–814. doi: 10.1016/0092-8674(83)90537-8. [DOI] [PubMed] [Google Scholar]
- Nathans J. Molecular biology of visual pigments. Annu Rev Neurosci. 1987;10:163–194. doi: 10.1146/annurev.ne.10.030187.001115. [DOI] [PubMed] [Google Scholar]
- Oprian D. D., Molday R. S., Kaufman R. J., Khorana H. G. Expression of a synthetic bovine rhodopsin gene in monkey kidney cells. Proc Natl Acad Sci U S A. 1987 Dec;84(24):8874–8878. doi: 10.1073/pnas.84.24.8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovchinnikov YuA, Abdulaev N. G., Bogachuk A. S. Two adjacent cysteine residues in the C-terminal cytoplasmic fragment of bovine rhodopsin are palmitylated. FEBS Lett. 1988 Mar 28;230(1-2):1–5. doi: 10.1016/0014-5793(88)80628-8. [DOI] [PubMed] [Google Scholar]
- Pedersen S. E., Ross E. M. Functional activation of beta-adrenergic receptors by thiols in the presence or absence of agonists. J Biol Chem. 1985 Nov 15;260(26):14150–14157. [PubMed] [Google Scholar]
- Plummer T. H., Jr, Elder J. H., Alexander S., Phelan A. W., Tarentino A. L. Demonstration of peptide:N-glycosidase F activity in endo-beta-N-acetylglucosaminidase F preparations. J Biol Chem. 1984 Sep 10;259(17):10700–10704. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strader C. D., Sigal I. S., Register R. B., Candelore M. R., Rands E., Dixon R. A. Identification of residues required for ligand binding to the beta-adrenergic receptor. Proc Natl Acad Sci U S A. 1987 Jul;84(13):4384–4388. doi: 10.1073/pnas.84.13.4384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryer L. Cyclic GMP cascade of vision. Annu Rev Neurosci. 1986;9:87–119. doi: 10.1146/annurev.ne.09.030186.000511. [DOI] [PubMed] [Google Scholar]
- Trimble R. B., Maley F. Optimizing hydrolysis of N-linked high-mannose oligosaccharides by endo-beta-N-acetylglucosaminidase H. Anal Biochem. 1984 Sep;141(2):515–522. doi: 10.1016/0003-2697(84)90080-0. [DOI] [PubMed] [Google Scholar]
- Vauquelin G., Bottari S., Kanarek L., Strosberg A. D. Evidence for essential disulfide bonds in beta1-adrenergic receptors of turkey erythrocyte membranes. Inactivation by dithiothreitol. J Biol Chem. 1979 Jun 10;254(11):4462–4469. [PubMed] [Google Scholar]
- Wald G. The molecular basis of visual excitation. Nature. 1968 Aug 24;219(5156):800–807. doi: 10.1038/219800a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto T., Bishop R. W., Brown M. S., Goldstein J. L., Russell D. W. Deletion in cysteine-rich region of LDL receptor impedes transport to cell surface in WHHL rabbit. Science. 1986 Jun 6;232(4755):1230–1237. doi: 10.1126/science.3010466. [DOI] [PMC free article] [PubMed] [Google Scholar]