Abstract
Induction of type I interferons is a fundamental cellular response to both viral and bacterial infection. The role of the transcription factor IRF3 is well established in driving this process. However, equally as important are cellular mechanisms for turning off type I interferon production in order to limit this response. In this respect, IRF3 has previously been shown to be targeted for ubiquitin-mediated degradation post-viral detection in order to turn off the IFN-β response. Here we provide evidence that the E3 ligase Ro52 (TRIM21) targets IRF3 for degradation post-pathogen recognition receptor activation. We demonstrate that Ro52 interacts with IRF3 via its C-terminal SPRY domain, resulting in the polyubiquitination and proteasomal degradation of the transcription factor. Ro52-mediated IRF3 degradation significantly inhibits IFN-β promoter activity, an effect that is reversed in the presence of the proteasomal inhibitor MG132. Specific targeting of Ro52 using shRNA rescues IRF3 degradation following polyI:C-stimulation of HEK293T cells, with a subsequent increase in IFN-β production. Additionally, shRNA targeting of murine Ro52 enhances the production of the IRF3-dependent chemokine RANTES following Sendai virus infection of murine fibroblasts. Collectively, this demonstrates a novel role for Ro52 in turning off and thus limiting IRF3-dependent type I interferon production by targeting the transcription factor for polyubiquitination and subsequent proteasomal degradation.
Keywords: Signal Transduction, Lipopolysaccharide, Molecular Biology
Introduction
Central to innate immune responses following viral and bacterial infection is the production of type I interferons (IFN-α and -β) and the establishment of an anti-viral or anti-bacterial state (1-3). Pathogen recognition receptors (PRRs)3, such as the transmembrane Toll-like receptors (TLRs) or the cytosolic PRRs (RIG-I, MDA-5), respond to viral or bacterial infection and activate the regulatory networks that coordinate the induction of type I IFN genes. A family of transcription factors, the IFN regulatory factors (IRFs), has gained much attention in this respect, as IRF3 and IRF7 have been demonstrated to be essential for type I IFN gene induction in response to pathogen recognition (4-6). However, overproduction of type I IFNs results in adverse pathogenic effects characteristic of many autoimmune disorders, such as systemic lupus erythematosus (SLE) (7). Thus, understanding the mechanisms that limit or down-regulate type I IFN production downstream of pathogen recognition is critical to protecting against such harmful effects.
One mechanism by which IFN-β production is turned off post-PRR stimulation is via polyubiquitination and subsequent degradation of the transcription factor IRF3 (8-10). IRF3 is a constitutively expressed member of the IRF family that regulates the primary induction of IFN-β in response to viral and bacterial infection downstream of TLR3, TLR4 and cytosolic PRRs (4, 5, 11, 12). Both TLR3 and TLR4 induce type I IFN production in a similar fashion, through the recruitment and activation of an adaptor protein TRIF (3-5, 11). This leads to the phosphorylation of IRF3 by IKKε and TBK1, followed by IRF3 nuclear translocation and induction of IFN-β transcription (13-15). IRF3 is also central to RIG-I and MDA-5 responses, which also involve IKKε and TBK1. The central role of IRF3 in mediating type I IFN induction in response to TLR3, TLR4 and intracellular PRRs would suggest that targeting it for ubiquitin-mediated degradation post-stimulation is a very efficient way of shutting off and limiting the type I IFN response. Early work supporting this demonstrated that the proteasomal inhibitor MG132 inhibited IRF3 degradation in response to viral infection (8). More recently, the peptide-prolyl isomerase Pin1 was shown to interact with IRF3 following polyI:C stimulation of 293T cells and promote polyubiquitination and proteasomal degradation of the transcription factor (9). Additionally, the involvement of a cullin-based ubiquitin ligase in Sendai virus-induced IRF3 degradation has been reported (10). Importantly, the E3 ligase responsible for targeting IRF3 has not yet been identified.
Ubiquitin-mediated proteasomal degradation regulates many biological events including cell cycle control, signal transduction, DNA repair and apoptosis (16, 17). Ubiquitination involves three enzymes: an E1 ubiquitin-activating enzyme, an E2 ubiquitin-conjugating enzyme, and an E3 ubiquitin ligase. It is the E3 ligase that provides the specificity in the ubiquitin process as it recruits both the E2-ubiquitin complex and the target protein, often resulting in polyubiquitination and proteasomal degradation by the 26S proteasome (18). The 52 members of the TRIM family are single-protein E3 ligases that have multiple roles in cell biology (19) and their substrate specificity is determined by the C-terminal SPRY domain (20, 21). Ro52 (TRIM21) was first described as a target for autoantibody production in SLE and Sjögren’s syndrome (22-24). Interestingly, TRIM family members, such as TRIM5α and TRIM25, have been shown to play important roles in anti-viral defenses (19, 25, 26). Indeed, recent work identifying IRF8 as a direct substrate for Ro52 (27), suggests a role for Ro52 in immune responses.
In this study we show that IRF3 is specifically targeted by Ro52 for ubiquitin-mediated degradation in order to negatively regulate the IFN-β promoter downstream of lipopolysacharride (LPS) and polyI:C stimulation and Sendai virus infection. In addition, we demonstrate that inhibiting Ro52 expression with shRNA results in enhanced TLR3-driven IFN-β production and Sendai virus-stimulated RANTES production. Taken together, our results demonstrate a novel role for Ro52 as a negative regulator of type I IFN induction downstream of pathogen recognition.
Materials and Methods
Cell culture
THP1 cells were cultured in RPMI 1640 supplemented with 10% FCS and 10 μg/ml gentamicin. Human embryonic kidney (HEK) 293 cells stably transfected with TLR3 (TLR3-293) or TLR4 (TLR4/MD2-293), HEK293T cells, HeLa cells and NIH 3T3 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% FCS and 10 μg/ml gentamicin. TLR3-293 and TLR4-293 cells were cultured in the presence of 500 μg/ml G418 (Sigma).
Plasmids and reagents
Flag-tagged pCMV-IRF3, Flag-tagged pCMV-IRF7, pEF-Bos-TRIF-Flag and the IFN-β promoter constructs were from Dr. Kate Fitzgerald (University of Massachusetts Medical School, Worcester, MA). Xpress™-tagged Ro52, Xpress™-tagged Ro52ΔExon1 and Xpress™-tagged Ro52ΔExon6 were gifts from Dr. David Rhodes (Cambridge Institute for Medical Research, Cambridge, UK) and hemagglutinin (HA)-ubiquitin from Dr. Andrew Bowie (School of Biochemistry with Immunology, Trinity College Dublin, Ireland). The NFκB-luciferase plasmid was a kind gift from Dr. R. Hofmeister (Universitat Regensburg, Regensburg, Germany). Primary antibodies used are anti-HA, anti-IRF3 (Santa Cruz Biotechnologies), anti-Flag (Sigma), anti-Xpress™ (Invitrogen), anti-polyubiquitin FK1 (Biomol) and anti-β-actin (Abcam). Polyclonal antibodies against Ro52 that were used in immunoprecipitation experiments were made by Sigma-Genosys and anti-Ro52 antibodies used in western blots were purchased from BioReagents, Cambridge, UK.
Luciferase reporter gene assays
HEK293T, TLR3-293 and TLR4/MD2-293 cells were transiently transfected for 18 hrs with 50 ng of the indicated reporter constructs and increasing amounts of a Ro52 construct (10 ng, 50 ng and 100 ng). HEK293T cells were also co-transfected with either a TRIF or IRF3 construct (50 ng). All transfections were performed using Metafectene™ (Biontex) according to the manufacturer’s recommendations. Following transfection, TLR3-293 and TLR4-293 cells were either untreated or stimulated with polyI:C (20 μg/ml) or LPS (1 μg/ml) for 6 hrs. Luciferase activity was standardised to Renilla luciferase plasmid activity to normalise for transfection efficiency.
Western blot and immunoprecipitation
Immunoblots were performed as described previously (28). Cells were lysed on ice in 1X radioimmune precipitation lysis buffer (1X PBS, 1% Nonidet P-40, 0.5% Na-deoxycholate, 0.1% SDS, 1 mM KF, 1 mM Na3VO4, 10 μg/ml leupeptin and 1 mM phenylmethylsulfonyl fluoride) followed by immunoprecipitation with either anti-HA agarose, or anti-Flag or anti-Ro52 bound to protein-G sepharose beads. For peptide pulldowns, lysates were incubated with 1 μg His-Ro52 bound to Ni2+-agarose beads (Qiagen). Immunoprecipitates were analysed by Western blot. Each blot is representative of 2-3 independent experiments.
Realtime PCR
RNA was extracted from cell cultures using TRIZOL™ (Invitrogen) and reverse transcribed to cDNA using Omniscript reverse transcriptase (Qiagen) according to manufacturer’s recommendations. Quantitative realtime PCR was performed using SYBR® Green Taq ReadyMix™ (Sigma) on an Applied Biosystems 7500 realtime PCR machine at an annealing temperature of 56°C. Realtime PCR data was analysed using the 2−ΔΔCt method (29).
RNA interference
A shRNA sequence targeting human Ro52 (30) was cloned into the pRNA-H1.1/neo plasmid vector (GenScript Corp.). Scrambled shRNA was similarly cloned as a negative control and used in parallel. 293T and TLR3-293 fibroblast cells were transfected with 1 μg of Ro52 shRNA or 1 μg of scrambled control shRNA, using Metafectene™ according to the manufacturer’s recommendations. Following 24 hrs transfection, cells were treated with polyI:C (25 μg/ml) for the indicated times. IFN-β and IRF-3 mRNA and IRF3 protein levels were determined as described. Murine NIH 3T3 fibroblast cells were stably transfected with 250 ng each of four shRNAs (Sigma) directed towards murine Ro52 or 1 μg of scrambled control shRNA using Metafectene™. Following 48 hrs Sendai virus infection, supernatants were collected from NIH 3T3 cells and RANTES production was measured by ELISA (R & D Systems) according to the manufacturer’s recommendations.
Results
Ro52 acts downstream of TLR4, TLR3 and RIG-I to negatively regulate activation of the IFN-β promoter
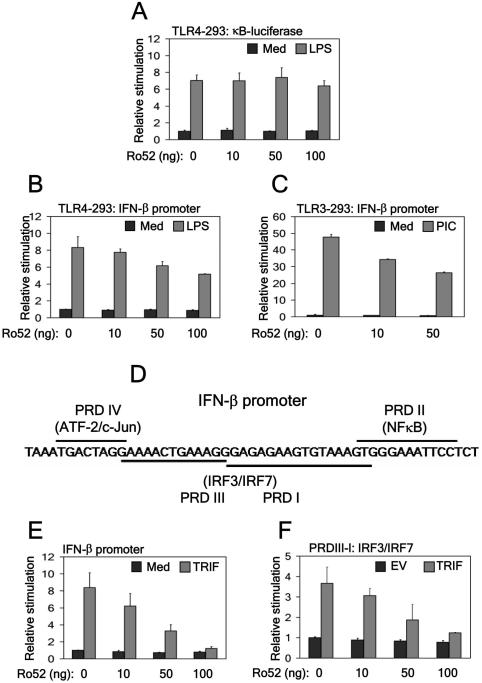
TRIM family members have been shown to be important in mediating anti-viral immunity (19, 25, 26). Given this and the putative role of Ro52 in regulating immune function (27, 30), we investigated the effect of Ro52 expression on TLR-driven NFκB- and IFN-β-reporter gene activity. Whilst transient transfection of TLR4/MD2-293 cells with increasing concentrations of Ro52 had no effect on the activation of the NFκB promoter in response to LPS stimulation (Fig. 1A), Ro52 dose-dependently inhibited TLR4-driven IFN-β-reporter gene activity (Fig. 1B). This result suggests that Ro52 acts by negatively regulating the IFN-β promoter following LPS stimulation, but is not involved in regulating NFκB. Ro52 inhibition of the IFN-β promoter was also observed following polyI:C stimulation of similarly transfected TLR3-293 cells (Fig. 1C).
Figure 1. Ro52 negatively regulates IFN-β promoter activity but not NFκB activation.
(a,b) TLR4/MD2-293 cells were transfected with either 50 ng (a) NFκB-dependent or (b) IFN-β-dependent reporter construct and a construct expressing Ro52 as shown. 18 hrs post-transfection cells were stimulated with 1 μg/ml LPS (6 hrs) and promoter activity measured. Med, medium. (c) TLR3-293 cells were transfected with 50 ng full-length IFN-β promoter and the indicated amounts of Ro52-expressing plasmid. 18 hrs post-transfection cells were stimulated with 20 μg/ml polyI:C (6 hrs) and promoter activity measured. (d) A diagrammatic representation of the transcription factor sites in the IFN-β promoter. (e) 293T cells were transfected with 50 ng full-length IFN-β promoter. Cells were co-transfected with 50 ng TRIF or empty vector (EV) control and increasing amounts of Ro52-expressing construct as indicated. (f) 293T cells were transfected with a reporter construct containing the IRF3 and IRF7 binding sites only. Cells were co-transfected with 50 ng TRIF or EV control and increasing amounts of Ro52-expressing construct as indicated. Cells were assayed for reporter gene activity 18 hrs post-transfection.
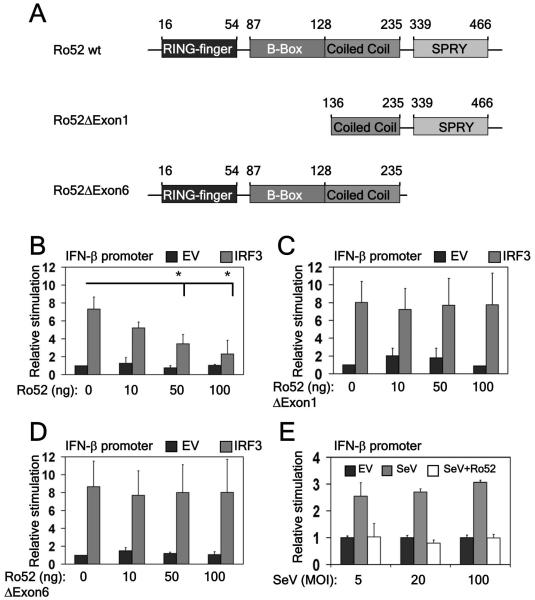
IFN-β has four regulatory cis-elements in its enhancer region: positive regulatory domains (PRDs) I-IV (Fig. 1D). NFκB and ATF-c-Jun bind to PRD II and IV, respectively, whilst IRF3/7 bind overlapping PRD I and III elements. Looking at the effect of Ro52 on TRIF-driven IFN-β promoter activity in 293T cells, we found that whilst Ro52 dose-dependently inhibited the full-length promoter (Fig. 1E), its effects were localised to PRD I/III (Fig. 1F) and not PRD II (data not shown), suggesting that Ro52 specifically regulates IRF3 activation. Thus, we assessed if Ro52 could inhibit IRF3-driven IFN-β promoter activity directly using the constructs shown in Figure 2A. Increasing amounts of Ro52 significantly inhibited the ability of IRF3 to induce the IFN-β promoter in a dose-dependent manner (Fig. 2B). Furthermore, plasmids encoding Ro52 mutants lacking either the N-terminal RING-finger domain or C-terminal SPRY domain were unable to inhibit IRF3-driven IFN-β promoter activity (Fig. 2C and 2D, respectively), indicating that both domains are functionally important for the negative effect of Ro52.
Figure 2. IRF3 activation of the IFN-β promoter is inhibited by Ro52.
(a) A schematic diagram of wildtype Ro52 and its mutants. (b,c,d) 293T cells were transfected with 50 ng full-length IFN-β promoter. Cells were co-transfected with 50 ng IRF3 or EV control and increasing amounts of constructs expressing either (b) wildtype Ro52, (c) Ro52ΔExon1 or (d) Ro52ΔExon6 as indicated. Cells were assayed for reporter gene activity 18 hrs post-transfection. *, p < 0.01 as determined by Student’s T test. (e) 293T cells were transfected with 50 ng full-length IFN-β promoter. Cells were co-transfected with 100 ng Ro52 or EV control and 18 hrs post-transfection cells were stimulated for 48 hrs with Sendai virus (SeV) at 5, 20 and 100 multiplicities of infection (MOI) and promoter activity was measured.
As the intracellular PRR RIG-I, which recognises Sendai virus, also signals through IRF3, we examined the effect of Ro52 on Sendai virus infection of 293T cells. Ro52 potently inhibited IFN-β promoter activation following infection of cells with Sendai virus confirming that RIG-I-regulated activation of IRF3 is also targeted by Ro52 (Fig. 2E).
Ro52 associates with IRF3
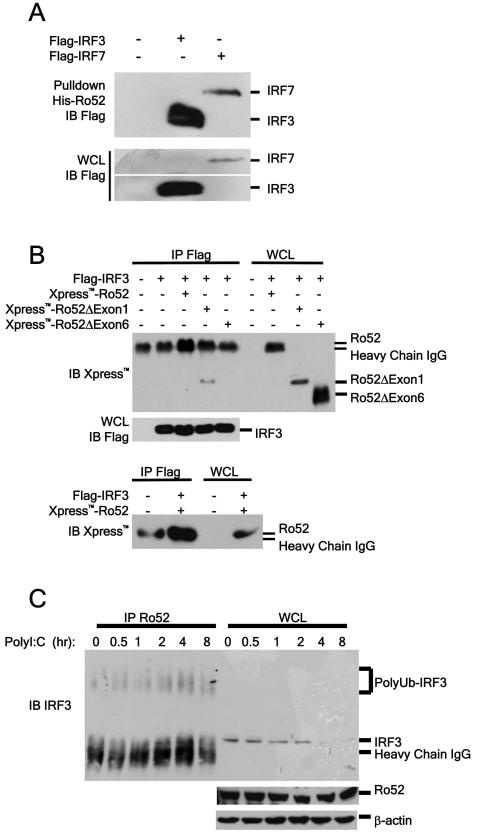
Our studies on the effects of Ro52 on the IFN-β promoter strongly indicated that the target for Ro52 was IRF3. To address this hypothesis we investigated the possibility that Ro52 and IRF3 interact. 293T cells were transfected with a construct expressing Flag-tagged IRF3. The ability of over-expressed IRF3 to interact with Ro52 was assessed in a pull-down experiment using recombinant His-tagged Ro52. Figure 3A demonstrates that Flag-IRF3 strongly associates with His-Ro52, suggesting that IRF3 may be an endogenous substrate for Ro52. Interestingly, Ro52 also associates with IRF7, which is important for the secondary type I IFN response following LPS stimulation.
Figure 3. Ro52 interacts with IRF3.
(a) 293T cells were transfected with constructs expressing 4 μg Flag-tagged IRF3 or IRF7. 18 hours post-transfection cell lysates were incubated with 1 μg recombinant Ro52 in a pull-down experiment. (b) 293T cells were transfected with 4 μg indicated constructs. 18 hours post-transfection Flag-associated proteins were immunoprecipitated from lysates with an anti-Flag antibody. Association of Ro52 and Ro52 mutants with Flag-tagged proteins was assessed by immunoblotting. The association between wildtype Ro52 and IRF3 is confirmed in the lower panel by migrating 50% of each sample, thus distinctly showing the increased intensity of the Ro52 band in comparison to the IgG heavy chain alone. (c) HeLa cells were stimulated with 25 μg/ml polyI:C for the indicated times. Ro52-associated proteins were immunoprecipitated from lysates with an anti-Ro52 antibody. Presence of Ro52-associated IRF3 was detected by immunoblotting.
To determine which domain of Ro52 is involved in the association with IRF3, plasmids expressing either full-length Ro52 or Ro52 lacking either the N-terminal RING-finger domain or C-terminal SPRY domain were used in co-expression studies with Flag-IRF3. As expected, full-length Ro52 interacted with Flag-IRF3 as shown by the increased intensity of the observed band compared with empty vector control (Fig. 3B, upper panel, lane 3). However as Ro52 consistently co-migrated with the heavy chain of the IgG molecule, we repeated the separation using 50% sample volume in order to make the association more distinct (Fig. 3B, lower panel). Importantly, whilst Flag-IRF3 was able to interact with Ro52 lacking the RING domain (Ro52ΔExon1), no association was observed between Flag-IRF3 and Ro52 lacking the C-terminal SPRY domain (Ro52ΔExon6) (Fig. 3B). This result indicates that the C-terminal SPRY domain of Ro52 is crucial for its interaction with IRF3.
We next looked at the ability of Ro52 and IRF3 to interact endogenously. HeLa cells were stimulated with polyI:C at specific time-points and cell lysates were incubated with anti-Ro52 antibody coupled to sepharose beads in an immunoprecipitation experiment. Importantly the endogenous association between Ro52 and IRF3 was stimulation dependent, with no apparent association under resting conditions, whereas a substantial increase in the association between the two proteins was observed at 2-4 hrs post-stimulation (Fig. 3C). This was accompanied by an increase in higher migrating molecular weight bands in the IRF3 immunoblot, indicative of increased polyubiquitination. Furthermore, accompanying the association between the two proteins, a decrease in total IRF3 levels was observed at 4-8 hrs post-stimulation. This suggests that polyI:C-induced degradation of IRF3 may be Ro52-dependent.
Ro52 promotes the ubiquitination and degradation of IRF3
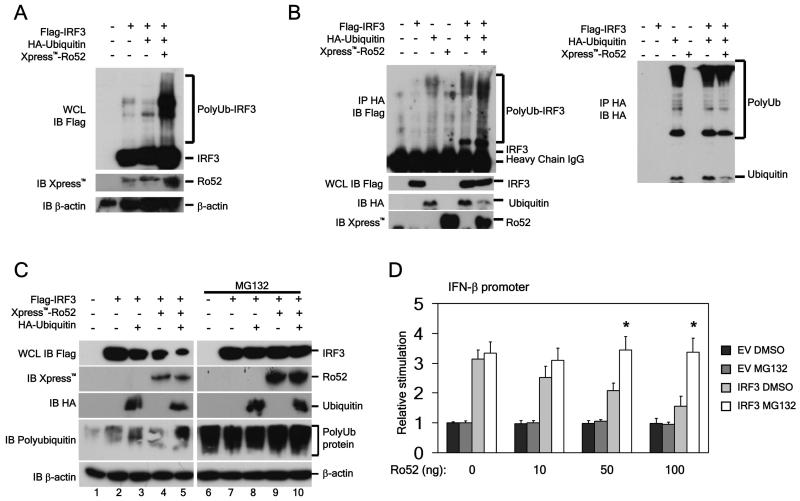
In our experimental system, IRF3 ubiquitination was observed following PRR stimulation, consistent with previous reports (data not shown). Studies have shown that Ro52 functions as an E3 ligase (23, 31, 32). Having first confirmed that Ro52 autoubiquitinates and thus has E3 ligase activity (data not shown), we next examined if the observed association between Ro52 and IRF3 was responsible for the observed ubiquitination of IRF3. In lysates from 293T cells, the presence of Xpress™-Ro52 and HA-ubiquitin increased the polyubiquitination of Flag-IRF3 compared to HA-ubiquitin alone, as observed by the presence of multiple high molecular mass bands (Fig. 4A). To confirm the ability of Ro52 to ubiquitinate IRF3, HA-ubiquitinated proteins were immunoprecipitated from 293T cells that had been transfected with HA-ubiquitin and Flag-IRF3 in the presence and absence of Ro52. As seen in Figure 4B, the presence of Ro52 markedly increased the polyubiquitination of IRF3. Total HA-ubiquitin modification was also examined and found to be similar in all lanes in which HA-ubiquitin was expressed, indicating that the increased IRF3 ubiquitination observed in the presence of Ro52 is not due to an increase in total ubiquitination. Taken together, these results indicate that Ro52 targets IRF3 for polyubiquitnation.
Figure 4. Ro52 promotes IRF3 ubiquitination and proteasomal degradation.
(a) 293T cells were transfected with 2 μg of the indicated constructs for 18 hrs and whole cell lysates were prepared. Presence of Flag-IRF3 and Xpress™-Ro52 were detected by immunoblotting. (b) 293T cells were transfected with 2 μg of the indicated constructs. 18 hrs post-transfection HA-ubiquitinated proteins were immunoprecipitated from lysates with an anti-HA antibody. Presence of HA-ubiquitinated Flag-IRF3 was detected by immunoblotting. Panel to the right shows total HA-ubiquitin modification detected by immunoblotting with an anti-HA antibody (c) 293T cells were transfected with 500 ng of the indicated constructs. 18 hrs post-transfection cells were treated with either 10 μM MG132 or vehicle control for 1 hr prior to lysis of the cells. Flag-tagged IRF3 expression in cell lysates was analysed by immunoblotting. (d) 293T cells were transfected with a reporter construct containing the IFN-β promoter. Cells were co-transfected with 50 ng IRF3 or empty vector (EV) control and increasing amounts of Ro52-expressing construct as indicated. Cells were treated with either 5 μM MG132 or vehicle control 1 hr prior to cell lysis. Cells were assayed for reporter gene activity 18 hrs post-transfection. *, p < 0.05 as determined by Student’s T test.
One consequence of polyubiquitination of proteins is subsequent proteasomal-mediated degradation. Consistent with a role for Ro52-mediated polyubiquitination of IRF3 in promoting its degradation we observed that IRF3 levels were reduced in 293T cells co-expressing Flag-IRF3 and Xpress-Ro52, (Fig. 4C, panel 1, lane 4). Furthermore, this effect was substantially increased when HA-ubiquitin was co-expressed (Fig. 4C, panel 1, lane 5). Pretreatment of cells with the proteasomal inhibitor MG132 completely rescued IRF3 degradation, confirming that the observed IRF3 degradation was proteasome-dependent (Fig. 4C, panel 1, lane 9-10). The ability of MG132 to inhibit proteasomal degradation is shown by the accumulation of total polyubiquitinated proteins (Fig. 4C, panel 4, lanes 6-10).
We next determined if Ro52-mediated IRF3 degradation had a direct effect on IFN-β promoter activity. Ro52 dose-dependently inhibited the IFN-β promoter when driven by IRF3 in 293T cells (Fig. 4D). However, inhibition of the proteasome with MG132 resulted in a significant rescue of IFN-β promoter activity, suggesting that Ro52-induced IRF3-degradation is responsible for inhibition of the IFN-β promoter. This corresponded with a dose-dependent decrease in the levels of IRF3 in the cells transfected with Ro52, which was rescued following MG132 treatment (data not shown)
Ro52 functions as a negative regulator of IFN-β signaling by degrading IRF3
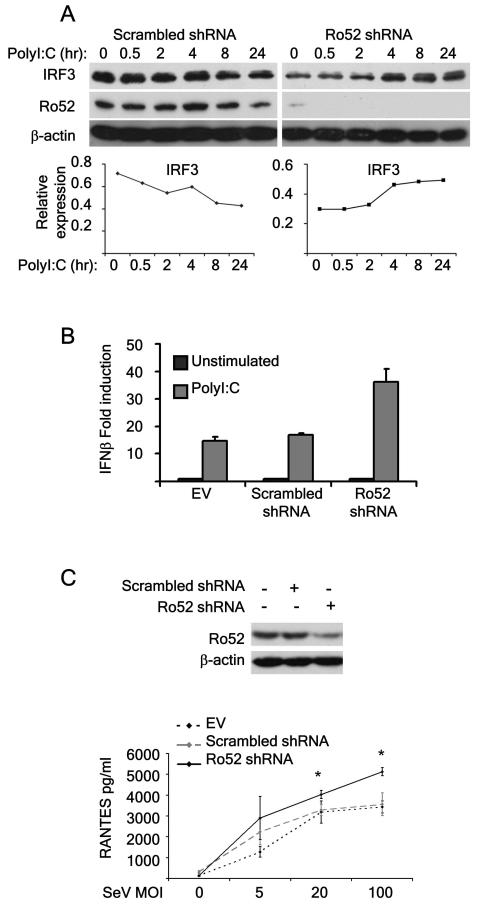
To determine the functional relevance of Ro52-mediated degradation of IRF3 and its corresponding effects on the IFN-β promoter, we targeted the expression of Ro52 in 293T cells using shRNA directed against human Ro52. As expected, polyI:C stimulation of cells treated with a non-target scrambled shRNA resulted in IRF3 degradation, an effect that could be seen by 8 hrs post-TLR3 stimulation. In contrast, knockdown of Ro52 using shRNA resulted in an accumulation of IRF3, consistent with a role for Ro52 as a negative regulator of IRF3 protein levels post-PRR stimulation (Fig. 5A). In addition, IRF3 degradation in response to PRR stimulation was rescued in Ro52-depleted NIH 3T3 cells compared to control cells (data not shown). We next measured the effects of Ro52 knockdown on IFN-β mRNA levels using real-time PCR. Whilst basal levels of the IFN-β gene were comparable between cells treated with Ro52 shRNA or scrambled shRNA control (data not shown), treatment of cells with polyI:C for 48 hours resulted in a 40-fold induction of the IFN-β gene in the presence of Ro52 shRNA compared with a 15-fold increase in gene induction in control cells (Fig. 5B). In contrast, there was no difference in IRF3 gene induction, suggesting that the effect of Ro52 observed in Figure 5A is specific to IRF3 protein (data not shown). Additional experiments confirmed these findings, using NIH 3T3 murine fibroblast cell lines stably transfected with either scrambled shRNA or Ro52 shRNA (Fig. 5C). Consistent with previous findings in 293T cells, following Sendai-virus infection, induction of the IFN-β-stimulated chemokine RANTES was significantly increased in Ro52-depleted cells compared to control cells (Fig. 5C).
Figure 5. Ro52 functions as a negative regulator of IFN-β and RANTES production post-PRR stimulation.
(a) 293T cells were transfected with either non-target scrambled shRNA or Ro52 shRNA for 24 hrs. Following transfection, cells were stimulated with 25 μg/ml polyI:C for the indicated times. Endogenous IRF3, Ro52 and β-actin expression in cell lysates was analysed by immunoblotting. IRF3 expression was quantitated densitometrically and graphed below the corresponding blots. (b) 293T cells were transfected with EV control, non-target scrambled shRNA or Ro52 shRNA for 48 hrs. Following transfection, cells were stimulated with 25 μg/ml polyI:C for 48 hrs. IFN-β mRNA induction was analysed by realtime PCR. (c) NIH 3T3 cells were stably transfected with non-target scrambled shRNA or Ro52 shRNA. Endogenous Ro52 and β-actin expression in cell lysates was analysed by immunoblotting. Cells were stimulated with Sendai virus for 48 hrs at 5, 20 and 100 MOI. RANTES expression in cell supernatants was determined by ELISA. *, p < 0.05 as determined by Student’s T test.
Collectively, these results indicate that loss of Ro52 in the cells results in an accumulation of the transcription factor IRF3 and subsequently an enhanced production of IFN-β. This implicates Ro52 in targeting IRF3 protein to turn off and limit IFN-β production post-pathogen detection by PRRs.
Discussion
In this study, we have identified Ro52 as an E3 ligase that acts to limit IFN-β production downstream of pathogen recognition receptors, specifically TLR3, TLR4 and RIG-I. Our results demonstrate that Ro52 achieves these effects by polyubiquitinating the transcription factor IRF3, thus targeting it for proteasomal-mediated degradation. By degrading IRF3, Ro52 prevents excessive production of IFN-β in response to pathogen detection.
Negative regulation of anti-viral pathways post-PRR activation via ubiquitination of key downstream components is an effective way to turn off and limit the production of type I IFNs. Indeed, targeted degradation of RIG-I by the E3 ligase RNF125 has recently been described and negatively regulates IFN-β production in response to infection of cells with Sendai virus (33). In addition, the anti-apoptotic protein A20 has been shown to be a potent inhibitor of RIG-I-mediated activation of both IRF3 and NFκB, an effect that requires its E3 ligase activity, and not its deubiquitinating activity (34, 35). We investigated a possible role for the E3 ligase Ro52 as a negative regulator of TLR-dependent pathways. Whilst Ro52 had no effect on TLR4-driven NFκB-dependent reporter gene activity, subsequent results indicated that Ro52 negatively regulated pathways promoting IFN-β production in response to TLR stimulation. Further analysis indicated that its target lay downstream of TRIF, with IRF3, and not NFκB, being a candidate target for Ro52. In addition, through the use of Ro52 mutants, we have shown that the N-terminal RING-finger domain is essential for the observed inhibition of IFN-β promoter activity, indicating that Ro52 may be acting as an E3 ligase in this pathway, targeting IRF3 for degradation.
Down-regulation of IRF3 activation by ubiquitin-mediated degradation is an efficient means to turn of IFN-β production, thus making IRF3 a prime target for viral immune evasion strategies (reviewed in 12, 36). In this context, bovine herpesvirus 1 infected cell protein 0 (bICP0) has been shown to act as an E3 ligase and promote IRF3 degradation in a proteasome-dependent manner, thus inhibiting the IFN-β promoter (37). Investigation into endogenous mechanisms targeting IRF3 to down-regulate type I IFN signaling downstream of pathogen detection has focussed on the involvement of a cullin-based ubiquitin ligase in the polyubiquitination and subsequent degradation of IRF3, however the E3 ligase responsible has not been identified (10).
Here we have identified Ro52 as an endogenous E3 ligase responsible for regulating IRF3 levels. Ro52 was found to associate with IRF3 via the C-terminal domain SPRY domain of Ro52, thought to be involved in protein-protein interactions (20, 21). We demonstrate here that the observed association of Ro52 with IRF3 promotes the polyubiquitination and subsequent proteasomal-mediated degradation of IRF3. In addition, Ro52 dose-dependently inhibits IRF3-driven IFN-β promoter activity and this effect is significantly reversed in the presence of the proteasomal inhibitor MG132, suggesting that inhibition of the IFN-β promoter is a direct consequence of Ro52-mediated degradation of IRF3. Critically, we have demonstrated that the observed association between Ro52 and IRF3 in over-expression studies also occurs endogenously and that it is stimulation dependent. Our results show that Ro52 associates with IRF3 2-4 hours post polyI:C-stimulation and that this is accompanied by a subsequent loss of IRF3 levels in the cells. Furthermore, targeting Ro52 with shRNA, and thus inhibiting Ro52-induced IRF3-degradation, results in enhanced TLR3-driven IFN-β production and Sendai virus-stimulated RANTES production, confirming our hypothesis that Ro52 negatively regulates IFN-β production post-PRR stimulation by targeting IRF3 for degradation.
As this manuscript was in preparation, Kong et al described a role for Ro52 in ubiquitinating the transcription factor IRF8, which positively regulates IL-12p40 production in murine macrophages (27). Like Kong et al, we identify a member of the IRF family, IRF3, as a target for Ro52. We also observed an interaction between Ro52 and IRF7, suggesting a global role for Ro52 in regulating the IRF family. Further work on the role of Ro52 in IRF7 signaling will be revealing as previous studies have shown enhanced IRF7 activity following ubiquitination by both TRAF6 and the Epstein-Barr virus oncoprotein LMP1 (38, 39).
Collectively our results demonstrate that the E3 ligase Ro52 targets IRF3 for polyubiquitination and proteasomal-mediated degradation. The overall function of this targeted degradation of IRF3 is to turn off or limit the production of IFN-β post-pathogen detection. In this context, our current focus is to determine how Ro52 is regulated post-PRR stimulation, specifically which E2 ligase is involved and if Ro52 is post-translationally modified. Interestingly, Ro52 is best known for its ability to act as an autoantigen in SLE and Sjögren’s syndrome. Whether there is a possible link between increased levels of Ro52 autoantibodies in patients with SLE and Sjögren’s syndrome, and Ro52 function as a regulator of IFN-β production, remains to be seen. In addition, Ro52 polymorphisms have been described that are associated with SLE (40), and linkage analysis reports have indicated chromosome 11p15.5, containing the Ro52 locus, as a susceptibility region for SLE (41, 42). Consequently, given its role in regulating IFN-β production described in this study, it is possible that Ro52 activity may be compromised in these autoimmune disorders, thus contributing to the increased production of type I IFNs associated with disease pathogenesis. Therefore, our findings have important implications for our understanding of mechanisms that regulate both antiviral immunity and autoimmunity.
Acknowledgements
We thank Prof. Jim Johnston and Kiva Brennan for helpful discussion and comments on the manuscript.
Footnotes
This work was funded by Science Foundation Ireland, the Health Research Board and by NIAID, NIH (AI067497 to K.A.F.).
- PRR
- Pathogen Recognition Receptor
- IRF
- interferon regulatory factor
- TRIF
- TIR domain containing adaptor inducing IFN-β
- LPS
- lipopolysacharride
- polyI:C
- polyinosinic:polycytidylic acid
- HA
- hemagglutinin
- PRD
- positive regulatory domain
References
- 1.Hertzog PJ, O’Neill LA, Hamilton JA. The interferon in TLR signaling: more than just antiviral. Trends Immunol. 2003;24:534–539. doi: 10.1016/j.it.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 2.Hoshino K, Kaisho T, Iwabe T, Takeuchi O, Akira S. Differential involvement of IFN-beta in Toll-like receptor-stimulated dendritic cell activation. Int Immunol. 2002;14:1225–1231. doi: 10.1093/intimm/dxf089. [DOI] [PubMed] [Google Scholar]
- 3.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 4.Doyle S, Vaidya S, O’Connell R, Dadgostar H, Dempsey P, Wu T, Rao G, Sun R, Haberland M, Modlin R, Cheng G. IRF3 mediates a TLR3/TLR4-specific antiviral gene program. Immunity. 2002;17:251–263. doi: 10.1016/s1074-7613(02)00390-4. [DOI] [PubMed] [Google Scholar]
- 5.Sakaguchi S, Negishi H, Asagiri M, Nakajima C, Mizutani T, Takaoka A, Honda K, Taniguchi T. Essential role of IRF-3 in lipopolysaccharide-induced interferon-beta gene expression and endotoxin shock. Biochem Biophys Res Commun. 2003;306:860–866. doi: 10.1016/s0006-291x(03)01049-0. [DOI] [PubMed] [Google Scholar]
- 6.Sato M, Suemori H, Hata N, Asagiri M, Ogasawara K, Nakao K, Nakaya T, Katsuki M, Noguchi S, Tanaka N, Taniguchi T. Distinct and essential roles of transcription factors IRF-3 and IRF-7 in response to viruses for IFN-alpha/beta gene induction. Immunity. 2000;13:539–548. doi: 10.1016/s1074-7613(00)00053-4. [DOI] [PubMed] [Google Scholar]
- 7.Banchereau J, Pascual V. Type I interferon in systemic lupus erythematosus and other autoimmune diseases. Immunity. 2006;25:383–392. doi: 10.1016/j.immuni.2006.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Lin R, Heylbroeck C, Pitha PM, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saitoh T, Tun-Kyi A, Ryo A, Yamamoto M, Finn G, Fujita T, Akira S, Yamamoto N, Lu KP, Yamaoka S. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol. 2006;7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- 10.Bibeau-Poirier A, Gravel SP, Clement JF, Rolland S, Rodier G, Coulombe P, Hiscott J, Grandvaux N, Meloche S, Servant MJ. Involvement of the IkappaB kinase (IKK)-related kinases tank-binding kinase 1/IKKi and cullin-based ubiquitin ligases in IFN regulatory factor-3 degradation. J Immunol. 2006;177:5059–5067. doi: 10.4049/jimmunol.177.8.5059. [DOI] [PubMed] [Google Scholar]
- 11.Kawai T, Takeuchi O, Fujita T, Inoue J, Muhlradt PF, Sato S, Hoshino K, Akira S. Lipopolysaccharide stimulates the MyD88-independent pathway and results in activation of IFN-regulatory factor 3 and the expression of a subset of lipopolysaccharide-inducible genes. J Immunol. 2001;167:5887–5894. doi: 10.4049/jimmunol.167.10.5887. [DOI] [PubMed] [Google Scholar]
- 12.Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem. 2007;282:15325–15329. doi: 10.1074/jbc.R700002200. [DOI] [PubMed] [Google Scholar]
- 13.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao SM, Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 14.McWhirter SM, Fitzgerald KA, Rosains J, Rowe DC, Golenbock DT, Maniatis T. IFN-regulatory factor 3-dependent gene expression is defective in Tbk1-deficient mouse embryonic fibroblasts. Proc Natl Acad Sci U S A. 2004;101:233–238. doi: 10.1073/pnas.2237236100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sharma S, tenOever BR, Grandvaux N, Zhou GP, Lin R, Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 16.Ben-Neriah Y. Regulatory functions of ubiquitination in the immune system. Nat Immunol. 2002;3:20–26. doi: 10.1038/ni0102-20. [DOI] [PubMed] [Google Scholar]
- 17.Wang J, Maldonado MA. The ubiquitin-proteasome system and its role in inflammatory and autoimmune diseases. Cell Mol Immunol. 2006;3:255–261. [PubMed] [Google Scholar]
- 18.Liu YC. Ubiquitin ligases and the immune response. Annu Rev Immunol. 2004;22:81–127. doi: 10.1146/annurev.immunol.22.012703.104813. [DOI] [PubMed] [Google Scholar]
- 19.Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays. 2005;27:1147–1157. doi: 10.1002/bies.20304. [DOI] [PubMed] [Google Scholar]
- 20.Rhodes DA, Trowsdale J. TRIM21 is a trimeric protein that binds IgG Fc via the B30.2 domain. Mol Immunol. 2007;44:2406–2414. doi: 10.1016/j.molimm.2006.10.013. [DOI] [PubMed] [Google Scholar]
- 21.Woo JS, Imm JH, Min CK, Kim KJ, Cha SS, Oh BH. Structural and functional insights into the B30.2/SPRY domain. Embo J. 2006;25:1353–1363. doi: 10.1038/sj.emboj.7600994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chan EK, Hamel JC, Buyon JP, Tan EM. Molecular definition and sequence motifs of the 52-kD component of human SS-A/Ro autoantigen. J Clin Invest. 1991;87:68–76. doi: 10.1172/JCI115003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Espinosa A, Zhou W, Ek M, Hedlund M, Brauner S, Popovic K, Horvath L, Wallerskog T, Oukka M, Nyberg F, Kuchroo VK, Wahren-Herlenius M. The Sjogren’s syndrome-associated autoantigen Ro52 is an E3 ligase that regulates proliferation and cell death. J Immunol. 2006;176:6277–6285. doi: 10.4049/jimmunol.176.10.6277. [DOI] [PubMed] [Google Scholar]
- 24.Wahren-Herlenius M, Muller S, Isenberg D. Analysis of B-cell epitopes of the Ro/SS-A autoantigen. Immunol Today. 1999;20:234–240. doi: 10.1016/s0167-5699(99)01458-9. [DOI] [PubMed] [Google Scholar]
- 25.Nisole S, Stoye JP, Saib A. TRIM family proteins: retroviral restriction and antiviral defence. Nat Rev Microbiol. 2005;3:799–808. doi: 10.1038/nrmicro1248. [DOI] [PubMed] [Google Scholar]
- 26.Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- 27.Kong HJ, Anderson DE, Lee CH, Jang MK, Tamura T, Tailor P, Cho HK, Cheong J, Xiong H, Morse HC, 3rd, Ozato K. Cutting Edge: Autoantigen Ro52 Is an Interferon Inducible E3 Ligase That Ubiquitinates IRF-8 and Enhances Cytokine Expression in Macrophages. J Immunol. 2007;179:26–30. doi: 10.4049/jimmunol.179.1.26. [DOI] [PubMed] [Google Scholar]
- 28.Doyle SL, Jefferies CA, O’Neill LA. Bruton’s tyrosine kinase is involved in p65-mediated transactivation and phosphorylation of p65 on serine 536 during NFkappaB activation by lipopolysaccharide. J Biol Chem. 2005;280:23496–23501. doi: 10.1074/jbc.C500053200. [DOI] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Ishii T, Ohnuma K, Murakami A, Takasawa N, Yamochi T, Iwata S, Uchiyama M, Dang NH, Tanaka H, Morimoto C. SS-A/Ro52, an autoantigen involved in CD28-mediated IL-2 production. J Immunol. 2003;170:3653–3661. doi: 10.4049/jimmunol.170.7.3653. [DOI] [PubMed] [Google Scholar]
- 31.Wada K, Kamitani T. Autoantigen Ro52 is an E3 ubiquitin ligase. Biochem Biophys Res Commun. 2006;339:415–421. doi: 10.1016/j.bbrc.2005.11.029. [DOI] [PubMed] [Google Scholar]
- 32.Sabile A, Meyer AM, Wirbelauer C, Hess D, Kogel U, Scheffner M, Krek W. Regulation of p27 degradation and S-phase progression by Ro52 RING finger protein. Mol Cell Biol. 2006;26:5994–6004. doi: 10.1128/MCB.01630-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Arimoto K, Takahashi H, Hishiki T, Konishi H, Fujita T, Shimotohno K. Negative regulation of the RIG-I signaling by the ubiquitin ligase RNF125. Proc Natl Acad Sci U S A. 2007;104:7500–7505. doi: 10.1073/pnas.0611551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lin R, Yang L, Nakhaei P, Sun Q, Sharif-Askari E, Julkunen I, Hiscott J. Negative regulation of the retinoic acid-inducible gene I-induced antiviral state by the ubiquitin-editing protein A20. J Biol Chem. 2006;281:2095–2103. doi: 10.1074/jbc.M510326200. [DOI] [PubMed] [Google Scholar]
- 35.Wang YY, Li L, Han KJ, Zhai Z, Shu HB. A20 is a potent inhibitor of TLR3- and Sendai virus-induced activation of NF-kappaB and ISRE and IFN-beta promoter. FEBS Lett. 2004;576:86–90. doi: 10.1016/j.febslet.2004.08.071. [DOI] [PubMed] [Google Scholar]
- 36.Ozato K, Tailor P, Kubota T. The interferon regulatory factor family in host defense: mechanism of action. J Biol Chem. 2007;282:20065–20069. doi: 10.1074/jbc.R700003200. [DOI] [PubMed] [Google Scholar]
- 37.Saira K, Zhou Y, Jones C. The infected cell protein 0 encoded by bovine herpesvirus 1 (bICP0) induces degradation of interferon response factor 3 and, consequently, inhibits beta interferon promoter activity. J Virol. 2007;81:3077–3086. doi: 10.1128/JVI.02064-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huye LE, Ning S, Kelliher M, Pagano JS. Interferon regulatory factor 7 is activated by a viral oncoprotein through RIP-dependent ubiquitination. Mol Cell Biol. 2007;27:2910–2918. doi: 10.1128/MCB.02256-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawai T, Sato S, Ishii KJ, Coban C, Hemmi H, Yamamoto M, Terai K, Matsuda M, Inoue J, Uematsu S, Takeuchi O, Akira S. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat Immunol. 2004;5:1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 40.Frank MB, Itoh K, Fujisaku A, Pontarotti P, Mattei MG, Neas BR. The mapping of the human 52-kD Ro/SSA autoantigen gene to human chromosome 11, and its polymorphisms. Am J Hum Genet. 1993;52:183–191. [PMC free article] [PubMed] [Google Scholar]
- 41.Gaffney PM, Kearns GM, Shark KB, Ortmann WA, Selby SA, Malmgren ML, Rohlf KE, Ockenden TC, Messner RP, King RA, Rich SS, Behrens TW. A genome-wide search for susceptibility genes in human systemic lupus erythematosus sib-pair families. Proc Natl Acad Sci U S A. 1998;95:14875–14879. doi: 10.1073/pnas.95.25.14875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, Tsao BP, Vyse TJ, Langefeld CD, Nath SK, Guthridge JM, Cobb BL, Mirel DB, Marion MC, Williams AH, Divers J, Wang W, Frank SG, Namjou B, Gabriel SB, Lee AT, Gregersen PK, Behrens TW, Taylor KE, Fernando M, Zidovetzki R, Gaffney PM, Edberg JC, Rioux JD, Ojwang JO, James JA, Merrill JT, Gilkeson GS, Seldin MF, Yin H, Baechler EC, Li QZ, Wakeland EK, Bruner GR, Kaufman KM, Kelly JA. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40:204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]