Abstract
During initiation of apoptosis, Bcl-2 family proteins regulate the permeability of mitochondrial outer membrane. BH3-only protein, tBid, activates pro-apoptotic Bax to release cytochrome c from mitochondria. tBid also activates anti-apoptotic Bcl-2 in the mitochondrial outer membrane, changing it from a single-spanning to a multi-spanning conformation that binds the active Bax and inhibits cytochrome c release. However, it is not known whether other mitochondrial proteins are required to elicit the tBid-induced Bcl-2 conformational alteration. To define the minimal components that are required for the functionally important Bcl-2 conformational alteration, we reconstituted the reaction using purified proteins and liposomes. We found that purified tBid was sufficient to induce a conformational alteration in the liposome-tethered, but not cytosolic Bcl-2, resulting in a multi-spanning form that is similar to the one found in the mitochondrial outer membrane of drug treated cells. Mutations that abolished tBid/Bcl-2 interaction also abolished the conformational alteration, demonstrating that a direct tBid/Bcl-2 interaction at the membrane is both required and sufficient to elicit the conformational alteration. Furthermore, active Bax also elicited the Bcl-2 conformational alteration. Bcl-2 mutants that displayed increased or decreased activity in the conformational alteration assay, showed corresponding activities in inhibiting pore formation by Bax in vitro, and in preventing apoptosis in vivo. Thus, there is a strong correlation between the direct interaction of membrane-bound Bcl-2 and tBid with activation of Bcl-2 in vitro and in vivo.
Proteins of the Bcl-2 family are key regulators of apoptosis that function either as promoters or inhibitors and that display homology in one to four short sequences termed Bcl-2 homology (BH)# motifs(1–3). Anti-apoptotic subfamily proteins such as Bcl-2 and Bcl-xL contain four BH motifs (BH1-4). Pro-apoptotic proteins are grouped into either multiple or single BH-motif subfamilies. The former subfamily includes the proteins Bax and Bak that display homology in BH1-3 motifs. The latter includes proteins such as Bid, Bim and Bad that are similar only in the limited BH3 motif. Despite the limited sequence homology, the three-dimensional structures determined for seven Bcl-2 family proteins including members from all three subfamilies are very similar, consisting of a hydrophobic core of one to three helices wrapped by five to eight amphipathic helices and their connecting loops. This structure is strikingly similar to the structure of the pore-forming domains of diphtheria toxin and E. coli colicins. In addition there is an obvious hydrophobic groove on the surface of all of the anti-apoptotic Bcl-2 family proteins (4–6).
From decades of extensive studies, two properties are well known to be shared by most, if not all, Bcl-2 family proteins: homo- or hetero-binding and pore formation in membranes. More experiments have been done on the homo- or hetero-binding of Bcl-2 family proteins, resulting in the following major conclusions. 1) The hetero-binding between different subfamily proteins is important for their function. Most BH3-only proteins have been proposed to bind to Bcl-2-like proteins following various death signals, either neutralizing the pro-survival function or inhibiting the anti-death activities of Bcl-2-like proteins. A few BH3-only proteins have also been shown to bind Bax-like proteins and activate their pro-death function(7–9). 2) Binding between two different subfamily proteins is mediated by different motifs. The interaction of a BH3-only protein and a Bcl-2-like protein is likely accomplished by the binding of the BH3-motif helix of the former into the hydrophobic groove of the latter(10–12). A similar interface may be formed in the complex between a Bax-like protein and a Bcl-2-like protein(13). 3) The interface for homo-binding of Bcl-2 is different from that for Bcl-xL. While a Bcl-2 dimer is formed asymmetrically by an acceptor surface including the BH1-3 hydrophobic groove and a donor surface including BH4 helix, a Bcl-xL dimer is formed symmetrically by inserting the C-terminal tail of one protomer into the hydrophobic groove of the other and vise versa(14,15). Interestingly, in healthy cells Bcl-2 is exclusively bound to membranes in a conformation in which the C-terminal tail is inserted into the lipid bilayer, and therefore not available for binding to the hydrophobic groove in the cytosolic domain of other Bcl-2 proteins(16,17). In contrast, a substantial fraction of Bcl-xL is found either in the cytosol or loosely bound to membranes in healthy cells. The C-terminal tail of the soluble Bcl-xL could therefore, be involved in homo-binding(18). During the induction of apoptosis Bcl-xL inserts into intracellular membranes. A recent crystallography study suggests that the membrane-bound Bcl-xL may swap helices 6–8 to form a symmetrical homo-dimer(19).
Since Bax can release cytochrome c from purified mitochondria, pore formation by Bax has been studied extensively. The following major conclusions have been reached. 1) tBid protein or BH3 peptide from Bid or Bim was required for Bax to release cytochrome c from purified mitochondria, and to release dextrans up to 2 MDa from mitochondrial outer membrane (MOM) vesicles or liposomes of MOM lipids (MOM-liposomes)(9,20). Pore formation is dependent on the mitochondrial specific lipid, cardiolipin, and can be inhibited by Bcl-xL. 2) The putative Bax pore that can release 2-MDa dextrans should have a radius more than 273 Å, suggesting that massive Bax oligomerization may be involved(21). 3) Oligomerization of Bax is required for Bax to permeabilize the MOM, thereby releasing proapoptotic factors such as cytochrome c and AIF that activate caspases and nucleases which eventually dismantle the cell(20,22–24). The oligomerization occurs after Bax is inserted into the membrane as a multispanning monomer with helices 5, 6 and 9 buried in the bilayer(25). The oligomerization of Bax-like proteins requires their BH3 motif and can be inhibited by Bcl-2-like proteins(26–29). 4) The BH3-activated Bax-like proteins can activate more Bax-like proteins to amplify the initial pro-death signal produced by BH3-only proteins(30,31).
Only a few studies have examined pore formation by Bcl-2 and Bcl-xL proteins, resulting in the following conclusions(19,32–36). 1) Both Bcl-2 and Bcl-xL form pores in anionic lipid containing liposomal membranes at extreme acidic pH. The pore-forming activity can be enhanced by a trans-negative membrane potential. 2) In planar lipid bilayers both proteins form ion-conducting channels that display a linear current-voltage relationship, following Ohm’s law. The channels have discrete conductances ranging from 20 to 90 pS for Bcl-2 and 80 to 276 pS for Bcl-xL, suggesting that they may have different structures. Unlike the pores in liposomal membranes that are only detectable at acidic pH, the channels formed in planar bilayers can be detected at both neutral and acidic pH. But the channels open more frequently at acidic pH. The channels are cation selective. 3) Deletion of helices 5 and 6 eliminates the channel activity for Bcl-2, and replacing helices 5 and 6 of Bcl-xL with those from Bax alters the channel property, suggesting that the two hydrophobic helices are required for channel formation by the two anti-apoptosis proteins. 4) During induction of apoptosis, Bcl-2 changes from the tail-anchored conformation to a multispanning one in which in addition to the C-terminal tail (helix 9) both helices 5 and 6 are inserted into the membrane(37). This conformational alteration can be induced in isolated mitochondria by tBid and is required for anti-Bax activity(29). Thus, only the conformationally altered Bcl-2 can form a stable complex with the active, multispanning Bax, thereby inhibiting its oligomerization and membrane permeabilization.
It is intriguing that both pro- and anti-apoptotic multi-BH motif proteins can be switched to a similar multispanning conformation by the BH3-only protein, tBid. Perhaps they form different structures in the membrane to fulfill their opposite functions. The multispanning Bax is known to form large oligomeric pores. Acidic pH induces the Bcl-2 pore formation in liposomal membranes, but the relationship between the acidic pH-induced pore formation and the tBid-induced conformational alteration remains elusive. Moreover, the correlation between the in vitro pore-forming activity and the in vivo anti-apoptotic activity of Bcl-2 has yet to be established. Finally, although tBid can induce the Bcl-2 conformational alteration in isolated mitochondria, the minimal components required for the alteration needs to be defined.
In this study we used the MOM-liposomes with encapsulated fluorescent dyes and purified cytosolic domain of Bcl-2 (Bcl-2ΔTM) to reconstitute the pore-forming process in vitro. We found that acidic pH induced a conformational alteration in soluble Bcl-2ΔTM, exposing a hydrophobic core helix to facilitate the membrane insertion and pore formation. In contrast, tBid did not induce the pore formation by the soluble Bcl-2ΔTM. However, when Bcl-2ΔTM was tethered to the liposome, it changed conformation and formed pores after interacting with tBid. Furthermore, the pore formed by the tBid-activated Bcl-2ΔTM is smaller than that formed by the tBid-activated Bax. Most importantly, the Bcl-2ΔTM inhibited the large pore formation by the activated Bax only when it was also activated by either tBid or tBid-activated Bax. Mutations that enhanced or reduced the tBid-mediated Bcl-2ΔTM activation in liposomes also enhanced or reduced Bcl-2 anti-apoptosis function in cells, respectively, thereby correlating the in vitro detected conformational alteration with the in vivo function.
EXPERIMENTAL PROCEDURES
Materials
All phospholipids and lipid analogs were purchased from Avanti Polar Lipids. Cascade Blue (CB), CB-labeled dextrans and rabbit anti-CB polyclonal antibody were from Molecular Probes. Peptides were synthesized by Global Peptide Services (Fort Collins, CO) and their sequences were as described (31).
Preparation of plasmids and proteins
Construction of pET22b(+)-based plasmid encoding His6-tagged Bcl-2ΔTM with either Gly154-Gly155 replaced by two alanines or Val159 replaced by an aspartate (designated G154/155A or V159D) was done using appropriate primers and overlapping PCR-based mutagenesis as described(38). Construction of pBabe/Hygromycin-based plasmid encoding each corresponding full-length Bcl-2 mutant was done by replacing the corresponding DNA segment with the one containing the mutation. The sequence of all the plasmids was verified by DNA sequencing. Expression and purification of the His6-tagged Bcl-2ΔTM and mutant, tBid and mutant with Met97-Asp98 replaced by two alanines (designated M97A/D98A), and Bax proteins were as described(14,39).
Preparation of liposomes
Liposomes of MOM lipid composition and with CB or CB-dextrans encapsulated were prepared by the extrusion method as described(31). The plain liposomes without fluorophores were prepared similarly except that no fluorophores were in the buffer A used for resuspension of the lipids and no gel filtration chromatography was needed after the extrusion. The liposomes containing lipophilic quenchers were prepared similarly except that 30 mol% of phosphatidylcholine (PC) was replaced by a doxyl-labeled PC [1-palmitoyl-2-stearoyl-(7-doxyl)-sn-glycero-3-PC, 7-doxyl-PC]. The Ni-chelating liposomes were prepared similarly except that 1 mM of Ni-chelating lipid analog, 1,2-dioleoyl-sn-glycero-3-{[N(5-amino-1-carboxypentyl)iminodiacetic acid]succinyl}(nickel salt), was added to 20 mM of MOM lipids before resuspension in buffer A.
Isolation of liposome-bound proteins or peptides
For Figure 2A or 3B, 400 or 200 nM Bcl-2ΔTM protein, respectively, was incubated with liposomes of 50 μM lipids (50 μM liposomes for short from here on) in 116 μl of buffer A of certain pH at 25 °C for 1–2 hr. For Figure 3D, 50 nM Bcl-2ΔTM, 20 nM tBid, and/or 50 nM Bax were incubated with 12.5 μM Ni-liposomes. When indicated, 5 mM EDTA or 200 mM imidazole was included either during or after the incubation. The samples were then subjected to sucrose gradient float-up centrifugation as described(39). For the sample containing EDTA or imidazole, same concentration of corresponding chemical was also presented in the sucrose gradient. Four 250-μl fractions were drawn, starting from the top of the gradient, resulting in fractions S1-S4. The pellet was re-suspended in 250 μl of buffer A, resulting in fraction P. The Bcl-2ΔTM proteins in these fractions were analyzed by SDS-PAGE and immunoblotting with sheep anti-Bcl-2 polyclonal antibody. The distribution of the liposomes among these fractions was estimated by scintillation counting of the 14C-PC of each fraction as described(31).
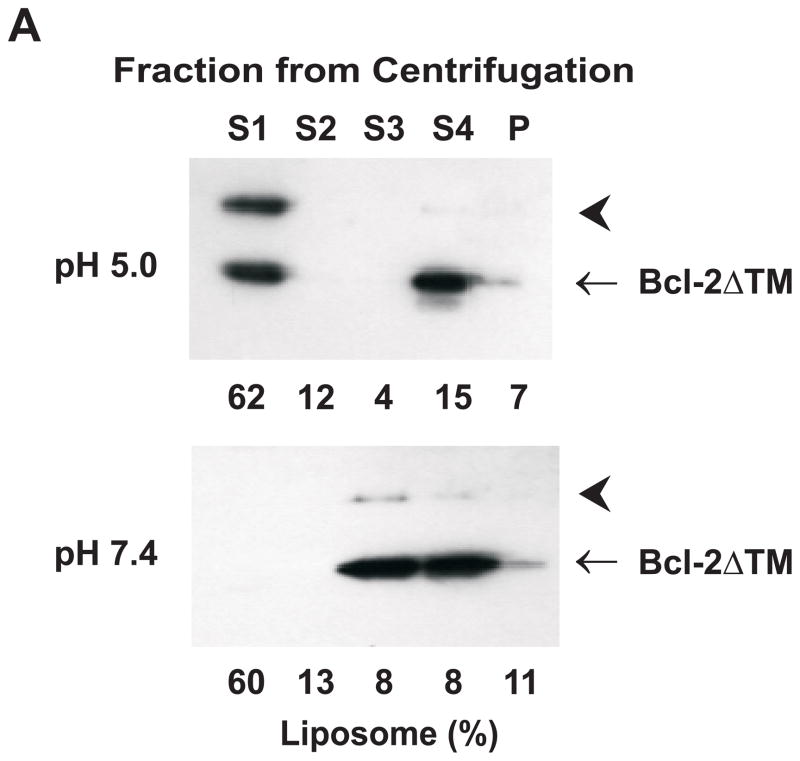
Fig. 2. Effect of pH on Bcl-2ΔTM insertion into liposomal membranes and pore formation.
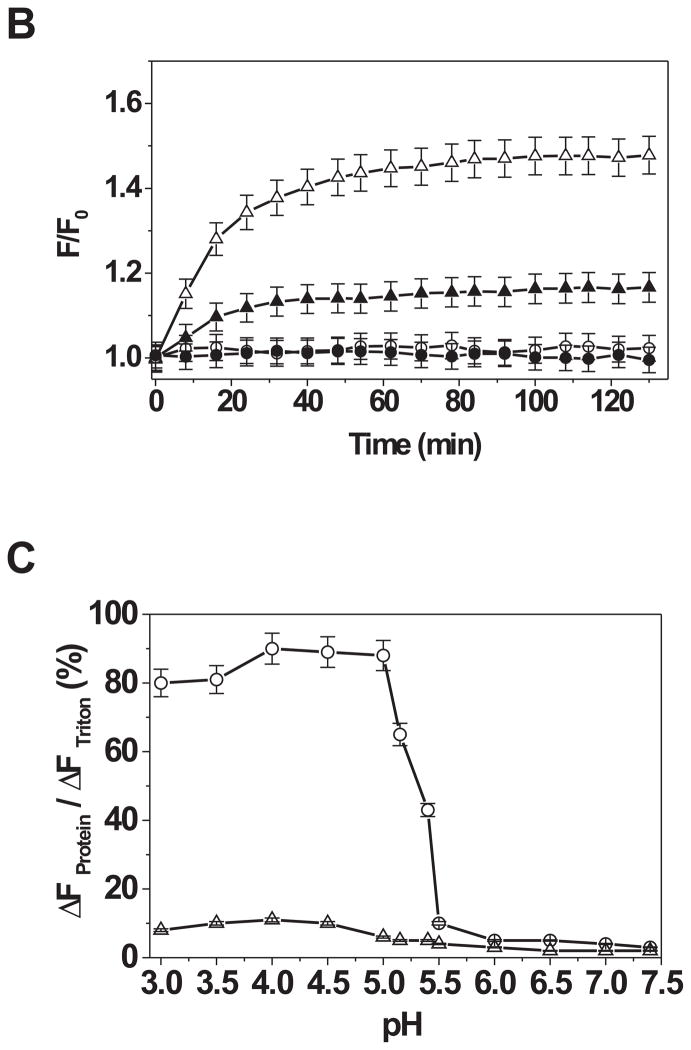
A. Data shown in the top or bottom panel are immunoblots of Bcl-2ΔTM in various fractions from a sucrose gradient float-up centrifugation after incubating the protein with liposomes at pH 5.0 or 7.4, respectively. The liposome-bound protein migrates into the upper supernatant fractions (S1–2), whereas the soluble protein remains in the bottom fractions (S3–4). The aggregated protein is in the pellet fraction (P). The distribution of liposomes in the gradient is indicated below each lane as the % of total liposomes. Data shown are representatives of three independent experiments. Arrowheads indicate a SDS-resistant Bcl-2ΔTM dimer. B. Insertion of helix 5 of Bcl-2ΔTM into liposomal membranes was monitored by the change of fluorescence intensity of the NBD labeled at Cys158 after mixing the protein and liposome at pH 7.4 (○) or 5.0 (△). The liposome contained 7-doxyl-PC that quenches the fluorescence if the NBD dye inserts into the bilayer was used in the parallel experiment at pH 7.4 (●) or 5.0 (▲). F0, the net intensity prior to addition of liposome; F, the net dilution-corrected intensities at indicated times after addition of liposome. Data shown are averages from two independent experiments with S.D. (error bars). C. Bcl-2ΔTM pore-formation in the liposomal membrane at various pH was monitored by the Bcl-2ΔTM-induced release of CB dyes from the liposomes after a 5-hr incubation. The release resulted in the quenching of CB fluorescence by the anti-CB antibody located outside the liposome. The extent of Bcl-2ΔTM-induced release is determined by the value of ΔFProtein/ΔFTriton. Data shown are averages from three independent experiments using Bcl-2ΔTM protein (○) or the buffer control (△) with S.D. (error bars).
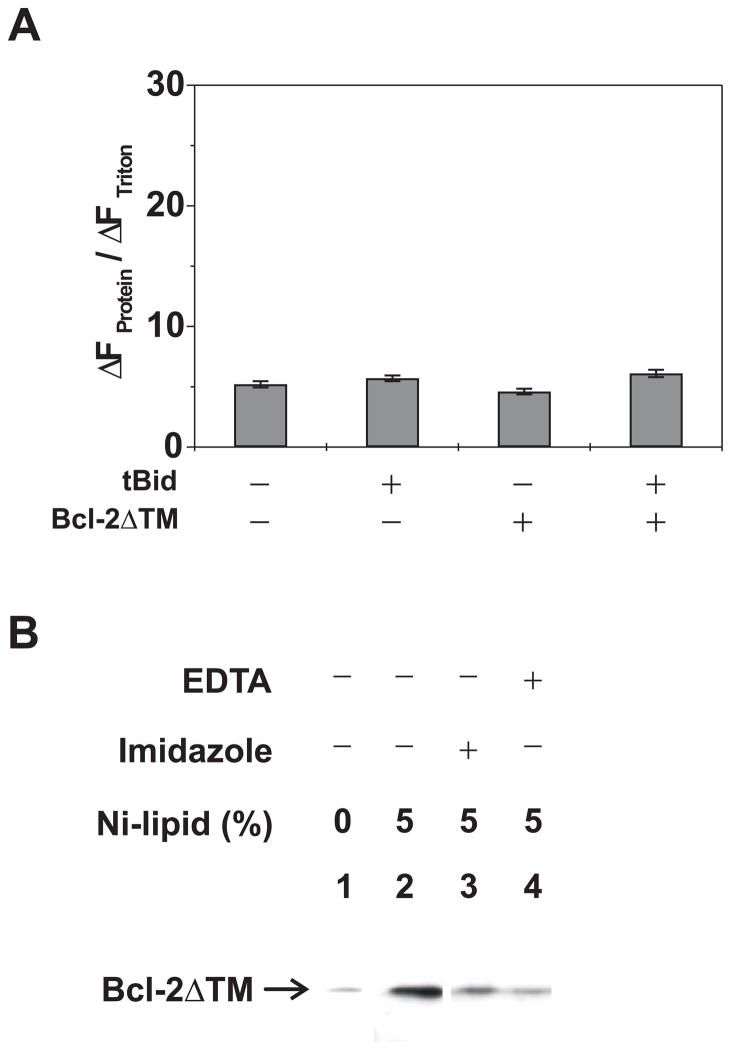
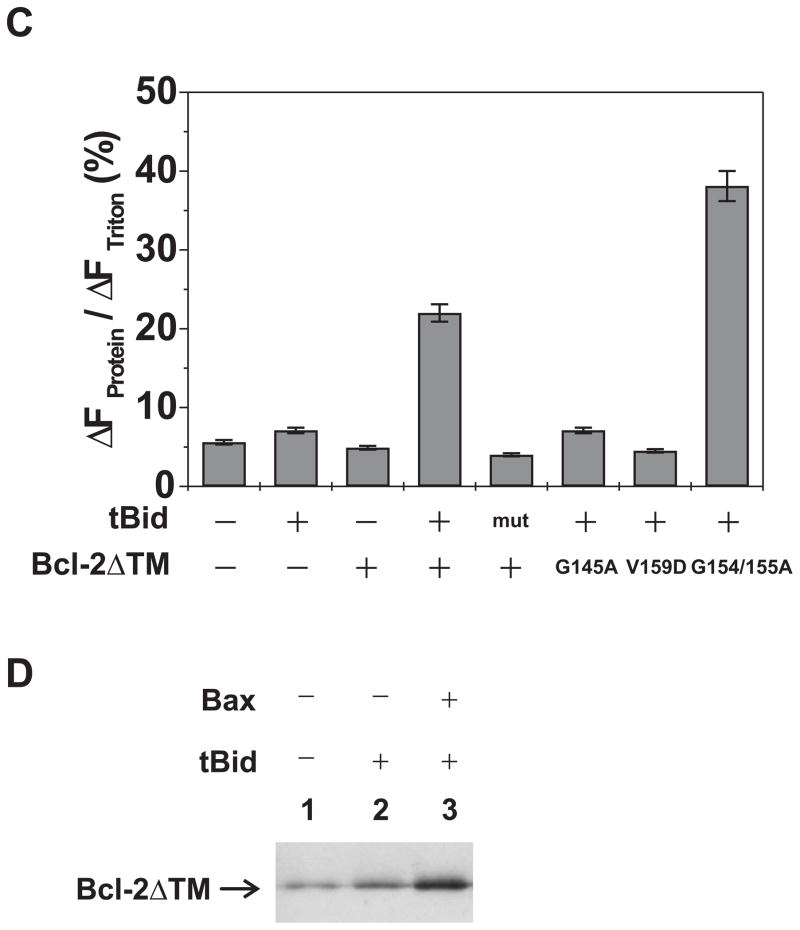
Fig. 3. Effect of tBid on the pore formation by soluble and membrane-tethered Bcl-2ΔTM.
A. Extent of CB dye release by the buffer control (minus tBid and Bcl-2), the soluble Bcl-2ΔTM and/or tBid at pH 7.4 was monitored as above. Data shown are averages of 2–3 independent experiments with S.D. (error bars). B. Binding of the C-terminally His6-tagged Bcl-2ΔTM with regular or Ni-liposome (with 0 or 5% Ni+-lipid, respectively) in the absence or presence of EDTA or imidazole was assayed by the float-up centrifugation. Data shown are immunoblots of the Bcl-2ΔTM that was recovered from S1 fraction after the centrifugation. Similar results were obtained in other two independent repeats. C. Extent of CB dye release by the buffer control, tBid or mutant (M97A/D98A, mut), and/or His6-tagged Bcl-2ΔTM or mutant (G145A, V159D or G154/155A) from Ni-liposome at pH 7.4 was monitored as above. Data shown are averages of 2–3 independent experiments with S.D. (error bars). D. Insertion of Bcl-2ΔTM into the membrane was monitored by the float-up centrifugation after incubating the protein with Ni-liposomes in the absence or presence of tBid and/or Bax and then treating with EDTA. Data shown are representative immunoblots of Bcl-2ΔTM bound to the liposomes in S1 fraction of the float-up centrifugation. Similar data were obtained in other two independent experiments.
Assay of CB or CB-dextran release from liposomes by fluorescence quenching
For Figure 2C, 50 μM liposomes with CB dyes encapsulated were mixed with 6 μg/ml anti-CB antibodies in 250 μl of buffer A of certain pH that was adjusted by varying the concentration of Na2HPO4 and citric acid. The initial emission intensity (F0) was determined after equilibrating the sample at 25 °C for 5 min. Purified Bcl-2ΔTM protein (400 nM) was then added. For other figures, if indicated, 12.5 μM liposomes with CB dyes or CB-dextrans were added, and after F0 was taken, 50 nM Bcl-2ΔTM or mutant protein, 20 nM tBid or mutant protein, and/or 50 nM Bax were added. The first fluorescence intensity measurement was started exactly 20 sec after the addition of the protein(s) and followed by multiple measurements in a predetermined time interval for 5 hr, resulting in multiple intermediate intensities (F). At the end of the time course 0.1% Triton X-100 was added and the final measurement was taken, resulting in the final intensity (Ft). The extent of CB or CB-dextran release is proportional to the extent of fluorescence quenching that is equal to ΔFProtein /ΔFTriton, where ΔFProtein = F0 − F, and ΔFTriton = F0 − Ft. All fluorescence intensities were measured using SLM-8100 fluorometer as described(31).
Collisional quenching of soluble NBD-labeled Bcl-2ΔTM by iodide ions
NBD (7-nitrobenz-2-oxa-1,3-diazole) labeling of Bcl-2ΔTM with a single Cys (Cys158) was done as described(40). The NBD fluorescence emission scan was carried out at 1 nm interval between 500 and 600 nm with the excitation at 468 nm. The spectrum was integrated to determine the intensity. To determine the pH effect on iodide ion accessibility to the NBD, the initial net fluorescence intensity (F0) for a sample with 100 nM NBD-Bcl-2ΔTM was determined in buffer A of indicated pH by subtracting the signal of a control without the NBD-Bcl-2ΔTM. The sample and control were titrated with aliquots of 1 M KI and 1 mM Na2S2O3. The intensities of sample and control were measured after each addition of KI/Na2S2O3 and corrected for dilution to obtain the net intensity (F). In parallel, another pair of sample and control was titrated with 1 M KCl and 1 mM Na2S2O3 for correcting the effect of ionic strength on the NBD fluorescence. In each experiment, the net change in fluorescence due to the quenching at each KI/KCl concentration was determined by the equation F0/F = (F0/F)KI/(F0/F)KCl, which was used to plot against the KI concentration to obtain the Stern-Volmer plot as described(40).
Fluorescence intensity of NBD-labeled Bcl-2ΔTM inserting into liposomal membranes
The initial fluorescence intensity (F0) was determined in absence of liposomes with 100 nM NBD-Bcl-2ΔTM in buffer A of indicated pH. Emission intensity (F) was monitored exactly 20 sec after addition of 100 μM liposomes to the sample, and continuously monitored until the intensity reached a plateau. The liposomes used are either the plain liposomes or the liposomes containing lipophilic quencher 7-doxyl-PC.
Other fluorescence methods
The tryptophan fluorescence intensity and anisotropy of Bcl-2ΔTM, and the anisotropy of carboxyfluorescein (CF)-labeled Bax H2 peptide were measured as described(31).
Assay Bcl-2 mutants in Rat-1MycERTAM cell line
Rat-1MycERTAM cell culture, retroviral infection and apoptosis induction were conducted as described(37,41,42). Briefly, the Rat-1 cells stably expressing Bcl-2, Bcl-2 mutant or vector were first treated with tamoxifen that induces the c-myc protooncogene activity and sensitizes the cells to apoptotic signals. The cells were then treated with either 6 μM etoposide for 12 and 18 hr or low serum (0.03% fetal bovine serum) for 24 and 48 hr. The cleavage of poly(ADP-ribose) polymerase (PARP) by caspases was used to assess the drug-induced apoptosis as described(29). The labeling of Cys158 in helix 5 by 4-acetamido-4′-[(iodoacetyl)amino]stilbene-2,2′-disulfonic acid (IASD), a bilayer-impermeant, sulfhydryl-specific reagent, was used to monitor the conformational alteration of Bcl-2 as described(29,37).
RESULTS
Acidic pH induces a conformational alteration in soluble Bcl-2ΔTM exposing the hydrophobic core yet retaining homo- and hetero-dimerization properties
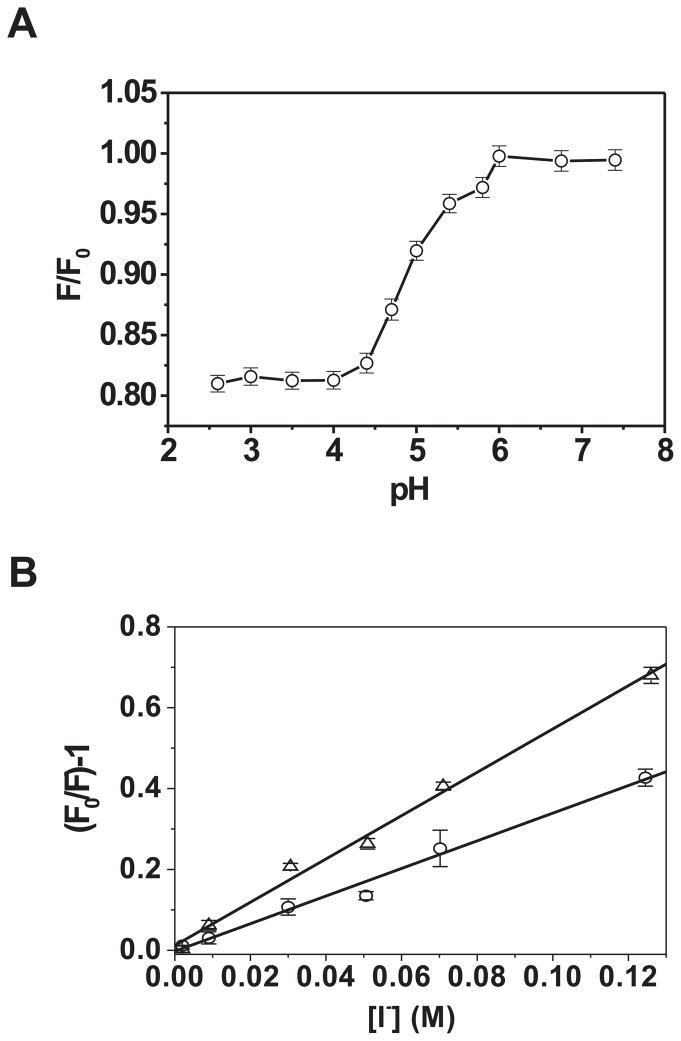
Acidic pH has been shown to induce Bcl-2ΔTM pore formation in membranes. The mechanism is unclear but it was suggested that acidic pH could induce a conformational alteration in the homologous Bcl-xL(43). We thus monitored the conformation of Bcl-2ΔTM in solutions of different pH by measuring tryptophan fluorescence of the protein. The tryptophan fluorescence provides information about the protein conformation since it is sensitive to the environment of tryptophan residues in the protein structure. Bcl-2ΔTM contains six tryptophan residues most of which are buried in the protein core at neutral pH(44). If reducing pH changes the protein conformation and exposes the tryptophan residues to water, the tryptophan fluorescence will decrease. As shown in Figure 1A, a pH lower than 6.0 significantly decreased the tryptophan fluorescence of Bcl-2ΔTM. About 50% of maximum fluorescence reduction was observed at pH 5.0, indicating that the protein is partially unfolded at the acidic pH required for the protein to form pores in membranes (see Fig. 2C). Of note, the tryptophan fluorescence itself is not sensitive to the range of pH change used here(45).
Fig. 1. Effect of pH on Bcl-2ΔTM conformation, homo-binding and hetero-binding with Bax peptide.
A. The pH-dependent conformational alteration of soluble Bcl-2ΔTM was assessed by monitoring the tryptophan fluorescence intensity in solutions of various pH. Data shown are averages of relative intensities (F/F0) from three independent measurements (○) with standard deviations (S.D.; error bars). F0, the intensity measured at pH 7.4; F, the intensity measured at other pH. B. Solvent exposure of the hydrophobic helix 5 of Bcl-2ΔTM was assessed by measuring I− quenching of fluorescence intensity of an NBD dye attached to Cys158 in helix 5. F0 is the net fluorescence intensity prior to addition of KI, and F the dilution-corrected fluorescence intensities after addition of indicated concentrations of KI. Data shown are averages of fluorescence quenching (F0/F-1) from two independent experiments conducted either at pH 7.4 (○) or 5.0 (△) with S.D. (error bars). The linear least-squares best-fit lines are also shown. C. Bcl-2ΔTM homo-binding in solution was monitored by measuring the tryptophan fluorescence anisotropy (rW) at various protein concentrations. Data shown are averages from three independent titrations conducted either at pH 7.4 (○) or 5.0 (△) with S.D. (error bars) and nonlinear least-squares best-fit curves (dash or solid) that were generated using the quadratic equation as described(31). D. Binding of Bax H2-H3 peptide to Bcl-2ΔTM at pH 5.0 was monitored by measuring the CF fluorescence anisotropy (rCF) of a CF-labeled Bax H2 peptide after titrating Bcl-2ΔTM. Data shown are averages from three independent titrations with S.D. (error bars), conducted either in the absence (□) or presence of an unlabeled Bax H2-H3 peptide (○) or its mutant (G67R) (△). Also shown are the nonlinear least-squares best-fit curves for the former two data sets that were generated using the cubic and quartic equations, respectively, as described(31).
To assess if the putative pore-forming helix 5 in the hydrophobic core of Bcl-2ΔTM is exposed to solvent by the acidic pH, we used a Bcl-2ΔTM derivative with an NBD fluorescent probe attached to the helix 5 via the sulfhydryl group of Cys158, the only cysteine in the protein. We monitored the quenching of the NBD fluorescence in a buffer of pH 5.0 or 7.4 by iodide ions. Iodide ions are charged and hence cannot diffuse into the protein hydrophobic core to collide with the NBD and quench its fluorescence, one thus expects to see more quenching for the more solvent-exposed probe(46). As shown in Figure 1B, more quenching was detected for the NBD-labeled Bcl-2ΔTM in the pH 5.0 buffer than that in the pH 7.4 buffer, indicating that the NBD at Cys158 in helix 5 is more exposed to solvent at the acidic pH. The NBD as well as tryptophan fluorescence data thus suggest that the conformational alteration of Bcl-2ΔTM induced by acidification elicits an exposure of the core helix 5 to solvent.
We recently reported that at neutral pH Bcl-2ΔTM associated with other Bcl-2ΔTM and with a peptide containing the sequence of Bax helices 2 and 3 (designated H2-H3 peptide) (31). Using the fluorescence anisotropy-based binding assay as described(31), we investigated whether the partially unfolded Bcl-2ΔTM at pH 5.0 retained similar binding properties. As shown in Figure 1C, the isotherm of Bcl-2ΔTM homo-binding determined at pH 5.0 was very similar to that determined at pH 7.4, indicating that the acidic pH-unfolded proteins interacted similarly to the folded proteins. The Bcl-2ΔTM protein also bound to the H2-H3 peptide in the pH 5.0 buffer with an isotherm similar to that obtained at neutral pH (compare Fig. 1D with the Fig. 4B in(31)). More importantly, a mutation in the peptide (G67R) that eliminated the binding at pH 7.4 also eliminated the binding at pH 5.0, suggesting that the partially unfolded protein still retains the ligand selectivity of the folded protein. These results indicate that the acidic pH-unfolded Bcl-2ΔTM protein still retains certain important properties of the folded protein.
Fig. 4. Effect of tBid, Bax and/or membrane-tethered Bcl-2ΔTM on pore formation.
Extent of CB-dextran release by the buffer control, tBid or mutant, Bax and/or His6-Bcl-2ΔTM or mutants from Ni-liposome was monitored as above. Data shown are averages of 2–3 independent experiments with S.D. (error bars).
Acidic pH induces insertion of helix 5 of Bcl-2ΔTM into liposomal membranes resulting in pore formation
Since the acidic pH induces the exposure of the hydrophobic core helix 5 to water, it may enhance binding of the protein to liposomal membranes. We monitored Bcl-2ΔTM binding with MOM-liposomes after an incubation at pH 5.0 or 7.4 using a sucrose gradient float-up centrifugation to separate the membrane-bound from the soluble and aggregated proteins(39). As shown in Figure 2A, a significant fraction of total Bcl-2ΔTM co-fractionated with the liposome after the incubation at pH 5.0. In contrast, liposome-bound protein was not detected after incubation at pH 7.4.
We used the Bcl-2ΔTM protein with an NBD attached to Cys158 in helix 5 to see if the helix is inserted into the liposomal membrane at the acidic pH since the NBD fluorescence will increase when the fluorophore is moved from aqueous milieu to hydrophobic bilayer(46). As shown in Figure 2B, the NBD fluorescence was increased by about 50% after the protein was incubated with the liposomes at pH 5.0. Furthermore, the fluorescence increase was quenched by a lipophilic quenching agent, 7-doxyl-PC, incorporated into the liposomal membrane, demonstrating a direct contact between the NBD and the hydrophobic core of lipid bilayer. As expected neither the increase nor the quenching of NBD fluorescence was observed when the incubation was done at pH 7.4 (Fig. 2B), consistent with the fact that Bcl-2ΔTM does not bind to the liposome at this pH (Fig. 2A). Therefore the acidic pH facilitates the membrane binding of Bcl-2ΔTM by partially unfolding the protein, exposing the hydrophobic core helix 5 to the liposome and eventually this helix inserts into the liposomal membrane.
To determine if the above Bcl-2ΔTM conformational alteration and helix 5 insertion results in pore formation in the liposomal membrane, we encapsulated fluorescent CB dyes into the MOM-liposome. Purified Bcl-2ΔTM protein was then incubated with the liposome in solutions of various pH. The Bcl-2ΔTM-dependent CB dye release from the liposome was monitored by quenching of the CB fluorescence by anti-CB antibody on the outside of the liposome. Since both CB dye and anti-CB antibody do not diffuse across the lipid bilayer spontaneously, quenching occurs only if the CB dye passes through the pore formed by Bcl-2ΔTM in the bilayer. In contrast the antibody is likely too large to pass through the pore. As shown in Figure 2C, no or very little Bcl-2ΔTM-dependent CB fluorescence quenching was observed when the protein was incubated with the liposome at pH 5.5 or higher, whereas maximal quenching was detected when the pH was lowered to 5.0 or less. Intermediate quenching was observed when the pH was between 5.0 and 5.5. Therefore pH of 5.0 or lower is required to induce a full pore-forming activity for Bcl-2ΔTM in the bilayer of MOM lipids, consistent with previous reports(32,33,35). Interestingly pH 5.0 also induces a conformational alteration in Bcl-2ΔTM that triggers insertion of the hydrophobic core helix 5 into the membrane (Fig. 1B and 2B), a reaction that most likely elicits pore formation.
tBid induces pore formation by membrane-tethered, but not soluble Bcl-2ΔTM
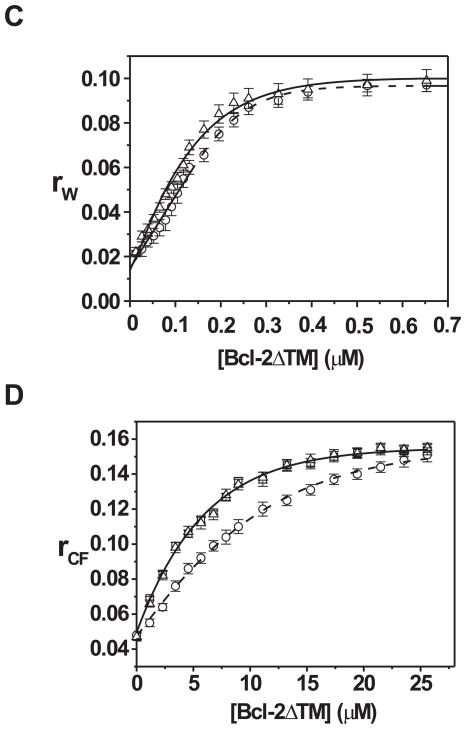
We recently reported that tBid can switch the Bcl-2 conformation at mitochondrion from the tail-anchored one to a conformation in which the helix 5 is inserted into the membrane(29). Since a similar helix 5 insertion also occurred during the acidic pH-induced Bcl-2ΔTM pore conformation in liposome, we tested if tBid induced Bcl-2ΔTM pore formation as well. However, when both tBid and Bcl-2ΔTM were incubated with the liposome with entrapped CB dyes at neutral pH, the fluorescence quenching was not increased compared to the buffer control or each protein alone (Fig. 3A). Therefore, tBid did not induce the pore formation by Bcl-2ΔTM.
One obvious difference between the Bcl-2 in mitochondria and the Bcl-2ΔTM in reactions containing liposomes is that the former is bound to the membrane via the C-terminal transmembrane sequence, whereas the latter is free in solution because it lacks the transmembrane sequence. To investigate the importance of membrane tethering in the tBid/Bcl-2 interaction, we tested whether Bcl-2ΔTM could be tethered to the liposomal membrane via a C-terminal His6-tag binding to a Ni-chelating lipid analog incorporated into the membrane. The His6-tagged Bcl-2ΔTM was incubated with either the Ni-chelating or regular liposomes at neutral pH and then subjected to the float-up centrifugation. As shown in Figure 3B, a large amount of the His6-tagged Bcl-2ΔTM was detected in the liposome-containing top fraction when the liposome contained the Ni-chelating lipid analog. Furthermore, the amount of His6-Bcl-2ΔTM associated with the Ni-liposome was greatly reduced if the incubation was done in presence of imidazole or EDTA. In contrast, the residual amount of His6-Bcl-2ΔTM bound to the regular liposomes was sensitive to neither imidazole nor EDTA treatment (data not shown). These results demonstrate that His6-Bcl-2ΔTM can be tethered to the Ni-liposome via His6-Ni interaction. Since the His6-tag is at the C-terminus of Bcl-2ΔTM and the Ni is at the surface of liposome, the liposome-tethered Bcl-2ΔTM topologically mimics the native mitochondrion-bound Bcl-2.
Using the liposome-tethered Bcl-2ΔTM we reinvestigated whether tBid can induce the pore formation. As shown in Figure 3C, neither Bcl-2ΔTM nor tBid alone released more CB dyes from the Ni-liposome than the buffer control did. In contrast, together they caused a significant dye release and hence the fluorescence quenching. A mutant tBid (M97A/D98A) that did not bind Bcl-2 also did not trigger the Bcl-2ΔTM pore formation(23,47). Similarly, a mutant Bcl-2ΔTM (G145A) that did not bind tBid also did not cause the dye release in the presence of tBid(47). Moreover, the mutation V159D shown previously to inhibit insertion of Bcl-2 helix 5 into membranes showed no function in the dye release assay(29). However, the hyperfunctional mutant Bcl-2ΔTM (G154A/155A; see Figure 5) triggered more dye release than Bcl-2ΔTM did. In the presence of tBid, more Bcl-2ΔTM became resistant to the EDTA treatment that removed the membrane-tethered Bcl-2 from the membrane (Fig. 3D), suggesting that tBid caused insertion of membrane-tethered Bcl-2ΔTM into the membrane. Together these results suggest that tBid interacts with the membrane-tethered Bcl-2ΔTM, causes the insertion of Bcl-2ΔTM and thereby elicits pores in the membrane. These results also demonstrate that tBid alone can induce the conformational alteration in the membrane-bound Bcl-2ΔTM, suggesting that no other mitochondrial proteins are required to mediate the tBid-induced Bcl-2 conformational alteration(29).
Fig. 5. Effect of mutations in helix 5 on the anti-apoptosis activity and conformational alteration for Bcl-2.
A. The anti-apoptotic activity of Bcl-2 or the G154/155A mutant was assayed after addition of etoposide for 12 (open bar) or 18 hr (shaded bar) or growth in low serum for 24 (open bar) or 48 hr (shaded bar) by measuring PARP cleavage. Rat-1MycERTAM cells were infected with packaged retroviral vector, or vector that expresses wild-type human Bcl-2 or Bcl-2-G154/155A, as indicated. Clones were selected that stably expressed equivalent amounts of the two proteins and then assayed for resistance to etoposide or serum starvation as indicated below the bars. Data shown are averages of three independent experiments with S.D. (error bars). B. The conformational alteration of Bcl-2 or the G154/155A mutant induced by the etoposide treatment or serum starvation was monitored by protection from IASD labeling assayed by isoelectric focusing followed by immunoblotting for human Bcl-2. Modification with IASD adds two negative charges at each cysteine modified and results in a discrete shift in mobility during isoelectric focusing. The cysteines available for modification in helix 5 and 9 are indicated as white triangles on diagram to the right of the panels. The migration position before IASD labeling (IASD, 0 min) indicates the migration position of the unlabeled protein (O). The migration position after labeling with detergent added to solubilize the membrane (Det) indicates the migration position of the protein labeled at both cysteines (**). Labeling of a single cysteine (Cys158 in helix 5) results in a band with intermediate migration (*; IASD, 10 min). In untreated cells Bcl-2 has not changed conformation and the major band (*) after IASD labeling represents protein in which Cys158 is labeled but Cys229 in helix 9 is protected unless detergent is added (Det). After treatment with etoposide or low serum the relative intensity of the unlabeled band (O) increases indicating protection of both cysteines from IASD. Addition of detergent renders both cysteines accessible to the reagent confirming that protection from IASD was due to insertion of the cysteines into the bilayer. The predicted topology of Bcl-2 with both cysteines protected from IASD is shown to the left of the panel with helices 9, 6 and 5 indicated.
When activated by tBid, Bcl-2ΔTM forms small pores, whereas Bax forms large pores
If the native mitochondrion-bound Bcl-2 forms pores in the MOM after it interacts with tBid, why don’t these pores release cytochrome c and other apoptotic proteins from the mitochondrial intermembrane space(29)? One possibility is that Bcl-2 forms pores that are smaller than these mitochondrial proteins. To asses whether the pore formed by the tBid-activated Bcl-2ΔTM is large enough to release cytochrome c, we incubated tBid and His6-Bcl-2ΔTM with Ni-liposomes that entrapped 10-kDa CB-dextrans, and monitored the release of CB-dextran by the fluorescence quenching assay. As shown in Figure 4, tBid and Bcl-2ΔTM together did not release more CB-dextrans than were released by the buffer control or each protein alone. Together with the data shown in Figure 3C, we demonstrated that after it was activated by tBid, the membrane-tethered Bcl-2ΔTM formed pores that can let 0.5-kDa CB dye but not 10-kDa CB-dextran pass through. Since the size of cytochrome c (12 kDa) is similar to that of 10-kDa CB-dextran, the Bcl-2 pore, if it forms in the native MOM after tBid treatment, is unlikely to release cytochrome c or other larger apoptotic mitochondrial proteins.
In contrast, tBid activated soluble Bax and released the 10-kDa or larger CB-dextrans from the liposomes (Fig. 4; data not shown), consistent with a previous report(20). Therefore, tBid induced the pore-forming activity of both anti-apoptotic Bcl-2 and pro-apoptotic Bax, but the consequences were different: the former formed a pore much smaller than cytochrome c, whereas the latter formed a pore much larger than cytochrome c.
The conformational alteration of membrane-tethered Bcl-2ΔTM can also be induced by active Bax, and is critical for Bcl-2 to inhibit active Bax
The Bcl-2 conformational alteration induced by tBid or another apoptotic stimulus is required for Bcl-2 to inhibit Bax-mediated cytochrome c release from mitochondria as we demonstrated recently(29). Since the Bcl-2ΔTM pore formation involved a similar conformational alteration and was induced by a similar stimulus, we tested whether the pore-forming conformer of Bcl-2ΔTM inhibits Bax pore formation. As shown in Figure 4, the membrane-tethered Bcl-2ΔTM significantly reduced the release of 10-kDa CB-dextran from the liposome by tBid-activated Bax. Interestingly, more EDTA-resistant Bcl-2ΔTM proteins were detected in the liposomes in the presence of both tBid and Bax than were observed when only tBid was added to liposomes with membrane-tethered Bcl-2ΔTM (Fig. 3D). This result suggests that tBid-activated Bax can also alter the conformation of membrane-tethered Bcl-2ΔTM.
To distinguish whether the inhibition of tBid-activated Bax is caused by the membrane-tethered or conformationally altered Bcl-2ΔTM (with Cys158 inserted into the bilayer), we employed a Bcl-2 mutant (V159D) that binds tBid but is defective in the conformational alteration induced by tBid(29). In accordance with our previous results in cells and isolated mitochondria, membrane-tethered Bcl-2ΔTM-V159D did not form pores in the liposome in response to added tBid (Fig. 3C). This mutant also exhibited reduced inhibition of pore formation by tBid-activated Bax (Fig. 4). In contrast, another Bcl-2ΔTM mutant (G154/155A) that had increased tBid-induced pore-forming activity provided a greater inhibition of pore formation by tBid-activated Bax (Fig. 3C and 4). These results suggest that the conformational alteration of Bcl-2-ΔTM induced by tBid is critical for Bcl-2-ΔTM to inhibit membrane permeabilization by Bax.
Since tBid-activated Bax also induced a conformational alteration of Bcl-2ΔTM (Fig. 3D), we tested whether this conformational alteration is also correlated with the Bcl-2ΔTM inhibition of Bax. For these experiments we used the tBid mutant (M97A/D98A) since it does not bind stably to Bcl-2 and did not lead to the membrane-tethered Bcl-2ΔTM adopting the pore conformation (Fig. 3C). However, this mutant activated Bax to permeabilize the membrane (Fig. 4), consistent with previous reports that the tBid mutant does not interact with Bcl-2 but interacts with Bax(23,47). The membrane-tethered Bcl-2ΔTM inhibited pore formation by the tBid mutant-activated Bax (Fig. 4). Moreover, addition of the mutant tBid and Bax increased the amount of Bcl-2-ΔTM that bound to liposomes in a fashion resistant to the addition of EDTA (data not shown), confirming that Bax activated by the mutant tBid induced the conformational alteration required for Bcl-2ΔTM to inhibit membrane permeabilization by Bax. In addition, the Bcl-2ΔTM mutants (V159D and G154/155A) that showed decreased and increased activities in the conformational alteration assays shown in Figure 3C also showed decreased and increased activities in inhibiting the tBid mutant-activated Bax, respectively (Fig. 4). Therefore, the conformational alteration of membrane-tethered Bcl-2ΔTM can be induced by either tBid or active Bax and plays a critical role in the inhibition of active Bax.
Conformational alteration activity in vitro and anti-apoptosis activity in vivo are correlated for two Bcl-2 mutants
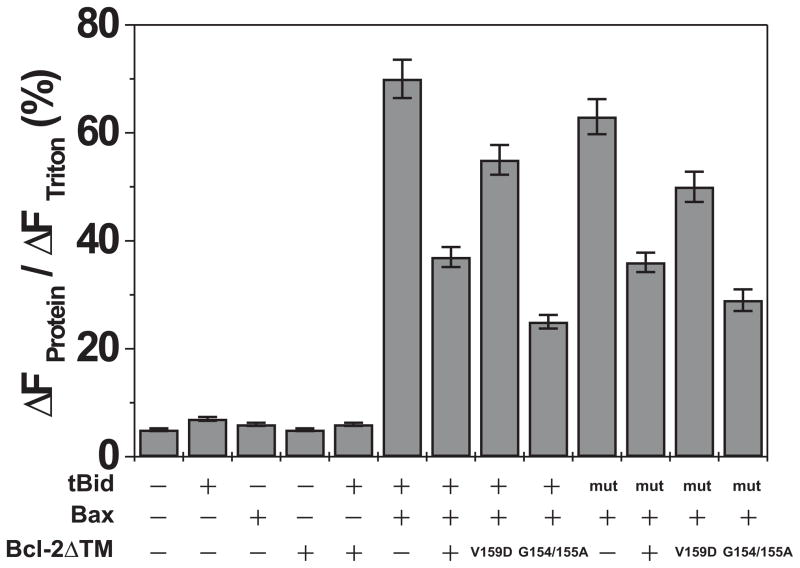
To assess further the physiological significance of the Bcl-2ΔTM conformational alteration in liposomes, we tested whether mutations in Bcl-2 that affect the tBid-induced pore formation in liposomes also affect anti-apoptosis activity in cells. Our previous study showed that the V159D mutation resulted in a hypo-functional Bcl-2 in cells(29). The V159D mutation abolished the pore-forming activity in liposomes (Fig. 3C) and inhibited the conformational alteration in both cells and isolated mitochondria(29), suggesting a positive correlation between the pore-forming activity, the conformational alteration and the anti-apoptosis function of Bcl-2.
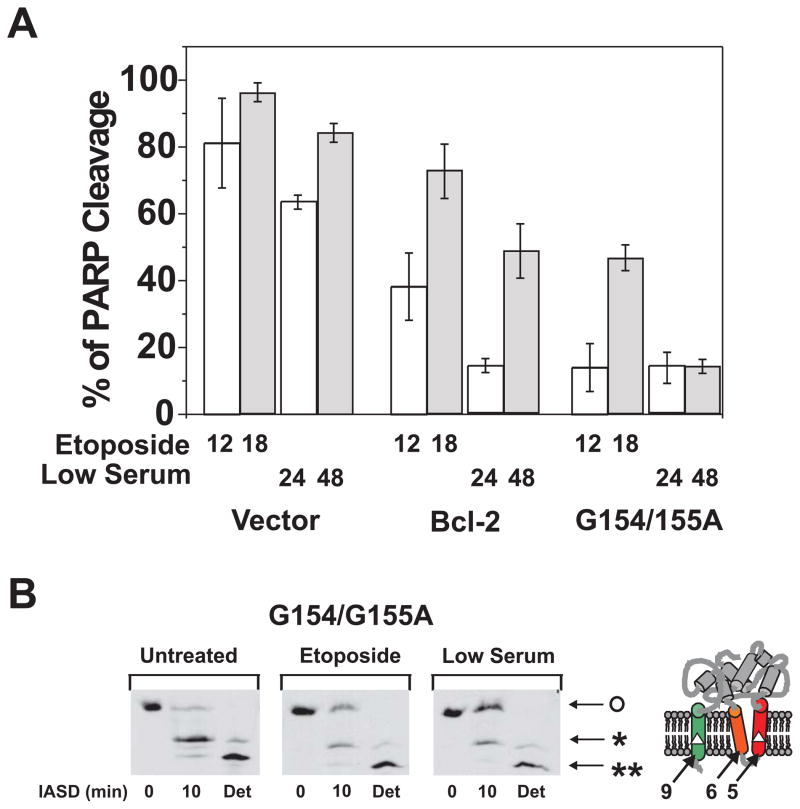
To further substantiate this correlation, we identified a Bcl-2 mutant, Bcl-2-G154/155A, that is hyper-active in forming pores in liposomes (Fig. 3C) and then assayed it for inhibition of apoptosis and for the accompanying change in conformation inside cells. To determine the anti-apoptosis activity of Bcl-2-G154/155A, it was expressed in Rat-1MycERTAM cells and then apoptosis was induced either by treating the cells with etoposide or by growth in low serum with activated c-myc as described(42). The anti-apoptosis activity Bcl-2-G154/155A was compared to wild-type Bcl-2 by measuring the extent of caspases-mediated PARP cleavage 12 or 18 hours after the cells were treated with etoposide, a DNA-damaging agent. The effects of growth in low serum with activated c-myc were assayed after 24 or 48 hours. Rat-1 cells expressing wild type Bcl-2 or empty vector were used as positive or negative control, respectively. As shown in Figure 5A, the mutant Bcl-2 had substantially higher anti-apoptotic activity (inhibition of PARP cleavage) than the wild type in both etoposide and low serum/activated myc-treated cells. We have shown elsewhere that in these cells the extent of PARP cleavage accurately reflects ongoing apoptosis measured as externalization of phosphatidylserine at the plasma membrane (Annexin V labeling) and chromatin condensation (Hoescht staining)(42).
Next, we monitored the conformational alteration of Bcl-2-G154/155A using the IASD labeling assay as described(29,37). As shown in Figure 5B, more protection of Cys158 in the mutant Bcl-2 from the labeling was observed in the membranes isolated from the etoposide-treated cells than the untreated cells, indicating that the mutant changed conformation in response to the apoptotic stimulus as reported previously for the wild type protein(29). The mutant displayed an even greater extent of conformational alteration (protection from labeling, indicated by o) and elevated anti-apoptosis activity in the Rat-1 cells cultured in the low serum media (Fig. 5A and 5B, low serum), suggesting that the conformation change correlates with anti-apoptotic activity for the mutant as reported previously for wild-type Bcl-2(29). Taken together the results from the hypo and hyper-functional mutants suggest a very strong correlation between the in vitro conformational alteration activity and the in vivo anti-apoptosis function of Bcl-2.
DISCUSSION
Whether native Bcl-2 forms pores in intracellular membranes is currently unknown. However, we previously found that native Bcl-2 changed conformation during the induction of apoptosis, resulting in insertion Cys158 of helix 5 into intracellular membranes, and that tBid could trigger the Bcl-2 conformational alteration in isolated mitochondria(29,37). Perhaps a direct interaction with tBid can trigger a conformational alteration in Bcl-2, promoting helix 5 insertion that may result in pore formation in intracellular membranes. Our data demonstrate that acidic pH increases the exposure of the internal helix 5 of Bcl-2ΔTM thereby promoting the insertion of the helix into the membrane. Acidic pH also induces some conformational alterations in Bcl-xLΔTM such as increasing solvent-exposure of hydrophobic sites while retaining the overall secondary structure(43), similar to what we observed for Bcl-2ΔTM. Therefore, it seems likely that the conformational alteration in Bcl-2 triggered by tBid involves exposure of helix 5 to promote integration into membranes.
However, tBid did not trigger the soluble Bcl-2ΔTM to form pores in the liposomal membrane. In addition to altering the conformation, acidic pH may also promote protonation of some ionizable residues such as glutamate, aspartate and histidine, as shown previously for the pore-forming diphtheria toxin and some membrane-inserting peptides(48–51). Notably there are four glutamates, one aspartate and one histidine in the helices 5 and 6 of Bcl-2. Protonation of these residues will reduce the free energy cost for insertion of the two putative pore-forming helices into the lipid bilayer. Thus, the combination of the conformational alteration and protonation of ionizable residues may account for the effect of low pH on insertion and pore formation by Bcl-2ΔTM.
Bcl-2ΔTM is the cytosolic domain of Bcl-2, and therefore it is not anchored onto membranes in our assay system using regular liposomes. Bcl-2 that is naturally anchored to an intracellular membrane via the C-terminal transmembrane sequence responds to interaction with tBid by inserting helix 5 into the membrane(29). Therefore, the additional effect of low pH compared to interaction with tBid may serve to promote membrane binding by Bcl-2ΔTM. When Bcl-2ΔTM was tethered to a liposomal membrane we have discovered that tBid directly interacts with the Bcl-2 cytosolic domain and alters its conformation. The conformationally altered Bcl-2 inserts the helix 5 into the membrane and forms small pores. Significantly, our results suggest that the major function of the constitutively transmembrane helix 9 in wild-type Bcl-2 may be only to hold the protein in close proximity to the membrane. Therefore, by using purified proteins and liposomes we have defined the minimal requirements for the Bcl-2 conformational alteration to be the membrane-bound Bcl-2 and tBid. Furthermore, since more conformationally altered Bcl-2 was generated in the presence of tBid and Bax than tBid alone, the tBid-activated Bax seems also able to induce the conformational alteration of Bcl-2.
We recently demonstrated that the Bcl-2 conformational alteration was required for Bcl-2 to inhibit apoptosis in cells and to inhibit active Bax in mitochondria(29), but we could not rule out the possibility that other mitochondrial proteins may be required either to induce the conformation change or for the conformationally altered Bcl-2 to inhibit Bax activity. Using the liposome system with minimal and defined components, we have shown that the conformationally altered Bcl-2 is sufficient and likely required to inhibit large pore formation by active Bax.
Based on this study and other studies(20,25,29,37), it is clear that tBid can independently activate both Bcl-2 and Bax changing them to what appears to be a similar multispanning transmembrane conformation. In the absence of the multispanning Bcl-2, the multispanning Bax forms large pores in the MOM to release cytochrome c and perhaps other mitochondrial proteins. In the presence of the multispanning Bcl-2, both multispanning proteins form hetero-complexes that do not permeabilize the mitochondrion. This suggests that the critical feature of BH3 mimetics with anti-Bcl-2 activity may be to inhibit the conformation change in Bcl-2 rather than simply blocking the hydrophobic binding surface on Bcl-2(52,53). It is interesting to speculate what structure the multispanning Bcl-2 may form in the MOM in the absence of the multispanning Bax. Our liposome data suggest that the multispanning Bcl-2 may form small pores in the MOM and this activity may intimately link to the anti-apoptosis activity. If the multispanning Bcl-2 indeed forms small pores in vivo, what would be the function of these pores? Are they pre-activated Bcl-2 traps waiting for active Bax? Do they affect the MOM permeability independent of inhibiting Bax? These intriguing questions about the functionally important multispanning Bcl-2 conformer must be addressed by future experiments before one can elucidate the molecular mechanism by which Bcl-2 inhibits apoptosis and develop better anti-Bcl-2 drugs for cancer therapy.
Acknowledgments
We thank Xiujun C. Zhang at Oklahoma Medical Research Foundation for making a structure-based prediction that certain mutations in helix 5 may affect the activity of Bcl-2.
Footnotes
The abbreviations used are: CB, Cascade Blue; CF, carboxyfluorescein; Bcl-2ΔTM, Bcl-2 lacking the C-terminal transmembrane sequence thus equivalent to the cytosolic domain; BH, Bcl-2 homology; 7-doxyl-PC, 1-palmitoyl-2-stearoyl-(7-doxyl)-sn-glycero-3-phosphotidylcholine; IASD, 4-acetamido-4′-[(iodoacetyl)amino]stilbene-2,2′-disulfonic acid; MOM, mitochondrial outer membrane; NBD, 7-nitrobenz-2-oxa-1,3-diazole; PARP, poly(ADP-ribose) polymerase; S.D., standard deviation.
This work was supported by American Heart Association Postdoctoral Fellowship 0420070Z (to ON), NIH grant GM062964 (to JL) and CIHR grant FRN 12517 (to DWA). DWA holds the Canada Research Chair in Membrane Biogenesis.
References
- 1.Danial NN, Korsmeyer SJ. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 2.Green DR, Kroemer G. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 3.Sharpe JC, Arnoult D, Youle RJ. Biochim Biophys Acta. 2004;1644:107–113. doi: 10.1016/j.bbamcr.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 4.Petros AM, Olejnicizak ET, Fesik SW. Biochim Biophys Acta. 2004;1644:83–94. doi: 10.1016/j.bbamcr.2003.08.012. [DOI] [PubMed] [Google Scholar]
- 5.Woo JS, Jung JS, Ha NC, Shin J, Kim KH, Lee W, Oh BH. Cell Death Differ. 2003;10:1310–1319. doi: 10.1038/sj.cdd.4401303. [DOI] [PubMed] [Google Scholar]
- 6.Day CL, Chen L, Richardson SJ, Harrison PJ, Huang DC, Hinds MG. J Biol Chem. 2005;280(6):4738–4744. doi: 10.1074/jbc.M411434200. [DOI] [PubMed] [Google Scholar]
- 7.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DCS. Mol Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 8.Cheng EH, Wei MC, Weiler S, Flavell JK, Mak TW, Lindsten T, Korsmeyer SJ. Mol Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 9.Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD. Mol Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Petros AM, Nettesheim DG, Wang Y, Olejnicizak ET, Meadows RP, Mack J, Swift K, Matayoshi ED, Zhang H, Thompson CB, Fesik SW. Protein Sci. 2000;9:2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu X, Dai S, Zhu Y, Marrack P, Kappler JW. Immunity. 2003;19:341–352. doi: 10.1016/s1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
- 12.Yan N, Gu L, Kokel D, Chai J, Li W, Han A, Chen L, Xue D, Shi Y. Mol Cell. 2004;15:999–1006. doi: 10.1016/j.molcel.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 13.Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW. Science. 1997;275(5302):983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z, Lapolla SM, Annis MG, Truscott M, Roberts GJ, Miao Y, Shao Y, Tan C, Peng J, Johnson AE, Zhang XC, Andrews DW, Lin J. J Biol Chem. 2004;279:43920–43928. doi: 10.1074/jbc.M406412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong SY, Gaume B, Lee YJ, Hsu YT, Ryu SW, Yoon SH, Youle RJ. EMBO J. 2004;23:2146–2155. doi: 10.1038/sj.emboj.7600225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen-Levy Z, Cleary ML. J Biol Chem. 1990;265:4929–4933. [PubMed] [Google Scholar]
- 17.Janiak F, Leber B, Andrews DW. J Biol Chem. 1994;269:9842–9849. [PubMed] [Google Scholar]
- 18.Hsu YT, Wolter KG, Youle RJ. Proc Nat Acad Sci USA. 1997;94:3668–3672. doi: 10.1073/pnas.94.8.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.O’Neill JW, Manion MK, Maguire B, Hockenbery DM. J Mol Biol. 2006;356(2):367–381. doi: 10.1016/j.jmb.2005.11.032. [DOI] [PubMed] [Google Scholar]
- 20.Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 21.Venturoli D, Rippe B. Am J Physiol Renal Physiol. 2005;288:605–613. doi: 10.1152/ajprenal.00171.2004. [DOI] [PubMed] [Google Scholar]
- 22.Antonsson B, Montessuit S, Lauper S, Eskes R, Martinou JC. Biochem J. 2000;345:271–278. [PMC free article] [PubMed] [Google Scholar]
- 23.Desagher S, Osen-Sand A, Nichols A, Eskes R, Montessuit S, Lauper S, Maundrell K, Antonsson B, Martinou JC. J Cell Biol. 1999;144(5):891–901. doi: 10.1083/jcb.144.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Arnoult D, Gaume B, Karbowski M, Sharpe JC, Cecconi F, Youle RJ. EMBO J. 2003;22:4385–4399. doi: 10.1093/emboj/cdg423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Annis MG, Soucie EL, Dlugosz PJ, Cruz-Aguado JA, Penn LZ, Leber B, Andrews DW. EMBO J. 2005;24:2096–2103. doi: 10.1038/sj.emboj.7600675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zha H, Aime-Sempe C, Sato T, Reed JC. J Biol Chem. 1996;271(13):7440–7444. doi: 10.1074/jbc.271.13.7440. [DOI] [PubMed] [Google Scholar]
- 27.Simonian PL, Grillot DA, Andrews DW, Leber B, Nunez G. J Biol Chem. 1996;271(50):32073–32077. doi: 10.1074/jbc.271.50.32073. [DOI] [PubMed] [Google Scholar]
- 28.Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, Adams JM, Huang DCS. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dlugosz PJ, Billen L, Annis MG, Zhu W, Zhang Z, Lin J, Leber B, Andrews DW. EMBO J. 2006;25:2287–2296. doi: 10.1038/sj.emboj.7601126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruffolo S, Shore GC. J Biol Chem. 2003;278:25039–25045. doi: 10.1074/jbc.M302930200. [DOI] [PubMed] [Google Scholar]
- 31.Tan C, Dlugosz PJ, Peng J, Zhang Z, Lapolla SM, Plafker SM, Andrews DW, Lin J. J Biol Chem. 2006;281:14764–14775. doi: 10.1074/jbc.M602374200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schendel SL, Xie Z, Montal MO, Matsuyama S, Montal M, Reed JC. Proc Natl Acad Sci USA. 1997;94:5113–5118. doi: 10.1073/pnas.94.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schlesinger PH, Gross A, Yin XM, Yamamoto K, Saito M, Waksman G, Korsmeyer SJ. Proc Nat Acad Sci USA. 1997;94:11357–11362. doi: 10.1073/pnas.94.21.11357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Minn AJ, Velez P, Schendel SL, Liang H, Muchmore SW, Fesik SW, Fill M, Thompson CB. Nature. 1997;385(6614):353–357. doi: 10.1038/385353a0. [DOI] [PubMed] [Google Scholar]
- 35.Kim KM, Giedt CD, Basanez G, O’Neil JW, Hill JJ, Han YH, Tzung SP, Zimmerberg J, Hockenbery DM, Zhang KYJ. Biochemistry. 2001;40:4911–4922. doi: 10.1021/bi002368e. [DOI] [PubMed] [Google Scholar]
- 36.Lam M, Bhat MB, Nunez G, Ma J, Distelhorst CW. J Biol Chem. 1998;273(28):17307–17310. doi: 10.1074/jbc.273.28.17307. [DOI] [PubMed] [Google Scholar]
- 37.Kim PK, Annis MG, Dlugosz PJ, Leber B, Andrews DW. Mol Cell. 2004;14:523–529. doi: 10.1016/s1097-2765(04)00263-1. [DOI] [PubMed] [Google Scholar]
- 38.Ho SN, Hunt HD, Horton RM, Pullen JK, Pease LR. Gene. 1989;77:51–59. doi: 10.1016/0378-1119(89)90358-2. [DOI] [PubMed] [Google Scholar]
- 39.Yethon JA, Epand RF, Leber B, Epand RM, Andrews DW. J Biol Chem. 2003;278:48935–48941. doi: 10.1074/jbc.M306289200. [DOI] [PubMed] [Google Scholar]
- 40.Shepard LA, Heuck AP, Hamman BD, Rossjohn J, Parker MW, Ryan KR, Johnson AE, Tweten RK. Biochemistry. 1998;37:14563–14574. doi: 10.1021/bi981452f. [DOI] [PubMed] [Google Scholar]
- 41.Zhu W, Cowie A, Wasfy GW, Penn LZ, Leber B, Andrews DW. EMBO J. 1996;15:4130–4141. [PMC free article] [PubMed] [Google Scholar]
- 42.Fiebig AA, Zhu W, Hollerbach C, Leber B, Andrews DW. BMC Cancer. 2006;6(1):213. doi: 10.1186/1471-2407-6-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xie Z, Schendel SL, Matsuyama S, Reed JC. Biochemistry. 1998;37:6410–6418. doi: 10.1021/bi973052i. [DOI] [PubMed] [Google Scholar]
- 44.Petros AM, Medek A, Nettesheim DG, Kim DH, Yoon HS, Swift K, Matayoshi ED, Oltersdorf T, Fesik SW. Proc Natl Acad Sci USA. 2001;98:3012–3017. doi: 10.1073/pnas.041619798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lakowicz JR. Principles of fluorescence spectroscopy. 2. Kluwer Academic/Plenum Publishers; New York: 1999. pp. 488–489. [Google Scholar]
- 46.Johnson AE. Traffic. 2005;6(12):1078–1092. doi: 10.1111/j.1600-0854.2005.00340.x. [DOI] [PubMed] [Google Scholar]
- 47.Wang K, Yin XM, Chao DT, Milliman CL, Korsmeyer SJ. Genes Dev. 1996;10:2859–2869. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- 48.O’Keefe DO, Cabiaux V, Choe S, Eisenberg D, Collier RJ. Proc Natl Acad Sci USA. 1992;89:6202–6206. doi: 10.1073/pnas.89.13.6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ren J, Sharpe JC, Collier RJ, London E. Biochemistry. 1999;38:976–984. doi: 10.1021/bi981576s. [DOI] [PubMed] [Google Scholar]
- 50.Lew S, Ren J, London E. Biochemistry. 2000;39:9632–9640. doi: 10.1021/bi000694o. [DOI] [PubMed] [Google Scholar]
- 51.Caputo GA, London E. Biochemistry. 2004;43:8794–8806. doi: 10.1021/bi049696p. [DOI] [PubMed] [Google Scholar]
- 52.O’Neill J, Manion M, Schwartz P, Hockenbery DM. Biochim Biophys Acta. 2004;1705(1):43–51. doi: 10.1016/j.bbcan.2004.09.004. [DOI] [PubMed] [Google Scholar]
- 53.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, Joseph MK, Kitada S, Korsmeyer SJ, Kunzer AR, Letai A, Li C, Mitten MJ, Nettesheim DG, Ng SC, Nimmer PM, O’Connor JM, Oleksijew A, Petros AM, Reed JC, Shen W, Tahir SK, Thompson CB, Tomaselli KJ, Wang B, Wendt MD, Zhang H, Fesik SW, Rosenberg SH. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]