Abstract
MSRA (EC 1.8.4.6) is a member of the methionine sulfoxide reductase family that can reduce methionine sulfoxide (MetO) in proteins. This repair function has been shown to protect cells against oxidative damage. In this study we have assembled the complete gene structure of msrA and identified the presence of two distinct putative promoters that generate three different transcripts. These transcripts were cloned by 5'RACE and code for three MSRA isoforms with different N-termini. The different forms of MSRA target to distinct intracellular regions. The main MSRA transcript (msrA1) had been previously shown to target the mitochondria. MsrA2 and 3 originate from a second promoter and target the cytosol and nuclei. In the monkey retina msrA message was detected mainly in the macular RPE-choroid region while its activity was measured mainly in the soluble fractions of fractionated neural retina and RPE-choroid. The MSRA protein is found throughout the retina but is especially abundant at the photoreceptor synapses, ganglion and Müller cells. Interestingly, MSRA was not detected in the mitochondria of the photoreceptor inner segments. The RPE in the peripheral retina shows very low levels of expression but the RPE in the macular region is strongly labeled. Targeted silencing of msrA message rendered cultured RPE cells more sensitive to oxidative damage suggesting a role for MSRA in RPE protection against oxidative stress. Collectively these data suggest MSRA may play an important role in protecting macular RPE from oxidative damage.
Keywords: retina, macula, methionine sulfoxide reductase, gene structure, mitochondria
1. Introduction
Methionine sulfoxide reductase (MSRA; EC 1.8.4.6) is an enzyme capable of reducing the highly oxidizeable surface exposed methionines in proteins back to methionine (Weissbach et al., 2002). This seems to be a critical process in reversing oxidative damage to proteins and may play an important role in protecting cells against oxidative damage. MSRA was initially discovered in Escherichia coli (Brot et al., 1981), and orthologs have been identified and cloned in many species (Weissbach et al., 2002; Lowther et al., 2000). MSRA is widely expressed in mammalian tissues (Moskovitz et al., 1996a) and was previously reported to target the mitochondria (Hansel et al., 2002) and the protein was detected in cytosolic fractions (Vougier et al., 2003). MSRA was shown to be stereospecific reducing the S enantiomer of methionine sulfoxide (Met(o)) in proteins (Sharov et al., 1999; Moskovitz et al., 2000). More recently, other members of the MSR family have been identified, called MSRB, which reduce the R enantiomer of Met(o) in proteins (Grimaud et al., 2001; Moskowitz, 2005). In addition to its protein repair function the MSR system may be acting to scavenge reactive oxygen species by way of methionine oxidation and reduction (Levine et al., 1999; Stadtman et al., 2002). Evidence for a role of MSRA in protecting cells against oxidative damage was first shown in Escherichia coli where MSRA mutants were more sensitive to H2O2 (Moskovitz et al., 1995; St. John et al., 2001). Studies in animals have supported these earlier studies. Mutant mice lacking msrA gene expression were found to have reduced lifespan, to be susceptible to oxidative stress and to have neurological defects (Moskovitz et al., 2001). Moreover, overexpression of MSRA in fruit fly neurons prolonged their lifespan and fertility as well as increased their resistance to paraquat-induced oxidative stress (Ruan et al., 2002). MSRA protected human lens cells against oxidative stress damage implicating MSRA in lens maintenance and cataractogenesis (Kantorow et al., 2004). These results suggest that MSRA expression may be important in the aging process and age-related diseases including eye diseases (see reviews by Berlett and Stadtman, 1997; Weissbach et al., 2005; Moskowitz, 2005; Petropoulos and Friguet, 2005).
The eye is an organ system particularly susceptible to aging diseases as demonstrated by the high incidence of cataracts (Hejtmancik and Kantorow, 2004), glaucoma (McKinnon, 2003) and age-related macular degeneration (Bonnel et al., 2003). Age-related macular degeneration (AMD) is a complex disease that involves genetic and environmental factors (Berger et al., 1999). This disease affects the macular region of the human retina and is responsible for the greatest number of cases of legal blindness among the elderly (Bonnel et al., 2003; Evans, 2001). Although the pathogenesis of AMD is not well understood, loss of function of the retinal pigment epithelium (RPE) supporting the macula is suspected of being responsible for photoreceptor loss. Oxidative damage has not been directly linked to AMD; however, its potential association with the aging process makes MSRA an interesting protein to study in the retina. MSRA has been found to be highly expressed in lens epithelium cells (Kantorow et al., 2004) and in the RPE (Moskovitz et al., 1996).
The macula is a retinal structure that among mammals is unique to primates. The cornea and the lens serve to focus light on the macular and this structure is responsible for the exquisite visual acuity enjoyed by primates (Oyster, 1999). The human macula is approximately 5 mm in diameter and at the center of this structure is the fovea. The fovea is composed entirely of cone photoreceptors and is completely dependent on the choroidal vasculature (Oyster, 1999). The neural macula is morphologically different from the neural peripheral retina in many ways. In the macula the photoreceptor density is greater as well as the number of supporting cells, especially the ganglion cells. There is also a greater ratio of cone to rod photoreceptors in the macula. The peripheral retina has a lower density of cone photoreceptors. In contrast, there are no obvious morphological changes in the RPE under the macula although the choriocapillaris is significantly denser providing a wider surface for exchange with the RPE. The structural and metabolic difference between these two areas of the retina may be the reason why the macula is more susceptible to aging and possible oxidative damage.
In this study, we have examined the expression, localization and activity of MSRA and its isoforms in monkey retina. We have elucidated the organization of the human gene and identified two putative promoters that express three different isoforms of msrA. We also provide evidence that MSRA is important for RPE resistance to oxidative stress damage.
2. Materials and methods
2.1. Monkey retinal tissue
Fresh eye tissue was obtained from rhesus monkeys (Macacca mulatta, 2–3 years old) through the courtesy of the Center for Biologics Research and Testing, U.S. Food and Drug Administration (Bethesda, MD). All animal studies were conducted in accordance with the NIH guidelines for the care and use of animal in research. Monkeys were euthanized and enucleated within minutes after death. The eyes were placed on ice or in ice-cold fixative. Macular and peripheral tissue was obtained using a 5 mm trephine. The neural retina and the RPE-choroid were carefully separated and the tissues were frozen at −80 °C before further processing. Each retinal sample was from a pool of 10 eyes.
2.2. 5' Rapid amplification of cDNA ends (5'RACE)
Rapid amplification of cDNA ends was performed on solid phase cDNA synthesized on magnetic beads (Dynal Inc, Oslo, Norway) as previously described (Rodriguez et al., 1994). The amplified fragments were cloned and sequenced.
2.3. DNA sequencing
Plasmids were purified (Plasmid Midi kit, Qiagen Inc., Valencia, CA) and sequenced using the BigDye Terminator v3.1 Cycle Sequencing kit (BigDye 3.1 kit, Applied Biosystems, Foster City, CA) and an Applied Biosystems Model 377 automate fluorescent DNA sequencer (Applied Biosystems, Foster City, CA).
2.4. Anti-MSRA antibodies
Anti-MSRA was obtained from Upstate LLC (Charlottesville, VA). The antibody is generated in rabbits immunized with a GST fusion protein containing amino acids 1–233 of the bovine MSRA as previously described (Moskovitz et al., 1996).
2.5. Cell culture
ARPE19 cells were purchased from American Type Culture Collection (Manassas, VA). The cells were cultured in DMEM/F12 (1:1) medium containing 10% fetal bovine calf serum (Atlanta Biological Inc., Atlanta, GA), 2 mM glutamine, 100 IU/ml penicillin, and 100 μg/ml streptomycin (all from Invitrogen Corp., Carlsbard, CA).
2.6. Preparation of GFP fusion constructs
The different msrAs were amplified from human retina cDNA using the forward primers 5′-AGCATGCTCTCGGCCACCCGGAGG-3′ (msrA1), 5′-AGCATGGCTGTATTTGGAATGGG (msrA2), 5′-AGAATGTGTTCAGAACCCAAACAT-3′ (msrA3), and the reverse primer 5′-CTTTTTTAATACCCACT GGGCAGG-3′. The PCR products were cloned into the pcDNA3.1/CT-GFP-TOPO vector from Invitrogen Corporation (Calrsbad, CA). The colonies were screened by PCR using the CMV-F primer (5′-CGCAAATGGGCGGTAGGCGTG-3′) and an msrA reverse primer (5′-CTGTTCTGTTGCCATTGACAT-3′) to ensure selection of properly oriented clones. All of the plasmids were sequenced to verify authenticity and orientation before use. The pDsRed2-Mito plasmid used as a mitochondrial marker was purchased from Clontech (Palo Alto, CA).
2.7. SDS-polyacrylamide gel electrophoresis and immunoblotting analyses
The samples (20 μg protein each) were separated in 12% NuPAGE Novex Bis-Tris Gels running in 1X NuPAGE MOPS SDS Running Buffer at room temperature for 50 min at 200 V. Protein samples were mixed with 4× NuPAGE LDS Sample Buffer and 10× NuPAGE Reducing Agent and incubated at 65 °C for 10 min. The protein electrophoresis reagents and apparatuses were purchased from Invitrogen/NOVEX (Carlsbad, CA). The gels were transferred overnight onto a PROTRAN-nitrocellulose membrane (Schleicher & Schuell BioScience Inc., Keene, NH) using a Trans-Blot electrophoresis apparatus (Bio-Rad, Hercules, CA). The transfer was performed in NuPAGE Transfer Buffer and 10% methanol at 20 V, 4 °C overnight. The membrane was equilibrated in 1× Tris-Buffered Saline (TBS)/Tween 20 for 15 min, and blocked in 1× TBS, pH 7.4, 5% Carnation nonfat milk, and 0.05% Tween 20, for 2 hr. Incubations with anti-MSRA antibodies (Upstate, Lake Placid, NY) at 1:500 dilutions were performed overnight at 4 °C. The blots were developed using anti-rabbit IgG HRP conjugated secondary antibody at a dilution of 1:20,000 (Pierce, Rockford, IL) and imaged on X-ray film using SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL) after a 10–30 sec. exposure.
2.8. Cell transfection and imaging
The msrA-GFP fusion constructs were transfected into ARPE19 cells using the Amaxa Nucleofector kit (Amaxa Inc, Gaithersburg, MD) according to the manufacturer's protocol. ARPE19 cells were plated in two-well Lab-Tek chamber slides (Nalge Nunc International, Naperville, IL) and monitored for fluorescence 24 hr after transfection. Cells were imaged live using a Nikon TE2000-U inverted fluorescent microscope.
2.9. Subcellular fractionation of monkey retina tissue
The monkey neural retina was separated from the RPE-choroid and homogenized in 50 ml of 10 mM HEPES buffer, pH 7.2, containing 5 mM MgCl2, 4% (w/v) sucrose and Complete Protease Inhibitor at 1 tablet per 50 ml buffer (Roche, Mannheim, Germany). The homogenate was subjected to a low speed (300×g) centrifugation to separate the nuclei (P1) and the remaining supernatant was centrifuged at high speed (27,000×g) to pellet the subcellular organelles (P2). The P2 pellet was subsequently subjected to a 0.5 M NaCl wash (P2-salt) and a 2% Triton X-100 wash (P2-detergent). The remaining pellet (P2-residue) was not further processed. The supernatant (S1) and the proteins extracted from the P2 pellet (P2 salt and P2 detergent) were placed at 4 °C or frozen for further analysis.
2.10. Immunohistochemistry
Vibrotome retina sections were blocked using 1:10 diluted goat serum in ICC buffer containing 0.5% BSA, 0.2% Tween-20, and 0.05% sodium azide for 4 hr at 4 °C. The tissue sections were then incubated with rabbit anti-MSRA antibody (1:50) overnight followed by additional washes with PBS. The secondary antibody—goat anti-rabbit Cy5 (Jackson ImmunoResearch Lab., Inc., West Grove, PA) was applied at 1:1000 dilution for 4 hr. Isolectin GS-IB4 AlexaFluor 488 conjugate from Grifonia simplicifolia (Molecular Probes, Eugene, OR) was used to stain capillary endothelium cell at 1:500 dilution. The nuclei were counterstained with 4′, 6′-diamino-2-phenylindole (DAPI; 1 μg/ml in PBS). The slides were mounted (GelMount; Biomeda Corp., Foster City, CA) and kept in the dark until viewing.
2.11. Confocal microscopy
Confocal microscopy was performed on a laser scanning confocal microscope (model SP2, with TCS software version 11.04; Leica Microsystems, Exton, PA) and a 40× objective. The DAPI staining was visualized by exciting with 351 and 364 nm lasers and collecting emissions between 400 and 500 nm. Green fluorescence (lipofuscin) and AlexaFluor 488 staining were visualized by exciting with a 488 nm laser beam and collecting emissions between 500 and 552 nm. Cy5 staining was visualized by exciting with the 633 nm laser beam and collecting emissions between 650 and 750 nm. Magnifications varied, and scale bars are, therefore, digitally included in some of the pictures.
2.12. Northern blots
Total RNA was purified from monkey tissues using the RNeasy kit from Qiagen Inc. (Valencia, CA). The gel was stained with SYB green II (Molecular Probes Inc., Eugene, OR) then scanned using a Typhoon 9410 variable mode imager (Amersham Biosciences, Piscataway, NJ). The gel was blotted and probed with a 706 base pair msrA1 PCR product generated with the following forward 5′-AGCATGCTCTCGGCCACCCGGAGG-3′ and reverse 5′-CTTTTTTAATACCCACTGGGCAGG-3′ primers. Relative quantification was performed by normalizing the radioactive signal to the 28S SYB green II signal using ImageQuant Software (Molecular Dynamics, Seattle, WA) as previously described (Spiess and Ivell, 1999).
2.13. MSR enzymatic assay
The assay for MSR activity was based on the reduction of 3H-N-acetylmet-R,S-sulfoxide (N-acetylmet(o)), using 15 mM DTT as the reducing system, and specific MSRA activity was determined as previously described (Brot et al., 1982; Marchetti et al., 2005).
2.14. Detection of MSRA mRNA post- siRNA transfection by RT-PCR
RPE cells were plated in six-well plates at a density of 500,000 cells per well and transfected as previously described (Kantorow et al., 2004). GAPDH was used as a control, and was amplified for 15, 20 and 30 PCR cycles to ensure linearity using an annealing temperature of 60 °C. The msrA transcripts were amplified for 33 PCR cycles with an annealing temperature of 56 °C. All products were formed linearly over the number of PCR cycles indicated and sequenced to ensure authenticity.
2.15. TBHP sensitivity of control and siRNA treated cultured RPE cells
ARPE19 cells were plated in 96-well plates at a density of 20,000 cells per well and mock-transfected or transfected with 0.2 μg of siRNA per well 24 hr after seeding. The msrA knockdown was performed as previously described (Kantorow et al., 2004) using the following sequences (5′-CAAAGUACAAAGGAAUUUAUU-3′) and (5′-UAAAUUCCUUUGUACUUUGUG-3′). At 48 hr post-transfection the cells were treated with increasing concentrations of tertiary-butyl hydroperoxide (TBHP) for 24 hr in serum-free supplemented medium and cell viability was monitored using the MTS assay as described by the manufacturer (Promega, Madison WI).
3. Results
3.1. Structure of human msrA gene and transcriptional control
Assembling of msrA ESTs from the public databases showed the presence of five distinct cDNAs. Three of these had ORFs corresponding to the known msrA and differing by their 5′ end sequence and two had short unrelated ORFs. In order to see if these transcripts were present in retina we performed 5′ RACE using human retina cDNA as previously described (Rodriguez et al., 1994). Three different 5′ ends were cloned which corresponded to three of the cDNAs we assembled from the GenBank ESTs. These three distinct cDNAs encoded three peptides differing in their N terminus sequences (Fig. 1). These three peptides and cDNAs were named msrA1, 2 and 3. The sequences are represented by ESTs CD244734, 18170218 for msrA1, BI599246, AL536061 for msrA2, and BX461653, F06683 for msrA3. The corrected and annotated sequences for msrA1-3 cDNAs have been submitted to GenBank the accession numbers are as follows: AY958429 for msrA1, AY958430 for msrA2 and AY958431 for msrA3.
Fig. 1.
Alignment of MSRA peptides, transcript assembly and gene map. (A) Alignment of MSRA ORF peptides by the Jostun-Hein method (B) Splicing of the msrA transcripts. The exons are numbered and the splice joints are depicted by broken lines. msrA2 and 3 share exon 2–1 but msrA3 skips exon 2. All transcripts are identical after exon 3.
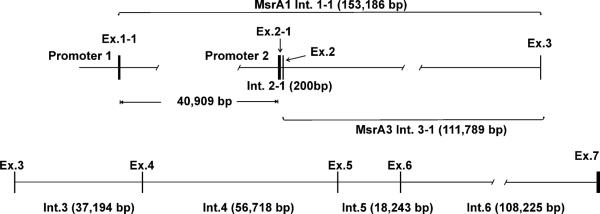
In order to verify the cDNAs found by 5′ RACE and the database ESTs, we assembled the gene sequence using genomic sequences available from GenBank with accession numbers AC034111, AC233085, AC079200, AC112673, and AC104964 which localized to human chromosome 8p23.1. These sequences and others assemble a longer contig of 741,580 bases. Comparing the cDNAs with the assembled gene sequence verified the three main transcripts found by 5′RACE and revealed two different transcription start sites 40,909 bases apart (Fig. 1, Table 1), suggesting two distinct promoters. The exons from all three transcripts showed consensus donor (AG) and acceptor (GT) splice sites (Fig. 1, Table 1). The two other transcripts mentioned above (represented by ESTs BX107824, AA44605, and BU663140, BG719999, respectively) that were found in the database but not by 5′RACE had nonconsensus splice sites. This suggests they may have been generated by aberrant splicing and were not studied further.
Table 1.
| Gene structure msrA1 | |||||||
|---|---|---|---|---|---|---|---|
| Exon # | Size (bp) | 5′ | 3′ | Intron # | Size (bp) | 5′ | 3′ |
| 1–1 | 390 | GAACTCGCCG.. | CCTGTAGCGG | 1–1 | 153 186 | gtaagcactg.. | ttttttctag |
| 3 | 69 | CCAAACATCA.. | GCTGTATTTG | 3 | 37 194 | gtaagatatc.. | cttttttaag |
| 4 | 120 | GAATGGGATG. | GTCTGCTCAG | 4 | 56 667 | gtaggaagaa.. | tttttccaag |
| 5 | 105 | AAAAAACTGG.. | CCGACCCAAG | 5 | 18 243 | gtagagtgat.. | gctttcctag |
| 6 | 107 | GTATGCGCCA.. | CTACCAAAAG | 6 | 108 225 | gtagggattg.. | gtccccacag |
| 7 | 742 | GTTCTTTCAG.. | CAATAATGCC | ||||
| Gene structure msrA2 | |||||||
|---|---|---|---|---|---|---|---|
| Exon # | Size (bp) | 5′ | 3′ | Intron # | Size (bp) | 5′ | 3′ |
| 2–1 | 488 | GAGAGAGAGA.. | TGTTCAGAAC | 2–1 | 200 | gtaagttttt..c | catcccccag |
| 2 | 75 | CAATTAGTGA.. | AACAAGAAAT | 2–2 | 111 514 | gtgagcttgg.. | ttttttctag |
| Gene structure msrA3 | |||||||
|---|---|---|---|---|---|---|---|
| Exon # | Size (bp) | 5′ | 3′ | Intron # | Size (bp) | 5′ | 3′ |
| 2–1 | 488 | GAGAGAGAGA.. | TGTTCAGAAC | 3–1 | 111 789 | gtaagttttt.. | ttttttctag |
Structure of the msrA gene showing the differences between the three main transcripts. After exon 3 all three msrAs mRNAs splice identically. The table shows the first 10 bases of the intron and exon junctions.
The msrA gene spans approximately 379 kb and seems to have two distinct promoters (Fig. 2, Table 1). The first promoter regulates the transcription of msrA1 and the second promoter regulates the transcription of msrA2 and 3. MsrA2 and 3 share exon 2–1 but msrA3 skips exon 2 (Table 1, Fig. 2) and forms a transcript with a slightly longer ORF than msrA2 (Fig. 1). The splicing after exon 3 is identical for all transcripts. The annotated gene sequence for msrA has been submitted to GenBank with accession number AY958432.
Fig. 2.
Scale map of the human msrA gene Scale map of the msrA gene (~379 kb) showing the intron and exon sizes and the splice distances of the first exons to exon 3 as well as the location of the putative promoters. msrA1 splices exon1–1 to exon 3 while msrA3 splices exon 2–1 to exon 3. msrA2 splices exon 2–1 to exon 2 then to exon 3.
3.2. Expression of msrA mRNA in monkey macula and peripheral retina
Northern blot analysis was performed on two different areas and four different tissues of the monkey retina. Total RNA was prepared from 5 mm trephine punches encompassing the fovea/macula and peripheral retina. The neural retina and the RPE/choroid tissue from each punch were separated and RNA prepared as described above. The blot was probed with an msrA specific probe generated by PCR. The relative levels of msrA in each tissue fraction were determined by normalizing the radioactivity signal to the SYB green II signal from the original gel as previously described (Spiess and Ivell, 1999) (Fig. 3). The results indicate that the ratio of msrA mRNA to total RNA is highest in the macular RPE/choroid fraction (MPC) than in the other tissues areas.
Fig. 3.
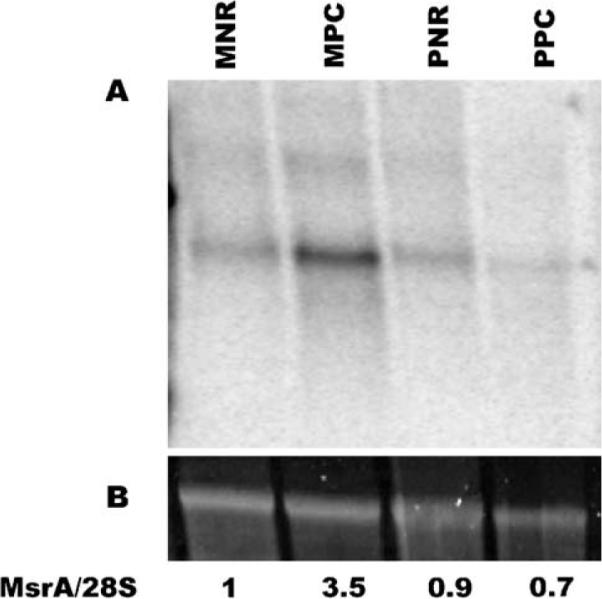
Northern blot analysis of msrA in macula versus peripheral retina. (A) Blot containing RNA from macular neural retina (MNR), macular pigment epithelium-choroid (MPC), peripheral neural retina (PNR), and peripheral pigment epithelium-choroid (PPC). The blot was probed with an msrA specific probe. Each lane contains approximately 5 μg of total RNA. For more details see Methods. (B) The 28S ribosomal RNA band stained with SYB green II. The relative ratio of msrA to 28S is shown below the lanes.
3.3. Detection of MSRA protein expression in monkey retina
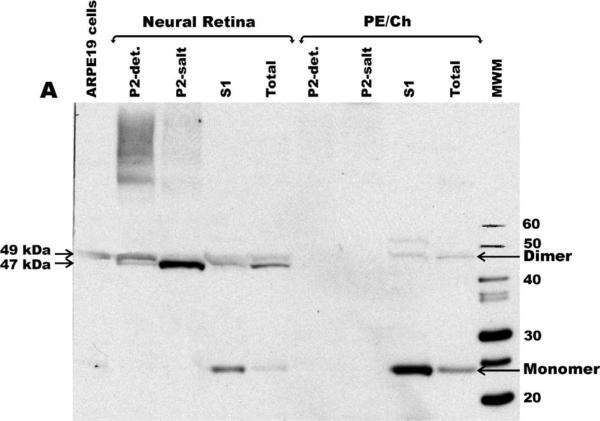
Fresh monkey retinas were dissected and the RPE and choroid (PE/Ch) tissues were separated from the neural retina tissue. Each tissue was then homogenized and the whole homogenate and subcellular fractions (prepared as described in Methods) were separated by SDS-PAGE followed by immunoblot analysis (Fig. 4). The S1 fractions contain soluble proteins while the P2 fractions contain cellular organelles and membrane associated proteins. The P2 fraction is subsequently extracted with 0.5 M NaCl (P2-salt) and then with 2% Triton X-100 (P2-det), see Methods. MSRA1 is normally detected as a ~26 kDa protein. However, in the unfractionated neural retina MSRA was detected mainly as a dimer in two bands of approximately 47 and 49 kDa (Fig. 4). Only a small amount of monomer was detected in the total unfractionated neural retina. In the fractionated neural retina MSRA was detected as a monomer mainly in the S1 fraction. In the P2-salt and P2-det fractions it was detected mainly as a dimer of 47 and 49 kDa respectively. In the unfractionated PE/Ch MSRA was detected both as a monomer and as a dimer. In the fractionated PE/Ch MSRA was detected in the S1 fraction mainly as a monomer and a small amount as the 49 kDa dimer. Very little MSRA was detected in the P2 fractions of the PE/Ch. In ARPE19 cultured cells MSRA was detected mainly as the 49 kDa band.
Fig. 4.
Immunoblot analysis of MSRA in the monkey retina and ARPE19 cells. MSRA in subfractionated monkey neural retina and retinal pigment epithelium-choriocapillaris (PE/Ch) tissues. The location of the monomer and dimers are shown with arrows.
3.4. MSRA enzymatic activity in monkey retina
MSR activity was measured on the same retina fractions used for immunoblot analysis (Table 2). The activity of MSRA and MSRB were determined as described in Methods. In the P2-detergent fractions the MSRA activity was corrected for the 50% inhibition caused by the 2% Triton X-100 used to extract the protein from the pellet. The MSRB activity was determined indirectly by inhibiting MSRA activity using p-tolyl-methylsulfoxide. In both neural retina and RPE-choroid tissues, most of the total MSR and MSRA activities were found in the S1 fractions. However, a significant amount of activity was also associated with the P2-det fraction suggesting interactions with organelles and membranes. The results are generally consistent with the immunoblot data.
Table 2.
MSRA activity in retinal fractions
| Fraction | Specific activity pmoles/mg | Total Protein (mg) | Total msr activity | Total MSRA activity | Relative% |
||
|---|---|---|---|---|---|---|---|
| MSRA | MSRB | ||||||
| Neural retina | S1-soluble | 241 | 36 | 8676 | 1995 | 23 | 77 |
| P2-0.5MNaCl | 99 | 6.3 | 624 | 405 | 65 | 35 | |
| P2-2%triton | 124 | 10.5 | 1302 | 885 | 68 | 32 | |
| RPE and choroid | S1-soluble | 180 | 13.5 | 2430 | 1871 | 77 | 23 |
| P2-0.5MNaCl | 122 | 3.9 | 476 | 190 | 40 | 60 | |
| P2-2%triton | 171 | 3.5 | 598 | 454 | 76 | 24 | |
The incubation conditions and assay are previously described (Brot et al., 1982; Marchetti et al., 2005) and in Methods. The relative amounts of MSRA and MSRB were determined using p-tolyl-methylsulfoxide as described in Methods. The % of MSRA and MSRB in the P2-det fractions have been corrected for the inhibition of the MSRA by the detergent present in these extracts. The pmoles formed are pmoles of N-acetyl-methionine.
3.5. Intracellular localization of MSRA1, 2 and 3
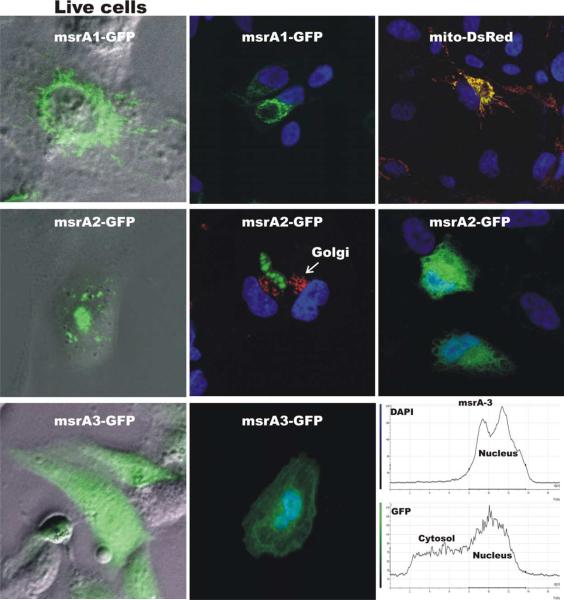
In order to determine the intracellular localization of the different forms of MSRA in RPE cells, green fluorescent protein (GFP) fusion constructs containing the MSRA in the N-terminus and GFP in the C-terminus were made using the Invitrogen pcDNA3.1/CT-GFP-TOPO vector. The msrAs were amplified from retina cDNA using different 5′ end primers reflecting the individual N-termini and a modified 3′ end primer that fused the msrA ORF with the GFP in the vector (see Methods). The three constructs were transfected into ARPE19 cells and the cells were photographed 24 hr later while still alive (Fig. 5). The cells were fixed and re-photographed by confocal microscopy after immunolocalization with known cellular markers (Fig. 5). As previously reported MSRA was localized to the mitochondria (Hansel et al., 2002) and cytosol (Vougier et al., 2003). This was also confirmed by co-transfection with pDsRed2-mito mitochondrial marker. MSRA2 localized to the cytosol in some cells and to unidentified vesicles. No localization was observed to lysosomes, endosomes, endoplasmic reticulum and golgi using specific markers (data not shown). MSRA3 localized to the cytosol and the nucleus.
Fig. 5.
Intracellular localization of MSRAs in ARPE19 cells. Image of msrA-GFP fusion constructs in transfected ARPE19 cells in the live cells and after fixation by confocal microscopy. MSRA1 demonstrated a clear mitochondrial localization in co-localizing with mito-DsRed. MSRA2 demonstrated two distinct localization patterns, a vesicular localization and a cytosolic localization. MSRA3 demonstrated a cytosolic and nuclear localization.
3.6. Immunohistochemical localization of MSRA in monkey macula and peripheral retina
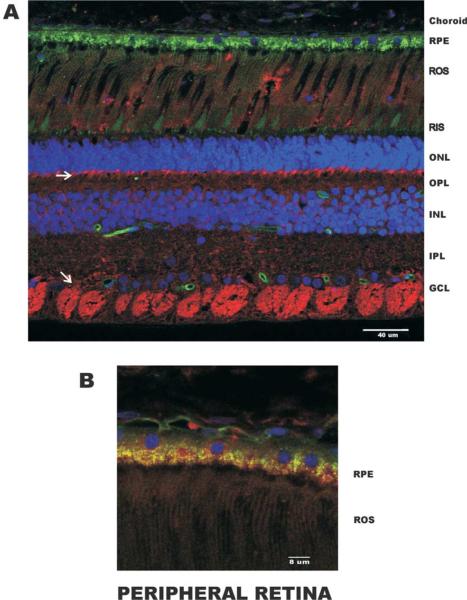
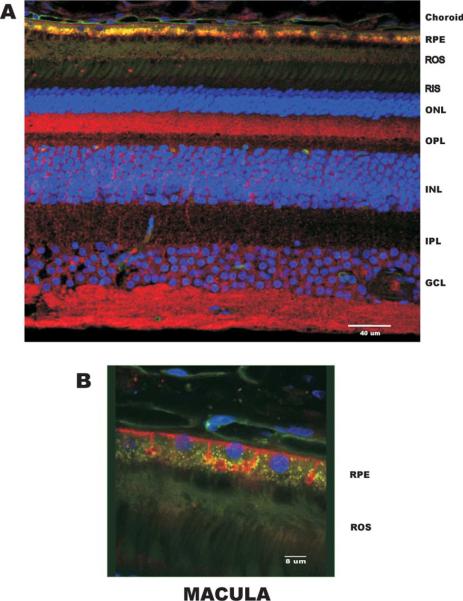
MSRA was localized in the monkey retina by confocal microscopy using 100 μm vibrotome sections (Figs. 6 and 7). The peripheral retina and macular region were examined. Capillaries were stained with isolectin B (Green Alexa 488) and the nuclei with DAPI (Blue). Anti-MSRA antibodies were localized using anti-rabbit Cy5 (Red)-conjugated secondary antibody. The image shown in Fig. 6(A) is a cross section of the peripheral retina which contains mostly rod photoreceptors and the number of ganglion cells is significantly lower than in the macula. The MSRA was localized throughout the retina but the most prominent labeling occurs in the ganglion cells and in the synapses between the photoreceptors and the bipolar and horizontal cells (see arrows, Fig. 6(A)). The peripheral RPE was lightly labeled in the apical side (Fig. 6(B)). The green fluorescence observed in the RPE is due to lipofuscin accumulation which is very common and pronounced in primates. There is some lipofuscin autofluorescence in the Cy5 channel but this was minimal and did not interfere with the stronger MSRA signal.
Fig. 6.
Localization of MSRA in peripheral retina. (A) Low magnification demonstrating the MSRA (Cy5, Red) localization across the peripheral retina. Staining was observed throughout the retina with the brightest staining in the retinal ganglion cells and in the rod outer segment synapses. Nuclei were stained with DAPI (Blue) and capillaries with Alexa 488-labeled isolectinB (Green). The green fluorescence in the RPE is the naturally occurring lipofuscin. (B) Higher magnification in the RPE. Choroid, choriocapillaris; RPE, retinal pigment epithelium; ROS, rod outer segments; RIS, rod inner segments; ONL, outer nuclear layer; OPL, outer plexiform layer; INL, inner nuclear layer; IPL, inner plexiform layer; GCL, ganglion cell layer.
Fig. 7.
Localization of MSRA in the macular region of the retina. (A) Low magnification demonstrating the MSRA (Cy5, Red) localization across the macular region. Staining was observed throughout the retina with the brightest staining in the retinal ganglion cells, Müller cells, photoreceptor synapses and RPE. (B) Localization in the RPE showing basal membrane staining. Labels are identical to Fig. 6.
In the macula (Fig. 7(A)) MSRA localized to similar areas as in the peripheral retina except for the RPE which was much more strongly labeled. In the macular RPE MSRA seems to associate with, but not exclusively to, the basal plasma membrane (Fig. 7(B)). The photoreceptor synapses are also strongly labeled in the macular region. These results are in agreement with the RNA blot data in Fig. 3.
3.7. Silencing of individual MSRA gene in retina cells results in loss of viability and decreased resistance to oxidative stress
Previously it has been shown that MSRA is required for human lens cell viability and provides oxidative stress resistance to cultured lens cells (Kantorow et al., 2004). To examine the role of MSRA on RPE cell viability and resistance to oxidative stress damage we used siRNA-mediated gene silencing to suppress the levels of the msrA transcripts from ARPE19 cells and subsequently measured the viability of the gene-silenced cells in the presence or absence of increasing TBHP concentrations (Fig. 8). The silencing of msrA resulted in increased sensitivity of the cultured RPE cells to TBHP as compared to mock transfected cell suggesting an essential role of MSRA in RPE cell resistance to oxidative stress. The effectiveness of the siRNA suppression of msrA mRNA was demonstrated by RT-PCR (Fig. 8, inset). The values were normalized to the untreated and untransfected control because an observed loss of cell viability after msrA RNA levels was reduced by siRNA treatment even without TBHP. Untransfected ARPE19 cells and mock transfected ARPE19 cells demonstrated similar TBHP sensitivity profiles (data not shown).
Fig. 8.
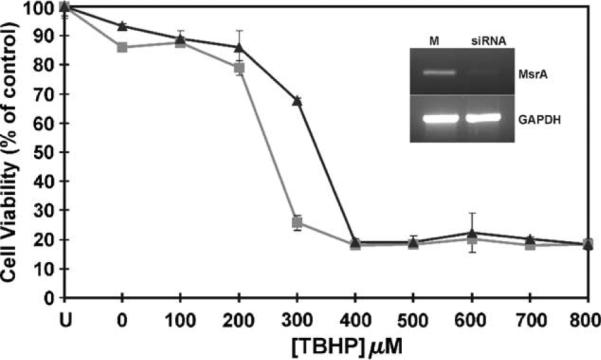
siRNA-mediated gene silencing of MSRA in RPE cells. Untransfected cells are represented by `U' siRNA transfected control cells not exposed to TBHP are indicated as 0. Mock transfected cells `M' are represented by solid squares (▴) and siRNA transfected cells are indicated by solid triangles (∎). Error bars represent standard deviations of four separate cell viability assays with the exception of the `U' value, which was obtained, based on two separate assays. X-axis shows TBHP increasing concentrations and Y-axis indicates cell viability as percent of untreated untransfected control. RT-PCR of ARPE19 cells 48 hr after transfection with mock and msrA siRNAs as described in Methods (see inset).
4. Discussion
The msrA gene structure is interesting for several reasons. The gene has uncommonly large introns (Fig. 2, Table 1) making its overall length very large (~379 kb). This is particularly dramatic when compared with the length of the mRNA (~1.6 kb) and coding regions (0.6–0.7 kb). Our 5′ RACE experiments identified three distinct 5′ ends originating from three different mRNA transcripts (Fig. 1) which match large numbers of EST sequences in GenBank. The matching of these 5′ untranslated sequences from the cDNAs to two regions in the genomic DNA sequences 40 kb apart (Fig. 2, Table 1) is very convincing evidence for the presence of two transcription regulatory regions or promoters.
The higher expression of msrA mRNA in the macular RPE is extremely interesting because this is the area of the RPE which is most likely under the greatest oxidative stress (Fig. 3). The RPE under the macula not only has to support a higher density of photoreceptors on its apical side and is more exposed to the blood by the higher density of the fenestrated choroidal capillaries on its basal side. Since most of the light that enters the eye is focused by the cornea and the lens on the fovea (the center of the macula), this area of the retina, with its greater density of photoreceptors, faces greater light intensity during normal daytime or ambient light conditions. It is in this area of the RPE where msrA mRNA is most abundant (Fig. 3) and suggests that MSRA may be extremely important for protecting the macular RPE from oxidative damage.
Another interesting aspect of MSRA in the retina is its size. MSRA is normally a 26 kDa protein but in the retina it is observed as a dimer at 47 and 49 kDa (Fig. 4). In some RPE-choroid preparation it can be observed as high as 120 kDa suggesting a tetramer (data not shown). This was reproduced with four different monkey retina preparations and in homogenization buffers with and without 5 mM DTT. This aggregation was also observed to some extent with human recombinant MSRA generated as a thioredoxin-fusion peptide in E. coli. Once purified the human recombinant MSRA also formed apparent dimers and tetramers although most of it remained in monomeric form (data not shown). In any event, the aggregated MSRA seems to be enzymatically active (Table 2). The MSRA activity measurements (Table 2) are in general agreement with the immunoblot data (Fig. 4). The nature of this aggregation is under further investigation.
In cultured human RPE cells (ARPE19) the three different MSRAs (Fig. 1) localized to distinct areas of the cell (Fig. 5). MSRA1 as previously described localized to the mitochondria (Hansel et al., 2002) and evidence of MSRA protein was also previously found in the cytosol (Vougier et al., 2003). MSRA2 localized mainly to unidentified vesicles and in some cells to the cytosol. This difference in localization for MSRA2 is unclear but may be related to the different stages in the cell cycle since these were not synchronized cultures. MSRA3 localized to the nucleus and the cytosol. MSRA3 distinctly associated with DAPI in the nuclei (Fig. 5). Thus the N-terminus sequence of the different MSRAs plays a very important role in the intracellular localization of MSRA. This observation coupled with the identification of two transcriptional control regions suggests that MSRA could specifically respond to stress in different cellular organelles. During preparation of this manuscript another study was published using mouse msrA which showed that N-terminal deletions targeted MSRA to the cytosol and nucleus (Kim and Gladyshev, 2005). This result is similar to what we have found in this study with the human transcripts (Fig. 5). However, our transcripts were determined directly from human RNA by 5′RACE and we have also determined their genomic origin. We do not have any evidence that the msrA2 and msrA3 transcripts are actually translated in the retina. Indirect evidence suggests that msrA1 is the predominant isoform in the retina. MsrA1 was the predominant transcript in the 5′RACE experiments and is also the predominant EST in the databases. Moreover, the immunohistochemistry staining (Figs. 6 and 7) is also mostly consistent with a mitochondrial staining. However, this is far from compelling evidence and the roles of these three different transcripts in the retina need to be further investigated.
In the monkey retina MSRA localized to all cells types (Figs. 6 and 7) with ganglion cells, Müller cells and the photoreceptor synapses most strongly labeled. This follows a general mitochondrial staining pattern (data not shown) except for the photoreceptor inner segments which are highly enriched in mitochondria but not stained. This suggests a differential MSRA expression between inner segment and synaptic mitochondrion. In the macula the photoreceptor synapses are much more numerous than in the peripheral retina explaining the broader labeled band in the outer plexiform layer (OPL, Fig. 7). In the peripheral retina the RPE was weakly labeled but in the macula the RPE was strongly labeled in the basal (choriocapillaris side) membrane.
A recent study has shown that overexpression of MSRA in cultured neuronal cells (PC12) protects them against oxidative damage due to hypoxia-reoxygenation (Yermolaieva et al., 2004). Similar results were also obtained with overexpression and siRNA knockdown in cultured lens cells (Kantorow et al., 2003). Another recent study demonstrated the expression of MSRA in primary cultured RPE cells and in human postmortem tissues (Sreekumar et al., 2005). Our results using siRNA knockdown of msrA in cultured RPE cells resulted in decreased resistance to TBHP-induced oxidative stress (Fig. 8). These results suggest that MSRA may be important in protecting RPE cells from oxidative damage and possibly in the maintenance of functional RPE proteins. Our results are consistent with these previous studies in other cell types (Kantorow et al., 2003; Yermolaieva et al., 2004) and in primary RPE cells (Sreekumar et al., 2005). Cultured ARPE19 cells and RPE cells in general are extremely robust. The fact that an MSRA knockdown has clearly shown an effect in their ability to resists TBHP suggests that MSRA may be an important part of their oxidative resistance mechanism.
In summary, the present data demonstrate high levels of MSRA expression and activity in the monkey retina. MSRA levels are enriched in the macular RPE suggesting an important role in the protection of RPE cells against oxidative stress damage. Thus, MSRA may be a potential therapeutical target for age-related macular degeneration.
4.1. Gene accession numbers
GenBank accession numbers: human msrA gene AY958432. The numbers for the human msrA cDNAs are msrA1 (AY958429), msrA2 (AY958430), msrA3 (AY958431).
Other numbers cited: ESTs for msrA1: CD244734, 18170218; ESTs for msrA2: BI599246, AL536061; ESTs for msrA3: BX461653, F06683; msrA gene: AC034111, AC023385, AC079200, AC112673, and AC104964.
Acknowledgements
We thank Dr Robert Fariss and Dr Maria Campos for their help in preparing the monkey vibrotome sections and with the confocal microscopy. We would also like to thank Ms Romina Gruber and Mr Catsim Fassassi for their help in preparing the msrA-GFP constructs. Special thanks to N. DeAmicis for the initial characterization of the siRNAs. This work was supported by the National Eye Institute Intramural research program, EY13022 (MK) and by a grant from the Center of Excellence in Biomedical and Marine Biotechnology (HW) contribution number 2005-11.
Glossary
Abbreviations
- RPE
retinal pigment epithelium
- CH
choroid or choriocapillaris
- ROS
rod outer segments
- RIS
rod inner segments
- ONL
outer nuclear layer
- OPL
outer plexiform layer
- INL
inner nuclear layer
- IPL
inner plexiform layer
- GCL
ganglion cell layer
- 5'RACE
Rapid amplification of cDNA ends
- MSRA
methionine sulfoxide reductase protein
- msrA
methionine sulfoxide reductase gene and mRNA transcripts
- MNR
macula neural retina
- MPC
macula RPE-choroid
- PNR
peripheral neural retina
- PPC
peripheral RPE-choroid
- TBHP
tertiary-butyl hydroperoxide
References
- Berger J, Fine SL, Maguire MG. Age-Related Macular Degeneration. Mosby, Inc.; St. Louis, MO: 1999. [Google Scholar]
- Berlett S, Stadtman ER. Protein oxidation in aging, disease, and oxidative stress. J. Biol. Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- Bonnel S, Mohand-Said S, Sahel J-A. The aging of the retina. Exp. Gerontol. 2003;38:825–831. doi: 10.1016/s0531-5565(03)00093-7. (review) [DOI] [PubMed] [Google Scholar]
- Brot N, Weissbach L, Werth J, Weissbach H. Enzymatic reduction of protein-bound methionine sulfoxide. Proc. Natl Acad. Sci. USA. 1981;78:2155–2158. doi: 10.1073/pnas.78.4.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brot N, Werth J, Koster D, Weissbach H. Reduction of N-acetyl methionine sulfoxide: a simple assay for peptide methionine sulfoxide reductase. Anal. Biochem. 1982;122:291–294. doi: 10.1016/0003-2697(82)90283-4. [DOI] [PubMed] [Google Scholar]
- Evans JR. Risk factors for age-related macular degeneration. Prog. Retin. Eye Res. 2001;20:227–253. doi: 10.1016/s1350-9462(00)00023-9. (review) [DOI] [PubMed] [Google Scholar]
- Grimaud R, Ezraty B, Mitchell JK, Lafitte D, Briand C, Derrick PJ, Barras FJ. Repair of oxidized proteins: identification of a new methionine sulfoxide reductase. J. Biol. Chem. 2001;276:48915–48920. doi: 10.1074/jbc.M105509200. [DOI] [PubMed] [Google Scholar]
- Hansel A, Kuschel L, Hehl S, Lemke C, Agricola H-J, Hoshi T, Heinemann SH. Mitochondrial targeting of the human peptide methionine sulfoxide reductase (MSRA), an enzyme involved in the repair of oxidized proteins. FASEB J. 2002;16:911–913. doi: 10.1096/fj.01-0737fje. [DOI] [PubMed] [Google Scholar]
- Hejtmancik JF, Kantorow M. Molecular genetics of age-related cataract. Exp. Eye Res. 2004;79:3–9. doi: 10.1016/j.exer.2004.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kantorow M, Hawse J, Cowell T, Benhamed S, Pizarro G, Reddy VN, Hejtmancik F. Methionine sulfoxide reductase A is important for lens cell viability and resistance to oxidative stress. Proc. Natl Acad. Sci. USA. 2004;101:9654–9659. doi: 10.1073/pnas.0403532101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H-Y, Gladyshev VN. Role of structural and functional elements of mouse methionine-S-sulfoxide reductase in its subcellular distribution. Biochemistry. 2005;44:8059–8067. doi: 10.1021/bi0501131. [DOI] [PubMed] [Google Scholar]
- Levine RL, Berlett BS, Moskovitz J, Mosoni L, Stadtman ER. Methionine residues may protect proteins from critical oxidative damage. Mech. Ageing Dev. 1999;107:323–332. doi: 10.1016/s0047-6374(98)00152-3. [DOI] [PubMed] [Google Scholar]
- Lowther WT, Brot N, Weissbach H, Honek JF, Matthews BW. Thiol-disulfide exchange is involved in the catalytic mechanism of peptide methionine sulfoxide reductase. Proc. Natl Acad. Sci. USA. 2000;97:6463–6468. doi: 10.1073/pnas.97.12.6463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetti MA, Pizarro GO, Sagher D, DeAmicis C, Brot N, Hejtmancik JF, Weissbach H, Kantorow M. Methionine sulfoxide reductases B1, B2, and B3 are present in the human lens and confer oxidative stress resistance to lens cells. Invest. Ophthalmol. Vis. Sci. 2005;46:2107–2112. doi: 10.1167/iovs.05-0018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKinnon SJ. Glaucoma: ocular alzheimer's disease? Front. Biosci. 2003;8:s1140–s1156. doi: 10.2741/1172. (Review) [DOI] [PubMed] [Google Scholar]
- Moskovitz J, Rahman MA, Strassman J, Yancey SO, Kushner SR, Brot N, Weissbach H. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J. Bateriol. 1995;177:502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Weissbach H, Brot N. Cloning the expression of a mammalian gene involved in the reduction of methionine sulfoxide residues in proteins. Proc. Natl Acad. Sci. USA. 1996a;93:2095–2099. doi: 10.1073/pnas.93.5.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Jenkins NA, Gilbert DJ, Copeland NG, Jursky F, Weissbach W, Brot N. Chromosomal localization of the mammalian peptide-methionine sulfoxide reductase gene and its differential expression in various tissues. Proc. Natl Acad. Sci. USA. 1996b;93:3205–3208. doi: 10.1073/pnas.93.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskovitz J, Poston JM, Berlett BS, Nosworthy NJ, Szczepanowski R, Stadtman ER. Identification and characterization of a putative active site for peptide methionine sulfoxide reductase (MsrA) and its substrate stereospecificity. J. Biol. Chem. 2000;275:14167–14172. doi: 10.1074/jbc.275.19.14167. [DOI] [PubMed] [Google Scholar]
- Moskovitz J, Bar-Noy S, Williams WM, Requena J, Berlett BS, Stadtman ER. Methionine sulfoxide reductase (MsrA) is a regulator of antioxidant defense and lifespan in mammals. Proc. Natl Acad. Sci. USA. 2001;98:12920–12925. doi: 10.1073/pnas.231472998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moskowitz J. Methionine sulfoxide reductases: ubiquitous enzymes involved in antioxidant defense, protein regulation, and prevention of aging-associated diseases. Biochim. Biophys. Acta. 2005;1703:213–219. doi: 10.1016/j.bbapap.2004.09.003. (review) [DOI] [PubMed] [Google Scholar]
- Oyster C. The Human Eye: Structure and Function. Sinauer Associates, Inc.; Sunderland, MA: 1999. [Google Scholar]
- Petropoulos I, Friguet B. Protein maintenance in aging and replicative senescence: a role for the peptide methionine sulfoxide reductases. Biochim. Biophys. Acta. 2005;1703:261–266. doi: 10.1016/j.bbapap.2004.08.018. (review) [DOI] [PubMed] [Google Scholar]
- Rodriguez IR, Mazuruk K, Schoen TJ, Chader GJ. Structural analysis of the human hydroxyindole-O-methyltransferase gene. Presence of two distinct promoters. J. Biol. Chem. 1994;269:31969–31977. [PubMed] [Google Scholar]
- Ruan H, Tang XD, Chen M-L, Joiner M-L, Sun G, Brot N, Weissbach H, Heineman SH, Iverson L, Wu C-F, Hoshi T. High-quality life extension by the enzyme peptide methionine sulfoxide reductase. Proc. Natl Acad. Sci. USA. 2002;99:2748–2753. doi: 10.1073/pnas.032671199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharov VS, Ferrington TC, Squier C, Schoneich C. Diaestereoselective reduction of protein-bound methionine sulfoxide by methionine sulfoxide reductase. FEBS Lett. 1999;445:247–250. doi: 10.1016/s0014-5793(99)00888-1. [DOI] [PubMed] [Google Scholar]
- Spiess AN, Ivell R. Normalization of RNA hybridization signals by means of SYBR green II-stained 28S or 18S ribosomal RNA and a phosphor imager. Biotechniques. 1999;26:46–48. doi: 10.2144/99261bm06. [DOI] [PubMed] [Google Scholar]
- Sreekumar PG, Kannan R, Yaung J, Spee CK, Ryan SJ, Hinton DR. Protection from oxidative stress by methionine sulfoxide reductases in RPE cells. Biochem. Biophys. Res. Commun. 2005;334:245–253. doi: 10.1016/j.bbrc.2005.06.081. [DOI] [PubMed] [Google Scholar]
- John G, Brot N, Ruan J, Erdjument-Bromage H, Tempst P, Weissbach H, Nathan C. Peptide methionine sulfoxide reductase from escherichia coli and mycobacterium tuberculosis protects bacteria against oxidative damage from reactive nitrogen intermediates. Proc. Natl Acad. Sci. USA. 2001;98:9901–9906. doi: 10.1073/pnas.161295398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman ER, Moskovitz J, Berlett BS, Levine RL. Cyclic oxidation and reduction of protein methionine residues is an important antioxidant mechanism. Mol. Cell. Biochem. 2002;234/235:3–9. [PubMed] [Google Scholar]
- Vougier S, Mary J, Friguet B. Subcellular localization of methionine sulphoxide reductase A (MsrA): evidence for mitochondrial and cytosolic isoforms in rat liver cells. Biochem. J. 2003;373:531–537. doi: 10.1042/BJ20030443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissbach H, Etienne F, Hoshi T, Heinemann SH, Lowther WT, Matthews BW, John G, Nathan C, Brot N. Peptide methionine sulfoxide reductase: structure, mechanism of action, and biological function. Arch. Biochem. Biophys. 2002;397:172–178. doi: 10.1006/abbi.2001.2664. [Review] [DOI] [PubMed] [Google Scholar]
- Weissbach H, Resnick L, Brot N. Methionine sulfoxide reductases: history and cellular role in protecting against oxidative damage. Biochim. Biophys. Acta. 2005;1703:203–212. doi: 10.1016/j.bbapap.2004.10.004. (review) [DOI] [PubMed] [Google Scholar]
- Yermolaieva O, Xu R, Schinstock C, Brot N, Weissbach H, Heinemann S, Hoshi T. Methionine sulfoxide reductase A protects neuronal cells against brief hypoxia/reoxygenation. Proc. Natl Acad. Sci. USA. 2004;101:1159–1164. doi: 10.1073/pnas.0308215100. [DOI] [PMC free article] [PubMed] [Google Scholar]