Abstract
Haemophilus ducreyi is an extracellular pathogen of human epithelial surfaces that resists human antimicrobial peptides (APs). The organism's genome contains homologs of genes sensitive to antimicrobial peptides (sap operon) in nontypeable Haemophilus influenzae. In this study, we characterized the sap-containing loci of H. ducreyi 35000HP and demonstrated that sapA is expressed in broth cultures and H. ducreyi-infected tissue; sapA is also conserved among both class I and class II H. ducreyi strains. We constructed a nonpolar sapA mutant of H. ducreyi 35000HP, designated 35000HPsapA, and compared the percent survival of wild-type 35000HP and 35000HPsapA exposed to several human APs, including α-defensins, β-defensins, and the cathelicidin LL-37. Unlike an H. influenzae sapA mutant, strain 35000HPsapA was not more susceptible to defensins than strain 35000HP was. However, we observed a significant decrease in the survival of strain 35000HPsapA after exposure to LL-37, which was complemented by introducing sapA in trans. Thus, the Sap transporter plays a role in resistance of H. ducreyi to LL-37. We next compared mutant strain 35000HPsapA with strain 35000HP for their ability to cause disease in human volunteers. Although both strains caused papules to form at similar rates, the pustule formation rate at sites inoculated with 35000HPsapA was significantly lower than that of sites inoculated with 35000HP (33.3% versus 66.7%; P = 0.007). Together, these data establish that SapA acts as a virulence factor and as one mechanism for H. ducreyi to resist killing by antimicrobial peptides. To our knowledge, this is the first demonstration that an antimicrobial peptide resistance mechanism contributes to bacterial virulence in humans.
Haemophilus ducreyi is the causative agent of chancroid, a genital ulcer disease that facilitates both the transmission and acquisition of HIV-1 (4, 45). An obligate human pathogen, H. ducreyi infects epithelial surfaces and primarily remains localized in the skin (49). During experimental human infection, polymorphonuclear leukocytes (PMNs) and macrophages are rapidly recruited to the bacterial entry site. The organism colocalizes with PMNs and macrophages throughout disease but remains extracellular (8, 10).
As part of the innate immune system's response to infection, PMNs, macrophages, and epithelial cells secrete antimicrobial peptides (APs). Most APs are small, cationic peptides that are secreted into the extracellular milieu and have both bactericidal and chemotactic properties (26). Cationic APs are attracted to the anionic bacterial cell membrane where they lyse the bacterial cell. Some APs specifically recruit macrophages, PMNs, T cells, and immature dendritic cells to the site of bacterial infection, serving as a bridge between innate and adaptive immunity (39, 40, 53).
In humans, the best-studied APs include the family of defensins, which are subdivided by structure into α- and β-defensins, and one human cathelicidin, LL-37. Defensins are cysteine- and arginine-rich β-sheet peptides that range from 29 to 47 amino acids in length (29). Defensins form three intramolecular disulfide bonds via six invariant cysteine residues. Both the positions of the cysteines and the pattern of disulfide bonding differ between the α- and β-defensins (17, 29). In contrast, cathelicidin LL-37 is devoid of cysteines and forms an α-helix (20).
H. ducreyi encounters several cellular sources of APs during human infection: keratinocytes constitutively express human β-defensin 1 (HBD-1) and upregulate expression of HBD-2, HBD-3, and LL-37 in response to inflammation (22, 23, 30, 54); PMNs secrete preformed α-defensins, including human neutrophil peptide 1 (HNP-1), HNP-2, HNP-3, and HNP-4, as well as HBD-4 and LL-37 during infection (16, 50); and macrophages secrete HBD-1, HBD-2, and LL-37 in response to inflammatory mediators (15, 31). Vaginal epithelial cells also constitutively express the α-defensin human defensin 5 (HD-5) (43). Dual staining for H. ducreyi and HNP-1, HNP-2, and HNP-3 demonstrate that H. ducreyi is exposed to APs at the papular, pustular, and ulcerative stages of disease (10; C. A. Townsend and M. E. Bauer, unpublished data). H. ducreyi resists the bactericidal effects of several human APs, including those predicted to be at the site of infection; this resistance has been observed in representative members of two phenotypic H. ducreyi classes (38, 51). The mechanism(s) by which H. ducreyi evades AP-mediated killing is unknown.
Bacterial pathogens have evolved many mechanisms to resist killing by APs, including enzymatic inactivation of APs, electrostatic repulsion by the addition of positively charged residues on the surface, and expression of transporters that remove APs before they can attack the cell membrane (27). To identify putative AP resistance factors in H. ducreyi, we examined the H. ducreyi genome (http://stdgen.northwestern.edu) for evidence of homology with previously described resistance strategies in other bacteria. Using a BLASTP search, we found that the H. ducreyi genome included homologs of the previously described transporter genes sensitive to antimicrobial peptides (sap genes) (3). The sap genes encode the Sap influx pump, which confers resistance to APs in several Gram-negative pathogens, including Salmonella enterica serovar Typhimurium, nontypeable Haemophilus influenzae, Proteus mirabilis, and Erwinia chrysanthemi (32, 35, 36, 41).
The Sap transporter consists of five proteins: SapB and SapC are permease proteins that form a pore in the bacterial inner membrane, SapD and SapF function as ATPase subunits of the transporter, and SapA is a periplasmic solute binding protein (34, 41). The nontypeable H. influenzae SapA protein has been shown to directly bind APs, including both LL-37 and HBD-3 (34). Once an AP enters the periplasm, SapA is thought to shuttle the AP to the SapBCDF channel, where the AP is transported into the cytoplasm, bypassing direct interaction between the AP and the cytoplasmic membrane, which would be the lethal event in AP attack (34, 41). Once in the cytoplasm, the fate of the AP is unknown, but presumably, it is degraded and its amino acids are recycled.
Sap transporters take up a variety of APs; among human APs tested, the S. enterica serovar Typhimurium Sap transporter confers resistance to crude extracts of human PMNs, and the nontypeable H. influenzae Sap transporter confers resistance to LL-37 and HBD-3 (19, 34, 35). Further, the nontypeable H. influenzae SapA protein has been shown to directly bind APs, including both LL-37 and HBD-3 (34).
Because Sap transporters protect other pathogens from APs, we hypothesized that the putative H. ducreyi Sap transporter system would confer resistance against APs and that the loss of the periplasmic component SapA would result in increased susceptibility to APs in vitro and loss of virulence in vivo. Herein, we describe the construction of an isogenic sapA insertion-deletion mutant to test this hypothesis and to define a mechanism for AP resistance of H. ducreyi.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains used in this study are listed in Table 1. H. ducreyi strains were grown on chocolate agar plates at 33°C with 5% CO2. Liquid cultures were grown with aeration at 33°C in gonococcal broth (15% proteose peptone [BD, Sparks, MD], 4% K2HPO4, 2% KH2PO4, 10% NaCl) supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 1% IsoVitaleX, and 50 μg of hemin (Aldrich Chemical Co., Milwaukee, WI) per ml (5). Escherichia coli strains were grown in Luria-Bertani (LB) broth at 37°C with aeration or on LB plates at 37°C (44).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Genotype or description and origin | Source or reference(s) |
|---|---|---|
| E. coli strains | ||
| HB101 | F thi-1 hsdS20(rB mB) supE44 recA13 ara-14 leuB6 proA2 lacY1 galK2 rpsL20 (strR) xyl-5 mtl-1 | Promega |
| TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) φ80lacZΔM15 ΔlacX74 recA1 araΔ139 Δ(ara-leu)7697 galU galK rpsL (strR) endA1 nupG | Invitrogen |
| H. ducreyi strains | ||
| 35000HP | Class I clinical isolate; human-passaged variant of strain 35000, Winnipeg, Canada, 1975 | 2, 21, 51 |
| HD183 | Class I clinical isolate; Singapore, 1982 | 9, 48 |
| HD188 | Class I clinical isolate; Kenya, 1982 | 9, 48 |
| 82-029362 | Class I clinical isolate; California, 1982 | 9, 48 |
| 6644 | Class I clinical isolate; Massachusetts, 1989 | 9, 48 |
| HD85-023233 | Class I clinical isolate; New York, 1985 | 9, 48 |
| CIP542-ATCC | Class II clinical isolate; Hanoi, Viet Nam, 1954 | 21, 51 |
| HMC112 | Class II clinical isolate; 1984 (location not reported) | 51 |
| 33921 | Class II clinical isolate; Kenya (yr not reported) | 9, 13 |
| DMC164 | Class II clinical isolate; Bangladesh (yr not reported) | 51 |
| MEB107 | 35000HPsapA::kan | This study |
| MEB121 | 35000HPsapA::kan/psapA | This study |
| Plasmids | ||
| pGEM-T Easy | TA cloning vector; Ampra | Promega |
| pSC-A | TA cloning vector; Ampr | Stratagene |
| pUC18K2 | Source of nonpolar Kanr cassette | 37 |
| pRSM2072 | H. ducreyi suicide vector | 12 |
| pLSSK | H. ducreyi shuttle vector | 52 |
| pMEB078 | 5′ end of sapA in pGEM-T Easy | This study |
| pMEB080 | 3′ end of sapA in pGEM-T Easy | This study |
| pMEB092 | 5′ end of sapA + Kanr cassette in pGEM-T Easy | This study |
| pMEB098 | 5′ end of sapA + Kanr cassette + 3′ end of sapA in pGEM-T Easy | This study |
| pMEB108 | sapA ORF in pGEM-T-Easy | This study |
| pMEB114 | Putative sap promoter in pSC-A | This study |
| pMEB115 | Putative sap promoter + sapA ORF in pGEM-T Easy | This study |
| pMEB120 | Putative sap promoter + sapA ORF in pLSSK | This study |
Ampr denotes resistance to ampicillin.
Nested RT-PCR.
RNA was isolated from mid-logarithmic cultures of H. ducreyi 35000HP and from a biopsy tissue specimen of H. ducreyi-infected skin obtained from a volunteer, using the Ultraspec reagent (Clontech, Mountainview, CA) as previously described (7). cDNA was synthesized using random hexamers (Clontech). Reverse transcriptase PCR (RT-PCR) was performed using HiFi Taq (Invitrogen, Carlsbad, CA) and the Sap Forward 2 and Sap Reverse 3 primers (Table 2). An annealing temperature of 60°C was used for 45 cycles of PCR amplification to produce a 569-bp product. Nested RT-PCR was performed using 2 μl of the previous reaction mixture as a template and the Sap Forward 6 and Sap Reverse 6 primers to amplify a 194-bp nested SapA RT-PCR product. Samples lacking template and samples lacking RT were included in all reaction mixtures.
TABLE 2.
Primers used in this study
| Primer | Construct or use(s) | Sequencea |
|---|---|---|
| 5′SapA Forward | pMEB078 | CATATCtctagaGCGGATAGCTTAATTTATTGCACCAG |
| 5′SapA Reverse | pMEB078 | CATATCggtaccCATATCctcgagGAATCTTTACTGTATATTCATTCGGTGCTG |
| 3′SapA Forward | pMEB080 | CATATCtctagaGCGGATAGCTTAATTTATTGCACCAG |
| 3′SapA Reverse | pMEB080 | TAAACGAACTTGCCCGAATGGCTC |
| SapAcompFor1 | pMEB108 | GATGTATCATTACTCACTAATATCCCTGCT |
| SapAcompRev1 | pMEB108 | CGGTCGAAATCCAACAGAACACAG |
| SapPromoterFor1 | pMEB114 | CCTTTAATTTGTTCTAAATACATAATGATCC |
| SapPromoterRev1 | pMEB114 | CATATCcatatgTCTTTTCGCCTTAATTTAAGC |
| Sap Forward 2 | Nested RT-PCR | CGGATAATTCGGTGTTGGCGCATT |
| Sap Reverse 3 | Nested RT-PCR | TGTGCACTCGACCTGTATGCGGAT |
| Sap Forward 5 | PCR of clinical isolates | GCATTTCTCATACTGCGGTGGCAA |
| Sap Forward 6 | Nested RT-PCR | AATGCGCATATTGAGCCATTCGGG |
| Sap Reverse 6 | Nested RT-PCR; PCR of clinical isolates | ATCCGAGCGGAATCCCGATAATCA |
Restriction enzyme sites are shown in lowercase type and underlined.
Survey of clinical isolates of H. ducreyi for sapA gene.
Chromosomal DNA was isolated from six class I clinical H. ducreyi isolates and four class II clinical H. ducreyi isolates (Table 1), as described previously (9). A 2,032-bp fragment, including sapA, was PCR amplified using primers Sap Forward 5 and Sap Reverse 6 (Table 2). A reaction mixture containing no template served as a negative control. Amplimers were used as templates for sequencing the sapA open reading frames (ORFs), performed by the DNA Sequencing Core Facility at the Indiana University School of Medicine. Sequences were aligned using the ClustalW algorithm in the Lasergene software package (DNASTAR, version 7).
Construction and complementation of a sapA mutant.
The primers used to generate constructs in this study are listed in Table 2. All plasmids were maintained in E. coli TOP10, and mutagenic and complementing plasmids were passed through E. coli HB101 before introduction into H. ducreyi (6). A 517-bp fragment containing the 5′ end of HD1230 (sapA) was PCR amplified from H. ducreyi 35000HP genomic DNA. The amplicon was TA cloned into pGEM-T Easy (Promega, Madison, WI) to generate pMEB078. A 489-bp fragment containing the 3′ end of HD1230 was PCR amplified from genomic DNA and TA cloned into pGEM-T Easy (Promega) to generate pMEB080. A nonpolar kanamycin resistance (Kanr) cassette from pUC18K2 (37) was inserted downstream of the 5-prime fragment of sapA in H. ducreyi MEB078 to generate pMEB092. The sapA′-Kanr fragment of pMEB092 was subcloned upstream of the 3-prime region of sapA in pMEB080, generating pMEB098. In the resulting mutagenic construct, 676 bp of sapA was replaced with the Kanr cassette. The sapA′-Kanr-′sapA fragment was subcloned into pRSM2072, which expresses lacZ and acts as a suicide vector in H. ducreyi (12), and introduced into strain 35000HP by electroporation. Kanamycin-resistant transformants were propagated on chocolate agar plates containing 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) in order to select for colonies in which the wild-type allele had been successfully replaced by the mutagenized allele (12). The sapA mutation was confirmed by digestion, PCR analysis, Southern blotting, and DNA sequencing (data not shown). H. ducreyi 35000HP and 35000HPsapA demonstrated similar growth rates in broth (data not shown). Quantitative RT-PCR (qRT-PCR) was used to verify that transcription of downstream genes was unaffected by the sapA mutation (data not shown).
To complement H. ducreyi 35000HPsapA in trans, we expressed sapA under the control of its native promoter in the H. ducreyi shuttle vector pLSSK (52). The sapA ORF was PCR amplified and TA cloned into pGEM-T Easy (Promega) to generate pMEB108. The 5′ primer used to amplify sapA inserted CAT bases immediately upstream of the sapA start codon to introduce an NdeI site (CATATG) at the 5′ end of the sapA clone. Using BPROM software (SoftBerry, Inc., Mount Kisco, NY), a putative promoter was identified starting 177 bp upstream of tyrR in an untranslated region. To place the putative promoter upstream of the sapA gene, sequence 237 bp upstream of tyrR was PCR amplified with primers that introduced an NdeI site at the 3′ end and TA cloned into pSC-A (Stratagene, Cedar Creek, TX) to generate pMEB114. The putative promoter region was excised from pMEB114, digested with NdeI, and ligated with NdeI-digested pMEB108 to generate pMEB115, in which the sapA ORF lies immediately downstream of the untranslated region upstream of tyrR. This construct was subcloned into pLSSK on a NotI fragment to produce pMEB120, which was introduced into mutant strain 35000HPsapA by electroporation. Transformants were selected on plates containing kanamycin and streptomycin; one transformant was selected and designated 35000HPsapA/psapA. RT-PCR was used to verify that the sapA gene was transcribed in mid-logarithmic cultures of strain 35000HPsapA/psapA (data not shown).
Sources of peptides.
Recombinant α- and β-defensins were purchased from PeproTech Inc. (Rocky Hill, NJ), Sigma-Aldrich (St. Louis, MO), Peptides International (Louisville, KY), and AnaSpec (San Jose, CA). Synthetic LL-37 was purchased from Phoenix Pharmaceuticals, Inc. (Belmont, CA).
96-Well AP bactericidal assay.
The 96-well AP bactericidal assay was performed as previously described (38). Briefly, bacteria were grown to mid-logarithmic phase and suspended in 10 mM sodium phosphate (pH 7.4) supplemented with 0.1% brain heart infusion broth (BD). Approximately 103 CFU of bacteria were mixed in duplicate with the indicated concentration of peptides in wells of a 96-well polypropylene plate (catalog no. 3790; Costar) and incubated for 1 h at 33°C. The concentration of bacteria remaining in the wells was determined by counting three replicate plates, and the results from duplicate wells were averaged. Survival in the presence of APs was calculated as a percentage of survival in control wells without APs. Because we have observed lot-to-lot variations in the level of AP activity, comparisons among bacterial strains for AP susceptibility were made only when the strains were assayed using the same lot of AP.
Assessment of virulence of the sapA mutant in human volunteers.
Human challenge studies with H. ducreyi were performed under the guidelines of the U.S. Department of Health and Human Services, the U.S. Food and Drug Administration, and the Institutional Review Board of Indiana University-Purdue University at Indianapolis. After giving informed consent for participation and HIV serology, six healthy, adult volunteers (4 males and 2 females; 3 white, 3 black; age range, 25 to 57 years; mean age ± standard deviation [SD], 44 ± 10 years) were inoculated on the upper arm with H. ducreyi 35000HP and 35000HPsapA and monitored to the clinical endpoint, exactly as described previously for parent-mutant trials in this model (1, 5, 24). Parameters compared between parent-inoculated and mutant-inoculated sites included rates of papule and pustule formation and day 1 lesion size.
There are gender and other host effects on susceptibility to disease progression in the human model (11, 46); subjects are inoculated with the parent and mutant strains and serve as their own controls for these effects. To account for within-subject correlation among inoculated sites, statistical analysis of papule and pustule formation rates utilized a logistic regression model with generalized estimating equations (GEE), as described previously (47). Ninety-five percent confidence intervals (95% CIs) were calculated using the GEE sandwich estimate for standard errors. Differences in the rates of papule or pustule formation between the two strains tested were considered statistically significant if P was <0.05. Statistical analysis of day 1 papule sizes was calculated using a two-tailed Student's t test.
RESULTS
Characterization of sap genes in H. ducreyi.
Homology searches showed that H. ducreyi 35000HP contains genes whose predicted products were highly homologous to five of the nontypeable H. influenzae 86-028NP Sap proteins (3), including SapA (44% identity; 67% similarity), SapB (46% identity; 67% similarity), SapC (52% identity; 73% similarity), SapD (65% identity; 82% similarity), and SapF (59% identity; 73% similarity). Nontypeable H. influenzae also contains a sixth Sap protein, SapZ, which is found in some but not all bacteria containing Sap transporters; the function of SapZ is unknown (35). We found no homolog of SapZ in H. ducreyi 35000HP.
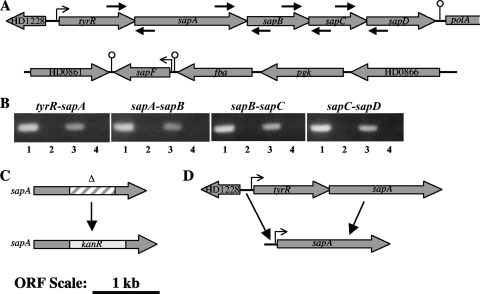
The locus containing sapA in H. ducreyi 35000HP is shown in Fig. 1A. A putative regulatory gene, tyrR, and a predicted promoter region were identified upstream of sapA, and homologs of sapBCD were identified downstream of sapA. A rho-independent terminator was predicted in the intergenic space between sapD and the next downstream gene, polA (55). Surprisingly, the sapA locus did not contain a sapF homolog; rather, the genome contained an unlinked sapF homolog that was flanked by predicted promoter and terminator regions, suggesting that sapF may be in a monocistronic operon (Fig. 1A). We used RT-PCR to map the operon structure of the H. ducreyi sapABCD locus and found that the four contiguous sap genes, along with the upstream tyrR, are cotranscribed in a single operon (Fig. 1B).
FIG. 1.
Analysis and mutagenesis of the sap genes in H. ducreyi 35000HP. (A) Genetic map of sap-containing loci. An operon containing tyrR and sapABCD was located between ORFs HD1228 and HD1236, and the unlinked sapF-containing locus was located between ORFs HD0861 and HD0866. Arrowheads show the direction of transcription, stalked arrows represent predicted promoters, and ball-and-stick structures indicate predicted stem-loop transcriptional terminators. Primer binding sites are indicated by small arrows. (B) Operon mapping of the sapABCD locus. RT-PCR was used to amplify mRNA across junctions of the indicated adjacent genes. Lanes 1, chromosomal DNA; lanes 2, no-template control; lanes 3, RNA samples with RT; lanes 4, RNA samples with no RT added. (C) Insertion/deletion mutagenesis of sapA. A 676-bp fragment of sapA (hatched box) was replaced with a 840-bp nonpolar Kanr cassette (gray box). This construct was inserted into the chromosome by allelic replacement as described in Materials and Methods. (D) Expression of sapA downstream of its native promoter for trans-complementation. sapA was cloned downstream of 237 bp of the untranslated sequence 5′ of tyrR for expression from the native promoter. The cloned 5′ sequence contained a predicted −35 and −10 region and ribosome binding site. Panels A, C, and D are drawn at the same scale.
The H. ducreyi Sap transporter genes are transcribed during human infection.
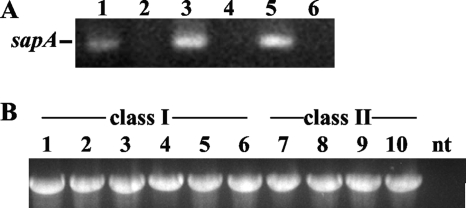
We recently completed a study using selective capture of transcribed sequences (SCOTS) to identify genes important for H. ducreyi virulence (7). With SCOTS, the competitive hybridization of tissue-derived and broth-derived sequences is used to identify genes that may be preferentially expressed in vivo (18). Among the in vivo-derived genes identified were homologs of H. influenzae sapA, sapB, and sapF (7). To confirm in vivo expression of sapA, we performed nested RT-PCR on RNA from an H. ducreyi-infected pustule of a human volunteer. Transcripts were amplified in both tissue-derived RNA and broth-derived RNA (Fig. 2A). These data confirm that H. ducreyi expresses sapA at the pustular stage of disease during experimental human H. ducreyi infection.
FIG. 2.
sapA is expressed in vivo and conserved among H. ducreyi strains. (A) Nested RT-PCR of sapA transcripts was performed on RNA from broth culture (lanes 3 and 4) or a pustule biopsy specimen from the human model (lanes 5 and 6). Controls were included in which no RT was added to reaction mixtures (lanes 4 and 6) or no template was used (lane 2). Genomic DNA (lane 1) served as a positive control. (B) PCR amplification of sapA-containing sequence from class I and class II clinical isolates of H. ducreyi (see Table 1). A 2.032-kbp fragment was amplified from genomic DNA of each strain using primers flanking the sapA ORF in H. ducreyi 35000HP. Lanes 1 to 6 contain class I strains as follows: lane 1, 35000HP; 2, HD183; 3, HD188; 4, 82-029362; 5, 6644; 6, HD85-023233. Lanes 7 to 10 contain class II strains as follows: lane 7, CIP542-ATCC; 8, HMC112; 9, 33921; 10, DMC64. nt, no-template control.
Identification of sap genes by SCOTS suggested that H. ducreyi may upregulate sap genes in vivo. H. ducreyi encodes one two-component regulator, a homolog of CpxRA (28). Although we identified a putative CpxR recognition motif upstream of tyrR, qRT-PCR analysis showed no difference in sapA expression between isogenic cpxR or cpxA mutants and their parent strain 35000HP (data not shown) (14, 28). We also used qRT-PCR to examine whether exposure to cathelicidin LL37 (0.06 or 1 μg/ml) signals H. ducreyi to upregulate sapA. Neither LL37 dose affected the level of sapA transcripts in strain 35000HP (data not shown).
The sapA gene is conserved among both class I and class II strains of H. ducreyi.
Two phenotypic classes of H. ducreyi have been described (51). Our previous studies demonstrated that both classes of H. ducreyi are resistant to human APs (38). We examined whether a panel of 10 clinical isolates harbored the sapA gene. Using primers flanking the sapA ORF from the H. ducreyi 35000HP sequence, we PCR amplified genomic DNAs of all clinical isolates tested; amplimers comigrating with the amplimer from strain 35000HP were identified (Fig. 2B). Sequencing confirmed the conservation of the full-length, 1,683-bp sapA ORF in all 10 isolates analyzed. In three class I strains, the sapA sequence was identical to sapA in the prototype class I strain 35000HP. However, in class I strains 6644 and 82-029362, we found a single G-to-A base pair change, which predicted an amino acid change from R to H at position 132 of the protein sequence. The sapA genes in four class II strains were identical to each other and differed at 12 bp from the 35000HP sapA ORF; these substitutions resulted in 4 amino acid changes in the predicted protein sequence, including S to A at position 26, A to V at position 183, V to A at position 428, and Y to D at position 510. The class II strains did not contain the single base pair change detected in two of the class I strains. Overall, the sequence data show that sapA is highly conserved in both classes of H. ducreyi, including strains from several geographic locations.
Construction and complementation of a sapA mutant in H. ducreyi.
To determine the role of SapA in AP resistance of H. ducreyi, we mutagenized sapA in H. ducreyi 35000HP by insertion of a nonpolar cassette (Fig. 1C). qRT-PCR showed that the transcription level of sapD, the most downstream gene within the operon, was unaffected by the mutagenesis procedure, confirming that the mutation in strain 35000HPsapA was nonpolar (data not shown). To confirm that phenotypic differences between strains 35000HP and 35000HPsapA were due solely to the loss of a functional sapA gene in the mutant, we cloned sapA downstream of its predicted native promoter sequence located upstream of tyrR (Fig. 1D) to trans-complement 350000HPsapA; the resulting strain was designated 35000HPsapA/psapA.
The H. ducreyi sapA gene confers resistance to the human cathelicidin LL-37.
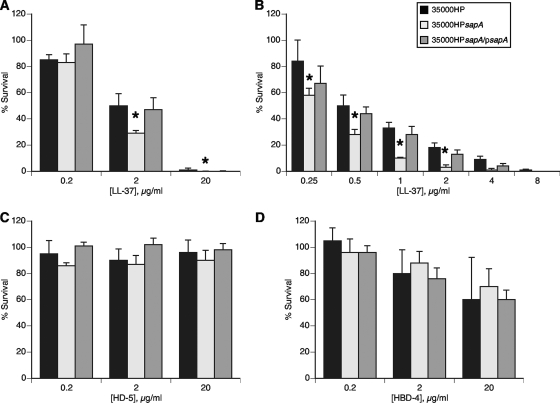
H. ducreyi is likely exposed to multiple sources of LL-37 during infection, and the bacterium is resistant to the bactericidal effects of this peptide (38). In a 96-well AP bactericidal assay with 10-fold serial dilutions of LL-37, H. ducreyi 35000HPsapA was significantly more sensitive than H. ducreyi 35000HP to LL-37 at all but the lowest concentration (Fig. 3A). Assays with 2-fold serial dilutions of LL-37 were performed to more specifically characterize the effect of the sapA mutation on H. ducreyi resistance to LL-37. Wild-type H. ducreyi demonstrated significantly greater levels of survival than the sapA mutant when challenged with 0.25 μg/ml to 2 μg/ml LL-37 (Fig. 3B). trans-Complementation with sapA expressed on pLSSK restored resistance to LL-37 (Fig. 3). Additional experiments showed that there were no differences in susceptibility of strains 35000HPsapA and 35000HPsapA transformed with vector pLSSK to LL-37 resistance (data not shown). These data suggest that the Sap transporter plays a role in protecting H. ducreyi from the bactericidal activity of LL-37.
FIG. 3.
H. ducreyi SapA confers resistance to the human cathelicidin LL-37, but not to human α- or β-defensins. Bactericidal assays comparing the percent survival of H. ducreyi 35000HP, 35000HPsapA, and 35000HPsapA/psapA exposed to the indicated concentrations of LL-37 (A and B), α-defensin HD-5 (C), or β-defensin HBD-4 (D). Data represent the means plus standard errors (error bars) for three independent assays. Percent survival values that are significantly different (P ≤ 0.01) from those of the parent strain are indicated by an asterisk.
The H. ducreyi sapA gene does not confer resistance to human defensins.
H. ducreyi is exposed to PMN-derived α-defensins at all stages of disease (10; data not shown), and H. ducreyi is likely exposed to the α-defensin HD-5 during vaginal and cervical infection (43). Similarly, H. ducreyi is exposed to multiple cellular sources of β-defensins at the site of infection. H. ducreyi is resistant to the bactericidal activity of these human α- and β-defensins (38), and the Sap transporter has been shown to contribute to β-defensin resistance in other bacteria. Thus, we examined the role of the Sap transporter in H. ducreyi resistance to these human defensins. In a 96-well AP bactericidal assay with 10-fold serial dilutions of peptide, there was no discernible difference in AP resistance levels of H. ducreyi 35000HP and 35000HPsapA exposed to the α-defensins HNP-1, HNP-2, and HD-5 (Fig. 3C and data not shown). Similarly, we found no significant difference in the rate of survival of wild-type H. ducreyi and the 35000HPsapA mutant after exposure to increasing concentrations of the β-defensins HBD-2, HBD-3, and HBD-4 (Fig. 3D and data not shown). These findings indicate that the H. ducreyi Sap transporter is not required for the bacterium's resistance to the α-defensins or β-defensins tested.
The H. ducreyi sapA mutant is partially attenuated for virulence in humans.
To determine whether SapA plays a role in disease, we tested H. ducreyi 35000HPsapA alongside 35000HP for virulence in the human model of H. ducreyi infection. Six healthy adults were challenged with H. ducreyi in two iterations (Table 3). In the first iteration, 3 volunteers were each inoculated with an estimated delivered dose of 81 CFU of H. ducreyi 35000HP at three sites on one arm and with estimated delivered doses of 42, 84, and 165 CFU of H. ducreyi 35000HPsapA at three sites on the other arm. Papules formed at all sites, and pustules formed at 6 of 9 parent-inoculated sites and at 5 of 9 mutant-inoculated sites (Table 3). In the second iteration, 3 volunteers received an estimated delivered dose of 97 CFU of strain 35000HP at each of three sites and estimated delivered doses of 47, 94, and 188 CFU of strain 35000HPsapA at three sites. Papules formed at all parent-inoculated sites and at 8 of 9 mutant-inoculated sites. Pustules formed at 6 of 9 parent-inoculated sites and at only 1 of 9 mutant-inoculated sites (Table 3).
TABLE 3.
Effect of sapA on human inoculation with H. ducreyia
| Volunteer (gender) | No. of days of observation | Strainb | Dose (CFU)c | No. of initial papules | No. of initial pustules | Final outcome of site |
||
|---|---|---|---|---|---|---|---|---|
| No. of papules | No. of pustules | No. of papules/pustules that resolved | ||||||
| 346 (male) | 6 | P | 81 | 3 | 3 | 3 | ||
| M | 42-165 | 3 | 2 | 2 | 1 | |||
| 347 (male) | 6 | P | 81 | 3 | 2 | 2 | 1 | |
| M | 42-165 | 3 | 3 | 3 | ||||
| 348 (male) | 7 | P | 81 | 3 | 1 | 1 | 1 | 1 |
| M | 42-165 | 3 | 0 | 3 | ||||
| 349 (female) | 6 | P | 97 | 3 | 3 | 3 | ||
| M | 47-188 | 3 | 1 | 1 | 2 | |||
| 350 (male) | 6 | P | 97 | 3 | 2 | 2 | 1 | |
| M | 47-188 | 3 | 0 | 3 | ||||
| 351 (female) | 7 | P | 97 | 3 | 1 | 1 | 2 | |
| M | 47-188 | 2 | 0 | 2 | ||||
Volunteers 346, 347, and 348 were inoculated in the first iteration; volunteers 349, 350, and 351 were inoculated in the second iteration. See the text for details.
P, parent strain 35000HP; M, mutant strain 35000HPsapA.
42-165, one dose each of 42, 84, and 165 CFU; 47-188, one dose each of 47, 94, and 188 CFU.
The two iterations were combined for a statistical comparison of the strains (see Materials and Methods). Results showed no differences between the papule formation rates at parent-inoculated sites (100% [95% CI, 60.6 to 100%]) and at mutant-inoculated sites (94.4% [95% CI, 83.9 to 99.9%]) (P = 0.14); day 1 papule sizes were also similar between the parent-inoculated sites (24.6 ± 14.1 mm2) and mutant-inoculated sites (26.1 ± 23.3 mm2) (P = 0.70). However, pustules formed at 66.7% (95% CI, 44.9 to 88.4%) of 18 parent-inoculated sites and at 33.3% (95% CI, 11.6 to 55.1%) of 18 mutant-inoculated sites (P = 0.007). Thus, although papule formation was not affected, expression of sapA had a significant effect on the ability of H. ducreyi to progress to pustule formation in humans. Although we are precluded by biosafety regulations from testing complemented mutants in humans, the data suggest that SapA contributes significantly to the virulence of H. ducreyi in humans.
Surface cultures taken daily during infection were positive for H. ducreyi at 67% of parent-inoculated and 44% of mutant-inoculated sites. At the clinical endpoint, 3 parent-inoculated and 3 mutant-inoculated sites were biopsied and cultured; H. ducreyi was recovered from 2 parent biopsy specimens and 3 mutant biopsy specimens. All colonies recovered from surface cultures (n = 370 from parent sites; n = 200 from mutant sites) and biopsy specimens (n = 71 from parent sites; n = 55 from mutant sites), as well as colonies from the parent (n = 62) and mutant (n = 72) inocula, were screened for susceptibility to Kan to confirm the presence of H. ducreyi 35000HP (Kan sensitive) or 35000HPsapA (Kanr). All colonies had the expected kanamycin susceptibility phenotype, suggesting no cross-contamination had occurred between parent-inoculated and mutant-inoculated sites.
DISCUSSION
Pathogenic mechanisms involved in the ability of H. ducreyi to survive in vivo are not well defined. Our previous research demonstrated that class I and class II H. ducreyi strains resist killing by several human APs and suggested that this phenotype may be an important survival mechanism during infection (38). In this study, we characterized the H. ducreyi Sap operon and generated an isogenic sapA mutant in H. ducreyi 35000HP. Using this mutant, we have demonstrated a role in vitro for SapA in H. ducreyi resistance to the cathelicidin LL-37; we also defined SapA as an important virulence factor of H. ducreyi in human disease. To our knowledge, this is the first study to show that an AP resistance mechanism contributes to bacterial virulence in humans.
Sap transporters have been shown to be required for AP resistance in various Gram-negative pathogens of both animal and plant hosts. In S. enterica serovar Typhimurium, sapABCDF mutants are more susceptible than the wild-type bacteria to the defensin melittin and to crude extracts of human granulocytes and contribute to virulence in mice (19, 41). In nontypeable H. influenzae, a sapA mutant confers resistance to the chinchilla beta defensin 1 (cBD-1) and to human APs HBD-3 and LL-37; the sapA mutant is also attenuated in a chinchilla model of infection (34, 35). A Sap transporter in the plant pathogen Erwinia chrysanthemi confers resistance to two classes of plant AP and contributes to virulence (32). McCoy et al. generated AP-susceptible mutants housing mutations within the sap operon of Proteus mirabilis; each of these mutants is susceptible to polymyxin B, and two are also susceptible to the synthetic protegrin IB-367 (36). Interestingly, while the Sap transporter plays a role in the AP resistance of many different bacteria, it does not confer AP resistance to all bacteria expressing sap genes. In Vibrio fischeri, a polar mutation within the sapABCDF operon does not confer resistance to any of the eight APs tested, including LL-37, but it does affect in vitro growth and in vivo colonization of the host (33). Taken together, these studies indicate that the Sap transporter has evolved distinct functionality within each bacterial species to meet the need of that particular pathogen.
The contributions of Sap transporters to AP resistance of H. ducreyi and H. influenzae are not identical. H. ducreyi 35000HPsapA showed increased sensitivity to the human cathelicidin but not to α- or β-defensins; in contrast, the H. influenzae sapA mutant affects resistance to both cathelicidin and β-defensins (34, 35). Additionally, although the effect of the H. ducreyi sapA mutation on LL-37 resistance was significant, the roughly 25% reduction in survival (Fig. 3) was not as profound as the 8-fold reduction reported in H. influenzae (35). The lack of observed Sap-mediated effects on defensins in H. ducreyi suggests that the H. ducreyi and H. influenzae Sap transporters may differ in their specificities for APs. Whether this difference would be at the level of the AP binding SapA or at the downstream step of transport through the Sap channel is unknown.
How Sap transporters function to protect the cell from AP attack is not clearly defined, although models have been proposed based on the homology of sap genes with genes of other ABC transporters (34, 41). Studies to compare the fate of APs in parent and isogenic sap mutant bacteria could shed light on the mechanism of Sap-mediated AP resistance. Because Sap is an uptake transporter, APs would be expected to be incorporated into the cell's cytoplasm or inner membrane, respectively, in parent and sap mutant bacteria, with differences being discerned by subcellular localization of the AP. In H. ducreyi, however, the modest difference in LL-37 susceptibility between strains 35000HP and 35000HPsapA suggests that we would observe only a minor difference in AP localization between the two strains, which is not likely to be informative. Thus, we did not pursue these experiments in our system.
Although the Sap transporter has a significant effect on LL-37 resistance in H. ducreyi, our findings also suggest that H. ducreyi expresses additional AP resistance mechanism(s) that function alongside the Sap transporter to thwart killing by defensins. Investigations with S. Typhimurium and Staphylococcus aureus have revealed the presence of several redundant resistance mechanisms within the same bacterial species (27). These redundant mechanisms seem to complement one another to achieve high-level resistance against a broad spectrum of APs. Sequence analysis of the H. ducreyi genome reveals the presence of regions of homology to other known AP resistance factors, suggesting that a similar dynamic may occur within H. ducreyi. We are currently examining the roles of these other putative resistance factors in H. ducreyi.
Our previous work examining H. ducreyi transcripts in vivo identified several sap genes and suggested that the sap operon may be upregulated during human infection. Neither the regulator nor an in vivo signal has been identified for upregulation of the sap operon. We examined the ability of LL-37 to act as a regulatory signal but observed no transcriptional response of sapA to LL-37. We also examined the effects of cpxRA genes, the only intact two-component regulator in H. ducreyi, and saw no effect on sapA expression. Interestingly, the first gene in the sap operon is a homolog of tyrR, the gene product of which regulates aromatic amino acid synthesis and transport in E. coli (42). Whether the sap operon is regulated by TyrR is under investigation.
Sequence analysis demonstrated that the sapA ORF is present in both class I and class II H. ducreyi strains. As has been shown with other H. ducreyi virulence factors, we found a minor variation in the ORFs of class I strains but relatively greater variation between sapA ORFs of class I and class II strains (9, 51). The conservation of SapA among class I and class II clinical isolates of H. ducreyi, coupled with the conserved AP resistance phenotype (38), suggests that SapA-mediated AP resistance may be a conserved mechanism in H. ducreyi. PCR amplification using primers specific to other genes in the sapABCD operon further suggested that the operon structure may be conserved among class I and class II strains (data not shown).
The human model of H. ducreyi infection is a powerful tool for discerning genes that contribute to H. ducreyi pathogenesis (25). In this study, we used the human challenge model to show that SapA contributes significantly to human disease caused by H. ducreyi. Although these studies do not demonstrate the mechanism for attenuation of the sapA mutant in vivo, our in vitro data suggest that SapA may be important in vivo for resisting LL-37-mediated killing. An AP resistance mechanism contributing to in vivo survival is consistent with our understanding of H. ducreyi pathogenesis, in that the organism is surrounded by LL-37-secreting cells during infection (8, 10). The partial resistance to LL-37 in vitro correlates with the partial attenuation in vivo. Future studies will include determining whether loss of the Sap channel proteins leads to a greater phenotype in vitro and in vivo.
Acknowledgments
We thank Ashley Lambert for technical assistance, Susan Ofner for statistical support, Sheila Ellinger for assistance with regulatory documents for the human trials, and X. Frank Yang for helpful discussions and critical review of the manuscript. We thank the volunteers who enrolled in the human challenge study.
This work was supported by National Institutes of Health (NIH) National Institute of Allergy and Infectious Disease (NIAID) Public Health Service grant R21 AI075008, Developmental Awards Program of the NIH NIAID Sexually Transmitted Infections and Topical Microbicide Cooperative Research Centers (STI-TM CRC) grants to the University of Washington (AI 31448) and Indiana University (AI 31494), and a grant from the Ralph W. and Grace M. Showalter Research Trust. The human challenge trials were supported by NIH NIAID Public Health Service grant U19 AI31494 and by the Indiana Clinical and Translational Sciences Institute and the Indiana Clinical Research Center (UL RR052761).
Editor: S. R. Blanke
Footnotes
Published ahead of print on 19 January 2010.
REFERENCES
- 1.Al-Tawfiq, J. A., K. R. Fortney, B. P. Katz, C. Elkins, and S. M. Spinola. 2000. An isogenic hemoglobin receptor-deficient mutant of Haemophilus ducreyi is attenuated in the human model of experimental infection. J. Infect. Dis. 181:1049-1054. [DOI] [PubMed] [Google Scholar]
- 2.Al-Tawfiq, J. A., A. C. Thornton, B. P. Katz, K. R. Fortney, K. D. Todd, A. F. Hood, and S. M. Spinola. 1998. Standardization of the experimental model of Haemophilus ducreyi infection in human subjects. J. Infect. Dis. 178:1684-1687. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Annan, N. T., and D. A. Lewis. 2005. Treatment of chancroid in resource-poor countries. Expert Rev. Anti Infect. Ther. 3:295-306. [DOI] [PubMed] [Google Scholar]
- 5.Banks, K. E., K. R. Fortney, B. Baker, S. D. Billings, B. P. Katz, R. S. Munson, Jr., and S. M. Spinola. 2008. The enterobacterial common antigen-like gene cluster of Haemophilus ducreyi contributes to virulence in humans. J. Infect. Dis. 197:1531-1536. [DOI] [PubMed] [Google Scholar]
- 6.Bauer, B. A., M. K. Stevens, and E. J. Hansen. 1998. Involvement of the Haemophilus ducreyi gmhA gene product in lipooligosaccharide expression and virulence. Infect. Immun. 66:4290-4298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bauer, M. E., K. R. Fortney, A. Harrison, D. M. Janowicz, R. S. Munson, Jr., and S. M. Spinola. 2008. Identification of Haemophilus ducreyi genes expressed during human infection. Microbiology 154:1152-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bauer, M. E., M. P. Goheen, C. A. Townsend, and S. M. Spinola. 2001. Haemophilus ducreyi associates with phagocytes, collagen, and fibrin and remains extracellular throughout infection of human volunteers. Infect. Immun. 69:2549-2557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer, M. E., C. A. Townsend, R. S. Doster, K. R. Fortney, B. W. Zwickl, B. P. Katz, S. M. Spinola, and D. M. Janowicz. 2009. A fibrinogen-binding lipoprotein contributes to the virulence of Haemophilus ducreyi in humans. J. Infect. Dis. 199:684-692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bauer, M. E., C. A. Townsend, A. R. Ronald, and S. M. Spinola. 2006. Localization of Haemophilus ducreyi in naturally acquired chancroidal ulcers. Microb. Infect. 8:2465-2468. [DOI] [PubMed] [Google Scholar]
- 11.Bong, C. T. H., J. Harezlak, B. P. Katz, and S. M. Spinola. 2002. Men are more susceptible to pustule formation than women in the experimental model of Haemophilus ducreyi infection. Sex. Transm. Dis. 29:114-118. [DOI] [PubMed] [Google Scholar]
- 12.Bozue, J. A., L. Tarantino, and R. S. Munson, Jr. 1998. Facile construction of mutations in Haemophilus ducreyi using lacZ as a counter-selectable marker. FEMS Microbiol. Lett. 164:269-273. [DOI] [PubMed] [Google Scholar]
- 13.Deneer, H. G., L. Slaney, I. W. Maclean, and W. L. Albritton. 1982. Mobilization of nonconjugative antibiotic resistance plasmids in Haemophilus ducreyi. J. Bacteriol. 149:726-732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Wulf, P., A. M. McGuire, X. Liu, and E. C. C. Lin. 2002. Genome-wide profiling of promoter recognition by the two-component response regulator CpxR-P in Escherichia coli. J. Biol. Chem. 277:26652-26661. [DOI] [PubMed] [Google Scholar]
- 15.Duits, L. A., B. Ravensbergen, M. Rademaker, P. S. Hiemstra, and P. H. Nibbering. 2002. Expression of β-defensin 1 and 2 mRNA by human monocytes, macrophages, and dendritic cells. Immunology 106:517-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Faurschou, M., O. E. Sörensen, A. H. Johnsen, J. Askaa, and N. Borregaard. 2002. Defensin-rich granules of human neutrophils: characterization of secretory properties. Biochim. Biophys. Acta 1591:29-35. [DOI] [PubMed] [Google Scholar]
- 17.Ganz, T. 2003. Defensins: antimicrobial peptides of innate immunity. Nat. Rev. Immunol. 3:710-720. [DOI] [PubMed] [Google Scholar]
- 18.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. U. S. A. 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Groisman, E. A., C. Parra-Lopez, M. Salcedo, C. J. Lipps, and F. Heffron. 1992. Resistance to host antimicrobial peptides is necessary for Salmonella virulence. Proc. Natl. Acad. Sci. U. S. A. 89:11939-11943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gudmundsson, G. H., B. Agerberth, J. Odeberg, T. Bergman, B. Olsson, and R. Salcedo. 1996. The human gene FALL39 and processing of the cathelin precursor to the antibacterial peptide LL-37 in granulocytes. Eur. J. Biochem. 238:325-332. [DOI] [PubMed] [Google Scholar]
- 21.Hammond, G. W., C. J. Lian, J. C. Wilt, and A. R. Ronald. 1978. Antimicrobial susceptibility of Haemophilus ducreyi. Antimicrob. Agents Chemother. 13:608-612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harder, J., J. Bartels, E. Christophers, and J.-M. Schröder. 2001. Isolation and characterization of human β-defensin-3, a novel human inducible peptide antibiotic. J. Biol. Chem. 276:5707-5713. [DOI] [PubMed] [Google Scholar]
- 23.Harder, J., U. Meyer-Hoffert, K. Wehkamp, L. Schwichtenberg, and J.-M. Schröder. 2004. Differential gene induction of human β-defensins (hBD-1, -2, -3, and -4) in keratinocytes is inhibited by retinoic acid. J. Investig. Dermatol. 123:522-529. [DOI] [PubMed] [Google Scholar]
- 24.Janowicz, D. M., K. R. Fortney, B. P. Katz, J. L. Latimer, K. Deng, E. J. Hansen, and S. M. Spinola. 2004. Expression of the LspA1 and LspA2 proteins by Haemophilus ducreyi is required for virulence in human volunteers. Infect. Immun. 72:4528-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Janowicz, D. M., S. Ofner, B. P. Katz, and S. M. Spinola. 2009. Experimental infection of human volunteers with Haemophilus ducreyi: fifteen years of clinical data and experience. J. Infect. Dis. 199:1671-1679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jenssen, H., P. Hamill, and R. E. Hancock. 2006. Peptide antimicrobial agents. Clin. Microbiol. Rev. 19:491-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kraus, D., and A. Peschel. 2006. Molecular mechanisms of bacterial resistance to antimicrobial peptides. Curr. Top. Microbiol. Immunol. 306:231-250. [DOI] [PubMed] [Google Scholar]
- 28.Labandeira-Rey, M., J. R. Mock, and E. J. Hansen. 2009. Regulation of expression of the Haemophilus ducreyi LspB and LspA2 proteins by CpxR. Infect. Immun. 77:3402-3411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lehrer, R. I. 2004. Primate defensins. Nat. Rev. Microbiol. 2:727-738. [DOI] [PubMed] [Google Scholar]
- 30.Liu, L., L. Wang, H. P. Jia, C. Zhao, H. H. Q. Heng, B. C. Schutte, P. B. McCray, Jr., and T. Ganz. 1998. Structure and mapping of the human β-defensin HBD-2 gene and its expression at sites of inflammation. Gene 222:237-244. [DOI] [PubMed] [Google Scholar]
- 31.Liu, P. T., S. Stenger, H. Li, L. Wenzel, B. H. Tan, S. R. Krutzik, M. T. Ochoa, J. Schauber, K. Wu, C. Meinken, D. L. Kamen, M. Wagner, R. Bals, W. Seteinmeyer, U. Zugel, R. L. Gallo, D. Eisenberg, M. Hewison, B. W. Hollis, J. S. Adams, B. R. Bloom, and R. L. Modlin. 2006. Toll-like receptor triggering of a vitamin D-mediated human antimicrobial response. Science 311:1770-1773. [DOI] [PubMed] [Google Scholar]
- 32.López-Solanilla, E., F. García-Olmedo, and P. Rodríguez-Palenzuela. 1998. Inactivation of the sapA to sapF locus of Erwinia chrysanthemi reveals common features in plant and animal bacterial pathogenesis. Plant Cell 10:917-924. [PMC free article] [PubMed] [Google Scholar]
- 33.Lupp, C., R. E. W. Hancock, and E. G. Ruby. 2002. The Vibrio fischeri sapABCDF locus is required for normal growth, both in culture and in symbiosis. Arch. Microbiol. 179:57-65. [DOI] [PubMed] [Google Scholar]
- 34.Mason, K. M., M. E. Bruggeman, R. S. Munson, Jr., and L. O. Bakaletz. 2006. The non-typeable Haemophilus influenzae Sap transporter provides a mechanism of antimicrobial peptide resistance and SapD-dependent potassium acquisition. Mol. Microbiol. 62:1357-1372. [DOI] [PubMed] [Google Scholar]
- 35.Mason, K. M., R. S. Munson, Jr., and L. O. Bakaletz. 2005. A mutation in the sap operon attenuates survival of nontypeable Haemophilus influenzae in a chinchilla model of otitis media. Infect. Immun. 73:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCoy, A. J., H. Liu, T. J. Falla, and J. S. Gunn. 2001. Identification of Proteus mirabilis mutants with increased sensitivity to antimicrobial peptides. Antimicrob. Agents Chemother. 45:2030-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Menard, R., P. J. Sansonetti, and C. Parsot. 1993. Nonpolar mutagenesis of the ipa genes defines IpaB, IpaC, and IpaD as effectors of Shigella flexneri entry into epithelial cells. J. Bacteriol. 175:5899-5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mount, K. L. B., C. A. Townsend, and M. E. Bauer. 2007. Haemophilus ducreyi is resistant to human antimicrobial peptides. Antimicrob. Agents Chemother. 51:3391-3393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Niyonsaba, F., H. Ogawa, and I. Nagaoka. 2004. Human β-defensin-2 functions as a chemotactic agent for tumour necrosis factor-α-treated human neutrophils. Immunology 111:273-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niyonsaba, F., H. Ushio, I. Nagaoka, K. Okumura, and H. Ogawa. 2005. The human β-defensins (-1, -2, -3, -4) and cathelicidin LL-37 induce IL-18 secretion through p38 and ERK MAPK activation in primary human keratinocytes. J. Immunol. 175:1776-1784. [DOI] [PubMed] [Google Scholar]
- 41.Parra-Lopez, C., M. T. Baer, and E. A. Groisman. 1993. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 12:4053-4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pittard, J., H. Camakaris, and J. Yang. 2005. The TyrR regulon. Mol. Microbiol. 55:16-26. [DOI] [PubMed] [Google Scholar]
- 43.Quayle, A. J., E. M. Porter, A. A. Nussbaum, Y. M. Wang, C. Brabee, K. P. Yip, and S. C. Mok. 1998. Gene expression, immunolocalization, and secretion of human defensin-5 in human female reproductive tract. Am. J. Pathol. 152:1247-1258. [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 45.Spinola, S. M., M. E. Bauer, and R. S. Munson, Jr. 2002. Immunopathogenesis of Haemophilus ducreyi infection (chancroid). Infect. Immun. 70:1667-1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spinola, S. M., C. T. H. Bong, A. L. Faber, K. R. Fortney, S. L. Bennett, C. A. Townsend, B. E. Zwickl, S. D. Billings, T. L. Humphreys, M. E. Bauer, and B. P. Katz. 2003. Differences in host susceptibility to disease progression in the human challenge model of Haemophilus ducreyi infection. Infect. Immun. 71:6658-6663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spinola, S. M., K. R. Fortney, B. P. Katz, J. L. Latimer, J. R. Mock, M. Vakevainen, and E. J. Hansen. 2003. Haemophilus ducreyi requires an intact flp gene cluster for virulence in humans. Infect. Immun. 71:7178-7182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spinola, S. M., G. E. Griffiths, J. A. Bogdan, and M. A. Menegus. 1992. Characterization of an 18,000-molecular-weight outer membrane protein of Haemophilus ducreyi that contains a conserved surface-exposed epitope. Infect. Immun. 60:385-391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Trees, D. L., and S. A. Morse. 1995. Chancroid and Haemophilus ducreyi: an update. Clin. Microbiol. Rev. 8:357-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Turner, J., Y. Cho, N.-N. Dinh, A. J. Waring, and R. I. Lehrer. 1998. Activities of LL-37, a cathelin-associated antimicrobial peptide of human neutrophils. Antimicrob. Agents Chemother. 42:2206-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.White, C. D., I. Leduc, B. Olsen, C. Jeter, C. Harris, and C. Elkins. 2005. Haemophilus ducreyi outer membrane determinants, including DsrA, define two clonal populations. Infect. Immun. 73:2387-2399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wood, G. E., S. M. Dutro, and P. A. Totten. 1999. Target cell range of Haemophilus ducreyi hemolysin and its involvement in invasion of human epithelial cells. Infect. Immun. 67:3740-3749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yang, D., O. Chertov, S. N. Bykovskaia, Q. Chen, M. J. Buffo, J. Shogan, M. Anderson, J. M. Schröder, J. M. Wang, O. M. Z. Howard, and J. J. Oppenheim. 1999. β-Defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525-528. [DOI] [PubMed] [Google Scholar]
- 54.Zhao, C., I. Wang, and R. I. Lehrer. 1996. Widespread expression of beta-defensin hBD-1 in human secretory glands and epithelial cells. FEBS Lett. 396:319-322. [DOI] [PubMed] [Google Scholar]
- 55.Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31:3406-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]