Abstract
Anaplasma and related Ehrlichia spp. are important tick-borne, Gram-negative bacterial pathogens of livestock and humans that cause acute infection and disease and can persist. Immunization of cattle with an Anaplasma marginale fraction enriched in outer membranes (OM) can provide complete protection against disease and persistent infection. Serological responses of OM vaccinees to the OM proteome previously identified over 20 antigenic proteins, including three type IV secretion system (T4SS) proteins, VirB9-1, VirB9-2, and VirB10. Subsequent studies showed that these three proteins also stimulated CD4+ T-cell responses in OM vaccinees. The T4SS, composed of a complex of proteins spanning the inner and outer membranes of certain bacteria, is an important virulence factor but is relatively unexplored as a vaccine target. The goal of this study was to determine if additional T4SS proteins are immunogenic for animals immunized with the protective OM fraction of A. marginale. T4SS proteins expressed by in vitro transcription and translation were screened for stimulating proliferation of T cells from OM vaccinees, and immunogenic proteins were expressed as recombinant proteins in Escherichia coli and their immunogenicity was verified. VirB2, a putative VirB7, VirB11, and VirD4 were immunogenic for OM vaccinees expressing several common major histocompatibility complex (MHC) class II haplotypes. VirB2 is encoded by multiple genes that share a conserved central region, and epitope mapping revealed T-cell epitopes in this region. The discovery of novel immunogenic T4SS proteins recognized by outbred individuals with common MHC haplotypes further justifies evaluating the T4SS as a potential vaccine candidate for pathogenic bacteria.
Anaplasma marginale is a tick-transmitted, Gram-negative intraerythrocytic bacterium in the class Alphaproteobacteria and order Rickettsiales (20). Infection of cattle by A. marginale results in ascending rickettsemia that peaks 4 to 5 weeks postinfection, resulting in fever, anemia, and if not controlled, death. Animals that control the infection remain persistently infected for life and serve as reservoirs for infection of naïve animals. Immunization with a fraction enriched for A. marginale outer membranes (OM) can induce complete protection against bacteremia and disease following experimental infection (13, 44, 52, 61, 70). Protective immunity correlates with CD4+ T-cell responses and T-cell-dependent immunoglobulin G2 (IgG2) responses to OM antigens (13). In contrast, equivalent levels of protection have not been afforded by immunization with several major surface proteins (MSPs), which included MSP1 and the highly abundant, immunodominant, and antigenically variable MSP2 and MSP3 (1, 10-12, 57, 60, 62, 63). This raises the question of which proteins or protein complexes in the outer membrane are protective in the face of these immunodominant surface proteins that continually undergo antigenic variation and result in immune evasion (59).
We recently identified over 20 novel, subdominant, immunogenic membrane proteins in the protective outer membrane fraction of A. marginale, among which were several members of the type IV secretion system (T4SS) (45-47). The immunogenic T4SS proteins were VirB2, formerly annotated as OrfX, which is encoded by multiple genes and was immunogenic for one of two animals tested (8, 9, 45, 47), VirB9, VirB10, and conjugal transfer protein (CTP) (46, 47). CTP is now designated VirB9-1, and VirB9 is designated VirB9-2 because of homology to the corresponding proteins in Anaplasma phagocytophilum (GenBank accession nos. YP_504712 and YP_50587). In certain bacteria, including the well-characterized Agrobacterium tumefaciens, approximately 12 proteins comprise the VirB/D T4SS complex, which spans the inner and outer membrane and functions in host cell adhesion/invasion and transfer of DNA and proteins directly to the eukaryotic host cell (5, 14-16). The identification of structurally conserved and immunogenic T4SS proteins in the protective outer membrane fraction, which contains an abundance of antigenically variant and immunodominant MSP2 and MSP3 proteins, suggested these might be good vaccine candidates.
The goal of this study was to determine if any other members of the T4SS are immunogenic in the OM-enriched fraction of A. marginale. Using an in vitro transcription and translation (IVTT) protein expression system (45), A. marginale T4SS proteins were expressed and screened in T-cell proliferation assays with lymphocytes from five OM vaccinees expressing different MHC class II haplotypes. The stimulatory proteins were then produced in Escherichia coli, with purified recombinant proteins tested to verify their immunogenicity. We found that among the newly tested proteins, a putative VirB7 homolog, VirB11, and VirD4 stimulated significant recall T-cell responses from immune animals. Furthermore, the immunogenicity of VirB2 was documented for an additional three OM-immunized cattle. T-cell epitopes in VirB2 were localized to the central region of this protein that is highly conserved among A. marginale VirB2 paralogs and with VirB2s from other members of the Anaplasmataceae. Discovery of these novel CD4+ T-cell antigens that comprise the T4SS further justifies assessing proteins that comprise the T4SS protein complex as vaccine candidates for A. marginale and other Gram-negative pathogens.
MATERIALS AND METHODS
Cattle used in this study.
Holstein cattle designated 04B90, 04B91, 04B92, 4848, and 5982 were immunized with purified A. marginale St. Maries strain OM, as previously described (13, 47). Immune sera used in this study were obtained before immunization and 2 weeks after four immunizations (04B90, 04B91, and 04B92), 2 weeks after five immunizations (5982), or 1 week after three immunizations (4848) with 60 μg OM resuspended in 1.3 ml phosphate-buffered saline (PBS; pH 7.0) containing 6 mg saponin. Sera were then frozen at −20°C. The bovine leukocyte antigen (BoLA) DRB3 alleles were determined by the PCR restriction fragment length polymorphism (PCR-RFLP) method as described previously (72). Further, each PCR product from exon 2 of each DRB3, DQA, and DQB gene was sequenced as described previously (53, 65). The nomenclature of bovine class II genes can be found at the following websites: http://www.projects.roslin.ac.uk/bola/bolahome.html and http://www.ebi.ac.uk/ipd/mhc/bola. BoLA-DRB3 haplotypes for the cattle in this study are as follows: animal 04B90, DRB3 *1101/*1501; animal 04B91, DRB3 *1201/*2703; animal 04B92, DRB3 *0201/*1201; animal 4848, DRB3 *1101/*1101; and animal 5982, DRB3 *1201/*1501. For simplicity, we refer to the RFLP analysis of DRB3 exon 2 designation for the corresponding allelic designations: 04B90, 22/16; 04B91, 8/23; 04B92, 8/7; 4848, 22/22; and 5982, 8/16. During the course of this study, all animals maintained memory/effector T-cell responses, which enabled the development of OM-specific short-term T-cell lines. Animals were used in this study in compliance with the Washington State University Institutional Animal Care and Use Committee.
Cloning and expression of Anaplasma marginale St. Maries strain T4SS genes by IVTT.
Fifteen T4SS genes were amplified by PCR with Accuprime pfx (Invitrogen, Carlsbad, CA) as either full-length genes for open reading frames (ORFs) less than 1.2 kb or as overlapping fragments of approximately 1.2 kb in size by using the primers listed in Table 1. PCR primers were also designed to incorporate a C-terminal sequence encoding the FLAG epitope DYKDDDDK as described previously (45). PCR products were purified using a High Pure 96 UF Cleanup Plate (Roche, Indianapolis, IN) and resuspended in 60 μl of double-distilled H2O. One microliter of each PCR product was used in a cloning reaction with pENTR-D-Topo (Invitrogen), and the mixture was transformed into Top10 E. coli cells. pENTR-D-Topo plasmid clones were sequenced using M13 forward and reverse sequencing primers (Invitrogen). The subsequent clones were used in an LR clonase II reaction with pEXP1-DEST (Invitrogen) to generate plasmids encoding N-terminally tagged 6× His proteins. pEXP1-DEST plasmid clones were sequenced using the T7 and pEXP1 sequencing primers. Approximately 600 ng of each pEXP1-DEST clone was used as a template in a 50-μl in vitro transcription/translation (IVTT) reaction using an RTS 100 E. coli HY kit (Roche, Mannheim, Germany) to achieve expression of cell-free recombinant protein.
TABLE 1.
Primers and protein annotations for expression of T4SS proteins
| Genome ID | Protein annotation | Forward primer (5′ to 3′) | Reverse primera (5′ to 3′) |
|---|---|---|---|
| AM030 | VirB2 | CACCATGGGCTTGCTAAAACTTAGATTCTTACTCATTGC | TCGGTTACTTTGGGCAGTGCC |
| AM097 | CTP | CACCATGGGCAAAAGGCTTTCATGGTTTGTGCTG | CCCACGTCCCCTTCTGGATG |
| AM757 | VirB8-2 | CACCATGGGCCGCGTACGGCCACAAAACAG | GTCTACGTATCTGTACGTATTGATAGTAAAC |
| AM810 | VirB6-4 | CACCATGGGCACACTCCTGCAGGAGAAAAGC | TTGAGAAGACCCACCCAGACC |
| AM810b | VirB6-4F1 | CACCATGGGCACACTCCTGCAGGAGAAAAGC | AAAGTCCGCCATGATGAGGGC |
| AM810b | VirB6-4F2 | CACCATGGGCCTTCTGAAGCTGTTTACAGGGGGC | ACCCTTCTTGAGATCCTTGATTATGTT |
| AM810b | VirB6-4F3 | CACCATGGGCAAGGTGATCGAGGAGAGAGAG | TATTCTATCCCGCACCTCGGG |
| AM810b | VirB6-4F4 | CACCATGGGCGCGCAAAAGGACGATGATGCG | CAGACCCCTATCCTCATCAAC |
| AM810b | VirB6-4F5 | CACCATGGGCGAGCCGCAGGAGGTTGAACCT | TTGAGAAGACCCACCCAGACC |
| AM811 | VirB6-3 | CACCATGGGCTTCAGGTTACTGCTCATTGCG | TTCCTTGTCTTGCTTTTCTCCTGC |
| AM811b | VirB6-3F1 | CACCATGGGCTTCAGGTTACTGCTCATTGCG | AATAACGGCAACCGCACCTTC |
| AM811b | VirB6-3F2 | CACCATGGGCGAGGACTGCACACGCTCC | CCTATCGCCATCTAGGTACCC |
| AM811b | VirB6-3F3 | CACCATGGGCGTGATCAAAATTACCAAGTACGATCTC | AGGCCCACCGAAATCAGTCACATC |
| AM811b | VirB6-3F4 | CACCATGGGCCCGCCAGGAGAAGAGAAGCTG | TTCCTTGTCTTGCTTTTCTCCTGC |
| AM812 | VirB6-2 | CACCATGGGCTTTTGTAACGCTTCTGCGCTGC | AAAGCTAGCCGGAGGATCAGC |
| AM812b | VirB6-2F1 | CACCATGGGCTTTTGTAACGCTTCTGCGCTGC | AGGATGCAATGTCTGTGTGCC |
| AM812b | VirB6-2F2 | CACCATGGGCGACGAGCTTGCAGCCAACGAA | TGCTAGCAGCTGAATCCACGTTTG |
| AM812b | VirB6-2F3 | CACCATGGGCCCGGACAGCTGGACCTTTTTTAAC | AAAGCTAGCCGGAGGATCAGC |
| AM813 | VirB6-1 | CACCATGGGCCGTAGAGCGGTCAGGGC | CGTCCCAGACGACCCTCC |
| AM813b | VirB6-1F1 | CACCATGGGCCGTAGAGCGGTCAGGGC | TACTGCCTTTGACTTCTCGCA |
| AM813b | VirB6-1F2 | CACCATGGGCGCCATTTCTGGGGCTGGA | AGTAAACATCAGCGACAGGAACGA |
| AM813b | VirB6-1F3 | CACCATGGGCCCCGCAATCCTCATAAACGCC | CGTCCCAGACGACCCTCC |
| AM814 | VirB4-1 | CACCATGGGCTTGAGACTCGGGAGAACTGCAG | CGCATTTCTAACCTTCTGGCAG |
| AM814b | VirB4-1F1 | CACCATGGGCTTGAGACTCGGGAGAACTGCAG | AACCGTCAGGTGATGGAGCCC |
| AM814b | VirB4-1F2 | CACCATGGGCTCTGCCGGAATGTTAGATGCG | CTCAAACCCAAAGACCCTGGA |
| AM814b | VirB4-1F3 | CACCATGGGCAGCGACCTAACTCCAGATGAC | CGCATTTCTAACCTTCTGGCAG |
| AM815 | VirB3 | CACCATGGGCTCGTCCGGTAGCGTAAAGAC | CATCACATCGTAAGAATTGGCAT |
| AM1053 | VirB4-2 | CACCATGGGCTCTTTCATAGATAGTTTTGTGCGC | CTGCGCGAGTTTATCGCAC |
| AM1053b | VirB4-2F1 | CACCATGGGCTCTTTCATAGATAGTTTTGTGCGC | TGAGCTTTCGTCGGTGGA |
| AM1053b | VirB4-2F2 | CACCATGGGCGGAGCAATTCTTGGCGTCAAG | CGAAAAAGCCCCCTGGGC |
| AM1053b | VirB4-2F3 | CACCATGGGCGAGATAGCTGTTGGGGTTGTG | CTGCGCGAGTTTATCGCAC |
| AM1312 | VirD4 | CACCATGGGCCACAGTAGCTCTAATCACATACGTAAC | CGGGTTGTCATCATCACGCTG |
| AM1312b | VirD4 F1 | CACCATGGGCCACAGTAGCTCTAATCACATACGTAAC | GGGGTTGGCCCACAGTTC |
| AM1312b | VirD4 F2 | CACCATGGGCTATCTGCTTGCAGTGCCG | CTGAGCGCTTTCTTCATGAGC |
| AM1312b | VirD4 F3 | CACCATGGGCATCGAATCTACGTATCCGATC | CGGGTTGTCATCATCACGCTG |
| AM1313 | VirB11 | CACCATGGGCTTGTGTGCGCATATGACAGC | ATCATTGCCTTGTGAACATTTAGTG |
| AM1314 | VirB10 | CACCATGGGCAGTTTAGGGATGTCAGACGAAACC | CCTACGCACCGCCTCCC |
| AM1314b | VirB10F1 | CACCATGGGCAGTTTAGGGATGTCAGACGAAACC | CCTAGAGCCTTTCGGTATCAT |
| AM1314b | VirB10F2 | CACCATGGGCACTTGGAGCACACTGGACGGC | CCTACGCACCGCCTCCC |
| AM1315 | VirB9 | CACCATGGGCAATTTCTATAAAAACTTTGCTTGCGTGC | AAGCACCGTATTCACTACTTCGAC |
| AM1316 | VirB8-1 | CACCATGGGCGGTATGTTCGGTTTCGGTAAGAAG | TAGAAATTCATCGTCTGCCCTATAG |
| AM306 | VirB7c | ATGAGATTTTCTAGGGTGGTGC | CCCAAGCCACCACTGGTTTACTc |
| BBOV_I003060 | MSA1d | GCCGATACTTCAATCGTCCTTCC | TGTACCCTGTTGTCCTTGGAGG |
Each reverse primer contains a leader sequence, CTATTTGTCGTCGTCGTCTTTATAGTC, encoding the FLAG epitope to be expressed on the C terminus.
Due to large gene size, fragments of AM810, AM811, AM812, AM813, AM1053, AM1312, and AM1314 were constructed along with the full-length counterparts.
Primers used to express recombinant VirB7 were constructed without the FLAG sequence.
Primers used to express recombinant B. bovis MSA1.
Protein quantity was determined by dot blot analysis and a monoclonal antibody (MAb) specific for the FLAG epitope (Sigma-Aldrich, St. Louis, MO), as previously described (45) with slight modifications. A HybriDot Manifold (BRL Life Technologies, Inc. Gaithersburg, MD) was used to spot a prewetted nitrocellulose membrane with 1 μl IVTT product diluted in 200 μl phosphate-buffered saline (PBS), pH 7.0. The membranes were washed in I-Block reagent (Applied Biosystems, Bedford, MA) containing 0.05% Tween 20 (Bio-Rad, Hercules, CA) (referred to here as I-Block) for 1 h, with a subsequent 1 h of incubation with either mouse anti-FLAG (Sigma-Aldrich) or anti-penta His MAb (Qiagen, Valencia, CA) diluted 1:10,000 in blocking reagent, and extensive washing. The membranes were then incubated for 1 h with alkaline phosphatase goat anti-mouse IgG conjugate (Applied Biosystems) diluted 1:10,000 in blocking reagent and washed. Antibody binding was detected with a Western-Star reagent system (Applied Biosystems). Dot blot densitometry was analyzed using AlphaEase FC version 4.0.0 software (Alpha Innotech, San Leandro, CA) to determine relative expression of each IVTT product. As only proteins expressing the C-terminal FLAG epitope were full length, protein concentrations were based on nanomolar levels of the FLAG epitope on recombinant VirB9 used as a standard and determined for 0.015 to 2 μg of protein. These ranged from 36.13 nM to 26.17 μM IVTT-expressed proteins in each reaction mixture, which was within the range of detection of IVTT-expressed proteins in our previous study (45).
Bead purification of IVTT proteins.
IVTT protein products were purified as previously described, with slight modifications (45). Briefly, 10 μl of each IVTT reaction mixture was bound to 2.5 μg of either anti-FLAG MAb or, for VirD4 in one experiment, anti-Penta His MAb (Qiagen) that was diluted in 440 μl A/G buffer (0.1 M Tris-HCl, 0.15 M NaCl [pH 7.4]), with a final concentration of 5 μg MAb/ml. The MAb-IVTT protein mixture was incubated at 4°C for 2 h with continuous rocking. For each reaction, 12.5 μl protein G carboxylate latex microspheres (Polysciences, Inc., Warrington, PA) were washed with 750 μl A/G buffer three times and pelleted after each wash at 16,000 × g for 5 min and resuspended in A/G buffer. The mixture was then incubated for 2 h at 4°C with continuous rocking. The final bead-MAb-protein matrixes were pelleted at 16,000 × g for 5 min and washed three times with 750 μl A/G buffer. Based on the manufacturers' specifications, protein G beads were diluted with RPMI medium to a final concentration of 2 to 10 μg protein G/ml so that each well for proliferation assays received 1 to 5 μg protein G beads/ml.
Recombinant protein expression and purification.
Recombinant T4SS proteins used in Western blotting and T-cell proliferation assays were expressed from pEXP1-DEST plasmid. Plasmids containing AM1313 (VirB11), the first (N-terminal) and second (middle) fragments of AM1312 (VirD4F1 and VirD4F2), full-length AM1312 (VirD4), and AM030 (VirB2) were transformed, according to the manufacturers' specifications, into BL-21 DE3 LysS One Shot chemically competent E. coli (Invitrogen). Individual colonies were selected and grown in LB broth with 50 μg/ml carbenicillin (LBC broth). The E. coli were then harvested, and plasmid DNA was obtained using a Wizard Plus SV Minipreps DNA purification system (Promega, Madison, WI). Upon sequence confirmation, E. coli containing the particular genes of interest was grown in 1 ml LBC broth overnight, harvested after centrifugation at 4,000 × g, and frozen at −20°C as a 50% LBC broth-glycerol stock.
To produce recombinant VirB2, VirB11, VirD4F1, and VirD4F2, bacteria were expanded in LBC broth overnight, induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 5 h, and then harvested after centrifugation at 4,000 × g for 20 min. The E. coli was resuspended in 6 M guanidine hydrochloride, 20 mM dibasic sodium phosphate, and 500 mM sodium chloride, pH 7.8, for 30 min, and then sonicated four times at 350 W for 45 s each time. Proteins were extracted from the E. coli lysate using nickel chelating resin from a ProBond purification system (Invitrogen). Recombinant VirB11 and VirD4F1 were purified using a Probond system two consecutive times, followed by dialysis into PBS using Slide-A-Lyzer dialysis cassettes with a 10-kDa molecular mass cutoff (Thermo Fisher Scientific, Rockford, IL). Recombinant VirB2 and VirD4F2 were purified once using a ProBond system, under denaturing conditions, and dialyzed in 10 mM Tris buffer at pH 7.8 containing 0.1% Triton X detergent (Bio-Rad) using Slide-A-Lyzer dialysis cassettes with a 10-kDa molecular mass cutoff, purified by immunoaffinity chromatography using an anti-FLAG agarose matrix (Sigma-Aldrich), and dialyzed against PBS. VirB9-1, VirB9-2, VirB10, and a negative-control protein from M07 strain Babesia bovis, merozoite surface antigen 1 (MSA1), and T4SS protein VirB7 were also expressed and purified for this study as previously described (43, 46), using gene-specific primers (Table 1). AM306 (putative virb7), was amplified from A. marginale St. Maries strain genomic DNA without a C-terminal FLAG epitope. For the msa1 construct, a C-terminal reverse primer was constructed to contain the FLAG epitope DYKDDDDK. Both msa1 and virb7 constructs were cloned into the pBAD/TOPO ThioFusion plasmid under the manufacturer's specifications (Invitrogen) and then sequenced, screened and purified using a ProBond system.
Protein purity was determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and Western blotting using 4 to 20% precast Tris-HCl polyacrylamide Ready Gels (Bio-Rad). One gel was stained with Coomassie brilliant blue, and the second was transferred to a nitrocellulose membrane for immunoblot analysis. The nitrocellulose membrane was blocked overnight in I-Block and probed with either mouse anti-FLAG MAb (Sigma-Aldrich) or mouse anti-Penta His MAb (Qiagen), both diluted 1:10,000 in I-Block, and incubated for 1 h. After an extensive washing step, the blots were incubated with alkaline phosphatase conjugated goat anti-mouse IgG antibody (Applied Biosystems), diluted 1:10,000 in I-Block, and incubated for 1 h. The nitrocellulose membranes were then washed with I-Block for 1 h and developed with a Western-Star chemiluminescent immunoblot detection system (Applied Biosystems). The purity of the recombinant proteins was determined after the comparison of bands between the Coomassie brilliant blue-stained gels and immunoblots. Protein concentrations were determined using a Quick Start Bradford assay (Bio-Rad), and then the proteins were frozen at −20°C.
Bioinformatic comparison of VirB7s.
A. marginale strain St. Maries putative VirB7 (AM306; GenBank accession no. AAV86397) amino acid sequence was aligned with Com7 from Campylobacter jejuni strain 81-176 (accession no. AAN46947), Com7 from Helicobacter pylori strain Shi170 (accession no. ACF17783), and VirB7 from Agrobacterium tumefaciens strain Bo542 (accession no. AAZ50524). Alignment was performed with ClustalW (AlignX) followed by manual editing.
VirB2 peptides and bioinformatic comparison of VirB2s.
Six peptides, with overlapping amino acids that span the entire sequence of VirB2 (AM030), were synthesized (Laboratory for Biotechnology and Bioanalysis I, Washington State University, Pullman, WA), diluted in PBS with or without dimethyl sulfoxide, as needed for solubility, to a concentration of 1 mg/ml, and frozen at −20°C. Representative VirB2 sequences were obtained from Anaplasmataceae genome sequences and aligned using ClustalW (22). The genes used to obtain the deduced amino acid sequences are as follows: for A. marginale strain St. Maries, AM030, AM044, AM065, AM077, AM082, AM210, AM717, AM723, AM989, AM1061, AM1149, and AM1253 (CP000030; reference 9); for A. phagocytophilum strain HZ, APH_1130-1134, and APH_1144 (CP000235; reference 21); for Ehrlichia canis strain Jake, ECAJ_0842 (CP000107); for Ehrlichia chaffeensis strain Arkansas, ECH_1042 (CP000236; reference 21); for E. ruminantium strain Welgevonden, Erum7990 (CR767821; reference 17); for Wolbachia pipientis wMel, WD_0651 (AE017196; reference 74); and for Neorickettsia sennetsu strain Miyayama, NSE_0770 (CP000237; reference 21).
Short-term T-cell lines specific for A. marginale OM proteins and proliferation assays.
Two-week CD4+ T-cell lines were established as previously described (45), with slight modifications. Briefly, peripheral blood mononuclear cells (PBMC) were isolated from A. marginale OM-immunized cattle (47, 52), plated at 4 × 106 cells/well in 24-well flat-bottom plates for 7 days with 5 μg OM, and incubated at 37°C with 5% CO2 in air. On day 7, the cells were harvested and replated at 0.7 × 106 cells/well with freshly harvested, irradiated (3,000 rads) autologous PBMC as a source of antigen presenting cells (APC) at 2 × 106 cells/well (resting). The cells were harvested after 7 days, washed repeatedly in complete RPMI, and tested in proliferation assays. Cell lines were tested for a proliferative response to A. marginale OM and T4SS antigens, expressed either by IVTT or as recombinant proteins in E. coli, or as VirB2 peptides. Cells were cultured in triplicate wells of 96-well U-bottom plates at 3 × 104 cells/well with 2 × 105 irradiated, autologous PBMC as a source of APC and incubated at 37°C in 5% CO2 in air with medium or various concentrations of antigen. Positive controls consisted of OM antigen and 10% T-cell growth factor (TCGF) and negative-control antigens consisted of uninfected red blood cell membranes (URBC), the no-DNA control IVTT reaction mixture incubated with beads, B. bovis MSA1, and B. bovis rhoptry-associated protein 1 (RAP-1) peptide P1 (54). Each antigen was tested at a final concentration of 1, 2.5, or 10 μg protein/ml, and IVTT beads were tested at 1 to 5 μg protein G beads/ml in 100-μl triplicate cultures. After 3 days, the cells were radiolabeled for 6 to 18 h with 0.25 μCi of [3H]thymidine (Dupont; New England Nuclear, Boston, MA) and harvested onto glass filters, and radionucleotide incorporation was measured with a Betaplate 1205 liquid scintillation counter (Wallac, Gaithersburg, MD). Proliferation is expressed as either mean counts per minute (CPM) ± 1 standard deviation (SD) for triplicate cultures or as a stimulation index (SI), determined as the mean CPM of cells cultured with antigen divided by the mean CPM of cells cultured with negative-control antigen. Proliferative responses to IVTT-expressed antigens were considered to be biologically significant if the SI was ≥2.0, and the mean CPM were ≥1,000. For other recombinant proteins and synthetic peptides, the statistical significance of the response to T4SS antigens compared to the response to negative control MSA1 or peptide RAP-1 P1, respectively, were determined using a one-tailed Student t test with a P value of <0.05 and the Bonferroni correction for multiple comparisons.
Surface phenotype analysis of T-cell lines.
Flow cytometry was used to characterize the composition of the T-cell lines, as previously described (46). Briefly, MAbs specific for bovine T-cell specific surface antigens CD8 (CACT80c), CD4 (CACT138), and γδ T-cell receptor (GB21A) were used to stain 2-week OM-stimulated T-cell lines. The MAbs were purchased from the Monoclonal Antibody Center, Washington State University, Pullman, WA.
Serological analysis of immune bovine sera.
Sera were obtained from the five OM-immunized cattle before immunization and after the last immunization, and from two cattle 41 days after experimental infection with the St. Maries strain by needle inoculation, and they were used to probe immunoblots of recombinant T4SS proteins VirB2, VirB9-1, VirB9-2, VirB7, VirB10, VirB11, and VirD4. B. bovis MSA-1 was used as a negative control. Recombinant proteins (1 μg), A. marginale homogenate (15 μg), or URBC (15 μg) were electrophoresed on SDS-PAGE gels and transferred to nitrocellulose membranes as described above, and bovine sera were diluted 1:100, incubated in I-Block as described above, and washed. The blots were then incubated with alkaline phosphatase-conjugated goat anti-bovine IgG antibody (Kirkegaard and Perry Laboratories, Gaithersburg, MD), diluted 1:10,000 in I-Block, and incubated for 1 h. The nitrocellulose membranes were washed, and they were developed with a Western-Star chemiluminescent immunoblot detection system.
Detection of VirB2, VirB7, VirB11, and VirD4 mRNA in A. marginale.
Total RNA was isolated from A. marginale (St. Maries strain)-infected bovine erythrocytes as described previously (10) and used for reverse transcription-PCR (RT-PCR) analysis to verify expression of transcripts encoding the newly identified T4SS antigens. Briefly, cDNA was obtained following reverse transcription using a iScript cDNA synthesis kit (Bio-Rad) following the manufacturer's specifications. cDNAs were amplified by PCR. Primers for amplifying VirB2 and VirB7 are the same as those listed in Table 1. For VirB11, the forward primer was 5′-CACCATGGGCTTGTGTGCGCATATGACAGC-3′ and the reverse primer was 5′-GATATCCCCGCTATCCAGGAGCTC-3′, and for VirD4, the forward primer was 5′-CCCTCTAGACTGGATTAGCAAAAAGCCC-3′ and the reverse primer was 5′-GCCTGCTGCTCTCCTTCTTCAGATGTAGGC-3′. PCRs consisted of 18 μl Supermix High Fidelity (Invitrogen), 5 μM (0.5 μl) each forward and reverse primer, and 1 μl of cDNA template, and amplification consisted of 35 cycles with a melting point of 94°C for 30 s, annealing temperature of 55°C for 1 min, and extension at 72°C for 2.5 min.
RESULTS
T-cell responses to T4SS proteins expressed by IVTT.
A. marginale T4SS proteins VirB9-1, VirB9-2, and VirB10 are recognized by both IgG and CD4+ T cells from cattle immunized with the protective OM vaccine (46, 47). The present study was designed to express and test the immunogenicity of additional predicted T4SS proteins that have homology to known T4SS proteins of other Gram-negative bacteria (9). In addition, the number of cattle tested was increased to five.
To rapidly express and screen the A. marginale T4SS proteins for stimulation of a recall T-cell response, a high-throughput IVTT expression system was employed (45). The primers used to express full-length genes or overlapping gene fragments encoding these proteins are listed in Table 1. Overlapping fragments were used for many of the larger open reading frames (ORFs) that were difficult to express as full-length proteins. As for all Rickettsiales pathogens, A. marginale has two VirB4 genes, multiple VirB6 genes, two VirB8 genes, and two VirB9 genes (9, 21, 32, 55). The A. marginale T4SS proteins were constructed to express a C-terminal FLAG epitope and an N-terminal 6× His tag, and they were affinity purified on anti-FLAG-protein G beads to ensure purification of the full-length protein. Only those proteins that expressed a C-terminal FLAG epitope, determined by dot blot analysis with a MAb specific for FLAG, were bead-affinity purified and tested for T-cell proliferation, with the exception of full-length VirD4 (Table 2). In one experiment, VirD4 that did not react with anti-FLAG MAb was affinity purified with anti-6× His beads, and protein was examined by Western blotting using anti-His MAb, which revealed two bands of approximately 30 and 50 kDa (the full-length protein is predicted to be ∼100 kDa; data not shown). Expression of three overlapping fragments of VirD4 (Table 1) was also attempted. However, only the first two (F1 and F2) could be expressed by IVTT. This was true as well for some of the VirB4, VirB6, and VirB8 protein fragments (Tables 1 and 2).
TABLE 2.
Proliferative responses to A. marginale T4SS protein antigens expressed by IVTT
| Proteina | Proliferation (SI) for animal no.b: |
||||
|---|---|---|---|---|---|
| 04B90 (22/16)c | 04B91 (8/23) | 04B92 (8/7) | 4848 (22/22) | 5982 (8/16) | |
| Expt 1 | |||||
| A. marginale OM | 44.6 | 80.7 | 740.8 | 104.1 | 409.8 |
| VirB9-1 (AM097) | 69.9 | 1.3 | 207.2 | 48.8 | 3.2 |
| VirB9-2 (AM1315) | 33.3 | 69.8 | 244.6 | 9.4 | 158.9 |
| VirB10 (AM1314) | 1.1 | 23.2 | 27.7 | 4.1 | 4.1 |
| VirB2 (AM030) | 15.5 | 0.6 | 24.9 | 3.2 | 33.5 |
| VirB3 (AM815) | 1.0 | 0.8 | 0.6 | 1.2 | 0.5 |
| VirB4-1 (AM814) F1 | 1.0 | 1.9 | 1.4 | 1.8 | 0.6 |
| VirB4-1 (AM814) F2 | 1.1 | 1.9 | 1.7 | 1.2 | 0.8 |
| VirB4-1 (AM814) F3 | 0.9 | 2.5 | 1.6 | 1.8 | 0.8 |
| VirB4-2 (AM1053) F1 | 2.1 | 0.6 | 0.8 | 1.4 | 0.4 |
| VirB4-2 (AM1053) F3 | 0.7 | 0.8 | 0.7 | 1.8 | 1.2 |
| VirB6 (AM810) F2 | 1.4 | 2.1 | 0.9 | 1.2 | 0.6 |
| VirB6 (AM811) F1 | 2.1 | 1.6 | 0.6 | 1.4 | 0.7 |
| VirB6 (AM811) F2 | 0.9 | NDd | 1.1 | 1.3 | 0.6 |
| VirB6 (AM812) F1 | 1.7 | 1.4 | 1.6 | 1.1 | 0.9 |
| VirB6 (AM812) F3 | 1.7 | 1.9 | 1.0 | 1.7 | 1.1 |
| VirB6 (AM813) F1 | 1.2 | 1.8 | 1.4 | 2.3 | 0.9 |
| VirB8-1 (AM1316) | 1.3 | ND | 2.2 | 1.6 | 0.6 |
| VirB11 (AM1313) | 1.4 | 6.5 | 3.7 | 1.4 | 0.5 |
| VirD4 (AM 1312) | 0.6 | 3.6 | 0.9 | 1.2 | 0.2 |
| VirD4 (AM1312) F1 | 1.0 | ND | 3.3 | 4.0 | 0.5 |
| VirD4 (AM1312) F2 | 0.7 | 1.1 | 0.9 | 3.3 | 0.7 |
| Expt 2 | |||||
| A. marginale OM | 7.2 | 8.4 | 12.9 | 61.4 | 27.6 |
| VirB9-1 (AM097) | 3.0 | 1.2 | 16.5 | 129.4 | 1.4 |
| VirB9-2 (AM1315) | ND | ND | 27.8 | 73.6 | 14.9 |
| VirB10 (AM1314) | 0.9 | 7.8 | 7.6 | 13.7 | 1.4 |
| VirB2/OrfX (AM030) | 0.9 | 1.2 | 7.4 | 3.3 | 8.1 |
| VirB3 (AM815) | 1.2 | 1.4 | 1.3 | 0.7 | 1.9 |
| VirB11 (AM1313) | 1.4 | 2.8 | 3.1 | 0.7 | 2.3 |
| VirD4 (AM1312) | 3.3 | 24.9 | 1.7 | 0.8 | 1.9 |
| VirD4 (AM1312) F1 | ND | ND | 1.3 | 9.5 | 0.9 |
| VirD4 (AM1312) F2 | ND | ND | 1.2 | 4.0 | 0.5 |
Proteins were expressed by IVTT, and dot blots were performed with anti-FLAG and anti-6× His MAb. Only proteins that expressed the C-terminal FLAG epitope were tested in proliferation assays, with the exception of VirD4. The IVTT-expressed VirD4, used in all but one experiment with 04B91, was truncated at the C terminus, and protein bands of ∼30 and 50 kDa were visualized on Western blots with anti-His MAb. The VirD4 protein used with animal 04B91 in experiment 1 was expressed with a FLAG epitope.
SI were derived by dividing the mean CPM of triplicate cultures of cells with antigen by the mean CPM of triplicate cultures of cells with medium (for A. marginale OM) or negative control IVTT antigen (for IVTT-expressed antigens). A. marginale OM were used at 1 μg protein/ml and bead-bound proteins from IVTT reaction mixtures were tested at a concentration of 1 or 2.5 μg protein G/ml, depending on the assay. Results in boldface type were considered significantly greater than those for medium or negative control antigen (URBC for A. marginale and bead-incubated negative-control no-DNA IVVT reaction for other antigens) where the SI is ≥2.0 and the mean CPM is ≥1,000.
The DRB3 RFLP haplotypes for each animal are indicated in parentheses.
ND, not determined.
Two-week T-cell lines established from PBMC of five OM-immunized cattle with different major histocompatibility complex (MHC) class II haplotypes represented broadly in Holstein cattle (67) were used to screen the IVTT-expressed T4SS proteins (Table 2). Flow cytometry showed that in these cell lines, 94 to 98% of the T cells expressed CD4, whereas only 1 to 6% expressed CD8 and 0.1% were γδ T cells (data not shown). Thus, the response to the T4SS proteins is mediated by CD4+ T cells. VirB9-1, VirB9-2, and VirB10 were included as positive-control antigens (45, 46). We also showed previously that VirB2 expressed by IVTT stimulated T cells from animal 04B90 but not animal 04B91 (45); however, additional animals had not been tested. In the present study, OM and VirB9-2 stimulated significant lymphocyte proliferation for all animals. Other antigens that stimulated specific proliferation of short-term T-cell lines in two or more animals in at least one assay are the following: VirB2 (animals 04B90, 04B92, 4848, and 5982), VirB9-1 (04B90, 04B92, 4848, and 5982), VirB10 (04B91, 04B92, 4848, 5982), VirB11 (04B91, 04B92, and 5982), and VirD4/VirD4F1/VirD4F2 (04B91 and 4848). Screening antigens by IVTT thus verified that known immunogenic T4SS proteins could be identified by this method but, importantly, showed that previously under-tested VirB2 and untested VirD4 and VirB11 proteins were able to stimulate T-cell responses from more than one OM-immunized animal.
Production of T4SS proteins.
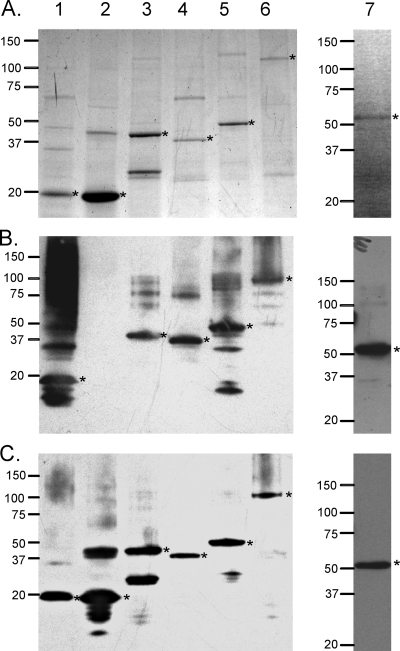
T4SS proteins that stimulated CD4+ T-cell responses from more than one animal were selected for expression as recombinant proteins in E. coli to verify their ability to induce recall T-cell proliferative responses from the OM vaccinees. VirB2, VirB9-1, VirB9-2, VirB10, VirB11, and VirD4 were expressed in E. coli and affinity purified. VirD4 was expressed as overlapping fragments F1 (amino acids [aa] 1 to 344) and F2 (aa 270 to 606), but F3 (aa 536 to 806) did not express. Therefore, full-length VirD4 (aa 1 to 806) was later expressed and purified in sufficient quantity for testing. After we had completed testing IVTT-expressed antigens, we identified a putative VirB7 homolog (Am306; discussed below) in the genome that was not originally annotated as VirB7, and expressed this as a His-tagged fusion protein (Table 1). VirB2, VirB7, VirB11, VirD4F1, VirD4F2, VirD4, and control MSA1 were stained with Coomassie blue to assess relative purity (Fig. 1A). The approximate molecular masses of the proteins were as follows: VirB2, 19 kDa; VirB7, 19 kDa; VirB11, 45 kDa; VirD4F1, 39 kDa; VirD4F2, 50 kDa; VirD4, 100 kDa; and MSA-1, 58 kDa. These bands correspond to the predicted molecular mass of each fusion protein. Immunoblots of these reacted with anti-FLAG MAb (Fig. 1B), with the exception of VirB7 lacking the FLAG epitope, and with anti-6× His MAb (Fig. 1C). VirB9-1, VirB9-2, and VirB10 were also of the predicted molecular masses, as previously described (reference 44 and data not shown).
FIG. 1.
Expression and relative purity of the T4SS recombinant proteins. Proteins were separated using SDS-PAGE and stained with Coomassie blue (A) or transferred to nitrocellulose membranes and probed with anti-FLAG MAb (B) or anti-6× His MAb (C), and bands were detected by chemiluminescence. Wells received 1.5 μg purified VirB2 (lane 1), VirB7 (lane 2), VirB11 (lane 3), VirD4F1 (lane 4), VirD4F2 (lane 5), or VirD4 (lane 6) or 2 μg MSA1 (lane 7). VirB2 and VirB7 are ∼19 kDa, VirB11 is ∼45 kDa, VirD4F1 is ∼39 kDa, VirD4F2 and MSA1 are ∼55 kDa, and VirD4 is ∼100 kDa, as indicated by asterisks.
Recognition of recombinant T4SS proteins by CD4+ T-cell lines from outer membrane-immunized cattle.
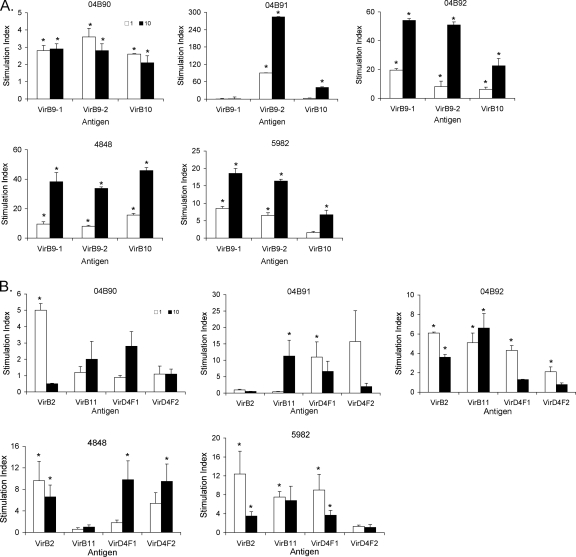
T-cell lines derived from PBMC of OM-immunized cattle were used in proliferation assays to further characterize the memory CD4+ T-cell response to newly expressed T4SS proteins as well as the known immunogenic T4SS proteins. VirB9-1, VirB9-2, and VirB10 stimulated significant proliferation of T cells of all five animals, with the exception of 04B91, which does not have T-cell responses to VirB9-1 (45, 46) (Fig. 2A). Four animals had significant T-cell proliferative responses against VirB2 (04B90, 04B92, 4848, and 5982) and VirD4F1 (04B91, 04B92, 4848, and 5982), three animals had significant responses to VirB11 (04B91, 04B92, and 5982), and two animals responded significantly to VirD4F2 (04B92 and 4848) at one or both antigen concentrations (Fig. 2B). With the exception of animal 5982, which did not respond to VirD4 proteins expressed by IVTT, the results are consistent with responses to IVTT-expressed proteins. Proliferation assays for each animal and antigen were performed at least twice with comparable results, with the exception of animal 04B91, which died unexpectedly and so not every protein could be tested repeatedly.
FIG. 2.
Short-term T-cell lines established from OM vaccinees proliferate in response to recombinant T4SS proteins. T cells from cattle 04B90, 04B91, 04B92, 4848, and 5982 were tested for proliferation to 1 (white bars) and 10 (black bars) μg/ml VirB9-1, VirB9-2, and VirB10 (A) or VirD4F1, VirD4F2, VirB11, and VirB2 (B). Responses are presented as the SI compared to negative control MSA1 ± 1 SD for triplicate cultures. Responses significantly greater than those for the same concentration of MSA1 are indicated by asterisks.
Many T4SSs encode small (∼50 aa) lipoproteins (VirB7 and related homologs) that interact with various scaffold components, particularly those close to the OM pore. For example, VirB7 of Agrobacterium tumefaciens binds and stabilizes VirB9 in the OM (25, 68), and VirB7 interacts with the C-terminal domain of VirB9 as revealed by nuclear magnetic resonance (NMR) (6) and cryo-electron microscopy (cryo-EM) (28) structures. While Gillespie et al. provided informatics evidence for a VirB7 homolog in all sequenced genomes of Rickettsia spp. (32), a virb7 gene has not yet been identified in A. marginale or annotated in any of the Anaplasmataceae genomes. However, recent analyses demonstrated similarities between Rickettsiaceae VirB7 and ComB7 proteins (32), and we therefore used this analysis as the basis to identify a VirB7 homolog in A. marginale. An alignment of the predicted amino acid sequences from A. marginale AM306 and virB7 genes from other Gram-negative pathogens, including Agrobacterium tumefaciens VirB7 and the VirB7 ortholog ComB7 from Helicobacter pylori and Campylobacter jejuni, is shown in Fig. 3. This revealed characteristics conserved across these lipoproteins. Therefore, the putative VirB7 was tested on T-cell lines from each of the five animals and was shown to stimulate significant responses in animals 04B90, 4848, and 5982 (Table 3).
FIG. 3.
Alignment of a putative A. marginale VirB7 with VirB7 homologs from other bacteria. The A. marginale strain St. Maries (AM_StM) putative VirB7 (AM306) amino acid sequence was aligned with Com7 from C. jejuni strain 81-176 (CJ_81-176) and H. pylori strain Shi170 (HP_Shi170) and with VirB7 from Agrobacterium tumefaciens strain Bo542 (AT_Bo542). Alignment was performed with ClustalW (AlignX) and was followed by manual editing. Residues identical in three or more sequences are shown as white text on a black background, while conserved residues are shown as black text on a gray background.
TABLE 3.
Proliferative response of T cells from outer membrane vaccinees to putative VirB7 and VirD4
| Antigen | Concn (μg/mg) | Proliferation (mean CPM ± 1 SD) for the following animala: |
||||
|---|---|---|---|---|---|---|
| 04B90 (22/16)b | 04B91 (8/23) | 04B92 (8/7) | 4848 (22/22) | 5982 (8/16) | ||
| MSA1 | 1 | 12,810 ± 2,725 | 237 ± 56 | 200 ± 117 | 737 ± 276 | 679 ± 289 |
| 10 | 9,788 ± 3,264 | 212 ± 27 | 159 ± 177 | 472 ± 120 | 646 ± 280 | |
| VirB7 | 1 | 28,676 ± 5,127 | 248 ± 24 | 104 ± 51 | 1,535 ± 434 | 668 ± 196 |
| 10 | 45,504 ± 7,990 | 572 ± 175 | 162 ± 81 | 5,134 ± 1,866 | 5,298 ± 596 | |
| VirD4 | 1 | 27,803 ± 4,777 | 610 ± 389 | 98 ± 21 | 3,301 ± 199 | 1,406 ± 304 |
| 10 | 41,763 ± 11,530 | 5,223 ± 2,334 | 608 ± 532 | 13,515 ± 5,780 | 3,587 ± 1,791 | |
Recombinant proteins were tested in a 3-day proliferation assay with short-term (2-week) cell lines that had been stimulated for 1 week with A. marginale OM and rested for 1 week. Results are presented as the mean CPM of triplicate cultures ± 1 SD, and those in boldface type are significantly greater than those for B. bovis MSA1.
MHC class II haplotypes are indicated in parentheses using the DRB3 RFLP nomenclature.
We additionally tested full-length VirD4 in the same assays, and animals 04B90, 04B91, 4848, and 5982 had significant responses to VirD4. The results with VirD4 are consistent with those using recombinant VirD4F1 and VirD4F2, except for animal 04B90, and the data suggest that T cells from animal 04B90 recognize an epitope in VirD4 that was not present in VirD4F1 or VirD4F2. These proliferation assays were performed at least twice with comparable results, with the exception of animal 04B91.
Unlike VirB9-2 and VirB10, the other immunogenic T4SS proteins were not recognized by every OM vaccinee, which is likely explained by MHC restriction, as immunization induced strong responses to OM in all animals (Table 2). For example, animals with DRB3 RFLP haplotypes 8/23 and 8/7 did not respond to VirB7, whereas animals with haplotypes 22/16, 22/22, and 8/16 did respond, suggesting that the response is mediated by a class II molecule associated with the 22 and 16 haplotypes but not with the 8, 7, or 23 haplotypes. Similar conclusions can be drawn for VirB11, which is likely presented by class II molecules associated with haplotype 8 but not 16 or 22, and for VirD4, which is likely presented by class II molecules associated with haplotypes 16, 22, and 23 but not 8 or 7. Additional experiments would need to be performed to identify the DR or DQ molecules that actually present epitopes from these proteins.
VirB2 T-cell epitopes are located in the central conserved region.
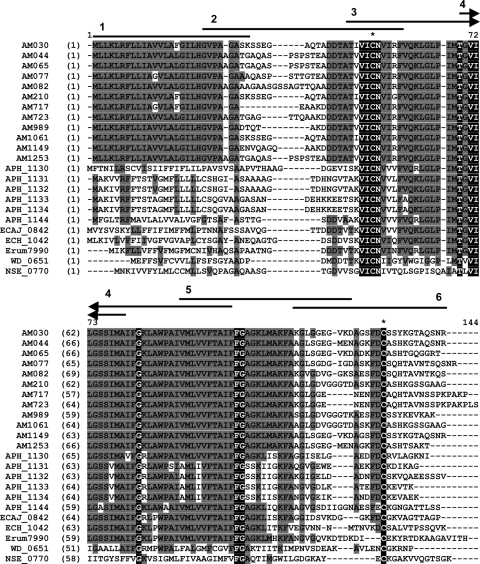
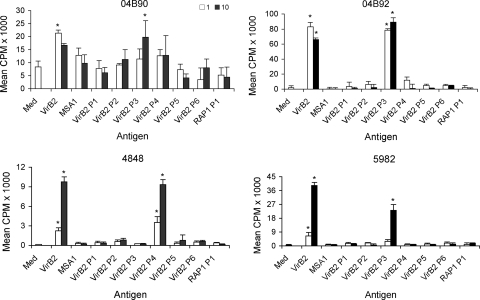
A. marginale has 12 copies of a gene annotated as orfx (9, 49) that are now considered orthologs of VirB2 in the Anaplasmataceae (8, 50). Comparison of the A. marginale VirB2 sequences AM030 to AM1263 with representative sequences across the family Anaplasmataceae reveals extensive conservation in the central portion of the VirB2 molecules of Anaplasma and Ehrlichia species, with less conservation when comparing Wolbachia and Neorickettsia proteins (Fig. 4). Interestingly, there are several residues completely conserved across the family, even when more representatives are included in the alignment (data not shown), including two cysteine residues. Comparison of the 12 A. marginale VirB2 sequences revealed two variable regions, one just after the predicted signal peptide and the second at the carboxy terminus of the protein. Some VirB2 proteins form a cyclic protein following enzymatic cleavage of the variable amino and carboxy termini (23), and it is thought that processed VirB2 proteins polymerize and line the entire periplasmic region of the T4SS channel (36). Thus, we were interested to determine if the T-cell epitope(s) would localize to the central conserved region of A. marginale VirB2, and furthermore, if multiple copies of VirB2 were expressed during infection, whether this central conserved region would also serve as a common antigen for all VirB2 variants. To identify the T-cell epitope-rich region, overlapping 30-amino-acid peptides spanning the sequence of VirB2 (AM030) were synthesized (Fig. 4 and Table 4) and tested for induction of a CD4+ T-cell response in OM-immunized cattle (Fig. 5). As expected, animal 04B91, which did not respond to VirB2, did not respond to any of the VirB2 peptides (data not shown). However, T-cell lines from animals 04B90, 04B92, and 5982 had significant responses to VirB2 peptide 3, whereas T-cell lines from animal 4848 responded significantly to peptide 4. These assays were performed at least twice with similar results, except for those from animal 04B91.
FIG. 4.
Alignment of Anaplasmataceae VirB2 predicted amino acid sequences. ClustalW alignment of 12 VirB2 sequences from A. marginale (AM), six (of eight) sequences from A. phagocytophilum (APH), and representative sequences from Ehrlichia canis (ECAJ), E. chaffeensis (ECH), E. ruminantium (Erum), Wolbachia pipientis wMel (WD), and Neorickettsia sennetsu (NSE) are shown. Identical residues in all sequences are shown as white text on a black background, while blocks of identical residues are shown as black text on a gray background. Conserved cysteine residues are indicated with an asterisk. The positions of A. marginale peptides 1 through 6 designed against the AM030 sequence are indicated as numbered horizontal lines above the sequences.
TABLE 4.
VirB2 (AM030) peptides used in proliferation assays
| Peptide | Amino acid position | Sequencea |
|---|---|---|
| 1 | 1-30 | MLLKLRFLLIAVVLAFGILHGVPAGASKSS |
| 2 | 21-50 | GVPAGASKSSEGAQTADDTATIVICNVIRF |
| 3 | 41-70 | TIVICNVIRFVQKLGLPIMTGVILGSSIMA |
| 4 | 61-90 | GVILGSSIMAIFGKLAWPAIVMLVVFTAIF |
| 5 | 81-110 | VMLVVFTAIFFGAGKLMAKFAKGLGGEGVK |
| 6 | 101-128 | AKGLGGEGVKDAGSFDCSSYKGTAQSNR |
Thirty amino acid peptides overlapping by 10 amino acids (underlined) were synthesized.
FIG. 5.
T-cell epitopes in VirB2 localize to the central conserved region. T-cells from animals 04B90, 04B92, 4848, and 5982 were tested for proliferation to 1 (white bars) and 10 (black bars) μg/ml of VirB2 and overlapping peptides. Responses are represented as the SI in relation to medium. Responses to antigen that are significantly greater than those to the medium or negative control MSA1 (for VirB2) or RAP-1 P1 (for VirB2 peptides) are indicated by asterisks.
Serologic responses to immunogenic T4SS proteins.
We also determined whether sera from cattle immunized with the OM-enriched fraction and from cattle experimentally infected with A. marginale St. Maries strain had detectable levels of antibody to the T4SS T-cell antigens by immunoblotting. Except for 5982, which only recognized VirB9-1 and VirB9-2, the immunized animals had IgG specific for VirB9-1, VirB9-2, and VirB10 (reference 46 and data not shown). When we tested VirB2, VirB7, VirB11, and VirD4, immune serum from animal 4848 recognized only VirB11, and the other sera were not convincingly positive for any of the these antigens (data not shown). When two postinfection sera were tested, VirB9-2 and VirB10 had the strongest reactivity, VirB9-1 was relatively weaker, and the other antigens tested were nonreactive (data not shown).
Transcription of immunogenic T4SS genes.
Because antibody responses to the newly tested T4SS proteins were not apparent in OM-immunized cattle, which would have provided additional indirect evidence for protein expression, RT-PCR was used to verify that genes encoding the novel immunogenic T4SS proteins were transcribed. Figure 6 shows that cDNAs encoding VirB2, VirB7, VirB11, and VirD4 were detected in A. marginale from infected bovine blood. Sequencing these cDNAs showed complete identity with the genomic sequences, verifying the identity of each amplicon.
FIG. 6.
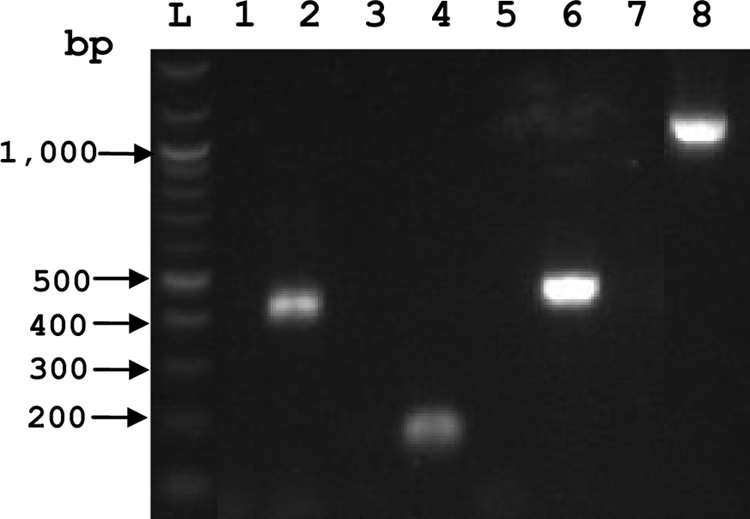
RT-PCR analysis of VirB2, VirB7, VirB11, and VirD4. RT-PCR was performed using RNA derived from A. marginale St. Maries strain-infected red blood cells to amplify T4SS cDNAs. PCR products for VirB2 (lanes 1 and 2), VirB7 (lanes 3 and 4), VirB11F1 (lanes 5 and 6), and VirD4F2 (lanes 7 and 8), performed without (lanes 1, 3, 5, and 7) or with (lanes 2, 4, 6, and 8) RT, are shown. L represents the 1-kb ladder. All RT-PCR products failed to produce bands and served as negative controls.
DISCUSSION
Immunization with immunodominant surface proteins, including antigenically variable MSP2 and MSP3, has not conferred protective immunity comparable to that achieved with outer membranes (13, 44, 52, 58, 59, 70). Our research is therefore focused on identifying nonvariant proteins in the protective outer membrane fraction of A. marginale that may induce protective immunity, either as individual proteins or as multiple proteins, such as those that comprise a molecular complex. Naturally occurring protein complexes also offer the potential for linked recognition of T- and B-cell epitopes on different, but associated, proteins in the bacterial membrane, which can generate effective antibody responses against surface-exposed proteins that have few or lack T-cell epitopes (48). The T4SS, required for invasion and intracellular survival of many Gram-negative bacterial pathogens, is a membrane protein complex that offers the potential for use as a vaccine target.
Very few studies other than ours have reported that T4SS proteins are antigenic, and to our knowledge, none have reported T-cell recognition of T4SS proteins. Antibody specific for VirB9 in dogs infected with Ehrlichia canis was detected (24), and more recently, antibody specific for VirB9-2 and VirB10 was detected in cattle experimentally or naturally infected with A. marginale in Brazil (4, 73). However, there is some evidence that type III secretion system (T3SS) proteins can induce protective immunity in several animal models with human pathogens (34, 38, 42, 69). Thus, the ability to elicit a protective response against proteins of the T3SS, which serves a function similar to that of the T4SS, suggests that disruption or blocking of the T4SS by immunization against selected T4SS proteins could interfere with the bacteria-host cell interaction required for bacterial survival.
The structure and function of the T4SS of rickettsial pathogens, including the Anaplasmataceae, have not been well defined (32). However, the importance of the T4SS for these pathogens, possibly as a functional virulence factor, is inferred by its retention and protein conservation among different rickettsial species that have undergone reductive evolution (32). In support of this, A. phagocytophilum AnkA, a protein with ankryin repeat domains that are typically found in eukaryotic proteins, localizes to the host cell nucleus in infected neutrophils (64) and can be secreted by the T4SS of Agrobacterium tumefaciens (44). Lin et al. (44) further showed that inhibiting AnkA by targeting AnkA-specific antibody to the neutrophil cytoplasm inhibited host cell infection. This probable role for the A. phagocytophilum T4SS in intracellular infection supports the applicability of a vaccine strategy targeting this conserved structure. Furthermore, although each immunogenic A. marginale T4SS protein was not recognized by all of the animals in our study, each protein was recognized by T cells from at least three animals with different, yet commonly occurring, MHC haplotypes found in at least 50% of Holstein cattle (67). This suggests it may be possible to develop a multivalent subunit vaccine that could target a large proportion of outbred animals.
Our studies also indicate the feasibility of immunizing against T4SS proteins to stimulate CD4+ T-cell and IgG responses that could inhibit T4SS function. For the T4SS proteins that we have identified as immunogenic for T cells, several are predicted to be surface exposed, where they could interact with neutralizing antibody. Alternatively, T4SS T-cell antigens that are not surface exposed could be physically associated with T4SS proteins that are, thereby stimulating T cells to provide help to augment antibody responses against the surface-exposed proteins. In A. marginale, the immunogenic T4SS proteins that are likely surface exposed are VirB2, VirB7, VirB9-1, VirB9-2, and VirB10. In some Gram-negative bacteria, VirB2 is the major component of the pilus apparatus and appears to require VirB5 to initiate pilus formation (7, 14, 18, 26, 37). However, in Helicobacter pylori, which lack true pili, a VirB2 ortholog was identified that was found to be surface exposed and associated with appendage-like structures on the outer membrane (3). VirB2 is essential for both substrate transfer (in the absence of a true pilus) and pilus formation (in the presence of VirB5) (27). As A. marginale lacks a VirB5 gene, we assume VirB2 plays a role in substrate transfer and is therefore likely exposed on the outer membrane.
VirB7 and VirB9 are covalently associated with each other (2), and together with VirB10, they aggregate to form a 14-oligomer core channel of the T4SS encoded on the pKM101 plasmid (28). Within this core channel structure, VirB7 and VirB9 are the major components of the cap of the core, which is inserted in the OM, whereas VirB10 forms the outer surface of the core below the cap (28). In Agrobacterium tumefaciens, VirB7 also interacts with VirB2 (14). When the composition of a complex of proteins formed by cross-linking outer membranes of intact A. marginale with nonmembrane permeable cross-linkers was studied, VirB10 was identified (52), suggesting that this protein is either surface exposed in A. marginale or tightly associated with surface-exposed proteins, such as VirB9 or VirB7. Antibody recognition of VirB9-2 and VirB10 (4, 73) as well as VirB9-1 (this study) in A. marginale-infected animals additionally supports their surface exposure. VirB9-1 of related A. phagocytophilum and E. chaffeensis were identified on the surface of these bacteria (31, 51).
VirB11 and VirD4 are not predicted to be OM proteins, but they could still play a role in linked recognition as T-cell antigens. VirB11 is an ATPase that may be responsible for the production of energy used to facilitate effector molecule translocation or the assembly of the T4SS complex (7, 16, 19, 35, 40, 66). It localizes to the bacterial cytoplasm or periplasmic space between the inner and outer membranes (28, 66) and was associated with VirB9 in assays defining protein-protein interactions (14). VirD4 is a coupling protein/ATPase that mediates the transfer of effector proteins and/or DNA across the periplasmic space and into the host cell (27, 33, 39).
Prior studies have shown that VirB2 undergoes posttranslational modification via cleavage at the N and C termini, and in Agrobacterium tumefaciens, the processed protein is circularized (23, 37, 41). We observed that the T-cell epitopes localize to the conserved central region of VirB2 that is shared by all 12 A. marginale VirB2 paralogs. In A. phagocytophilum, the VirB2 paralogs are differentially expressed in tick and mammalian cell cultures (50), yet for A. marginale, it is not known which of the VirB2 gene family members are expressed in vitro or in vivo. However, the significant T-cell responses to VirB2 (AM030) might be due to a relatively abundant amount of the conserved T-cell epitopes if multiple copies are expressed and/or if VirB2 multimers comprise a pilus-like apparatus. Additional studies are needed to answer these questions.
Compared with MSP2 and MSP3, the immunogenic T4SS proteins are considered subdominant antigens, as they induce weaker antibody responses and, in general, weaker T-cell responses (45-47). These subdominant antigens are of interest for vaccine development not only because they are antigenically conserved but also because in other disease models, protective immunity does not necessarily correlate with the immunodominance hierarchy of epitopes (29, 30, 56, 71, 75). Among the immunogenic T4SS proteins, VirB9-1, VirB9-2, and VirB10 are the most stimulatory, and both T cells and antibody recognize these proteins in the majority of OM vaccinees. VirB7, VirB11, and VirD4, which were generally not recognized by immune sera, may not be highly exposed to B cells after immunization with the OM-enriched fraction, whereas epitopes on these proteins would be presented by antigen-presenting cells to T cells.
The T4SS has conserved structure and function and is important for virulence and intracellular survival of several Gram-negative pathogens. The T4SS has not been well characterized for any of the Anaplasmataceae; however, since this protein complex is likely necessary for the survival and dissemination of A. marginale, vaccine development targeting this protein complex to stimulate a protective immune response is warranted. Furthermore, the predicted localization of several of the T4SS proteins to the OM of A. marginale further justifies targeting this protein complex as a vaccine candidate. Investigating the specific interactions among A. marginale T4SS proteins within the complex, and the immunogenic properties of T4SS proteins in relation to recognition of linked T-cell and B-cell epitopes, is needed to achieve this goal.
Acknowledgments
We thank Shelley Whidbee, Emma Karel, and Ralph Horn for excellent technical assistance and Heather Fallquist for providing A. marginale RNA.
This research was supported by National Institutes of Health (NIH) National Institutes of Allergy and Infectious Diseases (NIAID) grant AI053692 (W.C.B.) and the Intramural Research Program at NIH, NIAID (R.A.H., P.A.B., and J.E.L.). J.J.G. acknowledges support from NIAID awards R01AI017828 and R01AI59118 to Abdu Azad (University of Maryland) and contract HHSN266200400035C to Bruno Sobral (Virginia Bioinformatics Institute at Virginia Tech).
Editor: R. P. Morrison
Footnotes
Published ahead of print on 11 January 2010.
REFERENCES
- 1.Abbott, J. R., G. H. Palmer, K. A. Kegerreis, P. F. Hetrick, C. J. Howard, J. C. Hope, and W. C. Brown. 2005. Rapid and long-term disappearance of CD4+ T lymphocyte responses specific for Anaplasma marginale major surface protein-2 (MSP2) in MSP2 vaccinates following challenge with live A. marginale. J. Immunol. 174:6702-6715. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, L. B., A. V. Hertzel, and A. Das. 1996. Agrobacterium tumefaciens VirB7 and VirB9 form a disulfide-linked protein complex. Proc. Natl. Acad. Sci. U. S. A. 93:8889-8894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrzejewska, J., S. K. Lee, P. Olbermann, N. Lotzing, E. Katzowitsch, B. Linz, M. Achtman, C. I. Kado, S. Suerbaum, and C. Josenhans. 2006. Characterization of the pilin ortholog of the Helicobacter pylori type IV cag pathogenicity apparatus, a surface-associated protein expressed during infection. J. Bacteriol. 188:5865-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Araujo, F. R., C. M. Costa, C. A. Ramos, T. A. Farias, I. I. Souza, E. S. Melo, C. Elisei, G. M. Rosinha, C. O. Soares, S. P. Fragoso, and A. H. Fonseca. 2008. IgG and IgG2 antibodies from cattle naturally infected with Anaplasma marginale recognize the recombinant vaccine candidate antigens VirB9, VirB10, and elongation factor-Tu. Mem. Inst. Oswaldo Cruz 103:186-190. [DOI] [PubMed] [Google Scholar]
- 5.Baron, C., D. O'Callaghan, and E. Lanka. 2002. Bacterial secrets of secretion: EuroConference on the biology of type IV secretion processes. Mol. Microbiol. 43:1359-1365. [DOI] [PubMed] [Google Scholar]
- 6.Bayliss, R., R. Harris, L. Coutte, A. Monier, R. Fronzes, P. J. Christie, P. C. Driscoll, and G. Waksman. 2007. NMR structure of a complex between the VirB9/VirB7 interaction domains of the pKM101 type IV secretion system. Proc. Natl. Acad. Sci. U. S. A. 104:1673-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Berger, B. R., and P. J. Christie. 1994. Genetic complementation analysis of the Agrobacterium tumefaciens virB operon: virB2 through virB11 are essential virulence genes. J. Bacteriol. 176:3646-3660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brayton, K. A., M. J. Dark, and G. H. Palmer. 2008. Anaplasma, p. 85-116. In V. Nene and C. Kole (ed.), Genome mapping and genomics in animal-associated microbes. Springer-Verlag, Berlin, Germany.
- 9.Brayton, K. A., L. S. Kappmeyer, D. R. Herndon, M. J. Dark, D. L. Tibbals, G. H. Palmer, T. C. McGuire, and D. P. Knowles, Jr. 2005. Complete genome sequencing of Anaplasma marginale reveals that the surface is skewed to two superfamilies of outer membrane proteins. Proc. Natl. Acad. Sci. U. S. A. 102:844-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brayton, K. A., D. P. Knowles, T. C. McGuire, and G. H. Palmer. 2001. Efficient use of a small genome to generate antigenic diversity in tick-borne ehrlichial pathogens. Proc. Natl. Acad. Sci. U. S. A. 98:4130-4135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brayton, K. A., P. F. Meeus, A. F. Barbet, and G. H. Palmer. 2003. Simultaneous variation of the immunodominant outer membrane proteins, MSP2 and MSP3, during Anaplasma marginale persistence in vivo. Infect. Immun. 71:6627-6632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brayton, K. A., G. H. Palmer, and W. C. Brown. 2006. Genomic and proteomic approaches to vaccine candidate identification for Anaplasma marginale. Expert Rev. Vaccines 5:95-101. [DOI] [PubMed] [Google Scholar]
- 13.Brown, W. C., V. Shkap, D. Zhu, T. C. McGuire, W. Tuo, T. F. McElwain, and G. H. Palmer. 1998. CD4+ T-lymphocyte and IgG2 responses in calves immunized with Anaplasma marginale outer membranes and protected against homologous challenge. Infect. Immun. 66:5406-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cascales, E., and P. J. Christie. 2003. The versatile bacterial type IV secretion systems. Nat. Rev. Microbiol. 1:137-149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Celli, J., C. de Chastellier, D. M. Franchini, J. Pizzaro-Cerda, E. Moreno, and J. P. Gorvel. 2003. Brucella evades macrophage killing via VirB-dependent sustained interactions with the endoplasmic reticulum. J. Exp. Med. 198:545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Christie, P. J., K. Atmakuri, V. Krishnamoorthy, S. Jakubowski, and E. Cascales. 2005. Biogenesis, architecture, and function of bacterial type IV secretion systems. Annu. Rev. Microbiol. 59:451-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Collins, N. E., J. Liebenberg, E. P. de Villiers, K. A. Brayton, E. A. Pretorius, F. E. Faber, H. van Heerden, A. Josemans, M. van Kleef, H. C. Steyn, M. F. van Strijp, E. Zweygarth, F. Jongejan, J. C. Maillard, D. Berthier, M. Botha, F. Joubert, C. H. Corton, N. R. Thomson, M. T. Allsopp, and B. A. Allsopp. 2005. The genome of the heartwater agent Ehrlichia ruminantium contains multiple tandem repeats of actively variable copy number. Proc. Natl. Acad. Sci. U. S. A. 102:838-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dang, T. A., and P. J. Christie. 1997. The VirB4 ATPase of Agrobacterium tumefaciens is a cytoplasmic membrane protein exposed at the periplasmic surface. J. Bacteriol. 179:453-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.den Hartigh, A. B., H. G. Rolan, M. F. de Jong, and R. M. Tsolis. 2008. VirB3 to VirB6 and VirB8 to VirB11, but not VirB7, are essential for mediating persistence of Brucella in the reticuloendothelial system. J. Bacteriol. 190:4427-4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dumler, J. S., A. F. Barbet, C. P. Bekker, G. A. Dasch, G. H. Palmer, S. C. Ray, Y. Rikihisa, and F. R. Rurangirwa. 2001. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51:2145-2165. [DOI] [PubMed] [Google Scholar]
- 21.Dunning Hotopp, J. C., M. Lin, R. Madupu, J. Crabtree, S. V. Angiuoli, J. A. Eisen, R. Seshadri, Q. Ren, M. Wu, T. R. Utterback, S. Smith, M. Lewis, H. Khouri, C. Zhang, H. Niu, Q. Lin, N. Ohashi, N. Zhi, W. Nelson, L. M. Brinkac, R. J. Dodson, M. J. Rosovitz, J. Sundaram, S. C. Daugherty, T. Davidsen, A. S. Durkin, M. Gwinn, D. H. Haft, J. D. Selengut, S. A. Sullivan, N. Zafar, L. Zhou, F. Benahmed, H. Forberger, R. Halpin, S. Mulligan, J. Robinson, O. White, Y. Rikihisa, and H. Tettelin. 2006. Comparative genomics of emerging human ehrlichiosis agents. PLoS Genet. 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eddy, S. R. 1995. Multiple alignment using hidden Markov models. Proc. Int. Conf. Intell. Syst. Mol. Biol. 3:114-120. [PubMed] [Google Scholar]
- 23.Eisenbrandt, R., M. Kalkum, E. M. Lai, R. Lurz, C. I. Kado, and E. Lanka. 1999. Conjugative pili of IncP plasmids, and the Ti plasmid T pilus are composed of cyclic subunits. J. Biol. Chem. 274:22548-22555. [DOI] [PubMed] [Google Scholar]
- 24.Felek, S., H. Huang, and Y. Rikihisa. 2003. Sequence and expression analysis of virB9 of the type IV secretion system of Ehrlichia canis strains in ticks, dogs, and cultured cells. Infect. Immun. 71:6063-6067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fernandez, D., T. A. Dang, G. M. Spudich, X. R. Zhou, B. R. Berger, and P. J. Christie. 1996. The Agrobacterium tumefaciens virB7 gene product, a proposed component of the T-complex transport apparatus, is a membrane-associated lipoprotein exposed at the periplasmic surface. J. Bacteriol. 178:3156-3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Finberg, K. E., T. R. Muth, S. P. Young, J. B. Maken, S. M. Heitritter, A. N. Binns, and L. M. Banta. 1995. Interactions of VirB9, -10, and -11 with the membrane fraction of Agrobacterium tumefaciens: solubility studies provide evidence for tight associations. J. Bacteriol. 177:4881-4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fronzes, R., P. J. Christie, and G. Waksman. 2009. The structural biology of type IV secretion systems. Nat. Rev. Microbiol. 7:703-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fronzes, R., E. Schafer, L. Wang, H. R. Saibil, E. V. Orlova, and G. Waksman. 2009. Structure of a type IV secretion system core complex. Science 323:266-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gallimore, A., T. Dumrese, H. Hengartner, R. M. Zinkernagel, and H. G. Rammensee. 1998. Protective immunity does not correlate with the hierarchy of virus-specific cytotoxic T cell responses to naturally processed peptides. J. Exp. Med. 187:1647-1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gallimore, A., J. Hombach, T. Dumrese, H. G. Rammensee, R. M. Zinkernagel, and H. Hengartner. 1998. A protective cytotoxic T cell response to a subdominant epitope is influenced by the stability of the MHC class I/peptide complex and the overall spectrum of viral peptides generated within infected cells. Eur. J. Immunol. 28:3301-3311. [DOI] [PubMed] [Google Scholar]
- 31.Ge, Y., and Y. Rikihisa. 2007. Identification of novel surface proteins of Anaplasma phagocytophilum by affinity purification and proteomics. J. Bacteriol. 189:7819-7828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gillespie, J. J., N. C. Ammerman, S. M. Dreher-Lesnick, M. Sayeedur Rahman, M. J. Worley, J. C. Setubal, B. S. Sobral, and A. A. Azad. 2009. An anomalous type IV secretion system in Rickettsia is evolutionarily conserved. PLoS One 4:e4833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hamilton, C. M., H. Lee, P. L. Li, D. M. Cook, K. R. Piper, S. B. von Bodman, E. Lanka, W. Ream, and S. K. Farrand. 2000. TraG from RP4 and TraG and VirD4 from Ti plasmids confer relaxosome specificity to the conjugal transfer system of pTiC58. J. Bacteriol. 182:1541-1548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Haque, A., K. Chu, A. Easton, M. P. Steverns, E. E. Galyov, T. Atkins, R. Titball, and G. J. Bancroft. 2006. A live experimental vaccine against Burkolderia pseudomallei elicits CD4+ T cell-mediated immunity, priming T cells specific for 2 type III secretion system proteins. J. Infect. Dis. 194:1241-1248. [DOI] [PubMed] [Google Scholar]
- 35.Hoppner, C., A. Carle, D. Sivanesan, S. Hoeppner, and C. Baron. 2005. The putative lytic transglycosylase VirB1 from Brucella suis interacts with the type IV secretion system core components VirB8, VirB9 and VirB11. Microbiology 151:3469-3482. [DOI] [PubMed] [Google Scholar]
- 36.Jakubowski, S. J., J. E. Kerr, I. Garza, V. Krishnamoorthy, R. Bayliss, G. Waksman, and P. J. Christie. 2009. Agrobacterium VirB10 domain requirements for type IV secretion and T pilus biogenesis. Mol. Microbiol. 71:779-794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones, A. L., E. M. Lai, K. Shirasu, and C. I. Kado. 1996. VirB2 is a processed pilin-like protein encoded by the Agrobacterium tumefaciens Ti plasmid. J. Bacteriol. 178:5706-5711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jones, S. M., K. F. Griffin, I. Hodgson, and E. D. Williamson. 2003. Protective efficacy of a fully recombinant plague vaccine in the guinea pig. Vaccine 21:3912-3918. [DOI] [PubMed] [Google Scholar]
- 39.Krall, L., U. Wiedemann, G. Unsin, S. Weiss, N. Domke, and C. Baron. 2002. Detergent extraction identifies different VirB protein subassemblies of the type IV secretion machinery in the membranes of Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. U. S. A. 99:11405-11410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kutter, S., R. Buhrdorf, J. Haas, W. Schneider-Brachert, R. Haas, and W. Fischer. 2008. Protein subassemblies of the Helicobacter pylori Cag type IV secretion system revealed by localization and interaction studies. J. Bacteriol. 190:2161-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai, E. M., R. Eisenbrandt, M. Kalkum, E. Lanka, and C. I. Kado. 2002. Biogenesis of T pili in Agrobacterium tumefaciens requires precise VirB2 propilin cleavage and cyclization. J. Bacteriol. 184:327-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leary, S. E. C., E. D. Williamson, K. F. Griffin, P. Russell, S. M. Eley, and R. W. Titball. 1995. Active immunization with recombinant V antigen from Yersinia pestis protects mice against plague. Infect. Immun. 63:2854-2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Leroith, T., K. A. Brayton, J. B. Molloy, R. E. Bock, S. A. Hines, A. E. Lew, and T. F. McElwain. 2005. Sequence variation and immunologic cross-reactivity among Babesia bovis merozoite surface antigen 1 proteins from vaccine strains and vaccine breakthrough isolates. Infect. Immun. 73:5388-5394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lin, M., A. den Dulk-Ras, P. J. Hooykaas, and Y. Rikihisa. 2007. Anaplasma phagocytophilum AnkA secreted by type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell. Microbiol. 9:2644-2657. [DOI] [PubMed] [Google Scholar]
- 45.Lopez, J. E., P. A. Beare, R. A. Heinzen, J. Norimine, K. K. Lahmers, G. H. Palmer, and W. C. Brown. 2008. High-throughput identification of T-lymphocyte antigens from Anaplasma marginale expressed using in vitro transcription and translation. J. Immunol. Methods 332:129-141. [DOI] [PubMed] [Google Scholar]
- 46.Lopez, J. E., G. H. Palmer, K. A. Brayton, M. J. Dark, S. E. Leach, and W. C. Brown. 2007. Immunogenicity of Anaplasma marginale type IV secretion system proteins in a protective outer membrane vaccine. Infect. Immun. 75:2333-2342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lopez, J. E., W. F. Siems, K. A. Brayton, G. H. Palmer, T. C. McGuire, and W. C. Brown. 2005. Identification of novel antigenic proteins in a complex Anaplasma marginale outer membrane immunogen by mass spectrometry and genomic mapping. Infect. Immun. 73:8109-8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macmillan, H., J. Norimine, K. A. Brayton, G. H. Palmer, and W. C. Brown. 2008. Physical linkage of naturally complexed bacterial outer membrane proteins enhances immunogenicity. Infect. Immun. 76:1223-1229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meeus, P. F. M., K. A. Brayton, G. H. Palmer, and A. F. Barbet. 2003. Conservation of gene conversion mechanism in two distantly related paralogues of Anaplasma marginale. Mol. Microbiol. 47:633-643. [DOI] [PubMed] [Google Scholar]
- 50.Nelson, C. M., M. J. Herron, R. F. Felsheim, B. R. Schloeder, S. M. Grindle, A. O. Chavez, T. J. Kurtti, and U. G. Munderloh. 2008. Whole genome transcription profiling of Anaplasma phagocytophilum in human and tick host cells by tiling array analysis. BMC Genomics 9:364-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Niu, H., Y. Rikihisa, M. Yamaguchi, and N. Ohashi. 2006. Differential expression of VirB9 and VirB6 during the life cycle of Anaplasma phagocytophilum in human leukocytes is associated with differential binding and avoidance of lysosome pathway. Cell. Microbiol. 8:523-534. [DOI] [PubMed] [Google Scholar]
- 52.Noh, S., K. A. Brayton, W. C. Brown, J. Norimine, G. R. Munske, C. M. Davitt, and G. H. Palmer. 2008. Composition of the surface proteome of Anaplasma marginale and its role in protective immunity induced by outer membrane immunization. Infect. Immun. 76:2219-2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Norimine, J., and W. C. Brown. 2005. Intrahaplotype and interhaplotype pairing of bovine leukocyte antigen DQA and DQB molecules generate functional DQ molecules important for priming CD4+ T-lymphocyte responses. Immunogenetics 57:750-762. [DOI] [PubMed] [Google Scholar]
- 54.Norimine, J., J. Mosqueda, C. Suarez, G. H. Palmer, T. F. McElwain, G. Mbassa, and W. C. Brown. 2003. Stimulation of T-helper cell gamma interferon and immunoglobulin G responses specific for Babesia bovis rhoptry-associated protein 1 (RAP-1) or a RAP-1 protein lacking the carboxy-terminal repeat region is insufficient to provide protective immunity against virulent challenge. Infect. Immun. 71:5021-5032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ohashi, N., N. Zhi, Q. Lin, and Y. Rikihisa. 2002. Characterization and transcriptional analysis of gene clusters for a type IV secretion machinery in human granulocytic and monocytic ehrlichiosis agents. Infect. Immun. 70:2128-2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Oukka, M., J. C. Manuguerra, N. Livaditis, S. Tourdot, N. Riche, I. Vergnon, P. Cordopatis, and K. Kosmatopoulos. 1996. Protection against lethal viral infection by vaccination with nonimmunodominant peptides. J. Immunol. 157:3039-3045. [PubMed] [Google Scholar]
- 57.Palmer, G. H., A. F. Barbet, W. C. Davis, and T. C. McGuire. 1986. Immunization with an isolate-common surface protein protects cattle against anaplasmosis. Science 231:1299-1302. [DOI] [PubMed] [Google Scholar]
- 58.Palmer, G. H., and K. A. Brayton. 2007. Gene conversion is a convergent strategy for antigenic variation. Trends Parasitol. 23:408-413. [DOI] [PubMed] [Google Scholar]
- 59.Palmer, G. H., W. C. Brown, and F. R. Rurangirwa. 2000. Antigenic variation in the persistence and transmission of the ehrlichia Anaplasma marginale. Microbes Infect. 2:1-10. [DOI] [PubMed] [Google Scholar]
- 60.Palmer, G. H., and T. F. McElwain. 1995. Molecular basis for vaccine development against anaplasmosis and babesiosis. Vet. Parasitol. 57:233-253. [DOI] [PubMed] [Google Scholar]
- 61.Palmer, G. H., D. Munodzana, N. Tebele, T. Ushe, and T. F. McElwain. 1994. Heterologous strain challenge of cattle immunized with Anaplasma marginale outer membranes. Vet. Immunol. Immunopathol. 42:265-273. [DOI] [PubMed] [Google Scholar]
- 62.Palmer, G. H., S. M. Oberle, A. F. Barbet, W. L. Goff, W. C. Davis, and T. C. McGuire. 1988. Immunization of cattle with a 36-kilodalton surface protein induces protection against homologous and heterologous Anaplasma marginale challenge. Infect. Immun. 56:1526-1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Palmer, G. H., F. R. Ruringirwa, K. M. Kocan, and W. C. Brown. 1999. Molecular basis for vaccine development against Anaplasma marginale. Parasitol. Today 15:281-286. [DOI] [PubMed] [Google Scholar]
- 64.Park, J., K. J. Kim, K.-S. Hoi, D. J. Grab, and J. S. Dumler. 2004. Anaplasma phagocytophilum AnkA binds to granulocyte DNA and nuclear proteins. Cell. Microbiol. 6:743-751. [DOI] [PubMed] [Google Scholar]
- 65.Park, Y. H., Y. S. Joo, J. Y. Park, J. S. Moon, S. H. Kim, N. H. Kwon, J. S. Ahn, W. C. Davis, and C. J. Davies. 2004. Characterization of lymphocyte subpopulations and major histocompatibility complex haplotypes of mastitis-resistant and susceptible cows. J. Vet. Sci. 5:29-39. [PubMed] [Google Scholar]
- 66.Rashkova, S., G. M. Spudich, and P. J. Christie. 1997. Characterization of membrane and protein interaction determinants of the Agrobacterium tumefaciens VirB11 ATPase. J. Bacteriol. 179:583-591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Sharif, S., B. A. Mallard, B. N. Wilkie, J. M. Sargeant, H. M. Scott, J. C. Dekkers, and K. E. Leslie. 1999. Association of the bovine major histocompatibility complex DRB3 (BoLA-DRB3) with production traits in Canadian dairy cattle. Anim. Genet. 30:157-160. [DOI] [PubMed] [Google Scholar]
- 68.Spudich, G. M., D. Fernandez, X. R. Zhou, and P. J. Christie. 1996. Intermolecular disulfide bonds stabilize VirB7 homodimers and VirB7/VirB9 heterodimers during biogenesis of the Agrobacterium tumefaciens T-complex transport apparatus. Proc. Natl. Acad. Sci. U. S. A. 93:7512-7517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Swietnicki, W., B. S. Powell, and J. Goodin. 2005. Yersinia pestis Yop secretion protein F: purification, characterization, and protective efficacy against bubonic plague. Protein Expr. Purif. 42:166-172. [DOI] [PubMed] [Google Scholar]
- 70.Tebele, N., T. C. McGuire, and G. H. Palmer. 1991. Induction of protective immunity using Anaplasma marginale initial body membranes. Infect. Immun. 59:3199-3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.van der Most, R. G., A. Sette, C. Oseroff, J. Alexander, K. Murali-Krishna, L. L. Lau, S. Southwood, J. Sidney, R. W. Chesnut, M. Matloubian, and R. Ahmed. 1996. Analysis of cytotoxic T cell responses to dominant and subdominant epitopes during acute and chronic lymphocytic choriomeningitis virus infection. J. Immunol. 157:5543-5554. [PubMed] [Google Scholar]
- 72.van Eijk, M. J., J. A. Stewart-Haynes, and H. A. Lewin. 1992. Extensive polymorphism of the BoLA-DRB3 gene distinguished by PCR-RFLP. Anim. Genet. 23:483-496. [DOI] [PubMed] [Google Scholar]
- 73.Vidotto, M. C., E. J. Venancio, and O. Vidotto. 2008. Cloning, sequencing, and antigenic characterization of rVirB9 of Anaplasma marginale isolated from Parana State, Brazil. Gen. Mol. Res. 7:460-466. [DOI] [PubMed] [Google Scholar]
- 74.Wu, M., L. V. Sun, J. Vamathevan, M. Riegler, R. Deboy, J. C. Brownlie, E. A. McGraw, M. Martin, C. Esser, N. Ahmadinejad, C. Wiegand, R. Madupu, M. J. Beanan, L. M. Brikac, S. C. Daughtery, A. S. Durkin, J. F. Kolonay, W. C. Nelson, Y. Mohamoud, P. Lee, K. Berry, M. B. Young, T. Utterback, J. Weidman, W. C. Nierman, I. T. Paulsen, K. E. Nelson, H. Tettelin, S. L. O'Neill, and J. A. Eisen. 2004. Phylogenomics of the reproductive parasite Wolbachia pipentis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2:E69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ye, M., C. S. Morello, and D. H. Spector. 2002. Strong CD8 T-cell responses following coimmunization with plasmids expressing the dominant pp89 and subdominant M84 antigens of murine cytomegalovirus correlate with long-term protection against subsequent viral challenge. J. Virol. 76:2100-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]