Abstract
Speciation often involves the evolution of incompatible gene interactions that cause sterility or lethality in hybrids between populations. These so-called hybrid incompatibilities occur between two or more functionally divergent loci. We show that the Nup160 gene of the fruitfly Drosophila simulans is incompatible with a factor(s) on the D. melanogaster X chromosome, causing hybrid lethality. Nup160 encodes a nuclear pore complex protein and shows evidence of adaptive evolution. Furthermore, the protein encoded by Nup160 interacts directly with that of another hybrid lethality gene, Nup96, indicating that at least two lethal hybrid incompatibility genes have evolved as by-products of divergent coevolution among interacting components of the Drosophila nuclear pore complex.
As species diverge from one another, they accumulate genetic substitutions that function within their own genomic background but, when brought together in hybrids, can disrupt gametogenesis (causing hybrid sterility) or development (causing hybrid lethality) (1, 2). The evolution and genetics of these hybrid incompatibilities have been shown to follow specific rules. For instance, hybrid incompatibilities tend to accumulate gradually as species diverge (3); behave as partial recessives in hybrids (4); follow Haldane’s rule [i.e., the preferential sterility or inviability of hybrids of the heterogametc (XY or ZW) sex (5)]; and accumulate disproportionately on the X chromosome (i.e., the large X-effect) (6, 7). The molecular biology of hybrid incompatibilities has revealed that five of six hybrid incompatibility genes identified so far show signatures of recurrent adaptive evolution (8–13), suggesting that hybrid sterility and inviability generally evolve as incidental by-products of positive natural selection.
To test if this emerging molecular rule of speciation holds for additional hybrid incompatibility loci we performed a genetic screen for lethal hybrid incompatibilities between Drosophila melanogaster and D. simulans, two species that diverged ~3 Mya. As all hybrids between D. melanogaster and D. simulans are sterile, we performed a chromosomal deletion (deficiency) screen to identify recessive, lethal hybrid incompatibility factors in the D. simulans autosomal genome in the F1 generation [Fig. 1A; (14)]. By screening ~70% of the D. simulans autosomal genome with ~200 deficiencies, we identified 20 small regions that cause hybrid lethality when combined with a hemizygous D. melanogaster X chromosome (14). In one of these 20 regions, we mapped a new hybrid lethality gene using chromosomal deficiencies from D. melanogaster to a region that includes 21 genes (Fig. 1B; Table 1, Lines 1–5) and then performed single-locus complementation tests (Table S1). Only one lesion, PBac{RB}RfC38e00704, uncovers hybrid lethality (Table 1, line 6). To confirm that hybrid lethality is uncovered by the PBac{RB}RfC38e00704 mutation (a piggyBac transposon insertion) and not an unidentified linked mutation, we generated revertant chromosomes that were genetically identical to the original PBac{RB}RfC38e00704 chromosome except that the piggyBac insertion is precisely excised (Fig. S1). We recovered eight independent revertant chromosomes that lacked the piggyBac insertion (PBac{RB}RfC38e00704-R1, R2,…R8) and all were viable in hybrid males (Table S2), confirming that the piggyBac insertion unmasks hybrid lethality.
Figure 1.
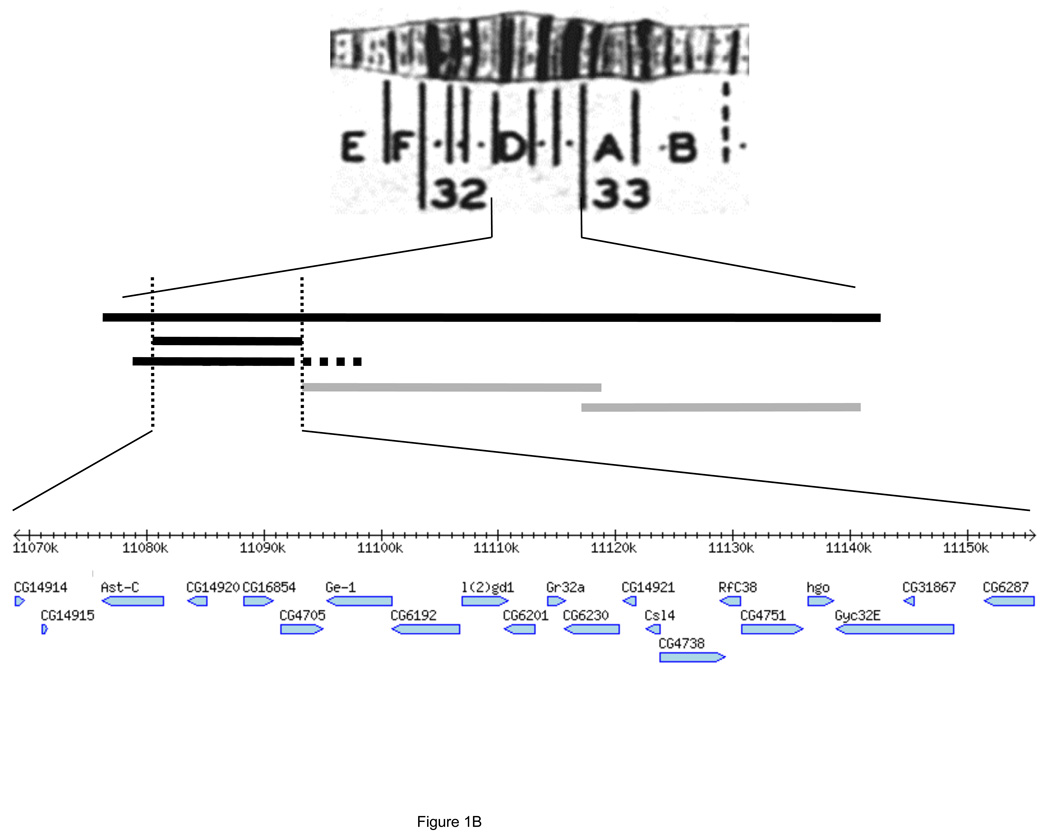
(A) Deficiency mapping recessive X-autosome hybrid lethals in F1 hybrids between D. melanogaster (mel, gray) and D. simulans (sim, white). The sex chromosomes (left, Y chromosome with hook) and one representative pair of autosomes (right) are shown, where Df = autosomal deficiency chromosome, and CyO = balancer chromosome. D. melanogaster Df/CyO females were crossed to D. simulans Lhr males; Lhr is a mutation that rescues ordinarily lethal hybrid males. Of the four zygotic genotypes produced, only Df-bearing hybrid males are forced to develop using only D. simulans material for the autosomal region exposed by the deficiency and only D. melanogaster material for the X chromosome. Thus, if only Df-bearing hybrid males die, we infer that a D. simulans autosomal region (red) is incompatible with D. melanogaster X chromosome, causing lethality (for more details see ref. (14)). Note that the hybrid lethal interactions identified in this screen would normally afflict advanced-generation (e.g., F2), but not F1, hybrids as both partners in these X-autosome incompatibilities are recessive. (B) Fine-scale mapping results in region 32D on chromosome arm 2L. Three deficiencies fail to complement hybrid lethality in males (black bars): Df(2L)FCK-20, Df(2L)Exel6027, and Df(2L)BSC36; two deficiencies complement hybrid lethality in males (gray bars): Df(2L)Exel6028 and Df(2L)Exel6029. Together, these deficiencies implicate one of 21 genes in hybrid lethality that maps to a 89.6-kb region in the cytological interval 32D02;32D05.
Table 1.
Interspecific complementation mapping of a hybrid lethal factor on D. simulans chromosome arm 2L
|
D. melanogaster maternal genotype |
Cytological breakpoints |
Hybrid females | Hybrid males | ||
|---|---|---|---|---|---|
| − / sim | CyO/sim | − / sim | CyO/sim | ||
| 1 Df(2L)FCK-20/CyO | 32D1;32F1-3 | 471 | 504 | 0 | 88 |
| 2 Df(2L)Exel6027/CyO | 32D2;32D5 | 338 | 296 | 1 | 261 |
| 3 Df(2L)BSC36/CyO | 32D1;32F3 | 134 | 137 | 0 | 76 |
| 4 Df(2L)Exel6028/CyO | 32D5;32E4 | 204 | 183 | 151 | 127 |
| 5 Df(2L)Exel6029/CyO | 32E4;32F2 | 265 | 274 | 254 | 192 |
| 6 PBac{RB}RfC38e00704/CyO | 32D4 | 359 | 306 | 0 | 219 |
| 7 C(1)A, y; PBac{RB}RfC38e00704/CyO | 32D4 | 0 | 93 | 114 | 120 |
To determine which gene(s) is disrupted by PBac{RB}RfC38e00704, we analyzed the genomic flanking sequence of the insertion (15) and found that the overlapping 3’-UTRs of two genes, CG4738 and RfC38, are disrupted (Fig. 1B). Of these two genes, CG4738 is the stronger candidate on the basis of three observations: DNA sequences are more diverged between species at CG4738 than RfC38 (58 fixed replacement differences versus one, respectively); disruption of RfC38mel is not generally associated with hybrid lethality as two other transposable element insertions that disrupt RfC38 but not CG4738, P{lacW}RfC38k13807 and PBac{WH}RfC38f07177, do not unmask hybrid lethality (Table S1, Lines 2 and 9); and CG4738 encodes the predicted protein sequence of the Drosophila homolog of the nucleoporin Nup160, a protein component of the nuclear pore complex. The latter finding is especially intriguing as we previously identified another nucleoporin, Nup96, as a hybrid lethality gene between these species (11).
While compelling, all three lines of evidence are circumstantial. We reasoned that if the D. simulans allele, Nup160sim, causes recessive hybrid lethality, then introduction of the D. melanogaster allele, Nup160mel, should rescue otherwise inviable hybrid males. To test this hypothesis, we cloned the Nup160mel cDNA into a vector that carries a w+ eye-color marker (conferring red eyes) and a GAL4-inducible UAS promoter (hereafter, we refer to this construct as P{UAS-Nup160mel}). We transformed P{UAS-Nup160mel} into D. simulans w; Lhr flies and recovered four independent stocks with autosomal insertions of the P{UAS-Nup160mel} construct. We then crossed D. melanogaster PBac{RB}RfC38e00704/CyO; P{Tub-Gal4}/TM3,Ser females carrying a ubiquitously expressed GAL4-driver of P{UAS-Nup160mel} expression to D. simulans w; Lhr P{UAS-Nup160mel} males. These crosses produce four male zygotic genotypes in equal proportions (1:1:1:1), all of which inherit the P{UAS-Nup160mel} construct. As expected, the two kinds of hybrid males inheriting CyO are viable (Table 2, Line 1), and hybrid males inheriting the PBac{RB}RfC38e00704 insertion but no induced Nup160mel expression (i.e., no P{Tub-Gal4}) are lethal (Table 2, Line 1). However, in contrast to their PBac{RB}RfC38e00704/sim; TM3/sim brothers, hybrid males inheriting the PBac{RB}RfC38e00704 insertion and induced Nup160mel expression, PBac{RB}RfC38e00704/sim; P{Tub-Gal4}/sim, are viable (Table 2, Line 1). Moreover, the ratio of the three viable classes of hybrid males is not significantly different from 1:1:1 (χ2 = 3.69, P = 0.158). These data show that ubiquitous expression of D. melanogaster Nup160 rescues the hybrid lethality caused by the insertion of PBac{RB}RfC38e00704 and that D. simulans Nup160 is a lethal hybrid incompatibility gene.
Table 2.
Ubiquitous expression of a D. melanogaster Nup160 transgene rescues hybrid lethality.
| Hybrid autosomal genotypes |
|||||
|---|---|---|---|---|---|
| Line | 2nd chromosome genotype: 3rd chromosome genotype: |
PBac{RB}RfC38e00704/sim; TM3, Ser/sim |
CyO/sim; TM3, Ser/sim |
PBac{RB}RfC38e00704/sim; P{Tub-Gal4}/sim |
Cyo/sim; P{Tub-Gal4}/sim |
| 1 | Hybrid males, Xmel/Ysim | 0 | 119 | 112 | 141 |
| 2 | Hybrid females, Xmel/Xsim | 191 | 204 | 224 | 219 |
Progeny counts are pooled results from four different D. simulans Lhr P{UAS-Nup160mel} stocks as similar results hold across replicates. All progeny from these crosses inherit Lhr and P{UAS-Nup160mel}, which, for simplicity, are not shown.
To confirm that Nup160sim is incompatible with a factor(s) on the D. melanogaster X chromosome, we switched the species origin of the X in hybrids. We constructed a D. melanogaster C(1)A, y/Y; PBac{RB}RfC38e00704/CyO stock, which possess two Xmel chromosomes fused to a single centromere that are transmitted together, and crossed females from this stock to D. simulans w; Lhr males. These crosses produced progeny with the same autosomal genotypes (and cytoplasm) as those in Fig. 1A, but hybrid males inherit their X from D. simulans whereas hybrid females inherit both X chromosomes from D. melanogaster (Fig. S2). [Attached-Xmel hybrid females also inherit a Ysim chromosome, but the Y does not determine sex in Drosophila and is not essential for viability in these species (16).] These crosses show that, whereas Xmel/Ysim; PBac{RB}RfC38e00704/sim hybrid males are lethal (Table 1, Line 6), Xsim/Ymel; PBac{RB}RfC38e00704/sim hybrid males are viable (Table 1, Line 7; Fisher’s Exact P < 10−40 ). We also found that, whereas Xmel/Xsim; PBac{RB}RfC38e00704/sim hybrid females are viable (Table 1, Line 6), attached-Xmel, C(1)A, y/Ysim; PBac{RB}RfC38e00704/sim, hybrid females are lethal (Table 1, Line 7; Fisher’s Exact P < 10−29). These results show, respectively, that Nup160sim causes hybrid lethality via an incompatible epistatic interaction with a factor(s) on the D. melanogaster X chromosome and that Nup160sim-dependent hybrid lethality occurs in both sexes.
To study the evolution of Nup160, we surveyed DNA sequence polymorphism and divergence within and between D. melanogaster and D. simulans from 12 isofemale lines of each species collected from Zimbabwe (see SOM). We tested the neutral mutation-drift model of protein evolution with the McDonald-Kreitman (MK) test (17). The MK test rejects the neutral hypothesis for Nup160 and instead reveals a highly significant excess of fixed replacement differences between species (Table 3, Line 1), consistent with recurrent adaptive protein evolution. To distinguish if adaptive evolution at Nup160 occurred during the history of one or both species, we polarized substitutions along lineages using sequence data from the outgroup species, D. yakuba, and contrasted polymorphic and fixed mutations in the D. melanogaster and D. simulans lineages separately. These lineage-specific MK tests showed that Nup160 has experienced adaptive evolution in both lineages (Table 3, line 2–3).
Table 3.
Recurrent adaptive evolution at the hybrid lethality gene, Nup160, in both D. melanogaster and D. simulans.
| Polymorphic |
Divergent |
Fisher's Exact | |||||
|---|---|---|---|---|---|---|---|
| R | S | R/S | R | S | R/S | P-value | |
| 1 D. melanogaster-D. simulans pooled | 27 | 154 | 0.175 | 58 | 64 | 0.906 | 9.3 × 10−10 |
| 2 D. melanogaster lineage | 10 | 48 | 0.208 | 19 | 33 | 0.576 | 0.0299 |
| 3 D. simulans lineage | 11 | 85 | 0.129 | 26 | 21 | 1.238 | 7.1 × 10−8 |
Despite its history of recurrent positive selection, we found no evidence for a recent selective sweep at Nup160 in either the D. melanogaster or D. simulans lineages. Selective sweeps should reduce local nucleotide diversity (18, 19) and shift the distribution of allele frequencies towards an excess of both rare variants (20) and high-frequency derived variants (21). We found that mean silent nucleotide diversity, π, at Nup160 is similar to other autosomal loci sampled from African populations of these species (22, 23): π = 0.0160 in D. melanogaster and π = 0.0335 in D. simulans. Moreover, coalescent simulations show that neither Tajima’s D (20) nor Fay and Wu’s H (21), two summaries of the allele frequency spectrum, deviate significantly from standard neutral equilibrium expectations in either species (D. melanogaster: D = −0.511 and H = −14.273; in D. simulans, D = 0.134 and H = 7.455; P ≥ 0.124 in all tests). These findings suggest that recurrent positive selection drove sequence evolution at Nup160 but that none of the selected substitutions occurred recently (i.e., within the last ~0.1Ne generations, where Ne is the effective population size).
Our current and previous (11) findings show that two rapidly evolving autosomal genes from D. simulans, Nup160sim and Nup96sim, are incompatible with a factor(s) on the D. melanogaster X chromosome. NUP160 and NUP96 are members of the NUP107 subcomplex, a subset of interacting nucleoporins that together form a stable, architectural component of the nuclear pore complex (NPC), the macromolecular structures that mediate all molecular traffic between the nucleus and cytoplasm (24). The structure, the function, and the interactions among particular nucleoporins seem largely conserved among eukaryotes (25). In addition to NPC functions, members of the NUP107 subcomplex also contribute to kinetochore function (26), cell cycle progression (27), and dosage compensation in Drosophila males (28). In D. melanogaster, three other Nup107 subcomplex genes (including Nup96) have histories of adaptive evolution, similar to that observed here for Nup160, whereas in D. simulans six others have histories of adaptive evolution (29). Interestingly, the components of the dosage compensation complex (30, 31) and its X chromosome binding sites have evolved by positive selection between D. melanogaster and D. simulans (32). However, it is unlikely that the rapid evolution of Nup160, and other members of the Nup107 subcomplex, reflects coevolution with the dosage compensation pathway or that hybrid inviability involves a disruption of dosage compensation in hybrids. For one, the rapid evolution of dosage compensation is largely limited to the D. melanogaster lineage (30–32) and thus cannot explain the especially rapid evolution of Nup160 and the other Nup107 subcomplex genes in D. simulans. For another, Nup160sim kills Xmel/Ysim hybrid males and Xmel.Xmel/Ysim hybrid females, excluding a loss-of-function disruption of dosage compensation— a male-specific problem— as the cause of hybrid lethality. Instead, the recurrent bouts of adaptation of Nup107 subcomplex genes are most likely driven by selection arising from evolutionary conflict involving pathogens, retrotransposons, or meiotic drive elements (29, 33).
As part of the NUP107 subcomplex, NUP160 and NUP96 interact directly (24), raising the possibility that Nup160sim and Nup96sim are incompatible with the same X-linked factor(s) from D. melanogaster. The D. melanogaster allele, Nup153mel, is a strong candidate X-linked partner for both Nup96sim and Nup160sim: Nup153 has a history of adaptive evolution (29), its protein interacts physically with the NUP107 subcomplex (34), and it is the only known interacting nucleoporin encoded on the X chromosome. However, it is not clear if Nup160 and Nup96 are involved in two distinct two-locus hybrid incompatibilities with the D. melanogaster X or if they are components of one complex hybrid incompatibility (i.e., one involving more than two loci). Theory predicts that complex hybrid incompatibilities should evolve more readily than simple hybrid incompatibilities (35), and there is the suggestion that complex hybrid incompatibilities may be typical (e.g., refs. (9, 36)). Regardless of whether nucleoporin-based hybrid lethality has a simple or complex basis, our study suggests that the adaptive coevolution of a large multi-protein complex may have given rise to multiple hybrid incompatibility genes. These findings suggest that divergent coevolution among the interacting partners of macromolecular complexes, particularly those prone to evolutionary conflicts, may drive the evolution of molecular incompatibilities that contribute to speciation.
Supplementary Material
References and Notes
- 1.Coyne JA, Orr HA. Speciation. Sunderland, Massachusetts: Sinauer; 2004. pp. [Google Scholar]
- 2.Dobzhansky T. Genetics and the Origin of Species. New York: Columbia University Press; 1937. p. 364. [Google Scholar]
- 3.Coyne JA, Orr HA. Evolution. 1989;43:362. doi: 10.1111/j.1558-5646.1989.tb04233.x. [DOI] [PubMed] [Google Scholar]
- 4.Turelli M, Orr HA. Genetics. 1995;140:389. doi: 10.1093/genetics/140.1.389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haldane JBS. Journal of Genetics. 1922;12:101. [Google Scholar]
- 6.Coyne JA, Orr HA. In: Speciation and Its Consequences. Otte D, Endler J, editors. Sunderland, MA: Sinauer Associates; 1989. pp. 180–207. [Google Scholar]
- 7.Presgraves DC. Trends in Genetics. 2008;24:336. doi: 10.1016/j.tig.2008.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barbash DA, Siino DF, Tarone AM, Roote J. Proceedings of the National Acadamy of Sciences. 2003;100 doi: 10.1073/pnas.0836927100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brideau NJ, et al. Science. 2006;314:1292. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- 10.Masly JP, Jones CD, Noor MAF, Locke J, Orr HA. Science. 2006;313:1448. doi: 10.1126/science.1128721. [DOI] [PubMed] [Google Scholar]
- 11.Presgraves DC, Balagopalan L, Abmayr SM, Orr HA. Nature. 2003;423:715. doi: 10.1038/nature01679. [DOI] [PubMed] [Google Scholar]
- 12.Ting C-T, Tsaur S-C, Wu M-L, Wu C-I. Science. 1998;282:1501. doi: 10.1126/science.282.5393.1501. [DOI] [PubMed] [Google Scholar]
- 13.Wittbrodt J, et al. Nature. 1989;341:415. doi: 10.1038/341415a0. [DOI] [PubMed] [Google Scholar]
- 14.Presgraves DC. Genetics. 2003;163:955. doi: 10.1093/genetics/163.3.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thibault ST, et al. Nature Genetics. 2004;36:283. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- 16.Ashburner M, Golic KG, Hawley RS. Drosophila: A Laboratory Handbook. Second Edition. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2005. ed. pp. [Google Scholar]
- 17.McDonald JH, Kreitman M. Nature. 1991;351:652. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- 18.Kaplan NL, Hudson RR, Langley CH. Genetics. 1989;123:887. doi: 10.1093/genetics/123.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maynard Smith J, Haigh J. Genetical Research. 1974;23:23. [PubMed] [Google Scholar]
- 20.Tajima F. Genetics. 1989;123:585. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fay JC, Wu C-I. Genetics. 2000;155:1405. doi: 10.1093/genetics/155.3.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andolfatto P. Molecular Biology and Evolution. 2001;18:279. doi: 10.1093/oxfordjournals.molbev.a003804. [DOI] [PubMed] [Google Scholar]
- 23.Hutter S, Li H, Beisswanger S, DeLorenzo D, Stephan W. Genetics. 2007;177:469. doi: 10.1534/genetics.107.074922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suntharalingam M, Wente SR. Developmental Cell. 2003;4:775. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- 25.Bapteste E, Charlebois RL, MacLeod D, Brochier C. Genome Biology. 2005;6:R85. doi: 10.1186/gb-2005-6-10-r85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zuccolo M, et al. EMBO Journal. 2007;26:1853. doi: 10.1038/sj.emboj.7601642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chakraborty P, et al. Developmental Cell. 2008;15:657. doi: 10.1016/j.devcel.2008.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendjan S, et al. Molecular Cell. 2006;21:811. doi: 10.1016/j.molcel.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 29.Presgraves DC, Stephan W. Molecular Biology and Evolution. 2007;24:306. doi: 10.1093/molbev/msl157. [DOI] [PubMed] [Google Scholar]
- 30.Levine MT, Holloway AK, Arshad U, Begun DJ. Genetics. 2007;177:1959. doi: 10.1534/genetics.107.079459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodriguez MA, Vermaak D, Bayes JJ, Malik HS. Proceedings of the National Academy of Sciences. 2007;104:15412. doi: 10.1073/pnas.0707445104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bachtrog D. Genetics. 2008;180:1123. doi: 10.1534/genetics.107.084244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Presgraves DC. BioEssays. 2007;29:386. doi: 10.1002/bies.20555. [DOI] [PubMed] [Google Scholar]
- 34.Vasu SK, et al. Journal of Cell Biology. 2001;155:339. doi: 10.1083/jcb.200108007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Orr HA. Genetics. 1995;139:1805. doi: 10.1093/genetics/139.4.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Barbash DA. Genetics. 2007;176:543. doi: 10.1534/genetics.107.072827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.We thank the Drosophila Genome Resource Center for cloning vectors and the D. melanogaster Nup160 cDNA; the Bloomington Stock Center and the Exelixis Stock Collection at Harvard Medical School for fly stocks; and V. Cattani, P. Gerard, C. Meiklejohn, A. Orr, A. Sweigart and two anonymous reviewers for comments. This work was supported by funds to DCP from NIH grant R01-GM079543, the University of Rochester, and the Radcliffe Institute for Advanced Study at Harvard University. Sequences have been deposited in GenBank under accession numbers FJ600378-FJ600401.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.