SUMMARY
Arginine kinase catalyzes reversible phosphoryl transfer between ATP and arginine, buffering cellular ATP concentrations. Structures of substrate-free and -bound enzyme have highlighted a range of conformational changes thought to occur during the catalytic cycle. Here, NMR is used to characterize the intrinsic backbone dynamics over multiple timescales. Relaxation dispersion indicates rigid-body motion of the N-terminal domain and flexible dynamics in the I182-G209 loop, both at milli-second rates commensurate with kcat, implying that either might be rate limiting upon catalysis. Lipari-Szabo analysis indicates backbone flexibility on the nanosecond timescale in the V308-V322 loop, while the rest of the enzyme is more rigid in this timescale. Thus, intrinsic dynamics are most prominent in regions that have been independently implicated in conformational changes. Substrate-free enzyme may sample an ensemble of different conformations, of which a subset are selected upon substrate binding, with critical active site residues appropriately configured for binding and catalysis.
INTRODUCTION
Arginine kinase (E.C. 2.7.3.3), a member of the phosphagen kinase family of enzymes, catalyzes reversible phosphoryl transfer between ATP and arginine, thus achieving cellular buffering of ATP levels in cells with high and/or variable energy demands (Ellington, 2001). It is chosen as a model for investigating enzyme dynamics due to its amenability to both high resolution crystallography and NMR spectroscopy.
The phosphagen kinase family of enzymes has been the focus of a large body of research resulting in significant advances in our understanding of bimolecular enzyme catalysis (Ellington, 2001). Steady-state enzyme kinetics have shown that arginine kinase undergoes a rapid equilibrium, random order bimolecular-bimolecular reaction, likely common to all phosphagen kinases, with a turnover number (kcat) in the range of 100 to 150 s−1 (Blethen, 1972; Gattis et al., 2004; Pruett et al., 2003). X-ray solution scattering experiments, which detected a decrease in the radius of gyration of arginine kinase upon substrate binding, were the first to indicate that substrate binding is accompanied by large conformational changes and raised the possibility that the change could be rate-limiting (Dumas and Janin, 1983).
X-ray crystal structures have provided the clearest view of the extent of conformational changes in Limulus polyphemus arginine kinase. Two structures of arginine kinase are available: one in a substrate-free, or ‘open’, form at 2.4 Å resolution, and one in a transition state analog complex, substrate-bound or ‘closed’, form at 1.2 Å resolution (Yousef et al., 2003; Yousef et al., 2002; Zhou et al., 1998). These structures show arginine kinase to be a monomeric, two-domain enzyme with a small, α-helical N-terminal domain (NTD) linked to a larger, primarily β-sheet C-terminal domain (CTD), with substrates binding in the cleft between these two domains. In the substrate-bound form, both N-terminal and C-terminal domains fold over the active site, along with a number of loops, analogous to a hand making a grabbing motion (Yousef et al., 2003).
Over the last decade, the concept of “dynamic domain” has been used increasingly to describe clusters of atoms whose conformational change can be approximated a common rotation/translation operator (Gerstein et al., 1994; Hayward, 1999; Hayward and Berendsen, 1998; Lee et al., 2003). A dynamic domain may include non-contiguous regions of linear sequence, so dynamic domains are an extension of the classical definition of domains as globular and linearly contiguous units. Analysis of substrate-free and transition state arginine kinase show the conformational differences can be approximated as rotations of three dynamic domains by up to 18° relative to a fixed domain with additional loop reconfigurations overlaid that bring catalytically important residues to the active site (Yousef et al., 2003). These changes may be involved in mediating substrate specificity, preventing wasteful hydrolysis of ATP, and precisely aligning substrates for phosphoryl transfer (Jencks, 1969; Jencks, 1975; Jencks and Mage, 1974; Lahiri et al., 2002).
There is growing evidence that enzymes bind substrates with a ‘conformational selection’ mechanism (Boehr et al., 2006; Boehr and Wright, 2008; Lange et al., 2008; Ma et al., 1999). In the absence of substrate, an equilibrium may exist between substrate-free states and ones that are closer to the substrate-bound conformation. Binding of substrates stabilizes the ligand-bound state, shifting the pre-existing equilibrium towards the ligand-bound form. Enzyme-substrate interactions in an ATP binding cassette (Wang et al., 2004), RNase A (Beach et al., 2005; Kovrigin and Loria, 2006), dihydrofolate reductase (Osborne et al., 2001) and other enzymes (Boehr et al., 2006), as well as antibody-antigen interactions (Bosshard, 2001), have been explained in terms of conformational selection. Conformational selection embodies statistical mechanics, in subtle contrast to Koshland’s deterministic view of induced-fit where the bound configuration is only attained when the enzyme binds a substrate molecule (Koshland Jr., 1958, 1994). The two mechanistic models represent extremes on a continuum, and there may be elements of both in the same system. Thus an excited state sampled via conformational selection may move closer to the ligand-bound state, but a substrate-initiated change may be needed to attain the final substrate-bound configuration (Sullivan and Holyoak, 2008). In this paper, we examine the timescales of backbone dynamics of an enzyme in the absence of substrates and correlate these findings with the structural changes and kinetics of enzyme turnover which occur in the presence of substrates.
RESULTS
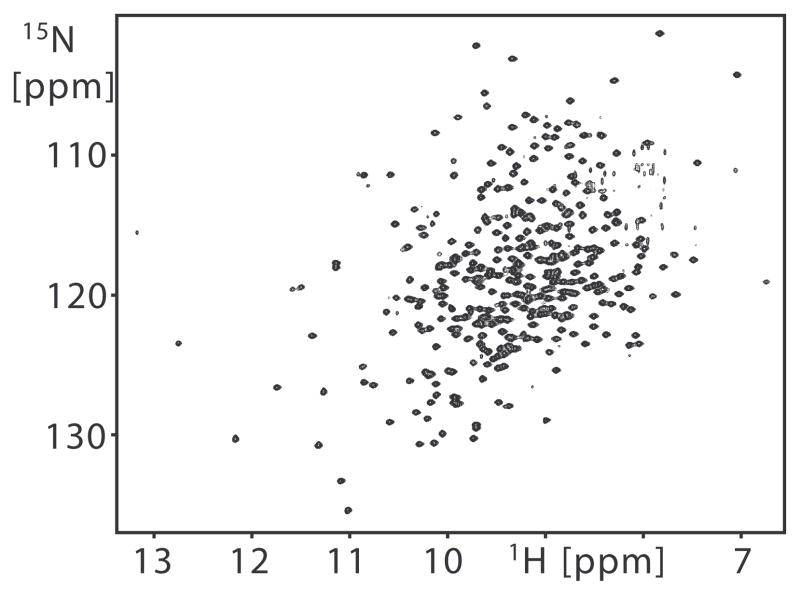
NMR 15N relaxation rate constants were measured for 303 of the 344 non-proline backbone amides in substrate-free arginine kinase. This included all assigned non-overlapping resonances (Davulcu et al., 2005). The NMR data are of high quality as illustrated by a 2D [15N,1H] TROSY spectrum of the enzyme (Figure 1). All relaxation rate constant measurements were detected on the slow relaxing spin transition using [15N,1H] TROSY detection schemes.
Figure 1.
2D [15N,1H] TROSY of arginine kinase recorded at 800 MHz for the shortest T1 delay measurement. The average signal-to-noise ratio of signals from the shortest relaxation delay time of T1 and T1rho measurement are 165/1 and 181/1 at 50.65 MHz, 202/1 and 200/1 at 60.78 MHz, and 370/1 and 250/1 at 81.04 MHz. The average signal-to-noise ratio from the {1H}15N NOE reference spectrum is 165/1.
Slow (μs-ms) Timescale Dynamics
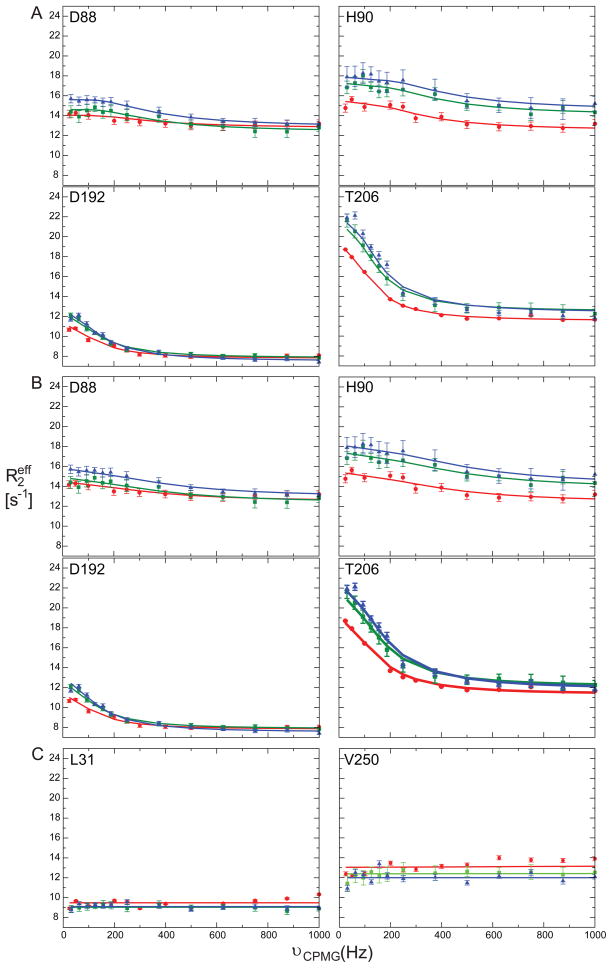
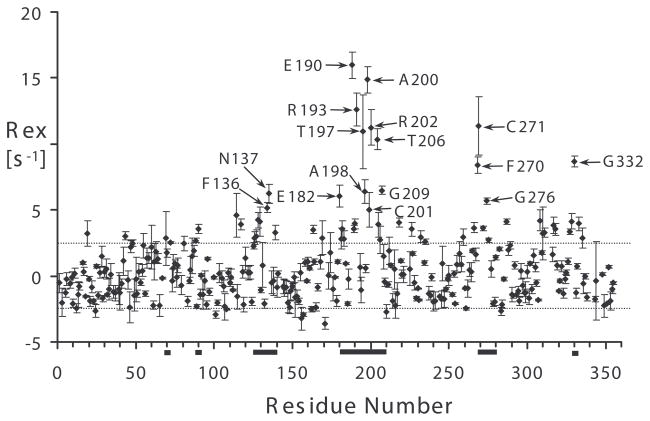
We began this study with a focus on identifying arginine kinase residues that exhibit μs-ms timescale motion. Main chain dynamics on this timescale were measured using two independent experimental approaches. Firstly, 15N relaxation dispersion identified 34 residues with Rex > 2 s−1 (Table 1). Representative 15N relaxation dispersion data at three static field strengths are shown in Figure 2. Secondly, independent Rex measurements from a complementary NMR relaxation experiment identified 17 residues with Rex above a conservative threshold of 5 s−1 (Figure 3 & Table 1) of which just F136 had not previously been identified (Wang et al., 2003). Thus, we identified a total of 35 residues with relaxation arising from chemical exchange (Rex) representing at least 12% of the protein.
Table 1.
Summary of two-state exchange parameters for individual and groups of residues in substrate-free arginine kinase.
| Individual Fits |
Collective Fits |
|||||||
|---|---|---|---|---|---|---|---|---|
| Residue | 2° | kex [s−1] | pB (%) | |Δω| [ppm] | kex [s−1] | pB (%) | |Δω| [ppm] | |
| NTD HINGE | D88 | α4 | 3200 ± 240 | 0.3 ± 0.1 | 1.9 ± 0.3 | 1930 ± 350 | 0.4 ± 0.1 | 3.2 ± 0.3 |
| H90 | α4 | 1020 ± 480 | 0.4 ± 0.1 | 4.5 ± 0.5 | 4.4 ± 0.6 | |||
| G92 | L4 | 1560 ± 760 | 0.3 ± 0.1 | 5.1 ± 0.6 | 3.4 ± 0.4 | |||
| NTD-CTD INTERFACE | D71 | L3 | * | * | * | 800 ± 100 | 1.1 ± 0.1 | 1.1 ± 0.1 |
| F136 | L6 | |||||||
| N137 | L6 | 1150 ± 160 | 1.9 ± 0.7 | 0.9 ± 0.6 | 1.7 ± 0.2 | |||
| F270 | L11 | 2830 ± 230 | 0.9 ± 0.8 | 2.3 ± 0.5 | 2.4 ± 0.4 | |||
| C271 | L11 | * | * | * | 0.9 ± 0.3 | |||
| N274 | L11 | 1100 ± 710 | 0.6 ± 0.1 | 3.6 ± 0.7 | 1.6 ± 0.2 | |||
| G276 | L11 | 1930 ± 340 | 1.4 ± 0.4 | 3.0 ± 0.6 | 3.8 ± 0.5 | |||
| G332 | L14 | 1330 ± 320 | 1.5 ± 2.1 | 1.5 ± 1.0 | 1.5 ± 0.1 | |||
| DYNAMIC DOMAIN 1 AND HINGE | R126 | β1 | 690 ± 220 | 0.3 ± 0.1 | 2.9 ± 0.3 | 790 ± 70 | 0.4 ± 0.1 | 2.2 ± 0.3 |
| C127 | β1 | 940 ± 550 | 0.2 ± 0.1 | 3.7 ± 0.6 | 1.9 ± 2.3 | |||
| R129 | β1 | 950 ± 450 | 0.3 ± 0.1 | 5.6 ± 0.4 | 5.6 ± 0.2 | |||
| V220 | β4 | 2360 ± 210 | 1.0 ± 0.1 | 1.3 ± 0.1 | 2.7 ± 3.0 | |||
| V222 | β4 | * | * | * | 1.7 ± 0.6 | |||
| R229 | β5 | 360 ± 350 | 0.3 ± 0.2 | 2.5 ± 0.4 | 1.7 ± 0.1 | |||
| Q234 | β5 | * | * | * | 1.0 ± 1.0 | |||
| SUBSTRATE BINDING LOOP L8 | I182 | α7 | 700 ± 420 | 0.5 ± 0.2 | 4.5 ± 0.4 | 790 ± 60 | 2.6 ± 0.6 | 0.8 ± 0.4 |
| D183 | α7 | * | * | * | * | |||
| D184 | α7 | 2730 ± 1760 | 0.5 ± 1.3 | 3.1 ± 4.5 | 0.6 ± 0.3 | |||
| H185 | α7 | 1810 ± 650 | 0.3 ± 0.1 | 5.1 ± 0.6 | 0.8 ± 0.4 | |||
| E190 | L8 | 990 ± 70 | 4.5 ± 0.6 | 1.6 ± 0.1 | 2.4 ± 0.8 | |||
| G191 | L8 | * | * | * | * | |||
| D192 | L8 | 760 ± 90 | 1.7 ± 0.4 | 1.3 ± 0.2 | 1.0 ± 0.2 | |||
| R193 | L8 | 1470 ± 390 | 0.5 ± 0.1 | 4.4 ± 0.4 | 1.0 ± 0.5 | |||
| T197 | L8 | 3660 ± 1360 | 0.5 ± 0.1 | 6.8 ± 1.3 | 1.1 ± 0.9 | |||
| A198 | L8 | 4190 ± 1160 | 0.0 ± 1.2 | 2.6 ± 1.6 | * | |||
| A200 | L8 | 1610 ± 440 | 0.7 ± 0.1 | 4.6 ± 0.5 | 1.3 ± 0.7 | |||
| C201 | L8 | 510 ± 230 | 1.3 ± 0.1 | 1.9 ± 0.3 | 1.2 ± 0.1 | |||
| R202 | L8 | 1300 ± 220 | 0.0 ± 0.0 | 0.8 ± 0.1 | 0.8 ± 0.6 | |||
| T206 | L8 | 570 ± 50 | 2.6 ± 0.2 | 1.7 ± 0.1 | 1.7 ± 0.2 | |||
| G207 | L8 | 1350 ± 140 | 0.0 ± 0.0 | 0.8 ± 0.1 | 0.7 ± 0.3 | |||
| R208 | L8 | 920 ± 180 | 0.6 ± 0.1 | 3.8 ± 0.2 | 1.1 ± 0.1 | |||
| G209 | β3 | 1450 ± 380 | 0.7 ± 0.2 | 3.3 ± 0.6 | 1.1 ± 0.2 | |||
Secondary structure of each residue is denoted with 2° as observed in the substrate-free crystal structure of arginine kinase. In all cases, fitting was performed assuming a two-state exchange model, where kex is the sum of forward and reverse rate constants, pB is the relative population of the higher energy state, and Δω is the chemical shift difference between the two states. Asterisks denote residues for which fitting did not converge.
Figure 2.
15N spin relaxation dispersion curves for selected residues with μs-ms timescale dynamics: D88, H90, D192, and T206. (a) Individual fit dispersion curves at 15N Larmor frequencies 60.78 (red circles), 72.05 (green squares), and 81.04 MHz (blue triangles). (b) Collective fit dispersion curves for same residues: D88 and H90 with G92 and D192 and T206 with 12 other residues (Table 1). (c) 15N spin relaxation dispersion curves for residues with no Rex.
Figure 3.
Chemical exchange in substrate-free arginine kinase identified from Rex = R2β – R12HzNz/2 – ηxy(κ − 1) + R1N/2. κ = trimmed mean of 1 + (R2β − R12HzNz/2)/ηxy = 1.43. For apparently non-exchanging residues, the average Rex is 0.05 ± 2.5 s−1, error is shown with dashed lines. The large error is partly due to residue-specific variation in the 15N CSA tensor magnitude and orientation. The solid bars represent polypeptide segments having residues with 15N spin relaxation dispersion. Error bars are from measurements in triplicate. Residues with Rex greater than 2 standard deviations (5 s−1) are labeled.
Residues with Rex are prevalent at four prominent locations in the protein (Figures 3 & 4 & Table 1): (1) D88-G92, within a hinge linking the N- and C-terminal domains; (2) D71, F136-N137, F270-G276 and G332 which all lie at the interface between the N- and C-terminal domains; (3) R126-L131 and V220-Q234, which largely constitute the hinges of dynamic domains 1 and 2 and (4) I182-G209, a substrate binding loop.
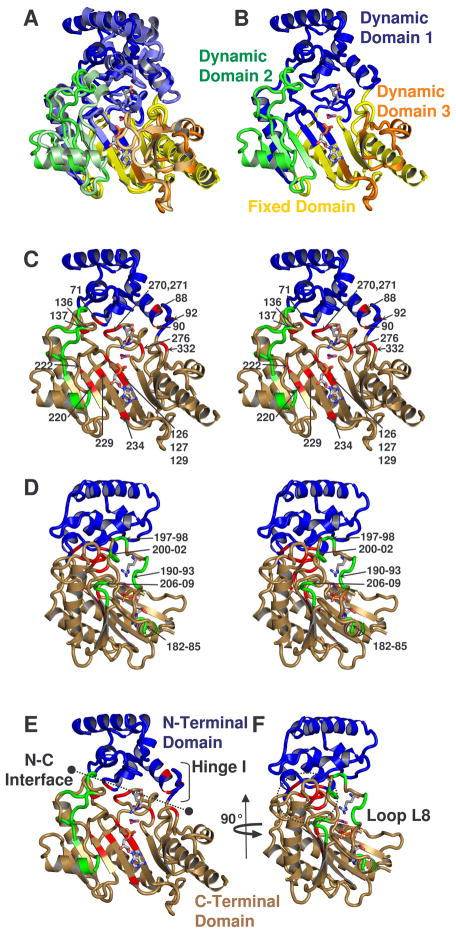
Figure 4.
(a) Comparison of the dynamic domains of substrate-free and -bound structures of arginine kinase (Yousef et al., 2003; Zhou et al., 1998). Dynamic domains 1, 2, and 3 are colored respectively blue, green and orange. The darker shade of each color is substrate-free and fixed domain is shown in yellow. (b) shows only substrate-free. (c – f) show amino acids with measurable Rex, colored red, with the exception of residues between 180-209, near loop L8, which are shown in green. The classical N- and C-terminal domains are shown blue and sand. c & d are stereographic pairs with loop 8 residues labeled in panel d, and all others labeled in panel c. Panels d & f are rotated 90° with respect to panels c & e. e shows the interface between N- and C-terminal domains and location of N-terminal domain hinge. In all panels, substrates ADP, NO3 anion, and arginine are shown from the -bound structure to guide the eye.
Resonances showing chemical exchange were fit to the Bloch-McConnell equation, and assuming two-state exchange, estimates of kex, populations pA and pB, and the chemical shift difference between states A and B, Δω, were obtained (Cavanagh et al., 1996; Palmer et al., 2001). Representative 15N spin relaxation dispersion curves are shown in Figure 2 and their fit exchange parameters are listed in Table 1. Conformational exchange rate constants, kex, of individual residues range from 400 to 4200 s−1 with significantly skewed pA/pB ratios. The fit 15N chemical shift difference, Δω, ranges from 0.8 to 5.6 ppm. Analysis for a small number of residues did not converge, likely the result of small Rex. Dispersion data from residues in spatially distinct locations of the enzyme were fit together assuming they share a common motion responsible for the chemical exchange with the same kex and relative populations pA and pB. The groupings and fit parameters are listed in Table 1. Further attempts to consolidate by fitting common parameters dynamic domain 1, its hinge, and flexible loop L8 were unsuccessful.
Fast (ns-ps) Timescale Dynamics
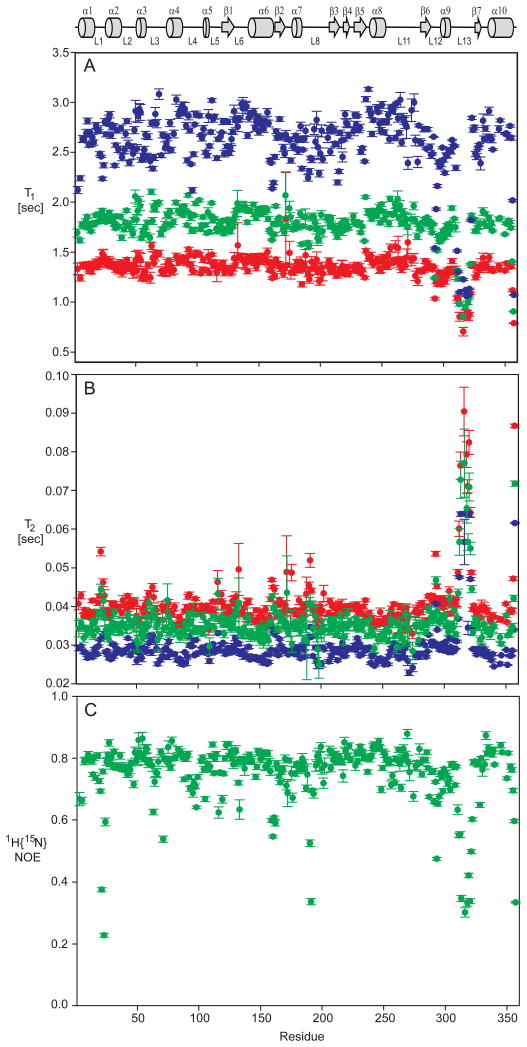
NMR spin relaxation measurements were recorded at three static field strengths, 15N ωo = 50.65, 60.78, and 81.04 MHz, and analysed with the Lipari-Szabo model-free approach, or extended model-free, providing ns-ps dynamics information (Clore et al., 1990; Lipari and Szabo, 1982a, b). 15N spin longitudinal (T1) and transverse (T2) relaxation time constants were measured at each field and the heteronuclear {1H}15N nuclear Overhauser effect (NOE) measured at 60.78 MHz only, yielding a total of seven measurements per residue. The data were recorded with high signal to noise (Figure 1), resulting in reliable T1, T2, and NOE values with most standard deviations < 2%. The relaxation time constants and NOE values are plotted in Figure 5 and listed in Supplemental Table 1. Mean and “trimmed” mean values, the subset used in the isotropic τm calculation, are listed in Table 2. The theoretical maximum NOE, in the rigid limit and τm = 23 ns, is 0.83 and only two residues are larger by more than 2 standard deviations, T269 and L333.
Figure 5.
15N NMR spin relaxation time constants (a) T1, (b) T2 and (c){1H}15N NOE for arginine kinase at 15N Larmor frequencies 50.65 (red), 60.78 (green), and 81.04 MHz (blue). Individual values are listed in Supplemental Table 1. Error bars on each measurement represent one standard deviation. The T1rho spin lock time at 50.65 and 60.78 MHz was shorter than optimal for the slow relaxing amide signals in loop L13 resulting in relatively large errors. A schematic diagram of arginine kinase secondary structure is shown at the top with α-helices shown as cylinders, β-strands as arrows, and loops and extended structure as lines.
Table 2.
Mean and trimmed mean 15N T1, T2, and heteronuclear {1H}15N NOE values for backbone amides of arginine kinase.
| Parameter | 50.65 MHz | 60.78 MHz | 81.04 MHz |
|---|---|---|---|
| T1 [s] | 1.35 ± 0.12 | 1.79 ± 0.17 | 2.61 ± 0.33 |
| Trimmed T1 [s] | 1.36 ± 0.06 | 1.82 ± 0.09 | 2.69 ± 0.16 |
| T2 [s] | 0.041 ± 0.007 | 0.036 ± 0.006 | 0.029 ± 0.005 |
| Trimmed T2 [s] | 0.039 ± 0.002 | 0.034 ± 0.002 | 0.029 ± 0.002 |
| Trimmed T1/T2 | 35.34 ± 3.25 | 53.08 ± 5.00 | 96.46 ± 9.49 |
| {1H}15N NOE | 0.75 ± 0.10 | ||
| Trimmed {1H}15N NOE | 0.79 ± 0.04 |
Values are reported as the all-residue or trimmed mean ± one standard deviation. Trimmed means are determined following criteria outlined in Tjandra et al., 1995 (Tjandra et al., 1995b).
{1H}15N NOE < 0.65 are excluded from trimmed {1H}15N NOE mean.
NMR spin relaxation measurements were fit to isotropic and axial-symmetric anisotropic rotational diffusion models using established methods (Tjandra et al., 1995a). Evidence derived from calculation and experiment strongly suggests that the axial-symmetric diffusion model is more appropriate. First, calculation of rotational diffusion anisotropy from NMR data with the substrate-free arginine kinase coordinates, assuming axial symmetric diffusion, resulted in a ratio of rotational diffusion rate constants, Dpar/Dperp of 0.86, which corresponds to an oblate ellipsoid (Yousef et al., 2003). The final diffusion tensor for 1M80.pdb was Dpar = 6.50 × 106 s−1 Dperp = 7.54 × 106 s−1, theta = 1.62 rad. and phi = 0.72 rad. Secondly, the calculated ratio of rotational diffusion rate constants for substrate-free arginine kinase using HYDRONMR was 0.80, consistent with the experimental results (Garcia de la Torre et al., 2000; Yousef et al., 2003). Finally, the consensus optimal value of τm and the average value of τm for the amide resonances of the individual residues fit to isotropic models both indicate that the value of the rotational correlation time is 23 ns. In contrast, the τm values for the amide nuclei in individual range between 21.0 – 25.7 ns, which is inconsistent with isotropic diffusion, but consistent with data acquired for a system that exhibits anisotropic rotational diffusion but fit to an isotropic model. We therefore proceed with subsequent analysis of the NMR relaxation data using an anisotropic model.
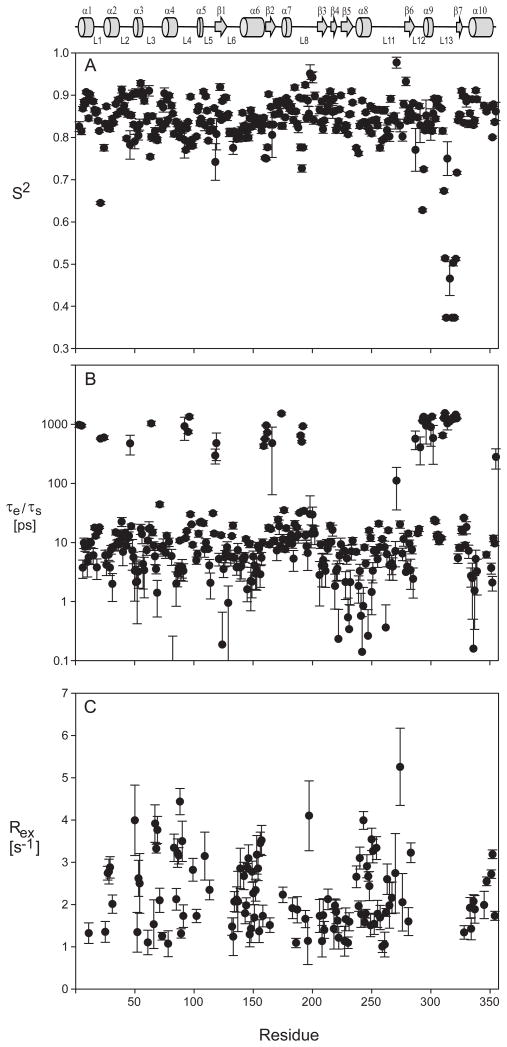
Following standard procedures and using axial symmetric diffusion, the ns-ps dynamics of each residue was analyzed and reported in Figure 6 and listed in Supplemental Table 2 (Clarkson et al., 2006; d’Auvergne and Gooley, 2003; Mandel et al., 1995). Model selection was guided by the χ2 error. Where χ2could not distinguish two or more models, the simplest was chosen. Models 3 (S2, Rex) & 4 (S2, τe, Rex) were excluded if the fit Rex was < 1 s−1. This cutoff was chosen since the experimental error of R2 is about 1 s−1. Residues V308 - V322, which are absent in the PDB coordinates, were necessarily fit using the isotropic model. Model selection results are summarized in Table 3. About 35% of the residues were best fit using models 3 & 4, indicating even more extensive μs-ms chemical exchange (Figure 6) than reported by relaxation dispersion. Alternative analysis using an isotropic diffusion model indicates a slightly higher fraction (40%) of residues best fit to models 3 & 4 (not shown). About 14% of the residues (43) were best fit to model 5 (S2f, S2s, τs) and are most abundant in loops and turns. Most residues best fit with model 5 have a slow time constant, aveτs = 1 ± 0.4 ns. It should be appreciated that the error in estimating the local correlation time is larger than that of other fitted parameters. Furthermore, in the current analysis, a single measurement of the {1H}15N NOE was used which decreases the precision of the estimate somewhat. Although the τs values are relatively large, because τm is also large (23 ns), the Lipari-Szabo decoupling approximation is not violated (Lipari and Szabo, 1982a, b; Vugmeyster et al., 2003). Nearly all residues best fit to model 2 (S2, τe) & 4 (S2, τe, Rex) have τe values < 20 ps. The aveS2 over all residues is 0.85 ± 0.06 and is significantly lower only for residues in loops L1, L12, and L13 (Figure 6 & Supplemental Table 2). Overall, the enzyme is relatively rigid on the ns-ps timescale with exception of three loops, including one, loop L13, which has previously been shown to be crucial for substrate binding.
Figure 6.
Lipari-Szabo model-free results for substrate-free arginine kinase dynamics. (a) Generalized order parameters, (b) τe/τs internal time constants and (c) Rex at 600 MHz. All fit parameters and model selections are listed in Supplemental Table 2.
Table 3.
Summary of Lipari-Szabo model selection for substrate-free arginine kinase.
| Model | Number of Residues | Percentage | |
|---|---|---|---|
| 1 | (S2) | 5 | 1.7 |
| 2 | (S2, τe) | 142 | 46.8 |
| 3 | (S2, Rex) | 7 | 2.3 |
| 4 | (S2, τe, Rex) | 106 | 35.0 |
| 5 | (S2f, S2s τs) | 43 | 14.2 |
| Total | 303 | 100.0 | |
DISCUSSION
Interpretation of the main chain dynamics will be discussed with reference to classical as well as “dynamic” domain structure, dynamic domains being more appropriate in describing the differences between substrate-free and -bound crystal structures. This is not to imply that the intrinsic dynamics observed by NMR in substrate-free form must necessarily be of the same magnitude or direction as the large conformational changes revealed in the substrate-bound crystallographic structures. Furthermore, NMR can reveal motions between open and closed states, some of which may be populated at levels too low to be detectable crystallographically. Despite these caveats, there is good correspondence between the locations of residues exhibiting chemical change or increased flexibility and prediction based upon the crystal structures.
The classical description of arginine kinase is as two domains: a small, α-helical N-terminal domain (NTD) spanning residues 1-90 and a larger, primarily β-sheet C-terminal domain (CTD) spanning residues 120-357, with the active site lying in the cleft between these domains. The domains are connected with residues 91-119, a region well-defined by crystallography but lacking regular α-helical or β-strand structure. Upon binding substrates, the two domains close over the active site. Closing of the two domains positions several substrate-binding and catalytically-important residues in the active site, underlying the importance of dynamics on turnover of the enzyme (Yousef et al., 2003; Zhou et al., 1998).
Arginine kinase can also be described as three dynamic domains that rotate independently relative to a fourth, fixed domain (Yousef et al., 2003). The overall structure of dynamic domains is shown in Figure 4a,b and compared to classical domains in Figure 4c–f. Dynamic domain 1 is an extended and larger version of the N-terminal domain, while dynamic domains 2 and 3 are primarily large loops (L8 and L13) of the classical C-terminal domain.
Rigid Body Motion of the N-terminal Domain
Between the substrate-free and bound states, the (classical) N-terminal domain of arginine kinase undergoes a significant rigid-body motion - a hinged rotation of the domain about residues 88-94 (Figure 4a,b) (Yousef et al., 2003; Zhou et al., 1998). Domain rotation also causes a flexing at the NTD-CTD interface (Figure 4). The open and closed forms of the enzyme show little difference in the internal structure of the NTD. The average composite torsion angle difference, defined as (Δφ2 + Δψ2)1/2, for residues 3-90 is only 10 degrees, and the backbone RMSD is < 1 Å compared to a 6 Å overall displacement of the N-terminal domain upon substrate-binding (Yousef et al., 2003; Zhou et al., 1998). The linker between the NTD and CTD also shows sizeable differences between the open and closed-form crystal structures at residues 92-94, with composite torsion angle changes of 15°, 18°, and 29°(Yousef et al., 2003; Zhou et al., 1998).
NMR relaxation dispersion identifies μs-ms timescale motion for multiple residues in the hinge and interface (Figure 4e,f). Their distribution is consistent with a rigid body motion with little change in internal structure: the NTD core is devoid of Rex with the exception of D71, located in the interface with the CTD (Figure 4). Assuming two-state exchange between open and closed forms, we fit exchange parameters for individual residues and groups with the Bloch-McConnell equation (Table 1). Fitting NMR relaxation dispersion for three individual hinge residues (D88, G90, G92) resulted in similar kex and pB values while fitting a single collective motion yields kex = 1930 ± 350 s−1 and pB = 0.4 ± 0.1%. The equilibrium is highly skewed; the approximate kopen and kclose rate constants are 1920 and 10 s−1, respectively. For eight NTD-CTD interface residues (D71, F136, N137, F270, C271, N274, G276, and G332) individual kex values range from 1200 to 2800 s−1 with skewed populations, pB < 2%, while a collective fit yields kex = 800 ± 100 s−1 and pB = 1.1 ± 0.1% (Table 1). Estimates of kopen and kclose are 790 s−1 and 10 s−1 respectively. The collective interface kex is about half that of the hinge. We note that the hinge and interface residues fit small relative population pB, which can lead to uncertainty in the exact parameter values fit. A collective fit of hinge and interface residues together was unsuccessful. In summary, the hinge and interface residues exhibit μs-ms timescale dynamics, with a highly skewed equilibrium pB ≈1%, kopen ≈ 790 – 1920 s−1, and kclose ≈ 10 s−1. The slower of the two rate constants is of the same order as kcat, and potentially rate-limiting.
Additional residues exhibiting Rex can be understood with the dynamic domain model (Figure 4a,b; Zhou et al., 1998; Yousef et al., 2003). R126, C127, R129, V220, V222, R229, and Q234 are located in the 49 residue dynamic domain 1 hinge or near the bottoms of the substrate binding pockets (Figure 4; (Yousef et al., 2003)). A collective fit for all dynamic domain 1 and hinge residues results in a reasonable kex = 990 ± 110 s−1 but very small pB (< 0.3%) and unreasonably large Δω and errors (not shown). Thus, even if only semi-quantitatively, NMR confirms μs-ms timescale dynamics of the NTD (or dynamic domain 1 and hinge 1) and demonstrates that the motions are intrinsic to substrate free arginine kinase, and not just substrate-gated.
On the ps-ns timescale, the entirety of the classical N-terminal domain or dynamic domain 1 and hinges are mostly rigid, with only a few locations showing increased flexibility (S2 < 0.8). The average order parameter, S2, for the NTD is 0.85 ± 0.04 and of the NTD loops, only L1 (residues S21-K24) shows significantly lower S2 values while L2 (N41-T49), and L3 (L61-A72) reveal only slightly below average S2 (supplemental Table 2; Figure 6). Loop L1 is surface exposed and distant from the active site. The average S2 of hinge residues is similar to the average of the NTD residues. Loop L4, G91-G105, of the NTD-CTD linker and loops L6, S130-L140, and L11, K256-M279, at the NTD-CTD interface also show very small decreases in S2, indicating some increased local flexibility.
Dynamics of Substrate Binding Loop L8, I182-G209
Substrate binding loop L8 changes conformation between open and closed structures to help close the entrance to the nucleotide binding site (Yousef et al., 2003; Zhou et al., 1998). The overall displacement of L8 is 5.4 Å, but internal to L8, the RMSD is only 1.4 Å (Figure 4). The average composite torsion angle change in loop L8 between open and closed forms of arginine kinase is 36°. Similar to the NTD, loop L8 has characteristics of a rigid body motion, but the larger composite torsion angle change and RMSD suggest a more complex motional model.
NMR relaxation dispersion shows a high fraction (60%) of loop L8 residues with Rex. Fitting dispersion for individual residues yields a range of kex from 500 – 2700 s−1 and skewed populations, pB < 4.5%; a collective fit of all seventeen residues yields kex = 790 ± 60 s−1, pB = 2.6 ± 0.6%, kopen ≈ 770 s−1, and kclose ≈ 20 s−1 (Table 1) and assumes the predominant conformer is the open state. The smaller of the two rate constants is on the same order as kcat.
NMR confirms μs-ms timescale dynamics in loop L8 are intrinsic to substrate free arginine kinase and not only substrate-gated. Backbone crystallographic B-factors for this loop in the structure of the unbound protein are elevated about two-fold. In contrast, B-factors for loop L8 in the substrate-bound form are not elevated, indicating relative ordering compared to the substrate free structure. Overall, the NMR results are consistent with the crystallographic B-factors. Loop L8 is involved with substrate nucleotide binding. The closed structure of the enzyme places loop L8 residue H185, along with H284, in a stacked arrangement with the purine ring of substrate ADP (Zhou et al., 1998). We thus speculate that the intrinsic motion present in loop L8 may sample the closed-form configuration, which is then selected with nucleotide binding.
The primary structure of loop L8 is highly conserved across the phosphagen kinase family. Crystal structures of substrate-free mitochondrial creatine kinase and transition state analog form of muscle creatine kinase show an identical B-factor pattern for the creatine kinase equivalent of loop L8 (Fritz-Wolf et al., 1996; Ohren et al., 2007). Furthermore, the results of hydrogen/deuterium exchange studies of creatine kinase are consistent with increased mobility in this loop (Mazon et al., 2004; Mazon et al., 2005). These correlations between arginine kinase and creatine kinase suggest that the phosphagen kinase family of enzymes has evolved not only in structure, but also in dynamics of this and other catalytically important regions.
On the ns-ps timescale, loop L8, and the whole of dynamic domain 2, appears relatively rigid (S2 = 0.85 ± 0.03) with exception of three residues E190, G191, D192 located near but not at the loop tip, which have S2 of 0.78, 0.73, and 0.78 respectively (Supplemental Table 2; Figure 4). Two of these residues, E190 and D192, exhibit measurable Rex and also best fit model 5 (S2f, S2s, τs) and their NOEs are smaller than average (Supplemental Table 2). This suggests a more complicated loop L8 motion.
Dynamics of Substrate Binding Loop L13, V308-V322
On the ns-ps timescale, the most prominent feature of motion in the enzyme is present in substrate binding loop L13, residues V308-V322 (Figures 5 & 6). Residues in the loop are best fit with model 5 (τs, S2s, S2f) resulting in a τs ≈ 1 ns (Table 3). As noted above, use of a single {1H}15N NOE measurement limits the precision of the local correlation time constant. The average S2 is 0.53, substantially lower than average (Supplemental Table 2). This increased flexibility is consistent with an absence of observable electron density for this region in the substrate-free arginine kinase crystal structure (Yousef et al., 2003). The correspondence of low order parameters and the absence of electron density suggest loop disorder is not an artifact of the crystalline state, since loop L13 becomes well-ordered in the substrate-bound complex, with only slightly elevated B-factors.
The conservation of elevated measures of thermal disorder in substrate-free crystal structures of phosphagen kinases from different phyla suggests a link to function. In substrate-bound arginine kinase, the side chains from two loop L13 residues directly contact substrate, R309 with nucleotide and E314 forming a salt link with substrate arginine (Zhou et al., 1998). R309 is highly conserved in the phosphagen kinase family and E314 is less conserved, aligning with a valine in creatine kinases. An E314V mutant, in the context of swapping the arginine and creatine kinase loops, resulted in only 35% loss of activity suggesting stabilization of the substrate-bound configuration involves interactions besides E314 (Azzi et al., 2004; Pruett et al., 2003). The structural, mutagenesis, and NMR studies suggest that the conserved primary structure of loop L13 across phosphagen kinases may reflect conservation of flexibility, as opposed to stabilization of substrate-bound state, as a necessary condition for catalysis.
Dynamics and Enzyme Catalysis
Through an increasing number of examples, it is becoming apparent that dynamics is important in the turnover of many enzymes (Boehr et al., 2006). General comments have been made about the slow turnover of phosphagen kinases being consistent with rate-limiting conformational change, but it is only now, with this work, that the specific mechanistic details are beginning to emerge. Particularly intriguing is the approximate correspondence between rate constants of conformational changes determined here by NMR (3 – 20 s−1) and the enzyme turnover rate, kcat =100 –150 s−1, measured earlier by experimental kinetics, suggesting that conformational changes might be rate limiting on enzyme turnover. The two regions with motions at corresponding timescales are loop L8 and the N-terminal domain (or dynamic domain 1), which moves as a (quasi-)rigid group. It is possible that the intrinsic motions are in the same direction, if not of the same magnitude as the changes seen between the substrate-free and -bound crystal structures, but the NMR analysis provides no direct evidence (Yousef et al., 2003; Zhou et al., 1998). The changes to loop L8 and the NTD contribute to the closing of the enzyme around the active site pocket. The most intriguing possibility is that turnover is gated by one or both of the motions. The difference between the slow rate constants of conformational exchange and turnover is not large, and might be brought even closer if the presence of substrates modulates dynamics. It is also possible that substrate-associated changes to loop L8 and NTD were required, and that evolutionary selection of residues that facilitate the conformational change could also have resulted in a loop that is intrinsically dynamic. In either case, it is possible that the ms timescale motions are part of an intrinsic conformational equilibrium whose populations are shifted by substrate binding towards a state required for reaction turnover (Boehr and Wright, 2008; Lange et al., 2008).
Loop L8 includes H185 that forms two interactions with the substrate nucleotide, stacking with the base, and hydrogen-bonding to the ribose (Zhou et al., 1998). While the rest of the loop is in the general area of the active site, this is the only direct interaction, and it is not proximal to where bond breakage/formation is catalyzed. The same is true for the NTD, for which the only direct substrate interaction is between the substrate-specificity loop and the end of the phosphagen substrate that is remote from the reactive atoms. Thus, if intrinsic dynamics, observed by NMR, are rate-limiting, their effects are exerted through substrate-binding or indirectly upon catalysis through propagated changes, rather than through direct influence on the catalytic chemistry.
The rate-limiting effect of protein dynamics on turnover has been shown for a number of other enzymes. Lid opening and closing has been shown to be rate-limiting upon catalysis in, for example, adenylate kinase (Wolf-Watz et al., 2004) and triose phosphate isomerase (Rozovsky et al., 2001). A single histidine residue has been implicated in coupling motions in RNase A to turnover (Watt et al., 2007). The transition from a closed to an occluded conformation in dihydrofolate reductase, marked by a loop motion, has been shown to be rate-limiting (Boehr et al., 2008). The motions in arginine kinase have analogs in these other systems. It is noteworthy that several motions are found together in arginine kinase: motion in a lid-like loop (L8), hinged domain motion (NTD), both of which are at turnover-commensurate rates, and the faster dynamics of loop L13 that folds over the active site. The presence of several modes of motions in arginine kinase, some of which might be turnover-limiting, and the conservation of the dynamic structural elements adds to a growing awareness that conformational dynamics might be more widespread and functionally relevant to enzymes than has been heretofore widely appreciated.
EXPERIMENTAL PROCEDURES
Sample Preparation
Uniformly 15N-enriched and 80% 2H-enriched Limulus polyphemus arginine kinase was prepared as previously described (Davulcu et al., 2005). The final enzyme sample was concentrated to 1.0 mM in a buffer containing 10 mM sodium citrate (pH=6.5), 0.5 mM dithiothreitol, 50 mM potassium chloride, 50 μm sodium azide, 90% H2O, and 10% D2O. Arginine kinase is stable for months at 4 or 20 °C, showing no signs of degradation or reduced enzymatic activity as judged by 2D [15N,1H] TROSY spectra and enzyme activity profiles over this same timescale (not shown).
NMR Spectroscopy
NMR data were recorded on Varian INOVA NMR spectrometers operating at 1H(15N) Larmor frequencies (ωo) of 499.8(50.65), 599.7(60.78), 719.9(72.95), or 799.7(81.04) MHz. Each spectrometer was equipped with a HCN triple resonance probe and a Z-axis pulsed field gradient. All relaxation experiments were performed at 25 ± 0.5 °C, determined using a methanol standard (Raiford et al., 1979). Proton chemical shifts were referenced to the DSS methyl protons at 0 ppm and 15N chemical shifts referenced indirectly (Markley et al., 1998). Amide 1H and 15N resonance assignments of arginine kinase have been reported (Davulcu et al., 2005).
15N T1 and heteronuclear {1H}15N NOE values were measured with relaxation experiments modified for TROSY detection (Farrow et al., 1994). T1ρ was measured using relaxation experiments described in Korzhnev et al., 2002 (Korzhnev et al., 2002); T2 values were calculated from T1 and T1ρ values. At 50.65 MHz, 15N T1 was measured using relaxation delays of 0.151, 0.503*, 1.006, 1.509, 2.012, and 3.017 s (“*” denoting measurements duplicated for error estimation); 15N T1ρ was measured using relaxation delays of 5, 10*, 25, 35, and 55 ms and a 15N spinlock field strength of 2.6 KHz. At 60.78 MHz, 15N T1 was measured using relaxation delays of 0.151, 0.503*, 1.006, 1.509, 2.012, and 4.5 s; 15N T1ρ was measured using relaxation delays of 5, 10*, 25, 35, and 55 ms, and a 15N spinlock field strength of 2.7 KHz. At 81.04 MHz, 15N T1 was measured using relaxation delays of 0.151*, 0.504, 1.010, 1.510, 2.010*, 3.020, and 3.527 s, 15N T1ρ was measured using relaxation delays of 5*, 15, 25*, 35, and 55 ms and a 15N spinlock field strength of 2.7 KHz. The steady-state {1H}15N NOE was measured at 60.78 MHz using a 1H saturation time of 11 s (5 times the longest 15N T1). {1H}15N NOE was calculated from the intensity ratio of an experiment recorded with and without 1H saturation during the mixing time. Peak intensity error was estimated from the rms base plane noise and {1H}15N NOE error from propagation of error in quadrature.
Slow timescale chemical exchange was quantified using two different experimental NMR approaches. The first approach made use of constant relaxation-time 15N spin CPMG relaxation dispersion with TROSY detection at three static fields, 15N ω0 = 60.78, 72.95 and 81.04 MHz (Loria et al., 1999). In these experiments, the CPMG field strength (υ) is varied between 0 and 1 KHz by adjustment of the CPMG refocusing delay time. Data were recorded using the following parameters: 15N ω0 = 60.78 MHz, constant relaxation time = 40 ms, υ = 0, 25, 50*, 100, 200, 250, 300, 400, 500, 625, 750, 875, and 1000 Hz; 15N ω0 = 72.95 and 81.04 MHz, constant transverse relaxation time of 32 ms, υ = 0*, 31.25, 62.50, 93.75, 125*, 156.25, 187.50, 250, 375, 500*, 625*, 750, 875, and 1000* Hz. The second approach is based on measurement of 15N spin R2β (TROSY), R2α (anti-TROSY), and longitudinal two-spin order R12HzNz rate constants, which are then used to estimate Rex (Wang et al., 2003). 15N spin R2α, R2β, and R12HzNz experiments were recorded at 15N ω0 = 60.78 MHz. Experiments were recorded in duplicate at 2τ = 5/JNH and, alternatively, the complete decay curves were recorded using relaxation time delays of 2τ = N/JNH with N = 0, 1, 2*, 3, 6, and 12 for R2β; N = 0, 1*, 2, 3 for R2α; τ = N/JNH with N = 0, 1, 4*, 10, 20, and 30 for R12HzNz. Rate constants were taken from two-parameter single exponential fits of peak intensities to relaxation delay times using the Levenberg-Marquardt algorithm. Rex and kappa were identical within measurement error, whether determined using a single 2τ = 5/JNH relaxation time or by recording a full decay curve using multiple relaxation time increments.
All 15N spin relaxation data were recorded with ~100 ms 1H acquisition time (t2max) and 75 ms 15N acquisition time (t1max) and processed with FELIX2004 to final matrices of 4096 (1H) × 1024 (15N) points. Peak intensities were measured with SPARKY (Goddard and Kneller). 15N spin relaxation time constants T1 and T1ρ were calculated from two-parameter single exponential fits of peak intensities to relaxation delay times using the Levenberg-Marquardt algorithm. The residual χ2 values of the single exponential fits were generally less than or equal to the number of fit data points, suggesting the data were well fit.
Slow (μs-ms) Timescale Dynamics Analysis
Chemical exchange is a prevalent phenomenon in NMR spectroscopy and provides valuable information on kinetic processes occurring on the μs – ms timescale. It results from alternating an NMR spin between different magnetic environments, or chemical states. Arginine kinase residues experiencing μs – ms chemical exchange were identified using the method described in Wang, 2003 (Wang et al., 2003) and using 15N R2 relaxation dispersion (Loria et al., 1999). For those residues with Rex, the former approach estimates the Rex value, while the latter depends upon analyzing the 15N R2 rate constant as a function of effective CPMG field strength, υ. Instead of recording a full relaxation decay curve at each CPMG field strength, a two-point 15N R2eff is determined using a common reference experiment (I0) and a single intensity value (Iυ): R2eff = (−1/τrelax) ln (Iυ/I0), where τrelax is the constant relaxation time period. In the absence of chemical exchange, R2eff will be independent of υ, while R2eff will vary with υ in the presence of appropriate timescale chemical exchange. A plot of R2eff versus υ generates a dispersion profile that can generally be fit to a simple, two-site exchange model when ΔR2eff = R2eff (υ→0) – R2eff (υ = 1 KHz) > 2 s−1. Values below this threshold could not be reliably identified or fit. Arginine kinase residues with relaxation dispersion of this criteria were fit with a numerical approximation of the Bloch-McConnell equation using software kindly provided by Drs. Lewis Kay and Dmitry Korzhnev (Cavanagh et al., 1996). In all cases, a two state exchange model was best-fit with the summed exchange rate constant kex = kAB + kBA, population of states A, pA, and B, pB, and their 15N chemical shift difference, Δω. No assumption was made regarding fast or slow exchange regime of kex relative to Δω.
Fast Timescale Model-Free Analyses
The Lipari-Szabo simple model-free formalism describes protein dynamics with two timescales: τm describes the rotational correlation time that reports on protein tumbling and τe describes relative rapid internal motions (Lipari and Szabo, 1982a, b). These motions are assumed to be decoupled from each other, and typically exhibit a ratio (τm/τe) greater than 10. Internal motions are described with the amplitude-dependent square of the generalized order parameter S2 and a ps-ns correlation time constant, τe. This simple model has been extended to describe situations where fast, S2f and τf, and slow, S2s and τs, internal motions are present (Clore et al., 1990). The generalized order parameter describing this situation is expressed as the product: S2 = S2f S2s. Model-free analysis can also include an exchange term, Rex, that adjusts the transverse relaxation rate constant to account for motions on the μs-ms timescale. The Lipari-Szabo spectral density functions are parameterized using a NH bond length of 1.04 Å and a 15N CSA of −170 ppm.
Selection of the appropriate rotational diffusion model for substrate-free arginine kinase was established by considering the prediction of hydrodynamic behavior of the molecule based on structural coordinate using HYDRONMR (Garcia de la Torre et al., 2000), and by conducting an analysis of the χ2 values derived from fits of the experimental NMR relaxation data using spectral density models for isotropic and axially-symmetric rotational diffusion. Both diffusion models were parameterized with an identical subset of residues that does not contain contributions from motion on multiple timescales. The residues excluded were those with {1H}15N NOE < 0.75, those showing evidence of conformational exchange from T1/T2 ratio analyses (Tjandra et al., 1995b), those with measured Rex, and those not in regular secondary structure elements. Using these criteria, 136 residues were retained for use in parameterization in the diffusion models. The isotropic diffusion model was determined from a least squares fit of the spectral density functions for T1/T2 ratios to the experimental data as described. This calculation was performed with a subroutine in the software relxn2.2 with the rvi interface (Boyer and Lee, 2008). The axially symmetric diffusion model was parameterized from the crystal structure of substrate free arginine kinase (PDB ID 1M80). This was accomplished by rotating the PDB coordinates to align the moments of inertia with the Cartesian axes and translated such that the enzyme center of mass was located at the origin. These manipulations were performed with PDBINTERTIA provided by Dr. Arthur G. Palmer (Columbia University). Direction cosines of amide bond vectors in the alignment frame were determined using software from Dr. Andrew L Lee (University of North Carolina).
The parameters S2, S2f, S2s, τe, τs and Rex were fit to five standard motional models: Model 1, S2 only; Model 2, S2 and τe; Model 3, S2 and Rex; Model 4, S2, τe and Rex; and Model 5, S2f, S2s and τs using the model-selection subroutine of relxn2.2 (Boyer and Lee, 2008). Model selection for each residue was guided by the χ2 statistic and the model with the lowest χ2 was usually selected (d’Auvergne and Gooley, 2003). For residues in which Rex was not observed in relaxation dispersion experiments, model 3 or 4 was only selected if the fit Rex was greater than the experimental error on R2, estimated as 1 s−1. Fits with χ2 values > 150 were not interpreted.
Supplementary Material
Acknowledgments
This research was supported by NIH (GM77643; M.S.C.), the NSF through the National High Magnetic Field Laboratory IHRP (5024-641-22 project 5045; J.J.S. & M.S.C.), and the American Heart Association (0415115B; O.D.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Azzi A, Clark SA, Ellington WR, Chapman MS. The role of phosphagen specificity loops in arginine kinase. Protein Sci. 2004;13:575–585. doi: 10.1110/ps.03428304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beach H, Cole R, Gill ML, Loria JP. Conservation of mus-ms enzyme motions in the apo- and substrate-mimicked state. J Am Chem Soc. 2005;127:9167–9176. doi: 10.1021/ja0514949. [DOI] [PubMed] [Google Scholar]
- Blethen SL. Kinetic Properties of the Arginine Kinase Isoenzymes of Limulus polyphemus. Archives of Biochemistry and Biophysics. 1972;149:244–251. doi: 10.1016/0003-9861(72)90319-0. [DOI] [PubMed] [Google Scholar]
- Boehr DD, Dyson HJ, Wright PE. An NMR perspective on enzyme dynamics. Chem Rev. 2006;106:3055–3079. doi: 10.1021/cr050312q. [DOI] [PubMed] [Google Scholar]
- Boehr DD, Dyson HJ, Wright PE. Conformational relaxation following hydride transfer plays a limiting role in dihydrofolate reductase catalysis. Biochemistry. 2008;47:9227–9233. doi: 10.1021/bi801102e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehr DD, Wright PE. Biochemistry. How do proteins interact? Science. 2008;320:1429–1430. doi: 10.1126/science.1158818. [DOI] [PubMed] [Google Scholar]
- Bosshard HR. Molecular recognition by induced fit: how fit is the concept? News Physiol Sci. 2001;16:171–173. doi: 10.1152/physiologyonline.2001.16.4.171. [DOI] [PubMed] [Google Scholar]
- Boyer JA, Lee AL. Monitoring aromatic picosecond to nanosecond dynamics in proteins via 13C relaxation: expanding perturbation mapping of the rigidifying core mutation, V54A, in eglin c. Biochemistry. 2008;47:4876–4886. doi: 10.1021/bi702330t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh J, Fairbrother WJ, Palmer AG, III, Skelton AJ. Protein NMR Spectroscopy. San Diego: Academic Press; 1996. [Google Scholar]
- Clarkson MW, Gilmore SA, Edgell MH, Lee AL. Dynamic coupling and allosteric behavior in a nonallosteric protein. Biochemistry. 2006;45:7693–7699. doi: 10.1021/bi060652l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clore GM, Szabo A, Bax A, Kay LE, Driscoll PC, Gronenborn AM. Deviations from the simple two-parameter model-free approach to the interpretation of Nitrogen-15 nuclear magnetic relaxation of proteins. J Am Chem Soc. 1990;112:4989–4991. [Google Scholar]
- d’Auvergne EJ, Gooley PR. The use of model selection in the model-free analysis of protein dynamics. J Biomol NMR. 2003;25:25–39. doi: 10.1023/a:1021902006114. [DOI] [PubMed] [Google Scholar]
- Davulcu O, Clark SA, Chapman MS, Skalicky JJ. Main chain (1)H, (13)C, and (15)N resonance assignments of the 42-kDa enzyme arginine kinase. J Biomol NMR. 2005;32:178. doi: 10.1007/s10858-005-6731-8. [DOI] [PubMed] [Google Scholar]
- Dumas C, Janin J. Conformational changes in arginine kinase upon ligand binding seen by small-angle X-ray scattering. FEBS Letters. 1983;153:128–130. [Google Scholar]
- Ellington WR. Evolution and Physiological Roles of Phosphagen Systems. Annual Review of Physiology. 2001;63:289–325. doi: 10.1146/annurev.physiol.63.1.289. [DOI] [PubMed] [Google Scholar]
- Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- Fritz-Wolf K, Schnyder T, Wallimann T, Kabsch W. Structure of Mitochondrial Creatine Kinase. Nature. 1996;381:341–345. doi: 10.1038/381341a0. [DOI] [PubMed] [Google Scholar]
- Furter R, Furter-Graves EM, Wallimann T. Creatine Kinase: The Reactive Cysteine Is Required for Synergism But Is Nonessential for Catalysis. Biochemistry. 1993;32:7022–7029. doi: 10.1021/bi00078a030. [DOI] [PubMed] [Google Scholar]
- Garcia de la Torre J, Huertas ML, Carrasco B. HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations. J Magn Reson. 2000;147:138–146. doi: 10.1006/jmre.2000.2170. [DOI] [PubMed] [Google Scholar]
- Gattis JL, Ruben E, Fenley MO, Ellington WR, Chapman MS. The active site cysteine of arginine kinase: structural and functional analysis of partially active mutants. Biochemistry. 2004;43:8680–8689. doi: 10.1021/bi049793i. [DOI] [PubMed] [Google Scholar]
- Gerstein M, Lesk AM, Chothia C. Structural mechanisms for domain movements in proteins. Biochemistry. 1994;33:6739–6749. doi: 10.1021/bi00188a001. [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG. SPARKY. Vol. 3. University of California; San Francisco: [Google Scholar]
- Hayward S. Structural principles governing domain motions in proteins. Proteins. 1999;36:425–435. [PubMed] [Google Scholar]
- Hayward S, Berendsen HJ. Systematic analysis of domain motions in proteins from conformational change: new results on citrate synthase and T4 lysozyme. Proteins. 1998;30:144–154. [PubMed] [Google Scholar]
- Jencks WP. Catalysis in Chemistry and Enzymology. New York: McGraw-Hill; 1969. [Google Scholar]
- Jencks WP. Binding energy, specificity, and enzymic catalysis: the circe effect. Adv Enzymol Relat Areas Mol Biol. 1975;43:219–410. doi: 10.1002/9780470122884.ch4. [DOI] [PubMed] [Google Scholar]
- Jencks WP, Mage MI. “Orbital Steering”, Entropy and Rate Accelerations. Biochemical and Biophysical Research Communications. 1974;57:887–892. doi: 10.1016/0006-291x(74)90629-9. [DOI] [PubMed] [Google Scholar]
- Korzhnev DM, Skrynnikov NR, Millet O, Torchia DA, Kay LE. An NMR experiment for the accurate measurement of heteronuclear spin-lock relaxation rates. J Am Chem Soc. 2002;124:10743–10753. doi: 10.1021/ja0204776. [DOI] [PubMed] [Google Scholar]
- Koshland DE., Jr Application of a theory of enzyme specificity to protein synthesis. Proceedings of the National Academy of Sciences, USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshland DE., Jr The key-lock theory and the induced-fit theory. Angew Chem Int Ed Engl. 1994;33:2375–2378. [Google Scholar]
- Kovrigin EL, Loria JP. Enzyme dynamics along the reaction coordinate: critical role of a conserved residue. Biochemistry. 2006;45:2636–2647. doi: 10.1021/bi0525066. [DOI] [PubMed] [Google Scholar]
- Lahiri SD, Wang PF, Babbitt PC, McLeish MJ, Kenyon GL, Allen KN. The 2.1 A Structure of Torpedo californica Creatine Kinase Complexed with the ADP-Mg2+-NO3−-Creatine Transition-State Analogue Complex. Biochemistry. 2002;41:13861–13867. doi: 10.1021/bi026655p. [DOI] [PubMed] [Google Scholar]
- Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KF, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- Lee RA, Razaz M, Hayward S. The DynDom database of protein domain motions. Bioinformatics. 2003;19:1290–1291. doi: 10.1093/bioinformatics/btg137. [DOI] [PubMed] [Google Scholar]
- Lipari G, Szabo A. Model-Free approach to the interpretation of Nuclear Magnetic Resonance Relaxation in Macromolecules. 1. Theory and Range of Validity. J Am Chem Soc. 1982a;104:4546–4559. [Google Scholar]
- Lipari G, Szabo A. Model-Free Approach to the Interpretation of Nuclear Magnetic Resonance Relaxation in Macromolecules. 2. Analysis of Experimental Results. J Am Chem Soc. 1982b;104:1559–4570. [Google Scholar]
- Loria JP, Rance M, Palmer AG., 3rd A TROSY CPMG sequence for characterizing chemical exchange in large proteins. J Biomol NMR. 1999;15:151–155. doi: 10.1023/a:1008355631073. [DOI] [PubMed] [Google Scholar]
- Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- Mandel AM, Akke M, Palmer AG., 3rd Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- Markley JL, Bax A, Arata Y, Hilbers CW, Kaptein R, Sykes BD, Wright PE, Wuthrich K. Recommendations for the presentation of NMR structures of proteins and nucleic acids. IUPAC-IUBMB-IUPAB Inter-Union Task Group on the Standardization of Data Bases of Protein and Nucleic Acid Structures Determined by NMR Spectroscopy. J Biomol NMR. 1998;12:1–23. doi: 10.1023/a:1008290618449. [DOI] [PubMed] [Google Scholar]
- Mazon H, Marcillat O, Forest E, Smith DL, Vial C. Conformational dynamics of the GdmHCl-induced molten globule state of creatine kinase monitored by hydrogen exchange and mass spectrometry. Biochemistry. 2004;43:5045–5054. doi: 10.1021/bi049965b. [DOI] [PubMed] [Google Scholar]
- Mazon H, Marcillat O, Forest E, Vial C. Local dynamics measured by hydrogen/deuterium exchange and mass spectrometry of creatine kinase digested by two proteases. Biochimie. 2005;87:1101–1110. doi: 10.1016/j.biochi.2005.05.012. [DOI] [PubMed] [Google Scholar]
- Ohren JF, Kundracik ML, Borders CL, Jr, Edmiston P, Viola RE. Structural asymmetry and intersubunit communication in muscle creatine kinase. Acta Crystallogr D Biol Crystallogr. 2007;63:381–389. doi: 10.1107/S0907444906056204. [DOI] [PubMed] [Google Scholar]
- Osborne MJ, Schnell J, Benkovic SJ, Dyson HJ, Wright PE. Backbone dynamics in dihydrofolate reductase complexes: role of loop flexibility in the catalytic mechanism. Biochemistry. 2001;40:9846–9859. doi: 10.1021/bi010621k. [DOI] [PubMed] [Google Scholar]
- Palmer AG, 3rd, Kroenke CD, Loria JP. Nuclear magnetic resonance methods for quantifying microsecond-to-millisecond motions in biological macromolecules. Methods Enzymol. 2001;339:204–238. doi: 10.1016/s0076-6879(01)39315-1. [DOI] [PubMed] [Google Scholar]
- Pruett PS, Azzi A, Clark SA, Yousef M, Gattis JL, Somasundaram T, Ellington WR, Chapman MS. The putative catalytic bases have, at most, an accessory role in the mechanism of arginine kinase. J Biol Chem. 2003;29:26952–26957. doi: 10.1074/jbc.M212931200. [DOI] [PubMed] [Google Scholar]
- Raiford DS, Fisk CL, Becker ED. Calibration of methanol and ethylene glycol nuclear magnetic resonance thermometers. Analytical Chemistry. 1979;51:2050–2051. [Google Scholar]
- Rozovsky S, Jogl G, Tong L, McDermott AE. Solution-state nmr investigations of triosephosphate isomerase active site loop motion: ligand release in relation to active site loop dynamics. J Mol Biol. 2001;310:271–280. doi: 10.1006/jmbi.2001.4673. [DOI] [PubMed] [Google Scholar]
- Sullivan SM, Holyoak T. Enzymes with lid-gated active sites must operate by an induced fit mechanism instead of conformational selection. Proc Natl Acad Sci U S A. 2008;105:13829–13834. doi: 10.1073/pnas.0805364105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Kawasaki Y, Furukohri T, Ellington WR. Evolution of phosphagen kinase. VI. Isolation, characterization and cDNA-derived amino acid sequence of lombricine kinase from the earthworm Eisenia foetida, and identification of a possible candidate for the guanidine substrate recognition site. Biochim Biophys Acta. 1997;1343:152–159. doi: 10.1016/s0167-4838(97)00128-3. [DOI] [PubMed] [Google Scholar]
- Tjandra N, Feller SE, Pastor RW, Bax A. Rotational diffusion anisotropy of human ubiquitin from 15N NMR relaxation. Journal of the American Chemical Society. 1995a;117:12562–12566. [Google Scholar]
- Tjandra N, Kuboniwa H, Ren H, Bax A. Rotational dynamics of calcium-free calmodulin studied by 15N-NMR relaxation measurements. Eur J Biochem. 1995b;230:1014–1024. doi: 10.1111/j.1432-1033.1995.tb20650.x. [DOI] [PubMed] [Google Scholar]
- Vugmeyster L, Raleigh DP, Palmer AG, 3rd, Vugmeister BE. Beyond the decoupling approximation in the model free approach for the interpretation of NMR relaxation of macromolecules in solution. J Am Chem Soc. 2003;125:8400–8404. doi: 10.1021/ja029480f. [DOI] [PubMed] [Google Scholar]
- Wang C, Karpowich N, Hunt JF, Rance M, Palmer AG. Dynamics of ATP-binding cassette contribute to allosteric control, nucleotide binding and energy transduction in ABC transporters. J Mol Biol. 2004;342:525–537. doi: 10.1016/j.jmb.2004.07.001. [DOI] [PubMed] [Google Scholar]
- Wang C, Rance M, Palmer AG., 3rd Mapping chemical exchange in proteins with MW > 50 kD. J Am Chem Soc. 2003;125:8968–8969. doi: 10.1021/ja035139z. [DOI] [PubMed] [Google Scholar]
- Watt ED, Shimada H, Kovrigin EL, Loria JP. The mechanism of rate-limiting motions in enzyme function. Proc Natl Acad Sci U S A. 2007;104:11981–11986. doi: 10.1073/pnas.0702551104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf-Watz M, Thai V, Henzler-Wildman K, Hadjipavlou G, Eisenmesser EZ, Kern D. Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat Struct Mol Biol. 2004;11:945–949. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- Yousef MS, Clark SA, Pruett PK, Somasundaram T, Ellington WR, Chapman MS. Induced fit in guanidino kinases-comparison of substrate-free and transition state analog structures of arginine kinase. Protein Sci. 2003;12:103–111. doi: 10.1110/ps.0226303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousef MS, Fabiola F, Gattis J, Somasundaram T, Chapman MS. Refinement of Arginine Kinase Transition State Analogue Complex at 1.2 Å resolution; mechanistic insights. Acta Crystallographica Section D: Biological Crystallography. 2002;58:2009–2017. doi: 10.1107/s0907444902014683. [DOI] [PubMed] [Google Scholar]
- Zhou G, Somasundaram T, Blanc E, Parthasarathy G, Ellington WR, Chapman MS. Transition state structure of arginine kinase: Implications for catalysis of bimolecular reactions. Proceedings of the National Academy of Sciences, USA. 1998;95:8449–8454. doi: 10.1073/pnas.95.15.8449. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.