Abstract
During their intraerythrocytic development, malaria parasites export hundreds of proteins to remodel their host cell. Nutrient acquisition, cytoadherence and antigenic variation are among the key virulence functions effected by this erythrocyte takeover. Proteins destined for export are synthesized in the endoplasmic reticulum (ER) and cleaved at a conserved (PEXEL) motif, which allows translocation into the host cell via an ATP-driven translocon called the PTEX complex. We report that plasmepsin V, an ER aspartic protease with distant homology to the mammalian processing enzyme BACE, recognizes the PEXEL motif and cleaves it at the correct site. This enzyme is essential for parasite viability and ER residence is essential for its function. We propose that plasmepsin V is the PEXEL protease and is an attractive enzyme for antimalarial drug development.
The human malaria parasite Plasmodium falciparum exports an estimated 200–300 proteins into the host erythrocyte1,2. In doing so, the parasite remodels the cytoskeleton and plasma membrane to create cytoadherence knobs, nutrient permeation pathways and altered erythrocyte mechanical stability3,4. Export of these effectors is dependent on a Plasmodium export element or PEXEL sequence, RxLxE/Q/D5,6. Proteins destined for export are cleaved after the conserved PEXEL leucine in the ER and mutation of the R or L residues attenuates cleavage and export7,8. Plasmepsin V (PM V) is an aspartic protease that has distant homology to mammalian BACE or beta-secretase9, an enzyme involved in the processing of amyloid precursor protein10. Both have a C-terminal extension that contains a hydrophobic membrane anchor sequence. An N-terminal aspartic protease “pro-domain” remains unprocessed in PM V9. PM V is expressed in intraerythrocytic P. falciparum parasites and has orthologs in other Plasmodium species. Phytophthoria infestans, the potato blight pathogen that has a similar export system11, has a homologous sequence in the database (PITG_02623.1). PM V has been localized to the ER9 and is therefore a candidate to be the PEXEL processing protease.
Role of the trans-membrane domain in PM V Localization
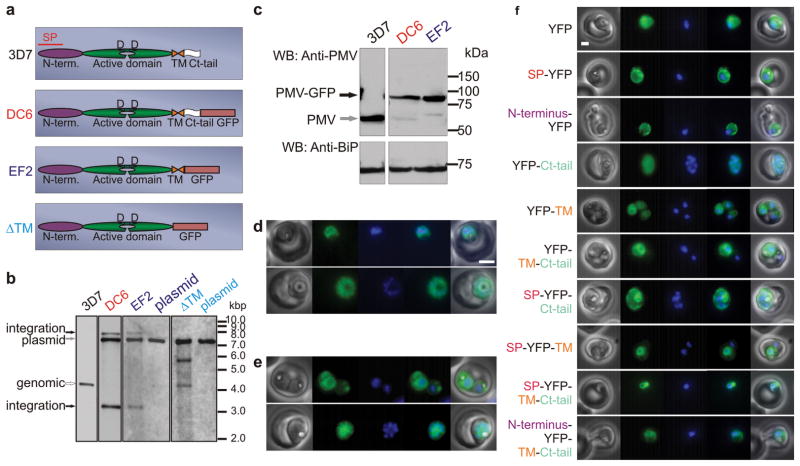
PM V lacks a classical ER retention signal. To identify the element responsible for localization and to assess the importance of ER residence for PM V function, we made sequential C-terminal truncation mutants (Fig. 1a). Single crossover homologous recombination into the endogenous locus was performed, introducing a GFP tag after a full-length C-terminus or in place of C-terminal sequence (Fig. 1b). Full-length PM V-GFP integrants (clone DC6) and integrants with deletion of the C-terminus downstream of the membrane-spanning segment (clone EF2) had no phenotype and retained ER targeting (Fig. 1c-e and Supplementary Fig. 2), whereas deletions involving the membrane anchor were lethal (Fig. 1b). Fusion of the TM region but not other portions of PM V was sufficient to target a reporter to the ER (Fig. 1f). Thus the TM sequence is important for ER localization and probably for cellular activity on its substrates as well.
Figure 1.
The transmembrane region of plasmepsin V confers ER localization. a) Schematic of PM V and C-terminal integrants. SP: signal peptide; N-term.: N-terminal domain; Ct: C-terminal; TM: transmembrane region. b) Southern blots of full-length (DC6) or C-terminal tail deletion (EF2) integrant clones and transmembrane region deletion transfectants (ΔTM). Parental strain (3D7) and plasmids are shown for reference. c) Western blot using anti-PM V antibody. BiP served as loading control. d and e) Live fluorescence images of DC6 and EF2, respectively. Left to right: phase, GFP, DAPI, fluorescence merge and total merge. Bar, 2 μm. f) Localization of PM V fragments (defined in a) fused to yellow fluorescent protein (YFP). Expression was episomal, using the HSP86 promoter. Panel order as in d and e.
PM V Essentiality
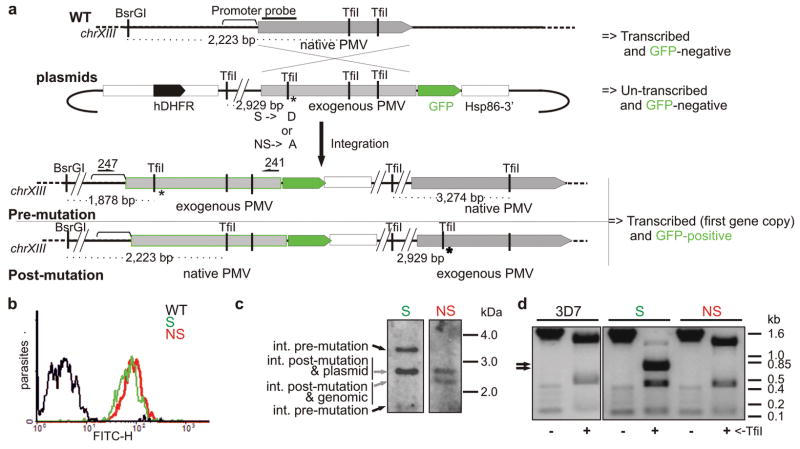
We further assessed essentiality of the PM V gene for intraerythrocytic parasites by using an allelic replacement approach12. An integration vector in which the first catalytic aspartate was modified by synonymous or non-synonymous mutation was transfected into parasites (Fig. 2a) and recombinants were obtained (Fig. 2b, c). Crossover into the endogenous PM V gene proved possible only when the aspartate codon was preserved (replacement via the synonymous mutation vector or downstream crossover bypassing the mutation with the non-synonymous mutation vector) (Fig. 2d). Non-synonymous alteration of the active site codon could not be achieved in four separate transfection experiments. These results support the notion that PM V has an essential function in the cell.
Figure 2.
Plasmepsin V is essential for intraerythrocytic parasite viability. a) Active site allelic replacement scheme. Integration vectors possessing a synonymous (S) or non-synonymous (NS) mutation in the Asp 108 codon and a new TfiI restriction site just upstream were transfected into parental strain 3D7. Possible outcomes for upstream crossover (pre-mutation) and downstream crossover (post-mutation) are drawn. Integrants were selected and assessed by PCR. Primers used for amplification (241 and 247) are marked. b) Flow cytometry of parasites. Parasites were assessed for GFP expression, an indication of integration at the endogenous locus. Parental strain 3D7 (WT) is shown as control. c) Southern blot of transfected parasite pools. At left are expected positions of possible products of the BsrGI/TfiI digest mapped in c. d) Screening of integrants by PCR and restriction digest. Arrows indicate a doublet (predicted 787 and 748 bp) resulting from restriction fragmentation of product from recombinants that have crossed over upstream of the active site codon.
Dominant-negative PM V Phenotype
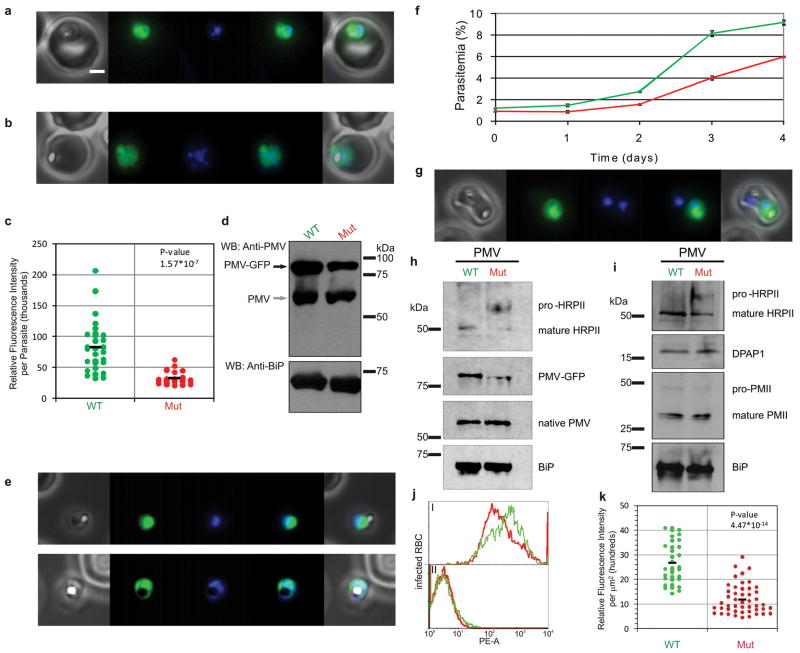
To investigate PM V function, we episomally expressed two GFP-tagged versions of PM V, one wild type and the other containing a D to A mutation in the active site aspartate 108 to render expressed protein catalytically dead. Both versions localized to the ER but the mutant had 3-fold reduced signal by immunofluorescence (Fig. 3a–c) and by western blot (Fig. 3d). Mutant enzyme-expressing parasites were frequently seen encased in erythrocyte ghosts (Fig. 3e), suggesting impaired host cell homeostasis. Indeed the mutant PM V-expressing culture grew more slowly than the wild-type PM V-expressing culture (Fig. 3f). Occasional cells expressing the mutant construct had intense signal similar to wild type; in such cases two parasites could be seen in a single erythrocyte, one of which was not fluorescent and had presumably lost or down-regulated the plasmid (Fig. 3g). Our interpretation of this result is that the plasmid-free parasite can export proteins normally into the shared red blood cell, overcoming the effect that the mutant PM V has on the neighboring parasite. To explore this further, we assessed processing of the exported histidine-rich protein II (HRPII) in the PM V-transfected cells (Fig. 3h, i). Unprocessed HRPII was barely detectable in wild-type PM V-expressing parasites. In contrast, unprocessed HRPII accumulated in mutated PM V-expressing parasites. Plasmepsin II and DPAP1, non-PEXEL containing proteins that use the secretory pathway but are then internalized instead of exported13,14, were processed normally (Fig. 3i). Export was assessed by immunofluorescence (Fig. 3j, k). The levels of host erythrocyte HRPII and another exported protein, RESA (ring-infected erythrocyte surface antigen), were diminished in the mutated PM V-expressing parasitized erythrocytes by 30–50%. These data suggest that episomal expression of catalytically dead PM V has a dominant-negative effect on parasite growth and on protein export. A survey of PEXEL gene essentiality estimated that about one-fourth are required for intraerythrocytic parasite growth15. Thus, perhaps 50–75 exported proteins are essential. Some will be required at near wild-type levels for optimal growth, while others will tolerate more drastic reduction without consequence. The 30–50% reduction in protein export is therefore about what would be expected, given the growth phenotype seen (Fig. 3f).
Figure 3.
Episomal expression of wild-type and catalytic mutant PM V. a,b) Immunofluorescence of wild-type and 108 D to A (mutant) PM V expressing lines, respectively. Left to right: phase, GFP, DAPI, merge. Bar, 2 μm. c) Fluorescence intensity of 52 transfected early trophozoites was measured. Mean relative fluorescence units were 82,559 and 32,059 for wild-type and mutant-transfected parasites, respectively. d) Western blot. BiP serves as a loading control. e) Mutant PM V-expressing parasites encased in erythrocyte ghosts. f) Growth curves. Asynchronous cultures episomally expressing wild-type (green) or mutant (red) PM V were monitored by flow cytometry for four days in triplicate (error bars indicate standard deviation). Mutant growth rate is reduced by 49.2+/−1.6 %. g) Double-infected erythrocyte containing a parasite expressing high levels of mutant PM V. h and i) Western blots of HRPII processing in parasites episomally expressing mutant or wild-type PM V. h: top - anti-HRPII; middle panels - anti-PM V; bottom - anti-BiP. Contrast and brightness enhanced slightly over the entire panel to bring out features seen on the original film. i: top - anti-HRPII; middle panels - antibodies against non-exported secretory proteins dipeptidyl peptidase I (DPAP1) and plasmepsin II (PMII); bottom - anti-BiP. j) Flow cytometry. Wild-type (green) and mutant (red) PM V-expressing parasites were fixed, treated with tetanolysin (5 min, 20 units/ml) to selectively permeabilize the erythrocyte compartment and exported RESA was quantified by flow cytometry with specific antibody. Control without primary antibody is shown below. Results are representative of 4 experiments. k) HRP II staining was inconsistent by the fixation procedure used in j, so HRPII erythrocyte fluorescence intensity (80 cells) was quantified. Mean relative fluorescence intensity per μm2 was 2671 and 1179 for wild-type and mutant, respectively.
Enzyme Activity and Specificity
To assess enzyme activity, PM V-GFP was detergent-solubilized from recombinant clone DC6 (see Fig. 1a, c) and enzyme was isolated using anti-GFP antibody. The enzyme was able to cleave a fluorogenic decapeptide based on the PEXEL motif from the exported HRPII (Fig. 4a, b). Pull-downs from the parental strain (3D7) with untagged PM V had no activity. When anti-PM V antibody was used for enzyme isolation, both tagged (DC6) and untagged (3D7) enzyme could be isolated and were active. A second PEXEL peptide based on PfEMP2 was also cleaved by isolated enzyme (Fig. 4b). Mutation of P1 Leu or P3 Arg abolishes export of PEXEL proteins10. Peptides with either of these residues changed to Ala were refractory to cleavage (Fig. 4b), confirming specificity of the enzyme for the PEXEL motif. Activity was seen between pH 5 and 7 (Fig. 4c). The pH of the mammalian ER has been measured to be 7.116. While we do not know the pH of the Plasmodium ER and cannot reproduce the cellular ionic environment in our in vitro assays, the measured activity is consistent with an ER function. Activity of the PM V active site mutant enzyme was undetectable (Fig. 4d). This result shows that PM V itself is the active protease, not an associated protein. Boddey and co-workers (Nature, this issue) have obtained active recombinant enzyme from E. coli, another indication that PM V is the protease in question. PM V but not control preparations, cleaved a full-length PEXEL-containing proprotein (Fig. 4e).
Figure 4.
Plasmepsin V activity. a) Substrate cleavage. PM V was isolated from parental (3D7) and PM V-GFP fusion (DC6) clones using anti-GFP (yellow and red, respectively) or anti-PM V (blue and green, respectively). A similar isolation without Ab (protein A) served as a control for non-specific binding (purple and cyan, respectively). Purified enzyme was incubated at pH 6.5 for 35 min with fluorogenic peptide (Anaspec) corresponding to the PEXEL motif for HRPII (DABCYL-LNKRLLHETQ-EDANS). Error bars indicate standard deviation of three activity curves. b) PM V specificity for PEXEL motif. Processing of fluorogenic peptides containing the PEXEL motifs of HRPII (blue) or PfEMP2 (red, DABCYL-RYVRILSETE-EDANS) are shown. Left - wild-type peptide (WT); middle - L to A mutant peptide; right - R to A mutant peptide. Inset: peptide sequences with mutated residues in black. Error bars as in a. c) pH dependence in Tris-Malate buffer. Two separate experiments are shown in different colors. Error bars as in a. d) Activity of active-site mutant PM V compared to wild type. PMV-GFP was isolated from 3D7 (untransfected mock isolation), wild-type- or mutant-PM V-GFP-transfected parasites using anti-GFP for purification. Activity on HRPII-PEXEL peptide is normalized to PM V protein content. Error bars as in a. e) Cleavage of pro-HRPII by isolated enzyme from d. p: pro-form; m: mature protein.
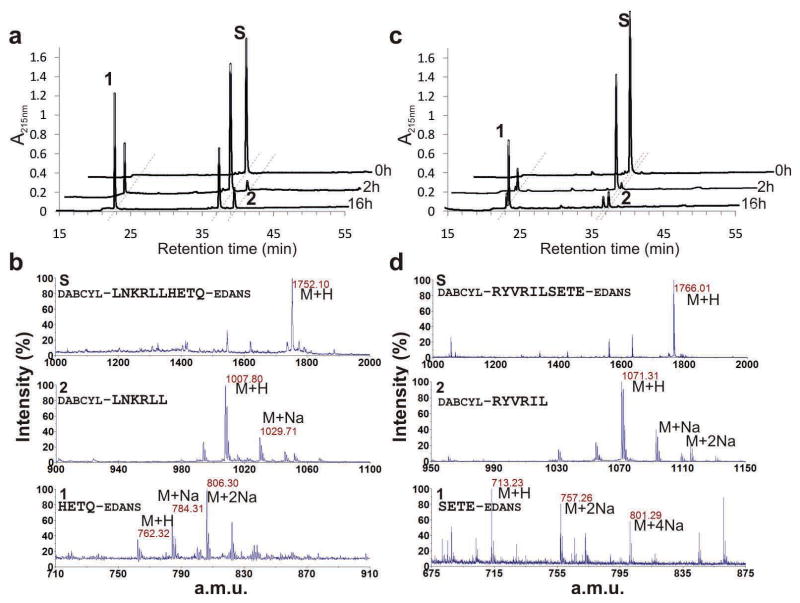
To confirm cleavage specificity, the products of the HRPII PEXEL peptide incubation with isolated WT PM V enzyme were fractionated by reverse phase HPLC (Fig. 5a) and analyzed by mass spectrometry (Fig. 5b). The fragments generated corresponded to proteolysis after the leucine that is the in vivo processing site. Similar results were obtained using the PfEMP2 peptide (Fig. 5c, d).
Figure 5.
Analysis of substrate cleavage. PM V was incubated with HRPII (a) or PfEMP2 (c) PEXEL peptides (DABCYL-LNKRLLHETQ-EDANS and DABCYL-RYVRILSETE-EDANS, respectively). Cleavage products were separated on a C18 column by reverse-phase HPLC. Back to front: incubation for 0, 2 and 16h. S, substrate peak. b,d) isolated products and substrates from a and c were analyzed by MALDI-TOF mass spectrometry. Ion peaks and sodium adducts are labeled. HRPII peptide product masses: calculated 762.51 and 1008.25; detected 762.32 and 1007.80. PfEMP2 peptide product masses: calculated 1071.30 and 713.72; detected 1071.31 and 713.23. Ions corresponding to alternative peptide bond cleavage were not detected.
PM V Interactions
We have shown that PM V is an essential ER protease in Plasmodium falciparum. Residence in the ER is necessary for its function, since deletion of the C-terminal tail had no effect on location or parasite viability while deletion of the transmembrane (TM) region rendered parasites non-viable (Fig. 1). Our data suggest that PM V is the enzyme that processes PEXEL-containing proteins to send them on their way for export into the host cell. It is not clear how PEXEL-containing proteins are recognized by the translocon in the parasitophorous vacuole17 when most of the PEXEL has been cleaved off by PM V in the ER. It is conceivable that the propeptide stays associated with the mature polypeptide during transport, but we favor a model (Supplementary Fig. 1) in which chaparones associate with PM V in the ER. Upon PEXEL cleavage, these chaparones receive the protein destined for export, usher it through the secretory pathway and then thread it through the translocon channel. In support of this, PM V pull-downs (Fig. 6) consistently identified an ER-resident HSP70 and HSP101 (a key translocon component17) as associated proteins. Much remains to be done to define the PM V-chaperone relationship but it is certainly plausible that chaperones could act in a complex or relay to shepherd proteins from ER to translocon for export.
Figure 6.
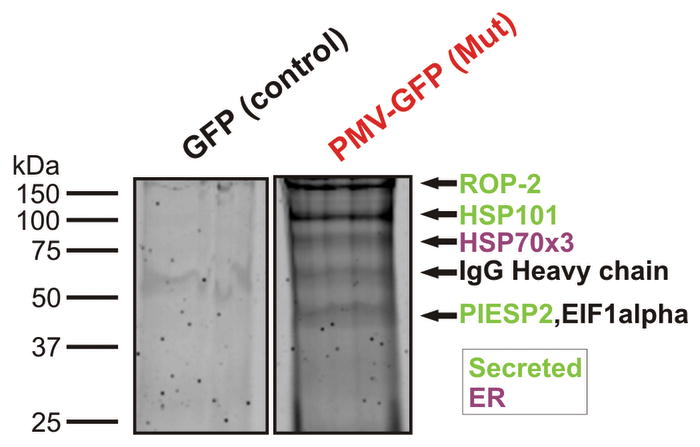
Proteomic analysis of PM V-associated proteins. Shown is a Coomassie-stained gel of an anti-GFP pulldown for the episomal GFP-expressing control strain (GFP) and the mutant PM V-GFP-expressing strain (Mut). Bands were excised, trypsinized and analyzed by MS-MS. The same analysis was also performed on the wild-type PM V-GFP strain, and on the parental 3D7 strain but using anti-PM V antibody. Shown are proteins for which at least four peptides were identified from the anti-PM V pulldown and three or more peptides from at least one of the two anti-GFP pulldowns of episomal PM V-expressing parasites. All proteins identified are tabulated in Supplementary Table 1. None of the proteins were detected in the GFP control pulldown.
Enzyme Inhibition
In vitro enzyme activity was partially inhibited by high micromolar concentrations of HIV protease inhibitors or pepstatin A (Supplementary Fig. 3a) but not by other classes of inhibitors. We tested a panel of protease inhibitors for ability to block processing of the PEXEL-containing exported protein HRPII but have not yet found a good inhibitor. BACE inhibitors had minimal effect, perhaps not surprising given the evolutionary distance between the two orthologs. Only HIV protease inhibitors had any effect and the blockade was partial (Supplementary Fig. 3b, c). Action on PM V is unlikely to be their primary effect since they kill cultured parasites in the single digit micromolar range18,19, while effects on protein export and on isolated PM V were observed at 50–200 micromolar concentrations.
We propose that plasmepsin V is the PEXEL protease. This enzyme recognizes a simple RxL motif on secretory proteins destined for export into the host erythrocyte. Since PM V cleaves the PEXEL sequence away from the mature protein, the simplest conclusion is that PM V is primarily responsible for the specificity of export. An xE/Q/D dipeptide at the N-terminus of mature exported proteins is also important for export though not for the cleavage itself8. Perhaps this polar residue comprises a secondary recognition element that interacts with the chaperone that will bring the protein to the translocon for export. It is very likely that the physical association of an escort system with PM V is needed to transfer the license for export. PM V appears then to be the gatekeeper for protein export. If potent inhibitors can be found, blocking the entire parasite virulence and intracellular survival program with one stroke will be a promising new strategy for combating this nefarious organism.
Methods
Techniques for parasite culture, 3′ end integrations and truncations and their analysis, allelic replacement, site-directed mutagenesis, fluorescence imaging, parasite extraction and western blotting, as well as flow cytometry growth monitoring have been previously described12,20. Parasite fluorescence intensity was measured blinded on random fields using Velocity 4 software (Improvision, Lexington, MA). For enzyme isolation, 50 ml of parasite culture at 2% hematocrit, 10% parasitemia was harvested and parasites freed by saponin treatment as described12. Cells were solubilized for 30 min in 0.5% Triton X-100 in PBS buffer and incubated with anti-GFP (3E6 -Invitrogen) or anti-PM V9 antibodies for 1 h at 4 C. Immune complexes were collected using protein A-Sepharose, washed extensively with PBS and isolated enzyme released in the activity buffer (see below) with 2 mM DTT. PM V enzyme activity assays were performed by incubation at 37° C in 50 mM Tris-Malate, pH 6.5, 50 mM NaCl, 0.05 % Triton X-100 and reaction progress monitored on a Biorad fluorimeter. Activity against pro-HRPII21 was assessed after 16h incubation by SDS-PAGE/anti-HRPII western blot. C18 reverse-phase liquid chromatography/MALDI mass spectrometry was performed as reported22. For pull-downs, enzyme was isolated as above except that solubilization was performed in RIPA buffer20. Immunoprecipitates were fractionated by SDS-PAGE and gel slices were analyzed by MS-MS after trypsinization23. Primers for allelic replacement diagnosis were 241: AATTCCTAGGAGAAACTTTTAAGAAGATATTTTCTTTTTCTCTATATTC and 247: AATTCCTAGGAGAAACTTTTAAGAAGATATTTTCTTTTTCTCTATATTC. PM V sequences for 3′ fusion constructs were, full-length: nt 280 to 1770; tail deletion: nt 280 to 1721; transmembrane region deletion: nt 280 to 1623. Mutagenesis oligos for the allelic replacements:
S, CGCAAAGAATTTCTTTGATTCTAGACACAGGTTCATCTTCGTTAAGTTTCCCGTG;
NS, CGCAAAGAATTTCTTTGATTCTAGCGACAGGTTCATCTTCGTTAAGTTTCCCGTG.
Supplementary Material
Acknowledgments
Supported by NIH grant AI-047798. We thank A. Cowman, B. Crabb and S. Lundquist for helpful suggestions, J. Adams and ATCC (MR4) for BiP antibody, D. Taylor for HRPII antibody, R. Anders for RESA antibody, A. Miller and M. Ndonwi for HRPII protein, P. Hruz for HIV protease inhibitors, J. Tang and S. Romeo for BACE inhibitors, W. Beatty for fluorescence analysis, S. Beverley for fluorimeter access, B. Vaupel for technical assistance and J. Turk for mass spectrometer access. Proteomics analysis was carried out at the ‘Fingerprints’ Proteomics Facility, College of Life Sciences, University of Dundee.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature. A figure summarising the main result of this paper is also included as SI.
Competing financial interests: none.
Reprints and permissions information is available at npg.nature.com/reprints and permissions.
Author Contributions: I.R. designed and executed most of the experiments and wrote the paper; S.B., V.M., T.B. and A.O. designed and executed experiments; D.G. designed experiments and wrote the paper.
References
- 1.Sargeant TJ, et al. Lineage-specific expansion of proteins exported to erythrocytes in malaria parasites. Genome Biol. 2006;7:R12. doi: 10.1186/gb-2006-7-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Ooij C, et al. The malaria secretome: from algorithms to essential function in blood stage infection. PLoS Pathogens. 2008;4:e1000084. doi: 10.1371/journal.ppat.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Haldar K, Mohandas N. Erythrocyte remodeling by malaria parasites. Current Opinion Hematol. 2007;14:203–209. doi: 10.1097/MOH.0b013e3280f31b2d. [DOI] [PubMed] [Google Scholar]
- 4.Maier AG, Cooke BM, Cowman AF, Tilley L. Malaria parasite proteins that remodel the host erythrocyte. Nature Reviews. 2009;7:341–354. doi: 10.1038/nrmicro2110. [DOI] [PubMed] [Google Scholar]
- 5.Hiller NL, et al. A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science. 2004;306:1934–1937. doi: 10.1126/science.1102737. [DOI] [PubMed] [Google Scholar]
- 6.Marti M, et al. Targeting malaria virulence and remodeling proteins to the host erythrocyte. Science. 2004;306:1930–1933. doi: 10.1126/science.1102452. [DOI] [PubMed] [Google Scholar]
- 7.Chang HH, et al. N-terminal processing of proteins exported by malaria parasites. Mol Biochem Parasitol. 2008;160:107–115. doi: 10.1016/j.molbiopara.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Boddey JA, Moritz RL, Simpson RJ, Cowman AF. Role of the Plasmodium export element in trafficking parasite proteins to the infected erythrocyte. Traffic. 2009;10:285–289. doi: 10.1111/j.1600-0854.2008.00864.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klemba M, Goldberg DE. Characterization of plasmepsin V, a membrane-bound aspartic protease homolog in the endoplasmic reticulum of Plasmodium falciparum. Mol Biochem Parasitol. 2005;143:183–191. doi: 10.1016/j.molbiopara.2005.05.015. [DOI] [PubMed] [Google Scholar]
- 10.Tang J. Memapsin 2. In: Barrett AJ, Rawlings ND, Woessner JF, editors. Handbook of Proteolytic Enzymes. Elsevier; 2004. [Google Scholar]
- 11.Bhattacharjee S, et al. The malarial host-targeting signal is conserved in the Irish potato famine pathogen. PLoS Pathogens. 2006;2:e50. doi: 10.1371/journal.ppat.0020050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Russo I, Oksman A, Vaupel B, Goldberg DE. A calpain unique to alveolates is essential in Plasmodium falciparum and its knockdown reveals an involvement in pre-S-phase development. Proc Natl Acad Sci USA. 2009;106:1554–1559. doi: 10.1073/pnas.0806926106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klemba M, Gluzman I, Goldberg DE. A Plasmodium falciparum dipeptidyl aminopeptidase I participates in vacuolar hemoglobin degradation. J Biol Chem. 2004;279:43000–43007. doi: 10.1074/jbc.M408123200. [DOI] [PubMed] [Google Scholar]
- 14.Klemba M, Beatty W, Gluzman I, Goldberg DE. Trafficking of plasmepsin II to the food vacuole of the malaria parasite Plasmodium falciparum. J Cell Biol. 2003;164:47–56. doi: 10.1083/jcb200307147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maier AG, et al. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell. 2008;134:48–61. doi: 10.1016/j.cell.2008.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim JH, et al. Noninvasive measurement of the pH of the endoplasmic reticulum at rest and during calcium release. Proc Natl Acad Sci USA. 1998;95:2997–3002. doi: 10.1073/pnas.95.6.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Koning-Ward TF, et al. A newly discovered protein export machine in malaria parasites. Nature. 2009;459:945–950. doi: 10.1038/nature08104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Skinner-Adams TS, et al. Antiretrovirals as antimalarial agents. J Infect Dis. 2004;190:1998–2000. doi: 10.1086/425584. [DOI] [PubMed] [Google Scholar]
- 19.Parikh S, et al. Antimalarial activity of human immunodeficiency virus type 1 protease inhibitors. Antimicrob Agents Chemother. 2005;49:2983–2985. doi: 10.1128/AAC.49.7.2983-2985.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Russo I, Oksman A, Goldberg DE. Fatty acid acylation regulates trafficking of the unusual Plasmodium falciparum calpain to the nucleolus. Mol Microbiol. 2009;72:229–245. doi: 10.1111/j.1365-2958.2009.06639.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sullivan DJ, Gluzman IY, Goldberg DE. Plasmodium hemozoin formation mediated by histidine-rich proteins. Science. 1996;271:219–222. doi: 10.1126/science.271.5246.219. [DOI] [PubMed] [Google Scholar]
- 22.Drew ME, et al. Plasmodium food vacuole plasmepsins are activated by falcipains. J Biol Chem. 2008;283:12870–12876. doi: 10.1074/jbc.M708949200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–8. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.