SUMMARY
Interactions among Bcl-2 family proteins mediated by Bcl-2 homology (BH) regions transform apoptosis signals into actions. The interactions between BH3 region-only proteins and multi-BH region proteins such as Bax and Bcl-2 have been proposed to be the dominant interactions required for initiating apoptosis. Experimental evidence also suggests that both homo- and hetero-interactions are mediated primarily by the BH3 regions in all Bcl-2 family proteins and contribute to commitment to or inhibition of apoptosis. We found that a peptide containing the BH3 helix of Bax was not sufficient to activate recombinant Bax to permeabilize mitochondria. However, an extended peptide containing the BH3 helix and additional downstream sequences activated Bax to permeabilize mitochondria and liposomes. Bcl-2 inhibited the membrane permeabilizing activity of peptide activated Bax. This activity of Bcl-2 was inhibited by the extended but not the BH3-only peptide despite both peptides binding to Bcl-2 with similar affinity. Further, membrane-bound Bax activation intermediates directly activated soluble Bax further permeabilizing the membrane. Bcl-2 inhibited Bax auto-activation. We therefore propose that Bax auto-activation amplifies the initial death signal produced by BH3-only proteins and that Bcl-2 functions as an inhibitor of Bax auto-activation.
INTRODUCTION
The dominant form of programmed cell death, apoptosis, plays an essential role in embryogenesis and tissue homeostasis of multi-cellular organisms by removing unwanted or damaged cells. Impaired regulation of apoptosis is implicated in a variety of diseases including cancer and stroke. Two kinds of signals can trigger apoptosis, death signals received by death receptors on cell surface and stress signals such as structural and genotypic damage. The latter apoptotic signals mainly route through mitochondria to provoke activation of caspases and nucleases that cleave critical proteins and DNAs thereby dismantling the cell(1,2).
Apoptosis is regulated by a complicated series of interactions between Bcl-2 family proteins. These interactions include hetero- and homo-interactions of proteins containing either multiple Bcl-2 homology (BH)* regions (BH1–3 or BH1–4) or a single (BH3) region. After receiving certain apoptotic stimuli, proapoptotic BH3-only proteins induce the release of cytochrome c and other proapoptotic proteins from mitochondria by triggering permeabilization of the mitochondrial outer membrane (MOM) by the multi-BH domain proteins Bax and Bak(3,4). One proposed mechanism of MOM permeabilization involves BH3 only protein induced conformational changes in Bax and/or Bak leading to their oligomerization in the MOM(5–7).
BH3 only proteins are found in a variety of different subcellular locations and share very limited sequence similarity, often only in the short BH3 region. There is insufficient sequence similarity in BH3 region sequences to permit unambiguous identification from sequence alone. It appears that BH3 region proteins act as sensors within the cell, reporting on a variety of subcellular defects. BH3 proteins are thought therefore to function to integrate signals from disparate cell signaling pathways and subcellular locations. Typically BH3-only proteins are classified as either a “sensitizer” that binds to and inhibits anti-apoptotic Bcl-2 family proteins including Bcl-2, Bcl-xL, Bcl-w, Mcl-1 and A1(8–13), or an “activator” that interacts with Bax and Bak proteins, transforming them from cytosolic and MOM-bound monomers to MOM-bound oligomers that permeabilize the membrane(10,14–21). However, this distinction is somewhat artificial as all of the activator BH3-only proteins characterized so far can also have sensitizer activity. Thus, depending on both the relative affinities and abundance of all the Bcl-2 family proteins in the cell a BH3-only protein may function as activator, sensitizer or both (13,15,17,22–26). Furthermore it appears that some functional interactions between BH3 only proteins and other Bcl-2 family proteins may be transient.
To map the various interactions and begin to deduce their relative importance in different apoptotic pathways several groups have measured the affinity of BH3 peptides derived from BH3-only proteins for binding to a variety of Bcl-2 family proteins(10,12,21). The common theme from these measurements is that most BH3 peptides bind preferentially to certain type of Bcl-2-like proteins, suggesting that selectivity exists between most BH3-only proteins and Bcl-2-like proteins. Two different views, however, have emerged from these studies mainly because the behavior of a few BH3 peptides derived from the “activators” Bid and Bim. The Bid and Bim BH3 peptides are more potent than other BH3 peptides in permeabilizing the MOM and inducing apoptosis. One view, based on the evidence that these peptides can directly activate Bax and Bak, is that MOM permeabilzation and cell demise require activation of pro-death Bax-like proteins. The other view, derived from the observation that these peptides can bind to most known pro-survival Bcl-2-like proteins, is that cell death will proceed only when most, if not all, pro-survival proteins are neutralized by Bid or Bim or by combinations of their more specialized BH3-only cousins.
In support the above neutralization model stable complexes or direct binding have been observed between various BH3 peptides and Bcl-2-like proteins (27,28). Evidence for direct binding of Bax and/or Bak by BH3-only proteins to activate Bax and Bak directly is scarce(9,20,29). A kiss and run model has been proposed to reconcile the data(17). Thus, the BH3-only proteins only transiently touch (kiss) the Bax and Bak proteins. Once the latter are activated, the former will leave (run) so the latter can homo-oligomerize in MOM. According to this model one BH3 protein can activate many multi-domain pro-apoptotic proteins through a series of transient interactions. Bcl-2-like proteins may disrupt the kiss by binding the BH3-only protein thereby inhibiting activation of Bax and Bak.
Understandably the above studies focused on the effects of peptides from BH3-only proteins, neglecting the potentially important contributions of homo-interactions in regulating commitment to apoptosis. It is well documented that both Bcl-2-like proteins and Bax-like proteins can form homo-oligomers in vitro and in vivo. Though much less work has been done on the homo-association of Bcl-2-like proteins(26), the homo-oligomerization of Bax and Bak has received more attention(7,19,30,31). However, most of these studies are focused on the effect of oligomerization on MOM permeability. Much less is known about the role of homo-interaction between Bax-like proteins in activating more Bax-like proteins, though the obvious fact is that the Bax BH3 region sequence and structure are not much different from that of BH3-only proteins. However in a recent study, Ruffolo and Shore demonstrated that Bak, activated by a BH3-only protein, can activate other Bak proteins via homo-interactions, thereby amplifying the signal from the BH3-only activator(25). This represents a third view about the apoptosis commitment step. Thus, after the initial activation events mediated by BH3-only activators, the activated Bax and Bak proteins may themselves function as sensitizers or activators or both.
To determine whether Bax auto-activation contributes to homo-oligomerization and to address the function(s) of Bax in the process leading toward MOM permeabilization, we studied various Bax peptides, Bax and Bcl-2 proteins in cell, mitochondrion, liposome, and solution-based systems. We found that the Bax BH3 peptide is not sufficient to activate Bax. Instead an extended peptide including both BH3 helix (helix 2) and a downstream helix (helix 3) (designated H2-H3 peptide) is required. Moreover, we demonstrated that while the extended peptide can interact with Bcl-2 and inhibit its function in solution, the primary target of the peptide in both mitochondrion and liposome-based systems is Bax, suggesting that the primary target of active Bax may be inactive Bax molecules. Auto-activation of Bax may also be related to the oligomerization of Bax proteins in the MOM that accompanies MOM permeabilization.
EXPERIMENTAL PROCEDURES
Materials
The following peptides containing helix 2 (H2) and/or 3 (H3) sequence of human Bax were synthesized by Global Peptide Services (Fort Collins, CO). Their Mr was verified by mass spectrometry, and purity was determined by analytical reverse phase HPLC to be > 90%. H2, 52-QDASTKKLSECLKRIGDELDS-72; and CF-H2, H2 with a carboxyfluorescein (CF) labeled at the N-terminus; H2-H3, 53-DASTKKLSECLKRIGDELDSNMELQRMI AAVDTD-86; CF-H2-H3, H2-H3 with a CF labeled at the N-terminus, and H2-H3 G67R with the underlined G (Gly) changed to R (Arg); and H3, 71-DSNMELQRMIAAVDTD-86. The numbers are the sequence numbers of the first and last residues in Bax protein sequence. Expression and purification of full-length human Bax and Bcl-2ΔTM proteins were as described(26,32). Phospholipids were purchased from Avanti Polar Lipids. Cascade Blue (CB) or tetramethylrhodamine labeled dextrans and an anti-CB antibody were from Molecular Probes.
Microinjection
Rat-1MycERTAM cells were cultured in DMEM for 16–24 hours prior to injection. The injection solution contains 500 μM peptide and 1 mg/ml tetramethylrhodamine labeled dextran (as injection marker) in microinjection buffer (70 mM KCl, 1 mM MgCl2 and 10 mM NaPO4, pH 7.2). Injections were done using an Eppendorf semiautomatic microinjection system (1X Injectman NI2 micromanipulator, 1X microscope adapter, and 1X Femtojet) with the pressure set at 100 Pa. Cells were filmed for 4 hours post-injection with images captured every 5 min using a Hamamatsu Orca AG charge-cooled digital camera. Throughout the experiment, cells were maintained in Leibovitz's L15 media supplemented with 10 mM HEPES (pH 7.2) at 37 °C using a Bioptechs Delta T dish mounted in a Bioptechs heated stage apparatus. Images were processed with the Openlab suite of imaging software (Improvision, Inc., Lexington, MA) and imported into Photoshop (version 8.0) for generating figures.
Assay of cytochrome c release from mitochondria
Cell free assays for cytochrome c release were performed as described(7) except that tBid protein was replaced by various Bax peptides, and the mitochondria were isolated from Rat-1MycERTAM cells either expressing or not expressing human Bcl-2(33).
Preparation of liposomes
Liposomes were made by the extrusion method as described but with the following modifications(34). Lipids were mixed in chroloform with a composition similar to that of Xenopus oocyte MOM as described(19). The lipid mixture was dried by nitrogen and vaccum, and re-suspended in 0.55 ml of buffer A (100 mM NaCl, 51 mM Na2HPO4, and 3.8 mM citric acid, pH 7.4) with 30 μM CB-labeled 3 or 10-kDa dextan. The total lipid concentration of this sample was 20 mM. In addition, trace of [14C]-labeled phosphatidylcholine was added into the sample to determine the lipid concentration in final liposome fractions from gel filtration chromatography. After extrusion, free CB-dextrans were separated from the liposomes with entrapped CB-dextrans by a gel filtration chromatography using a 0.7×50 cm Sephadex CL-2B column.
Assay of dextran release from liposomes by fluorescence quenching
Purified Bax protein (50 nM) was incubated with the liposomes with 6.25 μM lipids and encapsulated CB-dextrans in buffer A at 37 °C. The anti-CB antibody (6 μg/ml) was presented outside the liposomes to monitor the release of CB-dextrans from liposomes since the binding of antibody to CB-dextran quenches CB fluorescence. The Bax protein did not induce any fluorescence quenching, thereby the fluorescence intensity of above sample was taken as the initial intensity (F0). The H2, H2-H3 or H2-H3 G67R peptide was then titrated into the sample and the intensity (F) was measured again when the intensity became stable, typically one hour after each titration. Finally 0.1% of Triton X-100 was used to lysis the liposomes, releasing all CB-dextrans, and the final intensity (FT) was then measured. The extent of CB-dextran release caused by each addition of peptide is proportional to the extent of fluorescence quenching that is equal to (F0 – F) / (F0 – FT). Of note, all the peptides did not induce any CB-dextran release in absence of Bax protein even at the maximal concentration tested. The CB fluorescence intensity was measured using the SLM-8100 spectrofluorometer with excitation and emission wavelengths set at 385 and 417 nm, respectively, and bandpass at 4 nm. All measurements were done in 4 × 4 mm quartz microcells at 37 °C.
To assay the effect of Bcl-2ΔTM protein on above Bax/peptide-induced CB-dextran release, 500 nM of purified Bcl-2ΔTM protein was added and the F0 was measured prior to the addition of any peptides. Otherwise the experiments were done similarly as described above. To assay the effect of peptides on relieving Bcl-2ΔTM-mediated inhibition of CB-dextran release, the extent of CB-dextran release was determined first with a sample containing 50 nM Bax protein, 500 nM Bcl-2ΔTM protein and 10 μM H2-H3 peptide. The additional peptides were then titrated into the sample and their effect on the extent of CB-dextran release was determined as described above.
To assay the effect of liposome-bound Bax on activating soluble Bax to induce further release of CB-dextrans from the liposome, 6.25 μM of the liposome with encapsulated CB-dextrans were first incubated with 50 nM of Bax, 20 μM of H2-H3 peptide, and/or 500 nM of Bcl-2ΔTM at 37 °C for 40 min, and then subjected to sucrose step gradient float-up centrifugation as described(32). The resulting top fraction (250 μl) that contains proteoliposomes was incubated with anti-CB antibodies either in the absence or presence of freshly added 50 nM of Bax, 20 μM of H2-H3 peptide, and/or 500 nM of Bcl-2ΔTM. The CB fluorescence quenching was then followed for 6 hrs.
To determine if any H2-H3 peptides were carried over from the first incubation into the top fraction that was used for the second incubation, 40 nM CF-H2-H3 peptides were included in the first incubation. The CF emission intensity of the top fraction and other fractions was monitored as described below. The distribution of the peptide in the fractions was estimated based on the intensities of each fraction. The distribution of liposomes was determined by measuring 14C counts that were resulted from the trace of [14C]-phosphatidylcholine incorporated into the liposomes.
Assay the effect of Bax peptides on association of Bax and Bcl-2ΔTM proteins in solution
[35S] methionine-labeled Bax protein was synthesized using a rabbit reticulocyte lysate-based in vitro translation system as described(26). The resulted translation product (10 μl) was incubated with 2.5 μM of purified His6-tagged Bcl-2ΔTM protein either in absence or presence of various concentrations of H2 or H2-H3 peptide in buffer B (20 mM HEPES, pH 7.5, 100 mM KOAc, 5 mM Mg(OAc)2, 10% (v/v) glycerol, 0.25% (v/v) Triton X-100, and 5 mM imidazole) with a total volume of 13 μl at 4 °C for 1 hour. Ni2+-nitrilotriacetic acid-argarose [200 μl of 4% (v/v) suspension in buffer B] was added. After incubation at 4 °C for two hours, proteins bound to the Ni2+-beads were isolated and analyzed by SDS-PAGE and phosphorimaging as described(26).
Measurement of equilibrium binding of Bax peptides to Bcl-2ΔTM protein in solution by fluorescence anisotropy
Steady-state fluorescence measurements were done with SLM-8100 spectrofluorometer. All measurements were done at 25 °C in buffer A using 1 × 1 cm (for tryptophan fluorescence) or 4 × 4 mm (for CF fluorescence) and a bandpass of 4 nm. The excitation and emission wavelengths are 295 and 348 or 490 and 520 nm for tryptophan or CF fluorescence, respectively.
Steady-state anisotropy was measured in the L-format using Glan-Thompson prism polarizers in both the excitation and emission light passes. The emission intensity measured through a horizontal polarizer when a sample was excited by vertical plan-polarized light was designated as IVH. IHH, IHV and IVV were defined analogously. The component intensities of a fluorophore-free sample (without protein or CF-peptide) were subtracted from the corresponding component intensities of the sample containing the protein or CF-peptide to obtain the net intensities that were then used to calculate the anisotropy using the following equation.
| (Eq. 1) |
where the G-factor G = IHV/IHH.
To monitor the homo-association of Bcl-2ΔTM proteins, tryptophan anisotropy was measured after each titration of Bcl-2ΔTM protein (1 μl) into 2.5 ml of buffer A. To monitor the binding of a Bax peptide to Bcl-2ΔTM protein, CF anisotropy was measured after each titration of Bcl-2ΔTM protein (1 μl) into 250 μl of buffer A containing 10 nM of CF-labeled H2 peptide in absence or presence of 10 μM of competitor, unlabeled corresponding Bax peptide. All measurements were done when equilibrium was reached, typically 30 min after each titration.
Anisotropy data analysis
When Bcl-2ΔTM protein was titrated into the solution without any peptides, the following model binding equilibrium (Eq. 2) was used to derive the quadratic equation (Eq. 3) for data analysis (see Supplemental Data for the derivation of this and other equations).
| (Eq. 2) |
| (Eq. 3) |
where B represents the monomer, and B2 the dimer of Bcl-2ΔTM. Kd is the dissociation constant for the binding equilibrium, and b0 the concentration of total Bcl-2ΔTM in the sample after each titration. According to the additivity law for anisotropies and considering the intensity change upon the Bcl-2ΔTM dimerization (C. T. and J. L., unpublished data)(35), the fraction of Bcl-2ΔTM in the dimer form, fB, can be related to the measured tryptophan anisotropy (r) of the sample after each titration using
| (Eq. 4) |
where R is the ratio of the intensity of dimer to that of monomer Bcl-2ΔTM, and rM and rD the anisotropies of the monomer and dimer Bcl-2ΔTM, respectively.
When Bcl-2ΔTM protein was titrated into the sample containing CF-labeled H2 peptide, the above binding equilibrium (Eq. 2) competes with the following binding equilibrium (Eq. 5) resulting in the cubic equation (Eq. 6) for data analysis.
| (Eq. 5) |
| (Eq. 6) |
where L represents the labeled peptide, and BL the Bcl-2ΔTM/labled peptide complex. Kl is the dissociation constant for the binding equilibrium, and l0 the concentration of total L in the sample after each titration. Similar to the fB above, the fraction of L in the bound form, fl, can be related to the measured CF anisotropy (r1) of the sample after each titration using
| (Eq. 7) |
where R1 is the ratio of intensity of bound to free CF-peptide, and rF and rB the anisotropy of the free and bound CF-peptide, respectively.
When Bcl-2ΔTM protein was titrated into the sample containing the CF-peptide and the competitor peptide, the above two binding equilibriums (Eq. 2 and Eq. 5) compete with the following binding equilibrium (Eq. 8) resulting in the quartic equation (Eq. 9) for data analysis.
| (Eq. 8) |
| (Eq. 9) |
| (Eq. 10) |
| (Eq. 11) |
| (Eq. 12) |
| (Eq. 13) |
where U represents the unlabeled competitor peptide, and BU the Bcl-2ΔTM/unlabled peptide complex. Ku is the dissociation constant for the binding equilibrium, and u0 the concentration of total U in the sample after each titration.
The Kd value for Bcl-2ΔTM homo-association was first determined by combining three individual titration data sets and analyzing them simultaneously by nonlinear least squares fitting of the quadratic equation (Eq. 3) and the equation (Eq. 4) using the SCIENTIST fitting program(36). The fitting parameters other than the Kd are rM, rD and R. The b0 value was determined from the A280 value of the Bcl-2ΔTM stock used for the titration using ε280 of 43430 M−1cm−1(37). Similarly, the Kl value for CF-H2 binding to Bcl-2ΔTM was then determined as a fitting parameter by fitting the corresponding data sets with the cubic equation (Eq. 6) and the equation (Eq. 7). The other fitting parameters are rF, rB and R1. The l0 value was determined according to the concentration of CF-H2 peptide stock solution that was determined from the measured weight of dry peptide used for preparing the solution; the b0 value was determined as above; and Kd in the cubic equation (Eq. 6) used the value that was determined above for Bcl-2ΔTM homo-dimerization. Finally, the Ku values for various unlabeled Bax peptides binding to Bcl-2ΔTM were determined similarly by fitting the data with the quartic equation (Eq. 9) and the equation (Eq. 7). The other fitting parameters are rF, rB and R1. The u0 value was determined similarly as was the l0 value described above. Other parameters are as describe above or used the values of Kd and Kl determined above.
RESULTS AND DISCUSSION
A peptide of both BH3 and downstream regions of Bax triggers apoptosis in cells and activates Bax inducing cytochrome c release from mitochondria
The BH3 region of Bax containing amino acids 59–73 is required for Bax homo-interaction and death agonist activity(38). Previous studies using peptides containing Bax BH3 region have generated conflicting data. Particularly it was reported that a long Bax BH3 peptides can induce cytochrome c release from mitochondria by activating Bax directly(39,40), whereas a shorter peptide induced cytochrome c release by preventing Bax sequestration by Bcl-xL(41). Since activation of Bax by BH3-only proteins results in exposure of the BH3 region of Bax required for homo- and hetero-oligomerization(42), it is important to clarify the role of the Bax BH3 region peptide in Bax activation. This may provide answers to the important questions such as how cells can generate many active Bax molecules after initial activation of a few Bax molecules; why there is always a lag, from several hours to days, between the initial Bax activation and the first detectable mitochondrial permeabilization; and finally, what Bcl-2 does to interfere with the mitochondrial permeabilization during this lag period.
To examine the role of the Bax BH3 region separately from the rest of the protein, we studied two peptides containing amino acids 52–72 and 53–86 of Bax, corresponding to the shorter and longer peptides used in previous publications, respectively. By using both peptides in a single experimental system it should be possible to reveal the mechanisms responsible for the differences reported previously. The shorter peptide contains the intact BH3 helix (helix 2, amino acids 54–71) of Bax, while the longer peptide contains the BH3 helix plus residues in the down stream helix 3 (amino acids 74–81), and are here designated H2 or H2-H3 peptide, respectively(43).
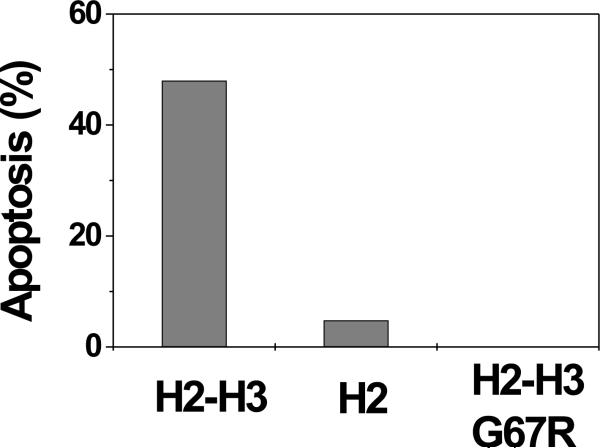
To examine the capacity of the peptides to trigger apoptosis, Rat-1MycERTAM cells were injected separately with equal molar amounts of each peptide and analyzed for 4 hrs by time-lapse video microscopy. Injection of the H2-H3 peptide resulted in morphological changes consistent with apoptosis including plasma membrane blebbing (Fig. 1A). By 4 hours post-injection, 57 of the 117 (48%) injected cells underwent apoptosis (Fig. 1B). In contrast, only 4 of the 83 (5%) cells receiving the H2 peptide underwent apoptosis, suggesting that this peptide is insufficient to trigger apoptosis. Furthermore, none of the 109 (0%) cells injected with the mutant H2-H3 peptide became apoptotic by 4 hours post-injection (Fig. 1B). Because the mutant peptide has Gly67 in the H2 region replaced by an Arg (G67R), these data demonstrate that the H2 region in the H2-H3 peptide is important for the apoptosis-inducing activity.
Fig. 1. Bax H2-H3 peptide but not H2 peptide triggers apoptosis in Rat-1MycERTAM cells.
A. Bax H2-H3 peptide was injected into cells together with a fluorescent injection marker. The first and last panels are fluorescent images showing the injected cells. All other panels are transmitted light images of a field containing injected and uninjected cells. The time (in min) that each image was captured post-injection is indicated in the upper right hand corner of each photomicrograph. An injected cell that underwent apoptosis is indicated by a white arrow. B. The numbers of apoptotic cells were counted 4 hours post-injection of either the H2-H3, H2 or H2-H3 G67R peptides. Data shown are the percentages of apoptotic cells among total injected cells.
To elucidate the mechanism by which the extended but not the short Bax BH3 peptide triggers apoptosis, we switched to in vitro system that contains defined components and is easier to manipulate. To assay membrane permeabilization the peptides were added alone or together with purified monomeric Bax to heavy membranes enriched with mitochondria isolated from Rat-1MycERTAM cells(33). When each peptide was added alone to the membranes, none caused a notable increase of cytochrome c release when compared with the minus peptide control (Fig. 2A, control mito). Therefore, neither peptide alone can induce the mitochondrial permeabilization, suggesting that other non-mitochondrial proteins are required for the observed pro-apoptotic activity for the H2-H3 peptide in Rat-1MycERTAM cells.
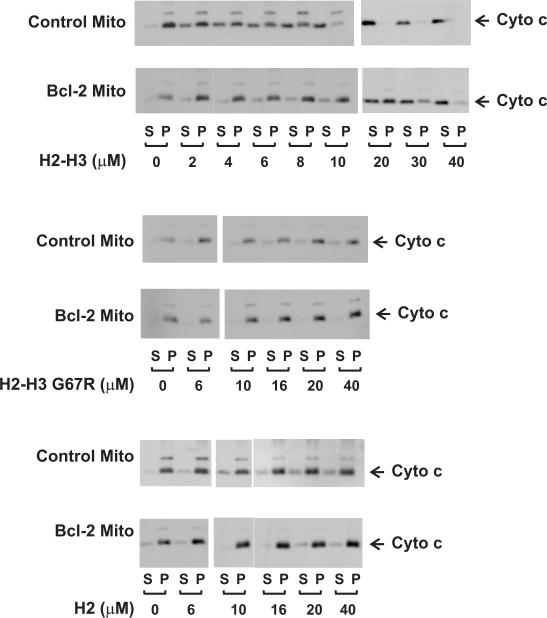
Fig. 2. Bax H2-H3 peptide activates Bax releasing cytochrome c from isolated mitochondria, a reaction inhibited by Bcl-2.
Mitochondria isolated from control or Bcl-2-expressing Rat-1MycERTAM cells were incubated with 0 (A) or 20 nM Bax (B) and various Bax peptides as indicated. The concentration of each peptide used in (A) was 25 μM, whereas in (B) was from 0 to 40 μM as indicated. Cytochrome c release was assayed by pelleting the mitochondria and visualizing the cytochrome c in the supernatant (S) and pellet (P) fractions by SDS-PAGE and immunoblotting. Data shown are representatives from three independent experiments in which each sample was analyzed in duplicate or triplicate. C. Cytochrome c release by H2-H3 peptide from the experiments shown in (B) was quantified using densitometry. Percentages of the release were obtained by dividing the intensity of the protein signal in S fraction with that in both S and P fractions of the same sample. D. Various concentrations of Bax were added with 1 μM H2-H3 peptide to the control or Bcl-2-expressing mitochondria. Cytochrome c release was analyzed as above.
When up to 40 μM of H2 peptide was added to the membranes together with 20 nM of monomeric Bax, the amount of cytochrome c recovered from the supernatants was only slightly increased compared to the control sample to which only the Bax was added (Fig. 2B, bottom panel, Control Mito). This suggests that the H2 peptide is not sufficient to activate the monomeric Bax to permeabilize the MOM, consistent with the microinjection and some but not all previous results(21,41,44). In accordance with the microinjection and previous results(39,40), addition of the H2-H3 peptide increased the level of cytochrome c release in a Bax-dependent fashion (Fig. 2B, top panel, Control Mito). Control peptides, H2-H3 G67R and H3 that contains only the helix 3 sequence increased cytochrome c release only slightly even at the highest concentration (40 μM) in parallel assays (Fig. 2B, middle panel, Control Mito; data not shown), further demonstrating that although the H2 (BH3) sequence is not sufficient for activating Bax, it is required.
To determine the effectiveness of the H2-H3 peptide, it was titrated into the samples with fixed amount of mitochondria and Bax in several independent experiments. The results from a typical experiment are shown in Figure 2B, top panel (Control Mito). The extent of cytochrome c release was quantified, and the results are shown in Figure 2C. The cytochrome c release was increased in a dose-dependent manner for the H2-H3 peptide with a maximum reached at about 20 μM of the peptide when 20 nM of Bax was present (diamonds). At this concentration the H2-H3 G67R, H2 or H3 peptide showed very limited activity (Fig. 2B, middle and bottom panels; data not shown). These results confirm that both H2 and H3 sequences are required for the peptide to effectively activate Bax, and indicate that a hydrophobic residue in the H2 region is important for this activity.
Mitochondrial permeabilization by peptide-activated Bax depends on the Bax to Bcl-2 ratio
Bcl-2 is known to bind Bax, preventing homo-oligomerization in the MOM and release of cytochrome c(22,26,31,38). Not surprisingly when the heavy membranes isolated from the Rat-1MycERTAM cells that express human Bcl-2 were used, cytochrome c release induced by the H2-H3 peptide-activated Bax was inhibited (Fig. 2B, top panel, Bcl-2 Mito), consistent with previous results(39). The concentration of exogenous human Bcl-2 expressed in the rat mitochondria used in these experiments was about 20 nM based on quantitative immunoblotting analysis (data not shown). Quantitative analysis showed that the cytochrome c release by up to 6 μM peptide from mitochondria in reactions containing and 20 nM Bax was mostly blocked by the Bcl-2 in mitochondria (Fig. 2C, triangles). Inhibition of cytochrome c release by the Bcl-2 can be overcome by higher concentrations of peptide. At concentration of 20 μM peptide cytochrome c release was about two-thirds that observed for mitochondria without the exogenous Bcl-2. When 40 μM of peptide was added, about 80% of the cytochrome c in mitochondria was released, an amount equal to the maximum obtained for mitochondria without the exogenous Bcl-2. In contrast, the H2-H3 G67R and H2 peptides that were ineffective in activation of Bax were also unable to overcome the inhibition by Bcl-2 even at the highest concentration tested (Fig. 2B, middle and bottom panels, Bcl-2 Mito). It was not a surprise to see that Bcl-2 blocked the minimal cytochrome c release caused by high concentrations of these relatively ineffective peptides (compare Control and Bcl-2 Mito in Fig. 2B, middle and bottom panels). However, it was a surprise to see that H2 peptide was unable to overcome the Bcl-2 inhibition on cytochrome c release, since previous studies suggested that it may inhibit Bax/Bcl-2 hetero-dimerization releasing active Bax from Bcl-2 to permeabilize mitochondria(44,45).
To compare the effectiveness of additional monomeric Bax in overcoming inhibition due to Bcl-2 with that of H2-H3 peptide, monomeric Bax was titrated into a heavy membrane sample with ~20 nM of exogenous Bcl-2 and 1 μM of H2-H3 peptide. Under these conditions cytochrome c release by 20 nM Bax is effectively inhibited by the Bcl-2 (Fig. 2D). To overcome fully the Bcl-2 mediated inhibition of cytochrome c release by the Bax an additional 60 nM Bax was needed (Fig. 2D). In contrast about 35 μM of additional H2-H3 peptide was required to achieve the same effect (Fig. 2C), indicating that activating more Bax is a more effective way to inhibit Bcl-2 than adding additional H2-H3 peptide. This also raised questions about how the H2-H3 peptide overrides Bcl-2 mediated inhibition of Bax. The peptide could neutralize Bcl-2 directly or indirectly through activation of more Bax. Furthermore, although the initial activation of Bax requires the H2-H3 peptide, does the peptide act on Bax directly or via other mitochondrial proteins? Finally, is the peptide required to activate all of the Bax needed to permeabilize the MOM? Perhaps, the initially activated Bax can active more Bax since it may contain the exposed H2-H3 motif(42). These questions are difficult to address in the complex mitochondrial system, we therefore switched to a simpler in vitro system using purified peptides/proteins and lipid vesicles. The vesicles were prepared from pure lipids according to the lipid composition of MOM determined previously(19).
The extended Bax peptide is sufficient to activate Bax to induce liposome permeabilization
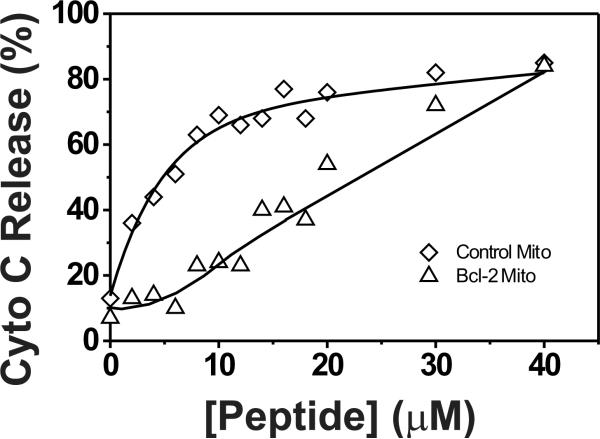
We and others have shown that monomeric Bax can be activated by tBid or the BH3 peptide of certain BH3-only proteins, permeabilizing liposomes of MOM lipids, an activity that can be inhibited by Bcl-xL or Bcl-xLΔTM, a Bcl-xL lacking the C-terminal transmembrane (TM) sequence(19,21,32). As expected, the H2-H3 peptide that activated Bax to permeabilize mitochondria also activated Bax to release fluorescent dextran molecules of 3 and 10 kDa from liposomes (Fig. 3A, diamonds; data not shown but see Fig. 6). Since in this simple system, only the peptide and Bax protein are required for the permeabilization of liposomes, this result demonstrates that in the presence of a lipid bilayer the peptide can activate Bax directly. This result also suggests that the activation of Bax by the peptide in mitochondria does not require other mitochondrial proteins or displacing pre-activated Bax from a Bcl-2-like protein. Quantitatively the data from the liposomes matched surprisingly well with those from the mitochondria, suggesting that the direct activation of Bax by the peptide and its consequence are likely the dominant mode (if not the only mode) of action for the peptide and Bax in the mitochondrial system. As expected the H2 and H2-H3 G67R peptides did not induce any Bax-mediated dextran release from liposomes even at a concentration of 320 μM, a concentration that is 16-time higher than that of H2-H3 peptide (20 μM) which can cause maximal dextran release via 50 nM of Bax (Fig. 3A, circles and squares; data not shown). These results provide further evidence that both H2 and H3 regions are required for Bax activation.
Fig. 3. Bax H2-H3 peptide activates Bax releasing dextrans from liposomes, a reaction inhibited by Bcl-2ΔTM.
A. Various Bax peptides were titrated into liposomes with encapsulated CB-labeled dextrans (Mr ~3 kDa) to activate 50 nM Bax in the absence (◊) or presence (Δ) of 500 nM Bcl-2ΔTM. Release of CB-dextrans from the liposomes by active Bax was monitored by anti-CB antibodies presented outside of the liposomes that would bind the released CB-dextrans and quench their fluorescence. The extent of fluorescence quenching after each titration (ΔFProtein) was measured and shown as a percentage of the maximal quenching observed after complete lysis of the liposomes by Triton X-100 (ΔFTriton). Data shown are means from three independent titrations using H2-H3 (◊ and Δ), H2(○) or H2-H3 G67R(□) peptide with standard deviations (S.D., error bars). B. H2-H3 (Δ), H2 (○) or H2-H3 G67R (□) peptide was titrated into a sample of CB-dextran encapsulated liposomes, 10 μM H2-H3 peptides, 50 nM Bax and 500 nM Bcl-2ΔTM. The extent of CB-dextran release was determined after each titration. Data shown are means from three independent titrations with S.D. (error bars).
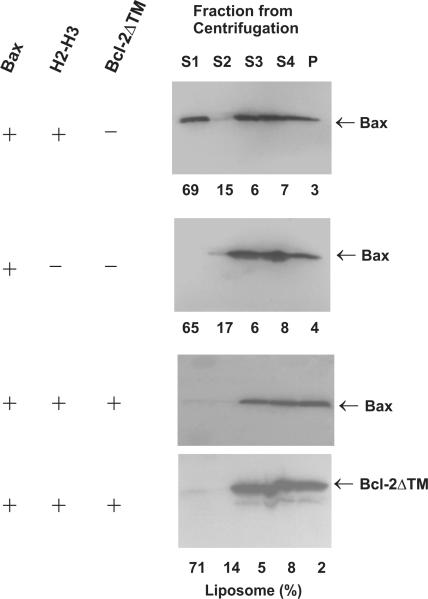
Fig. 6. Activation soluble Bax by membrane-bound Bax.
A. The time course of Bax-dependent CB-dextran release from liposomes was monitored by measuring the quenching of CB-fluorescence in the presence of Bax (□), Bax and H2-H3 peptide (◊), or Bax, H2-H3 peptide and Bcl-2ΔTM (Δ). Data shown are means from two or three independent experiments with S.D. (error bars). Since some S.D. values are very small, the corresponding error bars are inside the symbols. B. Liposomes were incubated with Bax, H2-H3 peptide and/or Bcl-2ΔTM for 40 min, and then subjected to sucrose gradient float-up centrifugation. Four fractions were removed from the supernatant resulting fractions S1–4 with S1 being the top most fraction. Bax or Bcl-2ΔTM proteins in the supernatant and pellet (P) fractions were analyzed by SDS-PAGE and immunoblotting using antibodies specific to Bax (top three panels) or Bcl-2 (bottom panel), respectively. The amount of liposomes in each fraction was determined by scintillation counting the 14C-phosphatidylcholine in the liposomes and shown as the percentage of the total liposomes under the blots. C. The amount of H2-H3 peptides in each fraction was determined by measuring the fluorescence intensity (F) of CF-H2-H3 peptides that were included in the sample during the 40-min incubation and shown as the percentage of the summed intensities of all fractions (FTotal). D. The liposomes incubated with both Bax and H2-H3 peptide for 40 min were isolated by the float-up centrifugation. The CB-dextran release from the resulted liposomes was monitored in the presence of Bax (□), Bax and Bcl-2ΔTM (Δ), or H2-H3 peptide (○). Data shown are means from three independent titrations with S.D. (error bars). E. The experiments were similar to that described in (D), except that the liposomes were incubated only with Bax for 40 min prior to the float-up centrifugation, and that the dextran release was measured in the presence of Bax (□) or Bax and H2-H3 peptide (◊).
Excess of H2-H3 peptide was required to activate Bax in both mitochondria and liposomes. Similar results were reported for other BH3 peptides before(21). One explanation is that the binding affinity of peptides to Bax may be low. In accordance we found that a carboxyfluorescein-labeled H2-H3 peptide did not bind Bax in solution in a anisotropy-based binding assay similar to that described below. When the binding assay was carried out in the presence of liposomes, an anisotropy increase was observed only when excess of Bax was titrated into the CF-labeled peptide, indicating that the binding affinity is low (C.T. and J.L., unpublished data).
Altering the ratio of peptide-activated Bax to Bcl-2 is sufficient to regulate liposome permeability
Although Bcl-xLΔTM was shown to be as potent as the full-length Bcl-xL as an inhibitor of active Bax in previous assays using MOM vesicles or liposomes(19), Bcl-2ΔTM has not been tested in similar assays. Furthermore, while some groups have reported that a recombinant Bcl-2ΔTM retains certain activities of full-length Bcl-2, consistent with early reports that Bcl-2ΔTM was an effective anti-apoptotic protein in cells(26,37,46), other studies have reported that Bcl-2ΔTM is mostly inactive(47,48). In our liposome system the recombinant Bcl-2ΔTM effectively inhibited the dextran release induced by the H2-H3 peptide-activated Bax (Fig. 3A, triangles), to a similar extent as reported for the Bcl-xLΔTM in a liposome system(19). Thus, in absence of Bcl-2ΔTM about 50% of dextrans were released by 50 nM of Bax protein and 10 μM of H2-H3 peptide. In presence of 500 nM of Bcl-2ΔTM, about 25% of dextrans were released by the same concentrations of Bax protein and peptide. When more peptides were titrated into the samples, the gap between the percentages of dextran release in samples with and without Bcl-2ΔTM became less. Eventually when 20 μM or more peptides were added, the extent of dextran release was indistinguishable for the two samples, demonstrating that higher concentrations of the peptide can overcome the inhibition of Bax-induced membrane permeabilization by Bcl-2ΔTM.
In liposome based assays excess Bcl-2ΔTM was required to inhibit the liposome permeabilization by active Bax. In contrast when mitochondria were used equal molar Bcl-2 effectively inhibited the active Bax mediated mitochondrion permeabilization. A major difference between the two systems is that in the latter experiments mitochondria were isolated with the Bcl-2 anchored in the membrane via the C-terminal TM sequence. In the liposome system Bcl-2ΔTM was purified from E. coli which necessitated the deletion of the TM sequence and resulted in a soluble protein that does not bind to liposomes. Therefore, it is not surprising that the specific activity of Bcl-2 synthesized and targeted to mitochondria in eukaryotic cells is higher than that of Bcl-2ΔTM added to liposomes.
Only peptides that activate Bax displace Bax from Bcl-2
One possible explanation for the discrepancy between the different published results for the H2 peptide is the H2 peptide is not sufficient to activate Bax but can release otherwise activated Bax from Bcl-2. In this scenario the H2 peptide would function similar to peptides from the BH3 region of sensitizer BH3-only proteins. To test this hypothesis 10 μM of H2-H3 peptide was added to reactions containing 50 nM of Bax and 500 nM of Bcl-2ΔTM. In such reactions the expectation is that H2-H3 peptide will activate some Bax which will then be inhibited by Bcl-2ΔTM. If subsequent additions of peptides to these reactions displace Bax from Bcl-2, the peptide should trigger dye release from liposomes. As shown in Figure 3B, addition of more H2-H3 peptide overcame the inhibition by Bcl-2 (triangles), whereas addition of either H2 or H2-H3 G67R peptide did not result in any additional dye release form liposomes (circles or squares, respectively). These results suggest that the peptides that do not activate Bax directly do not release activated Bax sequestered by Bcl-2.
Binding of Bax peptides to Bcl-2 does not correlate with inhibiting Bcl-2
It is unclear if the H2-H3 peptide inhibited Bcl-2 by direct binding or indirectly by activating Bax proteins that in turn bound to and inhibited Bcl-2. The expectation in the literature is that direct BH3 peptide binding correlates with inhibition. This presumption has been used to rank the substrate specificity of BH3 proteins from surface plasmon resonance data(12). To determine whether the H2-H3 peptide inhibits Bcl-2 directly or indirectly we measured the binding of the peptide, and the control peptides H2 and H2-H3 G67R, to Bcl-2ΔTM protein in solution at equilibrium using fluorescence. Bcl-2 family proteins are well known to form homo- and hetero-oligomers. In particular homo-oligomerization uses the hydrophobic groove also reported to bind BH3 peptides (26,49,50). In co-crystals of Bcl-2ΔTM and Bax H2-H3 peptide the same groove was occupied by the Bax H2-H3 peptide (S. Terzyan, X. C. Zhang, J. Peng, and J. Lin, unpublished data). We also observed that the Bcl-2ΔTM homo-oligomerization-dependent photocross-linking was reduced in the presence of the Bax peptide, suggesting that the two binding reactions, Bcl-2 homo-oligomerization and Bcl-2/Bax peptide hetero-dimerization, compete with each other (data not shown). To compare these interactions we must determine the binding affinity for Bcl-2 homo-oligomerization and for hetero-oligomerization with the various peptides.
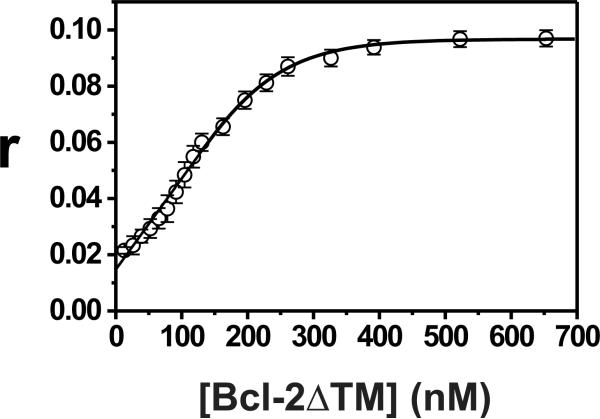
Bcl-2ΔTM contains six tryptophan residues that allow us to measure fluorescence anisotropy. Since the anisotropy is sensitive to the size of the molecule (35), the anisotropies of Bcl-2ΔTM monomer and oligomer in solution are expected to be different. As shown in Figure 4A (circles), the anisotropy of Bcl-2ΔTM was concentration dependent and increased as more protein was titrated into the sample, reaching a plateau when the concentration was 500 nM or higher. After the anisotropy measurement, the samples were subjected to gel filtration chromatography. Most Bcl-2ΔTM proteins were co-eluted with the protein standards of 25 and 43 kDa (data not shown), indicating that the proteins are mostly monomers and dimers in the concentration range used for titration. As expected the data can be fit very well using a linked equation set including the quadratic equation (Eq. 3) derived for the simple monomer-dimer equilibrium model (Supplemental Data) in a nonlinear least squares fitting (Fig. 4A, solid curve). The equilibrium dissociation constant for Bcl-2ΔTM homo-dimerization, determined as a fitting parameter, is equal to 0.25 μM (Kd, Table 1).
Fig. 4. Bcl-2 Δ TM homo-association and hetero-association with Bax peptides detected by fluorescence anisotropy.
A. Bcl-2ΔTM homo-association was monitored by measuring its Trp fluorescence anisotropy (r) after titrating the protein into a buffer. Data shown are means (ο) from three independent titrations with S.D. (error bars). Nonlinear least squares fitting of the quadratic equation (Eq. 3) to the titration data yielded the best fit line shown and the best fit parameters in Table 1. B. Binding of H2-H3 (○) or H2 (Δ) peptide to Bcl-2ΔTM in a buffer was monitored by measuring the CF fluorescence anisotropy of a CF-labeled H2 peptide after titrating Bcl-2ΔTM in absence (□) or presence of the corresponding peptide as the competitor. Data shown are means from three independent titrations with S.D. (error bars). Nonlinear least squares fitting of cubic or quartic equation (Eq. 6 or 9) to the titration data yielded the best fit lines shown and the best fit parameters in Table 1.
Table 1.
Bcl-2ΔTM homo- or hetero-association with various Bax peptidesf .
| Ligand | rFreea | rBoundb | Rc | Kd (μM)d |
|---|---|---|---|---|
| Bcl-2ΔTM | 0.015 ± 0.005 | 0.101 ± 0.009 | 1.9 ± 0.1 | 0.25 ± 0.05 |
| CF-H2 | 0.043 ± 0.004 | 0.15 ± 0.02 | 1.2 ± 0.1 | 2.4 ± 0.5 |
| H2-H3e | 0.043 ± 0.004 | 0.15 ± 0.02 | 1.2 ± 0.1 | 0.6 ± 0.2 |
| H2-H3 G67Re | 0.043 ± 0.004 | 0.15 ± 0.02 | 1.2 ± 0.1 | No binding |
| H2e | 0.043 ± 0.004 | 0.15 ± 0.02 | 1.2 ± 0.1 | 2.8 ± 0.6 |
The anisotropy of free ligand, i.e., Trp anisotropy of monomeric Bcl-2ΔTM in the homo-association (rM in Eq. 4), or CF anisotropy of free CF-H2 in the hetero-association (rF in Eq. 7).
The anisotropy of bound ligand, i.e., Trp anisotropy of dimeric Bcl-2ΔTM in the homo-association (rD in Eq. 4), or CF anisotropy of Bcl-2ΔTM-bound CF-H2 in the hetero-association (rB in Eq. 7).
The intensity ratio of bound to free ligand, i.e., the Trp intensity ratio of dimeric to monomeric Bcl-2ΔTM in the homo-association (R in Eq. 4), or the CF intensity ratio of Bcl-2ΔTM-bound to free CF-H2 in hetero-association (R1 in Eq. 7).
The equilibrium dissociation constant for the homo-association (Kd in Eq. 3), or the hetero-association with either CF-H2 (K1 in Eq. 6) or unlabeled H2-H3, H2-H3 G67R or H2 (Ku in Eq. 9–13).
CF-H2 is the additional ligand in the reaction, competing with the indicated ligand for binding to Bcl-2ΔTM.
We then determined the equilibrium dissociation constant for the binding of a carboxyfluorescein (CF)-labeled H2 peptide to Bcl-2ΔTM by titrating purified Bcl-2ΔTM protein into a solution containing the peptide and monitoring anisotropy of the CF (Fig. 4B, squares). The data can be fit very well using a linked equation set including the cubic equation (Eq. 6) derived from the two-binding equilibrium model (Supplemental Data) in a nonlinear least squares fitting (Fig. 4B, solid curve). The fitting yielded an equilibrium dissociation constant of 2.4 μM for CF-H2 binding to Bcl-2ΔTM (Kl, Table 1).
To determine the equilibrium dissociation constant for the unlabeled H2 or H2-H3 peptide binding to Bcl-2ΔTM, the above titration was done in the presence of the corresponding unlabeled peptide. Since the unlabeled peptide competed with the CF-peptide for binding to Bcl-2ΔTM, the fraction of CF-peptide bound to Bcl-2ΔTM and hence the anisotropy of CF was reduced (Fig. 4B, triangles or circles for presence of unlabeled H2 or H2-H3 peptide, respectively). The data can be fit very well using a linked equation set including the quartic equation (Eq. 9) derived from the three-binding equilibrium model (Supplemental Data) in nonlinear least squares fittings (Fig. 3B, dash and dash-dot curves). The resulting equilibrium dissociation constants are 2.8 and 0.6 μM for the H2 and H2-H3 peptides binding to Bcl-2ΔTM, respectively (Ku, Table 1). Thus, the H2 peptide binds to Bcl-2ΔTM with a 4.7-fold lower affinity than the H2-H3 peptide.
The H2 peptide binds Bcl-2ΔTM, yet cannot overcome the inhibition of active Bax by Bcl-2ΔTM in liposomal assays even at 32-fold higher concentration than that required for the H2-H3 peptide to inhibit Bcl-2ΔTM (Fig. 3B). Therefore, the inhibition of Bcl-2ΔTM by the H2-H3 peptide involves different binding interactions than those of H2 peptide-binding to Bcl-2ΔTM. Otherwise, a 4.7-fold increase in H2 peptide should also inhibit Bcl-2ΔTM. The fact that H2-H3 but not the H2 peptide inhibits Bcl-2 in cellular membranes (Fig. 2) argues that the two peptides also differ mechanistically when interacting with Bcl-2 on mitochondria.
The addition of mutant H2-H3 (G67R) to the competition assay had no effect on the observed Bcl-2ΔTM-dependent CF anisotropy change (data not shown). This result demonstrates that this mutant peptide does not compete with the CF-H2 peptide for binding of Bcl-2ΔTM, therefore does not bind to Bcl-2ΔTM (Table 1). If peptide-Bcl-2 binding alone was sufficient for the peptide to inhibit Bcl-2, then the H2 should inhibit Bcl-2 while the H2-H3 G67R peptide should not.
Inhibition of Bcl-2/Bax interaction by Bax peptides does not correlate with inhibition of Bcl-2
Another explanation for the ineffectiveness of H2 peptide to abolish the inhibition of Bax by Bcl-2 would be that only H2-H3 peptide inhibits Bcl-2/Bax hetero-dimerization. To test this, we monitored Bcl-2/Bax hetero-dimerization using a pull-down assay. Ni-beads were used to precipitate the Bcl-2ΔTM protein via a His6-tag at the C-terminus and the precipitates were examined for co-precipitation of in vitro translated, [35S]-labeled Bax protein. The non-ionic detergent Triton X-100 was added to induce Bax-Bcl-2 binding as described (26,51). Bax precipitated with the Ni-beads only when incubated with His6-tagged Bcl-2ΔTM, indicating that Bax binds to Bcl-2ΔTM (Fig. 5). When incubated with increasing amounts of either H2 or H2-H3 peptide, the amount of Bax bound to the Bcl-2ΔTM was decreased. Although the H2-H3 peptide is much more effective than the H2 peptide in inhibiting Bax/Bcl-2 binding, the shorter peptide can still inhibit the hetero-binding. Thus, 180 μM of H2 peptide produced a similar reduction of Bax/Bcl-2ΔTM binding as did 10 μM of H2-H3 peptide. In contrast 320 μM of H2 peptide did not overcome any Bcl-2ΔTM inhibition on Bax-mediated membrane permeabilization whereas 10 μM of H2-H3 peptide did completely (Fig. 3B), suggesting that the ineffectiveness of the shorter peptide in Bcl-2 inhibition in assays containing membranes must be explained by a mechanism other than inhibition of binding of Bax to Bcl-2.
Fig. 5. Inhibition of Bcl-2Δ TM/Bax hetero-association by Bax peptides.
In vitro synthesized, [35S]-labeled Bax was incubated with 2.5 μM of His6-tagged Bcl-2ΔTM protein in presence of various concentrations of H2-H3 or H2 peptide. The concentration of radioactive Bax was in nM range as estimated before(63). The radioactive Bax bound to Bcl-2ΔTM was detected in Ni2+-bound fractions by SDS-PAGE and phosphor-imaging. The Mr of standards is indicated on the left of the phosphor-image. Data shown are representatives from three independent experiments.
Bax auto-activation regulates MOM permeability
Our data cannot be explained by the two currently dominant models (described in the introduction above) about how Bcl-2 family proteins regulate MOM permeability. In contrast, our data are most consistent with a less prominent model put forward by Shore and colleagues(25) which argues that Bcl-2-like proteins preferentially bind and inhibit conformation-changed Bax-like proteins thereby preventing their auto-activation. In this way Bcl-2 like proteins intercept apoptotic signals initiated by BH3-only proteins. This model is based on the observation that an activated Bak mutant potently induced a conformation change in, and oligomerization of, non-activated Bak. Moreover, Bak auto-activation occurred even in the presence of Bcl-2, but Bcl-2 inhibited membrane permeabilization by preferentially binding to the conformation-changed Bak thereby blocking Bak oligomerization. In our experiments the H2-H3 peptide is sufficient to activate Bax in the presence of membranes. The H2-H3 is part of the Bax that is exposed after Bax is activated by BH3-only proteins or other stimuli(42), therefore, this result suggests that an active Bax could activate other Bax. Also adding additional Bax to the mitochondria that contain Bcl-2 effectively neutralized the inhibitory effect of Bcl-2 (Fig. 2D).
However, direct evidence for Bak or Bax auto-activation is still lacking. For example, in previous experiments Bak was activated using a mutant Bak in which the N-terminal 36 residues were deleted. This mutant may not mimic the native conformation-changed Bak at membranes. Similarly the H2-H3 peptide used in our assays to activate Bax may not mimic the natural Bax activation intermediate. Furthermore the presence of other proteins in cell and mitochondria-based assays precludes an unambiguous demonstration of direct auto-activation of Bak or Bax. Therefore, to test this model more rigorously we examined permeabilization of liposomes by recombinant Bax in vitro.
Bax auto-activation in liposomal membranes
Based on the above analysis, we predicted that the auto-activation of Bax might require two initiating events: activation by the H2-H3 peptide and insertion into the liposomal membrane. To follow the time course of active Bax-dependent membrane permeabilization fluorescent dextran release was monitored to determine when permeabilization starts and thus when the Bax begins to insert into the membrane. As shown in Figure 6A (diamonds), CB-dextrans (Mr ~10 kDa) were gradually released in the presence of Bax and H2-H3 peptide with a plateau reached after 12 hrs. No CB-dextran release was detected for 12 hrs or longer if only the Bax is present (Fig. 6A, squares), indicating that liposomes alone cannot activate Bax to induced permeabilization, consistent with our previous observation(32). As expected addition of Bcl-2ΔTM to the sample delayed dextran release caused by Bax and peptide (Fig. 6A, triangles).
To separate the membrane-bound active Bax from soluble monomeric Bax and inactive Bax aggregates we took advantage of the low density of proteoliposomes to isolate them by sucrose gradient centrifugation. In this assay the liposomes and molecules entrapped in or bound to the liposomes float up to the top fraction of sucrose gradients (32). As expected Bax protein was recovered with the liposomes in the top fractions of the gradient only after Bax was incubated with the liposomes and H2-H3 peptide for 40 minutes (compare the S1 fractions in the top two panels of Fig. 6B). At this time point dextran release is just beginning (Fig. 6A). As expected, Bcl-2ΔTM inhibited Bax binding to the membrane suggesting that one function of Bcl-2 is to inhibit Bax-membrane interaction. Unlike membrane bound Bax, the H2-H3 peptide does not remain bound to either the liposomes or membrane bound Bax since the CF-labeled H2-H3 peptide used as a tracer did not co-fractionate with Bax proteoliposomes (Fig. 6C, S1 fraction). The liposomes isolated from the top fractions of the gradient containing activated Bax (but not H2-H3 peptide) could be used to test whether membrane-bound Bax can activate recombinant monomeric Bax since they still contain fluorescent dextrans. Only ~10% of the entrapped CB-dextrans were release from the liposomes during the 40-min incubation (Fig. 6A) and although it is possible more dextrans were lost during the 3-hr centrifugation, there was still ample fluorescent signal entrapped in the isolated liposomes to monitor the release induced by added monomeric Bax (Fig. 6D, squares). In contrast release of CB-dextran was not detected for the control liposomes without bound Bax that were isolated from samples prepared from the 40-min incubation of Bax with liposomes but without the peptide (Fig. 6E, squares). Comparison of the data in Figures 6D and 6E clearly indicates that added monomeric Bax induced dextran release only from the liposomes with bound Bax. Control experiment in which buffer was added instead of recombinant Bax demonstrated that dextran release required the new monomeric Bax (data not shown). Dextran release was not due to further activation of the Bax proteins that were already bound to the liposomes as addition of H3-H3 peptide to liposomes with bound Bax (Fig. 6D, circles) did not result in liposome permeabilization. Addition of Bcl-2ΔTM inhibited the activation of monomeric Bax by membrane-bound Bax (Fig. 6D, triangles) suggesting that other function of Bcl-2 is to inhibit auto-activation of Bax.
Model and discussion
The data presented in this study support the following model. The dominant interactions among Bcl-2 family proteins after the initial activation of monomeric Bax by some apoptotic factors, e.g., BH3-only proteins, are Bax/Bax homotypic interaction and Bax/Bcl-2 heterotypic interaction. The former activates other monomeric Bax, triggering a cascade of Bax auto-activation. Sufficient numbers of activated Bax protein homo-oligomerize in the MOM permeabilizing the membrane to release cytochrome c and perhaps other pro-apoptosis proteins from mitochondria. When the ratio of Bcl-2 to active Bax is high at the MOM, Bcl-2 binds to active Bax preventing Bax auto-activation. When the ratio is low, the homo-association of Bax proteins becomes dominant leading to a full-scale Bax auto-activation and MOM permeabilization. BH3-only proteins may play some role during this post-Bax (initial) activation event, e.g., binding to Bcl-2 that may either prevent Bcl-2 from interacting with active Bax or release active Bax from Bcl-2, but they are not required.
It has been established that certain BH3-only proteins can activate Bax via their exposed BH3 motif. The H2 peptide as a bone fide BH3 peptide should function as some BH3-only protein in vitro. However, our results suggest that a longer H2-H3 peptide is required for activation of Bax in vitro. The H2 peptide is sufficient to displace Bax from Bcl-2 and therefore in the presence of some other stimulus may augment membrane permeabilization. We speculate that the H2-H3 peptide mimics Bax auto-activation. We reported previously that an interaction with the membrane surface triggers a conformational change in Bax that is required for BH3-only protein induced Bax oligomerization and membrane permeabilization(32). Consistent with this suggestion, our preliminary experiments indicate that the H2-H3 peptide only interacts with Bax in presence of liposomes (C.T. and J.L., unpublished data). Since the membrane-triggered conformational change in Bax is reversible and transient, it is conceivable that the first pool of Bax proteins being activated by BH3-only proteins or peptides is that loosely associated with mitochondria or endoplasmic reticulum. Such a pool of Bax proteins have been found in cells(52). This scenario also fits with the finding that BH3-only proteins such as tBid and Bim are translocated from cytosol to mitochondria during the initial phase of apoptosis induction, and insertion of the BH3-only proteins into the MOM may facilitate their interaction with Bax or Bak(18,28,29,53–56).
What happens next after the initial Bax activation? The active Bax proteins may form multi-spanning monomers in MOM that oligomerize to permeabilize the membrane as we demonstrated recently(7). The oligomerization step requires multiple Bax proteins co-localizing to a membrane site, a task that may not be accomplished by the initial Bax activation. Additional Bax activations may be needed generating the continuous movement of Bax from cytosol to mitochondria observed during apoptotic induction. Release of cytochrome c and other mitochondrial proteins remains undetectable until a threshold has passed, hours or days after the initial Bax translocation(57–60). Thus, the secondary Bax translocation may be a thermo-diffusion event that functions to replenish the empty pool of Bax that previously exists near the mitochondria, or an unknown secondary activation event. The newly arrived Bax proteins may be (further) activated by the mitochondria-bound tBid or Bim proteins, just like their predecessors. Alternatively or in addition, they may be activated by the previously-activated Bax. The latter is likely to occur during apoptosis as suggested by our finding that monomeric Bax can be activated by the H2-H3 peptide of Bax in the mitochondrial system and by the membrane-bound active Bax in the MOM-mimicking liposomal system. Interestingly in the multi-spanning monomeric Bax that we discovered recently as a Bax activation intermediate, both H2 and H3 sequences are predicted to be above the mitochondrial surface, available for interaction with another Bax(7). Consistent with this prediction, the liposome-bound Bax proteins that have the capacity to activate monomeric Bax include some monomeric Bax (data not shown). The auto-activation of Bax is not unique since a similar conclusion was reached based on more indirect measurements for Bak, a homolog of Bax that is constitutively bound to mitochondrion but also displays conformation change during apoptosis induction(25). Therefore, our data extend the model put forward by Shore and colleagues, by indicating that auto-activation of Bax-like proteins may be a commonly used mechanism to amplify the initial apoptotic signals. Indeed our data suggest that the major target of multi-BH domain pro-apoptotic proteins is multi-BH domain pro-apoptotic proteins. It remains to be determined whether activated Bak can activate cytoplasmic monomeric Bax.
The interaction between Bax and Bcl-2 was originally proposed to determine the cell's fate in the rheostat hypothesis originated by Korsmeyer(22). The time and location of the interaction has been modified after most Bax proteins were discovered to be localized in the cytosol of healthy cells(23,61). The current model is that Bax may interact with Bcl-2 only after an initial activation and the interaction takes place at the mitochondrial or endoplasmic reticular membrane. The function of this interaction is open for debate. One may view the interaction as a means to inhibit the Bax membrane permeabilization or pro-death activity. It can also be argued that it is a way to neutralize the Bcl-2 membrane protection or pro-survival activity. The fate of the Bax/Bcl-2 complex is also unknown. While we have suggested that Bcl-2 may act as a suicide inhibitor by binding to Bax and promoting its assembly into a dead-end complex(33), stable and direct Bax/Bcl-2 interaction in a membrane seems beyond current detection limit. For example, Bax and Bak homo-oligomers have been detected in mitochondrial membranes by chemical crosslinking, yet similar technique has failed to reveal any hetero-complexes formed between these death proteins and their survival counterparts such as Bcl-2 and BclxL(18,31,62). Therefore, the other possibility is that Bax/Bcl-2 complex may be reversible. In this scenario Bcl-2 may function as a competitive inhibitor by binding to Bax and preventing its assembly into a homo-complex. Our results that Bcl-2 can block small but not large numbers of active Bax-mediated membrane permeabilization (Fig. 2 and 3) is predicted by both models. That the Bcl-2 blocking is only temporary (Fig. 6A) seems to support a competitive binding model. But we noted that some (albeit very few) Bax molecules were localized in the membranes together with Bcl-2 after a short incubation (Fig. 6B, bottom two panels), and more Bax and Bcl-2 were inserted into the membrane as the incubation went longer (data not shown), suggesting that these Bax and Bcl-2 molecules may form some stable complexes in the membrane. Whether or not this is true, and if this is true, what is the function of these Bax/Bcl-2 complexes will be some of the most interesting subjects to investigate further.
Supplementary Material
Acknowledgements
We thank Arthur Johnson (Texas A&M University) for valuable suggestions to design fluorescence experiments, Paul Bock (Vanderbilt University) for SCIENTIST program to fit the anisotropy data, and Kathleen Kinnally (New York University) for H2-H3 peptide to test the binding to Bcl-2.
This work was supported by NIH grant GM062964 (to JL) and CIHR grant FRN 12517 (to DWA). DWA holds the Canada Research Chair in Membrane Biogenesis.
Footnotes
The abbreviations used are: CB, Cascade Blue; CF, carboxyfluorescein; Bcl-2ΔTM, Bcl-2 lacking the C-terminal transmembrane sequence; BH, Bcl-2 homology; MOM, mitochondrial outer membrane; Mr, relative molecular mass; S.D., standard deviation; TM, transmembrane.
REFERENCES
- 1.Danial NN, Korsmeyer SJ. Cell. 2004;116:205–219. doi: 10.1016/s0092-8674(04)00046-7. [DOI] [PubMed] [Google Scholar]
- 2.Green DR, Kroemer G. Science. 2004;305:626–629. doi: 10.1126/science.1099320. [DOI] [PubMed] [Google Scholar]
- 3.Wei MC, Zong WX, Cheng EH, Lindsten T, Panoutsakopoulou V, Ross AJ, Roth KA, MacGregor GR, Thompson CB, Korsmeyer SJ. Science. 2001;292:727–730. doi: 10.1126/science.1059108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zong W-X, Lindsten T, Ross AJ, MacGregor GR, Thompson CB. Genes Dev. 2001;15:1481–1486. doi: 10.1101/gad.897601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharpe JC, Arnoult D, Youle RJ. Biochim. Biophys. Acta. 2004;1644:107–113. doi: 10.1016/j.bbamcr.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 6.Lucken-Ardjomande S, Martinou J-C. J. Cell Sci. 2005;118:473–483. doi: 10.1242/jcs.01654. [DOI] [PubMed] [Google Scholar]
- 7.Annis MG, Soucie EL, Dlugosz PJ, Cruz-Aguado JA, Penn LZ, Leber B, Andrews DW. EMBO J. 2005;24:2096–2103. doi: 10.1038/sj.emboj.7600675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang E, Zha J, Jockel J, Boise LH, Thompson CB, Korsmeyer SJ. Cell. 1995;80:285–291. doi: 10.1016/0092-8674(95)90411-5. [DOI] [PubMed] [Google Scholar]
- 9.Cheng EH, Wei MC, Weiler S, Flavell JK, Mak TW, Lindsten T, Korsmeyer SJ. Mol. Cell. 2001;8:705–711. doi: 10.1016/s1097-2765(01)00320-3. [DOI] [PubMed] [Google Scholar]
- 10.Letai A, Bassik MC, Walensky LD, Sorcinelli MD, Weiler S, Korsmeyer SJ. Cancer Cell. 2002;2:183–192. doi: 10.1016/s1535-6108(02)00127-7. [DOI] [PubMed] [Google Scholar]
- 11.Wilson-Annan J, O'Reilly LA, Crawford SA, Hausmann G, Beaumont JG, Parma LP, Chen L, Lackmann M, Lithgow T, Hinds MG, Day CL, Adams JM, Huang DCS. J. Cell Biol. 2003;162:877–887. doi: 10.1083/jcb.200302144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen L, Willis SN, Wei A, Smith BJ, Fletcher JI, Hinds MG, Colman PM, Day CL, Adams JM, Huang DCS. Mol. Cell. 2005;17:393–403. doi: 10.1016/j.molcel.2004.12.030. [DOI] [PubMed] [Google Scholar]
- 13.Willis SN, Chen L, Dewson G, Wei A, Naik E, Fletcher JI, Adams JM, Huang DCS. Genes Dev. 2005;19:1294–1305. doi: 10.1101/gad.1304105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang K, Yin X-M, Chao DT, Milliman CL, Korsmeyer SJ. Genes Dev. 1996;10:2859–2869. doi: 10.1101/gad.10.22.2859. [DOI] [PubMed] [Google Scholar]
- 15.Desagher S, Osen-Sand A, Nichols A, Eskes R, Montessuit S, Lauper S, Maundrell K, Antonsson B, Martinou JC. J. Cell Biol. 1999;144(5):891–901. doi: 10.1083/jcb.144.5.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eskes R, Desagher S, Antonsson B, Martinou JC. Mol. Cell. Biol. 2000;20(3):929–935. doi: 10.1128/mcb.20.3.929-935.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perez D, White E. Mol. Cell. 2000;6:53–63. [PubMed] [Google Scholar]
- 18.Wei MC, Lindsten T, Mootha VK, Weiler S, Gross A, Ashiya M, Thompson CB, Korsmeyer SJ. Genes Dev. 2000;14:2060–2071. [PMC free article] [PubMed] [Google Scholar]
- 19.Kuwana T, Mackey MR, Perkins G, Ellisman MH, Latterich M, Schneiter R, Green DR, Newmeyer DD. Cell. 2002;111:331–342. doi: 10.1016/s0092-8674(02)01036-x. [DOI] [PubMed] [Google Scholar]
- 20.Cartron P-F, Gallenne T, Bougras G, Gautier F, Manero F, Vusio P, Meflah K, Vallette FM, Juin P. Mol. Cell. 2004;16:807–818. doi: 10.1016/j.molcel.2004.10.028. [DOI] [PubMed] [Google Scholar]
- 21.Kuwana T, Bouchier-Hayes L, Chipuk JE, Bonzon C, Sullivan BA, Green DR, Newmeyer DD. Mol. Cell. 2005;17:525–535. doi: 10.1016/j.molcel.2005.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Oltvai ZN, Milliman CL, Korsmeyer SJ. Cell. 1993;74(4):609–619. doi: 10.1016/0092-8674(93)90509-o. [DOI] [PubMed] [Google Scholar]
- 23.Hsu YT, Wolter KG, Youle RJ. Proc. Nat. Acad. Sci. USA. 1997;94:3668–3672. doi: 10.1073/pnas.94.8.3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuconati A, Degenhardt K, Sundararajan R, Anschel A, White E. J. Virol. 2002;76:4547–4558. doi: 10.1128/JVI.76.9.4547-4558.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruffolo S, Shore GC. J. Biol. Chem. 2003;278:25039–25045. doi: 10.1074/jbc.M302930200. [DOI] [PubMed] [Google Scholar]
- 26.Zhang Z, Lapolla SM, Annis MG, Truscott M, Roberts GJ, Miao Y, Shao Y, Tan C, Peng J, Johnson AE, Zhang XC, Andrews DW, Lin J. J. Biol. Chem. 2004;279:43920–43928. doi: 10.1074/jbc.M406412200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Petros AM, Nettesheim DG, Wang Y, Olejnicizak ET, Meadows RP, Mack J, Swift K, Matayoshi ED, Zhang H, Thompson CB, Fesik SW. Protein Sci. 2000;9:2528–2534. doi: 10.1110/ps.9.12.2528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu X, Dai S, Zhu Y, Marrack P, Kappler JW. Immunity. 2003;19:341–352. doi: 10.1016/s1074-7613(03)00234-6. [DOI] [PubMed] [Google Scholar]
- 29.Harada H, Quearry B, Ruiz-Vela A, Korsmeyer SJ. Proc. Natl. Acad. Sci. USA. 2004;101:15313–15317. doi: 10.1073/pnas.0406837101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Antonsson B, Montessuit S, Sanchez B, Martinou JC. J. Biol. Chem. 2001;276:11615–11623. doi: 10.1074/jbc.M010810200. [DOI] [PubMed] [Google Scholar]
- 31.Mikhailov V, Mikhailova M, Pulkrabek DJ, Dong Z, Venkatachalam MA, Saikumar P. J. Biol. Chem. 2001;276:18361–18374. doi: 10.1074/jbc.M100655200. [DOI] [PubMed] [Google Scholar]
- 32.Yethon JA, Epand RF, Leber B, Epand RM, Andrews DW. J. Biol. Chem. 2003;278:48935–48941. doi: 10.1074/jbc.M306289200. [DOI] [PubMed] [Google Scholar]
- 33.Kim PK, Annis MG, Dlugosz PJ, Leber B, Andrews DW. Mol. Cell. 2004;14:523–529. doi: 10.1016/s1097-2765(04)00263-1. [DOI] [PubMed] [Google Scholar]
- 34.Heuck AP, Tweten RK, Johnson AE. J. Biol. Chem. 2003;278:31218–31225. doi: 10.1074/jbc.M303151200. [DOI] [PubMed] [Google Scholar]
- 35.Lakowicz JR. Principles of fluorescence spectroscopy. 2nd Ed. Kluwer Academic/Plenum Publishers; New York: 1999. [Google Scholar]
- 36.Bock PE, Olson ST, Bjork I. J. Biol. Chem. 1997;272:19837–19845. doi: 10.1074/jbc.272.32.19837. [DOI] [PubMed] [Google Scholar]
- 37.Kim KM, Giedt CD, Basanez G, O'Neil JW, Hill JJ, Han Y-H, Tzung S-P, Zimmerberg J, Hockenbery DM, Zhang KYJ. Biochemistry. 2001;40:4911–4922. doi: 10.1021/bi002368e. [DOI] [PubMed] [Google Scholar]
- 38.Wang K, Gross A, Waksman G, Korsmeyer SJ. Mol. Cell. Biol. 1998;18:6083–6089. doi: 10.1128/mcb.18.10.6083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polster BM, Kinnally KW, Fiskum G. J. Biol. Chem. 2001;276:37887–37894. doi: 10.1074/jbc.M104552200. [DOI] [PubMed] [Google Scholar]
- 40.Polster BM, Robertson CL, Bucci CJ, Suzuki M, Fiskum G. Cell Death Differ. 2003;10:365–370. doi: 10.1038/sj.cdd.4401158. [DOI] [PubMed] [Google Scholar]
- 41.Moreau C, Cartron P-F, Hunt A, Meflah K, Green DR, Evan G, Vallette FM, Juin P. J. Biol. Chem. 2003;278:19426–19435. doi: 10.1074/jbc.M209472200. [DOI] [PubMed] [Google Scholar]
- 42.Cartron P-F, Arokium H, Oliver L, Meflah K, Manon S, Vallette FM. J. Biol. Chem. 2005;280:10587–10598. doi: 10.1074/jbc.M409714200. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki M, Youle RJ, Tjandra N. Cell. 2000;103:645–654. doi: 10.1016/s0092-8674(00)00167-7. [DOI] [PubMed] [Google Scholar]
- 44.Shangary S, Johnson DE. Biochemistry. 2002;41:9485–9495. doi: 10.1021/bi025605h. [DOI] [PubMed] [Google Scholar]
- 45.Diaz J-L, Oltersdorf T, Horne W, McConnell M, Wilson G, Weeks S, Garcia T, Fritz LC. J. Biol. Chem. 1997;272:11350–11355. doi: 10.1074/jbc.272.17.11350. [DOI] [PubMed] [Google Scholar]
- 46.Borner C, Martinou I, Mattmann C, Irmler M, Schaerer E, Martinou J-C, Tschopp J. J. Cell Biol. 1994;126:1059–1068. doi: 10.1083/jcb.126.4.1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alnemri ES, Robertson NM, Fernandes TF, Croce CM, Litwack G. Proc. Natl. Acad. Sci. USA. 1992;89:7295–7299. doi: 10.1073/pnas.89.16.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tanaka S, Saito K, Reed JC. J. Biol. Chem. 1993;268:10920–10926. [PubMed] [Google Scholar]
- 49.Petros AM, Medek A, Nettesheim DG, Kim DH, Yoon HS, Swift K, Matayoshi ED, Oltersdorf T, Fesik SW. Proc. Natl. Acad. Sci. USA. 2001;98:3012–3017. doi: 10.1073/pnas.041619798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sattler M, Liang H, Nettesheim D, Meadows RP, Harlan JE, Eberstadt M, Yoon HS, Shuker SB, Chang BS, Minn AJ, Thompson CB, Fesik SW. Science. 1997;275(5302):983–986. doi: 10.1126/science.275.5302.983. [DOI] [PubMed] [Google Scholar]
- 51.Hsu YT, Youle RJ. J. Biol. Chem. 1997;272(21):13829–13834. doi: 10.1074/jbc.272.21.13829. [DOI] [PubMed] [Google Scholar]
- 52.Goping IS, Gross A, Lavoie JN, Nguyen M, Jemmerson R, Roth K, Korsmeyer SJ, Shore GC. J. Cell Biol. 1998;143(1):207–215. doi: 10.1083/jcb.143.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li H, Zhu H, Xu CJ, Yuan J. Cell. 1998;94(4):491–501. doi: 10.1016/s0092-8674(00)81590-1. [DOI] [PubMed] [Google Scholar]
- 54.Luo X, Budihardjo I, Zou H, Slaughter C, Wang X. Cell. 1998;94(4):481–490. doi: 10.1016/s0092-8674(00)81589-5. [DOI] [PubMed] [Google Scholar]
- 55.Puthalakath H, Huang DC, O'Reilly LA, King SM, Strasser A. Mol. Cell. 1999;3:287–296. doi: 10.1016/s1097-2765(00)80456-6. [DOI] [PubMed] [Google Scholar]
- 56.Oh KJ, Barbuto S, Meyer N, Kim R-S, Collier RJ, Korsmeyer SJ. J. Biol. Chem. 2005;280:753–767. doi: 10.1074/jbc.M405428200. [DOI] [PubMed] [Google Scholar]
- 57.Goldstein JC, Waterhouse NJ, Juin P, Evan GI, Green DR. Nat. Cell. Biol. 2000;2:156–162. doi: 10.1038/35004029. [DOI] [PubMed] [Google Scholar]
- 58.Valentijn AJ, Metcalfe AD, Kott J, Streuli CH, Gilmore AP. J. Cell Biol. 2003;162:599–612. doi: 10.1083/jcb.200302154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rehm M, Dubmann K, Prehn JHM. J. Cell Biol. 2003;162:1031–1043. doi: 10.1083/jcb.200303123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Madesh MB, Antonsson SM, Srinivasula ES, Alnemri ES, Hajnoczky G. J. Biol. Chem. 2002;277:5651–5659. doi: 10.1074/jbc.M108171200. [DOI] [PubMed] [Google Scholar]
- 61.Wolter KG, Hsu KT, Smith CL, Nechushtan A, Xi XG, Youle RS. J. Cell Biol. 1997;139:1281–1292. doi: 10.1083/jcb.139.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mikhailov V, Mikhailova M, Degenhardt K, Venkatachalam MA, White E, Saikumar P. J. Biol. Chem. 2003;278:5367–5376. doi: 10.1074/jbc.M203392200. [DOI] [PubMed] [Google Scholar]
- 63.Kim PK, Janiak-Spens F, Trimble WS, Leber B, Andrews DW. Biochemistry. 1997;36(29):8873–8882. doi: 10.1021/bi970090t. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.