Abstract
Retinal membrane guanylyl cyclase (retGC) and Ca2+/Mg2+ -sensor proteins, GCAPs1, control the recovery of the photoresponse in vertebrate photoreceptors, through their molecular interactions that remain rather poorly understood and controversial. Here we have determined the main retGC isozyme (RetGC1):GCAP1 binding stoichiometry at saturation in cyto, using fluorescently labeled RetGC1 and GCAP1 co-expressed in HEK293 cells. In a striking manner, the equimolar binding of RetGC1 with GCAP1 in transfected HEK293 cells typical for the wild type RetGC1 was eliminated by a substitution, D639Y, in the kinase homology domain of RetGC1 found in a patient with a severe form of retinal dystrophy, Leber congenital amaurosis (LCA). Similar effect was observed with another LCA-related mutation, R768W, in the same domain of RetGC1. In contrast to the completely suppressed binding and activation of RetGC1 by Mg2+-liganded GCAP1, neither of these two mutations eliminated the GCAP1-independent activity of retGC stimulated by Mn2+. These results directly implicate the D639 (and possibly R768) -containing portion of the RetGC1 kinase homology domain in its primary recognition by the Mg2+-bound activator form of GCAP1.
Keywords: GCAP1, calcium-binding protein, guanylyl cyclase, retGC, retina, protein fluorescence, binding stoichiometry, Leber congenital amaurosis
Ca2+-sensitive membrane guanylyl cyclase (retGC) plays a key role in regulation of the photoresponse in a vertebrate retina (reviewed in ref. 1). Light absorption by visual pigments in rods and cones triggers activation of the transducin-PDE6 cascade thus reducing free cGMP levels in the outer segment and closing cGMP-gated channels. The interruption of Na+ and Ca2+ influx through the channels consequently causes hyperpolarization of the photoreceptors plasma membrane and interrupts the release of neuromediator from their synaptic terminals. Rods and cones recover from excitation and regain their light-sensitivity by both inactivating the PDE6 cascade and re-synthesizing cGMP by RetGC; the latter reaction is controlled via a Ca2+ feedback mechanism (2, 3) by Ca2+/Mg2+ binding proteins, GCAPs (guanylyl cyclase activating proteins) (4 – 7). Because Ca2+ influx stops in the light, its concentration in the outer segment falls nearly 10-fold (8, 9). In response to this change, GCAPs convert from their Ca2+-bound (inhibitor) state to the Mg2+-bound (activator) state and boost catalytic activity of retGC (10, 11). Two retGC isozymes, RetGC1 and retGC2 (12 –14) have been found in rods and cones. While RetGC1 is the main GCAP-regulated isozyme (6, 15) and is essential for the survival of cones (13), both isozymes are required for the normal photoresponse in rods (14). Two isoforms of GCAPs (GCAP1 and GCAP2), ubiquitous among vertebrates (5, 6, 16, 17), together provide the normal recovery speed in mouse rods (3, 11, 18), but additional GCAP isoforms have also been found in a variety of vertebrate species (19, 20). Despite significant progress in understanding how GCAPs mediate the Ca2+ feedback loop of retGC activity (11, 19), some key intricate processes involved in the GCAP/retGC interaction remain unclear, including the stoichiometry of the GCAP/retGC complexes and the interaction between amino acid residues in retGC and GCAPs responsible for the cyclase activation.
A number of mutations in RetGC1 that alter its activity and/or regulation by GCAPs have been linked to blinding disorders in humans. These mutations can be divided in two major types – the loss-of-function mutations that result in absence of RetGC1 activity and are frequent among patients with Leber Congenital Amaurosis (LCA) (21 – 26) and gain-of-function mutations associated with congenital dominant cone-rod dystrophy, CORD6 (27, 28) that decrease Ca2+ sensitivity of GCAP/RetGC1 complex, thus possibly leading to an abnormal increase of cGMP production in photoreceptors in the dark (29 – 31). Disease-related mutations that alter interactions of retGC and GCAPs can help not only explain the cause of congenital blindness, but also elucidate the mechanisms of GCAP/retGC interactions. In this paper, we present evidence that the stoichiometry of a GCAP1/RetGC1 complex produced in HEK293 cells is 1:1 and describe that two new mutations found in patients with LCA completely prevent formation of the RetGC1/GCAP1 complex by blocking GCAP1 docking on RetGC1.
EXPERIMENTAL PROCEDURES
Recombinant GCAP1
GFP-tagged bovine GCAP1 was expressed in HEK293 cells from a Clontech pQBI25-fN3 vector as described previously (32). Recombinant myristoylated wild type GCAP1 was expressed in E.coli and purified as described previously (33).
Screening for RetGC1 mutations in LCA patients
Three affected individuals whose DNA was analyzed in this study present with typical Leber congenital amaurosis (OMIM #204000). In the case of the D639Y mutation, two affected individuals carrying the mutation were siblings; their mother, who was also heterozygous for the mutation, exhibited subclinical changes in cone ERG, but was otherwise unremarkable. The R768W mutation was a simplex case and no further information is available on the patient or the family. DNA was isolated from peripheral blood samples obtained with informed consent by Dr. Jeremy Nathans (Johns Hopkins University, Baltimore, MD, USA) and Dr. Benedetto Falsini (Università del Cattolica Sacro Cuore, Rome, Italy).
PCR Optimization with Epicentre FailSafe system
Amplification reactions were optimized with FailSafe PCR 2X PreMixes A–L (Epicentre Technologies). Amplification reactions for each exon of the GC1 gene were done using the manufacturer’s protocol in 25 ul reaction volumes for 35 cycles with an initial denaturation step of 5 min. at 94°C, and a final extension for 7 min. at 72°C. Failsafe buffers F for exon 12, and buffer I for exon 9 were used. Annealing temperature was 55°C for exon 9 and 60°C for exon 12. Amplification of exon 9 was done with forward primer 5’-CCCACATTGCCCTGGGCAGA, reverse primer 5’-CCTGCCCCCAGGACGTCACC generating a 204 bp product; for exon 12 forward primer 5’-GGCAGCCTTTGTGTTCTGGG, reverse primer 5’-GTTGCTGACAAGCATTTGGG generating a 267 bp product.
Denaturing High Pressure Liquid Chromatography
Mutation detection was carried out using the Transgenomic WAVE DNA Fragment Analysis System. For run parameter optimization, each PCR fragment (200 ul of unpurified “wild type” PCR product) was denatured for 5 minutes at 95°C and gradually reannealed by decreasing sample temperature from 95 to 25°C over a period of 45 minutes. The reannealed products were analyzed by injecting 5 ul on a linear gradient of 65% buffer A (0.1 M TEAA, pH 7.0) and 35% buffer B (0.1 M TEAA, 25% acetonitrile) with a 2% change in slope per min. and total acquisition time of 18.8 min at a flow rate of 0.9 ml/min at 50°C. From retention time an optimized gradient was calculated to set peak elution at around 6 to 6.5 min. Additionally, three ul of PCR product was repeatedly injected for temperature titration over the range from 50 to 70°C using the optimized gradient. The optimized gradient ranged from 46 % – 57% buffer B with total acquisition time of 10 minutes and melting temperatures ranging from 60 – 68°C for all GC1 exons.
DNA Sequencing of PCR products
PCR products that displayed an altered migration were cycle sequenced using Sequitherm Excel II with end-labeled primers and analyzed on a Genomyx LR DNA Sequencer (Beckman Coulter). All products were sequenced on both DNA strands and more than one independent PCR product was sequenced for confirmation.
Generation of plasmids carrying engineered RetGC1 alleles
A pRCCMV plasmid carrying WT RetGC1 (34) was mutated using the Stratagene QuikChange Site-Directed Mutagenesis kit according to the manufacturer’s protocol. Plasmid sequence was verified on both strands for the entire GC1 cDNA contained in the plasmid.
Recombinant dsRed-tagged RetGC1 and dsRed-RetGC1-GCAP1GFP chimera
Fluorescently labeled RetGC1 was produced by inserting in RetGC1 cDNA a DNA fragment coding for a monomeric variant of red fluorescent protein (m-DsRed, Clontech), as described previously (32). The resultant construct, dsRed-RetGC1 pRCCMV, was used to express functional fluorescently tagged RetGC1 in HEK293 cells. To produce dsRed-RetGC1-GCAP1GFP "chimera", the GCAP1GFP sequence was PCR-amplified by PFU polymerase (Stratagene) from a DNA clone used for expression of GCAP1GFP in HEK293 cells (32) using a forward primer, 5'-AAAAAACGGCCGATGGGGAACATTATGAGCGGTAAGTCG-3', and a reverse primer, 5'-GCCAACGCGTCTAGATAGTCAATCGATG-3', thus adding EagI and XbaI restriction sites at the ends of the PCR product. The PCR product was then inserted into the EagI/XbaI sites of the dsRed-RetGC1 pRCCMV construct. All constructs were verified by automated DNA sequencing. Recombinant RetGC1 was expressed in HEK293 cells from a modified pRCCMV vector (Stratagene), as previously described (10).
Antibodies
Anti-RetGC1 antibodies were produced in rabbits against large recombinant fragments of human RetGC1 (34), Met747-Ser1052 (antibody RetGC1Cat) or Arg540-Asn815 (antibody RetGC1KHD), expressed in E. coli from pET15b vector (Novagen/Calbiochem). The IgG fraction was purified using Protein A-Sepharose (GE Healthcare). Anti-GCAP1 polyclonal antibody was produced as described in (32). Commercial antibodies against c-myc tag, dsRed, and GFP were purchased from Millipore, Clontech, and Invitrogen, respectively. Secondary peroxidase-conjugated antibodies were from Pierce/Thermo Scientific and Alexa Fluor 568 goat anti-rabbit antibody from Molecular Probes/Invitrogen.
Expression of RetGC1 in HEK293 cells
HEK293 cells were grown at 37°C, 5% CO2, in high-glucose Dulbecco's modified Eagle medium (DMEM, Invitrogen) supplemented with 10% fetal bovine serum (Invitrogen). To express RetGC1 and its mutants for the functional assay in vitro, HEK293 cells were transfected with 40 µg per 100-mm culture dish of pRCCMV plasmid containing wild type or mutant RetGC1 using the Ca2+-phosphate method (a Promega Profection protocol), and the membranes were harvested as previously described (31). In co-expression experiments, HEK293 cells were typically transfected with 40 µg/dish of an equimolar mixture of pRCCMV plasmids containing ΔRetGC1 and mutant RetGC1 using the same protocol. The expression level of each protein was determined by immunoblot using the rabbit polyclonal anti-RetGC1Cat antibody.
Co-expression of RetGC1 and GCAP1 in HEK 293 cells and confocal laser scanning microscopy
Cells were grown in standard glass cover-slip chambers (four 2-cm2 chambers per slide) in Dulbecco's modified Eagle medium supplemented with 10% fetal bovine serum, and were transfected with a mixture of expression constructs using the Ca2+-phosphate method. Routinely, a mixture of 3 µg of pRCCMV plasmid containing wild type or mutant RetGC1 cDNA and 0.02 µg of the GCAP1-GFP pQBI25fN3 plasmid (32) was used per chamber. In 24–32 hours we either directly viewed the live cells or fixed them with freshly prepared 4% paraformaldehyde in a standard Tris-buffered saline (TBS) at room temperature for subsequent immunostaining (32). Where indicated, the DNA in nuclei of the cells was counterstained for 10 min with 1 mM TO-PRO-3 iodide (Invitrogen) containing 20 µg/ml RNAse A added to the first wash following the incubation with the secondary antibody and then washed as described above.
The cells were viewed using an inverted Olympus IX81 microscope/FV1000 Spectral laser confocal system, and images were collected and analyzed using Olympus FluoView FV10-ASW software.
To determine the stoichiometry of GCAP1 binding to RetGC1 HEK293 the cells in the neighboring chambers of the same coverslip were transfected with 3 µg of pRCCMV plasmid containing monomeric dsRed-RetGC1-GCAP1GFP "chimera" cDNA or a mixture of 3 µg of dsRed-RetGC1 construct and 0.02 to 0.6 µg of the GCAP1-GFP pQBI25fN3. All cells were viewed and fluorescence intensity profiles were recorded at the same laser excitation and photomultiplier gain settings.
Immunoblotting and immunoprecipitation
HEK293 membrane fractions were washed in TBS, dissolved in a Laemmli SDS PAGE sample buffer, and aliquots were separated by 4–12% PAGE (Invitrogen) or 7% PAGE. Following electroblotting on Immobilon P membrane (Millipore), proteins were probed with antibodies and developed using a Pierce Femto Supersignal luminescent peroxidase substrate, according to the manufacturer’s protocol. The signal intensity was quantified by densitometry X-ray film as previously described (32) or using LuminousFX imaging system (FotoDyne, Inc.).
Immunoprecipitation
Ca. 30 µl of each membrane fraction containing expressed RetGC1 mutants was dissolved for 15 min at room temperature in 200 µl of Extraction Buffer (20 mM TrisHCl (pH 7.5), 1.5 % Triton X-100, 500 mM KCl, 10 mM NaCl, 1 mM Mg Cl2) supplemented with 0.5 mg/ml bovine serum albumin and centrifuged at 10,000 rpm × 5 min. The supernatant was collected, mixed with 5 µg of anti-Myc IgG, incubated for 30 min, and then mixed with 100 µl of protein A-Sepharose suspension equilibrated with Extraction Buffer (~20 µl set bed volume), and incubated for additional 15 min with constant agitation. The beads were then centrifuged at 2000 rpm × 2 min, washed in 1 ml of Extraction Buffer, collected at 2000 rpm × 2 min, and washed once in 50 mM Tris (pH6.8) containing 1 mM MgCl2. The beads collected by centrifugation were resuspend in 80 µl of 2× Laemmly SDS sample buffer and removed by centrifugation at 10,000 rpm ×1 min. The supernatant was analyzed by electrophoresis in 7% PAAG-SDS and immunoblotting using RetGC1Cat antibody.
Guanylyl cyclase activity
RetGC activity was assayed as previously described (10, 32). Briefly, the assay mixture (25 µl) contained 30 mM MOPS/KOH, pH 7.2, 60 mM KCl, 4 mM NaCl, 1 mM DTT, 1 mM free Mg2+, 2 mM Ca/EGTA buffer, 0.3 mM ATP, 4 mM cGMP, 1 mM GTP, 4 mM creatine phosphate, 0.5 U creatine phosphokinase, 1 µCi of [α-32P] GTP, 0.1 µCi of [8- 3H] cGMP, GCAP1 and HEK 293 cell membranes. The reaction mixture was incubated for 40 min at 30°C, stopped by heating for 2.5 min at 95°C, and aliquots were analyzed by TLC on fluorescent plastic-backed polyethylenimine cellulose plates (E.Merck) as described previously (10, 35). The data shown are representative from 3–4 independent experiments producing virtually identical results.
RESULTS AND DISCUSSION
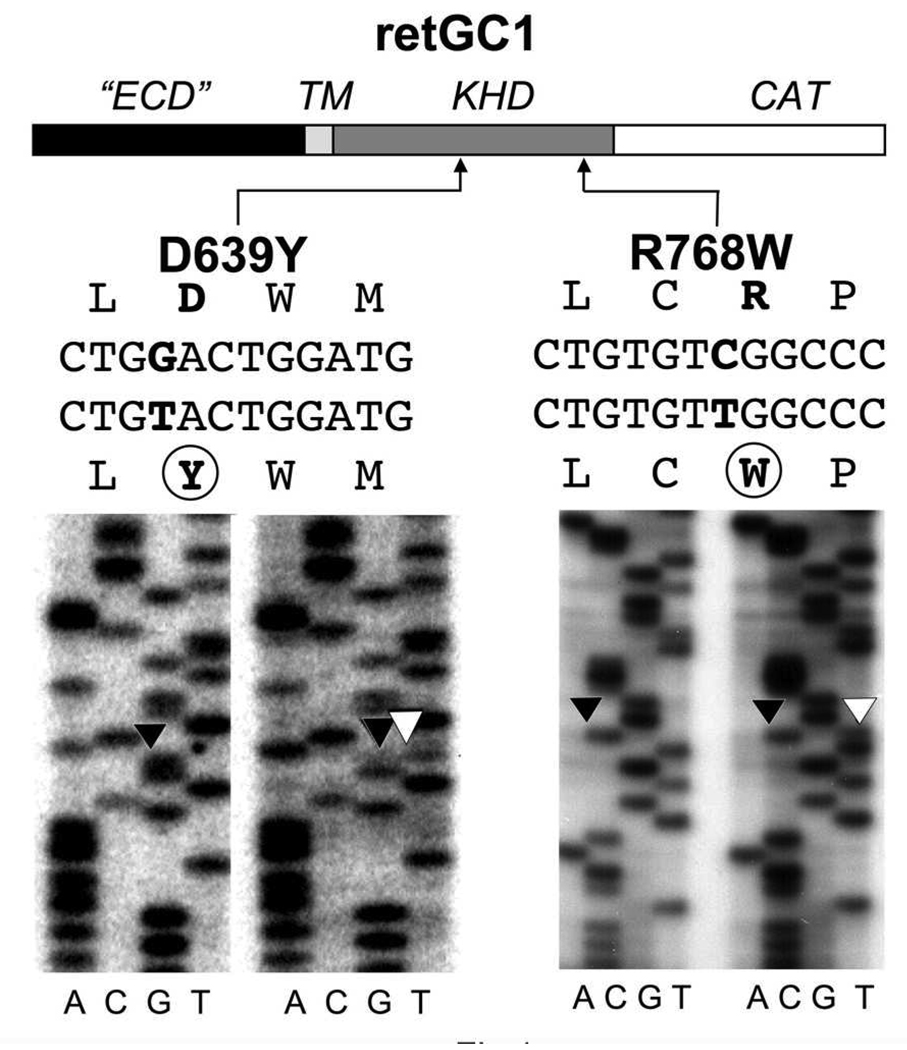
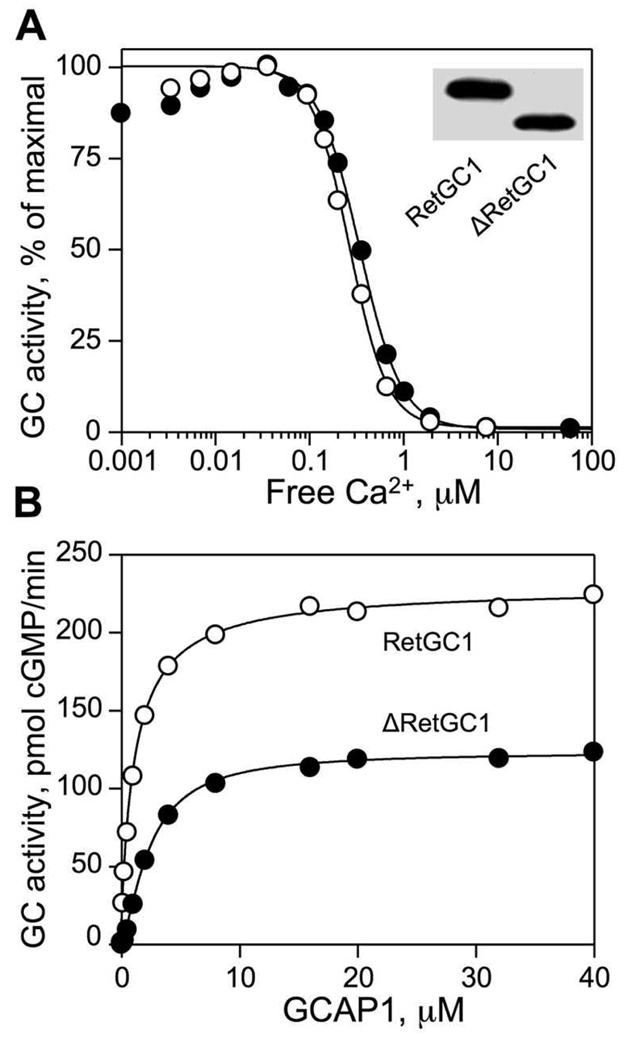
Mutations in the chromosome 17p13.1 GUCY2D locus encoding RetGC1 have been frequently found in patients with LCA (21, 22, 24, 25, 36). Therefore, we verified the sequence of RetGC1 in new LCA patients as a likely link to the onset of the disease. Screening results in DNA of two LCA patients revealed two mutations; D639Y, a novel sequence change, and R768W (37), both in the kinase-homology domain (KHD) of RetGC1 (Fig. 1). In each case, however, each mutant allele was heterozygous both in patients and in one of the parents (see Methods). This finding seemingly reduced the likelihood of a direct role of the two mutants alone in the development of the retinal degeneration, because the LCA symptoms evident in each patient were not observed in their parents. Yet, one parent carrier (D639Y) was shown to exhibit subclinical cone ERG changes and ERG changes have been reported previously in other LCA carriers (38). Thus, the possibility remained that the mutant RetGC1 could contribute to the LCA through additional, presently unclear, genetic interaction(s), if its normal function was altered by the mutations. Therefore, we tested the activity and the regulation of the mutant RetGC1 by GCAPs for any evidence for such abnormalities. We expressed the D639Y and the R768W RetGC1 mutants in HEK293 cells and tested their activation by purified recombinant GCAP1 in vitro (Fig. 2). As controls, we used wild type recombinant human RetGC1 and another RetGC1 mutant, R838S, that is linked to dominant cone-rod degeneration (27, 30) through increased affinity for Mg2+-bound GCAP1 (31) causing abnormal Ca2+ sensitivity of RetGC1. All variants of RetGC1 used in this assay were present in similar quantities (Fig. 2C) equalized by polyclonal anti-RetGC1Cat antibody staining. The antibody was produced against a large 30 kDa fragment of RetGC1 containing the catalytic domain (34) and therefore would unlikely be sensitive to single point mutations (of which the D639Y mutation was located >100 amino acid residues upstream of the region recognized by the antibody).
FIG. 1. Mutations identified in exon 9 and exon 12 of GUCY2D in LCA patients.
Analysis of exon 12 revealed a C➔T nucleotide change resulting in an R768W mutation. In exon 9 a G➔T nucleotide change results in a D639Y mutation. The sequence changes were not observed in over 100 control samples analyzed. For each sequencing reaction set, the control is on the left and the LCA patient sequence is on the right. The positions of the mutated amino acid residues in the primary structure are indicated in the schematics of the conventional RetGC1 domain map: ECD – extracellular domain; TM – transmembrane domain; KHD – kinase homology domain; CAT – catalytic domain.
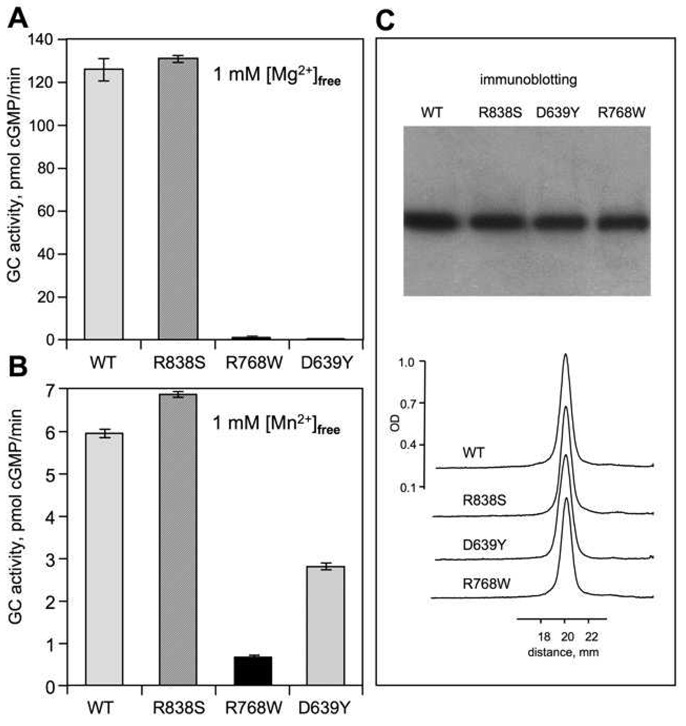
FIG. 2. Effect of D639Y and R768W mutations on GC activity of RetGC1.
A, B. Recombinant WT RetGC1 and its mutants present in HEK293 membrane fraction were assayed for guanylyl cyclase activity in the presence of 1 mM free Mg2+, 10 µM GCAP1 and 2 mM EGTA (A) or 5 mM Mn2+ (B) The activities are standardized for RetGC1 content in each assay, equalized with the wild type as a standard. The average of three independent measurements is shown. For other conditions of the assay see "Experimental Procedures". C. Immunoblotting probed with anti-RetGC1Cat antibody and analyzed by densitometry was used to equalize the RetGC1 content between the HEK293 membrane preparations.
Both the D639Y and the R768W mutations drastically changed the enzymatic properties of RetGC1 (Fig. 2). Whereas the wild type and the R838S RetGC1 demonstrated a robust stimulation by GCAP1, the D639Y RetGC1and R768W RetGC1 failed to show any activation even at saturating concentrations of wild type GCAP1 (Fig. 2A). The lack of activity was unlikely due to a non-specific inactivation of the mutant cyclase, because both the D639Y and the R768W RetGC1 displayed reduced, but readily detectable GCAP-independent basal activity in the presence of Mn2+ (Fig. 2B). The reduction in basal activity was by far less severe than the complete lack of the GCAP1-dependent activity, especially for the D639Y mutant, which retained nearly half the normal level of the basal activity. Therefore, the more likely explanation could be one of the following: i) the mutant cyclases were unable to bind GCAP1 or ii) the cyclase mutants were capable of binding GCAP1, but failed to increase their catalytic activity, because some potential secondary interactions in the GCAP1/RetGC1 complex failed to properly occur.
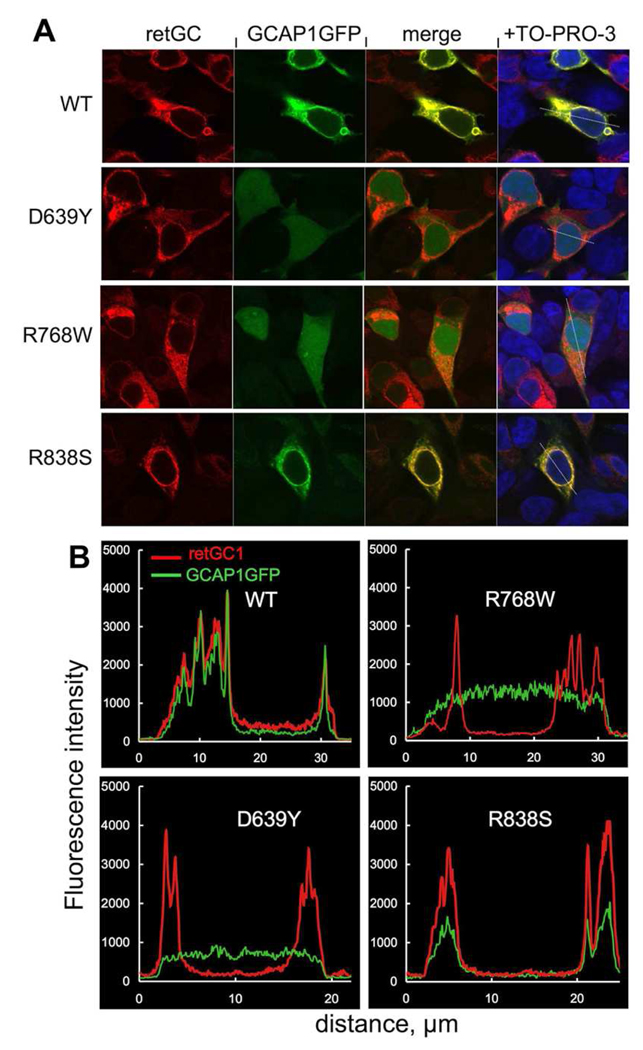
To assess the efficiency of GCAP1 binding to D639Y and R768WRetGC1, we utilized our recently described cell-based assay using RetGC1 and GCAP1-GFP co-expression in HEK293 cells (32) (Fig. 3). While GCAP1-GFP expressed alone always shows a diffuse uniform distribution throughout the cytoplasm and the nucleus, when co-expressed with RetGC1 it co-localizes with the cyclase in the membranes of the endoplasmic reticulum and to a much lesser extent in the plasma membrane, thus forming a characteristic “donuts and tennis rackets” pattern (32). Such co-localization pattern was observed in our experiments with both wild type and the R838S RetGC1 (Fig. 3A), with the across-the-cell profile of GCAP1 fluorescence matching that of the anti-RetGC1 immunofluorescence (Fig. 3B). In striking contrast, GCAP1-GFP co-expressed with either D639Y or R768W RetGC1 remained diffusely distributed through the entire cell, thus showing no evidence for binding to the cyclase. The levels of expression of both RetGC1 and GCAP1-GFP unavoidably vary between different cells, even in the same culture sample (32). Therefore, for more accurate semi-quantitative characterization of the cyclase ability to bind GCAP1 we determined the ratio between the maximal GFP fluorescence intensity in the whole cell and its maximal fluorescence intensity inside the nucleus (Fig. 4). For GCAP1-GFP expressed without the cyclase this ratio is close to 1.0, but sharply increases after co-transfection with RetGC1, reflecting GCAP1 depletion from the nucleus due to compartmentalization with the cyclase in the membranes (32). Whereas for both the wild type and the R838S RetGC1 the GCAP1 clearance between the membranes and the nucleus averaged from a random sample of cells from the same culture rose near 10-fold compared with the GCAP1-GFP expressed alone; it remained close to 1 for both the R639Y and the R768W mutants. The difference from the wild type and the R838S RetGC1 was statistically highly significant (ANOVA/Bonferroni post-hoc test, p<0.01, CL 99%). Hence, the lack of activity for both the R639Y and the R768W can be explained primarily by loss of their affinity for GCAP1.
FIG. 3. Cellular localization of GCAP1-GFP co-expressed with RetGC1 mutants.
A. Fluorescence of GCAP1- GFP and RetGC1 (anti-RetGC1KHD primary and Alexa Fluor 568 secondary antibody) in HEK293 co-expressing GCAP1-GFP and wild type (WT) or mutant (D639Y, R768W and R838S) RetGC1. The first two columns show GCAP1-GFP and Alexa Fluor 568 fluorescence, respectively. The third column shows the merged fluorescence images in the first two columns. Rightmost panels show the merged images counterstained with TO-PRO-3 (pseudo blue). The cells were fixed with 4% paraformaldehyde. The GFP fluorescence of GCAP1-GFP was excited at 488 nm and Alexa Fluor 568 antibody complex at 543 nm. For other details see "Experimental Procedures". B. Representative fluorescence intensity profiles recorded from the cells expressing GCAP1-GFP and wild type or mutant RetGC1 along the lines superimposed on the merged fluorescent images shown in the rightmost panels in A.
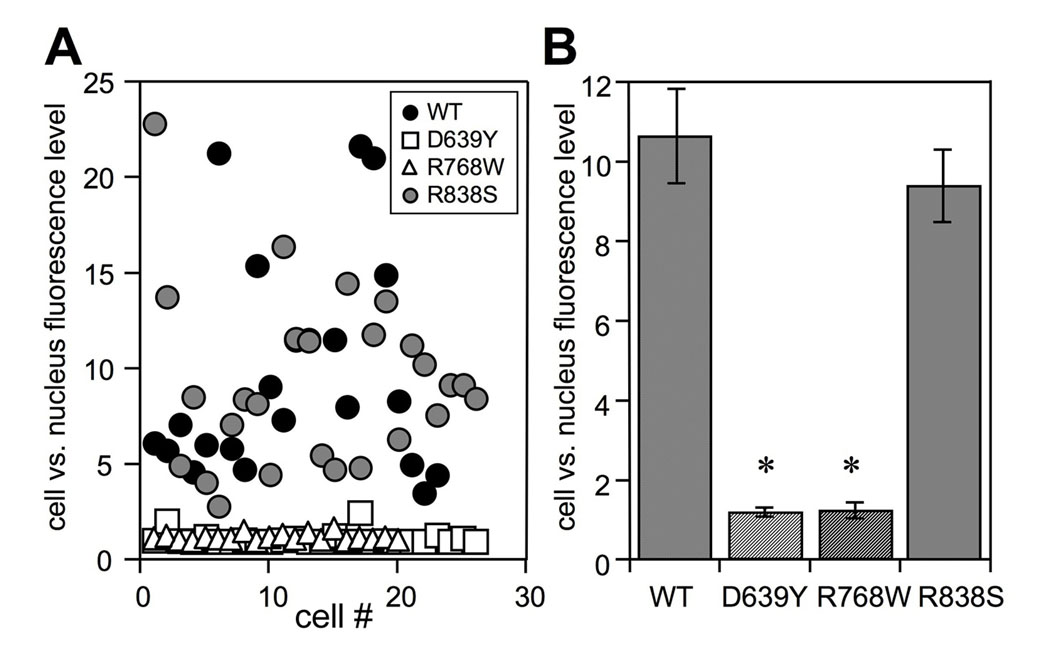
FIG. 4. Effect of RetGC1 mutations on GCAP1-GFP co-localization with RetGC1.
A. The cell:nucleus GCAP1-GFP fluorescence intensity ratio among the HEK293 cells that co-express GCAP1-GFP with WT (●), D639Y (□), R768W (Δ) and R838S ( ) RetGC1. The cell:nucleus fluorescence intensity ratio is the ratio between the maximal GFP fluorescence intensity in the whole cell versus its maximal fluorescence intensity inside the nucleus. Each data point represents a single cell. B. Average ratio (mean ± standard error, cell number) for GCAP1-GFP co-expressed with WT (10.6 ± 1.4, n = 25), D639Y (1.2 ± 0.1, n = 26), R768W (1.2 ± 0.04, n = 20) and R838S (9.3 ± 0.9, n = 26) RetGC1; *p<0.01 different from wild type.
) RetGC1. The cell:nucleus fluorescence intensity ratio is the ratio between the maximal GFP fluorescence intensity in the whole cell versus its maximal fluorescence intensity inside the nucleus. Each data point represents a single cell. B. Average ratio (mean ± standard error, cell number) for GCAP1-GFP co-expressed with WT (10.6 ± 1.4, n = 25), D639Y (1.2 ± 0.1, n = 26), R768W (1.2 ± 0.04, n = 20) and R838S (9.3 ± 0.9, n = 26) RetGC1; *p<0.01 different from wild type.
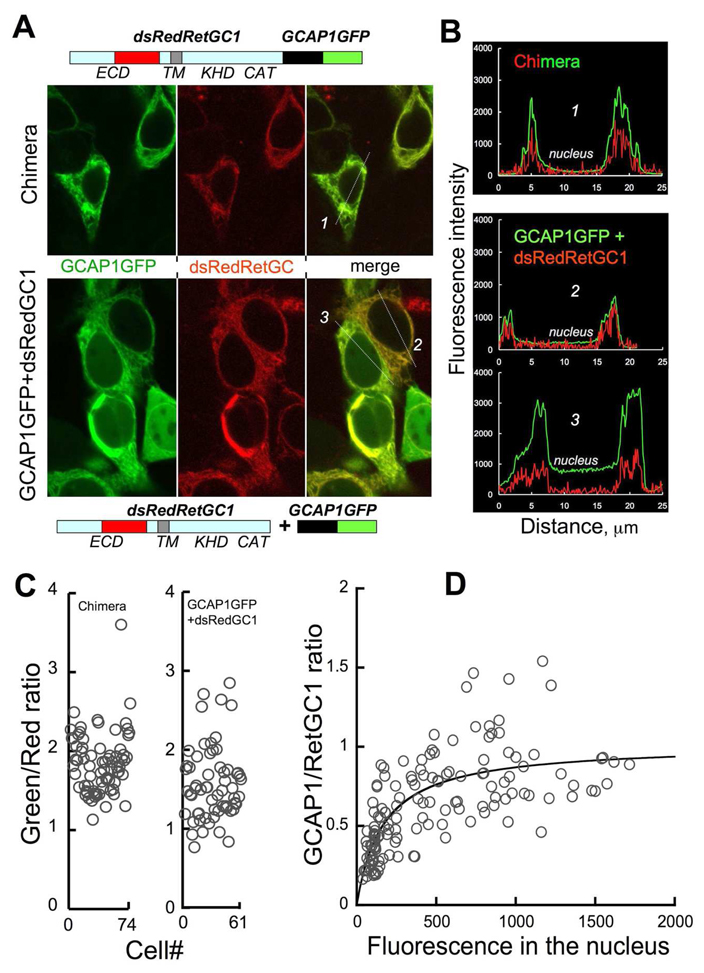
Does this mean that each mutant cyclase has just a smaller number of GCAP1 molecules associated with it or that GCAP1 simply does not bind to RetGC1? The answer depends on how many molecules of GCAP1 are actually associated with the cyclase at saturation – one or many. Although the levels of expression for RetGC1 and GCAPs are both estimated to be in the µM range (38), the exact stoichiometry between GCAP1 and RetGC1 in their complex is unknown. In order to resolve this question, we estimated the ratio between GCAP1 and RetGC1 using GCAP1-GFP and RetGC1 fluorescently tagged by replacing a portion of the cyclase ECD domain with monomeric dsRed red fluorescent protein (32). Neither of these modifications critically affected RetGC1 activity or its affinity for GCAP1 (32). To normalize the fluorescence of both tags in the membranes of co-transfected cells relative to a 1:1 molar ratio of GCAP1-GFP/dsRed-RetGC1 complex, we produced the “1:1 standard chimera” in which GCAP1-GFP was directly fused with the dsRedRetGC1 in a single ~170 kDa chimera protein that contained both fluorescent tags (Fig. 5A and Supplemental Fig. 1S). The ratio between the green versus the red tags fluorescence brightness for the "1:1 chimera" was calculated by dividing the average value of the green fluorescence in the cell by the average value of red fluorescence in the same region of the cell. The average ratio determined from many cells measured in this experiment was 1.87 ± 0.04 (mean ± SEM, n = 75). It also needs to be noted that there is a distinct cell-to-cell variability in the same cell culture, which could appear somewhat surprising to see in case of a single polypeptide containing both tagged proteins (Supplemental Fig. 1S). One possible explanation is that perhaps a small variation in the fluorescent tags folding or maturation exists between different cells in the same population. Therefore, a large number of cells were included in the analysis in order to average out the cell-to-cell variations.
FIG. 5. The stoichiometry of GCAP1:RetGC1 binding in HEK293 cells.
A. The eGFP-tagged GCAP1 and a monomeric dsRed-tagged RetGC1 were fused together as a single polypeptide containing both fluorescently tagged proteins (“1:1 chimera” standard); ECD-extracelular domain, TM – transmembrane domain, KHD - kinase homology domain, CAT – catalytic domain. The upper three panels show fluorescence of the eGFP and dsRed tags in the chimera-expressing HEK293 cells. The "1:1 chimera" displayed strictly membrane localization (primarily ER), indistinguishable from dsRed-tagged RetGC1 (32). The lower three panels show fluorescence of the eGFP and dsRed tags and their merged image in the cells that co-expressed a mixture of GCAP1GFP and dsRedRetGC1 cDNAs. B. An example of fluorescence intensity profiles recorded from the cells expressing the "1:1 chimera" (cell #1) or co-expressing individual GCAP1GFP and dsRedRetGC1 (cells #2 and #3) is shown in panel A. Subtracting GCAP1GFP fluorescence level in the nucleus (“free GCAP1GFP") from that in the endoplasmic reticulum yields the net fluorescence value for GCAP1GFP co-localized with dsRedRetGC1. C. The ratio between the GFP and the dsRed tags fluorescence in HEK293 cells that expressed "1:1 chimera" standard (left panel) or a mixture of the individual GCAP1GFP and dsRedRetGC1. In the latter case (right panel), the ratio was plotted for the cells where the GCAP1GFP fluorescence in the nucleus exceeded 10% of the proportional PMA range maximum (GFP levels >450, see panel B for an example). Each data point represents a separate cell. D. GCAP1:RetGC1 molar ratio calculated from the fluorescence ratio of their two tags divided by that of the “1:1 chimera” standard. The data points were fitted using a fractional saturation function, R = Rmax × F/(F + F1/2), where R is the molar ratio between membrane-associated GCAP1GFP and dsRed RetGC1; Rmax is the R value at saturation by GCAP1GFP; F is the GCAP1GFP fluorescence level in the nucleus (“free GCAPGFP”), F1/2 is the F value corresponding to half-saturation by GCAP1GFP. Each data point represents a separate cell. All measurements were conducted using the same settings for excitation and image acquisition.
We then measured the fluorescence of the two tags in the cells that co-expressed individual dsRedRetGC1 and GCAP1-GFP using the same settings for fluorescence excitation and image acquisition as we used for the “1:1 chimera” as a standard. In our previous experiments, the cell-based binding assay was normally conducted at the GCAP1-GFP:RetGC1 cDNAs ratio as low as 1:150 in order to prevent RetGC1 from saturation by GCAP1. Under those conditions, there was 10–20 molar excess of RetGC1 over GCAP1 (32). Consequently, almost all GCAP1 was associated with the membrane-bound RetGC1 and very little, if any, of the green fluorescence was detectable in the nuclei. However, in order to evaluate the stoichiometry for the GCAP1:RetGC1 complex, GCAP1-GFP had to be in an excess over RetGC1 such that the concentration of GCAP1-GFP in the cell that is not bound to the RetGC1 had to be high enough to saturate RetGC1. At the same time, we could not express a large excess of the tagged GCAP1 over RetGC1, because high background of the free GCAP1-GFP reduces the accuracy of the measurement in its membrane-associated fraction containing dsRed-tagged RetGC1. Therefore, we had to increase the proportion of the GCAP1 expressing construct in the co-transfection mixture until the relative fluorescence intensity of the free GCAP1-GFP in the nucleus became higher than ~25% of its maximal fluorescence in the cell (thus indicating that the GCAP/RetGC1 binding approached saturation), yet was still within the proportional range for PMU sensitivity. Since the free GCAP1-GFP is distributed evenly throughout the cell (32), the fluorescence intensity of GCAP1-GFP in the nuclei reflected the concentration of the free GCAP1-GFP in the cell. Therefore, subtracting GCAP1-GFP fluorescence level in the nucleus from that in the endoplasmic reticulum gives the net fluorescence value for GCAP1-GFP co-localized with dsRedRetGC1 and allows us to calculate the ratio between the two fluorescent tags in comparison with the “1:1 chimera” standard (Fig. 5C). For the wild type RetGC1 this ratio was 1.6 ± 0.06 (mean ± SEM, n = 61), which in comparison with the 1.87 ratio for the standard yields a GCAP1:RetGC1 ratio in the complex 0.86:1, strongly suggesting their 1:1 stoichiometry.
The fluorescence ratio between the two tags associated with the membranes versus the levels of the free GCAP1GFP fluorescence was also fitted with a fractional saturation curve (Fig. 5D). In comparison with the "1:1 standard chimera", the value of 1.01 was found for the molar ratio between the two proteins at saturation. We would also like to point out that the GFP and the dsRed tags in the chimera and in the GCAP1/RetGC complex are located on the opposite sides of the transmembrane domain of RetGC, hence, they are separated by the minimal distance equal to the thickness of the membrane (39) and very likely even farther apart, because GCAP1 in HEK293 cells is not directly anchored by the membrane bilayer surface (Fig. 3 and 4). Therefore, the efficiency of a fluorescence resonance energy transfer between the GFP and dsRed tags (40), which is inversely proportional to the 6th power of the distance between them, if any, should be very low and should not affect the results.
The important conclusion from those experiments was that wild type RetGC1 and GCAP1 formed an equimolar complex when produced in HEK293 cells. Since two catalytic domains are required to form the active center of the cyclase (29, 30), RetGC1 is most likely a dimer (although the existence of higher-order complexes cannot be completely excluded at this point). Therefore, our experiments argue that each dimer of RetGC1 must contain two GCAP1 molecules, one per each RetGC1 subunit. These results also argue that since both D639Y and R768W mutants failed to “anchor” GCAP1 to the membranes (Fig. 3), each of them was unable to bind and recruit the single molecule of GCAP1 required to produce their 1:1 complex, such as with the wild type RetGC1.
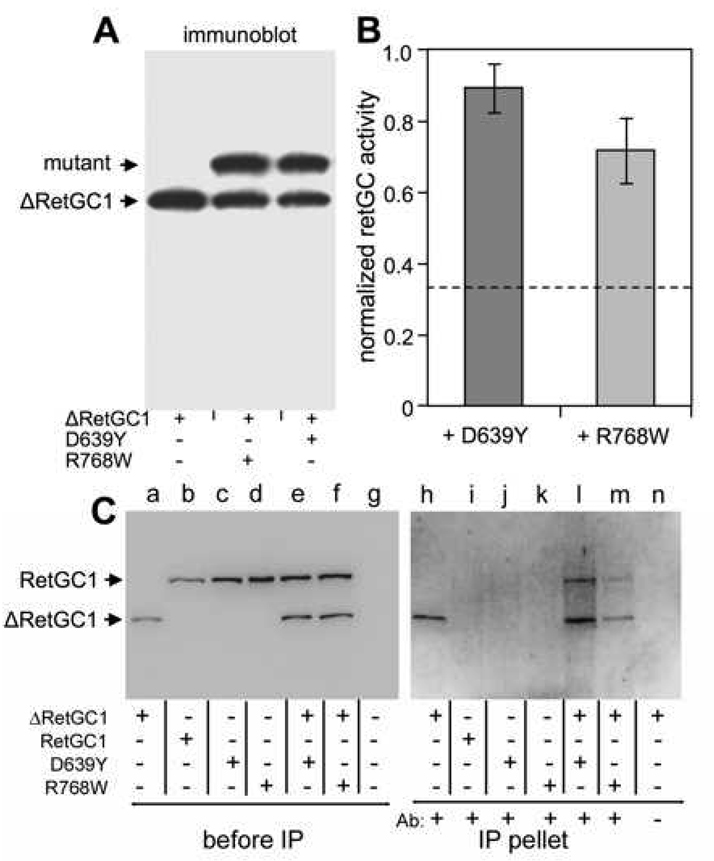
Since the RetGC1/GCAP1 complex is likely to be a heterotetramer consisting of two RetGC1 and two GCAP1 molecules, could the effect of both mutations in RetGC1 be dominant negative, with the mutant RetGC1 being able to abolish the activity of the wild type RetGC1 as a result of their interaction? We tested the activity of the wild type RetGC1 co-expressed with each mutant. In that case, however, it was challenging to reliably measure the ratio between the mutant and non-mutant cyclase from the immunoblotting; our antibodies produced against either the catalytic or the kinase-homology domain of the cyclase do not discriminate between the wild type and any of the mutants. Therefore, we removed a ~20-kDa fragment from the cyclase extracellular domain (the same region that can be substituted with the dsRed tag (32)) with a short peptide fragment containing EQKLISEEDL myc tag. That modification (ΔRetGC1) did not alter Ca2+/GCAP1 sensitivity of the cyclase regulation (Fig. 6A), only slightly reduced the maximal activity of the cyclase (Fig. 6A), and did not dramatically affect the apparent affinity of the shortened RetGC1 for GCAP1 in vitro (Fig. 6B). At the same time, it made the shortened “wild type” ΔRetGC1 easily distinguishable from the normal-size D639Y and R768W mutants when each mutant and the ΔRetGC1 were co-expressed in the same cells (Fig. 6A, inset).
FIG. 6. Properties of ΔRetGC1.
A. Ca2+ sensitivity of wild type RetGC1 and ΔRetGC1 stimulated by 10 µM recombinant GCAP1 in the presence of 1 mM free Mg2+. The data were fitted as fractional activity of RetGC1 by the function, Y=100% /(1 + ([Ca]f/[Ca]1/2) n’); [Ca]1/2 is the free Ca2+ concentration required for half-maximal inhibition of RetGC1, n’ is the cooperativity coefficient. Insert, western blot analysis of HEK293 membranes from the cells expressing wild type RetGC1 and ΔRetGC1 using RetGC1Cat antibody produced against the catalytic domain of RetGC1. B. Activation of wild type RetGC1 and ΔRetGC1 by purified GCAP1 in the presence of 1 mM EGTA and 1 mM free Mg2+. The data were fitted by the equation, A = Amax × [GCAP]n’/((K1/2)n’ + [GCAP]n’), where A is the activity of guanylyl cyclase in the assay, Amax is the maximal activity of guanylyl cyclase, [GCAP] is the concentration of GCAP1, K1/2 is the concentration of GCAP1 required for half-maximal activation of RetGC1, n’ is the cooperativity coefficient. The cyclase activity in each assay was equalized per RetGC1 content relative to the wild type. For other conditions of the assay see "Experimental Procedures".
We reasoned that if both ΔRetGC1 and one of each of the mutants are co-expressed at an equimolar ratio and are capable of forming a RetGC1 heterodimer, then on average 1/3 of the ΔRetGC1 produced in the culture was expected to remain a homodimer whereas 2/3 would remain to form a heterodimer with the mutant. Neither D639Y nor R768W alone display any GCAP1-stimulated activity (Fig. 2), therefore, if either mutation had a strong dominant negative effect, one could expect that only the 1/3 of the ΔRetGC1 remaining as the homodimer would retain its activity. In contrast, Fig. 7B demonstrates that the activity of ΔRetGC1 co-expressed with each mutant was only modestly reduced compared to the activity of ΔRetGC1 expressed alone and remained well above the level expected for a strong dominant negative effect (dashed line).
FIG. 7. Effects of the ΔRetGC1 Co-expression with D639Y and R768W mutants in HEK293 cells.
A. mmunoblotting of HEK293 membranes from the cells co-expressing ΔRetGC1 and D639Y or R768W mutants using antibodies produced against the catalytic domain of RetGC1. B. Guanylyl cyclase activity of ΔRetGC1 in HEK293 cells co-expressing ΔRetGC1 and D639Y or R768W mutants. The cyclase activity was normalized to that of ΔRetGC1 expressed alone. The average of three independent measurements is shown. C. Co-immunoprecipitation (co-IP) of the D639Y and R768W mutants with the Myc-tagged ΔRetGC1. Left panel (lanes a – g), immunoblotting of RetGC1 variants expressed in HEK293 membranes used in the co-IP experiments (no further normalization per RetGC1 content between the preparations was done for the experiments); lane g – non-transfected cells. The membranes were then dissolved in the extraction buffer and processed for immunoprecipitation with anti-Myc antibody as described in Experimental Procedures. Right panel (lanes h – n), the immunoblot of the fractions recovered from the beads after the immunoprecipitation (the blot was probed by RetGC1Cat antibody). The higher background in the right panel was due to longer exposure time needed to visualize the immunoprecipitated proteins recovered from the beads. Notice that both D639Y and R768W are precipitated when co-expressed with the ΔRetGC1 (lines l and m), but not when expressed separately (lines j, k). In the lane n, the anti-Myc antibody was omitted from the co-IP mixture as a negative control for the ΔRetGC1 precipitation in line h. All samples were processed simultaneously in the same experiment.
A possible explanation for the lack of dominant negative effect for both the D639Y and the R768W mutations in Fig. 7B could be merely their complete inability to dimerize with the wild type counterpart. However, the results shown in Fig. 7C demonstrate that both D639Y and R768W RetGC1 can co-immunoprecipitate with the ΔRetGC1 (lines l and m, compare with j and k) by the antibody that recognizes Myc tag incorporated into the shortened extracellular domain of the ΔRetGC1 (lanes a and h), absent from the full-length RetGC1 (lanes b–d and i–k). Since the immunoprecipitation in our experiments could only be conducted with a detergent-solubilized RetGC1, a procedure that completely inactivates its regulation by GCAPs (41 and our unpublished observations) and cannot provide sufficiently quantitative characterization of the dimerization efficiency, we cannot rule out that dimerization of the mutants with the normal RetGC1 is indeed affected. However, the fact that both RetGC1 mutants were clearly able to co-IP with the ΔRetGC1 in these experiments would rather argue that the mutant cyclase does not lack the ability to associate with the normal gene product in vivo.
To summarize, our present findings strongly argue that RetGC1 affected by either of the two LCA-associated mutations cannot bind GCAP1. At the same time, neither of the two mutations was able to abolish the activity of the normal RetGC1 when co-expressed in the same cell (and likely even despite forming a dimer with the unaffected cyclase). The lack of a strong dominant-negative effect for each mutant would appear consistent with one of the subjects in the original clinical study who carried heterozygous D639Y mutation not developing the severe loss of both rod and cone function typical of LCA (although, like with some other mutations in GUCY2D gene (42), had subclinical change in cone responses to light - Dr. Falsini, personal communication), despite that the primary binding (i.e., “docking”) of GCAP1 on the mutated RetGC1 molecule itself was completely abolished. Therefore, it is most likely that both mutants characterized in this study can affect photoreceptor viability leading to the LCA symptoms, but propagation of its potential effect would also require additional, currently unidentified, genetic interactions.
This study also demonstrates that our recently developed cell-based assay for GCAP1 binding to RetGC1 (32) allows identification of the residues involved in the GCAP1/RetGC1 docking event. They show that the docking of GCAP1 on the cyclase can be completely prevented by a single amino acid substitution, such as D639Y or R768W, affecting the KHD domain of RetGC1. A peptide containing D639 was previously found among the products of RetGC1 chemical cross-linking with GCAP1 in vitro (43), although it was not clear at that time whether that product reflected the primary binding of GCAP1 to the cyclase or their secondary regulatory interactions, which may potentially involve different sites (44). Together with the chemical cross-linking data (43), our functional experiments now directly implicate the region containing the D639 residue in the primary binding (“docking”) of GCAP1. In addition to that, the part of the RetGC1 KHD domain containing R768 was never before suspected to participate in the RetGC1/GCAP1 docking interactions. Therefore, using fluorescently tagged RetGC1 and GCAP1 in live cells opens a possibility for more accurate identification of the sites required for their docking.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dr. B. Falsini and Dr. J. Nathans for their generous gift of the original samples from the LCA patients used in this study.
This work was supported by the National Institutes of Health grants EY11522 (AMD), EY09924 and EY018143(SJP), and Foundation Fighting Blindness (SJP), Pennsylvania Department of Health, and Pennsylvania Lions Sight Conservation and Eye Research Foundation (AMD). A.M. Dizhoor is the Martin and Florence Hafter Professor of Pharmacology.
ABBREVIATIONS
- DIC
differential interference contrast
- EGTA
ethylene glycol-bis(2-aminoethylether)-N,N,N’,N’-tetraacetic acid
- GCAP
guanylyl cyclase activating protein
- GFP
green fluorescent protein
- LCA
Leber congenital amaurosis
- MOPS
4-morpholinopropanesulfonic acid
- RetGC
photoreceptor membrane guanylyl cyclase.
REFERENCES
- 1.Pugh EN, Jr, Duda T, Sitaramayya A, Sharma RK. Photoreceptor guanylate cyclases: a review. Biosci Rep. 1997;17:429–473. doi: 10.1023/a:1027365520442. [DOI] [PubMed] [Google Scholar]
- 2.Hodgkin AL, Nunn BJ. Control of light-sensitive current in salamander rods. J Physiol. 1988;403:439–471. doi: 10.1113/jphysiol.1988.sp017258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Burns ME, Mendez A, Chen J, Baylor DA. Dynamics of cyclic GMP synthesis in retinal rods. Neuron. 2002;36:81–91. doi: 10.1016/s0896-6273(02)00911-x. [DOI] [PubMed] [Google Scholar]
- 4.Koch KW, Stryer L. Highly cooperative feedback control of retinal rod guanylate cyclase by calcium ions. Nature. 1988;334:64–66. doi: 10.1038/334064a0. [DOI] [PubMed] [Google Scholar]
- 5.Gorczyca WA, Gray-Keller MP, Detwiler PB, Palczewski K. Purification and physiological evaluation of a guanylate cyclase activating protein from retinal rods. Proc Natl Acad Sci U S A. 1994;91:4014–4018. doi: 10.1073/pnas.91.9.4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dizhoor AM, Lowe DG, Olshevskaya EV, Laura RP, Hurley JB. The human photoreceptor membrane guanylyl cyclase, RetGC, is present in outer segments and is regulated by calcium and a soluble activator. Neuron. 1994;12:1345–1352. doi: 10.1016/0896-6273(94)90449-9. [DOI] [PubMed] [Google Scholar]
- 7.Peshenko IV, Dizhoor AM. Guanylyl cyclase-activating proteins (GCAPs) are Ca2+/Mg2+ sensors: implications for photoreceptor guanylyl cyclase (RetGC) regulation in mammalian photoreceptors. J Biol Chem. 2004;279:16903–16906. doi: 10.1074/jbc.C400065200. [DOI] [PubMed] [Google Scholar]
- 8.Gray-Keller MP, Detwiler PB. The calcium feedback signal in the phototransduction cascade of vertebrate rods. Neuron. 1994;13:849–861. doi: 10.1016/0896-6273(94)90251-8. [DOI] [PubMed] [Google Scholar]
- 9.Woodruff ML, Sampath AP, Matthews HR, Krasnoperova NV, Lem J, Fain GL. Measurement of cytoplasmic calcium concentration in the rods of wild-type and transducin knock-out mice. J Physiol. 2002;542:843–854. doi: 10.1113/jphysiol.2001.013987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peshenko IV, Dizhoor AM. Activation and inhibition of photoreceptor guanylyl cyclase by guanylyl cyclase activating protein 1 (GCAP-1): the functional role of Mg2+/Ca2+ exchange in EF-hand domains. J Biol Chem. 2007;282:21645–21652. doi: 10.1074/jbc.M702368200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makino CL, Peshenko IV, Wen XH, Olshevskaya EV, Barrett R, Dizhoor AM. A role for GCAP2 in regulating the photoresponse. Guanylyl cyclase activation and rod electrophysiology in GUCA1B knock-out mice. J Biol Chem. 2008;283:29135–29143. doi: 10.1074/jbc.M804445200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lowe DG, Dizhoor AM, Liu K, Gu Q, Spencer M, Laura R, Lu L, Hurley JB. Cloning and expression of a second photoreceptor-specific membrane retina guanylyl cyclase (RetGC), RetGC-2. Proc Natl Acad Sci U S A. 1995;92:5535–5539. doi: 10.1073/pnas.92.12.5535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang RB, Foster DC, Garbers DL, Fulle HJ. Two membrane forms of guanylyl cyclase found in the eye. Proc Natl Acad Sci U S A. 1995;92:602–606. doi: 10.1073/pnas.92.2.602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baehr W, Karan S, Maeda T, Luo DG, Li S, Bronson JD, Watt CB, Yau KW, Frederick JM, Palczewski K. The function of guanylate cyclase 1 and guanylate cyclase 2 in rod and cone photoreceptors. J Biol Chem. 2007;282:8837–8847. doi: 10.1074/jbc.M610369200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yang RB, Robinson SW, Xiong WH, Yau KW, Birch DG, Garbers DL. Disruption of a retinal guanylyl cyclase gene leads to cone-specific dystrophy and paradoxical rod behavior. J Neurosci. 1999;19:5889–5897. doi: 10.1523/JNEUROSCI.19-14-05889.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dizhoor AM, Olshevskaya EV, Henzel WJ, Wong SC, Stults JT, Ankoudinova I, Hurley JB. Cloning, sequencing, and expression of a 24-kDa Ca(2+)-binding protein activating photoreceptor guanylyl cyclase. J Biol Chem. 1995;270:25200–25206. doi: 10.1074/jbc.270.42.25200. J Biol Chem 270, 25200-6. [DOI] [PubMed] [Google Scholar]
- 17.Gorczyca WA, Polans AS, Surgucheva IG, Subbaraya I, Baehr W, Palczewski K. Guanylyl cyclase activating protein. A calcium-sensitive regulator of phototransduction. J Biol Chem. 1995;270:22029–22036. doi: 10.1074/jbc.270.37.22029. [DOI] [PubMed] [Google Scholar]
- 18.Mendez A, Burns ME, Sokal I, Dizhoor AM, Baehr W, Palczewski K, Baylor DA, Chen J. Role of guanylate cyclase-activating proteins (GCAPs) in setting the flash sensitivity of rod photoreceptors. Proc Natl Acad Sci U S A. 2001;98:9948–9953. doi: 10.1073/pnas.171308998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stephen R, Filipek S, Palczewski K, Sousa MC. Ca2+ -dependent regulation of phototransduction. Photochem Photobiol. 2008;84:903–910. doi: 10.1111/j.1751-1097.2008.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Imanishi Y, Li N, Sokal I, Sowa ME, Lichtarge O, Wensel TG, Saperstein DA, Baehr W, Palczewski K. Characterization of retinal guanylate cyclase-activating protein 3 (GCAP3) from zebrafish to man. Eur J Neurosci. 2002;15:63–78. doi: 10.1046/j.0953-816x.2001.01835.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Perrault I, Rozet JM, Calvas P, Gerber S, Camuzat A, Dollfus H, Chatelin S, Souied E, Ghazi I, Leowski C, Bonnemaison M, Le Paslier D, Frezal J, Dufier JL, Pittler S, Munnich A, Kaplan J. Retinal-specific guanylate cyclase gene mutations in Leber's congenital amaurosis. Nat Genet. 1996;14:461–464. doi: 10.1038/ng1296-461. [DOI] [PubMed] [Google Scholar]
- 22.Perrault I, Rozet JM, Gerber S, Ghazi I, Ducroq D, Souied E, Leowski C, Bonnemaison M, Dufier JL, Munnich A, Kaplan J. Spectrum of RetGC1 mutations in Leber's congenital amaurosis. Eur J Hum Genet. 2000;8:578–582. doi: 10.1038/sj.ejhg.5200503. [DOI] [PubMed] [Google Scholar]
- 23.Duda T, Venkataraman V, Goraczniak R, Lange C, Koch KW, Sharma RK. Functional consequences of a rod outer segment membrane guanylate cyclase (ROS-GC1) gene mutation linked with Leber's congenital amaurosis. Biochemistry. 1999;38:509–515. doi: 10.1021/bi9824137. [DOI] [PubMed] [Google Scholar]
- 24.Dharmaraj SR, Silva ER, Pina AL, Li YY, Yang JM, Carter CR, Loyer MK, El-Hilali HK, Traboulsi EK, Sundin OK, Zhu DK, Koenekoop RK, Maumenee IH. Mutational analysis and clinical correlation in Leber congenital amaurosis. Ophthalmic Genet. 2000;21:135–150. [PubMed] [Google Scholar]
- 25.Tucker CL, Ramamurthy V, Pina AL, Loyer M, Dharmaraj S, Li Y, Maumenee IH, Hurley JB, Koenekoop RK. Functional analyses of mutant recessive GUCY2D alleles identified in Leber congenital amaurosis patients: protein domain comparisons and dominant negative effects. Mol Vis. 2004;10:297–303. [PubMed] [Google Scholar]
- 26.Rozet JM, Perrault I, Gerber S, Hanein S, Barbet F, Ducroq D, Souied E, Munnich A, Kaplan J. Complete abolition of the retinal-specific guanylyl cyclase (retGC-1) catalytic ability consistently leads to leber congenital amaurosis (LCA) Invest Ophthalmol Vis Sci. 2001;42:1190–1192. [PubMed] [Google Scholar]
- 27.Kelsell RE, Gregory-Evans K, Payne AM, Perrault I, Kaplan J, Yang RB, Garbers DL, Bird AC, Moore AT, Hunt DM. Mutations in the retinal guanylate cyclase (RETGC-1) gene in dominant cone-rod dystrophy. Hum Mol Genet. 1998;7:1179–1184. doi: 10.1093/hmg/7.7.1179. [DOI] [PubMed] [Google Scholar]
- 28.Perrault I, Rozet JM, Gerber S, Kelsell RE, Souied E, Cabot A, Hunt DM, Munnich A, Kaplan J. A retGC-1 mutation in autosomal dominant cone-rod dystrophy. Am J Hum Genet. 1998;63:651–654. doi: 10.1086/301985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tucker CL, Woodcock SC, Kelsell RE, Ramamurthy V, Hunt DM, Hurley JB. Biochemical analysis of a dimerization domain mutation in RetGC-1 associated with dominant cone-rod dystrophy. Proc Natl Acad Sci U S A. 1999;96:9039–9044. doi: 10.1073/pnas.96.16.9039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramamurthy V, Tucker C, Wilkie SE, Daggett V, Hunt DM, Hurley JB. Interactions within the coiled-coil domain of RetGC-1 guanylyl cyclase are optimized for regulation rather than for high affinity. J Biol Chem. 2001;276:26218–26229. doi: 10.1074/jbc.M010495200. [DOI] [PubMed] [Google Scholar]
- 31.Peshenko IV, Moiseyev GP, Olshevskaya EV, Dizhoor AM. Factors that determine Ca2+ sensitivity of photoreceptor guanylyl cyclase. Kinetic analysis of the interaction between the Ca2+-bound and the Ca2+-free guanylyl cyclase activating proteins (GCAPs) and recombinant photoreceptor guanylyl cyclase 1 (RetGC-1) Biochemistry. 2004;43:13796–13804. doi: 10.1021/bi048943m. [DOI] [PubMed] [Google Scholar]
- 32.Peshenko IV, Olshevskaya EV, Dizhoor AM. Binding of guanylyl cyclase activating protein 1 (GCAP1) to retinal guanylyl cyclase (RetGC1). The role of individual EF-hands. J Biol Chem. 2008;283:21747–21757. doi: 10.1074/jbc.M801899200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peshenko IV, Dizhoor AM. Ca2+ and Mg2+ binding properties of GCAP-1. Evidence that Mg2+-bound form is the physiological activator of photoreceptor guanylyl cyclase. J Biol Chem. 2006;281:23830–23841. doi: 10.1074/jbc.M600257200. [DOI] [PubMed] [Google Scholar]
- 34.Laura RP, Dizhoor AM, Hurley JB. The membrane guanylyl cyclase, retinal guanylyl cyclase-1, is activated through its intracellular domain. J Biol Chem. 1996;271:11646–11651. doi: 10.1074/jbc.271.20.11646. [DOI] [PubMed] [Google Scholar]
- 35.Olshevskaya EV, Hughes RE, Hurley JB, Dizhoor AM. Calcium binding, but not a calcium-myristoyl switch, controls the ability of guanylyl cyclase-activating protein GCAP-2 to regulate photoreceptor guanylyl cyclase. J Biol Chem. 1997;272:14327–14333. doi: 10.1074/jbc.272.22.14327. [DOI] [PubMed] [Google Scholar]
- 36.Camuzat A, Rozet JM, Dollfus H, Gerber S, Perrault I, Weissenbach J, Munnich A, Kaplan J. Evidence of genetic heterogeneity of Leber's congenital amaurosis (LCA) and mapping of LCA1 to chromosome 17p13. Hum Genet. 1996;97:798–801. doi: 10.1007/BF02346192. [DOI] [PubMed] [Google Scholar]
- 37.Yzer S, Leroy BP, De Baere E, de Ravel TJ, Zonneveld MN, Voesenek K, Kellner U, Ciriano JP, de Faber JT, Rohrschneider K, Roepman R, den Hollander AI, Cruysberg JR, Meire F, Casteels I, van Moll-Ramirez NG, Allikmets R, van den Born LI, Cremers FP. Microarray-based mutation detection and phenotypic characterization of patients with Leber congenital amaurosis. Invest. Ophthalmol. Vis. Sci. 2006;47:1167–1176. doi: 10.1167/iovs.05-0848. [DOI] [PubMed] [Google Scholar]
- 38.Hwang JY, Lange C, Helten A, Hoppner-Heitmann D, Duda T, Sharma RK, Koch KW. Regulatory modes of rod outer segment membrane guanylate cyclase differ in catalytic efficiency and Ca2+-sensitivity. Eur J Biochem. 2003;270:3814–3821. doi: 10.1046/j.1432-1033.2003.03770.x. [DOI] [PubMed] [Google Scholar]
- 39.Heller H, Schaefer M, Schulten K. Molecular dynamics simulation of a bilayer of 200 lipids in the gel and in the liquid-crystal phases. J. Phys. Chem. 1993;97:8343–8360. [Google Scholar]
- 40.Van der Krogt GN, Ogink J, Ponsioen B, Jalink K. A comparison of donor-acceptor pairs for genetically encoded FRET sensors: application to the Epac cAMP sensor as an example. PLoS One. 2008;3:e1916. doi: 10.1371/journal.pone.0001916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koch KW. Purification and identification of photoreceptor guanylate cyclase. J Biol Chem. 1991;266:8634–8637. [PubMed] [Google Scholar]
- 42.Koenekoop RK, Fishman GA, Iannaccone A, Ezzeldin H, Ciccarelli ML, Baldi A, Sunness JS, Lotery AJ, Jablonski MM, Pittler SJ, Maumenee I. Electroretinographic abnormalities in parents of patients with Leber congenital amaurosis who have heterozygous GUCY2D mutations. Arch Ophthalmol. 2002;120:1325–1330. doi: 10.1001/archopht.120.10.1325. [DOI] [PubMed] [Google Scholar]
- 43.Krylov DM, Hurley JB. Identification of proximate regions in a complex of retinal guanylyl cyclase 1 and guanylyl cyclase-activating protein-1 by a novel mass spectrometry-based method. J Biol Chem. 2001;276:30648–30654. doi: 10.1074/jbc.M104121200. [DOI] [PubMed] [Google Scholar]
- 44.Duda T, Fik-Rymarkiewicz E, Venkataraman V, Krishnan R, Koch KW, Sharma RK. The calcium-sensor guanylate cyclase activating protein type 2 specific site in rod outer segment membrane guanylate cyclase type 1. Biochemistry. 2005;44:7336–7345. doi: 10.1021/bi050068x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.