Abstract
Photoreceptors, rods and cones are the most abundant cell type in the mammalian retina. However, the molecules that control their development are not fully understood. In studies of photoreceptor fate determination, we found that Blimp1 (Prdm1) is expressed transiently in developing photoreceptors. We analyzed the function of Blimp1 in the mouse retina using a conditional deletion approach. Developmental analysis of mutants showed that Otx2+ photoreceptor precursors ectopically express the bipolar cell markers Chx10 (Vsx2) and Vsx1, adopting bipolar instead of photoreceptor fate. However, this fate shift did not occur until the time when bipolar cells are normally specified during development. Most of the excess bipolar cells died around the time of bipolar cell maturation. Our results suggest that Blimp1 expression stabilizes immature photoreceptors by preventing bipolar cell induction. We conclude that Blimp1 regulates the decision between photoreceptor and bipolar cell fates in the Otx2+ cell population during retinal development.
Keywords: Blimp1 (Prdm1), Retina, Cell fate determination, Photoreceptor, Rod, Cone, Bipolar cell, Mouse
INTRODUCTION
The retina comprises seven primary cell types: rod and cone photoreceptors, amacrine cells, retinal ganglion cells (RGCs), horizontal cells, bipolar cells and Müller glia. These cells are formed from a common pool of retinal progenitor cells during development and are born (exit the cell cycle) in a characteristic, but overlapping, order (Livesey and Cepko, 2001; Rapaport et al., 2004). The molecular mechanisms that control retinal cell fate determination involve both cell-intrinsic and extrinsic factors (Livesey and Cepko, 2001).
Photoreceptors are the most numerous cells in the mammalian retina and are responsible for detecting light stimuli. Cones are born early in mouse retinal development, starting around embryonic day (E) 12 and finishing by parturition (Carter-Dawson and LaVail, 1979). Rods are born starting at ∼E13.5 in mice, with the bulk formed postnatally, finishing around postnatal day (P) 7 (Carter-Dawson and LaVail, 1979). Previous studies have identified several key transcription factors required for photoreceptor fate specification and differentiation. Otx2 is a homeodomain transcription factor that is expressed in early and mature photoreceptors and bipolar cells (Fossat et al., 2007; Nishida et al., 2003). Loss of Otx2 from the retina prevents the development of these cell types, the cells adopting amacrine fate instead (Nishida et al., 2003; Sato et al., 2007). These data show that Otx2 is required for both photoreceptor and bipolar cell fate specification. The Otx2 homolog Crx is expressed in the same domain as Otx2 (Chen et al., 1997; Furukawa et al., 1997; Nishida et al., 2003). Crx-null mice have the normal number of photoreceptors initially, but do not express rod or cone opsins, causing gradual photoreceptor degeneration (Furukawa et al., 1999). Thus, Crx is required for photoreceptor differentiation, but not fate specification. The transcription factor Nrl is the earliest rod-specific marker, which is expressed shortly after cell cycle exit (Akimoto et al., 2006; Mears et al., 2001). Loss of Nrl results in a one-to-one fate shift to cones, demonstrating the requirement for Nrl in rod specification (Mears et al., 2001). The nuclear hormone receptor Trβ2 (Thrβ – Mouse Genome Informatics) is an early cone-specific marker (Ng et al., 2001). Deletion of Trβ2 from mice does not affect cone specification, but affects the subtype of cone formed (Ng et al., 2001).
Bipolar cells are interneurons that relay information from photoreceptors to RGCs and are divided into ten subtypes in mouse (Wassle et al., 2009). Broadly, these are characterized as ON or OFF depending on whether they depolarize or hyperpolarize in response to stimuli, respectively (Wassle et al., 2009). These are further categorized into rod bipolars, all of which are of the ON type, and cone bipolars based on their presynaptic targets (Wassle et al., 2009). Bipolar cells are born relatively late in mouse development and nearly all are generated postnatally (Rapaport et al., 2004; Sidman, 1961; Young, 1985). Several transcription factors have been identified that regulate bipolar development, including Chx10 (Vsx2) and Vsx1. Chx10 is expressed in retinal progenitors and bipolar cells (Burmeister et al., 1996) and is necessary for bipolar generation as Chx10-null retinas have few, if any, bipolar cells (Burmeister et al., 1996; Green et al., 2003; Livne-Bar et al., 2006). Vsx1 is expressed in a subset of cone bipolar cells and is not required for bipolar cell genesis, but for differentiation (Chow et al., 2001; Chow et al., 2004; Clark et al., 2008; Ohtoshi et al., 2001; Ohtoshi et al., 2004).
Photoreceptors and bipolar cells are similar in that they both express Otx2. This raises the question of how the Otx2+ population chooses between these cell fates during development. This is likely to be mediated through differential expression of transcription factors within the Otx2+ population. We examined the role of the PR-SET domain zinc-finger transcription factor Blimp1 (Prdm1) in more detail, based on preliminary expression characterization in the retina (Chang and Calame, 2002; Hsiau et al., 2007; Wang et al., 2008). Blimp1 can recruit Groucho family co-repressors and histone-modifying machinery to repress transcription (Ancelin et al., 2006; Hayashi et al., 2007; Ren et al., 1999; Yu et al., 2000). Blimp1 (B-lymphocyte-induced maturation protein) is required for B-cell maturation into plasma cells (Shapiro-Shelef et al., 2003; Turner et al., 1994) and is also important for T-cell, skin, vibrissae, pharyngeal arch, muscle, neural crest, limb, heart and primordial germ cell development (John and Garrett-Sinha, 2009). Here, we determined that Blimp1 is transiently expressed in developing photoreceptors. We generated conditional loss-of-function mice and show that Blimp1 mutants have fewer photoreceptors and more bipolar cells than wild-type mice. Blimp1 mutants have normal numbers of Otx2+ cells during development, but most of them develop as bipolar cells instead of photoreceptors, providing evidence for a one-to-one fate shift. After the first postnatal week, many of these excess bipolar cells die, yielding an adult retina with ∼50% more bipolar cells than normal. Our results show that Blimp1 controls the choice between photoreceptor and bipolar cell fates during development by repressing bipolar cell-specific gene expression in Otx2+ cells.
MATERIALS AND METHODS
Mice
For wild-type Blimp1 characterization, tissues were collected from C57BL/6 mice. For conditional knockout mice (CKO), two different Cre lines were used: Foxg1-Cre (Hebert and McConnell, 2000) and αPax6-Cre-GFP (Marquardt et al., 2001). These lines were crossed to Blimp1Flox/Flox (Jackson strain 008100) (Shapiro-Shelef et al., 2003) mice to generate Cre::Blimp1Flox/+ mice. These mice were crossed to generate the two populations used for analysis: Cre::Blimp1Flox/+ controls and Cre::Blimp1Flox/Flox CKO mice. We were unable to obtain postnatal CKO mice using the Foxg1-Cre strain, presumably because homozygous deletion resulted in perinatal lethality. For the E15.5 time-point we used Foxg1-Cre to generate CKO and control mice. For all other time-points we used the αPax6-Cre-GFP line to generate animals. Both lines resulted in efficient Blimp1 deletion. However, we observed variation between mice in the extent of the peripheral retina that was affected in the αPax6-Cre-GFP CKO mice. Mice were genotyped for Blimp1Flox following a previously published protocol (Shapiro-Shelef et al., 2003) and for Cre with the primers 5′-TGCCAGGATCAGGGTTAAAG-3′ and 5′-TCCTTAGCGCCGTAAATCAA-3′. Mice were used in accordance with University of Washington IACUC approved protocols.
Human fetal tissue
Tissues from 74, 96, 115 and 163 day post-conception fetuses, without identifiers, were obtained from the University of Washington Birth Defects Laboratory. The donated fetal tissue was collected from non-diseased elective abortion procedures. Tissues were used and collected in accordance with University of Washington IRB approved protocols.
Immunohistochemistry and microscopy
Tissues of various stages were fixed in 2% paraformaldehyde for 15 minutes to 2 hours at room temperature. Prior to fixation, the lens was removed from postnatal eyes. Tissues were cryopreserved through 30% sucrose, frozen in OCT (Sakura, Torrance, CA, USA) and sectioned at 10 μm. We found that Blimp1, Trβ2 and NeuroD1 immunostaining was fixation sensitive. A shorter fixation time resulted in more robust staining. Sections were blocked with milk-block (the supernatant from a solution of 5% dried milk powder and 0.5% Triton X-100 in PBS spun for 15 minutes at 11,750 g in a microcentrifuge) for 2-4 hours at room temperature. Primary antibodies were diluted in milk-block solution and incubated with the sections overnight. The sections were washed in PBS and incubated with appropriate fluorescent conjugated secondary antibodies (Invitrogen, Carlsbad, CA, USA) or streptavidin conjugate (Invitrogen) diluted in milk-block solution for 2 hours along with DAPI to counterstain nuclei. The sections were washed and mounted for microscopy.
Primary antibodies were as follows: mouse anti-Blimp1 (1:100) (NB600-235, Novus, Littleton, CO, USA), rat anti-Blimp1 (1:100) (sc47732, Santa Cruz, Santa Cruz, CA, USA), goat anti-Brn3 (1:50) (sc6026, Santa Cruz), rabbit anti-calbindin D-28K (Calb1) (1:500) (ab1778, Millipore, Billerica, MA, USA), rabbit anti-cleaved caspase 3 (1:200) (559565, BD Biosciences, San Jose, CA, USA), sheep anti-Chx10 (1:200) (X1179P, Exalpha, Shirley, MA, USA), mouse anti-CRALBP (1:250) (ab15051, Abcam, Cambridge, MA, USA), rabbit anti-Crx (1:100) (sc30150, Santa Cruz), chicken anti-GFP (1:500) (ab13970, Abcam), goat anti-NeuroD1 (1:50) (sc1084, Santa Cruz), rabbit anti-Olig2 (1:250) (ab33427, Abcam), goat anti-Otx2-biotin (2.5 μg/ml) (BAF1979, R&D Systems, Minneapolis, MN, USA), rabbit anti-Otx2/Crx (1:1000) (a gift of Cheryl Craft, University of California, Los Angeles) (Zhu and Craft, 2000), rabbit anti-m-opsin (Opn1mw) (1:250) (ab5405, Millipore), goat anti-s-opsin (Opn1sw) (1:150) (sc14363, Santa Cruz), rabbit anti-Pax6 (1:500) (PRB-278P, Covance, Princeton, NJ, USA), mouse anti-PKC (1:250) (P5704, Sigma, St Louis, MO, USA), rabbit anti-recoverin (1:1000) (ab5585, Millipore), mouse anti-rhodopsin (1:500) (a gift of Robert Molday, University of British Columbia) (Laird and Molday, 1988), goat anti-Sox2 (1:100) (sc17320, Santa Cruz), rabbit anti-Sox9 (1:750) (ab5535, Millipore), rabbit anti-Trβ2 (1:500) (a gift of Douglas Forrest, NIDDK) (Ng et al., 2001) and rabbit anti-Vsx1 (1:250) (a gift of Ed Levine, University of Utah) (Clark et al., 2008). The rat Blimp1 antibody worked much better than the previously published mouse antisera (Chang and Calame, 2002) and was used for all the images shown. For Otx2 immunostaining we used fluorescent streptavidin conjugates instead of secondary antibodies because this resulted in a cleaner signal.
Images were captured using a Zeiss (Thornwood, NY, USA) LSM 510 confocal microscope or a Nikon (Melville, NY, USA) A1 confocal microscope. Images were used as single planes or as z-stack projections. Images were processed in Adobe Photoshop (San Jose, CA, USA).
RT-PCR
For relative Blimp1 expression, we collected retinas in Trizol (Invitrogen) from BL/6 mice from several time-points and made three pools of RNA. RNA was prepared following the Trizol protocol and cleaned up using the RNeasy Kit (Qiagen, Germantown, MD, USA) according to the manufacturer's instructions. Each pool of RNA was treated with DNase, reverse transcribed with Superscript II (Invitrogen), and then subjected to RT-PCR twice using a DNA Engine Opticon and Opticon Monitor software (BioRad, Hercules, CA, USA) with SYBR Green Master Mix (Applied Biosystems, Foster City, CA, USA). Primer sequences: for Blimp1, 5′-GAACCTGCTTTTCAAGTATGCTG-3′ and 5′-AGTGTAGACTTCACCGATGAGG-3′; for Gapdh, 5′-GGCATTGCTCTCAATGACAA-3′ and 5′-CTTGCTCAGTGTCCTTGCTG-3′. Critical threshold cycle values for Blimp1 were normalized to Gapdh values and relative expression was determined by normalizing each point to the average E12.5 value. Data were plotted using GraphPad Prism (GraphPad Software, La Jolla, CA, USA).
Blimp1-GFP plasmid construction and in vivo electroporation
Human BLIMP1 cDNA was acquired from Tom Maniatis (Ren et al., 1999) and subcloned into the pMES expression vector (Swartz et al., 2001) to generate pMES-BLIMP1 (Blimp1-GFP). The pMES backbone contains chicken β-actin and CMV-1E promoter and enhancer sequences followed by a multiple cloning site and IRES2-EGFP sequences. For control electroporations, unmodified pMES (GFP) was used. For electroporations, these plasmids were mixed (2 to 1) with pCAGGS (Niwa et al., 1991), a vector that robustly drives GFP expression, in order to better visualize transfected cells over time, as the pMES and pMES-BLIMP1 vectors were not as abundantly expressed after 4 days in neurons (data not shown). For electroporations, P1 mice were anesthetized and a small cut was made to pull the eyelids apart. A 30-gauge needle was used to make a hole in the vitreous near the limbus of one eye. A pulled-glass micropipette containing the plasmid mix was inserted through the hole and ∼1 μl (∼6 μg) of DNA injected. Tweezertrodes (BTX, Hollistan, MA, USA) were placed on either side of the head such that the positive electrode was opposite the injected eye. The eyes were electroporated with a BTX T-820 electroporator with the following parameters: 90 Volts, five pulses, 50 milliseconds per pulse. Eyes were collected 4 days later at P5 for histology. Approximately 80% of the electroporated eyes had transfected cells in the retina and ∼25% had large patches of transfected cells, which were used for counting.
Cell counting and statistics
For quantification of adult cell demographics in control and CKO mice, we manually counted immunostained nuclei from 600× magnification images. For controls, we counted from at least five fields from four retinas for each staining combination. For CKO mice, we counted at least five fields from eight retinas for each condition. At P7 and P10, we manually counted labeled cells from 400× images. At P7, we counted at least ten fields from four retinas for each genotype and staining condition. At P10, we counted at least five fields from four retinas for each genotype and condition. At E14.5, we manually counted at least five 400× fields from wild-type mice for each marker. For in vivo electroporated retinas, we counted nine fields from three retinas for both GFP- and Blimp1-GFP-transfected retinas. We conducted two-tailed unpaired t-tests to evaluate comparisons between genotypes and transfections using Microsoft Excel (Bellevue, WA, USA).
RESULTS
Blimp1 is expressed in developing photoreceptors
Previous experiments have shown that Blimp1 is expressed in the embryonic and early postnatal retina of rodents (Chang and Calame, 2002; Hsiau et al., 2007; Wang et al., 2008). Although its expression pattern was consistent with developing photoreceptors, this was not explicitly characterized. To more fully describe the Blimp1 expression domain during retinal development, we immunostained sections from multiple developmental time-points.
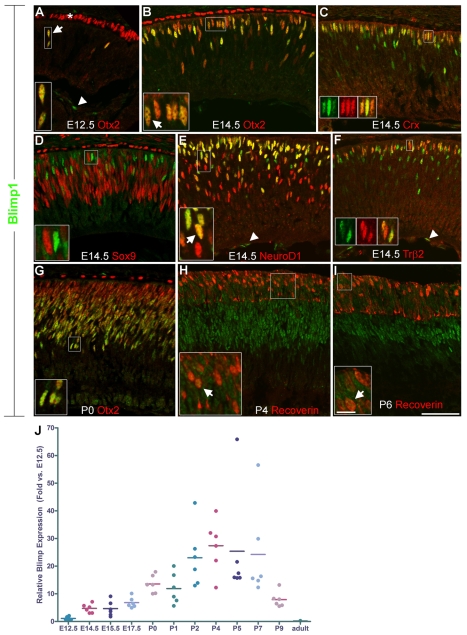
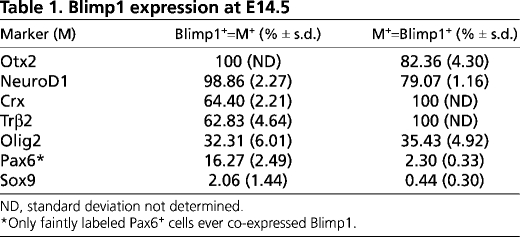
We first observed Blimp1 protein in the nucleus of cells in the central retina at E12. These Blimp1+ cells always co-expressed Otx2, a marker of developing photoreceptors and retinal pigmented epithelium (RPE) (Fossat et al., 2007; Nishida et al., 2003) (Fig. 1A). However, not all Otx2+ cells in the retina expressed Blimp1, nor did any Blimp1+ cells co-express Brn3 (Pou4f2 – Mouse Genome Informatics), an RGC marker (Xiang et al., 1995). We also observed that Blimp1 labeled vascular endothelial cells in the vitreous (Fig. 1A,E,F). At E14.5, the Blimp1 expression domain spanned from the central to peripheral retina, similar to what has been observed previously (Chang and Calame, 2002). The intensity of Blimp1 labeling was variable, but again, every Blimp1+ cell co-expressed Otx2 (Fig. 1B, Table 1). Crx is another photoreceptor-specific marker at E14.5 (Chen et al., 1997; Furukawa et al., 1997; Nishida et al., 2003). Every Crx+ cell co-expressed Blimp1 (Fig. 1C) and Otx2 (data not shown). However, there were more Blimp1+ cells than Crx+ cells and more Otx2+ cells than Blimp1+ cells (Table 1). Together, this showed that Blimp1 is expressed in developing photoreceptors and implied that Otx2 is expressed before Blimp1, which is expressed before Crx. Blimp1+ cells rarely co-expressed the progenitor marker Sox9 (Poche et al., 2008) (Fig. 1D, Table 1) and always co-expressed NeuroD1, which is expressed in postmitotic, developing photoreceptors and amacrine cells (Liu et al., 2008; Morrow et al., 1999) (Fig. 1E). Developing cones, labeled by Trβ2 (Ng et al., 2001), always co-expressed Blimp1, but accounted for only ∼60% of Blimp1+ cells at this stage (Fig. 1F, Table 1). This indicated that Blimp1 is expressed in both nascent postmitotic rods and cones at E14.5. We then examined the expression of Pax6 and Olig2, transcription factors that are expressed in both progenitors and postmitotic cells (de Melo et al., 2003; Nakamura et al., 2006). Pax6 labels progenitors weakly and postmitotic amacrines and RGCs intensely (de Melo et al., 2003). Although we observed no intensely labeled Blimp1+/Pax6+ cells, a small number of Blimp1+ cells expressed low levels of Pax6 (see Fig. S1 in the supplementary material; Table 1). Olig2 was expressed by ∼30% of Blimp1+ cells at E14.5 (see Fig. S1 in the supplementary material; Table 1). This implied that Olig2+ progenitors can give rise to photoreceptors and/or that Olig2 is transiently expressed in newly born photoreceptors. Transient retention of some progenitor markers is expected in newly postmitotic neurons. The small amount of progenitor marker overlap with Blimp1 implies that Blimp1 is an early marker for photoreceptor development.
Fig. 1.
Blimp1 expression in the developing mouse retina. (A) E12 retina stained for Blimp1 (green in all panels) and Otx2 (red). All Blimp1+ cells express Otx2 (arrow). Otx2 also labels the pigmented epithelium (asterisk). Blimp1 is also seen in vascular endothelial cells in the vitreous (arrowhead, as also in E,F). (B-F) E14.5 retinas stained for Blimp1. (B) All Blimp1+ cells co-express Otx2 (red) (arrow), but not all Otx2+ cells express Blimp1. (C) All Crx+ cells (red) co-express Blimp1, but not all Blimp1+ cells express Crx. (D) Few Blimp1+ cells co-express Sox9+ (red), a progenitor marker. (E) All Blimp1+ cells express NeuroD1 (red) (arrow), but not all NeuroD1+ cells express Blimp1. (F) All Trβ2+ cones express Blimp1. (G) At P0, all Blimp1+ cells co-express Otx2 (red). (H) At P4, Blimp1+ cells cover a large domain and many recoverin-labeled photoreceptors (red) co-express Blimp1 (arrow). (I) At P6, Blimp1 is seen only in the peripheral retina. Many recoverin-labeled (red) photoreceptors co-express Blimp1 (arrow). (J) Relative Blimp1 mRNA expression normalized to the E12.5 value. Shown are six measurements for each time-point; the bar represents the average. Expression starts at ∼E12 and is not seen in adult retina. Scale bars: 50 μm, except 10 μm for insets.
Table 1.
Blimp1 expression at E14.5
We next examined postnatal time-points for Blimp1 expression. At P0, Blimp1+ cells were broadly distributed throughout the retina and always co-expressed Otx2 (Fig. 1G). This pattern was similar at P4. Many recoverin-labeled photoreceptors (more mature) (Milam et al., 1993) co-expressed Blimp1 at P4 (Fig. 1H). Starting at P5, Blimp1 expression was lost from the central retina, and by P6 (Fig. 1I) and P7 expression was seen only in the peripheral retina. Expression was weaker in the outer retina, where photoreceptors were most mature (Fig. 1I). No immunostaining was seen in the adult retina (data not shown). Thus, the postnatal expression of Blimp1 was consistent with the genesis and maturation of photoreceptors (Carter-Dawson and LaVail, 1979). This transient Blimp1 expression pattern was also seen in the developing human retina (see Fig. S2 in the supplementary material). Lastly, we examined mRNA expression by quantitative RT-PCR, plotting relative expression normalized to E12 (Fig. 1J), the earliest time that Blimp1 was observed. The relative mRNA expression curve matched the immunostaining data, except that the mRNA peaks and ends slightly later. Thus, Blimp1 might also be regulated post-transcriptionally in the retina.
Blimp1 is needed for normal photoreceptor development
The transient expression of Blimp1 suggested that it plays a role in the development of photoreceptors. To test this, we examined Blimp1 loss-of-function mutants. Blimp1 knockout mice die in utero (Vincent et al., 2005), so we made conditional knockout (CKO) mice by crossing the previously generated Blimp1Flox strain (Shapiro-Shelef et al., 2003) with one of two strains that express Cre recombinase in retinal progenitors. The Foxg1-Cre strain expresses Cre recombinase in ∼50% of retinal progenitors during development, starting before the onset of Blimp1 expression (Hebert and McConnell, 2000). The αPax6-Cre-GFP strain expresses Cre in retinal progenitors in the peripheral part of the retina, starting before the onset of Blimp1 expression (Marquardt et al., 2001). This transgene is expressed in amacrine cells throughout the life of the animal (Marquardt et al., 2001). We crossed both Cre strains to Blimp1Flox mice to generate heterozygous controls (Cre::Blimp1Flox/+) and CKO mice (Cre::Blimp1Flox/Flox). Since Foxg1-Cre CKO mice did not survive to birth, we focused on αPax6-Cre-GFP CKO mice, which were viable.
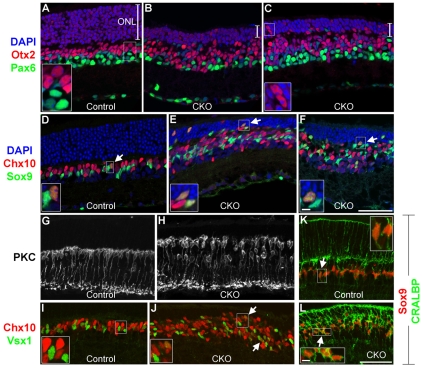
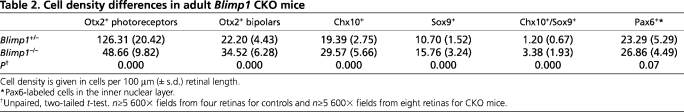
We first examined 3- to 4-week-old CKO mice, an age when all the cells in the retina have differentiated. The peripheral retinas of heterozygous controls and CKO mice were compared using various cell type-specific markers. It was immediately clear from nuclear counterstaining that the outer nuclear layer (ONL) of CKO mice contained fewer cells than control retinas (Fig. 2A-F). Moreover, the inner nuclear layer (INL) was thicker in CKO mice (Fig. 2A-F). This was consistent with a loss of photoreceptors and a gain of interneurons or glia. We then immunostained retinas for Otx2 to measure photoreceptors and bipolar cells. Mature photoreceptors have small nuclei that are stained near the edges, where euchromatin is preferentially packaged (Fossat et al., 2007; Solovei et al., 2009). Bipolar cells have larger, intensely stained nuclei located in the INL. Controls had normal Otx2 staining patterns, but CKO mice had far fewer Otx2+ photoreceptors and ∼50% more Otx2+ bipolar cells (Fig. 2A-C, Table 2). This reduction in photoreceptors affected rods and cones (see Fig. S3 in the supplementary material). The regular laminar organization of bipolar cells was lost in CKO mice and the outer plexiform layer (OPL) was disrupted. By contrast, the localization and number of Pax6+ cells in the INL did not differ between the strains (Fig. 2A-C, Table 2), nor were any differences seen in the number of RGCs or horizontal cells (data not shown). We stained retinas for Chx10 to label most bipolars (Burmeister et al., 1996) and observed ∼50% more Chx10+ bipolars in CKO mice (Fig. 2D-F, Table 2). Again, their laminar organization was disrupted and bipolar cell nuclei were often observed in the ONL (Fig. 2D-F). The increase in bipolar cells was confirmed with other markers, including protein kinase C (PKC), which labels rod bipolar cells (Greferath et al., 1990). Similar to the Otx2 and Chx10 staining results, there were more PKC+ rod bipolars in CKO mice and these bipolars showed disrupted lamination (Fig. 2G,H). Cone bipolars labeled for Vsx1 (Chow et al., 2001; Ohtoshi et al., 2001) were increased in CKO mice and showed disrupted laminar organization (Fig. 2I,J). Vsx1+ cells in CKO mice were more likely to co-express Chx10 than Vsx1+ cells in controls (Fig. 2I,J), but the relevance of this co-expression is unknown.
Fig. 2.
Blimp1 CKO mice have fewer photoreceptors and increased bipolars and glia. Heterozygous control and Blimp1 CKO peripheral retinal sections from P23-28 mice. (A-C) Sections stained for Otx2 (red), Pax6 (green) and with DAPI (blue). CKO sections (B,C) have a thinner outer nuclear layer (ONL) (bracket) and thicker inner nuclear layer (INL) compared with the control (A). There are more Otx2+ bipolar cells in CKO animals, but no change in the number of Pax6+ cells. (D-F) Sections stained for Chx10 (red), Sox9 (green) and with DAPI (blue). CKO mice (E,F) have more Chx10+ bipolars and Sox9+ glia than controls (D). CKO mice also have bipolars and glia in the ONL, along with increased numbers of Chx10+/Sox9+ glia (arrows). (G,H) PKC staining of control (G) and CKO (H) sections shows increased rod bipolars in CKO mice, along with disrupted laminar organization. (I,J) Chx10 (red) and Vsx1 (green) bipolar cell labeling of control (I) and CKO (J) sections. CKO mice have more of each bipolar population, more double-labeled cells (arrows), and have disrupted laminar organization, including cells located in the ONL. (K,L) Sox9 (red) and CRALBP (green) labeling shows increased Müller glia (arrows) in CKO mice. Scale bars: 50 μm, except 5 μm for insets.
Table 2.
Cell density differences in adult Blimp1 CKO mice
In addition to the increase in bipolar cells that we observed in CKO peripheral retina, there were also ∼50% more Sox9+ Müller glia (Poche et al., 2008) (Fig. 2D-F, Table 2). Sox9+ glial nuclei were not organized into a single row and were occasionally observed in the ONL of CKO retinas (Fig. 2D-F). We also observed an increase in the number of glia that co-expressed Sox9 and Chx10 (Hatakeyama et al., 2001; Rowan and Cepko, 2004) compared with the control (Table 2). The Müller glia-specific marker CRALBP (Rlbp1 – Mouse Genome Informatics) (Eisenfeld et al., 1985) co-localized, and increased proportionally, with Sox9 in CKO mice (Fig. 2K,L).
The loss of photoreceptors in adult CKO mice was about four times greater than the gains seen in bipolars and Müller glia (Table 2), raising the possibility of excess bipolar and glial genesis or photoreceptor cell death. We did not observe any proliferating cells or an increase in apoptotic cells in adult CKO retinas (data not shown), suggesting that all the cell population changes occurred during development.
Blimp1 represses bipolar fate in nascent photoreceptors
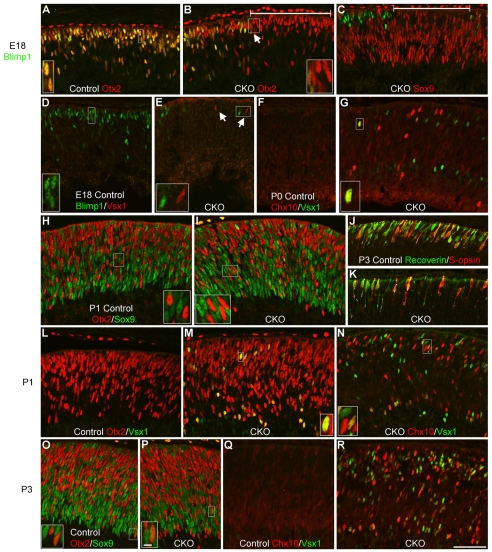
The presence of extra bipolar and Müller glia at the expense of photoreceptors in CKO mice suggested a role for Blimp1 in regulating photoreceptor fate choice. We examined CKO mice at earlier developmental stages for changes in photoreceptor markers. We first examined E15.5 Blimp1 CKO mice using the Foxg1-Cre line (for this age only). These mice had a broad loss of Blimp1 from the retina. Other than the loss of Blimp1, we did not observe any changes in Otx2, Sox9, Pax6, Trβ2, Crx or NeuroD1 (see Fig. S4 in the supplementary material). When we examined E18 CKO mice for the same markers, they appeared the same as in the controls (Fig. 3A-C). However, at this stage we observed a small number of Vsx1+ cells in Blimp1-deleted regions (Fig. 3D,E), which suggested precocious bipolar cell specification. The number of precocious Vsx1+ cells was even greater at P0 and P1 (Fig. 3D-G). At these stages, we detected cells that were intensely labeled for Chx10 (Fig. 3F,G,N), also indicative of precocious bipolar cell specification (Burmeister et al., 1996). The Vsx1+ and Chx10+ cells always co-expressed Otx2, consistent with bipolar fate choice (Fig. 3M and data not shown). At P1 and P3, we saw fewer recoverin- and S-opsin-labeled cells (Fig. 3J,K) in CKO than in control mice, consistent with a reduction in rod and cone differentiation. At P3, Blimp1 CKO mice had a large number of brightly labeled Chx10+ and Vsx1+ cells, whereas control mice still did not express these markers (Fig. 3Q,R). This suggested that Otx2+ cells were adopting bipolar fate at the expense of photoreceptor fate in CKO mice. At P1 and P3, expression of Otx2 and Sox9 was similar in CKO and control retinas (Fig. 3H,I,L,O,P). Together, these data show that Blimp1 is not required for the initial expression of photoreceptor markers (Otx2, Crx, Trβ2 and NeuroD1), but that it negatively regulates Chx10 and Vsx1 in the Otx2+ population.
Fig. 3.
Blimp1 CKO mice upregulate bipolar cell-specific genes. All panels show peripheral retina. (A-C) E18 control (A) and CKO (B,C) retina stained for Blimp1 (green). All Blimp1+ cells co-express Otx2 (red) in control and CKO mice (arrow). Despite the loss of Blimp1 (bracketed) in CKO sections, there is no effect on the expression of Otx2 (B) or Sox9 (red) (C). (D,E) E18 sections stained for Blimp1 (green) and Vsx1 (red). Control mice had no detectable Vsx1 (D), but a few Vsx1+ cells (arrows) are seen in Blimp1-deleted regions of CKO mice (E). (F,G) At P0, no intensely labeled Chx10+ (red) or Vsx1+ cells are seen in control sections (F), but they are present in Blimp1-deleted regions of CKO mice (G). (H,I) At P1, Otx2 (red) and Sox9 (green) have minimal overlap in control (H) and CKO (I) retinas and show similar expression patterns in both genotypes. (J,K) The early mature photoreceptor markers recoverin (green) and s-opsin (red) are reduced at P1 and P3 in CKO retinas (K) compared with controls (J). (L,M) At P1, control retinas (L) do not label for Vsx1 (green). In CKO sections (M), all Vsx1+ cells co-express Otx2 (red). (N) P1 CKO retina showing abundant staining for Chx10 (red) and Vsx1 (green). (O,P) P3 control (O) and CKO (P) retinas stained for Otx2 (red) and Sox9 (green). These markers had little overlap and a similar distribution in both genotypes. (Q,R) P3 control (Q) and CKO (R) sections stained for Chx10 (red) and Vsx1 (green) showing their absence in controls and abundance in CKO retinas. Scale bars: 50 μm, except 5 μm for insets.
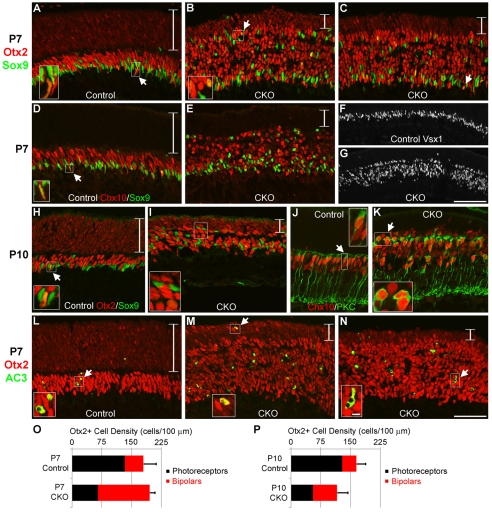
Next, we examined time-points at the end of retinogenesis. At P7, heterozygous controls had an Otx2+ ONL and brightly stained Otx2+ bipolar cells in the INL (Fig. 4A). These layers were noticeably separated by the OPL, with a few photoreceptors present on the wrong (inner) side of this border. By contrast, P7 CKO mice had numerous Otx2+ bipolar cells and few photoreceptors (Fig. 4B,C). The OPL was hard to discern in CKO retinas, but when observed, many Otx2+ bipolar cells were misplaced into the ONL. Blimp1 CKO retinas had increased numbers of Chx10+ and Vsx1+ bipolar cells compared with controls at P7 (Fig. 4D-G). When we counted the number of Otx2+ photoreceptors and bipolars, there were more bipolars and fewer photoreceptors in CKO than in control mice (Fig. 4O, Table 3). The decrease in photoreceptors was proportional to the increase in bipolar cells in CKO mice, yet there were no differences in the total number of Otx2+ cells between CKO mice and controls (Fig. 4O, Table 3). This implied that there was an approximately one-to-one fate shift between photoreceptors and bipolars. By P10, heterozygous control mice had laminated retinas with Otx2+ photoreceptors in the ONL and Otx2+ bipolars in the INL (Fig. 4H). Blimp1 CKO mice had a much thinner retina than controls, reminiscent of adult CKO mice (Fig. 4I). At P10, there were few photoreceptors, but also fewer bipolar cells than seen at P7 (Fig. 4I,P, Table 3). In contrast to P7 retinas, there were significantly fewer total Otx2+ cells in P10 CKO than control retinas (Fig. 4P, Table 3). At P10, there was excess PKC staining in CKO mice, consistent with the increase in bipolar cells (Fig. 4J,K). By contrast, PKC staining was not seen in the most peripheral parts of P7 control or CKO retinas, suggesting that the excess bipolar cells do not fully differentiate ahead of schedule (data not shown).
Fig. 4.
Otx2+ cells in Blimp1 CKO mice switch from photoreceptor to bipolar fate. All panels show peripheral retina. (A-C) P7 control (A) and CKO (B,C) retinas stained for Otx2 (red) and Sox9 (green). The small, fainter Otx2+ nuclei are photoreceptors and the large, bright nuclei are bipolar cells. The photoreceptor layer (bracketed) is much thinner in CKO mice and there is a proportional increase in the number of bipolar cells. In both control and CKO sections, several Sox9+ cells co-express Otx2 (arrows). CKO mice also have more Sox9+ cells (mostly glia), which are scattered throughout the retina. (D,E) P7 control (D) and CKO (E) sections stained for Chx10 (red) and Sox9 (green). CKO mice have more Chx10+ bipolars and Sox9+ cells. The arrow shows a double-labeled Müller cell. (F,G) P7 control (F) and CKO (G) retinas stained for Vsx1. There are many more Vsx1+ bipolars in CKO mice and their distribution is broader. (H,I) P10 control (H) and CKO (I) sections stained for Otx2 (red) and Sox9 (green). At this stage, the ONL (bracketed) is much thinner in CKO mice and there are fewer excess bipolars and glia than seen at P7. Otx2+/Sox9+ cells were detected (arrow). (J,K) P10 control (J) and CKO (K) mice stained for Chx10 (red) and PKC (green) to label bipolars. (J) All rod bipolars express PKC and Chx10 (arrow), forming a row in the INL. In CKO mice (K), there are more PKC- and Chx10-labeled bipolars (arrow). (L-N) P7 control (L) and CKO (M,N) retinas stained for Otx2 (red) and activated caspase 3 (AC3) (green). Most of the dying cells in controls and CKO mice are in the bipolar cell area rather than in the photoreceptor area (bracketed). CKO mice have ∼3-fold more dying cells than controls, some of which are still Otx2+ (arrows). (O,P) Otx2+ photoreceptor (black) and bipolar (red) cell density measured at P7 (O) and P10 (P). At P7 there is an approximately one-to-one fate shift in the CKO Otx2+ population. The error bars indicate the s.d. of the total Otx2+ cells counted. Scale bars: 50 μm, except 100 μm in F,G and 5 μm in insets.
Table 3.
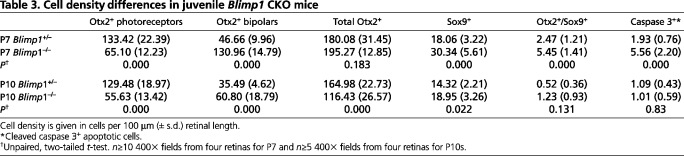
Cell density differences in juvenile Blimp1 CKO mice
The majority of Otx2+ cells in CKO mice adopted bipolar cell fate by P7. However, we also saw a smaller increase in the Sox9+ population at this stage (Table 3). At P7, in the control most of the Sox9+ cells were Müller glia, which formed a single layer in the INL (Fig. 4A). Blimp1 CKO mice had ∼2-fold more Sox9+ cells, which were spread throughout the area where bipolar cells are observed (Fig. 4B,C). Interestingly, both Blimp1 CKO and control retinas had a sizeable population of Otx2+/Sox9+ cells at P7 (Fig. 4A-C, Table 3). The ∼2-fold increase in Otx2+/Sox9+ cells paralleled the increase in Sox9+ cells in CKO mice (Table 3). What these double-labeled cells represent is unclear, but they might be newly formed Müller glia, raising the possibility that the excess glia seen in CKO retinas derived from Otx2+ cells. At P10, there are few, if any, progenitors left in the retina (Carter-Dawson and LaVail, 1979; Young, 1985). However, the presence of a small population of Otx2+/Sox9+ cells in both genotypes at this stage (Table 3) further suggested that these double-labeled cells were Müller glia.
The difference in the number of Otx2+ cells between P7 and P10 suggested that many of the fate-shifted bipolar cells were dying between these stages. We examined cell death at several time-points. At P1 and P3, no differences in activated caspase 3 labeling, a marker of apoptotic cells (Thornberry and Lazebnik, 1998), were seen between control and CKO mice (data not shown). However, at P7, we observed ∼3-fold more caspase 3+ cells in CKO mice as compared with control mice (Fig. 4L-N, Table 3). Most of the cell death was in the INL (Fig. 4M,N), consistent with the loss of bipolar cells and Müller glia. By P10, the amount of cell death was equivalent between genotypes (Table 3). Thus, many of the excess bipolar cells generated in the Blimp1 CKO mice died between P7 and P10, which correlated with the timing of bipolar maturation and the peak of normal cell death in this layer (Bramblett et al., 2004; Young, 1984).
Blimp1 overexpression inhibits bipolar cell fate choice
Blimp1 CKO mice have fewer photoreceptors and excess bipolar cells, suggesting that Blimp1 inhibits bipolar cell development. To determine whether Blimp1 is sufficient to inhibit bipolar cell genesis, we engineered expression plasmids to drive GFP or Blimp1-GFP in the retina at a time when bipolar versus photoreceptor fate decisions are being made. These constructs were injected into the vitreous of P1 eyes and electroporated into the retina. We examined the fate of transfected (GFP+) cells at P5, allowing 4 days for effects on bipolar versus photoreceptor fate to become apparent. At P5, most transfected cells co-expressed Otx2 in control (GFP) and Blimp1-GFP retinas (Fig. 5A,C) (see Table S1 in the supplementary material). Photoreceptors had smaller, faintly labeled nuclei, whereas bipolars and undifferentiated cells (nascent bipolars and photoreceptors) had bright, elongated Otx2 staining. Chx10 co-labeling was used to more definitively mark transfected bipolar cells (Fig. 5B,D). Transfected cells were scored for faint Otx2 (Otx2+), strong Otx2 (Otx2++), and Chx10 (Chx10+) expression (Fig. 5E). Blimp1-GFP-transfected cells adopted photoreceptor fate more readily (26% more) and bipolar (and undifferentiated) fate less frequently (24% fewer) than the GFP control (Fig. 5E). Thus, Blimp1 overexpression caused a shift in the fate distribution of the Otx2+ population, but in the opposite direction to that observed in Blimp1 CKO retinas. The inhibitory effect of Blimp1 was more pronounced on definitive, Chx10+ bipolar cells, which were reduced by 59% compared with GFP-transfected controls (Fig. 5E).
Fig. 5.
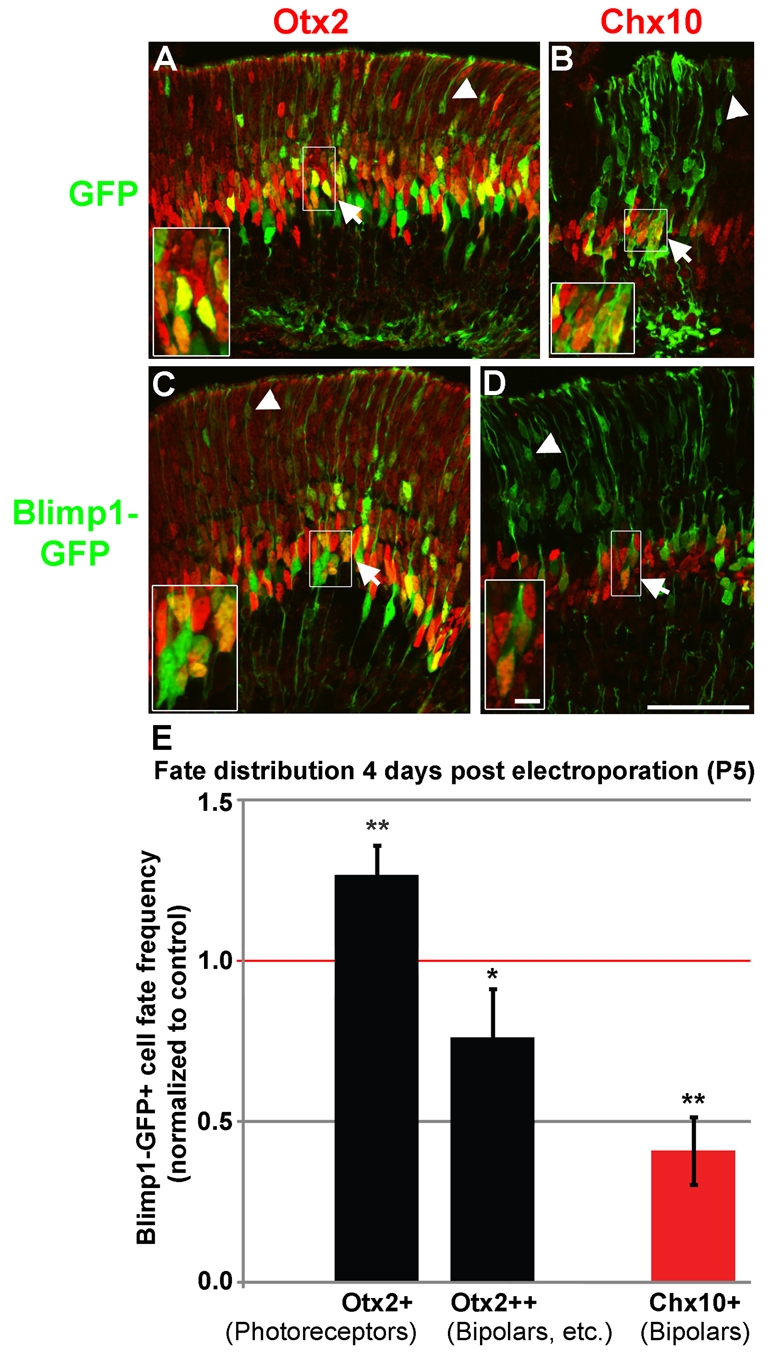
Blimp1 overexpression increases photoreceptor and decreases bipolar cell fate choice. (A-D) P1 retinas were electroporated in vivo with GFP control (A,B) or Blimp1-GFP (C,D) expression vectors and tissue collected at P5. (A,C) Transfected regions of control (A) and Blimp1-GFP (C) retinas stained for GFP (green) and Otx2 (red). The bright, elongated nuclei (termed Otx2++) represent bipolars and undifferentiated photoreceptors and bipolars (arrows), whereas the faint, small nuclei (Otx2+) mark photoreceptors (arrowheads). (B,D) Definitive bipolars in GFP control (B) or Blimp1-GFP (D) transfected retinas were labeled for Chx10 (red) (arrows). Arrowheads mark photoreceptors. (E) Relative frequency of cell fates observed in Blimp1-GFP-transfected cells normalized to control (1.0). At P5, Blimp1-GFP cells were more likely to be photoreceptors and less likely to be bipolar cells compared with controls. Error bars indicate the s.d. *, P<0.05; **, P<0.01; unpaired two-tailed t-tests. Scale bars: 50 μm, except 5 μm for insets.
DISCUSSION
We observed that the PR-SET domain zinc-finger transcription factor Blimp1 is expressed in developing photoreceptors in mammals. Genetic deletion of this transcription factor in the retina led to conversion of most of the photoreceptors into bipolar cells, although some may also have adopted Müller glial fate. As the Blimp1 CKO retinas matured, many of the fate-shifted cells were lost; however, there were still ∼50% more bipolars and glia than normal. Overexpression of Blimp1 promoted photoreceptor fate at the expense of bipolar cells. We conclude that during retinal development, Blimp1 regulates cell fate choice by inhibiting the `bipolar cell program' in nascent Otx2+ photoreceptors.
Previous experiments have shown that Blimp1 is likely to be expressed in developing photoreceptors (Chang and Calame, 2002; Hsiau et al., 2007; Wang et al., 2008). Our data are consistent with Blimp1 marking developing photoreceptors for the following reasons. First, Blimp1 expression starts at E12, the time when the first photoreceptors exit the cell cycle (Carter-Dawson and LaVail, 1979). Second, at all stages, Blimp1+ cells co-express Otx2, a marker of photoreceptors. Third, all Crx+ photoreceptors and Trβ2+ cones co-expressed Blimp1. Fourth, all Blimp1+ cells co-expressed NeuroD1, which is a marker of postmitotic photoreceptors and amacrines. Fifth, many recoverin+ photoreceptors co-express Blimp1 neonatally. Sixth, Blimp1 expression rarely coincided with markers of non-photoreceptor cell types. These criteria establish that Blimp1 is expressed in photoreceptors, although it remains possible that Blimp1 is produced transiently in all Otx2+ cells, including those that give rise to bipolar cells. A lineage-tracing study is needed to examine this possibility. Previous experiments have suggested that Otx2 expression precedes that of Crx (Nishida et al., 2003). At E14.5, all Crx+ cells expressed Blimp1 and both these populations co-expressed Otx2. This nested pattern suggests that the order of gene expression in nascent photoreceptors is Otx2, then Blimp1, and lastly Crx.
Conditional Blimp1 deletion resulted in retinas with fewer photoreceptors in the affected areas. A similar result has been reported in Blimp1 mutant fish, which have reduced expression of opsins (Wilm and Solnica-Krezel, 2005). It is unclear why some photoreceptors remain in Blimp1 CKO mice. It is possible that some cells in the affected regions did not lose Blimp1. Furthermore, although αPax6-Cre is expressed before Blimp1, it is possible that some cells did not delete Blimp1 until after it had been briefly expressed, stabilizing photoreceptor identity in this subset of cells. If only the latest born Otx2+ cells retained photoreceptor identity in Blimp1 CKO mice, the retina should contain only rods, as cones are not generated postnatally. However, both rods and cones were observed in CKO mice. Another possibility is that only postnatally generated Otx2+ cells convert to bipolars. This is unlikely because cones (early born) were also reduced in Blimp1 CKO mice. The cone reduction is seen early on (P1-P3), which argues against the possibility of selective cone death in adult CKO mice. Lastly, it is possible that another factor partially compensates for Blimp1 activity. In support of this, there are many Prdm family members in mice, most of which are expressed in the developing retina (J.A.B. and T.A.R., unpublished).
Blimp1 CKO mice had increased bipolar and Müller glial cells and, conversely, overexpression of Blimp1 resulted in fewer bipolar cells and increased photoreceptors. These experiments suggest that Blimp1 regulates cell fate during retinogenesis. Blimp1 has been shown to regulate fate choice in other systems. In B-cells, Blimp1 directly represses key transcription factors to allow plasma cell differentiation (Calame et al., 2003). In fish, Blimp1 acts as a switch between slow and fast twitch muscle types (Elworthy et al., 2008; von Hofsten et al., 2008). In primordial germ cells, Blimp1 is required to repress the somatic program, allowing germ cell specification in cells otherwise competent to adopt somatic fate (Ohinata et al., 2005; Vincent et al., 2005). In Blimp1 CKO mice, we observed a direct fate shift in the Otx2+ population, which is competent for photoreceptor and bipolar fates. Prior to birth, rod and cone fate specification was unaffected in Blimp1 CKO mice. By P1, there were fewer rods and cones, but no changes in Otx2 expression. In Blimp1 CKO mice, Otx2+ cells prematurely co-express the bipolar-specific markers Vsx1 and Chx10 (Fig. 6A,B) such that many previously generated rods and cones lose their photoreceptor identity in the absence of Blimp1. This indicates that Blimp1 is needed to stabilize, but not specify, photoreceptor fate during development (Fig. 6C). This also implies that photoreceptor identity is not permanently established at the time of cell cycle exit, but might require continued stabilization from the `bipolar cell program' until late in retinogenesis. Consistent with this hypothesis, Chx10 overexpression can directly repress the photoreceptor program (Dorval et al., 2006; Livne-Bar et al., 2006). It might be that Chx10 is needed to repress photoreceptor fate (Livne-Bar et al., 2006), whereas Blimp1 represses bipolar fate, allowing fate diversification in the postmitotic Otx2+ population. Our data suggest that Blimp1 represses Chx10 and Vsx1 expression, blocking bipolar development. Cells with high-level Chx10 and Vsx1 expression are not normally seen until P4 or later (Fig. 6A,B), well after many bipolar cells have exited the cell cycle (Morrow et al., 2008; Young, 1985). This raises the possibility that postmitotic Otx2+ cells remain plastic and do not commit to bipolar fate for up to several days after exiting the cell cycle.
Fig. 6.
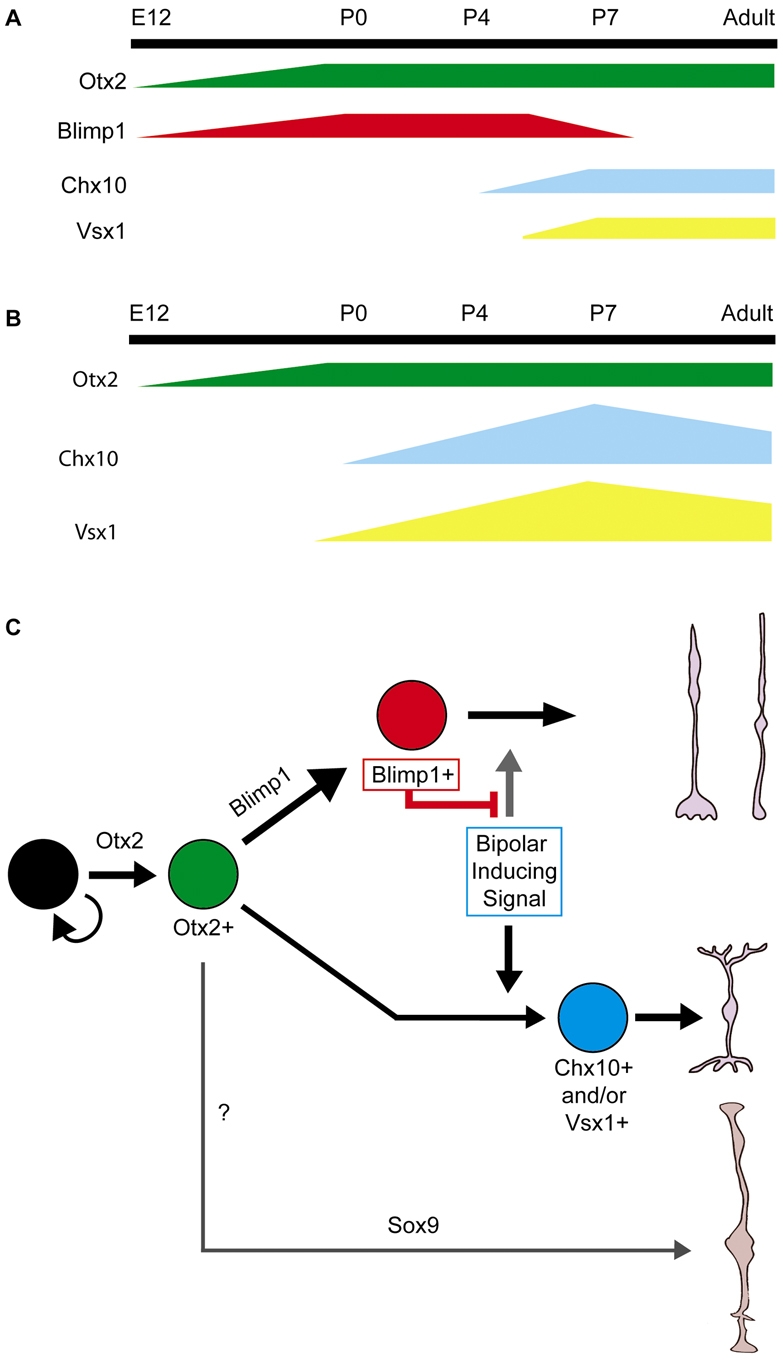
Blimp1 is required to stabilize, but not specify, photoreceptor fate during mouse retinal development. Cone photoreceptors are born from ∼E12 to P0, whereas rods are born from E13 to P7. (A) Transcription factor expression during normal development. Otx2 and Blimp1 are present in nascent photoreceptors starting at E12. Otx2 remains expressed in photoreceptors and bipolars, whereas Blimp1 is downregulated at ∼P7, the end of photoreceptor genesis. Chx10 and Vsx1 are present in bipolar cells starting at P4 and P5, respectively. (B) In Blimp1 mutants, Otx2 expression is unchanged. By contrast, Vsx1 expression is seen at E18.5 and intensely labeled Chx10+ cells are seen as early as P0. The number of Chx10+ and Vsx1+ bipolar cells peaks at P7 and many of the excess cells die by P10. (C) Blimp1 stabilizes photoreceptor fate from bipolar induction. Mitotic progenitors (black) give rise to Otx2+ postmitotic cells, which have competence for photoreceptor, bipolar and, possibly, Müller glial cell fates. Blimp1 expression labels developing photoreceptors. Around E18.5, a bipolar-inducing signal begins to act on the Otx2+ population. Blimp1 represses this signal, stabilizing photoreceptor fate. Otx2+ cells lacking Blimp1 adopt bipolar cell fate and express the markers Chx10 and/or Vsx1. It is unclear whether Blimp1 is produced in all Otx2+ cells, or just in those destined to become photoreceptors.
Blimp1 regulates the choice between photoreceptor and bipolar fates. Two other mouse mutants exhibit similarly extensive direct fate shifts in the retina. In Nrl-null mice, all rods become cones (Mears et al., 2001). In conditional Otx2 mutants, photoreceptors and bipolars are lost and these cells adopt amacrine cell fate (Nishida et al., 2003; Sato et al., 2007). In both of these mutants, many of the excess (fate shifted) cells are lost by apoptosis (Mears et al., 2001; Nishida et al., 2003; Sato et al., 2007). Similarly, many excess bipolar cells generated in Blimp1 CKO mice are lost around the time of bipolar cell maturation. It is unclear why these excess bipolar cells are lost. One possibility is that bipolar cells need trophic support from other neurons to mature and survive. The most likely sources of this support are photoreceptors (presynaptic) and RGCs (postsynaptic for most). In photoreceptor-degeneration mouse lines, bipolar cells change molecularly and physically, but survive (Punzo and Cepko, 2007). In mutants lacking RGCs, there are fewer bipolar cells, but most remain, suggesting that RGCs are not required for bipolar cell survival either (Brzezinski et al., 2005). Another possibility is that Müller glia are required to maintain bipolar cell numbers. In support of this, both bipolar and Müller glial numbers are increased by ∼50% in adult Blimp1 CKO mice. Thus, a 50% increase in glia might support a 50% increase in bipolar cells, but this remains to be demonstrated directly.
Although not as extensive as for bipolar cells, we also saw an increase in the number of Müller glia generated in CKO mice. This increase could also be the result of a direct fate shift of Otx2+ cells into glia. In support of this, we observed a ∼2-fold increase in both Sox9+ and Otx2+/Sox9+ cells in Blimp1 CKO mice at P7, suggesting that at least some glia derive from Otx2+ cells. We also observed a population of Otx2+/Sox9+ cells in adult CKO retinas (see Fig. S5 in the supplementary material). Nonetheless, it remains possible that the change in glial numbers is non-cell-autonomous. A lineage-tracing experiment is needed to distinguish these possibilities.
During retinal development a cohort of cells competent for cone, rod, bipolar and possibly glial fates exits the cell cycle and expresses Otx2 (Fig. 5C.) To achieve cell diversity in the Otx2+ population, a subset of these cells expresses Blimp1 (Fig. 5C). Blimp1 stabilizes photoreceptor fate by preventing bipolar-inducing signals from having an effect, in essence restricting the competence of this subset of Otx2+ cells to photoreceptors (Fig. 5C). The Otx2+ cells that do not express Blimp1 are competent to adopt bipolar and, perhaps, glial fates (Fig. 5C). Our data show that Blimp1 is the molecular switch that regulates photoreceptor versus bipolar fate choice during retinal development.
Supplementary Material
Acknowledgments
We thank Amy Weinmann, Emily Rowell and Christopher Wilson for advice and reagents; Ed Levine, Robert Molday and Cheryl Craft for sharing antibodies; the University of Washington Birth Defects Laboratory for providing tissues; Susan McConnell for Foxg1-Cre mice and Ruth Ashery-Padan for αPax6-Cre-GFP mice; Tom Maniatis and Huck-Hui Ng for BLIMP1 plasmids; and members of the Reh and Bermingham-McDonogh labs for helpful discussion, especially Sean Georgi and Mike Karl. J.A.B. was supported by NIH training grants T32-EY07031 and F32-EY19227. This research was supported by NIH grant R01 EY013475 to T.A.R. Deposited in PMC for release after 12 months.
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.043968/-/DC1
References
- Akimoto M., Cheng H., Zhu D., Brzezinski J. A., Khanna R., Filippova E., Oh E. C., Jing Y., Linares J. L., Brooks M., et al. (2006). Targeting of GFP to newborn rods by Nrl promoter and temporal expression profiling of flow-sorted photoreceptors. Proc. Natl. Acad. Sci. USA 103, 3890-3895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ancelin K., Lange U. C., Hajkova P., Schneider R., Bannister A. J., Kouzarides T., Surani M. A. (2006). Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 8, 623-630 [DOI] [PubMed] [Google Scholar]
- Bramblett D. E., Pennesi M. E., Wu S. M., Tsai M. J. (2004). The transcription factor Bhlhb4 is required for rod bipolar cell maturation. Neuron 43, 779-793 [DOI] [PubMed] [Google Scholar]
- Brzezinski J. A., Brown N. L., Tanikawa A., Bush R. A., Sieving P. A., Vitaterna M. H., Takahashi J. S., Glaser T. (2005). Loss of circadian photoentrainment and abnormal retinal electrophysiology in Math5 mutant mice. Invest. Ophthalmol. Vis. Sci. 46, 2540-2551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burmeister M., Novak J., Liang M. Y., Basu S, Ploder L., Hawes N. L., Vidgen D., Hoover F., Goldman D., Kalnins V. I., et al. (1996). Ocular retardation mouse caused by Chx10 homeobox null allele: impaired retinal progenitor proliferation and bipolar cell differentiation. Nat. Genet. 12, 376-384 [DOI] [PubMed] [Google Scholar]
- Calame K. L., Lin K. I., Tunyaplin C. (2003). Regulatory mechanisms that determine the development and function of plasma cells. Annu. Rev. Immunol. 21, 205-230 [DOI] [PubMed] [Google Scholar]
- Carter-Dawson L. D., LaVail M. M. (1979). Rods and cones in the mouse retina. II. Autoradiographic analysis of cell generation using tritiated thymidine. J. Comp. Neurol. 188, 263-272 [DOI] [PubMed] [Google Scholar]
- Chang D. H., Calame K. L. (2002). The dynamic expression pattern of B lymphocyte induced maturation protein-1 (Blimp-1) during mouse embryonic development. Mech. Dev. 117, 305-309 [DOI] [PubMed] [Google Scholar]
- Chen S., Wang Q. L., Nie Z., Sun H., Lennon G., Copeland N. G., Gilbert D. J., Jenkins N. A., Zack D. J. (1997). Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 19, 1017-1030 [DOI] [PubMed] [Google Scholar]
- Chow R. L., Snow B., Novak J., Looser J., Freund C., Vidgen D., Ploder L., McInnes R. R. (2001). Vsx1, a rapidly evolving paired-like homeobox gene expressed in cone bipolar cells. Mech. Dev. 109, 315-322 [DOI] [PubMed] [Google Scholar]
- Chow R. L., Volgyi B., Szilard R. K., Ng D., McKerlie C., Bloomfield S. A., Birch D. G., McInnes R. R. (2004). Control of late off-center cone bipolar cell differentiation and visual signaling by the homeobox gene Vsx1. Proc. Natl. Acad. Sci. USA 101, 1754-1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. M., Yun S., Veien E. S., Wu Y. Y., Chow R. L., Dorsky R. I., Levine E. M. (2008). Negative regulation of Vsx1 by its paralog Chx10/Vsx2 is conserved in the vertebrate retina. Brain Res. 1192, 99-113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Melo J., Qiu X., Du G., Cristante L., Eisenstat D. D. (2003). Dlx1, Dlx2, Pax6, Brn3b, and Chx10 homeobox gene expression defines the retinal ganglion and inner nuclear layers of the developing and adult mouse retina. J. Comp. Neurol. 461, 187-204 [DOI] [PubMed] [Google Scholar]
- Dorval K. M., Bobechko B. P., Fujieda H., Chen S., Zack D. J., Bremner R. (2006). CHX10 targets a subset of photoreceptor genes. J. Biol. Chem. 281, 744-751 [DOI] [PubMed] [Google Scholar]
- Eisenfeld A. J., Bunt-Milam A. H., Saari J. C. (1985). Localization of retinoid-binding proteins in developing rat retina. Exp. Eye Res. 41, 299-304 [DOI] [PubMed] [Google Scholar]
- Elworthy S., Hargrave M., Knight R., Mebus K., Ingham P. W. (2008). Expression of multiple slow myosin heavy chain genes reveals a diversity of zebrafish slow twitch muscle fibres with differing requirements for Hedgehog and Prdm1 activity. Development 135, 2115-2126 [DOI] [PubMed] [Google Scholar]
- Fossat N., Le Greneur C., Beby F., Vincent S., Godement P., Chatelain G., Lamonerie T. (2007). A new GFP-tagged line reveals unexpected Otx2 protein localization in retinal photoreceptors. BMC Dev. Biol. 7, 122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furukawa T., Morrow E. M., Cepko C. L. (1997). Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 91, 531-541 [DOI] [PubMed] [Google Scholar]
- Furukawa T., Morrow E. M., Li T., Davis F. C., Cepko C. L. (1999). Retinopathy and attenuated circadian entrainment in Crx-deficient mice. Nat. Genet. 23, 466-470 [DOI] [PubMed] [Google Scholar]
- Green E. S., Stubbs J. L., Levine E. M. (2003). Genetic rescue of cell number in a mouse model of microphthalmia: interactions between Chx10 and G1-phase cell cycle regulators. Development 130, 539-552 [DOI] [PubMed] [Google Scholar]
- Greferath U., Grunert U., Wassle H. (1990). Rod bipolar cells in the mammalian retina show protein kinase C-like immunoreactivity. J. Comp. Neurol. 301, 433-442 [DOI] [PubMed] [Google Scholar]
- Hatakeyama J., Tomita K., Inoue T., Kageyama R. (2001). Roles of homeobox and bHLH genes in specification of a retinal cell type. Development 128, 1313-1322 [DOI] [PubMed] [Google Scholar]
- Hayashi K., de Sousa Lopes S. M. C., Surani M. A. (2007). Germ cell specification in mice. Science 316, 394-396 [DOI] [PubMed] [Google Scholar]
- Hebert J. M., McConnell S. K. (2000). Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev. Biol. 222, 296-306 [DOI] [PubMed] [Google Scholar]
- Hsiau T. H., Diaconu C., Myers C. A., Lee J., Cepko C. L., Corbo J. C. (2007). The cis-regulatory logic of the mammalian photoreceptor transcriptional network. PLoS ONE 2, e643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- John S. A., Garrett-Sinha L. A. (2009). Blimp1: a conserved transcriptional repressor critical for differentiation of many tissues. Exp. Cell Res. 315, 1077-1084 [DOI] [PubMed] [Google Scholar]
- Laird D. W., Molday R. S. (1988). Evidence against the role of rhodopsin in rod outer segment binding to RPE cells. Invest. Ophthalmol. Vis. Sci. 29, 419-428 [PubMed] [Google Scholar]
- Liu H., Etter P., Hayes S., Jones I., Nelson B., Hartman B., Forrest D., Reh T. A. (2008). NeuroD1 regulates expression of thyroid hormone receptor 2 and cone opsins in the developing mouse retina. J. Neurosci. 28, 749-756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livesey F. J., Cepko C. L. (2001). Vertebrate neural cell-fate determination: lessons from the retina. Nat. Rev. Neurosci. 2, 109-118 [DOI] [PubMed] [Google Scholar]
- Livne-Bar I., Pacal M., Cheung M. C., Hankin M., Trogadis J., Chen D., Dorval K. M., Bremner R. (2006). Chx10 is required to block photoreceptor differentiation but is dispensable for progenitor proliferation in the postnatal retina. Proc. Natl. Acad. Sci. USA 103, 4988-4993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt T., Ashery-Padan R., Andrejewski N., Scardigli R., Guillemot F., Gruss P. (2001). Pax6 is required for the multipotent state of retinal progenitor cells. Cell 105, 43-55 [DOI] [PubMed] [Google Scholar]
- Mears A. J., Kondo M., Swain P. K., Takada Y., Bush R. A., Saunders T. L., Sieving P. A., Swaroop A. (2001). Nrl is required for rod photoreceptor development. Nat. Genet. 29, 447-452 [DOI] [PubMed] [Google Scholar]
- Milam A. H., Dacey D. M., Dizhoor A. M. (1993). Recoverin immunoreactivity in mammalian cone bipolar cells. Vis. Neurosci. 10, 1-12 [DOI] [PubMed] [Google Scholar]
- Morrow E. M., Furukawa T., Lee J. E., Cepko C. L. (1999). NeuroD regulates multiple functions in the developing neural retina in rodent. Development 126, 23-36 [DOI] [PubMed] [Google Scholar]
- Morrow E. M., Chen C. M., Cepko C. L. (2008). Temporal order of bipolar cell genesis in the neural retina. Neural Dev. 3, 2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K., Harada C., Namekata K., Harada T. (2006). Expression of olig2 in retinal progenitor cells. NeuroReport 17, 345-349 [DOI] [PubMed] [Google Scholar]
- Ng L., Hurley J. B., Dierks B., Srinivas M., Salto C., Vennstrom B., Reh T. A., Forrest D. (2001). A thyroid hormone receptor that is required for the development of green cone photoreceptors. Nat. Genet. 27, 94-98 [DOI] [PubMed] [Google Scholar]
- Nishida A., Furukawa A., Koike C., Tano Y., Aizawa S., Matsuo I., Furukawa T. (2003). Otx2 homeobox gene controls retinal photoreceptor cell fate and pineal gland development. Nat. Neurosci. 6, 1255-1263 [DOI] [PubMed] [Google Scholar]
- Niwa H., Yamamura K., Miyazaki J. (1991). Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene 108, 193-199 [DOI] [PubMed] [Google Scholar]
- Ohinata Y., Payer B., O'Carroll D., Ancelin K., Ono Y., Sano M., Barton S. C., Obukhanych T., Nussenzweig M., Tarakhovsky A., et al. (2005). Blimp1 is a critical determinant of the germ cell lineage in mice. Nature 436, 207-213 [DOI] [PubMed] [Google Scholar]
- Ohtoshi A., Justice M. J., Behringer R. R. (2001). Isolation and characterization of Vsx1, a novel mouse CVC paired-like homeobox gene expressed during embryogenesis and in the retina. Biochem. Biophys. Res. Commun. 286, 133-140 [DOI] [PubMed] [Google Scholar]
- Ohtoshi A., Wang S. W., Maeda H., Saszik S. M., Frishman L. J., Klein W. H., Behringer R. R. (2004). Regulation of retinal cone bipolar cell differentiation and photopic vision by the CVC homeobox gene Vsx1. Curr. Biol. 14, 530-536 [DOI] [PubMed] [Google Scholar]
- Poche R. A., Furuta Y., Chaboissier M. C., Schedl A., Behringer R. R. (2008). Sox9 is expressed in mouse multipotent retinal progenitor cells and functions in Muller glial cell development. J. Comp. Neurol. 510, 237-250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punzo C., Cepko C. (2007). Cellular responses to photoreceptor death in the rd1 mouse model of retinal degeneration. Invest. Ophthalmol. Vis. Sci. 48, 849-857 [DOI] [PubMed] [Google Scholar]
- Rapaport D. H., Wong L. L., Wood E. D., Yasumura D., LaVail M. M. (2004). Timing and topography of cell genesis in the rat retina. J. Comp. Neurol. 474, 304-324 [DOI] [PubMed] [Google Scholar]
- Ren B., Chee K. J., Kim T. H., Maniatis T. (1999). PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 13, 125-137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowan S., Cepko C. L. (2004). Genetic analysis of the homeodomain transcription factor Chx10 in the retina using a novel multifunctional BAC transgenic mouse reporter. Dev. Biol. 271, 388-402 [DOI] [PubMed] [Google Scholar]
- Sato S., Inoue T., Terada K., Matsuo I., Aizawa S., Tano Y., Fujikado T., Furukawa T. (2007). Dkk3-Cre BAC transgenic mouse line: a tool for highly efficient gene deletion in retinal progenitor cells. Genesis 45, 502-507 [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M., Lin K.-I., McHeyzer-Williams L. J., Liao J., McHeyzer-Williams M. G., Calame K. (2003). Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity 19, 607-620 [DOI] [PubMed] [Google Scholar]
- Sidman R. L. (1961). Histogenesis of mouse retina studied with thymidine H3. Structure of the Eye (ed. G. K. Smelser), 487-506 New York: Academic Press; [Google Scholar]
- Solovei I., Kreysing M., Lanctot C., Kosem S., Peichl L., Cremer T., Guck J., Joffe B. (2009). Nuclear architecture of rod photoreceptor cells adapts to vision in mammalian evolution. Cell 137, 356-368 [DOI] [PubMed] [Google Scholar]
- Swartz M. E., Eberhart J., Pasquale E. B., Krull C. E. (2001). EphA4/ephrin-A5 interactions in muscle precursor cell migration in the avian forelimb. Development 128, 4669-4680 [DOI] [PubMed] [Google Scholar]
- Thornberry N. A., Lazebnik Y. (1998). Caspases: enemies within. Science 281, 1312-1316 [DOI] [PubMed] [Google Scholar]
- Turner C. A., Jr, Mack D. H., Davis M. M. (1994). Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 77, 297-306 [DOI] [PubMed] [Google Scholar]
- Vincent S. D., Dunn N. R., Sciammas R., Shapiro-Shalef M., Davis M. M., Calame K., Bikoff E. K., Robertson E. J. (2005). The zinc finger transcriptional repressor Blimp1/Prdm1 is dispensable for early axis formation but is required for specification of primordial germ cells in the mouse. Development 132, 1315-1325 [DOI] [PubMed] [Google Scholar]
- von Hofsten J., Elworthy S., Gilchrist M. J., Smith J. C., Wardle F. C., Ingham P. W. (2008). Prdm1- and Sox6-mediated transcriptional repression specifies muscle fibre type in the zebrafish embryo. EMBO Rep. 9, 683-689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang D., Zhuang L., Gao B., Shi C. X., Cheung J., Liu M., Jin T., Wen X. Y. (2008). The Blimp-1 gene regulatory region directs EGFP expression in multiple hematopoietic lineages and testis in mice. Transgenic Res. 17, 193-203 [DOI] [PubMed] [Google Scholar]
- Wassle H., Puller C., Muller F., Haverkamp S. (2009). Cone contacts, mosaics, and territories of bipolar cells in the mouse retina. J. Neurosci. 29, 106-117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm T. P., Solnica-Krezel L. (2005). Essential roles of a zebrafish prdm1/blimp1 homolog in embryo patterning and organogenesis. Development 132, 393-404 [DOI] [PubMed] [Google Scholar]
- Xiang M., Zhou L., Macke J. P., Yoshioka T., Hendry S. H., Eddy R. L., Shows T. B., Nathans J. (1995). The Brn-3 family of POU-domain factors: primary structure, binding specificity, and expression in subsets of retinal ganglion cells and somatosensory neurons. J. Neurosci. 15, 4762-4785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young R. W. (1984). Cell death during differentiation of the retina in the mouse. J. Comp. Neurol. 229, 362-373 [DOI] [PubMed] [Google Scholar]
- Young R. W. (1985). Cell differentiation in the retina of the mouse. Anat. Rec. 212, 199-205 [DOI] [PubMed] [Google Scholar]
- Yu J., Angelin-Duclos C., Greenwood J., Liao J., Calame K. (2000). Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell. Biol. 20, 2592-2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Craft C. M. (2000). Modulation of CRX transactivation activity by phosducin isoforms. Mol. Cell. Biol. 20, 5216-5226 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.