Abstract
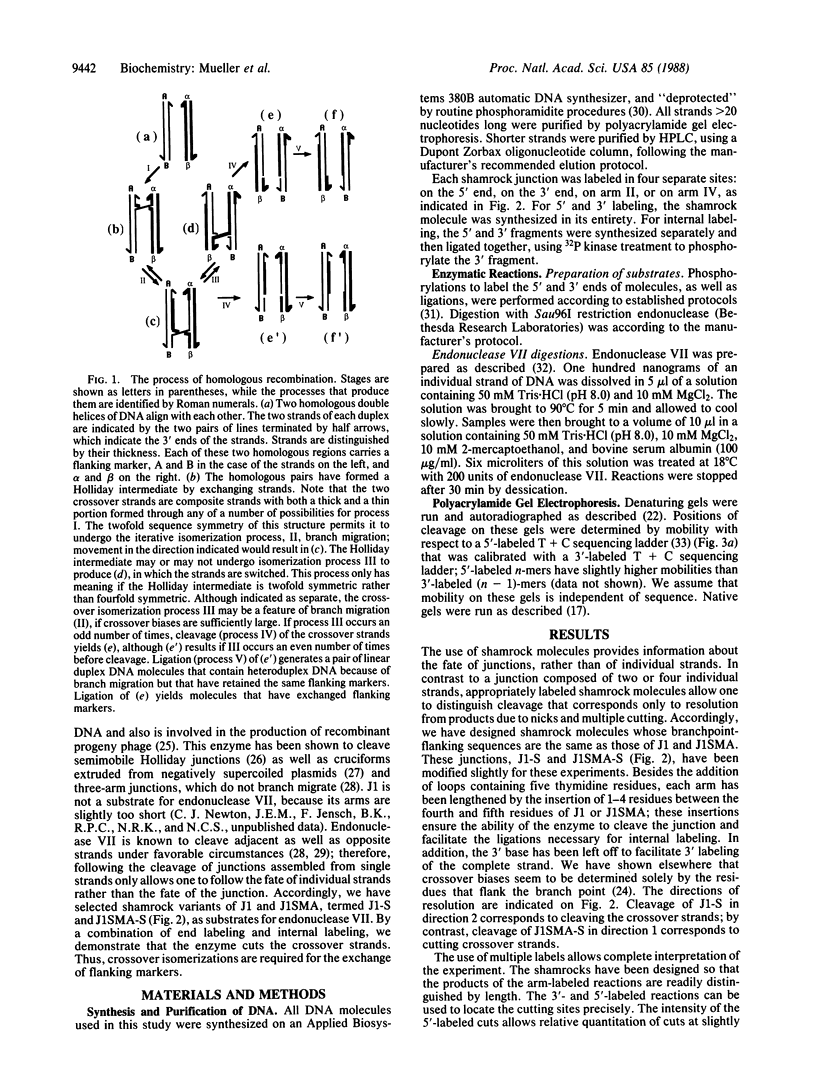
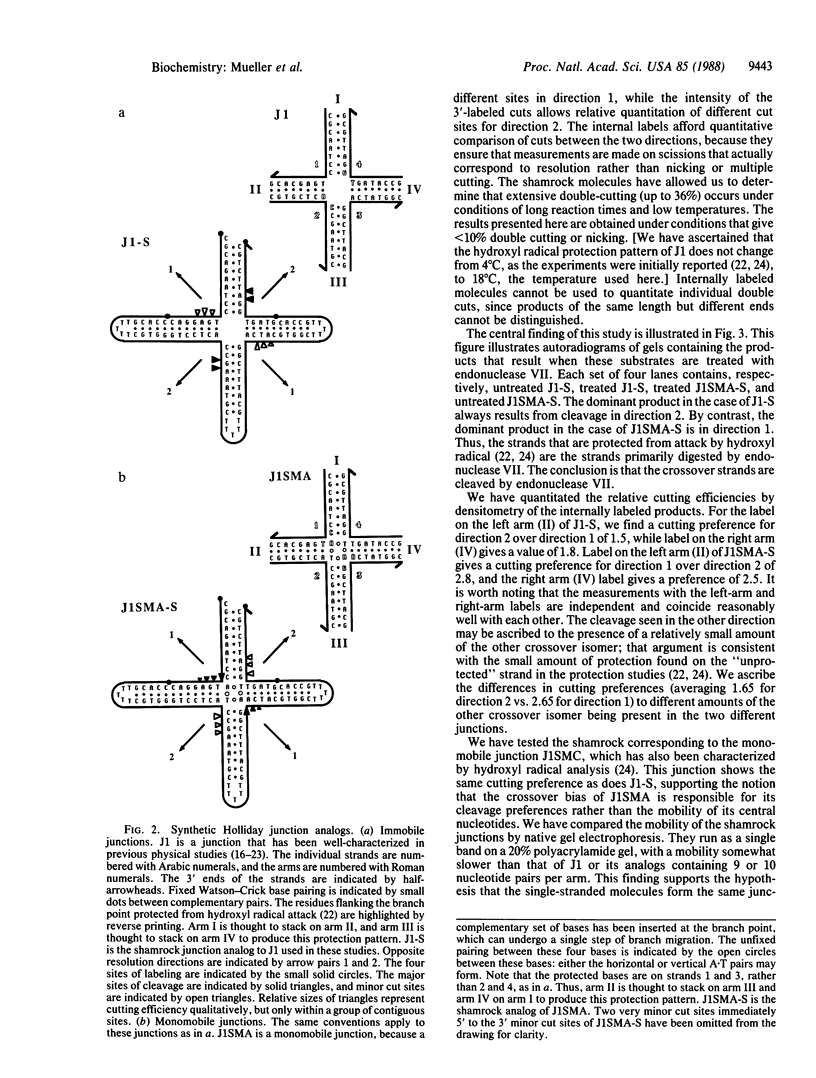
We have formed four-arm branched DNA junctions that contain no more than a single base pair of branch migratory freedom. Recently, we have shown that these Holliday junction analogs have twofold symmetric protection patterns in solution when probed with hydroxyl radicals: two opposite strands of one junction show extensive protection near the branch point, while the other pair of opposite strands is virtually as susceptible as a double helix. In a different junction, the hydroxyl radical protection pattern is reversed. These patterns suggest that a crossover-isomer bias exists in these molecules and that the protected strands form the crossover between helices. Here, we examine the cleavage pattern of these structures when they are resolved by T4 endonuclease VII. Junctions are formed from a single shamrock-shaped molecule, which contains 5', 3', or internal labels. The enzyme shows a preference for resolving these modified junctions at sites near those protected from hydroxyl radicals. This result suggests that only crossover strands in a Holliday junction are cleaved, and thus an odd number of crossover isomerizations must occur when flanking markers are exchanged.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Chen J. H., Churchill M. E., Tullius T. D., Kallenbach N. R., Seeman N. C. Construction and analysis of monomobile DNA junctions. Biochemistry. 1988 Aug 9;27(16):6032–6038. doi: 10.1021/bi00416a031. [DOI] [PubMed] [Google Scholar]
- Churchill M. E., Tullius T. D., Kallenbach N. R., Seeman N. C. A Holliday recombination intermediate is twofold symmetric. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4653–4656. doi: 10.1073/pnas.85.13.4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claverys J. P., Lacks S. A. Heteroduplex deoxyribonucleic acid base mismatch repair in bacteria. Microbiol Rev. 1986 Jun;50(2):133–165. doi: 10.1128/mr.50.2.133-165.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper J. P., Hagerman P. J. Gel electrophoretic analysis of the geometry of a DNA four-way junction. J Mol Biol. 1987 Dec 20;198(4):711–719. doi: 10.1016/0022-2836(87)90212-9. [DOI] [PubMed] [Google Scholar]
- Dickie P., McFadden G., Morgan A. R. The site-specific cleavage of synthetic Holliday junction analogs and related branched DNA structures by bacteriophage T7 endonuclease I. J Biol Chem. 1987 Oct 25;262(30):14826–14836. [PubMed] [Google Scholar]
- Dressler D., Potter H. Molecular mechanisms in genetic recombination. Annu Rev Biochem. 1982;51:727–761. doi: 10.1146/annurev.bi.51.070182.003455. [DOI] [PubMed] [Google Scholar]
- Hoess R., Wierzbicki A., Abremski K. Isolation and characterization of intermediates in site-specific recombination. Proc Natl Acad Sci U S A. 1987 Oct;84(19):6840–6844. doi: 10.1073/pnas.84.19.6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. L., Landy A. Resolution of synthetic att-site Holliday structures by the integrase protein of bacteriophage lambda. Nature. 1984 Oct 25;311(5988):721–726. doi: 10.1038/311721a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensch F., Kemper B. Endonuclease VII resolves Y-junctions in branched DNA in vitro. EMBO J. 1986 Jan;5(1):181–189. doi: 10.1002/j.1460-2075.1986.tb04194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemper B., Garabett M. Studies on T4-head maturation. 1. Purification and characterization of gene-49-controlled endonuclease. Eur J Biochem. 1981 Mar 16;115(1):123–131. [PubMed] [Google Scholar]
- Kitts P. A., Nash H. A. Homology-dependent interactions in phage lambda site-specific recombination. Nature. 1987 Sep 24;329(6137):346–348. doi: 10.1038/329346a0. [DOI] [PubMed] [Google Scholar]
- Kleff S., Kemper B. Initiation of heteroduplex-loop repair by T4-encoded endonuclease VII in vitro. EMBO J. 1988 May;7(5):1527–1535. doi: 10.1002/j.1460-2075.1988.tb02972.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilley D. M., Kemper B. Cruciform-resolvase interactions in supercoiled DNA. Cell. 1984 Feb;36(2):413–422. doi: 10.1016/0092-8674(84)90234-4. [DOI] [PubMed] [Google Scholar]
- Marky L. A., Kallenbach N. R., McDonough K. A., Seeman N. C., Breslauer K. J. The melting behavior of a DNA junction structure: a calorimetric and spectroscopic study. Biopolymers. 1987 Sep;26(9):1621–1634. doi: 10.1002/bip.360260912. [DOI] [PubMed] [Google Scholar]
- Meselson M. S., Radding C. M. A general model for genetic recombination. Proc Natl Acad Sci U S A. 1975 Jan;72(1):358–361. doi: 10.1073/pnas.72.1.358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meselson M. Formation of hybrid DNA by rotary diffusion during genetic recombination. J Mol Biol. 1972 Nov 28;71(3):795–798. doi: 10.1016/s0022-2836(72)80040-8. [DOI] [PubMed] [Google Scholar]
- Miyazaki J., Ryo Y., Minagawa T. Involvement of gene 49 in recombination of bacteriophage T4. Genetics. 1983 May;104(1):1–9. doi: 10.1093/genetics/104.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuuchi K., Kemper B., Hays J., Weisberg R. A. T4 endonuclease VII cleaves holliday structures. Cell. 1982 Jun;29(2):357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- Modrich P. DNA mismatch correction. Annu Rev Biochem. 1987;56:435–466. doi: 10.1146/annurev.bi.56.070187.002251. [DOI] [PubMed] [Google Scholar]
- Nunes-Düby S. E., Matsumoto L., Landy A. Site-specific recombination intermediates trapped with suicide substrates. Cell. 1987 Aug 28;50(5):779–788. doi: 10.1016/0092-8674(87)90336-9. [DOI] [PubMed] [Google Scholar]
- Robinson B. H., Seeman N. C. Simulation of double-stranded branch point migration. Biophys J. 1987 Apr;51(4):611–626. doi: 10.1016/S0006-3495(87)83386-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C., Kallenbach N. R. Design of immobile nucleic acid junctions. Biophys J. 1983 Nov;44(2):201–209. doi: 10.1016/S0006-3495(83)84292-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seeman N. C. Nucleic acid junctions and lattices. J Theor Biol. 1982 Nov 21;99(2):237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- Seeman N. C. Physical models for exploring DNA topology. J Biomol Struct Dyn. 1988 Apr;5(5):997–1004. doi: 10.1080/07391102.1988.10506445. [DOI] [PubMed] [Google Scholar]
- Sigal N., Alberts B. Genetic recombination: the nature of a crossed strand-exchange between two homologous DNA molecules. J Mol Biol. 1972 Nov 28;71(3):789–793. doi: 10.1016/s0022-2836(72)80039-1. [DOI] [PubMed] [Google Scholar]
- Thompson B. J., Camien M. N., Warner R. C. Kinetics of branch migration in double-stranded DNA. Proc Natl Acad Sci U S A. 1976 Jul;73(7):2299–2303. doi: 10.1073/pnas.73.7.2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wemmer D. E., Wand A. J., Seeman N. C., Kallenbach N. R. NMR analysis of DNA junctions: imino proton NMR studies of individual arms and intact junction. Biochemistry. 1985 Oct 8;24(21):5745–5749. doi: 10.1021/bi00342a009. [DOI] [PubMed] [Google Scholar]






