Abstract
Mass spectrometry plays a very visible role in biopharmaceutical industry, although its use in development, characterization and quality control of protein drugs is mostly limited to the analysis of covalent structure (amino acid sequence and post-translational modifications). Despite the centrality of protein conformation to biological activity, stability and safety of biopharmaceutical products, the expanding arsenal of mass spectrometry-based methods that are currently available to probe higher order structure and conformational dynamics of biopolymers did not enjoy until recently much attention in the industry. This is beginning to change as a result of recent work demonstrating the utility of these experimental tools for various aspects of biopharmaceutical product development and manufacturing. In this work we use a paradigmatic protein drug interferon β-1a as an example to illustrate the utility of mass spectrometry as a powerful tool not only to assess the integrity of higher order structure of a protein drug, but also to predict consequences of its degradation at a variety of levels.
Introduction
Pharmaceutical products based on biopolymers represent an important and rapidly growing part of the therapeutic arsenal of modern medicine (1). While a few of such medicines are based on polysaccharides (2) and nucleic acids (3), protein drugs (4, 5) constitute the largest fraction of this segment. Nearly 200 protein-based products have been already approved worldwide with nearly a thousand more either in clinical studies, or in various stages of the approval process (6). Protein therapeutics fundamentally differ from the traditional small-molecule medicines in many ways, perhaps the most obvious being the sheer size of the active pharmaceutical ingredients. The biopharmaceutical products range in size from several kDa (e.g., insulin) to nearly 1 MDa (e.g., botulinum toxin), vastly exceeding the molecular weight typical of small molecule drugs. This quantitative difference gives rise to an important qualitative distinction between the “traditional” small molecule medicines (where the covalent structure alone is the sole determinant of the three-dimensional structure and, ultimately, the therapeutic properties of the drug) and the protein pharmaceuticals (where the large physical size makes the multitude of non-covalent contacts not only inevitable, but in fact the defining element of their three-dimensional structure).
The unique three-dimensional organization of proteins, or higher order structure, is vital not only for their function, but also for many other aspects of their behavior. Proteins that are not folded properly are usually prone to aggregation both in vitro and in vivo, and are frequently a target for various degradation pathways both inside and outside the cell. Since a unique conformation is critically important for the ability of a protein drug to interact with its physiological targets, a failure to fold or maintain the native conformation at any time prior to or during administration would obviously have a negative impact on efficacy. Even partial unfolding that does not affect the structural elements of a protein drug critical for its function may have grave consequences, as such structurally compromised species are typically prone to aggregation (7). In addition to the negative impact on efficacy due to the protein drug loss, aggregation may trigger immune response, thereby adversely affecting the safety profile of the protein drug (8).
The central role played by higher order structure and conformational integrity in determining potency, stability and safety of protein therapeutics makes characterization of protein conformation a critical element for successful design, engineering, manufacturing and formulating of biopharmaceutical products. Furthermore, since the covalent structure alone does not define a protein drug, the ability to provide accurate and detailed characterization of protein conformation and dynamics will also be very important in establishing comparability (“sameness”) of protein drugs and the original approved drug products following a manufacturing process change. Recognition of the prominent role played by protein conformation and dynamics in establishing bioequivalence makes characterization of higher order structure of biopharmaceutical products especially important in light of the emergence of follow-on biologics (9) and the need to effectively regulate them (10, 11).
In the past several decades biophysics has amassed an impressive armamentarium of experimental techniques to probe various aspects of protein conformation and dynamics (12). In particular, X-ray crystallography and high-resolution NMR are able to provide in many cases detailed structure of proteins and their complexes, contributing knowledge that has been truly invaluable for the development of many bio- and small molecule pharmaceutical products (13). Unfortunately, these two powerful techniques have inherent limitations that often make their application to the analyses of biopharmaceutical products impractical. For example, X-ray crystallography by definition requires that the protein be crystallized prior to analysis. This makes it impossible to carry out direct examination of the protein drug conformation under relevant production/storage conditions (e.g., protein drug substance, product or dosing solution) or physiological conditions (e.g., mimicking the environment encountered by the protein post-administration). Furthermore, partially unfolded states are less likely to crystallize, thereby generating a bias against non-native conformations that may be present in solution. A sister technique that is capable of probing non-native ensembles of proteins directly in solution, small-angle X-ray scattering (14), does not provide detailed structural information.
Unlike X-ray crystallography, high-resolution NMR is capable of revealing intimate details of both higher order structure and conformational dynamics of proteins. However, it still is restricted in terms of the physical size of the protein systems it can handle routinely, placing the majority of biopharmaceutical products out of its reach. Therefore, it is not surprising that routine analyses of conformation and stability of protein drugs still rely on classical biophysical methods, such as various spectroscopic techniques (especially circular dichroism, fluorescence, UV-absorption and FTIR spectroscopy), light scattering, calorimetry, as well as analytical centrifugation and size exclusion chromatography (15-17). Most of these techniques have an advantage of being able to probe the conformational properties of macromolecular drugs in more relevant environments (i.e., those matching or closely mimicking formulations). However, they are typically focused on one particular aspect of higher order structure and fail to provide a detailed and comprehensive characterization of conformation and dynamics.
Mass spectrometry is another powerful analytical technique that is capable of characterizing biopolymer structure at a variety of levels, although most current applications of MS in the biopharmaceutical industry are focused on characterizing the covalent structure of proteins (amino acid sequence and post-translational modifications, PTMs). It is the ability to map both enzymatic and non-enzymatic PTMs with high sensitivity and unprecedented accuracy that made MS a staple in characterization of biopharmaceutical products (18, 19). However, in many cases there is no straightforward correlation between the presence or absence of certain PTMs within the protein molecule and its conformational stability or functional properties. Indeed, the consequences of PTMs in terms of stability and functional competence of biopharmaceutical products are highly context dependent and vary significantly from one system to another. While many PTMs are detrimental for the protein conformational stability (e.g., disulfide scrambling), some others appear to be fairly inconsequential, and can be easily tolerated by a protein without any noticeable impact on its function, stability or safety profile. At the same time, loss of native conformation may be triggered by a variety of factors, many of which have nothing to do with either enzymatic or non-enzymatic PTMs, and protein unfolding is not necessarily accompanied by alterations of its covalent structure.
Therefore, focusing protein drug characterization efforts on detecting PTMs may generate both false-positive and false-negative outcomes as far as conformation and stability are concerned. Furthermore, the complexity exhibited by many protein therapeutics in terms of both sheer size and structural heterogeneity makes precise mapping of all covalent alterations in many cases a very difficult, if not unfeasible, task. Clearly, there is a need in the biopharmaceutical industry to have new tools capable of detecting and characterizing changes in protein conformation that do not have to rely on techniques targeting covalent structure. Development of MS-based experimental strategies to probe higher order structure and dynamics of biopolymers, proteins in particular, has been a focal point of extensive research efforts, following the early recognition of the great potential of MS in this vast field nearly two decades ago (20-24). An impressive arsenal of MS techniques to probe non-covalent structure was developed as a result of these efforts (25), many of which proved highly successful in dealing with a variety of problems in biophysics and structural biology.
However, outside of academic laboratories, the embrace of these methodologies by industrial researchers has been somewhat limited until recently due to a variety of reasons. Some techniques, such as covalent cross-linking (26), have inherently low yields and therefore are not well suited for the specific needs of biopharmaceutical industry, where partial loss of structure must frequently be detected and characterized in a small fraction of proteins on the background of natively folded species. Methods that rely on direct ESI MS analysis of higher order structure, such as probing non-covalent assemblies (27-29) or analysis of protein ion charge state distributions (30), are frequently met with some skepticism due to the very nature of these measurements. Indeed, direct ESI MS measurements require that the proteins be placed into the so-called “electrospray-friendly” solvents (aqueous solutions of volatile salts), making structural characterization in a relevant environment (e.g., formulation buffer) nearly impossible. These limitations notwithstanding, probing non-covalent associations by direct ESI MS is increasingly used in the drug discovery process (31), including optimization of protein drugs (32). Charge state distribution analysis of protein ions in ESI MS is also beginning to enjoy recognition as a useful tool for comparability studies of related biopharmaceutical products (33).
Among MS-based techniques targeting protein higher order structure and dynamics, hydrogen/deuterium exchange (HDX) with MS detection (34-38) has demonstrated the greatest promise vis-à-vis conformational analysis of biopharmaceutical products. The technique is reliable, robust and sensitive, and is capable not only of detecting the presence of (partially) unfolded species in solution, but also localizing the protein segments with anomalous protection levels. This latter feature is particularly appealing as a means of mapping interaction sites, and is now actively evaluated for the purpose of optimizing the process of screening small molecule drug candidates (39-41). Furthermore, the ability of HDX MS to detect and characterize structurally compromised proteins on the background of the natively folded species in highly complex matrices makes it a very promising tool for conformational characterization of protein drugs (42, 43) and may open up new and exciting opportunities in drug formulation.
However, a complete embrace of this technique by the industry practitioners, as well as its better utilization, require that several questions be answered in order to better define its capabilities and limitations, particularly within the context of specific requirements of biopharmaceutical industry and regulatory agencies. For example, can MS-based methods provide information that cannot be obtained by classical methods commonly employed in the industry? How relevant is the information derived from MS characterization of protein drugs in terms of their conformational integrity, stability and functional competence? Can MS be used in comparability studies?
In this work we use an example of a glycoprotein interferon-β1a, the active pharmaceutical ingredient in several commercial biopharmaceutical products, in an attempt to address these questions. The work presented here clearly demonstrates that the value of MS extends far beyond mere detection of misfolded species. MS-based methods to probe higher order structure generate information that has high predictive value for analysis of properties and behavior of protein therapeutics. As such, these methodologies present an extremely valuable complement to the battery of “classic” biophysical techniques already employed in biopharmaceutical industry.
Detection of conformational changes in a biopharmaceutical product: alkylation of interferon as a model of protein drug degradation
Interferon-β1a (IFN) is a member of the type I interferon family, a group of homologous cytokines that display broad biological activity, including activation of anti-viral response, immunoregulation and anti-tumor activity (44, 45). IFN is the most widely prescribed disease-modifying therapy for multiple sclerosis (46), a chronic, progressive autoimmune disorder of the central nervous system (47). As is the case with many proteins, IFN has a problematic propensity to misfold, which leads to activity loss, aggregation and increased immunogenic response (8). This structure loss can be accelerated by a variety of factors, such as chemical modifications, surface binding, exposure to elevated temperatures, lyophilization, etc. Some non-enzymatic PTMs have been long suspected to act as likely triggers of misfolding, although establishing a firm correlation between such covalent modification events and the integrity of the protein higher order structure has proved difficult.
Recently, we demonstrated that both charge state distribution analysis of IFN ions in ESI MS and HDX MS readily provide evidence of IFN partial unfolding resulting from a single specific non-enzymatic PTM, alkylation of the sole free cysteine residue in IFN (Cys-17, see Figure 1) with N-ethylmaleimide (42). This non-enzymatic PTM dramatically reduces the activity of the protein (a 50-90% decrease of IFN anti-viral activity), which is likely to be mediated by changes in the protein conformation. However, classical biophysical techniques yielded mixed results when applied to detect this putative change: only size-exclusion chromatography (SEC) was able to make a clear distinction between the intact and NEM-alkylated forms of IFN, while spectroscopic techniques either failed to detect the conformational changes or reported differences that were not adequate for making a clear distinction (42).
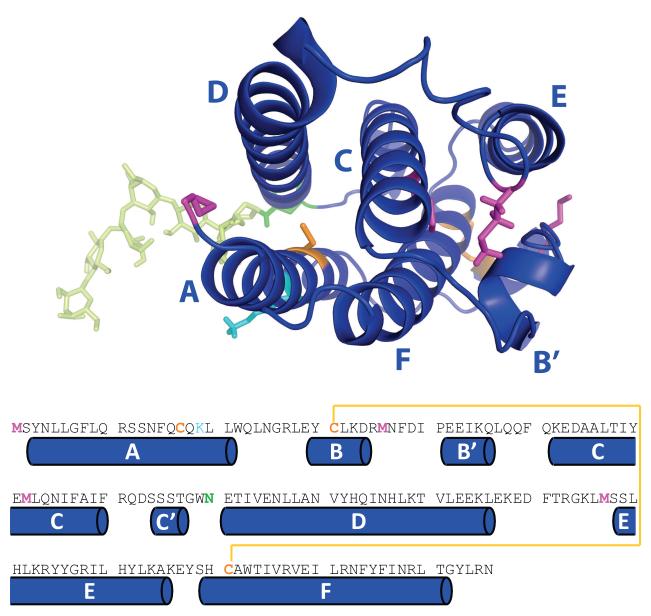
Figure 1.
Higher order structure (top) and amino acid sequence (bottom) of IFN with elements of secondary structure labeled according to a commonly accepted nomenclature (44). Colored residues are common targets of non-enzymatic or designer PTMs: orange, Cys-17 (alkylation, oxidation, formation of external disulfides, internal disulfide scrambling, replacement with Ser in interferon β1b, PEGylation via thiol-reactive group); yellow, Cys-31 and Cys-141 (disulfide scrambling); cyan, Lys-19 (glycation (86)); magenta, methionine residues (oxidation); and green, Asn-90 (variable glycosylation and deglycosylation).
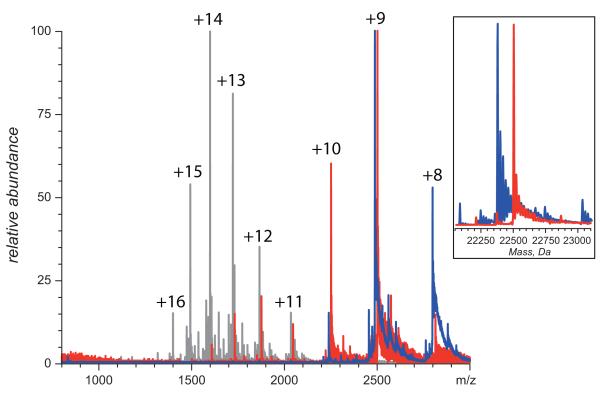
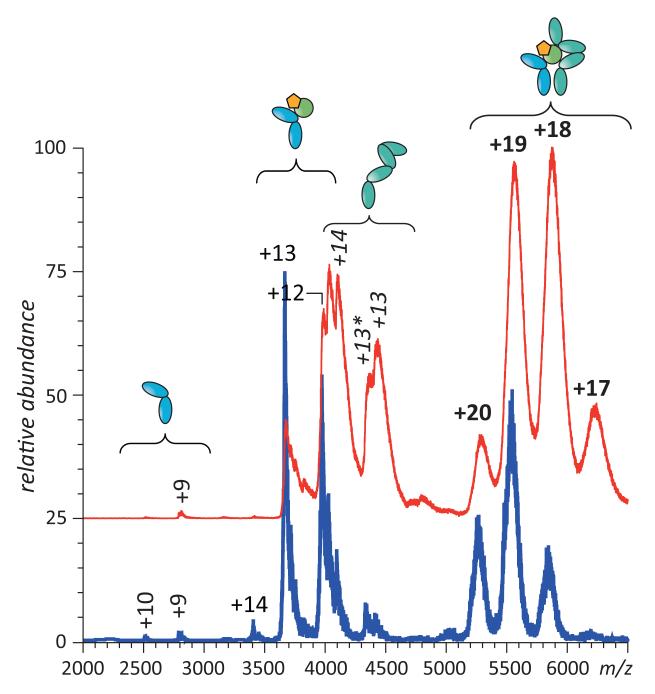
On the contrary, application of MS-based methods of probing higher order structure provided clear and unequivocal evidence that the native conformation of IFN was compromised as a result of NEM-alkylation. For example, direct ESI MS analysis of IFN and NEM-IFN following buffer exchange to an electrospray-friendly solvent reveals qualitative differences between the two forms of the protein. Ions of intact IFN exhibit a narrow charge state distribution with low charge density, as is expected for compact, tightly folded macromolecular species, while the charge state distribution of the NEM-IFN ions is clearly bimodal (Figure 2). Presence of the higher charge-density ions in ESI mass spectrum of NEM-IFN clearly signals that a fraction of the protein molecules fail to maintain the compact native structure. With some caution, it may even be possible to estimate the fractional concentrations of the (partially) unfolded and compact (tightly folded) protein species in solution based on the abundance of high- and low-charge density protein ions in ESI MS (48).
Figure 2.
ESI mass spectra of intact (blue) and NEM-alkylated IFN (red) buffer-exchanged to aqueous 100 mM ammonium acetate prior to MS analysis. The inset shows limited heterogeneity of IFN due to the presence of several glycoforms. Black trace shows an ESI mass spectrum of intact IFN acquired under strongly denaturing conditions (50% methanol, 6% acetic acid).
Although the presence of the NEM-alkylated residue within IFN can be easily deduced from the mass shift of NEM-IFN ions (see inset in Figure 2), the detection of partial unfolding here is based entirely on charge, rather than mass, measurements and, therefore, does not depend on the ability to detect any changes in covalent structure concurrently with (or prior to) the conformational analysis. In fact, this technique can detect changes in protein compactness triggered by non-enzymatic PTMs that do not alter the protein mass (e.g., disulfide scrambling).
It must be said that the extent of multiple charging of the NEM-IFN species representing the less compact conformations (charge states +11 through +14 in Figure 2) still implies the presence of some residual structure. Indeed, the average charge density of IFN ions in ESI mass spectrum acquired under denaturing conditions (black trace in Figure 2) is noticeably higher. This suggests that the solvent accessible surface area of “non-compact” conformers of NEM-IFN, whose presence under near-native conditions is revealed by the protein ion charge state distribution analysis, is lower compared to the fully denatured species of IFN (49, 50). However, this technique yields only a global measure of conformational “disorder” and, therefore, cannot provide any information that would allow these unfolding events to be localized within the protein structure.
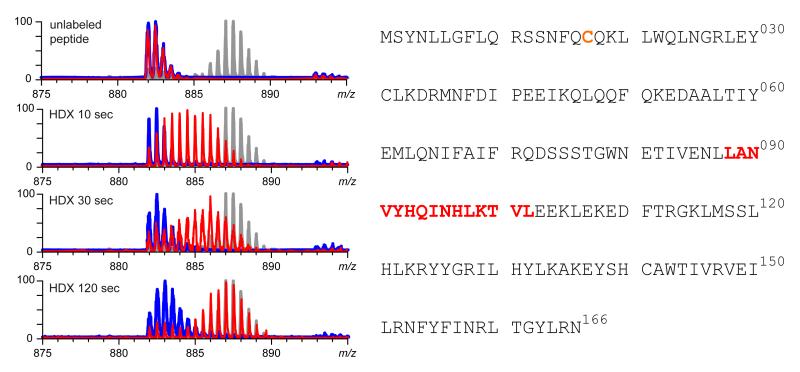
This gap can be easily filled by HDX MS, which provides local information on backbone dynamics when the exchange reaction in solution is supplemented by proteolysis under the slow exchange conditions and MS analysis. HDX MS easily identifies several segments in NEM-IFN whose stability is greatly compromised by alkylation of Cys-17 (42). One example is shown in Figure 3, where the evolution of the isotopic distribution of a peptic fragment [L88-L102] is traced over first 120 sec. of H/D exchange. Very slow uptake of deuterium is exhibited by the fragment derived from intact IFN (blue trace), while the protein alkylation results in a dramatic acceleration of the exchange kinetics of this segment. Presence of several such segments whose exchange kinetics is significantly altered by the alkylation event clearly signals a change in conformation and/or dynamics induced by the alkylation, consistent with the conclusions of the protein ion charge state distribution analysis (vide supra). Importantly, these measurements do not require that the protein be transferred to an electrospray-friendly buffer prior to the isotopic labeling, thereby allowing the conformational analysis to be carried out under relevant conditions (e.g., in the formulation buffer), since isolation of the protein drug material from the commercial product is often viewed as a step capable of altering its structural integrity (51)). Furthermore, the ability to localize the unfolding events within the protein sequence provides valuable information that can be used to predict the functional consequences of such partial unfolding. Just like the charge state distribution analysis, characterization of conformation and dynamics by HDX MS does not require any prior knowledge of the non-enzymatic PTMs that may be present in the protein drug. In fact, identification of all peptic fragments carried out prior to HDX MS analysis, is likely to detect most PTMs within the protein. Even if some of them escape the detection, this would obviously make no impact on the validity of the HDX MS analysis of protein conformation and dynamics.
Figure 3.
Evolution of isotopic distributions of peptic fragments [88-102] derived from intact (blue) and NEM-alkylated (red) IFN throughout the course of HDX. The endpoint of the exchange reaction is indicated with a gray trace (isotopic distribution of a fully exchanged peptide). Location of this peptide within the amino acid sequence of IFN is shown on the right. Adapted with permission from (42).
What do the results of the HDX MS conformational analysis tell us about the likely changes in the properties of the “degraded” protein drug?
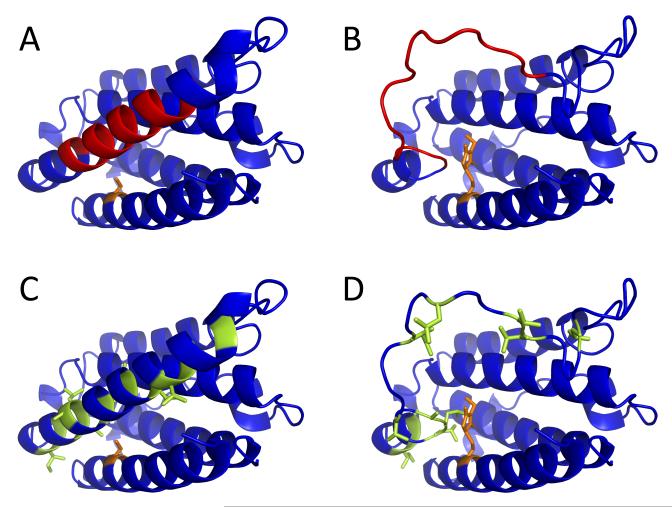
The detailed information on the backbone protection of IFN and NEM-IFN provided by HDX MS measurements allows the conformational properties of intact and alkylated forms of the protein to be compared directly. Perhaps the most important conclusion derived from the HDX MS work is the dramatic destabilization of helix D by the PTM event which affects a remote site in the amino acid sequence of this protein (42). Although this segment is very distant from the residue affected by alkylation (Cys-17) within the protein sequence, it is proximal to the Cys-17 side chain within the three-dimensional structure of the protein (Figure 4A).
Figure 4.
Location of the [88-102] segment (highlighted in red) within the crystal structure of IFN (1AU1, panel A) with respect to Cys-17 (highlighted in orange). Alkylation of Cys-17 inevitably leads to steric clashes within the native structure, which can be removed by unfolding of the helix D containing the [88-102] segment (panel B). Side chains of hydrophobic residues within helix D are sequestered in the protein interior (highlighted in green, panel C), but become exposed to solvent upon unfolding of this structural element (panel D).
Analysis of the crystal structure of IFN (PDB id 1AU1 (52)) makes it clear that modification of Cys-17 with NEM introduces a significant steric clash in the protein interior, which distorts helix packing, thereby adversely affecting the stability of the protein higher order structure. One extreme scenario of “relieving” this steric clash is presented in Figure 4B, where a total loss of secondary structure within helix D converts it to a flexible loop, thereby allowing the bulky NEM group to be accommodated within the protein structure. Another plausible scenario involves conversion of helix A to a flexible loop (this element of the secondary structure appears to be highly dynamic even in the absence of alkylation (42) and may also explain the possibility of forming a disulfide-linked IFN dimer despite sequestration of Cys-17 side chain in the protein interior (53)). Since the alkylation increases the dynamic character of both helices (A and D), NEM-IFN is likely to transiently sample conformations where either or both of these elements of secondary structure are compromised either through more frequent local structural fluctuations or via cooperative unfolding of the entire elements. We chose to present in Figure 4 the scenario where helix D melts, while helix A remains intact, due to its relevance for the mechanism of IFN deactivation (vide infra).
Even transient unfolding of either helix A or D (or both) would inevitably increase the aggregation propensity of IFN by exposing the hydrophobic residues, which are sequestered in the inter-helical interfaces in the native structure of IFN (Figure 4C and D). In fact, we have noticed diminished stability of NEM-IFN in solution compared to intact IFN in the course of our experimental work. These conformational changes at the IFN monomer level and the presence of aggregates in NEM-IFN solution are also likely to increase the likelihood of an immune response (8, 17).
Conformational changes detected by HDX MS: relevance for the functional competence of the protein drug. Verification by “native” ESI MS
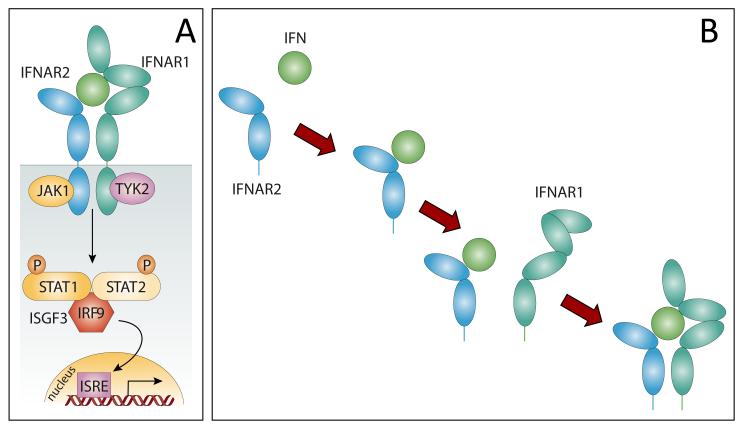
The analysis of the backbone flexibility maps of IFN and NEM-IFN also provides important clues regarding the molecular mechanism of inactivation of the modified form of IFN. IFN exerts its action by binding to the ectodomains of two transmembrane interferon receptors, IFNAR1 and IFNAR2 (Figure 5A). The assembly of this ternary complex in the extracellular space is required for bringing the cytoplasmic domains of IFNAR1 and IFNAR2 into close proximity. This latter step leads to activation of the JAK-STAT signaling pathway, which triggers a convoluted sequence of events that regulate the activity of interferon-stimulated response element (ISRE) in the nucleus (45, 54). IFNAR1 and IFNAR2 cannot interact directly with each other without IFN mediation, and the assembly of the ternary complex is believed to proceed via IFN binding to the high-affinity receptor IFNAR2, followed by recruitment of the low affinity receptor IFNAR1 (45), Figure 5B. The latter event is accompanied by a conformational change in the ectodomain of IFNAR1, which propagates to its cytoplasmic domain, thereby initializing signal transduction within the cell (55).
Figure 5.
A schematic representation of the JAK/STAT pathway activation by IFN initiated by its binding with the receptors IFNAR1 and IFNAR2 (A), adapted from (45); and the proposed sequence of events leading to the assembly of the ternary complex IFNAR2/IFN/IFNAR1 (B).
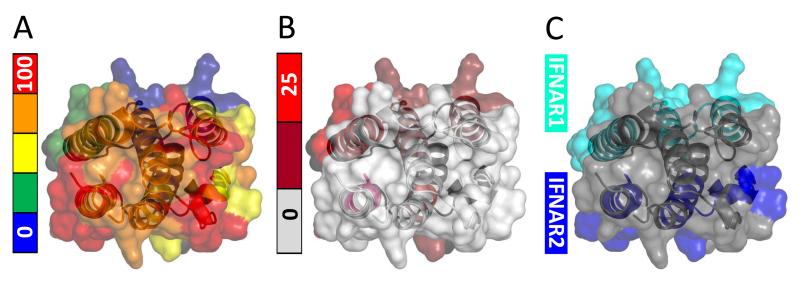
The IFNAR 1 and IFNAR2 binding interfaces revealed by mutagenesis (44, 56) are localized in two distinct parts of the IFN molecule (Figure 6C). Interestingly, comparison of this interface map with the IFN flexibility diagram deduced from the HDX MS measurements (Figure 6A) suggests that the protein segments forming the interface with the high-affinity receptor IFNAR2 are among the most dynamic (least protected) elements of the IFN structure. Although this observation may seem paradoxical, elevated polypeptide chain flexibility has been suggested to play an important role in protein interactions (57), and transient structural disorder was shown in the past to be an important facilitator of protein binding to small ligands (58) and other proteins (59).
Figure 6.
A: flexibility map of intact IFN generated by mapping the extent of deuterium incorporation in various peptic fragments (following 120 sec. HDX) onto the tertiary structure of the protein. B: regions of the protein exhibiting the difference in backbone protection between the intact and NEM-alkylated forms of IFN. C: receptor binding interfaces of IFN from earlier mutagenesis work (56).
Importantly, increased fluctuation within the receptor-binding interface region of human growth hormone (hGH) was recently linked directly to the increased free energy of the protein-receptor binding (60), which belongs to the same hematopoietic super-family that includes type I interferons (61). The two proteins (hGH and IFN) are highly homologous, so much so that hGH was used in earlier homology modeling of IFN and INF/IFNARs binding (61, 62). Helix 1 of hGH is noticeably more stable compared to its cousin helix A of IFN; however, introducing a structure-destabilizing mutation in helix 1 leads to a noticeable increase in the receptor binding affinity (60). Therefore, it should not be surprising that a critical element of IFN/IFNAR2 interface is one of the most dynamic segments of IFN.
If the dynamic character of helix A does in fact catalyze formation of the IFN/IFNAR2 complex, a modest flexibility increase within this element of IFN induced by alkylation of Cys-17 would be unlikely to disrupt the binding. Therefore, the first step in IFN/IFNARs ternary complex assembly at the cell surface is unlikely to be important for the activity loss of NEM-IFN. However, binding of IFN or NEM-IFN to IFNAR2 is almost certain to cement the structure of the IFN segments located in the interface region, similar to the stabilization of helix 1 observed upon hGH binding to its receptor (60). In this case, partial or complete melting of helix D would remain the only possible way to avoid the steric clash introduced by NEM-alkylation of Cys-17 (Figure 4B).
The next step in the ternary complex assembly involves association of the low-affinity receptor IFNAR1 with the IFN·IFNAR2 complex. The IFNAR1-binding interface of IFN maps on to the most stable region of the protein surface (Figure 6A and C), which is hardly surprising. Indeed, according to the accepted scenario, at this stage of the ternary complex assembly IFN serves as a binding template to which a flexible partner (IFNAR1) adapts (63). This consideration is particularly important within the context of the observation (deduced from the HDX MS measurements (42)) that the alkylation of Cys-17 results in significant destabilization of the IFNAR1-binding interface region of IFN (Figure 6B). The increase of IFN flexibility, which maps almost perfectly to the binding interface (compare panels B and C in Figure 6), is very likely to be highly detrimental to the IFN/IFNAR1 interaction.
The effect of alkylation is likely to be even more pronounced within the context of the ternary complex assembly, where the IFN/IFNAR1 binding occurs via recruitment of IFNAR1 to the IFN·IFNAR2 binary complex. As already mentioned, the expected stabilization of helix A in NEM-IFM following its association with IFNAR2 would result in additional stress on helix D due to the presence of a bulky NEM group sandwiched between them (Figure 4B). As a result, helix D is even less likely to be able to maintain a native-like structure within the NEM-IFN·IFNAR2 complex compared to NEM-IFN alone. Therefore, one should expect the stability of the ternary complex IFNAR1·NEM-IFN·IFNAR2 to be greatly diminished compared to IFNAR1·IFN·IFNAR2.
Analysis of the HDX MS data presented in this section suggests that the NEM-alkylation of IFN should impact its ability to bind the low affinity receptor and assemble a ternary complex to a much more significant extent compared to the effect (if any) on the IFN interaction with the high affinity receptor. A relatively easy and convenient way to assess the predictive value of these conclusions is provided by “native mass spectrometry,” a technique that relies on the unique ability of ESI to preserve non-covalent complexes upon their transition from solution to the gas phase (28). Here we apply this technique to monitor IFN interactions with its receptors and the effect exerted by NEM-alkylation on these processes.
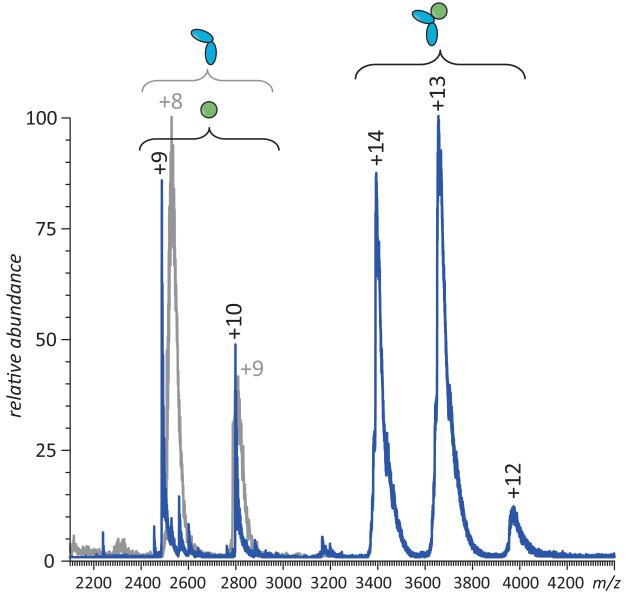
ESI MS of IFN/IFNAR2 mixture acquired under near-native conditions (aqueous solution of 100 mM ammonium acetate) using a hybrid quadrupole/time-of-flight MS (QStar-XL, ABI/Sciex, Toronto, Canada) provides clear evidence of a binary complex formation. The binding appears to be complete when the proteins are present in solution in the μM concentration range (e.g., no signal of free IFNAR2 is present in the mass spectrum alongside IFN or IFN·IFNAR2, as long as there is excess of IFN in the mixture, see Figure 7). This is consistent with the earlier results provided by SEC (64) and a low-nM binding constant estimated for this interaction (65). Importantly, the appearance of ESI mass spectra does not change when IFN is substituted with NEM-IFN, apart from small mass shifts due to alkylation (data not shown), consistent with the predictions generated in the course of the analysis of HDX MS data (vide supra).
Figure 7.
ESI MS of IFN/IFNAR2 interaction in the presence of excess IFN. Note that free IFNAR2 is absent from the spectrum (a reference mass spectrum of free IFNAR2 is shown in gray).
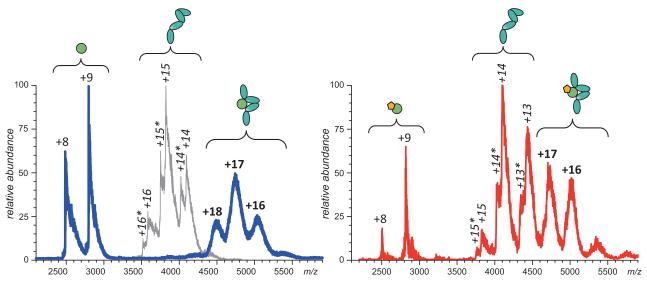
Analysis of the IFNAR1/IFN interaction might be somewhat trickier, since the binding energy is significantly weaker compared to the high affinity receptor (KD is estimated to be in the high sub-μM range (65)). As a result, earlier attempts to detect the IFNAR1·IFN using SEC failed (66). ESI MS appears to be a much more sensitive technique in this regard, demonstrating not only the formation of a 1:1 complex in solution, but also the absence of unbound IFNAR1 species (when excess IFN is present), as long as the protein concentrations in the mixture are maintained at least in the low μM range (Figure 8A). However, the ESI mass spectrum of the NEM-IFN/IFNAR1 mixture acquired under identical conditions clearly shows that the binding is affected by the alkylation (Figure 8B). Even though ionic signal of IFNAR1·IFN complex is present in the mass spectrum, contributions of both unbound NEM-IFN and free IFNAR1 are also prominent. Since both proteins are present in the mixture at low μM concentrations, the dissociation constant of the binary complex must be in the μM range or higher, an order of magnitude above that of the IFNAR1·IFN complex.
Figure 8.
A: ESI MS of IFN/IFNAR1 interaction in the presence of excess IFN. Note that free IFNAR1 is absent from the spectrum (a reference mass spectrum of free IFNAR1 is shown in gray). B: ESI MS of NEM-IFN/IFNAR1 interaction in the presence of excess NEM-IFN. Note that both free NEM-IFN and IFNAR1 are present in the spectrum alongside the NEM-IFN·IFNAR1 complex.
A very similar effect is exerted by IFN alkylation upon the stability of the ternary complex IFNAR1·IFN·IFNAR2. ESI MS provides strong evidence of complete binding in the IFNAR1/IFN/IFNAR2 mixture (Figure 9). Even though the ionic signals of some of the constituents of the ternary complex are observed alongside the ternary complex (e.g., IFNAR2 and IFN·IFNAR2 in the mass spectrum shown in Figure 9), the complementary components (e.g., IFNAR1·IFN and IFNAR1) are clearly absent. The situation changes very dramatically once the intact IFN is substituted with the alkylated form (top trace in Figure 9). Even though this spectrum provides unequivocal evidence that the ternary complex is formed in solution, the presence of the binary complex NEM-IFN·IFNAR2 alongside free IFNAR1 clearly suggests that the binding does not go to completion despite the fact that all participating proteins are present in solution at low μM concentrations.
Figure 9.
ESI MS of NEM-IFN interaction with IFNAR1 and IFNAR2 in the presence of excess NEM-IFN and IFNAR2. Note that both free IFNAR1 and a binary complex NEM-IFN·IFNAR2 are present in the spectrum alongside the ternary complex. Mixing intact IFN with the two receptors results in complete elimination of either free IFNAR1 or the binary complex IFN·IFNAR2 depending on the relative abundance of each of the three proteins (blue trace).
It is quite remarkable that native ESI MS confirms the conclusions drawn from the HDX MS experiments regarding the consequences of the PTM-triggered conformational changes for the ability of the protein to interact with its cognate receptors. Taken together, these two powerful techniques provide a means to determine the molecular basis of the protein drug inactivation following deleterious PTMs or other stress-induced degradation events.
Conformational analysis of the model protein: predictive value for identifying the “weak links” in the protein chain
Although alkylation of Cys-17 with NEM considered in the preceding sections was chosen only as a model of IFN degradation, analysis of the ensuing conformational changes provides important lessons regarding the likely consequences of many non-enzymatic PTMs that may affect IFN either by chance or by choice. Indeed, several covalent modification events that are known to occur under certain conditions target either Cys-17 or its immediate neighbors. For example, protein drug exposure to reactive oxygen species during manufacturing or storage frequently results in oxidation (67), one of the most common covalent modifications in biopharmaceutical products (68). The results of unreported work carried out in our laboratories suggest that controlled forced oxidation of IFN with H2O2 (which oxidizes three out of four methionine residues to sulfoxides and converts the side chain of Cys-17 initially to sulfenic acid, −SOH, a common but frequently unstable form of an oxidized thiol group (69)) does not result in a significant change of its biological activity measured by a standard anti-viral assay. However, more extensive oxidation converts Cys-17 to sulfinic acid (−SO2H), which appear to affect negatively the ability of IFN to bind the low-affinity receptor, IFNAR1, as revealed by native ESI MS measurements (data not shown). Once again, the physical size of the adduct formed on the Cys-17 side chain appears to be a factor destabilizing the critical element of the IFN/IFNAR1 interface, as was the case with NEM-alkylation.
Another deleterious non-enzymatic PTM that targets Cys-17 is disulfide scrambling, a phenomenon, which can also be detected and characterized using various mass spectrometry-based strategies (70). Although extensive efforts to observe scrambling under native conditions have been unsuccessful, IFN denaturation in the presence of reducing agents does lead to formation of IFN isomers with non-native disulfide bonds (i.e., Cys-17 is oxidized, while either Cys-31 or Cys-141 is reduced). We and others also demonstrated that under certain conditions Cys-17 may become involved in formation of an external disulfide bond, leading to formation of a covalent IFN dimer (53). An attempt to address this potential vulnerability of Cys-17 was made in the design of the earliest commercial IFN product, Betaseron™ by substituting this residue with a relatively inert serine (71). However, this form of IFN (interferon β1b) also lacks a carbohydrate chain. Glycosylation is often a critical element defining the stability of protein pharmaceuticals, and de-glycosylation is particularly damaging for IFN, where the carbohydrate chain shields a relatively hydrophobic patch on the protein surface from the solvent (72). As a result, the biological activity of the carbohydrate-free form of IFN is significantly lower than that of the wild type protein, while its aggregation propensity is much higher (72).
Interestingly, Cys-17 was also proposed in the past as a target for PEGylation, a “designer” PTM that is frequently used to enhance solubility and improve pharmacokinetic profile of protein drugs (73). PEGylation was shown to be beneficial to IFN, where it improved the pharmacokinetic profile of this protein by reducing its tendency to aggregate while maintaining its activity (74, 75). However, one commonly cited problem with the PEGylation of biopharmaceutical products is the difficulty in controlling the specificity of conjugation, which frequently generates an ensemble of protein molecules where the polymer chains are attached to different sites (76). Since there is only one free cysteine residue in the protein, a suggestion was made to increase the homogeneity of IFN PEGylation by using a thiol-reactive PEG chain (77, 78). While the initial report on this PEG-IFN conjugate was encouraging (77), no follow-up pre-clinical work was reported in the following decade, in contrast to other products of IFN PEGylation showing excellent results in pre-clinical investigation and entering clinical trials. This lack of action in such a dynamic field suggests that the use of a thiol-reactive PEG chain to specifically target Cys-17 met with insurmountable difficulties, most likely due to the instability of the conjugate, a dramatic reversal of the trend that is observed when solvent-exposed amino groups serve as PEGylation sites (74, 75). This example further highlights the value of HDX MS work that identifies Cys-17 as a rather troublesome site of IFN molecule.
HDX MS as a protein drug comparability tool: establishing equivalence at the higher order structure level
In the specific model of IFN degradation considered in the preceding sections, the backbone protection of the [88-102] segment serves as a convenient reporter of the protein conformational integrity. Because of the proximity of this segment in the 3D structure of IFN to Cys-17, one of the vulnerable elements of the protein, its utility as a reporter of protein degradation is likely to extend well beyond conformational changes triggered by NEM-alkylation. However, generally speaking it is not clear a priori which region(s) of the protein will have their stability diminished as a result of stress or degradation. Therefore, unbiased analysis of protein conformation and dynamics should focus not on a single element of the protein higher order structure, but rather catalog all conformational changes. Such analysis should cover the entire sequence of the protein (or at least as much as possible) and follow the exchange kinetics over extended periods of time. Capturing the exchange patterns on different time scales has a potential to be much more informative compared to simply taking a single snap-shot at an arbitrary selected time point (e.g., changes in protection patterns on shorter time scales may reveal compromised functional properties, while changes at longer time scale may betray the enhanced aggregation propensity of the protein).
Adaptation of this approach in the biopharmaceutical sector is likely to make a major impact on facilitating comparability studies of protein drugs, whose goal is to establish “sameness” of two (or more) related biopharmaceutical products. In the case of biopharmaceuticals, approval to a final product is given in the form of accepting the specific process of its manufacturing and formulation. However, the once unquestionable mantra “the process is the product” has been replaced by realization that in many cases post-approval changes to the process (e.g., change of a manufacturing site, protein production/purification protocol, and/or product formulation) can be dealt with by establishing the equivalence of the “new” product to the original one using “comparability” criteria (51), giving rise to the concept of “well characterized biological products” (79). The minimum set of comparability criteria includes molecular characterization, biological activity relevant for the therapeutic effect and relative bioavailability or “bioequivalence” (80). Although this approach will also play a role in regulation of the so-called biosimilars or follow-on biologics, “generic” versions of the innovator protein drugs facing patent expiration, more exhaustive studies are likely to be needed in this case (51).
In all of these situations the producer of the drug is required to prove to the regulatory agencies “sameness” of the drug to the original (approved) product. Effectively the manufacturer needs to prove drug equivalence (although the use of the term “identical” would be preferred, it is realized that in many cases this is unrealistic for biopharmaceuticals). The absence of alterations must be demonstrated at several levels, including covalent structure, purity, conformation (higher order structure), biological activity and clinical behavior in terms of toxicology and efficacy (including dosage) and immunogenicity. Of particular concern are possible changes in the higher order structure, including those triggered by environmental physical factors, whose detection is still viewed as lying out of the reach of current analytical methodology (as applied to biopharmaceuticals) (51). It is precisely in the area of conformational analysis that the MS-based experimental tools, especially HDX MS, can and should play an important role.
For example, MS-based comparability studies focusing on conformation and dynamics of two versions of the same protein drug may assess their three-dimensional structures and conformational dynamics by obtaining backbone protection maps using HDX MS. The goal of such an investigation would be to determine if there are any protein segments where a statistically significant difference in protection levels exists between the two forms (i.e., lies outside of the range of variability that is normally observed in conducting these experiments). Detection of such statistically significant differences in backbone protection patterns would immediately signal that the two forms of the protein drug are not equivalent at the molecular level and, therefore, do not satisfy one of the comparability criteria.
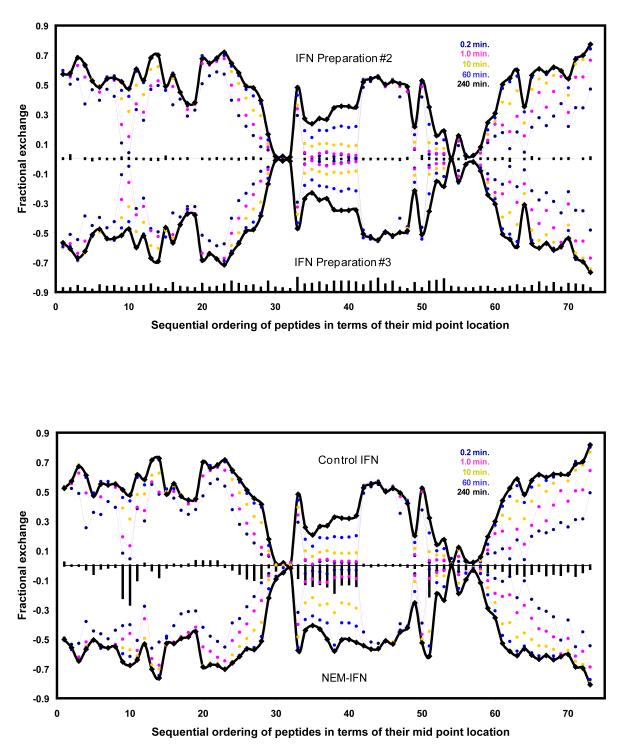
As mentioned above, the HDX MS data would have to be acquired at various time scales to enable detection of a variety of relevant dynamic events. This approach is illustrated in Figure 10A, where the two forms of IFN were produced using different growth media, a standard biological source medium (preparation 2) and a modified synthetic medium (preparation 3), which reduces the potential threat of contaminating the growth media with biologically active agents and provides more reproducible growth conditions for achieving higher product consistently. In this study, four independent HDX MS experiments were carried out to determine if there are any statistically significant differences in protection patterns between these two products. The results of this comparability study (Figure 10A) indicate that there are no protein segments exhibiting a statistically significant difference in backbone protection. This is in sharp contrast to the results of a similar comparability analysis carried out for the NEM-alkylated form of IFN (using intact IFN as a reference). In this latter case, several regions can be clearly identified where the differences between the two forms of the protein exceed the statistical variability (Figure 10B).
Figure 10.
HDX MS comparability studies of two different preparations of IFN grown in different culture media (A) and NEM-alkylated vs. intact IFN (B). The fractional exchange for each peptide at each HDX time point (plotted in different colors) is represented as a single point in a graph where the x-axis shows the sequential order of each peptic fragment in IFN sequence and the y-axis represents the fraction of exchanged amides for each peptide. To allow for better visualization, the data for the two samples are plotted in opposite directions (the so-called butterfly plot). Differences in total HDX levels (summations of all time points) for each peptide are plotted as vertical bars across the x-axis (the average standard deviation of these values is below 2% of the total exchange level). The vertical lines at the bottom of the graph A are scaled to represent the relative size of each peptic fragment.
Current challenges and future outlook
Biopharmaceutical products cover a very wide molecular weight range, and IFN is located in the lower half of this spectrum. While there are a large number of protein drugs that have either similar or smaller size, which would also benefit from the conformational analysis discussed in this article, development of reliable tools to probe conformation and dynamics of larger proteins is a more urgent task. Larger protein drugs (such as monoclonal antibodies) are much more complex, and as the size/complexity of any system increases, so does the probability that something may go wrong. This is certainly true in any branch of technology, and biotechnology is no exception to this rule. Of particular concern is the possibility of local unfolding events in large proteins, which can trigger aggregation, a highly undesirable process as far as protein therapeutics are concerned. An increase in size of a protein drug also inevitably translates into elevated frequency of non-enzymatic PTMs, which often makes precise mapping of these changes a gargantuan task. Therefore, availability of a robust, easy-to-use and reasonably high-throughput method to probe conformational integrity of such large systems directly would obviously be a boon to the biopharmaceutical industry.
Implementation of HDX MS for conformational analysis of large (>60 kDa) protein drugs faces several challenges. First, non-specificity of pepsin cleavage combined with the large physical size of the protein typically results in a large number of fragment peptides, which must be identified in order to extract meaningful HDX information. This problem is magnified by the frequent occurrence of glycosylation and disulfide bonds in protein drugs, which both multiply the sheer number of candidate peptides that fit a given mass and also make it more difficult to obtain meaningful sequence information using classical MS/MS approaches, such as collision-activated dissociation (CAD). In many cases, reliable peptide identification may be achieved only by using ultra-high resolution mass measurements (e.g., with FT ICR MS), applying electron-based ion fragmentation techniques (ECD or ETD), or using collision-energy dependent data collection in MS/MS experiments (MSE).
Furthermore, even complete identification of all observed peptic fragments does not necessarily result in complete sequence coverage of a large protein. Indeed, the necessity to minimize the sample handling time prior to MS analysis as a means to limit the extent of back-exchange typically translates to very short (and, therefore, crowded) LC runs, while utilization of more efficient chromatographic schemes (such as UPLC) may improve the separation, but results in more extensive back exchange (81). While MS detection itself (and particularly high-resolution MS) solves the problem of detection of multiple co-eluting peaks, signal suppression is likely to result in loss of some of the peptides, thereby leaving gaps in sequence coverage. Despite these difficulties, significant progress was made recently in this field, as demonstrated by successful use of HDX MS to probe conformational properties of larger protein therapeutics, such as 63 kDa β-glucocerebrosidase (41), 73 kDa (non-redundant mass) IgG (43), as well as our own recent work with transferrin (38, 82), an 80 kDa component of several therapies that are currently in pre-clinical evaluation.
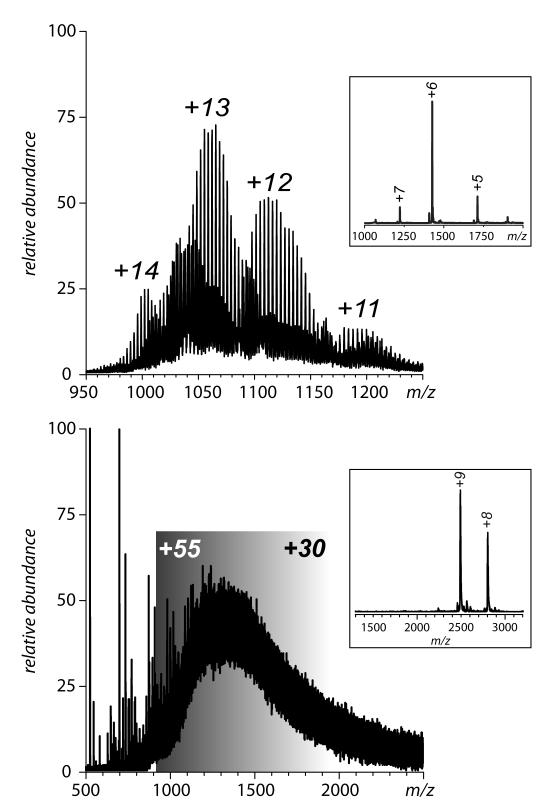
Another serious challenge to the characterization of protein drugs is posed by the highly heterogeneous character exhibited by a large fraction of these species. While glycosylation is an inherent (enzymatic) PTM that was originally the major source of heterogeneity in many of protein therapeutics, the second generation of biopharmaceuticals often employs “designer” PTMs to enhance their therapeutic properties. This can be achieved by either engineering elaborate and extensive glycosylation patterns (83), conjugating the protein drug to a synthetic polymer (73) or to a carrier protein (84). Although mass spectrometry certainly helps to appreciate the extent of heterogeneity of such engineered species (Figure 11), it also makes it very clear that the polydispersity of designer PTMs may present a significant barrier vis-à-vis characterization of conformation and dynamics of these complex systems. Indeed, while ESI MS allows a distinction to be made among modestly sized protein molecules conjugated to relatively short PEG chains of varying lengths (e.g., an 8 kDa ubiquitin conjugated to 5 kDa PEG, Figure 11A), individual species of larger proteins conjugated to longer PEG chains cannot be resolved. This is exemplified in Figure11B, which shows a mass spectrum of IFN conjugated to 20 kDa PEG. Nevertheless, some preliminary work carried out in our laboratories indicates that HDX MS can be successfully applied to probe conformational stability of these systems, and even ionic charge state distribution analysis may be possible following some modifications of the standard routine (49) in order to adapt it to continuous, rather than discrete, m/z distributions. Combination of ion chemistry (in the form of charge reduction) and separation (ion mobility) in the gas phase are also currently evaluated as a means to improve the analysis of these heterogeneous targets (85).
Figure 11.
Heterogeneity of protein-polymer conjugates exemplified by ESI MS of mono-PEGylated ubiquitin (A) and mono-PEGylated IFN (B). The average molecular weights of the PEG chains are 5 kDa (ubiquitin) and 20 kDa (IFN). Insets show mass spectra of unconjugated proteins.
Perhaps the ultimate success of mass spectrometry as a tool to probe conformation of protein drugs and its acceptance in this capacity will be determined not only by its ability to deal with very large and highly heterogeneous systems, but by its adaptability to the specific needs of the biopharmaceutical industry. Several important questions have to be addressed in order for mass spectrometry to become an integral part of the analytical routine in assessing conformation and dynamics of biopharmaceutical products. Can it probe integrity of the higher order structure in a true high-throughput fashion? Can the entire procedure (from sample handling to data processing) be automated and commercialized in a form of an easy-to-use “turn-key” instrument? What is the reproducibility and robustness of these measurements if they are carried out in different laboratories and/or using different LC-MS platforms? What is the sensitivity of this technique in detecting small conformational changes and/or alterations of the higher order structure that affect only a small fraction of the protein molecules? Addressing these questions will require extensive concerted efforts of academic and industrial researchers, but the end result is well worth it. Indeed, adoption of mass spectrometry in biopharmaceutical industry in this new role will not only become a boon to analytical characterization, but is also certain to greatly catalyze development of new and enhance existing potent therapies.
Acknowledgements
The authors thank Robert Smock (Molecular and Cellular Biology program, UMass-Amherst) for his help with modeling of partially unfolded protein structures, Adriana Zeledon (Molecular and Cellular Biology program, UMass-Amherst) for help in acquiring some of the data presented in the manuscript, Dr. Igor V. Chernushevich (ABI/Sciex, Toronto, Canada) for invaluable help with instrument set-up enabling native MS measurements, Drs. J. Paul Speir and Michael Easterling (Bruker Daltonics, Billerica, MA) for providing access to prototype instrumentation and Prof. John Engen (Barnett Institute at Northeastern University, Boston, MA and Waters Corp., Milford, MA) for providing access for one of the co-authors (D.H) to prototype equipment. The samples of ectodomains of IFNAR1 and IFNAR2 were provided by Dr. Darren Baker (Biogen IDEC) and Prof. Dr. Jacob Piehler (Fachbereich Biologie, Universität Osnabrück, Osnabrück, Germany).
This work was supported in part by grants from the National Institutes of Health R01 GM061666 and National Science Foundation CHE-0750389.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Knäblein J, editor. Modern Biopharmaceuticals: Design, Development and Optimization. Vol. 1-4. WILEY-VCH Verlag GmbH & Co.; Weinheim: 2005. [Google Scholar]
- 2.Volpi N. Therapeutic applications of glycosaminoglycans. Curr. Med. Chem. 2006;13:1799–1810. doi: 10.2174/092986706777452470. [DOI] [PubMed] [Google Scholar]
- 3.Alvarez-Salas LM. Nucleic Acids as Therapeutic Agents. Curr. Top. Med. Chem. 2008;8:1379–1404. doi: 10.2174/156802608786141133. [DOI] [PubMed] [Google Scholar]
- 4.Spada S, Walsh G. Directory of approved biopharmaceutical products. CRC Press; Boca Raton: 2005. [Google Scholar]
- 5.Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nat. Rev. Drug Discov. 2008;7:21–39. doi: 10.1038/nrd2399. [DOI] [PubMed] [Google Scholar]
- 6.Stolnik S, Shakesheff K. Formulations for delivery of therapeutic proteins. Biotechnol. Lett. 2009;31:1–11. doi: 10.1007/s10529-008-9834-y. [DOI] [PubMed] [Google Scholar]
- 7.Morozova-Roche L, Malisauskas M. A false paradise - mixed blessings in the protein universe: the amyloid as a new challenge in drug development. Curr. Med. Chem. 2007;14:1221–1230. doi: 10.2174/092986707780597989. [DOI] [PubMed] [Google Scholar]
- 8.Maas C, Hermeling S, Bouma B, Jiskoot W, Gebbink MFBG. A role for protein misfolding in immunogenicity of biopharmaceuticals. J. Biol. Chem. 2007;282:2229–2236. doi: 10.1074/jbc.M605984200. [DOI] [PubMed] [Google Scholar]
- 9.Hughes B. Gearing up for follow-on biologics. Nat. Rev. Drug Discov. 2009;8:181–181. doi: 10.1038/nrd2847. [DOI] [PubMed] [Google Scholar]
- 10.Frank RG. Regulation of follow-on biologics. N. Engl. J. Med. 2007;357:841–843. doi: 10.1056/NEJMp078095. [DOI] [PubMed] [Google Scholar]
- 11.Dudzinski DM, Kesselheim AS. Scientific and legal viability of follow-on protein drugs. N. Engl. J. Med. 2008;358:843–849. doi: 10.1056/NEJMhle0706973. [DOI] [PubMed] [Google Scholar]
- 12.Serdyuk IN, Zaccai NR, Zaccai G. Methods in molecular biophysics: structure, dynamics, function. Cambridge University Press; Cambridge, UK: 2006. [Google Scholar]
- 13.Scapin G. Structural biology and drug discovery. Curr. Pharm. Des. 2006;12:2087–2097. doi: 10.2174/138161206777585201. [DOI] [PubMed] [Google Scholar]
- 14.Petoukhov MV, Svergun DI. Analysis of X-ray and neutron scattering from biomacromolecular solutions. Curr. Opin. Struct. Biol. 2007;17:562–571. doi: 10.1016/j.sbi.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 15.Nguyen LT, Wiencek JM, Kirsch LE. Characterization methods for the physical stability of biopharmaceuticals. PDA J. Pharm. Sci. Technol. 2003;57:429–445. [PubMed] [Google Scholar]
- 16.Capelle MAH, Gurny R, Arvinte T. High throughput screening of protein formulation stability: Practical considerations. Eur. J. Pharm. Biopharm. 2007;65:131–148. doi: 10.1016/j.ejpb.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 17.Korotchkina LG, Ramani K, Balu-Iyer SV. Mol. Biol. Protein Fold., Pt. A. Elsevier Academic Press Inc; San Diego: 2008. Folding considerations for therapeutic protein formulations; pp. 255–270. [DOI] [PubMed] [Google Scholar]
- 18.Zhang Z, Pan H, Chen X. Mass spectrometry for structural characterization of therapeutic antibodies. Mass Spectrom. Rev. 2009;28:147–176. doi: 10.1002/mas.20190. [DOI] [PubMed] [Google Scholar]
- 19.Srebalus-Barnes CA, Lim A. Applications of mass spectrometry for the structural characterization of recombinant protein pharmaceuticals. Mass Spectrom. Rev. 2007;26:370–388. doi: 10.1002/mas.20129. [DOI] [PubMed] [Google Scholar]
- 20.Chowdhury SK, Katta V, Chait BT. Probing conformational changes in proteins by mass spectrometry. J. Am. Chem. Soc. 1990;112:9012–9013. [Google Scholar]
- 21.Loo JA, Loo RR, Udseth HR, Edmonds CG, Smith RD. Solvent-induced conformational changes of polypeptides probed by electrospray-ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1991;5:101–105. doi: 10.1002/rcm.1290050303. [DOI] [PubMed] [Google Scholar]
- 22.Katta V, Chait BT. Conformational changes in proteins probed by hydrogen-exchange electrospray-ionization mass spectrometry. Rapid Commun. Mass Spectrom. 1991;5:214–217. doi: 10.1002/rcm.1290050415. [DOI] [PubMed] [Google Scholar]
- 23.Smith RD, Lightwahl KJ, Winger BE, Loo JA. Preservation of noncovalent associations in electrospray ionization mass spectrometry - Multiply charged polypeptide and protein dimers. Org. Mass Spectrom. 1992;27:811–821. [Google Scholar]
- 24.Ganem B, Li YT, Henion JD. Detection of noncovalent receptor ligand complexes by mass spectrometry. J. Am. Chem. Soc. 1991;113:6294–6296. [Google Scholar]
- 25.Kaltashov IA, Eyles SJ. Studies of biomolecular conformations and conformational dynamics by mass spectrometry. Mass Spectrom. Rev. 2002;21:37–71. doi: 10.1002/mas.10017. [DOI] [PubMed] [Google Scholar]
- 26.Back JW, de Jong L, Muijsers AO, de Koster CG. Chemical cross-linking and mass spectrometry for protein structural modeling. J. Mol. Biol. 2003;331:303–313. doi: 10.1016/s0022-2836(03)00721-6. [DOI] [PubMed] [Google Scholar]
- 27.Hernandez H, Robinson CV. Determining the stoichiometry and interactions of macromolecular assemblies from mass spectrometry. Nat. Protoc. 2007;2:715–726. doi: 10.1038/nprot.2007.73. [DOI] [PubMed] [Google Scholar]
- 28.Heck AJR. Native mass spectrometry: a bridge between interactomics and structural biology. Nat. Meth. 2008;5:927–933. doi: 10.1038/nmeth.1265. [DOI] [PubMed] [Google Scholar]
- 29.Yin S, Loo JA. Mass spectrometry detection and characterization of noncovalent protein complexes. Methods Mol. Biol. 2009;492:273–282. doi: 10.1007/978-1-59745-493-3_16. [DOI] [PubMed] [Google Scholar]
- 30.Kaltashov IA, Abzalimov RR. Do ionic charges in ESI MS provide useful information on macromolecular structure? J. Am. Soc. Mass Spectrom. 2008;19:1239–1246. doi: 10.1016/j.jasms.2008.05.018. [DOI] [PubMed] [Google Scholar]
- 31.Hofstadler SA, Sannes-Lowery KA. Applications of ESI-MS in drug discovery: interrogation of noncovalent complexes. Nat. Rev. Drug Discov. 2006;5:585–595. doi: 10.1038/nrd2083. [DOI] [PubMed] [Google Scholar]
- 32.Atmanene C, Wagner-Rousset E, Malissard M, Chol B, Robert A, Corvaia N, Van Dorsselaer A, Beck A, Sanglier-Cianferani S. Extending mass spectrometry contribution to therapeutic monoclonal antibody lead optimization: Characterization of immune complexes using noncovalent ESI-MS. Anal. Chem. 2009;81:6364–6373. doi: 10.1021/ac9007557. [DOI] [PubMed] [Google Scholar]
- 33.Zamani L, Lindholm J, Ilag LL, Jacobsson SP. Discrimination among IgG1-κ monoclonal antibodies produced by two cell lines using charge state distributions in nanoESI-TOF mass spectra. J. Am. Soc. Mass Spectrom. 2009;20:1030–1036. doi: 10.1016/j.jasms.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 34.Hoofnagle AN, Resing KA, Ahn NG. Protein analysis by hydrogen exchange mass spectrometry. Annu. Rev. Biophys. Biomol. Struct. 2003;32:1–25. doi: 10.1146/annurev.biophys.32.110601.142417. [DOI] [PubMed] [Google Scholar]
- 35.Eyles SJ, Kaltashov IA. Methods to study protein dynamics and folding by mass spectrometry. Methods. 2004;34:88–99. doi: 10.1016/j.ymeth.2004.03.015. [DOI] [PubMed] [Google Scholar]
- 36.Konermann L, Tong X, Pan Y. Protein structure and dynamics studied by mass spectrometry: H/D exchange, hydroxyl radical labeling, and related approaches. J. Mass Spectrom. 2008;43:1021–1036. doi: 10.1002/jms.1435. [DOI] [PubMed] [Google Scholar]
- 37.Engen JR. Analysis of protein conformation and dynamics by hydrogen/deuterium exchange MS. Anal. Chem. 2009 doi: 10.1021/ac901154s. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kaltashov IA, Bobst CE, Abzalimov RR. H/D exchange and mass spectrometry in the studies of protein conformation and dynamics: Is there a need for a top-down approach? Anal. Chem. 2009;81:7892–7899. doi: 10.1021/ac901366n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Woods VL, Jr., Hamuro Y. High resolution, high-throughput amide deuterium exchange-mass spectrometry (DXMS) determination of protein binding site structure and dynamics: utility in pharmaceutical design. J. Cell. Biochem. 2001;37S:89–98. doi: 10.1002/jcb.10069. [DOI] [PubMed] [Google Scholar]
- 40.Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR. Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 2006;78:1005–1014. doi: 10.1021/ac051294f. [DOI] [PubMed] [Google Scholar]
- 41.Tropak MB, Kornhaber GJ, Rigat BA, Maegawa GH, Buttner JD, Blanchard JE, Murphy C, Tuske SJ, Coales SJ, Hamuro Y, Brown ED, Mahuran DJ. Identification of pharmacological chaperones for Gaucher disease and characterization of their effects on beta-glucocerebrosidase by hydrogen/deuterium exchange mass spectrometry. ChemBioChem. 2008;9:2650–2662. doi: 10.1002/cbic.200800304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bobst CE, Abzalimov RR, Houde D, Kloczewiak M, Mhatre R, Berkowitz SA, Kaltashov IA. Detection and characterization of altered conformations of protein pharmaceuticals using complementary mass spectrometry-based approaches. Anal. Chem. 2008;80:7473–7481. doi: 10.1021/ac801214x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Houde D, Arndt J, Domeier W, Berkowitz S, Engen JR. Characterization of IgG1 conformation and conformational dynamics by hydrogen/deuterium exchange mass spectrometry. Anal. Chem. 2009;81:2644–2651. doi: 10.1021/ac802575y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Whitty A, Karpusas M. The structure of human interferon-β-1a (Avonex™) and its relation to activity: a case study of the use of structural data in the arena of protein pharmaceuticals. In: Chasman DL, editor. Protein Structure: Determinantion, Analysis and Applications for Drug Discovery. Marcel Dekker; New York-Basel: 2003. pp. 483–519. [Google Scholar]
- 45.Borden EC, Sen GC, Uze G, Silverman RH, Ransohoff RM, Foster GR, Stark GR. Interferons at age 50: past, current and future impact on biomedicine. Nat. Rev. Drug Discov. 2007;6:975–990. doi: 10.1038/nrd2422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bermel RA, Rudick RA. Interferon-β treatment for multiple sclerosis. Neurotherapeutics. 2007;4:633–646. doi: 10.1016/j.nurt.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Noseworthy JH, Lucchinetti C, Rodriguez M, Weinshenker BG. Multiple sclerosis. N. Engl. J. Med. 2000;343:938–952. doi: 10.1056/NEJM200009283431307. [DOI] [PubMed] [Google Scholar]
- 48.Kuprowski MC, Konermann L. Signal response of coexisting protein conformers in electrospray mass spectrometry. Anal. Chem. 2007;79:2499–2506. doi: 10.1021/ac0620056. [DOI] [PubMed] [Google Scholar]
- 49.Dobo A, Kaltashov IA. Detection of multiple protein conformational ensembles in solution via deconvolution of charge state distributions in ESI MS. Anal. Chem. 2001;73:4763–4773. doi: 10.1021/ac010713f. [DOI] [PubMed] [Google Scholar]
- 50.Mohimen A, Dobo A, Hoerner JK, Kaltashov IA. A chemometric approach to detection and characterization of multiple protein conformers in solution using electrospray ionization mass spectrometry. Anal. Chem. 2003;75:4139–4147. doi: 10.1021/ac034095+. [DOI] [PubMed] [Google Scholar]
- 51.Davis GC, Beals JM, Johnson C, Mayer MH, Meiklejohn BI, Mitlak BH, Roth JL, Towns JK, Veenhuizen M. Recommendations regarding technical standards for follow-on biologics: comparability, similarity, interchangeability. Curr. Med. Res. Opin. 2009;25:1655–1661. doi: 10.1185/03007990903017313. [DOI] [PubMed] [Google Scholar]
- 52.Karpusas M, Nolte M, Benton CB, Meier W, Lipscomb WN, Goelz S. The crystal structure of human interferon β at 2.2-Å resolution. Proc. Natl. Acad. Sci. U. S. A. 1997;94:11813–11818. doi: 10.1073/pnas.94.22.11813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Orru S, Amoresano A, Siciliano R, Napoleoni R, Finocchiaro O, Datola A, De Luca E, Sirna A, Pucci P. Structural analysis of modified forms of recombinant IFN-β produced under stress-simulating conditions. Biol. Chem. 2000;381:7–17. doi: 10.1515/BC.2000.002. [DOI] [PubMed] [Google Scholar]
- 54.Claudinon J, Monier M-N, Lamaze C. Interfering with interferon receptor sorting and trafficking: Impact on signaling. Biochimie. 89:735–743. doi: 10.1016/j.biochi.2007.03.014. [DOI] [PubMed] [Google Scholar]
- 55.Strunk JJ, Gregor I, Becker Y, Li Z, Gavutis M, Jaks E, Lamken P, Walz T, Enderlein J, Piehler J. Ligand binding induces a conformational change in IFNAR1 that is propagated to its membrane-proximal domain. J. Mol. Biol. 2008;377:725–739. doi: 10.1016/j.jmb.2008.01.017. [DOI] [PubMed] [Google Scholar]
- 56.Runkel L, deDios C, Karpusas M, Betzenhauser M, Muldowney C, Zafari M, Benjamin CD, Miller S, Hochman PS, Whitty A. Systematic mutational mapping of sites on human interferon-β1a that are important for receptor binding and functional activity. Biochemistry. 2000;39:2538–2551. doi: 10.1021/bi991631c. [DOI] [PubMed] [Google Scholar]
- 57.Shoemaker BA, Portman JJ, Wolynes PG. Speeding molecular recognition by using the folding funnel: the fly-casting mechanism. Proc. Natl. Acad. Sci. U.S.A. 2000;97:8868–8873. doi: 10.1073/pnas.160259697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao H, Kaltashov IA. Transient structural disorder as a facilitator of protein-ligand binding: native H/D exchange-mass spectrometry study of cellular retinoic acid binding protein I. J. Am. Soc. Mass Spectrom. 2005;16:869–879. doi: 10.1016/j.jasms.2005.02.020. [DOI] [PubMed] [Google Scholar]
- 59.Griffith WP, Kaltashov IA. Protein conformational heterogeneity as a binding catalyst: ESI-MS study of hemoglobin H formation. Biochemistry. 2007;46:2020–2026. doi: 10.1021/bi062032q. [DOI] [PubMed] [Google Scholar]
- 60.Horn JR, Kraybill B, Petro EJ, Coales SJ, Morrow JA, Hamuro Y, Kossiakoff AA. The role of protein dynamics in increasing binding affinity for an engineered protein-protein interaction established by H/D exchange mass spectrometry. Biochemistry. 2006;45:8488–8498. doi: 10.1021/bi0604328. [DOI] [PubMed] [Google Scholar]
- 61.Seto MH, Harkins RN, Adler M, Whitlow M, Church WB, Croze E. Homology model of human interferon-α8 and its receptor complex. Protein Sci. 1995;4:655–670. doi: 10.1002/pro.5560040406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chill JH, Quadt SR, Levy R, Schreiber G, Anglister J. The human type I interferon receptor: NMR structure reveals the molecular basis of ligand binding. Structure. 2003;11:791–802. doi: 10.1016/s0969-2126(03)00120-5. [DOI] [PubMed] [Google Scholar]
- 63.Li Z, Strunk JJ, Lamken P, Piehler J, Walz T. The EM structure of a type I interferon-receptor complex reveals a novel mechanism for cytokine signaling. J. Mol. Biol. 2008;377:715–724. doi: 10.1016/j.jmb.2007.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Arduini RM, Strauch KL, Runkel LA, Carlson MM, Hronowski X, Foley SF, Young CN, Cheng W, Hochman PS, Baker DP. Characterization of a soluble ternary complex formed between human interferon-β-1a and its receptor chains. Protein Sci. 1999;8:1867–1877. doi: 10.1110/ps.8.9.1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jaitin DA, Roisman LC, Jaks E, Gavutis M, Piehler J, Van der Heyden J, Uze G, Schreiber G. Inquiring into the differential action of interferons (IFNs): an IFN-α2 mutant with enhanced affinity to IFNAR1 is functionally similar to IFN-β. Mol. Cell. Biol. 2006;26:1888–1897. doi: 10.1128/MCB.26.5.1888-1897.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kalie E, Jaitin DA, Podoplelova Y, Piehler J, Schreiber G. The stability of the ternary interferon-receptor complex rather than the affinity to the individual subunits dictates differential biological activities. J. Biol. Chem. 2008;283:32925–32936. doi: 10.1074/jbc.M806019200. [DOI] [PubMed] [Google Scholar]
- 67.Stadtman ER. Protein oxidation and aging. Free Radic. Res. 2006;40:1250–1258. doi: 10.1080/10715760600918142. [DOI] [PubMed] [Google Scholar]
- 68.Houde D, Kauppinen P, Mhatre R, Lyubarskaya Y. Determination of protein oxidation by mass spectrometry and method transfer to quality control. J. Chromatogr. A. 2006;1123:189–198. doi: 10.1016/j.chroma.2006.04.046. [DOI] [PubMed] [Google Scholar]
- 69.Eaton P. Protein thiol oxidation in health and disease: Techniques for measuring disulfides and related modifications in complex protein mixtures. Free Radic. Biol. Med. 2006;40:1889–1899. doi: 10.1016/j.freeradbiomed.2005.12.037. [DOI] [PubMed] [Google Scholar]
- 70.Gorman JJ, Wallis TP, Pitt JJ. Protein disulfide bond determination by mass spectrometry. Mass Spectrom. Rev. 2002;21:183–216. doi: 10.1002/mas.10025. [DOI] [PubMed] [Google Scholar]
- 71.Lin L. Betaseron. In: Brown F, Lubiniecki A, Murano G, editors. Symposium on the Characterization of Biotechnology Pharmaceutical Products; Washington, D.C.: Karger; 1995. pp. 97–104. [Google Scholar]
- 72.Runkel L, Meier W, Pepinsky RB, Karpusas M, Whitty A, Kimball K, Brickelmaier M, Muldowney C, Jones W, Goelz SE. Structural and functional differences between glycosylated and non-glycosylated forms of human interferon-beta (IFN-beta) Pharm. Res. 1998;15:641–649. doi: 10.1023/a:1011974512425. [DOI] [PubMed] [Google Scholar]
- 73.Veronese FM, Mero A. The impact of PEGylation on biological therapies. BioDrugs. 2008;22:315. doi: 10.2165/00063030-200822050-00004. [DOI] [PubMed] [Google Scholar]
- 74.Pepinsky RB, LePage DJ, Gill A, Chakraborty A, Vaidyanathan S, Green M, Baker DP, Whalley E, Hochman PS, Martin P. Improved pharmacokinetic properties of a polyethylene glycol-modified form of interferon-β-1a with preserved in vitro bioactivity. J. Pharmacol. Exp. Ther. 2001;297:1059–1066. [PubMed] [Google Scholar]
- 75.Baker DP, Lin EY, Lin K, Pellegrini M, Petter RC, Chen LL, Arduini RM, Brickelmaier M, Wen D, Hess DM, Chen L, Grant D, Whitty A, Gill A, Lindner DJ, Pepinsky RB. N-terminally PEGylated human interferon-β-1a with improved pharmacokinetic properties and in vivo efficacy in a melanoma angiogenesis models. Bioconjugate Chem. 2005;17:179–188. doi: 10.1021/bc050237q. [DOI] [PubMed] [Google Scholar]
- 76.Gaberc-Porekar V, Zore I, Podobnik B, Menart V. Obstacles and pitfalls in the PEGylation of therapeutic proteins. Curr. Opin. Drug Discov. Dev. 2008;11:242–250. [PubMed] [Google Scholar]
- 77.Roberts MJ, El Tayar N, McKenna S, Gorga J, Sawlivich W. Site-selective modification of human interferon beta. AAPS J. 2000;2-S1:140. [Google Scholar]
- 78.El-Tayar N, Roberts MJ, Harris M, Sawlivich W. Polyol-IFN-beta conjugate and composition containing same. Applied Research Systems, ARS Holdings; N.V., Curacao (NL), U.S.A.: 2004/0043002 A1 U.S. Patent. 2004
- 79.Wrotnowski C. Well-characterized biologicals: Trends and views. Genet. Eng. News. 2000;20:21–74. [Google Scholar]
- 80.Prync AES, Yankilevich P, Barrero PR, Bello R, Marangunich L, Vidal A, Criscuolo M, Benasayag L, Famulari AL, Dominguez RO, Kauffman MA, Diez RA. Two recombinant human interferon-beta 1a pharmaceutical preparations produce a similar transcriptional response determined using whole genome microarray analysis. Int. J. Clin. Pharmacol. Ther. 2008;46:64–71. doi: 10.5414/cpp46064. [DOI] [PubMed] [Google Scholar]
- 81.Wu Y, Engen JR, Hobbins WB. Ultra performance liquid chromatography (UPLC) further improves hydrogen/deuterium exchange mass spectrometry. J. Am. Soc. Mass Spectrom. 2006;17:163–167. doi: 10.1016/j.jasms.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 82.Bobst CE, Zhang M, Kaltashov IA. Existence of a noncanonical state of iron-bound transferrin at endosomal pH revealed by hydrogen exchange and mass spectrometry. J. Mol. Biol. 2009;388:954–967. doi: 10.1016/j.jmb.2009.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Sola RJ, Griebenow K. Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 2009;98:1223–1245. doi: 10.1002/jps.21504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Holzman DC. Whatever happened to immunotoxins? Research, and hope, are still alive. J. Natl. Cancer Inst. 2009;101:624–625. doi: 10.1093/jnci/djp110. [DOI] [PubMed] [Google Scholar]
- 85.Bagal D, Zhang H, Schnier PD. Gas-phase proton-transfer chemistry coupled with TOF mass spectrometry and ion mobility-MS for the facile analysis of poly(ethylene glycols) and PEGylated polypeptide conjugates. Anal. Chem. 2008;80:2408–2418. doi: 10.1021/ac7020163. [DOI] [PubMed] [Google Scholar]
- 86.Zheng X, Wu SL, Hancock WS. Glycation of interferon-beta-1b and human serum albumin in a lyophilized glucose formulation. Part III: application of proteomic analysis to the manufacture of biological drugs. Int. J. Pharm. 2006;322:136–145. doi: 10.1016/j.ijpharm.2006.06.038. [DOI] [PubMed] [Google Scholar]