Abstract
DNA double-strand breaks (DSBs) are a threat to cell survival and genome integrity. In addition to canonical DNA repair systems, DSBs can be converted to telomeres by telomerase. This process, herein termed telomere healing, endangers genome stability, since it usually results in chromosome arm loss. Therefore, cells possess mechanisms that prevent the untimely action of telomerase on DSBs. Here we report that Mec1, the ATR ortholog, couples the detection of DNA ends with the inhibition of telomerase. Mec1 inhibits telomere healing by phosphorylating Cdc13 on its S306 residue, a phosphorylation event that suppresses Cdc13 accumulation at DSBs. Conversely, telomere addition at accidental breaks is promoted by Pph3, the yeast protein phosphatase 4 (PP4). Pph3 is itself modulated by Rrd1, an activator of PP2A family phosphatases. Rrd1 and Pph3 oppose Cdc13 S306 phosphorylation and are necessary for the efficient accumulation of Cdc13 at DNA breaks. These studies therefore identify a mechanism by which the ATR family of kinases enforces genome integrity, and a process that underscores the contribution of Cdc13 to the fate of DNA ends.
Keywords: DNA double-strand breaks, telomerase, ATR, PP4, genome stability, Saccharomyces cerevisiae
A single unrepairable double-strand break (DSB) can lead to cell death (Sandell and Zakian 1993), and the inaccurate repair of DSBs is a major source of gross chromosomal rearrangements (GCRs) (Kolodner et al. 2002). In metazoans, GCRs can result in loss of heterozygosity at tumor suppressor loci or in the activation of proto-oncogenes (Lee and Myung 2009). Cells therefore possess an elaborate network of DNA damage repair and surveillance pathways that collaborate to minimize the deleterious impact of DSBs on chromosome integrity and viability.
DSBs are repaired by two main pathways: homologous recombination (HR) and nonhomologous end-joining (NHEJ) (Wyman and Kanaar 2006). In addition to these two repair processes, DSBs can be “repaired” by telomerase action at the break site, a phenomenon referred to as telomere healing or de novo telomere addition (Kramer and Haber 1993; Pennaneach et al. 2006). Since the essential function of telomeres is to protect chromosome ends from nucleolytic degradation, chromosome fusion, and the inappropriate engagement of checkpoint signaling (Lydall 2003), addition of telomere repeats onto a DSB results in the stabilization of the new chromosome end and allows resumption of cell cycling (Michelson et al. 2005). Therefore, rather than being a DNA repair process per se, telomere healing may be viewed more appropriately as a DSB tolerance pathway used at the cost of losing genetic information distal to the break, since the acentric fragment produced by the DSB is often lost or recombined. In order to protect genetic information, mechanisms are in place to restrict the action of telomerase at DSBs.
Telomerase is the specialized reverse transcriptase that solves the end-replication problem caused by discontinuous DNA replication. The telomerase enzyme is composed minimally of an RNA component that acts as a template and a reverse transcriptase catalytic subunit (for review, see Taggart and Zakian 2003; Smogorzewska and de Lange 2004). In budding yeast, telomerase catalyzes the polymerization of TG1–3 repeats at the 3′ end of chromosomes. Five genes are essential for this function. The first two are EST2, which codes for the reverse transcriptase subunit, and TLC1, which encodes the RNA template that guides telomere repeat polymerization. The next two are EST1 and EST3, which are ancillary subunits of the telomerase holoenzyme. The fifth gene, CDC13/EST4, codes for an OB fold-containing protein that directly binds the TG-rich ssDNA overhang (Nugent et al. 1996). Cdc13, in complex with Stn1 and Ten1, forms a chromosome end-capping complex sometimes referred to as CST (Cdc13–Stn1–Ten1) (Gao et al. 2007). In addition to this role in telomere protection, Cdc13 promotes telomere elongation via its interaction with Est1 (Evans and Lundblad 1999) and the lagging strand DNA replication apparatus (Qi and Zakian 2000). The interaction of Cdc13 with Est1 occurs via its telomerase recruitment domain (RD). Interestingly, the activity of the Cdc13RD is modulated by phosphorylation (Tseng et al. 2006; Li et al. 2009), pointing to the Cdc13RD as an important regulator of telomerase action.
Although telomere healing is a process that endangers genome integrity, only one mechanism that restricts telomerase action on DSBs has been characterized to date in budding yeast (or any other organism). This mechanism is underpinned by Pif1, a 5′–3′ helicase that acts as a potent inhibitor of telomere addition (at telomeres or at DSBs) (Schulz and Zakian 1994; Zhou et al. 2000). PIF1-deficient cells have elongated telomeres and a rate of telomere healing that is dramatically increased compared with wild-type cells, as measured by a GCR assay using a modified Chr V as a reporter (Myung et al. 2001). Pif1 inhibits telomere repeat polymerization by destabilizing the telomerase RNA–DNA hybrid (Boule et al. 2005). Interestingly, recent work has established that DNA damage signaling may direct the activity of Pif1 toward DSBs. Indeed, Pif1 is phosphorylated in response to DNA damage, likely by Dun1 and Rad53, and a phosphorylation-defective mutant of Pif1 (pif1-4A) displays an increased propensity to add telomere repeats on an HO-induced DSB (Makovets and Blackburn 2009). Finally, the activity of Pif1 is not restricted to telomeres (or DSBs), as Pif1 also participates in Okazaki fragment maturation and in the maintenance of the mitochondrial genome (Boule and Zakian 2006).
While telomere healing is observed in many eukaryotes, including humans (Harrington and Greider 1991; Morin 1991; Yu and Blackburn 1991; Kramer and Haber 1993; Sprung et al. 1999), the activity of Pif1 in inhibiting telomerase action may not be evolutionarily conserved. Indeed, deletion of the fission yeast Pif1 homolog Pfh1+ results in shorter, rather than longer, telomeres (Zhou et al. 2002). Moreover, there is debate as to whether the mammalian Pif1 homologs affect telomere length maintenance (Mateyak and Zakian 2006; Zhang et al. 2006; Snow et al. 2007). As these latter studies have not specifically examined telomere healing, it cannot be excluded that a role for mammalian Pif1 in the inhibition of telomere healing will be revealed. Nevertheless, it is likely that mechanisms other than those relying on Pif1 exist to inhibit telomerase action on DSBs.
Less is known about the gene activities that are involved specifically in de novo telomere formation, since most, if not all, genes that have been characterized to function in telomere healing also modulate telomere length homeostasis. However, in budding yeast, the Ku heterodimer (Yku70/Yku80) likely plays a unique role in telomere healing by virtue of the fact that Ku acts as a DNA end sensor during NHEJ and as a positive regulator of telomere elongation (Downs and Jackson 2004). Ku positively regulates telomere elongation largely via a specific interaction between the Yku80 subunit and a stem–loop located in the TLC1 RNA (Stellwagen et al. 2003).
Here, we report the identification of a mechanism by which the yeast ATR ortholog Mec1 restricts telomere repeat addition on accidental DSBs. We found that Mec1 inhibits telomere healing by suppressing the recruitment of Cdc13 to DSB sites. Mec1 does so by phosphorylating Cdc13 on its S306 residue. Conversely, Mec1-dependent phosphorylation of Cdc13 S306 is antagonized, likely at the site of DNA damage, by the action of Pph3, the yeast protein phosphatase 4 (PP4), and Rrd1, the yeast ortholog of human phosphotyrosyl phosphatase activator (PTPA) (Van Hoof et al. 2000). This study therefore uncovers a novel function for Mec1/ATR in promoting genome integrity via the inhibition of telomere healing, a potent source of genome rearrangements.
Results
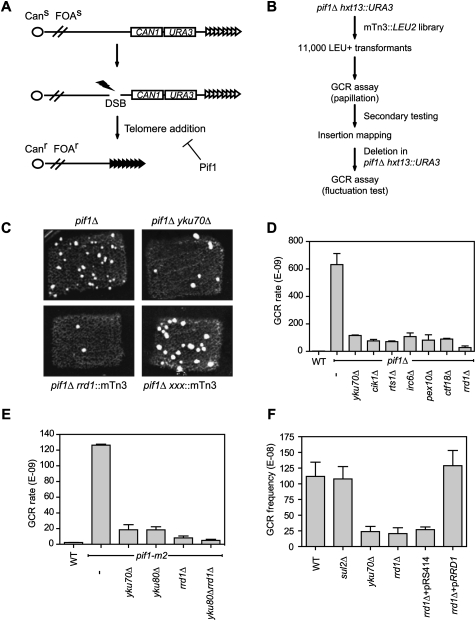
To identify genes that promote the healing of DSBs by telomerase, we employed a GCR assay developed by Kolodner and colleagues (Chen and Kolodner 1999). In this assay, the left arm of Chr V is engineered so that the URA3 gene is inserted at the HXT13 locus, ∼7.5 kb from CAN1 (Fig. 1A). Cells that undergo a GCR event that results in the simultaneous loss of URA3 and CAN1 are recovered by growth on media containing 5-fluoro-orotic acid (5-FOA) and canavanine (FC media).
Figure 1.
A genetic screen identifies Rrd1 as a factor important for de novo telomere formation. (A) Schematic representation of the Chr V GCR assay. The test strain (derivative of hxt13∷URA3) possesses two counterselectable markers (CAN1 and URA3) located near the telomeric end of Chr V–L. GCR events are identified by the simultaneous loss of CAN1 and URA3, which confer resistance to canavanine (Can) and 5-FOA (FOA). PIF1 inhibits telomere healing, and its deletion skews GCR events to the telomere healing pathway. (B) An outline of the transposon-based genetic screen carried out in this study. (C) Identification of telomere healing mutants by the papillation assay. Photographs were taken from the primary screen data. The top two panels show the papillation phenotypes of the control pif1Δ and pif1Δ yku70Δ strains, respectively. The bottom left panel shows a strain, later identified as a transposon disrupting the RRD1 gene. The bottom right panel displays a typical uncharacterized strain that scored negative by displaying a similar number of colonies as the pif1Δ control. (D) Quantitation of spontaneous GCR rates using the Chr V assay. The strains tested were derivatives of hxt13∷URA3 (WT), and, in the same background, pif1Δ, pif1Δ yku70Δ, pif1Δ cik1Δ, pif1Δ rts1Δ, pif1Δ irc6Δ, pif1Δ pex10Δ, pif1Δ ctf18Δ, and pif1Δ rrd1Δ. The data were obtained following a minimum of two independent fluctuation tests with 11 cultures. The data are presented as the mean ± SEM. (E) Quantitation of spontaneous GCR rates using the Chr V assay. The strains tested were derivatives of hxt13∷URA3 (WT), and, in the same background, pif1-m2, pif1-m2 yku70Δ, pif1-m2 yku80Δ, pif1-m2 rrd1Δ, and pif1-m2 yku80Δ rrd1Δ. The data were obtained following a minimum of two independent fluctuation tests with 11 cultures. The data are presented as the mean ± SEM. (F) Quantitation of MMS-induced frequency of GCR events using the Chr V assay. The strains tested were derivatives of hxt13∷URA3 (WT), and, in the same background, sul2Δ, yku70Δ, rrd1Δ, rrd1Δ <pRS414>, and rrd1Δ <pRRD1>. pRRD1 harbors the RRD1 gene under the control of its own promoter, in pRS414. The data are presented as the mean ± SEM (N = 3).
With this assay, we sought to identify mutations that reduce the high GCR rate observed in pif1Δ cells (Myung et al. 2001) using the scheme depicted in Figure 1B. The elevated GCR rate of pif1Δ cells provided two advantages. First, mutations that reduce GCR formation in pif1Δ cells most likely impact telomere healing, as nearly all of the GCR events produced by the pif1Δ mutation are telomere healing events. For example, deletion of genes encoding telomerase subunits or the Ku complex reduces the GCR rate of pif1Δ cells (Myung et al. 2001). Second, the high GCR rate of pif1Δ cells allowed for a facile evaluation of GCR rates by a papillation assay on plates, rather than by fluctuation analysis (see below).
We therefore transformed a pif1Δ hxt13∷URA3 strain with a yeast genomic DNA library containing random insertions of the bacterial minitransposon mTn3-lacZ/LEU2 (Kumar et al. 2002). Approximately 11,000 mTn-LEU2 transformants were screened, corresponding to ∼0.3× genome coverage (Kumar et al. 2002). To qualitatively estimate GCR rates, mTn-LEU2 transformants were patched and grown onto nonselective media containing 0.005% methyl methanesulfonate (MMS). MMS was included to stimulate the formation of GCR events (Myung and Kolodner 2003). The strains were then replica-plated onto FC plates to select for GCR events. On each plate, we included pif1Δ and pif1Δ yku70Δ strains as negative and positive controls, respectively (Fig. 1C; Supplemental Fig. S1A). Strains that displayed an apparent GCR rate lower than pif1Δ yku70Δ were selected for transposon insertion mapping. Using this scheme, we identified 69 transposon insertions in 57 genes (Supplemental Table S1). Notably, a total of seven insertions disrupting the TLC1 and EST2 genes were identified, indicating that our scheme could isolate genes required for telomere healing.
Since the suppression of the pif1Δ GCR rate in our strains might not necessarily be linked to the transposon insertion, we selected 18 candidate genes for gene disruption in the pif1Δ background. The GCR rate of the resulting strains was then determined by fluctuation analysis. From this analysis, we found that six gene deletions reduced the GCR rate of pif1Δ cells to a level lower than that of pif1Δ yku70Δ (Fig. 1D). These deletions affected the following genes: CIK1, which encodes a kinesin-associated protein required for karyogamy and mitotic spindle organization (Manning et al. 1999); RTS1, which codes for a B-type regulatory subunit of PP2A (Shu et al. 1997); IRC6, which codes for a protein of unknown function that displays increased levels of spontaneous Rad52 foci (Alvaro et al. 2007); PEX10, which encodes a peroxisomal membrane E3 ubiquitin ligase required for matrix protein import (Eckert and Johnsson 2003); CTF18, which codes for an Rfc1-like component of an alternative RFC complex required for sister chromatid cohesion (Mayer et al. 2001); and, finally, RRD1, which encodes the ortholog of human PTPA, a conserved PP2A activator (Fellner et al. 2003). Since the deletion of RRD1 was the most potent suppressor of GCR formation in the pif1Δ strain (Fig. 1C,D), we selected RRD1 for further analysis.
The pif1Δ mutation affects both the nuclear and mitochondrial forms of Pif1 (Schulz and Zakian 1994). Since the impaired mitochondrial function of pif1Δ cells might contribute to its high GCR rate, we first ascertained that the rrd1Δ mutation suppresses the GCR rate of pif1-m2, an allele that disrupts the nuclear function of Pif1. As shown in Figure 1E, the rrd1Δ mutation reduces the GCR rate of pif1-m2 cells to a level lower than that of Ku subunit deletion. Moreover, when rrd1Δ is combined with yku80Δ, we found that the double mutation had an additive reduction in the GCR rate of pif1-m2, indicating that RRD1 and YKU80 act in different pathways. Finally, since the GCR assay is based on selection on FC media, we confirmed that Rrd1 is not required for growth in the presence of 5-FOA and canavanine (Supplemental Fig. S1B).
Next, we examined whether the possible role of Rrd1 in promoting telomere healing was dependent on the pif1 mutations. To do so, we evaluated the frequency of GCR formation in PIF1 cells treated with a low dose of MMS to stimulate GCRs (Fig. 1F; Myung and Kolodner 2003). In addition to the rrd1Δ mutation, we included as a positive control the yku70Δ mutation, and, as an additional negative control, the deletion of an irrelevant gene, SUL2, to rule out any effect associated with the KanMX drug resistance cassette. Strikingly, rrd1Δ decreased the GCR frequency to the same extent as yku70Δ, indicating that the role of Rrd1 in GCR formation is independent of Pif1. Moreover, a plasmid harboring RRD1 under the control of its own promoter restored the frequency of GCR formation to wild-type levels, indicating that the reduction of GCR frequency in rrd1Δ cells is due to the loss of the Rrd1 protein (Fig. 1F).
Rrd1 facilitates telomere healing at DSBs with little or no telomere-like sequences
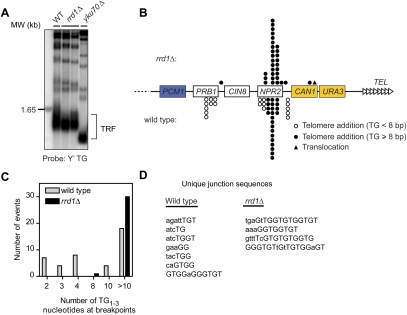
To date, most if not all genes that affect de novo telomere addition have also been found to influence telomere length homeostasis. We thus examined whether the rrd1Δ mutation affected telomere length maintenance by assessing the terminal restriction fragment (TRF) length. In contrast to yku70Δ cells, which have short telomeres, we observed that rrd1Δ cells maintain normal telomere length (Fig. 2A; Supplemental Fig. S2). In parallel, we also tested whether Rrd1 was required for telomere position effect, and found that rrd1Δ cells have normal telomeric silencing (Supplemental Fig. S2). These results imply that Rrd1 might not play a major function at telomeres, and that its role in telomere addition might instead be limited to DSBs.
Figure 2.
Rrd1 facilitates telomere healing at DSBs with no or short TG1–3 tracts. (A) rrd1Δ cells maintain normal telomere length. The TRFs of the indicated strains were detected by Southern blot with a Y′-TG probe targeting telomere sequences. (B) Schematic representation of the Chr V–L. The breakpoints of the GCR events must land between the essential gene PCM1 (blue) and the selection markers CAN1 and URA3 (yellow). Positions of independent telomere additions or translocations are indicated for wild-type (below) and rrd1Δ (above) strains. (C) Distribution of the number of TG1–3 residues found at the breakpoints of the telomere healing events described in B. (D) Breakpoint junction sites from telomere addition events recovered from wild-type or rrd1Δ strains. The events shown correspond to telomere addition events that occurred outside the NPR2 TG-rich hot spot. Nucleotide sequences are shown as 5′-to-3′ direction, and telomere repeat polymerization began following the last nucleotide indicated. Nucleotides conforming to a TG1–3 sequence are capitalized.
Next, we characterized the nature of the GCR events recovered in wild-type and rrd1Δ cells (following MMS treatment to stimulate GCR events). In the Chr V assay, GCR events must lie within a 12-kb region located between CAN1 and PCM1, the last essential gene on Chr V–L (Fig. 2B; Supplemental Fig. S3A). Under our experimental conditions, nearly all damage-induced GCR events mapped were telomere additions, except one translocation observed in rrd1Δ (Fig. 2B,C; Supplemental Fig. S3B). When the breakpoint sequences of the GCR events derived from wild-type and rrd1Δ cells were compared, a striking dichotomy was observed. In wild-type cells, telomere additions occurred at chromosomal sites that contained TG1–3 sequences that varied from 2 nucleotides (nt) to 23 nt in length (Fig. 2C,D). In contrast, in rrd1Δ cells, 30 out of 31 telomere additions occurred at sites that contain at least 11 nt of TG1–3 sequences (Fig. 2C,D). Therefore, Rrd1 appears to be essential for the conversion of DSBs into telomeres when DSBs contain <11nt of the telomere-like TG1–3 sequence. This phenotype is remarkably stronger than a similar phenotype imparted by the disruption of the Yku80–TLC1 RNA interaction (Stellwagen et al. 2003). Indeed, disruption of the Yku80–TLC1 interaction abrogates de novo telomere additions at DSBs that contain TG1–3 sequences of <4 nt (Stellwagen et al. 2003), compared with 11 nt for rrd1Δ cells. Interestingly, we note that the minimal site for Cdc13 binding to ssDNA is an 11-nt TG1–3 sequence (Lin and Zakian 1996; Nugent et al. 1996; Hughes et al. 2000). While it is not clear if Cdc13 can bind to this minimal site in vivo, a compelling possibility raised by these results is that binding of Cdc13 to a new chromosome end bypasses the need for Rrd1 during de novo telomere addition.
Rrd1 promotes telomere healing at HO-induced DSBs
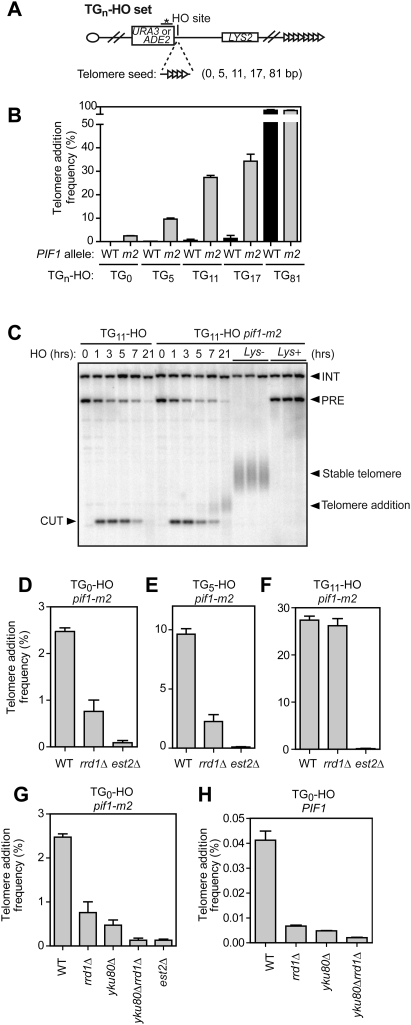
The GCR assay used above relies on the formation of spontaneous or MMS-induced DSBs. To rule out any possible participation of RRD1 in the generation of DSBs, and to have an experimental system where the role of Rrd1 in telomere healing can be studied more mechanistically, we sought to examine the contribution of Rrd1 to the formation of de novo telomeres at a DSB triggered by the HO endonuclease (Diede and Gottschling 1999; Mangahas et al. 2001). We therefore created a set of isogenic strains (the TGn-HO set) where, in addition to an HO site devoid of TG1–3 repeats, we inserted TG1–3 sequences of 5, 11, 17, or 81 base pairs (bp) in length adjacent to the HO site, yielding the TG0-HO, TG5-HO, TG11-HO, TG17-HO, and TG81-HO strains, respectively (Fig. 3A). The 11-nt sequence used in the TG11-HO strain corresponds to a minimal binding site for Cdc13 (Hughes et al. 2000). In addition, the RAD52 gene was deleted to eliminate recombination-mediated telomere elongation. Measurement of telomere addition frequency at the HO site was assessed primarily by the loss of the HO-distal marker LYS2 (counterselected by α-aminoadipate [αAA]). We also examined the kinetics of de novo telomere formation by Southern blotting, but we could only observe detectable levels of telomere repeat polymerization in pif1-m2 strains with at least 11 nt of telomere-like seed sequence adjacent to the HO site (Fig. 3C; data not shown). Interestingly, when we directly compared the genetic assay of telomere addition, which monitors the frequency of stable telomere formation, to the Southern blotting method, we found that the genetic assay was a more stringent readout of de novo telomere formation (see Supplemental Fig. S4A,B for a direct comparison). Last, we found during the course of developing these strains that telomere polymerization in the TG81-HO strain was so robust that Pif1 had only a marginal impact on either the kinetics of telomere addition (Supplemental Fig. S4C) or the frequency of stable telomere formation (Fig. 3B). The cleaved TG81-HO substrate might therefore more faithfully represent a short telomere than a DSB in this assay.
Figure 3.
A TGn-HO set of strains to study telomere healing at HO-induced DSBs. (A) Schematic representation of the modified Chr VII–L to generate the TGn-HO set of strains. The ADH4 locus was replaced by the ADE2 or URA3 gene followed by telomere seed sequences of different sizes (no telomeric seed, 5, 11, 17, and 81 bp of TG1–3 repeats), followed by the HO endonuclease cleavage site. The location of the probe used for Southern blotting (star on a bar) is also shown. (B) Quantitation of telomere addition frequency, as determnined by αAA resistance. The strains tested were PIF1 (WT; black bars) and pif1-m2 (gray bars) derivatives of strains TG0-HO, TG5-HO, TG11-HO, TG17-HO, and TG81-HO. The data are presented as the mean ± SEM (N ≥ 3). (C) The pif1-m2 mutation stimulates telomere addition in the TG11-HO strain. Cultures of wild-type and pif1-m2 derivatives of TG11-HO rad52Δ were arrested with nocodazole. HO expression was induced using galactose, and samples were taken at the indicated time points. In addition, Lys+ and Lys− derivatives of TG11-HO rad52Δ pif1-m2 were collected from the genetic assay based on αAA selection. A Southern blot analysis of EcoRV-cut genomic DNA probed with a section of the URA3 gene is shown. The band labeled PRE represents the EcoRV-digested fragment from Chr VII–L. After cleavage with HO, this fragment is converted into a new band (CUT). The URA3 probe also detects the ura3-52 locus (INT) and serves as a loading control. New telomere elongation forms a smear above the CUT band. (D–F) Quantitation of telomere addition frequency, as determnined by αAA resistance. The strains tested were wild-type (WT), rrd1Δ, and est2Δ derivatives of pif1-m2 TG0-HO (D), pif1-m2 TG5-HO (E), or TG11-HO (F). The data are presented as the mean ± SEM (N ≥ 3). (G) Quantitation of telomere addition frequency of wild-type, rrd1Δ, yku80Δ, rrd1Δ yku80Δ, and est2Δ derivatives of pif1-m2 TG0-HO. The data are presented as the mean ± SEM (N ≥ 3). (H) Quantitation of telomere addition frequency of wild-type, rrd1Δ, yku80Δ, and rrd1Δ yku80Δ derivatives of TG0-HO. The data are presented as the mean ± SEM (N ≥ 3).
As expected, telomere addition frequency in TG0-HO to TG17-HO (PIF1) strains was very low, ranging from 0.04% to 1% (Fig. 3B). However, when we introduced the pif1-m2 mutation, telomere healing frequency was greatly elevated in a telomerase-dependent manner in the TG0-HO to TG17-HO strains (Fig. 3B,D–F). We then deleted RRD1 in the TGn-HO pif1-m2 set of strains and calculated the frequency of telomere addition. Consistent with the GCR breakpoint mapping data, telomere healing events decreased by 70% and 77% in the rrd1Δ TG0-HO and rrd1Δ TG5-HO strains, respectively (Fig. 3D,E). In stark contrast, telomere addition frequency in strains with telomere-like seeds of 11 bp and higher was minimally affected by the rrd1Δ mutation (Fig. 3F; Supplemental Fig. S5A). The reduction in the frequency of telomere healing caused by the rrd1Δ mutation in the TG0-HO strain approached the reduction imparted by yku80Δ mutation (Fig. 3G), which was used as a positive control. Interestingly, if we combined the YKU80 deletion with rrd1Δ in TG0-HO cells, the frequency of telomere addition approached that of est2Δ cells (Fig. 3G). The role of RRD1 in promoting telomere healing at HO-induced breaks was independent of pif1 mutations, since deletion of RRD1 in the TG0-HO (PIF1) strain also greatly impaired telomere healing frequency (Fig. 3H). Together, these data suggest that Rrd1 and Yku80 may represent the two major pathways on which cells rely to recruit telomerase activity to DSBs.
Rrd1 promotes telomere healing via Pph3, the yeast PP4 catalytic subunit
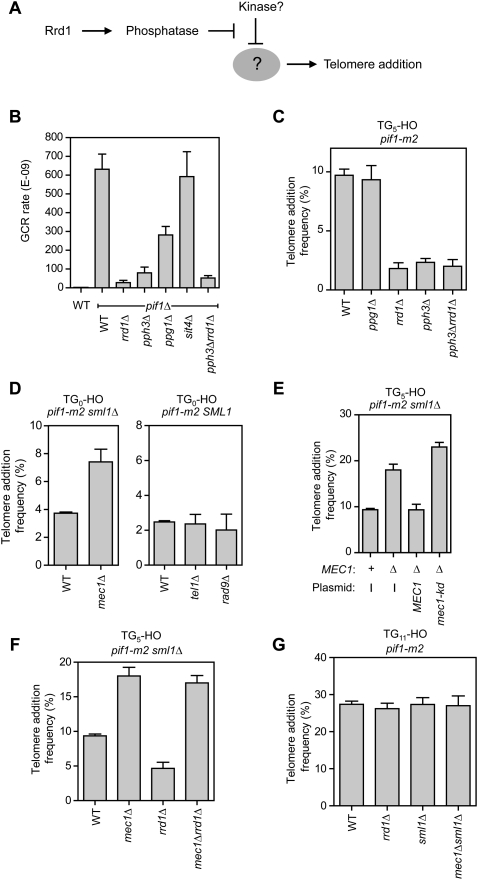
Rrd1 and Rrd2 are the two PTPA homologs in budding yeast. PTPA-like enzymes function in promoting the biogenesis of protein phosphatases of the PP2A superfamily. While Rrd1 and Rrd2 share some functions, Rrd1 was shown to interact primarily with the Pph3, Ppg1, and Sit4 phosphatase catalytic subunits, while Rrd2 appears to be more specific to the canonical yeast PP2A enzymes Pph21 and Pph22 (Van Hoof et al. 2005). The exact mechanism by which Rrd1 (or Rrd2) promotes the activity or biogenesis of PP2A-type phosphatases is not clear and is still a matter of debate. This issue notwithstanding, we sought to identify which phosphatase could be acting downstream from Rrd1 to promote telomere healing of DSBs (Fig. 4A). Since Pph3, Ppg1, and Sit4 interact physically with Rrd1, we therefore deleted their corresponding genes in the pif1Δ hxt13∷URA3 background, and measured the GCR rate of the resulting strains (Fig. 4B). We found that, of the three deletions, the pph3Δ mutation most severely reduced the GCR rate of pif1Δ cells, and did so to a level that approached that of the rrd1Δ mutation. Deletion of PPG1 reduced GCR rates to 45% of the wild-type levels, while deletion of SIT4 did not impact GCR rates. We then introduced the pph3Δ or ppg1Δ mutations into the TG5-HO strain and found that only the pph3Δ mutation reduced telomere healing to a level approaching that of rrd1Δ cells (Fig. 4C). Moreover, the rrd1Δ pph3Δ double mutant had a telomere addition frequency similar to that of the single pph3Δ or rrd1Δ mutants, indicating that Pph3 is the protein phosphatase that acts in conjunction with Rrd1 to promote telomere addition at DSBs.
Figure 4.
Mec1 and Rrd1–Pph3 form a phosphoregulatory circuit that controls telomere healing at DSBs. (A) Model of Rrd1 action, given its role as a PP2A-type phosphatase activator. Rrd1 might activate a phosphatase that antagonizes a phosphorylation event that inhibits telomere healing. (B) Quantitation of spontaneous GCR rates using the Chr V assay. The strains tested were derivatives of hxt13∷URA3 (WT), and, in the same background, pif1Δ, pif1Δ sul2Δ, pif1Δ, pif1Δ rrd1Δ, pif1Δ pph3Δ, pif1Δ ppg1Δ, pif1Δ sit4Δ, and pif1Δ rrd1Δ pph3Δ. The data were obtained following a minimum of two independent fluctuation tests with 11 cultures. The data are presented as the mean ± SEM. (C) Quantitation of telomere addition frequency of wild-type, ppg1Δ, rrd1Δ, pph3Δ, and rrd1Δ pph3Δ derivatives of pif1-m2 TG5-HO. The data are presented as the mean ± SEM (N ≥ 3). (D) Quantitation of telomere addition frequency of wild-type and mec1Δ derivatives of pif1-m2 sml1Δ TG0-HO (left panel), and wild-type, tel1Δ, and rad9Δ derivatives of pif1-m2 TG0-HO (right panel). SML1 was deleted to maintain the viability of mec1Δ. The data are presented as the mean ± SEM (N ≥ 3). (E) Quantitation of telomere addition frequency of wild-type and mec1Δ derivatives of pif1-m2 sml1Δ TG5-HO. The telomere addition frequency of mec1Δ derivatives containing plasmids that expressed either MEC1 or a mec1 kinase-dead allele (mec1-kd) is also shown. The data are presented as the mean ± SEM (N ≥ 3). (F) Quantitation of telomere addition frequency of wild-type, mec1Δ, rrd1Δ, and mec1Δ rrd1Δ derivatives of pif1-m2 sml1Δ TG5-HO. The data are presented as the mean ± SEM (N ≥ 3). (G) Quantitation of telomere addition frequency of wild-type, mec1Δ, rrd1Δ, sml1Δ, and mec1Δ sml1Δ derivatives of pif1-m2 TG11-HO. The data are presented as the mean ± SEM (N ≥ 3).
Mec1 suppresses telomere healing and is antagonized by Rrd1–Pph3
The latter result suggested that Rrd1, via Pph3, promotes the removal of a phosphate moiety that inhibits telomere healing (Fig. 4A). This possibility was tantalizing, since it hinted that telomerase action at DSBs is actively suppressed by phosphorylation. Strong candidates for kinases that might negatively regulate telomere healing were Mec1 and Tel1, the orthologs of ATR and ATM, since they are both activated by DSBs. We therefore deleted both genes in the TG0-HO strain and measured telomere healing frequencies. To circumvent the lethality of the MEC1 deletion, we also deleted SML1 (Zhao et al. 1998). Strikingly, the deletion of MEC1, but not of TEL1, stimulated the frequency of telomere additions (Fig. 4D). The impact of Mec1 on telomere healing is not due to its function in enforcing cell cycle arrest, as the experiments were carried out in nocodazole-arrested cells. In line with this idea, deletion of RAD9, which abolishes the DNA damage checkpoint, did not impact the frequency of telomere healing (Fig. 4D). Finally, we found that the kinase activity of Mec1 was critical to inhibit de novo telomere addition (Fig. 4E). Together, these data identified Mec1 as a strong candidate for the protein kinase that negatively regulates telomere healing, and whose action is antagonized by Rrd1 and Pph3.
A critical prediction to the above model is that the deletion of RRD1 should have no effect on the telomere healing frequency of mec1Δ cells, since the absence of inhibitory phosphorylation would render the dephosphorylation step irrelevant. Indeed, we observed in the TG5-HO background that the telomere frequency of rrd1Δ mec1Δ cells was as high as that of mec1Δ cells, suggesting that Rrd1 and Mec1 regulate the same substrate (Fig. 4F). The same result was observed with TG0-HO (Supplemental Fig. S5B). Interestingly, we also observed that MEC1 deletion did not impact telomere addition frequency on a DSB containing a TG11 seed (TG11-HO) (Fig. 4G). This latter result indicates that 11 nt of telomere-like sequence overcomes the Mec1-dependent inhibition of telomere healing, thereby reinforcing the idea that Rrd1–Pph3 and Mec1 operate within a phosphoregulatory loop.
Tethering Cdc13 to a DSB renders telomere addition insensitive to Mec1 and Rrd1
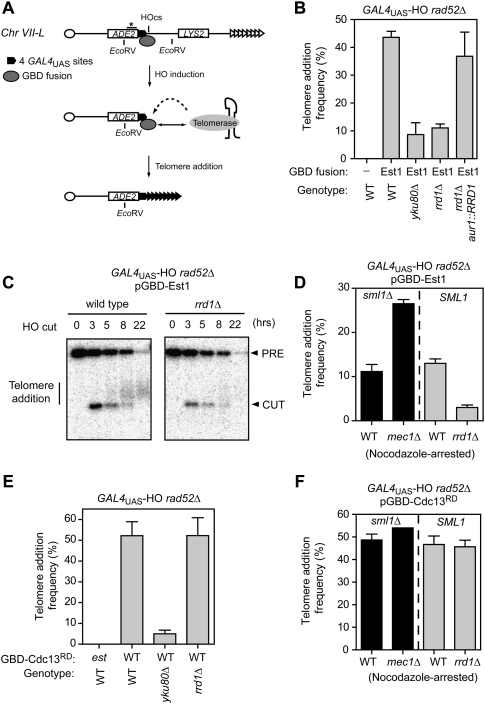
As an additional means to examine the contribution of the emerging Rrd1–Pph3/Mec1 network on de novo telomere formation, we took advantage of a system developed by Bianchi et al. (2004), where four copies of the GAL4UAS are placed adjacent to an HO endonuclease cleavage site, ectopically inserted on Chr VII (Fig. 5A). This system allows for the artificial delivery of Gal4 DNA-binding domain (GBD) fusion proteins to the vicinity of an HO-induced DSB. Telomere healing at this locus can be monitored by detecting the loss of LYS2, located distally to the HO cleavage site, or by Southern blotting (Fig. 5A; Supplemental Fig. S6A). In the absence of a GBD fusion protein, telomere healing is negligible, whereas it is greatly stimulated by expression of either GBD-Est1 or GBD-Cdc13 fusion proteins (Bianchi et al. 2004). This system was ideally suited to test whether the artificial recruitment of Est1 or Cdc13 to a DSB can override the regulatory network underpinned by Mec1 and Rrd1–Pph3.
Figure 5.
Tethering Cdc13 to a DSB overrides the Rrd1/Pph3-Mec1 regulatory network. (A) Schematic representation of the modified Chr VII–L in the GBD fusion assay. The restriction sites (EcoRV) and the probe location (star on a bar) used for Southern blotting are depicted. Telomere healing events stimulated by the GBD fusion can be detected via the loss of LYS2 and the retention of ADE2. (B) Quantitation of telomere addition frequency of wild-type (WT), yku80Δ, rrd1Δ, and rrd1Δ aur1∷RRD1 derivatives of the GAL4UAS rad52Δ strain in the presence of either an empty GBD-expressing plasmid (−) or the GBD-Est1 fusion (Est1). The data are presented as the mean ± SEM (N ≥ 3). (C) Wild-type and rrd1Δ cells were grown in galactose to induce HO expression, and samples were taken at the indicated time points. A Southern blot of EcoRV-cut genomic DNA was probed with a portion of the ADE2 gene. The band labeled PRE represents the EcoRV restriction-digested fragment from Chr VII–L. After HO cleavage, this fragment is converted into a new band (CUT). New telomere elongation appears as a smear above the CUT band. (D) Quantitation of telomere addition frequency of wild-type and mec1Δ derivatives of a GAL4UAS sml1Δ rad52Δ strain (black bars), and wild-type and rrd1Δ derivatives of a GAL4UAS rad52Δ strain (gray bars). All strains expressed the GBD-Est1 fusion. The experiments were carried out in nocodazole-arrested cells. The data are presented as the mean ± SEM (N ≥ 3). (E) Quantitation of telomere addition frequency of wild-type, yku80Δ, and rrd1Δ derivatives of a GAL4UAS rad52Δ strain expressing the GBD-Cdc13RD fusion. As a control, we also determined the telomere addition frequency of a GAL4UAS rad52Δ strain expressing the nonfunctional GBD-Cdc13RD-est fusion. The data are presented as the mean ± SEM (N ≥ 3). (F) Quantitation of telomere addition frequency of wild-type and mec1Δ derivatives of a GAL4UAS sml1Δ rad52Δ strain (black bars), and wild-type and rrd1Δ derivatives of a GAL4UAS rad52Δ strain (gray bars). All strains expressed the GBD-Cdc13RD fusion. The experiments were carried out in nocodazole-arrested cells. The data are presented as the mean ± SEM (N ≥ 3).
With this system, we first examined the impact of deleting RRD1 in a strain expressing the GBD-Est1 fusion protein. As a control, we also examined telomere healing in a strain carrying the yku80Δ mutation. As shown in Figure 5B, the rrd1Δ mutation reduced telomere healing stimulated by GBD-Est1 to a level similar to that of the yku80Δ mutation. This phenotype could be reversed by reintroducing RRD1 at another chromosome locus (Fig. 5B). The importance of Rrd1 for GBD-Est1-stimulated telomere healing could also be observed by Southern blotting, providing a direct demonstration of the importance of Rrd1 in telomere repeat polymerization at a DSB (Fig. 5C). Furthermore, deletion of MEC1 elevated the frequency of telomere addition in the presence of GBD-Est1, in line with the idea that Rrd1 opposes an inhibitory phosphorylation by Mec1 (Fig. 5D). Together, these results indicate that the tethering of Est1 to a DSB is not sufficient to override the requirement for Rrd1 during de novo telomere formation.
Next, we examined whether a GBD-Cdc13RD fusion could promote telomere addition on the cleaved GAL4UAS-HO substrate in a manner that is insensitive to Mec1 and Rrd1. In stark contrast to the situation observed with GBD-Est1, deletion of RRD1 had essentially no impact on GBD-Cdc13RD-stimulated de novo telomere formation (Fig. 5E). An identical result was seen when full-length Cdc13 was fused to the GBD (Supplemental Fig. S6B). Deletion of YKU80, which was used as a control for the assay, greatly reduced the frequency of telomere healing, consistent with the role of Yku80 in the recruitment of TLC1 to DNA ends (Fig. 5E). In addition, we found that deletion of MEC1 had only a marginal impact on the frequency of telomere healing stimulated by GBD-Cdc13RD (Fig. 5F). These results indicate that artificial recruitment of Cdc13 to a DSB bypasses the need for Rrd1/Pph3 during telomere healing, and renders this process insensitive to Mec1 action.
Cdc13 S306 phosphorylation by Mec1 inhibits telomere healing
The above results are consistent with a model in which Cdc13 is the critical target of Rrd1–Pph3 and Mec1. Cdc13 is a phosphoprotein that is subject to Mec1-, Tel1-, and Cdk1-dependent phosphorylation (Tseng et al. 2006; Smolka et al. 2007; Li et al. 2009). Tseng et al. (2006) identified the S306 residue on Cdc13 as a site that is predominantly phosphorylated in vitro by Mec1. Moreover, mutation of S306 to the nonphosphorylatable alanine residue does not impact telomere length homeostasis, even though this residue lies within the Cdc13RD (Tseng et al. 2006). Since we found that inhibition of telomere addition was Mec1-dependent but not Tel1-dependent, and since the Rrd1–Pph3 pathway does not promote telomere addition at native telomeres, the S306 residue was selected as a candidate for a Mec1 phosphorylation site that inhibits telomere healing.
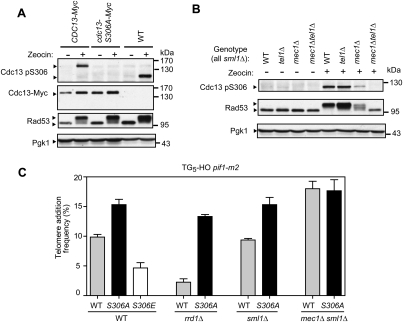
First, we determined whether the Cdc13 S306 site was phosphorylated in vivo in response to DNA damage. We raised a phosphospecific antibody against a phosphorylated Cdc13 S306 (pS306)-derived peptide (Supplemental Fig. S7). We found that induction of DSBs with zeocin, a radio-mimetic compound, resulted in robust Cdc13 S306 phosphorylation (Fig. 6A). We reached this conclusion based on the observation that pS306 immunoreactivity was abolished in a cdc13-S306A mutant, and by the fact that the mobility of the immunoreactive band is much slower in a strain expressing a Cdc13 protein fused to the 13 tandem copies of the Myc epitope tag (Cdc13-Myc). Importantly, deletion of MEC1 strongly reduced, but did not completely abolish, Cdc13 S306 phosphorylation (Fig. 6B), indicating that Mec1 is the main kinase that phosphorylates this S306 in response to DSBs. The residual S306 phosphorylation was due to Tel1, indicating that the yeast ATM homolog can also target Cdc13 S306 when Mec1 is disabled, albeit with a much lower efficiency. We also observed that Cdc13 S306 phosphorylation remained unaffected in strains carrying deletions of either RAD9 or RAD53 (Supplemental Fig. S7D). Together, these observations support the possibility that Mec1 directly phosphorylates the Cdc13 S306 residue. Moreover, we noted that S306 is phosphorylated in response to DSBs even in the context of the GBD-Cdc13RD fusion (Supplemental Fig. S8), indicating that the tethering of Cdc13 to a DSB does indeed override the impact of Mec1-dependent S306 phosphorylation on telomere healing.
Figure 6.
Cdc13 S306 phosphorylation by Mec1 inhibits telomere healing. (A) Western blot analysis of whole-cell extracts obtained from CDC13-Myc, cdc13-S306A-Myc, and wild-type (WT) strains, before and after DNA damage (zeocin) treatment. In the top panel, the blot was probed with the Cdc13 pS306 antibody. In the second panel, the blot was probed with a monoclonal antibody against the Myc epitope. The third panel was probed with a Rad53 antibody to control for the addition of zeocin. (Bottom panel) Pgk1 was used as a loading control. (B) Western blot analysis of whole-cell extracts obtained from sml1Δ, sml1Δ tel1Δ, sml1Δ mec1Δ, and sml1Δ mec1Δ tel1Δ strains, before and after DNA damage (zeocin) treatment. In the top panel, the blot was probed with the Cdc13 pS306 antibody. (Middle panel) Rad53 phosphorylation was used as a control for the addition of zeocin and Mec1 activity. (Bottom panel) Pgk1 was used as a loading control. (C) Quantitation of telomere addition frequency of wild-type (WT), cdc13-S306A, cdc13-S306E, rrd1Δ, cdc13-S306A rrd1Δ, sml1Δ, cdc13-S306A sml1Δ, mec1Δ sml1Δ, and cdc13-S306A mec1Δ sml1Δ derivatives of pif1-m2 TG5-HO. The data are presented as the mean ± SEM (N ≥ 3).
We next tested whether the Cdc13 S306 residue contributes to the inhibition of telomere healing. We introduced the cdc13-S306A mutation in the TG5-HO strain and estimated the frequency of new telomere formation. In a wild-type or sml1Δ background, the cdc13-S306A mutation increased de novo telomere formation, although to a level slightly lower than that of the mec1Δ mutation (Fig. 6C). Since the Cdc13S306A protein can be viewed as a constitutively “dephosphorylated” version of the Cdc13 S306 residue, the cdc13-S306A mutation should lead to an increase in telomere healing, even in the rrd1Δ or pph3Δ background. Indeed, we found that the deletion of RRD1 had only a minimal impact on the telomere healing frequency of the cdc13-S306A strain (Fig. 6C). Consistent with this latter result, we also observed that the cdc13-S306A mutation was entirely epistatic with the MEC1 deletion in terms of telomere healing (Fig. 6C).
To further test whether the phenotype associated with the cdc13-S306A mutation was due to defective phosphorylation, we also monitored the impact of the phosphomimic cdc13-S306E mutation on the frequency of de novo telomere formation in the TG5-HO strain. Consistent with Cdc13 S306 phosphorylation being inhibitory for telomere addition, we found that the cdc13-S306E strain displayed a lower frequency of telomere addition than the wild-type protein (Fig. 6C). Together, these results confirm that Cdc13 S306 phosphorylation by Mec1 inhibits telomere healing. While Cdc13 S306 represents the major site targeted by Mec1, our results suggest that an additional Mec1 site other than Cdc13 S306 plays a minor role in restricting telomere healing.
Cdc13 S306 phosphorylation inhibits its accumulation at DSBs
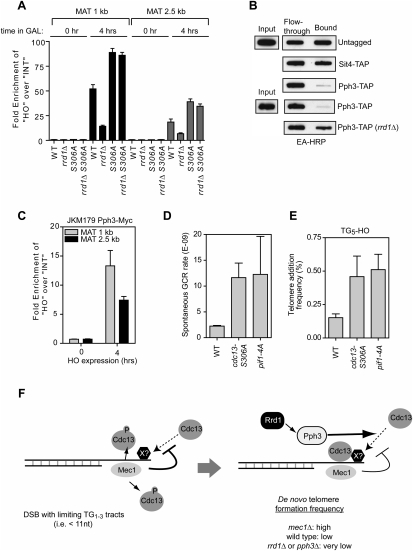
The observation that RRD1 deletion impairs de novo telomere formation on DSBs that contain a <11-nt telomeric seed (Figs. 2C, 4C), coupled to our observation that the tethering of Cdc13 to a DSB renders telomere healing insensitive to Mec1 action, strongly hinted that Cdc13 recruitment to DSBs is a step that is promoted by Rrd1/Pph3 and opposed by Mec1. To directly test this possibility, we monitored Cdc13-Myc accumulation at an irreparable DSB located at the MAT locus in a donorless strain (JKM179) by chromatin immunoprecipitation (ChIP) followed by quantitative real-time PCR. As shown in Figure 7A, 4 h after HO endonuclease induction, we observed a robust accumulation of Cdc13 in the vicinity (1 kb) of the DSB (52-fold over background). Moreover, we found that this accumulation was largely dependent on Rrd1, as Cdc13 recruitment to a DSB in the rrd1Δ strain was reduced by 73% to 14-fold over background. An identical trend was also observed when we examined Cdc13 accumulation 2.5 kb away from the DSB (Fig. 7A). We did not detect any difference between wild-type and rrd1Δ cells in terms of Cdc13 enrichment at the MAT locus prior DSB induction (Fig. 7A), nor did we detect a change in Cdc13 accumulation at telomeres during the course of the experiment (data not shown). Together, these observations indicate that Cdc13 accumulation at a DSB requires Rrd1.
Figure 7.
(A) ChIP of Cdc13-Myc to the MAT-HO locus in wild-type (WT), cdc13-S306A, rrd1Δ, and cdc13-S306A rrd1Δ derivatives of the JKM179 donorless strain. Relative enrichment at the HO locus (MAT 1 kb and 2.5 kb) over the negative control locus PDI1 is shown. The data are presented as the mean ± SEM (N = 3). (B) Cdc13 pS306 phosphatase assays with purified Pph3-TAP and Sit4-TAP. Detailed descriptions of the purification and assay are found in Supplemental Figure S9. After reactions, peptides were immobilized by slot-blotting, and the biotinylated peptides were detected using extravidin (EA) coupled to HRP. The eluate from an untagged strain was used as negative control. Loss of signal in the bound fraction indicates phosphatase activity. (C) ChIP of Pph3-Myc to the MAT-HO locus in the JKM179 donorless strain. Relative enrichment at the HO locus (MAT 1 kb and 2.5 kb) over the negative control locus PDI1 is shown. The data are presented as the mean ± SEM (N = 3). (D) Quantitation of spontaneous GCR rates using the Chr V assay. The strains tested were derivatives of hxt13∷URA3 (WT), and, in the same background, cdc13-S306A and pif1-4A. The data are presented as the mean ± SEM. (E) Quantitation of telomere addition frequency of wild-type (WT), cdc13-S306A, and Pif1-4A derivatives of TG5-HO. The data are presented as the mean ± SEM (N ≥ 3). (F) Model of the regulation of de novo telomere formation by Mec1 and Rrd1–Pph3. (Left panel) At DSBs that contain limited telomere-like seed sequences, ssDNA-bound Mec1 phosphorylates Cdc13 on its S306 residue to suppress Cdc13 accumulation at resected ends, possibly via the inhibition of a protein–protein interaction. (Right panel) However, in situations where a DSB is irreparable, cells can allow telomerase to act on a DSB substrate when all other DNA repair options have been exhausted. In this situation, the accumulation of Pph3 at the DSB site allows it to dephosphorylate Cdc13 locally in order to allow its recruitment to the break, which in turn promotes telomerase action. The new telomere is then masked from the DSB repair and signaling machinery, and promotes recovery from the checkpoint arrest.
Since our model predicts that the Cdc13S306A protein is largely insensitive to an inhibitory phosphorylation by Mec1, we next examined whether Cdc13S306A is recruited to DSBs more efficiently than the wild-type protein. Consistent with our telomere healing results, we found that Cdc13S306A accumulated at the DSB more avidly than the wild-type protein (89-fold over background for Cdc13S306A vs. 52-fold for the wild-type protein) (Fig. 7A). Critically, we finally tested whether the cdc13-S306A allele, which essentially encodes a constitutively dephosphorylated protein, can restore the accumulation of Cdc13 at DSBs in the absence of Rrd1. Indeed, as our model predicted, we found that deletion of RRD1 did not impair the accumulation of Cdc13S306A accumulation at a DSB (Fig. 7A). Together, these results strongly suggest that Mec1-dependent phosphorylation of the Cdc13 S306 residue inhibits its accumulation at DSBs, and that the Rrd1–Pph3 pathway acts to relieve this inhibition.
Pph3 can dephosphorylate Cdc13 pS306
Next, we determined whether Cdc13 pS306 can be a direct target of Pph3. The Pph3 phosphatase complex was purified from Pph3-TAP or Pph3-TAP rrd1Δ strains following IgG sepharose affinity chromatography and TEV site cleavage. As a control, we carried out a mock purification with extracts from an untagged strain. We also purified the Sit4 complex from a Sit4-TAP strain to test phosphatase selectivity toward Cdc13 pS306. Immunoblotting of the phosphatase catalytic subunits after TEV cleavage and elution from the IgG-agarose column showed that similar amounts of the Pph3-TAP and Sit4-TAP catalytic subunits were purified (Supplemental Fig. S9A). We then determined whether the purified phosphatase complexes were active against the generic phosphatase substrate 6,8-difluoro-4-methylumbelliferyl phosphate (DiFMUP). As shown in Supplemental Figure S9B, all three phosphatase complexes displayed equivalent activity toward DiFMUP, with Sit4 being slightly more active than the Pph3 complexes.
To examine pS306 phosphatase activity, we developed an assay based on the retention of phosphopeptides on Ga3+ immobilized metal affinity chromatography (IMAC) (Supplemental Fig. S9C). In this assay, dephosphorylation of the peptide results in loss of signal in the IMAC-bound fraction. The peptides used were biotinylated and detected with extravidin-coupled HRP (Supplemental Fig. S9D). Using this assay, we found that Pph3, but not the related Sit4 phosphatase, efficiently dephosphorylated a Cdc13 pS306-derived peptide (Fig. 7B), even though Sit4 displayed more activity toward DiFMUP (Supplemental Fig. S9B). We next examined whether Rrd1 was important for the activity of Pph3 toward pS306, and carried out phosphatase assays with Pph3 purified from either wild-type or rrd1Δ cells. We observed that, even though both Pph3 preparations were equally active toward DiFMUP (Supplemental Fig. S9B), Pph3 purified from rrd1Δ cells displayed a remarkable reduction in activity toward pS306 (Fig. 7B). These results suggest that Rrd1 positively regulates the activity of Pph3 toward the Cdc13 pS306 site.
Since Rrd1 can associate with Pph3 (Van Hoof et al. 2005), we next examined whether immunopurified Rrd1 was also associated with pS306 phosphatase activity. We immunoprecipitated Rrd1-Flag from wild-type and pph3Δ cells (Supplemental Fig. S9E), and first established that Rrd1-Flag immunoprecipitates had equivalent phosphatase activity against the generic DiFMUP substrate (Supplemental Fig. S9F). We then incubated equivalent amounts of Rrd1-Flag complexes with the pS306 peptide and found that Rrd1 was associated with pS306 phosphatase activity that was almost entirely Pph3-dependent (Supplemental Fig. S9G), further reinforcing the idea that Rrd1 and Pph3 act in the same pathway to dephosphorylate Cdc13 pS306. From these studies, it is clear that Pph3 can act as a pS306 phosphatase, at least in vitro.
Pph3 accumulates at the site of an HO-induced DSB
Although the latter results indicated that Pph3 can act as a selective Cdc13 pS306 phosphatase, we failed to detect a global increase in Cdc13 S306 phosphorylation in the absence of Rrd1 or Pph3 (data not shown). This observation hinted at the possibility that Rrd1–Pph3 might regulate a subpopulation of Cdc13 rather than the global levels of Cdc13 phosphorylation. We therefore explored the possibility that Pph3 might accumulate at DSB sites, where it could act on Cdc13. To do so, we tagged Pph3 with a 13xMyc epitope (Pph3-Myc) and analyzed its accumulation at an irreparable DSB at the MAT locus, as described above. We found that Pph3 accumulates at the chromatin surrounding a DSB (13.2-fold over background), indicating that Pph3 might target Cdc13 locally, in the region surrounding DSBs (Fig. 7C).
Cdc13 and Pif1 phosphorylation likely cooperate to limit telomere addition at DSBs
Finally, recent work by Makovets and Blackburn (2009) found that Pif1 phosphorylation by Rad53 and Dun1 provides an alternative means to limit telomere addition at accidental DSBs. Since most of our work examined the impact of Cdc13 phosphorylation in Pif1-deficient contexts, we determined the relative contribution of Cdc13 and Pif1 phosphorylation to the inhibition of telomere healing and the promotion of genome integrity. We therefore introduced the cdc13-S306A and pif1-4A mutations in the GCR reporter and TG5-HO strains and determined their impact on genome integrity. We found that the cdc13-S306A and pif1-4A mutations increased de novo telomere formation equivalently in either system, strongly suggesting that Cdc13 and Pif1 phosphorylation cooperate to limit the action of telomerase on DSBs (Fig. 7D,E).
Discussion
We conclude that reversible phosphorylation of Cdc13 by Mec1 safeguards nontelomeric DNA ends from the untimely action of telomerase. Mec1 phosphorylates the Cdc13 S306 residue to inhibit the accumulation of Cdc13 at DSBs (Fig. 7F). Counteracting Mec1 is Pph3, the yeast ortholog of PP4, which is itself regulated by Rrd1, the homolog of PTPA. This work therefore identifies a new function for the Mec1/ATR kinase in promoting genome integrity, which is to inhibit the conversion of DSBs into telomeres. Since Mec1 accumulates at resected DNA ends, the coupling of Mec1 activation with the inhibition of telomerase action is an elegant means to ensure that DSB repair is favored over telomere healing. Although Tel1, the ATM homolog, does not inhibit telomere healing, it nevertheless plays a critical role in preventing break-induced chromosome translocations, another class of GCR events (Lee et al. 2008). Mec1 and Tel1 therefore cooperate to enforce genome integrity by inhibiting different types of inappropriate chromosome transactions.
Interestingly, Pif1 phosphorylation in response to DNA damage has emerged recently as another means by which DNA damage signaling restricts telomerase action at DSBs (Makovets and Blackburn 2009). It is likely that Cdc13 and Pif1 phosphorylation cooperate to enforce genome integrity, and each mechanism might have evolved to restrict telomere addition under complementary circumstances. For example, we find that Mec1-dependent phosphorylation of Cdc13 is particularly important to suppress telomere addition at breaks that contain limited telomere-like seed sequences (Fig. 7F). Given the role of Pif1 in inhibiting the RNA–DNA hybrid formed between telomerase and its substrate (Boule et al. 2005), it is possible that Pif1 phosphorylation may be more important to limit telomere addition at DSBs that contain significant telomere seed sequences.
Because it is a threat to genome integrity, the conversion of DSBs to telomeres must be allowed only when all other DNA repair options have been exhausted. For this reason, the action of Mec1 on Cdc13 should dominate over the Rrd1–Pph3 pathway. In support of this possibility, we so far failed to see a convincing increase in the bulk Cdc13 pS306 signal in rrd1Δ and pph3Δ strains after the induction of DSBs. This is despite strong in vitro and genetic evidence that supports Pph3 as a selective Cdc13 pS306 phosphatase. Although other possibilities exist, the most parsimonious interpretation of our results is that Rrd1 and Pph3 affect only a small pool of phosphorylated Cdc13. Indeed, we observed that Pph3 accumulates at DSBs (Fig. 7C), raising the possibility that Pph3 specifically targets Cdc13 at the site of DNA damage. However, these data do not exclude that Pph3 activity is also regulated by DNA damage, and raise the intriguing question of what regulates the steady-state levels of Cdc13 S306 phosphorylation. Clearly, other mechanisms such as dephosphorylation or targeted protein degradation must be at play to remove the bulk of S306-phosphorylated Cdc13 after DSB repair.
How is Cdc13 targeted to DSBs, and how does S306 phosphorylation inhibit this interaction? We envision two possibilities. First, Cdc13, either alone or in the context of the CST complex, might harbor some intrinsic DNA end-binding capacity that allows it to be recruited to resected DNA ends. In this model, S306 phosphorylation would antagonize this DNA end-binding activity. Alternatively, the S306 residue is located within the Cdc13RD, which is an important domain of protein–protein interactions (Pennock et al. 2001; Petreaca et al. 2007). One possibility that we explored is that Cdc13 S306 phosphorylation might antagonize the Cdc13–Est1 interaction. However, deletion of EST1 did not impact Cdc13 recruitment to an HO break (Supplemental Fig. S10), strongly suggesting that the Cdc13–Est1 interaction is not what is affected by S306 phosphorylation. It will be important in the future to determine how Cdc13 is recruited to a DSB, and how S306 phosphorylation impacts this recruitment at the molecular level.
Finally, the identification of Rrd1 and Pph3 as positive regulators of telomere healing provides some clues toward understanding what differentiates a DSB from a short telomere. Indeed, we found that deletion of RRD1 produces a remarkable phenotype where telomere addition is severely impaired specifically at DSBs that contain <11 nt of TG1–3 sequence. Native telomere length homeostasis is unaffected in the rrd1Δ strain. Conversely, Mec1 inhibits telomere healing only at DSBs with <11 TG1–3 nucleotides. Therefore, our results suggest that 11 nt of TG1–3 repeats might be sufficient to give a break a telomeric nature. As this sequence corresponds to the minimal binding site for Cdc13, these results suggest that Cdc13 binding to a DNA end can play a critical role in determining the fate of a resected DNA end.
Materials and methods
Yeast strains and plasmids
The genotypes of all of the yeast strains used in this study are listed in Supplemental Table S2. Strains were constructed using standard genetic methods. For strains used in the telomere healing assays, the RAD52 gene was disrupted with pAB367, a gift from A. Bianchi. The pif1-m2 allele was generated with pVS31 from V. Zakian. All TGn-HO strains are derivatives of RMY241 (Michelson et al. 2005). The pif1-4A allele was introduced using pYT148, a gift from S. Makovets.
GCR assay
The measurement of GCR rates by fluctuation analysis was carried out as described (Chen and Kolodner 1999; Myung et al. 2001; Downey et al. 2006; Kanellis et al. 2007). All GCR rates were determined by fluctuation analysis at least two times using either five or 11 independent cultures. The measurement of damage-induced GCR was carried out as described previously (Myung and Kolodner 2003).
Telomere healing assays
Telomere healing assays were carried out essentially as described in Diede and Gottschling (1999), Bianchi et al. (2004), and Negrini et al. (2007), with some modifications. Strains were inoculated in 10 mL of SC-Leu media containing 2% glucose and grown to log phase. The cell cultures were then diluted to 25 or 50 mL of XY + 2% raffinose and grown overnight to a concentration of 5 × 106 to 8 × 106 cells per milliliter. For experiments where a G2/M arrest was needed, nocodazole (1.5 mg/mL) was added to a final concentration of 15 μg/mL. In the experiments where HO-induced DSBs were generated, cells were taken prior to HO induction to calculate plating efficiency. After 4 h of growth in the galactose medium, cultures were plated onto nonselective media (XY for the TGn-HO assay, and SC-Trp for the GBD fusion assay). After 2 d, cells were replica-plated to αAA plates to select for αAAR (Lys−) isolates. Telomere addition frequency was calculated using the following formula: [number of αAAR colonies/(colony-forming units plated − number of αAAS colonies)].
Southern blotting
The preparation of genomic DNA for Southern blotting was carried out as described previously (Kanellis et al. 2003). Genomic DNA was digested with XhoI (telomere blotting), SpeI (TG81-HO), or EcoRV (TGn-HO and GBD hybrid assays). The digested DNA were then probed with 32P-radiolabeled Y′-TG probe (telomere blotting), ADE2 probe (TG81-HO and GBD assays), URA3 probe (TGn-HO assay), or MCM3 probe (PFGE). All of the primers used to create these probes are available on request.
ChIP
Anti-Myc 9E10 (Santa Cruz Biotechnologies) antibody and Dynabeads pan-mouse IgG (Dynal) were used for the immunoprecipitations. Quantitative real-time PCR at MAT locus of JKM179 strains was performed as in Oza et al. (2009), with modifications. Quantitation of immunoprecipitated DNAs was achieved using an Applied Biosystems 7500 Fast Real-Time PCR system. Enrichment of DNA near the cut site (HO site) over an internal control DNA (INT) located within the PDI1 gene at ∼50 kb from the left arm of Chr III was determined after normalization against values obtained with input samples.
Acknowledgments
We thank R. Szilard, M. Downey, and S. Panier for comments on the manuscript. We thank B. Yoruk, W. Kuo, A. Smith, and D. Trudeau for assistance on the project. We are indebted to A. Bianchi, D. Shore, V. Zakian, V. Lundblad, D. Gottschling, T. Weinert, R. Kolodner, M. Tyers, A.C. Gingras, G. Chen, J. Rouse, C. Peterson, and S. Makovets for reagents and advice. W.Z. is an alumnus of the Excellence in Radiation Research for the 21st Century training program funded by CIHR. D.D. is the Thomas Kierans Chair in Mechanisms of Cancer Development and a Canada Research Chair (Tier 2) in Proteomics, Bioinformatics, and Functional Genomics. This work was supported by Canadian Cancer Society grant 018172 to D.D.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1869110.
Supplemental material is available at http://www.genesdev.org.
References
- Alvaro D, Lisby M, Rothstein R. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet. 2007;3:e228. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi A, Negrini S, Shore D. Delivery of yeast telomerase to a DNA break depends on the recruitment functions of Cdc13 and Est1. Mol Cell. 2004;16:139–146. doi: 10.1016/j.molcel.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Boule JB, Zakian VA. Roles of Pif1-like helicases in the maintenance of genomic stability. Nucleic Acids Res. 2006;34:4147–4153. doi: 10.1093/nar/gkl561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boule JB, Vega LR, Zakian VA. The yeast Pif1p helicase removes telomerase from telomeric DNA. Nature. 2005;438:57–61. doi: 10.1038/nature04091. [DOI] [PubMed] [Google Scholar]
- Chen C, Kolodner RD. Gross chromosomal rearrangements in Saccharomyces cerevisiae replication and recombination defective mutants. Nat Genet. 1999;23:81–85. doi: 10.1038/12687. [DOI] [PubMed] [Google Scholar]
- Diede SJ, Gottschling DE. Telomerase-mediated telomere addition in vivo requires DNA primase and DNA polymerases α and δ. Cell. 1999;99:723–733. doi: 10.1016/s0092-8674(00)81670-0. [DOI] [PubMed] [Google Scholar]
- Downey M, Houlsworth R, Maringele L, Rollie A, Brehme M, Galicia S, Guillard S, Partington M, Zubko MK, Krogan NJ, et al. A genome-wide screen identifies the evolutionarily conserved KEOPS complex as a telomere regulator. Cell. 2006;124:1155–1168. doi: 10.1016/j.cell.2005.12.044. [DOI] [PubMed] [Google Scholar]
- Downs JA, Jackson SP. A means to a DNA end: The many roles of Ku. Nat Rev Mol Cell Biol. 2004;5:367–378. doi: 10.1038/nrm1367. [DOI] [PubMed] [Google Scholar]
- Eckert JH, Johnsson N. Pex10p links the ubiquitin conjugating enzyme Pex4p to the protein import machinery of the peroxisome. J Cell Sci. 2003;116:3623–3634. doi: 10.1242/jcs.00678. [DOI] [PubMed] [Google Scholar]
- Evans SK, Lundblad V. Est1 and Cdc13 as comediators of telomerase access. Science. 1999;286:117–120. doi: 10.1126/science.286.5437.117. [DOI] [PubMed] [Google Scholar]
- Fellner T, Lackner DH, Hombauer H, Piribauer P, Mudrak I, Zaragoza K, Juno C, Ogris E. A novel and essential mechanism determining specificity and activity of protein phosphatase 2A (PP2A) in vivo. Genes & Dev. 2003;17:2138–2150. doi: 10.1101/gad.259903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- Harrington LA, Greider CW. Telomerase primer specificity and chromosome healing. Nature. 1991;353:451–454. doi: 10.1038/353451a0. [DOI] [PubMed] [Google Scholar]
- Hughes TR, Weilbaecher RG, Walterscheid M, Lundblad V. Identification of the single-strand telomeric DNA binding domain of the Saccharomyces cerevisiae Cdc13 protein. Proc Natl Acad Sci. 2000;97:6457–6462. doi: 10.1073/pnas.97.12.6457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellis P, Agyei R, Durocher D. Elg1 forms an alternative PCNA-interacting RFC complex required to maintain genome stability. Curr Biol. 2003;13:1583–1595. doi: 10.1016/s0960-9822(03)00578-5. [DOI] [PubMed] [Google Scholar]
- Kanellis P, Gagliardi M, Banath JP, Szilard RK, Nakada S, Galicia S, Sweeney FD, Cabelof DC, Olive PL, Durocher D. A screen for suppressors of gross chromosomal rearrangements identifies a conserved role for PLP in preventing DNA lesions. PLoS Genet. 2007;3:e134. doi: 10.1371/journal.pgen.0030134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolodner RD, Putnam CD, Myung K. Maintenance of genome stability in Saccharomyces cerevisiae. Science. 2002;297:552–557. doi: 10.1126/science.1075277. [DOI] [PubMed] [Google Scholar]
- Kramer KM, Haber JE. New telomeres in yeast are initiated with a highly selected subset of TG1–3 repeats. Genes & Dev. 1993;7:2345–2356. doi: 10.1101/gad.7.12a.2345. [DOI] [PubMed] [Google Scholar]
- Kumar A, Vidan S, Snyder M. Insertional mutagenesis: Transposon-insertion libraries as mutagens in yeast. Methods Enzymol. 2002;350:219–229. doi: 10.1016/s0076-6879(02)50965-4. [DOI] [PubMed] [Google Scholar]
- Lee SE, Myung K. Faithful after break-up: Suppression of chromosomal translocations. Cell Mol Life Sci. 2009;66:3149–3160. doi: 10.1007/s00018-009-0068-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Zhang Y, Lee SE. Saccharomyces cerevisiae ATM orthologue suppresses break-induced chromosome translocations. Nature. 2008;454:543–546. doi: 10.1038/nature07054. [DOI] [PubMed] [Google Scholar]
- Li S, Makovets S, Matsuguchi T, Blethrow JD, Shokat KM, Blackburn EH. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell-cycle progression. Cell. 2009;136:50–61. doi: 10.1016/j.cell.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JJ, Zakian VA. The Saccharomyces CDC13 protein is a single-strand TG1–3 telomeric DNA-binding protein in vitro that affects telomere behavior in vivo. Proc Natl Acad Sci. 1996;93:13760–13765. doi: 10.1073/pnas.93.24.13760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lydall D. Hiding at the ends of yeast chromosomes: Telomeres, nucleases and checkpoint pathways. J Cell Sci. 2003;116:4057–4065. doi: 10.1242/jcs.00765. [DOI] [PubMed] [Google Scholar]
- Makovets S, Blackburn EH. DNA damage signalling prevents deleterious telomere addition at DNA breaks. Nat Cell Biol. 2009;11:1383–1386. doi: 10.1038/ncb1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangahas JL, Alexander MK, Sandell LL, Zakian VA. Repair of chromosome ends after telomere loss in Saccharomyces. Mol Biol Cell. 2001;12:4078–4089. doi: 10.1091/mbc.12.12.4078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manning BD, Barrett JG, Wallace JA, Granok H, Snyder M. Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J Cell Biol. 1999;144:1219–1233. doi: 10.1083/jcb.144.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mateyak MK, Zakian VA. Human PIF helicase is cell cycle regulated and associates with telomerase. Cell Cycle. 2006;5:2796–2804. doi: 10.4161/cc.5.23.3524. [DOI] [PubMed] [Google Scholar]
- Mayer ML, Gygi SP, Aebersold R, Hieter P. Identification of RFC(Ctf18p, Ctf8p, Dcc1p): An alternative RFC complex required for sister chromatid cohesion in S. cerevisiae. Mol Cell. 2001;7:959–970. doi: 10.1016/s1097-2765(01)00254-4. [DOI] [PubMed] [Google Scholar]
- Michelson RJ, Rosenstein S, Weinert T. A telomeric repeat sequence adjacent to a DNA double-stranded break produces an anticheckpoint. Genes & Dev. 2005;19:2546–2559. doi: 10.1101/gad.1293805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morin GB. Recognition of a chromosome truncation site associated with α-thalassaemia by human telomerase. Nature. 1991;353:454–456. doi: 10.1038/353454a0. [DOI] [PubMed] [Google Scholar]
- Myung K, Kolodner RD. Induction of genome instability by DNA damage in Saccharomyces cerevisiae. DNA Repair (Amst) 2003;2:243–258. doi: 10.1016/s1568-7864(02)00216-1. [DOI] [PubMed] [Google Scholar]
- Myung K, Chen C, Kolodner RD. Multiple pathways cooperate in the suppression of genome instability in Saccharomyces cerevisiae. Nature. 2001;411:1073–1076. doi: 10.1038/35082608. [DOI] [PubMed] [Google Scholar]
- Negrini S, Ribaud V, Bianchi A, Shore D. DNA breaks are masked by multiple Rap1 binding in yeast: Implications for telomere capping and telomerase regulation. Genes & Dev. 2007;21:292–302. doi: 10.1101/gad.400907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nugent CI, Hughes TR, Lue NF, Lundblad V. Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science. 1996;274:249–252. doi: 10.1126/science.274.5285.249. [DOI] [PubMed] [Google Scholar]
- Oza P, Jaspersen SL, Miele A, Dekker J, Peterson CL. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes & Dev. 2009;23:912–927. doi: 10.1101/gad.1782209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pennaneach V, Putnam CD, Kolodner RD. Chromosome healing by de novo telomere addition in Saccharomyces cerevisiae. Mol Microbiol. 2006;59:1357–1368. doi: 10.1111/j.1365-2958.2006.05026.x. [DOI] [PubMed] [Google Scholar]
- Pennock E, Buckley K, Lundblad V. Cdc13 delivers separate complexes to the telomere for end protection and replication. Cell. 2001;104:387–396. doi: 10.1016/s0092-8674(01)00226-4. [DOI] [PubMed] [Google Scholar]
- Petreaca RC, Chiu HC, Nugent CI. The role of Stn1p in Saccharomyces cerevisiae telomere capping can be separated from its interaction with Cdc13p. Genetics. 2007;177:1459–1474. doi: 10.1534/genetics.107.078840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H, Zakian VA. The Saccharomyces telomere-binding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase α and the telomerase-associated est1 protein. Genes & Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA. Loss of a yeast telomere: Arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Schulz VP, Zakian VA. The Saccharomyces PIF1 DNA helicase inhibits telomere elongation and de novo telomere formation. Cell. 1994;76:145–155. doi: 10.1016/0092-8674(94)90179-1. [DOI] [PubMed] [Google Scholar]
- Shu Y, Yang H, Hallberg E, Hallberg R. Molecular genetic analysis of Rts1p, a B′ regulatory subunit of Saccharomyces cerevisiae protein phosphatase 2A. Mol Cell Biol. 1997;17:3242–3253. doi: 10.1128/mcb.17.6.3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A, de Lange T. Regulation of telomerase by telomeric proteins. Annu Rev Biochem. 2004;73:177–208. doi: 10.1146/annurev.biochem.73.071403.160049. [DOI] [PubMed] [Google Scholar]
- Smolka MB, Albuquerque CP, Chen SH, Zhou H. Proteome-wide identification of in vivo targets of DNA damage checkpoint kinases. Proc Natl Acad Sci. 2007;104:10364–10369. doi: 10.1073/pnas.0701622104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow BE, Mateyak M, Paderova J, Wakeham A, Iorio C, Zakian V, Squire J, Harrington L. Murine Pif1 interacts with telomerase and is dispensable for telomere function in vivo. Mol Cell Biol. 2007;27:1017–1026. doi: 10.1128/MCB.01866-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprung CN, Reynolds GE, Jasin M, Murnane JP. Chromosome healing in mouse embryonic stem cells. Proc Natl Acad Sci. 1999;96:6781–6786. doi: 10.1073/pnas.96.12.6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stellwagen AE, Haimberger ZW, Veatch JR, Gottschling DE. Ku interacts with telomerase RNA to promote telomere addition at native and broken chromosome ends. Genes & Dev. 2003;17:2384–2395. doi: 10.1101/gad.1125903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taggart AK, Zakian VA. Telomerase: What are the Est proteins doing? Curr Opin Cell Biol. 2003;15:275–280. doi: 10.1016/s0955-0674(03)00040-1. [DOI] [PubMed] [Google Scholar]
- Tseng SF, Lin JJ, Teng SC. The telomerase-recruitment domain of the telomere binding protein Cdc13 is regulated by Mec1p/Tel1p-dependent phosphorylation. Nucleic Acids Res. 2006;34:6327–6336. doi: 10.1093/nar/gkl786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Hoof C, Janssens V, De Baere I, de Winde JH, Winderickx J, Dumortier F, Thevelein JM, Merlevede W, Goris J. The Saccharomyces cerevisiae homologue YPA1 of the mammalian phosphotyrosyl phosphatase activator of protein phosphatase 2A controls progression through the G1 phase of the yeast cell cycle. J Mol Biol. 2000;302:103–120. doi: 10.1006/jmbi.2000.4062. [DOI] [PubMed] [Google Scholar]
- Van Hoof C, Martens E, Longin S, Jordens J, Stevens I, Janssens V, Goris J. Specific interactions of PP2A and PP2A-like phosphatases with the yeast PTPA homologues, Ypa1 and Ypa2. Biochem J. 2005;386:93–102. doi: 10.1042/BJ20040887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wyman C, Kanaar R. DNA double-strand break repair: All's well that ends well. Annu Rev Genet. 2006;40:363–383. doi: 10.1146/annurev.genet.40.110405.090451. [DOI] [PubMed] [Google Scholar]
- Yu GL, Blackburn EH. Developmentally programmed healing of chromosomes by telomerase in Tetrahymena. Cell. 1991;67:823–832. doi: 10.1016/0092-8674(91)90077-c. [DOI] [PubMed] [Google Scholar]
- Zhang DH, Zhou B, Huang Y, Xu LX, Zhou JQ. The human Pif1 helicase, a potential Escherichia coli RecD homologue, inhibits telomerase activity. Nucleic Acids Res. 2006;34:1393–1404. doi: 10.1093/nar/gkl029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Muller EG, Rothstein R. A suppressor of two essential checkpoint genes identifies a novel protein that negatively affects dNTP pools. Mol Cell. 1998;2:329–340. doi: 10.1016/s1097-2765(00)80277-4. [DOI] [PubMed] [Google Scholar]
- Zhou J, Monson EK, Teng S, Schulz VP, Zakian VA. Pif1p helicase, a catalytic inhibitor of telomerase in yeast. Science. 2000;289:771–774. doi: 10.1126/science.289.5480.771. [DOI] [PubMed] [Google Scholar]
- Zhou JQ, Qi H, Schulz VP, Mateyak MK, Monson EK, Zakian VA. Schizosaccharomyces pombe pfh1+ encodes an essential 5′ to 3′ DNA helicase that is a member of the PIF1 subfamily of DNA helicases. Mol Biol Cell. 2002;13:2180–2191. doi: 10.1091/mbc.02-02-0021. [DOI] [PMC free article] [PubMed] [Google Scholar]