Abstract
The mammalian brain is plastic in the sense that it shows a remarkable capacity for change throughout life. The contribution of neuronal activity to brain plasticity was first recognized in relation to critical periods of development, when manipulating the sensory environment was found to profoundly affect neuronal morphology and receptive field properties. Since then, a growing body of evidence has established that brain plasticity extends beyond development and is an inherent feature of adult brain function, spanning multiple domains, from learning and memory to adaptability of primary sensory maps. Here we discuss evolution of the current view that plasticity of the adult brain derives from dynamic tuning of transcriptional control mechanisms at the neuronal level, in response to external and internal stimuli. We then review the identification of “plasticity genes” regulated by changes in the levels of electrical activity, and how elucidating their cellular functions has revealed the intimate role transcriptional regulation plays in fundamental aspects of synaptic transmission and circuit plasticity that occur in the brain on an every day basis.
I. INTRODUCTION
Groundbreaking work in the 1960s and 1970s demonstrated that brain plasticity is shaped by sensory input during critical periods of development. Experimental manipulation of the sensory environment was shown to profoundly affect neuronal morphology and receptive field properties (120, 160, 274). Later studies demonstrated that brain plasticity is not limited to development but persists in adulthood and seems to be an inherent feature of everyday brain function, critical for learning and memory, and the adaptability of primary sensory maps (reviewed in Refs. 31, 114, 116, 180, 278).
The understanding of neuronal adaptability in response to the external environment is largely owed to the discovery in other cell systems of signal transduction pathways that couple extracellular cues to changes in transcriptional activity. The study of tumor viruses and their mechanism of action opened the door to our understanding of the cellular pathways that control cell growth and proliferation, and in the mature nervous system mediate neuronal responses to electrical activity.
The first molecules shown to translate extracellular signals to transcriptional programs that alter cell properties were transcription factors activated within minutes of stimulation by growth factors and encoded by the cellular counterparts of tumor virus oncogenes. Early studies of the Rous sarcoma virus (RSV) revealed that its transforming function resided in a single gene called src (25, 125, 210). In work for which they received the Nobel prize, Bishop and Varmus (210) showed that the viral src gene, v-src, was derived from a normal cellular protooncogene c-src. Clues to the machinery of intercellular signaling via growth factors and mitogens came when it was discovered that in normal cells protooncogenes function in signaling roles that regulate cell growth and proliferation (189, 256, 272). Cellular oncogenes were found to encode proteins that represent all major components of the growth factor response pathway: from the growth factor sis (42, 62, 133, 134) and growth factor receptor erbB (64, 218), to the small G protein ras (59, 189, 256) or nonre-ceptor tyrosine kinase src (25, 125, 210), and finally nuclear proteins, such as myc (3, 71, 105, 146), fos (55), or jun (26, 88, 202).
It was known that application of growth factors like platelet-derived growth factor (PDGF) leads to rapid activation of gene expression despite the presence of protein synthesis inhibitors such as cycloheximide. These rapid response genes were termed immediate-early genes (IEGs) (46). Nuclear run-off transcription assays after growth factor treatment revealed that the protoonco-genes c-fos and c-myc were among the IEGs (96, 190). Several facts implicated c-fos in regulation of gene expression. It was known to encode a nuclear protein (55) associated with chromatin and capable of binding DNA cellulose in vitro (226, 233). It turned out that c-fos and c-jun are specific members of inducible gene families whose products associate combinatorially to form dimeric complexes that function as transcriptional activators (54). These transcription factors, in turn, help induce expression of second wave genes, termed delayed early genes.
Studies of the protooncogene c-fos also pioneered the identification of cis- and trans-acting regulatory elements that play a role in controlling IEG induction (269). The first operationally defined regulatory cis-element, the serum response element (SRE), was initially mapped in the promoter region of the c-fos gene (91, 219, 260). The c-fos promoter was also found to contain a cAMP responsive element (CRE) (243), common to cAMP-regulated genes (185). The search for trans-acting protein factors that bind to CRE identified a bZip transcription factor termed CREB (CRE-binding protein) (111). The CREB protein family was found to comprise more than 10 members (102) able to heterodimerize in multiple combinations and associate with Fos and Jun as heterodimers, further fine-tuning the binding specificity of the complex (101). The CREB gene gives rise to three main activating isoforms: α (94), β (22), and Δ (111, 287). In addition, alternative splicing appears to be a common feature of CREB family genes and is used as a mechanism to generate both transcriptional activators and repressors. The gene encoding the CREB homolog CREM (cAMP-responsive element modulator) also shows IEG induction kinetics, is subject to cAMP-dependent activation (183), and is differentially spliced to encode both activators and repressors of cAMP-dependent transcription (reviewed in Ref. 152). The CREM products can heterodimerize with each other, as well as with CREB, adding further complexity to the regulation of cAMP-induced transcription (154).
In 1989, Gonzalez and Montminy (94) demonstrated that phosphorylation is a key mechanism in CREB regulation. They showed that elevated cAMP levels led to phosphorylation of CREB at a specific residue, serine-133. Phosphorylation of CREB at this particular residue is a prerequisite for its ability to trigger gene transcription in trans, as was first assessed for the somatostatin gene (94). The phosphorylated CREB protein associates with CBP (CREB-binding protein) (45) and mediates interaction with TFIIB, a major component of the transcription machinery of the RNA polymerase II complex (150). Termination of CREB signaling can be achieved by dephosphorylation through protein phosphatase 1 (PP-1), or PP-2a (201).
In parallel to in-depth studies of protooncogene IEGs, such as c-fos, other studies greatly expanded the pool of known inducible genes by administering different stimuli, such as mitogens and phorbol esters (4, 155, 161, 253), leading to the more general view that gene transcription in the nucleus is part of the cellular response program to alterations in signaling from outside the cell. An integral aspect of this cellular response program is the biphasic nature of transcriptional activation, with IEG induction closely followed by expression of a second wave of delayed early genes. Another important feature is that multiple signals and intracellular pathways can influence IEG transcriptional activation via an assortment of cis- and trans-acting elements, and nuclear IEGs themselves comprise a diverse array of factors that can potentially act combinatorially to differentially affect distinct second wave gene sets.
II. TRANSCRIPTIONAL ACTIVATION OF IMMEDIATE EARLY GENES IN THE NERVOUS SYSTEM
Generalization of these concepts to neurons started before they were fully understood and articulated, with demonstration that treatment of pheochromocytoma (PC12) cells with nerve growth factor (NGF) stimulates expression of c-fos (56, 95, 149). Surprisingly, in this case c-fos induction was associated with events related to cell differentiation rather than cell proliferation. This suggested that the role of IEGs and their activation of a late response transcriptional program could differ according to the differentiation and physiological state of the stimulated cell and was not necessarily restricted to proliferative cells. In addition, c-fos expression in PC12 cells could be induced by agents other than growth factors and mitogens, including depolarizing stimuli and voltage-gated calcium influx (187). These studies provided motivation for examining expression of c-fos and other IEGs in the brain, and their in vivo induction in response to various neural stimuli, most notably seizure paradigms.
Immunostaining revealed Fos-positive neurons throughout the mouse and rat brain (65, 186). Basal levels of c-fos expression could be robustly increased by administration of convulsants such as Metrazol (66, 186, 250) or picrotoxin (231), or by electrical kindling (67). The IEGs c-jun, zif/268, and junB were also found to be rapidly and coordinately activated in seizure paradigms (231). This led to the hypothesis that IEGs may play a part in normal neuronal function and that their increased levels were part of a programmed genomic response to intense stimulation analogous to the genomic response of nonneuronal cells to growth factors (231). However, while one could speculate that convulsant-induced transcription was merely an amplification of normal neuronal responses, it had yet to be shown that similar changes in gene expression could be detected after more natural, physiological levels of stimulation.
This critical point was subsequently addressed in the late 1980s. Several studies demonstrated that changes in IEG transcription factor mRNA levels could occur in response to physiological stimuli. For example, water deprivation resulted in Fos protein expression in hypothalamic neurons known to be activated by dehydration (232). Also, activation of small-diameter cutaneous sensory afferents by noxious heat or chemical stimulation rapidly induced c-Fos immunoreactivity (123) as well as zif/268 gene and protein expression (276) in the superficial layers of the spinal cord dorsal horn. Activation of low-threshold cutaneous afferents resulted in Fos-labeled cells with a different laminar distribution. Neither type of stimuli elicited Fos expression in the dorsal root ganglia, nucleus gracilis, or ventral horn (123). Similarly, baseline zif/268 mRNA levels in the visual cortex of rats was significantly increased by exposure to light after dark-adaptation and decreased after retinal action potential blockade by monocular intravitreal injection of TTX, whereas c-jun mRNA levels were unchanged by these treatments (277). Interestingly, zif/268 mRNA baseline levels were NMDA receptor dependent and dropped significantly throughout cortex and hippocampus upon treatment with the NMDA receptor antagonist MK-801, whereas c-fos mRNA levels were unaffected by NMDA receptor blockade.
These findings provided strong support for the idea that endogenous IEG expression in the brain is a result of normal synaptic activity and could therefore be influenced by neuronal stimulation or activity blockade in a pathway and stimulus specific manner. A side product of these developments was the realization that if IEG activation was part of a neuronal response program to natural stimuli, their expression could be used as a metabolic marker for mapping functional pathways at a cellular level (186, 232). There are excellent examples of successful implementation of this strategy (16, 83, 98, 241, 294), although there are concerns with strictly equating IEG expression with neuronal activity (reviewed in Refs. 100, 141).
III. TRANSCRIPTIONAL ACTIVATION AS THE BASIS FOR LONG-TERM SYNAPTIC PLASTICITY
The realization that synaptic activity induces gene transcription in mature neurons dawned as it was becoming clear that synaptic activity could also drive long-term changes in neuronal structure and function. This led to the proposal that electrical and chemical stimuli that produce long-term change in neurons, such as during learning and memory, act through a mechanism analogous to that of growth factors, namely, through second messenger pathways leading to transcriptional activation (reviewed in Refs. 21, 93).
It had been known for two decades that protein synthesis inhibitors could affect memory retention in mammals, first demonstrated in mice by intracerebral injection of the protein synthesis inhibitor puromycin (82). However, early studies monitoring behavioral performance could not separate the cellular and subcellular effects of the applied drugs from potential effects on aspects of behavior such as attention and motivation. Studies in Aplysia were the first to demonstrate that transcriptional activation and subsequent protein translation were critical for learning-related synaptic plasticity (184). In the intact animal, gently touching the siphon leads to its retraction, as well as gill retraction. This defense response could be sensitized by repeated stimulation such that sensitization could last for minutes (short-term sensitization, STS) to hours or days (long-term sensitization, LTS) (33, 39). The circuitry that controls this behavior in vivo involves sensory neurons from the siphon that connect to motor neurons in the gill. The sensory-motor-neuron synapses are modulated by facilitating interneurons driven by sensory neurons from the tail (86). Unlike STS, LTS is seen only after repeated stimulation and is mediated by serotonin (137). In this system serotonin acts as a neuromodulator and activates the cAMP-PKA pathway. Cocultured Aplysia sensory and motor neurons recapitulate the STS and LTS seen in vivo, with the long-term facilitation (LTF) of synaptic connections paralleling behavioral LTS in its requirement for multiple spaced applications of serotonin (184, 224). In an elegant study, Montarolo et al. (184) demonstrated that application of protein or RNA synthesis inhibitors to the Aplysia coculture system blocks LTF of sensorimotor connections, but has no effect on short-term facilitation (184). Application of the inhibitors during or immediately after stimulation with serotonin suppressed LTF, whereas administration before or after the stimulation was ineffective. This is in line with behavioral studies in mammals, where long-term memory is suppressed after administration of RNA or protein synthesis inhibitors during or immediately after learning (82, 182, 223; reviewed in Refs. 58, 108, 179). The Aplysia studies clearly demonstrated that long-term plasticity and its dependence on RNA and protein synthesis derives from cellular properties that are independent of the complex architecture or intact circuitry of the living animal.
Hints that transcriptional activation mediated by CREB is essential for synaptic plasticity also came from the Aplysia culture system, since bath application of cAMP could be used in place of serotonin to induce LTF (234). Administration of oligonucleotides containing CRE sequences resulted in selective loss of LTF, but not STF in this preparation (57), and repeated serotonin application was shown to induce CREB phosphorylation and activation of CREB-dependent gene transcription (136). Meanwhile, studies in PC12 cells revealed that the CRE element in the c-fos promoter was one and the same as the calcium-responsive element (CaRe), and CREB becomes phosphorylated at serine-133 in response to both membrane depolarization and elevation of intracellular Ca2+ levels, subsequently leading to transcriptional activation of the c-fos gene (243). This study also demonstrated that cAMP and Ca2+ signaling had synergistic effects on CREB-dependent c-fos transcription in vitro, suggesting that these two signaling pathways converge on the CREB protein to regulate activity-dependent transcription in neurons.
In vertebrates, studies of long-term synaptic plasticity at the cellular and synaptic level mainly focused on the hippocampus, a region of the brain known for its importance in learning and memory. In 1973 Bliss and Lømo (24) reported the phenomenon of long-term potentiation (LTP), whereby high-frequency stimulation of the perforant path produces enhanced transmission in the dentate gyrus lasting hours to weeks. Similar to behavioral memory, LTP in hippocampal slices has distinct temporal phases, an early phase lasting 1–3 h after tetanic stimulation, and a persistent late phase lasting at least 8 h (late LTP, L-LTP) (227). Like hippocampal-dependent learning, L-LTP in vitro and in vivo requires repetitive stimulation and is dependent on coincidence detection by the NMDA receptor (49, 188). These attributes and the fact that it can be induced in brain slices in vitro have contributed to the emergence of LTP as the premier synaptic model of memory in the hippocampus, and more generally as a model for activity-dependent synaptic plasticity in the vertebrate brain (23).
Two groups that first monitored the expression of IEGs in response to LTP (48, 276) found that high-frequency stimulation of the perforant path in rat hippocampus at an intensity and frequency sufficient for LTP induction also markedly increased zif/268 mRNA in the dentate gyrus. Other IEG mRNAs, such as c-fos, c-jun, and junB, were less consistently increased under the same experimental conditions, in line with earlier findings (63). While Cole et al. (48) found that stimulation of convergent inhibitory synaptic inputs known to block LTP also blocked IEG induction, Wisden et al. (276) using a similar protocol but slightly different placement of the stimulating electrode, saw a bilateral increase in mRNA levels of zif/268, c-fos, and c-jun alike. This finding indicated that upregulation of IEG mRNA is not always indicative of LTP (276). However, induction of LTP was always accompanied by concomitant zif/268 mRNA expression. The fact that zif/268 mRNA upregulation and LTP induction by high-frequency stimulation both require similar stimulus conditions, including activation of the NMDA receptor (48), argued that they were regulated by the same synaptic mechanisms. Consistent with this, induction of L-LTP in hippocampal slices produced either by tetanic stimulation or application of a membrane-permeable cAMP analog could be blocked by inhibitors of protein synthesis or transcriptional blockers (85, 118, 200). The similarity to the Aplysia system was further established with the demonstration that stimuli that induced L-LTP in CA1 of the hippocampus could also induce CRE-mediated gene expression (126) and CREB phosphorylation (60). L-LTP-induced gene expression could be blocked by PKA inhibitors or L-type Ca2+ channel blockers (117), again implicating CREB as a convergence point for both cAMP and Ca2+ signaling.
The first conclusive test of whether CREB was required for memory was performed in Drosophila, where genetic screens for learning and memory mutants identified several mutants with defects in genes encoding key enzymes that regulate intracellular cAMP levels (reviewed in Ref. 266). Inducible overexpression of a dominant negative truncated CREB protein containing the CRE-binding domain but lacking transactivation activity interfered with the formation of long-term memory if induced prior to training, without affecting short-term memory (290). Attempts to show the requirement for CREB in long-term memory using a knockdown approach in mice was complicated by the existence of multiple CREB isoforms (reviewed in Ref. 246). Initial results from analysis of CREBα Δ– knockout mice were consistent with the Aplysia and Drosophila studies in that short-term memory was unimpaired whereas long-term memory was severely compromised (28), despite upregulation of the CREB β-isoform in these mutants. In contrast, Gass et al. (90) found that CREBα Δ– knockout mice with a different genetic background did not show deficits in social transmission of food preference, nor in dentate gyrus or CA1 LTP in slice preparations. A strain in which all CREB isoforms were reduced showed clear behavioral deficits in the Morris water maze (90), although concerns remain that the mutant mice may be impaired in behavioral flexibility, rather than spatial memory. Another study used conditional CREB knockouts and found both hippocampal LTP and long-term depression (LTD) to be normal (12). Also, learning and memory in the water maze task were not grossly affected. Surprisingly, the mutant mice showed impairment in a conditioned taste aversion learning task, thought to be hippocampus independent. These studies cast a controversial light on the findings of CREB deletion work in mammals and call to mind that gene dosage, genetic background, and compensatory mechanisms (22, 122) may confound the interpretation of such findings.
Mouse knockout studies of other IEG transcription factors linked to activity-dependent gene transcription and synaptic plasticity further validated the view that the cellular underpinnings of activity-dependent synaptic plasticity and learning and memory were one and the same. Zif/268 mutant mice (259) that lack the Zif/268 protein were found to be defective in L-LTP, indicating that zif/268 mRNA expression is not just correlated with LTP, but is in fact required for its expression (135). Long-term memory was also impaired in zif/268 mutant mice when assessed by a variety of behavioral tests, including object recognition and spatial memory tasks. Genetic evidence for a role of c-Fos in memory formation came from c-fos knockout mice that displayed impaired spatial memory in a water maze task (213), although it could not be ruled out that this was partially due to developmental abnormalities in the c-fos mice (132).
Thus, over the course of 15 years, three model systems, Aplysia, Drosophila, and mouse, have contributed to our understanding of how behavioral plasticity, such as is manifested during learning and memory, derives from activity-dependent plasticity at the synaptic level and is mediated by transcriptional activation of IEG transcription factors. The transcription factor CREB was key to establishing this sequence and emerged as a potential convergence point for two major signaling pathways communicating extracellular signals to the neuronal nucleus: depolarization-induced Ca2+ signaling by ionotrophic glutamate receptors, and cAMP linked catecholamine or growth factor signaling. The importance of these pathways for activity-dependent neuronal function and malfunction cannot be overstated and has recently been comprehensively reviewed by Greenberg and colleagues (47, 80).
IV. SEARCH FOR ACTIVITY-REGULATED GENES
The above studies lent credence to the idea that in postmitotic neurons IEG activation is a normal response to synaptic activity and the first step towards long-term activity-dependent plasticity. However, they all focused on IEGs, such as c-fos, c-jun, or zif/268, identified in screens for rapid response to growth factors. It was clear that to fully understand the activity-dependent genetic programs utilized by neurons in the mammalian brain, one had to screen for genes that were induced by neuronal activity, rather than by growth factor stimulation.
In the early 1990s, several groups set out to identify genes that are regulated by changes in activity and could therefore be relevant to activity-dependent neuronal plasticity (196, 222, 283). While the differential screening and subtractive hybridization methods that constituted the state of the art at the time may seem outdated, these screens laid the conceptual groundwork for later screen design using more up-to-date methods. In addition, their combined output provided a significant fraction of the activity-regulated genes whose cellular function has been characterized in the context of synaptic plasticity, therefore affording us a first glimpse of the cellular machinery activated by IEGs in response to neuronal activity.
In screening for activity-regulated genes, as in any differential screen, a critical aspect of screen design is selection of the tissues for comparison. The greater the number of differences between two tissue sources, the more difficult it is to select from the cloned genes those that are relevant to the specific difference of interest. The more complex a tissue source, containing multiple cell types or multiple functional regions, the more differences will likely be detected. For these reasons it is best to compare cell populations that are as similar and homogeneous as possible. This consideration independently led all three groups to the hippocampus, a region associated with learning and memory, known to undergo cellular plasticity, and relatively uniform regarding cell type, compared with cortex or whole brain. The choice of adult rather than developing tissue circumvented the confounding influence of developmental change. They all used seizure to induce strong, synchronized neural activity with protocols previously shown to result in IEG induction in the brain. Many of the activity-regulated genes identified using the seizure protocol were later demonstrated to be induced by less severe stimulation (see Table 1), such as during LTP (110, 166, 196, 222, 263, 283, 285, 286), and by physiological stimuli, for example, visual input (30, 51, 157, 170, 195, 263, 283, 285), suggesting their induction was not specifically seizure related, but was likely to occur in the brain with day to day levels of activity.
TABLE 1.
IEG summary
| Protein | IEG | Stimulus Protocol | Plasticity Paradigm | Cellular Function |
|---|---|---|---|---|
| tPA | Yes | Seizure (222), LTP (222), learning (239) | L-LTP (13) | Extracellular matrix degradation (262), synaptic growth (13) |
| MHC I | No | Seizure (5l), sensory (51) | LTP (121), LTD (121) | Competitive structural refinement (121) |
|
Intracellular signaling | ||||
| SNK | Yes | Seizure (142), LTP (142), electroconvulsive therapy (199), epileptiform activity (238) | Synaptic scaling (236, 238) | Kinase, synapse weakening (236, 238) |
| RGS2 | Yes | Seizure (128), LTP (128), drugs (32, 128) | ND | G protein signaling (20, 128) |
| CPG16 | No | Seizure (196) | ND | S/T-kinase (110, 247), cortical architecture?, neuronal migration? |
| Rheb | Yes | Seizure (286), LTP (110, 286), sensory (195) | ND | Ras-Raf signaling (289) |
|
Extracellular signaling | ||||
| BDNF | No | Seizure (5, 74, 110, 131, 199 292), LTP (212), sensory (41) | LTP (78, 138, 148, 211) | Neurite outgrowth (15), neuronal survival (113), competitive structural refinement (34), axonal and dendritic arborization (129, 176–178) |
| COX-2 | Yes | Seizure (5,110,199, 283), LTP (283), neuronal activity (283), stress (283) | ND | Arachidonic acid metabolism (248), transcellular signaling (109) |
| Narp | Yes | Seizure (263), LTP (263), sensory (263) | ND | Pentraxin (263), neurite outgrowth (263), AMPAR clustering (203), excitatory synaptogenesis (181, 203, 245) |
| CPG15 | Yes | Seizure (196), diabetes (139), ischemia (104), axotomy (258), sensory (107, 157, 195) | ND | Dendritic outgrowth (198), axonal outgrowth (37), synapse maturation (37, 87), neuronal survival (221) |
| Arcadlin | Yes | Seizure (284), LTP (284) | ND | Cadherin (284, 288), endocytosis of N-cadherin (288), spine turnover (288) |
|
Synaptic machinery | ||||
| Arc | Yes | Seizure (166, 170), LTP (110, 166, 170), sensory (170) | Synaptic scaling (244), L-LTP (215), LTD (215), mGluR-LTD (209, 273), memory consolidation (215, 244) | AMPA receptor internalization (44, 215, 228) |
| Homer-1a | Yes | Seizure (30, 196), sensory (30, 195) | ND | mGluR clustering (30, 280), signaling (9, 77), and surface expression (8, 10, 229); coupling of mGluRs to the PSD (264), IP3R (265); TRPC1 gating (291); AMPAR cycling (169) |
| CPG2 | No | Seizure (196), sensory (195) | ND | Glutamate receptor endocytosis (53) |
ND, not determined. Reference numbers are given in parentheses. See text for definitions.
The Kuhl and Worley screens focused on IEGs by applying protein synthesis inhibitors prior to stimulation (222, 283). Kuhl and colleagues (222) identified, in addition to c-fos and zif/268, five independent genes upregulated by metrazole-induced seizures, that they termed brain activity dependent (BAD) 1 through 5. BAD1, BAD2, and BAD3 were novel genes not further examined. BAD4 encoded a secreted protein of unknown function, TIS21, also induced by NGF (29). BAD5 turned out to be the tissue-type plasminogen activator (tPA), an extracellular protease known to activate the protease plasmin by proteolytic cleavage of plasminogen (262). Further analysis showed that tPA mRNA is upregulated in electrophysiological paradigms that induce LTP in hippocampus and that tPA induction relies on the NMDA receptor (222). Consistent with a potential role in synaptic plasticity, tPA is induced in the cerebellum in response to learning (239), and tPA inhibitors suppress the expression of late LTP (13). Follow-up screens by the Kuhl group identified arg3.1, coding for a dendritically enriched protein (166) also identified by the Worley (170) and Nedivi groups (110).
The screen performed by Worley and co-workers proved even more fruitful thanks to the use of subtractive hybridization, yielding 15 novel IEGs (153), approximately half of which encoded transcription factors, including a novel zinc finger protein Egr3, a Zif/268 homolog (285). The MECS paradigm also identified a number of specifically upregulated IEGs with predicted cellular functions unrelated to transcriptional regulation, including COX-2, an inducible form of prostaglandin H synthase (283); Rheb, a novel Ras-related small GTP-binding protein (286, 289); Arc, a novel cytoskeleton-associated protein enriched in dendrites (170) (also termed Arg3.1 by Link et al., see above); Narp, a novel member of the pentraxin family of secreted lectins (263); Homer, a PDZ protein that binds metabotropic glutamate receptors (30) (also termed CPG22 by the Nedivi group, Ref. 195), and Arcadlin, a novel cadherin-like molecule (284). These are further discussed below.
The Nedivi screen also benefited from enrichment by subtractive hybridization and was further enhanced by a highly sensitive differential screening procedure (110, 196). In contrast to the above-mentioned screens, it was performed without protein synthesis inhibitors. As a result, this screen identified a large number of candidate plasticity genes (CPGs) induced in the hippocampal dentate gyrus in response to kainic acid seizure, including both IEGs and late response genes. The authors initially reported 52 genes, listing 17 with known cellular function, among them, the IEG transcription factors c-Jun, Zif/268, c-Fos, and CREM (196). Continued screening eventually expanded this number to 362 upregulated CPGs, 66 of them known, and 7 novel CPGs with motifs predicting their putative function (110). Among the candidate genes there was a clear representation of membrane-, vesicle-, and synapse-related proteins.
Even before characterization of the novel “plasticity” genes identified in these screens, perusing the cellular functions of known genes newly recognized in the context of a seizure response, provided some strong clues regarding the cellular response of neurons to synaptic activity. It became clear that the nervous system employs a wide array of genes in response to activity, with the potential of affecting multiple cellular properties, ranging from process outgrowth and rearrangement of the cytoskeleton and the extracellular matrix, to changes in neuronal excitability and modulation of synaptic strength.
The Nedivi and Worley screens also allowed a preliminary estimate of the number of genes that may be activity-regulated in the brain. Considering that CPGs comprised 5% of the screened clones, and a complexity of ~20–30,000 brain RNA species (158), the total number of activity-regulated genes was estimated at 500–1,000 (196). Exhaustive screening of the libraries prepared after MECS stimulation in the presence of protein synthesis inhibitors generated an estimate of 30–40 independent activity-regulated IEGs, of which perhaps 10–15 were transcription factors (153). These estimates likely represent lower bounds, limited by the finite sensitivity of the screens.
A conceptually different screen for activity-regulated genes in this early generation was not based on seizure activity, but rather on genes downregulated by activity blockade during development (51). This screen identified the immune molecule class I major histocompatibility complex (MHC) antigen as an activity-regulated gene and showed for the first time that the mRNAs for CD3ζ, a component of the MHC I receptor complex, and for β2-microglobulin, a cosubunit of the MHC I complex, are present in the central nervous system (CNS) (51). Hence, molecules formerly described as mediating cell-cell recognition in the immune system were shown to be regulated by activity in the central nervous system. In mutant mice lacking functional MHC I signaling, developmental refinement of connections between the retina and central targets is incomplete, hippocampal LTP is enhanced, and LTD is absent (121).
Approximately 10 years after these first screens, with the advent of microarray chip technology, a second wave of studies attempted to provide a more comprehensive view of transcriptional changes associated with enhanced activity. Since the microarray approach does not require the amount of tissue necessary for conventional differential screening or subtractive hybridization, it allows the use of less efficient, but more specific stimulation protocols, resulting in a greater diversity of approaches. Some chip screens were similar to the earlier differential screens, using hippocampal tissue after an in vivo seizure-like stimulus (5, 72, 84, 199), or hippocampal slices after an LTP-inducing stimulus (208). Other screens made use of cultured cortical neurons treated with different pharmacological agents (115, 147, 279). Whether the stimulus was seizure, LTP, NMDA application, or other pharmacological manipulations that affect activity, consensus groups were identified for many of these screens. Two common groups were those related to signal transduction and transcriptional activation. Effector gene categories were more varied between the different screens but did contain genes seen in earlier screens representing neuritogenesis, synaptic transmission, and cytoskeletal regulation. Interestingly, a genome-wide screen for seizure-induced transcriptional changes in the Drosophila nervous system also yielded a crop of transcripts, suggesting activity-dependent modulation of membrane excitability and synaptic transmission, as well as cytoskeletal architecture and synaptic growth (97).
Most of the above-mentioned screens partially validate their positives at the transcript level, but the significance of each individual hit to neuronal plasticity can only be assessed after a characterization of its cellular function in this context. This holds true for a second type of recent microarray screens that did not target the general category of activity-regulated genes, but focused on gene expression changes during developmental activity-dependent plasticity in the visual system (151, 172, 261). Interpretation of these data is even more complex due to the confounding effects of age-specific gene regulation, multiple developmental events occurring concurrently during the period examined, and the difference in outcome of the deprivation protocols used to manipulate activity. While there is some overlap with genes previously identified in response to more general seizure manipulations, especially in the transcription factor category, for the most part, the visual system development screens have yielded a pool of genes distinct from each other and from other screens for activity-regulated genes. It is too early to interpret the mechanistic significance of these differences.
Microarray screens have some major limitations: they are only capable of interrogating a predefined set of sequences, and they report relative, rather than absolute, abundance of a given transcript. In addition, because they are based on sequence-specific probe hybridization, they are particularly vulnerable to false positives due to probe cross-reactivity. Lately, newer techniques that are less susceptible to these problems are replacing gene expression microarray screens (for overview, see Ref. 50). Serial analyses of gene expression (SAGE) has been used to identify genes regulated in response to NGF in PC12 cells (7) and to identify candidate early target genes of JNK or AP-1 in Drosophila (76). SAGE technique has also been combined with chromatin immunoprecipitation (ChIP) for serial analysis of chromatin occupancy (SACO) in a genome-wide screen for CREB target sites (127). Comparing the SACO results with those obtained from affymetrix chips with and without cAMP stimulation, the authors validated their results and greatly expanded our view of the CREB regulon. Deep sequencing, a new, unbiased approach for identifying active transcripts (254), allows the detailed detection of multiple splice variants from a single gene, a goal notoriously difficult to achieve with conventional hybridization techniques. Although their application with respect to the activity-regulated gene program in the nervous system is yet to come, the advent of these powerful screening technologies holds promise for more comprehensive approaches to identification of activity-regulated genes.
In summary, forward genetic screens for activity-regulated genes have advanced our thinking regarding the cellular changes set in motion by synaptic activity. While signaling from the synapse to the nucleus utilizes traditional second messenger pathways similar to those activated by growth factors, inducing many of the same transcription factor IEGs, the cellular response to these two types of stimuli clearly diverges at the transcriptional level. Transcriptional programs initiated by synaptic activity are dominated by gene products that directly impinge on neuronal structure and function, with particular emphasis on proteins that promote process outgrowth and differentiation, or interface with the synaptic machinery.
V. IMPLEMENTATION OF ACTIVITY-DEPENDENT NEURONAL TRANSCRIPTIONAL PROGRAMS
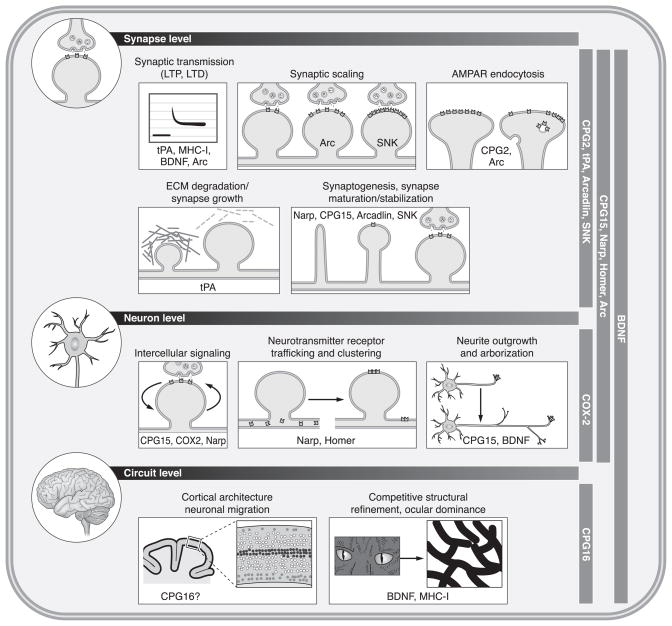
Forward genetic screens have yielded a large crop of activity-regulated genes with cellular functions described in nonneuronal contexts or in contexts of early neural developmental. While these gave a feel for what kind of cellular processes may be recruited in response to synaptic activation, few were followed up and studied mechanistically in the context of cellular or behavioral plasticity. Below we focus on activity-regulated genes that have been characterized sufficiently to yield insight into the cellular mechanisms recruited in response to altered levels of neuronal activity.
A. Intracellular Signaling Molecules
As might have been anticipated by analogy to the growth factor response pathway, two major classes of activity-regulated genes encode transcription factors and signaling proteins. In both classes there are “generic” members in common with the response pathways to extracellular stimuli unrelated to synaptic activity. Also identified were novel transcription factors and signaling proteins, predominantly kinases (168, 247), that seem unique to synaptic stimulation. We will not elaborate further on the role of activity-dependent transcription factors in synaptic plasticity, as this topic is comprehensively discussed in a number of excellent recent reviews (2, 47, 80). We discuss several candidate plasticity genes in the signaling molecule category newly investigated in the context of activity-dependent neuronal response pathways.
1. SNK
The serum-induced kinase SNK, also called Plk2, belongs to the family of pololike kinases, first identified in Drosophila (168, 255). In mammals, three genes encode pololike kinases that have been described in the context of proliferating cells, and their function in oncogenic transformation is well characterized (for review, see Ref. 69). In postmitotic neurons, SNK was found to be upregulated in response to various stimuli, including pharmacologically induced seizures, LTP-producing high-frequency stimulation (142), electroconvulsive seizure therapy (199), and epileptiform activity (238). Early studies showed that SNK is activity-regulated both at the mRNA and protein levels, with the most robust induction in hippocampus, amygdala, and the cerebral cortex. SNK protein is localized to soma and dendrites (142) and in particular to dendritic spines (6, 206).
While an early study demonstrated a direct in vitro interaction of SNK with the Ca2+-binding protein Cib (142), an integrin αIIb binding factor (192), the relevance of this interaction to activity-dependent plastic changes in the nervous system remains unclear. Evidence for a role in synaptic plasticity came from a study demonstrating SNK interaction with spine-associated Rap GTPase activating protein (SPAR) (206). SPAR is part of the postsynaptic scaffolding associated with PSD95, as well as the NMDA receptor (207), and was found to regulate dendritic spine morphology by reorganizing the F-actin cytoskeleton (207). SNK inhibits SPAR-mediated cytoskeletal reorganization, inducing loss of SPAR, as well as PSD95 and even presynaptic Bassoon clusters, suggesting it may play a role in weakening or possibly eliminating synaptic contacts (206). The molecular mechanism by which SNK phosphorylation of SPAR leads to SPAR loss is through association with the multisubunit E3 ubiquitin ligase complex SCF (β-TRCP), which directs SPAR towards proteasome-mediated degradation (6).
Two recent studies suggest a requirement for SNK in synaptic scaling. Disruption of SNK function using a dominant-negative approach abolished synaptic scaling in cultured hippocampal neurons (236). Eliminating CDK5 function has a similar outcome, since CDK5-mediated phosphorylation of SPAR is a prerequisite for its recognition by SNK (6, 236). Other experiments in organotypic slice cultures showed that SNK is required for the down-regulation of membrane excitability in pyramidal neurons. Thus SNK loss of function results in a persistent potentiation of synaptic strength which precludes the expression of LTP (238). Interestingly, SNK seems to regulate itself negatively, likely through autophosphorylation, and is therefore capable of limiting its own actions over time (144). Taken together, these data demonstrate that SNK is an activity-induced molecule that plays a role in homeostatic synaptic plasticity. The potential substrate specificity of the SNK kinase and the fact that not every spine head contains SPAR in detectable quantities (207) warrants consideration of a synaptic tagging-like mechanism for SNK action (237).
2. RGS2
The RGS gene family (regulators of G protein signaling) encodes proteins that accelerate the intrinsic GTPase activity of heterotrimeric G protein α-subunits, thus functioning to dampen G protein-mediated signal transduction (20). In contrast to other members of the mammalian RGS family, RGS2 is specifically induced by neuronal activity, with induction characteristics of an IEG (32, 128). Early studies using single turnover assays performed in solution indicated that RGS2 functions as a GTPase-activating protein (GAP) selectively for Gqα subunits. However, RGS2 also stimulates Giα when G protein is reconstituted in vesicles together with the muscarinic acetylcholine receptor (mAChR) (128). The same study showed that RGS2 inhibits both M1- and M2-dependent mAChR-mediated activation of mitogen-activated protein (MAP) kinase, consistent with its GAP activity for both Gqα and Giα subunits, and indicating how RGS2’s function may be integrated in activity-induced downstream signaling pathways.
Behavioral analyses of RGS2 knockout (KO) mice revealed increased anxiety and reduced male (but not female) aggression, although no other cognitive or motor deficits were observed (205). These findings were partially supported by a genetic trait analysis of outbred mice that identified the RGS2 locus as a quantitative trait gene influencing anxiety (282). Anatomically, RGS2 KO mice display reduced apical and basilar spine density in Golgi-Cox stained hippocampal slices and impaired basal electrical activity in hippocampus CA1 neurons seen as a reduction in the input-output relationship. Surprisingly, LTP in CA1 neurons of these mice was normal (205). A recent study looking more closely for circuit deficits that potentially underlie the behavioral anxiety seen in the RGS2 KO mice examined RGS2 KO neurons in culture and found that in the absence of RGS2 paired-pulse ratios were increased, while expression of RGS2 in either wild-type or RGS2−/− background led to reduced paired-pulse ratios (103). Pharmacological analyses demonstrated that this RGS2 action is specific to Gi/o trimeric α-subunits and that Gq pathways are not involved. Consistent with its effects on paired-pulse facilitation, the authors show that RGS2 increases presynaptic release probability by increasing local intracellular Ca2+ concentrations, likely by downregulating the Gi/o-mediated inhibition of presynaptic Ca2+ channel activity (103). Taken together, these studies establish a role for RGS2 in short-term synaptic plasticity, that may underlie its contribution to anxiety-and aggression-related behaviors.
B. Extracellular Signaling Molecules
Activity-dependent signaling is not confined within the cell. A significant fraction of the novel activity regulated genes examined in detail encode extracellular molecules that mediate intercellular communication. We are accustomed to thinking of extracellular signaling as mediated by diffusible molecules that act systemically or in controlled gradients across long distances. Some of the extracellular activity-dependent gene products, such as brain-derived neurotrophic factor (BDNF), clearly act in this manner too. However, an unusual aspect of this category as a whole is that their signaling can also be distinctly localized to the synaptic vicinity.
1. BDNF
BDNF was first isolated on the basis of its ability to support the survival of, and process outgrowth from, CNS neurons (15, 113). It was thought to act with other neurotrophins to promote neuronal survival and differentiation during nervous system development (reviewed in Ref. 249). However, expression of neurotrophins, including BDNF, in the adult brain, particularly in the hippocampus (11, 75, 112, 171), suggested they may play additional roles in regulating neuronal function (reviewed in Ref. 225). Shortly thereafter it was shown that transcription of the BDNF gene is strongly regulated by experimentally induced seizure (74, 131, 292), as well as by LTP (212) and physiological levels of activity such as exposure to light (41). Not surprisingly, BDNF also surfaced in screens for activity-regulated genes (5, 110, 199).
The potential relevance of BDNF function to synaptic plasticity was bolstered by studies manipulating its levels in vivo. Intracerebral infusion of BDNF or NT4/5, which share TrkB as their common receptor (130), inhibited formation of ocular dominance columns in the developing visual cortex of cats (34). Other known neurotrophins such as NGF and NT3 did not show these effects, hinting that the TrkB receptor specifically is involved in mediating competition-driven segregation of thalamocortical afferents into eye-specific domains. In a later complementary study, Cabelli et al. (35) reduced endogenous BDNF levels by infusing visual cortex at the peak of ocular dominance segregation with soluble fusion proteins that combine the TrKB receptor extracellular domain with IgG (TrkB-IgG receptor bodies) and act as scavengers of TrkB ligands (242). After intraocular injection of 3H-labeled proline and subsequent visualization by autoradiography, it became clear that ocular dominance stripes were absent near the infusion cannula (35).
Involvement of BDNF in the synaptic plasticity paradigm of LTP was first demonstrated in mice with targeted disruption of the BDNF gene. Hippocampal slices from BDNF knockout mice showed significantly reduced LTP in the CA1 region of the hippocampus (148, 211). Treatment of these slices with recombinant BDNF entirely rescued deficits in LTP as well as in basal synaptic transmission. Thus synaptic impairments in the knockout mice are not secondary to developmental defects but due to a direct requirement for BDNF during synaptic function and plasticity (211). These findings are consistent with studies showing that TrkB-IgG receptor bodies reduce the synaptic response to tetanic stimulation and diminish the magnitude of LTP in adult hippocampal slices, while exogenous BDNF promotes LTP induction in young slices (78). Additional studies showed that application of BDNF to hippocampal slices from young adults elicited a sustained enhancement of synaptic transmission that mimicked, but did not occlude, high-frequency induced LTP (138). These combined studies strongly indicate an acute role for BDNF in hippocampal synaptic plasticity.
Another aspect of neuronal function influenced by BDNF is structural plasticity of axonal and dendritic arbors. Application of BDNF to organotypic slice cultures of developing neocortex resulted in a rapid increase in length and complexity of pyramidal neuron dendrites (178). Conversely, removing endogenous BDNF (and NT4–5) in this system using TrkB-IgG receptor bodies showed the requirement for BDNF in the growth and maintenance of pyramidal cell dendritic arbors (177). Although evidence for its effect on axons is less extensive than for dendritic growth, application of BDNF has also been shown to enhance arborization of retinal axons in vitro (129). Inhibition of spontaneous neuronal activity, synaptic transmission, or L-type calcium channels all prevents the positive effect of BDNF on dendritic arborization (176), suggesting that BDNF may serve to link levels of synaptic activity with process outgrowth and activity-dependent structural plasticity.
2. Narp
Narp, neuronal activity-regulated pentraxin, is a member of the pentraxin family of secreted proteins. Consistent with the conservation of its Ca2+ and carbohydrate binding domains, Narp has the biochemical properties of a calcium-dependent lectin (263). Since pentraxins are structurally conserved across species (73), and the plant-derived lectin concanavalin A promotes neurite outgrowth in mammalian neurons (163), Narp was tested in a heterologous expression system where COS-1 cells transfected with a Narp expression construct were cocultured with cortical explants. Neurons, but not glia, from the explant migrated out to the surrounding area and exhibited exuberant process outgrowth (263). These effects of Narp were not apparent when explants were cultured on collagen, but were most obvious on a less supportive substrate, such as poly-L-ornithine. While it is possible that Narp is simply a better substrate for growth than poly-L-ornithine, its potency in promoting outgrowth is in the nanomolar range, unlike substrates such as NCAM or laminin, and more comparable to neurotrophins and growth factors (159). The growth-promoting effects of plant lectins, also effective at nanomolar concentrations, are attributed to binding of specific receptors on the neuronal surface (164). Like other lectins, Narp is multivalent, and its binding efficacy as well as physiological activity may be dependent on this multivalency (263).
Heterologous expression in HEK 293 cells revealed that secreted Narp binds to itself and forms large aggregates that can trap cotransfected GluR1 subunits (203). A closer look at Narp distribution in the mammalian nervous system shows it is predominantly localized to aspiny neurons, where it resides almost exclusively at GluR1-positive excitatory synapses on the shaft of dendrites, whereas it is largely absent from glutamatergic synapses on spiny neurons, such as pyramidal cells. Ultrastructural analyses detected Narp at both pre- and postsynaptic terminals and in the synaptic cleft as well (203). Native Narp in brain is part of a pentraxin complex with neuronal pentraxin 1 (NP1, described in Ref. 235). Narp and NP1 form quarternary complexes covalently linked by disulfide bonds. Whereas Narp is readily induced in neuronal activity paradigms (263), NP1 is constitutively expressed and is nonresponsive to elevated activity (281). Compared with Narp, NP1 has only minor synaptogenic activity (281). Thus Narp/NP1 ratios affect the synaptogenic efficacy of pentraxin complexes. Since Narp levels are directly coupled to synaptic activity, excitatory synaptogenesis is dynamically regulated by Narp through its contribution to pentraxin complex composition. A third member of the neuronal pentraxin family is the neuronal pentraxin receptor (NPR). NPR is physically associated with Narp and NP1, and like Narp and NP1 it binds AMPA receptors and contributes to synaptogenesis (61, 245). NPR possesses a transmembrane domain that likely tethers a pentraxin complex containing all three neuronal pentraxins to the cell membrane. Interestingly, both NP1 and NPR are axonally derived and are secreted exclusively by excitatory cell axons at their contact points with interneurons (181, 245). Overexpression of Narp in neurons leads to an increase in the number of excitatory but not inhibitory synapses (203). These data taken together support a model in which Narp facilitates excitatory synaptogenesis by acting as an extracellular aggregating factor for AMPA receptors, specifically on dendritic shafts (181, 203, 245).
However, in response to activation of group 1 metabotropic glutamate receptors, the extracellular metalloprotease TACE releases a soluble form of NPR and the other neuronal pentraxins from the transmembrane tether, allowing their trafficking to endosomes together with attached AMPA receptors, thereby contributing to the downregulation of synaptic AMPA receptors (43). It is possible that the role of nonactivity regulated neuronal pentraxins is to mediate complex turnover, allowing real time incorporation of activity-regulated Narp, thus linking synaptogenic efficacy with neuronal activity.
3. CPG15 (neuritin)
cpg15 (candidate plasticity gene 15 also known as Nrn1) was initially identified in the Nedivi screen as seizure regulated. cpg15 has since been shown to be induced by physiological stimuli such as light (157, 195) and whisker stimulation (107), as well as in response to chemically induced diabetes (139), ischemia (104), and axotomy (258). Interestingly, while cpg15 is an IEG regulated by the same signal transduction pathways, transcription factors, and promoter elements that regulate expression of transcription factors such as Fos (87), cpg15 encodes a small extracellular protein (191, 195, 198). The CPG15 protein contains both a signal sequence and a consensus sequence for a glycosylphosphatidylinositol (GPI) anchor, and in its processed form is attached to the cell surface (191, 198). In vitro functional characterization of CPG15 suggested that it promotes neurite outgrowth in cultured hippocampal neurons (191). In vivo studies, using virally mediated overexpression in the optical tectum of Xenopus leavis tadpoles followed by live imaging, revealed that CPG15 promotes dendritic outgrowth in a non-cell-autonomous manner (198). Using the same system, Cantallops et al. (37) showed that CPG15 expression significantly increased the growth rate of retinal axons and promoted synaptic maturation by recruitment of functional AMPA receptors to synapses. Electrophysiological recording showed that while NMDA receptor-mediated currents remained unchanged in response to increased CPG15 levels, AMPA receptor currents were significantly increased and failure rates at hyperpolarizing potentials were decreased, potentially due to AMPA receptor recruitment to otherwise silent, immature synapses (37).
CPG15 protein is concentrated in axon tracts (197) and is distributed inside the axon as well as on the axon surface (36). Depolarization by KCl and kainate rapidly increases surface expression of CPG15, suggesting that its delivery to the axon membrane can be rapidly regulated by neural activity (36). These data support a model in which activity plays a dual role in CPG15 action, increasing its expression levels, and promoting delivery of axonally trafficked protein to the surface where it acts to locally modulate dendritic and axonal elaboration, and synaptic maturation.
CPG15 has no sequence homology with traditional neurotrophin ligands and is unusual as a growth factor in its action as a membrane-bound ligand. It does share with neurotrophins an ability to act both as a neuronal growth and differentiation factor and as a survival factor. Consistent with its identification as an activity-regulated gene, cpg15 expression is contemporaneous with critical periods for activity-dependent plasticity (52, 195). However, cpg15 is also expressed during early brain development before neuronal differentiation and circuit formation, suggesting a different role at this stage (221). Indeed, soluble CPG15 purified from the culture media in a heterologous expression system can rescue both cortical progenitors and differentiated cortical neurons from starvation-induced apoptosis in vitro (221). CPG15 overexpression in vivo during early development expands the cortical progenitor pool by preventing apoptosis, resulting in an expanded cortical plate and heterotypic cell masses in the ventricular zone. Conversely, RNAi-mediated CPG15 knockdown leads to a shrunken cortical plate with fewer neurons as a result of decreased progenitor survival (221). It is still unclear whether the soluble form of CPG15 harvested from culture media in vitro exists in vivo as a biologically active molecule, and whether CPG15’s distinct roles as a survival or differentiation factor are mediated by the soluble versus membrane-bound form.
4. Arcadlin
Arcadlin (activity-regulated cadherin-like protein), found by the Worley group to be upregulated in rat hippocampus after MECS or tetanic stimulation of the perforant path, was the first activity-regulated gene identified from the cadherin superfamily (284). Northern blot analyses showed expression of the arcadlin gene to be brain specific, with highest levels in hippocampus, cortex, and the striatum. The deduced amino acid sequence predicts a single pass transmembrane domain, an extracellular domain with six identified cadherin repeats, and an intracellular COOH terminus. The protein is localized at the soma, and at synapses it colocalizes with the marker synaptophysin. The extracellular domain of Arcadlin is most homologous to the human protocadherin 8, and similar to other cadherins, Arcadlin possesses Ca2+-dependent homophilic binding mediated by this domain. The intracellular portion of Arcadlin, however, seems to be unique among the cadherin family.
Anti-Arcadlin antibody application to acute hippocampal slice preparations suppressed synaptic transmission and impaired LTP induction by tetanic stimulation (284). While these initial results indicated a role for Arcadlin in synaptic transmission, the molecular mechanisms were not elucidated until recently, when Yasuda et al. (288) demonstrated that Arcadlin can regulate dendritic spine numbers by inducing endocytosis of N-cadherin. Using a variety of techniques, their study elegantly demonstrated that Arcadlin and N-cadherin colocalize at synapses and that Arcadlin diminishes the adhesive properties of N-cadherin. Furthermore, the authors show that the molecular pathway for activity-induced internalization of N-cadherin is regulated by Arcadlin through the following sequence of events: synaptic levels of Arcadlin are increased by activity, increasing homophilic interaction of extracellular Arcadlin domains. These interactions activate TAO2β kinase, which is constitutively bound to the intracellular domain of Arcadlin. TAO2β is a MAPKKK which is upstream of p38 MAPK. p38 MAPK then feeds back to phosphorylate a key residue of TAO2β that is required for N-cadherin endocytosis. Hippocampal neurons from Arcadlin-deficient mice show a significantly larger number of spines, perhaps due to increased adhesivity as a result of reduced N-cadherin endocytosis (288). These findings identify the synaptic adhesive apparatus as a target of activity-dependent synaptic remodeling and suggest that Arcadlin may be a negative regulator of synapse and spine numbers through its effect on N-cadherin internalization.
5. Cyclooxygenase 2
Cyclooxygenase 2 (COX-2) first identified by the Worley lab as an inducible immediate-early gene (283) also surfaced in other seizure screens (5, 110, 199). Cyclooxygenases catalyze the first step in prostaglandin synthesis (248) and are the pharmacological target of nonsteroidal anti-inflammatory drugs (reviewed in Ref. 173). The COX-2 substrate arachidonic acid is generated in the striatum in response to NMDA receptor activation (68) and in the hippocampus after synaptic stimulation that induces LTP (275). Hence, increased synaptic activity not only upregulates COX2 mRNA, but also increases availability of the enzyme’s substrate. Prostaglandin H2, the product of COX-2 catalysis, serves as the precursor for further prostaglandin synthesis, but also for the production of prostacyclins and thromboxanes (38, 173, 248), all of which are diffusible molecules that can cross the plasma membrane and therefore spread a local signal to influence neighboring cells (109). It is of particular interest that COX-2 immunostaining is seen in dendritic spines (140), suggesting it can produce diffusible signaling molecules that act locally at the synapse.
C. Regulation of the Synaptic Machinery
While both intra- and extracellular activity-regulated signaling molecules impinge on the synapse, the third category of activity-regulated genes encode proteins that are integral parts of the synaptic machinery.
1. Arc
Arc (activity-regulated cytoskeleton-associated protein) was first described in 1995 as a nontranscription factor IEG upregulated in response to seizure activity (170). At the same time, another group characterized an activity-regulated gene with a 3.1-kb transcript (arg3.1) that was later found to be identical to arc (166). Robust arc/arg3.1 mRNA upregulation is seen as early as 30 min after induction (166, 170), and recent work has identified a synaptic activity-responsive element (SARE) through study of the arc/arg3.1 promoter region (143). It is represented by a ~100-bp element located 5 kb upstream of the transcription initiation site and harbors four boxes that differentially respond to CREB, MEF2, SRF, and TCF to regulate SARE activation (143).
Both initial studies identified homologies between Arc and α-spectrin, characterized its rapid induction in response to neuronal activity, and demonstrated the dendritic localization of arc/arg3.1 mRNA in the hippocampus (166, 170). While dendritic mRNA localization had been seen for other transcripts, e.g., MAP-2 (89), this feature of arc expression is unique among the IEGs (251). Taking advantage of the precise laminar targeting of perforant path projections in the middle molecular layer of the hippocampal dentate gyrus, Steward et al. (251) showed that direct activation of this pathway induces arc expression, and accumulation of newly synthesized arc mRNA and Arc protein, selectively in the synaptically activated dendritic laminae (251). The local accumulation of both transcript and protein gave rise to the idea that arc/arg3.1 mRNA might be locally translated in dendrites, a speculation that was confirmed in hippocampal slices (273). Later, it became evident that arc/arg3.1 mRNA targeting to specific sites of synaptic activation requires NMDA receptor activation (252). Importantly, this study demonstrated that the induction of arc/arg3.1 mRNA and its targeting to regions of synaptic activity are separable, since MECS induced arc/arg3.1 mRNA was evenly distributed in the molecular layer, but when MECS was followed by region-specific synaptic input, the induced transcript accumulated specifically in the region of elevated activity.
The arc/arg3.1 transcript is one of few naturally occurring mRNAs where the STOP codon of the open reading frame is not located in the last exon. Hence, upon splicing of the transcript, an exon-junction-complex (EJC) tag labels the mRNA at a position 3′ to the STOP codon, triggering nonsense-mediated decay of the message (92). This suggests that not only transcriptional activation of arc/arg3.1 but also its translation is tightly regulated, possibly leading to a single pulse of Arc protein translation in activated terminals and enabling the system to react sensitively to sudden changes in local neuronal activity.
The consequences of Arc protein loss of function were first studied by administration of arc/arg3.1 anti-sense oligo-deoxynucleotides via a chronically implanted infusion cannula in the hilus of the fascia dentate (99). These and other experiments revealed that Arc is required for maintenance of LTP and memory consolidation, but not short-term memory (for review, see Ref. 268). These findings were reinforced by studies of an arc/arg3.1 knockout mouse. In these mice, LTP is compromised: while the early phase of LTP is enhanced, the late phase is essentially absent. The mice show disrupted memory consolidation during spatial learning, fear conditioning, conditioned taste aversion (which is hippocampus independent), and object recognition, although short-term memory was intact (215). A more recent study established a role for Arc in Pavlovian fear conditioning in the lateral amygdala (216). Together, these findings indicate that Arc is required for consolidation of enduring synaptic plasticity and memory storage.
The finding that Arc is necessary for expression of L-LTP and establishment of certain forms of memory led to examination of its role in another form of neuronal plasticity, synaptic scaling (244). In wild-type neurons, surface expression of postsynaptic AMPA-type glutamate receptors is upregulated upon chronic blockade of activity and downregulated under conditions of elevated activity to globally and homeostatically compensate for changes in general activity levels, a process termed synaptic scaling (for review, see Ref. 267). In arc/arg3.1 knockout neurons, synaptic scaling is absent (244). The effect on surface glutamate receptors seems to be specific for GluR1, since surface GluR2 levels are unchanged in arc/arg3.1 knockout neurons (244).
Screening for Arc interacting proteins with the yeast-2-hybrid system identified two critical components of the endocytic machinery, dynamin 2 and endophilin 3 (44). Both proteins coimmunoprecipitate with Arc from transfected HEK cells and from forebrain preparations. In HeLa cells, they colocalize on functional early endosomes. Fine-mapping of the binding sites showed that Arc binds to dynamin 2 and endophilin 3 via distinct domains. Amino acids 89–100 in Arc are necessary and sufficient for binding to endophilin 3 (44), and these same amino acids are also required for mediating the decrease in AMPA receptor currents (228), as is the motif for dynamin binding (44), further arguing for an endocytosis-based mechanism of Arc function. Analysis in neurons showed that Arc associates with endophilin and dynamin on early endosomes as well. arc/arg3.1 knockout neurons show increased AMPA receptor surface expression and reduced endocytosis (44) in line with findings in the arc/arg3.1 knockout mouse (215).
Since AMPA receptors have been demonstrated to undergo constitutive internalization that is regulated by synaptic activity (70, 162), glutamate receptor internalization has been suggested as a fundamental molecular mechanism to reduce synaptic strength during synaptic plasticity, leading to LTD (174). Two studies presented evidence for a direct role for Arc in AMPA receptor internalization (44, 228). Virally mediated overexpression of green fluorescent protein (GFP)-tagged Arc in CA1 pyramidal neurons in hippocampal slices reduced the amplitude of AMPA receptor-mediated synaptic currents. This was a result of GluR2/3 containing AMPA receptor removal from the synaptic membrane (228). siRNA directed against arc/arg3.1 could block the Arc-mediated reduction in AMPA currents, as did the deletion of the Arc region known to interact with endophilin and required for clathrin-mediated endocytosis. The interaction between the tail domain of GluR2 and the general endocytic adaptor AP2 can be suppressed using a specific peptide which competes for receptor tail binding (156). When delivered intracellularly through the recording pipette, this peptide abolishes the Arc-induced reduction of AMPA currents. Finally, Arc expression occludes NMDA-dependent LTD. These data suggest that Arc activates synaptic mechanisms similar to LTD, namely, the removal of functional AMPA-type glutamate receptors from the synapse (156, 228).
More recent studies shed light on Arc’s role in AMPA-receptor endocytosis during mGluR-mediated LTD. Two major mechanisms of long-term synaptic depression are known: one mediated through the ionotropic NMDA receptors, the other by metabotropic G protein-coupled mGluRs. Both scenarios lead to a loss of AMPA-type glutamate receptors in the postsynaptic membrane, but employ different molecular mechanisms (204). In an elegant study, Waung et al. (273) reported that brief activation of mGluRs led to local dendritic Arc synthesis and to a persistent increase of GluR1 endocytosis rate. Furthermore, overexpression of GFP-Arc occludes further mGluR-induced decrease of surface GluR1, arguing that Arc and mGluR-LTD employ the same molecular mechanism. Finally, the authors show that endogenous Arc is required for mGluR-mediated AMPA receptor endocytosis and the expression of mGluR-LTD (273).
Another study examining the same pathway showed that arc/arg3.1 translation is induced within minutes of mGluR activation and that this induction is essential for mGluR-LTD (209). Arc translation in response to mGluR activity requires the translation elongation factor eEF2 kinase, a CaM kinase that binds mGluR and dissociates upon its activation, whereupon it phosphorylates eEF2. Phosphorylation of eEF2 by eEF2K leads to a decrease in global translation, but an increase in specific Arc translation. Thus Park et al. (209) link local Arc translation to a key regulatory component of the translational machinery. Indeed, eEF2K knockouts show impaired mGluR-LTD, showing that eEF2K and Arc act in the same molecular pathway. Interestingly, synaptic scaling is normal in eEF2K knockouts, suggesting that Arc’s function in this plasticity paradigm (244) involves a molecular mechanism distinct from eEFK2 function. The fragile X mental retardation protein (FMRP) has also been shown to bind to arc/arg3.1 mRNA (124, 293) and is hypothesized to inhibit its translation (17). In concordance with this model, the authors observe a dependence of mGluR-LTD upon Arc in fmr1 knockout slices, arguing that the otherwise distinct signaling pathways for eEF2K and FMRP converge on Arc (209).
The product of an activity-regulated gene, Arc, has been shown as essential for the formation of long-term memories and the expression of both L-LTP and mGluR-LTD. It regulates AMPA receptor surface expression both in an acute way, and in homeostatic forms of plasticity such as synaptic scaling. While its molecular mode of action is being unraveled, we are also beginning to understand how arc/arg3.1 mRNA localization and translation are themselves tightly regulated.
2. Homer
Homer-1a was first described as an IEG regulated by synaptic activity (30) that binds to group 1 metabotropic glutamate receptors (mGluRs). It was subsequently shown to be a member of a family of Homer-related proteins derived from three distinct genes (Homer 1–3) that each generate several splice variants (280). Like Homer-1a, all Homer proteins bind group 1 mGluRs, yet Homer-1a is the only IEG among them. Homer-1a is also distinct from other Homer proteins in that it does not contain a COOH-terminal coiled-coil domain that allows for leucine zipper-mediated dimerization and is hence unable to oligomerize (27, 280). Homer proteins are synaptically localized (264, 280) and contain a proline-rich NH2-terminal EVH1 domain that is critical for their specific binding to G protein-linked metabotropic glutamate receptors mGluR1α and mGluR5 (19, 30, 264) that are coupled to phospholipase C and thereby regulate phosphoinositide hydrolysis (194, 214). The same EVH domain apparently binds other Homer ligands. Binding partners of particular interest for synaptic function are the inositide trisphosphate receptor (IP3R) (265), the TRPC1 cation channel (291), and the PSD scaffolding molecule Shank (264).
Consistent with their multimeric nature, Homer complexes can be coimmunoprecipitated from brain with both mGluR1α and the IP3R, suggesting that Homer couples these receptors in a higher order signaling complex (265). Similarly, Homer facilitates physical association between the TRPC1 channel and the IP3R (291). Shank binds Homer through a single EVH binding site that is distinct from its GKAP binding domain, thus bridging between these two proteins (264). Since GKAP had been described as part of the NMDA receptor anchoring and signaling complex in the postsynaptic density (PSD) (193), the Homer-Shank interaction potentially links NMDA receptors to both mGluRs and IP3Rs. Using heterologous expression in COS-7 cells, Tu et al. (265) showed that Shank mediates attachment of Homer to PSD95/GKAP clusters and that mGluR5 clustering relies on the presence of both Homer and Shank. Importantly, Homer self-association and Shank binding are mediated by different portions of Homer, potentially allowing for assembly of a multivalent adapter complex (265).
The IP3R is located in the ER specialization of the spine apparatus in close proximity to the PSD (270) and functions as an intracellular store calcium channel that can be activated by IP3 (which can be produced through mGluR signaling and phospholipase C activation; reviewed in Ref. 230). In turn, release of intracellular Ca2+ stores by ER IP3Rs regulates entry of extracellular Ca2+ through plasma membrane TRPC channels (145, 291). Homer proteins thus physically couple intracellular IP3R signaling to the surface mGluRs that activate them, and to surface TRPC channels that they activate themselves. In addition, Homer proteins in this large signaling complex also link mGluR signaling to NMDA receptor signaling cascades via their association with Shank and GKAP, potentially linking IP3 receptor-mediated Ca2+ emission to general synaptic transmission. Elegantly, the Homer-linked signaling complex can be dynamically regulated by Homer-1a which can bind either mGluRs or IP3R, but does not contain a cross-linking domain that would allow their association within the same complex. Thus Homer 1a effectively functions as an activity-regulated dominant negative that can terminate mGluR/IP3R interactions (265, 280) as well as IP3R/TRPC1 interactions (291) and perhaps disassociate them from the NMDA signaling complex at the PSD.
While the above functions may imply that Homer is a “mere” scaffolding molecule, there is also evidence that Homer can act as an intracellular agonist of G protein-coupled mGluR1α and mGluR5 (77). Apparently these receptors can shift from an inactive to an active conformation even in the absence of agonist (217). Ango et al. (9) showed that constitutive (agonist-independent) activity of these receptors is controlled by Homer proteins and that disruption of the Homer/mGluR interaction leads to constitutively active mGluR1 and mGluR5 receptors (9). Using a pharmacological approach, they further demonstrated that Homer-1a directly competes with Homer-3 for mGluR1α and triggers constitutive activity of the receptor. Thus, in this case, too, Homer-1a functions as an endogenous, activity-regulated dominant negative that dynamically competes with constitutive Homer isoforms to regulate mGluR function.
Additionally, Homer proteins may play a role in trafficking and surface expression of group I mGluRs, although this role has not been entirely clarified. In cerebellar neurons where endogenous Homer-1b and -1c are absent, mGluR5 receptors display restricted somatic localization. Cotransfection of Homer-1b and -1c results in dendritic targeting of Homer-mGluR5 complexes (8). Furthermore, Homer-1b expression prevents trafficking of mGluRs from the ER to the cell surface in heterologous systems (229) and suppresses their surface expression in dendrites (10). In contrast, mGluR surface expression is positively regulated by expression of Homer-1a (10, 229).
A recent paper also shows a role for Homer in mediating a physical link between the PSD and the endocytic zone adjacent to it, perhaps facilitating capture and maintenance of a proximal pool of cycling AMPA receptors (169). Although understanding of all Homer functions is still preliminary, clearly, the identification of Homer-1a as an activity-regulated gene product has opened a window into the inner workings of the basic synaptic machinery and its regulation by synaptic activity.
3. CPG2
CPG2, candidate plasticity gene 2, was identified as the product of an activity-regulated transcript (195, 196). Similar to other activity-regulated transcripts, it seems to be the only activity-regulated product of a multitranscript gene. However, as a splice variant of the massive syne-1 gene, it gives rise to a protein that is unrelated structurally or functionally to its siblings transcribed from the same gene (53). CPG2 is expressed exclusively in brain, predominantly in hippocampus, neocortex, striatum, and cerebellum. In mature cultured hippocampal neurons, CPG2 immunoreactivity is punctate and mainly localizes adjacent to PSD95 clusters, a marker for excitatory postsynaptic sites (53).
Immunoelectron microscopic studies have shown that CPG2 resides in the postsynaptic compartment, adjacent to the PSD and in close vicinity of clathrin-coated pits and clathrin-coated vesicles. Clathrin-mediated endocytosis is believed to underlie glutamate receptor internalization (18, 40, 175, 271). The finding that CPG2 colocalizes with clathrin light chain in the postsynaptic endocytic zone lateral to the PSD prompted Cottrell et al. (53) to investigate the role of CPG2 in glutamate receptor internalization. RNAi-mediated knockdown of CPG2 showed that both constitutive and activity-induced AMPA-type glutamate receptor internalization were impaired in the absence of CPG2. Under the same conditions, a fourfold increase of clathrin-coated vesicles was observed, some of which carried the NR1 subunit of the NMDA receptor (53). Spine head size was also affected by CPG2 levels. Under conditions where CPG2 was overexpressed, spine head areas were increased by 7%, upon CPG2 knockdown they were decreased by 18%.
Given that CPG2 is located adjacent to F-actin in spine heads and features a spectrin repeat and coiled-coil domains, it has been speculated that CPG2 may act as a nexus connecting components of the endocytic machinery to the actin cytoskeleton (53). However, it is equally possible that CPG2 fulfills multiple functions, and that variations in spine head size seen upon altered CPG2 levels are not secondary to altered membrane internalization resulting from defective endocytosis, but are direct effects of CPG2 on the spine cytoskeleton.
Neurons may express CPG2 after elevated synaptic activity to prompt removal of AMPA- and NMDA-type glutamate receptors from the synapse as a homeostatic mechanism to counteract the effect of elevated synaptic activity. It is not clear whether CPG2 acts globally as a mechanism of synaptic scaling on all synapses of a given neuron, or whether synaptic tagging directs it to specific synapses that undergo subsequent glutamate receptor internalization.
D. Activity-Regulated Genes Coordinating Inhibitory Synapse Formation
In addition to orchestrating functional and structural changes at the excitatory synapse, recent work shows that the activity-dependent transcription program also coordinates the formation of inhibitory synapses, and likely underlies the maturation of inhibitory circuits.
In an elegant study, Lin et al. (165) used microarrays to identify activity-regulated genes that respond to membrane depolarization. Out of more than 300 potential hits, one candidate gene was responsive only to Ca2+ influx, showed neuron-specific expression, and is expressed coincidentally with inhibitory synapse development. The study further shows that this protein, the bHLH-PAS transcription factor Npas4, regulates the development of inhibitory synapses by orchestrating the expression of a large set of genes that controls formation and/or maintenance of GABAergic synapses, among them BDNF. Notably, Npas4 binds to sites in the BDNF promoter region (165) responsible for its activity-dependent transcription (1, 167, 257). Furthermore, BDNF knockdown partially attenuates Npas4-induced elevation of GABAergic synapses, suggesting that Npas4 regulation of BDNF transcription is at least partially responsible for its effect on inhibitory synapse numbers.
Another study from the same lab analyzed the targets of the transcriptional activator MEF2 on a genome-wide scale (81). In neurons, MEF2 responds to neurotrophins and synaptic Ca2+ influx (79) and has been shown to restrict excitatory synapse numbers in certain brain regions (14, 79, 220, 240). Using a microarray approach, the new study by Flavell et al. (81) identifies 182 activity-dependent MEF2 target genes that are implicated in various aspects of synapse function, among them several genes that encode proteins with a structural or functional role at the inhibitory synapse. Together with the findings related to Npas4, which also seems to be a direct target of MEF2 transcription (81), these data demonstrate that the neuronal activity-regulated gene program orchestrates inhibitory synapse formation and function, and likely underlies earlier observations of activity-dependent maturation of inhibitory circuits in the brain (106, 119).
VI. SUMMARY
It has been commonly accepted that during development environmental cues act through specific receptors to influence a cell’s transcriptional profile and hence its fate. The idea that the environment similarly affects transcription as a means to alter the response properties of mature differentiated cells is newly emerging. In the brain in particular, perhaps because of the paucity of cell proliferation after birth, the view of a hard-wired structure has long dominated the field.
Studies of protooncogene IEGs such as c-fos introduced the general concept that gene transcription in the nucleus is part of the cellular response program to alterations in signaling from outside the cell. Integral to this concept was the biphasic nature of transcriptional activation, with IEG induction closely followed by a second wave of delayed-early gene expression. Stimulation of IEG expression by NGF in nonproliferating PC12 cells suggested that this biphasic response could differ according to the differentiation and physiological state of the stimulated cell and was not necessarily restricted to proliferative cells. Furthermore, IEG expression in culture could be induced by agents other than growth factors and mitogens, including depolarizing stimuli and voltage-gated calcium influx. In vivo, IEGs were shown to be responsive to physiological stimuli, providing strong support for the idea that endogenous IEG expression in the brain is a result of normal synaptic activity and can therefore be influenced by neuronal stimulation or activity blockade in a pathway and stimulus specific manner. At the same time it was becoming clear that synaptic activity can also drive long-term changes in neuronal structure and function. These concepts were unified with the proposal that electrical and chemical stimuli that produce long-term change in neurons, such as during learning and memory, act through mechanisms analogous to those of growth factors, namely, through second messenger pathways that lead to transcriptional activation.
Forward genetic screens aimed at identifying genes induced by neuronal activity rather than by growth factor stimulation proved essential to understanding the activity-dependent genetic programs utilized by neurons in the mammalian brain on an everyday basis. Characterization of activity-regulated genes identified in these screens has advanced our knowledge of the cellular changes set in motion by synaptic activity. While signaling from the synapse to the nucleus utilizes traditional second messenger pathways, similar to those activated by growth factors and inducing many of the same transcription factor IEGs, the cellular response program activated by these two types of stimuli diverges at the transcriptional level. Transcriptional programs initiated by synaptic activity are dominated by gene products that directly impinge on neuronal structure and function, with particular emphasis on proteins that promote process outgrowth and differentiation or interface with synaptic machinery (see Fig. 1).
FIG. 1.
Activity-regulated genes affect multiple cellular functions at different organizational levels. Boxed sketches depict specific biological processes and show the activity-regulated gene products involved. The boxes are arranged according to the level of organization they represent: top, synaptic level; middle, cellular level; bottom, systems level. Vertical bars on the right indicate which levels each activity-regulated gene product spans with respect to its mechanism of operation.
Given the large number of candidates identified in screens for activity-regulated genes, many of which remain uncharacterized in a neuronal context, we are only beginning to fathom the complexity of processes regulated by synaptic activity in the brain. Yet, despite our incomplete understanding, several themes are beginning to emerge surrounding the role of activity-regulated genes in brain function. Neurons react to changes in levels of synaptic activity with a complex response that takes place on multiple levels and at different time points. Neuronal plasticity, Hebbian or homeostatic, is a consequence of potentially overlapping and interacting actions of a wide number of genes. Furthermore, multiple signals and intracellular pathways can influence transcriptional activation via an assortment of cis- and trans-acting elements, and nuclear IEGs themselves make up a diverse array of factors that can potentially act in a multitude of combinations to differentially affect distinct second-wave gene sets.
The large number of genes induced by neuronal activity, and their individual functions suggest that “plasticity genes” are not a unique regulatory gene set whose action is layered on top of constitutive cellular and synaptic genes; rather, they constitute the backbone of the synaptic and cellular machinery and their regulated expression is a means to dynamically tune everyday neuronal function. In addition, regulation of activity-dependent genes is tightly controlled not only at the transcriptional level, but also at the levels of translation and localization, and perhaps at the levels of mRNA and protein half-life, allowing fine spatial and temporal control of the machinery they oversee.
C-fos, c-jun, CREB, CREM, zif/268, tPA, Rheb, RGS2, CPG16, COX-2, Narp, BDNF, CPG15, Arcadlin, Homer-1a, CPG2, and Arc likely constitute only the tip of the iceberg in the fascinating array of cellular mechanisms our brains use to bring about remarkable functions such as learning and memory, as well as cognitive abilities like thought and reasoning.
Acknowledgments
We thank Jeff Hoch, Robert Weinberg, and members of the Nedivi lab for helpful comments on the manuscript.
GRANTS
We acknowledge funding by the National Eye Institute, the Picower Institute Innovation Fund, the American Federation for Aging Research, and the Deutsche Forschungsgemeinschaft.
References
- 1.Aid T, Kazantseva A, Piirsoo M, Palm K, Timmusk T. Mouse and rat BDNF gene structure and expression revisited. J Neurosci Res. 2007;85:525–535. doi: 10.1002/jnr.21139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alberini CM. Transcription factors in long-term memory and synaptic plasticity. Physiol Rev. 2009;89:121–145. doi: 10.1152/physrev.00017.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alitalo K, Ramsay G, Bishop JM, Pfeifer SO, Colby WW, Levinson AD. Identification of nuclear proteins encoded by viral and cellular myc oncogenes. Nature. 1983;306:274–277. doi: 10.1038/306274a0. [DOI] [PubMed] [Google Scholar]
- 4.Almendral JM, Sommer D, Macdonald-Bravo H, Burckhardt J, Perera J, Bravo R. Complexity of the early genetic response to growth factors in mouse fibroblasts. Mol Cell Biol. 1988;8:2140–2148. doi: 10.1128/mcb.8.5.2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altar CA, Laeng P, Jurata LW, Brockman JA, Lemire A, Bullard J, Bukhman YV, Young TA, Charles V, Palfreyman MG. Electroconvulsive seizures regulate gene expression of distinct neurotrophic signaling pathways. J Neurosci. 2004;24:2667–2677. doi: 10.1523/JNEUROSCI.5377-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ang XL, Seeburg DP, Sheng M, Harper JW. Regulation of postsynaptic RapGAP SPAR by Polo-like kinase 2 and the SCFbeta-TRCP ubiquitin ligase in hippocampal neurons. J Biol Chem. 2008;283:29424–29432. doi: 10.1074/jbc.M802475200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Angelastro JM, Klimaschewski L, Tang S, Vitolo OV, Weissman TA, Donlin LT, Shelanski ML, Greene LA. Identification of diverse nerve growth factor-regulated genes by serial analysis of gene expression (SAGE) profiling. Proc Natl Acad Sci USA. 2000;97:10424–10429. doi: 10.1073/pnas.97.19.10424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ango F, Pin JP, Tu JC, Xiao B, Worley PF, Bockaert J, Fagni L. Dendritic and axonal targeting of type 5 metabotropic glutamate receptor is regulated by homer1 proteins and neuronal excitation. J Neurosci. 2000;20:8710–8716. doi: 10.1523/JNEUROSCI.20-23-08710.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ango F, Prezeau L, Muller T, Tu JC, Xiao B, Worley PF, Pin JP, Bockaert J, Fagni L. Agonist-independent activation of metabotropic glutamate receptors by the intracellular protein Homer. Nature. 2001;411:962–965. doi: 10.1038/35082096. [DOI] [PubMed] [Google Scholar]
- 10.Ango F, Robbe D, Tu JC, Xiao B, Worley PF, Pin JP, Bockaert J, Fagni L. Homer-dependent cell surface expression of metabotropic glutamate receptor type 5 in neurons. Mol Cell Neurosci. 2002;20:323–329. doi: 10.1006/mcne.2002.1100. [DOI] [PubMed] [Google Scholar]
- 11.Ayer-LeLievre C, Olson L, Ebendal T, Seiger A, Persson H. Expression of the beta-nerve growth factor gene in hippocampal neurons. Science. 1988;240:1339–1341. doi: 10.1126/science.2897715. [DOI] [PubMed] [Google Scholar]
- 12.Balschun D, Wolfer DP, Gass P, Mantamadiotis T, Welzl H, Schutz G, Frey JU, Lipp HP. Does cAMP response element-binding protein have a pivotal role in hippocampal synaptic plasticity and hippocampus-dependent memory? J Neurosci. 2003;23:6304–6314. doi: 10.1523/JNEUROSCI.23-15-06304.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baranes D, Lederfein D, Huang YY, Chen M, Bailey CH, Kandel ER. Tissue plasminogen activator contributes to the late phase of LTP and to synaptic growth in the hippocampal mossy fiber pathway. Neuron. 1998;21:813–825. doi: 10.1016/s0896-6273(00)80597-8. [DOI] [PubMed] [Google Scholar]
- 14.Barbosa AC, Kim MS, Ertunc M, Adachi M, Nelson ED, Mc-Anally J, Richardson JA, Kavalali ET, Monteggia LM, Bassel-Duby R, Olson EN. MEF2C, a transcription factor that facilitates learning and memory by negative regulation of synapse numbers and function. Proc Natl Acad Sci USA. 2008;105:9391–9396. doi: 10.1073/pnas.0802679105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barde YA, Edgar D, Thoenen H. Purification of a new neurotrophic factor from mammalian brain. EMBO J. 1982;1:549–553. doi: 10.1002/j.1460-2075.1982.tb01207.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barth AL, Gerkin RC, Dean KL. Alteration of neuronal firing properties after in vivo experience in a FosGFP transgenic mouse. J Neurosci. 2004;24:6466–6475. doi: 10.1523/JNEUROSCI.4737-03.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bear MF, Huber KM, Warren ST. The mGluR theory of fragile X mental retardation. Trends Neurosci. 2004;27:370–377. doi: 10.1016/j.tins.2004.04.009. [DOI] [PubMed] [Google Scholar]
- 18.Beattie EC, Carroll RC, Yu X, Morishita W, Yasuda H, von Zastrow M, Malenka RC. Regulation of AMPA receptor endocytosis by a signaling mechanism shared with LTD. Nat Neurosci. 2000;3:1291–1300. doi: 10.1038/81823. [DOI] [PubMed] [Google Scholar]
- 19.Beneken J, Tu JC, Xiao B, Nuriya M, Yuan JP, Worley PF, Leahy DJ. Structure of the Homer EVH1 domain-peptide complex reveals a new twist in polyproline recognition. Neuron. 2000;26:143–154. doi: 10.1016/s0896-6273(00)81145-9. [DOI] [PubMed] [Google Scholar]
- 20.Berman DM, Gilman AG. Mammalian RGS proteins: barbarians at the gate. J Biol Chem. 1998;273:1269–1272. doi: 10.1074/jbc.273.3.1269. [DOI] [PubMed] [Google Scholar]
- 21.Berridge M. Second messenger dualism in neuromodulation and memory. Nature. 1986;323:294–295. doi: 10.1038/323294a0. [DOI] [PubMed] [Google Scholar]
- 22.Blendy JA, Kaestner KH, Schmid W, Gass P, Schutz G. Targeting of the CREB gene leads to up-regulation of a novel CREB mRNA isoform. EMBO J. 1996;15:1098–1106. [PMC free article] [PubMed] [Google Scholar]
- 23.Bliss TV, Collingridge GL. A synaptic model of memory: long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- 24.Bliss TV, Lomo T. Long-lasting potentiation of synaptic transmission in the dentate area of the anaesthetized rabbit following stimulation of the perforant path. J Physiol. 1973;232:331–356. doi: 10.1113/jphysiol.1973.sp010273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bolen JB, Thiele CJ, Israel MA, Yonemoto W, Lipsich LA, Brugge JS. Enhancement of cellular src gene product associated tyrosyl kinase activity following polyoma virus infection and transformation. Cell. 1984;38:767–777. doi: 10.1016/0092-8674(84)90272-1. [DOI] [PubMed] [Google Scholar]
- 26.Bos TJ, Monteclaro FS, Mitsunobu F, Ball AR, Jr, Chang CH, Nishimura T, Vogt PK. Efficient transformation of chicken embryo fibroblasts by c-Jun requires structural modification in coding and noncoding sequences. Genes Dev. 1990;4:1677–1687. doi: 10.1101/gad.4.10.1677. [DOI] [PubMed] [Google Scholar]
- 27.Bottai D, Guzowski JF, Schwarz MK, Kang SH, Xiao B, Lanahan A, Worley PF, Seeburg PH. Synaptic activity-induced conversion of intronic to exonic sequence in Homer 1 immediate early gene expression. J Neurosci. 2002;22:167–175. doi: 10.1523/JNEUROSCI.22-01-00167.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bourtchuladze R, Frenguelli B, Blendy J, Cioffi D, Schutz G, Silva AJ. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell. 1994;79:59–68. doi: 10.1016/0092-8674(94)90400-6. [DOI] [PubMed] [Google Scholar]
- 29.Bradbury A, Possenti R, Shooter EM, Tirone F. Molecular cloning of PC3, a putatively secreted protein whose mRNA is induced by nerve growth factor and depolarization. Proc Natl Acad Sci USA. 1991;88:3353–3357. doi: 10.1073/pnas.88.8.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brakeman PR, Lanahan AA, O’Brien R, Roche K, Barnes CA, Huganir RL, Worley PF. Homer: a protein that selectively binds metabotropic glutamate receptors. Nature. 1997;386:284–288. doi: 10.1038/386284a0. [DOI] [PubMed] [Google Scholar]
- 31.Buonomano DV, Merzenich MM. Cortical plasticity: from synapses to maps. Annu Rev Neurosci. 1998;21:149–186. doi: 10.1146/annurev.neuro.21.1.149. [DOI] [PubMed] [Google Scholar]
- 32.Burchett SA, Volk ML, Bannon MJ, Granneman JG. Regulators of G protein signaling: rapid changes in mRNA abundance in response to amphetamine. J Neurochem. 1998;70:2216–2219. doi: 10.1046/j.1471-4159.1998.70052216.x. [DOI] [PubMed] [Google Scholar]
- 33.Byrne JH. Cellular analysis of associative learning. Physiol Rev. 1987;67:329–439. doi: 10.1152/physrev.1987.67.2.329. [DOI] [PubMed] [Google Scholar]
- 34.Cabelli RJ, Hohn A, Shatz CJ. Inhibition of ocular dominance column formation by infusion of NT-4/5 or BDNF. Science. 1995;267:1662–1666. doi: 10.1126/science.7886458. [DOI] [PubMed] [Google Scholar]
- 35.Cabelli RJ, Shelton DL, Segal RA, Shatz CJ. Blockade of endogenous ligands of trkB inhibits formation of ocular dominance columns. Neuron. 1997;19:63–76. doi: 10.1016/s0896-6273(00)80348-7. [DOI] [PubMed] [Google Scholar]
- 36.Cantallops I, Cline HT. Rapid activity-dependent delivery of the neurotrophic protein CPG15 to the axon surface of neurons in intact Xenopus tadpoles. Dev Neurobiol. 2008;68:744–759. doi: 10.1002/dneu.20529. [DOI] [PubMed] [Google Scholar]
- 37.Cantallops I, Haas K, Cline HT. Postsynaptic CPG15 promotes synaptic maturation and presynaptic axon arbor elaboration in vivo. Nat Neurosci. 2000;3:1004–1011. doi: 10.1038/79823. [DOI] [PubMed] [Google Scholar]
- 38.Capdevila JH, Falck JR, Estabrook RW. Cytochrome P450 and the arachidonate cascade. FASEB J. 1992;6:731–736. doi: 10.1096/fasebj.6.2.1537463. [DOI] [PubMed] [Google Scholar]
- 39.Carew TJ, Hawkins RD, Kandel ER. Differential classical conditioning of a defensive withdrawal reflex in Aplysia californica. Science. 1983;219:397–400. doi: 10.1126/science.6681571. [DOI] [PubMed] [Google Scholar]
- 40.Carroll RC, Beattie EC, Xia H, Luscher C, Altschuler Y, Nicoll RA, Malenka RC, von Zastrow M. Dynamin-dependent endocytosis of ionotropic glutamate receptors. Proc Natl Acad Sci USA. 1999;96:14112–14117. doi: 10.1073/pnas.96.24.14112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Castren E, Zafra F, Thoenen H, Lindholm D. Light regulates expression of brain-derived neurotrophic factor mRNA in rat visual cortex. Proc Natl Acad Sci USA. 1992;89:9444–9448. doi: 10.1073/pnas.89.20.9444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chiu IM, Reddy EP, Givol D, Robbins KC, Tronick SR, Aaronson SA. Nucleotide sequence analysis identifies the human c-sis protooncogene as a structural gene for platelet-derived growth factor. Cell. 1984;37:123–129. doi: 10.1016/0092-8674(84)90307-6. [DOI] [PubMed] [Google Scholar]
- 43.Cho RW, Park JM, Wolff SB, Xu D, Hopf C, Kim JA, Reddy RC, Petralia RS, Perin MS, Linden DJ, Worley PF. mGluR1/5-dependent long-term depression requires the regulated ectodomain cleavage of neuronal pentraxin NPR by TACE. Neuron. 2008;57:858–871. doi: 10.1016/j.neuron.2008.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chowdhury S, Shepherd JD, Okuno H, Lyford G, Petralia RS, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 interacts with the endocytic machinery to regulate AMPA receptor trafficking. Neuron. 2006;52:445–459. doi: 10.1016/j.neuron.2006.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chrivia JC, Kwok RP, Lamb N, Hagiwara M, Montminy MR, Goodman RH. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- 46.Cochran BH, Reffel AC, Stiles CD. Molecular cloning of gene sequences regulated by platelet-derived growth factor. Cell. 1983;33:939–947. doi: 10.1016/0092-8674(83)90037-5. [DOI] [PubMed] [Google Scholar]
- 47.Cohen S, Greenberg ME. Communication between the synapse and nucleus in neuronal development, plasticity, and disease. Annu Rev Cell Dev Biol. 2008;24:183–209. doi: 10.1146/annurev.cellbio.24.110707.175235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cole AJ, Saffen DW, Baraban JM, Worley PF. Rapid increase of an immediate early gene messenger RNA in hippocampal neurons by synaptic NMDA receptor activation. Nature. 1989;340:474–476. doi: 10.1038/340474a0. [DOI] [PubMed] [Google Scholar]
- 49.Collingridge GL, Kehl SJ, McLennan H. The antagonism of amino acid-induced excitations of rat hippocampal CA1 neurones in vitro. J Physiol. 1983;334:19–31. doi: 10.1113/jphysiol.1983.sp014477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Colvis CM, Pollock JD, Goodman RH, Impey S, Dunn J, Mandel G, Champagne FA, Mayford M, Korzus E, Kumar A, Renthal W, Theobald DE, Nestler EJ. Epigenetic mechanisms and gene networks in the nervous system. J Neurosci. 2005;25:10379–10389. doi: 10.1523/JNEUROSCI.4119-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Corriveau RA, Huh GS, Shatz CJ. Regulation of class I MHC gene expression in the developing and mature CNS by neural activity. Neuron. 1998;21:505–520. doi: 10.1016/s0896-6273(00)80562-0. [DOI] [PubMed] [Google Scholar]
- 52.Corriveau RA, Shatz CJ, Nedivi E. Dynamic regulation of cpg15 during activity-dependent synaptic development in the mammalian visual system. J Neurosci. 1999;19:7999–8008. doi: 10.1523/JNEUROSCI.19-18-07999.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cottrell JR, Borok E, Horvath TL, Nedivi E. CPG2: a brain- and synapse-specific protein that regulates the endocytosis of glutamate receptors. Neuron. 2004;44:677–690. doi: 10.1016/j.neuron.2004.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Curran T, Franza BR., Jr Fos and Jun: the AP-1 connection. Cell. 1988;55:395–397. doi: 10.1016/0092-8674(88)90024-4. [DOI] [PubMed] [Google Scholar]
- 55.Curran T, Miller AD, Zokas L, Verma IM. Viral and cellular fos proteins: a comparative analysis. Cell. 1984;36:259–268. doi: 10.1016/0092-8674(84)90219-8. [DOI] [PubMed] [Google Scholar]
- 56.Curran T, Morgan JI. Superinduction of c-fos by nerve growth factor in the presence of peripherally active benzodiazepines. Science. 1985;229:1265–1268. doi: 10.1126/science.4035354. [DOI] [PubMed] [Google Scholar]
- 57.Dash PK, Hochner B, Kandel ER. Injection of the cAMP-responsive element into the nucleus of Aplysia sensory neurons blocks long-term facilitation. Nature. 1990;345:718–721. doi: 10.1038/345718a0. [DOI] [PubMed] [Google Scholar]
- 58.Davis HP, Squire LR. Protein synthesis and memory: a review. Psychol Bull. 1984;96:518–559. [PubMed] [Google Scholar]
- 59.DeFeo-Jones D, Tatchell K, Robinson LC, Sigal IS, Vass WC, Lowy DR, Scolnick EM. Mammalian and yeast ras gene products: biological function in their heterologous systems. Science. 1985;228:179–184. doi: 10.1126/science.3883495. [DOI] [PubMed] [Google Scholar]
- 60.Deisseroth K, Bito H, Tsien RW. Signaling from synapse to nucleus: postsynaptic CREB phosphorylation during multiple forms of hippocampal synaptic plasticity. Neuron. 1996;16:89–101. doi: 10.1016/s0896-6273(00)80026-4. [DOI] [PubMed] [Google Scholar]
- 61.Dodds DC, Omeis IA, Cushman SJ, Helms JA, Perin MS. Neuronal pentraxin receptor, a novel putative integral membrane pentraxin that interacts with neuronal pentraxin 1 and 2 and taipoxin-associated calcium-binding protein 49. J Biol Chem. 1997;272:21488–21494. doi: 10.1074/jbc.272.34.21488. [DOI] [PubMed] [Google Scholar]
- 62.Doolittle RF, Hunkapiller MW, Hood LE, Devare SG, Robbins KC, Aaronson SA, Antoniades HN. Simian sarcoma virus onc gene, v-sis, is derived from the gene (or genes) encoding a platelet-derived growth factor. Science. 1983;221:275–277. doi: 10.1126/science.6304883. [DOI] [PubMed] [Google Scholar]
- 63.Douglas RM, Dragunow M, Robertson HA. High-frequency discharge of dentate granule cells, but not long-term potentiation, induces c-fos protein. Brain Res. 1988;464:259–262. doi: 10.1016/0169-328x(88)90033-2. [DOI] [PubMed] [Google Scholar]
- 64.Downward J, Yarden Y, Mayes E, Scrace G, Totty N, Stockwell P, Ullrich A, Schlessinger J, Waterfield MD. Close similarity of epidermal growth factor receptor and v-erb-B oncogene protein sequences. Nature. 1984;307:521–527. doi: 10.1038/307521a0. [DOI] [PubMed] [Google Scholar]
- 65.Dragunow M, Peterson MR, Robertson HA. Presence of c-fos-like immunoreactivity in the adult rat brain. Eur J Pharmacol. 1987;135:113–114. doi: 10.1016/0014-2999(87)90767-9. [DOI] [PubMed] [Google Scholar]
- 66.Dragunow M, Robertson HA. Generalized seizures induce c-fos protein(s) in mammalian neurons. Neurosci Lett. 1987;82:157–161. doi: 10.1016/0304-3940(87)90121-2. [DOI] [PubMed] [Google Scholar]
- 67.Dragunow M, Robertson HA. Kindling stimulation induces c-fos protein(s) in granule cells of the rat dentate gyrus. Nature. 1987;329:441–442. doi: 10.1038/329441a0. [DOI] [PubMed] [Google Scholar]
- 68.Dumuis A, Sebben M, Haynes L, Pin JP, Bockaert J. NMDA receptors activate the arachidonic acid cascade system in striatal neurons. Nature. 1988;336:68–70. doi: 10.1038/336068a0. [DOI] [PubMed] [Google Scholar]
- 69.Eckerdt F, Yuan J, Strebhardt K. Polo-like kinases and oncogenesis. Oncogene. 2005;24:267–276. doi: 10.1038/sj.onc.1208273. [DOI] [PubMed] [Google Scholar]
- 70.Ehlers MD. Reinsertion or degradation of AMPA receptors determined by activity-dependent endocytic sorting. Neuron. 2000;28:511–525. doi: 10.1016/s0896-6273(00)00129-x. [DOI] [PubMed] [Google Scholar]
- 71.Eisenman RN, Tachibana CY, Abrams HD, Hann SR. V-myc-and c-myc-encoded proteins are associated with the nuclear matrix. Mol Cell Biol. 1985;5:114–126. doi: 10.1128/mcb.5.1.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Elliott RC, Miles MF, Lowenstein DH. Overlapping microarray profiles of dentate gyrus gene expression during development- and epilepsy-associated neurogenesis and axon outgrowth. J Neurosci. 2003;23:2218–2227. doi: 10.1523/JNEUROSCI.23-06-02218.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Emsley J, White HE, O’Hara BP, Oliva G, Srinivasan N, Tickle IJ, Blundell TL, Pepys MB, Wood SP. Structure of pentameric human serum amyloid P component. Nature. 1994;367:338–345. doi: 10.1038/367338a0. [DOI] [PubMed] [Google Scholar]
- 74.Ernfors P, Bengzon J, Kokaia Z, Persson H, Lindvall O. Increased levels of messenger RNAs for neurotrophic factors in the brain during kindling epileptogenesis. Neuron. 1991;7:165–176. doi: 10.1016/0896-6273(91)90084-d. [DOI] [PubMed] [Google Scholar]
- 75.Ernfors P, Ibanez CF, Ebendal T, Olson L, Persson H. Molecular cloning and neurotrophic activities of a protein with structural similarities to nerve growth factor: developmental and topographical expression in the brain. Proc Natl Acad Sci USA. 1990;87:5454–5458. doi: 10.1073/pnas.87.14.5454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Etter PD, Narayanan R, Navratilova Z, Patel C, Bohmann D, Jasper H, Ramaswami M. Synaptic and genomic responses to JNK and AP-1 signaling in Drosophila neurons. BMC Neurosci. 2005;6:39. doi: 10.1186/1471-2202-6-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Fagni L, Worley PF, Ango F. Sci STKE 2002. 2002. Homer as both a scaffold and transduction molecule; p. RE8. [DOI] [PubMed] [Google Scholar]
- 78.Figurov A, Pozzo-Miller LD, Olafsson P, Wang T, Lu B. Regulation of synaptic responses to high-frequency stimulation and LTP by neurotrophins in the hippocampus. Nature. 1996;381:706–709. doi: 10.1038/381706a0. [DOI] [PubMed] [Google Scholar]
- 79.Flavell SW, Cowan CW, Kim TK, Greer PL, Lin Y, Paradis S, Griffith EC, Hu LS, Chen C, Greenberg ME. Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science. 2006;311:1008–1012. doi: 10.1126/science.1122511. [DOI] [PubMed] [Google Scholar]
- 80.Flavell SW, Greenberg ME. Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system. Annu Rev Neurosci. 2008;31:563–590. doi: 10.1146/annurev.neuro.31.060407.125631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Flavell SW, Kim TK, Gray JM, Harmin DA, Hemberg M, Hong EJ, Markenscoff-Papadimitriou E, Bear DM, Greenberg ME. Genome-wide analysis of MEF2 transcriptional program reveals synaptic target genes and neuronal activity-dependent polyadenylation site selection. Neuron. 2008;60:1022–1038. doi: 10.1016/j.neuron.2008.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Flexner JB, Flexner LB, Stellar E. Memory in mice as affected by intracerebral puromycin. Science. 1963;141:57–59. doi: 10.1126/science.141.3575.57. [DOI] [PubMed] [Google Scholar]
- 83.French PJ, O’Connor V, Jones MW, Davis S, Errington ML, Voss K, Truchet B, Wotjak C, Stean T, Doyere V, Maroun M, Laroche S, Bliss TV. Subfield-specific immediate early gene expression associated with hippocampal long-term potentiation in vivo. Eur J Neurosci. 2001;13:968–976. doi: 10.1046/j.0953-816x.2001.01467.x. [DOI] [PubMed] [Google Scholar]
- 84.French PJ, O’Connor V, Voss K, Stean T, Hunt SP, Bliss TV. Seizure-induced gene expression in area CA1 of the mouse hippocampus. Eur J Neurosci. 2001;14:2037–2041. doi: 10.1046/j.0953-816x.2001.01818.x. [DOI] [PubMed] [Google Scholar]
- 85.Frey U, Huang YY, Kandel ER. Effects of cAMP simulate a late stage of LTP in hippocampal CA1 neurons. Science. 1993;260:1661–1664. doi: 10.1126/science.8389057. [DOI] [PubMed] [Google Scholar]
- 86.Frost WN, Castellucci VF, Hawkins RD, Kandel ER. Monosynaptic connections made by the sensory neurons of the gill- and siphon-withdrawal reflex in Aplysia participate in the storage of long-term memory for sensitization. Proc Natl Acad Sci USA. 1985;82:8266–8269. doi: 10.1073/pnas.82.23.8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Fujino T, Lee WC, Nedivi E. Regulation of cpg15 by signaling pathways that mediate synaptic plasticity. Mol Cell Neurosci. 2003;24:538–554. doi: 10.1016/s1044-7431(03)00230-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gao M, Morgan I, Vogt PK. Differential and antagonistic effects of v-Jun and c-Jun. Cancer Res. 1996;56:4229–4235. [PubMed] [Google Scholar]
- 89.Garner CC, Tucker RP, Matus A. Selective localization of messenger RNA for cytoskeletal protein MAP2 in dendrites. Nature. 1988;336:674–677. doi: 10.1038/336674a0. [DOI] [PubMed] [Google Scholar]
- 90.Gass P, Wolfer DP, Balschun D, Rudolph D, Frey U, Lipp HP, Schutz G. Deficits in memory tasks of mice with CREB mutations depend on gene dosage. Learn Mem. 1998;5:274–288. [PMC free article] [PubMed] [Google Scholar]
- 91.Gilman MZ, Wilson RN, Weinberg RA. Multiple protein-binding sites in the 5′-flanking region regulate c-fos expression. Mol Cell Biol. 1986;6:4305–4316. doi: 10.1128/mcb.6.12.4305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Giorgi C, Yeo GW, Stone ME, Katz DB, Burge C, Turrigiano G, Moore MJ. The EJC factor eIF4AIII modulates synaptic strength and neuronal protein expression. Cell. 2007;130:179–191. doi: 10.1016/j.cell.2007.05.028. [DOI] [PubMed] [Google Scholar]
- 93.Goelet P, Castellucci VF, Schacher S, Kandel ER. The long and the short of long-term memory–a molecular framework. Nature. 1986;322:419–422. doi: 10.1038/322419a0. [DOI] [PubMed] [Google Scholar]
- 94.Gonzalez GA, Montminy MR. Cyclic AMP stimulates somatostatin gene transcription by phosphorylation of CREB at serine 133. Cell. 1989;59:675–680. doi: 10.1016/0092-8674(89)90013-5. [DOI] [PubMed] [Google Scholar]
- 95.Greenberg ME, Greene LA, Ziff EB. Nerve growth factor and epidermal growth factor induce rapid transient changes in protooncogene transcription in PC12 cells. J Biol Chem. 1985;260:14101–14110. [PubMed] [Google Scholar]
- 96.Greenberg ME, Ziff EB. Stimulation of 3T3 cells induces transcription of the c-fos protooncogene. Nature. 1984;311:433–438. doi: 10.1038/311433a0. [DOI] [PubMed] [Google Scholar]
- 97.Guan Z, Saraswati S, Adolfsen B, Littleton JT. Genome-wide transcriptional changes associated with enhanced activity in the Drosophila nervous system. Neuron. 2005;48:91–107. doi: 10.1016/j.neuron.2005.08.036. [DOI] [PubMed] [Google Scholar]
- 98.Guthrie KM, Anderson AJ, Leon M, Gall C. Odor-induced increases in c-fos mRNA expression reveal an anatomical “unit” for odor processing in olfactory bulb. Proc Natl Acad Sci USA. 1993;90:3329–3333. doi: 10.1073/pnas.90.8.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Guzowski JF, Lyford GL, Stevenson GD, Houston FP, Mc-Gaugh JL, Worley PF, Barnes CA. Inhibition of activity-dependent arc protein expression in the rat hippocampus impairs the maintenance of long-term potentiation and the consolidation of long-term memory. J Neurosci. 2000;20:3993–4001. doi: 10.1523/JNEUROSCI.20-11-03993.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Guzowski JF, Timlin JA, Roysam B, McNaughton BL, Worley PF, Barnes CA. Mapping behaviorally relevant neural circuits with immediate-early gene expression. Curr Opin Neurobiol. 2005;15:599–606. doi: 10.1016/j.conb.2005.08.018. [DOI] [PubMed] [Google Scholar]
- 101.Hai T, Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc Natl Acad Sci USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hai TW, Liu F, Coukos WJ, Green MR. Transcription factor ATF cDNA clones: an extensive family of leucine zipper proteins able to selectively form DNA-binding heterodimers. Genes Dev. 1989;3:2083–2090. doi: 10.1101/gad.3.12b.2083. [DOI] [PubMed] [Google Scholar]
- 103.Han J, Mark MD, Li X, Xie M, Waka S, Rettig J, Herlitze S. RGS2 determines short-term synaptic plasticity in hippocampal neurons by regulating Gi/o-mediated inhibition of presynaptic Ca2+ channels. Neuron. 2006;51:575–586. doi: 10.1016/j.neuron.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 104.Han Y, Chen X, Shi F, Li S, Huang J, Xie M, Hu L, Hoidal JR, Xu P. CPG15, a new factor upregulated after ischemic brain injury, contributes to neuronal network re-establishment after glutamate-induced injury. J Neurotrauma. 2007;24:722–731. doi: 10.1089/neu.2006.0174. [DOI] [PubMed] [Google Scholar]
- 105.Hann SR, Abrams HD, Rohrschneider LR, Eisenman RN. Proteins encoded by v-myc and c-myc oncogenes: identification and localization in acute leukemia virus transformants and bursal lymphoma cell lines. Cell. 1983;34:789–798. doi: 10.1016/0092-8674(83)90535-4. [DOI] [PubMed] [Google Scholar]
- 106.Hanover JL, Huang ZJ, Tonegawa S, Stryker MP. Brain-derived neurotrophic factor overexpression induces precocious critical period in mouse visual cortex. J Neurosci. 1999;19:RC40. doi: 10.1523/JNEUROSCI.19-22-j0003.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Harwell C, Burbach B, Svoboda K, Nedivi E. Regulation of cpg15 expression during single whisker experience in the barrel cortex of adult mice. J Neurobiol. 2005;65:85–96. doi: 10.1002/neu.20176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Hernandez PJ, Abel T. The role of protein synthesis in memory consolidation: progress amid decades of debate. Neurobiol Learn Mem. 2008;89:293–311. doi: 10.1016/j.nlm.2007.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hertting G, Seregi A. Formation and function of eicosanoids in the central nervous system. Ann NY Acad Sci. 1989;559:84–99. doi: 10.1111/j.1749-6632.1989.tb22600.x. [DOI] [PubMed] [Google Scholar]
- 110.Hevroni D, Rattner A, Bundman M, Lederfein D, Gabarah A, Mangelus M, Silverman MA, Kedar H, Naor C, Kornuc M, Hanoch T, Seger R, Theill LE, Nedivi E, Richter-Levin G, Citri Y. Hippocampal plasticity involves extensive gene induction and multiple cellular mechanisms. J Mol Neurosci. 1998;10:75–98. doi: 10.1007/BF02737120. [DOI] [PubMed] [Google Scholar]
- 111.Hoeffler JP, Meyer TE, Yun Y, Jameson JL, Habener JF. Cyclic AMP-responsive DNA-binding protein: structure based on a cloned placental cDNA. Science. 1988;242:1430–1433. doi: 10.1126/science.2974179. [DOI] [PubMed] [Google Scholar]
- 112.Hofer M, Pagliusi SR, Hohn A, Leibrock J, Barde YA. Regional distribution of brain-derived neurotrophic factor mRNA in the adult mouse brain. EMBO J. 1990;9:2459–2464. doi: 10.1002/j.1460-2075.1990.tb07423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hofer MM, Barde YA. Brain-derived neurotrophic factor prevents neuronal death in vivo. Nature. 1988;331:261–262. doi: 10.1038/331261a0. [DOI] [PubMed] [Google Scholar]
- 114.Hofer SB, Mrsic-Flogel TD, Bonhoeffer T, Hubener M. Lifelong learning: ocular dominance plasticity in mouse visual cortex. Curr Opin Neurobiol. 2006;16:451–459. doi: 10.1016/j.conb.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 115.Hong SJ, Li H, Becker KG, Dawson VL, Dawson TM. Identification and analysis of plasticity-induced late-response genes. Proc Natl Acad Sci USA. 2004;101:2145–2150. doi: 10.1073/pnas.0305170101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Hooks BM, Chen C. Critical periods in the visual system: changing views for a model of experience-dependent plasticity. Neuron. 2007;56:312–326. doi: 10.1016/j.neuron.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 117.Huang YY, Kandel ER. Recruitment of long-lasting and protein kinase A-dependent long-term potentiation in the CA1 region of hippocampus requires repeated tetanization. Learn Mem. 1994;1:74–82. [PubMed] [Google Scholar]
- 118.Huang YY, Li XC, Kandel ER. cAMP contributes to mossy fiber LTP by initiating both a covalently mediated early phase and macromolecular synthesis-dependent late phase. Cell. 1994;79:69–79. doi: 10.1016/0092-8674(94)90401-4. [DOI] [PubMed] [Google Scholar]
- 119.Huang ZJ, Kirkwood A, Pizzorusso T, Porciatti V, Morales B, Bear MF, Maffei L, Tonegawa S. BDNF regulates the maturation of inhibition and the critical period of plasticity in mouse visual cortex. Cell. 1999;98:739–755. doi: 10.1016/s0092-8674(00)81509-3. [DOI] [PubMed] [Google Scholar]
- 120.Hubel DH, Wiesel TN. Ferrier lecture. Functional architecture of macaque monkey visual cortex. Proc R Soc Lond B Biol Sci. 1977;198:1–59. doi: 10.1098/rspb.1977.0085. [DOI] [PubMed] [Google Scholar]
- 121.Huh GS, Boulanger LM, Du H, Riquelme PA, Brotz TM, Shatz CJ. Functional requirement for class I MHC in CNS development and plasticity. Science. 2000;290:2155–2159. doi: 10.1126/science.290.5499.2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Hummler E, Cole TJ, Blendy JA, Ganss R, Aguzzi A, Schmid W, Beermann F, Schutz G. Targeted mutation of the CREB gene: compensation within the CREB/ATF family of transcription factors. Proc Natl Acad Sci USA. 1994;91:5647–5651. doi: 10.1073/pnas.91.12.5647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Hunt SP, Pini A, Evan G. Induction of c-fos-like protein in spinal cord neurons following sensory stimulation. Nature. 1987;328:632–634. doi: 10.1038/328632a0. [DOI] [PubMed] [Google Scholar]
- 124.Iacoangeli A, Rozhdestvensky TS, Dolzhanskaya N, Tournier B, Schutt J, Brosius J, Denman RB, Khandjian EW, Kindler S, Tiedge H. On BC1 RNA and the fragile X mental retardation protein. Proc Natl Acad Sci USA. 2008;105:734–739. doi: 10.1073/pnas.0710991105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Iba H, Takeya T, Cross FR, Hanafusa T, Hanafusa H. Rous sarcoma virus variants that carry the cellular src gene instead of the viral src gene cannot transform chicken embryo fibroblasts. Proc Natl Acad Sci USA. 1984;81:4424–4428. doi: 10.1073/pnas.81.14.4424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Impey S, Mark M, Villacres EC, Poser S, Chavkin C, Storm DR. Induction of CRE-mediated gene expression by stimuli that generate long-lasting LTP in area CA1 of the hippocampus. Neuron. 1996;16:973–982. doi: 10.1016/s0896-6273(00)80120-8. [DOI] [PubMed] [Google Scholar]
- 127.Impey S, McCorkle SR, Cha-Molstad H, Dwyer JM, Yochum GS, Boss JM, McWeeney S, Dunn JJ, Mandel G, Goodman RH. Defining the CREB regulon: a genome-wide analysis of transcription factor regulatory regions. Cell. 2004;119:1041–1054. doi: 10.1016/j.cell.2004.10.032. [DOI] [PubMed] [Google Scholar]
- 128.Ingi T, Krumins AM, Chidiac P, Brothers GM, Chung S, Snow BE, Barnes CA, Lanahan AA, Siderovski DP, Ross EM, Gilman AG, Worley PF. Dynamic regulation of RGS2 suggests a novel mechanism in G-protein signaling and neuronal plasticity. J Neurosci. 1998;18:7178–7188. doi: 10.1523/JNEUROSCI.18-18-07178.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Inoue A, Sanes JR. Lamina-specific connectivity in the brain: regulation by N-cadherin, neurotrophins, and glycoconjugates. Science. 1997;276:1428–1431. doi: 10.1126/science.276.5317.1428. [DOI] [PubMed] [Google Scholar]
- 130.Ip NY, Stitt TN, Tapley P, Klein R, Glass DJ, Fandl J, Greene LA, Barbacid M, Yancopoulos GD. Similarities and differences in the way neurotrophins interact with the Trk receptors in neuronal and nonneuronal cells. Neuron. 1993;10:137–149. doi: 10.1016/0896-6273(93)90306-c. [DOI] [PubMed] [Google Scholar]
- 131.Isackson PJ, Huntsman MM, Murray KD, Gall CM. BDNF mRNA expression is increased in adult rat forebrain after limbic seizures: temporal patterns of induction distinct from NGF. Neuron. 1991;6:937–948. doi: 10.1016/0896-6273(91)90234-q. [DOI] [PubMed] [Google Scholar]
- 132.Johnson RS, Spiegelman BM, Papaioannou V. Pleiotropic effects of a null mutation in the c-fos protooncogene. Cell. 1992;71:577–586. doi: 10.1016/0092-8674(92)90592-z. [DOI] [PubMed] [Google Scholar]
- 133.Johnsson A, Betsholtz C, von der Helm K, Heldin CH, Westermark B. Platelet-derived growth factor agonist activity of a secreted form of the v-sis oncogene product. Proc Natl Acad Sci USA. 1985;82:1721–1725. doi: 10.1073/pnas.82.6.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Johnsson A, Heldin CH, Wasteson A, Westermark B, Deuel TF, Huang JS, Seeburg PH, Gray A, Ullrich A, Scrace G. The c-sis gene encodes a precursor of the B chain of platelet-derived growth factor. EMBO J. 1984;3:921–928. doi: 10.1002/j.1460-2075.1984.tb01908.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Jones MW, Errington ML, French PJ, Fine A, Bliss TV, Garel S, Charnay P, Bozon B, Laroche S, Davis S. A requirement for the immediate early gene Zif268 in the expression of late LTP and long-term memories. Nat Neurosci. 2001;4:289–296. doi: 10.1038/85138. [DOI] [PubMed] [Google Scholar]
- 136.Kaang BK, Kandel ER, Grant SG. Activation of cAMP-responsive genes by stimuli that produce long-term facilitation in Aplysia sensory neurons. Neuron. 1993;10:427–435. doi: 10.1016/0896-6273(93)90331-k. [DOI] [PubMed] [Google Scholar]
- 137.Kandel ER, Schwarz JH, Jessel TM. Principles of Neural Science. 4. New York: McGraw-Hill; 2000. Cellular mechanisms of learning and the biological basis of individuality; pp. 1247–1279. [Google Scholar]
- 138.Kang H, Schuman EM. Long-lasting neurotrophin-induced enhancement of synaptic transmission in the adult hippocampus. Science. 1995;267:1658–1662. doi: 10.1126/science.7886457. [DOI] [PubMed] [Google Scholar]
- 139.Karamoysoyli E, Burnand RC, Tomlinson DR, Gardiner NJ. Neuritin mediates nerve growth factor-induced axonal regeneration and is deficient in experimental diabetic neuropathy. Diabetes. 2008;57:181–189. doi: 10.2337/db07-0895. [DOI] [PubMed] [Google Scholar]
- 140.Kaufmann WE, Worley PF, Pegg J, Bremer M, Isakson P. COX-2, a synaptically induced enzyme, is expressed by excitatory neurons at postsynaptic sites in rat cerebral cortex. Proc Natl Acad Sci USA. 1996;93:2317–2321. doi: 10.1073/pnas.93.6.2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Kaufmann WE, Yamagata K, Andreasson KI, Worley PF. Rapid response genes as markers of cellular signaling during cortical histogenesis: their potential in understanding mental retardation. Int J Dev Neurosci. 1994;12:263–271. doi: 10.1016/0736-5748(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 142.Kauselmann G, Weiler M, Wulff P, Jessberger S, Konietzko U, Scafidi J, Staubli U, Bereiter-Hahn J, Strebhardt K, Kuhl D. The polo-like protein kinases Fnk and Snk associate with a Ca2+-and integrin-binding protein and are regulated dynamically with synaptic plasticity. EMBO J. 1999;18:5528–5539. doi: 10.1093/emboj/18.20.5528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Kawashima T, Okuno H, Nonaka M, Adachi-Morishima A, Kyo N, Okamura M, Takemoto-Kimura S, Worley PF, Bito H. Synaptic activity-responsive element in the Arc/Arg3.1 promoter essential for synapse-to-nucleus signaling in activated neurons. Proc Natl Acad Sci USA. 2009;106:316–321. doi: 10.1073/pnas.0806518106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Kelm O, Wind M, Lehmann WD, Nigg EA. Cell cycle-regulated phosphorylation of the Xenopus polo-like kinase Plx1. J Biol Chem. 2002;277:25247–25256. doi: 10.1074/jbc.M202855200. [DOI] [PubMed] [Google Scholar]
- 145.Kim JY, Zeng W, Kiselyov K, Yuan JP, Dehoff MH, Mikoshiba K, Worley PF, Muallem S. Homer 1 mediates store- and inositol 1,4,5-trisphosphate receptor-dependent translocation and retrieval of TRPC3 to the plasma membrane. J Biol Chem. 2006;281:32540–32549. doi: 10.1074/jbc.M602496200. [DOI] [PubMed] [Google Scholar]
- 146.Kingston RE, Baldwin AS, Jr, Sharp PA. Regulation of heat shock protein 70 gene expression by c-myc. Nature. 1984;312:280–282. doi: 10.1038/312280a0. [DOI] [PubMed] [Google Scholar]
- 147.Kitamura C, Takahashi M, Kondoh Y, Tashiro H, Tashiro T. Identification of synaptic activity-dependent genes by exposure of cultured cortical neurons to tetrodotoxin followed by its withdrawal. J Neurosci Res. 2007;85:2385–2399. doi: 10.1002/jnr.21391. [DOI] [PubMed] [Google Scholar]
- 148.Korte M, Carroll P, Wolf E, Brem G, Thoenen H, Bonhoeffer T. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc Natl Acad Sci USA. 1995;92:8856–8860. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Kruijer W, Schubert D, Verma IM. Induction of the protooncogene fos by nerve growth factor. Proc Natl Acad Sci USA. 1985;82:7330–7334. doi: 10.1073/pnas.82.21.7330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Kwok RP, Lundblad JR, Chrivia JC, Richards JP, Bachinger HP, Brennan RG, Roberts SG, Green MR, Goodman RH. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature. 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- 151.Lachance PE, Chaudhuri A. Microarray analysis of developmental plasticity in monkey primary visual cortex. J Neurochem. 2004;88:1455–1469. doi: 10.1046/j.1471-4159.2003.02274.x. [DOI] [PubMed] [Google Scholar]
- 152.Lamas M, Monaco L, Zazopoulos E, Lalli E, Tamai K, Penna L, Mazzucchelli C, Nantel F, Foulkes NS, Sassone-Corsi P. CREM: a master-switch in the transcriptional response to cAMP. Philos Trans R Soc Lond B Biol Sci. 1996;351:561–567. doi: 10.1098/rstb.1996.0055. [DOI] [PubMed] [Google Scholar]
- 153.Lanahan A, Worley P. Immediate-early genes and synaptic function. Neurobiol Learn Mem. 1998;70:37–43. doi: 10.1006/nlme.1998.3836. [DOI] [PubMed] [Google Scholar]
- 154.Laoide BM, Foulkes NS, Schlotter F, Sassone-Corsi P. The functional versatility of CREM is determined by its modular structure. EMBO J. 1993;12:1179–1191. doi: 10.1002/j.1460-2075.1993.tb05759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Lau LF, Nathans D. Identification of a set of genes expressed during the G0/G1 transition of cultured mouse cells. EMBO J. 1985;4:3145–3151. doi: 10.1002/j.1460-2075.1985.tb04057.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Lee SH, Liu L, Wang YT, Sheng M. Clathrin adaptor AP2 and NSF interact with overlapping sites of GluR2 and play distinct roles in AMPA receptor trafficking and hippocampal LTD. Neuron. 2002;36:661–674. doi: 10.1016/s0896-6273(02)01024-3. [DOI] [PubMed] [Google Scholar]
- 157.Lee WC, Nedivi E. Extended plasticity of visual cortex in dark-reared animals may result from prolonged expression of cpg15-like genes. J Neurosci. 2002;22:1807–1815. doi: 10.1523/JNEUROSCI.22-05-01807.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Lein ES, Hawrylycz MJ, Ao N, Ayres M, Bensinger A, Bernard A, Boe AF, Boguski MS, Brockway KS, Byrnes EJ, Chen L, Chen TM, Chin MC, Chong J, Crook BE, Czaplinska A, Dang CN, Datta S, Dee NR, Desaki AL, Desta T, Diep E, Dolbeare TA, Donelan MJ, Dong HW, Dougherty JG, Duncan BJ, Ebbert AJ, Eichele G, Estin LK, Faber C, Facer BA, Fields R, Fischer SR, Fliss TP, Frensley C, Gates SN, Glattfelder KJ, Halverson KR, Hart MR, Hohmann JG, Howell MP, Jeung DP, Johnson RA, Karr PT, Kawal R, Kidney JM, Knapik RH, Kuan CL, Lake JH, Laramee AR, Larsen KD, Lau C, Lemon TA, Liang AJ, Liu Y, Luong LT, Michaels J, Morgan JJ, Morgan RJ, Mortrud MT, Mosqueda NF, Ng LL, Ng R, Orta GJ, Overly CC, Pak TH, Parry SE, Pathak SD, Pearson OC, Puchalski RB, Riley ZL, Rockett HR, Rowland SA, Royall JJ, Ruiz MJ, Sarno NR, Schaffnit K, Shapovalova NV, Sivisay T, Slaughterbeck CR, Smith SC, Smith KA, Smith BI, Sodt AJ, Stewart NN, Stumpf KR, Sunkin SM, Sutram M, Tam A, Teemer CD, Thaller C, Thompson CL, Varnam LR, Visel A, Whitlock RM, Wohnoutka PE, Wolkey CK, Wong VY, Wood M. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 159.Lemmon V, Burden SM, Payne HR, Elmslie GJ, Hlavin ML. Neurite growth on different substrates: permissive versus instructive influences and the role of adhesive strength. J Neurosci. 1992;12:818–826. doi: 10.1523/JNEUROSCI.12-03-00818.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.LeVay S, Stryker MP, Shatz CJ. Ocular dominance columns and their development in layer IV of the cat’s visual cortex: a quantitative study. J Comp Neurol. 1978;179:223–244. doi: 10.1002/cne.901790113. [DOI] [PubMed] [Google Scholar]
- 161.Lim RW, Varnum BC, Herschman HR. Cloning of tetradecanoyl phorbol ester-induced “primary response” sequences and their expression in density-arrested Swiss 3T3 cells and a TPA non-proliferative variant. Oncogene. 1987;1:263–270. [PubMed] [Google Scholar]
- 162.Lin JW, Ju W, Foster K, Lee SH, Ahmadian G, Wyszynski M, Wang YT, Sheng M. Distinct molecular mechanisms and divergent endocytotic pathways of AMPA receptor internalization. Nat Neurosci. 2000;3:1282–1290. doi: 10.1038/81814. [DOI] [PubMed] [Google Scholar]
- 163.Lin SS, Levitan IB. Concanavalin A alters synaptic specificity between cultured Aplysia neurons. Science. 1987;237:648–650. doi: 10.1126/science.3603045. [DOI] [PubMed] [Google Scholar]
- 164.Lin SS, Levitan IB. Concanavalin A: a tool to investigate neuronal plasticity. Trends Neurosci. 1991;14:273–277. doi: 10.1016/0166-2236(91)90136-i. [DOI] [PubMed] [Google Scholar]
- 165.Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Link W, Konietzko U, Kauselmann G, Krug M, Schwanke B, Frey U, Kuhl D. Somatodendritic expression of an immediate early gene is regulated by synaptic activity. Proc Natl Acad Sci USA. 1995;92:5734–5738. doi: 10.1073/pnas.92.12.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Liu QR, Lu L, Zhu XG, Gong JP, Shaham Y, Uhl GR. Rodent BDNF genes, novel promoters, novel splice variants, and regulation by cocaine. Brain Res. 2006;1067:1–12. doi: 10.1016/j.brainres.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 168.Llamazares S, Moreira A, Tavares A, Girdham C, Spruce BA, Gonzalez C, Karess RE, Glover DM, Sunkel CE. Polo encodes a protein kinase homolog required for mitosis in Drosophila. Genes Dev. 1991;5:2153–2165. doi: 10.1101/gad.5.12a.2153. [DOI] [PubMed] [Google Scholar]
- 169.Lu J, Helton TD, Blanpied TA, Racz B, Newpher TM, Weinberg RJ, Ehlers MD. Postsynaptic positioning of endocytic zones and AMPA receptor cycling by physical coupling of dynamin-3 to Homer. Neuron. 2007;55:874–889. doi: 10.1016/j.neuron.2007.06.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Lyford GL, Yamagata K, Kaufmann WE, Barnes CA, Sanders LK, Copeland NG, Gilbert DJ, Jenkins NA, Lanahan AA, Worley PF. Arc, a growth factor and activity-regulated gene, encodes a novel cytoskeleton-associated protein that is enriched in neuronal dendrites. Neuron. 1995;14:433–445. doi: 10.1016/0896-6273(95)90299-6. [DOI] [PubMed] [Google Scholar]
- 171.Maisonpierre PC, Belluscio L, Friedman B, Alderson RF, Wiegand SJ, Furth ME, Lindsay RM, Yancopoulos GD. NT-3, BDNF, and NGF in the developing rat nervous system: parallel as well as reciprocal patterns of expression. Neuron. 1990;5:501–509. doi: 10.1016/0896-6273(90)90089-x. [DOI] [PubMed] [Google Scholar]
- 172.Majdan M, Shatz CJ. Effects of visual experience on activity-dependent gene regulation in cortex. Nat Neurosci. 2006;9:650–659. doi: 10.1038/nn1674. [DOI] [PubMed] [Google Scholar]
- 173.Malhotra S, Shafiq N, Pandhi P. COX-2 inhibitors: a CLASS act or Just VIGORously promoted. Med Gen Med. 2004;6:6. [PMC free article] [PubMed] [Google Scholar]
- 174.Malinow R, Malenka RC. AMPA receptor trafficking and synaptic plasticity. Annu Rev Neurosci. 2002;25:103–126. doi: 10.1146/annurev.neuro.25.112701.142758. [DOI] [PubMed] [Google Scholar]
- 175.Man HY, Lin JW, Ju WH, Ahmadian G, Liu L, Becker LE, Sheng M, Wang YT. Regulation of AMPA receptor-mediated synaptic transmission by clathrin-dependent receptor internalization. Neuron. 2000;25:649–662. doi: 10.1016/s0896-6273(00)81067-3. [DOI] [PubMed] [Google Scholar]
- 176.McAllister AK, Katz LC, Lo DC. Neurotrophin regulation of cortical dendritic growth requires activity. Neuron. 1996;17:1057–1064. doi: 10.1016/s0896-6273(00)80239-1. [DOI] [PubMed] [Google Scholar]
- 177.McAllister AK, Katz LC, Lo DC. Opposing roles for endogenous BDNF and NT-3 in regulating cortical dendritic growth. Neuron. 1997;18:767–778. doi: 10.1016/s0896-6273(00)80316-5. [DOI] [PubMed] [Google Scholar]
- 178.McAllister AK, Lo DC, Katz LC. Neurotrophins regulate dendritic growth in developing visual cortex. Neuron. 1995;15:791–803. doi: 10.1016/0896-6273(95)90171-x. [DOI] [PubMed] [Google Scholar]
- 179.McGaugh JL. Memory–a century of consolidation. Science. 2000;287:248–251. doi: 10.1126/science.287.5451.248. [DOI] [PubMed] [Google Scholar]
- 180.Merzenich MM, Recanzone GH, Jenkins WM, Grajski KA. Adaptive mechanisms in cortical networks underlying cortical contributions to learning and nondeclarative memory. Cold Spring Harb Symp Quant Biol. 1990;55:873–887. doi: 10.1101/sqb.1990.055.01.082. [DOI] [PubMed] [Google Scholar]
- 181.Mi R, Tang X, Sutter R, Xu D, Worley P, O’Brien RJ. Differing mechanisms for glutamate receptor aggregation on dendritic spines and shafts in cultured hippocampal neurons. J Neurosci. 2002;22:7606–7616. doi: 10.1523/JNEUROSCI.22-17-07606.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Mizumori SJ, Rosenzweig MR, Bennett EL. Long-term working memory in the rat: effects of hippocampally applied anisomycin. Behav Neurosci. 1985;99:220–232. doi: 10.1037//0735-7044.99.2.220. [DOI] [PubMed] [Google Scholar]
- 183.Molina CA, Foulkes NS, Lalli E, Sassone-Corsi P. Inducibility and negative autoregulation of CREM: an alternative promoter directs the expression of ICER, an early response repressor. Cell. 1993;75:875–886. doi: 10.1016/0092-8674(93)90532-u. [DOI] [PubMed] [Google Scholar]
- 184.Montarolo PG, Goelet P, Castellucci VF, Morgan J, Kandel ER, Schacher S. A critical period for macromolecular synthesis in long-term heterosynaptic facilitation in Aplysia. Science. 1986;234:1249–1254. doi: 10.1126/science.3775383. [DOI] [PubMed] [Google Scholar]
- 185.Montminy MR, Sevarino KA, Wagner JA, Mandel G, Goodman RH. Identification of a cyclic-AMP-responsive element within the rat somatostatin gene. Proc Natl Acad Sci USA. 1986;83:6682–6686. doi: 10.1073/pnas.83.18.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Morgan JI, Cohen DR, Hempstead JL, Curran T. Mapping patterns of c-fos expression in the central nervous system after seizure. Science. 1987;237:192–197. doi: 10.1126/science.3037702. [DOI] [PubMed] [Google Scholar]
- 187.Morgan JI, Curran T. Role of ion flux in the control of c-fos expression. Nature. 1986;322:552–555. doi: 10.1038/322552a0. [DOI] [PubMed] [Google Scholar]
- 188.Morris RG, Anderson E, Lynch GS, Baudry M. Selective impairment of learning and blockade of long-term potentiation by an N-methyl-D-aspartate receptor antagonist, AP5. Nature. 1986;319:774–776. doi: 10.1038/319774a0. [DOI] [PubMed] [Google Scholar]
- 189.Mulcahy LS, Smith MR, Stacey DW. Requirement for ras protooncogene function during serum-stimulated growth of NIH 3T3 cells. Nature. 1985;313:241–243. doi: 10.1038/313241a0. [DOI] [PubMed] [Google Scholar]
- 190.Muller R, Bravo R, Burckhardt J, Curran T. Induction of c-fos gene and protein by growth factors precedes activation of c-myc. Nature. 1984;312:716–720. doi: 10.1038/312716a0. [DOI] [PubMed] [Google Scholar]
- 191.Naeve GS, Ramakrishnan M, Kramer R, Hevroni D, Citri Y, Theill LE. Neuritin: a gene induced by neural activity and neurotrophins that promotes neuritogenesis. Proc Natl Acad Sci USA. 1997;94:2648–2653. doi: 10.1073/pnas.94.6.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Naik UP, Patel PM, Parise LV. Identification of a novel calcium-binding protein that interacts with the integrin alphaIIb cytoplasmic domain. J Biol Chem. 1997;272:4651–4654. doi: 10.1074/jbc.272.8.4651. [DOI] [PubMed] [Google Scholar]
- 193.Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, Weinberg RJ, Worley PF, Sheng M. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- 194.Nakanishi S. Metabotropic glutamate receptors: synaptic transmission, modulation, and plasticity. Neuron. 1994;13:1031–1037. doi: 10.1016/0896-6273(94)90043-4. [DOI] [PubMed] [Google Scholar]
- 195.Nedivi E, Fieldust S, Theill LE, Hevron D. A set of genes expressed in response to light in the adult cerebral cortex and regulated during development. Proc Natl Acad Sci USA. 1996;93:2048–2053. doi: 10.1073/pnas.93.5.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Nedivi E, Hevroni D, Naot D, Israeli D, Citri Y. Numerous candidate plasticity-related genes revealed by differential cDNA cloning. Nature. 1993;363:718–722. doi: 10.1038/363718a0. [DOI] [PubMed] [Google Scholar]
- 197.Nedivi E, Javaherian A, Cantallops I, Cline HT. Developmental regulation of CPG15 expression in Xenopus. J Comp Neurol. 2001;435:464–473. doi: 10.1002/cne.1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Nedivi E, Wu GY, Cline HT. Promotion of dendritic growth by CPG15, an activity-induced signaling molecule. Science. 1998;281:1863–1866. doi: 10.1126/science.281.5384.1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Newton SS, Collier EF, Hunsberger J, Adams D, Terwilliger R, Selvanayagam E, Duman RS. Gene profile of electroconvulsive seizures: induction of neurotrophic and angiogenic factors. J Neurosci. 2003;23:10841–10851. doi: 10.1523/JNEUROSCI.23-34-10841.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Nguyen PV, Abel T, Kandel ER. Requirement of a critical period of transcription for induction of a late phase of LTP. Science. 1994;265:1104–1107. doi: 10.1126/science.8066450. [DOI] [PubMed] [Google Scholar]
- 201.Nichols M, Weih F, Schmid W, DeVack C, Kowenz-Leutz E, Luckow B, Boshart M, Schutz G. Phosphorylation of CREB affects its binding to high and low affinity sites: implications for cAMP induced gene transcription. EMBO J. 1992;11:3337–3346. doi: 10.1002/j.1460-2075.1992.tb05412.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Nishimura T, Vogt PK. The avian cellular homolog of the oncogene jun. Oncogene. 1988;3:659–663. [PubMed] [Google Scholar]
- 203.O’Brien RJ, Xu D, Petralia RS, Steward O, Huganir RL, Worley P. Synaptic clustering of AMPA receptors by the extracellular immediate-early gene product Narp. Neuron. 1999;23:309–323. doi: 10.1016/s0896-6273(00)80782-5. [DOI] [PubMed] [Google Scholar]
- 204.Oliet SH, Malenka RC, Nicoll RA. Two distinct forms of long-term depression coexist in CA1 hippocampal pyramidal cells. Neuron. 1997;18:969–982. doi: 10.1016/s0896-6273(00)80336-0. [DOI] [PubMed] [Google Scholar]
- 205.Oliveira-Dos-Santos AJ, Matsumoto G, Snow BE, Bai D, Houston FP, Whishaw IQ, Mariathasan S, Sasaki T, Wakeham A, Ohashi PS, Roder JC, Barnes CA, Siderovski DP, Penninger JM. Regulation of T cell activation, anxiety, and male aggression by RGS2. Proc Natl Acad Sci USA. 2000;97:12272–12277. doi: 10.1073/pnas.220414397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Pak DT, Sheng M. Targeted protein degradation and synapse remodeling by an inducible protein kinase. Science. 2003;302:1368–1373. doi: 10.1126/science.1082475. [DOI] [PubMed] [Google Scholar]
- 207.Pak DT, Yang S, Rudolph-Correia S, Kim E, Sheng M. Regulation of dendritic spine morphology by SPAR, a PSD-95-associated RapGAP. Neuron. 2001;31:289–303. doi: 10.1016/s0896-6273(01)00355-5. [DOI] [PubMed] [Google Scholar]
- 208.Park CS, Gong R, Stuart J, Tang SJ. Molecular network and chromosomal clustering of genes involved in synaptic plasticity in the hippocampus. J Biol Chem. 2006;281:30195–30211. doi: 10.1074/jbc.M605876200. [DOI] [PubMed] [Google Scholar]
- 209.Park S, Park JM, Kim S, Kim JA, Shepherd JD, Smith-Hicks CL, Chowdhury S, Kaufmann W, Kuhl D, Ryazanov AG, Huganir RL, Linden DJ, Worley PF. Elongation factor 2 and fragile X mental retardation protein control the dynamic translation of Arc/Arg3.1 essential for mGluR-LTD. Neuron. 2008;59:70–83. doi: 10.1016/j.neuron.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Parker RC, Varmus HE, Bishop JM. Expression of v-src and chicken c-src in rat cells demonstrates qualitative differences between pp60v-src and pp60c-src. Cell. 1984;37:131–139. doi: 10.1016/0092-8674(84)90308-8. [DOI] [PubMed] [Google Scholar]
- 211.Patterson SL, Abel T, Deuel TA, Martin KC, Rose JC, Kandel ER. Recombinant BDNF rescues deficits in basal synaptic transmission and hippocampal LTP in BDNF knockout mice. Neuron. 1996;16:1137–1145. doi: 10.1016/s0896-6273(00)80140-3. [DOI] [PubMed] [Google Scholar]
- 212.Patterson SL, Grover LM, Schwartzkroin PA, Bothwell M. Neurotrophin expression in rat hippocampal slices: a stimulus paradigm inducing LTP in CA1 evokes increases in BDNF and NT-3 mRNAs. Neuron. 1992;9:1081–1088. doi: 10.1016/0896-6273(92)90067-n. [DOI] [PubMed] [Google Scholar]
- 213.Paylor R, Johnson RS, Papaioannou V, Spiegelman BM, Wehner JM. Behavioral assessment of c-fos mutant mice. Brain Res. 1994;651:275–282. doi: 10.1016/0006-8993(94)90707-2. [DOI] [PubMed] [Google Scholar]
- 214.Pin JP, Duvoisin R. The metabotropic glutamate receptors: structure and functions. Neuropharmacology. 1995;34:1–26. doi: 10.1016/0028-3908(94)00129-g. [DOI] [PubMed] [Google Scholar]
- 215.Plath N, Ohana O, Dammermann B, Errington ML, Schmitz D, Gross C, Mao X, Engelsberg A, Mahlke C, Welzl H, Kobalz U, Stawrakakis A, Fernandez E, Waltereit R, Bick-Sander A, Therstappen E, Cooke SF, Blanquet V, Wurst W, Salmen B, Bosl MR, Lipp HP, Grant SG, Bliss TV, Wolfer DP, Kuhl D. Arc/Arg3.1 is essential for the consolidation of synaptic plasticity and memories. Neuron. 2006;52:437–444. doi: 10.1016/j.neuron.2006.08.024. [DOI] [PubMed] [Google Scholar]
- 216.Ploski JE, Pierre VJ, Smucny J, Park K, Monsey MS, Overeem KA, Schafe GE. The activity-regulated cytoskeletal-associated protein (Arc/Arg3.1) is required for memory consolidation of pavlovian fear conditioning in the lateral amygdala. J Neurosci. 2008;28:12383–12395. doi: 10.1523/JNEUROSCI.1662-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Prezeau L, Gomeza J, Ahern S, Mary S, Galvez T, Bockaert J, Pin JP. Changes in the carboxyl-terminal domain of metabotropic glutamate receptor 1 by alternative splicing generate receptors with differing agonist-independent activity. Mol Pharmacol. 1996;49:422–429. [PubMed] [Google Scholar]
- 218.Privalsky ML, Ralston R, Bishop JM. The membrane glycoprotein encoded by the retroviral oncogene v-erb-B is structurally related to tyrosine-specific protein kinases. Proc Natl Acad Sci USA. 1984;81:704–707. doi: 10.1073/pnas.81.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Prywes R, Roeder RG. Purification of the c-fos enhancer-binding protein. Mol Cell Biol. 1987;7:3482–3489. doi: 10.1128/mcb.7.10.3482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Pulipparacharuvil S, Renthal W, Hale CF, Taniguchi M, Xiao G, Kumar A, Russo SJ, Sikder D, Dewey CM, Davis MM, Greengard P, Nairn AC, Nestler EJ, Cowan CW. Cocaine regulates MEF2 to control synaptic and behavioral plasticity. Neuron. 2008;59:621–633. doi: 10.1016/j.neuron.2008.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Putz U, Harwell C, Nedivi E. Soluble CPG15 expressed during early development rescues cortical progenitors from apoptosis. Nat Neurosci. 2005;8:322–331. doi: 10.1038/nn1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Qian Z, Gilbert ME, Colicos MA, Kandel ER, Kuhl D. Tissue-plasminogen activator is induced as an immediate-early gene during seizure, kindling and long-term potentiation. Nature. 1993;361:453–457. doi: 10.1038/361453a0. [DOI] [PubMed] [Google Scholar]
- 223.Quinton EE, Kramarcy NR. Memory impairment correlates closely with cycloheximide dose and degree of inhibition of protein synthesis. Brain Res. 1977;131:184–190. doi: 10.1016/0006-8993(77)90041-5. [DOI] [PubMed] [Google Scholar]
- 224.Rayport SG, Schacher S. Synaptic plasticity in vitro: cell culture of identified Aplysia neurons mediating short-term habituation and sensitization. J Neurosci. 1986;6:759–763. doi: 10.1523/JNEUROSCI.06-03-00759.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Reichardt LF. Neurotrophin-regulated signalling pathways. Philos Trans R Soc Lond B Biol Sci. 2006;361:1545–1564. doi: 10.1098/rstb.2006.1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Renz M, Verrier B, Kurz C, Muller R. Chromatin association and DNA binding properties of the c-fos protooncogene product. Nucleic Acids Res. 1987;15:277–292. doi: 10.1093/nar/15.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Reymann KG, Malisch R, Schulzeck K, Brodemann R, Ott T, Matthies H. The duration of long-term potentiation in the CA1 region of the hippocampal slice preparation. Brain Res Bull. 1985;15:249–255. doi: 10.1016/0361-9230(85)90147-9. [DOI] [PubMed] [Google Scholar]
- 228.Rial Verde EM, Lee-Osbourne J, Worley PF, Malinow R, Cline HT. Increased expression of the immediate-early gene arc/arg3.1 reduces AMPA receptor-mediated synaptic transmission. Neuron. 2006;52:461–474. doi: 10.1016/j.neuron.2006.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Roche KW, Tu JC, Petralia RS, Xiao B, Wenthold RJ, Worley PF. Homer 1b regulates the trafficking of group I metabotropic glutamate receptors. J Biol Chem. 1999;274:25953–25957. doi: 10.1074/jbc.274.36.25953. [DOI] [PubMed] [Google Scholar]
- 230.Ross WN, Nakamura T, Watanabe S, Larkum M, Lasser-Ross N. Synaptically activated Ca2+ release from internal stores in CNS neurons. Cell Mol Neurobiol. 2005;25:283–295. doi: 10.1007/s10571-005-3060-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Saffen DW, Cole AJ, Worley PF, Christy BA, Ryder K, Baraban JM. Convulsant-induced increase in transcription factor messenger RNAs in rat brain. Proc Natl Acad Sci USA. 1988;85:7795–7799. doi: 10.1073/pnas.85.20.7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sagar SM, Sharp FR, Curran T. Expression of c-fos protein in brain: metabolic mapping at the cellular level. Science. 1988;240:1328–1331. doi: 10.1126/science.3131879. [DOI] [PubMed] [Google Scholar]
- 233.Sambucetti LC, Curran T. The Fos protein complex is associated with DNA in isolated nuclei and binds to DNA cellulose. Science. 1986;234:1417–1419. doi: 10.1126/science.3491427. [DOI] [PubMed] [Google Scholar]
- 234.Schacher S, Castellucci VF, Kandel ER. cAMP evokes long-term facilitation in Aplysia sensory neurons that requires new protein synthesis. Science. 1988;240:1667–1669. doi: 10.1126/science.2454509. [DOI] [PubMed] [Google Scholar]
- 235.Schlimgen AK, Helms JA, Vogel H, Perin MS. Neuronal pentraxin, a secreted protein with homology to acute phase proteins of the immune system. Neuron. 1995;14:519–526. doi: 10.1016/0896-6273(95)90308-9. [DOI] [PubMed] [Google Scholar]
- 236.Seeburg DP, Feliu-Mojer M, Gaiottino J, Pak DT, Sheng M. Critical role of CDK5 and Polo-like kinase 2 in homeostatic synaptic plasticity during elevated activity. Neuron. 2008;58:571–583. doi: 10.1016/j.neuron.2008.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Seeburg DP, Pak D, Sheng M. Polo-like kinases in the nervous system. Oncogene. 2005;24:292–298. doi: 10.1038/sj.onc.1208277. [DOI] [PubMed] [Google Scholar]
- 238.Seeburg DP, Sheng M. Activity-induced Polo-like kinase 2 is required for homeostatic plasticity of hippocampal neurons during epileptiform activity. J Neurosci. 2008;28:6583–6591. doi: 10.1523/JNEUROSCI.1853-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Seeds NW, Williams BL, Bickford PC. Tissue plasminogen activator induction in Purkinje neurons after cerebellar motor learning. Science. 1995;270:1992–1994. doi: 10.1126/science.270.5244.1992. [DOI] [PubMed] [Google Scholar]
- 240.Shalizi A, Gaudilliere B, Yuan Z, Stegmuller J, Shirogane T, Ge Q, Tan Y, Schulman B, Harper JW, Bonni A. A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science. 2006;311:1012–1017. doi: 10.1126/science.1122513. [DOI] [PubMed] [Google Scholar]
- 241.Sharp JW, Sagar SM, Hisanaga K, Jasper P, Sharp FR. The NMDA receptor mediates cortical induction of fos and fos-related antigens following cortical injury. Exp Neurol. 1990;109:323–332. doi: 10.1016/s0014-4886(05)80023-8. [DOI] [PubMed] [Google Scholar]
- 242.Shelton DL, Sutherland J, Gripp J, Camerato T, Armanini MP, Phillips HS, Carroll K, Spencer SD, Levinson AD. Human trks: molecular cloning, tissue distribution, and expression of extracellular domain immunoadhesins. J Neurosci. 1995;15:477–491. doi: 10.1523/JNEUROSCI.15-01-00477.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Sheng M, McFadden G, Greenberg ME. Membrane depolarization and calcium induce c-fos transcription via phosphorylation of transcription factor CREB. Neuron. 1990;4:571–582. doi: 10.1016/0896-6273(90)90115-v. [DOI] [PubMed] [Google Scholar]
- 244.Shepherd JD, Rumbaugh G, Wu J, Chowdhury S, Plath N, Kuhl D, Huganir RL, Worley PF. Arc/Arg3.1 mediates homeostatic synaptic scaling of AMPA receptors. Neuron. 2006;52:475–484. doi: 10.1016/j.neuron.2006.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Sia GM, Beique JC, Rumbaugh G, Cho R, Worley PF, Huganir RL. Interaction of the N-terminal domain of the AMPA receptor GluR4 subunit with the neuronal pentraxin NP1 mediates GluR4 synaptic recruitment. Neuron. 2007;55:87–102. doi: 10.1016/j.neuron.2007.06.020. [DOI] [PubMed] [Google Scholar]
- 246.Silva AJ, Kogan JH, Frankland PW, Kida S. CREB and memory. Annu Rev Neurosci. 1998;21:127–148. doi: 10.1146/annurev.neuro.21.1.127. [DOI] [PubMed] [Google Scholar]
- 247.Silverman MA, Benard O, Jaaro H, Rattner A, Citri Y, Seger R. CPG16, a novel protein serine/threonine kinase downstream of cAMP-dependent protein kinase. J Biol Chem. 1999;274:2631–2636. doi: 10.1074/jbc.274.5.2631. [DOI] [PubMed] [Google Scholar]
- 248.Smith WL, Marnett LJ, DeWitt DL. Prostaglandin and thromboxane biosynthesis. Pharmacol Ther. 1991;49:153–179. doi: 10.1016/0163-7258(91)90054-p. [DOI] [PubMed] [Google Scholar]
- 249.Snider WD. Functions of the neurotrophins during nervous system development: what the knockouts are teaching us. Cell. 1994;77:627–638. doi: 10.1016/0092-8674(94)90048-5. [DOI] [PubMed] [Google Scholar]
- 250.Sonnenberg JL, Macgregor-Leon PF, Curran T, Morgan JI. Dynamic alterations occur in the levels and composition of transcription factor AP-1 complexes after seizure. Neuron. 1989;3:359–365. doi: 10.1016/0896-6273(89)90260-2. [DOI] [PubMed] [Google Scholar]
- 251.Steward O, Wallace CS, Lyford GL, Worley PF. Synaptic activation causes the mRNA for the IEG Arc to localize selectively near activated postsynaptic sites on dendrites. Neuron. 1998;21:741–751. doi: 10.1016/s0896-6273(00)80591-7. [DOI] [PubMed] [Google Scholar]
- 252.Steward O, Worley PF. Selective targeting of newly synthesized Arc mRNA to active synapses requires NMDA receptor activation. Neuron. 2001;30:227–240. doi: 10.1016/s0896-6273(01)00275-6. [DOI] [PubMed] [Google Scholar]
- 253.Sukhatme VP, Cao XM, Chang LC, Tsai-Morris CH, Stamenkovich D, Ferreira PC, Cohen DR, Edwards SA, Shows TB, Curran T, et al. A zinc finger-encoding gene coregulated with c-fos during growth and differentiation, after cellular depolarization. Cell. 1988;53:37–43. doi: 10.1016/0092-8674(88)90485-0. [DOI] [PubMed] [Google Scholar]
- 254.Sultan M, Schulz MH, Richard H, Magen A, Klingenhoff A, Scherf M, Seifert M, Borodina T, Soldatov A, Parkhomchuk D, Schmidt D, O’Keeffe S, Haas S, Vingron M, Lehrach H, Yaspo ML. A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science. 2008;321:956–960. doi: 10.1126/science.1160342. [DOI] [PubMed] [Google Scholar]
- 255.Sunkel CE, Glover DM. polo, a mitotic mutant of Drosophila displaying abnormal spindle poles. J Cell Sci. 1988;89:25–38. doi: 10.1242/jcs.89.1.25. [DOI] [PubMed] [Google Scholar]
- 256.Sweet RW, Yokoyama S, Kamata T, Feramisco JR, Rosenberg M, Gross M. The product of ras is a GTPase and the T24 oncogenic mutant is deficient in this activity. Nature. 1984;311:273–275. doi: 10.1038/311273a0. [DOI] [PubMed] [Google Scholar]
- 257.Tao X, Finkbeiner S, Arnold DB, Shaywitz AJ, Greenberg ME. Ca2+ influx regulates BDNF transcription by a CREB family transcription factor-dependent mechanism. Neuron. 1998;20:709–726. doi: 10.1016/s0896-6273(00)81010-7. [DOI] [PubMed] [Google Scholar]
- 258.Tetzlaff JE, Huppenbauer CB, Tanzer L, Alexander TD, Jones KJ. Motoneuron injury and repair: new perspectives on gonadal steroids as neurotherapeutics. J Mol Neurosci. 2006;28:53–64. doi: 10.1385/jmn:28:1:53. [DOI] [PubMed] [Google Scholar]
- 259.Topilko P, Schneider-Maunoury S, Levi G, Trembleau A, Gourdji D, Driancourt MA, Rao CV, Charnay P. Multiple pituitary and ovarian defects in Krox-24 (NGFI-A, Egr-1)-targeted mice. Mol Endocrinol. 1998;12:107–122. doi: 10.1210/mend.12.1.0049. [DOI] [PubMed] [Google Scholar]
- 260.Treisman R. Transient accumulation of c-fos RNA following serum stimulation requires a conserved 5′ element and c-fos 3′ sequences. Cell. 1985;42:889–902. doi: 10.1016/0092-8674(85)90285-5. [DOI] [PubMed] [Google Scholar]
- 261.Tropea D, Kreiman G, Lyckman A, Mukherjee S, Yu H, Horng S, Sur M. Gene expression changes and molecular pathways mediating activity-dependent plasticity in visual cortex. Nat Neurosci. 2006;9:660–668. doi: 10.1038/nn1689. [DOI] [PubMed] [Google Scholar]
- 262.Tsirka SE, Rogove AD, Bugge TH, Degen JL, Strickland S. An extracellular proteolytic cascade promotes neuronal degeneration in the mouse hippocampus. J Neurosci. 1997;17:543–552. doi: 10.1523/JNEUROSCI.17-02-00543.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Tsui CC, Copeland NG, Gilbert DJ, Jenkins NA, Barnes C, Worley PF. Narp, a novel member of the pentraxin family, promotes neurite outgrowth and is dynamically regulated by neuronal activity. J Neurosci. 1996;16:2463–2478. doi: 10.1523/JNEUROSCI.16-08-02463.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, Doan A, Aakalu VK, Lanahan AA, Sheng M, Worley PF. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- 265.Tu JC, Xiao B, Yuan JP, Lanahan AA, Leoffert K, Li M, Linden DJ, Worley PF. Homer binds a novel proline-rich motif and links group 1 metabotropic glutamate receptors with IP3 receptors. Neuron. 1998;21:717–726. doi: 10.1016/s0896-6273(00)80589-9. [DOI] [PubMed] [Google Scholar]
- 266.Tully T. Physiology of mutations affecting learning and memory in Drosophila–the missing link between gene product and behavior. Trends Neurosci. 1991;14:163–164. doi: 10.1016/0166-2236(91)90096-d. [DOI] [PubMed] [Google Scholar]
- 267.Turrigiano GG. The self-tuning neuron: synaptic scaling of excitatory synapses. Cell. 2008;135:422–435. doi: 10.1016/j.cell.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Tzingounis AV, Nicoll RA. Arc/Arg3.1: linking gene expression to synaptic plasticity and memory. Neuron. 2006;52:403–407. doi: 10.1016/j.neuron.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 269.Verma IM, Sassone-Corsi P. Protooncogene fos: complex but versatile regulation. Cell. 1987;51:513–514. doi: 10.1016/0092-8674(87)90115-2. [DOI] [PubMed] [Google Scholar]
- 270.Villa A, Sharp AH, Racchetti G, Podini P, Bole DG, Dunn WA, Pozzan T, Snyder SH, Meldolesi J. The endoplasmic reticulum of Purkinje neuron body and dendrites: molecular identity and specializations for Ca2+ transport. Neuroscience. 1992;49:467–477. doi: 10.1016/0306-4522(92)90111-e. [DOI] [PubMed] [Google Scholar]
- 271.Wang YT, Linden DJ. Expression of cerebellar long-term depression requires postsynaptic clathrin-mediated endocytosis. Neuron. 2000;25:635–647. doi: 10.1016/s0896-6273(00)81066-1. [DOI] [PubMed] [Google Scholar]
- 272.Waterfield MD, Scrace GT, Whittle N, Stroobant P, Johnsson A, Wasteson A, Westermark B, Heldin CH, Huang JS, Deuel TF. Platelet-derived growth factor is structurally related to the putative transforming protein p28sis of simian sarcoma virus. Nature. 1983;304:35–39. doi: 10.1038/304035a0. [DOI] [PubMed] [Google Scholar]
- 273.Waung MW, Pfeiffer BE, Nosyreva ED, Ronesi JA, Huber KM. Rapid translation of Arc/Arg3.1 selectively mediates mGluR-dependent LTD through persistent increases in AMPAR endocytosis rate. Neuron. 2008;59:84–97. doi: 10.1016/j.neuron.2008.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Wiesel TN, Hubel DH. Single-cell responses in striate cortex of kittens deprived of vision in one eye. J Neurophysiol. 1963;26:1003–1017. doi: 10.1152/jn.1963.26.6.1003. [DOI] [PubMed] [Google Scholar]
- 275.Williams JH, Errington ML, Lynch MA, Bliss TV. Arachidonic acid induces a long-term activity-dependent enhancement of synaptic transmission in the hippocampus. Nature. 1989;341:739–742. doi: 10.1038/341739a0. [DOI] [PubMed] [Google Scholar]
- 276.Wisden W, Errington ML, Williams S, Dunnett SB, Waters C, Hitchcock D, Evan G, Bliss TV, Hunt SP. Differential expression of immediate early genes in the hippocampus and spinal cord. Neuron. 1990;4:603–614. doi: 10.1016/0896-6273(90)90118-y. [DOI] [PubMed] [Google Scholar]
- 277.Worley PF, Cole AJ, Murphy TH, Christy BA, Nakabeppu Y, Baraban JM. Synaptic regulation of immediate-early genes in brain. Cold Spring Harb Symp Quant Biol. 1990;55:213–223. doi: 10.1101/sqb.1990.055.01.023. [DOI] [PubMed] [Google Scholar]
- 278.Xerri C. Imprinting of idiosyncratic experience in cortical sensory maps: neural substrates of representational remodeling and correlative perceptual changes. Behav Brain Res. 2008;192:26–41. doi: 10.1016/j.bbr.2008.02.038. [DOI] [PubMed] [Google Scholar]
- 279.Xiang G, Pan L, Xing W, Zhang L, Huang L, Yu J, Zhang R, Wu J, Cheng J, Zhou Y. Identification of activity-dependent gene expression profiles reveals specific subsets of genes induced by different routes of Ca2+ entry in cultured rat cortical neurons. J Cell Physiol. 2007;212:126–136. doi: 10.1002/jcp.21008. [DOI] [PubMed] [Google Scholar]
- 280.Xiao B, Tu JC, Petralia RS, Yuan JP, Doan A, Breder CD, Ruggiero A, Lanahan AA, Wenthold RJ, Worley PF. Homer regulates the association of group 1 metabotropic glutamate receptors with multivalent complexes of homer-related, synaptic proteins. Neuron. 1998;21:707–716. doi: 10.1016/s0896-6273(00)80588-7. [DOI] [PubMed] [Google Scholar]
- 281.Xu D, Hopf C, Reddy R, Cho RW, Guo L, Lanahan A, Petralia RS, Wenthold RJ, O’Brien RJ, Worley P. Narp and NP1 form heterocomplexes that function in developmental and activity-dependent synaptic plasticity. Neuron. 2003;39:513–528. doi: 10.1016/s0896-6273(03)00463-x. [DOI] [PubMed] [Google Scholar]
- 282.Yalcin B, Willis-Owen SA, Fullerton J, Meesaq A, Deacon RM, Rawlins JN, Copley RR, Morris AP, Flint J, Mott R. Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nat Genet. 2004;36:1197–1202. doi: 10.1038/ng1450. [DOI] [PubMed] [Google Scholar]
- 283.Yamagata K, Andreasson KI, Kaufmann WE, Barnes CA, Worley PF. Expression of a mitogen-inducible cyclooxygenase in brain neurons: regulation by synaptic activity and glucocorticoids. Neuron. 1993;11:371–386. doi: 10.1016/0896-6273(93)90192-t. [DOI] [PubMed] [Google Scholar]
- 284.Yamagata K, Andreasson KI, Sugiura H, Maru E, Dominique M, Irie Y, Miki N, Hayashi Y, Yoshioka M, Kaneko K, Kato H, Worley PF. Arcadlin is a neural activity-regulated cadherin involved in long term potentiation. J Biol Chem. 1999;274:19473–19479. doi: 10.1074/jbc.274.27.19473. [DOI] [PubMed] [Google Scholar]
- 285.Yamagata K, Kaufmann WE, Lanahan A, Papapavlou M, Barnes CA, Andreasson KI, Worley PF. Egr3/Pilot, a zinc finger transcription factor, is rapidly regulated by activity in brain neurons and colocalizes with Egr1/zif268. Learn Mem. 1994;1:140–152. [PubMed] [Google Scholar]
- 286.Yamagata K, Sanders LK, Kaufmann WE, Yee W, Barnes CA, Nathans D, Worley PF. rheb, a growth factor- and synaptic activity-regulated gene, encodes a novel Ras-related protein. J Biol Chem. 1994;269:16333–16339. [PubMed] [Google Scholar]
- 287.Yamamoto KK, Gonzalez GA, Menzel P, Rivier J, Montminy MR. Characterization of a bipartite activator domain in transcription factor CREB. Cell. 1990;60:611–617. doi: 10.1016/0092-8674(90)90664-z. [DOI] [PubMed] [Google Scholar]
- 288.Yasuda S, Tanaka H, Sugiura H, Okamura K, Sakaguchi T, Tran U, Takemiya T, Mizoguchi A, Yagita Y, Sakurai T, De Robertis EM, Yamagata K. Activity-induced protocadherin arcadlin regulates dendritic spine number by triggering N-cadherin endocytosis via TAO2beta and p38 MAP kinases. Neuron. 2007;56:456–471. doi: 10.1016/j.neuron.2007.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289.Yee WM, Worley PF. Rheb interacts with Raf-1 kinase and may function to integrate growth factor- and protein kinase A-dependent signals. Mol Cell Biol. 1997;17:921–933. doi: 10.1128/mcb.17.2.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Yin JC, Del Vecchio M, Zhou H, Tully T. CREB as a memory modulator: induced expression of a dCREB2 activator isoform enhances long-term memory in Drosophila. Cell. 1995;81:107–115. doi: 10.1016/0092-8674(95)90375-5. [DOI] [PubMed] [Google Scholar]
- 291.Yuan JP, Kiselyov K, Shin DM, Chen J, Shcheynikov N, Kang SH, Dehoff MH, Schwarz MK, Seeburg PH, Muallem S, Worley PF. Homer binds TRPC family channels and is required for gating of TRPC1 by IP3 receptors. Cell. 2003;114:777–789. doi: 10.1016/s0092-8674(03)00716-5. [DOI] [PubMed] [Google Scholar]
- 292.Zafra F, Hengerer B, Leibrock J, Thoenen H, Lindholm D. Activity dependent regulation of BDNF and NGF mRNAs in the rat hippocampus is mediated by non-NMDA glutamate receptors. EMBO J. 1990;9:3545–3550. doi: 10.1002/j.1460-2075.1990.tb07564.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293.Zalfa F, Giorgi M, Primerano B, Moro A, Di Penta A, Reis S, Oostra B, Bagni C. The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell. 2003;112:317–327. doi: 10.1016/s0092-8674(03)00079-5. [DOI] [PubMed] [Google Scholar]
- 294.Zangenehpour S, Chaudhuri A. Patchy organization and asymmetric distribution of the neural correlates of face processing in monkey inferotemporal cortex. Curr Biol. 2005;15:993–1005. doi: 10.1016/j.cub.2005.04.031. [DOI] [PubMed] [Google Scholar]