Abstract
Cation diffusion facilitator (CDF) proteins are a recently discovered family of cation efflux transporters that might play an essential role in metal homeostasis and tolerance. Here, we describe the identification, characterization, and localization of PtdMTP1, a member of the CDF family from the hybrid poplar Populus trichocarpa × Populus deltoides. PtdMTP1 is expressed constitutively and ubiquitously, although at low levels. Heterologous expression in yeast showed that PtdMTP1 was able to complement the hypersensitivity of mutant strains to Zn but not to other metals, including Cd, Co, Mn, and Ni. PtdMTP1 fused to green fluorescent protein localized to the vacuolar membrane both in yeast and in plant cells, consistent with a function of PtdMTP1 in zinc sequestration. Overexpression of PtdMTP1 in Arabidopsis confers Zn tolerance. We show that PtdMTP1, when expressed in yeast and Arabidopsis, forms homooligomers, a novel feature of CDF members. Oligomer formation is disrupted by reducing agents, indicating possible disulfide bridge formation. PtdMTP1 also contains a conserved Leu zipper motif. Although not necessary for oligomer formation, Leu residues within this motif are required for PtdMTP1 functional activity.
INTRODUCTION
Transition elements such as Fe, Co, Ni, Cu, and Zn play a wide variety of roles in biology as enzyme cofactors and must be absorbed from the soil by plants. However, either naturally or as a result of human activity, these metals can be present at potentially toxic concentrations. A number of other metal ions with no known biological function—including Cd, Pb, and Hg—also are potentially highly toxic for plants. Two basic strategies for decreasing the toxicity of metals are apparent: chelation and efflux from the cytosol, either into the apoplast or by intracellular sequestration. A number of cation transporter families have emerged recently and been classified in plants (Mäser et al., 2001), and although it is apparent that some (e.g., the Zrt-, Irt-like protein [ZIP] family) are involved in the high-affinity uptake of metals for nutritional purposes, overexpression of a member of another family (the cation diffusion facilitator [CDF] family) can lead to decreased metal toxicity and enhanced accumulation (van der Zaal et al., 1999).
Interest regarding transporters that facilitate the accumulation of potentially toxic metals has centered on their potential exploitation in phytoremediation. Phytoremediation is an emerging technology potentially effective and applicable to a number of different contaminants and site conditions (Lasat, 2002; Pilon-Smits and Pilon, 2002). The practical use of many well-known hyperaccumulators, such as Thlaspi caerulescens, for metal phytoremediation might be limited because they are slow growing and produce little biomass. The use of larger plants that are not currently classified as hyperaccumulators can compensate for somewhat lower accumulation factors in aboveground organs with greater biomass production and high transpiration rates. Poplar, which has emerged as a model system for genomic approaches to wood formation and tree physiology (Sterky et al., 1998; Taylor, 2002; Kohler et al., 2003), also is a good candidate for phytoremediation purposes (Gordon et al., 1998; Rugh et al., 1998; Pilon-Smits and Pilon, 2002; Stanton et al., 2002; Di Baccio et al., 2003). Thus, an ideal plant for phytoremediation purposes would combine high biomass production and superior capacity for pollutant accumulation and tolerance.
Among essential metals, zinc plays critical roles in a wide variety of biochemical processes; therefore, intracellular zinc concentrations must be maintained at adequate levels to support cell growth (Gaither and Eide, 2001). Recent reviews have discussed the basis for the mechanisms of Zn tolerance in plants and proposed that transport-mediated sequestration can contribute greatly to Zn tolerance (Williams et al., 2000; Clemens, 2001; Clemens et al., 2002b; Lasat, 2002). Gaither and Eide (2001) reported the major advances that have been made in the last decade through the discovery of two families of zinc transporters and their regulators in eukaryotes: the ZIP and CDF families. Members of the CDF family have been implicated in the metal tolerance mechanisms of a range of organisms and are found in all biological kingdoms (Paulsen and Saier, 1997). ZRC1 and COT1 in Saccharomyces cerevisiae localize to the vacuole membrane and are thought to contribute to the storage of Zn and Co ions, respectively (Li and Kaplan, 1998; MacDiarmid et al., 2000, 2002; Miyabe et al., 2001). MSC2, a third CDF member from S. cerevisiae, was shown to affect the cellular distribution of zinc, particularly the zinc content of nuclei (Li and Kaplan, 2001). ZHF in Schizosaccharomyces pombe is localized in the endoplasmic reticulum/nuclear envelope and plays an important role in cellular zinc homeostasis by mediating the transport of zinc into the endoplasmic reticulum (Clemens et al., 2002a). In yeast, overexpression of metal-tolerance proteins (MTPs; which are members of the CDF family) from Thlaspi goesingense was shown to confer resistance to Cd, Co, Ni, and Zn, possibly as a result of transport into the vacuole (Persans et al., 2001). Arabidopsis ZAT (also known as AtMTP1; Mäser et al., 2001) is expressed in all organs of the plant, and overexpression of ZAT can lead to enhanced zinc resistance and accumulation in roots (van der Zaal et al., 1999). Despite this recent progress, no localization has been assigned to zinc transporters of the CDF family in plants. Recently, a Mn transporter belonging to this family was shown to be located on the tonoplast (Delhaize et al., 2003).
Here, we report the identification of poplar MTP1 (PtdMTP1), which encodes a protein with features typical of CDF proteins. PtdMTP1 was identified in a poplar EST database (Kohler et al., 2003), and experiments were performed to analyze its functional properties. Heterologous expression of PtdMTP1 in various yeast mutants was shown to confer resistance specifically to Zn, possibly as a result of transport into the vacuole. Fusion proteins of PtdMTP1 and green fluorescent protein (GFP) were shown to be localized to the vacuolar membrane of yeast, onion epidermal, and Arabidopsis root cells, consistent with a function for PtdMTP1 in Zn sequestration. Moreover, overexpression of PtdMTP1 in Arabidopsis confers Zn tolerance. We found that PtdMTP1 possesses key biochemical features: the expected CDF signature and Leu zipper motifs, both of which are necessary for its functional activity. We also demonstrate that PtdMTP1 forms oligomers that are disrupted by reducing agents.
RESULTS
Molecular Analysis of a Poplar Zinc Transporter
To examine the molecular basis of Zn homeostasis in poplar, we searched the EST database of Populus trichocarpa × Populus deltoides roots (Kohler et al., 2003) for sequence homology with known Zn transporters of the ZAT/MTP/CDF family. We identified a cDNA homologous with plant and yeast zinc transporters. We named the poplar homolog PtdMTP1 (Populus trichocarpa × deltoides metal tolerance protein), in accord with the suggestion in a recent review (Mäser et al., 2001) and the new nomenclature given by Delhaize et al. (2003). The PtdMTP1 cDNA corresponds to a 393-residue protein (43.5 kD). Analysis of predicted sequence from the cDNA showed 73.7% identity with Arabidopsis ZAT (now known as AtMTP1; Mäser et al., 2001). The hydropathy profile of PtdMTP1, generated with the Kyte and Doolittle (1982) algorithm (http://www.expasy.org/cgi-bin/protscale.pl), predicts six hydrophobic domains of sufficient length to be considered potential membrane-spanning domains, with a long loop separating transmembrane domains IV and V (data not shown). PtdMTP1 possesses the most conserved region of the CDF family proteins, which encompasses transmembrane spans I to IV. Within this region, the CDF family–specific signature sequence (Paulsen and Saier, 1997), which begins with the fully conserved Ser-80 and continues just past the fully conserved Asp-93, was identified. A hydrophilic, His-rich stretch, to which zinc probably binds (Bloss et al., 2002), extends from His-182 to His-232.
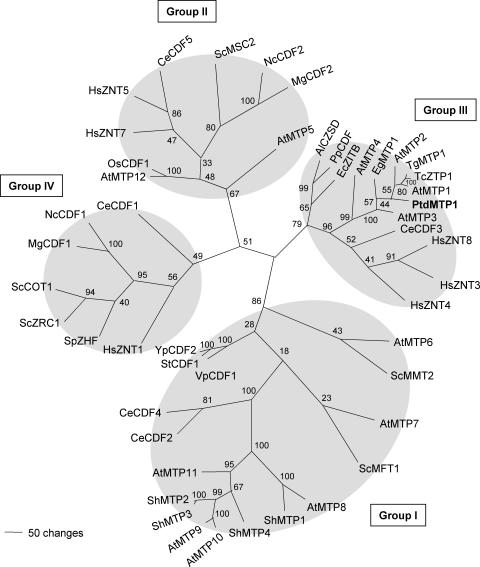
A phylogenetic tree for the CDF family proteins is shown in Figure 1. This tree is a revised and extended version of that presented by Mäser et al. (2001), and it includes four additional CDF members that have emerged from a more detailed study of the Arabidopsis genome. PtdMTP1 is most closely related to group III of the CDF transporters (including AtMTP1), as defined by Gaither and Eide (2001). Group II is a distant group that includes AtMTP12 and an Oryza sativa CDF, the mammalian ZNT5 and ZNT7, and fungal CDFs from S. cerevisiae (MSC2), Neurospora crassa (U07262), and Magnaporthe grisea (10493). Other important plant CDF members from Arabidopsis (AtMTP8 to AtMTP11) and Stylosanthes hamata (ShMTP1 to ShMTP4) clearly cluster as a very distant and separate group, which would form part of group I defined by Gaither and Eide (2001). This subgroup includes the more distantly related plant CDF members AtMTP6 and AtMTP7 (previously named AtMTPc1 and AtMTPc4, respectively; Mäser et al., 2001) as well as bacterial, animal, and yeast CDF members. Group IV includes the well-characterized yeast CDF members ScZRC1 and ScCOT1, three other fungal CDFs from S. pombe (ZHF), N. crassa (U03145), and M. grisea (03634), and animal CDFs.
Figure 1.
An Unrooted, Parsimony-Based Tree of the CDF Gene Family.
The tree was generated using PAUP 4.0b10 (D. Swofford, Smithsonian Institution, Washington, DC) after sequence alignment with CLUSTAL X (Thompson et al., 1997). Bootstrap values are indicated (1000 replicates, full heuristic search option). Accession numbers are given at the end of Methods. Arabidopsis gene names follow the new nomenclature used by Delhaize et al. (2003).
PtdMTP1 Is Expressed Constitutively
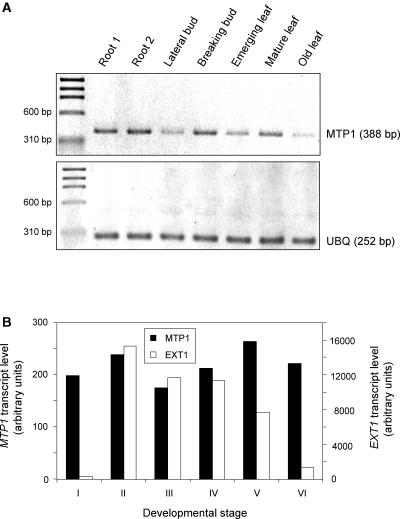
Reverse transcription (RT)–PCR analyses demonstrated that PtdMTP1 was expressed constitutively in most poplar tissues, as shown in Figure 2A. The highest level of PtdMTP1 transcript was detected in mature leaves and roots. We further measured the expression of PtdMTP1 by RT-PCR in root and leaf from poplar seedlings grown axenically and found that transcript level was not affected by exposure to low or high concentrations of Zn (0.038, 0.38, and 3.8 mM) for short or extended periods (3 to 5 days) (data not shown). The additional metals Mn, Cd, and Ni also were checked, and we found no response to these either. Transcript profiling experiments to study poplar adventitious root development revealed that PtdMTP1 was expressed constitutively, although at low levels, as shown in Figure 2B. Expression was independent of root development stage, in contrast to that of the extensin gene EXT1. The low expression level also was reflected by the low abundance of the PtdMTP1 clone in a Populus trichocarpa × deltoides root cDNA library (just 1 in >7000 ESTs [Kohler et al., 2003]; http://mycor.nancy.inra.fr/poplardb/index.html) and in the Populus spp.cDNA libraries (8 in >95,151 ESTs from various tissues; http://poppel.fysbot.umu.se). Fortuitously, in other cDNA array projects performed to study drought stress, root colonization by the ectomycorrhizal fungus Paxillus involutus, and leaf infection by the rust fungus Melampsora larici-populina, we found that PtdMTP1 expression was not affected significantly (data not shown).
Figure 2.
Expression Levels of PtdMTP1 in Poplar.
(A) Transcript levels in different tissues of mature poplar were estimated by reverse transcription–PCR as described in Methods. The expression level of PtdMTP1 was compared with that of UBIQUITIN (UBQ). Experiments were repeated three times, and representative gels are shown.
(B) Transcript levels during the development of adventitious poplar roots were estimated from cDNA arrays as described in Methods. Developmental stages are as follows: I, bark tissues of dormant cuttings; II, root primordium; III, root callus; IV, emerging roots; V, primary roots; and VI, lateral root tips. The expression level of PtdMTP1 was compared with that of EXTENSIN (EXT1), a highly expressed and strongly regulated gene. Experiments were performed in triplicate, and representative data are shown.
Expression of PtdMTP1 in Yeast Confers Tolerance to Zn but Not to Cd, Co, Mn, or Ni
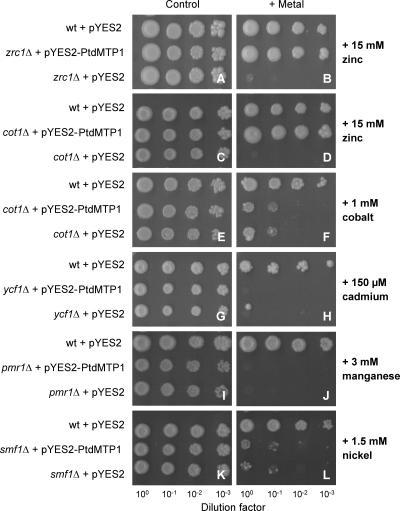
To characterize PtdMTP1 further, PtdMTP1 cDNA was expressed in S. cerevisiae mutant strains that are unable to grow in high concentrations of various metals, and growth was monitored on either control or metal-supplemented medium. Figure 3 shows that the wild-type strain was able to grow at high Zn concentrations (Figure 3B), whereas deletion of the ZRC1 gene, which encodes a transporter that sequesters Zn into the vacuole, rendered the mutant highly sensitive to Zn (Kamizono et al., 1989; Li and Kaplan, 1998). The Zn-sensitive phenotype of the zrc1Δ mutant was fully complemented by PtdMTP1. cot1Δ mutants are hypersensitive to both Zn and Co, because COT1 mediates the efflux of both ions into the vacuole (Conklin et al., 1992; Li and Kaplan, 1998; Lyons et al., 2000). Interestingly, although PtdMTP1 complemented the cot1Δ mutant phenotype when grown on Zn (Figures 3C and 3D), PtdMTP1 failed to complement when grown on Co (Figures 3E and 3F). We further transformed the ycf1Δ, pmr1Δ, and smf1Δ mutant strains, which are unable to grow on Cd, Mn, and Ni, respectively. YCF1 is an ABC transporter that confers Cd tolerance through the transport of Cd conjugates into the vacuole (Szczypka et al., 1994; Li et al., 1997). PMR1 is the yeast secretory pathway pump responsible for high-affinity transport of Mn2+ and Ca2+ into the Golgi and confers Mn tolerance by effectively removing Mn from the cytoplasm (Ton et al., 2002). SMF1 functions in the cellular accumulation of Mn, and the smf1Δ mutant was shown to be Ni sensitive (Supek et al., 1996). Transformation with PtdMTP1 did not restore the growth of ycf1Δ on 0.15 mM Cd (Figures 3G and 3H), of pmr1Δ on 3 mM Mn (Figures 3I and 3J), or of smf1Δ on 1.5 mM Ni (Figures 3K and 3L). The data obtained with the five mutant strains suggest that PtdMTP1 specifically transports Zn but not Co, Cd, Mn, or Ni.
Figure 3.
Complementation of Yeast Mutants on Selective Media.
S. cerevisiae mutant strains were transformed with the empty vector pYES2 or with pYES2-PtdMTP1. Wild-type (wt) cells (strain BY4741) also were transformed with pYES2 as a control. Yeast cultures were adjusted to OD = 1.0, and 2 μL of serial dilutions (from left to right in each panel) were spotted on SD medium without extra metal ([A], [C], [E], [G], [I], and [K]) or supplemented with 15 mM Zn ([B] and [D]), 1 mM Co (F), 150 μM Cd (H), 3 mM Mn (J), or 1.5 mM Ni (L). Plates were incubated for 6 days at 30°C.
PtdMTP1 Partially Rescues the Growth of Yeast Vacuolar Acidification Mutants under Zn Stress
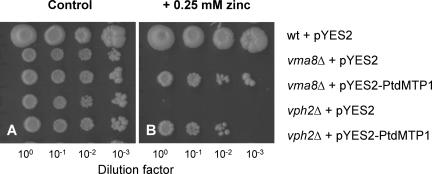
Yeast vacuolar acidification mutants are unable to acidify their vacuoles and have increased sensitivity to some heavy metals, possibly because proton-driven antiport on the vacuolar membrane is required for the sequestration of metals into the lumen (Ramsay and Gadd, 1997). Therefore, to gain insight into the dependence of PtdMTP1 on a proton gradient, two vacuole acidification mutants (vma8Δ and vph2Δ) were transformed with PtdMTP1 and colonies were assayed subsequently for growth at different Zn concentrations. The VMA8 gene encodes a subunit of the catalytic domain of the vacuolar-type H+-ATPase, and the VPH2 protein is required for the biogenesis of a functional vacuolar ATPase (Bachhawat et al., 1993; Graham et al., 1995). Figure 4 shows that vma8Δ and vph2Δ are highly sensitive to Zn (no growth at 0.25 mM Zn) compared with the wild type (growth up to 15 mM; Figure 3). Interestingly, the growth of both acidification mutants was partially rescued when cells were transformed with PtdMTP1. These PtdMTP1-expressing transformants were able to grow at 0.25 mM Zn (Figure 4) and also partially at 0.5 mM Zn (data not shown).
Figure 4.
Partial complementation of Yeast Vacuolar Acidification Mutants.
For yeast transformation and growth conditions, see legend to Figure 3. Yeast growth was assayed on SD medium without extra metal (A) or supplemented with 0.25 mM Zn (B), with serial dilutions from left to right in each panel. wt, wild type.
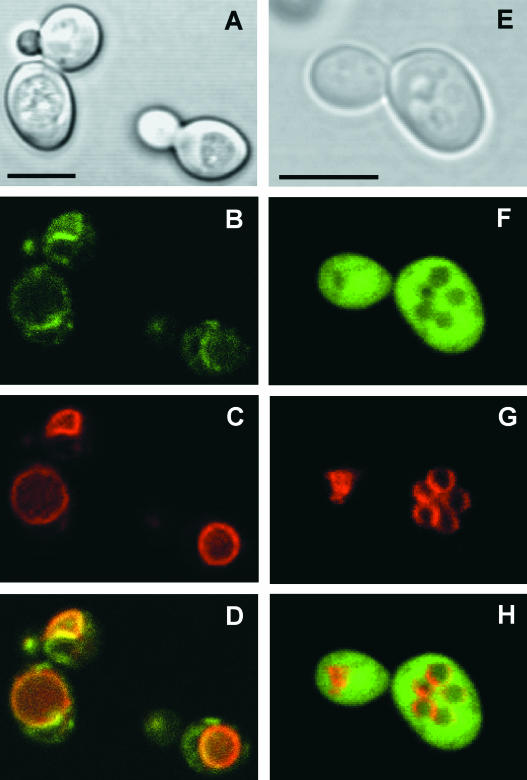
PtdMTP1:GFP Is Targeted to the Vacuolar Membrane in Yeast Cells
Phenotypic suppression by PtdMTP1 of the zrc1Δ phenotype indicates a putative vacuolar membrane localization of the transporter in yeast cells. We constructed the PtdMTP1:GFP fusion protein to determine the intracellular localization of PtdMTP1 more precisely. Because the PtdMTP1:GFP fusion gene complemented the zinc sensitivity of the zrc1Δ mutant (data not shown), the fusion protein was confirmed to retain its original function. The bright-field image in Figure 5A shows the clear presence of vacuoles in zrc1Δ yeast cells. At the standard image acquisition settings used for GFP visualization, autofluorescence from cells transformed with the untagged PtdMTP1 was absent (data not shown), so all detectable fluorescence in the transformants was GFP specific. Figure 5B shows that, when exponentially growing cells were analyzed by confocal laser scanning microscopy, green fluorescence resulting from PtdMTP1:GFP showed a ring-like pattern in the cells. We speculated that this ring-like pattern might reflect vacuolar membrane location. We then stained the vacuolar membrane with the lipophilic dye FM4-64 to identify the location of this organelle (Vida and Emr, 1995). After 60 min of internalization, FM4-64 was found exclusively at the vacuolar membrane in transformed S. cerevisiae cells (Figure 5C). Figure 5D shows that superposition of the GFP and FM4-64 images yields a coincident staining pattern, demonstrating that PtdMTP1 was localized to the vacuolar membrane. When the zrc1Δ mutant was transformed with the vector carrying only the GFP, the fluorescence was found throughout the cytosol (Figure 5F) and was not coincident with the staining of the vacuoles (Figures 5G and 5H).
Figure 5.
Vacuolar Membrane Localization of the PtdMTP1:GFP Fusion Protein in Yeast, Viewed by Confocal Laser Scanning Microscopy.
Cells were visualized after 24 h of induction. Four images from the same cells are shown: bright-field image (A), GFP fluorescence from a PtdMTP1:GFP-expressing strain (B), FM4-64 staining of vacuolar membranes (C), and a pseudocolored merged image with GFP in green and FM4-64 in red (D). (E) to (H) are as for (A) to (D) except for (F) and (H), which show GFP fluorescence from nonfused GFP. Bars = 5 μm.
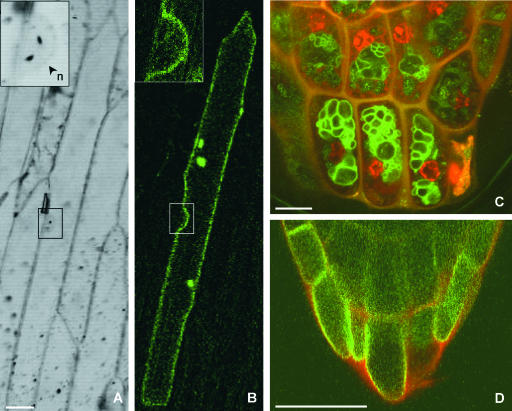
PtdMTP1 Localizes to the Vacuolar Membrane of Plant Cells
Prediction by TargetP (Emanuelsson et al., 2000) indicated no preferred location of PtdMTP1. Therefore, we tested experimentally whether the vacuolar location of the transporter in yeast cells reflects the location in the plant. The PtdMTP1:GFP fusion protein was expressed transiently in living onion epidermal cells (Scott et al., 1999), which then were subjected to analysis by confocal laser scanning microscopy. The bright-field image in Figure 6A shows that, although onion epidermal cells have a large vacuole that effectively contours the cell, a clear separation from the cell periphery was seen in the region of the nucleus (inset). The corresponding fluorescence image in Figure 6B shows a clear localization of GFP to the indented region, rather than the cell periphery, unambiguously demonstrating a vacuolar location. Membrane-lined transvacuolar strands of cytoplasm spanning the cell also produced fluorescence, another typical feature of a vacuolar membrane location (data not shown).
Figure 6.
Vacuolar Membrane Localization of PtdMTP1:GFP in Planta.
(A) and (B) Transient expression of PtdMTP1:GFP in onion epidermal cells. Bar = 50 μm.
(A) Bright-field image along the middle plane of a cell showing the nucleus (n) and invagination of the vacuolar membrane (inset).
(B) GFP fluorescence in the same cell concentrated to the vacuolar membrane, which follows the cell contour except in the perinuclear region (inset).
(C) and (D) Vacuolar localization of PtdMTP1:GFP in Arabidopsis. The red fluorescence is caused by cell walls and nuclei stained with propidium iodide. Bars = 10 μm.
(C) Root tips of transgenic Arabidopsis expressing PtdMTP1:GFP.
(D) Control cells expressing AtMGT1:GFP, a transporter located in the plasma membrane.
Transgenic lines of Arabidopsis expressing PtdMTP1:GFP were used to confirm the vacuolar location of PtdMTP1 found by transient expression in onion cells. In elongated root cells, the large central vacuole usually occupies most of the cell volume, and the vacuolar and plasma membranes are closely juxtaposed. Conversely, cells in the root tip region contain several small immature vacuoles, so we focused on the examination of GFP fluorescence in root tips. Figure 6C shows the presence of numerous intracellular globular localization patterns in those cells, strongly suggesting a vacuolar location. The defined peripheral location of the AtMGT1:GFP fusion protein, as described previously by Li et al. (2001), is shown in Figure 6D as a control for plasma membrane localization.
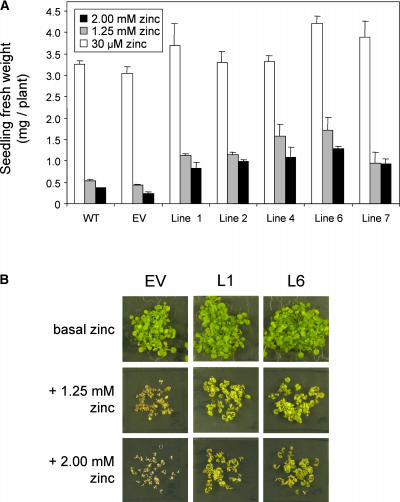
PtdMTP1 Confers Zn2+ Tolerance in Planta
The vacuolar membrane location of PtdMTP1 and its complementation of the Zn sensitivity phenotype of the yeast zrc1Δ mutants suggest that PtdMTP1 might provide Zn tolerance in planta. To address this possibility, we overexpressed PtdMTP1 in transgenic Arabidopsis plants under the control of the 35S promoter. All lines were phenotypically normal when grown on plates or in soil. Figure 7A shows that five representative homozygous lines performed significantly better at high Zn concentrations than did the controls. Two independently transformed lines that overexpressed PtdMTP1 were selected for further study. Figure 7B illustrates the increased Zn tolerance of these two lines when grown on plates in medium supplemented with excess Zn2+ ions, compared with plants transformed with an empty vector grown under the same conditions. At 0.5 mM Zn, root development was nearly nonexistent in wild-type plants and in plants transformed with the empty vector, whereas it was only partially inhibited in lines expressing PtdMTP1 (data not shown). The two transgenic lines expressing PtdMTP1 produced more biomass than the control lines (untransformed plants and plants transformed with the empty vector), which grew poorly and finally died at 1.25 mM Zn (Figure 7B). These findings demonstrate that PtdMTP1 is capable of conferring Zn tolerance in planta.
Figure 7.
Overexpression of PtdMTP1 Confers Zn Tolerance to Arabidopsis.
(A) Plant fresh weight of Arabidopsis lines expressing PtdMTP1 on control half-strength MS medium (containing 30 μM Zn) or supplemented with 1.25 or 2 mM Zn. Plants were harvested after 3 weeks of growth on plates. Lines 1, 2, 4, 6, and 7 are independent homozygous lines expressing PtdMTP1. EV indicates a homozygous line transformed with the empty vector pART27. WT refers to untransformed Arabidopsis wild type. Data are means of 28 to 42 plants, and vertical bars represent standard errors.
(B) Zn-tolerance test of two representative Arabidopsis lines expressing PtdMTP1 and a control line (EV) transformed with the empty vector after 3 weeks of growth on control (basal), 1.25 mM, or 2 mM Zn plates.
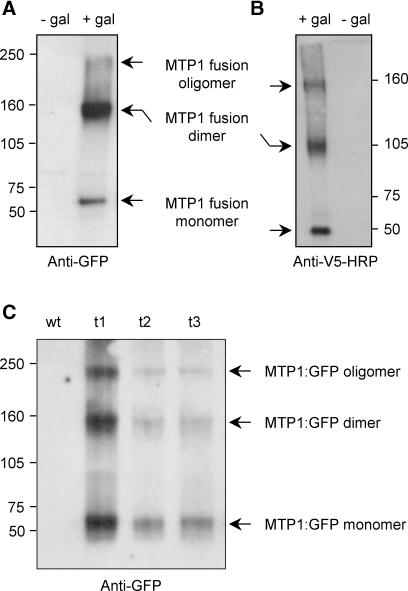
PtdMTP1 Forms Oligomers That Are Sensitive to Reducing Agents
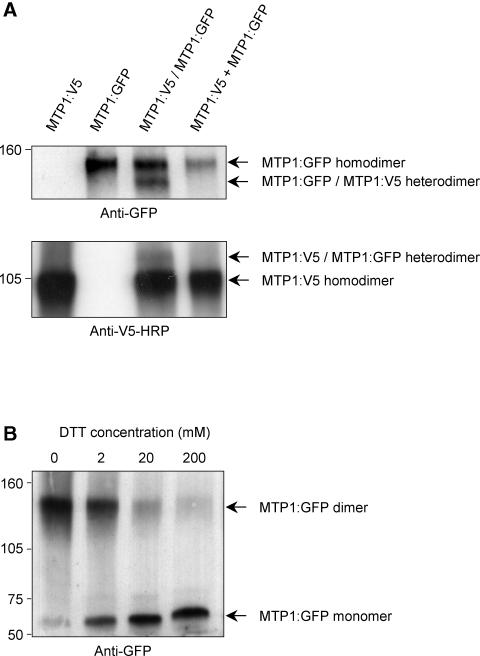
Size estimation by SDS-PAGE using metal blots indicated that AtZAT might form dimers when present in membranes (Bloss et al., 2002). To address whether PtdMTP1 forms oligomers, we used protein gel blot analysis. We expressed the PtdMTP1:GFP fusion protein in yeast cells and solubilized the membrane proteins with n-dodecyl β-d-maltoside. The total molecular mass of the fusion protein was 73 kD. Figure 8A shows the presence of two bands of ∼60 and 140 kD on protein gel blots when using GFP antibodies, with a third band running at ∼220 kD. Although these data imply that a substantial fraction of PtdMTP1, when expressed in yeast cells, exists in the form of dimers, it remained possible that PtdMTP1 forms heterodimers with an unknown protein of similar size and thus migrated as an apparent homodimer. To test this possibility, we generated a PtdMTP1:V5 fusion protein (49.5 kD) and performed similar nonreducing SDS-PAGE experiments. As shown in Figure 8B, PtdMTP1:V5 migrated as two bands of ∼50 and 100 kD, suggesting the presence of both monomeric and dimeric forms and consequently refuting the heterodimeric formation of PtdMTP1:GFP with another unknown protein of ∼80 kD. A third band running at ∼150 kD also was observed (Figure 8B). Figure 8C demonstrates that oligomer formation also occurred in planta. Therefore, whether expressed in planta or in yeast, PtdMTP1 proteins are likely to be present at least as dimers; nevertheless, the presence of higher molecular mass complexes cannot be excluded and are indicated in Figure 8.
Figure 8.
Oligomerization of PtdMTP1 Expressed in Yeast and Arabidopsis.
S. cerevisiae expressing either PtdMTP1:GFP (A) or PtdMTP1:V5 (B) were induced or not with 2% (w/v) galactose (gal). (C) shows the expression of PtdMTP1:GFP in Arabidopsis control plants (wt) or three transgenic lines (t1 to t3) overexpressing PtdMTP1:GFP. Proteins were extracted and separated on 5% (w/v) SDS-PAGE gels under nonreducing conditions and analyzed further using immunoblotting methods. The positions of the PtdMTP1:GFP and PtdMTP1:V5 monomers, dimers, and oligomers are indicated. The antibodies used for detection are noted below each gel. Numbers beside the gels indicate molecular masses in kilodaltons.
To confirm the presence of oligomers, cotransformation of yeast cells with both PtdMTP1:V5 and PtdMTP1:GFP constructs was performed. Because the two tagged species have significantly different molecular masses, heterodimeric and homodimeric chimeric proteins are expected to migrate differentially on gels of low acrylamide concentration. In one approach, anti-GFP antibodies were used to detect PtdMTP1 proteins. Figure 9A shows that in addition to the homodimeric species running with an apparent molecular mass of 140 kD, a band with a lower mass of ∼120 kD also was detected, indicating the presence of heterodimeric PtdMTP1:V5/PtdMTP1:GFP complexes. Complementary data were obtained when the converse experiment was performed; that is, anti-V5 antibodies revealed the presence of a heterodimeric complex, which although appearing fainter, had a higher molecular mass compared with that of the homodimeric complex. We also considered the possibility that PtdMTP1 complexes could be formed during the extraction procedure. To test this possibility, we mixed cells that expressed either PtdMTP1:GFP alone or PtdMTP1:V5 alone and then processed the mixture for protein gel blot analysis as described above. Heterodimers were not observed when the blot was probed with anti-GFP antibodies or with anti-V5 antibodies (Figure 9A). To determine whether PtdMTP1 dimerization occurred through disulfide bridges, extracts containing the PtdMTP1:GFP dimers were treated with the reducing agent DTT before gel loading. As shown in Figure 9B, increasing DTT concentration in the sample led to increasing dissociation of the dimers. At 20 mM DTT, almost complete dissociation was observed. Similar results were found when extracts containing PtdMTP1:V5 were used as well as with the reducing agents β-mercaptoethanol and tributylphosphine (data not shown).
Figure 9.
PtdMTP1 Forms Dimers.
(A) Coexpression of PtdMTP1:GFP and PtdMTP1:V5 fusion proteins in yeast shows dimer formation. As indicated above lanes 1 to 3 (from left), proteins were extracted from cells expressing only PtdMTP1:V5, only PtdMTP1:GFP, or both (PtdMTP1:GFP/PtdMTP1:V5). The right lane was loaded with proteins from cells expressing only PtdMTP1:GFP mixed with cells expressing only PtdMTP1:V5 (PtdMTP1:GFP + PtdMTP1:V5).
(B) Disruption of the PtdMTP1:GFP dimer by DTT. Samples were treated with increasing concentrations of DTT for 1 h before loading.
Cells were processed for immunodetection as described in the legend to Figure 8. The positions of homodimers and heterodimers are indicated. The antibodies used for detection are noted below each gel. Numbers beside the gels indicate molecular masses in kilodaltons.
PtdMTP1 Contains a Leu Zipper Motif That Is Necessary for Function
The discovery that PtdMTP1 forms multimers possibly stabilized by S-S bridges encouraged us to examine the possibility that other motifs are involved in functional aspects of protein–protein interaction. The Scanprosite interface (www.expasy.ch/cgi-bin/scanprosite) enabled the identification of a Leu zipper (LZ) motif in the C-terminal PtdMTP1 sequence from residues Leu-293 to Leu-314. Figure 10 shows that this LZ domain is conserved among plant CDF proteins. The most notable feature of the LZ is the presence of a repeating pattern of Leu residues at every seventh position (LX6)n. More generally, however, the overall architecture of coiled coils is defined by a 4,3 hydrophobic repeat in the primary amino acid sequence. Thus, the hydrophobic positions a and d of the sequence (abcdefg)n fall on the same face of an α-helix. In some cases, the Leu residues are replaced by Ile or Val and can include “skips” and “hiccups” (Leung and Lassam, 1998). This explains the unsuccessful searches for LZ motifs in some other CDF proteins (e.g., AtMTP1) with search programs that are designed to detect only the conventional (LX6)n motif, even though the motifs are effectively present (Figure 10).
Figure 10.
Conservation of a LZ Motif in Plant CDFs.
Numbers in parentheses correspond to the first residue in each sequence. Gray-shaded residues indicate conserved “d” positions of the Leu/Ile/Val heptad repeats. Sequences were aligned as described in the legend to Figure 1. Accession numbers are given at the end of Methods.
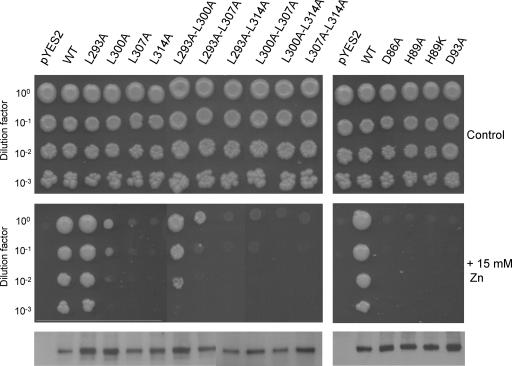
To determine the role of the conserved LZ in the oligomeric structure and biological activity of PtdMTP1:GFP, the heptadic Leu residues 293, 300, 307, and 314 were changed individually or in pairwise combination to Ala residues. Ala was chosen as a substituting residue for Leu because (1) Ala has a strong propensity to form an α-helix and its substitution has minimal disruptive effects on the α-helical structure characteristic of a LZ domain in proteins, and (2) Ala lacks the bulky hydrophobic side chain of Leu, which is believed to be the major hydrophobic force for a LZ domain to facilitate protein interaction (Luo et al., 1999). Figure 11 (bottom gel) shows that all of the mutant proteins were expressed at levels identical to that of the wild-type protein. Substitution of one or two of the four Leu residues did not alter the size of the fusion protein oligomer, as determined by gel electrophoresis, nor was the subcellular localization of the mutant PtdMTP1:GFP proteins changed. However, as shown in Figure 11, although the L293A substitution had no apparent effect on function, the L314A substitution completely abolished the functional complementation of the zrc1Δ mutant by PtdMTP1 on 15 mM Zn. Other single substitutions (L300A and L307A) showed intermediate patterns: the reduction in yeast growth at high Zn concentration was marked but not fully abolished. Double Leu substitution mutants confirmed the patterns observed with the single Leu mutants, with a slight additional effect in growth inhibition, especially for the L300A-L307A mutant (Figure 11).
Figure 11.
Evaluation of the Functionality of PtdMTP1 Mutants by Yeast Complementation Assay.
Yeast zrc1Δ mutants were transformed with the empty vector pYES2, with pYES2-PtdMTP1, or with various mutant pYES2-PtdMTP1 vectors. Cultures were adjusted to OD = 1.0, and 2 μL of serial dilutions were spotted on SD medium without extra metal (top panels) or supplemented with 15 mM Zn (middle panels). Plates were incubated for 6 days at 30°C. Protein gel blot analysis of microsomes of transformed mutants with anti-GFP antibodies is shown in the bottom panel. Cells were processed for immunodetection as described in the legend to Figure 8. WT, wild type.
Residue Substitutions in the CDF Motif Abolish Yeast Complementation
PtdMTP1 contains the CDF signature, which begins with Ser-80 and continues just past the fully conserved Asp-93. We created four mutants (D86A, H89A, H89K, and D93A) by site-directed mutagenesis of either semi (D86 and H89) or fully (D93) conserved residues. All of the mutant proteins were expressed at levels identical to that of the wild-type protein (Figure 11, bottom gel), still formed dimers, and were localized on the vacuolar membrane (data not shown). However, as shown in Figure 11, all four mutants failed to complement the Zn hypersensitivity of zrc1Δ cells, as shown by the complete abolition of yeast growth on 15 mM Zn.
DISCUSSION
The novel transporter from the hybrid poplar Populus trichocarpa × deltoides, PtdMTP1, is a member of the CDF family. The CDF family can be subdivided into three (van der Zaal et al., 1999; Gaither and Eide, 2001) or four (Mäser et al., 2001) groups. Following the classification given by Gaither and Eide (2001), PtdMTP1 is most closely related to group-III CDF members (Figure 1). Heterologous expression of PtdMTP1 in yeast lacking either of the vacuolar cytoplasmic efflux carriers ZRC1 or COT1 complements only the zinc sensitivity of these yeast strains, strongly suggesting that PtdMTP1 is able to selectively transport zinc ions into the yeast vacuole. These properties reflect those of AtZAT (AtMTP1), which when reconstituted in proteoliposomes was shown to transport Zn but not Co (Bloss et al., 2002). However, the AtZAT gene did not rescue the zinc sensitivity of a zrc1Δ single mutant or a zrc1Δ cot1Δ double mutant strain of S. cerevisiae, but it did rescue that of a zrc1Δ mutant strain of S. pombe (Bloss et al., 2002).
A critical question in establishing the physiological function of CDF transporters relates to their cellular location. Table 1 shows for a number of CDF family members a range of disparate locations, including on most intracellular membranes and the plasma membrane. Thus, ShMTP1, a S. hamata CDF member that specifically transports Mn, was found recently to be targeted to the vacuolar membrane in planta but localized to endoplasmic reticulum membranes when expressed in yeast (Delhaize et al., 2003). One of the homologous mammalian zinc resistance transporters, ZNT1, has been localized to the plasma membrane and reported to function in Zn efflux (Palmiter and Findley, 1995). A second, related mammalian zinc resistance transporter, ZNT2, is located on intracellular membranes and appears to facilitate the vesicular sequestration of Zn (Palmiter et al., 1996a). The reported localization of MSC2, a CDF family member from S. cerevisiae, in the endoplasmic reticulum/nucleus and the higher nuclear zinc content of msc2Δ cells suggested a requirement for transporter-facilitated exchange of zinc between the cytosol and the nucleus (Li and Kaplan, 2001). ZHF, a CDF member from S. pombe, is hypothesized to mediate the transport of zinc into the endoplasmic reticulum and the nuclear envelope in fission yeast (Clemens et al., 2002a). By contrast, the S. cerevisiae CDF transporter ZRC1 has been localized to the yeast vacuole membrane (Li and Kaplan, 1998; MacDiarmid et al., 2000, 2002; Miyabe et al., 2001). The present study demonstrates the targeting of PtdMTP1:GFP fusion proteins to the vacuolar membranes of yeast, living onion epidermal cells, and Arabidopsis cells, effectively confirming the physiological role of the transporter in vacuolar Zn sequestration.
Table 1.
Fully Localized Eukaryotic Members of the CDF Family
| Gene | Substrate | Organism | Localization | Reference |
|---|---|---|---|---|
| MTP1 | Zn | Hybrid poplar | Vacuole | Present study |
| MTP1 | Mn | S. hamata | Vacuole | Delhaize et al. (2003) |
| ZNT7 | Zn | H. sapiens | Golgi | Kirschke and Huang (2003) |
| ZHF1 | Zn, Co, Cd | S. pombe | Endoplasmic reticulum | Clemens et al. (2002a) |
| ZNT6 | Zn | H. sapiens | Golgi | Huang et al. (2002) |
| MCS2 | Zn | S. cerevisiae | Endoplasmic reticulum/nucleus | Li and Kaplan (2001) |
| ZRC1 | Zn, Cd | S. cerevisiae | Vacuole | Li and Kaplan (1998) |
| COT1 | Co, Zn | S. cerevisiae | Vacuole | Li and Kaplan (1998) |
| ZNT3 | Zn | H. sapiens | Vesicles | Palmiter et al. (1996b) |
| ZNT2 | Zn | H. sapiens | Endosome/lysosome | Palmiter et al. (1996a) |
| ZNT1 | Zn | H. sapiens | Plasma membrane | Palmiter and Findley (1995) |
In eukaryotes, two families of transporters have been implicated in zinc transport. ZIP family members play prominent roles in cytosolic zinc import, transporting zinc from outside the cell (Pence et al., 2000) or from within an intracellular compartment (Gaither and Eide, 2001). The second group of transporters, the CDF family, transports zinc in a direction opposite that of the ZIP transporters, promoting cytosolic zinc efflux. Chardonnens et al. (1999) reported evidence that transport across the vacuolar membrane plays an essential role in naturally selected zinc-tolerant Silene vulgaris. van der Zaal et al. (1999) suggested that increased Zn transport into the vacuoles of Zn-tolerant species such as S. vulgaris might be attributable to a ZAT-like protein, although data on ZAT localization were lacking. The present study demonstrates that the ZAT homolog PtdMTP1 is located at the vacuolar membrane, which is consistent with a role for PtdMTP1 in zinc sequestration. Zinc transporters—either of the ZIP or CDF families—have not previously been identified plant vacuolar membranes.
PtdMTP1 mRNA was expressed constitutively throughout the hybrid poplar used in this study and was not expressed differentially by varying the Zn concentrations. Similarly, RNA gel blot experiments in Arabidopsis indicated that ZAT mRNA was expressed throughout the plant at uniform but low levels and was not induced by increasing the Zn concentrations (van der Zaal et al., 1999). Expression of the Ni transporter MTP1 from T. goesingense failed to increase during Ni exposure, although in this MTP1-related Ni hyperaccumulator, MTP1 is much more highly expressed than in nonaccumulator species, presumably conferring enhanced ability to accumulate Ni ions within vacuoles (Persans et al., 2001). Under metal stress, the regulation of CDF transporters might occur post-transcriptionally, or these transporters might simply perform a housekeeping function.
To date, no clear picture has emerged regarding whether plant CDF transporters might energize metal transport through coupling to antiport with H+. Bloss et al. (2002) showed that the rate and extent of Zn uptake in proteoliposomes containing purified and reconstituted ZAT1 (AtMTP1) were unaffected by the presence of a protonmotive force, indicating the probable absence of H+ coupling. By contrast, the failure of the Mn transporter ShMTP1 to complement the Mn-sensitive phenotype of yeast mutants defective in vacuolar acidification led Delhaize et al. (2003) to conclude that this CDF transporter is H+ coupled. We adopted a similar approach, expressing PtdMTP1 in yeast vacuolar ATPase mutants in which ATP-driven H+ pumping is severely defective (vph2Δ; Bachhawat et al., 1993) or in which vacuolar ATPase activity is abolished completely (vma8Δ; Graham et al., 1995). Our results demonstrate the complementation of the Zn-sensitive mutant phenotype, albeit not to the extent to which complementation occurred when PtdMTP1 was expressed in a zrc1Δ background. It is possible, then, that PtdMTP1, like ZAT1, is not H+ coupled, although these complementation results should be interpreted with caution. The possible presence of PtdMTP1 on nonvacuolar endomembranes in yeast (Figure 5D) might confer a partial complementation phenotype if acidification of compartments bounded by such membranes is not vacuolar ATPase dependent. This conclusion raises the question of whether PtdMTP1 actively sequesters Zn in the vacuolar lumen through energized transport or whether the capacity to confer Zn tolerance arises through the formation of a passive Zn permeation pathway that provides access to the additional intracellular volume afforded by the vacuolar lumen. Nevertheless, a clear physiological role in conferring Zn tolerance in planta is indicated by the ability of plants that overexpress PtdMTP1 to survive and grow at high Zn concentrations that are lethal to control plants.
Previous studies involving metal blots have suggested that AtZAT might form dimers (Bloss et al., 2002), although direct evidence for this contention has been lacking. The protein gel blots presented here show that the dominant extractable form of PtdMTP1, whether expressed in yeast or plants, is oligomeric. Further experiments with other CDF members and other species are needed to determine whether CDF oligomerization is a common feature of plant CDFs and is present in members of other kingdoms. Although it is clear that PtdMTP1 can form homooligomers, the possibility of heterooligomer formation with other CDF members cannot be excluded where CDF proteins are encoded by multigene families. Stable oligomer formation in PtdMTP1 might be dependent, at least in part, on the formation of one or more disulfide bonds, because the addition of DTT resulted in a shift toward the monomeric form.
Interestingly, we discovered a LZ motif in PtdMTP1. The LZ domain is a helical structure that forms coiled coils and was identified originally as a highly conserved motif mediating the binding of transcription factors to DNA (Landschulz et al., 1988). The role of LZ domains in promoting the homodimerization and heterodimerization of transcription factors has been well documented (Turner and Tjian, 1989). LZ domains also have been shown to mediate protein–protein interactions (Surks et al., 1999) and intraprotein oligomerization (Simmerman et al., 1996) and also were implicated in the formation of macromolecular complexes (Marx et al., 2001). The finding of a LZ motif in PtdMTP1 encouraged us to examine its functional importance. Our results indicate that the conserved Leu residues are not necessary for oligomer formation but are required for the functional activity of PtdMTP1. Analogous findings have been reported for other systems. Thus, triple Leu-to-Ala substitutions on the paramyxovirus fusion proteins demonstrated that the conserved Leu residues in the LZ motif were not necessary for oligomer formation but were required for the fusogenic ability of the protein (Reitter et al., 1995). Subunit assembly of animal Shaker K+ channels does not depend on the Leu heptad repeat, even though substitutions of the Leu residues in the repeat produced large effects on the voltage dependence of conductance and prepulse activation curves (McCormack et al., 1991). It seems likely that the LZ in PtdMTP1 mediates subtle interactions that are required for the function of the multimeric complex.
It appears that the fourth and last Leu residue (Leu-314) of the LZ motif is particularly critical for the function of PtdMTP1. Interestingly, this final Leu residue of the heptad repeat was found to be highly conserved among plant CDFs (Figure 10). Studies of the AtZAT protein have revealed that the C-terminal putative intracellular region is critical for activity, because truncation just after the last transmembrane domain was not able to confer Zn resistance or Zn accumulation in Arabidopsis (van der Zaal et al., 1999). This truncation removed the final two residues of the LZ domain (Val-318 and Leu-319) and, although not precluding an important role for other residues at the C terminus, it is intriguing to speculate that the removal of Leu-319 alone likely would have abolished function.
PtdMTP1 possesses the CDF family–specific signature sequence (Paulsen and Saier, 1997) that begins with Ser-80 and continues just past Asp-93. Substitution with Ala of any of the semi (Asp-86 or His-89) or fully (Asp-93) conserved residues within this sequence yielded a nonfunctional protein that was unable to rescue zrc1Δ mutant yeast. Because the mutated amino acids are charged residues, it remained possible that the mutation abolished a salt bridge. However, in the case of His-89, the H89K mutant also was nonfunctional; thus, disruption of a salt bridge seems unlikely. Therefore, this motif might be essential for in vivo transport activity of the protein but is not required for oligomerization or for tonoplast localization of PtdMTP1. The demonstration of oligomerization abilities and the identification of a LZ motif provide a new foundation for understanding the structure and function of the CDF family, proteins that could be important in phytoremediation technologies.
To breed plants with superior phytoremediation potential, one possible strategy is to enhance the biomass productivity of species that are hyperaccumulators. An alternative strategy is to enhance metal accumulation in high-biomass species (Pilon-Smits and Pilon, 2002). Indeed, the combination of fast-growing and high-biomass-producer characteristics together with the metal hyperaccumulation trait would be an invaluable tool in phytoremediation technology. Hybrid poplars have been grown primarily for raw material production, but the utility of poplar now extends to the treatment of wastewater, nutrient removal from agriculture runoff, and phytoremediation of industrial landfills (Stanton et al., 2002). It was demonstrated recently that poplar pants have the potential to be used for plantations in Zn-contaminated soils (Di Baccio et al., 2003). Because vacuolar sequestration is clearly a process to target to improve metal tolerance (Küpper et al., 1999; Persans et al., 2001; Clemens et al., 2002b), poplar species/clones exhibiting high zinc accumulation–like characteristics could be screened using MTP1 protein as a suitable marker with a view to improving the potential of this gene for Zn phytoremediation. Poplar clones overexpressing PtdMTP1 also could be used for phytoremediation technologies.
METHODS
Plant Growth Conditions
Hybrid poplar (Populus trichocarpa × Populus deltoides cv Beaupré) was grown in a greenhouse in a controlled environment as described previously (Kohler et al., 2003). Arabidopsis thaliana plants (ecotype Columbia) were grown in Klasman Substrate No. 1 compost (Klasmann-Deilmann, Geest, Germany) in a greenhouse at 23°C with a 16-h photoperiod. Alternatively, seeds were sterilized and grown at 22°C with a 16-h photoperiod on agar plates containing sterile MS medium (Murashige and Skoog, 1962).
For Zn resistance assays of Arabidopsis lines expressing PtdMTP1 and lines transformed with the empty vector pART27, homozygous T3 seeds were sown on plates containing half-strength MS medium supplemented with various concentrations of ZnSO4. Wild-type plants were also used as a control. After 3 weeks of growth, plant fresh weights were determined.
Cloning of PtdMTP1, cDNA Array, and Reverse Transcriptase–PCR Analysis
Standard techniques were used for DNA preparation (Sambrook and Russell, 2001). A cDNA library was made from the adventitious root system of 2-month-old rooted cuttings as described by Kohler et al. (2003). The pTriplex2 phagemid clones in Escherichia coli were obtained using the mass excision protocol according to the manufacturer's instructions (Clontech, Erembodegem, Belgium), and DNA sequencing on randomly chosen bacterial clones was performed as described by Kohler et al. (2003). Generation and analysis of cDNA arrays were performed as reported elsewhere (Lacourt et al., 2002; Kohler et al., 2003). Developing root tissues of cuttings were harvested at six different stages (dormant bark tissues, root primordia, root calli, emerging roots, primary roots, and lateral root tips), frozen in liquid N2, and stored at −80°C for RNA isolation. Photographs of the different developmental stages are available at http://mycor.nancy.inra.fr/poplardb/index.html. EST database searches were performed using Basic Local Alignment Search Tool (BLAST; Altschul et al., 1990).
Total RNA isolation was performed with the RNeasy Plant Mini kit (Qiagen, Courtabœuf, France) from ∼100 mg of frozen material. An average of 40 μg of total RNA per 100 mg of fresh material was isolated and stored in diethyl pyrocarbonate–treated water at −70°C until further use. The integrity and size distribution were checked by agarose gel electrophoresis. Reverse transcription (RT) reactions were performed from 1 μg of RNA using the enzyme Omniscript (Qiagen) as recommended by the manufacturer. RT reactions were performed for 60 min at 37°C, and RT products were used immediately in PCR or kept frozen until use. Two microliters of RT products were amplified by PCR using the primers MTP1FP1 (5′-CCTCCTCTTCGATTCTTGGA-3′) and MTPRP1 (5′-TCAAGCACAAGCACATGGAT-3′) in the following conditions: 95°C for 1 min followed by 35 cycles at 95°C for 5 s, 65°C for 5 s, and 68°C for 6 min using an Eppendorf Mastercycler (Eppendorf, Le Pecq, France). The suitability of the extracted RNA for RT-PCR amplification was checked by performing RT-PCR control experiments with ubiquitin using the primers UBQFP1 (5′-GCACCTCTGGCAGACTACAAA-3′) and UBQRP1 (5′-TAACAGCCGCTCCAAACAGT-3′) in the same amplification conditions.
Yeast Strains, Media, and Transformation
The yeast strains used for the heterologous expression of PtdMTP1 were BY4741 (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0), zrc1Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 ZRC1::kanMX4), cot1Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 COT1::kanMX4), ycf1Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 YCF1::kanMX4), pmr1Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 PMR1::kanMX4), smf1Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 SMF1::kanMX4), vma8Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 VMA8::kanMX4), and vph2Δ (MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 VPH2::kanMX4). The seven mutants used derived from the parental strain BY4741 and were all obtained from Euroscarf (http://www.uni-frankfurt.de/fb15/mikro/euroscarf/). Growth was in yeast potato dextrose or in synthetic defined (SD) medium with 2% (w/v) glucose or galactose, supplemented as necessary (Sherman, 1991). For metal tolerance assays, yeast was grown in liquid SD medium (with 2% [w/v] galactose) until OD = 1, and drop assays were performed on SD plates containing different concentrations of Zn, Cd, Co, Mn, or Ni. Yeast cells were transformed as described elsewhere (Gietz et al., 1992). The PtdMTP1 cDNA was subcloned from the pTriplex2 clone into the pYES2 plasmid (Invitrogen, Paisley, UK) using MTP1f (5′-CCCCCAAGCTTATGGAAGCACAAAATCCTCA-3′) and MTP1r (5′-CCCCGGATCCCTAACGCTCTATCTGGATGGTT-3′) primers, introducing HindIII and BamHI sites (sites underlined).
Site-Directed Mutagenesis
Mutant PtdMTP1 plasmid clones were generated by oligonucleotide-directed PCR mutagenesis. The desired amino acid codon changes were incorporated into a PCR-amplified fragment using the 5′ flanking primer MTP1f and the 3′ flanking primer MTP1r and a series of mutagenic primer pairs (Table 2). The resulting mutant HindIII-BamHI fragments were excised and ligated back into the pYES2 vector. To generate single amino acid mutants, pYES2-PtdMTP1 was used as a template for the PCR. Double Leu-to-Ala substitution mutants were generated by the same process as the single mutants except that the PCR templates were plasmids containing corresponding single Leu-to-Ala codon substitutions instead of pYES2-PtdMTP1. The presence of targeted mutations in all plasmid constructs was verified by DNA sequencing.
Table 2.
Oligonucleotides for Amino Acid Substitution by PCR Mutagenesis
| Name a | Positions b | Sequence (5′ to 3′) c | Mutation d |
|---|---|---|---|
| Mut1f | 241-270 | CTGGCAATCTTGACTgcTGCTGCACATTTG | D86A |
| Mut1r | 270-241 | CAAATGTGCAGCAgcAGTCAAGATTGCCAG | |
| Mut2f | 250-279 | TTGACTGATGCTGCAgcTTTGCTTTCAGAT | H89A |
| Mut2r | 279-250 | ATCTGAAAGCAAAgcTGCAGCATCAGTCAA | |
| Mut3f | 250-279 | TTGACTGATGCTGCAaAgTTGCTTTCAGAT | H89K |
| Mut3r | 279-250 | ATCTGAAAGCAAcTtTGCAGCATCAGTCAA | |
| Mut4f | 262-291 | GCACATTTGCTTTCAGcTGTTGCAGGCTTT | D93A |
| Mut4r | 291-262 | AAAGCCTGCAACAgCTGAAAGCAAATGTGC | |
| Mut5f | 865-894 | CTGATCTGCACCgcAATCTTTTCTGTTATA | L293A |
| Mut5r | 894-865 | TATAACAGAAAAGATTgcGGTGCAGATCAG | |
| Mut6f | 886-915 | TCTGTTATAGTTgcGGGTACAACAATCAAA | L300A |
| Mut6r | 915-886 | TTTGATTGTTGTACCCgcAACTATAACAGA | |
| Mut7f | 907-936 | ACAATCAAAATGgcACGGAACATACTTGAA | L307A |
| Mut7r | 936-907 | TTCAAGTATGTTCCGTgcCATTTTGATTGT | |
| Mut8f | 928-957 | ATACTTGAAGTCgcGATGGAGAGCACTCCA | L314A |
| Mut8r | 957-928 | TGGAGTGCTCTCCATCgcGACTTCAAGTAT |
Two partially complementary primers (f and r) were used in each PCR mutagenesis for the introduction of amino acid codon substitutions.
The region of the PtdMTP1 sequence in each oligonucleotide is marked by the positions of the 5′ and 3′ nucleotides separated by a dash. The position of the 5′ oligonucleotide is shown first for each oligonucleotide. All positions are numbered in reference to the first nucleotide of the PtdMTP1 coding sequence.
Uppercase letters indicate that the sequence is exactly the same as the wild-type PtdMTP1 DNA sequence. Lowercase letters indicate the mismatched nucleotides designed for specific mutations.
The designated amino acid substitutions resulting from each mutagenic primer pair.
Construction of GFP and V5 Fusions for Expression in Yeast
To construct PtdMTP1:GFP, the GFP (S65T) cDNA was amplified from the plasmid PFA6a-GFP65T-kanMX6 with the primers GFPf (5′-CCCCTCTAGAATTAACAGTAAAGGAGAAGAACTTTTCA-3′) and GFPr (5′-CCCCTCTAGACTATTTGTATAGTTCATCCATGCC-3′), introducing a XbaI site at both ends (sites underlined), and cloned at the XbaI site in the pYES2 plasmid to generate a new plasmid called pYES2-GFP. Then, a PtdMTP1 fragment, lacking the stop codon of the PtdMTP1 cDNA, was amplified with the primers MTP1f and MTP1rnostop (5′-CCCCGGATCCCCACGCTCTATCTGGATGGTTACAT-3′) (BamHI site underlined) and cloned into the HindIII-BamHI–digested pYES2-GFP plasmid. PtdMTP1:V5 was constructed by inserting the HindIII-BamHI fragment of PtdMTP1 into pYES6/CT (Invitrogen, Carlsbad, CA). Mutant PtdMTP1 clones also were fused in frame with GFP or V5 into the pYES2-GFP or pYES6/CT plasmid, respectively. As a control, the expression of nontargeted GFP in yeast was achieved by transforming cells with pYES2 in which only the full-length GFP cDNA was cloned.
Transient Expression of PtdMTP1 in Onion Cells
The intracellular localization of PtdMTP1 was determined by monitoring the transient expression of a PtdMTP1:GFP translational fusion product in onion epidermal cells after DNA particle bombardment. The coding region of mGFP5, a green fluorescent protein modified for plants (Haseloff et al., 1997), was fused in frame to the 3′ terminus of full-length PtdMTP1 cDNA using a three-step PCR procedure. First, a mGFP5 fragment was amplified by PCR from the plasmid pCAMBIA1303 using the primers GFP5f (5′-AAAGGAGAAGAACTTTTCACTGGAGTTGTC-3′) and GFP5r (5′-AAAATCTAGATTTGTATAGTTCATCCATGC-3′), introducing a XbaI site at the 3′ end of the fragment (site underlined). In a second step, a fragment consisting of the full-length PtdMTP1 cDNA (stop codon removed) plus the first 21 nucleotides of mGFP5 was amplified by PCR from the plasmid pYES2-PtdMTP1 with the primer pair MTP1 gfpf (5′-AAAACTCGAGATGGAAGCACAAAATCCTCA-3′) and MTP1 gfpr (5′-AGTGAAAAGTTCTTCTCCTTTACGCTCTATCTGGATGGTTACATG-3′), introducing a XhoI site at the 5′ end of the fragment (site underlined). DNA fragments were gel purified, mixed in a PCR mix lacking primers, denatured at 94°C for 1 min, annealed at 55°C for 1 min, elongated at 68°C for 3 min, and subsequently subjected to 25 PCR cycles after the addition of the MTP1 gfpf and GFP5r primers. The resulting XhoI-XbaI fragment was cloned between the 35S promoter of Cauliflower mosaic virus and the octopine synthase terminator in the XhoI-XbaI–digested pART7 plasmid (Gleave, 1992). The construct was coated onto gold particles (0.6 μm) and delivered into onion cells with a Helios Gene Gun System (Bio-Rad Laboratories, Hercules, CA). The bombardment parameters were as follows: discharge pressure of 150 p.s.i., and distance to target tissue of 3.5 cm. Onion cells were placed onto MS agar plates before bombardment and incubated at 22°C for 24 h after particle delivery.
Generation of Transgenic Arabidopsis Expressing PtdMTP1 or PtdMTP1:GFP
For PtdMTP1 and PtdMTP1:GFP overexpression in Arabidopsis, expression cassettes present in pART7 (see above) were isolated as NotI fragments and cloned in NotI-digested pART27 (Gleave, 1992). The resulting plasmids were used to transform Agrobacterium tumefaciens strain GV3101. Transformants were selected for spectinomycin resistance and were used to transform Arabidopsis by the floral-dip method (Clough and Bent, 1998). Plant transformants were selected on MS agar plates containing 50 mg/L kanamycin. Subsequent generations also were selected on kanamycin to identify homozygous lines. Arabidopsis transgenic plants expressing AtMGT1:GFP (Li et al., 2001) were the kind gift of L. Li and S. Luan (University of California, Berkeley).
Cell Labeling and Confocal Microscopy
Yeast vacuolar membranes were selectively stained with the red fluorescent probe FM4-64 (Molecular Probes, Leiden, The Netherlands). This lipophilic styryl dye has been reported to stain yeast vacuolar membranes selectively (Vida and Emr, 1995). Cells were incubated with FM4-64 for 30 min at 30°C, washed with SD medium, and incubated 1 h before observation by confocal microscopy. These incubation times are sufficient for the dye to reach the vacuolar membrane with no residual plasma membrane or endosomal staining (Humair et al., 2001). Staining of root cell walls and nuclei was achieved by incubating roots in 100 μg/L propidium iodide.
The GFP fluorescence of yeast and plant cells was visualized by confocal laser scanning microscopy (MRC-1000 [Bio-Rad] or Meta 510 [Zeiss, Jena, Germany]). Control cells did not show any green or red fluorescence for the settings at which images usually were collected. Digital images were recorded using the confocal microscope operating system software. For final processing, digital images were pseudocolored using Adobe Photoshop 4.0 (Mountain View, CA), with red attributed to FM4-64 or propidium iodide and green attributed to GFP, to correspond to real red and green colors. An image with transmitted light also was collected when needed using the confocal microscope.
Protein Extraction
Yeast cells were collected by centrifugation at 3000g for 5 min. Cell lysates were obtained by vortexing cells at full speed for 2 min with glass beads (420 to 600 μm; Sigma, Dorset, UK) in ice-cold lysis buffer containing 100 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM EDTA, 10% (v/v) glycerol, 100 μg/mL phenylmethylsulfonyl fluoride, 1 μg/mL leupeptin, and 10 μg/mL pepstatin A. Lysates were centrifuged at 3,000g for 5 min at 4°C to pellet cell debris, and beads and the supernatants were centrifuged subsequently at 12,000g for 45 min at 4°C. Pellets (membrane fraction) were resuspended subsequently in ice-cold lysis buffer containing 2 mg/mL n-dodecyl β-d-maltoside and incubated for 2 h on ice with occasional mixing. The extracts were centrifuged at 13,000g for 5 min to remove insoluble material.
To obtain proteins of microsome fractions from plants expressing PtdMTP1:GFP, 3-week-old shoot tissues were ground in the presence of liquid nitrogen and homogenized subsequently with Tris buffer, pH 7.5, that contained 5 mM EGTA, 0.25% (v/v) Tween 20, 10 mM phenylmethylsulfonyl fluoride, and a cocktail of protease inhibitors (EDTA-free protease inhibitor cocktail tablets; Roche Diagnostics, Mannheim, Germany). The extracts were centrifuged first at 3,600g for 5 min to remove debris and subsequently at 13,000g for 15 min to pellet the microsome fraction. Finally, the proteins were homogenized in the extraction buffer. Even if an equivalent number of cells or mass of plant tissue was processed in each extraction, protein quantitation was performed with the Bio-Rad protein assay to standardize the extracts for protein concentration.
Protein Gel Blot Analysis
Protein extracts were mixed with sample buffer (60 mM Tris-HCl, pH 6.8, 25% [v/v] glycerol, 2% [w/v] SDS, and 0.0125% [v/v] bromophenol blue) either with (1 to 200 mM) or without the reducing agent DTT and subjected to SDS-PAGE (Laemmli, 1970), and proteins were transferred to polyvinylidene difluoride membranes (Bio-Rad). Blots were probed either with mouse anti-V5-HRP antibodies (Invitrogen) (1:5000) or with rabbit GFP antiserum (Invitrogen) (1:5000). In the latter case, goat anti-rabbit peroxidase-conjugated antibodies (Calbiochem-Novabiochem, Bad Soden, Germany) (1:10,000) were used as the secondary reagent. Proteins were revealed by enhanced chemiluminescence using the ECL protein gel blot detection kit (Amersham Pharmacia Biotech, Freiburg, Germany) according to the manufacturer's instructions.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Michel Chalot, michel.chalot@scbiol.uhp-nancy.fr.
Accession Numbers
The GenBank accession numbers for the sequences described in this article are as follows: Populus trichocarpa × deltoides: PtdMTP1, AY450453; Eucalyptus grandis: EgMTP1, AAL25646; Arabidopsis thaliana: AtMTP1, NP_182203; AtMTP2, NP_191753; AtMTP3, NP_191440; AtMTP4, NP_180502; AtMTP5, NP_187817; ATMTP6, NP_182304; AtMTP7, NP_564594; AtMTP8, NP_191365.1; AtMTP9, NP_178070.1; AtMTP10, NP_173081; AtMTP11, NP_181477; and AtMTP12, NP_178539; Oryza sativa: OsCDF1, BAC24961; Stylosanthes hamata: ShMTP1, AY181256; ShMTP2, AY181257; ShMTP3, AY181258; and ShMTP4, AY181259; Thlaspi caerulescens: TcZTP1, AAK69428; Thlaspi goesingense: TgMTP1, AAK91869; Saccharomyces cerevisiae: ScZRC1, NP_013970; ScCOT1, NP_014961; ScMSC2, NP_010491; ScMMT2, NP_015100; and ScMFT1, NP_013649; Schizosaccharomyces pombe: SpZHF, NP_593645; Neurospora crassa: NcCDF2, EAA32880.1; and NcCDF1, EAA34958; Magnaporthe grisea: MgCDF1, 03634; and MgCDF2, 10493; Homo sapiens: HsZNT1, Q9Y6M5; HsZNT3, Q99726; HsZNT4, O14863; HsZNT5, AAM09099.1; HsZNT7, NP_598003.1; and HsZNT8, NP_776250.1; Caenorhabditis elegans: CeCDF1, NP_509095.1; CeCDF2, NP_498611.1; CeCDF3, NP_499691.1; CeCDF4, NP_509279.2; and CeCDF5, NP_740931.1; Vibrio parahaemolyticus: VpCDF1, NP_799235.1; Pseudomonas putida: PpCDF, NP_742196.1; Escherichia coli: EcZITB, Q8X400; Alcaligenes sp CT14: AlCZSD, P94178; Salmonella typhimurium: StCDF1, NP_462942.1; and Yersinia pestis: YpCDF2, NP_667404.1. Other accession numbers are CA825068 (UBQ) and CB833313 (EXT1).
Acknowledgments
We thank Legong Li and Sheng Luan (University of California, Berkeley) for providing Arabidopsis transgenic plants expressing AtMGT1 fused to GFP and Catherine Jauniaux (Institut de Biologie et de Médecine Moléculaires, Gosselies, Belgium) for providing the PFA6a-GFP65T-kanMX6 plasmid. We acknowledge Chris Winefield (University of York, UK) for the gift of pART7 and pART27 plasmids and for helpful discussions. We also thank Peter O'Toole (University of York) for providing help with confocal microscopy, Sylvain Jeandroz (University H. Poincaré, Nancy, France) for helping with phylogenetic analysis, Valérie Legué (University H. Poincaré) for providing access to microscopy and image-analysis facilities, Pascal Frey (Institut National de la Recherche Agronomique [INRA], Nancy, France) for the gift of poplar trees, and Jared Cartwright, Viktor Filatov, Frans Maathuis (University of York), and Jean Pierre Jacquot (University H. Poincaré) for stimulating discussions. The contributions of Jeremy Couturier and Régis Belleville are gratefully acknowledged. A.K. was supported by postdoctoral fellowships from the INRA and the Région de Lorraine. The present investigation was supported by grants from the INRA (Programmes Functional Genomics of Poplar and LIGNOME) and from the European Union, Framework Programme 5 (Metallophytes project).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.017541.
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Bachhawat, A.K., Manolson, M.F., Murdock, D.G., Garman, J.D., and Jones, E.W. (1993). The VPH2 gene encodes a 25 kDa protein required for activity of the yeast vacuolar H+-ATPase. Yeast 9, 175–184. [DOI] [PubMed] [Google Scholar]
- Bloss, T., Clemens, S., and Nies, D.H. (2002). Characterization of the ZAT1p zinc transporter from Arabidopsis thaliana in microbial model organisms and reconstituted proteoliposomes. Planta 214, 783–791. [DOI] [PubMed] [Google Scholar]
- Chardonnens, A.N., Koevoets, P.L., van Zanten, A., Schat, H., and Verkleij, J.A. (1999). Properties of enhanced tonoplast zinc transport in naturally selected zinc-tolerant Silene vulgaris. Plant Physiol. 120, 779–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens, S. (2001). Molecular mechanisms of plant metal tolerance and homeostasis. Planta 212, 475–486. [DOI] [PubMed] [Google Scholar]
- Clemens, S., Bloss, T., Vess, C., Neumann, D., Nies, D.H., and Zur Nieden, U. (2002. a). A transporter in the endoplasmic reticulum of Schizosaccharomyces pombe cells mediates zinc storage and differentially affects transition metal tolerance. J. Biol. Chem. 277, 18215–18221. [DOI] [PubMed] [Google Scholar]
- Clemens, S., Palmgren, M.G., and Krämer, U. (2002. b). A long way ahead: Understanding and engineering plant metal accumulation. Trends Plant Sci. 7, 309–315. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Conklin, D.S., McMaster, J.A., Culbertson, M.R., and Kung, C. (1992). COT1, a gene involved in cobalt accumulation in Saccharomyces cerevisiae. Mol. Cell. Biol. 12, 3678–3688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delhaize, E., Kataoka, T., Hebb, D.M., White, R.G., and Ryan, P.R. (2003). Genes encoding proteins of the cation diffusion facilitator family that confer manganese tolerance. Plant Cell 15, 1131–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Baccio, D., Tognetti, R., Sebastiani, L., and Vitagliano, C. (2003). Responses of Populus deltoides × Populus nigra (Populus × euramericana) clone I-214 to high zinc concentrations. New Phytol. 159, 443–452. [DOI] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Gaither, L.A., and Eide, D.J. (2001). Zinc transporters and their regulation. Biometals 14, 251–270. [DOI] [PubMed] [Google Scholar]
- Gietz, D., St. Jean, A., Woods, R.A., and Schiestl, R.H. (1992). Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 20, 1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave, A.P. (1992). A versatile binary vector system with a T-DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207. [DOI] [PubMed] [Google Scholar]
- Gordon, M., Choe, N., Duffy, J., Ekuan, G., Heilman, P., Muiznieks, I., Ruszaj, M., Shurtleff, B.B., Strand, S., Wilmoth, J., and Newman, L.A. (1998). Phytoremediation of trichloroethylene with hybrid poplars. Environ. Health Perspect. 106, 1001–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham, L.A., Hill, K.J., and Stevens, T.H. (1995). VMA8 encodes a 32-kDa V1 subunit of the Saccharomyces cerevisiae vacuolar H+-ATPase required for function and assembly of the enzyme complex. J. Biol. Chem. 270, 15037–15044. [DOI] [PubMed] [Google Scholar]
- Haseloff, J., Siemering, K.R., Prasher, D.C., and Hodge, S. (1997). Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. USA 94, 2122–2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, L., Kirschke, C.P., and Gitschier, J. (2002). Functional characterization of a novel mammalian zinc transporter, ZNT6. J. Biol. Chem. 277, 26389–26395. [DOI] [PubMed] [Google Scholar]
- Humair, D., Hernandez Felipe, D., Neuhaus, J.M., and Paris, N. (2001). Demonstration in yeast of the function of BP-80, a putative plant vacuolar sorting receptor. Plant Cell 13, 781–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamizono, A., Nishizawa, M., Teranishi, Y., Murata, K., and Kimura, A. (1989). Identification of a gene conferring resistance to zinc and cadmium ions in the yeast Saccharomyces cerevisiae. Mol. Gen. Genet. 219, 161–167. [DOI] [PubMed] [Google Scholar]
- Kirschke, C.P., and Huang, L. (2003). ZNT7, a novel mammalian zinc transporter, accumulates zinc in the Golgi apparatus. J. Biol. Chem. 278, 4096–4102. [DOI] [PubMed] [Google Scholar]
- Kohler, A., Delaruelle, C., Martin, D., Encelot, N., and Martin, F. (2003). The poplar root transcriptome: Analysis of 7000 expressed sequence tags. FEBS Lett. 542, 37–41. [DOI] [PubMed] [Google Scholar]
- Küpper, H., Zhao, F.J., and McGrath, S.P. (1999). Cellular compartmentation of zinc in leaves of the hyperaccumulator Thlaspi caerulescens. Plant Physiol. 119, 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Lacourt, I., Duplessis, S., Abba, S., Bonfante, P., and Martin, F. (2002). Isolation and characterization of differentially expressed genes in the mycelium and fruit body of Tuber borchii. Appl. Environ. Microbiol. 68, 4574–4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Landschulz, W.H., Johnson, P.F., and McKnight, S.L. (1988). The leucine zipper: A hypothetical structure common to a new class of DNA binding proteins. Science 240, 1759–1764. [DOI] [PubMed] [Google Scholar]
- Lasat, M.M. (2002). Phytoextraction of toxic metals: A review of biological mechanisms. J. Environ. Qual. 31, 109–120. [PubMed] [Google Scholar]
- Leung, I.W., and Lassam, N. (1998). Dimerization via tandem leucine zippers is essential for the activation of the mitogen-activated protein kinase kinase kinase, MLK-3. J. Biol. Chem. 273, 32408–32415. [DOI] [PubMed] [Google Scholar]
- Li, L., and Kaplan, J. (1998). Defects in the yeast high affinity iron transport system result in increased metal sensitivity because of the increased expression of transporters with a broad transition metal specificity. J. Biol. Chem. 273, 22181–22187. [DOI] [PubMed] [Google Scholar]
- Li, L., and Kaplan, J. (2001). The yeast gene MSC2, a member of the cation diffusion facilitator family, affects the cellular distribution of zinc. J. Biol. Chem. 276, 5036–5043. [DOI] [PubMed] [Google Scholar]
- Li, L., Tutone, A.F., Drummond, R.S., Gardner, R.C., and Luan, S. (2001). A novel family of magnesium transport genes in Arabidopsis. Plant Cell 13, 2761–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z.S., Lu, Y.P., Zhen, R.G., Szczypka, M., Thiele, D.J., and Rea, P.A. (1997). A new pathway for vacuolar cadmium sequestration in Saccharomyces cerevisiae: YCF1-catalyzed transport of bis(glutathionato)cadmium. Proc. Natl. Acad. Sci. USA 94, 42–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, Z., Matthews, A.M., and Weiss, S.R. (1999). Amino acid substitutions within the leucine zipper domain of the murine coronavirus spike protein cause defects in oligomerization and the ability to induce cell-to-cell fusion. J. Virol. 73, 8152–8159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyons, T.J., Gasch, A.P., Gaither, L.A., Botstein, D., Brown, P.O., and Eide, D.J. (2000). Genome-wide characterization of the Zap1p zinc-responsive regulon in yeast. Proc. Natl. Acad. Sci. USA 97, 7957–7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDiarmid, C.W., Gaither, L.A., and Eide, D. (2000). Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 19, 2845–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDiarmid, C.W., Milanick, M.A., and Eide, D.J. (2002). Biochemical properties of vacuolar zinc transport systems of Saccharomyces cerevisiae. J. Biol. Chem. 277, 39187–39194. [DOI] [PubMed] [Google Scholar]
- Marx, S.O., Reiken, S., Hisamatsu, Y., Gaburjakova, M., Gaburjakova, J., Yang, Y.M., Rosemblit, N., and Marks, A.R. (2001). Phosphorylation-dependent regulation of ryanodine receptors: A novel role for leucine/isoleucine zippers. J. Cell Biol. 153, 699–708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mäser, P., et al. (2001). Phylogenetic relationships within cation transporter families of Arabidopsis. Plant Physiol. 126, 1646–1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCormack, K., Tanouye, M.A., Iverson, L.E., Lin, J.W., Ramaswami, M., McCormack, T., Campanelli, J.T., Mathew, M.K., and Rudy, B. (1991). A role for hydrophobic residues in the voltage-dependent gating of Shaker K+ channels. Proc. Natl. Acad. Sci. USA 88, 2931–2935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyabe, S., Izawa, S., and Inoue, Y. (2001). The Zrc1 is involved in zinc transport system between vacuole and cytosol in Saccharomyces cerevisiae. Biochem. Biophys. Res. Commun. 282, 79–83. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Palmiter, R.D., Cole, T.B., and Findley, S.D. (1996. a). ZNT-2, a mammalian protein that confers resistance to zinc by facilitating vesicular sequestration. EMBO J. 15, 1784–1791. [PMC free article] [PubMed] [Google Scholar]
- Palmiter, R.D., Cole, T.B., Quaife, C.J., and Findley, S.D. (1996. b). ZnT-3, a putative transporter of zinc into synaptic vesicles. Proc. Natl. Acad. Sci. USA 93, 14934–14939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmiter, R.D., and Findley, S.D. (1995). Cloning and functional characterization of a mammalian zinc transporter that confers resistance to zinc. EMBO J. 14, 639–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paulsen, I.T., and Saier, M.H., Jr. (1997). A novel family of ubiquitous heavy metal ion transport proteins. J. Membr. Biol. 156, 99–103. [DOI] [PubMed] [Google Scholar]
- Pence, N.S., Larsen, P.B., Ebbs, S.D., Letham, D.L., Lasat, M.M., Garvin, D.F., Eide, D., and Kochian, L.V. (2000). The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc. Natl. Acad. Sci. USA 97, 4956–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persans, M.W., Nieman, K., and Salt, D.E. (2001). Functional activity and role of cation-efflux family members in Ni hyperaccumulation in Thlaspi goesingense. Proc. Natl. Acad. Sci. USA 98, 9995–10000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon-Smits, E., and Pilon, M. (2002). Phytoremediation of metals using transgenic plants. Crit. Rev. Plant Sci. 21, 439–456. [Google Scholar]
- Ramsay, L.M., and Gadd, G.M. (1997). Mutants of Saccharomyces cerevisiae defective in vacuolar function confirm a role for the vacuole in toxic metal ion detoxification. FEMS Microbiol. Lett. 152, 293–298. [DOI] [PubMed] [Google Scholar]
- Reitter, J.N., Sergel, T., and Morrison, T.G. (1995). Mutational analysis of the leucine zipper motif in the Newcastle disease virus fusion protein. J. Virol. 69, 5995–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugh, C.L., Senecoff, J.F., Meagher, R.B., and Merkle, S.A. (1998). Development of transgenic yellow poplar for mercury phytoremediation. Nat. Biotechnol. 16, 925–928. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., and Russell, D.W. (2001). Molecular Cloning: A Laboratory Manual, 3rd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Scott, A., Wyatt, S., Tsou, P.L., Robertson, D., and Allen, N.S. (1999). System for plant cell biology: GFP imaging in living onion epidermal cells. Biotechniques 26, 1128–1132. [DOI] [PubMed] [Google Scholar]
- Sherman, F. (1991). Getting started with yeast. Methods Enzymol. 28, 3–20. [DOI] [PubMed] [Google Scholar]
- Simmerman, H.K., Kobayashi, Y.M., Autry, J.M., and Jones, L.R. (1996). A leucine zipper stabilizes the pentameric membrane domain of phospholamban and forms a coiled-coil pore structure. J. Biol. Chem. 271, 5941–5946. [DOI] [PubMed] [Google Scholar]
- Stanton, B., Eaton, J., Johnson, J., Rice, D., Schuette, B., and Moser, B. (2002). Hybrid poplar in the Pacific Northwest: The effects of market-driven management. J. For. 100, 28–33. [Google Scholar]
- Sterky, F., et al. (1998). Gene discovery in the wood-forming tissues of poplar: Analysis of 5,692 expressed sequence tags. Proc. Natl. Acad. Sci. USA 95, 13330–13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supek, F., Supekova, L., Nelson, H., and Nelson, N. (1996). A yeast manganese transporter related to the macrophage protein involved in conferring resistance to mycobacteria. Proc. Natl. Acad. Sci. USA 93, 5105–5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surks, H.K., Mochizuki, N., Kasai, Y., Georgescu, S.P., Tang, K.M., Ito, M., Lincoln, T.M., and Mendelsohn, M.E. (1999). Regulation of myosin phosphatase by a specific interaction with cGMP-dependent protein kinase I alpha. Science 286, 1583–1587. [DOI] [PubMed] [Google Scholar]
- Szczypka, M.S., Wemmie, J.A., Moye-Rowley, W.S., and Thiele, D.J. (1994). A yeast metal resistance protein similar to human cystic fibrosis transmembrane conductance regulator (CFTR) and multidrug resistance-associated protein. J. Biol. Chem. 269, 22853–22857. [PubMed] [Google Scholar]
- Taylor, G. (2002). Populus: Arabidopsis for forestry. Do we need a model tree? Ann. Bot. 90, 681–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Gibson, T.J., Plewniak, F., Jeanmougin, F., and Higgins, D.G. (1997). The CLUSTAL_X Windows interface: Flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton, V.K., Mandal, D., Vahadji, C., and Rao, R. (2002). Functional expression in yeast of the human secretory pathway Ca2+,Mn2+-ATPase defective in Hailey-Hailey disease. J. Biol. Chem. 277, 6422–6427. [DOI] [PubMed] [Google Scholar]
- Turner, R., and Tjian, R. (1989). Leucine repeats and an adjacent DNA binding domain mediate the formation of functional cFos-cJun heterodimers. Science 243, 1689–1694. [DOI] [PubMed] [Google Scholar]
- van der Zaal, B.J., Neuteboom, L.W., Pinas, J.E., Chardonnens, A.N., Schat, H., Verkleij, J.A., and Hooykaas, P.J. (1999). Overexpression of a novel Arabidopsis gene related to putative zinc-transporter genes from animals can lead to enhanced zinc resistance and accumulation. Plant Physiol. 119, 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vida, T.A., and Emr, S.D. (1995). New vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J. Cell Biol. 128, 779–792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, L.E., Pittman, J.K., and Hall, J.L. (2000). Emerging mechanisms for heavy metal transport in plants. Biochim. Biophys. Acta 1465, 104–126. [DOI] [PubMed] [Google Scholar]