Abstract
Auxin plays a role in nearly every aspect of a plant's life. Signals from the developmental program, physiological status, and encounters with other organisms all converge on the auxin pathway. The molecular mechanisms facilitating these interactions are diverse; yet, common themes emerge. Auxin can be regulated by modulating rates of biosynthesis, conjugation, and transport, as well as sensitivity of a cell to the auxin signal. In this article, we describe some well-studied examples of auxin's interactions with other pathways.
Auxin controls many processes in plants—from development to pathogenesis. Regulation of its synthesis, transport, and conjugation helps generate context-specific responses.
It has often been said—sometimes in joyful wonderment, sometimes in teeth-grinding frustration—auxin does everything. Although the major molecular details of the auxin pathway are largely known, many questions remain about how this one simple signaling molecule is responsible for directing so many diverse responses. The focus of this article is to try to illustrate several themes of how auxin acts in concert with other pathways to trigger specific cellular events in time and space.
One potentially useful analogy in trying to understand auxin's complicated roles is to think of it as money. Auxin does not have much intrinsic value—it stores very little energy or raw materials. However, like paper currency, it has great symbolic value, as an easily circulated means of facilitating transactions in the dynamic economy of plant life. As with currency, the amount, form, and location of auxin affects which transactions are possible. Other factors, such as what commodities are available in a given time and place, constrain which transactions auxin can facilitate. For instance, you cannot usually buy a goldfish at a shoe store, and it is quite challenging to purchase an ice cream sundae at 8 am.
Here, we describe some of the ways in which the developmental program, the abiotic environment, the circadian clock, other hormones, and other organisms modify the auxin response. Auxin can be regulated by modulating rates of biosynthesis, conjugation, and transport, as well as sensitivity of a cell to the auxin signal. In addition, the dominant pathway of biosynthesis, the composition and levels of IAA conjugates, the direction of auxin transport, and the downstream consequences of sensing auxin can all be radically altered by the cellular milieu. These context-specific networks help shape the outcome (transactions) of cellular exposure to auxin. Each of the networks impinging on the cellular response to auxin could easily fill a long article (or entire collection) on their own. For brevity's sake, only a handful of examples will be described highlighting some of the molecular mechanisms by which other signaling networks intersect with the auxin pathway.
THE DEVELOPMENTAL PROGRAM
Auxin has been implicated in a disconcertingly large number of developmental events. Its role in embryonic polarity, organ initiation, and stem cell maintenance have all been extensively reviewed (De Smet and Jurgens 2007; Bowman and Floyd 2008; Vanneste and Friml 2009). In many of these cases, a general model can be discerned. First, auxin accumulates in a subset of cells, triggering a change in cell fate. This is followed by establishment of a graded distribution of auxin, often setting up an axis. Finally, this axis is used, sometimes in combination with other signals, to establish zones of cell identity. In these scenarios, auxin acts both to initiate early events and subsequently to refine the locations of specific programs.
The relationship between auxin and the developmental program can be viewed as bidirectional. A genetically encoded developmental program pushes auxin into some cells and the auxin changes which genes are expressed in these cells. Part of the cellular response to auxin is to change auxin biosynthesis, conjugation, transport, and response. This then further alters the cellular environment. If auxin is cellular currency, the developmental program could be said to act like the government. By directing money to some programs and by levying or refunding taxes to certain groups or entities, governmental bodies exert a strong influence on where money flows. Within the cell, the suite of transcription factors are analogous to elected officials. Importantly, there is also a strong feedback from the auxin flux on which transcription factors are present. Similarly, how money is spent or saved often determines who gets elected. A few recent examples from the developing Arabidopsis fruit illustrate the coordination between auxin and the developmental program.
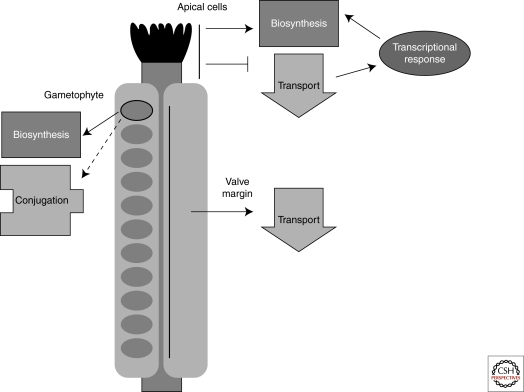
Auxin maxima are crucial for several steps in the development of the Arabidopsis gynoecium (Balanza et al. 2006; Ostergaard 2009). For example, auxin levels must be high for proper development of the apical tissues called the stigma and the style (Fig. 1). Current models propose that auxin levels are manipulated through coordinated regulation of SHORT INTERNODE (SHI)/STYLISH (STY) transcription factor family-induced local auxin biosynthesis and the relative efficiency of auxin transport away from sites of synthesis (Sohlberg et al. 2006; Alvarez et al. 2009; Trigueros et al. 2009). The balance of production and transport determines the timing and location of style and stigmatic tissue development. Expression of STY1 and STY2 become restricted to the apical end of the gynoecium 1–2 days before stigmatic papillae become visible. An inducible STY1 causes rapid induction of the gene encoding the auxin biosynthetic enzyme YUCCA4 and a concomitant increase in free auxin levels (Sohlberg et al. 2006). This auxin maximum in the ring of cells near the apex of gynoecium primordia triggers activation of the NGATHA (NGA) genes (Alvarez et al. 2009; Trigueros et al. 2009). These in turn alter the suite of transcription factors expressed in these cells, promoting apical cell identity. The NGA family also induces the SHI/STY genes, providing a positive‐feedback loop. Thus, the location and timing of the auxin maxima is genetically programmed and is required to trigger the next step in development.
Figure 1.
Many developmental events require modulation of auxin. In the Arabidopsis fruit (gynoecium), auxin maxima, minima, and gradients are used to define the location and timing of crucial developmental milestones. In the apical tissues, a suite of transcription factors acts in a positive‐feedback loop to produce a local auxin pool. This spatially precise auxin accumulation triggers a change in the transcriptional network, leading to the development of style and stigma. More basal tissues also use auxin during development. Auxin is actively transported out of a thin stripe of cells at the valve margin. The change in localization of auxin transporters needed for creating this auxin minima is caused by developmentally regulated expression of the transcription factor IND. Finally, in the female gametophyte, cell fate is highly correlated with position within the embryo sac. An auxin gradient—high at the micropylar end (near the egg cell) and low at the chalazal end—is essential for conveying this positional information. Coordinated regulation of biosynthesis and perhaps conjugation establish this pattern. Solid arrows represent strongly supported links, whereas dashed arrows indicate possible connections.
The absence of auxin is also a potent signal, as shown in the development of the valve margin separation layer where Arabidopsis fruits open for seed dispersal (Fig. 1). In wild-type plants, there is a marked reduction in auxin response (and likely auxin levels) in these boundary cells (Sorefan et al. 2009). The gene encoding the transcription factor INDEHISCENT (IND) is expressed in a thin stripe of cells at the boundary of the valves (Liljegren et al. 2004). IND is both necessary and sufficient to create these auxin minima and to direct the subsequent development of the boundary separation layer. IND regulates auxin transport away from these cells by directly regulating the expression of PINOID and related kinases (Sorefan et al. 2009). These kinases are able to direct the localization of PIN-FORMED (PIN) auxin efflux carriers (Friml et al. 2004; Robert and Offringa 2008). This is perhaps the first described case of a developmental program pushing a signaling molecule away from a subset of cells as a specific developmental trigger.
Recent work in the female gametophyte provides a striking example for the action of auxin gradients as positional cues on cell fate programs (Fig. 1). Several lines of evidence suggest that location within the syncitial gametophyte is a major determinant of egg cell fate (Pagnussat et al. 2009). By examining and manipulating auxin response in the embryo sac, researchers strongly implicated a gradient of auxin—highest at the micropylar end and lowest at the chalazal end—as a morphogenic factor directing cell fate (Pagnussat et al. 2009). In contrast to sporophytic tissue, where graded auxin levels have thus far largely been attributed to balanced biosynthesis and transport, the gametophytic gradient appears to be maintained via regulated biosynthesis and conjugation/degradation. How this gradient is interpreted remains to be determined.
ABIOTIC ENVIRONMENT
The modulation of auxin in response to abiotic changes in the environment is heavily documented, including a well-studied role for auxin in tropic responses to gravity (Morita and Tasaka 2004) and light (Holland et al. 2009), a connection between auxin and response to salt stress (Wang et al. 2009), and an interaction between auxin and high temperature (Gray et al. 1998; Koini et al. 2009). Just as the distribution of money by the government is affected by lobbyists and special interest groups, the developmental program is strongly influenced by information from the abiotic environment. Among these various lobbies, light is perhaps the most powerful, and its intimate relationship with auxin regulation is among the most complex. Here, we focus on light's influence over auxin as an example of how auxin integrates environmental information into the developmental program. During light/dark transitions, plants change auxin responsiveness by regulating auxin signaling components. In contrast, when optimizing light access in crowded environments, plants appear to act primarily by altering auxin biosynthesis and transport.
Light is perceived by a number of photoreceptors. Blue light is perceived mainly by cryptochromes and phototropins, whereas red and far/red light is sensed by phytochromes. All three classes of photoreceptors have been linked with the auxin response (Chen et al. 2004; Li and Yang 2007; Salisbury et al. 2007; Holland et al. 2009). Here, we focus on the phytochromes, as one example. When the family of red/far-red light receptor proteins known as the phytochromes are stimulated by light, the equilibrium of the phytochrome pool shifts toward the active form. Activated phytochromes move into the nucleus and facilitate the degradation of the PHYTOCHROME INTERACTING FACTOR (PIF) and PHYTOCHROME INTERACTING FACTOR-LIKE (PIL) proteins and help stabilize other transcription factors such as LONG HYPOCOTYL 5 (HY5) (Bae and Choi 2008). The degradation of HY5 in the dark requires the ubiquitin ligase CONSTITUTIVE PHOTOMORPHOGENIC 1 (COP1) and the multi-protein CONSTITUTIVE PHOTOMORPHOGENIC 9 (COP9) signalosome (CSN) (Cluis et al. 2004).
In the light, HY5 accumulates and drives transcription associated with the light program. Plants deficient in HY5 show many hyper-auxin response phenotypes, such as faster growing lateral roots, higher number of lateral root primordia, and altered lateral root emergence. These phenotypes depend on the auxin receptor TRANSPORT INHIBITOR RESPONSE 1 (TIR1). When the hy5 mutation is combined with a knockout allele of HY5 HOMOLOG (HYH), the de-repression of auxin signaling is more severe (Sibout et al. 2006). Transcriptome analysis of hy5 mutants reveals a widespread reduction in the expression level of many early auxin response genes, including the auxin transcriptional repressors Aux/IAAs. Consistent with this finding, HY5 can directly bind the promoter of AXR2/IAA7 in vitro. Overexpression of AXR2 rescues the hy5 long hypocotyl phenotype (Cluis et al. 2004). A genome-wide survey of DNA bound by HY5 revealed binding sites in the promoters of nine Aux/IAA genes and six ARF genes (Lee et al. 2007). Interestingly, phyA can phosphorylate many Aux/IAAs in vitro (Colon-Carmona et al. 2000), although the relevance of this phosphorylation is still unknown. Thus, light strongly reduces cellular response to auxin by simultaneously increasing expression of the Aux/IAAs and perhaps by stabilizing the encoded proteins (Fig. 2A).
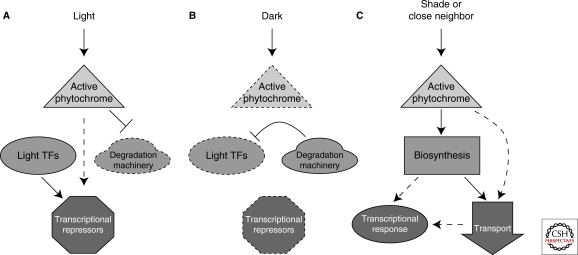
Figure 2.
The auxin pathway is affected by the presence and quality of light. (A) Light stimulation reduces sensitivity to auxin. Activated phytochrome promotes the accumulation of auxin transcriptional repressors. Repression of degradation machinery, including the COP9 signalosome, results in stabilization of transcription factors such as HY5. (B) In the dark, active degradation of light transcription factors results in reduced accumulation of auxin transcriptional repressors and high auxin sensitivity. (C) In shaded conditions or close proximity to a neighboring plant, increased auxin biosynthesis and transport result in an increased transcriptional response that contributes to the shade avoidance syndrome. Dotted lines indicate decreased levels of proteins or complexes.
Although light blocks auxin transactions in the cell, the absence of light promotes them (Fig. 2B). When light levels are low, positive regulators of light signaling such as HY5 and HYH are degraded, leading to lower levels of auxin repressors. Mutations in many CSN subunits result in plants showing a light-grown phenotype when grown in the dark (Schwechheimer et al. 2001). Such mutants also show decreased apical dominance, dwarfism, and decreased root growth inhibition by auxin. Antisense lines with reduced expression of the CSN5 subunit of the CSN have lower expression levels of Aux/IAA genes and increased Aux/IAA stability, similar to what is observed in low auxin conditions. The explanation for these effects is that the CSN directly facilitates the formation of SCFTIR1 complexes responsible for auxin-induced degradation of the Aux/IAAs. Another negative regulator of auxin transcription, AUXIN RESONSE FACTOR2 (ARF2), is repressed in the dark (Li et al. 2004). Reduced levels of Aux/IAAs and repressor ARFs would together elevate auxin sensitivity in dark-grown plants.
In addition to absolute presence or absence of light, light quality can also change the auxin balance (Fig. 2C). Appropriate response to changing light conditions is an essential part of a plant's daily routine. Light quality can be strongly altered by the proximity and positioning of nearby plants. Transcriptional studies show that auxin-related genes make up the largest group of genes up-regulated by shade (Devlin et al. 2003) and auxin signaling mutants are impaired in their shade-avoidance responses (Vandenbussche et al. 2003). Free IAA accumulates in shaded plants through a process that requires an enzyme that catalyzes an early step in the auxin biosynthetic pathway known as SHADE AVOIDANCE 3 (SAV3) (Tao et al. 2008) or TRYPTOPHAN AMINOTRANSERASE 1 (TAA1) (Stepanova et al. 2008). Plants deficient in SAV3/TAA1 show shorter hypocotyls in shaded conditions (Tao et al. 2008). SAV3/TAA1 is highly expressed in the leaves in a similar pattern as the auxin-responsive reporter DR5::GUS. In shaded conditions, DR5::GUS activity moves into the hypocotyl. This shift from leaves to stem is blocked by pretreatment with the polar auxin transport inhibitor naphthylphthalamic acid (NPA). This is consistent with previous evidence for the involvement of polar auxin transport during hypocotyl elongation in response to far-red light (Jensen et al. 1998). It is still unclear how the light signal regulates this change in transport pattern.
Shade avoidance also requires the PHYTOCHROME RAPIDLY REGULATED (PAR) genes up-regulated shortly after experiencing shaded conditions. PAR1 and PAR2 encode transcription factors that negatively regulate shade avoidance response (Roig-Villanova et al. 2007). PAR1 and PAR2 locate to the nucleus and repress the expression of a subset of auxin responsive genes including SAUR15 and SAUR68. This same ability of negatively regulating auxin-induced transcription during shade-avoidance was seen for the transcription factor ATHB4, another PAR gene (Sorin et al. 2009). Although the precise role of these negative regulators is unclear, auxin appears to be the target of both positive and negative control.
CIRCADIAN CLOCK
Complementing the guidance from the developmental program and the environment, plants use the circadian system to regulate auxin response. There is emerging evidence that the clock modulates auxin metabolism, transport, and signaling, determining how much auxin is available and how readily auxin can be perceived at different times of the day (Fig. 3). In effect, the circadian system sets the operating hours during which specific transactions may be conducted.
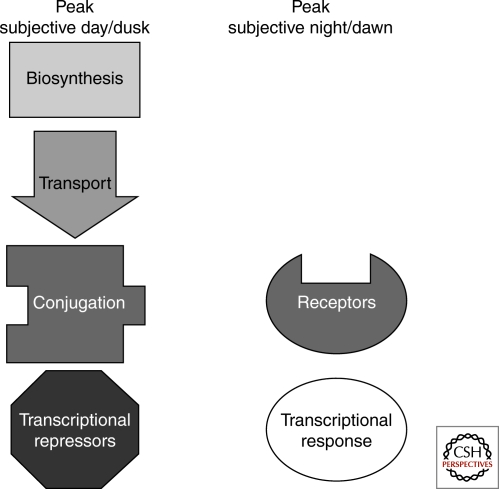
Figure 3.
Plants use the circadian system to regulate auxin response. Circadian regulation has been observed at every level of the auxin pathway. In continuous conditions, expression of genes involved in biosynthesis, transport, conjugation, and transcriptional repression peaks during subjective day or dusk. Conversely, receptor gene expression and the auxin transcriptional response peaks during subjective night or dawn. Thus, transcriptional and growth effects show peak sensitivity to exogenous auxin treatment during subjective night.
Decapitation experiments show that circadian controlled rhythmic elongation of the Arabidopsis inflorescence stem requires auxin transport from the apex (Jouve et al. 1999). Rhythmic growth can be restored by adding exogenous auxin to the tip of a decapitated stem. If polar auxin transport is blocked with NPA, rhythmic growth stops. Circadian fluctuations of IAA-aspartic acid conjugates in the stem suggest that the clock may control availability of auxin by regulated de-conjugation in combination with polar auxin transport. In a genome-wide study of clock-regulated genes, a large overrepresentation of auxin-related genes were identified (Covington and Harmer 2007). Genes in this group are involved in auxin biosynthesis, conjugation, transport, and signaling. In addition, more than half of highly auxin-induced genes are regulated by the clock. Neither elevated rates of auxin biosynthesis nor transport is required for these auxin-induced transcriptional oscillations, as treatment with auxin does not affect the phase or amplitude of an auxin-responsive reporter and NPA could cause slight phase delays but no change in amplitude. Timing of maximum induction of transcription by exogenous auxin is perfectly aligned with the time of peak endogenous auxin responses for untreated plants. This gating of auxin perception seems to extend to auxin physiological responses as well. Elongation of the hypocotyl under constant light in response to auxin was only induced when the auxin was applied during subjective night.
Unlike the bidirectional relationship observed with the developmental program, auxin does not play a major role in clock activity. Although the SCF complex is involved in both auxin signal transduction and generating circadian rhythms, turnover of the core clock component TOC1 is not affected by auxin (Harmon et al. 2008). Auxin applied at a range of concentrations is not able to reset the phase of the clock (Covington and Harmer 2007). Very high concentrations can dampen oscillations of clock reporter genes and produce slightly longer periods under constant conditions; however, near normal rhythms persist.
INTERACTIONS WITH OTHER HORMONES
Other hormones intersect with the auxin pathway at many levels. Cytokinin, brassinosteroids, gibberellins, jasmonic acid, ethylene, abscisic acid, and strigolactones all act at least in part through modifying auxin. If auxin is cellular currency, we could stretch our analogy (perhaps to the breaking point) to say that other hormones are responsible for setting up the various types of shops into which auxin might flow, each stocking its own merchandise. Here, we present a small number of examples from this rapidly growing area of research.
The molecular details of the classical antagonism of cytokinin and auxin action in differentiation are becoming clear. In root meristems, auxin produced locally and transported from the shoot promotes cell division and stem cell identity (Dello Ioio et al. 2008; Chapman and Estelle 2009). Cytokinin acts to promote differentiation. It does this by altering levels of auxin transport and response (Fig. 4) (Dello Ioio et al. 2008; Chapman and Estelle 2009). Cytokinin exerts this effect by up-regulating SHY2, a member of the Aux/IAA family, repressors of the transcriptional response to auxin. Among the genes negatively regulated by high levels of SHY2 are the PIN auxin transporters and IPT5, encoding a cytokinin biosynthetic enzyme. So, as cytokinin levels increase, SHY2 reduces auxin flow into meristematic cells while increasing the levels of auxin required to turn on genes needed for stem cell identity. At the same time, SHY2 likely causes a reduction in the local levels of cytokinin, leading to a negative‐feedback loop. Such a loop combined with local production of auxin at the root tip may be sufficient to establish the spatial architecture observed in the root—with stem cells at the tip, followed by cells transitioning into the differentiation program further away from the auxin pool.
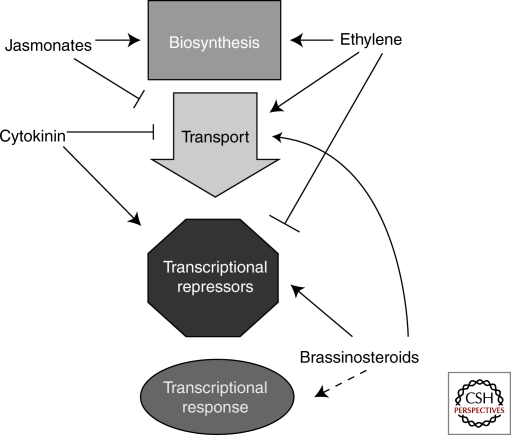
Figure 4.
Other hormones impact auxin levels and response. Jasmonates, cytokinin, ethylene, and brassinosteroids all modulate cellular auxin levels through changes in biosynthesis and transport. Cytokinin, ethylene, and brassinosteroids also alter how sensitive a cell is to the auxin available by manipulating levels of repressors of auxin transcription. Brassinosteroids likely interact with transcriptional activators as well, although the precise mechanism is still unknown.
In many cases, a large number of hormones may be coordinating auxin responses (Fig. 4). In the root apical meristem, auxin levels are also being modulated by jasmonic acid (Sun et al. 2009). In addition to down-regulating levels of auxin transporters, jasmonates increase expression of ASA1. ASA1 is an enzyme in tryptophan biosynthesis, a precursor to auxin. The size of the auxin pool and rate of transport laterally away from the tip directly determine the location and density of lateral root primordia. There is also new evidence that IAA or JA conjugates to tryptophan act as potent antiauxins (Staswick 2009). How these compounds interfere with auxin response is currently unknown, although preliminary in vitro studies suggest that they are not competing with free IAA for TIR1 binding. Ethylene is also acting with auxin in the root. It inhibits root growth in part by modulating auxin levels and transport (Ruzicka et al. 2007; Stepanova et al. 2007; Stepanova et al. 2008; Yoo et al. 2009). In the root tip, ethylene induces expression of several genes important for auxin biosynthesis, including ASA1, enhancing local levels of auxin (Stepanova et al. 2008). Ethylene also up-regulates PIN2 and AUX1, likely facilitating auxin transport into the elongation and differentiation zones (Ruzicka et al. 2007; Stepanova et al. 2007). The increase in auxin levels is proposed to inhibit cell growth and further sensitize cells to the effects of ethylene. Like light, ethylene stimulates turnover of the repressor ARF2, likely further promoting auxin response in some cells (Li et al. 2004).
In addition to joining the fray in the root, brassinosteroids act on many aspects of auxin biology throughout the plant (Hardtke et al. 2007) (Fig. 4). In the presence of both hormones, many responses are greatly enhanced; whereas in the absence of brassinosteroids, auxin transcriptional responses are severely reduced (Nakamura et al. 2003; Bao et al. 2004; Nemhauser et al. 2004; Vert et al. 2008). How brassinosteroids are connected to the auxin pathway is an area of active investigation. It is known that prolonged exposure to brassinosteroids increases the polar transport of auxin (Bao et al. 2004; Li et al. 2005) and that brassinosteroids regulate the expression of many early auxin response genes (Nakamura et al. 2003; Goda et al. 2004; Nemhauser et al. 2004; Vert et al. 2005). These include Aux/IAA and PIN genes, likely leading to changes in the dynamics of auxin transcriptional responses and transport. Like light and ethylene, brassinosteroids appear to act on ARF2, a repressor ARF thought to compete with activator ARFs for cis-regulatory elements (Vert et al. 2008).
In roots, auxin regulates both the expression and intracellular localization of the putative transcription factor BREVIS RADIX (BRX) (Scacchi et al. 2009). brx mutants have short roots and impaired auxin responses (Mouchel et al. 2006). Both of these phenotypes can be rescued by application of brassinosteroids or by constitutively elevating expression of auxin-responsive genes through introduction of a loss-of-function mutation in hy5 (Mouchel et al. 2006; Scacchi et al. 2009). Nuclear-localized BRX is required for cells to maintain normal levels of CONSTITUTIVE PHOTOMORPHOGENIC DWARF (CPD), a brassinosteroid biosynthetic enzyme. The strong loss of auxin responsiveness caused by reduced CPD expression in brx mutants further emphasizes the requirement of brassinosteroids for normal auxin responses. Treatment of wild-type roots with the polar auxin transport inhibitor NPA produces roots closely resembling those of brx mutants. Moreover, in low auxin conditions, BRX appears to colocalize with PIN1 at the plasma membrane. Together, these findings suggest that brassinosteroids are needed for normal auxin transport and that this transport is required for normal auxin response. Determining how transport feeds back on response and how brassinosteroids fit into this picture remain to be determined.
One potential insight into the brassinosteroid-auxin connection comes from the study of embryonic patterning. In the presence of high auxin levels at the tips of embryonic cotyledons, an ARF transcription factor MONOPTEROS (MP) directly induces expression of the transcription factor DORNROSCHEN (DRN) (Cole et al. 2009). In vitro and in vivo interaction studies showed that DRN interacts with the transcription factor BIM1 (Chandler et al. 2009), known to enhance brassinosteroid transcriptional responses (Yin et al. 2005). Loss of BIM1 causes similar patterning defects as loss of DRN, suggesting that this interaction is required for DRN function (Chandler et al. 2009). These and yet-to-be-described interactions among transcription factors on promoters of shared target genes are a compelling model to explain the interdependency observed between the two pathways.
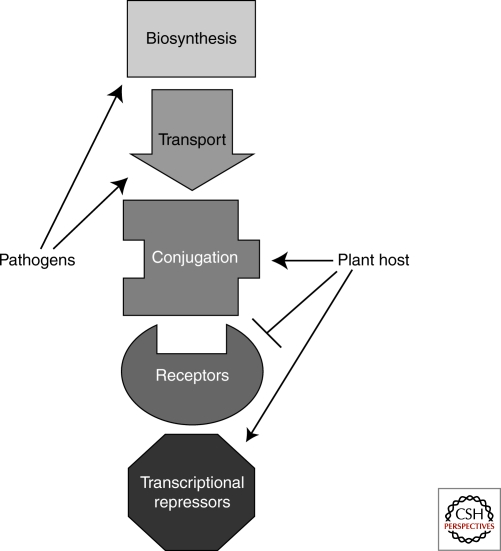
PATHOGENS
If auxin acts as a currency driving the cellular economy, it is not surprising that many pathogens use auxin to aid infection and disease. Pathogens manipulate their hosts' auxin pathway at many stages throughout growth and development and in diverse ways (Fig. 5). They can synthesize their own auxin (Ali et al. 2009), increase native auxin biosynthesis (Chen et al. 2007), reroute auxin transport (Grunewald et al. 2009), or alter auxin signal transduction (Padmanabhan et al. 2008). In this way, pathogens act as counterfeiters flooding cells with their auxin knockoffs or as greedy speculators manipulating the currency market for their own enrichment.
Figure 5.
The auxin pathway is a target of pathogen attack. High auxin levels are correlated with increased disease development. Pathogens increase their chances of success through increased auxin biosynthesis and altered auxin transport. Plant hosts defend against this strategy by conjugating free auxin, inhibiting receptor production, and promoting accumulation of auxin transcriptional repressors. In counter-offensive strategies, pathogens target the host's ability to control auxin response.
An association between increased disease development and auxin accumulation is well supported in diverse plant–pathogen interactions (Wang et al. 2007; Ding et al. 2008). In part, this may reflect auxin's ability to activate expansins that soften and degrade the cell wall (Ding et al. 2008). For example, the bacterial pathogen Pseudomonas syringae uses the type III effector AvrRpt2 to increase auxin content in host cells (Chen et al. 2007). Expressing AvrRpt2 in plants is sufficient to increase levels of free IAA. This effect can be enhanced by inoculating AvrRpt2-expressing transgenic plants with either virulent or avirulent P. syringae strains. How AvrRpt2 alters auxin levels is still not known, but the effect of this increase is clear. Whether auxin levels are manipulated by infection with the bacteria or by treatment of wild-type seedlings with exogenous auxin, the bacteria's ability to multiply within host tissues is positively correlated with auxin levels.
Pathogens can also redirect transport to increase auxin in particular cells. To exploit a host's resources, parasitic nematodes must establish nematode feeding sites, creating cysts of multiple cells that release nutrients easily and can spread to adjacent cells. Degrading the cell wall using auxin is an important part of this process (Grunewald et al. 2009). Shortly after exposure to the model parasitic nematode Heterodera schachtii, expression of the auxin transporters PIN3 and PIN4 are induced at the site of nematode attack (Grunewald et al. 2009). PIN3 localization is changed to orient the flow of auxin toward the sites adjacent to the initially parasitized plant cells. Conversely, the expression of PIN1 and PIN4 is strongly repressed after nematode infection. The current model suggests that down-regulating PIN1 and PIN4 prevents auxin efflux from the nematode feeding sites while PIN3 and PIN4 are repositioned to push auxin into adjacent cells priming them for feeding site expansion. This hypothesis is supported by the fact that PIN1 knockouts show a decrease in cyst number, whereas double mutants such as pin1pin3 and pin1pin4 show an increased proportion of small cysts compared with wild-type.
Because high auxin levels aid in pathogen success, it is not surprising that many components of the plant's immune system—both basal and R-gene-mediated—are geared toward controlling auxin during an attack. Plants treated with flg22, the conserved amino-terminal peptide from bacterial flagellin, produce the microRNA miR393, thought to target TIR1 (Navarro et al. 2006). Plants overexpressing miR393 show increased Aux/IAA stabilization and decreased transcription of auxin-responsive genes. Down-regulation of auxin response is accompanied by an increased ability to fight off infection by virulent Pto DC3000 as shown in tir1 mutants or miR393 overexpressing plants. Moreover, infection of resistant rice plants by Xanthomonas oryzae pv oryzae causes rapid up-regulation of GH3.8, an auxin conjugating enzyme. Overexpression of GH3.8 results in plants with less free auxin, higher levels of IAA-amino acid conjugates, and reduced auxin induced transcriptional response. These plants, like many resistant plants carrying R genes, are able to suppress the auxin-induced expression of expansins early after pathogen inoculation.
Systemic acquired resistance allows plants to anticipate future pathogen attacks and is under the control of salicylic acid (Durrant and Dong 2004). Salicylic acid promotes disease resistance in part although repression of the auxin pathway (Wang et al. 2007). Salicylic acid down-regulates auxin transporters and receptors leading to decreased expression of auxin responsive genes. At the same time, salicylic acid up-regulates IAA conjugating enzymes and increases the stability of Aux/IAA repressors. Consistent with these findings, accumulation of salicylic acid prevents expression of auxin reporters following fungal infection. When salicylic acid is not allowed to accumulate or when salicylic acid-targeted Aux/IAAs are knocked-out, fungal disease development is enhanced.
One major difference between disease resistant and susceptible plants may be their ability to detect and prevent acute spikes of auxin activity at the onset of infection. However, if auxin responses are suppressed for prolonged periods, the reward of limiting pathogen spread may come at the expense of host growth. This may explain the observed trade-offs between plant productivity and disease resistance (Durrant and Dong 2004). Plants may be further constrained in their ability to limit pathogen access to auxin, because auxin plays a critical role in beneficial interactions between plants and microbes, especially during colonization (Contreras-Cornejo et al. 2009; Schafer et al. 2009). Auxin spending is risky business. It may lead to great growth and success, or leave the plant open to exploitation and ruin.
CONCLUDING REMARKS
Biologists know a tremendous amount about auxin—how levels can be manipulated, how its presence (or absence) can trigger specific cellular programs, and, increasingly, how auxin plays a central role in integrating diverse pathways directing plant life. An informal survey of auxin research over the last 20 years reveals some striking trends. Most obvious is that interest in this pathway remains strong. There has been an approximately eightfold increase in the number of research articles where the term “auxin” appears between 1989 and 2009. In addition, a clear shift in the direction of research can be observed. In 1989, approximately 60% of papers described a core auxin signaling component—genes involved in auxin metabolism, signaling, or transport. Approximately 40% were primarily concerned with connecting auxin with a developmental or physiological program (including other hormones and biotic interactions). By 1999, the relative proportions of papers on core auxin components versus those relating auxin to other pathways had been reversed. This switch appears permanent. More than 70% of papers published in the first half of 2009 are primarily concerned with how auxin interacts with other pathways. Areas of particularly dramatic gains are connections with other hormones (∼5% in 1989 vs. ∼20% in 2009) and the role of auxin in biotic interactions (∼10% in 1989 vs. ∼20% in 2009). This reflects the logical progression of a field: initiating exploration close to the starting point, then using these initial findings to explore more distant areas. From another vantage, this trend likely reflects the development of sufficiently sophisticated tools to tackle more complex problems. Either way, it is clear that the future of auxin research, like studies of the global economy, is only going to get more focused on connections.
ACKNOWLEDGMENTS
We are indebted to Robert Cleland, Cristina Walcher, Andrew Chen, and Matthew Offenbacher for careful reading of this manuscript and stimulating discussions about the centrality of auxin in plant life. The idea that auxin might act as cellular money was first proposed to JLN by Niko Geldner several years ago. Any fault for its exuberant elaboration here is entirely our own. J.L.S. is supported by the University of Washington Royalty Research Fund, the National Science Foundation Graduate Research Fellowship Program, and the Seattle Chapter of the Achievement Rewards for College Scientists Foundation. Funding for work on auxin in our lab is provided by the University of Washington Royalty Research Fund and National Science Foundation IOS-0919021.
Footnotes
Editors: Mark Estelle, Dolf Weijers, Karin Ljung, and Ottoline Leyser
Additional Perspectives on Auxin Signaling available at www.cshperspectives.org
REFERENCES
- Ali B, Sabri AN, Ljung K, Hasnain S 2009. Auxin production by plant associated bacteria: Impact on endogenous IAA content and growth of Triticum aestivum L. Lett Appl Microbiol 48:542–547 [DOI] [PubMed] [Google Scholar]
- Alvarez JP, Goldshmidt A, Efroni I, Bowman JL, Eshed Y 2009. The NGATHA distal organ development genes are essential for style specification in Arabidopsis. Plant Cell 21:1373–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bae G, Choi G 2008. Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59:281–311 [DOI] [PubMed] [Google Scholar]
- Balanza V, Navarrete M, Trigueros M, Ferrandiz C 2006. Patterning the female side of Arabidopsis: The importance of hormones. J Exp Bot 57:3457–3469 [DOI] [PubMed] [Google Scholar]
- Bao F, Shen J, Brady SR, Muday GK, Asami T, Yang Z 2004. Brassinosteroids interact with auxin to promote lateral root development in Arabidopsis. Plant Physiol 134:1624–1631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman JL, Floyd SK 2008. Patterning and polarity in seed plant shoots. Ann Rev Plant Biol 59:67–88 [DOI] [PubMed] [Google Scholar]
- Chandler JW, Cole M, Flier A, Werr W 2009. BIM1, a bHLH protein involved in brassinosteroid signalling, controls Arabidopsis embryonic patterning via interaction with DORNROSCHEN and DORNROSCHEN-LIKE. Plant Mol Biol 69:57–68 [DOI] [PubMed] [Google Scholar]
- Chapman EJ, Estelle M 2009. Cytokinin and auxin intersection in root meristems. Genome Biol 10:210 [Google Scholar]
- Chen M, Chory J, Fankhauser C 2004. Light signal transduction in higher plants. Annu Rev Genet 38:87–117 [DOI] [PubMed] [Google Scholar]
- Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN 2007. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci 104:20131–20136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cluis CP, Mouchel CF, Hardtke CS 2004. The Arabidopsis transcription factor HY5 integrates light and hormone signaling pathways. Plant J 38:332–347 [DOI] [PubMed] [Google Scholar]
- Cole M, Chandler J, Weijers D, Jacobs B, Comelli P, Werr W 2009. DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136:1643–1651 [DOI] [PubMed] [Google Scholar]
- Colon-Carmona A, Chen DL, Yeh KC, Abel S 2000. Aux/IAA proteins are phosphorylated by phytochrome in vitro. Plant Physiol 124:1728–1738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras-Cornejo HA, Macias-Rodriguez L, Cortes-Penagos C, Lopez-Bucio J 2009. Trichoderma virens, a plant beneficial fungus, enhances biomass production and promotes lateral root growth through an auxin-dependent mechanism in Arabidopsis. Plant Physiol 149:1579–1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Harmer SL 2007. The circadian clock regulates auxin signaling and responses in Arabidopsis. PLoS Biol 5:e222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Smet I, Jurgens G 2007. Patterning the axis in plants–auxin in control. Curr Opin Genetics Dev 17:337–343 [DOI] [PubMed] [Google Scholar]
- Dello Ioio R, Nakamura K, Moubayidin L, Perilli S, Taniguchi M, Morita MT, Aoyama T, Costantino P, Sabatini S 2008. A genetic framework for the control of cell division and differentiation in the root meristem. Science 322:1380–1384 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Yanovsky MJ, Kay SA 2003. A genomic analysis of the shade avoidance response in Arabidopsis. Plant Physiol 133:1617–1629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S 2008. Activation of the indole-3-acetic acid-amido synthetase GH3–8 suppresses expansin expression and promotes salicylate- and jasmonate-independent basal immunity in rice. Plant Cell 20:228–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durrant WE, Dong X 2004. Systemic acquired resistance. Annu Rev Phytopathol 42:185–209 [DOI] [PubMed] [Google Scholar]
- Friml J, Yang X, Michniewicz M, Weijers D, Quint A, Tietz O, Benjamins R, Ouwerkerk PB, Ljung K, Sandberg G, et al. 2004. A PINOID-dependent binary switch in apical-basal PIN polar targeting directs auxin efflux. Science 306:862–865 [DOI] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S 2004. Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134:1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Ostin A, Sandberg G, Romano CP, Estelle M 1998. High temperature promotes auxin-mediated hypocotyl elongation in Arabidopsis. Proc Natl Acad Sci 95:7197–7202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, Cannoot B, Friml J, Gheysen G 2009. Parasitic nematodes modulate PIN-mediated auxin transport to facilitate infection. PLoS Pathog 5:e1000266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardtke CS, Dorcey E, Osmont KS, Sibout R 2007. Phytohormone collaboration: Zooming in on auxin-brassinosteroid interactions. Trends Cell Biol 17:485–492 [DOI] [PubMed] [Google Scholar]
- Harmon F, Imaizumi T, Gray WM 2008. CUL1 regulates TOC1 protein stability in the Arabidopsis circadian clock. Plant J 55:568–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland JJ, Roberts D, Liscum E 2009. Understanding phototropism: From Darwin to today. J Exp Bot 60:1969–1978 [DOI] [PubMed] [Google Scholar]
- Jensen PJ, Hangarter RP, Estelle M 1998. Auxin transport is required for hypocotyl elongation in light-grown but not dark-grown Arabidopsis. Plant Physiol 116:455–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jouve L, Gaspar T, Kevers C, Greppin H, Degli Agosti R 1999. Involvement of indole-3-acetic acid in the circadian growth of the first internode of Arabidopsis. Planta 209:136–142 [DOI] [PubMed] [Google Scholar]
- Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA 2009. High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19:408–413 [DOI] [PubMed] [Google Scholar]
- Lee J, He K, Stolc V, Lee H, Figueroa P, Gao Y, Tongprasit W, Zhao H, Lee I, Deng XW 2007. Analysis of transcription factor HY5 genomic binding sites revealed its hierarchical role in light regulation of development. Plant Cell 19:731–749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li QH, Yang HQ 2007. Cryptochrome signaling in plants. Photochem Photobiol 83:94–101 [DOI] [PubMed] [Google Scholar]
- Li H, Johnson P, Stepanova A, Alonso JM, Ecker JR 2004. Convergence of signaling pathways in the control of differential cell growth in Arabidopsis. Dev Cell 7:193–204 [DOI] [PubMed] [Google Scholar]
- Li L, Xu J, Xu ZH, Xue HW 2005. Brassinosteroids stimulate plant tropisms through modulation of polar auxin transport in Brassica and Arabidopsis. Plant Cell 17:2738–2753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liljegren SJ, Roeder AH, Kempin SA, Gremski K, Ostergaard L, Guimil S, Reyes DK, Yanofsky MF 2004. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell 116:843–853 [DOI] [PubMed] [Google Scholar]
- Morita MT, Tasaka M 2004. Gravity sensing and signaling. Curr Opin Plant Biol 7:712–718 [DOI] [PubMed] [Google Scholar]
- Mouchel CF, Osmont KS, Hardtke CS 2006. BRX mediates feedback between brassinosteroid levels and auxin signalling in root growth. Nature 443:458–461 [DOI] [PubMed] [Google Scholar]
- Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S 2003. Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol 133:1843–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD 2006. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312:436–439 [DOI] [PubMed] [Google Scholar]
- Nemhauser JL, Mockler TC, Chory J 2004. Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biology 2:E258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostergaard L 2009. Don't 'leaf' now. The making of a fruit. Curr Opin Plant Biol 12:36–41 [DOI] [PubMed] [Google Scholar]
- Padmanabhan MS, Kramer SR, Wang X, Culver JN 2008. Tobacco mosaic virus replicase-auxin/indole acetic acid protein interactions: Reprogramming the auxin response pathway to enhance virus infection. J Virol 82:2477–2485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagnussat GC, Alandete-Saez M, Bowman JL, Sundaresan V 2009. Auxin-dependent patterning and gamete specification in the Arabidopsis female gametophyte. Science 324:1684–1689 [DOI] [PubMed] [Google Scholar]
- Robert HS, Offringa R 2008. Regulation of auxin transport polarity by AGC kinases. Curr Opin Plant Biol 11:495–502 [DOI] [PubMed] [Google Scholar]
- Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portoles S, Rodriguez-Concepcion M, Martinez-Garcia JF 2007. Interaction of shade avoidance and auxin responses: A role for two novel atypical bHLH proteins. EMBO J 26:4756–4767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzicka K, Ljung K, Vanneste S, Podhorska R, Beeckman T, Friml J, Benkova E 2007. Ethylene regulates root growth through effects on auxin biosynthesis and transport-dependent auxin distribution. Plant Cell 19:2197–2212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salisbury FJ, Hall A, Grierson CS, Halliday KJ 2007. Phytochrome coordinates Arabidopsis shoot and root development. Plant J 50:429–38 [DOI] [PubMed] [Google Scholar]
- Scacchi E, Osmont KS, Beuchat J, Salinas P, Navarrete-Gomez M, Trigueros M, Ferrandiz C, Hardtke CS 2009. Dynamic, auxin-responsive plasma membrane-to-nucleus movement of Arabidopsis BRX. Development 136:2059–2067 [DOI] [PubMed] [Google Scholar]
- Schafer P, Pfiffi S, Voll LM, Zajic D, Chandler PM, Waller F, Scholz U, Pons-Kuhnemann J, Sonnewald S, Sonnewald U, et al. 2009. Manipulation of plant innate immunity and gibberellin as factor of compatibility in the mutualistic association of barley roots with Piriformospora indica. Plant J 59:461–474 [DOI] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng XW 2001. Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292:1379–1382 [DOI] [PubMed] [Google Scholar]
- Sibout R, Sukumar P, Hettiarachchi C, Holm M, Muday GK, Hardtke CS 2006. Opposite root growth phenotypes of hy5 versus hy5 hyh mutants correlate with increased constitutive auxin signaling. PLoS Genet 2:e202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohlberg JJ, Myrenas M, Kuusk S, Lagercrantz U, Kowalczyk M, Sandberg G, Sundberg E 2006. STY1 regulates auxin homeostasis and affects apical-basal patterning of the Arabidopsis gynoecium. Plant J 47:112–123 [DOI] [PubMed] [Google Scholar]
- Sorefan K, Girin T, Liljegren SJ, Ljung K, Robles P, Galvan-Ampudia CS, Offringa R, Friml J, Yanofsky MF, Ostergaard L 2009. A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature 459:583–586 [DOI] [PubMed] [Google Scholar]
- Sorin C, Salla-Martret M, Bou-Torrent J, Roig-Villanova I, Martinez-Garcia JF 2009. ATHB4, a regulator of shade avoidance, modulates hormone response in Arabidopsis seedlings. Plant J 59:266–277 [DOI] [PubMed] [Google Scholar]
- Staswick PE 2009. The tryptophan conjugates of jasmonic and indole-3-acetic acids are endogenous auxin inhibitors. Plant Physiol 150:1310–1321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanova AN, Robertson-Hoyt J, Yun J, Benavente LM, Xie DY, Dolezal K, Schlereth A, Jurgens G, Alonso JM 2008. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell 133:177–191 [DOI] [PubMed] [Google Scholar]
- Stepanova AN, Yun J, Likhacheva AV, Alonso JM 2007. Multilevel interactions between ethylene and auxin in Arabidopsis roots. Plant Cell 19:2169–2185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Xu Y, Ye S, Jiang H, Chen Q, Liu F, Zhou W, Chen R, Li X, Tietz O, et al. 2009. Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell 21:1495–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao Y, Ferrer JL, Ljung K, Pojer F, Hong F, Long JA, Li L, Moreno JE, Bowman ME, Ivans LJ, et al. 2008. Rapid synthesis of auxin via a new tryptophan-dependent pathway is required for shade avoidance in plants. Cell 133:164–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trigueros M, Navarrete-Gomez M, Sato S, Christensen SK, Pelaz S, Weigel D, Yanofsky MF, Ferrandiz C 2009. The NGATHA genes direct style development in the Arabidopsis gynoecium. Plant Cell 21:1394–1409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbussche F, Vriezen WH, Smalle J, Laarhoven LJ, Harren FJ, Van Der Straeten D 2003. Ethylene and auxin control the Arabidopsis response to decreased light intensity. Plant Physiol 133:517–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanneste S, Friml J 2009. Auxin: A trigger for change in plant development. Cell 136:1005–1016 [DOI] [PubMed] [Google Scholar]
- Vert G, Nemhauser JL, Geldner N, Hong F, Chory J 2005. Molecular mechanisms of steroid hormone signaling in plants. Ann Rev Cell Develop Biol 21:177–201 [DOI] [PubMed] [Google Scholar]
- Vert G, Walcher CL, Chory J, Nemhauser JL 2008. Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci 105:9829–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Li K, Li X 2009. Auxin redistribution modulates plastic development of root system architecture under salt stress in Arabidopsis thaliana. J Plant Physiol 166:1637–1645 [DOI] [PubMed] [Google Scholar]
- Wang D, Pajerowska-Mukhtar K, Culler AH, Dong X 2007. Salicylic acid inhibits pathogen growth in plants through repression of the auxin signaling pathway. Curr Biol 17:1784–1790 [DOI] [PubMed] [Google Scholar]
- Yin Y, Vafeados D, Tao Y, Yoshida S, Asami T, Chory J 2005. A new class of transcription factors mediates brassinosteroid-regulated gene expression in Arabidopsis. Cell 120:249–259 [DOI] [PubMed] [Google Scholar]
- Yoo SD, Cho Y, Sheen J 2009. Emerging connections in the ethylene signaling network. Trends Plant Sci 14:270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]