Abstract
The tomato transcription factor Pti4, an ethylene-responsive factor (ERF), interacts physically with the disease resistance protein Pto and binds the GCC box cis element that is present in the promoters of many pathogenesis-related (PR) genes. We reported previously that Arabidopsis plants expressing Pti4 constitutively express several GCC box–containing PR genes and show reduced disease symptoms compared with wild-type plants after inoculation with Pseudomonas syringae pv tomato or Erysiphe orontii. To gain insight into how genome-wide gene expression is affected by Pti4, we used serial analysis of gene expression (SAGE) to compare transcripts in wild-type and Pti4-expressing Arabidopsis plants. SAGE provided quantitative measurements of >20,000 transcripts and identified the 50 most highly expressed genes in Arabidopsis vegetative tissues. Comparison of the profiles from wild-type and Pti4-expressing Arabidopsis plants revealed 78 differentially abundant transcripts encoding defense-related proteins, protein kinases, ribosomal proteins, transporters, and two transcription factors (TFs). Many of the genes identified were expressed differentially in wild-type Arabidopsis during infection by Pseudomonas syringae pv tomato, supporting a role for them in defense-related processes. Unexpectedly, the promoters of most Pti4-regulated genes did not have a GCC box. Chromatin immunoprecipitation experiments confirmed that Pti4 binds in vivo to promoters lacking this cis element. Potential binding sites for ERF, MYB, and GBF TFs were present in statistically significantly increased numbers in promoters regulated by Pti4. Thus, Pti4 appears to regulate gene expression directly by binding the GCC box and possibly a non-GCC box element and indirectly by either activating the expression of TF genes or interacting physically with other TFs.
INTRODUCTION
Complex signaling networks that involve protein kinase cascades, transcription factors, other regulatory proteins, and pathogenesis-related (PR) genes play a central role in plant responses to pathogen attack (Yang et al., 1997; Tena et al., 2001; Cheong et al., 2002; Pedley and Martin, 2003). Many transcription factor (TF) genes are induced by pathogen infection or hormones associated with defense signaling (Kranz et al., 1998; Chen et al., 2002; Mysore et al., 2002). TFs also are activated at the post-translational level by phosphorylation or by binding to other regulatory proteins, including other TFs (Solano et al., 1998; Zhang et al., 1999; Després et al., 2000; Gu et al., 2000). TFs bind specific cis elements present in the promoters of many defense-related genes, thereby activating their expression and contributing to the plant's ability to overcome disease (Singh et al., 2002). Families of defense-related TFs and the cis elements they bind are conserved in divergent plant species (Rushton and Somssich, 1998; Rushton et al., 2002). The major TF families that have roles in defense are WRKY, ERF, bZIP, and MYB (Rushton and Somssich, 1998; Riechmann and Ratcliffe, 2000; Singh et al., 2002). These also constitute the major TF families encoded by the Arabidopsis genome (Riechmann and Ratcliffe, 2000).
WRKY proteins bind to the W box sequence found in the promoters of pathogen-responsive genes (e.g., parsley PR-1, potato PR-10a, and tobacco chitinase CHN50) (Després et al., 1995; Yang et al., 1997; Rushton and Somssich, 1998; Eulgem et al., 2000). W boxes are present in the promoter of PR1, the hallmark gene associated with the induction of systemic acquired resistance in Arabidopsis (Maleck et al., 2000). WRKY proteins have been characterized in diverse plant species (Arabidopsis, parsley, and tobacco) (Eulgem et al., 2000). The bZIP family of TFs include TGA and GBF factors and have been characterized in tobacco, soybean, and Arabidopsis (Singh et al., 2002). They bind to the as-1 and G box cis elements, respectively. The as-1 element has been shown to be responsive to the defense signaling molecules salicylic acid and jasmonate and has been identified in the promoters of glutathione S-transferase genes and on the 35S promoter of Cauliflower mosaic virus (Yang et al., 1997). Some TGA family members in Arabidopsis have been shown to interact with NPR1, a key regulator in the salicylic acid defense signaling pathway (Zhang et al., 1999; Després et al., 2000). The G box is a ubiquitous element, and it has been proposed that it functions in concert with neighboring cis elements in regulating gene expression related to different functions, including pathogen attack (Kim et al., 1992; Menkens et al., 1995). Finally, the MYB family of TFs is a very large family with a subset of genes that play a role in the defense response (Kranz et al., 1998). MYB family members bind to several different cis element sequences (Martin and Paz-Ares, 1997).
The ethylene-responsive factor (ERF) TF family is unique to plants and has been identified in many plant species (Ohme-Takagi et al., 2000; Riechmann and Ratcliffe, 2000). The expression of many ERF genes is induced by pathogen infection and ethylene. ERFs bind the GCC box (GCCGCC) element that is present in the promoters of many PR genes (Ohme-Takagi and Shinshi, 1995; Solano et al., 1998; Fujimoto et al., 2000; Ohme-Takagi et al., 2000; Gu et al., 2002). Individual members of the ERF family have been shown to be either positive or negative regulators of transcription (Fujimoto et al., 2000; Ohta et al., 2000, 2001). ERFs also are known to be involved in resistance to various pathogens. Overexpression of ERF1 in Arabidopsis increased resistance to necrotrophic fungi, and overexpression of TsiI in tobacco and pepper increased resistance to viral, bacterial, and oomycete pathogens (Park et al., 2001; Berrocal-Lobo et al., 2002; Shin et al., 2002). ERF genes that are expressed differentially during pathogen infection include tomato Pti4, Pti5, and Pti6, soybean GmEREBP1, and Arabidopsis AtERF1, AtERF13, and AtERF14 (Thara et al., 1999; Gu et al., 2000; Mazarei et al., 2002; Onate-Sanchez and Singh, 2002). Interestingly, the TsiI and Pti4 genes also are induced by hormones associated with defense responses (salicylic acid and jasmonic acid) and by wounding (Park et al., 2001; Gu et al., 2002).
The tomato ERF Pti4 was isolated, along with two other ERFs, Pti5 and Pti6, from a yeast two-hybrid screen by virtue of its interaction with the Pto kinase (Zhou et al., 1997). Pto specifies gene-for-gene resistance against Pseudomonas syringae pv tomato (Pst) strains that express the avirulence protein AvrPto or AvrPtoB (Martin et al., 1993; Kim et al., 2002). Pti4 was shown to bind the GCC box in vitro and to regulate the expression of several GCC box–containing genes in vivo (Gu et al., 2002). Phosphorylation of Pti4 by the Pto kinase enhances Pti4 binding capacity to the GCC box (Gu et al., 2000). Expression of Pti4 is induced in tomato leaves upon inoculation with Pseudomonas strains, by wounding, and by exposure to ethylene, salicylic acid, or jasmonic acid (Thara et al., 1999; Gu et al., 2000; Mysore et al., 2002). Although Pti4 was identified based on its interaction with Pto, the expression of Pti4 in leaves is induced equally well by inoculation with either avirulent or virulent Pseudomonas strains (Gu et al., 2000). Expression of Pti4 in Arabidopsis caused the activation of several PR genes and increased resistance to the fungal pathogen Erysiphe orontii and tolerance to the bacterial pathogen Pst strain DC3000 (Gu et al., 2002). An independent study of Arabidopsis plants expressing Pti4 identified a set of defense-related genes whose expression was affected by Pti4 (Wu et al., 2002). Based on these collective observations, we propose that Pti4 plays an important regulatory role in the expression of defense genes and that its activity is enhanced by phosphorylation (Gu et al., 2002).
To further explore the role that Pti4 plays in regulating gene expression, we first used serial analysis of gene expression (SAGE) (Zhang et al., 1997) to profile transcripts in wild-type and Pti4-containing Arabidopsis lines. SAGE is an “open-architecture” form of expression profiling that has been shown to be highly quantitative and capable of detecting small differences in gene expression (Zhang et al., 1997). Our study identified a small number of genes whose transcript abundance either increased or decreased in plants expressing Pti4 compared with wild-type plants. To determine if, as expected, Pti4 bound the promoters of these genes, we used a Pti4 antibody for chromatin immunoprecipitation (ChIP) assays (Johnson et al., 2001). Surprisingly, although the promoters of many of these genes were bound by Pti4 in vivo, most lacked a GCC box. Thus, our observations suggest that ERFs such as Pti4 regulate gene expression both directly and indirectly, probably in combination with other TFs.
RESULTS
SAGE Retrieved Tags Corresponding to 3314 Arabidopsis Genes
To identify genes that are upregulated or downregulated by Pti4, we used SAGE to determine differences in transcript accumulation between wild-type and Pti4-expressing Arabidopsis plants. SAGE is based on two principles: (1) a short sequence tag of 9 to 11 bp contains enough information to identify a unique transcript, provided that it is derived from a defined location within the transcript; and (2) many transcript tags can be concatenated into a single molecule, facilitating the sequencing and analysis of tags (Velculescu et al., 1995; www.sagenet.org/). SAGE profiling has been used widely and successfully in mammalian and yeast systems but relatively little for expression profiling in plants (Matsumura et al., 1999; Lorenz and Dean, 2002; Jung et al., 2003; Lee and Lee, 2003).
The SAGE profile of a tissue sample is determined by counting the number of individual tags from a constructed library of tags and identifying the genes that correspond to the tags. To minimize artifacts, we isolated RNA from vegetative tissues derived from 30 to 35 4-week-old plants from each of the wild-type and Pti4-expressing Arabidopsis lines. Four-week-old plants were chosen because we have shown previously that these plants display increased resistance to E. orontii and tolerance to Pst (Gu et al., 2002). We then constructed two SAGE libraries from these mRNA samples and sequenced the concatenated-tag inserts from 658 clones from each library (see Methods). SAGE analysis software (Velculescu et al., 1997) was used to extract the 10-bp tag sequences from each sequenced clone. Analysis of the data revealed that we had isolated tags from 27,925 possible transcripts (an average of 21 tags per clone; Table 1). SAGE tags that were identified only once might be attributable to PCR or sequencing errors; therefore, we limited our further analysis to the 20,213 tags that were retrieved at least twice. By comparing these tags with Arabidopsis sequence databases, we found that they corresponded to 3314 unique genes (Table 1). Of these 3314 unique genes, 2204 genes were represented in both libraries, whereas 501 and 609 genes were present uniquely in the wild-type and Pti4 libraries, respectively.
Table 1.
Summary of SAGE Analysis
| Variable | Wild-Type Library | Pti4 Library | Total |
|---|---|---|---|
| Total tags a | 13,710 (9,976) | 14,215 (10,237) | 27,925 (20,213) |
| Genesb | 2,705 | 2,813 | 3,314 |
Number of tags obtained from the wild-type and Pti4 libraries. The number of tags that were retrieved at least two times in the libraries are shown in parentheses.
Number of different genes represented by the tags. Tags that were retrieved at least two times, shown in parentheses above, were used to determine the number of unique genes represented in the entire SAGE project.
Previous RNA gel blot analysis showed that several GCC box–containing defense-related genes are expressed constitutively in the Pti4-expressing Arabidopsis line (Gu et al., 2002). Tags from four of these genes—PDF1.2, PR2, PR3, and PR4—were retrieved from our SAGE libraries. There were 34 copies of the tag sequence corresponding to PDF1.2 (tag 65) in the Pti4 library and none in the wild-type library (Table 2). Four PR4 tags (tag 854) were present in the Pti4 library, and none were present in the wild-type library (Table 2). For both PR2 and PR3, we found two tags in the Pti4 library and none in the wild-type library. These results correspond extremely well to our previous RNA gel blot analysis of wild-type and Pti4-expressing plants (Gu et al., 2002) and provided an initial validation of the SAGE data.
Table 2.
Genes That Are Expressed Differentially in Pti4-Expressing Plants Compared with Wild-Type Plants
| SAGE Tag Identifier a | Tag Sequence b | Tag Count c
|
Fold d | GenBank Accession Number |
Corresponding Gene e | Promoter f | |
|---|---|---|---|---|---|---|---|
| Wild Type |
Pti4 | ||||||
| Environmental/pathogen response (PR and antimicrobial genes, and drought- and salt-responsive genes) g | |||||||
| 65 | CATATTTCTG | 0 | 34 | 33.1 | T04323 | Antifungal protein PDF1.2 | P |
| 174 | TGTCAAGGAG | 3 | 11 | 3.6 | U40399 | Cytosolic cyclophilin ROC3 | |
| 212 | TCGTGTTTGG | 2 | 10 | 4.9 | U42724 | Chloroplast stromal cyclophilin ROC4 | P |
| 243 | GAGGAACTAA | 1 | 10 | 9.7 | X78584 | Drought-induced Di19 | P |
| 356 | GTAGTGACCA | 1 | 7 | 6.8 | X98189 | Peroxidase ATP1a | P |
| 523 | GTGCATTTGG | 1 | 5 | 4.9 | AF003728 | Salt-induced plasma membrane intrinsic protein SIMIP h |
P |
| 642 | ATGGAATGCT | 0 | 5 | 4.9 | AA395048 | Thylakoid lumen rotamase (cyclophilin) | P |
| 643 | GATTATAATG | 0 | 5 | 4.9 | D89051 | Drought-induced putative sugar transporter ERD6 | |
| 854 | CTTGTTTCGG | 0 | 4 | 3.9 | U01880 | Hevein-like protein precursor (PR-4) h | P |
| 617 | ACCGGACACA | 5 | 1 | −5.1 | AV527586 | Putative protein, disease resistance protein family (LRR) h |
P |
| 634 | CAACTCCTCA | 6 | 0 | −6.2 | BE038927 | Major latex protein (MLP)-related | P |
| Signal transduction (transcription factors, kinases, signaling molecules) | |||||||
| 352 | GGACGTGCCG | 0 | 8 | 7.8 | X91259 | Similar to putative lectin | P |
| 423 | AAGAAGTTTT | 1 | 6 | 5.8 | U27698 | Putative calreticulin AtCRTLh | P |
| 427 | GACAACCTGA | 1 | 6 | 5.8 | AJ242970 | General transcription factor BTF3b homologh | P |
| 844 | AATCGCGTCA | 0 | 4 | 3.9 | AA598125 | Receptor-protein kinase-like proteinh | P |
| 274 | GCACAAACAA | 9 | 2 | −4.6 | At5g02030 i | Homeodomain protein | |
| 347 | AGTTGTTTTT | 7 | 2 | −3.6 | AB013886 | Transcription factor RAV1h | P |
| 503 | ACTCTTTTAA | 6 | 1 | −6.2 | AF285106 | CBL-interacting protein kinase CIPK6 h | P |
| 832 | CGAGGAAGCA | 5 | 0 | −5.1 | Y12710 | Shaggy-related protein kinase, ASK-GAMMA | |
| Cell cycle, growth, and proliferation | |||||||
| 104 | AAGATTAAGG | 5 | 16 | 3.1 | U41998 | Actin2 | |
| 646 | GGAAAGAACT | 0 | 5 | 4.9 | AY058193 | Expansin At-EXP6 | |
| Detoxification | |||||||
| 639 | AAGAACGGAC | 0 | 5 | 4.9 | Z25705 | Putative cytochrome P450 | |
| 842 | AAAACTCGGT | 0 | 4 | 3.9 | U37697 | Glutathione reductase h | P |
| 636 | TCTTATGTCA | 6 | 0 | −6.2 | AV556265 | Putative M-type thioredoxin | P |
| Fatty acid, phospholipid, and isoprenoid metabolism | |||||||
| 645 | GCTGCAAACC | 0 | 5 | 4.9 | AB007799 | NADH-cytochrome b5 reductase | P |
| 208 | CAGGTTGTGG | 10 | 3 | −3.4 | AV547952 | Putative acetyl-CoA synthetase | |
| 620 | GAGGCCAAGG | 5 | 1 | −5.1 | AJ010713 | β-Ketoacyl-CoA synthase family (FIDDLEHEAD) h | |
| 631 | TTTAAGATAT | 5 | 1 | −5.1 | AF159801 | Lipid transfer protein ltp4h | |
| Energy metabolism and photosynthesis | |||||||
| 189 | GATTGAAGTT | 2 | 11 | 5.4 | BT002151 | Putative Thr synthase | |
| 321 | GGTGAAATTT | 1 | 8 | 7.8 | L44582 | Vacuolar H+-pumping ATPase ava-p2 | P |
| 422 | AAATTGATCT | 1 | 6 | 5.8 | X97484 | Putative phosphate transporter h | P |
| 637 | AACAACAAAA | 0 | 5 | 4.9 | AV547395 | Amino acid permease AAP2 | |
| 640 | AGCGTTCTCC | 0 | 5 | 4.9 | Z29881 | Putative fructose 1,6-bisphosphatase | P |
| 4 | AAGGTGTGGC | 200 | 146 | −1.4 | BE039376 | Rubiscoj small subunit 2b precursor | |
| 8 | CTTGTGATGG | 155 | 111 | −1.4 | X14212 | Rubisco activase | |
| 17 | TTTGTACAAA | 57 | 37 | −1.6 | AF134120 | Chlorophyll a/b binding protein Lhca2 | |
| 20 | TTCTCTATGT | 55 | 32 | −1.8 | AF134124 | Chlorophyll a/b binding protein Lhcb2 | |
| 27 | AAAGCTTTCT | 46 | 25 | −1.9 | AJ245629 | Putative photosystem I subunit III precursor | |
| 31 | TTTCTATAAA | 40 | 23 | −1.8 | X55970 | Photosystem II 10-kD polypeptide | |
| 91 | ATCATTCGTG | 18 | 7 | −2.6 | AA586203 | Transketolase-like protein | |
| 110 | TTACCTTTCT | 15 | 6 | −2.6 | AJ245631 | Photosystem I subunit VI precursor | |
| 112 | TACTTACATT | 17 | 4 | −4.4 | AV559242 | Putative glycolate oxidase | |
| 318 | TTTATTTTTC | 9 | 1 | −9.2 | S74719 | Sedoheptulose-1,7-bisphosphatase | P |
| 508 | TTTGTATTCT | 6 | 1 | −6.2 | AV557139 | H+-transporting ATPase-like protein h | |
| 834 | CTTTGTGATG | 5 | 0 | −5.1 | AA598190 | Similar to Rubisco activase | P |
| 836 | GCTATACAAA | 5 | 0 | −5.1 | AI993835 | Vacuolar H+-transporting ATPase chain E | |
| 838 | GTACAGCGCC | 5 | 0 | −5.1 | N96033 | Putative dihydroxyacetone kinase | |
| Ribosomal proteins | |||||||
| 355 | CTGGGAAAAA | 1 | 7 | 6.8 | AV549039 | 50S ribosomal protein L27 | P |
| 357 | TCCTTCAAGA | 1 | 7 | 6.8 | BE038325 | Ribosomal protein S13-like | P |
| 419 | AAGAGCCGAG | 0 | 7 | 6.8 | Y09635 | 50S ribosomal protein L24, chloroplast precursor | P |
| 647 | GTTCGTTGAG | 0 | 5 | 4.9 | AY063876 | Putative plastid ribosomal protein L34 precursor | |
| 209 | GTGAGACTTG | 10 | 3 | −3.4 | AV554596 | 40S ribosomal protein-like | |
| 275 | GACGTATTGA | 10 | 1 | −10.3 | AI100025 | Putative ribosomal protein L18 | P |
| 317 | GGACCACCAC | 9 | 1 | −9.2 | AA728511 | 60S ribosomal protein L10A | P |
| 351 | TGTACTTTGT | 8 | 1 | −8.2 | AI100051 | Ribosomal protein S1 | |
| 506 | GTTTTATATA | 6 | 1 | −6.2 | T43594 | 60S ribosomal protein L38-like protein h | P |
| 511 | TGTCTTAGCT | 7 | 0 | −7.2 | AA597822 | Putative 60S ribosomal protein L21 | P |
| 833 | CTTCCGTGTT | 5 | 0 | −5.1 | L28828 | Ribosomal protein S11 | |
| Protein processing/fate | |||||||
| 635 | CTTTTTAAGG | 6 | 0 | −6.2 | AA712419 | ATP-dependent Clp protease ATP binding subunit (ClpC1) |
P |
| Membrane proteins | |||||||
| 125 | GTTTCGCCGA | 14 | 5 | −2.9 | AF097648 | Phosphate/triose phosphate translocator precursor | |
| 210 | TGAATTTGTA | 11 | 2 | −5.6 | Y08061 | Endomembrane-associated protein | P |
| 242 | TGTGATGATC | 10 | 2 | −5.1 | AA728495 | Water channel–like protein | P |
| Other categories | |||||||
| 30 | TGTAGCTCAG | 0 | 63 | 60.8 | AF187951 | Octopine synthase 3′ terminator region from Pti4 and nptII gene cassettes |
|
| 244 | ATGGTGATTA | 2 | 9 | 4.4 | AF195896 | Arabinogalactan protein AGP15 | |
| 512 | AAGCAACTCT | 0 | 6 | 5.9 | AF017074 | RNA polymerase I, II, and III 16.5-kD subunit | P |
| 509 | CAAAAAAAAA | 7 | 0 | −7.2 | AY042804 | Putative RNA binding protein | |
| 837 | GCTCCGCTCC | 5 | 0 | −5.1 | AV566796 | Putative glycine dehydrogenase | |
| 841 | TTTCGTCTTG | 5 | 0 | −5.1 | AV557387 | Putative xyloglucan endotransglycosylase | |
| Unknown proteins | |||||||
| 173 | CATTTGGATT | 3 | 11 | 3.6 | AV536971 | Unknown protein | |
| 211 | AGAATGGTTG | 2 | 10 | 4.9 | AI099719 | Unknown protein | |
| 425 | CCTTGATGTT | 1 | 6 | 5.8 | R90105 | Unknown protein h | P |
| 648 | TGAAAGTTGT | 0 | 5 | 4.9 | AY045836 | Unknown protein | |
| 19 | ATAGAACCTT | 52 | 35 | −1.5 | BE038487 | Unknown protein | |
| 70 | AATTGGAATG | 23 | 11 | −2.2 | AY065156 | Unknown protein | |
| 171 | AGAAGAAGCC | 15 | 0 | −15.4 | At5g40700 i | Unknown protein | |
| 418 | CAATTAGAGT | 7 | 1 | −7.2 | AV534110 | Unknown protein | |
| 829 | AAGGAGTTGT | 5 | 0 | −5.1 | AY070394 | Unknown protein | |
| 830 | ACAAAATTTT | 5 | 0 | −5.1 | AY035176 | Unknown protein | |
SAGE tag identifier corresponding to a particular gene.
The 10-bp SAGE tag sequence, excluding the 5′ adjacent NlaIII site (CATG).
Number of tags retrieved from the wild-type and Pti4 libraries.
Fold increase or fold decrease of tag numbers observed in Pti4 plants compared with wild-type plants. Genes with decreased tag numbers in Pti4-expressing plants are designated with a minus sign.
The gene corresponding to the tag. For each functional category, the genes with increased tag numbers in Pti4-expressing plants are shown first, followed by the genes with decreased tag numbers in Pti4-expressing plants. Within these categories, tags are shown in order of SAGE tag identifiers.
P indicates that the promoter of the gene was analyzed (see Table 3).
The genes are grouped into different functional categories.
The difference in tag number between the wild-type and Pti4 libraries for these genes was significant at 0.05 ≤ P ≤ 0.15. For all other genes, the difference in tag number was significant at P ≤ 0.05.
The TAIR locus name. GenBank or EST clones containing the SAGE tag could not be identified for these genes.
Rubisco, ribulose-1,5-bisphosphate carboxylase/oxygenase.
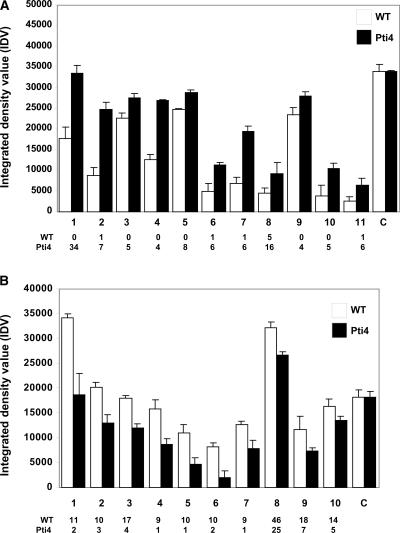
We also used reverse transcription PCR (RT-PCR) to attempt to verify the expression differences in a subset of 33 genes (see supplemental data online). New populations of plants (20 to 25 for each line), independent of the plants used for SAGE, were grown for these experiments, and RNA was isolated from pooled vegetative tissues. Twenty-one of the genes were found to have transcript abundances by RT-PCR that correlated well with the tag numbers observed by SAGE (Figures 1A and 1B; see also supplemental data online). Of the other 12 genes, we were unable to detect differences between wild-type and Pti4-expressing plants by RT-PCR for 10, whereas the PCR analyses for the other 2 genes failed (no products were observed). Thus, of the 35 genes we were able to analyze independently of SAGE (4 by RNA gel blot analysis and 31 by RT-PCR), 25 of them (71%) supported our SAGE results. Previous reports involving mammalian systems also found a good correlation between data obtained by SAGE and RT-PCR (Leerkes et al., 2002; Menssen and Hermeking, 2002). The lack of an even better correlation between these two techniques probably is the result of the inherent differences between them: SAGE determines actual transcript numbers, whereas RT-PCR depends on amplification. It is not possible, based on our data, to conclude which technique provides a better estimate of actual transcript abundance.
Figure 1.
RT-PCR Supports the Use of SAGE for Arabidopsis Gene Expression Profiling.
Differences in transcript abundances between wild type (WT) and Pti4-expressing (Pti4) Arabidopsis were measured by reverse transcription PCR. Values shown represent averages of three reactions plus standard deviations. Integrated density values (IDV) of PCR products are shown on the y axis. Gene names are shown on the x axis. C represents a control from the Lhcb5 gene (SAGE tag 12) that was found to have proportionately equal tag numbers in the two SAGE libraries (see Methods). The tag abundance for each gene from the SAGE analysis is shown at bottom.
(A) Genes with tag numbers increased in the Pti4 line: 1, PDF1.2; 2, peroxidase ATP1a; 3, putative sugar transporter ERD6; 4, PR4; 5, putative lectin; 6, putative calreticulin; 7, BTF3b homolog; 8, actin2; 9, glutathione reductase; 10, putative fructose 1,6-bisphosphatase; and 11, unknown protein (SAGE tags 65, 356, 643, 854, 352, 423, 427, 104, 842, 640, and 425, respectively).
(B) Genes with tag numbers decreased in the Pti4 line: 1, endomembrane-associated protein; 2, 40S ribosomal protein–like; 3, putative glycolate oxidase; 4, sedoheptulose bisphosphatase; 5, putative ribosomal protein L18; 6, water channel–like protein; 7, ribosomal protein L10A; 8, putative photosystem I subunit III precursor; 9, transketolase-like protein; and 10, phosphate/triose phosphate translocator precursor (SAGE tags 210, 209, 112, 318, 275, 242, 317, 27, 91, and 125, respectively).
Wu et al. (2002) have reported expression profiling of a Pti4-expressing line using an Affymetrix chip, and Lorenzo et al. (2003) profiled a line expressing another ERF factor, ERF1. Four genes were identified in both studies as being induced by ERF overexpression (i.e., genes encoding a putative endochitinase, basic endochitinase [PR3], glucanase precursor, and lipoxygenase). Of these four genes, our SAGE analysis identified only PR3 as being induced in our Pti4-expressing plants. Therefore, we used RT-PCR to test directly whether these genes were induced in our Pti4-expressing line (we could not test the putative endochitinase because it was annotated with the accession number of the BAC clone [AC002333], which contains several endochitinase genes). We observed the induction of genes that encode PR3 and the glucanase precursor, whereas we were unable to detect any lipoxygenase transcripts (see supplemental data online). The results for two of the three genes tested agree with our SAGE data; we found PR3 to be induced, and no SAGE tags were recovered for lipoxygenase. In the case of the glucanase precursor, we observed induction by RT-PCR but did not recover a SAGE tag for this gene.
SAGE Identified the 50 Most Highly Expressed Genes in Arabidopsis Vegetative Tissues
One tangential, but interesting, finding derived from our initial SAGE data analysis was the identification of the 50 most highly expressed genes in wild-type Arabidopsis vegetative tissues (see supplemental data online). Not surprisingly, 36 of the genes (72%) encode proteins related to the photosynthetic apparatus. The other genes encode proteins involved in energy metabolism, protein synthesis, defense, or cell structure. The 19th most highly expressed gene is annotated as an unknown protein. We retrieved very similar numbers of tags from both libraries for the majority (86%) of these genes. Seven of the top 50 genes, with all but one encoding photosynthesis-related proteins, had tag numbers that were statistically significantly decreased in the Pti4 library compared with the wild-type library (tags 4, 8, 17, 19, 20, 27, and 31; see below).
A recent article on the analysis of cold-stressed Arabidopsis leaves using SAGE also reported the most highly expressed genes in wild-type, untreated leaf tissue (Jung et al., 2003). Of their 45 most highly abundant SAGE tags, we observed 26 among our top 50 genes (see supplemental data online). The application of Massively Parallel Signature Sequencing (MPSS), another open-architecture expression profiling method, also was reported recently for Arabidopsis shoots and other tissues (http://dbixs001.dbi.udel.edu/MPSS4/java.html). We compared the MPSS signatures corresponding to the most highly expressed genes in shoots with the most highly expressed genes identified by our SAGE analysis (see supplemental data online). Of the top 30 genes identified by each method, 14 were the same. The lack of even more overlap between our observations and the other studies might be attributable to differences in the tissues used (3-week-old plants grown in continuous light [Jung et al., 2003]; 2-week-old leaves [MPSS study]; and 4-week-old aboveground vegetative tissues [this study]).
Expression of Pti4 Alters the Transcript Accumulation of 2% of Arabidopsis Genes Expressed in Vegetative Tissues
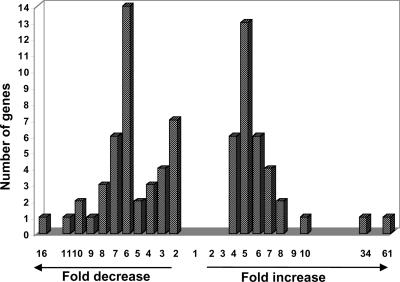
To increase confidence in the observed transcript differences, we focused on those 1139 genes (of the 3314 total) that produced at least four tags in the combined wild-type and Pti4 libraries. The possible statistical significance of the different number of each of these 1139 tags in the two libraries was determined, taking into account the total number of tags retrieved from each of the libraries (Table 1) (Audic and Claverie, 1997). From this analysis, we identified just 63 genes with tag differences between the Pti4 and wild-type libraries that were significant at P ≤ 0.05. An additional 15 tags that corresponded to genes with likely roles in defense and that had P values of ≤0.15 also were considered further (Table 2). The distribution of the fold increase or fold decrease of these 78 genes is shown in Figure 2. In cases in which the P value was ≤0.15, the differences would have been significant at the 0.05 level if either of the libraries had a tag number that differed by just 1 or 2. For simplicity, we refer to these 78 genes as either IPT genes (tag number increased in the Pti4 line compared with the wild-type line) or as DPT genes (tag number decreased in the Pti4 line compared with the wild-type line).
Figure 2.
Frequency Distribution of Fold Differences in Gene Expression Observed in Pti4-Expressing Arabidopsis Plants.
The fold differences in tag numbers are shown for the 78 genes that were found to have significantly different tag numbers between the Pti4-expressing lines and wild-type Arabidopsis (see Table 2). The level of fold difference in tag number is shown on the x axis. The number of genes with different levels of fold increase or decrease in tag number is shown on the y axis.
The 78 IPT and DPT genes identified above were classified into functional categories to gain insight into the overall biological processes that might be affected by the expression of Pti4 (Table 2). Many genes were induced (and a few repressed) that encode proteins involved in responses to pathogens, drought/salt stress, or toxins and in cell growth and proliferation (e.g., PDF1.2, PR4, drought-induced Di19, peroxidase ATP1a, actin2, cyclophilin ROC3, and CIPK). In general, genes coding for proteins involved in energy metabolism, photosynthesis, lipid metabolism, and membrane integrity were repressed by the expression of Pti4 (e.g., transketolase-like protein, chlorophyll a/b binding proteins, and lipid transfer protein Ltp4). Genes that encode TFs, kinases, and ribosomal proteins were both induced and repressed. There were two genes of unknown function that showed lower transcript accumulation in Pti4-expressing plants than in wild-type plants (tags 171 and 418; Table 2).
The Transcript Abundance of Some IPT and DPT Genes Is Altered in Wild-Type Arabidopsis in Response to Pathogen Infection
In tomato, Pti4 transcript abundance increases as early as 30 min after inoculation of susceptible leaves with a virulent Pst strain and is followed, at 2 h after inoculation, by expression of the GCC box–containing genes GluB and Osm (Gu et al., 2000). Because Pti4-expressing Arabidopsis plants show reduced disease symptoms when inoculated with bacterial or fungal pathogens, we hypothesized that many genes whose expression is altered in these plants will be associated with the plant defense response. To test this hypothesis, we determined if the expression of 13 IPT and 7 DPT genes was altered upon infection of Arabidopsis leaves with a virulent bacterial pathogen.
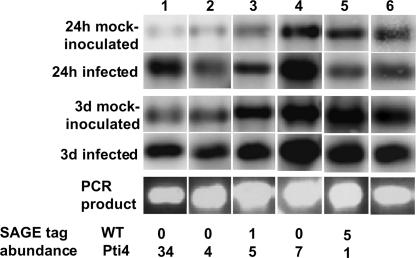
Arabidopsis plants (30 to 35 per line) were dipped into a suspension of Pst strain DC3000, tissue was harvested 24 h and 3 days later, and “reverse” RNA gel blots were used to compare transcripts in infected and uninfected leaf tissues (Figure 3). Transcripts from 4 of the 13 IPT genes were increased upon Pseudomonas infection: PDF1.2, PR4, the SIMIP gene encoding a salt-induced membrane intrinsic protein, and the 50S ribosomal gene L24. Transcripts from two of the seven DPT genes were decreased upon pathogen infection: L21, a 60S ribosomal protein (not shown in Figure 2), and the FIDDLEHEAD gene, which encodes a member of the β-ketoacyl-CoA synthase family (Figure 3). Of the remaining genes, transcripts were not detectable for five, whereas for another nine, transcript accumulation was not observably different between the infected and uninfected tissues. Because of possible cross-hybridization to related transcripts, we cannot exclude the possibility that some of these nine genes are expressed differentially during pathogen infection. In summary, the expression of at least 40% (6 of 15) of the IPT and DPT transcripts detectable by this method was altered in Arabidopsis leaves during Pseudomonas infection. This observation supports an association with the plant defense response for many of the IPT/DPT genes.
Figure 3.
Transcript Abundance of IPT and DPT Genes Is Similar in Pti4-Expressing and Pseudomonas-Infected Arabidopsis Plants.
Wild-type (WT) Arabidopsis plants were inoculated with Pst strain DC3000. Radiolabeled cDNA prepared from RNA isolated at the indicated time points was used to hybridize to PCR products of genes that were expressed differentially in Pti4-expressing Arabidopsis. Lanes are as follows: 1, PDF1.2 (tag 65); 2, PR4 (tag 854); 3, SIMIP (tag 523); 4, ribosomal gene L24 (tag 419); 5, FIDDLEHEAD (tag 620); and 6, H+-pumping ATPase subunit, which served as a control. A representative gel showing the abundance of the PCR products in each lane is shown. The tag abundance for each gene from the SAGE analysis is shown at the bottom.
Promoters of IPT and DPT Genes Are Enriched for Three cis Elements Associated with Defense-Related Gene Expression
Because Pti4, like many ERFs, is known to bind and regulate expression from promoters that contain the GCC box (Thara et al., 1999; Gu et al., 2002), we expected this sequence to be enriched in the IPT and DPT promoters. We chose a subset of IPT and DPT genes from each functional category in Table 2 to obtain 35 representative genes for promoter analysis. cis elements responsible for promoter activity in Arabidopsis are usually within 800 bp of the promoter, and Arabidopsis 5′ untranslated regions generally are short (<150 bp) (Maleck et al., 2000). Therefore, we retrieved sequences 1 kb upstream of the annotated ATG site from the TAIR database. Unexpectedly, only 8 of the 35 promoters had a GCC box (one promoter had two GCC boxes; Table 3). We then examined the 35 promoters for other cis elements that have been reported to play roles in defense- and stress-related gene expression (the G box [Kim et al., 1992; Menkens et al., 1995]; the DRE box DRE box [Baker et al., 1994][Stockinger et al., 1997]; MYB boxes [Martin and Paz-Ares, 1997; Kranz et al., 1998]; the W box W box [Rushton and Somssich, 1998]; and the as-1 element [Niggeweg et al., 2000a, 2000b]; details of our analysis are provided in the supplemental data online). Two elements, the GCC box and the Myb1 box, were found to be present in significantly increased numbers (P ≤ 0.05) in the IPT and DPT promoters (Table 3; see also supplemental data online). The G box also was present at slightly greater than the expected frequency (P ≤ 0.07). As a control, we retrieved 50 random promoters from the Arabidopsis genome sequence (most of these had no apparent role in defense) and examined their cis elements. All of the cis elements from this arbitrary set were present in statistically expected quantities (Table 3).
Table 3.
Analysis of Defense-Related cis Elements in the IPT and DPT Promoters
| Name and Sequence of cis Element |
Corresponding Transcription Factor a |
Observed Number in IPT/DPT Promoters |
Statistically Expected Number in 35 Promoters |
P1b | Observed Number in Random Promoters |
Statistically Expected Number in 50 Promoters |
P2c |
|---|---|---|---|---|---|---|---|
| GCC box (GCCGCC) | ERF | 9 | 1.7 | 5.60 E-05 | 3 | 2.3 | 4.17 E-01 |
| G box (CACGTG) | GBF | 10 | 5.9 | 7.52 E-02 | 8 | 8.4 | 5.99 E-01 |
| DRE box (CCGAC) | CBF | 19 | 20.7 | 6.78 E-01 | 22 | 29.6 | 9.38 E-01 |
| Myb1 (GTTAGTT) | MYB1 | 15 | 7.1 | 6.57 E-03 | 4 | 10.2 | 9.91 E-01 |
| Myb2 ([C/A]TCC[T/A]ACC | MYB2 | 4 | 3.0 | 3.49 E-01 | 6 | 4.3 | 2.57 E-01 |
| Myb3 (TAAC[C/G]GTT) | MYB3 | 3 | 2.8 | 5.34 E-01 | 2 | 4.0 | 9.10 E-01 |
| W box (TTGACC) | WRKY | 11 | 13.3 | 7.74 E-01 | 25 | 19.0 | 1.08 E-01 |
| as-1 (TGACG) | TGA | 29 | 32.6 | 7.62 E-01 | 44 | 46.6 | 6.70 E-01 |
Thirty-five promoters were analyzed for the presence of cis elements commonly found in defense-related genes. A set of 50 random promoters retrieved from the Arabidopsis genome sequence was analyzed as a control.
Transcription factor that binds to the cis element. ERF, ethylene response factor; GBF, G box binding factor; CBF, DRE box binding factor; MYB, Myb box binding factor (the nomenclature Myb/MYB1, -2, and -3 is used to distinguish between the different cis element sequences and their corresponding transcription factors); WRKY, W box binding protein; TGA, as-1 binding protein (Menkens et al., 1995; Martin and Paz-Arez, 1997; Jaglo-Ottosen et al., 1998; Rushton and Somssich, 1998; Niggeweg et al., 2000a, 2000b).
The P value (P1) for the probability of finding at least n copies of a cis element in 35 promoters. Values shown in boldface are statistically significant at P ≤ 0.05 (for the GCC and Myb1 boxes) and P ≤ 0.08. The P value for the G box in the IPT/DPT promoters was 1 order of magnitude higher than its P value in the random promoters; therefore, we considered it to be statistically significant.
The P value (P2) for the probability of finding at least n copies of a cis element in 50 promoters.
ChIP Analysis Reveals Different Classes of Promoters Bound by Pti4
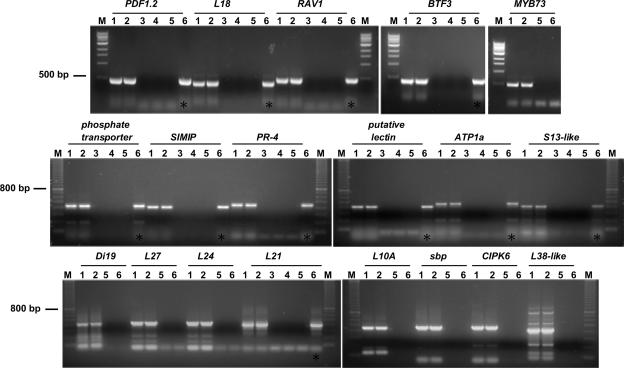
Our promoter analysis suggested three possible scenarios for the direct involvement of Pti4 in gene expression: (1) Pti4 binds the GCC box (or a non-GCC box element) that is present in some of the IPT/DPT promoters; (2) Pti4 activates the expression of TF genes whose products bind and regulate the IPT/DPT promoters; and (3) Pti4 associates physically with other TFs that bind non-GCC box elements in the IPT/DPT promoters. To examine these scenarios, we performed ChIP experiments using (uninoculated) wild-type and Pti4-expressing plants (Figure 4). Antibodies to Pti4 were raised against a peptide sequence that was not similar to any protein from Arabidopsis (see Methods). In preliminary experiments, we found that for all 18 promoters tested (see below), no PCR products were amplified from wild-type Arabidopsis chromatin extracts immunoprecipitated with Pti4 antibody (Figure 4, lane 5). Thus, the Pti4 antibody did not cross-react with any ERF proteins in wild-type (uninoculated) Arabidopsis. Leaves from uninoculated Pti4-expressing Arabidopsis plants were fixed with formaldehyde, and a Pti4 antibody was used to immunoprecipitate Pti4-DNA complexes from chromatin extracts (see Methods for details). Primer pairs specific to 18 different IPT or DPT promoters were used to amplify corresponding PCR products from the immunoprecipitate.
Figure 4.
Pti4 Interacts in Vivo with Both GCC Box– and Non-GCC Box–Containing Promoter Sequences.
Gels show results from the 18 promoters that were analyzed in the ChIP assay (see Table 4 for summary). The names of the promoters analyzed are given above the gels; MYB73 was used as a negative control. Lanes contain PCR products from promoter-specific primers obtained from genomic DNA or chromatin extract. Inputs for PCR amplification were as follows: 1, genomic DNA from wild-type Arabidopsis; 2, genomic DNA from Pti4-expressing Arabidopsis; 3, chromatin immunoprecipitated with preimmune serum from Pti4-expressing Arabidopsis; 4, chromatin immunoprecipitated with Pti4 antibody without cross-linking from Pti4-expressing Arabidopsis; 5, chromatin immunoprecipitated with Pti4 antibody from wild-type Arabidopsis; and 6, chromatin immunoprecipitated with Pti4 antibody after cross-linking from Pti4-expressing Arabidopsis. The presence of a PCR product in lane 6 (asterisks) indicates binding by Pti4. A 1-kb DNA size ladder (M) from New England Biolabs (Beverly, MA; top gels) and a 100-bp ladder from Amersham (middle and bottom gels) are labeled.
Eleven of the 18 promoters tested immunoprecipitated with Pti4 (Table 4). Four of these promoters, including one from a BTF3 TF gene, contain a GCC box. Another three promoters that were not bound by Pti4 also contain a GCC box, thus indicating that the presence of the GCC box alone in a promoter is insufficient for Pti4 binding and that the DNA context in which it lies might be important. For the seven promoters that were bound by Pti4 but lacked a GCC box in the 1-kb region we analyzed, we searched an additional 1.5 kb of upstream sequence for possible GCC boxes (i.e., a total of 2.5 kb upstream from the ATG). In only one of these promoters (S13 ribosomal-like protein; SAGE tag 357) did we observe a GCC box (at 1.4 kb from the ATG). However, among these seven promoters, we found three that have a Myb1 box and one with a G box (the PDF1.2 promoter contains both a GCC box and a Myb1 box; Table 4). Both of these cis elements, as reported above, are present in significantly increased numbers in the IPT/DPT promoters (Table 3).
Table 4.
ChIP Analysis of in Vivo Interactions of Pti4 and IPT/DPT Promoters
| Tag Identifier | GenBank Accession Number |
Corresponding Gene | Fold a | Bound by Pti4 b |
GCC Box c |
G Box c | Myb1 Box c |
Models d |
|---|---|---|---|---|---|---|---|---|
| Transcripts increased by Pti4 | ||||||||
| 65 | T04323 | Antifungal protein PDF1.2 | 33.1 | + | + | − | + | A, G |
| 243 | X78584 | Drought-induced Di19 | 9.7 | − | − | − | − | D, E |
| 352 | X91259 | Similar to putative lectin | 7.8 | + | − | − | − | B, F |
| 355 | AV549039 | 50S ribosomal protein L27 | 6.8 | − | − | − | − | D, E |
| 356 | X98189 | Peroxidase ATP1a | 6.8 | + | + | − | − | A |
| 357 | BE038325 | Ribosomal protein S13-like | 6.8 | + | − | − | + | B, F |
| 419 | Y09635 | 50S ribosomal protein L24, chloroplast precursor | 6.8 | − | + | − | − | D, E |
| 422 | X97484 | Putative phosphate transporter | 5.8 | + | − | − | + | B, F |
| 427 | AJ242970 | General transcription factor BTF3 | 5.8 | + | + | − | − | C |
| 523 | AF003728 | Salt-induced plasma membrane intrinsic protein SIMIP | 4.9 | + | − | − | + | B, F |
| 854 | U01880 | Hevein-like protein precursor (PR-4) | 3.9 | + | + | − | − | A |
| Transcripts reduced by Pti4 | ||||||||
| 275 | AI100025 | Putative ribosomal protein L18 | −10.3 | + | − | − | − | B, F |
| 317 | AA728511 | 60S ribosomal protein L10A | −9.2 | − | + | − | − | D, E |
| 318 | S74719 | Sedoheptulose-1,7-bisphosphatase | −9.2 | − | − | + | − | D, E |
| 347 | AB013886 | Transcription factor RAV1 | −3.6 | + | − | + | − | B, F |
| 503 | AF285106 | CBL-interacting protein kinase CIPK6 | −6.2 | − | + | + | − | D, E |
| 506 | T43594 | 60S ribosomal protein L38-like protein | −6.2 | − | − | − | − | D, E |
| 511 | AA597822 | Putative 60S ribosomal protein L21 | −7.2 | + | − | − | − | B, F |
Promoters of 18 genes were analyzed to determine if they were bound in vivo by Pti4.
Fold increase or fold decrease of tag numbers observed in Pti4 plants compared with wild-type plants. Genes with decreased tag numbers in Pti4-expressing plants are designated with a minus sign.
Promoters bound (+) or not bound (−) by Pti4 are shown.
The presence (+) or absence (−) of a GCC box, a G box, or a Myb1 box in the promoters is shown.
The models to which each promoter might correspond are indicated (see Figure 5).
DISCUSSION
Previous characterization of a Pti4-expressing Arabidopsis line showed that, along with exhibiting reduced disease symptoms in response to two pathogens, it also constitutively expressed several pathogenesis-related genes (Gu et al., 2002). To understand which genes are regulated by Pti4 and to gain insight into the mechanism of regulation, we combined the experimental approaches of SAGE and ChIP. SAGE identified >20,000 transcripts corresponding to >3000 unique genes. By applying stringent statistical tests, we identified just 78 IPT/DPT genes (2%) whose transcripts accumulated differentially in the Pti4-expressing line compared with the wild type. Differential expression of 25 of these genes (of 35 examined) was verified independently by RT-PCR or RNA gel blot analysis. As a step toward testing whether these genes actually play a role in defense, we examined the expression of 20 of them during infection of Arabidopsis leaves by Pst strain DC3000. Six of the 15 genes (40%) for which transcripts were detectable were expressed differentially during challenge with this virulent pathogen, thus supporting a potential defense-related role for them. Our expectation that the IPT/DPT promoters would contain the GCC box was not realized: only a minority of the promoters examined contained this cis element. ChIP analysis confirmed that Pti4 likely binds the GCC box in vivo but also suggested that Pti4 might bind a non-GCC box element or interact with other TFs that bind the IPT/DPT promoters. Collectively, our results elucidated both the specific genes regulated by Pti4 and the mechanism by which this regulation occurs.
Three Defense-Related cis Elements Are Enriched in the IPT/DPT Promoters
A statistical analysis of 35 IPT/DPT promoters indicated that two cis elements were present at significantly greater than expected frequencies (GCC and Myb1 elements); the G box also was enriched in the IPT/DPT promoters, but at a lower significance level. Although we had expected to find the GCC box in most of the IPT/DPT promoters, in fact, it occurred in only eight of them (23%; twice in one promoter). However, this number was significantly higher than the 1.7 expected in a set of random promoters, and along with the known specificity of Pti4 for binding the GCC box, this finding further supports a role for this element in Pti4-mediated gene expression. Of the seven GCC-containing promoters we analyzed by ChIP, four were bound by Pti4 and three were not. ERFs have been implicated in both transcriptional activation and repression (Fujimoto et al., 2000), so it is interesting that all of the GCC box–containing promoters that were bound by Pti4 were from IPT genes. Based on this observation, Pti4 appears to be mainly, or exclusively, a transcriptional activator of GCC box–containing genes. The fact the Pti4 did not bind three GCC box–containing promoters suggests that the nucleotide context in which the GCCGCC element lies might influence the binding of Pti4. We examined this possibility by comparing the nucleotides adjacent to the GCC box in the seven promoters described above. Although no consensus motif was apparent, we did observe that the sequences 3′ to the GCC box in promoters bound by Pti4 had a higher average pyrimidine content (72.5%) compared with the comparable region in the promoters not bound by Pti4 (the pyrimidine average was 42.5% in those promoters). The sequences 5′ of the GCC box showed no nucleotide bias in either promoter set. Whether this 3′ nucleotide composition difference influences the binding of Pti4 will require further investigation.
The two other elements present in greater than expected frequencies in the IPT/DPT promoters (Myb1 and G boxes) have both been reported previously to be associated with plant defense responses. For example, the DNA binding site that we call Myb1 (GTTAGTT) is present in the promoter of the well-characterized HR-related gene HSR203J of tobacco and contributes to the regulatory function of that promoter (Pontier et al., 2001). Individual genes of the MYB family in Arabidopsis are induced by bacterial infection and during the hypersensitive response (Kranz et al., 1998; Daniel et al., 1999). The G box (CACGTG) plays a role in gene expression in response to various stresses (Menkens et al., 1995). Transcription factors that bind this cis element belong to the bZIP superfamily of proteins and have been called GBF (G box binding factors). Earlier studies have shown synergism between the GCC box and the G box (Hart et al., 1993), and an ERF from Arabidopsis was shown to bind a bZIP factor in a protein–protein interaction screen (Buttner and Singh, 1997). The fact that the Myb1 and G box cis elements are enriched in the IPT/DPT promoters might further support a defense- or stress-related role for these genes and raises the possibility that Pti4 regulates gene expression synergistically with TFs that bind these elements. Neither bZIP nor MYB-like genes were identified as being transcribed differentially in the Pti4-expressing plants (but see below), and this indicates that, if Pti4 interacts with proteins expressed by such genes, it relies on their basal levels of expression.
Five cis elements with reported roles in defense-related gene expression were not enriched in the IPT/DPT promoters; therefore, it is unlikely that these elements play an important role in Pti4-mediated gene expression. In total, of the 35 IPT/DPT promoters we studied in detail, 10 contained none of the three cis elements we found to be present in significantly increased numbers in the IPT/DPT promoters; presumably, they have novel elements that are involved in Pti4-mediated expression. These 10 promoters were scanned using the motif-searching program MEME (Bailey and Elkan, 1994), but we were unable to identify any single cis element that is shared by a majority of these promoters. Our stringent criteria for deriving the 78 IPT/DPT genes from the >3000 SAGE genes identified strongly indicates that their promoters are regulated in some manner by the activity of Pti4. By isolating TFs that bind IPT/DPT promoters but that lack known cis elements, it should be possible to explore further the mechanisms by which they are regulated by Pti4.
Mechanisms by Which Pti4 Might Regulate Gene Expression
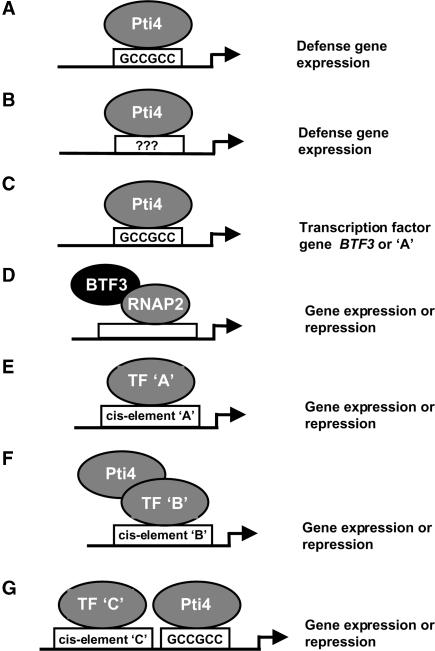
We used ChIP to examine the potential in vivo binding by Pti4 of 18 of the IPT/DPT promoters. Pti4 was shown to bind 11 of these promoters, with just 4 of them having a GCC box. Thus, our data support both a direct and a potentially indirect role for Pti4 in the regulation of gene expression. We show several mechanistic possibilities for the regulation of gene expression by Pti4 in Figure 5. In model A, Pti4 binds the GCC box directly to activate the expression of defense-related genes. Examples of this might be the genes that encode PDF1.2, peroxidase ATP1a, and PR4 (Table 4). However, because the GCC box is not present in many IPT/DPT promoters, it is possible that another novel element, yet to be determined, is bound by Pti4 (model B). In a third possibility, shown as model C, Pti4 binds the GCC box present in the promoter of a TF, and this TF then plays a role in modulating expression from other IPT/DPT genes. We identified one TF gene, BTF3, whose transcript was increased in the Pti4-expressing plants and that has a GCC box–containing promoter bound by Pti4. BTF3 TFs were purified originally from HeLa cells and play a role in the initiation of transcription in several eukaryotes by forming a complex with RNA polymerase II (Zheng et al., 1990). As illustrated in model D, we speculate that a Pti4-mediated increase in BTF3 might play a role in activating the expression of other IPT/DPT genes. Model E provides another mechanistic explanation for the IPT/DPT promoters that lack any previously known cis elements or that contain a GCC box but are not bound by Pti4. These promoters might be bound directly by TFs whose gene expression is altered by Pti4 or might be regulated indirectly by these TFs, as proposed for BTF3.
Figure 5.
Models for Pti4-Mediated Gene Expression.
Pti4 is postulated to regulate the expression of defense genes both directly via the GCC box (or a non-GCC box element) and indirectly via other transcription factors. Shown first are three “direct binding” models. In model A, Pti4 interacts directly with the GCC box to activate the expression of various defense-related genes. Model B shows that Pti4 might also bind directly to a currently unknown, non-GCC box element (shown as ???). In model C, Pti4 binds the GCC box present in the promoters of the BTF3 TF gene or another TF gene, designated TF A. In the next two “stepwise activation” models, the BTF3 protein associates with RNA polymerase II (RNAP2) to regulate gene expression (model D) or the TF A protein from model C binds the cognate cis element (A) present in the promoter of a defense-related gene (model E). In the “cooperative activation” model F, Pti4 interacts physically with another basally expressed transcription factor (TF B), and this interaction facilitates the expression of genes whose promoter contains a cis element bound directly by TF B. Finally, in the “tandem binding” model G, gene expression occurs upon binding of the GCC box by Pti4 and of another non-GCC box by a non-ERF TF. See Discussion for further details.
Two other possibilities might account for our ChIP observations. Model F (Figure 5) depicts a scenario in which non-GCC box–containing IPT/DPT promoters are associated with Pti4 and have either another known defense-related cis element or some other previously undescribed element. Four promoters in the former category are genes that encode a putative phosphate transporter, a salt-induced plasma membrane intrinsic protein, a ribosomal protein, and a TF, RAV1. All but the RAV1 gene are induced in the Pti4-expressing plants. The RAV1 promoter has a G box, and because this element is enriched significantly in the IPT promoters, it is plausible that the expression of the RAV1 gene is inhibited by a GBF-like TF in concert with the Pti4 protein (Figure 5F). Interestingly, we did identify a putative bZIP family TF gene (tag 644; GCAGAGTTGG) with increased tag numbers in the Pti4-expressing line; unfortunately, however, it was one of five tags that matched two different Arabidopsis genes, so we cannot be sure that its expression is truly altered. Nevertheless, it is possible that Pti4 interacts physically with a bZIP factor to alter the expression of some of the IPT/DPT genes. One precedent for such a model is the interaction between the bZIP factor OBF4 (TGA4) and the Arabidopsis ERF AtEBP (Buttner and Singh, 1997). Finally, our data support a model (Figure 5G) in which some IPT/DPT promoters are regulated by the synergistic binding of both Pti4 and another defense-related TF. The PDF1.2 promoter that contains both a GCC box and a Myb1 box might represent an example of such a promoter.
Some IPT/DPT Genes Likely Contribute to the Plant Defense Response
A central assumption in our work is that some of the genes whose expression is altered by Pti4 play roles in defense responses. This assumption is based on several previous observations: (1) Pti4 interacts with the Pto kinase, a disease resistance protein; (2) Pti4 binds the GCC box present in many defense-related genes; (3) the Pti4 gene itself is induced in both compatible and incompatible plant–pathogen interactions; and (4) the expression of Pti4 in Arabidopsis causes the upregulation of several known PR genes and inhibits the formation of disease symptoms by two very different pathogens. We found that of 15 genes tested that produced detectable transcripts, the expression of 6 was altered during Pseudomonas infection in a similar manner, as determined by our SAGE results. The remaining nine genes detected transcripts of similar abundance in both Pti4-expressing and wild-type plants. The expression differences of three of these nine genes would potentially be obscured by cross-hybridization with other closely related family members in Arabidopsis (S. Chakravarthy, unpublished data). Therefore, it is possible that the expression of an even greater percentage of IPT/DPT genes is altered by Pseudomonas infection.
The identities of many IPT/DPT genes suggest a role for them in defense. For example, the PDF1.2 and PR4 genes have been shown to be responsive to infection by fungal pathogens and are involved in the ethylene/jasmonate-dependent disease signaling pathways (Thomma et al., 1998, 1999). Other IPT/DPT genes are known to be activated under salt or dehydration stress (SIMIP, Di19, and ERD6) (Gosti et al., 1995; Kiyosue et al., 1998; Pih et al., 1999). We observed genes involved in calcium signaling: a putative calreticulin and a calcineurin B–like protein-interacting protein kinase (CIPK6). Various members of both classes of proteins have been proposed to play roles in defense and stress responses (Wyatt et al., 2002; Kim et al., 2003). We also identified an ASK-GAMMA kinase; members of the ASK family have been implicated in the wound response (Dornelas et al., 1998). Two TF genes, BTF3 and RAV1, were identified that have not been implicated previously in defense (Zheng et al., 1990; Kagaya et al., 1999). Finally, it should be noted that CIPK6, ASK-GAMMA, and RAV1 are DPT genes. Therefore, these genes may act normally to inhibit defense- or stress-related responses.
Many of the IPT/DPT genes are similar to the recently described APR genes, whose expression changes during the incompatible interaction between tomato and Pseudomonas (Mysore et al., 2002). Similar to what we observed in the Pti4-expressing plants, the transcript abundance of large numbers of genes associated with photosynthesis and energy metabolism was decreased in the tomato–Pseudomonas interaction, whereas PR transcripts were increased (Mysore et al., 2002). We did not determine whether the expression of any of the IPT/DPT genes is altered during infection with E. orontii. However, this is possible given the increased resistance to this pathogen observed in Pti4-expressing plants (Gu et al., 2002).
Several IPT/DPT genes had no obvious role in defense. Eleven photosynthesis-related genes were identified, and the expression of 10 of these was decreased. There were 11 ribosomal protein genes and 8 energy metabolism–related genes identified, and they showed both increased and decreased expression. The overall spectrum of the IPT/DPT genes, and the expression of some of these in response to pathogen infection in Arabidopsis, suggest that Pti4 causes a global diversion of gene expression toward defense response and away from cellular responses that are, at least temporarily, dispensable.
A previous study by Wu et al. (2002) compared global gene expression between wild-type and Pti4-expressing Arabidopsis using oligonucleotide-based microarrays (Affymetrix chip) and observed 28 genes that exhibited >2.5-fold induction in Pti4 plants. There is almost no overlap between these genes and our IPT/DPT genes, except for basic endochitinase (PR3), which they found to be induced 3.6-fold in Pti4 plants (we observed more PR3 tags in our Pti4 SAGE library: two tags compared with none in the wild type; SAGE tag 2205 [data not shown]). Surprisingly, these authors observed no enhanced expression of PDF1.2, the most highly induced gene in our study (SAGE tag 65). Some of the genes present on their microarray occur in clustered gene families, and because they were cited using a BAC clone accession number instead of the gene accession number, it was not possible to compare our results directly with theirs (i.e., for the transporter and calreticulin genes; SAGE tags 643 and 423). However, we did perform RT-PCR to determine if three genes identified by Wu et al. were induced in our Pti4-expressing plants. We found that glucanase precursor gene was induced and reconfirmed the induction of PR3 but could not detect lipoxygenase (see supplemental data online). It is possible that greater overlap between our IPT/DPT genes and the genes found by Wu et al. (2002) would have been observed if we had sequenced more clones from our SAGE libraries. However, it also is possible that the lack of overlap is the result of the use of different promoters (Wu et al. [2002] used tCUP, and we used 35S Cauliflower mosaic virus) and the fact that the authors of the earlier study used seedlings whereas our experiments were performed on vegetative tissue from 4-week-old plants. Interestingly, although it is not discussed explicitly by Wu et al. (2002), they also observed that only a small percentage of the genes identified in their study (6 of 28) possessed GCC boxes in their promoters.
ETHYLENE RESPONSE FACTOR1 (ERF1) from Arabidopsis was shown recently to play a role in integrating signals from the ethylene and jasmonate signaling pathways (Lorenzo et al., 2003). The authors showed that ERF1 plays a role in the activation of PDF1.2 in Arabidopsis. Overexpression of ERF1 in Arabidopsis led to the activation of several genes, many of which were defense-related genes, including endochitinase, cytochrome P450, pre-hevein-like gene, and peroxidase. Similar genes were found among our IPT genes, but given that cross-hybridization among homologous sequences occurs in microarrays, it is not possible to confirm if they are identical to our IPT genes. Most of the genes activated by ERF1, however, were not observed in our study. This is not surprising, because the amino acid sequences of Pti4 and ERF1 are very different and likely regulate the expression of distinct sets of genes in Arabidopsis.
In previous work, we and others showed that Pti4-expressing Arabidopsis plants display a partial triple-response phenotype (Gu et al., 2002; Wu et al., 2002). Several genes have been shown to be involved in ethylene biosynthesis and signaling related to the triple response, including ETO1, EIN2, CTR1, AUX1, HLS1, and EIR1 (Roman et al., 1995; Kieber, 1997). We did not recover tags corresponding to any of these genes from our SAGE libraries. It is possible that some of the kinases and TFs identified in our study might play a yet uncharacterized role in the triple response. For example, the RAV1 TF (SAGE tag 347) has an AP2-like DNA binding domain and may be involved in ethylene-mediated responses (Kagaya et al., 1999).
SAGE Is a Powerful Tool for Profiling Plant Transcriptomes
This is one of the first reports of the application of SAGE to Arabidopsis expression profiling. The technique is used widely in yeast, microbial, and animal systems (www.sagenet.org/) and has been applied successfully in rice, Arabidopsis, and loblolly pine (Matsumura et al., 1999; Lorenz and Dean, 2002; Jung et al., 2003; Lee and Lee, 2003). Compared with cDNA microarrays, SAGE offers the advantages of being an open-architecture, highly quantitative method, with the ability to detect rare transcripts and transcripts from individual members of a closely related gene family. Most current microarray technologies do not reliably detect genes expressed at low levels, and potential cross-hybridization among members of gene families renders microarrays less sensitive than SAGE (Menssen and Hermeking, 2002; Wan et al., 2002). The accuracy and reliability of SAGE improves with the size of the library obtained and depends on the quality of sequencing, whereas sequencing and annotation errors also lead to discrepancies in microarray data analysis (Wan et al., 2002). Because of the large amount of sequencing needed for SAGE analysis, the samples generally are not replicated (Matsumura et al., 1999; see also publications provided at www.sagenet.org/). However, we prepared each tag library from a large number of plants (30 to 35); thus, they represent a population average. In addition, the rigorous statistical analyses of tag number differences that are available produce highly reliable data (Audic and Claverie, 1997). SAGE also is inexpensive relative to microarrays and requires none of the specialized equipment needed for microarray fabrication and scanning.
We found a reasonable, but not perfect, correlation between our SAGE data and RT-PCR results. We were able to verify the differential expression of 25 genes (of 35 tested) using RT-PCR and RNA gel blot analysis. RT-PCR can detect low-abundance transcripts, but it depends on transcript amplification and is subject to other variables (i.e., we observed that differences between wild-type and Pti4 samples as detected by RT-PCR became less distinct with increasing cycle number and also depended on the dilution of the cDNA used). As mentioned above, SAGE also detects low-abundance transcripts, but it does not rely on the amplification of specific transcripts and derives tags representing actual transcript numbers. In summary, although SAGE, microarrays, and RT-PCR all yield useful information about differential gene expression, the results obtained with these methods do not always correlate perfectly with each other as a result of the inherent differences in the techniques (Ishii et al., 2000; Leerkes et al., 2002; Menssen and Hermeking, 2002). Further studies comparing these techniques are required before conclusions can be made about which one most accurately measures true transcript numbers.
We found two minor impediments in our SAGE analysis. The first was that in five cases individual tags corresponded to two different genes. Given the theory underlying SAGE, such an occurrence is very unusual. It is possible that sequencing errors present in the EST databases accounted for this observation (we eliminated these five tags from further analysis). The second observation was that 18 tags did not match any annotated open reading frames or ESTs in the Arabidopsis databases. These tags might correspond to unannotated open reading frames, or again, they might be caused by sequencing errors in the database, because in each case the tags differed by just a single nucleotide from an EST or annotated open reading frame (we also set these tags aside). Despite these minor problems, we found the SAGE protocol relatively simple to implement, the tag numbers reliable as judged by comparison with RNA gel blot and RT-PCR results, and the data analysis straightforward.
The identification of a set of promoters that are bound by Pti4 but that lack the GCC box provides interesting new leads to follow in our attempt to understand the role of this ERF in the regulation of gene expression. We anticipate that these promoters will be useful in yeast one-hybrid experiments to determine if Pti4 binds directly to a non-GCC box element and also to identify other plant proteins that bind the IPT/DPT promoters. Based on the models presented in Figure 5, we predict that some DNA binding proteins that bind the IPT/DPT promoters will interact physically with Pti4 and that their activity will be enhanced.
METHODS
Plant Material
Wild-type and Pti4-expressing Arabidopsis thaliana ecotype Col-0 plants (Gu et al., 2002) were grown in a light room at 22 to 23°C under a 16-h photoperiod. The aboveground vegetative portions of 4-week-old plants were harvested for serial analysis of gene expression (SAGE) profiling and chromatin immunoprecipitation experiments. For pathogen infection, plants were grown in 4-inch pots that were covered with mesh so that the seedlings could grow through the mesh. Approximately 30 to 35 plants were grown per pot. Three- to 4-week-old plants were used for pathogen infection.
Pathogen Infection
Arabidopsis plants were inoculated with Pseudomonas syringae pv tomato strain DC3000. Overnight cultures of the bacterium grown in Kings medium B (Martin et al., 1993) were washed twice in 10 mM MgCl and resuspended in 10 mM MgCl with 0.04% Silwet at 106 colony-forming units/mL. The plants were dipped into the suspension for 30 s. Control plants were mock-inoculated in a solution of 10 mM MgCl with 0.04% Silwet. Leaf tissue was harvested at 24 h and 3 days after inoculation. Four pots each were treated with the pathogen or the control solution. RNA was isolated from leaves pooled from the four pots.
RNA Isolation
RNA isolation was performed using the hot-phenol method as described previously (Gu et al., 2000), with an additional step of phenol-chloroform extraction and precipitation with sodium acetate and ethanol at the end. mRNA was isolated from total RNA using the PolyATract mRNA Isolation System I (Promega, Madison, WI). Five micrograms of poly(A)+ RNA was used for double-stranded cDNA synthesis with a cDNA synthesis kit (Stratagene, La Jolla, CA).
SAGE Protocol
SAGE procedures were performed according to the originally described protocol (Velculescu et al., 1995, 1997) up to and including step 8 (ligation of tags to form ditags). Gel-purified primers were obtained from Integrated DNA Technologies (Coralville, IA). Restriction and modification enzymes used were obtained from the manufacturers specified in the detailed SAGE protocol (version 1.0c; obtained from V.E. Velculescu, Johns Hopkins Oncology Center and Howard Hughes Medical Institute, Baltimore, MD) (Velculescu et al., 1997). The cDNA was digested with NlaIII and captured on streptavidin-coated magnetic beads (Dynal Biotech, Lake Success, NY). Linkers were ligated to captured cDNA ends, and SAGE tags adjacent to the linkers were released with BsmFI digestion. Two pools of tags were separately blunt-ended and ligated to linkers 1 and 2 (Velculescu et al., 1995), and ditags were formed by blunt-end ligation of the two pools.
The ditag amplification step of the SAGE procedure (step 9) was performed with the following modifications based on those described by Matsumura et al. (1999). Ditags were amplified with 5′ biotinylated linker-specific primers 1 (5′-TCTAACGATGTACGGGGACA-3′) and 2 (5′-TACAACTAGGCTTAATAGGGACA-3′). This resulted in ∼67-bp ditag PCR products. PCR amplifications (50-μL reactions) were performed on 0.2-mL 96-well PCR plates (Eppendorf, Westbury, NY) with an Eppendorf Mastercycler Gradient PCR machine. A master mix was prepared that contained, for each PCR well, 0.65 units of AmpliTaq Gold and 1× AmpliTaq Gold Buffer (Applied Biosystems, Foster City, CA), 1.5 mM MgCl2, 0.2 mM each deoxynucleotide triphosphate, 70 ng each of biotinylated primers 1 and 2, and 1 μL of a 1:200 dilution of ditag ligation product (from step 8) as a template. The PCR conditions were 95°C for 2 min followed by 33 cycles of 30 s at 94°C, 30 s at 60°C, and 1 min at 72°C. After a 5-min incubation at 70°C, the plates were stored on ice until they were processed. Eight 96-well plates were processed for each SAGE library constructed. The PCR products from each 96-well plate were pooled in a 30-mL Corex tube, extracted and precipitated as described in the detailed protocol (step 10), and resuspended in 102 μL of LoTE (3 mM Tris-HCl and 0.2 mM EDTA, pH 7.5). Two microliters was used for dot quantitation (an ∼5- to 7-μg PCR product was obtained from each 96-well plate), the remaining 100 μL was run on a 12% polyacrylamide gel, and the 67-bp bands were gel-purified as described in the SAGE protocol.
After extraction and precipitation, the amplified ditag was resuspended in 47 μL of LoTE. Two microliters was quantitated, and the remaining 45 μL (∼2 μg of gel-purified ditag from each 96-well plate) was digested with NlaIII. Biotinylated primer sequence was removed with streptavidin-magnetic beads, and ditags were again gel-purified as described in the version 1.0c protocol. The purified 26-bp ditags were extracted, precipitated, and ligated to form concatemers. Concatemers from 400 bp to 3 kb were size-selected on an 8% polyacrylamide gel, gel-purified, spun through a Spin-X microcentrifuge tube (Fisher, Suwanee, GA), precipitated, and cloned into SphI-digested pZERO (Invitrogen, Carlsbad, CA). Colonies that grew on LB-Zeocin plates (Invitrogen, Carlsbad, CA) were screened for plasmids with inserts >400 bp by colony PCR. Desired colonies were grown overnight in low-salt LB with Zeocin added, 96-well plasmid preparations (Qiagen, Valencia, CA) were performed for each, and the plasmids were sequenced with M13 forward primer on an ABI 3700 automated sequencer (Applied Biosystems) at the Cornell Biotechnology Sequencing Center. Seven 96-well plates (658 colonies, with two control wells on each plate) were prepared from each library.
SAGE Data Analysis and Tag-to-Gene Assignment
Sequences were analyzed with the SAGE software program (Velculescu et al., 1995). The total tags counted in this project were identified from both unique ditags and duplicated ditags that were counted only once to eliminate potential PCR bias in quantitation. The number of tags of a particular sequence present in the wild-type and Pti4 libraries was counted. Using these data, fold increase and fold decrease values for every tag in the Pti4 library compared with the wild-type library were calculated. Fold increase was calculated as the percentage of a tag's occurrence in the Pti4 library divided by the percentage of its occurrence in the wild-type library. Fold decrease was calculated as the percentage of a tag's occurrence in the wild-type library divided by the percentage of its occurrence in the Pti4 library. When necessary, a tag count of zero was changed to one to avoid division by zero. To determine if the difference in tag numbers between the two libraries for each tag sequence was statistically significant, we used the method described by Audic and Claverie (1997). This method is a rigorous statistical test to calculate the significance of “digital gene expression profiles” like those generated by SAGE. Given the total size of each library (total tag number), the significance of the difference in tag number between the two populations for every sequence was calculated and the tags with P values of ≤0.05 were identified.
Gene assignments for tags were made through progressive rounds of database searches. Tag sequences of 14 bp, consisting of the 10-bp tag sequence plus the 4-bp NlaIII restriction site, were submitted as queries for Basic Local Alignment Search Tool (BLAST) searches in the Arabidopsis genome sequences available in the TAIR (http://www.arabidopsis.org), TIGR (http://www.TIGR.org/tdb), and GenBank nonredundant and EST databases. Criteria used in the searches were as follows: the tag had to (1) match perfectly to the gene sequence, (2) occur in the correct orientation in the gene, and (3) be in the expected position (i.e., downstream of the farthest NlaIII site within the gene's transcript).
Reverse Transcription PCR
Total RNA from wild-type and Pti4-expressing Arabidopsis tissues (pooled from 20 to 25 plants that were grown independently of the plants used for the SAGE libraries) was used for first-strand cDNA synthesis at a concentration of 125 ng/μL with oligo(dT) primer and Superscript II (Invitrogen). Serial dilutions of the reaction mixture (1.25, 2.5, 5, 10, 25, and 100×) were prepared, and 1 μL of each dilution was taken for PCR with individual primer pairs to determine the linear concentration range of PCR for each primer pair (see supplemental data online). The final PCRs were performed using 1 (or 2) μL of the appropriate dilution. All PCRs were in 25 (or 50) μL volume using Taq polymerase (Fisher). Reaction parameters differed for each gene: thermocycling conditions were 94°C for 2 min followed by 22, 25, or 28 cycles of 94°C for 1 min, 51 or 56°C for 1 min, and 72°C for 2 min, with a final polymerization step at 72°C for 7 min. Reactions were performed in triplicate and analyzed by running 5 to 7 μL of the reaction on 1.4% agarose gels. The intensity of the PCR bands was determined using the Spot Densitometry feature of the AlphaEase program (Alpha Innotech, San Leandro, CA). The sequences of the primers used for PCR are given in the supplemental data online.
As a control, the gene encoding chlorophyll a/b binding protein, Lhcb5, was used (SAGE tag 12; see supplemental data online). When adjusted for the different tag numbers derived from each library, Lhcb5 tags were equal in the wild-type and Pti4 SAGE libraries (wild type, 58 of 13,310; Pti4, 62 of 14,215). In individual RT-PCR experiments, minor differences were observed in the abundance of Lhcb5 transcripts between wild-type and Pti4 samples. This finding was attributed to the slightly different efficiency of the wild-type and Pti4 reverse transcription reactions and errors in the dilution of cDNA. Therefore, we used the differences to normalize PCR product density values between samples including Lhcb5, as shown in Figures 1A and 1B.
Reverse RNA Gel Blot Analysis
EST clones corresponding to selected genes were obtained from the ABRC DNA Stock Center (Columbus, OH), and the cDNA inserts were amplified using M13 forward and reverse primers. PCR products were separated by gel electrophoresis and sandwich-blotted onto two Hybond-N+ membranes to generate identical duplicate blots (Sambrook et al., 1989). An H+-pumping ATPase subunit was used as a control. Ten micrograms of total RNA from pathogen- or mock-inoculated tissue was used for first-strand cDNA synthesis in a 20-μL reaction containing 500 ng of oligo(dT) primer, 0.5 mM each dTTP, dGTP, dCTP, and radiolabeled 32P-dATP (3000 Ci/mmol), and Superscript II (Invitrogen). The mRNA was removed with RNase H before hybridization. Hybridization was performed in tubes at 56°C for 16 h. Solutions for hybridization and washing were as recommended by the manufacturer of the membrane (Amersham Biosciences, Piscataway, NJ). Treatments and exposure times for inoculated and control samples were identical. Signals were visualized by phosphorimaging.
Promoter Analysis
Thirty-five IPT and DPT promoter sequences (1-kb sequence upstream of the annotated ATG site) were retrieved from the Arabidopsis genome sequence at TAIR. For the analysis of a set of random promoters, the genome sequence at TIGR (BAC tiling path) was scanned, and 50 promoter sequences were retrieved. A motif-searching program was written in Perl to perform sequence searches (M. D'Ascenzo and G. Martin, unpublished data). The program searched in the forward direction only using Perl Regular Expressions. Therefore, motifs were translated to the reverse complement to effectively search for the motif on both strands of the promoter.
The probability of the occurrence of each nucleotide was calculated from a random set of 20 promoters to obtain the nucleotide composition of promoter sequences. It was found to be 0.34 for A, 0.34 for T, 0.15 for G, and 0.18 for C. These values were used to calculate the probability (p) of finding a given motif (e.g., GCCGCC) in a DNA sequence. The probability (P) of seeing at least n copies of a motif in 35 (or 50) promoters of 1000 bp, looking on both strands, was calculated using the formula P(x ≥ n) = 1 − i=0n−1Σ e−λ × (λi/ i!), where n is the observed number of motifs and λ is the expected number of motifs of that sequence, or λ = 2 × (1000 − L + 1) × 35 (or 50) × p, where p is calculated as stated above and L is the length of the motif. The value P was considered as the p value to determine the significance of the cis element, and values of ≤0.05 were considered statistically significant.
Chromatin Immunoprecipitation
Two grams of leaf tissue of wild-type and Pti4-expressing Arabidopsis was fixed for 15 min in 1% formaldehyde (or left unfixed as a non-cross-linked control). Leaves were rinsed extensively with water, dried, frozen in liquid nitrogen, and stored at −80°C until further processing. Tissues were ground in IP buffer (0.1% SDS, 1% Triton X-100, 50 mM Hepes, pH 7.9, 150 mM NaCl, and 1 mM EDTA) supplemented with protease inhibitors (1 μg/mL aprotinin, 1 μg/mL leupeptin, 1 μg/mL pepstatin, 0.5 mM phenylmethylsulfonyl fluoride, and 1 mM benzamidine). The lysate was centrifuged for 5 min at 14,000 rpm and 4°C in a microcentrifuge before filtering through siliconized glass wool. The chromatin was sonicated to yield DNA fragments between 0.5 and 1.3 kb in size. After sonication, a 20-μL aliquot was set aside as the input material and stored at −20°C until further processing. The chromatin solution was split into two 500-μL aliquots and combined with 20 μL of protein A–Sepharose beads (Bio-Rad, Hercules, CA) that had been preadsorbed with 10 μL of preimmune serum or 10 μL of Pti4 antibodies. (Pti4-specific antibodies were raised against the peptide RSSVMQVGCQIEQLTGVHQL, which is at the C terminus and outside the ERF domain of Pti4. This peptide bears no similarity to any annotated protein from Arabidopsis [Y. Gu and G.B. Martin, unpublished data]). After an overnight incubation at 4°C on a rotation wheel, the beads were washed three times with IP buffer and twice with TE, pH 8.0. The immunoprecipitated material was released from the beads by heating at 65°C for 15 min in 200 μL of TE supplemented with 1% SDS. Forty microliters of immunoprecipitate or 20 μL of input was combined with 80 μL (immunoprecipitate) or 100 μL (input) of TE and 1% SDS and incubated at 65°C for 6 h. After precipitation with isopropanol, sodium acetate, and 2 μg of glycogen, the DNA was resuspended in 20 μL of TE (immunoprecipitate) or 200 μL of TE (input). After a brief centrifugation, 1 μL of the input was added to 29 μL of TE.
PCR Analyses and Sequencing after Chromatin Immunoprecipitation
PCR amplifications were performed in a volume of 50 μL with 50 pmol of each primer, 0.2 mM deoxynucleotide triphosphates, and 5 units of Taq polymerase (Amersham Biosciences, Baie D'Urfé, Québec, Canada) along with ∼2 μL of immunoprecipitated DNA or diluted input. The actual amount of DNA used was adjusted so that the input DNA from wild-type and Pti4 Arabidopsis gave similar amounts of amplified product. The volume of immunoprecipitate then was adjusted accordingly. Under these conditions, the amount of PCR product was proportional to the amount of template DNA added, as determined by serial dilutions of the input DNA (data not shown). The primer pairs used were specific to the gene of interest. The cycling conditions were 5 min of initial denaturation at 95°C followed by 35 cycles of 30 s at each temperature (95, 55, and 72°C). For sequencing, the PCR fragments were generated as described above except that Taq polymerase was replaced by the proofreading enzyme ExTaq (PanVera, Madison, WI). The PCR fragments were gel-excised and sequenced directly. Sequences of the primers used for PCR are shown in the supplemental data online.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Gregory B. Martin, gbm7@cornell.edu.
Accession Numbers
The GenBank accession numbers for the sequences mentioned in this article are as follows: BE038487 (an unknown protein); AY093268 (a putative bZIP family TF gene); and L44582 (a H+-pumping ATPase subunit).
Supplementary Material
Acknowledgments
We thank Rasmus Nielsen (Department of Biological Statistics and Computational Biology, Cornell University) for help with the statistical analysis of the promoters and the ABRC for EST clones. We thank Pete Pascuzzi and Kerry Pedley for helpful discussions and critical review of the manuscript. This work was supported, in part, by National Science Foundation Grant 00-90402 (to G.B.M.), by the National Research Council, Plant Biotechnology Institute core program (to P.R.F.), and by the National Science and Engineering Research Council discovery grant program (to C.D.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.017574.
Footnotes
Online version contains Web-only data.
References
- Audic, S., and Claverie, J.M. (1997). The significance of digital gene expression profiles. Genome Res. 7, 986–995. [DOI] [PubMed] [Google Scholar]
- Bailey, T.L., and Elkan, C. (1994). Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 2, 28–36. [PubMed] [Google Scholar]
- Baker, S.S., Wilhelm, K.S., and Thomashow, M.F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo, M., Molina, A., and Solano, R. (2002). Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. Plant J. 29, 23–32. [DOI] [PubMed] [Google Scholar]
- Buttner, M., and Singh, K.B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. USA 94, 5961–5966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14, 559–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheong, Y.H., Chang, H.S., Gupta, R., Wang, X., Zhu, T., and Luan, S. (2002). Transcriptional profiling reveals novel interactions between wounding, pathogen, abiotic stress, and hormonal responses in Arabidopsis. Plant Physiol. 129, 661–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel, X., Lacomme, C., Morel, J.B., and Roby, D. (1999). A novel myb oncogene homologue in Arabidopsis thaliana related to hypersensitive cell death. Plant J. 20, 57–66. [DOI] [PubMed] [Google Scholar]
- Després, C., DeLong, C., Glaze, S., Liu, E., and Fobert, P.R. (2000). The Arabidopsis NPR1/NIM1 protein enhances the DNA binding activity of a subgroup of the TGA family of bZIP transcription factors. Plant Cell 12, 279–290. [PMC free article] [PubMed] [Google Scholar]
- Després, C., Subramaniam, R., Matton, D.P., and Brisson, N. (1995). The activation of the potato PR-10a gene requires the phosphorylation of the nuclear factor PBF-1. Plant Cell 7, 589–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornelas, M.C., Lejeune, B., Dron, M., and Kreis, M. (1998). The Arabidopsis SHAGGY-related protein kinase (ASK) gene family: Structure, organization and evolution. Gene 212, 249–257. [DOI] [PubMed] [Google Scholar]
- Eulgem, T., Rushton, P.J., Robatzek, S., and Somssich, I.E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. [DOI] [PubMed] [Google Scholar]
- Fujimoto, S.Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box-mediated gene expression. Plant Cell 12, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosti, F., Bertauche, N., Vartanian, N., and Giraudat, J. (1995). Abscisic acid-dependent and -independent regulation of gene expression by progressive drought in Arabidopsis thaliana. Mol. Gen. Genet. 246, 10–18. [DOI] [PubMed] [Google Scholar]
- Gu, Y.Q., Wildermuth, M.C., Chakravarthy, S., Loh, Y.T., Yang, C., He, X., Han, Y., and Martin, G.B. (2002). Tomato transcription factors Pti4, Pti5, and Pti6 activate defense responses when expressed in Arabidopsis. Plant Cell 14, 817–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu, Y.Q., Yang, C., Thara, V.K., Zhou, J., and Martin, G.B. (2000). Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell 12, 771–786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart, C.M., Nagy, F., and Meins, F., Jr. (1993). A 61 bp enhancer element of the tobacco β-1,3-glucanase B gene interacts with one or more regulated nuclear proteins. Plant Mol. Biol. 21, 121–131. [DOI] [PubMed] [Google Scholar]
- Ishii, M., Hashimoto, S., Tsutsumi, S., Wada, Y., Matsushima, K., Kodama, T., and Aburatani, H. (2000). Direct comparison of GeneChip and SAGE on the quantitative accuracy in transcript profiling analysis. Genomics 68, 136–143. [DOI] [PubMed] [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces COR genes and enhances freezing tolerance. Science 280, 104–106. [DOI] [PubMed] [Google Scholar]
- Johnson, C., Boden, E., Desai, M., Pascuzzi, P., and Arias, J. (2001). In vivo target promoter-binding activities of a xenobiotic stress-activated TGA factor. Plant J. 28, 237–243. [DOI] [PubMed] [Google Scholar]
- Jung, S.H., Lee, J.Y., and Lee, D.H. (2003). Use of SAGE technology to reveal changes in gene expression in Arabidopsis leaves undergoing cold stress. Plant. Mol. Biol. 52, 553–567. [DOI] [PubMed] [Google Scholar]
- Kagaya, Y., Ohmiya, K., and Hattori, T. (1999). RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 27, 470–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieber, J.J. (1997). The ethylene response pathway in Arabidopsis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 277–296. [DOI] [PubMed] [Google Scholar]
- Kim, K.N., Cheong, Y.H., Grant, J.J., Pandey, G.K., and Luan, S. (2003). CIPK3, a calcium sensor–associated protein kinase that regulates abscisic acid and cold signal transduction in Arabidopsis. Plant Cell 15, 411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, S.R., Choi, J.L., Costa, M.A., and An, G. (1992). Identification of G-box sequence as an essential element for methyl jasmonate response of potato proteinase inhibitor II promoter. Plant Physiol. 99, 627–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y.J., Lin, N.C., and Martin, G.B. (2002). Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109, 589–598. [DOI] [PubMed] [Google Scholar]
- Kiyosue, T., Abe, H., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). ERD6, a cDNA clone for an early dehydration-induced gene of Arabidopsis, encodes a putative sugar transporter. Biochim. Biophys. Acta 1370, 187–191. [DOI] [PubMed] [Google Scholar]
- Kranz, H.D., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16, 263–276. [DOI] [PubMed] [Google Scholar]
- Lee, J.Y., and Lee, D.H. (2003). Use of serial analysis of gene expression technology to reveal changes in gene expression in Arabidopsis pollen undergoing cold stress. Plant Physiol. 132, 517–529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leerkes, M.R., Caballero, O.L., Mackay, A., Torloni, H., O'Hare, M.J., Simpson, A.J., and de Souza, S.J. (2002). In silico comparison of the transcriptome derived from purified normal breast cells and breast tumor cell lines reveals candidate upregulated genes in breast tumor cells. Genomics 79, 257–265. [DOI] [PubMed] [Google Scholar]
- Lorenz, W.W., and Dean, J.F. (2002). SAGE profiling and demonstration of differential gene expression along the axial developmental gradient of lignifying xylem in loblolly pine (Pinus taeda). Tree Physiol. 22, 301–310. [DOI] [PubMed] [Google Scholar]
- Lorenzo, O., Piqueras, R., Sanchez-Serrano, J.J., and Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K.A., Dangl, J.L., and Dietrich, R.A. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. [DOI] [PubMed] [Google Scholar]
- Martin, C., and Paz-Ares, J. (1997). MYB transcription factors in plants. Trends Genet. 13, 67–73. [DOI] [PubMed] [Google Scholar]
- Martin, G.B., Brommonschenkel, S.H., Chunwongse, J., Frary, A., Ganal, M.W., Spivey, R., Wu, T., Earle, E.D., and Tanksley, S.D. (1993). Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262, 1432–1436. [DOI] [PubMed] [Google Scholar]
- Matsumura, H., Nirasawa, S., and Terauchi, R. (1999). Technical advance: Transcript profiling in rice (Oryza sativa L.) seedlings using serial analysis of gene expression (SAGE). Plant J. 20, 719–726. [DOI] [PubMed] [Google Scholar]
- Mazarei, M., Puthoff, D.P., Hart, J.K., Rodermel, S.R., and Baum, T.J. (2002). Identification and characterization of a soybean ethylene-responsive element-binding protein gene whose mRNA expression changes during soybean cyst nematode infection. Mol. Plant-Microbe Interact. 15, 577–586. [DOI] [PubMed] [Google Scholar]
- Menkens, A.E., Schindler, U., and Cashmore, A.R. (1995). The G-box: A ubiquitous regulatory DNA element in plants bound by the GBF family of bZIP proteins. Trends Biochem. Sci. 20, 506–510. [DOI] [PubMed] [Google Scholar]
- Menssen, A., and Hermeking, H. (2002). Characterization of the c-MYC-regulated transcriptome by SAGE: Identification and analysis of c-MYC target genes. Proc. Natl. Acad. Sci. USA 99, 6274–6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore, K.S., Crasta, O.R., Tuori, R.P., Folkerts, O., Swirsky, P.B., and Martin, G.B. (2002). Comprehensive transcript profiling of Pto- and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 32, 299–315. [DOI] [PubMed] [Google Scholar]
- Niggeweg, R., Thurow, C., Kegler, C., and Gatz, C. (2000. a). Tobacco transcription factor TGA2.2 is the main component of as-1-binding factor ASF-1 and is involved in salicylic acid- and auxin-inducible expression of as-1-containing target promoters. J. Biol. Chem. 275, 19897–19905. [DOI] [PubMed] [Google Scholar]
- Niggeweg, R., Thurow, C., Weigel, R., Pfitzner, U., and Gatz, C. (2000. b). Tobacco TGA factors differ with respect to interaction with NPR1, activation potential and DNA-binding properties. Plant Mol. Biol. 42, 775–788. [DOI] [PubMed] [Google Scholar]
- Ohme-Takagi, M., and Shinshi, H. (1995). Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 7, 173–182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohme-Takagi, M., Suzuki, K., and Shinshi, H. (2000). Regulation of ethylene-induced transcription of defense genes. Plant Cell Physiol. 41, 1187–1192. [DOI] [PubMed] [Google Scholar]
- Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohta, M., Ohme-Takagi, M., and Shinshi, H. (2000). Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J. 22, 29–38. [DOI] [PubMed] [Google Scholar]
- Onate-Sanchez, L., and Singh, K.B. (2002). Identification of Arabidopsis ethylene-responsive element binding factors with distinct induction kinetics after pathogen infection. Plant Physiol. 128, 1313–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, J.M., Park, C.J., Lee, S.B., Ham, B.K., Shin, R., and Paek, K.H. (2001). Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2-type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13, 1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pedley, K.P., and Martin, G.B. (2003). Molecular basis of bacterial speck disease resistance in tomato mediated by the Pto pathway. Annu. Rev. Phytopathol. 41, 215–243. [DOI] [PubMed] [Google Scholar]
- Pih, K.T., Kabilan, V., Lim, J.H., Kang, S.G., Piao, H.L., Jin, J.B., and Hwang, I. (1999). Characterization of two new channel protein genes in Arabidopsis. Mol. Cells 9, 84–90. [PubMed] [Google Scholar]
- Pontier, D., Balague, C., Bezombes-Marion, I., Tronchet, M., Deslandes, L., and Roby, D. (2001). Identification of a novel pathogen-responsive element in the promoter of the tobacco gene HSR203J, a molecular marker of the hypersensitive response. Plant J. 26, 495–507. [DOI] [PubMed] [Google Scholar]
- Riechmann, J.L., and Ratcliffe, O.J. (2000). A genomic perspective on plant transcription factors. Curr. Opin. Plant Biol. 3, 423–434. [DOI] [PubMed] [Google Scholar]
- Roman, G., Lubarsky, B., Kieber, J.J., Rothenberg, M., and Ecker, J.R. (1995). Genetic analysis of ethylene signal transduction in Arabidopsis thaliana: Five novel mutant loci integrated into a stress response pathway. Genetics 139, 1393–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J., Reinstadler, A., Lipka, V., Lippok, B., and Somssich, I.E. (2002). Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell 14, 749–762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rushton, P.J., and Somssich, I.E. (1998). Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1, 311–315. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Shin, R., Park, J.M., An, J.M., and Paek, K.H. (2002). Ectopic expression of Tsi1 in transgenic hot pepper plants enhances host resistance to viral, bacterial, and oomycete pathogens. Mol. Plant-Microbe Interact. 15, 983–989. [DOI] [PubMed] [Google Scholar]
- Singh, K., Foley, R.C., and Onate-Sanchez, L. (2002). Transcription factors in plant defense and stress responses. Curr. Opin. Plant Biol. 5, 430–436. [DOI] [PubMed] [Google Scholar]
- Solano, R., Stepanova, A., Chao, Q., and Ecker, J.R. (1998). Nuclear events in ethylene signaling: A transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stockinger, E.J., Gilmour, S.J., and Thomashow, M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tena, G., Asai, T., Chiu, W.L., and Sheen, J. (2001). Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4, 392–400. [DOI] [PubMed] [Google Scholar]
- Thara, V.K., Tang, X., Gu, Y.Q., Martin, G.B., and Zhou, J.M. (1999). Pseudomonas syringae pv tomato induces the expression of tomato EREBP-like genes Pti4 and Pti5 independent of ethylene, salicylate and jasmonate. Plant J. 20, 475–483. [DOI] [PubMed] [Google Scholar]
- Thomma, B., Eggermont, K., Penninckx, I., Mauch-Mani, B., Vogelsang, R., Cammue, B.P.A., and Broekaert, W.F. (1998). Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proc. Natl. Acad. Sci. USA 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomma, B.P., Eggermont, K., Tierens, K.F., and Broekaert, W.F. (1999). Requirement of functional ethylene-insensitive 2 gene for efficient resistance of Arabidopsis to infection by Botrytis cinerea. Plant Physiol. 121, 1093–1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velculescu, V.E., Zhang, L., Vogelstein, B., and Kinzler, K.W. (1995). Serial analysis of gene expression. Science 270, 484–487. [DOI] [PubMed] [Google Scholar]
- Velculescu, V.E., Zhang, L., Zhou, W., Vogelstein, J., Basrai, M.A., Bassett, D.E., Jr., Hieter, P., Vogelstein, B., and Kinzler, K.W. (1997). Characterization of the yeast transcriptome. Cell 88, 243–251. [DOI] [PubMed] [Google Scholar]
- Wan, J., Dunning, F.M., and Bent, A.F. (2002). Probing plant-pathogen interactions and downstream defense signaling using DNA microarrays. Funct. Integr. Genomics 2, 259–273. [DOI] [PubMed] [Google Scholar]
- Wu, K., Tian, L., Hollingworth, J., Brown, D.C., and Miki, B. (2002). Functional analysis of tomato Pti4 in Arabidopsis. Plant Physiol. 128, 30–37. [PMC free article] [PubMed] [Google Scholar]
- Wyatt, S.E., Tsou, P.L., and Robertson, D. (2002). Expression of the high capacity calcium-binding domain of calreticulin increases bioavailable calcium stores in plants. Transgenic Res. 11, 1–10. [DOI] [PubMed] [Google Scholar]
- Yang, Y., Shah, J., and Klessig, D.F. (1997). Signal perception and transduction in plant defense responses. Genes Dev. 11, 1621–1639. [DOI] [PubMed] [Google Scholar]
- Zhang, L., Zhou, W., Velculescu, V.E., Kern, S.E., Hruban, R.H., Hamilton, S.R., Vogelstein, B., and Kinzler, K.W. (1997). Gene expression profiles in normal and cancer cells. Science 276, 1268–1272. [DOI] [PubMed] [Google Scholar]
- Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. USA 96, 6523–6528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng, X.M., Black, D., Chambon, P., and Egly, J.M. (1990). Sequencing and expression of complementary DNA for the general transcription factor BTF3. Nature 344, 556–559. [DOI] [PubMed] [Google Scholar]
- Zhou, J., Tang, X., and Martin, G.B. (1997). The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 16, 3207–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.