1. Introduction
The evidence for impairment in the ubiquitin proteasome system (UPS) in Parkinson Disease (PD) is mounting and becoming increasingly more convincing. However, it is presently unclear whether UPS dysfunction is a cause or result of PD pathology, a crucial distinction which impedes both the understanding of disease pathogenesis and the development of effectual therapeutic approaches. Thus recent findings specifically regarding the role of the UPS in PD are discussed within this review, and offer new insight and provide direction for future research to conclusively resolve this debate.
2. Parkinson disease (PD)
PD is a progressive neurodegenerative disease clinically characterized by bradykinesia, gait disturbances, resting tremor, muscular rigidity, and postural instability [1]. Pathological hallmarks of the disease include loss of dopaminergic neurons in the substantia nigra (SN), as well as the presence of eosinophilic cytoplasmic inclusions and dystrophic neurites in remaining neurons, first described by Friederich Heinrich Lewy in 1912 and termed Lewy bodies (LB) and Lewy neurites (LN) in his honor [2]. The identification of α-synuclein as the major, filamentous protein component of LBs [3], in addition to the linkage of missense mutations (A53T, A30P, E46K) and genomic duplication and triplication of the α-synuclein gene with autosomal dominant PD [4-8], is indicative of a key role for α-synuclein in disease pathogenesis. However, the detection of LBs in clinically normal individuals upon postmortem analysis, frequently called incidental Lewy body disease (iLBD), brings into question the pathological significance of α-synuclein aggregation. Utilizing a unique brain donation program to control for the inherent biases associated with more conventional case control studies, a recent population-based study estimates the prevalence of synuclein pathology in people over 70 years of age is approximately 37%, with synuclein burden a poor predictor of clinical status/diagnosis [9]. Despite these findings, Dickson and colleagues demonstrate that iLBD cases exhibit a decrement in tyrosine hydroxylase, a marker of dopaminergic and noradrenergic neurons and a characteristic feature of PD, in both striatal and epicardial nerve fibers that is intermediate to control and PD patients [10]. The authors conclude that the absence of parkinsonian symptoms is the result of a subthreshold-level of pathology, thus further solidifying the pathogenic role of α-synuclein in PD progression.
3. Ubiquitin proteasome system (UPS)
The UPS regulates the degradation of key regulatory proteins that control signal transduction, cell cycle progression, apoptosis, as well as cellular differentiation [11]. In addition to involvement in these processes, the UPS also degrades misfolded and damaged proteins, thus collectively implicating the UPS in a wide range of conditions, including neurodegenerative diseases, cancer, inflammation, and autoimmunity [12, 13]. Given the detrimental consequences of unregulated protein degradation, the UPS utilizes a class of enzymes to covalently link ubiquitin polypeptide chains to proteins, marking those proteins as substrates for the proteasome and allowing for targeted and selective degradation (reviewed in [14, 15]). Initially, the carboxyl end of ubiquitin is activated in an ATP-dependent process by the ubiquitin-activating enzyme (E1), which results in a highly reactive ubiquitin thiolester that is transferred to a ubiquitin-carrier protein (E2). The E3 class of enzymes, which are also called ubiquitin protein ligases, recognize and bind proteins to be marked for degradation, subsequently catalyzing the transfer of ubiquitin chains from the E2 to lysine residues on protein substrates, which can serve as a signal for proteasome-mediated degradation.
The proteasome is a large, multisubunit complex containing a common proteolytic core, the 20S proteasome, which is composed of 28 subunits arranged in four, heptameric rings (reviewed in [15, 16]. The two outer rings are each composed of seven alpha-type subunits (α1-α7), while the two inner rings each contain seven beta-type subunits (β1-β7). The proteolytic activity is enclosed within the inner rings, with only β1, β2, and β5 subunits possessing caspase-like, trypsin-like, and chymotrypsin-like cleavage specificity, respectively [17, 18]. These active sites have been shown to allosterically regulate one another through substrate binding or cleavage, leading to a proposed model in which single polypeptide chains are successively hydrolyzed by a structured and coordinated activation of these catalytic subunits [19, 20]. However, Liu and colleagues present evidence that a disordered polypeptide loop, such as a β-hairpin structure, can also be permitted entry into the inner canal of the 20S proteasome, allowing for the endoproteolytic cleavage and partial degradation of unstructured proteins that is not dependent upon ubiquitination [21]. This describes a novel function of the proteasome, liberating active peptides from precursor proteins, as well as correcting folding defects in internal domains of large proteins.
The activity of the 20S proteasome is modulated by a variety of regulators, including the 19S/PA700 complex, PA200, as well as PA28 α/β and PA28γ [22-24]. The most common regulator, the 19S/PA700 complex, contains six AAA-family ATPases and is capable of binding both ends of the 20S proteasome in an ATP-dependent manner, forming the 26S proteasome, which is involved in the degradation of ubiquitinated proteins [20, 25, 26]. Given that only the 19S/PA700 complex possesses ATPase activity and binds to polyubiquitin chains, alternative regulators of the 20S proteasome are believed to modulate ubiquitin-independent functions of the proteasome. However, hybrid proteasomes have also been described, in which the 19S/PA700 and PA28α/β complexes bind opposite ends of the 20S proteasome [27]. The specific function of these hybrid proteolytic complexes is unclear, and studies evaluating the cellular localization of the 20S proteasome, which has been detected in both nuclear and cytosolic compartments, have failed to distinguish between free and bound 20S proteasomes [28, 29]. A recent study has further investigated the modulation of 20S proteasome activity and/or localization, demonstrating that 20S proteasomes associated with PA28γ complexes are localized to nuclear speckles and implicated in the intranuclear trafficking of SR proteins [30]. Additional research of this nature will be needed to more fully characterize the precise cellular functions of these alternate proteasome-regulator complexes, as well as to decipher the specific physiological signals that regulate proteasome-regulator composition.
4. UPS and PD
The evaluation of human postmortem brain tissue has provided a considerable amount of evidence implicating proteasomal dysfunction in PD pathogenesis. Using enzymatic assays to measure proteasome activity, a significant decrement in chymotrypsin-like, trypsin-like, and caspase-like activity was detected in the SN of PD patients when compared to age-matched controls [31-34]. However, no deficits in proteasomal activity were detected in extranigral regions, and Furukawa and colleagues actually observed an increase in proteasomal activity in unaffected regions, specifically the cerebral cortex and striatum, of PD patients compared to age-matched controls [31]. In line with these findings, immunoblotting and histological techniques revealed a decrease in subunits of the 20S proteasome and the PA700/19S complex in the SN of PD patients, while protein levels where unchanged or increased in extranigral brain regions [31, 32, 35]. In addition, the accumulation of ubiquitinated proteins, heat shock proteins/chaperones, and components of the UPS within LBs provides further support for a central role of UPS dysfunction in the etiopathogenesis of PD [36-41]. However, these findings must be interpreted with caution, as the above-mentioned studies do not take into account neuronal loss, nor do they identify the affected cell type (i.e. neuronal vs glial).
The link between proteasomal inhibition and the pathogenesis of PD was further solidified by the demonstration that treatment with the proteasomal inhibitor lactacystin dose-dependently leads to the degeneration and the formation of synuclein and ubiquitin-positive inclusions in rat ventral mesencephalic primary neurons [42, 43]. In vivo, McNaught and colleagues reveal that systemic administration of proteasomal inhibitors in Sprague-Dawley rats produced both a behavioral and pathological phenotype reminiscent of PD [44]. In addition to the progressive nature of the motor impairment exhibited by treated rats, administration of dopamine agonists alleviated behavioral symptoms. Postmortem analysis revealed loss of dopamine in the striatum, as well as neuronal loss and the presence of eosinophilic, synuclein/ubiquitin-positive inclusions in remaining neurons of the SN [44, 45]. However, this model has since been viewed with great scrutiny due to the inability of different laboratories to replicate these findings [46-49]. Although two additional laboratories were able to replicate dopaminergic cell loss following systemic administration of proteasome inhibitors, only Zeng and associates detected the presence of synuclein aggregates in the SN, while neither group observed a progressive motor impairment [50, 51]. It is hypothesized that extraneous variables due to differences in formulation of the proteasomal inhibitors, strain background differences in treated rats and mice, as well as environmental factors, could account for this variability in findings. However, the extensive variability in consequences of in vivo proteasomal inhibition casts significant doubt on the utility of this approach as an accurate model of PD.
Despite the failure of in vivo administration of proteasome inhibitors to consistently produce a parkinsonian phenotype, an exciting new report from Bedford and associates provides striking evidence establishing a link between 26S proteasome dysfunction and the development of α-synuclein neuropathology [52]. In this study, Bedford and colleagues develop and characterize a novel mouse model expressing a conditional deletion of the Rpt2/PSMC1 subunit, an ATPase of the 19S regulatory complex, spatially restricted to neurons of the forebrain, or a second model in which the Rpt2/PSMC1 subunit is ablated in TH-positive neurons. As the Rpt2/PSMC1 subunit is required for both the assembly and activity of the 26S proteasome, conditional knockdown of Rpt2/PSMC1 expression produced a specific impairment of 26S proteasome activity, while 20S proteasome activity was unaffected. Intriguingly, synuclein and ubiquitin-positive inclusions resembling LBs were observed in either neurons of the forebrain region or the nigrostriatal pathway, with the localization of pathology coincident with Rpt2/PSMC1 knockdown, and thus 26S dysfunction [52]. Although no motor impairment or parkinsonian phenotype is reported in this study, genetic ablation of Rpt2/PSMC1 in the forebrain did produce a learning deficit, as well as progressive neurodegeneration of forebrain regions. As restriction of Rpt2/PSMC1 knockdown to TH-positive neurons is particularly relevant to PD pathology, it is disappointing that autonomic dysfunction leading to premature death by 1 month of age prevents a full behavioral assessment of these mice [52].
The relationship between UPS impairment and sporadic PD has also been strengthened by a number of in vitro studies demonstrating a decrease in proteasome activity following exposure to pesticides and environmental toxins linked to PD, including rotenone, paraquat, and maneb [53-55]. Consistent with in vitro findings, the in vivo administration of rotenone led to a reduction in proteasome activity specifically in the ventral midbrain of rats [53]. Intriguingly, utilization of osmotic minipumps to continually deliver the PD-linked toxin MPTP to mice for one month produced a PD-like phenotype, including depletion of striatal dopamine levels and neuronal loss in both the SN and locus coeruleus, which was accompanied by the formation of α-synuclein and ubiquitin-positive inclusions [56]. These mice also exhibited a decrease in proteolytic activity of the proteasome in striatal extracts as assessed by enzymatic assays, as well as a progressive decline in motor activity that was rescued by administration of dopamine agonists. Surprisingly, when experiments were replicated in mice lacking α-synuclein, neuronal loss, behavioral impairments, and the formation of ubiquitin-positive inclusions were alleviated [56]. Perhaps most telling was the demonstration that impairments in proteolytic activity following MPTP administration were also alleviated in the absence of α-synuclein, suggesting that α-synuclein exacerbates the deleterious effects of PD-linked environmental toxins on UPS function. Furthermore, these findings imply that α-synuclein, and possibly UPS dysfunction, is critically involved in the manifestation of a PD phenotype.
The demonstration that MPTP treatment alters proteasomal activity has also been replicated in non-human primates [57]. Specifically, both proteolytic activity and expression of proteasomal subunits is decreased in extracts from the SN of MPTP-treated marmoset monkeys similarly to alterations observed in PD patients, though synuclein pathology, neuronal loss, and behavioral impairments were not assessed in this cohort of monkeys. However, an earlier study performed by Kowall and colleagues revealed an initiation of α-synuclein aggregation upon MPTP treatment in baboons, with regrettably no evaluation of either proteasomal activity or expression performed in this study [58]. Thus the precise involvement of α-synuclein pathology and UPS impairment in MPTP-linked PD and parkinsonism remains to be more conclusively established.
5. Genetic links to PD and association with UPS
Although the majority of PD cases are sporadic, a number of genetic loci have been identified and linked to the inheritance of familial PD. The relationship between these genes is still presently unclear, as is the connection between familial-linked genes and the etiology of idiopathic PD. However, the clinical and pathophysiological similarities between familial and idiopathic forms of PD suggest they may share a common pathogenic mechanism [59]. Given the considerable evidence implicating a central role for UPS impairment in the development and progression of sporadic PD, it is intriguing that a number of genetic mutations linked to PD are also involved in the regulation of UPS function. The direct and indirect relationship(s) between these PD-linked genes and modulation of the UPS will be discussed below.
5.1. α-Synuclein
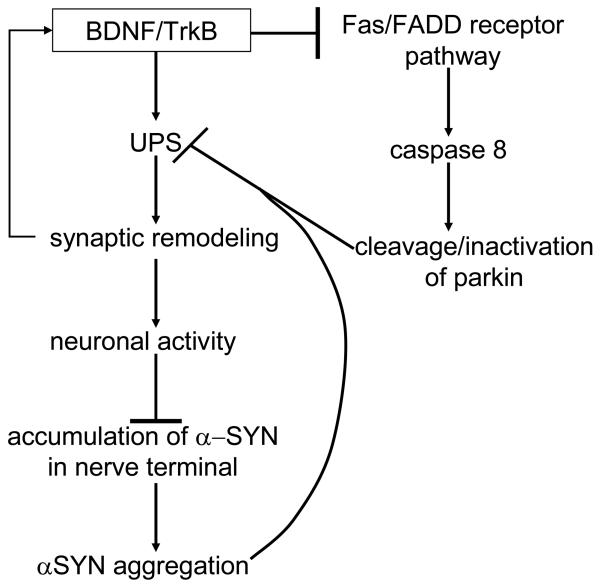
α-synuclein is a natively unfolded presynaptic protein initially cloned from the electric lobe of Torpedo californica [60]. Although the function of α-synuclein is still unknown, it adopts an α-helical structure upon binding to phospholipids [61], and has been shown to modulate synaptic transmission through the regulation of synaptic vesicle recycling and the compartmentalization of neurotransmitters [62-66]. In addition, Fortin and colleagues have demonstrated that lipid rafts are required for the presynaptic localization of α-synuclein, and further that both synaptic localization and membrane association of α-synuclein are modulated by neuronal activity [67, 68]. These findings, in concert with evidence that BDNF-TrkB signaling acts upstream of the UPS to regulate the expression level of key synaptic proteins in response to neuronal activity, could have significant implications for PD pathogenesis [69]. In particular, given the suspected link between PD and UPS dysfunction, a local impairment of the UPS within the synapse theoretically could promote the accumulation of ubiquitinated proteins irrespective of BDNF-TrkB signaling, thereby preventing BDNF-TrkB-mediated synaptic remodeling and leading to a decrease in neuronal activity. A reduction in neuronal activity would not only decrease BDNF expression and synaptic release [70-73], but based on the findings of Fortin and associates, would also be expected to increase the amount of membrane-bound α-synuclein localized to the synapse [67, 68]. Taking into consideration the higher propensity of membrane-bound synuclein to aggregate and seed the aggregation of the more abundant, cytosolic form of α-synuclein [74], a decrease in neuronal activity and subsequent increase in membrane-bound synuclein, further exacerbated by a decrement in BDNF expression, could effectively establish a pathogenic, positive-feedback mechanism linking neuronal activity and UPS function with synuclein aggregation (Figure 1). In addition, the demonstration by Dluzen and colleagues that targeted deletion of a BDNF allele potentiates the age-dependent decline in nigrostriatal dopaminergic function in mice provides a potential explanation for susceptibility of the nigrostriatal dopamine system to synuclein pathology with aging [75].
Figure 1.
A putative mechanism by which the UPS acts downstream of BDNF/TrkB signaling to ultimately regulate α-synuclein aggregation. According to this hypothetical model, synuclein-mediated inhibition of the UPS interferes with stimulatory effects of BDNF on synaptic activity, as well as the inhibitory influence of BDNF signaling on parkin cleavage and inactivation by the Fas/FADD death receptor pathway.
5.1.1. Modulation of aggregation potential of α-synuclein
The precipitating basis of α-synuclein aggregation in synucleinopathies is controversial, though broadly speculated to arise from an increase in α-synuclein protein expression (either through gene triplication or altered transcriptional or translational activities), excessive posttranslational modifications (including phosphorylation, ubiquitination, oxidation, nitration, truncation), or through increased interaction with other proteins, all of which could modulate the propensity of α-synuclein to fibrillize [76-88]. In addition, the negatively-charged C-terminus of α-synuclein, which has also been shown to bind dopamine derivatives [89], appears to act as a negative regulator of aggregation [82, 84, 90]. Thus it is highly likely that posttranslational modifications to this region, including phosphorylation, ubiquitination, oxidation, nitration, and truncation [77, 78, 91], influence the propensity of α-synuclein to aggregate.
Critical evaluation of the various α-synuclein species observed in LBs demonstrates that α-synuclein is selectively and extensively phosphorylated at Ser129 (pSer129) in these lesions, and further that this is the predominant modification of α-synuclein in LBs [77, 92, 93]. In addition to phosphorylation at Ser129, α-synuclein in LBs is also N-terminally acetylated and ubiquitinated, as well as C-terminally truncated [92]. Given that both normal and diseased brains contain trace amounts of soluble α-synuclein pSer129, as well as species that are truncated at Asp119, it is believed these forms of α-synuclein are generated through normal metabolism [92]. However, in postmortem brain tissue from synucleinopathy patients, the majority of pSer129 is detected in insoluble fractions. Because the main ubiquitinated α-synuclein species found in LBs is also pSer129 α-synuclein, it is hypothesized that an excess of pSer129 may actually serve as the priming event which ultimately culminates in the formation of LBs. Anderson and colleagues further posit that pSer129 may serve as a signal for proteolysis, supported by their observation that all α-synuclein species truncated at Tyr133 were also pSer129 [92].
Given the potential ramifications of modulating phosphorylation at Ser129, a number of laboratories have investigated prospective kinases that phosphorylate this site, leading to the identification of casein kinase 1 and 2, as well as G-protein coupled receptor kinases [94-97]. In support of a pathogenic role of pSer129, overexpression of α-synuclein and GRK5 (G-protein coupled receptor kinase 5), which colocalize in LBs, promotes GRK5-mediated phosphorylation at Ser129 and leads to the formation of soluble oligomers and aggregates of α-synuclein [94]. In addition, pSer129 has also been shown to increase the propensity of α-synuclein to aggregate following exposure to mitochondrial or oxidative stressors [98, 99]. In contrast, Paleologou and colleagues demonstrate that in vitro phosphorylation of α-synuclein at Ser129 inhibits fibrillogenesis, but does not perturb the overall conformation of synuclein and its ability to adopt α-helical conformations upon membrane-binding to synthetic vesicles [96]. Perhaps most importantly, Paleologou and associates reveal a discrepancy in the structural and aggregation properties of α-synuclein phosphorylated in vitro and the phosphorylation mimics S129E, S129D [96], which could explain the inconsistent findings evaluating the consequences of α-synuclein phosphorylation. Specifically, Gorbatyuk and coworkers report that the phosphorylation mimic S129D is protective against dopaminergic cell loss when injected into the SN of rats, while Chen and Feany demonstrate an enhanced toxicity associated with α-synuclein phosphorylation in Drosophila [76, 100]. However, Chen and Feany substantiate their findings by demonstrating that both the phosphomimetic and kinase-phosphorylated wild-type synuclein produce a similar phenotype [76]. In addition, Chen and Feany observe an inverse correlation between α-synuclein phosphorylation and aggregation potential, which is consistent with the report by Paleologou and colleagues [76, 96].
In addition to phosphorylation, α-synuclein present in LBs is also ubiquitinated [77, 92]. Recently, the RING-type E3 ubiquitin ligase SIAH (seven in absentia homolog) has been shown to interact with and monoubiquitinate α-synuclein in vitro and in vivo, thereby increasing the propensity of α-synuclein to aggregate [101, 102]. Although there was no difference in the ability of SIAH to monoubiquitinate wild-type or mutant α-synuclein, significantly more inclusions were observed in cells overexpressing the A53T mutant [102]. This suggests that despite an increased tendency of α-synuclein to aggregate upon SIAH-mediated monoubiquitination, additional factors further modulate this tendency. In addition, ubiquitination of α-synuclein by SIAH increases cell susceptibility to proteasome impairment and promotes apoptotic cell death, suggesting that SIAH activity plays a crucial role in determining the toxicity of α-synuclein under conditions of proteasome dysfunction [101, 102].
An additional substrate of SIAH, synphilin-1, is a synuclein-interacting protein that colocalizes with α-synuclein in LBs [103-105]. Intriguingly, overexpression of synphilin-1 inhibits proteasomal function, and also leads to the formation of ubiquitinated cytoplasmic inclusions positive for both synphilin-1 and α-synuclein [103, 105]. SIAH-mediated ubiquitination has been shown to target synphilin-1 for degradation by the UPS [104], though phosphorylation of synphilin-1 on serine 556 by GSK3β prevents SIAH-mediated ubiquitination and the subsequent degradation of synphilin-1 [106]. Prevention of this phosphorylation by either GSK3β inhibition or mutation of the phospho-residue (S556A) promotes the formation of synphilin-positive ubiquitinated inclusions that colocalize with increased expression of the UPS reporter, GFPμ, which may indicate that phosphorylation determines inhibitory potential of synphilin-1 on proteasome activity [106]. However, the effect of synphilin-1 phosphorylation or ubiquitination on ability to bind and interact with α-synuclein could be a confounding variable in these studies, in particular with the discovery that α-synuclein is also a substrate for SIAH [101, 102].
Although Anderson and colleagues state that truncated α-synuclein does not appear to be highly enriched in LBs in comparison to pSer129 α-synuclein [92], earlier studies report that approximately 15% of α-synuclein in LBs is truncated [34, 107, 108]. Based on these earlier findings, as well as the demonstration that truncated human α-synuclein (amino acid residues 1-120) fibrillizes faster than either wild-type or mutant protein [84, 90, 109], a mouse model was generated expressing truncated human α-synuclein (1-120) on a synuclein null background [88]. Surprisingly, synuclein-positive inclusions were detected in dopaminergic neurons in the substantia nigra and olfactory bulb, and decrements in striatal dopamine levels correlated with motor impairment [88]. The susceptibility of dopaminergic neurons to synuclein toxicity could be explained by the observation that dopamine has been shown to inhibit α-synuclein fibrillization in vitro, leading to the proposal that either dopamine or its metabolites kinetically stabilize oligomeric α-synuclein intermediates [89, 110-113]. This hypothesis is supported by a significantly higher α-synuclein oligomer to monomer ratio in the SN compared to cortical tissue of symptomatic A53T mice [114]. In addition, Mazzulli and associates demonstrate that increasing catechol levels in SH-SY5Y cells that overexpress mutant A53T α-synuclein by cotransfecting with tyrosine hydroxylase prevents the formation of insoluble α-synuclein aggregates and increases the concentration of soluble oligomers [114]. As the amino acid residues 125-129 in the C-terminus of α-synuclein have been shown to be required for catechol-mediated inhibition of synuclein aggregation [89, 115], it is interesting that a mouse model overexpressing truncated wild-type α-synuclein (1-120), which lacks the catechol-interaction site, develops insoluble synuclein aggregates in dopaminergic neurons [88].
Alternatively, a decrease in degradation of the α-synuclein protein could serve as the basis for pathogenic overexpression. To characterize the degradative pathway for α-synuclein, Bennett and colleagues demonstrate that both wild-type and mutant A53T α-synuclein are substrates of the proteasome in SH-SY5Y neuroblastoma cells [116]. However, Ancolio and associates were unable to observe proteasome-mediated degradation of either wild-type or mutant synuclein in HEK293 cells, though an effect of calpain inhibition on synuclein expression was also not detected [117], despite the fact that a number of investigators have observed calpain-mediated cleavage of α-synuclein [83, 118, 119]. Given the natively unfolded structure of α-synuclein, it has now been shown that wild-type α-synuclein can be degraded by a ubiquitin-independent proteasome pathway [21, 120, 121]. On the other hand, A53T α-synuclein exhibits a 50% longer half-life compared to wild-type, suggesting mutant A53T α-synuclein is not degraded as efficiently by the proteasome [116]. In addition, the metal-catalyzed oxidation of α-synuclein enhanced the formation of oligomeric and protofibrillar forms, simultaneously preventing mature fibril formation [120]. Although monomeric α-synuclein was degraded in a proteasome-dependent manner, the degradation of oxidized oligomeric α-synuclein was completely prevented [120].
Given that α-synuclein adopts an α-helical conformation upon binding to membranes, and further that natively unfolded α-synuclein has been reported to be a substrate for the proteasome [21, 120, 121], Liu and colleagues demonstrate that only unbound, cytosolic α-synuclein is a substrate for the proteasome, while membrane-bound, α-helical α-synuclein is not degraded by the proteasome [122]. This suggests that loss of vesicular or membrane-binding ability due to mutation [123] or oxidative damage [81, 124] would enhance 20S-mediated cleavage of α-synuclein, increasing the generation of truncated fragments. In vitro, proteasome-mediated cleavage of α-synuclein yielded three predominant fragments, including 1-119, 1-110, and 1-83, which all adopted a random coil conformation indistinguishable from the full-length protein [122]. Cotransfection of full-length and truncated variants of α-synuclein in SH-SY5Y cells increased cell vulnerability to oxidative stress, and induced aggregation of full-length protein, with A53T mutants aggregating more rapidly than parallel combinations of wild-type protein [122]. Thus Liu and associates speculate that proteolytic activity of the proteasome produces highly amyloidogenic α-synuclein fragments via partial degradation of cytosolic protein, which induces the aggregation of full-length α-synuclein [122]. These hybrid aggregates further impair proteasome activity, exacerbating the accumulation of truncated and full-length α-synuclein deposits, creating a vicious cycle of cytotoxicity.
5.1.2. Evaluation of α-synuclein-mediated UPS impairment
A direct inhibitory effect of synuclein on UPS activity has been reported, though the effects of overexpression, mutation, aggregation, and posttranslational modifications of the synuclein protein on proteasome function are still under debate due to conflicting reports, most likely resultant from methodological differences in sample preparation and analysis. Tanaka and colleagues demonstrated an impairment in proteasome activity in dopaminergic PC12 cells following the overexpression of wild-type α-synuclein, with an even greater inhibitory effect observed following overexpression of mutant A30P α-synuclein [125]. In contrast, Martin-Clemente and associates failed to detect an alteration in proteasome activity in stably transfected PC12 cells with EYFP-tagged α-synuclein constructs, including wild-type, as well as the PD-linked mutants A30P and A53T [126]. However, the authors fail to demonstrate a significant increase in exogenous synuclein expression over basal levels, and instead report significant overexpression of synuclein based on EYFP immunoreactivity, which is not a valid comparison [126].
An inhibitory effect on proteolytic activity mediated by overexpressed wild-type α-synuclein has since been replicated in dopaminergic N27 cells and MG63 (osteosarcoma) cells [127, 128], but not in CHO (ovarian) cells [129], which may indicate that effects of α-synuclein are cell-type specific. In support of this, Petrucelli and associates observed proteasomal impairment following the overexpression of mutant A53T and A30P α-synuclein, but not wild-type, in M17 neuroblastoma cells [130]. In addition, Chen and colleagues observed an inhibition of proteasome-mediated degradation of short-lived proteins in yeast cells overexpressing wild-type α-synuclein, with an even greater impairment exhibited by cells expressing mutant A30P α-synuclein [131]. Although there was no change in proteolytic activity of immunoprecipitated 20S proteasomes from cells overexpressing either wild-type or A30P α-synuclein, proteasome subunit composition and interaction with regulatory proteins was significantly altered by both wild-type and A30P α-synuclein overexpression [131]. These results may indicate that α-synuclein does not directly inhibit the active/catalytic site of the proteasome, but instead exerts an inhibitory effect through modulation of proteasome activity. A recent report from the Sudhof laboratory supports this hypothesis, demonstrating a dramatic alteration in the expression of proteasomal subunits in spinal cord from symptomatic A30P α-synuclein mice [132]. However, there was significant neuronal loss and gliosis observed in the spinal cord from symptomatic A30P mice, and as biochemical assessment of proteasome subunit expression does not differentiate between cell types, it is most likely that neuronal loss coupled with the significant increase in activated glia accounts for the changes in proteasomal subunit expression. In addition, a recent report by Emmanouilidou and coworkers detected a significant impairment in proteasome activity in the cortex of A53T α-synuclein mice, though expression of proteasomal subunits remained unchanged [133].
The effect of α-synuclein aggregation on proteasome activity has also been evaluated, with groups consistently reporting a greater proteasomal impairment in the presence of aggregated compared to monomeric α-synuclein [134-136]. Although both monomeric and aggregated α-synuclein have been shown to bind the S6'/TBP1 (Tat binding protein 1) subunit of the 19S/PA700 proteasome complex [135, 137], only aggregated α-synuclein inhibits ubiquitin-dependent and independent 26S proteasomal activity [135]. Zhang and colleagues have also demonstrated that α-synuclein protofibrils inhibit the ubiquitin-independent degradation of unstructured proteins by the 26S proteasome, though monomers and dimers have no effect on the proteolysis of these substrates [136]. In contrast, ubiquitin-dependent 26S proteasome activity is slightly inhibited by monomeric and dimeric α-synuclein, while protofibrillar α-synuclein potently inhibits the degradation of polyubiquitinated proteins. Given that α-synuclein protofibrils bind the 19S/PA700 regulatory complex of the 26S proteasome, as well as p21 (an unstructured proteasomal substrate) and K48-linked polyubiquitin chains, it is proposed that α-synuclein protofibrils inhibit 26S proteasome activity by interfering with substrate translocation into the proteasome core, achieved through direct interactions with the proteasome, as well as through the sequestration of proteasomal substrates [133, 136].
Given that, α-synuclein, Aβ, polyglutamine proteins, prion protein, and other amyloidogenic proteins adopt a similar structure upon oligomerization, it is hypothesized that these proteins also share similar pathogenic effects [138]. In agreement with this, impaired proteasomal function is observed in parallel with the first appearance of soluble Aβ oligomers in the triple transgenic mouse model of AD (3× Tg-AD), while proteasome activity is restored at a time point when soluble Aβ oligomers are converted into insoluble aggregates [139]. This suggests that soluble oligomeric Aβ species, and not the monomeric or fibrillar form of Aβ, inhibits proteasomal activity [139]. UPS impairment has also been observed in cell culture and animal models overexpressing mutant huntingtin protein [129, 140-144], and consistent with data proposing a protective effect of aggregation due to sequestration of toxic species, treatment with a compound that increases inclusion formation prevents huntingtin-mediated proteasome inhibition [129]. In addition, the abnormal prion conformer (PrPsc) inhibits the 26S proteasome in vitro, while either preincubation with an oligomer antibody or heat denaturation of PrPsc alleviated this inhibitory effect, indicating a specific conformation of an oligomeric PrPsc intermediate mediates the proteasomal inhibitory effect [145]. Proteasome activity was also significantly decreased in cells exposed to prion-infected mouse brain homogenates, as well as in brain regions exhibiting significant prion neuropathology in mice infected with PrPsc, establishing a solid link between UPS impairment and neurodegeneration associated with prion infection [145].
Based upon the above findings, as well as the lack of direct in vivo evidence of a link between α-synuclein pathology and UPS dysfunction, our laboratory has generated a transgenic mouse model expressing the proteasomal reporter GFPμ [140]. In vitro, expression of GFPμ is dose-dependently increased in the presence of the proteasome inhibitor MG132, illustrating the sensitivity of GFPμ to perturbations in proteasomal function [130]. In addition, cotransfection of GFPμ and mutant A53T or A30P α-synuclein leads to an upregulation of GFPμ, demonstrating the inhibitory effects of mutant α-synuclein on UPS activity [130]. Thus we are crossing GFPμ transgenic mice with mutant A53T α-synuclein mice [146], which will allow us to monitor effects of α-synuclein pathology on UPS function in vivo by evaluating bigenic mice at various time points. This model will also allow us to determine selective vulnerability of specific cell populations to synuclein-mediated perturbations in proteasome activity. Ultimately, following the initial characterization of GFPμ × A53T mice, it is anticipated this model can be utilized to develop novel therapeutic approaches to preclude inhibitory effects of α-synuclein on UPS function.
5.2. Parkin
Mutations in the E3 ubiquitin ligase parkin cause early onset PD with an autosomal recessive inheritance pattern [147, 148]. Although various mutations in the parkin gene have been linked to PD, including missense, nonsense, frameshift point mutations, exon deletions and duplications, to date there is no noticeable variation in clinical manifestation between the different mutations [148, 149]. Neurodegenerative changes are also relatively similar in sporadic PD and early onset PD caused by parkin mutations, with both types exhibiting neuronal loss and gliosis that is primarily restricted to the brainstem. However, LBs are not typically observed in patients with parkin-linked PD, though the significance of this observation is still under speculation [150-153].
Despite the lack of LB formation, PD patients with parkin mutations do show an accumulation of parkin substrates [154-156], which would suggest that α-synuclein is not a substrate for parkin. However, α-synuclein and parkin do colocalize in LBs [41]. In addition, under basal conditions, parkin and α-synuclein have been shown to associate and colocalize to the cytosol and neuritic processes [157, 158]. Inhibition of proteasome activity in cells coexpressing α-synuclein and parkin led to a decrease in parkin solubility accompanied by the formation of inclusions positive for both α-synuclein and parkin, while knockdown of α-synuclein increased parkin solubility under conditions of proteasomal impairment [158]. Given that proteasome inhibition also increases parkin expression [158], and that autoubiquitination of parkin promotes its degradation by the proteasome [159-162], it is possible that the level of expression and/or ubiquitination of parkin may regulate its association with α-synuclein. These alterations in parkin expression and/or ubiquitination may be further exacerbated by the accumulation of α-synuclein, which could contribute to the pathogenesis of PD by promoting a decrease in parkin solubility and in turn, compromising neural function. Mutations in α-synuclein have also been shown to more efficiently stimulate parkin aggregation in c.elegans [163] and neuroblastoma cells [158]. Thus it is hypothesized that mutations associated with familial parkinsonism in combination with exposure to cellular stressors, including proteolytic or oxidative stress, might facilitate the pathological interactions between α-synuclein and parkin.
The effects of PD-linked mutations in parkin also appear to universally result from alterations in parkin solubility and intracellular localization, possibly due to misfolding of the mutant protein [54, 164, 165]. Wang and colleagues demonstrate that exposure to PD-linked environmental toxins, as well as oxidative or proteolytic stressors, promotes the depletion of soluble, functional parkin, which correlates with reduced proteasomal activity and increased cell death [54]. In addition, Chung and associates observed S-nitrosylated parkin in both mice exposed to MPTP and human postmortem brain tissue from PD and diffuse LBD patients, which was negatively correlated with parkin function [166]. Given that parkin has also been shown to become increasingly more insoluble with age [167], and that risk for PD increases with age, it would appear that loss of functional parkin is a major, pathogenic mechanism. In particular taking into consideration that parkin has been shown to exert a significant neuroprotective effect against various toxic insults, including manganese-induced cell death, α-synuclein toxicity, proteasomal dysfunction, Pael-R and P38/JTv-1 accumulation, kainate-induced excitotoxicity, mitochondrial-dependent apoptosis, MPP+/rotenone-induced cell death, ER stress, and dopamine-mediated toxicity [54, 130, 154, 155, 168-175].
In Drosophila, deletion of the parkin gene leads to the progressive degeneration of dopaminergic neurons in the central nervous system (CNS), a phenotype which is exacerbated by loss of glutathione-S-transferase [176], a gene which is actually upregulated by the PD-linked gene DJ-1 in response to oxidative stress [177]. Confirming the specificity of this effect, the neurodegenerative phenotype in parkin mutants was suppressed by overexpression of glutathione-Stransferase [176], which suggests that loss of functional parkin increases sensitivity to oxidative stress. However, given that Drosophila do not express endogenous α-synuclein or Pael-R, it may not be possible to fully appreciate the consequences of alterations in parkin function in this model.
The selective vulnerability of dopaminergic neurons in PD could potentially be explained by the demonstration that dopamine can covalently modify parkin in vitro, decreasing parkin solubility and E3 ligase activity [178]. In support of these in vitro results, LaVoie and colleagues also observed a decrease in parkin solubility in PD patients, as well as the presence of catechol-modified parkin in the SN [178]. In addition, embryonic dopaminergic neurons are particularly sensitive to deprivation of the growth factors GDNF or BDNF, activating a novel, apoptotic pathway in their absence [179]. Cell death was shown to be independent of mitochondria, though caspase activation was still required, as treatment with a caspase inhibitor, and more specifically inhibition of caspase 8, prevented cell death mediated by GDNF/BDNF withdrawal. Yu and coworkers were also able to suppress cell death induced by GDNF/BDNF deprivation by inhibiting Fas or FADD (Fas-associated protein with death domain), which is an adaptor required for Fas-mediated activation of caspase 8, thus implicating the death receptor pathway in this phenomenon [179]. Intriguingly, Kahns and associates demonstrate that while caspases 1, 3, and 8 cleave parkin, both caspase 1 and 8 directly cleave parkin without requiring activation of the effector caspase 3, suggesting that death receptor activation and inflammatory stress promote the cleavage and inactivation of parkin [180]. Thus given the suspected link between depletion of neurotrophic factors and PD pathogenesis [181-183], in addition to the detection of cleaved parkin fragments in LBs isolated from the SN of PD patients [41, 184], it is possible that caspase 8-mediated cleavage and resultant inactivation of parkin may play a pathogenic role in PD progression, and additionally establishes a potential mechanism for the selective vulnerability of dopaminergic neurons in PD (Figure 1).
5.3. DJ-1
Mutations in DJ-1, an antioxidant, redox-sensitive molecular chaperone [185-188], are linked to rare forms of autosomal recessive, early-onset PD [189, 190]. PD-linked mutations in DJ-1 include missense, truncation, and splice site mutations, as well as large deletions, suggesting that loss of DJ-1 function leads to neurodegeneration [190-192]. Of particular relevance to PD neuropathology, DJ-1 inhibits the oligomerization and toxicity of mutant A53T α-synuclein, while a decrease in DJ-1 expression facilitates the aggregation of α-synuclein [177, 193]. Knockdown or deletion of DJ-1 also increases susceptibility to proteasome inhibition in vitro [194, 195]. Through the modulation of glutamate cysteine ligase expression, the rate-limiting enzyme in glutathione synthesis, wild-type DJ-1, but not the PD-linked mutant L166P, is protective against oxidative stress, while blocking glutathione synthesis abolishes the protective effect of DJ-1 [177]. Loss of DJ-1 function is further implicated in PD pathogenesis by the observation that glutathione levels are decreased in the SN in early stages of PD [196], as well as the demonstration that depletion of glutathione leads to an age-related neurodegeneration of the nigrostriatal pathway by oxidation-dependent inhibition of mitochondrial complex I [197-200].
Recently, a surprising link between expression levels of proteasome subunits and DJ-1 was identified. In nontransgenic mice, treatment with antioxidants enhances the expression of 20S and 19S proteasome subunits, as well as proteasome activity [201]. However, no induction of proteasome activity or expression of proteasome subunits is observed in mice lacking the transcription factor Nrf2 (nuclear factor erythroid 2-related factor). Further, activity of the promoter regulating expression of the 20S β5 subunit, PSMB5, is increased with either overexpression of Nrf2 or exposure to antioxidants [201]. Given that Nrf2 is a component of the transcription complex that binds to the cis-acting element ARE (antioxidant response element), which regulates the expression of proteins that are protective against oxidative stress (e.g. glutathione S-transferases, glutamyl cysteine ligase, and NADPH quinone oxidoreductase) [202-205], the proximal promoter of PSMB5 was evaluated and an ARE subsequently identified [201]. Under normal conditions, Nrf2 is sequestered in the cytosol by the actin-binding protein Keap1 [206]. Exposure to antioxidants causes the dissociation of Nrf2 from Keap1, allowing for the nuclear translocation of Nrf2 and transcription of ARE genes. Amazingly, DJ-1 actually binds and stabilizes Nrf2, preventing the interaction with Keap1 and decreasing ubiquitination and subsequent proteasome-dependent degradation of Nrf2 [207, 208]. In the absence of DJ-1, Nrf2 is unstable and rapidly degraded, leading to a decrease in transcription of ARE genes [207].
These findings could explain the dopaminergic cell loss and motor dysfunction observed in DJ-1 deficient mice upon exposure to the PD-associated herbicide paraquat [209]. Following treatment with paraquat, Yang and coworkers report a decrease in Nrf2, as well as the 19S ATPase Rpt6 and 20S β5 subunits in the ventral midbrain of DJ-1 knockout mice, while no pathological abnormalities were detected in wild-type mice treated with paraquat [209]. In addition, proteasome activity in the ventral midbrain was decreased by 30% in DJ-1 deficient mice treated with paraquat when compared to the saline-treated group, which was accompanied by an increase in protein ubiquitination. Thus DJ-1 appears to be crucial for the survival of dopaminergic neurons, in particular under conditions of cellular stress.
5.4. LRRK2 (Leucine-rich repeat kinase 2
LRRK2, which is detected in both LBs and granular α-synuclein deposits believed to represent LB precursors [210, 211], was originally linked to the PARK8 locus in a large Japanese family [212]. This linkage was subsequently questioned by the discovery Family SK, carriers of the most common LRRK2 mutation (G2019S) [213], which clinically present with a slowly progressive parkinsonism, though no LBs or synuclein pathology was observed in any brain region evaluated upon autopsy [214]. Further adding to the complexity, a recent case history details a patient carrying the G2019S LRRK2 mutation with a history of slowly-progressive PD, though histological assessment revealed nigral degeneration in the absence of both α-synuclein and tau pathology [215]. However, numerous Marinesco bodies, which are spherical eosinophilic ubiquitin-positive intranuclear inclusions, were observed in both the SN and locus coeruleus [215]. Given that proteasome inhibition in vitro can lead to aberrations in ubiquitin immunoreactivity reminiscent of Marinesco body formation [216], it is possible that mutations in LRRK2 may disrupt UPS function, though an explanation for the pleomorphic pathology associated with LRRK2 mutations is still unknown.
A more direct link between LRRK2 and the UPS was established by Smith and associates in their discovery of an interaction between LRRK2 and parkin, but not DJ-1, α-synuclein, or tau, consistent with observations by Rajput and coworkers [214, 217]. Interestingly, overexpression of LRRK2 led to the formation of inclusions in a small fraction of cells, which was exacerbated by the coexpression of parkin [217]. Although LRRK2 mutants displayed a similar cellular localization, ability to associate with parkin, and tendency to aggregate in comparison to the wild-type protein, all mutants evaluated (R1441C, Y1699C, G2019S) increased cell death, which could not be prevented by the coexpression of parkin [217]. As LRRK2 activity increases the autoubiquitination of parkin [217], thus promoting the proteasome-mediated degradation of parkin [159-162], and mutations in LRRK2 are predicted to increase kinase activity [218, 219], it is possible that toxicity attributed to LRRK2 mutations could result from the abnormal modulation of parkin levels and/or activity. In addition, both wild-type and mutant LRRK2 were recently shown to interact with heat shock protein 90 (hsp90), and dissociation of the LRRK2-hsp90 complex promotes the UPS-mediated degradation of LRRK2 [219, 220], suggesting that proteasome dysfunction could also lead to an increase in LRRK2 expression.
5.5. PINK1 (PTEN-induced putative kinase 1)
PINK1, a highly conserved kinase that is localized to the inner and outer mitochondrial membranes [221-224], has been identified as the gene locus for PARK6-linked PD, which is characterized by an earlier age of onset than sporadic PD [224]. The most common PD-associated mutation in PINK1 (C1366T) decreases mRNA transcript levels by 80-90% [225], while the mutations G309D, L437P, G386A and G409V have all been shown to reduce kinase activity in vitro [226, 227]. These findings implicate a loss of function or deficiency of the PINK1 protein in PD pathogenesis, leading to the development and evaluation of PINK1 knockout models. Intriguingly, knockout of the PINK1 gene in Drosophila led to defects in mitochondrial morphology and degeneration of dopaminergic neurons, a phenotype which was rescued by parkin overexpression [228, 229]. As deletion of the parkin gene produced a similar phenotype [230, 231] that could not likewise be reversed by overexpression of PINK1 [228, 229], and deletion of both the parkin and PINK1 genes does not lead to an exacerbated phenotype, it is believed that PINK1 is upstream of parkin in a signaling cascade that regulates mitochondrial function and integrity [228, 229].
The effects of PINK1 deficiency on mitochondrial function and morphology have since been replicated in both human and mouse-derived dopaminergic neurons [232]. The deletion of PINK1 also leads to an increased activation of the mitochondrial cell-death pathway, as well as an elevation of ROS levels [232]. Conversely, the overexpression of PINK1 is protective against proteasomal inhibition and staurosporine-induced apoptosis, decreasing both cytochrome c release and caspase 3 activation [233, 234]. The overexpression of PINK1 has also been shown to prevent abnormal depolarization of the mitochondrial membrane in response to UPS dysfunction [235, 236]. The link between PINK1 and the UPS has been further established by Muqit and colleagues, demonstrating an enhanced cleavage and recruitment of PINK1 to aggresomes under conditions of proteasomal impairment, with both wild-type and mutant PINK1 displaying a similar tendency to aggregate [222]. As PINK1 is detected in approximately 5-10% of LBs in PD [221], the finding that PINK1 colocalizes with parkin, synphilin-1, and α-synuclein in aggresomes in the presence of the proteasomal inhibitor MG132 provides additional support for a central role of UPS dysfunction in LB formation [222].
6. Conclusion
Thus it is becoming increasingly clear that genetic links to PD either promote UPS dysfunction, or interfere with the normal compensatory response(s) that occur to minimize toxicity from proteasomal impairment. However, although this review evaluates the existing data from the viewpoint that a decrement in UPS function is central to the pathogenesis of PD, it is possible that UPS function itself is actually modulated in response to a central impairment in an alternate system, such as the mitochondrial or lysosomal/autophagic pathway. Further confounding this issue, UPS impairment compounded by an inefficient upregulation of autophagy or mitochondrial protein quality control mechanisms may ultimately yield a similar phenotype to that observed by lysosomal or mitochondrial dysfunction exacerbated by proteasomal inhibition. Clarification of this issue will assist in the identification of specific targets that can be modulated, as it may be possible to rescue neuronal function through the augmentation of parallel and intersecting pathways, ultimately enhancing the repertoire of therapeutic agents available to treat PD and other neurodegenerative conditions characterized by abnormal protein aggregation.
Acknowledgements
This work was supported by the Mayo Clinic Foundation and funding by NIA R01-AG-026251-01, NINDS NS55698, and NIA AG17216 and NINDS NS59363J1.
References
- 1.Olanow CW, Watts RL, Koller WC. An algorithm (decision tree) for the management of Parkinson's disease (2001): treatment guidelines. Neurology. 2001;56(11 Suppl 5):S1–S88. doi: 10.1212/wnl.56.suppl_5.s1. [DOI] [PubMed] [Google Scholar]
- 2.Olanow CW, Perl DP, DeMartino GN, McNaught KS. Lewy-body formation is an aggresome-related process: a hypothesis. Lancet Neurol. 2004;3(8):496–503. doi: 10.1016/S1474-4422(04)00827-0. [DOI] [PubMed] [Google Scholar]
- 3.Spillantini MG, Crowther RA, Jakes R, Hasegawa M, Goedert M. alpha-Synuclein in filamentous inclusions of Lewy bodies from Parkinson's disease and dementia with lewy bodies. Proc Natl Acad Sci U S A. 1998;95(11):6469–6473. doi: 10.1073/pnas.95.11.6469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, et al. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997;276(5321):2045–2047. doi: 10.1126/science.276.5321.2045. [DOI] [PubMed] [Google Scholar]
- 5.Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S, et al. Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet. 2004;364(9440):1167–1169. doi: 10.1016/S0140-6736(04)17103-1. [DOI] [PubMed] [Google Scholar]
- 6.Kruger R, Kuhn W, Muller T, Woitalla D, Graeber M, Kosel S, et al. Ala30Pro mutation in the gene encoding alpha-synuclein in Parkinson's disease. Nat Genet. 1998;18(2):106–108. doi: 10.1038/ng0298-106. [DOI] [PubMed] [Google Scholar]
- 7.Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J, et al. alpha-Synuclein locus triplication causes Parkinson's disease. Science. 2003;302(5646):841. doi: 10.1126/science.1090278. [DOI] [PubMed] [Google Scholar]
- 8.Zarranz JJ, Alegre J, Gomez-Esteban JC, Lezcano E, Ros R, Ampuero I, et al. The new mutation, E46K, of alpha-synuclein causes Parkinson and Lewy body dementia. Ann Neurol. 2004;55(2):164–173. doi: 10.1002/ana.10795. [DOI] [PubMed] [Google Scholar]
- 9.Zaccai J, Brayne C, McKeith I, Matthews F, Ince PG. Patterns and stages of alpha-synucleinopathy: Relevance in a population-based cohort. Neurology. 2008;70(13):1042–1048. doi: 10.1212/01.wnl.0000306697.48738.b6. [DOI] [PubMed] [Google Scholar]
- 10.Dickson DW, Fujishiro H, DelleDonne A, Menke J, Ahmed Z, Klos KJ, et al. Evidence that incidental Lewy body disease is pre-symptomatic Parkinson's disease. Acta Neuropathol. 2008;115(4):437–444. doi: 10.1007/s00401-008-0345-7. [DOI] [PubMed] [Google Scholar]
- 11.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 12.Heinemeyer W, Kleinschmidt JA, Saidowsky J, Escher C, Wolf DH. Proteinase yscE, the yeast proteasome/multicatalytic-multifunctional proteinase: mutants unravel its function in stress induced proteolysis and uncover its necessity for cell survival. Embo J. 1991;10(3):555–562. doi: 10.1002/j.1460-2075.1991.tb07982.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz AL, Ciechanover A. The ubiquitin-proteasome pathway and pathogenesis of human diseases. Annu Rev Med. 1999;50:57–74. doi: 10.1146/annurev.med.50.1.57. [DOI] [PubMed] [Google Scholar]
- 14.Giasson BI, Lee VM. Are ubiquitination pathways central to Parkinson's disease? Cell. 2003;114(1):1–8. doi: 10.1016/s0092-8674(03)00509-9. [DOI] [PubMed] [Google Scholar]
- 15.McNaught KS, Olanow CW, Halliwell B, Isacson O, Jenner P. Failure of the ubiquitin-proteasome system in Parkinson's disease. Nat Rev Neurosci. 2001;2(8):589–594. doi: 10.1038/35086067. [DOI] [PubMed] [Google Scholar]
- 16.Kopp F, Hendil KB, Dahlmann B, Kristensen P, Sobek A, Uerkvitz W. Subunit arrangement in the human 20S proteasome. Proc Natl Acad Sci U S A. 1997;94(7):2939–2944. doi: 10.1073/pnas.94.7.2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dick TP, Nussbaum AK, Deeg M, Heinemeyer W, Groll M, Schirle M, et al. Contribution of proteasomal beta-subunits to the cleavage of peptide substrates analyzed with yeast mutants. J Biol Chem. 1998;273(40):25637–25646. doi: 10.1074/jbc.273.40.25637. [DOI] [PubMed] [Google Scholar]
- 18.Kisselev AF, Akopian TN, Castillo V, Goldberg AL. Proteasome active sites allosterically regulate each other, suggesting a cyclical bite-chew mechanism for protein breakdown. Mol Cell. 1999;4(3):395–402. doi: 10.1016/s1097-2765(00)80341-x. [DOI] [PubMed] [Google Scholar]
- 19.Navon A, Goldberg AL. Proteins are unfolded on the surface of the ATPase ring before transport into the proteasome. Mol Cell. 2001;8(6):1339–1349. doi: 10.1016/s1097-2765(01)00407-5. [DOI] [PubMed] [Google Scholar]
- 20.Voges D, Zwickl P, Baumeister W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu Rev Biochem. 1999;68:1015–1068. doi: 10.1146/annurev.biochem.68.1.1015. [DOI] [PubMed] [Google Scholar]
- 21.Liu CW, Corboy MJ, DeMartino GN, Thomas PJ. Endoproteolytic activity of the proteasome. Science. 2003;299(5605):408–411. doi: 10.1126/science.1079293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeMartino GN, Slaughter CA. The proteasome, a novel protease regulated by multiple mechanisms. J Biol Chem. 1999;274(32):22123–22126. doi: 10.1074/jbc.274.32.22123. [DOI] [PubMed] [Google Scholar]
- 23.Ortega J, Heymann JB, Kajava AV, Ustrell V, Rechsteiner M, Steven AC. The axial channel of the 20S proteasome opens upon binding of the PA200 activator. J Mol Biol. 2005;346(5):1221–1227. doi: 10.1016/j.jmb.2004.12.049. [DOI] [PubMed] [Google Scholar]
- 24.Rechsteiner M, Hill CP. Mobilizing the proteolytic machine: cell biological roles of proteasome activators and inhibitors. Trends Cell Biol. 2005;15(1):27–33. doi: 10.1016/j.tcb.2004.11.003. [DOI] [PubMed] [Google Scholar]
- 25.Coux O, Tanaka K, Goldberg AL. Structure and functions of the 20S and 26S proteasomes. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 26.Smith DM, Benaroudj N, Goldberg A. Proteasomes and their associated ATPases: a destructive combination. J Struct Biol. 2006;156(1):72–83. doi: 10.1016/j.jsb.2006.04.012. [DOI] [PubMed] [Google Scholar]
- 27.Cascio P, Call M, Petre BM, Walz T, Goldberg AL. Properties of the hybrid form of the 26S proteasome containing both 19S and PA28 complexes. Embo J. 2002;21(11):2636–2645. doi: 10.1093/emboj/21.11.2636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reits EA, Benham AM, Plougastel B, Neefjes J, Trowsdale J. Dynamics of proteasome distribution in living cells. Embo J. 1997;16(20):6087–6094. doi: 10.1093/emboj/16.20.6087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wojcik C, DeMartino GN. Intracellular localization of proteasomes. Int J Biochem Cell Biol. 2003;35(5):579–589. doi: 10.1016/s1357-2725(02)00380-1. [DOI] [PubMed] [Google Scholar]
- 30.Baldin V, Militello M, Thomas Y, Doucet C, Fic W, Boireau S, et al. A novel role for PA28gamma-proteasome in nuclear speckle organization and SR protein trafficking. Mol Biol Cell. 2008;19(4):1706–1716. doi: 10.1091/mbc.E07-07-0637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Furukawa Y, Vigouroux S, Wong H, Guttman M, Rajput AH, Ang L, et al. Brain proteasomal function in sporadic Parkinson's disease and related disorders. Ann Neurol. 2002;51(6):779–782. doi: 10.1002/ana.10207. [DOI] [PubMed] [Google Scholar]
- 32.McNaught KS, Belizaire R, Isacson O, Jenner P, Olanow CW. Altered proteasomal function in sporadic Parkinson's disease. Exp Neurol. 2003;179(1):38–46. doi: 10.1006/exnr.2002.8050. [DOI] [PubMed] [Google Scholar]
- 33.McNaught KS, Jenner P. Proteasomal function is impaired in substantia nigra in Parkinson's disease. Neurosci Lett. 2001;297(3):191–194. doi: 10.1016/s0304-3940(00)01701-8. [DOI] [PubMed] [Google Scholar]
- 34.Tofaris GK, Razzaq A, Ghetti B, Lilley KS, Spillantini MG. Ubiquitination of alpha-synuclein in Lewy bodies is a pathological event not associated with impairment of proteasome function. J Biol Chem. 2003;278(45):44405–44411. doi: 10.1074/jbc.M308041200. [DOI] [PubMed] [Google Scholar]
- 35.McNaught KS, Belizaire R, Jenner P, Olanow CW, Isacson O. Selective loss of 20S proteasome alpha-subunits in the substantia nigra pars compacta in Parkinson's disease. Neurosci Lett. 2002;326(3):155–158. doi: 10.1016/s0304-3940(02)00296-3. [DOI] [PubMed] [Google Scholar]
- 36.Auluck PK, Chan HY, Trojanowski JQ, Lee VM, Bonini NM. Chaperone suppression of alpha-synuclein toxicity in a Drosophila model for Parkinson's disease. Science. 2002;295(5556):865–868. doi: 10.1126/science.1067389. [DOI] [PubMed] [Google Scholar]
- 37.Ii K, Ito H, Tanaka K, Hirano A. Immunocytochemical co-localization of the proteasome in ubiquitinated structures in neurodegenerative diseases and the elderly. J Neuropathol Exp Neurol. 1997;56(2):125–131. doi: 10.1097/00005072-199702000-00002. [DOI] [PubMed] [Google Scholar]
- 38.Lennox G, Lowe J, Morrell K, Landon M, Mayer RJ. Anti-ubiquitin immunocytochemistry is more sensitive than conventional techniques in the detection of diffuse Lewy body disease. J Neurol Neurosurg Psychiatry. 1989;52(1):67–71. doi: 10.1136/jnnp.52.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lowe J, McDermott H, Landon M, Mayer RJ, Wilkinson KD. Ubiquitin carboxyl-terminal hydrolase (PGP 9.5) is selectively present in ubiquitinated inclusion bodies characteristic of human neurodegenerative diseases. J Pathol. 1990;161(2):153–160. doi: 10.1002/path.1711610210. [DOI] [PubMed] [Google Scholar]
- 40.McNaught KS, Shashidharan P, Perl DP, Jenner P, Olanow CW. Aggresome-related biogenesis of Lewy bodies. Eur J Neurosci. 2002;16(11):2136–2148. doi: 10.1046/j.1460-9568.2002.02301.x. [DOI] [PubMed] [Google Scholar]
- 41.Schlossmacher MG, Frosch MP, Gai WP, Medina M, Sharma N, Forno L, et al. Parkin localizes to the Lewy bodies of Parkinson disease and dementia with Lewy bodies. Am J Pathol. 2002;160(5):1655–1667. doi: 10.1016/S0002-9440(10)61113-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McNaught KS, Mytilineou C, Jnobaptiste R, Yabut J, Shashidharan P, Jennert P, et al. Impairment of the ubiquitin-proteasome system causes dopaminergic cell death and inclusion body formation in ventral mesencephalic cultures. J Neurochem. 2002;81(2):301–306. doi: 10.1046/j.1471-4159.2002.00821.x. [DOI] [PubMed] [Google Scholar]
- 43.Rideout HJ, Lang-Rollin IC, Savalle M, Stefanis L. Dopaminergic neurons in rat ventral midbrain cultures undergo selective apoptosis and form inclusions, but do not up-regulate iHSP70, following proteasomal inhibition. J Neurochem. 2005;93(5):1304–1313. doi: 10.1111/j.1471-4159.2005.03124.x. [DOI] [PubMed] [Google Scholar]
- 44.McNaught KS, Perl DP, Brownell AL, Olanow CW. Systemic exposure to proteasome inhibitors causes a progressive model of Parkinson's disease. Ann Neurol. 2004;56(1):149–162. doi: 10.1002/ana.20186. [DOI] [PubMed] [Google Scholar]
- 45.McNaught KS, Olanow CW. Proteasome inhibitor-induced model of Parkinson's disease. Ann Neurol. 2006;60(2):243–247. doi: 10.1002/ana.20936. [DOI] [PubMed] [Google Scholar]
- 46.Bove J, Zhou C, Jackson-Lewis V, Taylor J, Chu Y, Rideout HJ, et al. Proteasome inhibition and Parkinson's disease modeling. Ann Neurol. 2006;60(2):260–264. doi: 10.1002/ana.20937. [DOI] [PubMed] [Google Scholar]
- 47.Hawlitschka A, Haas SJ, Schmitt O, Weiss DG, Wree A. Effects of systemic PSI administration on catecholaminergic cells in the brain, adrenal medulla and carotid body in Wistar rats. Brain Res. 2007;1173:137–144. doi: 10.1016/j.brainres.2007.07.072. [DOI] [PubMed] [Google Scholar]
- 48.Kordower JH, Kanaan NM, Chu Y, Suresh Babu R, Stansell J, 3rd, Terpstra BT, et al. Failure of proteasome inhibitor administration to provide a model of Parkinson's disease in rats and monkeys. Ann Neurol. 2006;60(2):264–268. doi: 10.1002/ana.20935. [DOI] [PubMed] [Google Scholar]
- 49.Manning-Bog AB, Reaney SH, Chou VP, Johnston LC, McCormack AL, Johnston J, et al. Lack of nigrostriatal pathology in a rat model of proteasome inhibition. Ann Neurol. 2006;60(2):256–260. doi: 10.1002/ana.20938. [DOI] [PubMed] [Google Scholar]
- 50.Schapira AH, Cleeter MW, Muddle JR, Workman JM, Cooper JM, King RH. Proteasomal inhibition causes loss of nigral tyrosine hydroxylase neurons. Ann Neurol. 2006;60(2):253–255. doi: 10.1002/ana.20934. [DOI] [PubMed] [Google Scholar]
- 51.Zeng BY, Bukhatwa S, Hikima A, Rose S, Jenner P. Reproducible nigral cell loss after systemic proteasomal inhibitor administration to rats. Ann Neurol. 2006;60(2):248–252. doi: 10.1002/ana.20932. [DOI] [PubMed] [Google Scholar]
- 52.Bedford L, Hay D, Devoy A, Paine S, Powe DG, Seth R, et al. Depletion of 26S proteasomes in mouse brain neurons causes neurodegeneration and Lewy-like inclusions resembling human pale bodies. J Neurosci. 2008;28(33):8189–8198. doi: 10.1523/JNEUROSCI.2218-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Betarbet R, Canet-Aviles RM, Sherer TB, Mastroberardino PG, McLendon C, Kim JH, et al. Intersecting pathways to neurodegeneration in Parkinson's disease: effects of the pesticide rotenone on DJ-1, alpha-synuclein, and the ubiquitin-proteasome system. Neurobiol Dis. 2006;22(2):404–420. doi: 10.1016/j.nbd.2005.12.003. [DOI] [PubMed] [Google Scholar]
- 54.Wang C, Ko HS, Thomas B, Tsang F, Chew KC, Tay SP, et al. Stress-induced alterations in parkin solubility promote parkin aggregation and compromise parkin's protective function. Hum Mol Genet. 2005;14(24):3885–3897. doi: 10.1093/hmg/ddi413. [DOI] [PubMed] [Google Scholar]
- 55.Wang XF, Li S, Chou AP, Bronstein JM. Inhibitory effects of pesticides on proteasome activity: implication in Parkinson's disease. Neurobiol Dis. 2006;23(1):198–205. doi: 10.1016/j.nbd.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 56.Fornai F, Schluter OM, Lenzi P, Gesi M, Ruffoli R, Ferrucci M, et al. Parkinson-like syndrome induced by continuous MPTP infusion: convergent roles of the ubiquitin-proteasome system and alpha-synuclein. Proc Natl Acad Sci U S A. 2005;102(9):3413–3418. doi: 10.1073/pnas.0409713102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zeng BY, Iravani MM, Lin ST, Irifune M, Kuoppamaki M, Al-Barghouthy G, et al. MPTP treatment of common marmosets impairs proteasomal enzyme activity and decreases expression of structural and regulatory elements of the 26S proteasome. Eur J Neurosci. 2006;23(7):1766–1774. doi: 10.1111/j.1460-9568.2006.04718.x. [DOI] [PubMed] [Google Scholar]
- 58.Kowall NW, Hantraye P, Brouillet E, Beal MF, McKee AC, Ferrante RJ. MPTP induces alpha-synuclein aggregation in the substantia nigra of baboons. Neuroreport. 2000;11(1):211–213. doi: 10.1097/00001756-200001170-00041. [DOI] [PubMed] [Google Scholar]
- 59.Hardy J, Cookson MR, Singleton A. Genes and parkinsonism. Lancet Neurol. 2003;2(4):221–228. doi: 10.1016/s1474-4422(03)00350-8. [DOI] [PubMed] [Google Scholar]
- 60.Maroteaux L, Campanelli JT, Scheller RH. Synuclein: a neuron-specific protein localized to the nucleus and presynaptic nerve terminal. J Neurosci. 1988;8(8):2804–2815. doi: 10.1523/JNEUROSCI.08-08-02804.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Davidson WS, Jonas A, Clayton DF, George JM. Stabilization of alpha-synuclein secondary structure upon binding to synthetic membranes. J Biol Chem. 1998;273(16):9443–9449. doi: 10.1074/jbc.273.16.9443. [DOI] [PubMed] [Google Scholar]
- 62.Abeliovich A, Schmitz Y, Farinas I, Choi-Lundberg D, Ho WH, Castillo PE, et al. Mice lacking alpha-synuclein display functional deficits in the nigrostriatal dopamine system. Neuron. 2000;25(1):239–252. doi: 10.1016/s0896-6273(00)80886-7. [DOI] [PubMed] [Google Scholar]
- 63.Cabin DE, Shimazu K, Murphy D, Cole NB, Gottschalk W, McIlwain KL, et al. Synaptic vesicle depletion correlates with attenuated synaptic responses to prolonged repetitive stimulation in mice lacking alpha-synuclein. J Neurosci. 2002;22(20):8797–8807. doi: 10.1523/JNEUROSCI.22-20-08797.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Murphy DD, Rueter SM, Trojanowski JQ, Lee VM. Synucleins are developmentally expressed, and alpha-synuclein regulates the size of the presynaptic vesicular pool in primary hippocampal neurons. J Neurosci. 2000;20(9):3214–3220. doi: 10.1523/JNEUROSCI.20-09-03214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yavich L, Jakala P, Tanila H. Abnormal compartmentalization of norepinephrine in mouse dentate gyrus in alpha-synuclein knockout and A30P transgenic mice. J Neurochem. 2006;99(3):724–732. doi: 10.1111/j.1471-4159.2006.04098.x. [DOI] [PubMed] [Google Scholar]
- 66.Yavich L, Tanila H, Vepsalainen S, Jakala P. Role of alpha-synuclein in presynaptic dopamine recruitment. J Neurosci. 2004;24(49):11165–11170. doi: 10.1523/JNEUROSCI.2559-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fortin DL, Nemani VM, Voglmaier SM, Anthony MD, Ryan TA, Edwards RH. Neural activity controls the synaptic accumulation of alpha-synuclein. J Neurosci. 2005;25(47):10913–10921. doi: 10.1523/JNEUROSCI.2922-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fortin DL, Troyer MD, Nakamura K, Kubo S, Anthony MD, Edwards RH. Lipid rafts mediate the synaptic localization of alpha-synuclein. J Neurosci. 2004;24(30):6715–6723. doi: 10.1523/JNEUROSCI.1594-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Jia JM, Chen Q, Zhou Y, Miao S, Zheng J, Zhang C, et al. Brain-derived neurotrophic factor-tropomyosin-related kinase B signaling contributes to activity-dependent changes in synaptic proteins. J Biol Chem. 2008;283(30):21242–21250. doi: 10.1074/jbc.M800282200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bozzi Y, Pizzorusso T, Cremisi F, Rossi FM, Barsacchi G, Maffei L. Monocular deprivation decreases the expression of messenger RNA for brain-derived neurotrophic factor in the rat visual cortex. Neuroscience. 1995;69(4):1133–1144. doi: 10.1016/0306-4522(95)00321-9. [DOI] [PubMed] [Google Scholar]
- 71.Hardingham GE, Fukunaga Y, Bading H. Extrasynaptic NMDARs oppose synaptic NMDARs by triggering CREB shut-off and cell death pathways. Nat Neurosci. 2002;5(5):405–414. doi: 10.1038/nn835. [DOI] [PubMed] [Google Scholar]
- 72.Isackson PJ, Huntsman MM, Murray KD, Gall CM. BDNF mRNA expression is increased in adult rat forebrain after limbic seizures: temporal patterns of induction distinct from NGF. Neuron. 1991;6(6):937–948. doi: 10.1016/0896-6273(91)90234-q. [DOI] [PubMed] [Google Scholar]
- 73.Zafra F, Castren E, Thoenen H, Lindholm D. Interplay between glutamate and gamma-aminobutyric acid transmitter systems in the physiological regulation of brain-derived neurotrophic factor and nerve growth factor synthesis in hippocampal neurons. Proc Natl Acad Sci U S A. 1991;88(22):10037–10041. doi: 10.1073/pnas.88.22.10037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lee HJ, Choi C, Lee SJ. Membrane-bound alpha-synuclein has a high aggregation propensity and the ability to seed the aggregation of the cytosolic form. J Biol Chem. 2002;277(1):671–678. doi: 10.1074/jbc.M107045200. [DOI] [PubMed] [Google Scholar]
- 75.Dluzen DE, McDermott JL, Anderson LI, Kucera J, Joyce JN, Osredkar T, et al. Age-related changes in nigrostriatal dopaminergic function are accentuated in +/− brain-derived neurotrophic factor mice. Neuroscience. 2004;128(1):201–208. doi: 10.1016/j.neuroscience.2004.06.032. [DOI] [PubMed] [Google Scholar]
- 76.Chen L, Feany MB. Alpha-synuclein phosphorylation controls neurotoxicity and inclusion formation in a Drosophila model of Parkinson disease. Nat Neurosci. 2005;8(5):657–663. doi: 10.1038/nn1443. [DOI] [PubMed] [Google Scholar]
- 77.Fujiwara H, Hasegawa M, Dohmae N, Kawashima A, Masliah E, Goldberg MS, et al. alpha-Synuclein is phosphorylated in synucleinopathy lesions. Nat Cell Biol. 2002;4(2):160–164. doi: 10.1038/ncb748. [DOI] [PubMed] [Google Scholar]
- 78.Giasson BI, Duda JE, Murray IV, Chen Q, Souza JM, Hurtig HI, et al. Oxidative damage linked to neurodegeneration by selective alpha-synuclein nitration in synucleinopathy lesions. Science. 2000;290(5493):985–989. doi: 10.1126/science.290.5493.985. [DOI] [PubMed] [Google Scholar]
- 79.Giasson BI, Forman MS, Higuchi M, Golbe LI, Graves CL, Kotzbauer PT, et al. Initiation and synergistic fibrillization of tau and alpha-synuclein. Science. 2003;300(5619):636–640. doi: 10.1126/science.1082324. [DOI] [PubMed] [Google Scholar]
- 80.Giasson BI, Uryu K, Trojanowski JQ, Lee VM. Mutant and wild type human alpha-synucleins assemble into elongated filaments with distinct morphologies in vitro. J Biol Chem. 1999;274(12):7619–7622. doi: 10.1074/jbc.274.12.7619. [DOI] [PubMed] [Google Scholar]
- 81.Hodara R, Norris EH, Giasson BI, Mishizen-Eberz AJ, Lynch DR, Lee VM, et al. Functional consequences of alpha-synuclein tyrosine nitration: diminished binding to lipid vesicles and increased fibril formation. J Biol Chem. 2004;279(46):47746–47753. doi: 10.1074/jbc.M408906200. [DOI] [PubMed] [Google Scholar]
- 82.Li W, West N, Colla E, Pletnikova O, Troncoso JC, Marsh L, et al. Aggregation promoting C-terminal truncation of alpha-synuclein is a normal cellular process and is enhanced by the familial Parkinson's disease-linked mutations. Proc Natl Acad Sci U S A. 2005;102(6):2162–2167. doi: 10.1073/pnas.0406976102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mishizen-Eberz AJ, Norris EH, Giasson BI, Hodara R, Ischiropoulos H, Lee VM, et al. Cleavage of alpha-synuclein by calpain: potential role in degradation of fibrillized and nitrated species of alpha-synuclein. Biochemistry. 2005;44(21):7818–7829. doi: 10.1021/bi047846q. [DOI] [PubMed] [Google Scholar]
- 84.Murray IV, Giasson BI, Quinn SM, Koppaka V, Axelsen PH, Ischiropoulos H, et al. Role of alpha-synuclein carboxy-terminus on fibril formation in vitro. Biochemistry. 2003;42(28):8530–8540. doi: 10.1021/bi027363r. [DOI] [PubMed] [Google Scholar]
- 85.Norris EH, Giasson BI, Ischiropoulos H, Lee VM. Effects of oxidative and nitrative challenges on alpha-synuclein fibrillogenesis involve distinct mechanisms of protein modifications. J Biol Chem. 2003;278(29):27230–27240. doi: 10.1074/jbc.M212436200. [DOI] [PubMed] [Google Scholar]
- 86.Singleton A, Gwinn-Hardy K. Parkinson's disease and dementia with Lewy bodies: a difference in dose? Lancet. 2004;364(9440):1105–1107. doi: 10.1016/S0140-6736(04)17117-1. [DOI] [PubMed] [Google Scholar]
- 87.Smith WW, Margolis RL, Li X, Troncoso JC, Lee MK, Dawson VL, et al. Alpha-synuclein phosphorylation enhances eosinophilic cytoplasmic inclusion formation in SH-SY5Y cells. J Neurosci. 2005;25(23):5544–5552. doi: 10.1523/JNEUROSCI.0482-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tofaris GK, Garcia Reitbock P, Humby T, Lambourne SL, O'Connell M, Ghetti B, et al. Pathological changes in dopaminergic nerve cells of the substantia nigra and olfactory bulb in mice transgenic for truncated human alpha-synuclein(1-120): implications for Lewy body disorders. J Neurosci. 2006;26(15):3942–3950. doi: 10.1523/JNEUROSCI.4965-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Norris EH, Giasson BI, Hodara R, Xu S, Trojanowski JQ, Ischiropoulos H, et al. Reversible inhibition of alpha-synuclein fibrillization by dopaminochrome-mediated conformational alterations. J Biol Chem. 2005;280(22):21212–21219. doi: 10.1074/jbc.M412621200. [DOI] [PubMed] [Google Scholar]
- 90.Serpell LC, Berriman J, Jakes R, Goedert M, Crowther RA. Fiber diffraction of synthetic alpha-synuclein filaments shows amyloid-like cross-beta conformation. Proc Natl Acad Sci U S A. 2000;97(9):4897–4902. doi: 10.1073/pnas.97.9.4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Hashimoto M, Hsu LJ, Xia Y, Takeda A, Sisk A, Sundsmo M, et al. Oxidative stress induces amyloid-like aggregate formation of NACP/alpha-synuclein in vitro. Neuroreport. 1999;10(4):717–721. doi: 10.1097/00001756-199903170-00011. [DOI] [PubMed] [Google Scholar]
- 92.Anderson JP, Walker DE, Goldstein JM, de Laat R, Banducci K, Caccavello RJ, et al. Phosphorylation of Ser-129 is the dominant pathological modification of alpha-synuclein in familial and sporadic Lewy body disease. J Biol Chem. 2006;281(40):29739–29752. doi: 10.1074/jbc.M600933200. [DOI] [PubMed] [Google Scholar]
- 93.Waxman EA, Giasson BI. Specificity and regulation of casein kinase-mediated phosphorylation of alpha-synuclein. J Neuropathol Exp Neurol. 2008;67(5):402–416. doi: 10.1097/NEN.0b013e3186fc995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Arawaka S, Wada M, Goto S, Karube H, Sakamoto M, Ren CH, et al. The role of G-protein-coupled receptor kinase 5 in pathogenesis of sporadic Parkinson's disease. J Neurosci. 2006;26(36):9227–9238. doi: 10.1523/JNEUROSCI.0341-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Okochi M, Walter J, Koyama A, Nakajo S, Baba M, Iwatsubo T, et al. Constitutive phosphorylation of the Parkinson's disease associated alpha-synuclein. J Biol Chem. 2000;275(1):390–397. doi: 10.1074/jbc.275.1.390. [DOI] [PubMed] [Google Scholar]
- 96.Paleologou KE, Schmid AW, Rospigliosi CC, Kim HY, Lamberto GR, Fredenburg RA, et al. Phosphorylation at Ser-129 but not the phosphomimics S129E/D inhibits the fibrillation of alpha-synuclein. J Biol Chem. 2008;283(24):16895–16905. doi: 10.1074/jbc.M800747200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Pronin AN, Morris AJ, Surguchov A, Benovic JL. Synucleins are a novel class of substrates for G protein-coupled receptor kinases. J Biol Chem. 2000;275(34):26515–26522. doi: 10.1074/jbc.M003542200. [DOI] [PubMed] [Google Scholar]
- 98.Liu C, Fei E, Jia N, Wang H, Tao R, Iwata A, et al. Assembly of lysine 63-linked ubiquitin conjugates by phosphorylated alpha-synuclein implies Lewy body biogenesis. J Biol Chem. 2007;282(19):14558–14566. doi: 10.1074/jbc.M700422200. [DOI] [PubMed] [Google Scholar]
- 99.Sugeno N, Takeda A, Hasegawa T, Kobayashi M, Kikuchi A, Mori F, et al. Serine 129 phosphorylation of alpha-synuclein induces unfolded protein response-mediated cell death. J Biol Chem. 2008;283(34):23179–23188. doi: 10.1074/jbc.M802223200. [DOI] [PubMed] [Google Scholar]
- 100.Gorbatyuk OS, Li S, Sullivan LF, Chen W, Kondrikova G, Manfredsson FP, et al. The phosphorylation state of Ser-129 in human alpha-synuclein determines neurodegeneration in a rat model of Parkinson disease. Proc Natl Acad Sci U S A. 2008;105(2):763–768. doi: 10.1073/pnas.0711053105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Lee JT, Wheeler TC, Li L, Chin LS. Ubiquitination of alpha-synuclein by Siah-1 promotes alpha-synuclein aggregation and apoptotic cell death. Hum Mol Genet. 2008;17(6):906–917. doi: 10.1093/hmg/ddm363. [DOI] [PubMed] [Google Scholar]
- 102.Rott R, Szargel R, Haskin J, Shani V, Shainskaya A, Manov I, et al. Monoubiquitylation of alpha-synuclein by seven in absentia homolog (SIAH) promotes its aggregation in dopaminergic cells. J Biol Chem. 2008;283(6):3316–3328. doi: 10.1074/jbc.M704809200. [DOI] [PubMed] [Google Scholar]
- 103.Lee G, Junn E, Tanaka M, Kim YM, Mouradian MM. Synphilin-1 degradation by the ubiquitin-proteasome pathway and effects on cell survival. J Neurochem. 2002;83(2):346–352. doi: 10.1046/j.1471-4159.2002.01136.x. [DOI] [PubMed] [Google Scholar]
- 104.Liani E, Eyal A, Avraham E, Shemer R, Szargel R, Berg D, et al. Ubiquitylation of synphilin-1 and alpha-synuclein by SIAH and its presence in cellular inclusions and Lewy bodies imply a role in Parkinson's disease. Proc Natl Acad Sci U S A. 2004;101(15):5500–5505. doi: 10.1073/pnas.0401081101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Marx FP, Soehn AS, Berg D, Melle C, Schiesling C, Lang M, et al. The proteasomal subunit S6 ATPase is a novel synphilin-1 interacting protein--implications for Parkinson's disease. Faseb J. 2007;21(8):1759–1767. doi: 10.1096/fj.06-6734com. [DOI] [PubMed] [Google Scholar]
- 106.Avraham E, Szargel R, Eyal A, Rott R, Engelender S. Glycogen synthase kinase 3beta modulates synphilin-1 ubiquitylation and cellular inclusion formation by SIAH: implications for proteasomal function and Lewy body formation. J Biol Chem. 2005;280(52):42877–42886. doi: 10.1074/jbc.M505608200. [DOI] [PubMed] [Google Scholar]
- 107.Baba M, Nakajo S, Tu PH, Tomita T, Nakaya K, Lee VM, et al. Aggregation of alpha-synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. Am J Pathol. 1998;152(4):879–884. [PMC free article] [PubMed] [Google Scholar]
- 108.Campbell BC, McLean CA, Culvenor JG, Gai WP, Blumbergs PC, Jakala P, et al. The solubility of alpha-synuclein in multiple system atrophy differs from that of dementia with Lewy bodies and Parkinson's disease. J Neurochem. 2001;76(1):87–96. doi: 10.1046/j.1471-4159.2001.00021.x. [DOI] [PubMed] [Google Scholar]
- 109.Crowther RA, Jakes R, Spillantini MG, Goedert M. Synthetic filaments assembled from C-terminally truncated alpha-synuclein. FEBS Lett. 1998;436(3):309–312. doi: 10.1016/s0014-5793(98)01146-6. [DOI] [PubMed] [Google Scholar]
- 110.Cappai R, Leck SL, Tew DJ, Williamson NA, Smith DP, Galatis D, et al. Dopamine promotes alpha-synuclein aggregation into SDS-resistant soluble oligomers via a distinct folding pathway. Faseb J. 2005;19(10):1377–1379. doi: 10.1096/fj.04-3437fje. [DOI] [PubMed] [Google Scholar]
- 111.Conway KA, Rochet JC, Bieganski RM, Lansbury PT., Jr Kinetic stabilization of the alpha-synuclein protofibril by a dopamine-alpha-synuclein adduct. Science. 2001;294(5545):1346–1349. doi: 10.1126/science.1063522. [DOI] [PubMed] [Google Scholar]
- 112.Li J, Zhu M, Manning-Bog AB, Di Monte DA, Fink AL. Dopamine and L-dopa disaggregate amyloid fibrils: implications for Parkinson's and Alzheimer's disease. Faseb J. 2004;18(9):962–964. doi: 10.1096/fj.03-0770fje. [DOI] [PubMed] [Google Scholar]
- 113.Xu J, Kao SY, Lee FJ, Song W, Jin LW, Yankner BA. Dopamine-dependent neurotoxicity of alpha-synuclein: a mechanism for selective neurodegeneration in Parkinson disease. Nat Med. 2002;8(6):600–606. doi: 10.1038/nm0602-600. [DOI] [PubMed] [Google Scholar]
- 114.Mazzulli JR, Mishizen AJ, Giasson BI, Lynch DR, Thomas SA, Nakashima A, et al. Cytosolic catechols inhibit alpha-synuclein aggregation and facilitate the formation of intracellular soluble oligomeric intermediates. J Neurosci. 2006;26(39):10068–10078. doi: 10.1523/JNEUROSCI.0896-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mazzulli JR, Armakola M, Dumoulin M, Parastatidis I, Ischiropoulos H. Cellular oligomerization of alpha-synuclein is determined by the interaction of oxidized catechols with a C-terminal sequence. J Biol Chem. 2007;282(43):31621–31630. doi: 10.1074/jbc.M704737200. [DOI] [PubMed] [Google Scholar]
- 116.Bennett MC, Bishop JF, Leng Y, Chock PB, Chase TN, Mouradian MM. Degradation of alpha-synuclein by proteasome. J Biol Chem. 1999;274(48):33855–33858. doi: 10.1074/jbc.274.48.33855. [DOI] [PubMed] [Google Scholar]
- 117.Ancolio K, Alves da Costa C, Ueda K, Checler F. Alpha-synuclein and the Parkinson's disease-related mutant Ala53Thr-alpha-synuclein do not undergo proteasomal degradation in HEK293 and neuronal cells. Neurosci Lett. 2000;285(2):79–82. doi: 10.1016/s0304-3940(00)01049-1. [DOI] [PubMed] [Google Scholar]
- 118.Dufty BM, Warner LR, Hou ST, Jiang SX, Gomez-Isla T, Leenhouts KM, et al. Calpain-cleavage of alpha-synuclein: connecting proteolytic processing to disease-linked aggregation. Am J Pathol. 2007;170(5):1725–1738. doi: 10.2353/ajpath.2007.061232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Kim HJ, Lee D, Lee CH, Chung KC, Kim J, Paik SR. Calpain-resistant fragment(s) of alpha-synuclein regulates the synuclein-cleaving activity of 20S proteasome. Arch Biochem Biophys. 2006;455(1):40–47. doi: 10.1016/j.abb.2006.08.019. [DOI] [PubMed] [Google Scholar]
- 120.Cole NB, Murphy DD, Lebowitz J, Di Noto L, Levine RL, Nussbaum RL. Metal-catalyzed oxidation of alpha-synuclein: helping to define the relationship between oligomers, protofibrils, and filaments. J Biol Chem. 2005;280(10):9678–9690. doi: 10.1074/jbc.M409946200. [DOI] [PubMed] [Google Scholar]
- 121.Tofaris GK, Layfield R, Spillantini MG. alpha-synuclein metabolism and aggregation is linked to ubiquitin-independent degradation by the proteasome. FEBS Lett. 2001;509(1):22–26. doi: 10.1016/s0014-5793(01)03115-5. [DOI] [PubMed] [Google Scholar]
- 122.Liu CW, Giasson BI, Lewis KA, Lee VM, Demartino GN, Thomas PJ. A precipitating role for truncated alpha-synuclein and the proteasome in alpha-synuclein aggregation: implications for pathogenesis of Parkinson disease. J Biol Chem. 2005;280(24):22670–22678. doi: 10.1074/jbc.M501508200. [DOI] [PubMed] [Google Scholar]
- 123.Jensen PH, Nielsen MS, Jakes R, Dotti CG, Goedert M. Binding of alpha-synuclein to brain vesicles is abolished by familial Parkinson's disease mutation. J Biol Chem. 1998;273(41):26292–26294. doi: 10.1074/jbc.273.41.26292. [DOI] [PubMed] [Google Scholar]
- 124.Andersen JK. Oxidative stress in neurodegeneration: cause or consequence? Nat Med. 2004;10(Suppl):S18–25. doi: 10.1038/nrn1434. [DOI] [PubMed] [Google Scholar]
- 125.Tanaka Y, Engelender S, Igarashi S, Rao RK, Wanner T, Tanzi RE, et al. Inducible expression of mutant alpha-synuclein decreases proteasome activity and increases sensitivity to mitochondria-dependent apoptosis. Hum Mol Genet. 2001;10(9):919–926. doi: 10.1093/hmg/10.9.919. [DOI] [PubMed] [Google Scholar]
- 126.Martin-Clemente B, Alvarez-Castelao B, Mayo I, Sierra AB, Diaz V, Milan M, et al. alpha-Synuclein expression levels do not significantly affect proteasome function and expression in mice and stably transfected PC12 cell lines. J Biol Chem. 2004;279(51):52984–52990. doi: 10.1074/jbc.M409028200. [DOI] [PubMed] [Google Scholar]
- 127.Fujita M, Sugama S, Nakai M, Takenouchi T, Wei J, Urano T, et al. alpha-Synuclein stimulates differentiation of osteosarcoma cells: relevance to down-regulation of proteasome activity. J Biol Chem. 2007;282(8):5736–5748. doi: 10.1074/jbc.M606175200. [DOI] [PubMed] [Google Scholar]
- 128.Sun F, Anantharam V, Latchoumycandane C, Kanthasamy A, Kanthasamy AG. Dieldrin induces ubiquitin-proteasome dysfunction in alpha-synuclein overexpressing dopaminergic neuronal cells and enhances susceptibility to apoptotic cell death. J Pharmacol Exp Ther. 2005;315(1):69–79. doi: 10.1124/jpet.105.084632. [DOI] [PubMed] [Google Scholar]
- 129.Bodner RA, Outeiro TF, Altmann S, Maxwell MM, Cho SH, Hyman BT, et al. Pharmacological promotion of inclusion formation: a therapeutic approach for Huntington's and Parkinson's diseases. Proc Natl Acad Sci U S A. 2006;103(11):4246–4251. doi: 10.1073/pnas.0511256103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Petrucelli L, O'Farrell C, Lockhart PJ, Baptista M, Kehoe K, Vink L, et al. Parkin protects against the toxicity associated with mutant alpha-synuclein: proteasome dysfunction selectively affects catecholaminergic neurons. Neuron. 2002;36(6):1007–1019. doi: 10.1016/s0896-6273(02)01125-x. [DOI] [PubMed] [Google Scholar]
- 131.Chen Q, Thorpe J, Keller JN. Alpha-synuclein alters proteasome function, protein synthesis, and stationary phase viability. J Biol Chem. 2005;280(34):30009–30017. doi: 10.1074/jbc.M501308200. [DOI] [PubMed] [Google Scholar]
- 132.Gallardo G, Schluter OM, Sudhof TC. A molecular pathway of neurodegeneration linking alpha-synuclein to ApoE and Abeta peptides. Nat Neurosci. 2008;11(3):301–308. doi: 10.1038/nn2058. [DOI] [PubMed] [Google Scholar]
- 133.Emmanouilidou E, Stefanis L, Vekrellis K. Cell-produced alpha-synuclein oligomers are targeted to, and impair, the 26S proteasome. Neurobiol Aging. 2008 doi: 10.1016/j.neurobiolaging.2008.07.008. [DOI] [PubMed] [Google Scholar]
- 134.Lindersson E, Beedholm R, Hojrup P, Moos T, Gai W, Hendil KB, et al. Proteasomal inhibition by alpha-synuclein filaments and oligomers. J Biol Chem. 2004;279(13):12924–12934. doi: 10.1074/jbc.M306390200. [DOI] [PubMed] [Google Scholar]
- 135.Snyder H, Mensah K, Theisler C, Lee J, Matouschek A, Wolozin B. Aggregated and monomeric alpha-synuclein bind to the S6′ proteasomal protein and inhibit proteasomal function. J Biol Chem. 2003;278(14):11753–11759. doi: 10.1074/jbc.M208641200. [DOI] [PubMed] [Google Scholar]
- 136.Zhang NY, Tang Z, Liu CW. alpha-Synuclein protofibrils inhibit 26 S proteasome-mediated protein degradation: understanding the cytotoxicity of protein protofibrils in neurodegenerative disease pathogenesis. J Biol Chem. 2008;283(29):20288–20298. doi: 10.1074/jbc.M710560200. [DOI] [PubMed] [Google Scholar]
- 137.Ghee M, Fournier A, Mallet J. Rat alpha-synuclein interacts with Tat binding protein 1, a component of the 26S proteasomal complex. J Neurochem. 2000;75(5):2221–2224. doi: 10.1046/j.1471-4159.2000.0752221.x. [DOI] [PubMed] [Google Scholar]
- 138.Kayed R, Head E, Thompson JL, McIntire TM, Milton SC, Cotman CW, et al. Common structure of soluble amyloid oligomers implies common mechanism of pathogenesis. Science. 2003;300(5618):486–489. doi: 10.1126/science.1079469. [DOI] [PubMed] [Google Scholar]
- 139.Tseng BP, Green KN, Chan JL, Blurton-Jones M, Laferla FM. Abeta inhibits the proteasome and enhances amyloid and tau accumulation. Neurobiol Aging. 2007 doi: 10.1016/j.neurobiolaging.2007.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Bence NF, Sampat RM, Kopito RR. Impairment of the ubiquitin-proteasome system by protein aggregation. Science. 2001;292(5521):1552–1555. doi: 10.1126/science.292.5521.1552. [DOI] [PubMed] [Google Scholar]
- 141.Bennett EJ, Bence NF, Jayakumar R, Kopito RR. Global impairment of the ubiquitin-proteasome system by nuclear or cytoplasmic protein aggregates precedes inclusion body formation. Mol Cell. 2005;17(3):351–365. doi: 10.1016/j.molcel.2004.12.021. [DOI] [PubMed] [Google Scholar]
- 142.Bennett EJ, Shaler TA, Woodman B, Ryu KY, Zaitseva TS, Becker CH, et al. Global changes to the ubiquitin system in Huntington's disease. Nature. 2007;448(7154):704–708. doi: 10.1038/nature06022. [DOI] [PubMed] [Google Scholar]
- 143.Jana NR, Zemskov EA, Wang G, Nukina N. Altered proteasomal function due to the expression of polyglutamine-expanded truncated N-terminal huntingtin induces apoptosis by caspase activation through mitochondrial cytochrome c release. Hum Mol Genet. 2001;10(10):1049–1059. doi: 10.1093/hmg/10.10.1049. [DOI] [PubMed] [Google Scholar]
- 144.Verhoef LG, Lindsten K, Masucci MG, Dantuma NP. Aggregate formation inhibits proteasomal degradation of polyglutamine proteins. Hum Mol Genet. 2002;11(22):2689–2700. doi: 10.1093/hmg/11.22.2689. [DOI] [PubMed] [Google Scholar]
- 145.Kristiansen M, Deriziotis P, Dimcheff DE, Jackson GS, Ovaa H, Naumann H, et al. Disease-associated prion protein oligomers inhibit the 26S proteasome. Mol Cell. 2007;26(2):175–188. doi: 10.1016/j.molcel.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 146.Lee MK, Stirling W, Xu Y, Xu X, Qui D, Mandir AS, et al. Human alpha-synuclein-harboring familial Parkinson's disease-linked Ala-53 --> Thr mutation causes neurodegenerative disease with alpha-synuclein aggregation in transgenic mice. Proc Natl Acad Sci U S A. 2002;99(13):8968–8973. doi: 10.1073/pnas.132197599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Hattori N, Shimura H, Kubo S, Wang M, Shimizu N, Tanaka K, et al. Importance of familial Parkinson's disease and parkinsonism to the understanding of nigral degeneration in sporadic Parkinson's disease. J Neural Transm Suppl. 2000;60:101–116. doi: 10.1007/978-3-7091-6301-6_6. [DOI] [PubMed] [Google Scholar]
- 148.Lucking CB, Durr A, Bonifati V, Vaughan J, De Michele G, Gasser T, et al. Association between early-onset Parkinson's disease and mutations in the parkin gene. N Engl J Med. 2000;342(21):1560–1567. doi: 10.1056/NEJM200005253422103. [DOI] [PubMed] [Google Scholar]
- 149.Mata IF, Lockhart PJ, Farrer MJ. Parkin genetics: one model for Parkinson's disease. Hum Mol Genet. 2004;13(Spec No 1):R127–133. doi: 10.1093/hmg/ddh089. [DOI] [PubMed] [Google Scholar]
- 150.Hayashi S, Wakabayashi K, Ishikawa A, Nagai H, Saito M, Maruyama M, et al. An autopsy case of autosomal-recessive juvenile parkinsonism with a homozygous exon 4 deletion in the parkin gene. Mov Disord. 2000;15(5):884–888. doi: 10.1002/1531-8257(200009)15:5<884::aid-mds1019>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 151.Mori H, Kondo T, Yokochi M, Matsumine H, Nakagawa-Hattori Y, Miyake T, et al. Pathologic and biochemical studies of juvenile parkinsonism linked to chromosome 6q. Neurology. 1998;51(3):890–892. doi: 10.1212/wnl.51.3.890. [DOI] [PubMed] [Google Scholar]
- 152.Takahashi H, Ohama E, Suzuki S, Horikawa Y, Ishikawa A, Morita T, et al. Familial juvenile parkinsonism: clinical and pathologic study in a family. Neurology. 1994;44(3 Pt 1):437–441. doi: 10.1212/wnl.44.3_part_1.437. [DOI] [PubMed] [Google Scholar]
- 153.van de Warrenburg BP, Lammens M, Lucking CB, Denefle P, Wesseling P, Booij J, et al. Clinical and pathologic abnormalities in a family with parkinsonism and parkin gene mutations. Neurology. 2001;56(4):555–557. doi: 10.1212/wnl.56.4.555. [DOI] [PubMed] [Google Scholar]
- 154.Imai Y, Soda M, Inoue H, Hattori N, Mizuno Y, Takahashi R. An unfolded putative transmembrane polypeptide, which can lead to endoplasmic reticulum stress, is a substrate of Parkin. Cell. 2001;105(7):891–902. doi: 10.1016/s0092-8674(01)00407-x. [DOI] [PubMed] [Google Scholar]
- 155.Staropoli JF, McDermott C, Martinat C, Schulman B, Demireva E, Abeliovich A. Parkin is a component of an SCF-like ubiquitin ligase complex and protects postmitotic neurons from kainate excitotoxicity. Neuron. 2003;37(5):735–749. doi: 10.1016/s0896-6273(03)00084-9. [DOI] [PubMed] [Google Scholar]
- 156.Shimura H, Schlossmacher MG, Hattori N, Frosch MP, Trockenbacher A, Schneider R, et al. Ubiquitination of a new form of alpha-synuclein by parkin from human brain: implications for Parkinson's disease. Science. 2001;293(5528):263–269. doi: 10.1126/science.1060627. [DOI] [PubMed] [Google Scholar]
- 157.Choi P, Golts N, Snyder H, Chong M, Petrucelli L, Hardy J, et al. Co-association of parkin and alpha-synuclein. Neuroreport. 2001;12(13):2839–2843. doi: 10.1097/00001756-200109170-00017. [DOI] [PubMed] [Google Scholar]
- 158.Kawahara K, Hashimoto M, Bar-On P, Ho GJ, Crews L, Mizuno H, et al. alpha-Synuclein aggregates interfere with Parkin solubility and distribution: role in the pathogenesis of Parkinson disease. J Biol Chem. 2008;283(11):6979–6987. doi: 10.1074/jbc.M710418200. [DOI] [PubMed] [Google Scholar]
- 159.Choi P, Ostrerova-Golts N, Sparkman D, Cochran E, Lee JM, Wolozin B. Parkin is metabolized by the ubiquitin/proteosome system. Neuroreport. 2000;11(12):2635–2638. doi: 10.1097/00001756-200008210-00006. [DOI] [PubMed] [Google Scholar]
- 160.Kahle PJ, Haass C. How does parkin ligate ubiquitin to Parkinson's disease? EMBO Rep. 2004;5(7):681–685. doi: 10.1038/sj.embor.7400188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Snyder H, Wolozin B. Pathological proteins in Parkinson's disease: focus on the proteasome. J Mol Neurosci. 2004;24(3):425–442. doi: 10.1385/JMN:24:3:425. [DOI] [PubMed] [Google Scholar]
- 162.Zhang Y, Gao J, Chung KK, Huang H, Dawson VL, Dawson TM. Parkin functions as an E2-dependent ubiquitin- protein ligase and promotes the degradation of the synaptic vesicle-associated protein, CDCrel-1. Proc Natl Acad Sci U S A. 2000;97(24):13354–13359. doi: 10.1073/pnas.240347797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Springer W, Hoppe T, Schmidt E, Baumeister R. A Caenorhabditis elegans Parkin mutant with altered solubility couples alpha-synuclein aggregation to proteotoxic stress. Hum Mol Genet. 2005;14(22):3407–3423. doi: 10.1093/hmg/ddi371. [DOI] [PubMed] [Google Scholar]
- 164.Schlehe JS, Lutz AK, Pilsl A, Lammermann K, Grgur K, Henn IH, et al. Aberrant folding of pathogenic Parkin mutants: aggregation versus degradation. J Biol Chem. 2008;283(20):13771–13779. doi: 10.1074/jbc.M707494200. [DOI] [PubMed] [Google Scholar]
- 165.Wang C, Tan JM, Ho MW, Zaiden N, Wong SH, Chew CL, et al. Alterations in the solubility and intracellular localization of parkin by several familial Parkinson's disease-linked point mutations. J Neurochem. 2005;93(2):422–431. doi: 10.1111/j.1471-4159.2005.03023.x. [DOI] [PubMed] [Google Scholar]
- 166.Chung KK, Thomas B, Li X, Pletnikova O, Troncoso JC, Marsh L, et al. S-nitrosylation of parkin regulates ubiquitination and compromises parkin's protective function. Science. 2004;304(5675):1328–1331. doi: 10.1126/science.1093891. [DOI] [PubMed] [Google Scholar]
- 167.Pawlyk AC, Giasson BI, Sampathu DM, Perez FA, Lim KL, Dawson VL, et al. Novel monoclonal antibodies demonstrate biochemical variation of brain parkin with age. J Biol Chem. 2003;278(48):48120–48128. doi: 10.1074/jbc.M306889200. [DOI] [PubMed] [Google Scholar]
- 168.Darios F, Corti O, Lucking CB, Hampe C, Muriel MP, Abbas N, et al. Parkin prevents mitochondrial swelling and cytochrome c release in mitochondria-dependent cell death. Hum Mol Genet. 2003;12(5):517–526. doi: 10.1093/hmg/ddg044. [DOI] [PubMed] [Google Scholar]
- 169.Higashi Y, Asanuma M, Miyazaki I, Hattori N, Mizuno Y, Ogawa N. Parkin attenuates manganese-induced dopaminergic cell death. J Neurochem. 2004;89(6):1490–1497. doi: 10.1111/j.1471-4159.2004.02445.x. [DOI] [PubMed] [Google Scholar]
- 170.Imai Y, Soda M, Takahashi R. Parkin suppresses unfolded protein stress-induced cell death through its E3 ubiquitin-protein ligase activity. J Biol Chem. 2000;275(46):35661–35664. doi: 10.1074/jbc.C000447200. [DOI] [PubMed] [Google Scholar]
- 171.Jiang H, Ren Y, Zhao J, Feng J. Parkin protects human dopaminergic neuroblastoma cells against dopamine-induced apoptosis. Hum Mol Genet. 2004;13(16):1745–1754. doi: 10.1093/hmg/ddh180. [DOI] [PubMed] [Google Scholar]
- 172.Ko HS, von Coelln R, Sriram SR, Kim SW, Chung KK, Pletnikova O, et al. Accumulation of the authentic parkin substrate aminoacyl-tRNA synthetase cofactor, p38/JTV-1, leads to catecholaminergic cell death. J Neurosci. 2005;25(35):7968–7978. doi: 10.1523/JNEUROSCI.2172-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ledesma MD, Galvan C, Hellias B, Dotti C, Jensen PH. Astrocytic but not neuronal increased expression and redistribution of parkin during unfolded protein stress. J Neurochem. 2002;83(6):1431–1440. doi: 10.1046/j.1471-4159.2002.01253.x. [DOI] [PubMed] [Google Scholar]
- 174.Tsai YC, Fishman PS, Thakor NV, Oyler GA. Parkin facilitates the elimination of expanded polyglutamine proteins and leads to preservation of proteasome function. J Biol Chem. 2003;278(24):22044–22055. doi: 10.1074/jbc.M212235200. [DOI] [PubMed] [Google Scholar]
- 175.Yang Y, Nishimura I, Imai Y, Takahashi R, Lu B. Parkin suppresses dopaminergic neuron-selective neurotoxicity induced by Pael-R in Drosophila. Neuron. 2003;37(6):911–924. doi: 10.1016/s0896-6273(03)00143-0. [DOI] [PubMed] [Google Scholar]
- 176.Whitworth AJ, Theodore DA, Greene JC, Benes H, Wes PD, Pallanck LJ. Increased glutathione S-transferase activity rescues dopaminergic neuron loss in a Drosophila model of Parkinson's disease. Proc Natl Acad Sci U S A. 2005;102(22):8024–8029. doi: 10.1073/pnas.0501078102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Zhou W, Freed CR. DJ-1 up-regulates glutathione synthesis during oxidative stress and inhibits A53T alpha-synuclein toxicity. J Biol Chem. 2005;280(52):43150–43158. doi: 10.1074/jbc.M507124200. [DOI] [PubMed] [Google Scholar]
- 178.LaVoie MJ, Ostaszewski BL, Weihofen A, Schlossmacher MG, Selkoe DJ. Dopamine covalently modifies and functionally inactivates parkin. Nat Med. 2005;11(11):1214–1221. doi: 10.1038/nm1314. [DOI] [PubMed] [Google Scholar]
- 179.Yu LY, Saarma M, Arumae U. Death receptors and caspases but not mitochondria are activated in the GDNF- or BDNF-deprived dopaminergic neurons. J Neurosci. 2008;28(30):7467–7475. doi: 10.1523/JNEUROSCI.1877-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kahns S, Kalai M, Jakobsen LD, Clark BF, Vandenabeele P, Jensen PH. Caspase-1 and caspase-8 cleave and inactivate cellular parkin. J Biol Chem. 2003;278(26):23376–23380. doi: 10.1074/jbc.M300495200. [DOI] [PubMed] [Google Scholar]
- 181.Chauhan NB, Siegel GJ, Lee JM. Depletion of glial cell line-derived neurotrophic factor in substantia nigra neurons of Parkinson's disease brain. J Chem Neuroanat. 2001;21(4):277–288. doi: 10.1016/s0891-0618(01)00115-6. [DOI] [PubMed] [Google Scholar]
- 182.Siegel GJ, Chauhan NB. Neurotrophic factors in Alzheimer's and Parkinson's disease brain. Brain Res Brain Res Rev. 2000;33(23):199–227. doi: 10.1016/s0165-0173(00)00030-8. [DOI] [PubMed] [Google Scholar]
- 183.Weinreb O, Amit T, Bar-Am O, Youdim MB. Induction of neurotrophic factors GDNF and BDNF associated with the mechanism of neurorescue action of rasagiline and ladostigil: new insights and implications for therapy. Ann N Y Acad Sci. 2007;1122:155–168. doi: 10.1196/annals.1403.011. [DOI] [PubMed] [Google Scholar]
- 184.Shimura H, Hattori N, Kubo S, Yoshikawa M, Kitada T, Matsumine H, et al. Immunohistochemical and subcellular localization of Parkin protein: absence of protein in autosomal recessive juvenile parkinsonism patients. Ann Neurol. 1999;45(5):668–672. doi: 10.1002/1531-8249(199905)45:5<668::aid-ana19>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 185.Taira T, Saito Y, Niki T, Iguchi-Ariga SM, Takahashi K, Ariga H. DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep. 2004;5(2):213–218. doi: 10.1038/sj.embor.7400074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Takahashi K, Taira T, Niki T, Seino C, Iguchi-Ariga SM, Ariga H. DJ-1 positively regulates the androgen receptor by impairing the binding of PIASx alpha to the receptor. J Biol Chem. 2001;276(40):37556–37563. doi: 10.1074/jbc.M101730200. [DOI] [PubMed] [Google Scholar]
- 187.Xu J, Zhong N, Wang H, Elias JE, Kim CY, Woldman I, et al. The Parkinson's disease-associated DJ-1 protein is a transcriptional co-activator that protects against neuronal apoptosis. Hum Mol Genet. 2005;14(9):1231–1241. doi: 10.1093/hmg/ddi134. [DOI] [PubMed] [Google Scholar]
- 188.Zhong N, Kim CY, Rizzu P, Geula C, Porter DR, Pothos EN, et al. DJ-1 transcriptionally up-regulates the human tyrosine hydroxylase by inhibiting the sumoylation of pyrimidine tract-binding protein-associated splicing factor. J Biol Chem. 2006;281(30):20940–20948. doi: 10.1074/jbc.M601935200. [DOI] [PubMed] [Google Scholar]
- 189.Abou-Sleiman PM, Healy DG, Quinn N, Lees AJ, Wood NW. The role of pathogenic DJ-1 mutations in Parkinson's disease. Ann Neurol. 2003;54(3):283–286. doi: 10.1002/ana.10675. [DOI] [PubMed] [Google Scholar]
- 190.Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science. 2003;299(5604):256–259. doi: 10.1126/science.1077209. [DOI] [PubMed] [Google Scholar]
- 191.Clark LN, Afridi S, Mejia-Santana H, Harris J, Louis ED, Cote LJ, et al. Analysis of an early-onset Parkinson's disease cohort for DJ-1 mutations. Mov Disord. 2004;19(7):796–800. doi: 10.1002/mds.20131. [DOI] [PubMed] [Google Scholar]
- 192.Hague S, Rogaeva E, Hernandez D, Gulick C, Singleton A, Hanson M, et al. Early-onset Parkinson's disease caused by a compound heterozygous DJ-1 mutation. Ann Neurol. 2003;54(2):271–274. doi: 10.1002/ana.10663. [DOI] [PubMed] [Google Scholar]
- 193.Shendelman S, Jonason A, Martinat C, Leete T, Abeliovich A. DJ-1 is a redox-dependent molecular chaperone that inhibits alpha-synuclein aggregate formation. PLoS Biol. 2004;2(11):e362. doi: 10.1371/journal.pbio.0020362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Martinat C, Shendelman S, Jonason A, Leete T, Beal MF, Yang L, et al. Sensitivity to oxidative stress in DJ-1-deficient dopamine neurons: an ES-derived cell model of primary Parkinsonism. PLoS Biol. 2004;2(11):e327. doi: 10.1371/journal.pbio.0020327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Yokota T, Sugawara K, Ito K, Takahashi R, Ariga H, Mizusawa H. Down regulation of DJ-1 enhances cell death by oxidative stress, ER stress, and proteasome inhibition. Biochem Biophys Res Commun. 2003;312(4):1342–1348. doi: 10.1016/j.bbrc.2003.11.056. [DOI] [PubMed] [Google Scholar]
- 196.Sian J, Dexter DT, Lees AJ, Daniel S, Agid Y, Javoy-Agid F, et al. Alterations in glutathione levels in Parkinson's disease and other neurodegenerative disorders affecting basal ganglia. Ann Neurol. 1994;36(3):348–355. doi: 10.1002/ana.410360305. [DOI] [PubMed] [Google Scholar]
- 197.Burwell LS, Nadtochiy SM, Tompkins AJ, Young S, Brookes PS. Direct evidence for S-nitrosation of mitochondrial complex I. Biochem J. 2006;394(Pt 3):627–634. doi: 10.1042/BJ20051435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Chinta SJ, Kumar MJ, Hsu M, Rajagopalan S, Kaur D, Rane A, et al. Inducible alterations of glutathione levels in adult dopaminergic midbrain neurons result in nigrostriatal degeneration. J Neurosci. 2007;27(51):13997–14006. doi: 10.1523/JNEUROSCI.3885-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Chinta SJ, Rajagopalan S, Butterfield DA, Andersen JK. In vitro and in vivo neuroprotection by gamma-glutamylcysteine ethyl ester against MPTP: relevance to the role of glutathione in Parkinson's disease. Neurosci Lett. 2006;402(12):137–141. doi: 10.1016/j.neulet.2006.03.056. [DOI] [PubMed] [Google Scholar]
- 200.Hsu M, Srinivas B, Kumar J, Subramanian R, Andersen J. Glutathione depletion resulting in selective mitochondrial complex I inhibition in dopaminergic cells is via an NO-mediated pathway not involving peroxynitrite: implications for Parkinson's disease. J Neurochem. 2005;92(5):1091–1103. doi: 10.1111/j.1471-4159.2004.02929.x. [DOI] [PubMed] [Google Scholar]
- 201.Kwak MK, Wakabayashi N, Greenlaw JL, Yamamoto M, Kensler TW. Antioxidants enhance mammalian proteasome expression through the Keap1-Nrf2 signaling pathway. Mol Cell Biol. 2003;23(23):8786–8794. doi: 10.1128/MCB.23.23.8786-8794.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Itoh K, Chiba T, Takahashi S, Ishii T, Igarashi K, Katoh Y, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236(2):313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 203.Kwak MK, Itoh K, Yamamoto M, Sutter TR, Kensler TW. Role of transcription factor Nrf2 in the induction of hepatic phase 2 and antioxidative enzymes in vivo by the cancer chemoprotective agent, 3H-1, 2-dimethiole-3-thione. Mol Med. 2001;7(2):135–145. [PMC free article] [PubMed] [Google Scholar]
- 204.Thimmulappa RK, Mai KH, Srisuma S, Kensler TW, Yamamoto M, Biswal S. Identification of Nrf2-regulated genes induced by the chemopreventive agent sulforaphane by oligonucleotide microarray. Cancer Res. 2002;62(18):5196–5203. [PubMed] [Google Scholar]
- 205.Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc Natl Acad Sci U S A. 1997;94(10):5361–5366. doi: 10.1073/pnas.94.10.5361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999;13(1):76–86. doi: 10.1101/gad.13.1.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Clements CM, McNally RS, Conti BJ, Mak TW, Ting JP. DJ-1, a cancer- and Parkinson's disease-associated protein, stabilizes the antioxidant transcriptional master regulator Nrf2. Proc Natl Acad Sci U S A. 2006;103(41):15091–15096. doi: 10.1073/pnas.0607260103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Stewart D, Killeen E, Naquin R, Alam S, Alam J. Degradation of transcription factor Nrf2 via the ubiquitin-proteasome pathway and stabilization by cadmium. J Biol Chem. 2003;278(4):2396–2402. doi: 10.1074/jbc.M209195200. [DOI] [PubMed] [Google Scholar]
- 209.Yang W, Chen L, Ding Y, Zhuang X, Kang UJ. Paraquat induces dopaminergic dysfunction and proteasome impairment in DJ-1-deficient mice. Hum Mol Genet. 2007;16(23):2900–2910. doi: 10.1093/hmg/ddm249. [DOI] [PubMed] [Google Scholar]
- 210.Alegre-Abarrategui J, Ansorge O, Esiri M, Wade-Martins R. LRRK2 is a component of granular alpha-synuclein pathology in the brainstem of Parkinson's disease. Neuropathol Appl Neurobiol. 2008;34(3):272–283. doi: 10.1111/j.1365-2990.2007.00888.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Perry G, Zhu X, Babar AK, Siedlak SL, Yang Q, Ito G, et al. Leucine-rich repeat kinase 2 colocalizes with alpha-synuclein in Parkinson's disease, but not tau-containing deposits in tauopathies. Neurodegener Dis. 2008;5(34):222–224. doi: 10.1159/000113708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Funayama M, Hasegawa K, Kowa H, Saito M, Tsuji S, Obata F. A new locus for Parkinson's disease (PARK8) maps to chromosome 12p11.2-q13.1. Ann Neurol. 2002;51(3):296–301. doi: 10.1002/ana.10113. [DOI] [PubMed] [Google Scholar]
- 213.Gilks WP, Abou-Sleiman PM, Gandhi S, Jain S, Singleton A, Lees AJ, et al. A common LRRK2 mutation in idiopathic Parkinson's disease. Lancet. 2005;365(9457):415–416. doi: 10.1016/S0140-6736(05)17830-1. [DOI] [PubMed] [Google Scholar]
- 214.Rajput A, Dickson DW, Robinson CA, Ross OA, Dachsel JC, Lincoln SJ, et al. Parkinsonism, Lrrk2 G2019S, and tau neuropathology. Neurology. 2006;67(8):1506–1508. doi: 10.1212/01.wnl.0000240220.33950.0c. [DOI] [PubMed] [Google Scholar]
- 215.Gaig C, Marti MJ, Ezquerra M, Rey MJ, Cardozo A, Tolosa E. G2019S LRRK2 mutation causing Parkinson's disease without Lewy bodies. J Neurol Neurosurg Psychiatry. 2007;78(6):626–628. doi: 10.1136/jnnp.2006.107904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Beach TG, Walker DG, Sue LI, Newell A, Adler CC, Joyce JN. Substantia nigra Marinesco bodies are associated with decreased striatal expression of dopaminergic markers. J Neuropathol Exp Neurol. 2004;63(4):329–337. doi: 10.1093/jnen/63.4.329. [DOI] [PubMed] [Google Scholar]
- 217.Smith WW, Pei Z, Jiang H, Moore DJ, Liang Y, West AB, et al. Leucine-rich repeat kinase 2 (LRRK2) interacts with parkin, and mutant LRRK2 induces neuronal degeneration. Proc Natl Acad Sci U S A. 2005;102(51):18676–18681. doi: 10.1073/pnas.0508052102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Gloeckner CJ, Kinkl N, Schumacher A, Braun RJ, O'Neill E, Meitinger T, et al. The Parkinson disease causing LRRK2 mutation I2020T is associated with increased kinase activity. Hum Mol Genet. 2006;15(2):223–232. doi: 10.1093/hmg/ddi439. [DOI] [PubMed] [Google Scholar]
- 219.West AB, Moore DJ, Biskup S, Bugayenko A, Smith WW, Ross CA, et al. Parkinson's disease-associated mutations in leucine-rich repeat kinase 2 augment kinase activity. Proc Natl Acad Sci U S A. 2005;102(46):16842–16847. doi: 10.1073/pnas.0507360102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Wang L, Xie C, Greggio E, Parisiadou L, Shim H, Sun L, et al. The chaperone activity of heat shock protein 90 is critical for maintaining the stability of leucine-rich repeat kinase 2. J Neurosci. 2008;28(13):3384–3391. doi: 10.1523/JNEUROSCI.0185-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Gandhi S, Muqit MM, Stanyer L, Healy DG, Abou-Sleiman PM, Hargreaves I, et al. PINK1 protein in normal human brain and Parkinson's disease. Brain. 2006;129(Pt 7):1720–1731. doi: 10.1093/brain/awl114. [DOI] [PubMed] [Google Scholar]
- 222.Muqit MM, Abou-Sleiman PM, Saurin AT, Harvey K, Gandhi S, Deas E, et al. Altered cleavage and localization of PINK1 to aggresomes in the presence of proteasomal stress. J Neurochem. 2006;98(1):156–169. doi: 10.1111/j.1471-4159.2006.03845.x. [DOI] [PubMed] [Google Scholar]
- 223.Silvestri L, Caputo V, Bellacchio E, Atorino L, Dallapiccola B, Valente EM, et al. Mitochondrial import and enzymatic activity of PINK1 mutants associated to recessive parkinsonism. Hum Mol Genet. 2005;14(22):3477–3492. doi: 10.1093/hmg/ddi377. [DOI] [PubMed] [Google Scholar]
- 224.Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, et al. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004;304(5674):1158–1160. doi: 10.1126/science.1096284. [DOI] [PubMed] [Google Scholar]
- 225.Grunewald A, Breedveld GJ, Lohmann-Hedrich K, Rohe CF, Konig IR, Hagenah J, et al. Biological effects of the PINK1 c.1366C>T mutation: implications in Parkinson disease pathogenesis. Neurogenetics. 2007;8(2):103–109. doi: 10.1007/s10048-006-0072-y. [DOI] [PubMed] [Google Scholar]
- 226.Beilina A, Van Der Brug M, Ahmad R, Kesavapany S, Miller DW, Petsko GA, et al. Mutations in PTEN-induced putative kinase 1 associated with recessive parkinsonism have differential effects on protein stability. Proc Natl Acad Sci U S A. 2005;102(16):5703–5708. doi: 10.1073/pnas.0500617102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Sim CH, Lio DS, Mok SS, Masters CL, Hill AF, Culvenor JG, et al. C-terminal truncation and Parkinson's disease-associated mutations down-regulate the protein serine/threonine kinase activity of PTEN-induced kinase-1. Hum Mol Genet. 2006;15(21):3251–3262. doi: 10.1093/hmg/ddl398. [DOI] [PubMed] [Google Scholar]
- 228.Clark IE, Dodson MW, Jiang C, Cao JH, Huh JR, Seol JH, et al. Drosophila pink1 is required for mitochondrial function and interacts genetically with parkin. Nature. 2006;441(7097):1162–1166. doi: 10.1038/nature04779. [DOI] [PubMed] [Google Scholar]
- 229.Park J, Lee SB, Lee S, Kim Y, Song S, Kim S, et al. Mitochondrial dysfunction in Drosophila PINK1 mutants is complemented by parkin. Nature. 2006;441(7097):1157–1161. doi: 10.1038/nature04788. [DOI] [PubMed] [Google Scholar]
- 230.Greene JC, Whitworth AJ, Kuo I, Andrews LA, Feany MB, Pallanck LJ. Mitochondrial pathology and apoptotic muscle degeneration in Drosophila parkin mutants. Proc Natl Acad Sci U S A. 2003;100(7):4078–4083. doi: 10.1073/pnas.0737556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Pesah Y, Pham T, Burgess H, Middlebrooks B, Verstreken P, Zhou Y, et al. Drosophila parkin mutants have decreased mass and cell size and increased sensitivity to oxygen radical stress. Development. 2004;131(9):2183–2194. doi: 10.1242/dev.01095. [DOI] [PubMed] [Google Scholar]
- 232.Wood-Kaczmar A, Gandhi S, Yao Z, Abramov AS, Miljan EA, Keen G, et al. PINK1 is necessary for long term survival and mitochondrial function in human dopaminergic neurons. PLoS ONE. 2008;3(6):e2455. doi: 10.1371/journal.pone.0002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Petit A, Kawarai T, Paitel E, Sanjo N, Maj M, Scheid M, et al. Wild-type PINK1 prevents basal and induced neuronal apoptosis, a protective effect abrogated by Parkinson disease-related mutations. J Biol Chem. 2005;280(40):34025–34032. doi: 10.1074/jbc.M505143200. [DOI] [PubMed] [Google Scholar]
- 234.Wang HL, Chou AH, Yeh TH, Li AH, Chen YL, Kuo YL, et al. PINK1 mutants associated with recessive Parkinson's disease are defective in inhibiting mitochondrial release of cytochrome c. Neurobiol Dis. 2007;28(2):216–226. doi: 10.1016/j.nbd.2007.07.010. [DOI] [PubMed] [Google Scholar]
- 235.Abou-Sleiman PM, Muqit MM, McDonald NQ, Yang YX, Gandhi S, Healy DG, et al. A heterozygous effect for PINK1 mutations in Parkinson's disease? Ann Neurol. 2006;60(4):414–419. doi: 10.1002/ana.20960. [DOI] [PubMed] [Google Scholar]
- 236.Valente EM, Salvi S, Ialongo T, Marongiu R, Elia AE, Caputo V, et al. PINK1 mutations are associated with sporadic early-onset parkinsonism. Ann Neurol. 2004;56(3):336–341. doi: 10.1002/ana.20256. [DOI] [PubMed] [Google Scholar]