An Arabidopsis plastid triose phosphate isomerase mutant was reduced in transcript, protein, and activity, resulting in a severely stunted plant at the transition from heterotrophic to autotrophic growth. Accumulation of dihydroxyacetone phosphate and methylglyoxal to toxic levels is proposed as a cause of this phenotype, as exogenous application of these metabolites mimicked the phenotype.
Abstract
During postgerminative seedling establishment, reserves stored during seed filling are mobilized to provide energy and carbon for the growing seedling until autotrophic growth is possible. A plastid isoform of triose phosphate isomerase (pdTPI) plays a crucial role in this transition from heterotrophic to autotrophic growth. A T-DNA insertion in Arabidopsis thaliana pdTPI resulted in a fivefold reduction in transcript, reduced TPI activity, and a severely stunted and chlorotic seedling that accumulated dihydroxyacetone phosphate (DHAP), glycerol, and glycerol-3-phosphate. Methylglyoxal (MG), a by-product of DHAP, also accumulated in the pdtpi mutant. Wild-type seed sown in the presence of any of these four metabolites resulted in a phenocopy of this pdtpi mutant, although MG and DHAP were the most effective based upon dosage. These metabolites (except MG) are by-products of triacylglycerol mobilization and precursors for glycerolipid synthesis, suggesting that lipid metabolism may also be affected. Lipid profiling revealed lower monogalactosyl but higher digalactosyl lipids. It is unclear whether the change in lipid composition is a direct or indirect consequence of the pdtpi mutation, as ribulose-1,5-bis-phosphate carboxylase/oxygenase expression, chloroplast morphology, and starch synthesis are also defective in this mutant. We propose that DHAP and MG accumulation in developing plastids delays the transition from heterotrophic to autotrophic growth, possibly due to MG toxicity.
INTRODUCTION
During Arabidopsis thaliana seed development, large quantities of carbon, nitrogen, and other minor nutrients are stored from the mother plant (Graham, 2008). Starch accumulates transiently, eventually being converted to triacylglycerol (TAG), accounting for up to 45% of the dry mass of the mature Arabidopsis seed (O'Neill et al., 2003). These reserves are used to fuel postgerminative seedling establishment until chloroplast and leaf development are complete and photosynthesis can begin. Germination initiates with release from dormancy and seed imbibition and is finished when the radicle emerges through the seed coat (Bewley, 1997). Establishment involves the transition from a heterotrophic to a photoautotrophic seedling. In wild-type) Arabidopsis seedlings, germination is typically complete after 24 h and is largely driven by the metabolism of storage reserves other than lipids, while seed oil is used for subsequent seedling establishment (Penfield et al., 2005; Cernac et al., 2006).
Following germination, TAG is broken down and carbon used to support seedling growth (Bewley and Black, 1985). The initial step in this process is catalyzed by TAG lipase (Eastmond, 2006), which hydrolyzes TAG to yield free fatty acids and glycerol in a 3:1 molar ratio. Free fatty acids are then transferred to the glyoxysome and activated to acyl-CoAs for subsequent catabolism by β -oxidation to yield acetyl-CoA (Graham and Eastmond, 2002). Most of the acetyl-CoA produced is ultimately converted to sugars by the glyoxylate cycle and gluconeogenesis (Bunkelmann and Trelease, 1996). Radiolabeling studies have established that glycerol is also converted to sucrose in germinating seeds (Beevers, 1956). Glycerol is first phosphorylated to glycerol-3-phosphate (G-3-P) by glycerol kinase (Eastmond, 2004) and is then converted to dihydroxyacetone phosphate (DHAP) by a G-3-P dehydrogenase (Lin, 1976, 1977). The DHAP generated is available for glycolysis or converted to hexoses by gluconeogenesis (Beevers, 1956).
A genetic study has demonstrated that TAG mobilization plays an essential role for early seedling development. SUGAR-DEPENDENT1 (SDP1) encodes a patatin domain TAG lipase that initiates storage oil breakdown in germinating Arabidopsis seeds (Eastmond, 2006). Although not significantly impaired during germination, the sdp1 mutant does exhibit a much slower rate of postgerminative growth than the wild type. This phenotype can be rescued by applying an alternate carbon source, such as sucrose. Once sdp1 mutant seedlings develop photosynthetic activity, however, they grow normally and are indistinguishable from the wild type throughout the rest of the life cycle.
Genetic studies of a number of Arabidopsis mutants have also shown that fatty acid catabolism by β -oxidation is essential for embryo development, seedling establishment, and breakdown of stored TAG (Hayashi, et al., 1998; Cornah et al., 2004; Eastmond et al., 2000; Fulda et al., 2004; Adham et al., 2005; Pinfield-Wells et al., 2005; Rylott et al., 2006). Details of the pathway of glycerol catabolism in plants are also beginning to emerge (Eastmond, 2004). From genetic screenings of glycerol-insensitive (gli) mutants, GLI1 has been shown to encode glycerol kinase, which initiates glycerol catabolism by converting it into G-3-P. Following germination, gli1 mutant seedlings transiently accumulate glycerol derived from the breakdown of TAG and are more resistant to hyperosmotic stress, salt and oxidative stress, freezing, and desiccation (Eastmond, 2004).
After germination, seedling establishment requires a transition from heterotrophic to autotrophic growth to sustain plant growth once storage reserves are used. This likely involves multiple plastid biosynthetic pathways. In plants, triose phosphate isomerase (TPI; EC 5.3.1.1) is involved in several metabolic pathways operating during this transition, including glycolysis, gluconeogenesis, and Calvin cycle. In this article, we present findings demonstrating the importance of a plastid isoform of TPI (pdTPI) in Arabidopsis. A T-DNA insertion mutant (pdtpi) in intron 1 significantly reduces gene expression and plastid TPI activity, resulting in a severely stunted plant that does not reach reproductive maturity. Reduction of plastid TPI activity results in the buildup of glycerol, G-3-P, DHAP, and MG following seed germination. We demonstrate that the high concentrations of these intermediates impair seedling establishment and chloroplast development. These observations demonstrate that pdTPI plays an essential role in seedling establishment and chloroplast development in Arabidopsis.
RESULTS
At2g21170 Gene Encodes Two Plastid TPI Splice Variants
The Arabidopsis genome contains two genes annotated as TPI: At2g21170 and At3g55440. One of these genes, At2g21170, is predicted to encode a plastid-targeted protein based upon an extra 65 amino acids at the N terminus (see Supplemental Figure 1A online), which organellar prediction algorithms suggest is a plastid targeting peptide. An extensive query of the public Arabidopsis EST databases revealed two splice variants for this putative plastid TPI mRNA (At2g21170.1 and At2g21170.2). Unlike At2g21170.1, At2g21170.2 has a 27-bp deletion that spans the second exon. To determine tissue-specific expression, a pair of primers was designed to amplify a 122-bp fragment from At2g21170.1 and a 95-bp fragment from At2g21170.2. The At2g21170.1 form was expressed in roots, leaves, stems, flowers, and siliques, with highest expression in leaves and flowers (see Supplemental Figure 2 online). By contrast, At2g21170.2 was weakly detected only in roots. Differential expression patterns for these two splice variants suggest organ-specific roles for these gene products.
The Gene Product for At2g21170.1 Localizes to Plastids
To experimentally verify the subcellular localization of pdTPI, organelle fractionation was performed with a transgenic line overexpressing the gene product of At2g21170.1 in frame with green fluorescent protein (GFP) at the 3 ′ end. Developing leaves were homogenized and fractionated into 5k g, 20k g, and 100k g pellets and the 100k g supernatant. Distribution of plastids and mitochondria within these fractions was estimated by immunoblot analyses using marker antibodies, plastid isoforms of pyruvate dehydrogenase (PDH) E1 α subunit, and mitochondrion pyruvate dehydrogenase α subunit, respectively (Mooney et al., 2002). Antibodies to GFP were used to detect pdTPI-GFP fusion protein expression. Immunoblot results reveal that pdTPI localizes to the 5k g chloroplast fraction (see Supplemental Figure 1B online). The chloroplast localization of pdTPI was further supported by in vivo imaging of GFP-tagged pdTPI (see Supplemental Figure 1C online).
Mutation in pdTPI Results in Stunted Growth and Abnormal Chloroplast Development
Two T-DNA insertion lines (Salk-003991 and Salk-106805) were identified for the cytoTPI gene locus At3g55440; however, homozygous lines showed no visible phenotype, and gene transcription was unaffected (see Supplemental Figure 3 online). Three T-DNA insertion lines were identified for gene At2g21170 (see Supplemental Figure 4A online). Homozygous T-DNA insertion lines were verified by PCR using a combination of T-DNA left border primer and gene-specific primers. One example is shown in Supplemental Figure 4B online for the Salk-152526 T-DNA insertion line. Further sequence analysis indicated that the T-DNA left border has a 91-bp deletion. PCR conducted using a T-DNA right border primer and a gene-specific primer failed to amplify any band from genomic DNA, possibly due to T-DNA right border deletion during the insertion.
Gene expression was not affected in either the Salk-022963 or the CS829061 homozygous T-DNA lines but was significantly attenuated in the Salk-152526 line (see Supplemental Figure 4C online). This line also produced a severely stunted growth phenotype and was renamed plastid TPI mutant or pdtpi. Homozygous T-DNA lines Salk-022963 and CS829061 did not show any abnormal growth phenotype.
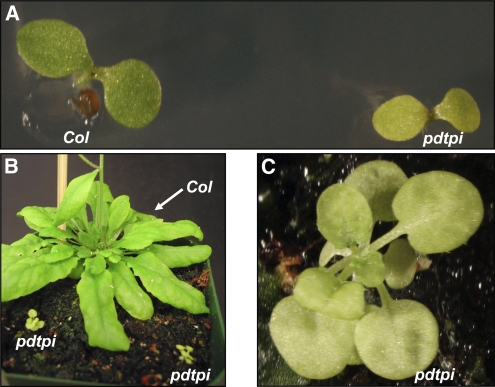
To further characterize this growth phenotype, pdtpi mutant and wild-type seeds were germinated on Murashige and Skoog (MS) plates. The pdtpi mutant seeds germinated normally, with a 97.6% germination rate (n = 320) compared with a 98.3% germination rate for the wild type (n = 366). Phenotypic differences between wild-type and pdtpi seedlings were observed beginning at 3 to 4 d postgermination. Mutant seedlings displayed pale cotyledons and shorter primary roots compared with wild-type seedlings (Figure 1A). At 10 d postgermination, wild-type seedlings were well established with multiple true green leaves and an extensive root system, while pdtpi seedlings had developed only a single, pale-green true leaf and did so at a much slower rate. At 10 weeks postgermination, wild-type plants began to develop reproductive shoots. The pdtpi plants, however, had diminutive pale-green leaves and a lower overall biomass compared with the wild type (Figures 1B and 1C). The pdtpi mutant plants were unable to transition into the reproductive stage prior to senescence.
Figure 1.
Phenotype of the pdtpi Mutant.
(A) Wild-type and pdtpi mutant plants germinated on MS plates for 5 d.
(B) Plants grown on soil for 10 weeks under a long-day photoperiod (14 h light/ 10 h dark).
(C) An enlarged view of the pdtpi mutant shown in (B).
The pdtpi heterozygous mutant was backcrossed three times with Columbia wild type, and the F1 population always displayed wild-type phenotype. The F2 population, however, showed a 3:1 segregation of green wild-type seedlings to pale-green mutants (216 wild type to 66 pdtpi mutants) when germinated on MS media, suggesting that pdtpi is a recessive, single gene mutation. The genotyping of 32 individual pale-green seedlings from the F2 population demonstrated that all are homozygous for the T-DNA insertion, indicating that the phenotype is tightly linked with the T-DNA insertion.
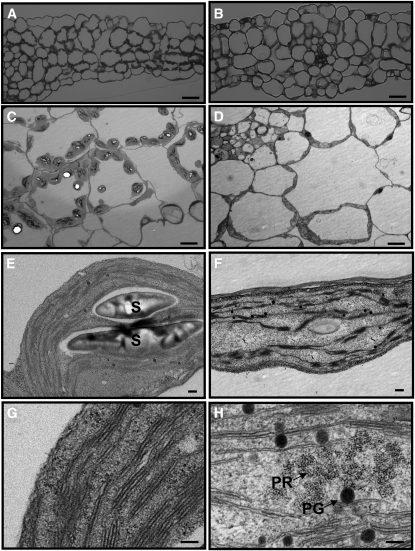
Cotyledons from 5-d-old seedlings were examined by light and electron microscopy. No apparent difference in cell size was observed between pdtpi and the wild type (Figures 2A and 2B). The number of chloroplasts per cell was much lower in pdtpi compared with the wild type (9.5 ± 3.0 chloroplasts versus 4.4 ± 0.9 for the wild type and pdtpi [n = 23], respectively) (Figures 2A and 2B). The reduced number and size of chloroplasts may account for the pale-green color of the pdtpi cotyledon. Examination of chloroplast membrane ultrastructure revealed portions of pdtpi chloroplasts contain prolamella bodies, a structure that typically exists in etioplasts (Figure 2H). Etioplasts are found in white or pale-yellow etiolated leaves and dark-adapted, green tissues. The plastoglobule, a lipoprotein subcompartment, is significantly larger in the mutant compared with the wild type (Figure 2H). In comparison, wild-type chloroplasts have developed stacked grana and unstacked stroma thylakoid membranes (Figure 2G). These observations suggest that chloroplast differentiation in pdtpi is delayed during the early stages of the greening process. In accordance with its changing structure, pdtpi accumulates fewer starch granules in the chloroplast stroma compared with the wild type (Figures 2E and 2F), suggesting that photosynthetic capacity in pdtpi is compromised.
Figure 2.
Chloroplast Structures of the Wild Type and the pdtpi Mutant.
Panels show leaf and chloroplast structure of Columbia wild type ([A], [C], [E], and [G]) and pdtpi mutant ([B], [D], [F], and [H]). Plants were grown on MS plate for 5 d under continuous light and were then harvested for microscopy. Bars = 40 μ m in (A) and (B), 10 μ m in (C) and (D), 0.2 μ m in (E) and (F), and 100 nm in (G) and (H). Starch grains (S), plastoglobule (PG), and prolamellar body (PR) are indicated.
(A) and (B) Leaf structure observed under a light microscope (magnification × 500).
(C) to (H) Chloroplast ultrastructure from transmission electron microscopy.
Exogenous Glycolytic Intermediates Do Not Rescue pdtpi Phenotype
TPI catalyzes the reversible isomerization of DHAP to glyceraldehyde-3-phosphate (GAP), a reaction that allows DHAP to enter glycolysis for energy production. Reduced TPI expression therefore could result in a shortage of carbon intermediates and consequently the mutant phenotype. To test this possibility, both wild-type and pdtpi seeds were germinated on MS agar plates supplemented with carbohydrates and glycolytic pathway intermediates, such as sucrose, glucose, fructose-1,6-bisphosphate, DHAP, GAP, and 3-phosphoglyceric acid (PGA). Seeds were germinated under continuous light for 5 d, and then the phenotype (i.e., color of the cotyledon and the length of the primary root) was scored. Surprisingly, the pdtpi phenotype could not be rescued by exogenously supplying any of these compounds (see Supplemental Figure 5A online). DHAP, at 1.0 mM, not only failed to mitigate the pdtpi phenotype but also strongly inhibited root elongation both in the wild type and in the pdtpi mutant (see Supplemental Figure 5A online).
To differentiate the transition from heterotrophic to autotrophic growth, wild-type and pdtpi seeds were initially germinated in MS media under darkness for 5 d, transferred to MS plates supplemented with different glycolytic intermediates (as described above), and grown an additional 2 d under continuous white light. Interestingly, dark-grown pdtpi seedlings were indistinguishable from wild-type etiolated seedlings, both having elongated hypocotyls, small, folded cotyledons, and a short root—all features typical of skotomorphogenesis (see Supplemental Figure 5B online, left). Upon transition to light, pdtpi etiolated seedlings began to initiate cotyledon expansion and greening at a slower pace than the wild type. In wild-type seedlings, the cotyledon opened, expanded, and then greened within 2 d. By contrast, the cotyledon of pdtpi mutant seedlings opened normally but then failed to expand and to initiate greening at the same rate as the wild type. The primary roots of pdtpi mutant seedlings also did not elongate at the same rate as wild-type seedlings upon transition to light (see Supplemental Figure 5B online). Exogenous sucrose, glucose, fructose-1,6-bisphosphate, DHAP, GAP, or PGA failed to rescue the pdtpi capacity to initiate cotyledon greening. Interestingly, wild-type seedlings grown on MS plates containing 1.0 mM DHAP showed normal greening of the cotyledon but slower cotyledon expansion and primary root elongation compared with wild-type seedlings grown on MS plates without addition of any intermediates. These observations support the notion that the pdtpi mutant is capable of mobilizing storage reserves in dark-adapted conditions but upon transition to autotrophic growth begins to show the chlorotic, delayed growth phenotype.
TPI Activity Is Reduced in pdtpi, Resulting in the Accumulation of Glycerol, G-3-P, and DHAP
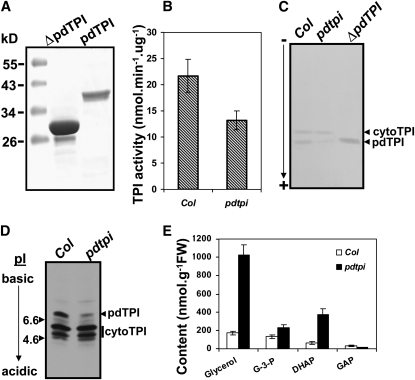
To experimentally demonstrate that gene At2g27710 encodes an enzyme with TPI activity, the open reading frame was cloned into a bacterial expression vector and recombinant protein expressed and purified under native condition from Escherichia coli (Figure 3A). In one construct, the N-terminal 59 amino acids, encoding a putative plastid targeting peptide, was deleted ( Δ pdTPI); the other construct expressed the full-length open reading frame. Both purified recombinant proteins showed TPI activity using an enzyme-linked spectrophotometric assay (388 ± 32.2 nmol min−1 μ g−1 for pdTPI and 469 ± 43.9 nmol min−1 μ g−1 for Δ pdTPI) with values close or exceeding those reported previously (Tomlinson and Turner, 1979; Tang et al., 1999).
Figure 3.
TPI Activity Was Reduced and Metabolic Intermediate Pools Were Altered in the pdtpi Mutant.
Plants were germinated on MS agar plates under continuous light for 5 d, and shoots were harvested for activity assays in solution (B) and in native gels (C) and for immunoblotting (D) and metabolite analysis (E). Values were determined to be significantly different by t test with P values of 0.02 in (B) and 0.03, 0.03, 0.04, and 0.04 in (E) for glycerol, G-3-P, DHAP, and GAP, respectively.
(A) Full-length (pdTPI) and N-terminal truncated ( Δ pdTPI) proteins were expressed and purified from E. coli as activity assay controls.
(B) TPI activity in the wild type (Col) and pdtpi mutant.
(C) TPI activity staining in PAGE gel under nondenaturing conditions. Native Δ pdTPI recombinant protein was loaded as a control.
(D) Isoelectric immunoblotting with anti-cytoTPI antibody. Twenty micrograms of total protein was loaded per lane. Plastid and cytosolic TPI bands are noted.
(E) Quantification of metabolites in wild type (Col; open bars) and pdtpi mutant (closed bars). Four biological replicates were used for TPI activity assay and metabolite quantification in the wild type and mutant; data are presented as the mean ± se.
Using this assay, total TPI activity was quantified from wild-type and pdtpi 5-d-old seedlings. Since the pdtpi mutant did not show a clear phenotype under darkness (see Supplemental Figure 5 online), it was difficult to differentiate the mutant from the wild type when the mutant was propagated as a heterozygote; thus, the light-grown tissues were chosen to perform the enzyme activity assay and subsequent metabolite comparison. TPI activity in pdtpi was ∼60% of the level measured in the wild type (Figure 3B). Lower TPI activity in pdtpi is likely due to reduced plastid TPI expression, and this was verified by TPI activity staining using native gels (Figure 3C). Both TPI isoforms migrated toward the anode, although pdTPI migrated faster than cytoTPI as reported previously (Pichersky and Gottlieb, 1984; Dorion et al., 2005). To further verify that pdTPI protein was reduced in the mutant, total protein was isoelectrically separated for quantitative immunoblotting. The predicted pI values for pdTPI and cytoTPI are 7.98 and 5.17, respectively, indicating that these two proteins can be isoelectrophoretically resolved. Immunoblotting with antibodies raised against recombinant cytoTPI revealed three isoelectric bands in the wild type and three bands in pdtpi, one strongly reduced. The two equally expressed bands were more acidic and, therefore, likely cytoTPI (Figure 3C), which was previously shown to migrate as a doublet (Ito et al., 2003). The basic protein band is likely pdTPI since it was located very close to its pI range (Figure 3D) and it was reduced by ∼75%.
Since the TPI enzyme catalyzes the interconversion of DHAP and GAP, it is possible the concentrations of these metabolites were altered in pdtpi. Glycerol and G-3-P concentrations may also be affected as these by-products of TAG breakdown are metabolized to DHAP. A comparison of wild-type and pdtpi 5-d seedlings revealed glycerol, G-3-P, and DHAP concentrations were all higher in the mutant, 4.9-, 0.7-, and 5-fold, respectively. By contrast, GAP levels in pdtpi were 40% of those in the wild type (Figure 3E).
Accumulation of DHAP Is Deleterious to Plant Growth
The aforementioned experiments demonstrate that (1) DHAP levels are significantly higher in pdtpi mutant seedlings (Figure 3E), (2) the addition of DHAP to wild-type seedlings results in an inhibition of root elongation and cotyledon expansion at a concentration of 1 mM, and (3) exogenously supplied GAP, and other glycolytic intermediates, are unable to rescue the pdtpi phenotype and are not deleterious to growth (see Supplemental Figure 5 online). These observations suggest that the pdtpi phenotype could be the result of DHAP accumulation.
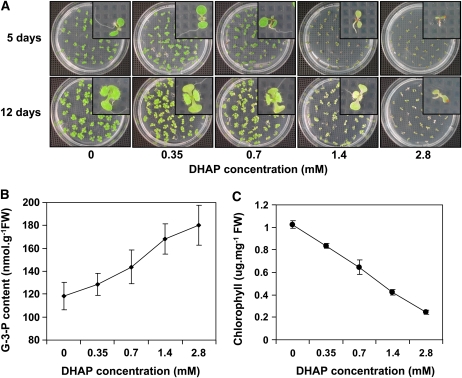
To test this, wild-type seeds were germinated on MS plates supplied with concentrations of DHAP ranging from 0.35 to 2.8 mM and then germinated under continuous white light. At 5 d postgermination, seedlings grown on plates containing <0.7 mM DHAP showed no significant difference from wild-type seedlings without DHAP treatment (Figure 4A, top). In the presence of 1.4 mM DHAP, wild-type seeds could germinate, but seedling growth was noticeably inhibited. The seedlings showed discoloration of the cotyledon and a shortened primary root, a phenotype similar to pdtpi. Interestingly, in vivo G-3-P levels in 5-d-old seedlings increased with exogenous application of DHAP (Figure 4B). Twelve days postgermination, the pale-green phenotype became stronger at concentrations as low as 0.35 mM (Figure 4A, bottom). Quantification of leaf chlorophyll content of 12-d-old seedlings indicated an inverse relationship between leaf chlorophyll content and DHAP concentration applied to the growth media (Figure 4C).
Figure 4.
Germinating Wild-Type Seed in DHAP-Enriched MS Medium Results in a Phenocopy of the pdtpi Mutant.
(A) Wild-type seeds were sown for 5 and 12 d on MS plates supplemented with a concentration series of DHAP. Insets show individual seedlings.
(B) G-3-P content of 5-d-old seedlings grown on MS media plus DHAP.
(C) Chlorophyll content of 12-d-old seedlings grown on MS plates plus varying concentrations of DHAP. Data are presented as the mean ± se from three biological replicates.
In Addition to DHAP, Exogenous Glycerol and G-3-P Can Also Mimic the pdtpi Phenotype
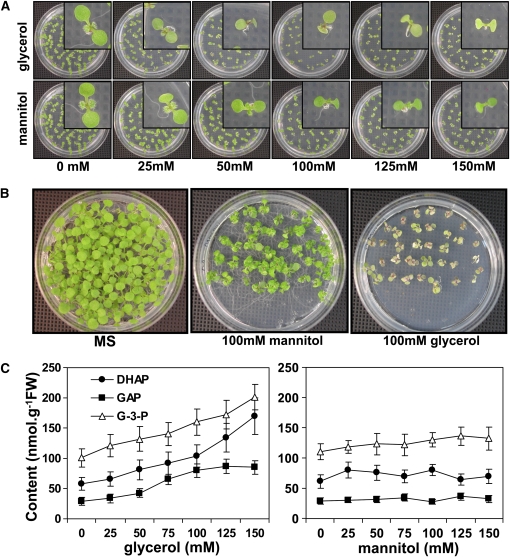
One of the products of TAG mobilization in germinating seed is glycerol. Glycerol breakdown proceeds by phosphorylation of glycerol to G-3-P followed by oxidation to DHAP by G-3-P dehydrogenase. To test if these intermediates are also toxic to plant cells, wild-type seeds were germinated for 5 d on MS media supplemented with different concentrations of glycerol (25 to 150 mM); MS plates with the same concentration of mannitol served as an osmotic control. In the presence of 25 mM glycerol, germinating wild-type seedlings accumulated slightly less chlorophyll and elongated the primary root at a slower rate than wild-type seedlings germinated on regular MS media (Figure 5A, top). The intensity of the pale-green phenotype also increased with increasing concentrations of glycerol present in the growth media (Figure 5A, top). The effect of glycerol on leaf greening and primary root elongation was most evident in seedlings grown for 12 d on MS media containing glycerol (Figure 5B). This phenotype was not due to osmotic stress since seedlings sown on 100 mM mannitol did not show chlorotic leaves and short primary roots compared with seedlings grown on glycerol (Figure 5B). Quantification of G-3-P, DHAP, and GAP from 5-d-old seedlings revealed that glycerol application raised the levels of these metabolites, especially when >100 mM glycerol (Figure 5C, left). Mannitol application had no effect on the levels of these three metabolites (Figure 5C, right).
Figure 5.
Wild-Type Seed Grown on MS Plates with the Addition of Glycerol Mimics the pdtpi Mutant Phenotype Due to the Accumulation of DHAP.
(A) Wild-type seed grown on MS medium plus 0 to 125 mM glycerol for 5 d. As an osmotic control, seeds were sown on equal concentrations of mannitol. Insets show individual seedlings.
(B) Wild-type seedlings grown for 8 d on MS plus 100 mM glycerol or mannitol (or neither; MS).
(C) Quantification of DHAP (closed circles), G-3-P (open triangles), and GAP (closed squares) from 5-d-old wild-type seedlings grown on various concentrations of glycerol or mannitol. Data are presented as the mean ± se from biological triplicate analyses. Values were determined to be significantly different from control by t test with P values of 0.04, 0.009, and 0.03 for DHAP, GAP, and G-3-P, respectively.
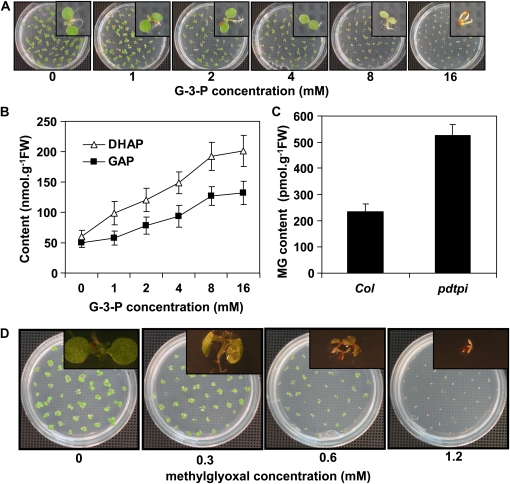
The effect of G-3-P on seedling growth was also examined. Wild-type seeds were germinated for 5 d on MS media supplemented with variable concentrations of G-3-P. G-3-P had no affect on seedling germination; however, primary root elongation and cotyledon greening were both inhibited beginning at 1 mM G-3-P (Figure 5A), and the severity of this phenotype increased with higher concentrations of G-3-P. Also, in vivo levels of DHAP and GAP in germinating seedlings increased with exogenous G-3-P concentrations (Figure 5B).
Methylglyoxal Content Is Significantly Higher in the pdtpi Mutant
Since DHAP can be converted into methylglyoxal (MG) by nonenzymatic phosphate elimination, MG content in the wild type and pdtpi was quantified from seedlings. The level of MG in pdtpi mutant was double that in the wild type (Figure 6C). Subsequent MG feeding experiments to the wild-type seeds indicated that MG is highly toxic to germinating seedlings (Figure 6D). Cotyledon chlorophyll biosynthesis was inhibited with as little as 0.3 mM, and root elongation was completely blocked by 0.6 mM MG (Figure 6D).
Figure 6.
Wild-Type Seed Grown on MS Plates with the Addition of G-3-P or MG Mimics the pdtpi Mutant Phenotype.
(A) Wild-type seed were grown on G-3-P–enriched MS plates for 5 d under continuous white light. Insets show individual seedlings.
(B) Quantification of DHAP (open triangles) and GAP (closed squares) in 5-d-old wild-type seedlings treated with G-3-P as shown in (A).
(C)The pdtpi mutant contains higher MG than the wild type. Values were determined to be significantly different by t test (P value of 0.03).
(D) Wild-type seeds were grown on MS plates with the addition of 0 to 1.2 mM MG. All data are presented as the mean ± se of from three biological replicates.
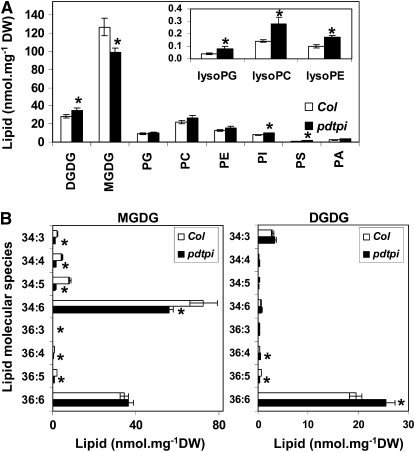
Galactolipid Levels Are Altered in the pdtpi Mutant
Glycerol metabolism is at the interface of glycerolipid turnover and synthesis. This is particularly true for germinating seed, as storage glycerolipids must be broken down for carbon availability, but membrane glycerolipids must also be synthesized for the developing seedling. Lipid composition analysis of the pdtpi mutant was therefore performed to determine if the growth phenotype impacted lipid synthesis. Five-day-old seedlings germinated on MS agar plate under light were harvested for lipid extraction. Quantitative lipid profiling revealed that total lipid content is slightly reduced in pdtpi (201 nmol mg−1 DW ± 12.8 se, n = 4) compared with the wild type (211 ± 13.6). Monogalactosyldiacylglycerol (MGDG) MGDG accounted for the biggest loss in pdtpi, dropping from 127.0 ± 9.7 nmol mg−1 in the wild type to 99.3 ± 4.5 in the mutant. At same time, Digalactosyldiacylglycerol (DGDG) was significantly higher in the mutant: 28.4 ± 1.9 nmol mg−1 in the wild type and 34.7 ± 2.5 in the mutant. Other lipids, including phosphatidyglycerol, phosphatidycholine, phosphatidylethanolamine, phosphatidylinositol, phosphatidylserine, phosphatidic acid, lysoPG, lysoPC, and lysoPE, were higher in the mutant (Figure 7A). Due to these lipid changes in pdtpi, the ratio of the nonbilayer-forming thylakoid lipid MGDG to the bilayer-forming thylakoid lipids phosphatidylglycerol, sulfolipid, and DGDG decreased from 3.2 to 2.1 (see Supplemental Data Set 1 online). In addition, MGDG shifted to lower levels of fatty acyl unsaturation, indicated by the strong reduction of MGDG 34:6 (Figure 7B); meanwhile, unsaturated acyl chains increased in DGDG, indicated by the strong increase in DGDG 36:6 and 38:6 (Figure 7B; see Supplemental Data Set 1 online).
Figure 7.
Downregulation of pdTPI Alters the Lipid Composition of Chloroplast Membranes.
Lipids were isolated from 5-d-old seedlings grown on MS plates and were analyzed by lipidomic profiling. Data are presented as the mean ± se from four biological replicates. Values marked with asterisks were determined to be significantly different from wild-type values by t test; the P values for total lipid in (A) are 0.048 (DGDG), 0.036 (MGDG), 0.33 (phosphatidyglycerol), 0.12 (phosphatidycholine), 0.096 (phosphatidylethanolamine), 0.020 (phosphatidylinositol), 0.014 (phosphatidylserine), 0.091 (phosphatidic acid), 0.021 (lysoPG), 0.015 (lysoPC), 0.0028 (lysoPE). The P values for MGDG in (B), left panel, are 0.0014 (34:3), 0.00044 (34:4), 0.00056 (34:5), 0.050 (34:6), 0.015 (36:3), 0.0015 (36:4), 0.00036 (36:5), and 0.26 (36:6). The P values for DGDG in (B), right panel, are 0.12 (34:3), 0.12 (34:4), 0.092 (34:5), 0.24 (34:6), 0.22 (36:3), 0.029 (36:4), 0.0042 (36:5), and 0.018 (36:6).
(A) Total glycerolipids in leaves of the wild-type (Col; open bars) and the pdtpi mutant (closed bars).
(B) Acyl composition of major chloroplast lipids MGDG (left panel) and DGDG (right panel).
Since MGDG is the precursor for DGDG synthesis, this raises the possibility that MGDG reduction in pdtpi resulted from enhanced conversion into DGDG. However, the increase in DGDG is much smaller compared with the decrease in MGDG in the mutant. In addition, decrease in the MGDG species resulted from reduced activity of the prokaryotic pathway of galactolipid biosynthesis, including 34:6, 34:5, 34:4, 34:3, 34:2, and 34:1 species. By contrast, the increase in DGDG was mostly due to 36:6 and 38:6 acyl species, which originate from the eukaryotic pathway of galactolipid biosynthesis, while DGDG 34:1 and 34:2, which are produced through the prokaryotic pathway, are >60% lower in the mutant compared with the wild type (see Supplemental Data Set 1 online). These data indicate the prokaryotic pathway of galactolipid biosynthesis is downregulated while the eukaryotic pathway is enhanced.
pdtpi Mutant Phenotype Was Complemented with cDNA Transgene
To determine if transgenic expression of At2g21170.1 cDNA could complement pdtpi, we constructed transgenic lines in the pdtpi heterozygous background by expressing the coding sequence of pdTPI cDNA under the control of a 35S promoter in a binary vector harboring the phosphinothricin acetyltransferase herbicide selection marker. T1 plants were PCR genotyped using a pair of primers of the P2/P3 combination (see Supplemental Figure 6 online). These primers, which are located on the exons of both sides of the original T-DNA insertion site, amplify a 114-bp fragment from the introduced pdTPI cDNA and a 523-bp fragment if the genotypes are pdTPI/pdTPI or pdTPI/pdtpi; the 523-bp fragment will disappear from homozygous pdtpi/pdtpi (see Supplemental Figure 6A online, top). A combination of LBa1 and P3 primers was used to distinguish pdTPI/pdTPI and pdTPI/pdtpi genotypes (see Supplemental Figure 6A online, bottom). By applying this method, 13 wild-type-like T1 transgenic plants were genotyped. Of these, five lines were pdTPI/pdTPI, six lines were pdTPI/pdtpi, and two lines were pdtpi/pdtpi. The phenotype of heterozygous lines 3 and 4 and homozygous line 6 was further observed in the T2 generation. After germination of the T2 seeds in soil, plants were first selected by spraying BASTA herbicide, individual wild-type-like plants were genotyped using the same strategy as described above, and homozygous plants with pdtpi/pdtpi locus were identified. The appearance of these plants through a complete life cycle was indistinguishable from wild-type plants grown under the same conditions (see Supplemental Figure 6B online). These genotyping results demonstrate that the pdtpi mutant phenotype is rescued by a pdTPI cDNA transgene and that At2g21170 is the mutant gene responsible for the observed phenotype.
DISCUSSION
pdTPI Encodes a Plastid TPI in Arabidopsis
The At2g21170 gene encodes an authentic plastid TPI based on five different experiments. First, when At2g21170 expression was reduced, total TPI activity was concomitantly 40% lower (Figure 3B). Second, recombinant At2g21170.1 protein showed strong TPI activity in vitro (Figure 3A). Third, in accordance with reduced TPI activity in the mutant in vivo levels of TPI substrates (DHAP and GAP) were altered; DHAP levels increased, while GAP decreased (Figure 3E). Fourth, the abundance and activity of a specific isoform of TPI was reduced (Figures 3B and 3C). Finally, subcellular localization analyses with a C-terminal GFP fusion demonstrated that the protein product of At2g21170 localized to chloroplasts (see Supplemental Figure 1 online).
Disruption of Triose and Glycerol Catabolism during Early Seedling Establishment Results in the Buildup of Toxic Metabolites and the pdtpi Mutant Phenotype
The most visible phenotype of the pdtpi mutant includes dwarfed stature, pale-green leaves, and reduced primary roots. One possible explanation for this is that pdTPI is required for glycerol metabolism during early seedling establishment. This is supported by the accumulation of glycerol and glycerol breakdown products G-3-P and DHAP, all of which are toxic to germinating seedlings at high concentrations. The loss of TPI function in other systems, including yeast, Kluyveromyces lactis, and humans (Shi et al., 2005; Capitanio et al., 2002; Oláh et al., 2005), also has been reported to lead to the significant buildup of DHAP. DHAP accumulation has been shown to trigger glycerol production in K. lactis, inositol auxotrophy in yeast (Capitanio et al., 2002; Shi et al., 2005), and multisystem disorders in human (Schneider et al., 1965).
Physiological evidence for the toxicity of DHAP is indirectly provided by chemical feeding experiments. Exogenous application of sucrose, glucose, F1, 6-P, GAP, or PGA did not mitigate the pdtpi mutant phenotype (see Supplemental Figure 5 online), suggesting the mutant phenotype is not the result of a limiting metabolite but rather the accumulation of a toxic metabolite. In support of this, feeding with glycerol, G-3-P, or DHAP in the wild type produced a phenotype similar to the pdtpi mutant (Figures 4 to 6). Plants that were supplied with exogenous glycerol or G-3-P showed elevated DHAP levels in vivo. Since DHAP was found to be more toxic than glycerol or G-3-P, buildup of this metabolite in germinating Arabidopsis seeds is a possible cause of the pdtpi phenotype. Presumably, DHAP buildup occurs within the plastid, since a functional cytosolic TPI would reduce DHAP levels to equilibrium with GAP. This premise is supported by abnormal chloroplast development and a defective transition from heterotrophic to photoautotrophic growth in the mutant.
Glycerol, applied exogenously to plants at high concentrations, is known to inhibit growth (Hippman and Heinz, 1976; Leegood et al., 1988; Aubert et al., 1994). The effect of glycerol on germinating seeds was used to isolate the glycerol insensitive mutant gli1, which encodes a glycerol kinase, the first enzyme in glycerol catabolism (Eastmond, 2004). It has been speculated that glycerol toxicity is a consequence of its catabolism (Aubert et al., 1994). One hypothesis suggests that phosphorylation of exogenously applied glycerol leads to the depletion of orthophosphate (Leegood et al., 1988). However, exogenously applied G-3-P is more effective than glycerol at mimicking the pdtpi mutant phenotype, suggesting that orthophosphate depletion is not likely the major cause of glycerol toxicity. Rather, our observations suggest that DHAP buildup at the early seedling stage is the major cause of the mutant phenotype since seed sown in glycerol and G-3-P accumulate DHAP (Figures 5 and 6). Alternatively, a nonenzymatic by-product of DHAP could also be responsible for this phenotype. DHAP can be converted into MG by nonenzymatic phosphate elimination (Ahmed et al., 2005). Interestingly, seed germination in the presence of MG revealed that it is more toxic than DHAP (Figure 6D). For example, to phenocopy the pdtpi mutant in 5-d-old germinating wild-type seedlings, the minimum exogenous concentrations of glycerol, G-3-P, DHAP, and MG required are ∼150, 8, 1.4, and 0.6 mM, respectively (Figures 4 to 6). Therefore, the toxicity for germinating seedlings from highest to lowest is MG>DHAP>G-3-P>glycerol, in reverse order of glycerol catabolism. In human cancer cells, MG can inactivate G-3-P dehydrogenase, although this has not been confirmed in plants. Since MG reacts with free amino groups of Lys and Arg residues of proteins forming glycation end products, its targets could be multiple. Our data suggest that MG toxicity also plays an important role in the pdtpi mutant phenotype.
Considerations for Metabolic Feeding to Germination Seeds
Metabolic feeding with heterotrophic organisms, such as E. coli and yeast, has historically been very successful at characterizing mutants and elucidating metabolic pathways. However, metabolic feeding to phototrophic organisms, especially germinating seedlings, presents some challenges. First, germinating seedlings are in transition from heterotrophic to phototrophic growth and therefore possess both properties. Unlike heterotrophic organisms, germinating seedlings have their own energy reserve instead of being dependent on exogenous carbon, so seed storage mobilization must also be taken into account when metabolic feeding experiments are performed on germinating seedlings. This is especially true if the metabolite being tested is also a metabolic intermediate. Second, germinating seedlings contain rapidly dividing and elongating tissues undergoing dramatic morphological changes, such as root growth, stem elongation, and leaf expansion. Exogenous application of metabolites first requires these compounds to be delivered into the contacting tissue and transported to distant tissues, such as the leaf. Also, cells may differ in their relative permeability to these compounds. Moreover, although the roots must have the capacity to absorb these compounds, once into the cell these compounds could potentially be metabolized and converted into other metabolites since they are intermediates. For these reasons, a higher concentration of metabolic intermediates was employed for these feeding studies, compared with the physiological concentrations of each compound in vivo. If the feeding compounds are toxic to germinating seedlings, one would expect that the roots would be first affected since they directly contact the media. Our feeding experiments indeed support this notion (Figures 4 to 6). For example, root elongation was inhibited by 25 mM glycerol, 1 mM G-3-P, 0.7 mM DHAP, and 0.3 mM MG, while the cotyledon only showed a marginal phenotype at these concentrations (Figures 4 to 6).
Reduced Expression of pdTPI Disrupts Metabolic Pathways in Chloroplasts
The biochemical basis for DHAP and MG toxicity in plants, as demonstrated here, is uncertain. In Saccharomyces cerevisiae, a tpi1 mutant was unable to grow in the absence of inositol (Shi et al., 2005) possibly because DHAP is an inhibitor for myo-inositol-3 phosphate synthase, the rate-limiting enzyme in inositol biosynthesis (Migaud and Frost, 1996). To test the possibility that plants might share a similar mechanism, wild-type and pdtpi seeds were germinated on MS plates with the addition of myo-inositol ranging from 25 μ M to 125 mM. However, myo-inositol application was unable to rescue the pdtpi phenotype (see Supplemental Figure 7 online), which likely excludes the possibility that myo-inositol depletion is the link between DHAP buildup and the mutant phenotype. A slightly higher phosphatidylinositol lipid content in the pdtpi mutant supports this conclusion (Figure 7).
Glycolysis and glycerol catabolism take place in the cytoplasm and involve a cytoTPI. It is therefore surprising that a functional cytoTPI cannot reduce DHAP levels in the pdtpi mutant. During the transition from heterotrophy to autotrophy, the plastid shuttles carbon from the cytoplasm to the plastid for various pathways, including fatty acid, amino acid, and chlorophyll biosynthesis. Isolated barley (Hordeum vulgare) leaf etioplasts were shown to import DHAP for use both as a carbon source and for energy production (Batz et al., 1992). It was proposed that DHAP uptake is mediated by a translocator with properties similar to those of the triose phosphate translocator (TPT; Batz et al., 1992). Interestingly, a recent proteomic study of proplastid envelope membranes identified TPT as well as several putative translocators (Bräutigam and Weber, 2009). The TPT translocator is widely accepted to transport photosynthesis-produced triose phosphate from chloroplast to the cytoplasm (Riesmeier et al., 1993; Schneider et al., 2002). Expression of TPT in proplastids but the inability of DHAP to equilibrate in this pdTPI mutant may suggest the possibility of substrate preference or regulation for this translocator, which could also be direction dependent.
Under circumstances where DHAP is imported to the plastid from the cytosol, reduced expression of pdTPI would prevent the conversion of DHAP to GAP in the chloroplast and result in DHAP accumulation and reduced GAP in the chloroplast, as shown in Figure 3. Total cellular GAP (cytosolic plus plastid) was reduced ∼60% in the pdtpi mutant (Figure 3E). Since a functional cytoTPI would equilibrate cytosolic DHAP and GAP, the plastid GAP pool in the pdtpi mutant is probably >60% reduced. However, additional experiments are necessary to determine if the increase in DHAP pool size is principally within the plastid, since the cytoTPI appears to have a higher affinity for GAP than DHAP (Tomlinson and Turner, 1979; Tang et al., 1999). This metabolic imbalance could profoundly affect the development of other biochemical pathways in a plastid transitioning to a mature chloroplast (Figure 8). First, GAP and pyruvate are the direct substrates for chlorophyll biosynthesis in the plastid (Lichtenthaler, 1999); reduced supply of GAP likely leads to reduced chlorophyll biosynthesis and the chlorotic developmental phenotype (Figure 1). Second, GAP is required for regenerating ribulose 1,5-bisphosphate for the Calvin cycle. The limitation of GAP and the surplus of DHAP would limit ribulose 1,5-bisphosphate production, suppressing the Calvin cycle (Figure 8). In support of this, the pdtpi mutant ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit was reduced (see Supplemental Figure 8A online), an indication that photosynthetic capacity was suppressed in the mutant. Reduced photosynthetic activity would ultimately limit starch production and photoautotrophic development. Interestingly, exogenously supplied GAP failed to rescue the pdtpi phenotype (see Supplemental Figure 5 online); this might result from the following: (1) GAP cannot be directly transported into the chloroplast (Weber et al., 2005), (2) germinating seedlings ineffectively absorb exogenously supplied GAP, or (3) GAP was metabolized either by cytosolic glycolysis or the gluconeogenic pathway before reaching the chloroplast stroma.
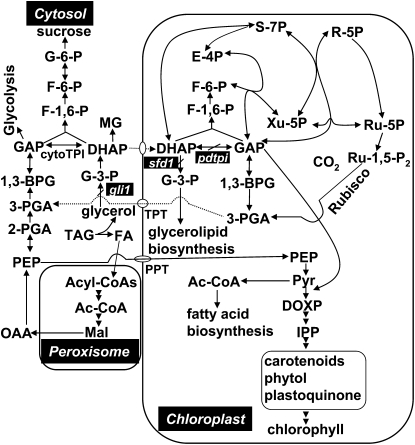
Figure 8.
Metabolic Pathways Affected by a Mutation in pdTPI during Postgerminative Seedling Transition from Heterotrophic to Photoautotrophic Development.
For comparison, the figure shows related mutants, gli1, encoding a glycerol kinase, and sfd1 (for suppressor of fatty acid desaturase deficiency1), encoding a putative plastid DHAP reductase involved in G-3-P supply for glycerolipid synthesis. G-6-P, d-glucose-6-phosphate; F-6-P, d-fructose-6-phosphate; F-1,6-P, fructose-1,6-bisphosphate; 1,3-BPG, 1,3-diphosphateglycerate; 3-PGA, 3-phosphoglycerate; 2-PGA, 2-phosphoglycerate; PEP, phosphoenolpyruvate; Pyr, pyruvate; FA, fatty acid; Ac-CoA: acetyl-CoA; Mal, malate; OAA, oxaloacetate; DOXP, 1-deoxy-d-xylulose-5-P; IPP, isopentenyl diphosphate; E-4P, erythrose-4-phosphate; R-5P, ribose-5-phosphate; Ru-1,5-P2, ribulose-1,5-bisphosphate; Ru-5P: ribulose-5-phosphate; S-7P, sedoheptulose-7-phosphate; Xu-5P, xylulose-5-phosphate; PPT, phosphoenolpyruvate/phosphate translocator.
Lastly, fatty acid content was only slightly reduced (see Supplemental Data Set 1 online) in the pdtpi mutant, possibly because a functional phosphoenolpyruvate (PEP) translocator delivers PEP from the cytosol to the chloroplast for acetyl-CoA production (Fischer et al., 1997; Figure 8), the precursor for de novo fatty acid biosynthesis. However, the polar lipid profile was significantly affected (Figure 7). This might be due to delayed photosynthetic development or the possibly that DHAP directly affects lipid biosynthetic activities in the chloroplast. Imported PEP cannot replace the GAP in the plastid, possibly due to the absence of enolase and phosphoglycerate mutase (Prabhakar et al., 2009), and suggests that glycolysis does not operate in reverse in plastids from developing seedlings.
Conclusions
Postgerminative seedlings produce DHAP through glycerol catabolism, which is a by-product of lipid mobilization during seedling establishment. A portion of this DHAP is likely transported into developing chloroplast through a translocator. Once into the chloroplast, DHAP is converted into GAP, catalyzed by pdTPI. DHAP and GAP are precursors for multiple metabolic pathways in the chloroplast. These pathways play critical roles in the transition of a heterotrophic etioplast into a photoautotrophic chloroplast. The loss of function of pdTPI compromises these biosynthetic pathways with a concurrent buildup of DHAP. Since exogenous DHAP, and its glycerol catabolic precursors, are capable of suppressing seedling growth in a manner similar to the pdTPI mutant, it is plausible that DHAP either directly or indirectly is responsible for this phenotype. This is supported by the observation that although this mutant phenotype can be mimicked, it cannot be rescued by exogenous metabolic intermediates. Interestingly, a significant portion of this DHAP is apparently converted into MG by nonenzymatic phosphate elimination. MG was previously reported to react with free amino groups of Lys and Arg residues of proteins forming glycation end products, which could adversely affect other metabolic pathways or cellular processes. The pdtpi mutant phenotype is likely the result of the combined accumulation of DHAP and MG and disruption of plastid intermediary metabolism.
METHODS
Plant Materials
T-DNA insertion lines were ordered from the ABRC. Seeds were sown in a 1:1 mixture of water-saturated vermiculite and peat moss-enriched soil and grown in a growth chamber (14-h-light/10-h-dark cycle, 23 ° C day/20 ° C night, 50% humidity, and light intensity of 200 μ mol m−2 s−1).
Chemical Feeding
Chemical feeding was performed on half-strength MS agar plates (0.21% [w/v] micro and macro elements [Research Products International], 0.025% [w/v] MES [Fisher Scientific], 0.0056% [w/v] Gamborg's B5 vitamin mixture [Research Products International], 0.8% [w/v] plant agar [Research Products International], pH 5.7, and KOH) at specified concentrations. Sucrose, d-glucose, and myo-inositiol were obtained from Research Products International, and glycerol was purchased from Fisher Scientific. All other chemicals were purchased from Sigma-Aldrich.
Recombinant pdTPI Protein Expression and Purification
To produce full-length and mature pdTPI, two set of N-terminal primers were designed: 5 ′ -CACCATGGCAGCTACCTCTCTCA-3 ′ and 5 ′ -CACCATGGCTGGATCCGAAAAGTT-3 ′, respectively. The same C-terminal primer was used for amplification: 5 ′ -TCAAGCAGCAACTTTCTTCGACGT-3 ′. Pfu-amplified cDNAs were directly cloned into the Topo pET200 vector (Invitrogen). Constructs were introduced into Escherichia coli BL21 cells and protein expression induced with 1 mM isopropyl- β -d-thiogalactopyranoside. Expressed proteins were purified by nickel-NTA agarose (Qiagen) under native conditions.
Enzyme Assays
Five-day-old seedlings grown on agar plates were ground into powder under liquid nitrogen. Extraction buffer (50 mM Tris-HCl, pH 8.0, 0.5 M sorbitol, and 2 mM EDTA) was added, and tissue was reground until completely thawed. Homogenates were centrifuged at 13,000g for 5 min at 4 ° C, and supernatants were transferred into fresh tubes. Total protein was quantified using a Coomassie Brilliant Blue dye binding assay with chicken γ -globulin as the standard (Bio-Rad). TPI activity was determined according to the procedure of Ito et al. (2003) by a coupled assay with α -glycerophosphate dehydrogenase. The TPI activity also was assayed on native PAGE gel (Dorion et al., 2005) and stained as described by Pichersky and Gottlieb (1984).
Metabolite Quantitation
For DHAP and GAP quantitation, samples were prepared according to Häusler et al. (2000) and enzymatically measured according to Stitt et al. (1989). For G-3-P quantitation, samples were prepared according to Jelitto et al. (1992) and enzymatically assayed as described previously (Wei et al., 2004). For glycerol quantification, sample extraction, derivatization, and gas chromatography–mass spectrometry analysis were all performed according to Roessner et al. (2000). A glycerol standard curve was made under the same experimental conditions to quantify glycerol content from plant extracts. For MG quantification, sample extraction and derivatization was conducted as described by Chaplen et al. (1996), and gas chromatography–mass spectrometry analysis was performed according to Du et al. (2008).
Polar Lipid Profiling
Lipid extraction and quantitative profiling were performed according to Devaiah et al. (2006), except that analyses of MGDG and DGDG were performed in the same solvent mixture as used for the phospholipid analysis and with different scan parameters for galactolipids. Neutral loss scans in positive ion mode were used to detect the [M + NH4] + ions of MGDG (NL179.06) and DGDG (NL 341.11). Collision energies, using nitrogen as the collision gas (gas pressure set at 2 arbitrary units) were 21 V for MGDG and 24 V for DGDG. Declustering potentials were 90 V, entrance potentials 10 V, and exit potentials 23 V for both MGDG and DGDG.
PCR, RT-PCR, and Immunoblotting
Total RNA was isolated from 5-d-old Arabidopsis thaliana seedlings grown on MS media using the RNeasy plant mini kit (Qiagen). M-MLV (Promega) was used to synthesize first-strand cDNA using decamers and 2 μ g of total RNA at 37 ° C for 2 h. Subsequent PCR reactions were conducted on 0.5 μ L of the RT reaction mixture. The primer sequences are P1, 5 ′ -GAGCACAACCACCAACCCTCT-3 ′, P2, 5 ′ -TCCTCTACCCGTCTCGTTTCT-3 ′, P3, 5 ′ -CCCGTTACACTTCCAGTTTCC-3 ′, LBa1, 5 ′ -TGGTTCACGTAGTGGGCCATCG-3 ′, S21170F, 5 ′ -CATCGCTCCCCCAGAGGTGT-3 ′, and S21170R, 5 ′ -GCACTGTTGAGATCGGAGATA-3 ′.
To discriminate pdTPI from cytoTPI protein, total protein was isolated from 5-d-old MS plate-grown seedlings and separated in an IEF gel by following the method described by Anderson and Peck (2008); protein was then transferred to nitrocellulose membrane and probed with anti-cytoTPI antibody. Anti-cytoTPI antibody cross-reacted with pdTPI protein, which was confirmed using purified recombinant cytoTPI and pdTPI protein (see Supplemental Figure 8B online).
Light and Transmission Electron Microscopy Analysis
Samples were fixed in 2% (v/v) glutaraldehyde and 2% (v/v) paraformaldehyde in 100 mM sodium cacodylate buffer-NaOH, pH 7.4, overnight at 4 ° C, rinsed three times with 0.01 M 2-mercaptoethanol, 0.1 M sodium cacodylate buffer, and 0.17 M sucrose, pH 7.4 (20 min for each rinse). Samples were postfixed with 1% (w/v) osmium tetroxide in 100 mM sodium cacodylate buffer, rinsed three times (5 min each) with water, and dehydrated through a graded series of acetone (20%, 50%, 70%, 90%, and three times 100% [v/v]). After infiltration through a graded acetone/Epon/Spurr's epoxy resin and polymerization at 60 ° C for 24 h, the polymerized blocks were sectioned on a Leica Ultracut UCT ultramicrotome. Samples for light microscope slides were sectioned onto glass slide at 2.5 μ m thickness. Samples for transmission electron microscopy were sectioned at ∼85 nm using an ultra 35 ° diatome diamond knife, and the thin sections were collected on 200 mesh copper thin bar grids. Grids were then poststained with 5% aqueous uranyl acetate and Sato's triple lead stain. Stained samples were viewed in the JEOL 1400 transmission electron microscope, and digital images collected using the Gatan 894 Ultrascan 1000 digital camera (4k × 4k).
Transgenic Complementation of the pdtpi Mutant and Subcellular Localization
pdtpi was genetically complemented by introducing the pdTPI cDNA. The pdTPI coding sequence (At2g21170.1) was PCR amplified using a pair of primers (5 ′ -CCGCTCGAGTCAGAAATGGCAGCTACCTCTCTCA-3 ′ and 5 ′ -GGAATTCAAGCAGCAACTTTCTTCGACGT-3 ′ ) containing XhoI and EcoRI restriction sites. The PCR fragment was digested by XhoI and EcoRI and cloned into the same sites of pEZT-NL vector (Carnegie Institute, Washington DC), in frame with a C-terminal GFP. The construct was introduced into Agrobacterium tumefaciens EHA105 R26C9 and then transformed into pdtpi mutant plants by vacuum infiltration (Koncz et al., 1984).
To verify subcellular localization, transgenic Arabidopsis leaf expressing pdTPI-GFP was harvested and homogenized, and subcellular protein fractions were prepared as described by Bao et al. (2003). Protein from each fraction was resolved by SDS-PAGE and transferred to nitrocellulose membranes for antibody probing. Polyclonal antibodies to plastid PDH E1 α were used to determine plastid enrichment, and monoclonal antibodies to the mitochondrial PDH α -subunit were used to assay mitochondria enrichment (Miernyk and Thelen, 2008). For GFP subcellular localization, T1 transgenic plants were screened using a Zeiss LSM 510 META NLO two-photon point scanning confocal microscope, under the mode of linear unmixing to effectively subtract the autofluorescence from the original signal.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL or Arabidopsis Genome Initiative data libraries under the following accession numbers: pdTPI (gi145360176, At2g21170), cytoTPI (gi145339534, At3g55440), Petunia hybrida TPI (gi602589), Solanum chacoense TPI (gi38112661), Coptis japonica TPI (gi340520), Oryza sativa TPI (gi169820 and gi115440976), Triticum aestivum TPI (gi11124571), Glycine max TPI (gi77540215), Phaseolus vulgaris TPI (gi57283984), Secale cereale TPI (gi407524), Hordeum vulgare (gi1785947), and Pteris vittata (gi84626306). T-DNA insertion lines used were SALK_152526 (pdtpi), SALK_022963, CS829061, SALK_003991, and SALK_106806.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. pdTPI Protein Localizes to the Chloroplast.
Supplemental Expression of Alternatively Spliced pdTPI Transcripts.
Supplemental CytoTPI Mutant Identification.
Supplemental Characterization of the pdtpi Mutant.
Supplemental Metabolite Augmentation Does Not Rescue the pdtpi Mutant Phenotype.
Supplemental Complementation of the pdtpi Mutant by Transgenic Expression of pdTPI cDNA.
Supplemental The pdtpi Mutant Cannot Be Rescued by the Application of Myo-Inositiol.
Supplemental Rubisco Large Subunit Is Downregulated in the pdtpi Mutant and Anti-cytoTPI Polyclonal Antibody Cross-Reacts with pdTPI Protein.
Supplemental Data Set 1. Lipidomics Profiling of the Wild Type and pdtpi Mutant.
Supplementary Material
Acknowledgments
We thank the Electron Microscopy Core Facility at the University of Missouri for assistance in preparing and imaging specimens for this research, the Richard Jeannotte and the Kansas Lipidomics Research Center for lipid profiling, and the Arabidopsis Stock Center for providing T-DNA insertion lines. Financial support was provided by a Life Science Postdoctoral Fellowship (to M.C.) from the University of Missouri and National Science Foundation-Plant Genome Research Grant DBI-0332418. We thank Melody Kroll for editing this manuscript. We also thank the three anonymous reviewers for helpful suggestions and comments.
References
- Adham A.R., Zolman B.K., Millius A., Bartel B. (2005). Mutations in Arabidopsis acyl-CoA oxidase genes reveal distinct and overlapping roles in β -oxidation. Plant J. 41: 859–874 [DOI] [PubMed] [Google Scholar]
- Ahmed N., Dobler D., Dean M., Thornalley P.J. (2005). Peptide mapping identifies hotspot site of modification in human serum albumin by methyglyoxal involved in ligand binding and esterase activity. J. Biol. Chem. 280: 5724–5732 [DOI] [PubMed] [Google Scholar]
- Anderson J.C., Peck S.C. (2008). A simple and rapid technique for detecting protein phosphorylation using one-dimensional isoelectric focusing gels and immunoblot analysis. Plant J. 55: 881–885 [DOI] [PubMed] [Google Scholar]
- Aubert S., Gout E., Bligny R., Douce R. (1994). Multiple effects of glycerol on plant cell metabolism. J. Biol. Chem. 269: 21420–21427 [PubMed] [Google Scholar]
- Bao X., Thelen J.J., Bonaventure G., Ohlrogge J.B. (2003). Characterization of cyclopropane fatty-acid synthase from Sterculia foetida. J. Biol. Chem. 278: 12846–12853 [DOI] [PubMed] [Google Scholar]
- Batz O., Scheibe R., Neuhaus H.E. (1992). Transport processes and corresponding changes in metabolite levels in relation to starch synthesis in barley (Hordeum vulgare L.) etioplasts. Plant Physiol. 100: 184–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beevers H. (1956). Utilization of glycerol in the tissues of the germinating castor bean. Plant Physiol. 31: 440–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley J.D. (1997). Seed germination and dormancy. Plant Cell 9: 1055–1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bewley J.D., Black M. (1985). Seeds: Physiology of Development and Germination. (New York: Plenum Publishing; ). [Google Scholar]
- Bräutigam A., Weber A. (2009). Proteomic analysis of the proplastid envelop membrane provides novel insights into small molecular and protein transport across proplastid membrane. Mol. Plant 2: . [DOI] [PubMed] [Google Scholar]
- Bunkelmann J., Trelease R.N. (1996). Ascorbate peroxidase: A prominent membrane protein in oilseed glyoxysomes. Plant Physiol. 110: 589–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capitanio D., Merico A., Ranzi B.M., Compagno C. (2002). Effects of the loss of triose phosphate isomerase activity on carbon metabolism in Kluyveromyces lactis.. Res. Microbiol. 153: 593–598 [DOI] [PubMed] [Google Scholar]
- Cernac A., Andre C., Hoffmann-Benning S., Benning C. (2006). WRI1 is required for seed germination and seedling establishment. Plant Physiol. 141: 745–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaplen F.W.R., Fahl W.E., Cameron D.C. (1996). Detection of methylglyoxal as a degradation product of DNA and nucleic acid components treated with strong acid. Anal. Biochem. 236: 262–269 [DOI] [PubMed] [Google Scholar]
- Cornah J.E., Germain V., Ward J.L., Beale M.H., Smith S.M. (2004). Lipid utilization, gluconeoenesis and seedling growth in Arabidopsis mutants lacking the glyoxylate cycle enzyme malate synthase. J. Biol. Chem. 279: 42916–42923 [DOI] [PubMed] [Google Scholar]
- Devaiah S.P., Roth M.R., Baughman E., Li M., Tamura P., Jeannotte R., Welti R., Wang X. (2006). Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a PHOSPHOLIPASE D α 1 knockout mutant. Phytochemistry 67: 1907–1924 [DOI] [PubMed] [Google Scholar]
- Dorion S., Parveen, Jeukens J., Matton D.P., Rivoal J. (2005). Cloning and characterization of a cytosolic isoform of triosephosphate isomerase developmentally regulated in potato leaves. Plant Sci. 168: 183–194 [Google Scholar]
- Du Z., Clery R.A., Hammond C.J. (2008). Volatile organic nitrogen-containing constituents in ambrette seed Abeloschus moschatus Medik (Malvaceae). J. Agric. Food Chem. 56: 7388–7392 [DOI] [PubMed] [Google Scholar]
- Eastmond P.J. (2004). Glycerol-insensitive Arabidopsis mutants: gli1 seedlings lack glycerol kinase, accumulate glycerol and are more resistant to abiotic stress. Plant J. 37: 617–625 [DOI] [PubMed] [Google Scholar]
- Eastmond P.J. (2006). SUGAR-DEPENDENT1 encodes a patain domain triacylglycerol lipase that initiates storage oil breakdown in germinating Arabidopsis seeds. Plant Cell 18: 665–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastmond P.J., Hooks M.A., Williams D., Lange P., Bechtold N., Sarrobert C., Nussaume L., Graham I.A. (2000). Promoter trapping of a novel medium-chain acyl-CoA oxidase, which is induced transcriptionally during Arabidopsis seed germination. J. Biol. Chem. 275: 34375–34381 [DOI] [PubMed] [Google Scholar]
- Fischer K., Kammerer N., Gutersohn M., Arbinger B., Weber A., Hausler R.E., Flügge U.I. (1997). A new class of plastidic phosphate translocators: a putative link between primary and secondary metabolism by the phosphoenolpyrute/phosphate antiporter. Plant Cell 9: 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda M., Schnurr J., Abbadi A., Heinz E., Browse J. (2004). Peroxisomal Acyl-CoA synthetase activity is essential for seedling development in Arabidopsis thaliana. Plant Cell 16: 394–405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham I.A. (2008). Seed storage oil mobilization. Annu. Rev. Plant Biol. 59: 115–142 [DOI] [PubMed] [Google Scholar]
- Graham I.A., Eastmond P.J. (2002). Pathways of straight and branched chain fatty acid catabolism in higher plants. Prog. Lipid Res. 41: 156–181 [DOI] [PubMed] [Google Scholar]
- Häusler R.E., Fischer K.L., Flügge U.I. (2000). Determination of low-abundant metabolites in plant extracts by NAD(P)H fluorescence with a microtiter plate reader. Anal. Biochem. 281: 1–8 [DOI] [PubMed] [Google Scholar]
- Hayashi M., Toriyama K., Kondo M., Nishimura M. (1998). 2,4-Dichlorophenoxybutrric acid-resistant mutants of Arabidopsis have defects in glyoxysomal fatty acid β -oxidation. Plant Cell 10: 183–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hippman H., Heinz E. (1976). Glycerol kinase in leaves. Z. Pflanzenphysiol. 79: 408–418 [Google Scholar]
- Ito H., Iwabuchi M., Ogawa K.I. (2003). The sugar-metabolic enzymes aldolase and triose-phosphate isomerase are targets of glutathionylation in Arabidopsis thaliana: detection using biotinylated glutathione. Plant Cell Physiol. 44: 655–660 [DOI] [PubMed] [Google Scholar]
- Jelitto T., Sonnewald U., Willmitzer L., Hajirezeai M., Stitt M. (1992). Inorganic pyrophosphate content and metabolites in potato and tobacco plants expressing E. coli pyrophosphatase in their cytosol. Planta 188: 238–244 [DOI] [PubMed] [Google Scholar]
- Koncz C., Kreuzaler F., Kalman Z., Schell J. (1984). A simple method to transfer, integrate and study expression of foreign genes, such as chicken ovalbumin and alpha-actin in plant tumors. EMBO J. 3: 1029–1037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leegood R.C., Labata C.A., Huber S.C., Neuhaus H.E., Stitt M. (1988). Phosphate sequestration by glycerol and its effects on photosynthetic carbon assimilation in leaves. Planta 176: 117–126 [DOI] [PubMed] [Google Scholar]
- Lichtenthaler H.K. (1999). The 1-deoxy-D-xylulose-5 phosphate pathway of isoprenoid biosynthesis in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50: 47–65 [DOI] [PubMed] [Google Scholar]
- Lin E.C. (1976). Glycerol dissimilation and its regulation in bacteria. Annu. Rev. Microbiol. 30: 535–578 [DOI] [PubMed] [Google Scholar]
- Lin E.C. (1977). Glycerol utilization and its regulation in mammals. Annu. Rev. Biochem. 46: 765–795 [DOI] [PubMed] [Google Scholar]
- Miernyk J.A., Thelen J.J. (2008). Biochemical approachs for discovering protein-protein interactions. Plant J. 53: 597–609 [DOI] [PubMed] [Google Scholar]
- Migaud M.E., Frost J.W. (1996). Elaboration of a general strategy for inhibition of myo-Inositol 1-phosphate synthase: active site interactions of analogues possessing oxidized reaction centers. J. Am. Chem. Soc. 118: 495–501 [Google Scholar]
- Mooney B.P., Miernyk J.A., Randall D.D. (2002). The complex fate of α -ketoacids. Annu. Rev. Plant Biol. 53: 357–375 [DOI] [PubMed] [Google Scholar]
- O'Neill C.M., Gill S., Hobbs D., Morgan C., Bancroft I. (2003). Natural variation for seed oil composition in Arabidopsis thaliana. Phytochemistry 64: 1077–1090 [DOI] [PubMed] [Google Scholar]
- Oláh J., Orosz F., Puskás L.G., Hackler L.J., Horányi M., Polgár L., Hollán S., Ovádi J. (2005). Triosephosphate isomerase deficiency: Consequence of an inherited mutation at mRNA, protein and metabolic levels. Biochem. J. 392: 675–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Graham S., Graham I.A. (2005). Storage reserve mobilization in germinating oilseeds: Arabidopsis as a model system. Biochem. Soc. Trans. 33: 380–383 [DOI] [PubMed] [Google Scholar]
- Pichersky E., Gottlieb L.D. (1984). Plant triose phosphate isomerase. Plant Physiol. 74: 340–347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinfield-Wells H., Rylott E.L., Gilday A.D., Graham S., Job K., Larson T.R., Graham I.A. (2005). Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. Plant J. 43: 861–872 [DOI] [PubMed] [Google Scholar]
- Prabhakar V., Löttgert T., Gigolashvili T., Bell K., Flügge U., Häusler R. (2009). Molecular and functional characterization of the plastid-localized phosphoenolpyruvate enolase (ENO1) from Arabidopsis thaliana.. FEBS Lett. 583: 983–991 [DOI] [PubMed] [Google Scholar]
- Riesmeier J.W., Flügge U., Schulz B., Heineke D., Heldt H., Willmitzer L., Frommer W. (1993). Antisense repression of the chloroplast triose phosphate translocator affects carbon partitioning in transgenic potato plants. Proc. Natl. Acad. Sci. USA 90: 6160–6164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessner U., Wagner C., Kopka J., Trethewey R.N., Willmitzer L. (2000). Simultanous analysis of metabolites in potato tuber by gas chromatography-mass spectrometry. Plant J. 23: 131–142 [DOI] [PubMed] [Google Scholar]
- Rylott E.L., Eastmond P.J., Gilday A.D., Slocombe S.P., Larson T.R., Baker A., Graham I.A. (2006). The Arabidopsis thaliana multifunctional protein gene (MFP2) of peroxisomal β -oxidation is essential for seedling establishment. Plant J. 45: 930–941 [DOI] [PubMed] [Google Scholar]
- Schneider A., Häusler R.E., Kolukisaoglu Ü., Kunze R., Graaff E.V.D., Schwacke R., Catoni E., Desimone M., Flügge U. (2002). An Arabidopsis thaliana knock-out mutant of the chloroplast triose phosphate/phosphate translocator is severely compromised only when starch synthesis, but not starch mobilization is abolished. Plant J. 32: 685–699 [DOI] [PubMed] [Google Scholar]
- Schneider A.S., Valentine W.N., Hattori M., Heins H.L., Jr (1965). Hereditary hemolytic anemia with triosephosphate isomerase deficiency. N. Engl. J. Med. 272: 229–235 [DOI] [PubMed] [Google Scholar]
- Shi Y.H., Vaden D.L., Ju S.L., Ding D.B., Geiger J.H., Greenberg M.L. (2005). Genetic perturbation of glycolysis results in inhibition of de novo inositol biosynthesis. J. Biol. Chem. 280: 41805–41810 [DOI] [PubMed] [Google Scholar]
- Stitt M., Lilley R.M., Gerhardt R., Heldt H.W. (1989). Metabolite levels in specific cells and subcellular compartments of plant leaves. Methods Enzymol. 174: 518–522 [Google Scholar]
- Tomlinson J.D., Turner J.F. (1979). Pea seed triose phosphate isomerase. Phytochemistry 18: 1959–1962 [Google Scholar]
- Tang G.L., Wang Y.F., Bao J.S., Chen H.B. (1999). Overexpression in Excherichia coli and characterization of the chloroplast triosephosphate isomerase from spinach. Protein Expr. Purif. 16: 432–439 [DOI] [PubMed] [Google Scholar]
- Weber A., Schwacke R., Flügge U. (2005). Solute transporters of the plastid envelope membrane. Annu. Rev. Plant Biol. 56: 133–164 [DOI] [PubMed] [Google Scholar]
- Wei Y., Shen W., Dauk M., Wang F., Selvaraj G., Zou J. (2004). Targeted gene disruption of glycerol-3-phosphate dehydrogenase in Colletotrichum gloeosporioides reveals evidence that glycerol is a significant transferred nutrient from host plant to fungal pathogen. J. Biol. Chem. 279: 429–435 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.