To determine how microtubule (MT) nucleation and nuclear migration are controlled in multinucleated hyphae we deleted genes encoding MTOC subunits and AgStu2. The novel phenotypes we observed in these mutants compared with analogous deletions in budding yeast allowed us to assign functions to the two types of cMTs that we observe in A. gossypii.
Abstract
In the multinucleate fungus Ashbya gossypii, cytoplasmic microtubules (cMTs) emerge from the spindle pole body outer plaque (OP) in perpendicular and tangential directions. To elucidate the role of cMTs in forward/backward movements (oscillations) and bypassing of nuclei, we constructed mutants potentially affecting cMT nucleation or stability. Hyphae lacking the OP components AgSpc72, AgNud1, AgCnm67, or the microtubule-stabilizing factor AgStu2 grew like wild- type but showed substantial alterations in the number, length, and/or nucleation sites of cMTs. These mutants differently influenced nuclear oscillation and bypassing. In Agspc72Δ, only long cMTs were observed, which emanate tangentially from reduced OPs; nuclei mainly moved with the cytoplasmic stream but some performed rapid bypassing. Agnud1Δ and Agcnm67Δ lack OPs; short and long cMTs emerged from the spindle pole body bridge/half-bridge structures, explaining nuclear oscillation and bypassing in these mutants. In Agstu2Δ only very short cMTs emanated from structurally intact OPs; all nuclei moved with the cytoplasmic stream. Therefore, long tangential cMTs promote nuclear bypassing and short cMTs are important for nuclear oscillation. Our electron microscopy ultrastructural analysis also indicated that assembly of the OP occurs in a stepwise manner, starting with AgCnm67, followed by AgNud1 and lastly AgSpc72.
INTRODUCTION
Microtubule-organizing centers (MTOCs) are a structurally diverse class of organelles involved in cell division, intracellular trafficking, cytoplasmic organization, and motility. Defined by the localization of γ-tubulin and its associated proteins, the function of MTOCs in nucleation and anchorage of microtubules is highly conserved in all eukaryotes. Centrosomes are the primary MTOC in metazoans, whereas nuclear-associated spindle pole bodies (SPBs) function as MTOCs in fungi. In the budding yeast Saccharomyces cerevisiae, the SPB serves as the sole site of microtubule nucleation, forming both nuclear microtubules involved in spindle assembly and chromosome segregation and cytoplasmic microtubules (cMTs) involved in nuclear migration and spindle positioning (reviewed in Jaspersen and Winey, 2004). The multinucleate filamentous fungus Ashbya gossypii also contains nuclear-associated SPBs as its only MTOC (Lang et al., 2010).
Structural analysis of the multilayered A. gossypii SPB from wild-type cells by using electron microscopy (EM) revealed considerable similarity to the SPB of S. cerevisiae (Lang et al., 2010). However, structural differences with functional implications where found at the cytoplasmic side of the A. gossypii SPB. Two types of cMTs emerge from a spherical outer plaque (OP), one type in perpendicular and the other type in tangential orientations. The perpendicular cMTs are generally short and make connections with the cortex, similar to budding yeast cMTs in G1 cells (Carminati and Stearns, 1997; Shaw et al., 1997). The tangential class of cMTs is not found in budding yeast. These very long cMTs bypass other nuclei in the multinucleated hyphae. A model for the role of both types of cMTs in nuclear movements was proposed in Lang et al., 2010 based on EM and live cell imaging experiments. Long tangential cMTs were predicted to be important for long-range nuclear bypassing events and the shorter cortex-connected cMTs were predicted to play a role in short-range nuclear oscillations. To test this model, it is necessary to find and investigate mutants with altered cMT nucleation and nuclear migration behavior. Based on the evolutionary relationship between A. gossypii and S. cerevisiae (Dietrich et al., 2004), it is highly likely that both organisms use similar proteins to nucleate and anchor microtubules. Therefore, genes deleted for analysis of cMT function in multinucleated A. gossypii hyphae were selected based on extensive knowledge of S. cerevisiae SPBs.
In budding yeast, three SPB substructures are directly involved in binding of the γ-tubulin complex and thus microtubule nucleation. The γ-tubulin complex is tethered to the nuclear face of the SPB, known as the inner plaque (IP), by Spc110 and nucleates the intranuclear microtubules that form the spindle (Kilmartin and Goh, 1996; Spang et al., 1996; Knop and Schiebel, 1997; Nguyen et al., 1998; Pereira et al., 1998). A different γ-tubulin complex binding protein, Spc72, is used for tethering Tub4, Spc97, and Spc98 to the cytoplasmic side of the SPB. The site of Spc72 binding and cMT nucleation changes throughout the cell division. During most of the cell cycle, Spc72 is present at the SPB OP but during G1 phase it can be associated with the half-bridge or bridge region (Rout and Kilmartin, 1990; Knop et al., 1997; Knop and Schiebel, 1998). Other components required for formation of the OP and anchoring of cMTs include the intermediate layer (IL)1 to OP spacer Cnm67 and the OP protein Nud1 (Brachat et al., 1998; Elliott et al., 1999; Gruneberg et al., 2000; Schaerer et al., 2001; Muller et al., 2005). In contrast to Spc72, which can be found associated with the half-bridge or OP depending on the cell cycle stage, Cnm67 and Nud1 are localized exclusively to the OP of the SPB (Brachat et al., 1998; Adams and Kilmartin, 1999; Pereira et al., 1999).
Although most SPB components are essential for viability in budding yeast due to their role in SPB duplication and bipolar spindle formation, deletion analysis showed that components of the outer layers of the SPB are not absolutely required for viability in all strain backgrounds (Brachat et al., 1998; Soues and Adams, 1998; Hoepfner et al., 2000, 2002). However, cnm67Δ or spc72Δ cells often lack a nucleus or carry two or more nuclei due to defects in nuclear migration and spindle positioning, processes that both require cMTs. Survival of cnm67Δ cells, which lack an OP and the cMTs normally formed at this structure, results from a rescue pathway involving microtubule nucleation from the half-bridge (Brachat et al., 1998; Hoepfner et al., 2000). Certain temperature-sensitive mutants such as nud1-2 share a similar phenotype to cnm67Δ cells (Gruneberg et al., 2000), suggesting that it is also essential for OP formation and microtubule nucleation. Analysis of NUD1 is complicated by the fact that this SPB component serves as a scaffold for a signaling pathway that monitors spindle positioning and controls mitotic exit, know as the mitotic exit network (MEN; reviewed in Hoyt, 2000; Pereira and Schiebel, 2001; Stegmeier and Amon, 2004). Segregation of chromosomes to the daughter cell is critical in budding yeast, so deletion of NUD1 or most MEN components results in lethality. The SPB may also be a loading or storage site of Stu2, an essential, conserved microtubule-plus end binding protein of the XMAP215/Dis1 family that regulates both nuclear and cMT dynamics (Kosco et al., 2001; Severin et al., 2001; Pearson et al., 2003; van Breugel et al., 2003; Usui et al., 2003; Al-Bassam et al., 2006).
A. gossypii carries syntenic homologues for these SPB genes (Table 1). However, some of the encoded orthologues have <20% sequence identity, and it is unclear whether and how differences in the primary sequence translate into changes in SPB structure and cMT nucleation or anchorage needed to coordinate movements of nuclei in a multinucleated cytoplasm. To better understand which type of cMT controls nuclear oscillation or nuclear bypassing in A. gossypii, we constructed and analyzed deletions of A. gossypii genes orthologous to SPB genes of budding yeast. We found that some deletions had an analogous phenotype in A. gossypii as they did in S. cerevisiae, whereas several other deletions resulted in unexpected and novel phenotypes, which allowed us to assign functions to the two types of cMTs and to present the ultrastructure of SPBs and attached cMTs in Agspc72Δ, Agstu2Δ, Agcnm67Δ and Agnud1Δ mutants.
Table 1.
Amino acid sequence comparison of SPB components
| S. cerevisiae SPB componenta | S. cerevisiae SPB localization | Role in SPB function in S. cerevisiae | A. gossypii orthologue | Protein length in amino acids (S.c./A.g.)b | % identityb |
|---|---|---|---|---|---|
| Tub4 | γ-Tubulin complex | Microtubule nucleation | AgTub4 | 473/470 | 56 |
| Spc97 | γ-Tubulin complex | Microtubule nucleation | AgSpc97 | 823/836 | 30 |
| Spc98 | γ-Tubulin complex | Microtubule nucleation | AgSpc98 | 846/845 | 41 |
| Spc72 | OP, HB | γ-tubulin complex binding protein | AgSpc72 | 622/795 | 22 |
| Nud1 | OP | MEN signaling | AgNud1 | 851/757 | 26 |
| Cnm67 | IL1, OP | Spacer, anchors OP to CP | AgCnm67 | 581/862 | 18 |
| Spc42 | CP, IL2 | Structural SPB core | AgSpc42 | 363/314 | 28 |
| Spc29 | CP | Structural SPB core | AgSpc29 | 253/293 | 20 |
| Cmd1 | CP | Spc110 binding protein | AgCmd1 | 147/147 | 95 |
| Spc110 | CP to IP | γ-Tubulin complex binding protein | AgSpc110 | 944/852 | 26 |
| Mps2 | SPB periphery | SPB insertion | AgMps2 | 387/338 | 19 |
| Ndc1 | SPB periphery | SPB insertion | AgNdc1 | 655/591 | 36 |
| Bbp1 | SPB periphery | SPB core to HB membrane linker | AgBbp1 | 385/353 | 26 |
| Nbp1 | SPB periphery | SPB core to HB membrane linker | AgNbp1 | 319/328 | 28 |
| Cdc31 | HB | SPB duplication | AgCdc31 | 161/172 | 71 |
| Sfi1 | HB | SPB duplication | AgSfi1 | 946/991 | 24 |
| Kar1 | HB | SPB duplication | AgKar1 | 433/330 | 23 |
| Mps3 | HB | SPB duplication | AgMps3 | 682/616 | 35 |
a SPB components are defined as proteins required to maintain the structural integrity of the organelle (Jaspersen and Winey, 2004).
b Based on gene information in the S. cerevisiae and A. gossypii databases. Percentage of identity was determined along the entire length as described in Materials and Methods.
MATERIALS AND METHODS
A. gossypii Media and Growth Conditions
A. gossypii media and culturing are described in Ayad-Durieux et al. (2000) and Wendland et al. (2000), and strains are listed in Supplemental Table S1.
Plasmid and Strain Construction
Plasmids generated and used in this study are described below. All DNA manipulations were carried out according to Sambrook and Russell (2001) with Escherichia coli DH5αF' as host (Hanahan, 1983). Polymerase chain reaction (PCR) amplification was performed using standard methods with Taq DNA polymerase, Expand High Fidelity PCR system, or the Expand Long Template PCR system (Roche Diagnostics, Indianapolis, IN). Oligonucleotides are listed in Supplemental Table S2 and were synthesized by Microsynth (Balgach, Switzerland).
A. gossypii deletion mutants were made using the PCR-based one-step gene targeting approach with heterologous selection markers (Wendland et al., 2000). The deletion cassettes were amplified from a plasmid containing the GEN3 (Wendland et al., 2000) cassette that mediates resistance to G418, using ‘gene name’_NS1/F2 oligonucleotide pairs. Correct integration of the deletion cassettes was verified with oligonucleotide primer pairs ‘gene name’_A1/Gen2_A2 (N terminus) and ‘gene name’_A4/Gen2_A3 (C terminus). Three independent transformants were characterized for each mutant. Transformation of multi-nucleate mycelium leads to heterokaryotic cells, which contain a mixture of transformed and wild-type nuclei. For subsequent analysis, homokaryotic mycelia were obtained by isolating and growing single spores. To evaluate phenotypes of lethal mutants or mutants with sporulation deficiency, spores from heterokaryotic mycelium were germinated and analyzed under selective conditions (200 μg/ml G418; Sigma-Aldrich, St. Louis, MO).
Fluorescence Microscopy and Image Processing
DNA (Hoechst) and immunofluorescence stainings were performed as described previously (Ayad-Durieux et al., 2000; Gladfelter et al., 2006). Rat anti-α-tubulin (YOL1/34; Serotec, Oxford, United Kingdom) was used at a 1:25 dilution and Alexa Fluor 568 goat anti-rat immunoglobulin (IgG; Invitrogen, Carlsbad, CA) at a 1:200 dilution.
An Axioplan2 microscope equipped with the objectives Plan-Apochromat 100×/1.40 numerical aperture (NA) Oil differential interference contrast (DIC) and Plan-Apochromat 63×/1.40 NA Oil DIC (Carl Zeiss, Feldbach, Switzerland) and appropriate filters (Carl Zeiss and Chroma Technology, Brattleboro, VT) was used for microscopy. The light source for fluorescence microscopy was either a 75-W XBO lamp (Osram, Augsburg, Germany), controlled by a MAC2000 shutter and filter wheel system (Ludl Electronics, Hawthorne, NY) or a Polychrome V monochromator (TILL Photonics, Gräfelfing, Germany). Images were acquired at room temperature using a cooled charge-coupled device camera (CoolSNAP HQ; Photometrics, Tucson, AZ) with MetaMorph 6.2r5 software (Molecular Devices, Sunnyvale, CA). Out-of-focus shading references were used for DIC image acquisitions.
For time-lapse image acquisition, a glass slide was covered with 1 ml of A. gossypii minimal medium containing 1% agarose. Once the medium had solidified, either small pieces of mature mycelium from the border of 3-d-old A. gossypii colonies or young mycelia cultured in liquid medium were spotted onto the slides. Seventy microliters of liquid minimal medium was added to the mycelia before cells were covered with a coverslip and incubated for at least 1 h before image acquisition. For still images, multiple planes with a distance between 0.3 and 1 μm in the z-axis were taken.
Image processing was performed with MetaMorph 6.2r5 software. Z-stacks were deconvolved with Nearest Neighbor and compressed by maximum or average projection with Stack Arithmetic. Brightness and contrast were adjusted using Scale Image. Images were colored and overlaid by using Overlay Images and exported from MetaMorph as 8-bit grayscale or RGB TIFF files. Z-Stacks and time-lapse picture series were converted to QuickTime H.264 movies with Quicktime Player Pro (Apple Computer, Cupertino, CA).
Transmission Electron Microscopy
Spores were grown for 10–14 h in liquid AFM to give rise to small mycelia containing no >100 nuclei. Samples were frozen on the Leica EM-Pact at ∼ 2050 bar and then transferred under liquid nitrogen into 2% osmium tetroxide/0.1% uranyl acetate/acetone and transferred to an automated freeze-substitution apparatus (AFS) (Leica, Wetzler, Germany). The freeze substitution protocol was as follows: −90°C for 16 h, up 4°C/h for 7 h, −60°C for 19 h, up 4°C/h for 10 h, and −20°C for 20 h. Samples were removed from the AFS and placed in the refrigerator for 4 h and then allowed to incubate at room temperature for 1 h. Samples went through three changes of acetone over 1 h and were removed from the planchettes. Then, they were embedded in acetone/Epon mixtures to final 100% Epon over several days in a stepwise procedure as described previously (McDonald, 1999). We cut 60-nm serial thin sections on a UC6 ultramicrotome (Leica), stained them with uranyl acetate and Sato's lead, and imaged them on a Spirit transmission electron microscope (FEI Technai, Hillsboro, OR). Serial section images were aligned using AutoAligner (Bitplane, Zurich, Switzerland). For the AgNUD1 deletion and some samples of the AgCNM67 deletion, mycelium of the border of 3-d-old colonies was frozen and subsequently treated as described above. We could not detect any differences in the SPB structure between either method of sample preparation.
Bioinformatic Analysis
Nuclear localization signal (NLS) search was performed with PredictNLS (http://cubic.bioc.columbia.edu/predictNLS/) and Prosite (http://www.expasy.org/prosite/PS50079). Protein alignments were performed with sequences retrieved from the Ashbya Genome Database (http://agd.vital-it.ch/; Gattiker et al., 2007) and the Saccharomyces Genome Database (http://www.yeastgenome.org/) by using the EMBOSS Pairwise Alignment Algorithms (Blosum62 Matrix, gap open 10, gap extend 0.5).
RESULTS
Essential and Nonessential SPB Components in A. gossypii
We searched the A. gossypii genome for homologues of genes that encode components of the evolutionary related budding yeast SPB. Sequence analysis revealed that the A. gossypii genome encodes syntenic homologues for all 18 mitotic S. cerevisiae SPB components (Table 1). Most orthologous proteins share only 20–40% identity. Notable exceptions are Tub4 (γ-tubulin) and the calcium-binding proteins Cmd1 (calmodulin) and Cdc31 (centrin), which are 56, 95, and 71% identical, respectively. These proteins are conserved components of many MTOCs, including SPBs and centrosomes (Jaspersen and Winey, 2004).
Given the structural similarities between the A. gossypii and S. cerevisiae SPB (Lang et al., 2010) but the relatively low level of sequence conservation of its constituent parts, we were interested to test functional conservations using a gene deletion approach. We asked whether elimination of core cytoplasmic components had similar effects on SPB structure, microtubule nucleation, and nuclear migration in multinucleated hyphae as in budding yeast cells or whether some deletions caused unexpected phenotypes due to the evolutionary adaptation of mechanisms of nuclear migration in both systems. A priori, it cannot be excluded, that an orthologue is essential in S. cerevisiae but nonessential in A. gossypii, so deletion analysis of SPB components in A. gossypii could result in structural and functional insight of mutant SPBs not possible in budding yeast. Specifically, we created deletion mutants in components of the γ-tubulin complex, the half-bridge, and the OP because these SPB substructures have well-documented roles in cMT organization as well as nuclear migration and positioning in S. cerevisiae (Rose and Fink, 1987; Geissler et al., 1996; Spang et al., 1996; Knop et al., 1997; Brachat et al., 1998; Chen et al., 1998; Pereira et al., 1999; Gruneberg et al., 2000; Hoepfner et al., 2000, 2002; Usui et al., 2003). In our analysis of A. gossypii SPB deletion mutants, we mainly focused on nuclear migration dynamics, formation of cMTs, and the structure of the mutant SPBs with the aim of providing a mechanistic model for long-range nuclear migration within multinucleate hyphae.
We found that A. gossypii genes encoding components of the γ-tubulin complex (AgTUB4, AgSPC97, and AgSPC98) are all essential. Visualization of nuclei in germinated spores lacking AgTUB4, AgSPC97, or AgSPC98 by using histone H4-green fluorescent protein (AgH4-GFP) revealed a clear nuclear division defect: nuclear density was decreased ∼10-fold compared with wild-type (n > 100) and nuclei were elongated or fragmented (Figure 1, A and B). The γ-tubulin complex probably nucleates microtubules at both the IP and OP of the SPB. Because cMTs are dispensable for hyphal growth of A. gossypii (Lang et al., 2010), the lethality of these mutants is probably due to defects in spindle microtubule nucleation at the IP rather than nucleation of cMTs at the OP.
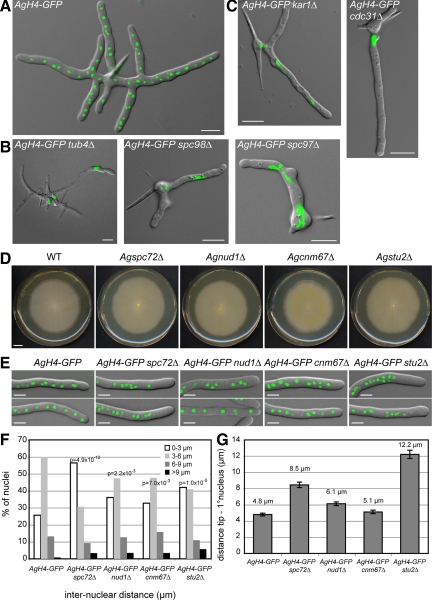
Figure 1.
Deletion of A. gossypii SPB components. Overlays of DIC image and AgH4-GFP signals from cells grown for ∼12 h at 30°C. (A) A typical wild-type mycelium at this stage contains multiple branches and 50–100 nuclei. (B) Terminal phenotype of mutants lacking components of the γ-tubulin complex. (C) Terminal phenotype of mutants lacking half-bridge components. Some mutants shown in B and C arrest as small mycelium with up to 12 branches containing a few nuclei and did not arrest as germlings with one nucleus or two nuclei. This is due to a maternal effect, where remnants of the wild-type protein are packed into mutant spores (Gladfelter et al., 2006). In A. gossypii, targeted deletions are performed with young mycelium containing ∼20 haploid nuclei that initially generate heterokaryotic strains: mixtures of nuclei carrying either the wild type or the deletion allele. On starvation, haploid mononucleate spores are produced in the heterokaryotic hyphae, and a substantial part of the mutant spores contain wild-type (maternal) protein, which can account for early growth and nuclear division in otherwise fatal deletions. Bars, 10 μm. (D) Radial growth of wild-type and deletion mutants on solid medium during 7 d of incubation at 30°C. Bar, 1 cm. (E) Overlays of DIC image and AgH4-GFP signals from hyphae showing nuclear distributions in wild-type and mutant strains. Bars, 5 μm. (F) Distribution of distances between adjacent nuclei was plotted using distance measurements between the nuclei in the first 50 μm of at least 20 different hyphae for each strain (n > 200 for each strain). The center of the GFP signal was used as the central point of the nucleus. Confidence values (p) for the χ2 test were calculated for each data set between wild-type and mutant distributions. (G) Distance between the hyphal tip and the first nucleus was determined. Error bars represent the SE of the mean, n > 60 for each strain.
During G1 phase of the S. cerevisiae cell cycle, cMTs are nucleated on the half-bridge in a Kar1-dependent manner (Byers and Goetsch, 1975; Pereira et al., 1999). The ability to form microtubules at the half-bridge is essential for survival of SPB OP mutants such as cnm67Δ (Brachat et al., 1998; Hoepfner et al., 2000). In A. gossypii, hyphae lacking the half-bridge component AgKar1 stopped growing as small mycelium containing up to 10 nuclei (Figure 1C). Thus, AgKAR1 is an essential gene. Dependent on the remnant AgKar1 proteins in mutant spores, SPBs can still duplicate two to three times, permitting a few nuclear divisions. The fact that deletion of AgCDC31, encoding another half-bridge component presumably not involved in cMT organization, resulted in the same terminal phenotype (cessation of growth as small mycelium; Figure 1C), suggests that lethality in cells lacking AgKAR1 is due to a general SPB duplication defect rather than a specific effect on cMTs.
Deletions of AgSPC72, AgNUD1, AgCNM67, and AgSTU2 did not affect radial growth of colonies, a sensitive method to test for reduced hyphal growth (Figure 1D). S. cerevisiae orthologues of the first three genes encode OP components, the orthologue of AgSTU2 encodes an OP-associated MT-stabilizing protein, and deletions of these genes are either lethal or cause reduced growth in S. cerevisiae (Brachat et al., 1998; Chen et al., 1998; Knop and Schiebel, 1998; Soues and Adams, 1998; Adams and Kilmartin, 1999; Hoepfner et al., 2000, 2002; Usui et al., 2003; Al-Bassam et al., 2006). Even though polar growth was not affected in the four A. gossypii deletions, alterations in nuclear distributions were noted in each (Figure 1, E–G).
The deletions are expected to alter the formation of microtubules at the cytoplasmic side of SPBs and microtubule stability thus affecting nuclear mobility. We therefore monitored the migration of H4-GFP labeled nuclei in wild-type and the four deletions, quantified the frequencies of forward and backward movements (oscillations) and bypassing of nuclei, visualized MTs in vitro by immunofluorescence microscopy and in vivo using GFP-AgTub1 and finally used EM to determine the structure of SPBs in the mutants and their ability to nucleate perpendicular and tangential cMTs. Because of the complexity of this analysis we have documented the nuclear mobilities for all mutants in one figure and one table to allow for direct comparisons (Figure 2; Table 2), and we compiled separately for each mutant images of cMTs and SPBs (Figures 3–6). In the following paragraphs, we present and discuss the data for each of the four mutants individually, starting each time with a functional description of the S. cerevisiae orthologue. The structural and functional results of our studies will be summarized at the end (Figures 7–9).
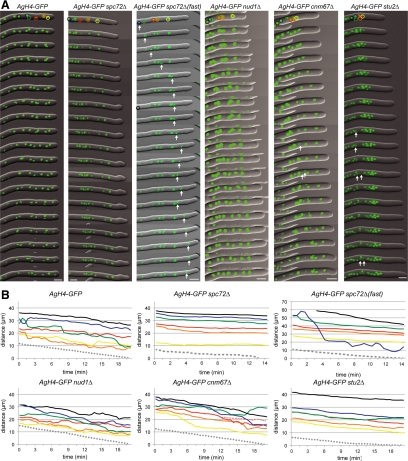
Figure 2.
Time-lapse analysis of nuclear migration in different mutants. (A) Overlays of DIC and AgH4-GFP signals from time-lapse video imaging of wild-type (Supplemental Movie S1), Agspc72Δ (Supplemental Movie S2), Agnud1Δ (Supplemental Movie S4), Agcnm67Δ (Supplemental Movie S5), and Agstu2Δ (Supplemental Movie S6) hyphae. Images were captured every 30 s, and 1-min interval frames are shown. Migration of the first six nuclei were tracked and are shown in the schematic. In Agspc72Δ(fast) (Supplemental Movie S3), images from each 30-s time point are shown for the first 7 min followed by 1-min interval frames; the rapidly moving nucleus is indicated by an arrow. Nuclei undergoing mitosis are indicated by arrows in Agcnm67Δ and Agstu2Δ. Bars, 5 μm. (B) Positions of the first six nuclei in each hyphae and the hyphal tip (gray dotted line) were tracked throughout each time course and are plotted. In wild-type, the nuclei were observed to undergo bypassing and oscillations. In Agspc72Δ(fast), one nucleus (blue) moves rapidly toward the tip, thereby bypassing four other nuclei in <5 min and traveling distances up to 8.9 μm within a 30-s interval (1.5–2 min). The other nuclei in this hypha as well as in the other Agspc72Δ and Agstu2Δ mutants move toward the tip with the cytoplasmic stream without undergoing any bypassing or oscillation.
Table 2.
Nuclear oscillations and bypassing in OP and Agstu2Δ mutants
| Straina | Distance migrated (μm)/30 s |
Nuclear oscillationsb |
Nuclear bypassing |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Avg. | max | n | pc value | Forward movement | Backward movement | Events | Total nuclei | % | |
| Wild-type | 0.51 | 2.82 | 1264 | 101 | 41 | 15 | 48 | 31 | |
| spc72Δ | 0.35 | 1.39 | 1177 | <0.001 | 48 | 0 | 4 | 42 | 10 |
| (2.12) | (8.91) | (82)d | <0.001 | (8)e | |||||
| nud1Δ | 0.45 | 2.3 | 861 | <0.01 | 81 | 28 | 8 | 36 | 22 |
| cnm67Δ | 0.56 | 3.11 | 770 | <0.05 | 138 | 28 | 17 | 25 | 68 |
| stu2Δ | 0.36 | 1.23 | 240 | <0.001 | 9 | 0 | 0 | 32 | 0 |
a Background AgH4-GFP.
b The movements of the five most apical nuclei in four different hyphae (1 hypha for stu2Δ) were followed >40 or >30-s time intervals. Forward or backward displacements ≥0.75 μm (approximately half the diameter of a nucleus) within 30 s were scored.
c Statistical significance between mutant and wild-type values was calculated using an unpaired Students t test, and the resulting p value is shown. We also compared different mutant data sets with each other and all average values are highly statistically significant except for the slight difference in the average values from Agspc72Δ and Agstu2Δ mutants (p > 0.05).
d A subpopulation of nuclei in Agspc72Δ mutants undergo fast, long-range bypassing (see Figure 2). The numbers in brackets concern these nuclei, which were excluded when the average of parameters was determined.
e Each bypassed nucleus was counted.
Figure 3.
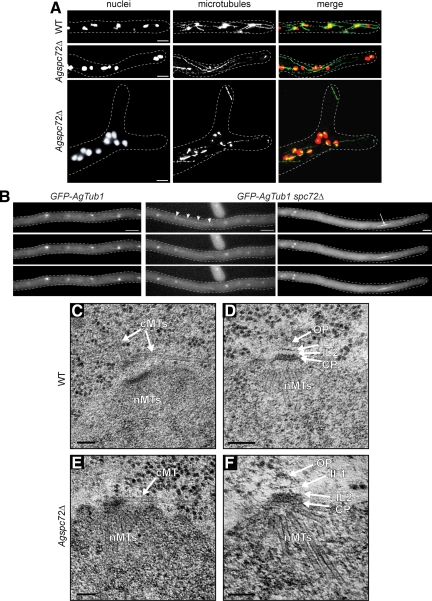
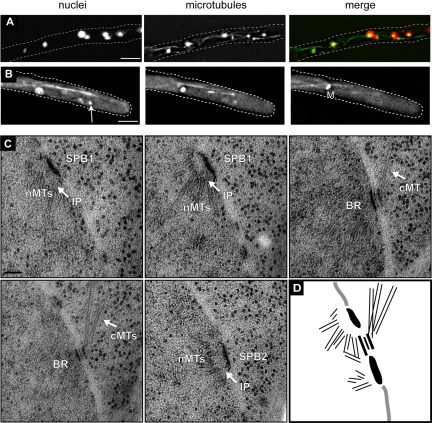
Nuclear movement, cMTs and SPB structure in Agspc72Δ. (A) Wild-type and Agspc72Δ mutants were stained with Hoechst to visualize DNA and anti-α-tubulin antibodies to detect microtubules. In the bottom image of Agspc72Δ, a cluster of nuclei can be seen at a branch site. These hyphae lack short cMTs, whereas long cMTs that extend along the growth axis are still present. A detached microtubule can be seen in the upper tip region. Bars, 5 μm. (B) Representative, deconvolved images of a Z-stack of a wild-type and two Agspc72Δ hyphae expressing GFP-AgTUB1. The complete stacks are available as Supplemental Movies S7, S8, and S9. Long and short cMTs can be seen emerging from bright foci that represent the SPBs in wild-type, but Agspc72Δ SPBs often lack associated cMTs. Arrowheads point to a long cMT in Agspc72Δ, which may facilitate bypassing. An arrow indicates an anaphase spindle. Bar, 5 μm. (C) EM image of SPB-attached microtubules in wild-type hyphae. Bar, 100 nm. The SPB is associated with nuclear microtubules (nMTs) at the IP and a tangential and perpendicular microtubule at its OP (arrows). (D) SPB structure in wild-type. Bar, 100 nm. A central plaque (CP) plus IL1 and IL2 are marked by arrows. A small amount of amorphous material could also be detected above IL1 and is part of the outer plaque (OP) (Lang et al., 2010). (E) EM image of SPB-attached microtubules in Agspc72Δ. Bar, 100 nm. The SPB is associated with nMTs at the IP and a tangential microtubule close to the central plaque (arrow). In this and other thin sections, we never observed perpendicular microtubules. (F) SPB structure in Agspc72Δ. Bars, 100 nm. A CP plus IL1 and IL2 are marked by arrows. In some cases, a small amount of amorphous material could also be detected above IL1, which could be part of the OP. However, the size of this OP remnant was substantially reduced compared with a wild-type OP (Lang et al., 2010).
Figure 4.
SPBs of Agnud1Δ lack an OP and nucleate cMTs from the bridge. (A) Agnud1Δ mutants were stained with Hoechst to visualize DNA and anti-α-tubulin antibodies to detect microtubules. Long cMTs that extend along the growth axis are still present and also a few short cMTs were detected. Bar, 5 μm. (B) Representative, deconvolved images of a Z-stack from Agnud1Δ hypha expressing GFP-AgTUB1. The complete stack is available as Supplemental Movie S10. Long and short cMTs can be seen emerging form both sides of a mitotic spindle (M) and from bright foci that represent the SPBs. An arrow points to a SPB lacking cMTs. Bar, 5 μm. (C and D) EM images of Agnud1Δ mutants. Bars, 100 nm. (C) Serial section images of a single SPB in an Agnud1Δ mutant. The central plaque (CP) and IL1 and IL2 were observed, whereas the outer plaque could not be detected in any section. (D) Serial section images of duplicated side-by-side SPBs (SPB1 and SPB2) connected by a bridge (BR) in the Agnud1Δ mutant. Nuclear microtubules (nMTs) as well as two cMTs can be seen. The cMTs seem to emerge from the bridge region. (E) Schematic summarizing the five serial sections images of D.
Figure 5.
cMT nucleation and SPB structure in Agcnm67Δ hyphae. (A) Agcnm67Δ hypha stained with Hoechst to visualize DNA and anti-α-tubulin antibodies to detect microtubules. Long cMTs that extend along the growth axis are present and a few short cMTs were detected. Bar, 5 μm. (B) Representative, deconvolved images of a Z-stack of an Agcnm67Δ hypha expressing GFP-AgTub1. The complete stack is available as Supplemental Movie S11. Long and short cMTs can be seen emerging form both sides of a mitotic spindle (M) and from bright foci that represent the SPBs. An arrow points to a SPB lacking cMTs. Bar, 5 μm. (C) Serial sections of duplicated side-by-side SPBs (SPB1 and SPB2) connected by a bridge (BR), which nucleates three cMTs in an Agcnm67Δ mutant. Nuclear microtubules (nMTs) are formed at the SPB IP. Images were aligned using AutoAligner to produce the schematic in D. Bars, 100 nm.
Figure 6.
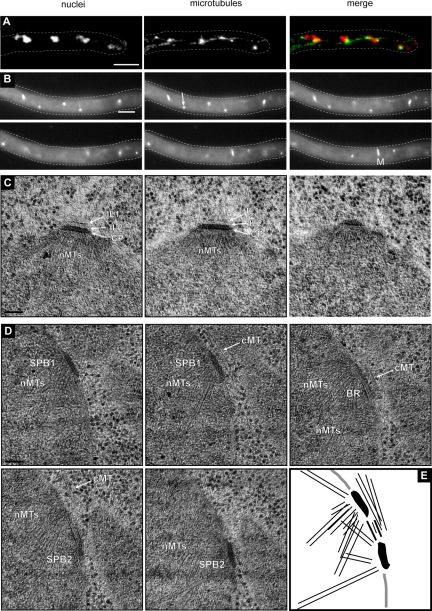
Microtubule stability and SPB structure in Agstu2Δ mutants. (A) Agstu2Δ hypha stained with Hoechst to visualize DNA and anti-α-tubulin antibodies to detect microtubules. Only short cMTs were detected. Bar, 5 μm. (B) Representative, deconvolved images of a Z-stack of an Agstu2Δ hypha expressing GFP-AgTUB1. The complete stack is available as Supplemental Movie S12. (C) EM image of a SPB from Agstu2Δ showing OP, IL2, IL1, and central plaque (CP) layers as well as nuclear microtubules (nMTs). (D) Serial sections of an Agstu2Δ SPB also show the layered SPB structure and nMTs as in A. In addition, three very short cMTs are visible, which have both perpendicular and tangential orientations with the SPB. Arrows point to “flared” microtubule ends frequently seen by EM in Agstu2Δ mutants. Bars, 100 nm.
Figure 7.
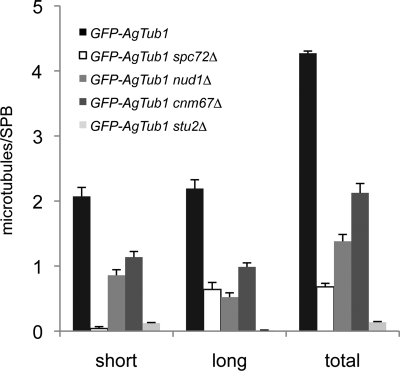
Quantitation of short and long cMTs in hyphae expressing GFP-AgTUB1. The number of short (≤5-μm) and long (>5-μm) cMTs per SPB was quantitated in wild-type and mutant hyphae expressing GFP-AgTUB1, and average values observed for both classes of microtubules are presented along with a combined total. Error bars indicate SE of the mean (n = 77, 39, 46, 92, and 58 SPBs for GFP-AgTUB1, GFP-AgTUB1 spc72Δ, GFP-AgTUB1 nud1Δ, GFP-AgTUB1 cnm67Δ, and GFP-AgTUB1 stu2Δ, respectively). Note that only a minor fraction of the long cMTs in GFP-AgTUB1 spc72Δ and GFP-AgTUB1 nud1Δ are >12 μm, whereas in GFP-AgTUB1 cnm67Δ mutants, a substantial fraction is >12 μm.
Figure 8.
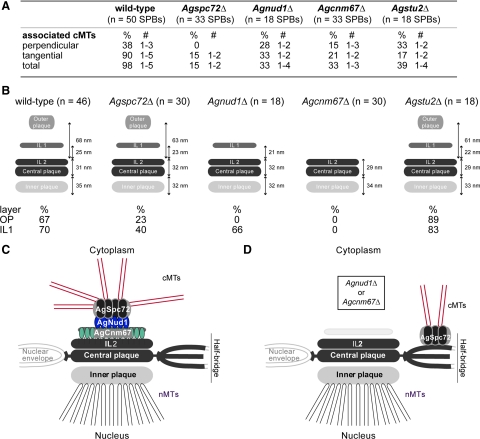
Comparison of A. gosspyii wild-type and mutant SPB structure and cMT nucleation based on EM analysis. (A) Quantitation of cMTs that emerge per SPB in wild-type and mutants based on EM analysis. (B) Schematics of wild-type and mutant SPBs depicting the observed layers and averaged distances between the layers, based on measurements shown in Supplemental Table S3. The percentage of SPBs in which the OP and IL1 were seen is indicated below. (C) Model of the A. gossypii SPB based on SPB morphology we observed in different deletion mutants. The layered SPB of A. gossypii is composed of AgCnm67 in IL1, which recruits AgNud1. Next, AgSpc72 binds to form the OP and tether the γ-tubulin complex that nucleates cMTs. (D) In the absence of AgCNM67 or AgNUD1, cMTs are formed from the bridge region (Brachat et al., 1998; Gruneberg et al., 2000; Hoepfner et al., 2000). Studies in budding yeast lead us to hypothesize that AgSpc72 and the γ-tubulin complex relocalize here (Knop and Schiebel, 1998), at least in these mutants. In Agnud1Δ, IL1 remains, whereas it is lost in Agcnm67Δ.
Figure 9.
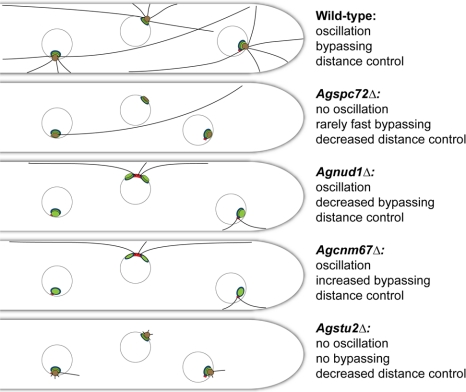
Model of the cytoplasmic sides of SPBs and attached cMTs in wild-type and mutant hyphae of A. gossypii. The four deletion mutants presented here have altered cMT arrays compared with wild-type, and these alterations can explain most of the observed changes in nuclear mobility and in distance control to the hyphal tip assuming that microtubules can exert pulling forces upon cortical contacts, which may be transient, e.g., causing oscillations, or more stable, e.g., for nuclear bypassing driven by long microtubules. See Discussion for details.
Agspc72Δ Lacks Nuclear Oscillations but Occasionally Shows Bypassing
Spc72 serves as the cytoplasmic γ-tubulin complex anchor in S. cerevisiae (Knop and Schiebel, 1998; Pereira et al., 1999) and a deletion of the ScSPC72 gene is lethal or yields in some strains slowly growing colonies with bi- and anucleated cells (Soues and Adams, 1998; Hoepfner et al., 2002; Usui et al., 2003). Based on analysis in budding yeast, it is likely that deletion of AgSPC72 also prevents anchoring of cMTs at the OP. Previously, we showed that nuclear migration within the multinucleate A. gossypii hyphae can be broken down into four types of movement: rotation; forward/backward oscillation and bypassing of nuclei, which are cMT-dependent processes; and cotransport with the cytoplasmic stream, the only observable nuclear movement in the absence of cMTs (Lang et al., 2010). Because of this later microtubule-independent mechanism of tip-directed nuclear movement, it seemed likely that deletion of AgSPC72 will not be lethal but would affect cMT-dependent nuclear mobility.
Indeed, Agspc72Δ mutants were viable and, as shown above, displayed radial colony growth like wild-type even though nuclei were less evenly distributed and formed small clusters (Figure 1, D–F). We monitored the movements of the first five nuclei in four growing hyphae of Agspc72Δ for a total of 800 half-min time intervals. Alternating forward/backward movements were not found, and only slow forward movements were observed that rarely exceeded the growth speed of the hyphae (Figure 2, A and B; Table 2; and Supplemental Movies S1 and S2). The one exception was in a rapidly forward moving nucleus able to bypass four nuclei as documented in the image series and the graph labeled AgH4-GFPspc72Δ(fast) in Figure 2, A and B (see also Supplemental Movie S3). Further investigation of 70 nuclei in 10 hyphae revealed that three of these highly motile nuclei, which could move with maximum speed of 8.9 μm in 30 s, 3 times faster than the maximum speed in wild type (Table 2). Thus, Agspc72Δ hyphae seem to contain two populations of nuclei: a subpopulation of highly mobile nuclei able to undergo long-range bypassing and the majority of nuclei, in which nuclear migration is controlled by cytoplasmic streaming and not by cMTs.
Next, we examined microtubules by immunostaining with antibodies to α-tubulin and by live cell imaging of hyphae expressing substoichiometric levels of GFP-labeled α-tubulin (GFP-AgTub1) in addition to wild-type α-tubulin. Both methods can be used for visualizing cMTs in A. gossypii but with some limitations (Gladfelter et al., 2006; Lang et al., 2010). The very thin cMTs of A. gossypii are refractory to fixation and are destabilized by more than a substoichiometric concentration of GFP-AgTub1. Compared with wild type, nuclei in Agspc72Δ hyphae lack short cMTs; but, surprisingly, a few long cMTs were observed attached or detached from nuclei (Figures 3, A and B, and 7; and Supplemental Movies S7–S9).
We used serial section EM to analyze the direction in which the few remaining cMTs emanate from Agspc72Δ mutant SPBs because cMTs emerge from wild-type SPBs in a perpendicular and a tangential direction (Figure 3C) as described previously (Lang et al., 2010). EM analysis of 33 Agspc72Δ mutant SPBs showed no perpendicular cMTs but one or two tangential cMTs attached to the OP region of five mutant SPBs (Figures 3E and 8A). This lack of perpendicular (short) cMTs and the low frequency of tangential (long) cMTs are consistent with our immunofluorescence and live cell imaging data; in GFP-AgTUB1 spc72Δ cells, only 31% (12/39) of the SPBs were associated with cMTs, and virtually all of these were long cMTs >5 μm (Figures 3B and 7 and Supplemental Movies S8 and S9). The few nuclei that contain SPBs attached to long tangential cMTs are probably those that rapidly migrate through the Agspc72Δ hyphae, thereby bypassing other nuclei. Together, this indicates that short perpendicular cMTs are probably important for oscillatory movements and long tangential cMTs for nuclear bypassing.
The serial EM sections also revealed structural differences between wild-type and mutant SPBs (Figures 3, D and F, and 8B). A clearly distinguishable OP and IL1 was seen in only 23 and 40% of Agspc72Δ SPBs, respectively, compared with 67 and 70% of wild type (Agspc72Δ, n = 30; wild type, n = 46; we excluded SPBs in this analysis if serial section images were not available). Other layers of the SPB were unaffected by deletion of AgSPC72.
Deletion of AgNUD1 Results in Loss of the OP but Not Nuclear Dynamics
The budding yeast gene ScNUD1 encodes an important component of the OP. Deletion of the gene is lethal, probably due to the role of Nud1 as a signaling scaffold for the essential MEN (reviewed in Hoyt, 2000; Pereira and Schiebel, 2001; Stegmeier and Amon, 2004). However, some viable point mutants of this gene were characterized and display disturbed organization of cMTs (Gruneberg et al., 2000), but the role of ScNud1 in OP structure could not be determined.
Surprisingly, the Agnud1Δ deletion mutant is viable displaying wild-type radial growth on agar (Figure 1D). Nuclear spacing was similar as in wild-type hyphae, although a tendency to form clusters was noted, as concluded from the increased (11%) number of nuclei with <3-μm distance to their nearest neighbor (Figure 1F). Forward/backward movements and bypassing of Agnud1Δ nuclei were observed as in wild-type (Figure 2, A and B, and Supplemental Movie S4). Examination of 800 half-min time intervals revealed a slight but statistically significant (p < 0.01) decrease in nuclear oscillation and a clear decrease in bypassing frequencies compared with wild type (Table 2). Interestingly, Agnud1Δ nuclei seemed to have a larger diameter than wild-type or other mutant nuclei we examined (Figure 2A). The reason for the difference is currently unknown but could be due to a delay in the nuclear division cycle or an increase in ploidy.
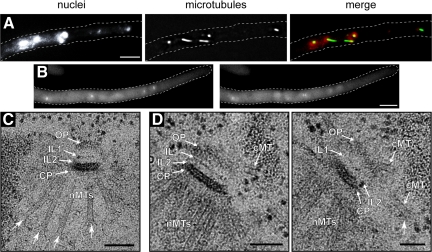
Hyphae lacking AgNUD1 contained both long and short cMTs, and analysis of GFP-AgTub1 images revealed that 89% (41/46) of the Agnud1Δ SPBs nucleated both types of cMTs (Figures 4, A and B, and 7; and Supplemental Movie S10). However, very long cMTs >12 μm were seldom observed in Agnud1Δ mutants. Unlike Agspc72Δ mutants, in which cMTs are often detached from nuclei (Figure 3A), we did not detect detached microtubules in the cytoplasm of >20 Agnud1Δ hyphae examined.
Analysis of 18 mutant SPBs by EM revealed a complete lack of the OP and a surprising arrangement of SPB-associated cMTs (Figures 4, C–E, and 8, A and B). Serial sections revealed that all duplicated mutant SPBs (n = 6 of 18) had cMTs attached to the bridge structure, approximately half in a perpendicular and half in a tangential direction (Figures 4D and 8A). We therefore conclude that A. gossypii SPBs that lack an OP can nucleate and apparently stably anchor cMTs from the bridge, presumably through AgSpc72. Furthermore, because nuclear oscillation and bypassing are still observed in Agnud1Δ mutants despite a reduced number of cMTs, microtubule anchorage, rather than the number of nucleated cMTs, is probably a key factor for nuclear migration dynamics in A. gossypii.
Deletion of AgCNM67 Does Not Impair Nuclear Oscillation or Bypassing
In budding yeast, Cnm67 attaches the OP to IL2, and cells lacking Cnm67 have no OP. This is lethal in some strains or leads to reduced colony growth in other strains because cells with no, two or many nuclei form due to a defect in nuclear positioning at the bud neck before anaphase (Brachat et al., 1998; Hoepfner et al., 2000; Schaerer et al., 2001). Based on these data from budding yeast, we anticipated an AgCNM67 deletion to be viable but have nuclear migration defects. It is possible that Agcnm67Δ mutants would be able to nucleate cMTs from the bridge/half-bridge as we observed in Agnud1Δ and has been observed in cells lacking ScCNM67 (Brachat et al., 1998).
We found that Agcnm67Δ colonies grow like wild-type and that nuclear spacing was only mildly affected (Figure 1, D–G). Agcnm67Δ nuclei were still able to undergo oscillation and bypassing (Figure 2, A and B, and Supplemental Movie S5). After nuclear oscillation and bypassing in 20 mutant nuclei for a total of 770 half-min intervals revealed a doubled frequency in bypassing events compared with wild-type, concomitant with increased forward and decreased backward movements (Table 2). Whereas the changed ratio in forward and backward movements, as defined in Table 2, can in part be explained by a slightly faster growth speed of the investigated Agcnm67Δ hyphae (Lang et al., 2010), the increased frequency in bypassing is probably a result of changes in cMT organization in this mutant.
Immunostaining with antibodies to α-tubulin and Z-stacks of GFP-AgTub1 labeled microtubules showed a reduced number of short and long cMTs attached to Agcnm67Δ nuclei compared with wild-type nuclei (Figures 5, A and B, and 7; and Supplemental Movie S11). The Z-stacks also revealed that 78% (72/92) of the Agcnm67Δ SPBs nucleated short, long, and very long (>12 μm) cMTs. Examination of 33 mutant SPB structures by EM showed a complete lack of the OP and IL1; but for 11 SPBs, cMTs emanated either from a half-bridge or a bridge structure in both perpendicular and tangential directions (Figures 5, C and D, and 8, A and B). This confirms that A. gossypii SPBs lacking an OP and an IL1 can nucleate and anchor cMTs from a bridge and presumably also half-bridge. It also shows that, in the presence of very long cMTs and a reduced number of short cMTs per SPB, the frequency in nuclear bypassing can increase. It would be interesting to test whether the half-bridge component AgKar1 participates in this rescue mechanism for cMT nucleation in the absence of an OP. But because AgKAR1 is essential (see above), we could not examine the phenotype of Agcnm67Δ kar1Δ or Agnud1Δ kar1Δ double mutants.
AgSTU2 Is Required for Nuclear Oscillation and Bypassing
Although Stu2 is not considered to be a core SPB component in budding yeast in the sense that it is required to maintain the structural integrity of the complex (Jaspersen and Winey, 2004), its localization to the SPB and microtubule plus-ends, biochemical and genetic interactions with SPB OP and γ-tubulin components, microtubule binding domain, and regulation of microtubule dynamics made it a leading candidate to participate in cMT organization (Wang and Huffaker, 1997; Chen et al., 1998; Kosco et al., 2001; Severin et al., 2001; Pearson et al., 2003; Usui et al., 2003; van Breugel et al., 2003; Al-Bassam et al., 2006), and thus control nuclear migration dynamics in A. gossypii. The A. gossypii genome contains a single XMAP215-like gene, AgSTU2, that is a syntenic homologue of S. cerevisiae STU2. AgStu2 and ScStu2 are 32% identical.
If AgStu2 is needed for cMT growth or stability, then we would expect to see short or absent microtubules in Agstu2Δ, which should manifest itself in defects in cMT-dependent forms of nuclear migration. Surprisingly, deletion of AgSTU2 resulted in viable cells that grew on agar plates similar to wild-type (Figure 1C). Labeling of Agstu2Δ nuclei with H4-GFP showed that nuclear spacing is affected: hyphae displayed enlarged distances from the leading nucleus to the tip region as well as decreased distances between adjacent nuclei (Figure 1, E–G). Nuclei in Agstu2Δ hyphae only migrated forward with the cytoplasmic stream (Figure 2, A and B, and Supplemental Movie S6). In 200 half-min intervals, no backward movement and no nuclear bypassing were observed (Table 2), indicating a lack of cMTs.
Analysis of microtubules by immunostaining with anti-α-tubulin antibodies showed some Agstu2Δ nuclei associated with very short cMTs, whereas long cMTs were absent (Figure 6A). Quantitation of microtubule length from immunofluorescence images revealed an average cMT length of 200 ± 56 nm (n = 31 cMTs/51 SPBs) in Agstu2Δ mutants compared with 1912 ± 93 nm (n = 275 cMTs/75 SPBs) in wild-type hyphae (data not shown). We also examined microtubules by live cell imaging using GFP-AgTub1 and found that cMTs were virtually undetectable (Figures 6B and 7 and Supplemental Movie S12). It is likely that GFP-AgTub1 leads to slightly less stable microtubules in vivo, which when combined with Agstu2Δ, further decreases cMT stability. Thus, the cMT number and length observed by immunofluorescence is very likely correct.
Unlike the other mutant SPBs we examined by EM, all known SPB layers were observed in Agstu2Δ and their dimensions are very similar to wild-type SPBs (Figures 6, C and D, and 8B). Even though the OP structure seems to be preserved, serial section EM analysis confirmed the significant reduction in the number of cMTs nucleated at Agstu2Δ SPBs: only 39% of the mutant OPs (n = 7 of 18) compared with 98% wild-type (n = 49 of 50) SPBs were associated with cMTs (Figure 8A). The cMTs we observed by EM were rather short but showed both perpendicular and tangential association to the SPB (Figures 6D and 8A). We also noticed that most of the non-SPB–associated microtubule ends in Agstu2Δ seemed to exhibit a “flared” or “peeling” structure, which has been observed previously in other EM studies and is indicative of shortening microtubules (Byers et al., 1978; Mandelkow et al., 1991; O'Toole et al., 1999). This phenotype was rarely detected in wild-type cells or other mutants we examined. From our analysis of Agstu2Δ, we conclude that cMTs nucleated in this mutant are too short to induce nuclear oscillations or even bypassing. The fact that Agstu2Δ is able to form both perpendicular and tangential attachments even though we never observed cMTs > 200 nm in this mutant (Figure 8, A and B; n = 60 cMTs) suggests that something other than microtubule length determines the orientation of cMTs. The short cMTs and normal SPB structure suggest that AgStu2 functions in plus-end microtubule dynamics and does not have a structural role at the SPB.
DISCUSSION
From our EM analysis of SPB structure in Agspc72Δ, Agnud1Δ and Agcnm67Δ mutants, we propose that the A. gossypii organelle is assembled in a stepwise manner. By analogy to S. cerevisiae SPB assembly, binding of AgCnm67 drives assembly of AgNud1, followed by AgSpc72 and the γ-tubulin complex to form the OP (Figure 8C). In wild-type hyphae, an intact OP is essential for nucleation and anchorage of cMTs involved in nuclear oscillations and bypassing. AgStu2 also plays an important role in the formation of cMTs and thus nuclear movements, although it is probably not involved in the structural maintenance of the SPB.
Role of cMTs in Nuclear Oscillations and Bypassing
A major goal of this work was to test the model for cMT-dependent nuclear movements proposed in our previous publication (Lang et al., 2010). Shown schematically in Figure 9, we proposed that arrays of short perpendicular and long tangential cMTs emanate from nuclear SPBs in the multinucleate hyphae of A. gossypii. Each of the three nuclei shown in the wild-type hypha is associated with short and long cMTs growing in apical and subapical directions from its OP. The most apical nucleus makes close contact to the growing tip via several cMTs. The other two nuclei are connected with the hyphal cortex via short cMTs and, based on preliminary data obtained with plus-end–labeled cMTs, also via their long cMTs (Lang et al., 2010; Grava and Philippsen, unpublished observations). We hypothesized that growth and shrinkage of cMTs provides pulling and pushing forces for short-range nuclear oscillations and that long-range movements during nuclear bypassing are probably achieved by pulling forces of the long cMTs when the cortex connection of short cMTs is reduced or absent. The four deletion mutants that we have analyzed here each have unique alterations in their cMT arrays compared with wild-type (summarized in Figures 7 and 8) that allowed us to test various aspects of this model. Based on the changes in cMT organization that we observed, we can explain most of the corresponding changes in nuclear mobility and in distance control in the mutants. In addition, although the A. gossypii SPB structure and core proteins are similar to S. cerevisiae, our analysis of the deletion mutants revealed unanticipated phenotypes that differed considerably from those described previously in budding yeast (Rose and Fink, 1987; Geissler et al., 1996; Spang et al., 1996; Knop et al., 1997; Brachat et al., 1998; Chen et al., 1998; Pereira et al., 1999; Gruneberg et al., 2000; Hoepfner et al., 2000, 2002; Usui et al., 2003), shedding light on the mechanism of cMT anchorage, SPB assembly, and the evolution of conserved cytoskeletal proteins in multinucleated hyphae.
Formation of the OP and cMT Nucleation: AgSpc72
In Agspc72Δ mutants, the majority of nuclei do not nucleate cMTs, therefore, they lack cMT-dependent movements and migrate with the cytoplasmic stream (Figure 9). The lack of cMTs in Agspc72Δ is similar to Scspc72Δ mutants, which form only very short and unstable cMTs (Soues and Adams, 1998; Hoepfner et al., 2002). In a few rare instances, we observed long tangential microtubules and an associated OP by EM in Agspc72Δ mutants; it seems that the long cMT can pull the attached nucleus over long distances, thereby rapidly bypassing several nuclei. The fact that short cortex-associated cMTs that may interfere with bypassing are also absent in Agspc72Δ hyphae could explain the high speed of these rare bypassing events. The lack of short cMTs also accounts for the increased distance of the most apical nucleus to the tip.
However, the question of how any cMTs are nucleated and attached at such a mutant SPB is unclear because Agspc72Δ mutants are lacking the presumed cytoplasmic anchor for the γ-tubulin complex. Due to the fact that budding yeast Stu2 binds to Spc72 and the γ-tubulin complex through sequences in its N terminus and stimulates polymerization of pure tubulin in vitro (Usui et al., 2003; van Breugel et al., 2003; Al-Bassam et al., 2006), AgStu2 was a candidate to help anchor cMTs to the SPB in the absence of AgSpc72, but our experimental data do not support this hypothesis. Another candidate for anchoring the γ-tubulin complex at the OP in the absence of AgSpc72 is AgSpc110. In S. cerevisiae, Spc110 tethers γ-tubulin to the SPB IP but it can function in cMT nucleation if it is artificially tethered to the OP (Knop and Schiebel, 1998). Normally, Spc110 is targeted to the nucleus by an N-terminal nuclear localization sequence (Adams and Kilmartin, 1999). This domain is not conserved in AgSpc110, and other nuclear localization motifs were not detected (Lang and Jaspersen, unpublished observations), so how AgSpc110 enters the nucleus is unknown. A portion of AgSpc110 might remain in the cytoplasm. The A. gossypii genome also carries multiple no homology in bakers yeast (NOHBY) genes that could play a role in SPB functions specific to multinucleate fungi, including nucleation of different classes of cMTs (Brachat et al., 2003).
Loss of AgNUD1 and AgCNM67 Reveals a Conserved Rescue Mechanism for cMT Nucleation at the SPB
In comparison with hyphae lacking AgSpc72, the absence of AgCnm67 or AgNud1 results in a radical change in SPB structure and microtubule organization: the OP is lost and cMTs are nucleated from the bridge/half bridge (Figure 8D). However, nuclear oscillation and bypassing are still observed in both mutants, indicating that bridge/half-bridge-nucleated cMTs are sufficient to direct typical nuclear movements within hyphae (Figure 9). In both mutants the total number of cMTs per nucleus is reduced approximately twofold compared with wild-type; therefore, anchorage to the SPB rather than the total number of nucleated cMTs seems to be a critical factor controlling both nuclear oscillations and bypassing. The increased frequency in bypassing we observed in Agcnm67Δ mutants compared with wild-type might be due to the relatively high abundance of very long cMTs concomitant with an overall decreased number of cMTs, which could physically obstruct rapid long-range movement of a large nucleus or reduce the tethering force that resists bypassing. Due to the larger size of Agnud1Δ nuclei, their movement is probably more restricted so a decrease in bypassing was observed.
Nucleation of microtubules from the half-bridge/bridge seems to be an evolutionary conserved rescue mechanism in the absence of an OP because it is also observed in budding yeast cnm67Δ and nud1 mutants (Brachat et al., 1998; Adams and Kilmartin, 1999; Gruneberg et al., 2000). Nucleation of microtubules from the half-bridge also occurs in wild-type budding yeast cells during G1 phase of the cell cycle (Byers and Goetsch, 1975). Although half-bridge microtubules are rarely observed in asynchronous cultures of S. cerevisiae, they can be easily visualized in cells treated with the mating pheromone α-factor because this class of cMTs is essential for the nuclear migration and fusion after mating (karyogamy). During our EM analysis of A. gossypii wild-type SPBs, we could not find evidence of cMT nucleation from the half-bridge simply because this structure was only observed twice (Lang et al., 2010). A. gossypii is not known to undergo nuclear fusions, so half-bridge–emanating microtubules may have been lost during evolution.
Perhaps one of our most unexpected findings was that Agnud1Δ mutants are viable, because most nud1 mutants in S. cerevisiae arrest cell division in anaphase due to the role of Nud1 in MEN at the SPB OP (reviewed in Hoyt, 2000; Pereira and Schiebel, 2001; Stegmeier and Amon, 2004). Our finding that AgNUD1 is not essential suggests that the MEN is either nonessential in A. gossypii or does not depend on SPB recruitment for its activation. Due to the growth mode of A. gossypii, in which segregation of chromosomes, an SPB, and other cellular material into the daughter cell is not critical for survival, we suspect that positioning of the nucleus and spindle is probably not under tight checkpoint control as it is in S. cerevisiae. Indeed, preliminary experiments with some A. gossypii MEN genes show that they are not essential (Finlayson and Philippsen, unpublished observations). Although Nud1 is one of the few core SPB components with recognizable homologues in other species, the functional conservation among these proteins is poor, aside from their role in astral microtubule formation. Given our findings, Nud1 orthologues may have a rapidly evolving activity during cell division.
Regulation of cMTs by AgStu2
Of all the deletions we analyzed, Agstu2Δ has the greatest effect on cMT length, which explains the complete lack of nuclear oscillations and bypassing in this mutant and the 2.5-fold increase in the distance from the tip to the first nucleus compared with wild-type. Although Agspc72Δ and Agstu2Δ mutants are similar in terms of their effects on nuclear dynamics, the mechanisms responsible for cMT loss in Agstu2Δ and Agspc72Δ mutants are very different, as shown by our EM analysis and depicted in Figure 9. AgSpc72 is most likely a structural component of the SPB; therefore, its elimination results in changes in SPB morphology and cMT nucleation, whereas AgStu2 is a microtubule-associated protein, which affects MT formation/elongation but not SPB structure. Unlike Agspc72Δ nuclei that are occasionally able to form very long cMTs and undergo rapid bypassing, we did not observe any bypassing in Agstu2Δ hyphae because no long cMTs are able to form. The number of short cMTs was also dramatically reduced, but we were able to detect these forming in both perpendicular and tangential orientations. Most likely, AgStu2 affects cMT polymerization as it does in many other eukaryotes, including budding and fission yeast, Aspergillus nidulans, and metazoans (reviewed in Gard et al., 2003). Curiously, neither nuclear migration nor positioning was affected in A. nidulans hyphae lacking the single XMAP215/Stu2 orthologue alpA, despite a dramatic reduction in the number and dynamics of cMTs (Enke et al., 2007). Perhaps nuclear dynamics is controlled through different mechanisms in filamentous fungi that use a single MTOC like A. gossypii versus multiple MTOCs like A. nidulans (Konzack et al., 2005; Lang et al., 2010).
We were also surprised by the fact that deletion of AgSTU2 did not result in loss of viability as it does in budding yeast. ScSTU2 is essential due to its role in nuclear microtubule dynamics and bipolar spindle assembly; it also has a nonessential function in cMT polymerization and nuclear positioning (Wang and Huffaker, 1997; Chen et al., 1998; Kosco et al., 2001; Severin et al., 2001; Pearson et al., 2003; Usui et al., 2003; van Breugel et al., 2003; Al-Bassam et al., 2006). It is unclear why AgSTU2 is not required for mitotic spindle formation as it is in budding yeast. Alterations in cell cycle regulation, the mechanism of spindle assembly, or both could account for this difference.
CONCLUSIONS
In conclusion, the multinucleate growth mode of A. gossypii has resulted in different demands on the microtubule cytoskeleton that have driven evolution of SPB components to fit its unique life-style. Because nuclear migration into a bud once every cell cycle does not apply in A. gossypii hyphae, cMTs and SPB OP components are not critical for cell viability but do function in nuclear oscillations and bypassing. One newly evolved cytoskeletal element are tangential long cMTs, which drive nuclear bypassing, whereas the conventional short cMTs, which make contacts with the cell cortex, are important for nuclear oscillations as summarized in Figure 9. Our functional analysis of A. gossypii SPB genes has allowed us to better understand the adaptive properties of the cytoskeleton involved in growth and development of many eukaryotic cells.
Supplementary Material
ACKNOWLEDGMENTS
We thank Géraldine Kässlin and Shanon Seger for help in strain construction and Dominic Hoepfner for guidance in the early stage of this project. We are grateful to Jenny Friederichs, Teri Johnson, and Fengli Guo for assistance with EM and to Katie Perko for help with AutoAligner. We acknowledge the advice and suggestions of Trisha Davis, Chad Pearson and the Philippsen and Jaspersen laboratories. This work was supported by Swiss National Science Foundation grant 3100A0-112688 (to P. P). S.L.J. is supported by a March of Dimes Basil O'Connor Award and the Stowers Institute for Medical Research.
Abbreviations used:
- cMT
cytoplasmic microtubule
- EM
electron microscopy
- IL
intermediate layer
- IP
inner plaque
- MEN
mitotic exit network
- MTOC
microtubule-organizing center
- OP
outer plaque
- SPB
spindle pole body.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/10.1091/mbc.E09-07-0555) on January 6, 2010.
REFERENCES
- Adams I. R., Kilmartin J. V. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J. Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Bassam J., van Breugel M., Harrison S. C., Hyman A. Stu2p binds tubulin and undergoes an open-to-closed conformational change. J. Cell Biol. 2006;172:1009–1022. doi: 10.1083/jcb.200511010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayad-Durieux Y., Knechtle P., Goff S., Dietrich F., Philippsen P. A PAK-like protein kinase is required for maturation of young hyphae and septation in the filamentous ascomycete Ashbya gossypii. J. Cell Sci. 2000;113:4563–4575. doi: 10.1242/jcs.113.24.4563. [DOI] [PubMed] [Google Scholar]
- Brachat A., Kilmartin J. V., Wach A., Philippsen P. Saccharomyces cerevisiae cells with defective spindle pole body outer plaques accomplish nuclear migration via half-bridge-organized microtubules. Mol. Biol. Cell. 1998;9:977–991. doi: 10.1091/mbc.9.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brachat S., Dietrich F. S., Voegeli S., Zhang Z., Stuart L., Lerch A., Gates K., Gaffney T., Philippsen P. Reinvestigation of the Saccharomyces cerevisiae genome annotation by comparison to the genome of a related fungus: Ashbya gossypii. Genome Biol. 2003;4:R45. doi: 10.1186/gb-2003-4-7-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Goetsch L. Behavior of spindles and spindle plaques in the cell cycle and conjugation of Saccharomyces cerevisiae. J. Bacteriol. 1975;124:511–523. doi: 10.1128/jb.124.1.511-523.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byers B., Shriver K., Goetsch L. The role of spindle pole bodies and modified microtubule ends in the initiation of microtubule assembly in Saccharomyces cerevisiae. J. Cell Sci. 1978;30:331–352. doi: 10.1242/jcs.30.1.331. [DOI] [PubMed] [Google Scholar]
- Carminati J. L., Stearns T. Microtubules orient the mitotic spindle in yeast through dynein-dependent interactions with the cell cortex. J. Cell Biol. 1997;138:629–641. doi: 10.1083/jcb.138.3.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X. P., Yin H., Huffaker T. C. The yeast spindle pole body component Spc72p interacts with Stu2p and is required for proper microtubule assembly. J. Cell Biol. 1998;141:1169–1179. doi: 10.1083/jcb.141.5.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrich F. S., et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science. 2004;304:304–307. doi: 10.1126/science.1095781. [DOI] [PubMed] [Google Scholar]
- Elliott S., Knop M., Schlenstedt G., Schiebel E. Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc. Natl. Acad. Sci. USA. 1999;96:6205–6210. doi: 10.1073/pnas.96.11.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enke C., Zekert N., Veith D., Schaaf C., Konzack S., Fischer R. Aspergillus nidulans Dis1/XMAP215 protein AlpA localizes to spindle pole bodies and microtubule plus ends and contributes to growth directionality. Eukaryot. Cell. 2007;6:555–562. doi: 10.1128/EC.00266-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard D. L., Becker B. E., Romney S. J. MAPing the eukaryotic tree of life: structure, function, and evolution of the MAP215/Dis1 family of microtubule-associated proteins. Int. Rev. Cytol. 2003;239:179–271. doi: 10.1016/S0074-7696(04)39004-2. [DOI] [PubMed] [Google Scholar]
- Gattiker A., Rischatsch R., Demougin P., Voegeli S., Dietrich F. S., Philippsen P., Primig M. Ashbya Genome Database 3. 0, a cross-species genome and transcriptome browser for yeast biologists. BMC Genomics. 2007;8:9. doi: 10.1186/1471-2164-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler S., Pereira G., Spang A., Knop M., Soues S., Kilmartin J., Schiebel E. The spindle pole body component Spc98p interacts with the gamma-tubulin-like Tub4p of Saccharomyces cerevisiae at the sites of microtubule attachment. EMBO J. 1996;15:3899–3911. [PMC free article] [PubMed] [Google Scholar]
- Gladfelter A. S., Hungerbuehler A. K., Philippsen P. Asynchronous nuclear division cycles in multinucleated cells. J. Cell Biol. 2006;172:347–362. doi: 10.1083/jcb.200507003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U., Campbell K., Simpson C., Grindlay J., Schiebel E. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 2000;19:6475–6488. doi: 10.1093/emboj/19.23.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hoepfner D., Brachat A., Philippsen P. Time-lapse video microscopy analysis reveals astral microtubule detachment in the yeast spindle pole mutant cnm67. Mol. Biol. Cell. 2000;11:1197–1211. doi: 10.1091/mbc.11.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D., Schaerer F., Brachat A., Wach A., Philippsen P. Reorientation of mispositioned spindles in short astral microtubule mutant spc72Delta is dependent on spindle pole body outer plaque and Kar3 motor protein. Mol. Biol. Cell. 2002;13:1366–1380. doi: 10.1091/mbc.01-07-0338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyt M. A. Exit from mitosis: spindle pole power. Cell. 2000;102:267–270. doi: 10.1016/s0092-8674(00)00031-3. [DOI] [PubMed] [Google Scholar]
- Jaspersen S. L., Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu. Rev. Cell Dev. Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- Kilmartin J. V., Goh P. Y. Spc110p: assembly properties and role in the connection of nuclear microtubules to the yeast spindle pole body. EMBO J. 1996;15:4592–4602. [PMC free article] [PubMed] [Google Scholar]
- Knop M., Pereira G., Geissler S., Grein K., Schiebel E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Schiebel E. Spc98p and Spc97p of the yeast gamma-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M., Schiebel E. Receptors determine the cellular localization of a gamma-tubulin complex and thereby the site of microtubule formation. EMBO J. 1998;17:3952–3967. doi: 10.1093/emboj/17.14.3952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konzack S., Rischitor P. E., Enke C., Fischer R. The role of the kinesin motor KipA in microtubule organization and polarized growth of Aspergillus nidulans. Mol. Biol. Cell. 2005;16:497–506. doi: 10.1091/mbc.E04-02-0083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosco K. A., Pearson C. G., Maddox P. S., Wang P. J., Adams I. R., Salmon E. D., Bloom K., Huffaker T. C. Control of microtubule dynamics by Stu2p is essential for spindle orientation and metaphase chromosome alignment in yeast. Mol. Biol. Cell. 2001;12:2870–2880. doi: 10.1091/mbc.12.9.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang C., Grava S., van den Hoorn T., Trimble R., Philippsen P., Jaspersen S. L. Mobility, microtubule nucleation and structure of MTOCs in multinucleated hyphae of Ashbya gossypii. Mol. Biol. Cell. 2010;21:18–28. doi: 10.1091/mbc.E09-01-0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandelkow E. M., Mandelkow E., Milligan R. A. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J. Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald K. High-pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol. Biol. 1999;117:77–97. doi: 10.1385/1-59259-201-5:77. [DOI] [PubMed] [Google Scholar]
- Muller E. G., Snydsman B. E., Novik I., Hailey D. W., Gestaut D. R., Niemann C. A., O'Toole E. T., Giddings T. H., Jr., Sundin B. A., Davis T. N. The organization of the core proteins of the yeast spindle pole body. Mol. Biol. Cell. 2005;16:3341–3352. doi: 10.1091/mbc.E05-03-0214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T., Vinh D. B., Crawford D. K., Davis T. N. A genetic analysis of interactions with Spc110p reveals distinct functions of Spc97p and Spc98p, components of the yeast gamma-tubulin complex. Mol. Biol. Cell. 1998;9:2201–2216. doi: 10.1091/mbc.9.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Toole E. T., Winey M., McIntosh J. R. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae. Mol. Biol. Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearson C. G., Maddox P. S., Zarzar T. R., Salmon E. D., Bloom K. Yeast kinetochores do not stabilize Stu2p-dependent spindle microtubule dynamics. Mol. Biol. Cell. 2003;14:4181–4195. doi: 10.1091/mbc.E03-03-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Grueneberg U., Knop M., Schiebel E. Interaction of the yeast gamma-tubulin complex-binding protein Spc72p with Kar1p is essential for microtubule function during karyogamy. EMBO J. 1999;18:4180–4195. doi: 10.1093/emboj/18.15.4180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Knop M., Schiebel E. Spc98p directs the yeast gamma-tubulin complex into the nucleus and is subject to cell cycle-dependent phosphorylation on the nuclear side of the spindle pole body. Mol. Biol. Cell. 1998;9:775–793. doi: 10.1091/mbc.9.4.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira G., Schiebel E. The role of the yeast spindle pole body and the mammalian centrosome in regulating late mitotic events. Curr. Opin. Cell Biol. 2001;13:762–769. doi: 10.1016/s0955-0674(00)00281-7. [DOI] [PubMed] [Google Scholar]
- Rose M. D., Fink G. R. KAR1, a gene required for function of both intranuclear and extranuclear microtubules in yeast. Cell. 1987;48:1047–1060. doi: 10.1016/0092-8674(87)90712-4. [DOI] [PubMed] [Google Scholar]
- Rout M. P., Kilmartin J. V. Components of the yeast spindle and spindle pole body. J. Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Russell D. W. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schaerer F., Morgan G., Winey M., Philippsen P. Cnm67p is a spacer protein of the Saccharomyces cerevisiae spindle pole body outer plaque. Mol. Biol. Cell. 2001;12:2519–2533. doi: 10.1091/mbc.12.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severin F., Habermann B., Huffaker T., Hyman T. Stu2 promotes mitotic spindle elongation in anaphase. J. Cell Biol. 2001;153:435–442. doi: 10.1083/jcb.153.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw S. L., Yeh E., Maddox P., Salmon E. D., Bloom K. Astral microtubule dynamics in yeast: a microtubule-based searching mechanism for spindle orientation and nuclear migration into the bud. J. Cell Biol. 1997;139:985–994. doi: 10.1083/jcb.139.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soues S., Adams I. R. SPC72: a spindle pole component required for spindle orientation in the yeast Saccharomyces cerevisiae. J. Cell Sci. 1998;111:2809–2818. doi: 10.1242/jcs.111.18.2809. [DOI] [PubMed] [Google Scholar]
- Spang A., Geissler S., Grein K., Schiebel E. gamma-Tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J. Cell Biol. 1996;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegmeier F., Amon A. Closing mitosis: the functions of the Cdc14 phosphatase and its regulation. Annu. Rev. Genet. 2004;38:203–232. doi: 10.1146/annurev.genet.38.072902.093051. [DOI] [PubMed] [Google Scholar]
- Usui T., Maekawa H., Pereira G., Schiebel E. The XMAP215 homologue Stu2 at yeast spindle pole bodies regulates microtubule dynamics and anchorage. EMBO J. 2003;22:4779–4793. doi: 10.1093/emboj/cdg459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Breugel M., Drechsel D., Hyman A. Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. J. Cell Biol. 2003;161:359–369. doi: 10.1083/jcb.200211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J. P., Huffaker T. C. Stu2p: a microtubule-binding protein that is an essential component of the yeast spindle pole body. J. Cell Biol. 1997;139:1271–1280. doi: 10.1083/jcb.139.5.1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland J., Ayad-Durieux Y., Knechtle P., Rebischung C., Philippsen P. PCR-based gene targeting in the filamentous fungus Ashbya gossypii. Gene. 2000;242:381–391. doi: 10.1016/s0378-1119(99)00509-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.