Phospholipid flipping across cellular membranes contributes to vesicle biogenesis in eukaryotes and involves flippases (P4-ATPases). However, the minimal composition of the flippase machinery remains to be determined. We demonstrate that cellular targeting and lipid specificity of P4-ATPases require the α-subunit but are independent of the β-subunit.
Abstract
Members of the P4 subfamily of P-type ATPases are believed to catalyze flipping of phospholipids across cellular membranes, in this way contributing to vesicle biogenesis in the secretory and endocytic pathways. P4-ATPases form heteromeric complexes with Cdc50-like proteins, and it has been suggested that these act as β-subunits in the P4-ATPase transport machinery. In this work, we investigated the role of Cdc50-like β-subunits of P4-ATPases for targeting and function of P4-ATPase catalytic α-subunits. We show that the Arabidopsis P4-ATPases ALA2 and ALA3 gain functionality when coexpressed with any of three different ALIS Cdc50-like β-subunits. However, the final cellular destination of P4-ATPases as well as their lipid substrate specificity are independent of the nature of the ALIS β-subunit they were allowed to interact with.
INTRODUCTION
A fundamental cellular process that requires a dynamic regulation of transbilayer lipid arrangements is the biogenesis of endocytic and secretory vesicles. Recently, accumulating evidence has pointed to an important role in this process for P4-ATPases, which form one of the five subfamilies of P-type ATPases (Tang et al., 1996; Axelsen and Palmgren, 1998; Palmgren and Harper, 1998). P-type ATPases constitute a large family of membrane pumps that are transiently autophosphorylated at a conserved aspartate residue, hence the designation P-type. Evidence for an important role of P4-ATPases in membrane vesiculation has mainly come from genetic studies (Graham, 2004). For example, 1) inactivation of a Saccharomyces cerevisiae P4-ATPase, DRS2, rapidly blocks formation of a clathrin-dependent class of post-Golgi secretory vesicles carrying exocytic cargo (Gall et al., 2002); 2) S. cerevisiae Δdnf1Δdnf2 mutant cells show a cold-sensitive defect in endocytosis (Pomorski et al., 2003); 3) conditional alleles of the essential yeast P4-ATPase Neo1p perturb ADP-ribosylation factor–dependent vesicle formation from endosomes (Hua and Graham, 2003; Wicky et al., 2004); 4) loss of ALA3, a Golgi-resident P4-ATPase in Arabidopsis thaliana, causes a defect in the production of slime vesicles containing polysaccharides and enzymes for secretion, and impairs growth of roots and shoots (Poulsen et al., 2008a); and finally; 5) the Caenorhabditis elegans P4-ATPase TAT-1 is required for yolk uptake in oocytes and for an early step of fluid-phase endocytosis in the intestine (Darland-Ransom et al., 2008; Ruaud et al., 2009).
At least three models can be proposed to explain how P4-ATPases contribute to vesicle formation. One model is that P4-ATPases directly catalyze an inward directed phospholipid translocation across the lipid bilayer, which creates an imbalance in phospholipid numbers between the two leaflets. In turn, this causes an inward bending of the membrane leading to budding and vesicle formation, which is stabilized by recruitment of coat proteins (e.g., clathrin or COPII proteins). Consistent with this hypothesis, insertion of exogenous phospholipids in the exoplasmic leaflet of the plasma membrane (PM), and their subsequent translocation to the cytosolic leaflet by a lipid flippase causes dramatic shape changes of red blood cells (Seigneuret and Devaux, 1984; Daleke and Huestis, 1985). Similarly, lipid flipping can provoke the formation of endocytic-like vesicles (Muller et al., 1994) and accelerates endocytosis (Farge et al., 1999). An unresolved question with this model is the “giant substrate” problem. All other P-type ATPases characterized so far, e.g., Na+/K+- and Ca2+-ATPases (Ebashi and Ebashi, 1962; Jorgensen et al., 2003), pump small cations and three-dimensional (3D) structures of such pumps show that the transported ions are occluded in the center of the transmembrane part of the protein, from where they have alternating access to either side of the membrane (Toyoshima et al., 2000; Morth et al., 2007; Pedersen et al., 2007; Shinoda et al., 2009). Consequently, it is unclear how this mechanism can be utilized to transport a phospholipid across the membrane bilayer.
A second model is that P4-ATPases contribute to vesicle budding by serving as scaffold proteins that recruit coat proteins (e.g., clathrin or COPII) to the membrane through protein–protein interactions and/or by increasing the concentration of specific lipids in the cytosolic leaflet, which in turn could be required for binding of coat proteins. This model is supported by the observation that S. cerevisiae Drs2p directly interacts with the ADP-ribosylation factor (ARF) activator, Gea2p (Chantalat et al., 2004). The latter protein is a GTP-exchange factor that regulates recruitment of ARF, adapter protein-1 (AP-1) and clathrin coat proteins to membranes of the trans-Golgi. However, a recent study shows that membrane association of ARF, AP-1, and clathrin does not require Drs2p (Liu et al., 2008).
In a third model, it is hypothesized that P4-ATPases, in view of their overall similarity to cation-transporting P-type ATPases, are cation pumps that only contribute indirectly to lipid flipping (Axelsen and Palmgren, 2001; Kuhlbrandt, 2004). According to the model, P4-ATPases pump cations to generate a transmembrane electrochemical gradient in secretory and endocytic pathway membrane structures. In turn, a second transport protein operating by a symport mechanism energized by this gradient drives a fundamental process in vesicle budding, which could be lipid translocation. This protein could in principle be a subunit of the flippase complex (Axelsen and Palmgren, 2001; Kuhlbrandt, 2004). Prime candidates for such accessory proteins would be the recently identified Cdc50-like β-subunits of P4-ATPases (Katoh and Katoh, 2004; Saito et al., 2004; Paulusma et al., 2008; Poulsen et al., 2008a). Because Cdc50 proteins lack any similarity to known transporters, it has recently been suggested that transmembrane flipping might require cooperation between both subunits and occur at the interface between a P4- ATPase and its Cdc50 binding partner, an arrangement in which Cdc50 proteins would contribute directly to the transport specificity of the complex (Coleman et al., 2009; Puts and Holthuis, 2009; Zhou and Graham, 2009). Alternatively, creation of a high-affinity phospholipid binding site may require Cdc50-induced conformational changes in the membrane domain of P4-ATPases, analogous to the role of the β-subunit in the oligomeric Na+/K+-ATPase (Geering, 2001; Puts and Holthuis, 2009).
In this work, we aimed at investigating closer the role of Cdc50-like β-subunits of P4-ATPases for targeting and function of P4-ATPases in higher eukaryotes using the plant Arabidopsis as a model organism. In this plant, P4-ATPases are encoded for by ALA genes and Cdc50p-like β-subunits by ALIS genes (Gomés et al., 2000; Poulsen et al., 2008a). First, we identified a novel P4-ATPase, ALA2, in Arabidopsis. We found that it only gained functionality while coexpressed with an ALIS protein, and in its absence ALA2 never exited the endoplasmic reticulum (ER) in planta. However, the final cellular destination of ALA2 as well as its lipid substrate specificity was not affected by the ALIS protein it was allowed to interact with. Further, when compared with ALA2, the related P4-ATPase ALA3, that also requires an ALIS protein for ER export, shows a different subcellular location and lipid specificity, which similarly was unaffected by the accompanying ALIS protein. We conclude that targeting signals and lipid specificity determinants of P4-ATPases reside in the ALA catalytic α-subunit. This would suggest that plant Cdc50p-like proteins mainly play a role in functional maturation and ER export of P4-ATPases, but do not affect the nature of the transported phospholipid and the final localization of the putative flippase complex.
MATERIALS AND METHODS
DNA Cloning and Sequence Analysis
The ALA2 (At5g44240) cDNA was isolated by PCR amplification using a cDNA library of size-fractionated (3–6 kb) cDNAs (Kieber et al., 1993) as template. Primers were designed on the basis of information contained in the P-type ATPase database (http://biobase.dk/∼axe/Patbase.html). All fragments were amplified using Phusion High-Fidelity DNA polymerase (Finnzymes, Espoo, Finland) and cloned into the pJET1 blunt-end cloning vector (Fermentas, Hanover, MD) for sequencing. Full sequencing of the cDNA clone revealed four point mutations causing amino acid changes in the protein sequence, which were subsequently corrected by site-directed mutagenesis giving rise to four silent mutations. The corrected clone was used as a template in new PCRs in which the full-length cDNA sequence was amplified without additions or where a sequence corresponding to a hemagglutinin (HA) epitope (YPYDVPDYA) was included at the N-terminal end. The PCR products were cloned into the Gateway compatible vector pENTR/D-TOPO (Invitrogen, Carlsbad, CA) using the pENTR/D-TOPO cloning kit. Previously, among the five ALIS genes in Arabidopsis, we successfully cloned ALIS1, ALIS3, and ALIS5 (Poulsen et al., 2008a). HA:ALA2 was cloned into a modified version of yeast plasmid pRS423GAL1–10 and its derivatives containing RGSH6:ALIS gene fusions (Poulsen et al., 2008a) using the Gateway technology. Employing an overlapping PCR strategy an HA-tagged mutant version of ALA2, ala2D381A, was generated and cloned in a similar manner into pENTR/D-TOPO and the Gateway-compatible yeast plasmids.
To obtain a green fluorescent protein (GFP)-tagged version of ALA2 for tobacco infiltration, untagged ALA2 was transferred into plasmid pMDC43 (Curtis and Grossniklaus, 2003) using the Gateway technology. Likewise, for generating C-terminal fusions of ALIS1, ALIS3, and ALIS5 to yellow fluorescent protein (YFP), the corresponding genes were transferred from pENTR/D-TOPO clones (Poulsen et al., 2008a) to plant binary plasmid pEarleyGate 104 (Earley et al., 2006) using the Gateway technology.
Topology predictions for ALA2 were carried out using the TMHMM (http://www.cbs.dtu.dk/services/TMHMM/) and TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) servers. Sequence identity/homology scores were calculated using CrustalW (http://www.ebi.ac.uk/Tools/clustalw2/index.html).
Yeast Strains and Media
Functional complementation and lipid translocation assays were carried out employing S. cerevisiae mutant strain ZHY709 (MATα his3 leu2 ura3 met15 dnf1Δ dnf2Δ drs2::LEU2; Hua et al., 2002), and strain BY4741 (MATα his3 leu2 ura3 met15; EUROSCARF) as wild type. Cells were grown at 30°C in standard rich medium with glucose (YPD) or galactose (YPG), or rich synthetic SD or SG media (Rose and Broach, 1990) containing yeast synthetic dropout medium supplement without histidine (Sigma-Aldrich; see also Supplemental Figure 1). Heavy metal-containing media were generated by incorporation of 200 μM CoCl2 or 2 mM ZnCl2. Solid media were added 2% agar (Villalba et al., 1992). Papuamide B (Flintbox, Lynsey Huxham; www.flintbox.com/), duramycin (Sigma-Aldrich, St. Louis, MO), and miltefosine (Calbiochem, La Jolla, CA) were added to rich synthetic SD or SG media to the indicated concentrations.
Yeast Transformation and Growth
Yeast cells were transformed by the lithium acetate method (Gietz and Woods, 2002). Transformants were incubated in liquid SG medium for 4 h and then diluted with water to 0.1, 0.01, and 0.001 OD600. Drops (5 μl) were spotted on plates and incubated at 20°C for 6–8 d or at 30°C for 2–3 d as indicated. All experiments were repeated independently at least three times.
Yeast Membrane Preparation and Protein Immunodetection
Total cellular membranes for protein expression analysis were prepared as previously described (Villalba et al., 1992). Quantification of proteins content and Western blot analysis were carried out as described (Poulsen et al., 2008a). For sucrose-density fractionation, fresh yeast transformants were inoculated in 25 ml selective SD media and grown for 24 h at 28°C under 160 rpm shaking. Cells were harvested (1,500 × g, 5 min, 24°C), washed twice with water, and inoculated into 500 ml YPG media. Cultures were grown to 1–1.2 OD600 at 28°C under 160 rpm shaking. Cells (400 ml) were harvested by centrifugation (1500 × g, 4°C, 5 min), and washed in 25 ml ice-cold lysis buffer (10 mM Tris-HCl, pH 7.5, 1 mM EDTA, 0.6 M sorbitol, 1 μg/ml pepstatin A, 0.1 mM phenylmethylsulfonyl fluoride, 1 mM dithiothreitol). After centrifugation, cells were resuspended in 4 ml of lysis buffer, and 2-ml aliquots were vortexed with glass beads (1.2 × g; 200 μm; Sigma-Aldrich). The cell lysate was clarified by centrifugation (1000 × g, 10 min, 4°C), and PM-enriched membranes, devoid of mitochondria and most of the endomembranes, were subsequently collected by centrifugation (9000 × g, 30 min, 4°C). Pellets were homogenized in 500 μl lysis buffer and centrifuged (1000 × g, 10 min, 4°C) before loading onto two-step sucrose gradients: 7.6 ml 43% (wt/wt) sucrose on top of 3.8 ml 53% (wt/wt) sucrose prepared in lysis buffer. After centrifugation (120,000 × g, 17 h, 4°C), 1.2-ml fractions were collected from the top. Equal volumes per fraction were used for Western blot analysis. Sucrose was quantified in each fraction using a PAL1 refractometer (Atago, Kirkland, WA).
Lipid Translocation Assays and Flow Cytometry
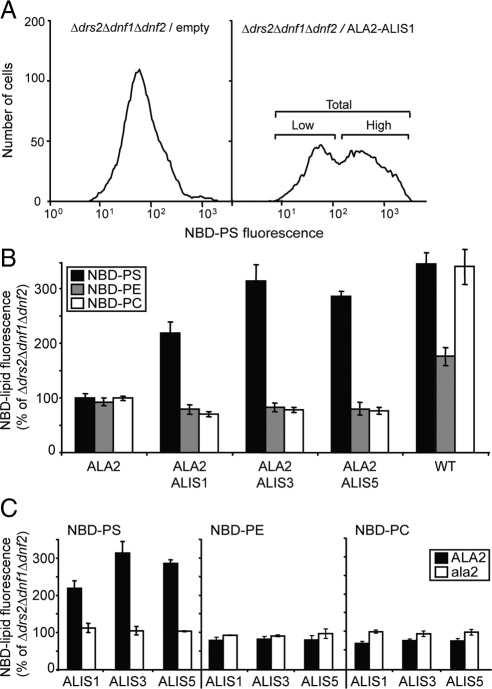
1-Palmitoyl-2-[6-(7-nitrobenz-2-oxa-1,3-diazol-4-yl)amino]hexanoyl-sn-glycero-3-phosphocholine (NBD-PC), phosphoethanolamine (NBD-PE), and phosphoserine (NBD-PS) were from Avanti Polar Lipids (Birmingham, AL). All NBD-lipid stocks (10 mM) were prepared in DMSO. Uptake experiments were performed essentially as previously described (Poulsen et al., 2008a) except that cells were grown in selective rich synthetic medium to midlogarithmic phase (0.5 1.0 OD600/ml). Cells (10 OD600/ml) were incubated in the same medium with 60 μM NBD-lipids at 30°C with periodic mixing. After 30 min, cells were washed twice in ice-cold medium containing 4% (wt/vol) bovine serum albumin to extract NBD lipids from the cell surface. Flow cytometry of NBD-labeled cells was performed on a Becton Dickinson FACSCalibur (San Jose, CA) equipped with an argon laser using Cell Quest software. One microliter of 1 mg/ml propidium iodide in water was added to 1 × 107 cells in 1 ml buffer (137 mM NaCl, 10 mM Na2HPO4, 2 mM NaH2PO4, pH 7.4) just before analysis. Thirty thousand cells were analyzed without gating during the acquisition. Live cells were selected based on forward/side-scatter gating and propidium iodide exclusion. A histogram of the green fluorescence (NBD) of living cells was used to calculate the mean fluorescence intensity of total cells (see Figure 2; Supplemental Figure 1).
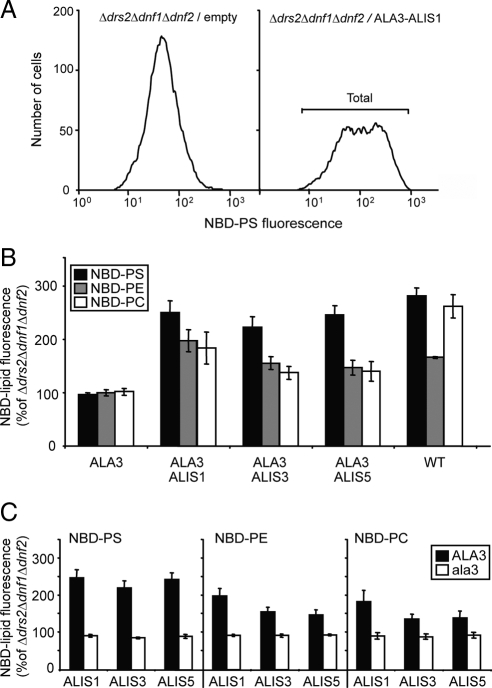
Figure 2.
Coexpression of ALA2 and ALIS proteins complements the lipid uptake defect of the Δdrs2Δdnf1Δdnf2 yeast mutant. Yeast mutant cells expressing different protein combinations and wild-type cells were labeled with 1-palmitoyl-NBD lipids and then washed and analyzed by flow cytometry. Accumulation of NBD lipids was expressed as percentage of fluorescence intensity relative to control Δdrs2Δdnf1Δdnf2 mutant cells. (A) Coexpression of ALA2 and ALIS1 resulted in two populations of cells with low and high NBD-PS uptake, respectively. Representative histograms are shown. For quantitative analysis of lipid uptake the fluorescence intensity of the total population was determined. (B) ALA2 specifically promoted NBD-PS internalization in the presence of ALIS proteins. (C) The catalytically inactive mutant ala2D381A failed to promote NBD-PS internalization. (B and C) Results are averages ± SE from three independent experiments. One hundred percent corresponds to 45 ± 13 arbitrary units (NBD-PS), 31 ± 7 arbitrary units (NBD-PE), and 32 ± 6 arbitrary units (NBD-PC). WT, wild type; ala2, ala2D381A.
Transient Expression in Tobacco Epidermal Leaf Cells
Agrobacterium tumefaciens strain C58C1 (Koncz and Schell, 1986) was transformed by electroporation, and transformants were selected on YEP plates (1% yeast extract, 2% peptone, 1.5% agar) containing 25 μg/ml gentamicin and 50 μg/ml kanamycin or 50 μg/ml spectinomycin as required. Transformants were either directly used for infiltration or resuspended in a 15% glycerol solution, frozen in liquid nitrogen, and kept at −80°C until needed. Transient expression in tobacco epidermal cells was carried out as described (Sparkes et al., 2006) using 3-wk old Nicotiana benthamiana plants. To facilitate high expression of recombinant proteins Agrobacterium strains carrying the different constructs were coinfiltrated with a strain carrying the p19 gene encoding the viral p19 protein that specifically inhibits plant posttranscriptional gene silencing (Voinnet et al., 2003). When several constructs were coinfiltrated, all Agrobacterium strains mixed were used to the same final concentration (0.03 OD600). Expression was visualized 4–5 d after infiltration.
Confocal Microscopy
A Leica TCS SP2/MP spectral confocal laser scanning microscope (Leica Microsystems, Heidelberg, Germany) with a 63×/1.2 NA water immersion objective was used as previously described (Poulsen et al., 2008a). Both GFP and YFP were excited at 488 nm, and emission spectra were recorded between 495 and 510 nm for GFP (green channel) and 530 and 545 nm for YFP (magenta channel). Sequential scanning between lines was used to follow both fluorescent proteins at once. No fluorescence bleed-through was detected under our experimental conditions (not shown).
RESULTS
Arabidopsis ALA2 Encodes a Novel P4-ATPase
The family of P4-ATPases in the model plant Arabidopsis comprises 12 members, named ALA1–ALA12 (Gomès et al., 2000). ALA1, ALA2, and ALA3 are the most divergent phylogenetically, whereas ALA4 to ALA12 group closely together. ALA1 has been partially characterized and shown to be involved in chilling tolerance in Arabidopsis (Gomès et al., 2000), whereas ALA3 seems to be required for vesicle formation in actively secreting cells at the plant root tip (Poulsen et al., 2008a) and for normal development of plant trichomes (Zhang and Oppenheimer, 2009). When expressed in the presence of different ALIS proteins, ALA3 is capable of complementing yeast mutants carrying deletions in P4-ATPases (Poulsen et al., 2008a). None of the other 10 members of the P4-ATPase family in Arabidopsis has been investigated so far. In this work, we aimed at studying ALA2 for the first time.
An Arabidopsis cDNA fragment containing the full-length ALA2 gene (At5g44240) was amplified from a cDNA library from 3-d-old hypocotyls (Kieber et al., 1993). This cDNA corresponds to the gene contained in the genomic clone AB005239 (accession no. AF419611). ALA2 encodes a protein with 1107 amino acid residues and a molecular weight of ∼124 kDa, predicted to have 10 transmembrane spanning segments and containing the conserved domains of the P-type ATPase superfamily and the characteristic motifs of the P4-ATPase subfamily (Axelsen and Palmgren, 1998). ALA2 shares 27 and 29% sequence identity with ALA1 and ALA3, respectively, and 27% identity with Drs2p.
ALA2 in Combination with an ALIS Functionally Complements an S. cerevisiae P4-ATPase Mutant
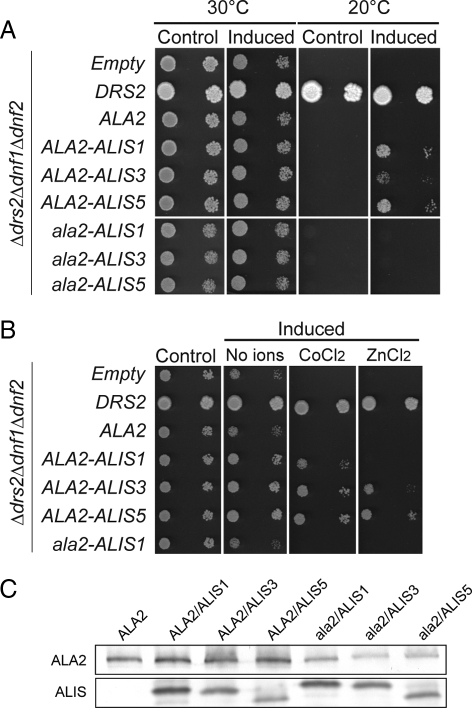
To investigate if ALA2 encodes a functional P4-ATPase, the gene was expressed in a S. cerevisiae mutant strain lacking three endogenous P4-ATPases (Δdrs2Δdnf1Δdnf2), which displays a cold-sensitive phenotype (Hua et al., 2002). In our system, expression of heterologous genes is carried out under the control of a bidirectional galactose-inducible promoter. Thus, glucose-grown yeast cells containing the desired plasmids will show no expression of the heterologous proteins, whereas culturing the yeast cells with galactose as a carbon source will result in heterologous protein overexpression. The bidirectionality of the promoter also allows for overexpression of two heterologous proteins at the same time. When transformed with a plasmid containing ALA2 alone, the yeast was unable to grow on galactose plates at 20°C (Figure 1A). Recently, we showed that functional complementation of this yeast strain by ALA3, is dependent on the presence of an ALIS gene (Poulsen et al., 2008a). When ALA2 was expressed in combination with ALIS1, ALIS3, or ALIS5, the yeast was able to grow to different extents below the restrictive temperature (Figure 1A). Among these, ALIS1 and ALIS5 were the most efficient at complementing the yeast cold-sensitive phenotype. A mutant version of ALA2 in which the aspartate residue being phosphorylated as part of the catalytic reaction is replaced by an alanine residue (ala2D381A) was not capable of rescuing the yeast cold-sensitive phenotype (Figure 1A), supporting that a catalytically active pump is required for functionality.
Figure 1.
ALA2 in combination with ALIS genes functionally complement the cold- and heavy metal–sensitive phenotype of a Δdrs2Δdnf1Δdnf2 yeast mutant. The triple yeast mutant Δdrs2Δdnf1Δdnf2 expressed ALA2 alone or in combination with ALIS genes under the control of a bidirectional galactose-inducible promoter. Yeast DRS2 and an empty vector were used as positive and negative controls, respectively. ALA2 was tagged with the hemagglutinin (HA) epitope and ALIS with the RGSH6 epitope. ALA2 expressed in concert with ALIS allows growth of the cold-sensitive yeast strain at 20°C, whereas the catalytically inactive mutant, ala2D381A, in combination with ALIS, did not support growth at 20°C. (A) Yeast cells were dropped on rich media at 30°C and at the restrictive temperature 20°C. (B) Yeast cells dropped on minimal media without metals or containing 200 μM CoCl2 or 2 mM ZnCl2. (C) Western blot analysis of membranes from yeast overexpressing HA:ALA2 or HA:ala2D381A in concert with RGSH6:ALIS proteins. Control, cells grown on glucose plates to repress expression; Induced, cells grown on galactose plates to induce overexpression; Empty, yeast cells transformed with an empty plasmid control; ala2, ala2D381A.
Next, we carried out Western blot analysis on total membranes isolated from yeast overexpressing epitope-tagged ALA2 alone or in combination with different ALIS proteins (Figure 1C). The epitope-tagged ALA2 was in all cases immunodecorated with an anti-HA antibody, indicating that the inability of ALA2 to complement the yeast cold-sensitive phenotype in the absence of an ALIS is not derived from lack of expression. ALA2 expression levels were found to be comparable in the presence of the different ALIS proteins, whereas ALIS3 was expressed at levels lower than ALIS1 and higher than ALIS5. This suggests that the differences in complementation of the cold-sensitive phenotype are not related to protein expression levels. Expression of the catalytically inactive mutant form ala2D381A seems to be lower than ALA2 expression in all cases, suggesting that lack of complementation by the mutant protein might be due to low expression. However, a catalytically inactive ALA3, analogous to the ala2D381A mutant used in this work, is unable to complement the cold-sensitive phenotype of the Δdrs2Δdnf1Δdnf2 mutant strain even when expressed at similar levels to active ALA3 (Poulsen et al., 2008a). This supports the hypothesis that a catalytically active ALA2 would be required for functional complementation of the yeast mutant phenotype.
Single Δdrs2 and double Δdnf1Δdnf2 mutant strains are unable to grow on elevated concentrations of Zn2+ and Co2+ ions (Siegmund et al., 1998; Pomorski et al., 2003), and, although the nature of this defect is unknown, it has been taken as evidence for overlapping physiological functions between Drs2p and members of the Dnf subfamily. ALA2 alone was unable to complement the heavy-metal–sensitive phenotype of Δdrs2Δdnf1Δdnf2 (Figure 1B). However, coexpression of ALA2 with any ALIS allowed growth on 200 μM CoCl2. Interestingly, in contrast to results obtained when testing complementation of the cold-sensitive phenotype, the combination of ALA2 and ALIS1 presented a slower growth on CoCl2-containing plates and was unable to restore growth on plates containing 2 mM ZnCl2 (Figure 1B).
ALA2 in Combination with an ALIS Promotes Flipping of a Fluorescent Analogue of PS
The Δdrs2Δdnf1Δdnf2 mutant yeast strain presents a low background flippase activity at the PM, which allows us to test the capacity of overexpressed proteins to promote internalization of fluorescent NBD-labeled lipids. This approach has recently been used in the characterization of the lipid flipping activity of the ALA3/ALIS1 complex with different NBD lipids (Poulsen et al., 2008a). Herein, we used palmitoyl-NBD lipids that only differ in their headgroup to characterize the lipid transport activity of ALA2. We chose ALIS1 as an example of a β-subunit, because this protein promotes lipid transport in the presence of ALA3 (Poulsen et al., 2008a). Coexpression of ALA2 with ALIS1 in the yeast triple mutant background resulted in a population of cells with increased internalization of NBD-PS, but not -PC or -PE (Figure 2, A and B). ALA2 alone was unable to promote internalization of NBD-lipids (Figure 2B). Likewise, the mutant protein ala2D381A lacking the ATPase activity in combination with ALIS1 was unable to support NBD-lipid translocation (Figure 2C), which confirms that P4-ATPase activity is a requirement for lipid internalization at the PM.
All ALA2/ALIS Combinations Result in the Same Lipid Transport Specificity
It has been suggested that P4-ATPase β-subunits could have a role in determining the transport specificity of the protein complex (Lenoir et al., 2007; Puts and Holthuis, 2009). The fact that ALA2 was capable of complementing the yeast mutant phenotype in the presence of three different members of the ALIS family allowed us to test this hypothesis. Coexpression of ALA2 in yeast with ALIS3 and ALIS5 resulted in internalization of NBD-PS to levels similar to those observed before for ALIS1 (Figure 2, B and C). No internalization was observed for NBD-PE or -PC. These results suggest that the specificity of lipid transport by ALA2 is not dependent on the nature of the coexpressed β-subunit or alternatively, that all β-subunits used in this work generate the same lipid specificity.
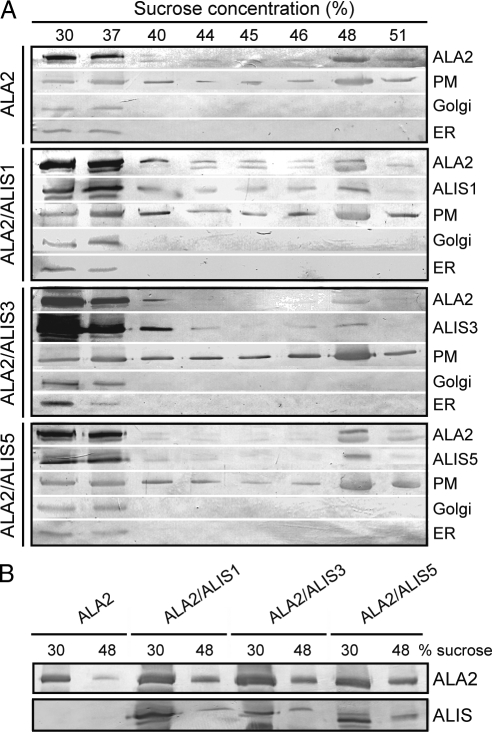
Because the lipid transport assay requires a flippase activity at the PM, we tested whether ALA2 was present in this membrane. PM-enriched yeast membranes expressing ALA2 alone or in the presence of an ALIS protein were subjected to discontinuous sucrose gradient fractionation followed by Western blot analysis. The ER marker protein dolichol-1-phosphate-mannose synthase 1 (Dpm1p) was restricted to the lower sucrose concentrations, whereas the PM proton ATPase Pma1p was enriched at fractions containing 48% sucrose (Figure 3A). Although ALA2 and the ALIS proteins were heavily represented in ER fractions, as expected for overexpressed heterologous proteins, a significant amount reached the PM in all cases (Figure 3, A and B).
Figure 3.
ALA2 reaches the plasma membrane when expressed in yeast. (A) PM-enriched membranes from yeast expressing HA:ALA2 or HA:ALA2 together with RGSH6:ALIS1, RGSH6:ALIS3, or RGSH6:ALIS5 were subjected to discontinuous sucrose density gradient fractionation. (B) Fractions corresponding to 30 and 48% sucrose were analyzed in parallel for each ALA2/ALIS combination to compare protein levels. Western blots were probed using the following antibodies: anti-Pma1p, PM; anti-Sed5p, Golgi apparatus; anti-Dpm1p, ER; anti-HA, ALA2; and anti-RGSH6, ALIS.
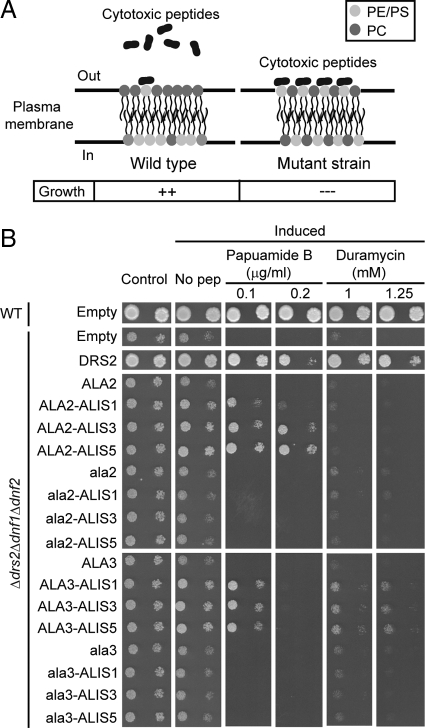
ALA2/ALIS Combinations Regulate Plasma Membrane Distribution of PS in Yeast Cells
Even when fluorescently labeled lipids are internalized, this needs not be the case for naturally occurring lipids. We therefore investigated the capacity of ALA2 and ALA2/ALIS combinations to promote the transport of natural lipids in the Δdrs2Δdnf1Δdnf2 triple mutant yeast strain exhibiting an aberrant exposure of PE and PS at the cell surface (Pomorski et al., 2003; Chen et al., 2006). Such abnormal lipid distribution at the PM can be detected using duramycin and papuamide B, which are cytolytic peptides that in order to exert their cytotoxicity require binding to cell surface–exposed PE and PS, respectively (Märki et al., 1991; Parsons et al., 2006). Thus, wild-type yeast that confines PS and PE in the cytoplasmic leaflet of the PM is less sensitive to peptide-induced cytolysis compared with the triple yeast mutant (Figure 4A). Yeast Δdrs2Δdnf1Δdnf2 cells expressing ALA2 alone or in combination with an ALIS protein were spotted on selective plates containing different concentrations of cytotoxic peptides (Figure 4B). A wild-type strain transformed with an empty plasmid was capable of growing normally on media containing 0.2 μg/ml papuamide B and 2.5 mM duramycin, whereas Δdrs2Δdnf1Δdnf2 cells transformed with an empty plasmid or overexpressing ALA2 alone were highly sensitive to both the PS- and the PE-binding peptide. Expression of ALA2 in combination with an ALIS protein reduced the sensitivity of cells toward papuamide B but not to duramycin. These results suggest that ALA2 in combination with any ALIS promotes inward translocation of natural PS, but not PE, across cellular membranes. ALA2/ALIS3 and ALA2/ALIS5 combinations promoted a better growth on papuamide B than ALA2/ALIS1 (Figure 4B). Interestingly, ALA2 in combination with ALIS3 or ALIS5 was also more potent in supporting yeast growth on elevated Cobalt and Zinc concentrations than when expressed with ALIS1 (Figure 1B). However, coexpression of ALA2 with ALIS1 or ALIS5 was more effective at rescuing the cold-sensitive phenotype of the Δdrs2Δdnf1Δdnf2 mutant strain (Figure 1A), and this could not be directly correlated to protein expression levels (Figure 1C) or cellular distribution (Figure 3, A and B). Whether this differences reflect physiologically related properties of ALA2/ALIS complexes remains to be determined.
Figure 4.
ALA2 as well as ALA3 in combination with ALIS proteins regulate lipid distribution in the PM of yeast. (A) Cytolytic, lipid-binding peptides allow probing of distribution of natural lipids. Wild-type yeast confines PS and PE in the cytoplasmic leaflet of the PM and is less sensitive to peptide-induced cytolysis compared with yeast mutants deficient in P4-ATPases. (B) Δdrs2Δdnf1Δdnf2 yeast cells were transformed with ALA2 and ALA3 alone or in combination with ALIS genes and dropped on plates containing PS-binding papuamide B or PE-binding duramycin. Yeast DRS2 and an empty vector were used as positive and negative controls, respectively. Control, nonexpressing cells; Induced, overexpressing yeast; No pep, no peptide included; WT, wild type; ala2, ala2D381A; ala3, ala3D413A.
Yeast wild-type strains can take up miltefosine, a toxic lyso-PC analog, and are unable to grow in the presence of this lysolipid, whereas any strain lacking Dnf1p and Dnf2p (like the Δdrs2Δdnf1Δdnf2 mutant) is capable of growing on moderate amounts of this drug (Pérez-Victoria et al., 2006; Riekhof and Voelker, 2009). Under our experimental conditions, the triple mutant strain was resistant to 2.5 μg/ml miltefosine, whereas no growth could be detected for the wild-type strain under these conditions (Supplemental Figure 2). All strains expressing ALA2/ALIS combinations or Drs2p were able to grow in the presence of miltefosine, suggesting that neither of them is capable of transporting lysolipid across the PM of yeast cells.
ALA2 Is Retained in the Plant ER in the Absence of a β-Subunit
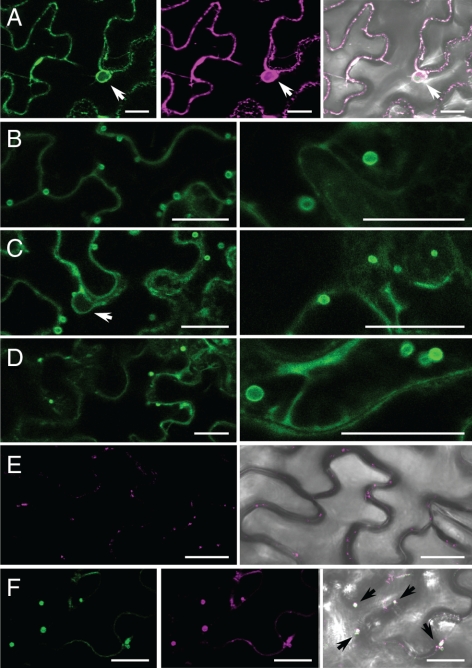
To study the physiological importance of ALA2, we decided to investigate its subcellular localization in planta. For this purpose, a GFP N-terminally tagged version of the protein was generated and transiently expressed in N. benthamiana epidermal leaf cells. In tobacco, the vacuole occupies most of the cell volume, implying that all other membrane compartments are detected along the periphery of the cells (see Supplemental Figure 3). When GFP:ALA2 was expressed in this system, the fluorescent signal could be detected in membrane structures following the contour of the cell, surrounding the nucleus and stretching into the cytoplasm, which is typical for ER membranes (Figure 5A). To test whether the observed localization indeed corresponds to the ER, a YFP containing a HDEL ER-retention signal (YFP:HDEL; Brandizzi et al., 2002) was coexpressed with GFP:ALA2. The YFP and GFP fluorescent signals colocalized (Figure 5A), indicating that GFP:ALA2 is indeed retained in the ER.
Figure 5.
ALA2 leaves the ER and localizes to vesicular structures in the presence of ALIS. GFP:ALA2 was expressed in tobacco leaf epidermal cells alone or in the presence of ALIS. (A) When expressed alone, GFP:ALA2 colocalized with an ER-retained YFP:HDEL: left, GFP fluorescence; middle, YFP fluorescence; right, fluorescent signal overlay on the bright-field image. (B–D) On coexpression with an untagged ALIS, GFP:ALA2 was localized to vesicular structures: (B) coexpression with ALIS1; (C) coexpression with ALIS3; and (D) coexpression with ALIS5. In each case, two different magnifications are shown; (E) Prevacuolar compartment visualized by expression of the fusion protein BP-80:YFP containing the targeting signals for a plant homologue of the yeast receptor protein Vsp10p: left, YFP fluorescence; right, overlay of YFP signal on a bright-field image. (F) GFP:ALA2 expressed in the presence of ALIS colocalized with BP-80:YFP in aberrant prevacuolar compartment structures. Coexpression with ALIS3 is shown: left, GFP fluorescence; middle, YFP fluorescence; right, overlay of the fluorescent signals on the bright-field image. Arrows indicate the position of nuclei when visible. Bar, 25 μm.
ALA2 Coexpressed with an ALIS Exits the ER to the Prevacuolar Compartment
To study the effect of the β-subunits on trafficking of ALA2, the GFP:ALA2 gene fusion was expressed in leaves of N. benthamiana in the presence of an untagged ALIS protein. In addition to the ER, the GFP fluorescent signal could now be localized in highly mobile vesicular structures of 1–4-μm diameter (Figure 5, B–D), and, in some cases, in structures that resemble the vacuolar membrane (cf. with Supplemental Figure 3D). The number of vesicular structures varied from cell to cell and between experiments, but these bodies were always associated with expression of ALA2/ALIS combinations, indicating that all ALIS proteins tested cause ALA2 to exit the ER and accumulate in similar organelles.
Vesicular structures similar to those apparent in Figure 5 were not observed in the absence of ALA2 (Figure 5E), but have been described in tobacco cells as aberrant prevacuolar compartment (PVC), that results from defects in the cell vesicular machinery (Kotzer et al., 2004; Tse et al., 2004; Wang et al., 2009). To test this hypothesis, we repeated the leaf infiltration with the combination of GFP:ALA2 and untagged ALIS3, this time in the presence of the PVC marker YFP:BP80 construct (Tse et al., 2004). Fluorescent signals for GFP:ALA2 and YFP:BP80 colocalized, indicating a PVC localization of GFP:ALA2 (Figure 5F).
ALA3 in Combination with an ALIS Generates Transport of NBD-PE, -PS, and -PC
To compare the properties of ALA2 with those of ALA3 (Poulsen et al., 2008a), we tested lipid internalization by ALA3 under the same conditions used before for ALA2. Overexpression of ALA3 with any of the ALIS proteins promoted the internalization of not only NBD-PS, but also -PE and -PC (Figure 6, A and B). These results suggest that the specificity of lipid transport of plant P4-ATPase/β-subunit complexes is determined by the nature of the P4-ATPase catalytic α-subunit.
Figure 6.
Coexpression of ALA3 and ALIS proteins complements the lipid uptake defect of the Δdrs2Δdnf1Δdnf2 yeast mutant. Yeast mutant cells expressing different protein combinations and wild-type cells were labeled with 1-palmitoyl-NBD lipids and then washed and analyzed by flow cytometry. Accumulation of NBD lipids was expressed as percentage of fluorescence intensity relative to control Δdrs2Δdnf1Δdnf2 mutant cells. (A) Coexpression of ALA3 and ALIS1 resulted in cells displaying a broad distribution of NBD-PS uptake. Representative histograms are shown. For quantitative analysis of lipid uptake the fluorescence intensity of the total population was determined. (B) ALA3 facilitated the internalization of NBD-PS, -PE, and -PC in the presence of ALIS proteins; (C) The catalytically inactive mutant ala3D413A failed to promote NBD-lipid internalization. Results in B and C are averages ± SE from five independent experiments. One hundred percent corresponds to 51 ± 5 arbitrary units (NBD-PS), 40 ± 5 arbitrary units (NBD-PE), and 42 ± 4 arbitrary units (NBD-PC). WT, wild type; ala3, ala3D413A.
ALA3/ALIS Combinations Regulate PM Asymmetry in Yeast Cells
All ALA3/ALIS combinations reduced the sensitivity of cells to both papuamide B and duramycin, whereas yeast strains transformed with a mutant form of ALA3, ala3D413A, displayed comparable sensitivity to the empty vector control (Figure 4B). On the basis of these results, we conclude that ALA3 with any ALIS indeed promotes transport of natural PS and PE. Notably, coexpression of ALA3 with any ALIS reduced the sensitivity of cells to papuamide B to a lesser extent compared with the results obtained for ALA2, in agreement with the results obtained for the translocation assays with fluorescent lipids.
Similarly to ALA2, ALA3/ALIS combinations did not increase miltefosine sensitivity (Supplemental Figure 2), suggesting that ALA3 is also not capable of transporting the lysolipid.
ALA3 Requires a β-Subunit for Exit from the ER
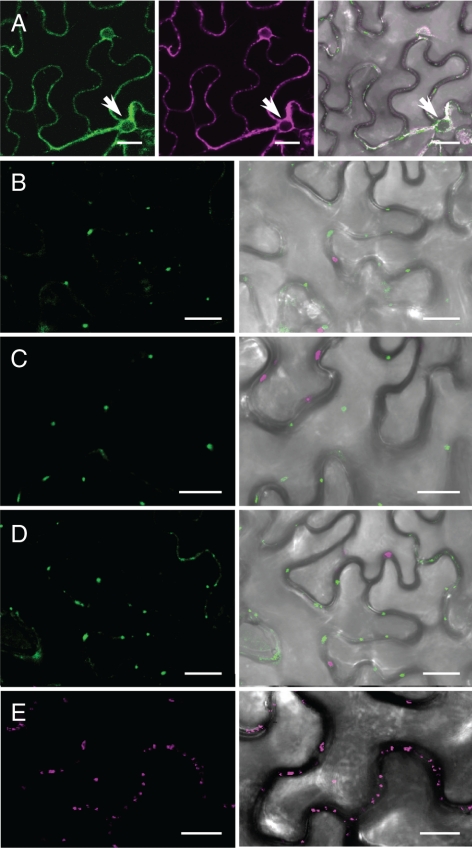
When expressed alone in N. tabacum, ALA3 can apparently exit the ER (Poulsen et al., 2008a). However, expression of a GFP:ALA3 fusion in N. benthamiana together with the ER-retained YFP:HDEL construct, resulted in colocalization of the fluorescent signals (Figure 7A), indicating that GFP:ALA3 is retained in the ER in this tobacco species. This result could be explained assuming that N. tabacum is equipped with a Cdc50-like protein that allows for ER exit of the heterologous Arabidopsis ALA3. In contrast, when the GFP:ALA3 construct was expressed in the presence of untagged ALIS1, ALIS3, or ALIS5, fluorescence could be detected in punctuated structures resembling Golgi bodies (Figure 7, B–D). Based on the expression pattern on a YFP-tagged version of the Golgi marker sialyl transferase (ST:YFP; Saint-Jore et al., 2002), these punctuate structures most likely represent the Golgi apparatus (Figure 7E; Poulsen et al., 2008a).
Figure 7.
ALA3 leaves the ER and localizes to Golgi-like structures in the presence of ALIS. GFP:ALA3 was expressed in tobacco leaf epidermal cells alone or in the presence of ALIS. (A) When expressed alone, GFP:ALA3 colocalized with an ER-retained YFP:HDEL: left, GFP fluorescence; middle, YFP fluorescence; right, fluorescent signal overlay on the bright-field image. (B–D) On coexpression with untagged ALIS, GFP:ALA3 was localized to Golgi-like structures: (B) coexpression with ALIS1; (C) coexpression with ALIS3; and (D) coexpression with ALIS5. In each case: left GFP fluorescence; right, overlay (including autofluorescence, magenta) of GFP signal on a bright-field image. (E) Golgi bodies visualized by expression of a Golgi localized sialyl transferase fusion to YFP (ST:YFP). Arrows indicate the position of nuclei when visible. Bar, 25 μm.
Localization of ALIS Proteins Is Determined by the Coexpressed P4-ATPase
At this point, we wondered whether ALIS proteins are able to exit the ER in the absence of a coexpressed P4-ATPase. To test this, YFP-tagged versions of the different ALIS genes were transiently expressed in N. benthamiana epidermal cells, together with GFP:HDEL encoding an ER-retained fusion protein (Hawes et al., 2001). Both fluorescent signals were found to colocalize in all cases, indicating that the β-subunits expressed alone are retained in the ER (Figure 8A; Supplemental Figures 4A and 5A).
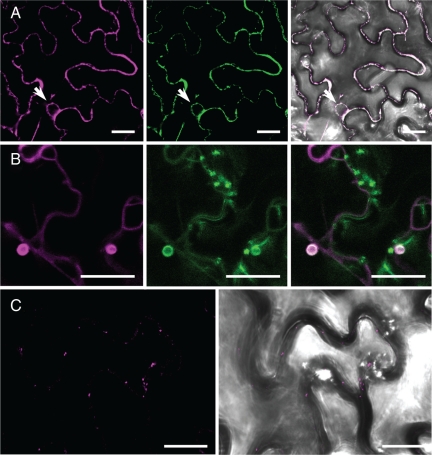
Figure 8.
ALIS1 leaves the ER and localizes to different compartments in the presence of ALA2 and ALA3, respectively. A C-terminal fusion of ALIS1 to YFP was expressed in tobacco leaf epidermal cells alone or in combination with ALA2 and ALA3. (A) When expressed alone, ALIS1:YFP colocalized with an ER-retained GFP:HDEL: left, YFP fluorescence; middle, GFP fluorescence; right, overlay image of YFP and GFP fluorescent signals on the bright-field image. (B) When expressed with GFP:ALA2, ALIS1:YFP colocalized to vesicular structures: left, YFP fluorescence; middle, GFP fluorescence; right, overlay of YFP and GFP fluorescent signals. (C) On coexpression with an untagged ALA3, ALIS1:YFP was detected in Golgi-resembling structures: left, YFP fluorescence; right, overlay of YFP signal on the bright-field image. Arrows indicate the position of nuclei when visible. Bar, 25 μm.
To study the effect of an ALA protein on ALIS localization in the cell, we coexpressed ALIS:YFP constructs with either GFP:ALA2 or GFP:ALA3. In combination with GFP:ALA2, YFP-tagged ALIS1, ALIS3, and ALIS5 all localized to large vesicular structures (Figure 8B; Supplemental Figures 4B and 5B). In the case of ALA3, coexpression of tagged ALIS3 and ALIS proteins appeared to be toxic for the cells. Therefore, we coexpressed ALIS:YFP fusions together with an untagged version of ALA3 (Figure 8C; Supplemental Figures 4C and 5C). The fluorescent signals for YFP could then be detected in punctuate structures resembling Golgi bodies as shown for ALA3. The different localization for ALIS proteins in the presence of ALA2 or ALA3 suggests that the P4-ATPase determines the final intracellular localization of the protein complex after exit from the ER.
DISCUSSION
Defining the minimal composition of the P4-ATPase dependent flippase machinery is a major challenge in cell biology. In this work we tested whether Cdc50 related β-subunits of P4-ATPases add novel features to the P4-ATPase catalytic subunit. We show that Arabidopsis P4-ATPases ALA2 and ALA3, which are putative aminophospholipid flippases, are retained in the ER when expressed in planta in the absence of an ALIS Cdc50-like β-subunit. After coexpression with any of three different ALIS β-subunits they leave the ER from where ALA2 travels to the endosomal system and ALA3 to the Golgi apparatus. Both ALA proteins are catalytically inactive in the absence of an ALIS protein but, in the presence of any ALIS β-subunit, ALA2 and ALA3 both promote transmembrane flipping of PS whereas ALA3 shows a broader transport specificity. This holds true both for labeled lipid probes as well as for natural lipids. We conclude that intracellular targeting signals and lipid specificity determinants reside in the catalytic ALA subunit.
P4-ATPases have been suggested to contribute indirectly to lipid flipping, e.g., by generating an electrochemical gradient in various membrane systems that drives specific lipid flippases. However, this is difficult to reconcile with our results showing that different ALA proteins when expressed in a heterologous host can generate flipping activities with different lipid specificities. Therefore, our results are in line with a direct role of P4-ATPases in lipid translocation as recently suggested (Coleman et al., 2009; Zhou et al., 2009).
It has previously been proposed that Cdc50-like proteins may help determine the substrate specificity of the flippase complex. It has been argued that the yeast trans-Golgi P4-ATPases Drs2p and Dnf3p, which exhibit different translocation profiles (Alder-Baerens et al., 2006), interact with different Cdc50 homologues (Cdc50p and Crf1p, respectively; Saito et al., 2004; Furuta et al., 2007). Furthermore, the yeast plasma membrane P4-ATPases Dnf1p and Dnf2p, which have the same substrate specificity (Pomorski et al., 2003), both interact with Lem3p (Saito et al., 2004; Furuta et al., 2007). However, these are only circumstantial pieces of evidence and do not produce a causative link between the nature of the subunits and the lipid specificity of the protein complex.
A higher number of P4-ATPase isoforms compared with Cdc50p homologues has been identified in most organisms, which argues against the notion that Cdc50-like proteins add specific features to P4-ATPase catalytic subunits. This imbalanced ratio is striking in multicellular organisms, because humans have 14 P4-ATPases and only three Cdc50 proteins (Paulusma and Oude Elferink, 2005), whereas in Arabidopsis 12 P4-ATPase isoforms (Gomés et al., 2000) and five subunits (Poulsen et al., 2008a) are present. No interaction partner among the Cdc50p homologues has thus far been found for yeast Neo1p, indicating that some P4-ATPases may act without a Cdc50p homologue. Lem3p interacts and sustains functionality of Dnf1p and Dnf2p (Saito et al., 2004; Furuta et al., 2007). In multicellular organisms several Cdc50p homologues have been shown capable of activating the same P4-ATPase (Poulsen et al., 2008a; Paulusma et al., 2008), which supports the notion that one Cdc50p isoform can interact with several P4-ATPases.
In light of the results presented in this study, it seems unlikely that Cdc50-like proteins directly participate in lipid binding and flipping. However, we cannot exclude the possibility that they support the transport function of the ALA subunit or play other role(s) in catalysis of the putative flippase complex. Indeed, three lines of evidence indicate that Cdc50 subunits directly participate in the P4-ATPase catalytic mechanism. First, separation of Drs2p from its binding partner Cdc50p affects its ability to form a phosphoenzyme intermediate (Lenoir et al., 2009). Second, association of the Cdc50 subunit with the Drs2p ATPase fluctuates during the reaction cycle, with the strongest interaction occurring at or near a point where the enzyme would be loaded with phospholipid ligand (Lenoir et al., 2009). Third, if Cdc50-like proteins are indispensable for catalytic function of P4-ATPases, mutation of Cdc50-like genes should also block phospholipid transport and phenocopy mutations in P4-ATPase genes. Indeed, mutations in members of the Cdc50 protein family produce such phenotypes. In yeast, this family consists of three members: Cdc50p, Crf1p, and Lem3p. CDC50 was identified in a screen for cold-sensitive, cell-division-cycle mutants, but how cdc50 causes a cold-sensitive block in the G1-to-S transition is unknown (Saito et al., 2004). At low temperatures, Δcdc50 mutant cells exhibit depolarization of cortical actin patches and mislocalization of polarity regulators, such as Bni1p and Gic1p, in a manner similar to the Δdrs2 mutant (Saito et al., 2004). LEM3 (previously ROS3) was recovered in two unbiased, genetic screens for yeast mutants that either aberrantly expose endogenous phosphatidylethanolamine on the cell surface (Kato et al., 2002) or are resistant to a toxic phosphatidylcholine analogue (Hanson et al., 2003). Deletion of LEM3 causes a defect in the translocation of NBD-PC and -PE across the plasma membrane and in this way phenocopies deletion mutants in the P4-ATPases dnf1 dnf2 (Pomorski et al., 2003).
Besides P4-ATPases, another subfamily of P-type ATPases is known to have additional subunits, namely the P2C subfamily of Na+/K+- and H+/K+-ATPases. With only 20–30% overall sequence identity, the three Na+/K+-ATPase β-subunit isoforms and one H+/K+-ATPase β-subunit are much less conserved than the α-subunits. Yet all share the same basic overall structure: a short cytosolic N-terminal tail, followed by a single membrane span and a large C-terminal ectodomain with multiple glycosylation sites and disulfide bonds. As pointed out previously (Poulsen et al., 2008b), Cdc50 proteins mimic a fusion between the β- and γ-subunits of the Na+/K+-ATPase in terms of polypeptide chain lengths and membrane topology. Both Cdc50 proteins and β-subunits are heavily N-glycosylated and contain highly conserved, disulfide bridge-forming cysteine residues (Geering, 2001; Kato et al., 2002; Lenoir et al., 2009; Shinoda et al., 2009).
The β-subunit of P2C-ATPases is required for the correct membrane insertion, functional maturation, and ER export of the α-subunit (Gottardi and Caplan, 1993; Geering, 2001). The γ-subunit is not required for function, but has a role in fine-tuning Na+/K+-ATPase activity in a tissue-specific manner (Geering, 2008). Similarly, Cdc50-like proteins required for P4-ATPase stability and export of the P4-ATPase from the ER (Paulusma et al., 2008; Saito et al., 2004; Chen et al., 2006; Furuta et al., 2007; this work). In this respect, Cdc50 proteins and P2C-ATPase β-subunits appear to be functionally similar. Hence, it seems that Cdc50 proteins function as chaperones responsible for the proper maturation of P4-ATPases, rather than serving as translocases or symporters themselves. P2C-ATPase β-subunits also modulate intrinsic kinetic parameters of the Na+/K+ pump (Geering, 2008), and a similar role for Cdc50-like proteins in P4-ATPases cannot be excluded.
In sum, the combination of cell biological and genetic data presented here demonstrate that Cdc50-like proteins are essential for ER exit of plant P4-ATPases and in this way show a striking similarity to the role of the β-subunit in P2C-ATPases. A surprising finding has been that Cdc50-like proteins do not contribute with specific ligands for intracellular targeting and lipid specificity and that these properties reside in the catalytic P4-ATPase subunit. These results mark an important step forward in understanding how P4-ATPases have been adapted to contribute to lipid flipping across membranes. Understanding the physiological roles of diverse plant P4-ATPase complexes remains a major challenge for the future.
Supplementary Material
ACKNOWLEDGMENTS
The authors acknowledge Prof. Chris Hawes (Oxford Brookes University), Dr. Federica Brandizzi (Michigan State University), and Prof. Liwen Jiang (The Chinese University of Hong Kong) for providing DNA constructs for colocalization experiments. This work was supported by the Danish National Research Foundation (M.G.P), The Danish Governments Globalisation Fund (L.R.P.), the Carlsberg Foundation (T.G.P.), and the Deutsche Forschungsgemeinschaft Grant Po748/10 (T.G.P.).
Abbreviations used:
- ER
endoplasmic reticulum
- HA
hemagglutinin
- NBD
7-nitrobenz-2-oxa-1,3-diazol-4-yl
- PC
phosphatidylcholine
- PE
phosphatidylethanolamine
- PM
plasma membrane
- PS
phosphatidylserine.
Footnotes
This article was published online ahead of print in MBC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E09-08-0656) on January 6, 2010.
REFERENCES
- Alder-Baerens N., Lisman Q., Luong L., Pomorski T., Holthuis J. C. Loss of P4 ATPases Drs2p and Dnf3p disrupts aminophospholipid transport and asymmetry in yeast post-Golgi secretory vesicles. Mol. Biol. Cell. 2006;17:1632–1642. doi: 10.1091/mbc.E05-10-0912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsen K. B., Palmgren M. G. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 1998;46:84–101. doi: 10.1007/pl00006286. [DOI] [PubMed] [Google Scholar]
- Axelsen K. B., Palmgren M. G. Inventory of the superfamily of P-type ion pumps in Arabidopsis. Plant Physiol. 2001;126:696–706. doi: 10.1104/pp.126.2.696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandizzi F., Fricker M., Hawes C. A greener world: the revolution in plant bioimaging. Nat. Rev. Mol. Cell Biol. 2002;3:520–530. doi: 10.1038/nrm861. [DOI] [PubMed] [Google Scholar]
- Chantalat S., Park S. K., Hua Z., Liu K., Gobin R., Peyroche A., Rambourg A., Graham T. R., Jackson C. L. The Arf activator Gea2p and the P-type ATPase Drs2p interact at the Golgi in Saccharomyces cerevisiae. J. Cell Sci. 2004;117:711–722. doi: 10.1242/jcs.00896. [DOI] [PubMed] [Google Scholar]
- Chen S., Wang J., Muthusamy B. P., Liu K., Zare S., Andersen R. J., Graham T. R. Roles for the Drs2p-Cdc50p complex in protein transport and phosphatidylserine asymmetry of the yeast plasma membrane. Traffic. 2006;7:1503–1517. doi: 10.1111/j.1600-0854.2006.00485.x. [DOI] [PubMed] [Google Scholar]
- Coleman J. A., Kwok C. M., Molday R. S. Localization, purification and functional reconstitution of the P4-ATPase, ATP8A2, a phosphatidylserine flippase in photoreceptor disc membranes. J. Biol. Chem. 2009;284:32670–32679. doi: 10.1074/jbc.M109.047415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis M. D., Grossniklaus U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 2003;133:462–469. doi: 10.1104/pp.103.027979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daleke D. L., Huestis W. H. Incorporation and translocation of aminophospholipids in human erythrocytes. Biochemistry. 1985;24:5406–5416. doi: 10.1021/bi00341a019. [DOI] [PubMed] [Google Scholar]
- Darland-Ransom M., Wang X., Sun C. L., Mapes J., Gengyo-Ando K., Mitani S., Xue D. Role of C. elegans TAT-1 protein in maintaining plasma membrane phosphatidylserine asymmetry. Science. 2008;320:528–531. doi: 10.1126/science.1155847. [DOI] [PubMed] [Google Scholar]
- Earley K. W., Haag J. R., Pontes O., Opper K., Juehne T., Song K., Pikaard C. S. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- Ebashi F., Ebashi S. Removal of calcium and relaxation in actomyosin systems. Nature. 1962;28:378–379. doi: 10.1038/194378a0. [DOI] [PubMed] [Google Scholar]
- Farge E., Ojcius D. M., Subtil A., Dautry-Varsat A. Enhancement of endocytosis due to aminophospholipid transport across the plasma membrane of living cells. Am. J. Physiol. 1999;276:C725–C733. doi: 10.1152/ajpcell.1999.276.3.C725. [DOI] [PubMed] [Google Scholar]
- Furuta N., Fujimura-Kamada K., Saito K., Yamamoto T., Tanaka K. Endocytic recycling in yeast is regulated by putative phospholipid translocases and the Ypt31p/32p-Rcy1p pathway. Mol. Biol. Cell. 2007;18:295–312. doi: 10.1091/mbc.E06-05-0461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gall W. E., Geething N. C., Hua Z., Ingram M. F., Liu K., Chen S. I., Graham T. R. Drs2p dependent formation of exocytic clathrin-coated vesicles in vivo. Curr. Biol. 2002;12:1623–1627. doi: 10.1016/s0960-9822(02)01148-x. [DOI] [PubMed] [Google Scholar]
- Geering K. The functional role of β-subunits in oligomeric P-type ATPases. J. Bioenerg. Biomembr. 2001;33:425–438. doi: 10.1023/a:1010623724749. [DOI] [PubMed] [Google Scholar]
- Geering K. Functional roles of Na,K-ATPase subunits. Curr. Opin. Nephrol. Hypertens. 2008;17:526–532. doi: 10.1097/MNH.0b013e3283036cbf. [DOI] [PubMed] [Google Scholar]
- Gietz R. D., Woods R. A. Transformation of yeast by lithium acetate/single-stranded carrier DNA/polyethylene glycol method. Methods Enzymol. 2002;350:87–96. doi: 10.1016/s0076-6879(02)50957-5. [DOI] [PubMed] [Google Scholar]
- Gomès E., Jakobsen M. K., Axelsen K. B., Geisler M., Palmgren M. G. Chilling tolerance in Arabidopsis involves ALA1, a member of a new family of putative aminophospholipid translocases. Plant Cell. 2000;12:2441–2454. [PMC free article] [PubMed] [Google Scholar]
- Gottardi C. J., Caplan M. J. Delivery of Na+,K+-ATPase in polarized epithelial cells. Science. 1993;260:552–554. doi: 10.1126/science.8386395. [DOI] [PubMed] [Google Scholar]
- Graham T. R. Flippases and vesicle-mediated protein transport. Trends Cell Biol. 2004;14:670–677. doi: 10.1016/j.tcb.2004.10.008. [DOI] [PubMed] [Google Scholar]
- Hanson P. K., Malone L., Birchmore J. L., Nichols J. W. Lem3p is essential for the uptake and potency of alkylphosphocholine drugs, edelfosine and miltefosine. J. Biol. Chem. 2003;278:36041–36050. doi: 10.1074/jbc.M305263200. [DOI] [PubMed] [Google Scholar]
- Hawes C., Saint-Jore C., Martin B., Zheng H. Q. ER confirmed as the location of mystery organelles in Arabidopsis plants expressing GFP! Trends Plant Sci. 2001;6:245–246. doi: 10.1016/s1360-1385(01)01980-x. [DOI] [PubMed] [Google Scholar]
- Hua Z., Fatheddin P., Graham T. R. An essential subfamily of Drs2p-related P-type ATPases is required for protein trafficking between Golgi complex and endosomal/vacuolar system. Mol. Biol. Cell. 2002;13:3162–3177. doi: 10.1091/mbc.E02-03-0172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hua Z., Graham T. R. Requirement for Neo1p in retrograde transport from the Golgi complex to the endoplasmic reticulum. Mol. Biol. Cell. 2003;14:4971–4983. doi: 10.1091/mbc.E03-07-0463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorgensen P. L., Hakansson K. O., Karlish S. J. Structure and mechanism of Na,K-ATPase: functional sites and their interactions. Annu. Rev. Physiol. 2003;65:817–849. doi: 10.1146/annurev.physiol.65.092101.142558. [DOI] [PubMed] [Google Scholar]
- Kato U., Emoto K., Fredriksson C., Nakamura H., Ohta A., Kobayashi T., Murakami-Murofushi K., Kobayashi T., Umeda M. A novel membrane protein, Ros3p, is required for phospholipid translocation across the plasma membrane in Saccharomyces cerevisiae. J. Biol. Chem. 2002;277:37855–37862. doi: 10.1074/jbc.M205564200. [DOI] [PubMed] [Google Scholar]
- Katoh Y., Katoh M. Identification and characterization of CDC50A, CDC50B and CDC50C genes. Oncol. Rep. 2004;12:939–943. [PubMed] [Google Scholar]
- Kieber J. J., Rothenberg M., Roman G., Feldmann K. A., Ecker J. R. CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell. 1993;72:427–441. doi: 10.1016/0092-8674(93)90119-b. [DOI] [PubMed] [Google Scholar]
- Koncz C., Schell J. The promoter of TL-DNA gene 5 controls the tissue-specific expression of chimeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 1986;204:383–396. [Google Scholar]
- Kotzer A. M., Brandizzi F., Neuman U., Paris N., Moore I., Hawes C. AtRabF2b (Ara7) acts on the vacuolar trafficking pathway in tobacco leaf epidermal cells. J. Cell Sci. 2004;117:6377–6389. doi: 10.1242/jcs.01564. [DOI] [PubMed] [Google Scholar]
- Kuhlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- Lenoir G., Williamson P., Holthuis J. C. On the origin of lipid asymmetry: the flip side of ion transport. Curr. Opin. Chem. Biol. 2007;11:654–661. doi: 10.1016/j.cbpa.2007.09.008. [DOI] [PubMed] [Google Scholar]
- Lenoir G., Williamson P., Puts C. F., Holthuis J. C. Cdc50p plays a vital role in the ATPase reaction cycle of the putative aminophospholipid transporter Drs2p. J. Biol. Chem. 2009;284:17956–17967. doi: 10.1074/jbc.M109.013722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu K., Surendhran K., Nothwehr S. F., Graham T. R. P4-ATPase requirement for AP-1/clathrin function in protein transport from the trans-Golgi network and early endosomes. Mol. Biol. Cell. 2008;19:3526–3535. doi: 10.1091/mbc.E08-01-0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Märki F., Hänni E., Fredenhagen A., van Oostrum J. Mode of action of the lanthionine-containing peptide antibiotics duramycin, duramycin B and C, and cinnamycin as indirect inhibitors of phospholipase A2. Biochem. Pharmacol. 1991;42:2027–2035. doi: 10.1016/0006-2952(91)90604-4. [DOI] [PubMed] [Google Scholar]
- Morth J. P., Pedersen B. P., Toustrup-Jensen M. S., Sørensen T. L., Petersen J., Andersen J. P., Vilsen B., Nissen P. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- Muller P., Pomorski T., Herrmann A. Incorporation of phospholipid analogues into the plasma membrane affects ATP-induced vesiculation of human erythrocyte ghosts. Biochem. Biophys. Res. Commun. 1994;199:881–887. doi: 10.1006/bbrc.1994.1311. [DOI] [PubMed] [Google Scholar]
- Palmgren M. G., Harper J. F. Pumping with plant P-type ATPases. J. Exp. Bot. 1998;50:883–893. [Google Scholar]
- Parsons A. B., et al. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- Paulusma C. C., Folmer D. E., Ho-Mok K. S., de Waart D. R., Hilarius P. M., Verhoeven A. J., Oude Elferink R. P. ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane lipid flippase activity. Hepatology. 2008;47:268–278. doi: 10.1002/hep.21950. [DOI] [PubMed] [Google Scholar]
- Paulusma C. C., Oude Elferink R. P. The type 4 subfamily of P-type ATPases, putative aminophospholipid translocases with a role in human disease. Biochim. Biophys. Acta. 2005;1741:11–24. doi: 10.1016/j.bbadis.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Pedersen B. P., Buch-Pedersen M. J., Morth J. P., Palmgren M. G., Nissen P. Crystal structure of the plasma membrane proton pump. Nature. 2007;450:1111–1114. doi: 10.1038/nature06417. [DOI] [PubMed] [Google Scholar]
- Pérez-Victoria F. J., Sánchez-Cañete M. P., Castanys S., Gamarro F. Phospholipid translocation and miltefosine potency require both L. donovani miltefosine transporter and the new protein LdRos3 in Leishmania parasites. J. Biol. Chem. 2006;281:23766–23775. doi: 10.1074/jbc.M605214200. [DOI] [PubMed] [Google Scholar]
- Pomorski T., Lombardi R., Riezman H., Devaux P. F., van Meer G., Holthuis J. C. Drs2p-related P-type ATPases Dnf1p and Dnf2p are required for phospholipid translocation across the yeast plasma membrane and serve a role in endocytosis. Mol. Biol. Cell. 2003;14:1240–1254. doi: 10.1091/mbc.E02-08-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen L. R., López-Marqués R. L., McDowell S. C., Okkeri J., Licht D., Schulz A., Pomorski T., Harper J. F., Palmgren M. G. The Arabidopsis P4-ATPase ALA3 localizes to the golgi and requires a β-subunit to function in lipid translocation and secretory vesicle formation. Plant Cell. 2008a;20:658–676. doi: 10.1105/tpc.107.054767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulsen L. R., López-Marqués R. L., Palmgren M. G. Flippases: still more questions than answers. Cell. Mol. Life Sci. 2008b;65:3119–3125. doi: 10.1007/s00018-008-8341-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puts C. F., Holthuis J. C. Mechanism and significance of P4 ATPase-catalyzed lipid transport: Lessons from a Na+/K+-pump. Biochim. Biophys. Acta. 2009;1791:603–611. doi: 10.1016/j.bbalip.2009.02.005. [DOI] [PubMed] [Google Scholar]
- Riekhof W. R., Voelker D. R. The yeast plasma membrane P4-ATPases are major transporters for lysophospholipids. Biochim. Biophys. Acta. 2009;1791:620–627. doi: 10.1016/j.bbalip.2009.02.013. [DOI] [PubMed] [Google Scholar]
- Rose A. B., Broach J. R. Propagation and expression of cloned genes in yeast: 2-microns circle-based vectors. Methods Enzymol. 1990;185:234–279. doi: 10.1016/0076-6879(90)85024-i. [DOI] [PubMed] [Google Scholar]
- Ruaud A. F., Nilsson L., Richard F., Larsen M. K., Bessereau J. L., Tuck S. The C. elegans P4-ATPase TAT-1 regulates lysosome biogenesis and endocytosis. Traffic. 2009;10:88–100. doi: 10.1111/j.1600-0854.2008.00844.x. [DOI] [PubMed] [Google Scholar]
- Saint-Jore C. M., Evins J., Batoko H., Brandizzi F., Moore I., Hawes C. Redistribution of membrane proteins between the Golgi apparatus and endoplasmic reticulum in plants is reversible and not dependent on cytoskeletal networks. Plant J. 2002;29:661–678. doi: 10.1046/j.0960-7412.2002.01252.x. [DOI] [PubMed] [Google Scholar]
- Saito K., Fujimura-Kamada K., Furuta N., Kato U., Umeda M., Tanaka K. Cdc50p, a protein required for polarized growth, associates with the Drs2p P-type ATPase implicated in phospholipid translocation in Saccharomyces cerevisiae. Mol. Biol. Cell. 2004;15:3418–3432. doi: 10.1091/mbc.E03-11-0829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seigneuret M., Devaux P. F. ATP-dependent asymmetric distribution of spin-labeled phospholipids in the erythrocyte membrane: relation to shape changes. Proc. Natl. Acad. Sci. USA. 1984;81:3751–3755. doi: 10.1073/pnas.81.12.3751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinoda T., Ogawa H., Cornelius H., Toyoshima C. Crystal structure of the sodium-potassium pump at 2.4 Å resolution. Nature. 2009;459:446–450. doi: 10.1038/nature07939. [DOI] [PubMed] [Google Scholar]
- Siegmund A., Grant A., Angeletti C., Malone L., Nichols J. W., Rudolph H. K. Loss of Drs2p does not abolish transfer of fluorescence-labeled phospholipids across the plasma membrane of Saccharomyces cerevisiae. J. Biol. Chem. 1998;273:34399–34405. doi: 10.1074/jbc.273.51.34399. [DOI] [PubMed] [Google Scholar]
- Sparkes I. A., Runions J., Kearns A., Hawes C. Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 2006;1:2019–2025. doi: 10.1038/nprot.2006.286. [DOI] [PubMed] [Google Scholar]
- Tang X., Halleck M. S., Schlegel R. A., Williamson P. A subfamily of P-type ATPases with aminophospholipid transporting activity. Science. 1996;272:1495–1497. doi: 10.1126/science.272.5267.1495. [DOI] [PubMed] [Google Scholar]
- Toyoshima C., Nakasako M., Nomura H., Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 A resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- Tse Y. C., Mo B., Hillmer S., Zhao M., Lo S. W., Robinson D. G., Jiang L. Identification of multivesicular bodies as prevacuolar compartments in Nicotiana tabacum BY-2 cells. Plant Cell. 2004;16:672–693. doi: 10.1105/tpc.019703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba J. M., Palmgren M. G., Berberián G. E., Ferguson C., Serrano R. Functional expression of plant plasma membrane H+-ATPase in yeast endoplasmic reticulum. J. Biol. Chem. 1992;267:12341–12349. [PubMed] [Google Scholar]
- Voinnet O., Rivas S., Mestre P., Baulcombe D. An enhanced transient expression system in plans based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003;33:949–956. doi: 10.1046/j.1365-313x.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- Wang J., Cai Y., Miao Y., Lam S. K., Jiang L. Wortmannin induces homotypic fusion of plant prevacuolar compartments. J. Exp. Bot. 2009;60:3075–3083. doi: 10.1093/jxb/erp136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicky S., Schwarz H., Singer-Kruger B. Molecular interactions of yeast Neo1p, an essential member of the Drs2 family of aminophospholipid translocases, and its role in membrane trafficking within the endomembrane system. Mol. Cell. Biol. 2004;24:7402–7418. doi: 10.1128/MCB.24.17.7402-7418.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Oppenheimer D. G. Irregular Trichome Branch 2 (ITB2) encodes a putative aminophospholipid translocase that regulates trichome branch elongation in Arabidopsis. Plant J. 2009;60:195–206. doi: 10.1111/j.1365-313X.2009.03954.x. [DOI] [PubMed] [Google Scholar]
- Zhou X., Graham T. R. Reconstitution of phospholipid translocase activity with purified Drs2p, a type-IV P-type ATPase from budding yeast. Proc. Natl. Acad. Sci. USA. 2009;106:16586–16591. doi: 10.1073/pnas.0904293106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.