Abstract
Signatures of natural selection occur throughout the human genome and can be detected at the sequence level. We have re-sequenced ABCE1, a host candidate gene essential for HIV-1 capsid assembly, in European- (n=23) and African-descent (Yoruban; n=24) reference populations for genetic variation discovery. We identified an excess of rare genetic variation in Yoruban samples, and the resulting Tajima’s D was low (−2.27). The trend of excess rare variation persisted in flanking candidate genes ANAPC10 and OTUD4, suggesting that this pattern of positive selection can be detected across the 184.5kb examined on chromosome 4. Because of ABCE1’s role in HIV-1 replication, we re-sequenced the candidate gene in three small cohorts of HIV-1-infected or resistant individuals. We were able to confirm the excess of rare genetic variation among HIV-1 positive African-American individuals (n=53; Tajima’s D = −2.34). These results highlight the potential importance of ABCE1’s role in infectious diseases such as HIV-1.
Keywords: ABCE1, African-Americans, single nucleotide polymorphisms, HIV-1
Introduction
The candidate gene ABCE1, located on chromosome 4q311, is a member of the ATP binding-cassette (ABC) family. ABCE1 has two main isoforms and is widely expressed1. Unlike other ABC family members, ABCE1 maintains an ATP-binding cassette lacking a transmembrane domain2. The product of ABCE1 was first described as an inhibitor of ribonuclease (RNase) L3, but more recently it has been shown to be involved in eukaryotic translational initiation4–9, and is essential in the assembly of immature HIV-1 capsids10, 11.
In human populations, little is known about the patterns of genetic variation in ABCE1. To date, four variation discovery efforts have been published for ABCE112–14; however, none provide a comprehensive reference of common variation across multiple populations or across both intronic and exonic sequence. Given the potential role ABCE1 has on HIV-1 replication, we, as part of the Program for Genomic Applications SeattleSNPs, re-sequenced both ABCE1 and the flanking sequence in 23 Centre d’Etude du Polymorphisme Humain samples (CEPHs) and 24 Yoruban samples using standard dye terminator sequencing technology15 to identify single nucleotide polymorphisms for future genetic association studies. In a parallel study approved by the University of Washington’s Human Subjects Review Committee, we also re-sequenced ABCE1 in three small populations ascertained for features of HIV-1 resistance or disease progression. In both the variation discovery dataset and the HIV-1 infection-related population dataset, we observed an excess of rare genetic variation in ABCE1 that extends to its neighboring genes ANAPC10 and OTUD4. These data suggest that the genomic region containing ABCE1 may have been a target for positive selection that has affected present day genetic diversity in human populations.
Results and Discussion
We successfully re-sequenced 30,773bp of ABCE1 (all introns and exons) as well as 1,870bp and 2,073bp of 5′ and 3′ flanking sequence, respectively, in 23 European-descent (CEPH) and 24 African-descent (Yoruban) samples. We identified 125 SNPs in these 47 reference DNA samples: 40 SNPs in the European-descent and 93 SNPs in the African-descent discovery panels (Table 1). Nucleotide diversity (π) was lower in the European- and African-descent discovery panels (5.5×10−4 and 7.6×10−4, respectively; Table 1) compared to the average nucleotide diversity based on 180 candidate genes re-sequenced in similar DNA discovery panels (7.0×10−4 and 9.0×10−4, respectively)16. In other words, based on 180 genes, one SNP is expected per 1,100 bps and 1,435 bps in African- and European-descent populations, respectively, compared to our observed rate of one SNP per 1,315 bps and 1,818 bps, respectively, in ABCE1.
Table 1.
Number individuals re-sequenced, number of SNPs (S), nucleotide diversity (π and θ), and Tajima’s D for ABCE1.
| Cohort/Samples | # individuals | S (# with MAF >5%) | π(×10−4) (θ) | Tajima’s D |
|---|---|---|---|---|
| CEPH | 23 | 40 (14) | 5.5 (7.0) | −1.36 |
| Yoruban | 24 | 93 (27) | 7.6 (34.5) | −2.27 |
| HIV-1 positive African-Americans1 | 53 | 170 (21) | 9.6 (95.4) | −2.34 |
| HIV-1 positive, non-progressing2 EA AA |
25 3 |
34 (17) 11 (11) |
5.9 (5.1) 3.8 (0.7) |
−0.77 −0.54 |
| Exposed, seronegatives3 | 10 | 24 (13) | 5.3 (3.1) | −0.82 |
University of Washington/Center for AIDS research (UW/CFAR) cohort: This cohort consists of HIV-infected individuals cared for at the University of Washington HIV clinics who were invited to donate their blood to the UW/CFAR HIV specimen repository for use in HIV-related virology, immunology, and disease pathogenesis studies. All 53 DNA samples are from African-Americans and the majority from men (62%). At enrollment, the average age of participants was 39 years (range: 20–51 years), the length of HIV-1 infection and treatment information was unknown, the mean viral load was 4.0 log10 copies/ml (range: 2.7–5.7 log10 copies/ml), and the mean CD4+ T-cell count was 341 cells/μl (range: 2–974 cells/μl). In follow-up exams, these participants had a mean viral load of 2.6 log10 copies/ml (range: 1.5–5.4 log10 copies/ml) and a mean CD4+ T-cell count of 409 cells/μl (range: 15–1439 cells/μl). 37 of the 53 participants (70%) eventually began antiretroviral therapy (ART), with a mean follow-up length of 6 years (range: 0–14 years).
Long-term non-progressor (LTNP) cohort: These volunteers were HIV-1 infected individuals who had a documented HIV-1 seropositivity for ≥ 10 years and CD4+ T-cell counts that were either ≥ 600 cells/μL or > 500 cells/μL, with a slope that was either zero or positive during the two years prior to enrollment. All of the 28 DNA samples sequenced were from male volunteers, while 25 of the 28 samples (89%) were from European-Americans and the remaining (11%) were from African-Americans. At enrollment, this subgroup of volunteers had the following characteristics: the average age was 41 years (range: 30–52 years), the average length of HIV-1 infection was 14 years (range: 10–20 years), the mean viral load was 3.3 log10 copies/ml (range: 2.7–5.1 log10 copies/ml), the mean CD4+ T-cell count was 819 cells/μl (range: 364–1372 cells/μl), and all individuals were ART naïve. In follow-up exams, these individuals had a mean viral load of 2.6 log10 copies/ml (range: 1.5–5.3 log10 copies/ml), a mean CD4+ T-cell count of 675 cells/μl (range: 262–1724 cells/μl). In addition, 9 of the individuals (32%) began ART during the follow-up examination period, with as the mean length of 18 years (range, 12–27 years) for the period between HIV-1 infection and the start of ART. The mean length of follow-up was 7 years for the 28 individuals (range: 1–11 years).
HIV-1 high-risk exposed seronegatives (ES) cohort: These volunteers were predominantly European-American men having sex with men (MSM). The ES cohort, their enrollment criteria, and the study procedures associated with this cohort have been previously described33. All 10 individuals were European-American and 7 (70%) were male. The mean age of the individuals was 34 years (range: 24–53 years).
Abbreviations: African-American (AA), Centre d’Etude du Polymorphisme Humain (CEPH), European-American, minor allele frequency (MAF)
In addition to the lower-than-expected nucleotide diversity, we observed low Tajima’s D statistics17, 18, a common test for deviation from the expected rate of mutation and genetic drift in the absence of selection, for both the European- and African-descent discovery panels (−1.36 and −2.27, respectively; Table 1). Based on 323 candidate genes re-sequenced in a similar DNA discovery panel, the average Tajima’s D is 0.14 and −0.49 in European- and African-descent populations, respectively16, 19. Lower Tajima’s D statistics have been observed for European-descent populations in candidate genes such as TRPV620, 21; however, lower Tajima’s D statistics have not yet been reported for African-descent populations for any one re-sequenced candidate gene19. The Tajima’s D statistic observed here is 2.65 standard deviations from the mean Tajima’s D statistic calculated for 323 candidate genes in African-descent samples, indicating that the pattern of ABCE1 genetic diversity is an extreme outlier and adding weight to the suggestive evidence of positive selection22.
Recent genome-wide screens for extreme Tajima’s D statistics based on Perlegen23, 24 data identified several genomic regions as extreme outliers in African-descent populations25; These analyses did not identify the region of chromosome 4 containing ABCE1 as an outlier of this statistic for either population. This is likely due to the ascertainment bias toward high-frequency alleles present in the Perlegen dataset 24, 26. Carlson and colleagues25 noted that this bias is evident from the higher mean value for Tajima’s D for 178 candidate genes in the Perlegen data (0.94 for African-descent) compared to the SeattleSNPs data (−0.54 for African-descent). For ABCE1, the Perlegen dataset only contains data for nine SNPs in for 23 African-American samples in contrast to the 93 SNPs in the 24 Yoruban samples described here (http://gvs.gs.washington.edu/GVS/). Of the nine SNPs in the Perlegen dataset, three are common (>5% minor allele frequency) with all three in high linkage disequilibrium with one another (r2=1 for all pair-wise combinations). Perlegen23, like HapMap27, is biased towards common variation, and this bias can impact statistics such as Tajima’s D which aims to summarize the natural allele frequency distribution of variations in the gene or region of interest.
Our variation discovery dataset, in contrast to the genome-wide datasets such as Perlegen23 and HapMap27, is based on re-sequencing that is less likely to be influenced by ascertainment bias28. The extreme low Tajima’s D statistics signal detected in this re-sequencing data may be due to positive selection but the possibility of population expansion cannot be formally excluded in this dataset. By comparison, it has been postulated that the signature of selection in and around TRPV6 in European-descent populations is more likely to be due to positive selection than population demography based on extensive simulations21. Also, that signature is observed only among European-descent populations, which suggests local adaptation21. Despite the uncertainty in the interpretation of the Tajima’s D statistic for ABCE1, it is notable that other host candidate genes associated with HIV-1, such as CCR5 (Tajima’s D = 2.2 in Europeans29) and APOBEC3G30, exhibit genetic signatures of natural selection in human populations. Indeed, pathogens in general are thought to have had a major impact on the landscape of the host genome over the course of human history31, 32.
Based on the preliminary evidence that ABCE1 may be subject to positive selection, and given its putative involvement in HIV-1 replication, we expanded our re-sequencing efforts to include clinical samples collected from volunteers enrolled in three separate study cohorts from the Seattle area: African-American HIV-1 positive individuals (n=53), HIV-1 positive, long-term non-progressing individuals (n=28), and HIV-1 high-risk seronegative individuals (n=10)33, 34. Overall, the pattern of genetic diversity in these populations was similar to that observed for the variation discovery dataset. That is, Tajima’s D is similar between the African-American HIV-1 infected individuals and the African population re-sequenced for SNP discovery, −2.34 and −2.27, respectively, (Table 1). The other patient populations also have negative Tajima’s D statistics that are consistent with a trend of to an excess of rare alleles despite the small samples sizes.
In addition to similar patterns of overall genetic diversity, both the HIV-1 infection-related populations and the SNP discovery panels have similar patterns of intronic and exonic diversity (Tables 1 and 2). Overall, we identified almost twice as many SNPs among the African-American HIV-1 infected individuals compared with the Yoruban sample (170 vs. 93), but this was expected given the sample size for the HIV-1 infected individuals was twice as large as the SNP discovery set (Table 1). In fact, of the SNPs not in common between these two sample sets, ~90% were rare (<5% minor allele frequency). Of the common SNPs not shared between the two African-descent samples, six were found only in the CEPH variation discovery and the African-American HIV-1 positive individuals datasets but not among the Yoruban sample dataset, suggesting that these variations represent a genetic admixture typical of African-descent populations ascertained in the United States35. Among the common SNPs shared between the two African-descent populations, only one (intronic rs34492893) was significantly less frequent among HIV-1 positive African-American samples (MAF=5%) compared to the samples from presumably healthy Yorubans (MAF=15%; p=0.05; Table 3). No differences in allele frequencies were observed when the CEPH data were compared with the data from the European-descent individuals of the long-term non-progressing cohort (data not shown). It is possible that the allele frequency difference observed for rs34492893 is due to statistical fluctuation from small sample sizes and/or to differences in European admixture in African-Americans and Yorubans36. Additional tests of association with larger sample sizes are needed to confirm this potential difference between HIV-1 positive and general population samples of African-descent.
Table 2. SNPs in ABCE1 exons.
Location and frequency of the SNP is given for each sample that was re-sequenced for variation discovery.
| Site1 (rs number) | Nucleotide | Amino Acid (position) | Sample (Frequency) |
|---|---|---|---|
| 8026 (rs34265438) | G>A | Lys to Lys (27) | CEPH (0.02) |
| 12707 (−)2 | A>T | Gly to Gly (110) | African-American HIV-1 positive (0.03) |
| 15787 (rs35406092) | G>A | Glu to Glu (242) | African-American HIV-1 positive (0.01) Yoruban (0.05) |
| 26775 (rs35729907) | A>G | Leu toLeu (467) | Yoruban (0.04) |
| 26799 (rs34070877)2 | C>T | Cys to Cys (475) | African-American HIV-1 positive (0.07) Yoruban (0.06) |
| 31200 (rs34305988) | C>G | N/a | Yoruban (0.02) |
| 31222 (−) | A>G | N/a | African-American HIV-1 positive (0.03) |
| 31291 (−) | C>T | N/a | African-American HIV-1 positive (0.02) |
| 31703 (rs35984115) | G>A | N/a | African-American HIV-1 positive (0.02) Yoruban (0.03) |
| 31810 (−) | A>T | N/a | African-American HIV-1 positive (0.01) |
| 31865 (−) | −/+ | N/a | African-American HIV-1 positive (0.01) |
| 32165 (rs17019897) | C>G | N/a | African-American HIV-1 positive (0.01) Yoruban (0.05) |
| 32304 (rs34634683) | G>A | N/a | African-American HIV-1 positive (0.03) Yoruban (0.02) |
Table 3. ABCE1 SNP allele frequency comparisons between Yorubans and African-American HIV-1 infected individuals.
Common tagSNPs were identified in the Yoruban dataset for SNPs with MAF≥0.05 in ABCE1 using the Genome Variation Server (gvs.gs.washington.edu/GVS/) at r2>0.80.
| SNP rs number (minor allele) | SNP location | Yoruban MAF (n=48) | African-American HIV-1 infected individuals MAF (n=106) |
|---|---|---|---|
| rs959304 (A) | Intron | 0.09 | 0.05 |
| rs34291406 (T) | Intron | 0.09 | 0.05 |
| rs17019887 (C) | Intron | 0.08 | 0.08 |
| rs35406092 (A) | Synonymous | 0.05 | 0.00 |
| rs34766684 (C) | Intron | 0.08 | 0.10 |
| rs34776612 (T) | Intron | 0.07 | 0.06 |
| rs34070877 (T) | Synonymous | 0.06 | 0.07 |
| rs34492893† (G) | Intron | 0.15 | 0.05 |
| rs34935304 (T) | Intron | 0.09 | 0.00 |
| rs35207405 (A) | Intron | 0.06 | 0.00 |
| rs35407026 (T) | Intron | 0.19 | 0.11 |
| rs35665369 (T) | Intron | 0.07 | 0.00 |
| rs36114072 (−) | Intron | 0.05 | 0.00 |
n = number of chromosomes
Abbreviations: minor allele frequency (MAF)
The minor allele frequency in Yorubans is higher compared with African-American HIV-1 infected individuals (p=0.05; Fisher’s exact test).
For exonic diversity, no nonsynonymous variation was identified in re-sequencing any of the patient populations or the SNP discovery panels. We identified a total of five synonymous SNPs and eight diallelic variants in the untranslated regions (Table 2). All identified exonic variations had minor allele frequencies of 7% or less in their respective samples.
The genetic profile of ABCE1 observed among African-descent populations suggests positive selection, and it is intriguing to speculate that the gene’s function is associated with this selection event. It is possible, however, that ABCE1 is contained within the region that exhibits the signature of selection but is not the locus under selection. To better define the boundaries of the signature of selection on chromosome 4, we merged the SeattleSNPs discovery dataset with the NIEHS Environmental Genome Project discovery dataset37 and surveyed the genetic diversity of a 184.5kb region that contains the candidate genes ANAPC10, ABCE1, and OTUD4 in 12 overlapping Yoruban samples (Figure 1). ANAPC10 (anaphase promoting complex subunit 10) has been reported essential for mitosis38, 39. Recent expression studies suggest that ANAPC10 is highly expressed in glioblastoma endothelial cells compared with normal brain and other tissues40. Interestingly, the neighboring gene OTUD4 (OTU domain containing 4), was identified in a screen for HIV-associated chimeric provirus-host gene transcripts 41. Tajima’s D for this region in the Yoruban samples was low (−1.72), suggesting the signature for positive selection is contained within this expanded region surveyed. This region also exhibits relatively low levels of linkage disequilibrium (Figure 2), a finding which may be expected given the excess of rare variation.
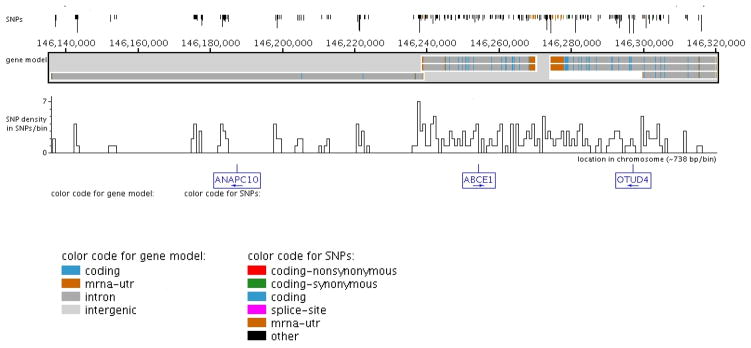
Figure 1. Location and density of SNPs discovered across ANAPC10, ABCE1, and OTUD4 on chromosome 4 the Yoruban population.
Candidate genes were re-sequenced in 12 Yoruban DNA samples (NA18502, NA19238, NA18504, NA18870, NA1855, NA19137, NA19201, NA19200, NA19203, NA19223, NA19153, and NA19144) for variation discovery. All ABCE1 and OTUD4 introns and exons were targeted for re-sequencing while ANAPC10 exons and representative intronic sequence was targeted for variation discovery. The presence of a SNP and its frequency are denoted by the vertical lines at the top of the figure generated by the Genome Variation Server (http://gvs.gs.washington.edu/GVS/). The lines are color coded to correspond with the gene model for each candidate gene (for example, orange represents the untranslated region of a gene). The graph below the gene model represents the SNP density per ~738 basepairs across this genomic region. Candidate genes and their direction are labeled at the bottom of the figure. All DNA variations were deposited into dbSNP and GenBank (accession numbers DQ304649, DQ148409, and DQ427109)
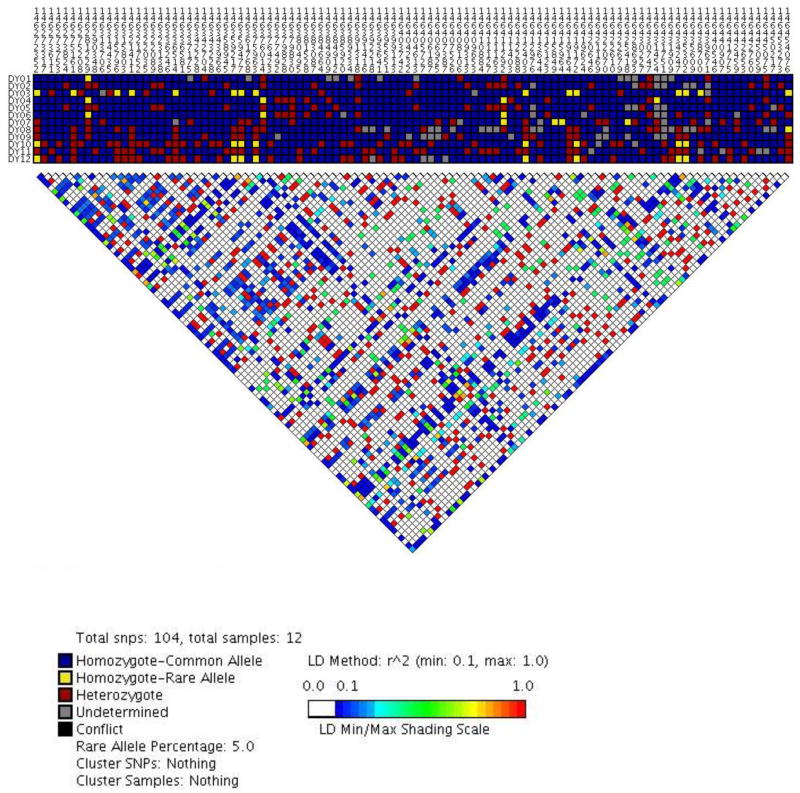
Figure 2. Patterns of linkage disequilibrium (r2) in Yoruban Africans (n=12) across ANAPC10, ABCE1, and OTUD4.
Common SNPs (minor allele frequency >5%) are numbered across the top of the figure, and samples are numbered to the left side of the figure. SNPs are numbered according to their chromosomal position based on NCBI Build 36. Each square represents the individual’s genotype for a specific SNP, and each square is color-coded so that blue represents homozygosity for the common allele, red represents heterozygosity, and yellow represents being homozygosity for the rare allele. Gray represents missing data.
Conclusions
Based on our variation discovery efforts, we show here that the genomic region containing ABCE1 has an excess of rare variation, resulting in a signature of natural selection in African-descent populations. The product of ABCE1 is highly conserved across species and is essential for life4. However, given that our data suggest positive selection across ABCE1 and surrounding genomic regions in African-descent compared to European-descent populations, it is possible that some factor separate from basic biological conservation is responsible for this signature. It is intriguing to speculate on the trigger of the selection event, given ABCE1’s role in HIV-1 assembly. Notably, an ABCE1 insertion/deletion variant (rs9333571) has been reported as associated with reduced HIV-1 replication13. This genetic variant was rare in its respective cohort (MAF=1%) and, as such, not identified in our small cohorts. Rare genetic variations such as this indel will require deep re-sequencing efforts in patient populations to identify associations with complex phenotypes42. These rare variations will likely be missed by current genome-wide association studies, which rely on common variation and linkage disequilibrium to detect associations for variations not directly genotyped in the experiment43.
The advent of HIV-1/AIDS is likely too recent in human history to have left a detectable footprint in present day populations, and we cannot formally exclude the possibility that the signature we are observing is due to recent population expansion. Further work with larger cohorts is needed to better understand potential sources of ABCE1 positive selection and to better characterize any possible association of rare variation within the ABCE1 gene in African-Americans with HIV-1 infection.
Acknowledgments
We would like to thank Dr. Jaisri Lingappa (University of Washington) for constructive comments, Devon Livingston-Rosanoff (VIDI) for technical assistance and Reneé Ireton for technical editing of the report. The work was funded by NIH grants 1 R21 A1073115-01 (JL), U01 HL66682 (DAN), U01 HL66642 (DAN), N01 ES-15478 (DAN), AI47806 (MJM), AI35605 (MJM), and AI057005 (MJM). ECS was supported by T32 grants AI007140 and GM0726 from National Institutes of Health, funding from the Seattle Chapter of ARCS (Achievement Rewards for College Scientists), and the Poncin Scholarship Fund. M.J.M. is a recipient of the Burroughs Wellcome Clinical Scientist Award for Translational Research. This publication/presentation/grant proposal was made possible with help from the University of Washington Center for AIDS Research (CFAR), an NIH funded program (P30 AI027757), which is supported by the following NIH Institutes and Centers (NIAID, NCI, NIMH, NIDA, NICHD, NHLBI, and NCCAM).
Footnotes
The authors have no conflict of interest to report.
Reference List
- 1.Aubry F, Mattei M-G, Barque J-P, Galibert F. Chromosomal localization and expression pattern of the RNase L inhibitor gene. FEBS Letters. 1996;381:135–9. doi: 10.1016/0014-5793(96)00099-3. [DOI] [PubMed] [Google Scholar]
- 2.Dean M, Annilo T. EVOLUTION OF THE ATP-BINDING CASSETTE (ABC) TRANSPORTER SUPERFAMILY IN VERTEBRATES*. Annual Review of Genomics and Human Genetics. 2005 September 1;6(1):123–42. doi: 10.1146/annurev.genom.6.080604.162122. [DOI] [PubMed] [Google Scholar]
- 3.Bisbal C, Martinand C, Silhol M, Lebleu B, Salehzada T. Cloning and characterization of a RNase L inhibitor. J Biol Chem. 1995;270(22):13308–17. doi: 10.1074/jbc.270.22.13308. [DOI] [PubMed] [Google Scholar]
- 4.Chen Zq, Dong J, Ishimura A, Daar I, Hinnebusch AG, Dean M. The Essential Vertebrate ABCE1 Protein Interacts with Eukaryotic Initiation Factors. J Biol Chem. 2006 March 17;281(11):7452–7. doi: 10.1074/jbc.M510603200. [DOI] [PubMed] [Google Scholar]
- 5.Zhao Z, Sheps JA, Ling V, Fang LL, Baillie DL. Expression Analysis of ABC Transporters Reveals Differential Functions of Tandemly Duplicated Genes in Caenorhabditis elegans. Journal of Molecular Biology. 2004 November 19;344(2):409–17. doi: 10.1016/j.jmb.2004.09.052. [DOI] [PubMed] [Google Scholar]
- 6.Yarunin A, Panse VG, Petfalski E, Dez C, Tolervey D, Hurt EC. Functional link between ribosome formation and biogenesis of iron-sulfur proteins. EMBO J. 2005;24:580–8. doi: 10.1038/sj.emboj.7600540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Estevez AM, Haile S, Steinbnchel M, Quijada L, Clayton C. Effects of depletion and overexpression of the Trypanosoma brucei ribonuclease L inhibitor homologue. Molecular and Biochemical Parasitology. 2004 January;133(1):137–41. doi: 10.1016/j.molbiopara.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 8.Dong J, Lai R, Nielsen K, Fekete CA, Qiu H, Hinnebusch AG. The Essential ATP-binding Cassette Protein RLI1 Functions in Translation by Promoting Preinitiation Complex Assembly. J Biol Chem. 2004 October 1;279(40):42157–68. doi: 10.1074/jbc.M404502200. [DOI] [PubMed] [Google Scholar]
- 9.Kispal G, Sipos K, Lange H, et al. Biogenesis of cytosolic ribosomes requires the essential iron-sulphur protein RLi1p and mitochondria. EMBO J. 2005;24(3):589–98. doi: 10.1038/sj.emboj.7600541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zimmerman C, Klein KC, Kiser PK, et al. Identification of a host protein essential for assembly of immature HIV-1 capsids. Nature. 2002;415:88–92. doi: 10.1038/415088a. [DOI] [PubMed] [Google Scholar]
- 11.Dooher JE, Lingappa JR. Conservation of a stepwise, energy-sensitive pathway invovling HP68 for assembly of primate lentivirus capsids in cells. J Virol. 2004;78(4):1645–56. doi: 10.1128/JVI.78.4.1645-1656.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shea PR, Ishwad CS, Bunker CH, Patrick AL, Kuller LH, Ferrell RE. RNASEL and RNASEL-inhibitor variation and prostate cancer risk in Afro-Caribbeans. Prostate. 2008;68(4):354–9. doi: 10.1002/pros.20687. [DOI] [PubMed] [Google Scholar]
- 13.Bleiber G, May M, Martinez R, et al. Use of a Combined Ex Vivo/In Vivo Population Approach for Screening of Human Genes Involved in the Human Immunodeficiency Virus Type 1 Life Cycle for Variants Influencing Disease Progression. J Virol. 2005 October 15;79(20):12674–80. doi: 10.1128/JVI.79.20.12674-12680.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Iida A, Saito S, Sekine A, et al. Catalog of 605 single-nucleotide polymorphisms (SNPs) among 13 genes encoding human ATP-binding cassette transporters: ABCA4, ABCA7, ABCA8, ABCD1, ABCD3, ABCD4, ABCE1, ABCF1, ABCG1, ABCG2, ABCG4, ABCG5, and ABCG8. J Hum Genet. 2002;47(6):285–310. doi: 10.1007/s100380200041. [DOI] [PubMed] [Google Scholar]
- 15.Crawford DC, Carlson CS, Rieder MJ, Carrington DP, Yi Q. Haplotype diversity across 100 candidate genes for inflammation, lipid metabolism, and blood pressure regulation in two populations. Am J Hum Genet. 2004;74:610. doi: 10.1086/382227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Crawford DC, Akey DT, Nickerson DA. The patterns of natural variation in human genes. Annual Review of Genomics and Human Genetics. 2005;6(1):287–312. doi: 10.1146/annurev.genom.6.080604.162309. [DOI] [PubMed] [Google Scholar]
- 17.Tajima F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics. 1989;123:585. doi: 10.1093/genetics/123.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartl DL, Clark AG. Principles of Population Genetics. 3. Sunderland: Sinauer Associates, Inc; 1997. Random Genetic Drift; pp. 267–313. [Google Scholar]
- 19.Nickerson DA, Rieder MJ. SeattleSNPs Variation Discovery Resource. Website. 2009 June 17; Available at: URL: http://pga.gs.washington.edu/
- 20.Akey JM, Swanson WJ, Madeoy J, Eberle M, Shriver MD. TRPV6 exhibits unusual patterns of polymorphism and divergence in worldwide populations. Hum Mol Genet. 2006 July 1;15(13):2106–13. doi: 10.1093/hmg/ddl134. [DOI] [PubMed] [Google Scholar]
- 21.Akey JM, Eberle MA, Rieder MJ, Carlson CS, Shriver MD. Population history and natural selection shape patterns of genetic variation in 132 genes. PLoS Biol. 2004;2:e286. doi: 10.1371/journal.pbio.0020286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akey JM. Constructing genomic maps of positive selection in humans: Where do we go from here? Genome Research. 2009 May;19(5):711–22. doi: 10.1101/gr.086652.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinds DA, Stuve LL, Nilsen GB, et al. Whole-Genome Patterns of Common DNA Variation in Three Human Populations. Science. 2005 February 18;307(5712):1072–9. doi: 10.1126/science.1105436. [DOI] [PubMed] [Google Scholar]
- 24.Kelley JL, Madeoy J, Calhoun JC, Swanson W, Akey JM. Genomic signatures of positive selection in humans and the limits of outlier approaches. Genome Res. 2006 August;16(8):980–9. doi: 10.1101/gr.5157306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carlson CS, Thomas DJ, Eberle MA, et al. Genomic regions exhibiting positive selection identified from dense genotype data. Genome Research. 2005 November 1;15(11):1553–65. doi: 10.1101/gr.4326505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Biswas S, Akey JM. Genomic insights into positive selection. Trends Genet. 2006 August;22(8):437–46. doi: 10.1016/j.tig.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 27.The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clark AG, Hubisz MJ, Bustamante CD, Williamson SH, Nielsen R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Research. 2005 November 1;15(11):1496–502. doi: 10.1101/gr.4107905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bamshad MJ, Mummidi S, Gonzalez E, et al. A strong signature of balancing selection in the 5ΓǦ cis -regulatory region of CCR5. Proceedings of the National Academy of Sciences of the United States of America. 2002 August 6;99(16):10539–44. doi: 10.1073/pnas.162046399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sawyer SL, Emerman M, Malik HS. Ancient adaptive evolution of the primate antiviral DNA-editing enzyme APOBEC3G. PLoS Biol. 2004;2(9):E275. doi: 10.1371/journal.pbio.0020275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frodsham AJ, Hill AVS. Genetics of infectious diseases. Human Molecular Genetics. 2004 October 1;13(suppl2):R187–R194. doi: 10.1093/hmg/ddh225. [DOI] [PubMed] [Google Scholar]
- 32.Sirugo G, Hennig BJ, Adeyemo AA, et al. Genetic studies of African populations: an overview on disease susceptibility and response to vaccines and therapeutics. Hum Genet. 2008;123(6):557–98. doi: 10.1007/s00439-008-0511-y. [DOI] [PubMed] [Google Scholar]
- 33.Goh WC, Markee RE, Akridge RE, et al. Protection against human immunodeficiency virus type 1 infection in persons with repeated exposure: evidence for T cell immunity in the absence of inherited CCR5 coreceptor defects. J Infect Dis. 1999;179:548–57. doi: 10.1086/314632. [DOI] [PubMed] [Google Scholar]
- 34.Speelmon EC, Livingston-Rosanoff D, Desbien AL, et al. Impaired viral entry cannot explain reduced CD4+ T cell susceptibility to HIV type 1 in certain highly exposed individuals. AIDS Res Hum Retroviruses. 2008;24(11):1415–27. doi: 10.1089/aid.2007.0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Parra EJ, Marcini A, Akey J, et al. Estimating African American Admixture Proportions by Use of Population-Specific Alleles. Am J Hum Genet. 1998 December;63(6):1839–51. doi: 10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yaeger R, vila-Bront A, Abdul K, et al. Comparing Genetic Ancestry and Self-Described Race in African Americans Born in the United States and in Africa. Cancer Epidemiol Biomarkers Prev. 2008 June 1;17(6):1329–38. doi: 10.1158/1055-9965.EPI-07-2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Livingston RJ, von NA, Jegga A, Crawford DC, Carlson CS. Pattern of sequence variation across 213 environmental response genes. Genome Res. 2004;14:1821. doi: 10.1101/gr.2730004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wendt KS, Vodermaier HC, Jacob U, et al. Crystal structure of the APC10/DOC1 subunit of the human anaphase-promoting complex. Nat Struct Biol. 2001;8(9):784–8. doi: 10.1038/nsb0901-784. [DOI] [PubMed] [Google Scholar]
- 39.Grossberger R, Gieffers C, Zachariae W, et al. Characterization of the DOC1/APC10 Subunit of the Yeast and the Human Anaphase-promoting Complex. J Biol Chem. 1999 May 14;274(20):14500–7. doi: 10.1074/jbc.274.20.14500. [DOI] [PubMed] [Google Scholar]
- 40.Beaty RM, Edwards JB, Boon K, Siu IM, Conway JE, Riggins GJ. PLXDC1 (TEM7) is identified in a genome-wide expression screen of glioblastoma endothelium. J Neurooncol. 2007;81(3):241–8. doi: 10.1007/s11060-006-9227-9. [DOI] [PubMed] [Google Scholar]
- 41.Raineri I, Senn HP. HIV-1 promotor insertion revealed by selective detection of chimeric provirus-host gene transcripts. Nucl Acids Res. 1992 December 11;20(23):6261–6. doi: 10.1093/nar/20.23.6261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Curtin K, Iles MM, Camp NJ. Identifying rarer genetic variations for common complex diseases: diseased versus neutral discovery panels. Ann Hum Genet. 2009;73(1):54–60. doi: 10.1111/j.1469-1809.2008.00483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eberle MA, Ng PC, Kuhn K, et al. Power to detect risk alleles using genome-wide tag SNP panels. PLoS Genet. 2007;3(10):1827–37. doi: 10.1371/journal.pgen.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]