Abstract
Ca2+ signalling in neurons through calmodulin (CaM) has a prominent function in regulating synaptic vesicle trafficking, transport, and fusion. Importantly, Ca2+–CaM binds a conserved region in the priming proteins Munc13-1 and ubMunc13-2 and thus regulates synaptic neurotransmitter release in neurons in response to residual Ca2+ signals. We solved the structure of Ca2+4–CaM in complex with the CaM-binding domain of Munc13-1, which features a novel 1-5-8-26 CaM-binding motif with two separated mobile structural modules, each involving a CaM domain. Photoaffinity labelling data reveal the same modular architecture in the complex with the ubMunc13-2 isoform. The N-module can be dissociated with EGTA to form the half-loaded Munc13/Ca2+2–CaM complex. The Ca2+ regulation of these Munc13 isoforms can therefore be explained by the modular nature of the Munc13/Ca2+–CaM interactions, where the C-module provides a high-affinity interaction activated at nanomolar [Ca2+]i, whereas the N-module acts as a sensor at micromolar [Ca2+]i. This Ca2+/CaM-binding mode of Munc13 likely constitutes a key molecular correlate of the characteristic Ca2+-dependent modulation of short-term synaptic plasticity.
Keywords: calcium, calmodulin, Munc13, neurotransmitter release, short-term plasticity
Introduction
Information transfer between neurons occurs predominantly through the release of neurotransmitter at synapses. This process starts with the transport and docking of neurotransmitter-filled vesicles to the active zone at the plasma membrane (PM), where they undergo maturation. This event is known as priming (Rettig and Neher, 2002; Südhof, 2004; Wojcik and Brose, 2007) and represents a major rate-limiting step in regulated exocytosis, particularly during phases of high synaptic activity (Augustin et al, 1999; Rosenmund et al, 2002; Varoqueaux et al, 2002). Only the primed vesicles can fuse to the PM in response to a Ca2+ trigger and release their content to the extracellular synaptic cleft. The priming and fusion processes depend on the formation of the ternary SNARE complex by the proteins syntaxin-1, SNAP-25, and synaptobrevin (VAMP2) (Jahn and Scheller, 2006).
In contrast to constitutive exocytosis, SNARE complex-dependent neurotransmitter release is regulated by multiple signalling pathways, involving numerous proteins as well as secondary messengers such as Ca2+ and diacylglycerol (DAG) (Südhof, 2004; Wojcik and Brose, 2007). A family of proteins essential in this regard consists of the mammalian homologs of Caenorhabditis elegans UNC13, Munc13-1, Munc13-2, and Munc13-3, which are essential regulators of synaptic vesicle priming (Augustin et al, 1999; Rosenmund et al, 2002; Varoqueaux et al, 2002). Munc13 isoforms are differentially expressed in the brain, where they are specifically localized in the active zone. Upon DAG/phorbol ester (PE) binding, these proteins bind to the PM, which regulates their fundamental function in synaptic vesicle priming and neurotransmitter release (Betz et al, 1998; Brose and Rosenmund, 2002; Rhee et al, 2002; Silinsky and Searl, 2003). Their action has been reported to involve the remodelling of syntaxin-1, leading to fusion-competent SNARE complexes (Richmond et al, 2001; Guan et al, 2008).
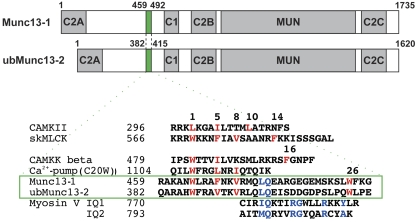
Munc13s are multidomain proteins sharing a C-terminal highly conserved region (Figure 1), consisting of two C2 domains, a DAG-binding C1 domain, and an MUN domain, which is essential for their function and responsible for the interaction with key binding partners like syntaxin-1 (Betz et al, 1997; Basu et al, 2005; Stevens et al, 2005). On the other hand, the N-terminal part differs in the three isoforms. Munc13-1 and ubMunc13-2 present a third N-terminal C2 domain and a unique calmodulin (CaM)-binding domain, which does not match any previously known CaM recognition motif and leads to the regulation of these proteins by CaM (Junge et al, 2004). CaM binds Munc13-1 and ubMunc13-2 in a Ca2+-dependent manner through this CaM recognition motif, resulting in the increase of priming activity and of the size of the pool of fusion competent, ready-releasable vesicles. Thus, the activation of the CaM/Munc13 complex by residual Ca2+ represents a molecular correlate for the phenomenon of Ca2+-dependent vesicle pool refilling and short-term synaptic plasticity (Junge et al, 2004).
Figure 1.
Domain structure of Munc13-1 and ubMunc13-2. The CaM-binding domains are highlighted in green. The expansion shows the primary sequence alignment of these CaM-binding domains with peptides belonging to different classes of CaM recognition motifs. The hydrophobic residues anchoring each motif to Ca2+–CaM are highlighted in red; the apo-CaM IQ-binding motif is highlighted in blue.
Like synaptic vesicle fusion itself, synaptic vesicle priming is strongly dependent on changes in presynaptic Ca2+ concentrations [Ca2+]i, albeit with a different (i.e. linear) Ca2+ concentration dependence. Depending on the synapse type under examination, increases in the residual presynaptic Ca2+ concentration can accelerate synaptic vesicle priming by a factor of 10–30 (Neher and Sakaba, 2008). Munc13s are key priming proteins and thought to also have an important function in the Ca2+-dependent acceleration of the priming reaction. Indeed, Ca2+–CaM binding to Munc13-1 and ubMunc13-2 strongly accelerates synaptic vesicle priming in hippocampal neurons (Junge et al, 2004). Moreover, studies in chromaffin cells showed that ubMunc13-2 is activated by two Ca2+-dependent processes. In a slow activation mode at low Ca2+ concentrations (∼100 nM), ubMunc13-2 acts as a priming switch. This activation mode is apparently independent of Ca2+–CaM binding and may involve the C2B domain of ubMunc13-2. In a fast activation mode at high Ca2+ concentrations (>5 μM), ubMunc13-2 is activated in a Ca2+–CaM-dependent manner and accelerates chromaffin granule recruitment and maturation during stimulation (Zikich et al, 2008).
CaM is a ubiquitous eukaryotic protein that has a prominent function in the integration of Ca2+ in signal transduction processes. It is composed of two homologous domains, each consisting of two Ca2+ binding helix-loop-helix motifs known as EF-hand motifs. On Ca2+ binding, a conformational change in the EF-hand motifs exposes a hydrophobic cleft (Kuboniwa et al, 1995; Zhang et al, 1995) that binds amphiphilic α-helices (Crivici and Ikura, 1995; Bayley et al, 1996; Ikura, 1996). This property, in addition to the flexibility of the linker joining the two domains, confers on CaM an extraordinary versatility with regard to its macromolecular interactions and the resulting regulatory mechanisms (Hoeflich and Ikura, 2002; Vetter and Leclerc, 2003). Several structures of CaM in complex with target peptides have been solved, leading to the definition of various Ca2+–CaM-binding motifs (Figure 1), which all consist of an amphiphilic α-helix with hydrophobic side chains facing one side at specific positions (Rhoads and Friedberg, 1997). Recent studies also revealed that a ‘half-activated' state of CaM, where two Ca2+ ions are bound to the CaM C-terminal domain at relatively low [Ca2+]i (<1 μM), is sufficient to activate some targets (Shifman et al, 2006; Edlich et al, 2007; Dick et al, 2008).
The understanding of the molecular mechanism underlying the regulation of Munc13-1 and ubMunc13-2 by Ca2+–CaM would provide further insight into the complex regulation of neurotransmitter release, especially into the link between the Ca2+ signals and synaptic vesicle priming. In the study described here, we characterized the structure of complexes formed by CaM and the CaM-binding domains of Munc13-1 and ubMunc13-2 by means of NMR and photoaffinity labelling (PAL). We show that Munc13-1 and ubMunc13-2 feature a novel (1-5-8-26) CaM recognition motif, forming CaM complexes that combine two independent modules connected by flexible linkers. This modular architecture allows a stepwise formation of (i) an intermediate Munc13/Ca2+2–CaM species and (ii) the fully loaded Munc13/Ca2+4–CaM complex. The fast interconversion between these two species confers on the Munc13/CaM complexes the ability to act as efficient Ca2+-sensors at μM (∼5 μM) Ca2+ concentrations, providing an explanation for the described function of Munc13/CaM in short-term synaptic plasticity (Junge et al, 2004).
Results
Munc13-1 features an unusually extended CaM-binding motif: Munc13-1459−492
The CaM-binding site in Munc13-1 was originally ascribed to the segment R459–A478 (Junge et al, 2004). To investigate the presence of additional interactions, we elongated this peptide towards the C-terminus and studied the effect on CaM by means of NMR titrations using the HSQC correlation (which displays signals of directly bound N and H atoms) of 15N-labelled Ca2+–CaM as a fingerprint. Interestingly, addition of the R459–A478 peptide only affected N–HN signals from the C-terminal CaM domain, whereas the longer peptide R459–G492, containing the hydrophobic segment L488–W489–F490, produced additional chemical shift changes in the N-terminal CaM domain (data not shown). This indicates additional hydrophobic interactions between the N-terminal CaM domain and the segment L488–F490. Further elongation of the peptide (i.e. R459–K511) did not produce any significant changes either in the HSQC spectrum of Ca2+–CaM or of the peptide (data not shown), indicating that the segment R459–G492 constitutes the CaM-binding site in Munc13-1. This segment interacts simultaneously with both CaM domains, although its sequence does not resemble any other known CaM target motifs (Figure 1).
NMR study of the Munc13-1459−492/CaM interactions
The interactions leading to the formation of the complex between CaM and Munc13-1459−492 were first studied by means of NMR titrations, using HSQC spectra as protein fingerprints. The studies were carried out under different conditions, to delineate the Ca2+ dependence of the binding. Titrations were carried out both in the absence and in the presence of an excess of Ca2+. A titration of the Ca2+-saturated complex with EGTA was also performed. The results of these experiments reveal the existence of three distinct species along the interaction pathway between the two proteins:
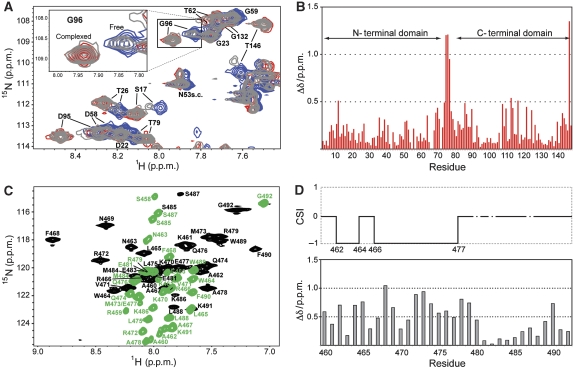
High-affinity Munc13-1459−492/Ca2+4–CaM complex. The titration of 15N-labelled Ca2+4–CaM with Munc13-1459−492 largely affected the resonances of both the C- and N-terminal domain of Ca2+4–CaM, inducing the presence of two sets of resonances (i.e. those corresponding to the free and bound states) for most of the amino acids (Figure 2A and B). Once a 1:1 stoichiometry was reached, only one set of resonances remained (corresponding to the bound state) and no changes in the HSQC spectrum were observed after further addition of Munc13-1459−492. This confirms the previously reported 1:1 stoichiometry of the Munc13-1459−492/Ca2+4–CaM complex (Junge et al, 2004; Dimova et al, 2006). Moreover, the observation of two sets of resonances for Ca2+4–CaM at sub-stoichiometric concentrations of Munc13-1459−492 indicates that the complex has a high affinity and a dissociation constant (Kd) in the nanomolar range, assuming a diffusion controlled on-rate. A similar behaviour was observed in the spectra of 13C,15N-labelled Munc13-1459−492, where the Munc13-1459−492/Ca2+4–CaM complex displays a much larger degree of chemical shifts dispersion than the free peptide (Figure 2C and D), indicating the formation of a secondary structure element upon Ca2+4–CaM binding. Indeed, the observation of a negative Hα chemical shift index indicates that Munc13-1459−492 features an α-helix spanning residues 462 to 477 when bound to Ca2+4–CaM (Figure 2D). The stronger chemical shift perturbations observed in these two titrations (i.e. those above 0.25 p.p.m.) spread along the whole Ca2+4–CaM and Munc13-1459−492 sequences (Figure 2B and D). However, for Ca2+4–CaM, the C-terminal domain and the linker region were more affected than the N-terminal domain, whereas in Munc13-1459−492, the N-terminal region displayed the strongest effects. This indicates an antiparallel arrangement in the complex, involving both CaM domains and the whole Munc13-1459−492 molecule.
Figure 2.
Binding of Munc13-1459−492 to Ca2+4–CaM followed by HSQC spectra. (A) Titration of 15N-labelled Ca2+4–CaM with Munc13-1459−492. The reference spectrum of Ca2+4–CaM is shown in blue, whereas the spectrum with 0.6:1 Munc13-1459−492/Ca2+4–CaM concentration ratio is shown in grey, and the saturated 1:1 spectrum is shown in red. (B) Amide chemical shift perturbations in Ca2+4–CaM upon complex formation with Munc13-1459−492. (C) Overlay of the 15N–1H-HSQC spectra of Munc13-1459−492 either free (green) or bound to Ca2+4–CaM (black). The free form of Munc13-1459−492 was not soluble in aqueous solution and could only be measured in 8 M urea. (D) Amide chemical shift perturbations in Munc13-1459−492 upon complex formation with Ca2+4–CaM. The upper part shows the secondary chemical shift index (CSI) for the Hα resonances of Munc13-1459−492, which suggests the formation of an α-helix (CSI=−1) spanning segment A462–E477; missing CSI for residues 480, 482 (both glycines), and S487 (no Hα assignment) are indicated by discontinuous lines.
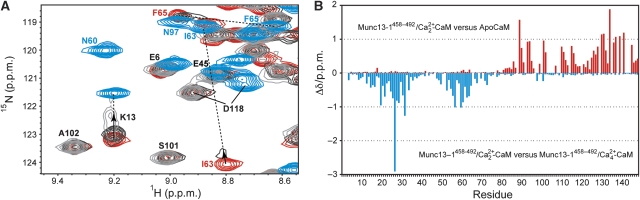
Half-loaded Munc13-1459−492/Ca2+2–CaM complex. As it is well known that the CaM N-terminal domain has a lower affinity for Ca2+ than the C-terminal domain, we followed the effects of Ca2+ removal from the Munc13-1459−492/Ca2+4–CaM complex. For this purpose, the 15N-labelled Munc13-1459−492/Ca2+4–CaM sample was titrated with EGTA up to a final concentration of 50 mM. In the course of the titration, the amide resonances corresponding to the N-terminal CaM domain migrated towards the chemical shift positions corresponding to apo-CaM (e.g. K13, Figure 3A). This was accompanied by line broadening, which was especially severe in the case of signals with larger chemical shift differences between the two species (especially in the 15N dimension). These resonances eventually disappeared (e.g. I63). On the other hand, signals from the C-terminal domain remained essentially unaffected (e.g. D118). At lower magnetic field (400 MHz instead of 600 MHz) and temperature (27°C instead of 35°C), the titration shows a discrete change of the chemical shifts, and the intensive line broadening was not observed, allowing the evaluation of the chemical shifts of the Munc13-1459−492/Ca2+2–CaM species for almost all N-terminal resonances. Indeed, the obtained values resemble those of apo-CaM (Figure 3B) in the N-terminal domain. This indicates that EGTA addition led to dissociation of the Ca2+ ions and Munc13-1459−492 from the N-terminal domain and that the resulting species (i.e. Munc13-1459−492/Ca2+2–CaM) represents an intermediate state that combines structural features of the Munc13-1459−492/Ca2+4–CaM complex (i.e. the C-terminal CaM domain and the segment of Munc13-1459−492 bound to it) and apo-CaM (i.e. the free N-terminal CaM domain). The observation of line broadening at 35°C indicates intermediate exchange on the NMR timescale between the Munc13-1459−492/Ca2+2–CaM intermediate complex and the fully loaded complex.
Figure 3.
Titration of the 15N-labelled Munc13-1459−492/Ca2+4–CaM complex with EGTA. (A) Overlay of a section of the HSQC spectra of Munc13-1459−492/Ca2+4–CaM in the presence of 10 mM Ca2+ (red) and after addition of 10 mM EGTA (dark grey) and 50 mM EGTA (light grey) with the reference spectrum from apo-CaM (cyan). The signals from the C-terminal CaM domain remained unaffected (e.g. D118), whereas those from the N-terminal CaM domain migrate towards the apo-CaM position and simultaneously experience line broadening (e.g. K13). (B) Overview of N–HN chemical shift perturbations of the intermediate Munc13-1459−492/Ca2+2–CaM complex (i.e. after addition of 50 mM EGTA in the same way as in (A), but at 27°C and 400 MHz instead of 35°C and 600 MHz) relative to apo-CaM (upper plot) and the Munc13-1459−492/Ca2+4–CaM complex (lower plot).
Low-affinity Munc13-1459−492/apo-CaM interaction. The titration of Munc13-1459−492 to 15N-labelled apo-CaM revealed an interaction in the intermediate to fast exchange regime on the NMR timescale, as inferred from the line broadening seen for several N–HN cross peaks and continuous, concentration-dependent chemical shift changes upon addition of peptide (Supplementary Figure S1A). The affected N–HN resonances were mapped to the C-terminal domain of apo-CaM, indicating that only this domain interacts with Munc13-1459−492 in the absence of Ca2+ (Supplementary Figure S1A). Furthermore, these N–HN resonances tend to adopt chemical shifts similar to those in the Ca2+-bound forms of the complex, which indicates an opening of the C-terminal CaM domain on peptide binding. This behaviour reflects the well-studied cooperativity in Ca2+ and peptide binding to CaM (Peersen et al, 1997; Mirzoeva et al, 1999). On the basis of the progressive chemical shift changes on peptide addition in the HSQC of apo-CaM, we obtained a dissociation constant (Kd) of around 4 mM, which explains the observed line broadening. This interaction might result from the presence of an LQ segment in the sequence of the CaM-binding domain (i.e. L475–Q476), which could partially mimic an IQ motif (Figure 1), a standard target of apo-CaM. However, the rest of the sequence is rather different from that of standard IQ motifs, possibly explaining the low affinity in the formation of this species (Jurado et al, 1999). This interaction may have a function in the pre-formation of the Ca2+-bound species.
Altogether, these experiments show that the Munc13-1/CaM interaction is characterized by a combination of two distinct binding modes: a high-affinity interaction of the C-terminal CaM domain with a helical segment at the N-terminal part of Munc13-1459−492, and a second interaction, of lower affinity, between the N-terminal CaM domain and the C-terminal end of Munc13-1459−492.
Solution structure of the Munc13-1459−492/Ca2+4–CaM complex
Next, we determined the structure of the complex between Ca2+4–CaM and Munc13-1459−492. The summary of the Nuclear Overhauser Enhancement (NOE) distance restraints is presented in Table 1. On average, there are >10 NOEs per amino-acid residue. The number of NOE restraints is significantly larger in the C-terminal domain of Ca2+4–CaM than in the N-terminal domain, seemingly the result of a less tight binding of Munc13-1459−492 to the N-terminal domain and consequent line broadening of some resonances from intermediate exchange. For the same reason, some resonances in the N-terminal domain remained unassigned. However, the amount of experimental data was sufficient to obtain a high-quality structure for the entire complex, with backbone root mean square deviations (RMSD) of 1.21 and 0.90 Å for the N- and C-terminal parts, respectively. Moreover, the structure is in good agreement with the N–HN residual dipolar coupling (RDC) data, as reflected by the Q-factors (Bax and Grishaev, 2005) of 0.03 and 0.04 for the N- and C-terminal CaM, respectively, calculated for experimental RDCs resulting from a steric alignment of CaM with phages (15 mg/ml Pf1 solution).
Table 1.
NMR and refinement statistics for the Munc13-1459−492/CaM complex
| NMR geometric restraints | Nterma | Ctermb |
|---|---|---|
| Total distance restraints | 658 | 1276 |
| Intra-residue | 252 | 364 |
| Inter-residue | 329 | 705 |
| Sequential (∣i−j∣=1) | 183 | 300 |
| Medium range (∣i−j∣⩽4) | 82 | 264 |
| Long range (∣i−j∣⩾5) | 64 | 141 |
| Intermolecular | 9 | 137 |
| Hydrogen bondsc | 28 | 29 |
| Ca2+-protein restraintsd | 12 | 12 |
| Total dihedral restraints | ||
| φ | 42 | 48 |
| ψ | 45 | 49 |
| Total 1D1H−15N residual dipolar couplings | 44 | 62 |
| Structure statistics | ||
| Violations (RMSD±s.d.) | ||
| Distance restraints (Å) | 0.044±0.002 | |
| Dihedral restraints (deg) | 0.75±0.12 | |
| 1D1H−15N RDC from the phage sample (Hz) | 0.71±0.05 | |
| Deviation from idealized geometry | ||
| Bond lengths (Å) | 0.0039±0.0001 | |
| Bond angles (deg) | 0.70±0.02 | |
| Ramachandran statisticse,f | All residues | Nterm and Cterm |
| Residues in most favoured regions | 88.4% | 88.7% |
| Residues in additional allowed regions | 9.2% | 9.8% |
| Residues in generously allowed regions | 1.4% | 1.2% |
| Residues in disallowed regions | 0.9% | 0.3% |
| Average RMSD (Å)f | Nterm | Cterm |
| Heavy atoms | 1.79±0.27 | 1.50±0.22 |
| Backbone atoms | 1.21±0.24 | 0.90±0.20 |
| aDefined as residues 1–78 of CaM and residues 483–492 of Munc13-1459−492. | ||
| bDefined as residues 79–148 of CaM and residues 459–482 of Munc13-1459−492. | ||
| cHydrogen bonds were identified on the basis of strong protection factors. Two restraints per hydrogen bond were included in the calculations (dHN−0⩽2.0Å and dN−O⩽3.0 Å). | ||
| dThe Ca2+–O distances were restrained to 2.333–2.503 Å. | ||
| eRamachandran statistics were obtained using the PROCHECK NMR software ([37]). | ||
| fNterm is defined as residues 5–74 of CaM and 488–489 of Munc13-1459−492, Cterm is defined as residues 84–146 of CaM and 459–479 of Munc13-1459−492. | ||
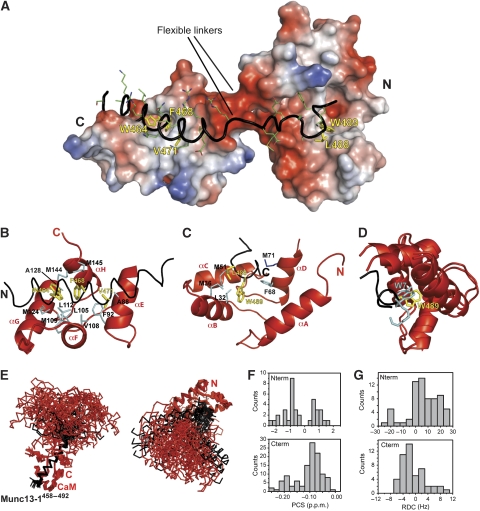
As already suggested by the NMR titrations, the structure of this complex consists of two separate modules each formed by a CaM domain and a segment of Munc13-1459−492, which are connected by two unstructured linkers (i.e. one from each protein) (Figure 4A). The C-terminal module (Figure 4B) consists of a helix spanning the segment S459–A478 of Munc13-1459−492 attached to the hydrophobic cleft of the C-terminal CaM domain. The hydrophobic side chains of residues W464, F468, and V471 from Munc13-1459−492, located in relative positions 1-5-8 (Figure 1), anchor this helix to the C-terminal CaM domain, thereby interacting with several hydrophobic side chains of CaM. The indole ring of W464 lies between the side chains of M124 and M144. This agrees with our recent finding that M124 and M144 of CaM are the sites of photoincorporation of shorter, Munc13-1459−479-derived photoprobes carrying a photoreactive amino acid para-benzoyl-Phe (Bpa) moiety in place of W464 (Dimova et al, 2009). Here, we used the longer, Munc13-1459−492-derived photoprobes (Figure 5A) and confirmed the contact of W464 with M124 and M144 (data not shown). The observed 1-5-8 arrangement of the N-terminal part of Munc13-1459−492 with CaM is the same as in the C20W/CaM complex (Elshorst et al, 1999), where the peptide interacts only with the C-terminal CaM domain. The structures of the C-terminal CaM domain in the two complexes are similar, but not identical (RMSD of 2.18 Å). The contact surface areas are also similar, reaching 992±33 Å2 in the C-terminal module of Munc13-1459−492/Ca2+4–CaM and 738±44 Å2 in the C20W/CaM complex.
Figure 4.
Munc13-1459−492/Ca2+4–CaM structure. (A) Overall binding mode. The backbone and side chains of Munc13-1459−492 are shown as black ribbon and sticks, respectively, whereas Ca2+4–CaM is represented as electrostatic potential surface. The complex features two modules: an amphiphilic α-helical 1-5-8 motif bound to the CaM C-terminal domain and the Trp in position 26 attached to the CaM N-terminal domain. The side chains of the anchoring residues are highlighted as yellow sticks. (B, C) Structures of the C- and N-modules of the Munc13-1459−492/Ca2+4–CaM complex, respectively. Segments of the backbone of Munc13-1459−492 are shown as black ribbon, while the anchor residues are highlighted as in (A). CaM is shown as red cartoon, with the side chains contacting the hydrophobic anchors of Munc131459−492 represented as blue lines. (D) Superposition of the N-terminal modules of the Munc13-1459−492/Ca2+4–CaM and the W-7/Ca2+4–CaM complexes. The N-terminal CaM domains are shown in red. A segment of Munc13-1459−492 is shown in black with the aromatic side chain of W489 highlighted in yellow. The W-7 inhibitor is shown in cyan. (E) Ensemble of the 20 lowest-energy structures of the Munc13-1459−492/Ca2+4–CaM complex aligned on the CaM C-terminal part of the interaction (left) and on the N-terminal part of the interaction (right). CaM and Munc13-1459−492 are shown in red and black, respectively. (F) Histograms of the CaM N–HN PCSs from the C- (lower histogram) and N-terminal CaM domain (upper histogram), measured using the terbium-loaded Munc13-1459−492/Ca2+4-N60D–CaM complex. (G) Histograms of the CaM N–HN RDCs from the C- (lower histogram, measured using the terbium-loaded Munc13-1459−492/Ca2+4-N60D–CaM complex at 900 MHz) and N-terminal CaM domain (upper histogram, calculated from the PCSs tensor in F).
Figure 5.
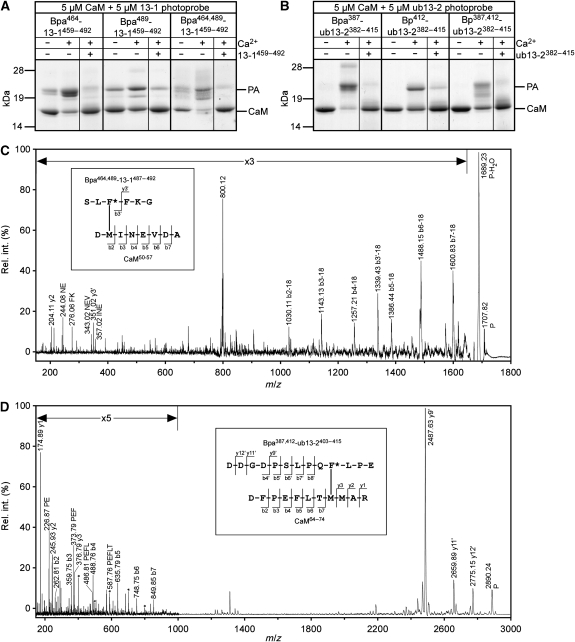
Photoaffinity labelling of CaM with Munc13-derived photoprobes. (A, B) Coomassie-stained SDS gels showing the photoadduct (PA) formation of different Munc13-1459−492- (A) and ubMunc13-2382−415-derived (B) photoprobes in the absence and presence of excess Ca2+ (1 mM). Photoprobe binding was considered specific as photoadduct formation is suppressed in the presence of a 10-fold molar excess of wild-type competitor. Mass spectrometry (not shown) revealed that only unlabelled CaM and 1:1 photoadduct species were present even in the samples showing photoadduct double bands, which were most likely due to conformational effects on electrophoretic mobility. The 1:2 photoadduct species detected to some extent at an apparent molecular mass of 28 kDa were probably related to the artificially high Ca2+ concentration of 1 mM, as these artefacts were not observed under the Ca2+ conditions (<100 nM) used for the generation of photoadducts to be structurally characterized by mass spectrometry (data not shown). (C) Fragment ion mass spectrum of the cross-linked peptide consisting of CaM50−57 and Bpa464,489-13-1487−492. P denotes the precursor ion ([M+H]+calc=1707.79) and primed ion designators indicate fragmentation in the photoprobe chain. After an immediate loss of water (P-H2O), a conclusive N-terminal b-ion series is formed, demonstrating that the sequence CaM52−57 (INEVDA) was not modified. As modification of D50 can be ruled out on the basis of the AspN cleavage specificity, M51 remains as the only possible site of photoincorporation (see inset, F* encodes Bpa). The dominant signal at m/z=800.12 is most likely related to Bpa464,489-13-1487−492 and may be explained by concurrent release and radicalic rearrangement of the benzophenone moiety under post-source decay conditions (Leite et al, 2003). (D) Fragment ion mass spectrum of the cross-linked peptide consisting of CaM64−74 and Bpa387,412-ub13-2403−415. P denotes the precursor ion ([M+H]+calc=2890.30) and primed ion designators indicate fragmentation in the photoprobe chain. The detection of a C-terminal y- (y1–y3) and an N-terminal b-ion series (b2–b7) revealed M71 as the site of photoincorporation (see inset, F* encodes Bpa). Within the enlarged region m/z<1000, fragment ions derived from cleavages in the photoprobe chain are only marked with an asterisk for the sake of clarity. The high abundance of the y9′ ion at m/z=2487.63 is in agreement with facile amide bond cleavages N-terminal of Proline and C-terminal of Aspartate.
The N-terminal module of the interaction is a novel feature in Ca2+–CaM complexes, as only L488 and W489 (occupying positions 25 and 26 relative to the 1-5-8 motif of the C-terminal module) are buried in the N-terminal domain of Ca2+4–CaM. These amino acids, especially W489, are embedded in the hydrophobic cleft of the CaM N-terminal domain, where they engage in van der Waals contacts with several hydrophobic side chains (Figure 4C). The contact surface area is only 359±33 Å2, almost identical to that of the N-terminal part of the W-7/CaM complex (307±23 Å2), where the chloronaphthalene ring of W-7 is also buried in the N-terminal CaM domain (Osawa et al, 1998). Moreover, the tryptophan ring of W489 and the chloronaphthalene ring of W-7 display very similar positions on superimposition of the N-terminal CaM domains from both complex structures (Figure 4D).
We performed PAL studies to verify this binding mode at physiological concentrations, given the low micromolar concentration of CaM in brain (Xia and Storm, 2005). In this case, PAL data also sustain the N-terminal module of the structure. When we performed PAL of CaM with Bpa489-13-1459−492, a Munc13-1459−492-derived photoprobe carrying the Bpa moiety in place of W489, we also observed a specific Ca2+-dependent photoadduct formation (Figure 5A) like in the PAL experiments with Bpa464-13-1459−492. The somewhat lower photoadduct yield reflects the looser binding of Munc13-1 to the N-terminal domain of CaM than to the C-terminal domain. To identify contact sites of W489 in CaM, we labelled CaM with the bifunctional photoprobe Bpa464,489-13-1459−492 (Figure 5A) and structurally characterized the photoadducts according to our workflow (Dimova et al, 2009). Trypsin digestion followed by HPLC/MALDI-TOF-MS confirmed the contact sites of W464 (M124 and M144 of CaM, see above) and revealed that the site of photoincorporation of Bpa489 into CaM is located within the tryptic peptide CaM38−74 (data not shown). As the corresponding cross-linked peptide consisting of the tryptic fragments CaM38−74 and Bpa464,489-13-1487−492 has a mass of 4.9 kDa and is therefore not available for mass spectrometric sequencing, it was isolated by HPLC and truncated further by AspN treatment. Thereby, the site of photoincorporation was narrowed down to CaM50−57 and MS/MS sequencing of the cross-linked peptide revealed a linkage between Bpa489 of the photoprobe and Met51 of CaM (Figure 5C). This finding was also confirmed for the monofunctional photoprobe Bpa489-13-1459−492 (data not shown). Thus, in the bound state, W489 contacts M51 of CaM, as readily visualized in the NMR structure (Figure 4C).
The lack of NOEs between the two domains together with missing sequential NOEs in the linker region of the two domains indicates that the two domains are flexible with respect to each other. Indeed, the calculated structural ensemble shows a high degree of inter-domain flexibility, sampling several different inter-domain orientations (Figure 4E). This inter-domain flexibility is confirmed by the N–HN pseudocontact shifts (PCSs) and RDCs measured using terbium-loaded N60D-CaM. First, we observed that the measured N- and C-terminal PCSs behave similarly to those reported for free terbium-loaded N60D-CaM (Bertini et al, 2004), where very large PCSs (up to 2.3 p.p.m.) were measured for the N-terminal domain (which carries the terbium ion), whereas the C-terminal domain showed smaller (<0.2 p.p.m.) PCSs (Figure 4F). This distribution confirms the presence of an extended conformation. Furthermore, although the PCSs of the N-terminal domain fit well to the determined structure (Q=0.23), the PCSs of the C-terminal domain could not be fitted assuming a single overall conformation, which again supports the presence of inter-domain motion (data not shown). To assess the extent of the motion, we first calculated the N–HN RDCs expected for the N-terminal domain given the alignment tensor (Da=−17.3 Hz and Rh=0.5) obtained by fitting the experimental PCSs to the structure of this domain. This procedure results in RDCs values that are independent of the experimental errors and of the incompleteness of the data caused by the extensive paramagnetic line broadening in the N-terminal domain. The N–HN RDCs of the N-terminal domain, containing the paramagnetic centre, depend only on bond orientations and are insensitive to inter-domain motion. Subsequently, we compared the RDCs calculated for the N-terminal domain to the RDCs measured for the C-terminal domain. The quality of the C-terminal domain experimental RDCs was assessed by fitting them to the structure of the domain, which resulted in a very good Q factor of 0.32 (Da=4.1 Hz and Rh=0.4). The experimental values measured for the C-terminal domain are approximately four times smaller than the RDCs calculated from the PCSs of the N-terminal domain (Figure 4G), indicating that the RDCs of the C-terminal domain result from averaging over different orientations of this domain with respect to the N-terminal domain, which contains the paramagnetic center and therefore largely determines the alignment tensor. From the relative size of the RDCs of the N- and C-terminal domains, we can infer an order parameter S of 0.25 for the inter-domain motion. Such consistent reduction of the size of the RDCs belonging to one lobe of a multidomain protein showing inter-domain flexibility has been previously reported for free CaM (Bertini et al, 2004); however, in this study, the RDCs of the C-terminal domain of CaM were scaled down by a factor of 8 with respect to those of the terbium-loaded N-terminal domain (S=0.12). The reduced flexibility (S=0.25 for the Munc13-1459−492/Ca2+4–CaM complex compared with S=0.12 for free CaM) is plausible as the inter-domain motion of CaM in complex with Munc13-1 is restricted by two linkers (one CaM linker and one Munc13-1 linker), whereas in the free CaM it is restricted by only one linker.
ubMunc13-2 features the same CaM recognition mechanisms as Munc13-1
The CaM recognition motifs of Munc13-1 and ubMunc13-2 are highly homologous (45% identity, 66% similarity, Figure 1). Moreover, the hydrophobic residues critical for CaM binding (i.e. at positions 1-5-8-26) are also present in ubMunc13-2. This raised the question as to whether the recognition motif found for Munc13-1 is the same for ubMunc13-2. With regard to the 1-5-8 arrangement, our recent cross-linking study with shorter model peptides already indicated that Munc13-1459−479 and ubMunc13-2382−402 share the same binding site in the C-terminal domain of CaM (Dimova et al, 2009). Here, we reproduced this finding with the longer, ubMunc13-2382−415-based photoprobes and found that the corresponding W387 of ubMunc13-2382−415 also contacts M124 and M144 of CaM (data not shown). More importantly, PAL experiments using Bpa412-ub13-2382−415 confirmed the interaction through the conserved Trp residue in position 26 relative to the 1-5-8 motif, with a photoadduct yield comparable to that of the Munc13-1 variant (Figure 5B). Using the bifunctional photoprobe Bpa387,412-ub13-2382−415 and our analytical strategy, we identified a cross-linked peptide consisting of the proteolytic fragments CaM64−74 and Bpa387,412-ub13-2403−415, indicating that the C-terminus of ubMunc13-2382−415 binds the N-terminal CaM domain. MS/MS sequencing of the cross-linked peptide revealed a linkage between Bpa412 of the photoprobe and M71 of CaM (Figure 5D), a finding that was also confirmed for the monofunctional photoprobe Bpa412-ub13-2382−415 (data not shown). Thus, in the bound state, W412 of ubMunc13-2382−415 contacts M71 of CaM, a residue that forms part of the hydrophobic cleft of the N-terminal CaM domain. This indicates that the ubMunc13-2381−413/CaM complex shares its overall topology and consequently the modular structure with the homologous Munc13-1459−492/CaM complex.
NMR titration of 15N-labelled Ca2+4–CaM with ubMunc13-2382−415 displayed the same behaviour as seen with Munc13-1459−492 (Supplementary Figure S2A), namely the appearance of two sets of resonances, before saturation was reached. This confirms the formation of a high-affinity ubMunc13-2382−415/Ca2+4–CaM complex. The titration of this complex with EGTA also led to a dramatic effect on the resonances from the N-terminal CaM domain (Supplementary Figure S2B), whereas those from the C-terminal domain remained unaffected. As in the case of the Munc13-1459−492/CaM complex, this indicates the formation of a half-loaded ubMunc13-2382−415/Ca2+2–CaM species after the N-terminal CaM domain releases Ca2+ and the peptide. We also observed a gradual migration of the resonances from the N-terminal CaM domain towards those of the intermediate species accompanied by line broadening. This indicates that also the ubMunc13-2382−415/CaM complex rapidly interconverts between the half loaded and the fully loaded species. Altogether our data confirm that ubMunc13-2 and Munc13-1 share a common pattern of interactions with CaM (Figure 6).
Figure 6.
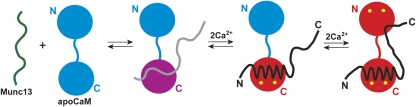
Scheme of the interactions between Munc13-1459−492 and CaM. Apo- and Ca2+-bound states of the CaM domains are represented in cyan and red, respectively, whereas the C-terminal CaM domain in the intermediate Munc13-1459−492/apo-CaM complex is represented in magenta. Ca2+ ions are shown in yellow. The free, apo-CaM-bound and Ca2+-CaM-bound peptides are shown in green, grey and black, respectively. The first step involves the low-affinity binding of apo-CaM (cyan) to the peptide (grey), followed by the partial activation after two Ca2+ ions bind the C-terminal CaM domain and the full activation, where the C-terminal tail of the peptide binds the Ca2+-loaded N-terminal CaM domain.
Discussion
Our results show that Munc13-1 and ubMunc13-2 share a novel 1-5-8-26 CaM recognition motif. The most prominent feature of this motif a is conserved tryptophan residue at position 26 in addition to the hydrophobic residues at positions 1, 5, and 8, which makes the Munc13 peptides the longest CaM recognition motifs among the peptides described to date. The high-resolution NMR structure of the Munc13-1459−492/Ca2+4–CaM complex reveals a modular architecture, where the C-module consists of an amphiphilic α-helix of Munc13-1 anchored to the C-terminal CaM domain by the hydrophobic residues in positions 1-5-8, whereas the N-module consists of tryptophan in position 26 embedded in the hydrophobic cleft of the N-terminal CaM domain. PAL data show contacts of W464 in position 1 of the 1-5-8-26 motif to CaM residues M124 and M144, and W489 in position 26 to the CaM residue M51, which are all in good agreement with the NMR structure. In addition, PAL data on ubMunc13-2 show a similar contact pattern for the W464/W489 equivalents W387/W412, that is W387-M124/M144 and W412-M71, indicating the presence of the same recognition motif for CaM in this Munc13 isoform.
A hallmark of this complex is the extended and flexible nature of the linkers connecting the two modules. This was revealed by the lack of inter-domain NOEs and further confirmed by a significant difference in the magnitude of paramagnetically induced RDCs and PCSs of one of the modules with respect to the other, qualitatively but not quantitatively comparable to earlier studies performed on free CaM (Bertini et al, 2004). Although numerous reports show that Ca2+–CaM interacts with its binding partner in an ‘extended' conformation (Elshorst et al, 1999; Larsson et al, 2001; Schumacher et al, 2001; Drum et al, 2002; Bouvier et al, 2003), to our knowledge, this is the first structure where two separated structural elements, connected by flexible linkers, are formed in a Ca2+–CaM/peptide complex.
A small difference in the intermolecular contacts of the N-module of the complex was found between Munc13-1 and Munc13-2 in the PAL analysis. Although in Munc13-1 W489 contacts M51, in ubMunc13-2 the equivalent W412 contacts M71. This indicates a different orientation of these tryptophans within the hydrophobic pocket of the N-terminal CaM domain, probably as a result of variations in the primary sequence of the linkers. Notable is the presence of two proline residues in ubMunc13-2 that are missing in Munc13-1 (Figure 1), as well as the replacement of L488 in Munc13-1 by Q411 in ubMunc13-2. This may influence the flexibility in the linkers connecting the two modules and affect the binding of the N-module.
The structural data presented here lead us to revise the conclusions drawn on the basis of a PAL-derived model of shorter Munc13-1 and ubMunc13-2 peptides, that is Munc13-1459−479 and ubMunc13-2382−402, in complex with Ca2+4–CaM (Dimova et al, 2009). The peptides used in this earlier study lack the C-terminal part of the 1-5-8-26 CaM-binding motif and therefore bind primarily to the C-terminal CaM domain. The models obtained in this study show the involvement of the residues in position 1-5-8 in the interaction with the C-terminal CaM domain. However, mainly due to the models used (i.e. nNOS- and skMLCK-CaM complexes) as templates for the structure calculation, these earlier Munc13/CaM models (Dimova et al, 2009) displayed a wrapped-around conformation, which is in disagreement with our extended and dynamic NMR structure. Indeed, the PAL data on the longer Munc13 peptides presented here support the open conformation obtained in the NMR structure.
NMR titration experiments revealed the existence of various molecular species along the interaction pathway of CaM with both Munc13-1 and ubMunc13-2 (Figure 6). The titration of Ca2+4–CaM with either Munc13-1459−492 or ubMunc13-2382−415 revealed the formation of high-affinity 1:1 complexes, as shown earlier (Junge et al, 2004). EGTA titrations led to the Ca2+ removal from the N-terminal module and the observation of half-loaded Munc13/Ca2+2–CaM complexes. In these species, the C-module remained intact, whereas the N–terminal module was completely dissociated, as indicated by N–HN chemical shifts similar to those of apo-CaM. The observation of intermediate exchange on the NMR timescale indicates that these intermediate species interconvert rapidly into the fully loaded Munc13/Ca2+4–CaM complex. Altogether, these results establish a function of the C-terminal CaM domain in the formation of a high-affinity complex with Munc13-1 and ubMunc13-2 (the C-module), whereas the lower affinity of the N-module allows these Munc13/CaM complexes to interconvert readily between half and fully loaded species, thereby acting as very efficient Ca2+ sensors.
Studies on both chromaffin cells and glutamatergic synapses of hippocampal neurons showed that the main consequence of Ca2+–CaM binding to Munc13-1 and ubMunc13-2 is an increase in chromaffin granule or synaptic vesicle priming (Junge et al, 2004; Zikich et al, 2008). In chromaffin cells, an increase of [Ca2+]i from resting levels of 50 nM to 10 μM causes a concomitant increase of chromaffin granule priming rates by at least one order of magnitude (Neher, 2006). Interestingly, Ca2+–CaM stimulates chromaffin granule maturation and recruitment through its binding to ubMunc13-2 only at high Ca2+ concentrations (Zikich et al, 2008). This phenomenon likely represents the physiological correlate of the properties that we observed in the N-module of the Munc13/CaM complexes studied here, which interconvert readily between half and fully loaded species at higher (μM) Ca2+ concentrations. Such an action of the N-terminal CaM domain as Ca2+-sensor finds analogy in the case of the Ca2+–CaM-dependent voltage-gated Ca2+ channel, where at CaM-saturating Ca2+ concentrations (∼10 μM) the CaM N-terminal domain mediates a spatial Ca2+ selectivity rather than the global Ca2+ selectivity mediated by the C-terminal domain of Ca2+–CaM (Dick et al, 2008).
A somewhat different scenario may arise in glutamatergic synapses. To date, the Ca2+ concentration dependence of the synaptic vesicle-priming rate in glutamatergic hippocampal neurons has not been studied in detail due to methodological limitations. However, in the calyx of Held, which is a specialized glutamatergic synapse in the auditory brainstem, the synaptic vesicle priming rate is linearly dependent on [Ca2+]i within a concentration range of 0–1 μM [Ca2+]i. At resting [Ca2+]i in the range of some 20–50 nM, priming rates of 0.2 vesicles/ms are observed, which accelerate with increasing [Ca2+]i (Ca2+-EC50≈300–800 nM) to an estimated maximum of 10 vesicles/ms that is reached at 2–5 μM [Ca2+]i (Hosoi et al, 2007; Neher and Sakaba, 2008). This Ca2+-dependent increase of synaptic vesicle priming rates in the calyx of Held is known to be dependent on CaM, even at low [Ca2+]i in the range of 100–500 nM (Sakaba and Neher, 2001); in addition, Munc13s are thought to be directly involved in this CaM-dependent regulation of vesicle priming rates (Junge et al, 2004; Neher and Sakaba, 2008). Assuming that this is indeed the case, the characteristics of the Ca2+ dependence of vesicle priming in the calyx of Held synapse, with an onset at about 100 nM [Ca2+]i and an EC50 for Ca2+ of 200–800 nM, would be best compatible with the notion that formation of the half-loaded Munc13-1459−492/Ca2+2–CaM complex is sufficient to activate Munc13-1 in glutamatergic synapses.
Ca2+–CaM has a multitude of targets in essentially every eukaryotic cell. Consequently, earlier studies on the function of Ca2+–CaM in regulated secretion using CaM inhibitors have often been difficult to interpret due to the pleiotropic effects of the corresponding CaM inhibitors. In consideration of this methodological problem, most recent studies on the role of specific Ca2+–CaM interactions with individual target proteins have used genetic tools to specifically perturb Ca2+–CaM binding by the target protein of interest. This study further challenges the suitability of some of the most frequently used CaM inhibitors for functional studies in complex cellular systems. As outlined in detail above, we show that the C-terminal and N-terminal CaM domains can bind different sites in Munc13-1 with strikingly different affinity—nanomolar in the case of the C-terminal domain and micromolar in the case of the N-terminal domain—and other CaM targets may use similar binding modes. Interestingly, the chloronaphtalene derivative W-7, which is one of the most widely used CaM inhibitors, has an IC50 of around 18 μM (Schweitzer, 1987). It forms a 2:1 complex with Ca2+4–CaM, with one W-7 molecule bound to each CaM domain, as revealed by the NMR structure of its complex with Ca2+4–CaM (Osawa et al, 1998). Considering the high IC50 of W-7 to CaM and the high stability of the C-module of the Munc13/CaM complexes, it is plausible that, depending on the applied concentration of W-7, only the N-terminal CaM domain would be targeted by this inhibitor. This would in turn lead to only partial inhibitory effects and consequent problems with their interpretation.
Although the molecular mechanisms of the Munc13/CaM regulation is not completely understood yet, the sequential character of the interactions between CaM and Munc13-1 and ubMunc13-2, which is favoured by the modular architectures of their complexes, provides a good explanation of the Munc13-mediated regulation of neurotransmitter release by Ca2+ and CaM. Possibly, CaM could directly or indirectly regulate other Munc13 functions. The complete mapping of such regulation networks might also help to identify the origin of the different Ca2+-responses in the various Munc13 isoforms.
Materials and methods
Protein and peptide purification
All unlabelled proteins were expressed in Escherichia coli BL21 (DE3) grown in LB medium at 37°C. The uniformly 15N or 15N, 13C-labelled proteins were expressed in E. coli BL21 (DE3) grown in M9 minimal medium supplemented either with 15NH4Cl or with 15NH4Cl and [13C6] glucose (Cambridge Isotope Laboratories) as the sole nitrogen and carbon sources. CaM was purified as described elsewhere (Guerini et al, 1984; Haberz et al, 2006). The N60D-CaM mutant was generated by site-directed mutagenesis using the QuickChange kit (Stratagene). For the recombinant expression and purification of Munc13-1459−492, the cDNA encoding this fragment was cloned into a pGEX 2T vector (GE Healthcare)C-terminal to the Glutathion S-transferase tag with a thrombin cleavage site in the linker region. This construct was co-expressed with CaM. The cells were disrupted by sonication and the supernatant was applied to a Glutathione-Sepharose (Pharmacia) affinity chromatography column. The Munc13-1459−492 fragment was released by on-column cleavage with thrombin (Sigma) and further purified by reverse phase HPLC. The same procedure was used for the longer Munc13-1459−511 fragment. The identity of all proteins was verified by ESI-mass spectrometry.
NMR measurements and spectral assignments
15N-labelled samples of apo- or Ca2+–CaM for titrations were prepared by dialysing CaM against either an EDTA-containing buffer (20 mM Bis-Tris, 150 mM KCl, 5 mM EDTA) or a CaCl2-containing buffer (20 mM Bis-Tris, 150 mM KCl and 10 mM CaCl2, pH 6.8). The selectively labelled Munc13-1459−492/Ca2+4–CaM samples used for the structural determination were prepared by dissolving 1 equivalent of Ca2+4–CaM and 1.2 equivalents of Munc13-1459−492 in 250 μl of buffer (20 mM Bis-Tris, 150 mM KCl and 10 mM CaCl2, pH 6.8) either in a 90% H2O, 10% D2O mixture or in 99.99% D2O. The final concentration of Munc13-1459−492/Ca2+4–CaM was ∼1 mM.
All NMR experiments were carried out at 35°C on Bruker DRX spectrometers equipped with z-gradient cryoprobes operating at different fields (600–900 MHz). The spectra were processed using Felix97 (Accelrys) or NMRpipe (Delaglio et al, 1995) and analysed using SPARKY (Goddard and Kneller, 1999).
Each partner in the complex was assigned separately by preparing two samples, where one partner was uniformly 15N/13C labelled and the second one contained isotopes at natural abundance. Backbone 1H, 13C, and 15N chemical shift assignments of the subscript for the Munc13-1459−492/Ca2+–CaM complex were obtained from 3D triple resonance 1H–15N NOESY-HSQC, HNCO, HN(CA)CO, HNCACB, and CBCA(CO)NH experiments, and side-chain resonance assignments were obtained with 3D H(CC)(CO)NH, (H)CC(CO)NH, HCCH-TOCSY, and 13C-edited NOESY-HSQC experiments. The assignments are deposited in the BMRB data bank under accession number 15470.
1H–15N RDCs were measured using a spin-state selective excitation scheme (Ottiger et al, 1998), in (i) a steric alignment with phages (15 mg/ml Pf1 solution) at 600 MHz or (ii) for 15N-labelled N60D-CaM (Bertini et al, 2003) loaded selectively with terbium, at 900 MHz. The singular value decomposition algorithm from PALES (Zweckstetter and Bax, 2000) was used for the RDC analysis. 1H and 15N PCSs were evaluated from HSQC spectra of the terbium-loaded 15N-labelled N60D-CaM sample.
Chemical shift perturbations were calculated from the backbone amide 15N (Δδ15N) and 1H (Δδ1H) chemical shift changes using the expression Δδ=[(Δδ1HN)2 + (0.15Δδ15N)2]1/2 (Craik and Wilce, 1997).
Structure calculation
Backbone φ and ψ dihedral restraints were derived from the 1H, 13C, and 15N backbone chemical shifts using the program TALOS (Cornilescu et al, 1999). Only TALOS predictions that have >10 matches were used as restraints, applying a minimal standard deviation range of 10°. Dihedral restraints, 1H–13C/15N HSQC-NOESY spectra and 1H, 13C, 15N chemical shifts were used as input for the ATNOS/CANDID software (Herrmann et al, 2002, 2002b) to generate a first ensemble of structures. This early ensemble was used to complete the assignment of the 1H–13C/15N HSQC-NOESY spectra manually and led to a final set of about 1300 distance restraints. Dihedral angles and distance restraints were then used in the torsion angle MD protocol of CYANA (Güntert, 2003) to generate an ensemble of 100 structures of the Munc13-1459−492/CaM complex. Hydrogen bond restraints were also added during this step, based on strong protection factors obtained from H/D exchange experiments, correlating with the secondary structure prediction.
Structure refinement with the RDC values measured by steric alignment (15 mg/ml Pf1) was performed using Xplor-NIH (Schwieters et al, 2003) and the protein-2.0 force field using a protocol described elsewhere (Lee et al, 2007). Ca2+ ions were added subsequently, based on the canonical Ca2+-binding sites of CaM. For each Ca2+, six distance restraints were defined between the protein and the ion. The compatibility of the Ca2+-binding restraints with the force field parameters and the experimental restraints (dihedral angle, distance restraints, and RDC) was carefully checked.
The 20 structures with the lowest NOE energy were selected as the final ensemble of structures describing the Munc13-1459−492/CaM complex in solution. Experimental restraints and structural statistics are summarized in Table 1. Coordinates of the final ensemble were deposited in the Brookhaven Protein Data Bank under the accession number 2KDU. The lowest energy structure from the final ensemble is considered as the most representative.
PAL and structural characterization of photoadducts
Munc13-1459−492- and ubMunc13-2382−415-derived photoprobes were synthesized using Bpa as described earlier (Jahn et al, 2002; Dimova et al, 2006). In Munc13-1459−492, W464, W489, or both were substituted with Bpa to generate monofunctional (Bpa464-13-1459−492, Bpa489-13-1459−492) and bifunctional (Bpa464,489-13-1459−492) photoprobes. Similarly, W387, W412, or both were replaced in ubMunc13-2382−415 to obtain Bpa387-ub13-2382−415, Bpa412-ub13-2459−492, and Bpa387, 412-ub13-2459−492. PAL reactions using recombinant CaM were carried out as described earlier and monitored by gel electrophoresis and protein mass spectrometry (Dimova et al, 2006). For the structural characterization of photoadducts by mass spectrometry, a recently introduced analytical workflow based on isotopically labelled CaM was applied (Dimova et al, 2009). Briefly, PAL reaction mixtures were subjected to in-solution trypsin digestion and subsequently analysed by HPLC/MALDI-TOF-MS to identify and sequence cross-linked peptides. When the exceptionally large tryptic CaM fragment, CaM38−74 was involved in photoadduct formation, the cross-linked peptides were isolated by HPLC and further digested with endoprotease AspN (Roche) to make them available for mass spectrometric sequencing by HPLC/MALDI-TOF-MS.
Supplementary Material
Supplementary Information
Review Process File
Acknowledgments
We thank Gerhard Wolff, Kerstin Overkamp, and Dr Volker Klaukien for HPLC and peptide synthesis; Karin Giller for technical assistance; Dr Joachim Krebs for the CaM construct; Dr Christophe Farès and Dr Vinesh Vijayan for assistance in NMR measurements; Dr Pierre Montaville for scientific discussions; and the Max-Planck Society as well as the Fonds der Chemischen Industrie (to CG) for funding. FRC did his PhD in the framework of the Molecular Biology IMPRS in Göttingen.
Footnotes
The authors declare that they have no conflict of interest.
References
- Augustin I, Rosenmund C, Südhof TC, Brose N (1999) Munc13-1 is essential for fusion competence of glutamatergic synaptic vesicles. Nature 400: 457–461 [DOI] [PubMed] [Google Scholar]
- Basu J, Shen N, Dulubova I, Lu J, Guan R, Guryev O, Grishin NV, Rosenmund C, Rizo J (2005) A minimal domain responsible for Munc13 activity. Nat Struct Mol Biol 12: 1017–1018 [DOI] [PubMed] [Google Scholar]
- Bax A, Grishaev A (2005) Weak alignment NMR: a hawk-eyed view of biomolecular structure. Curr Opin Struct Biol 15: 563–570 [DOI] [PubMed] [Google Scholar]
- Bayley PM, Findlay WA, Martin SR (1996) Target recognition by calmodulin: dissecting the kinetics and affinity of interaction using short peptide sequences. Protein Sci 5: 1215–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I, Del Bianco C, Gelis I, Katsaros N, Luchinat C, Parigi G, Peana M, Provenzali A, Zoroddu MA (2004) Experimentally exploring the conformational space sampled by domain reorientation in calmodulin. Proc Natl Acad Sci USA 101: 6841–6846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini I, Gelis I, Katsaros N, Luchinat C, Provenzali A (2003) Tuning the affinity for lanthanides of calcium binding proteins. Biochemistry 42: 8011–8021 [DOI] [PubMed] [Google Scholar]
- Betz A, Asheri U, Rickmann M, Augustin I, Neher E, Sudhof TC, Rettig J, Brose N (1998) Munc13-1 is a presynaptic phorbol ester receptor that enhances neurotransmitter release. Neuron 21: 123–126 [DOI] [PubMed] [Google Scholar]
- Betz A, Okamoto M, Brose N (1997) Direct interaction of the rat unc-13 homologue Munc13-1 with the N-terminus of syntaxin. J Biol Chem 272 2520–2526 [DOI] [PubMed] [Google Scholar]
- Bouvier D, Vanhaverbeke C, Simorre J-P, Arlaud GJ, Bally I, Forge V, Margolis RL, Gans P, Kleman J-P (2003) Unusual Ca2+-calmodulin binding interactions of the microtubule-associated protein F-STOP) Biochemistry 42: 11484–11493 [DOI] [PubMed] [Google Scholar]
- Brose N, Rosenmund C (2002) Move over protein kinase C, you've got company: alternative cellular effectors of diacylglycerol and phorbol esters. J Cell Sci 115: 4399–4411 [DOI] [PubMed] [Google Scholar]
- Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13: 289–302 [DOI] [PubMed] [Google Scholar]
- Craik DJ, Wilce JA (1997) Studies of protein-ligand interactions by NMR. Methods Mol Biol 60: 195–232 [DOI] [PubMed] [Google Scholar]
- Crivici A, Ikura M (1995) Molecular and structural basis of target recognition by calmodulin. Annu Rev Biophys Biomol Struct 24: 85–116 [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) Nmrpipe: a multidimensional spectral processing system based on Unix pipes. J Biomol NMR (6) 277–293 [DOI] [PubMed] [Google Scholar]
- Dick IE, Tadross MR, Liang H, Tay L-H, Yang W, Yue DT (2008) A modular switch for spatial Ca2+ selectivity in the calmodulin regulation of CaV channels. Nature 451: 830–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimova K, Kalkhof S, Pottratz I, Ihling C, Rodriguez-Castaneda F, Liepold T, Griesinger C, Brose N, Sinz A, Jahn O 2009. Structural insights into the Calmodulin–Munc13 interaction obtained by cross-linking and mass spectrometry. Biochemistry 48: 5908–5921 [DOI] [PubMed] [Google Scholar]
- Dimova K, Kawabe H, Betz A, Brose N, Jahn O (2006) Characterization of the Munc13-calmodulin interaction by photoaffinity labeling. Biochim Biophys Acta 1763: 1256–1265 [DOI] [PubMed] [Google Scholar]
- Drum CL, Yan S-Z, Bard J, Shen Y-Q, Lu D, Soelaiman S, Grabarek Z, Bohm A, Tang W-J (2002) Structural basis for the activation of anthrax adenylyl cyclase exotoxin by calmodulin. Nature 415: 396–402 [DOI] [PubMed] [Google Scholar]
- Edlich F, Maestre-Martínez M, Jarczowski F, Weiwad M, Moutty MC, eseviæ M, Jahreis G, Fischer G, Lücke C (2007) A novel calmodulin-Ca2+ target recognition activates the Bcl-2 regulator FKBP38. J Biol Chem 282: 36496–36504 [DOI] [PubMed] [Google Scholar]
- Elshorst B, Hennig M, Försterling H, Diener A, Maurer M, Schulte P, Schwalbe H, Griesinger C, Krebs J, Schmid H, Vorherr T, Carafoli E (1999) NMR solution structure of a complex of calmodulin with a binding peptide of the Ca2+ pump. Biochemistry 38: 12320–12332 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Kneller DG (1999) SPARKY 3 San Francisco: University of California [Google Scholar]
- Guan R, Dai H, Rizo J (2008) Binding of the Munc13-1 MUN domain to membrane-anchored SNARE complexes. Biochemistry 47: 1474–1481 [DOI] [PubMed] [Google Scholar]
- Guerini D, Krebs J, Carafoli E (1984) Stimulation of the purified erythrocyte Ca2+-ATPase by tryptic fragments of calmodulin) J Biol Chem 259: 15172–15177 [PubMed] [Google Scholar]
- Güntert P (2003) Automated NMR protein structure calculation) Prog Nucl Magn Reson Spectrosc 44: 33–96 [Google Scholar]
- Haberz P, Rodríguez-Castañeda F, Junker J, Becker S, Leonov A, Griesinger C (2006) Two new chiral EDTA-based metal chelates for weak alignment of proteins in solution) Org Lett 8: 1275–1278 [DOI] [PubMed] [Google Scholar]
- Herrmann T, Güntert P, Wüthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA) J Mol Biol 319: 209–227 [DOI] [PubMed] [Google Scholar]
- Herrmann T, Güntert P, Wüthrich K (2002b) Protein NMR structure determination with automated NOE-identification in the NOESY spectra using the new software ATNOS) J Biomol NMR 24: 171–189 [DOI] [PubMed] [Google Scholar]
- Hoeflich KP, Ikura M (2002) Calmodulin in action: diversity in target recognition and activation mechanisms) Cell 108: 739–742 [DOI] [PubMed] [Google Scholar]
- Hosoi N, Sakaba T, Neher E (2007) Quantitative analysis of calcium-dependent vesicle recruitment and its functional role at the calyx of Held synapse) J Neurosci 27: 14286–14298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikura M (1996) Calcium binding and conformational response in EF-hand proteins) Trends Biochem Sci 21: 14–17 [PubMed] [Google Scholar]
- Jahn O, Eckart K, Brauns O, Tezval H, Spiess J (2002) The binding protein of corticotropin-releasing factor: ligand-binding site and subunit structure) Proc Natl Acad Sci USA 99: 12055–12060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn R, Scheller RH (2006) SNAREs—engines for membrane fusion) Nat Rev Mol Cell Biol 7: 631–643 [DOI] [PubMed] [Google Scholar]
- Junge HJ, Rhee J-S, Jahn O, Varoqueaux F, Spiess J, Waxham MN, Rosenmund C, Brose N (2004) Calmodulin and Munc13 form a Ca2+ sensor/effector complex that controls short-term synaptic plasticity) Cell 118: 389–401 [DOI] [PubMed] [Google Scholar]
- Jurado LA, Chockalingam PS, Jarrett HW (1999) Apocalmodulin) Physiol Rev 79: 661–682 [DOI] [PubMed] [Google Scholar]
- Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A (1995) Solution structure of calcium-free calmodulin) Nat Struct Biol 2: 768–776 [DOI] [PubMed] [Google Scholar]
- Larsson G, Schleucher J, Onions J, Hermann S, Grundström T, Wijmenga SS (2001) A novel target recognition revealed by calmodulin in complex with the basic helix-loop-helix transcription factor SEF2-1/E2-2) Protein Sci 10: 169–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmann JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR) J Biomol NMR 8: 477–486 [DOI] [PubMed] [Google Scholar]
- Lee D, Walsh JD, Yu P, Markus MA, Choli-Papadopoulou T, Schwieters CD, Krueger S, Draper DE, Wang Y-X (2007) The structure of free L11 and functional dynamic of L11 in free, L11-rRNA(58nt) binary and L11-rRNA(58nt)-thiostrepton ternary complexes) J Mol Biol 367: 1007–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite JF, Dougherty DA, Lester HA, Shahgholi M (2003) Investigation of apparent mass deviations in electrospray ionization tandem mass spectrometry of a benzophenone-labeled peptide) Rapid Commun Mass Spectrom 17: 1677–1684 [DOI] [PubMed] [Google Scholar]
- Mirzoeva S, Weigand S, Lukas TJ, Shuvalova L, Anderson WF, Watterson DM (1999) Analysis of the functional coupling between calmodulin's calcium binding and peptide recognition properties) Biochemistry 38: 3936–3947 [DOI] [PubMed] [Google Scholar]
- Neher E (2006) A comparison between exocytic control mechanisms in adrenal chromaffin cells and a glutamatergic synapse) Pflugers Arch 453: 261–268 [DOI] [PubMed] [Google Scholar]
- Neher E, Sakaba T (2008) Multiple roles of calcium ions in the regulation of neurotransmitter release) Neuron 59: 861–872 [DOI] [PubMed] [Google Scholar]
- Osawa M, Swindella MB, Tanikawa J, Tanaka T, Mase T, Furuya T, Ikura M (1998) Solution structure of calmodulin-w-7 complex: the basis of diversity in molecular recognition) J Mol Biol 276: 165–176 [DOI] [PubMed] [Google Scholar]
- Ottiger M, Delaglio F, Bax A (1998) Measurement of J and dipolar couplings from simplified two-dimensional NMR spectra) J Magn Res 131: 373–378 [DOI] [PubMed] [Google Scholar]
- Peersen OB, Madsen TS, Falke JJ (1997) Intermolecular tuning of calmodulin by target peptides and proteins: differential effects on Ca2+ binding and implications for kinase activation) Protein Sci 6: 794–807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rettig J, Neher E (2002) Emerging roles of presynaptic proteins in Ca++-triggered exocytosis) Science 298: 781–785 [DOI] [PubMed] [Google Scholar]
- Rhee J-S, Betz A, Pyott S, Reim K, Varoqueaux F, Augustin I, Hesse D, Südhof TC, Takahashi M, Rosenmund C, Brose N (2002) β Phorbol ester- and diacylglycerol-induced augmentation of transmitter release is mediated by Munc13s and not by PKCs) Cell 108: 121–133 [DOI] [PubMed] [Google Scholar]
- Rhoads AR, Friedberg F (1997) Sequence motifs for calmodulin recognition) FASEB J 11: 331–340 [DOI] [PubMed] [Google Scholar]
- Richmond JE, Weimer RM, Jorgensen EM (2001) An open form of syntaxin bypasses the requirement for UNC-13 in vesicle priming) Nature 412: 338–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenmund C, Sigler A, Augustin I, Reim K, Brose N, Rhee JS (2002) Differential control of vesicle priming and short-term plasticity by Munc13 isoforms) Neuron 33: 411–424 [DOI] [PubMed] [Google Scholar]
- Sakaba T, Neher E (2001) Calmodulin mediates rapid recruitment of fast-releasing synaptic vesicles at a calyx-type synapse) Neuron 32: 1119–1131 [DOI] [PubMed] [Google Scholar]
- Schumacher MA, Rivard AF, Bächinger HP, Adelman JP (2001) Structure of the gating domain of a Ca2+-activated K+ channel complexed with Ca2+/calmodulin) Nature 410: 1120–1124 [DOI] [PubMed] [Google Scholar]
- Schweitzer E (1987) Coordinated release of ATP and ACh from cholinergic synaptosomes and its inhibition by calmodulin antagonists) J Neurosci 7: 2948–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM (2003) The Xplor-NIH NMR molecular structure determination package) J Magn Reson 160: 65–73 [DOI] [PubMed] [Google Scholar]
- Shifman JM, Choi MH, Mihalas S, Mayo SL, Kennedy MB (2006) Ca2+/calmodulin-dependent protein kinase II (CaKII) is activated by calmodulin with two bound calciums) Proc Natl Acad Sci USA 103: 13968–13973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silinsky EM, Searl TJ (2003) Phorbol esters and neurotransmitter release: more than just protein kinase C?) Br J Pharmacol 138: 1191–1201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens DR, Wu Z-X, Matti U, Junge HJ, Schirra C, Becherer U, Wojcik SM, Brose N, Rettig J (2005) Identification of the minimal protein domain required for priming activity of Munc13-1) Curr Biol 15: 2243–2248 [DOI] [PubMed] [Google Scholar]
- Südhof TC (2004) The synaptic vesicle cycle) Annu Rev Neurosci 27: 509–547 [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Sigler A, Rhee J-S, Brose N, Enk C, Reim K, Rosenmund C (2002) Total arrest of spontaneous and evoked synaptic transmission but normal synaptogenesis in the absence of Munc13-mediated vesicle priming) Proc Natl Acad Sci USA 99: 9037–9042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter SW, Leclerc E (2003) Novel aspects of calmodulin target recognition and activation) Eur J Biochem 270: 404–414 [DOI] [PubMed] [Google Scholar]
- Wojcik SM, Brose N (2007) Regulation of membrane fusion in synaptic excitation-secretion coupling: speed and accuracy matter) Neuron 55: 11–24 [DOI] [PubMed] [Google Scholar]
- Xia Z, Storm DR (2005) The role of calmodulin as a signal integrator for synaptic plasticity) Nat Rev Neurosci 6: 267–276 [DOI] [PubMed] [Google Scholar]
- Zhang M, Tanaka T, Ikura M (1995) Calcium-induced conformational transition revealed by the solution structure of apo-Calmodulin) Nat Struct Biol 2: 758–767 [DOI] [PubMed] [Google Scholar]
- Zikich D, Mezer A, Varoqueaux F, Sheinin A, Junge HJ, Nachliel E, Melamed R, Brose N, Gutman M, Ashery U (2008) Vesicle priming and recruitment by ubMunc13-2 are differentially regulated by calcium and calmodulin) J Neurosci 28: 1949–1960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zweckstetter M, Bax A (2000) Prediction of sterically induced alignment in a dilute liquid crystalline phase: aid to protein structure determination by NMR) J Am Chem Soc 122: 3791–3792 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Information
Review Process File