Abstract
This paper describes the development of laser desorption 7.87 eV vacuum ultraviolet postionization mass spectrometry (LDPI-MS) to detect antibiotics within intact bacterial colony biofilms. As >99% of the molecules ejected by laser desorption are neutrals, vacuum ultraviolet (VUV) photoionization of these neutrals can provide significantly increased signal compared to detection of directly emitted ions. Postionization with VUV radiation from the molecular fluorine laser single photon ionizes laser desorbed neutrals with ionization potentials below the 7.87 eV photon energy. Antibiotics with structures indicative of sub-7.87 eV ionization potentials were examined for their ability to be detected by 7.87 eV LDPI-MS. Tetracycline, sulfadiazine, and novobiocin were successfully detected neat as dried films physisorbed on porous silicon oxide substrates. Tetracycline and sulfadiazine were then detected within intact Staphylococcus epidermidis colony biofilms, the former with LOD in the micromolar concentration range.
Keywords: Mass spectra, Imaging, Bacteria, Biofilm, Matrix-assisted laser desorption/ionization time of flight mass spectrometry
1. Introduction
Staphylococcus epidermidis is a common bacteria that naturally resides on human skin and often forms biofilms associated with catheter and other hospital infections [1]. Treatment of biofilm infections are hindered by the limited ability of antibiotics to inhibit or kill biofilms [2] compared with the same microbial strain in planktonic culture. Uncovering the mechanisms of biofilm resistance to antibiotics requires imaging MS methods to probe antibiotics and their catabolites within intact biofilms.
MALDI-MS is commonly performed directly on biological tissue for imaging MS of molecular species [3-5]. MALDI-MS has also been used to detect clinically relevant concentrations of drugs in tissue [5, 6]. However, MALDI imaging of biofilms is limited by various experimental effects [7], including the difficulty of forming ions from such a high salt environment while preserving the biofilm's structural integrity.
As >99% of the molecules ejected by laser desorption are neutrals, vacuum ultraviolet (VUV) photoionization of these neutrals can provide significantly increased signal compared to detection of directly emitted ions. [8]. Postionization of laser desorbed neutrals with 7.87 eV photons from a molecular fluorine laser will single photon ionize various low ionization potential compounds including secondary, tertiary, and aromatic amines as well as various fused ring aromatics, tryptophan containing peptides, and pharmaceuticals [9-11]. Selectivity for specific higher ionization potential species can be achieved by derivatizing a molecular analyte with an appropriate chemical tag to lower the ionization potential of the resultant complex below 7.87 eV, thereby permitting postionization with the fluorine laser [12]. Prior work demonstrated that tagging with 6-aminoquinolyl-N-hydroxysuccinimidyl carbamate allowed detection of a known quorum sensing peptide within intact Bacillus subtilis biofilms by laser desorption 7.87 eV VUV postionization mass spectrometry (LDPI-MS) [7].
This paper describes the development of 7.87 eV LDPI-MS to detect antibiotics within intact bacterial biofilms. The experimental configuration of LDPI-MS is essentially that of a MALDI-MS with a molecular fluorine laser added for postionization. Five antibiotics with structures indicative of sub-7.87 eV ionization potentials were examined for their ability to be detected by 7.87 eV LDPI-MS without chemical derivatization. Tetracycline, sulfadiazine, and novobiocin were successfully detected neat as dried films physisorbed on porous silicon oxide substrates, then examined within intact S. epidermidis bacterial biofilms. Azithromycin and coumermycin A1 were found to not be detectable neat by 7.87 eV LDPI-MS and were not examined further.
Previous work with another instrument showed that LDPI-MS can be used for imaging of spatial distributions of quorum sensing peptides within intact biofilms [7]. Ongoing experimental upgrades will allow imaging MS on the instrument employed here.
2. Materials and Methods
A. Laser Desorption Postionization Mass Spectrometer (LDPI-MS)
A custom built LDPI-MS was constructed with both linear and reflectron TOF mass spectrometers. The instrument was described in detail previously [12], but has since undergone some changes that are described here. The major parts of the LDPI-MS are the load lock for sample introduction; the main chamber which contains the pulsed ion optics, steering plates, Einzel lens, flight tube, and detectors; the 355 nm Nd:YAG desorption laser; and the postionization laser. Separate dual microchannel plate detectors were used in linear (R.M. Jordan Co., Grass Valley, CA) and reflectron modes (Advanced Performance Detector, Burle, Sturbridge, MA). Data is recorded and processed using a four channel digital oscilloscope with a bandwidth of 1 GHz (TDS5104B series; Tektronix, Portland, OR).
Vacuum was maintained using three turbo pumps backed by mechanical pumps. The base pressures in the instrument were 1 × 10-6 Torr in the load lock, 7 × 10-9 Torr in the main chamber base and 1 × 10-9 Torr in the TOF tube. Typical operating pressures in the main chamber with a sample loaded was 9 × 10-8 Torr.
Sample desorption was achieved with a pulsed Nd:YAG laser (355 nm, 10 Hz, ∼4 ns pulse length, Minilite II, Continuum, Santa Clara, CA). The desorption laser power was varied from 0.8 to 11 MW/cm2 depending upon sample conditions to maximize desorbed neutral signal while minimizing direct ion formation (prior to postionization). The laser desorbed neutrals were postionized using a molecular fluorine laser (157 nm, 10 Hz, 10 ns pulse length, Optex Pro, Lambda Physik, Ft Lauderdale, FL) running with a gas mixture consisting of 0.075% F2 gas in helium and fired 3 μs after the desorption laser pulse. A MgF2 plano-convex lens (35.3 cm focal length at 180 nm) focused the 6 × 3 mm2 postionization laser beam about 2 mm above the sample to a power density of ∼1 MW/cm2. Each displayed mass spectrum was the average of 64 laser shots collected on a single spatial location on the sample.
The total path length in linear mode was 1.7 m with maximum resolution achieved of 500 at m/z 720 for a 5400 eV acceleration voltage. In order to increase resolution and signal to noise ratio an ion mirror was constructed of 40 plates separated by 0.635 cm and connected using vacuum compatible 1 MΩ resistors (ITT Power Solutions, West Springield, MA). The ion mirror contained one grid on the last plate (nickel mesh, 117.6 wires/inch, 88.6% transmission, InterNet Inc., internetmesh.net, Minneapolis, MN). Voltage applied to the back of the ion mirror was ∼1.3 times greater than the acceleration voltage in linear mode. The total path length in reflectron mode was 2.4 m with maximum resolution (m/Δm) of 1200 achieved at m/z 397. Efforts are underway to further improve the reflectron mass resolution.
B. Colony Biofilm Growth and Treatment
Colony biofilm growth and treatment generally followed previously published procedures [13-15] A freeze-dried bacterial pellet of Staphylococcus epidermidis ATCC35984 cells (ATCC, Manassus, VA) was rehydrated in 5 mL of tryptic soy broth (Fisher Scientific) in a 15 mL test tube, incubated at 37° C in a water bath, and agitated with a magnetic stir bar to provide oxygenation to planktonic solution for 12 to 18 hours. A new 5 mL of tryptic soy broth solution was prepared, inoculated with 100 μL of stock bacterial solution, and incubated for an additional 12 to 18 hours with agitation. Petri dishes were prepared with 25 mL of tryptic soy agar (Fisher Scientific) each with four ultraviolet sterilized polycarbonate membranes (Millipore, 0.20 μm pore size, 25 mm diameter, Fisher Scientific), which provided structural integrity to the biofilms. Each membrane was inoculated with 10 μL of cultured bacterial solution, and incubated for 71 hours with the transfer of biofilm membranes to fresh agar plates every 24 hours. Biofilm membranes were transferred to agar doped with the desired antibiotic (Sigma-Aldrich) at the given concentration and incubated for the last hour of growth.
The colony biofilm still attached to the polycarbonate membrane was transferred to a silicon substrate and anchored with carbon tape, thereby preventing the biofilm from cracking due to wrinkling or buckling of the membrane. Biofilms were treated with two 50 μL washes of 1:1 acetonitrile/deionized water solution each followed by 15 minutes of drying. The biofilms were then dried in a desiccator for 6 to 12 hours. The biofilm on the membrane along with the underlying silicon substrate were secured to a stainless steel sample holder with carbon tape for introduction to the vacuum system.
Cultures for biofilm cell counts and viability studies were prepared as noted above with the following modifications: after exposure to tetracycline doped agar plates, the biofilm containing membranes were harvested into 9 mL of phosphate buffer solution. The bacterial cells were recovered from the membranes by vigorously vortexing for one minute. The recovered biofilm samples were homogenized and diluted using previously reported techniques [13-15]. The minimal inhibitory concentration of tetracycline for planktonic S. epidermidis cells was ∼0.38 μg/ml (0.86 μM) as determined using antibiotic gradient test strips (Etest, AB BIODISK, Solna, Sweden) and the manufacturer's guidelines. All samples were run in triplicate and the standard deviations reported as errors.
C. Scanning Electron Microscopy
A scanning electron microscope (Model S-3000N, Hitachi) with a tungsten electron source operating at 15 keV electron energy under high vacuum was used obtain images of the colony biofilms. Biofilm samples were treated the same as described above for LDPI-MS except that they were not transferred to agar with antibiotic. The biofilms were then fixed with 2.5% glutaraldehyde in sterilized deionized water, dehydrated by treatment with a gradient series of ethanol:water solutions, dried overnight in a desiccator, and coated with platinum/palladium alloy [16]. Scanning electron micrographs indicated an average S. epidermidis microbe diameter of ∼0.8 μm and verified an absence of contamination by other microbes (not shown).
3. Results and Discussion
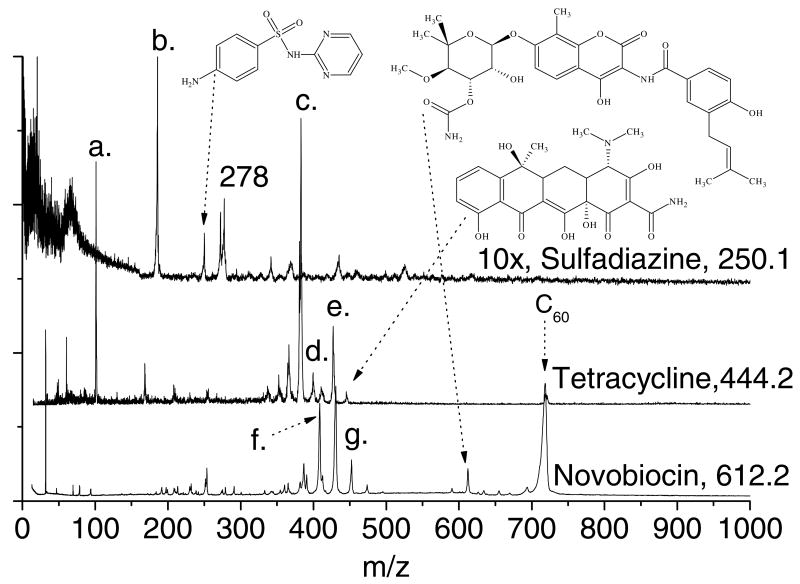
Figure 1 displays the laser desorption postionization mass spectra (LDPI-MS) of tetracycline, sulfadiazine, and novobiocin deposited on separate porous silicon oxide substrates [17]. The antibiotics were laser desorbed at 355 nm with 0.8 MW/cm2 power density, postionized with 7.87 eV vacuum ultraviolet radiation, then detected in linear TOF mode. Concurrent spectra of each antibiotic were also recorded at the same laser desorption power densities but with the postionization laser beam blocked (data not shown). None of the laser desorption only spectra showed any significant ion formation above ∼m/z 100, indicating that the spectra in Figure 1 represented postionization of laser desorbed neutrals. Furthermore, each sample was tested for and found to lack postionization signal from evaporated species in the absence of laser desorption.
Figure 1.
Laser desorption 7.87 eV vacuum ultraviolet postionization mass spectra (LDPI-MS) of neat sulfadiazine, tetracycline, and novobiocin physisorbed on porous silicon oxide.
LDPI-MS of novobiocin, tetracycline, and sulfadiazine all displayed intact radical cations at m/z 612, 444, and 250, respectively, of the undissociated parent (structures displayed for each compound in Figure 1 with arrows connecting the chemical structures to the parent ion peaks). Tetracycline displayed several prominent fragments, two of which were readily identified as the parent after loss of hydroxyl or dimethyl amino groups to form [M-OH]+ at m/z 427 (labeled d. on Figure 1) and [M-NMe2]+ at m/z 400 (labeled e.), respectively. The most intense peak occurred at m/z 383 (labeled c.) and resulted from loss of both groups to form [M-OHNMe2]+. Other characteristic tetracycline fragment ions appeared at m/z 410 due to loss of a hydroxyl from the m/z 427 fragment and m/z 357 due to loss of an amide group from the m/z 400 fragment. The peak at m/z 365 was not identified, but presumably resulted from a complex rearrangement. Sulfadiazine displayed major fragments at m/z 182 and 183 (labeled b.) which correspond to loss of C3H4N2 and C3H3N2, respectively. High mass resolution spectra of this peak supported the presence of two components with slightly higher intensity for m/z 183 than m/z 182 (data not shown). Novobiocin displayed fragments at m/z 451 (labeled g.), 433/434, and 408 (labeled f) which resulted from cleavage at the secondary amine where single photon ionization was expected to initiate, with charge retention on the fragment containing the fused ring.
Several peaks in the antibiotic spectra of Figure 1 were assigned to calibrant compounds or impurities. The m/z 101 peak (labeled a.) on the tetracycline spectrum was due to triethylamine vapor leaked into the instrument. The m/z 720 peak labeled C60 in the novobiocin and tetracycline spectra was due to the addition of C60 to the antibiotic sample prior to analysis. Both triethylamine and C60 were used to perform mass calibration and tuning of the instrument. Sulfadiazine displayed a prominent peak at m/z 278 and several lower intensity peaks at higher masses due to impurities (presumably present in the original compound as purchased) or clustering.
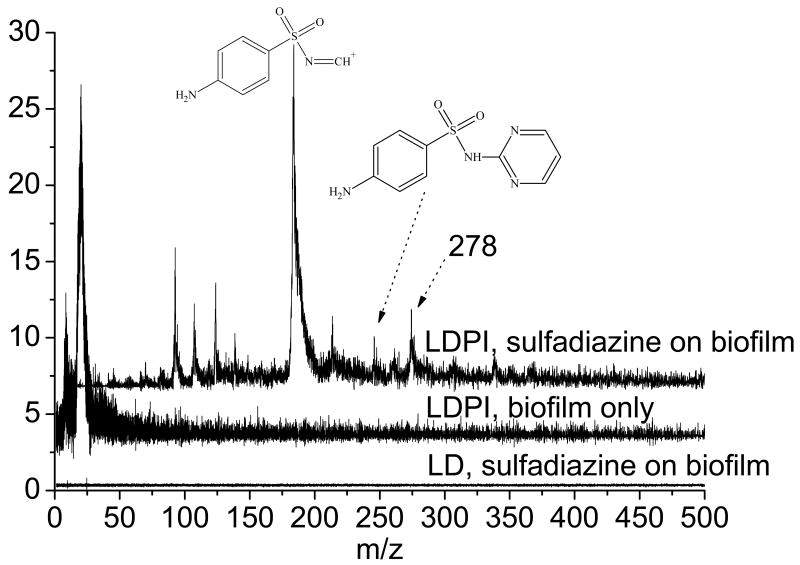
LDPI-MS was used to detect these antibiotics within intact biofilms without significant interference from any of their other chemical constituents. Figure 2 (top trace) displays LDPI-MS of sulfadiazine on S. epidermidis biofilm at 11 mM concentration, prepared by transferring a viable biofilm to an agar plate prepared with this concentration of the antibiotic and allowing the biofilm to equilibrate with the agar plate for one hour. The biofilm sample was then removed from the agar plate, dried overnight, fixed with acetonitrile:water washes, and placed in the load lock. The sample was laser desorbed using ∼3 MW/cm2 power density, postionized, and then detected in reflectron mode. The parent ion of sulfadiazine at m/z 250, its major fragment at m/z 182/183, and the peak at m/z 278 all appeared in the LDPI-MS of the biofilm. No signal above m/z 50 was seen for the LDPI-MS of the biofilm without sulfadiazine (middle trace of Figure 2). Furthermore, no signal at all was seen without postionization of the biofilm with sulfadiazine (bottom trace of Figure 2), indicating that MALDI-like events did not occur here. These results support the argument that of all the neutrals laser desorbed from the biofilm, only tetracycline had a sufficiently low ionization potential to undergo single photon ionization at 7.87 eV. This selectivity is an important feature of LDPI-MS and will enable spatially resolved analysis of these antibiotics.
Figure 2.
LDPI-MS of (top trace) 11 mM sulfadiazine in a Staphylococcus epidermidis colony bacterial biofilm and (middle trace) the biofilm without any added antibiotic. (Bottom trace) laser desorption only MS (without postionization) of sulfadiazine in biofilm.
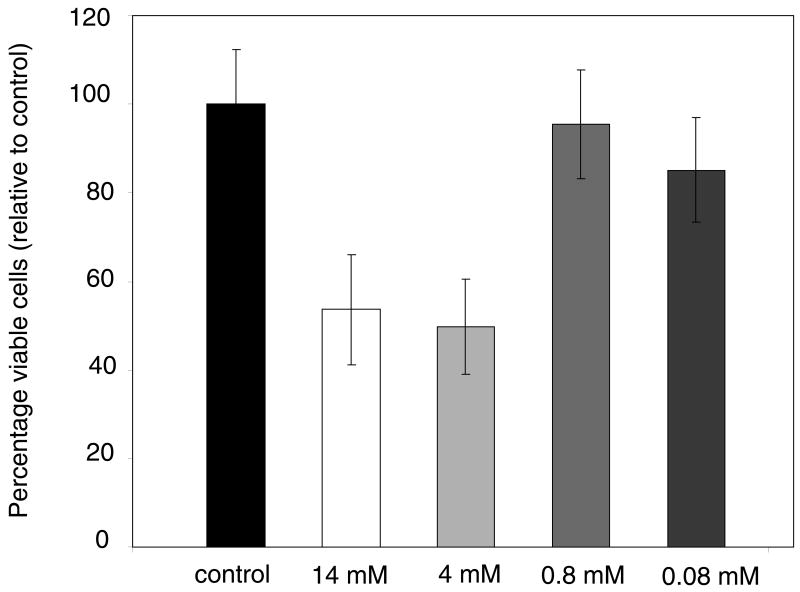
Antibiotic detection with biofilms should ideally be performed at inhibitory concentrations for the time of antibiotic exposure in a given trial. Viable S. epidermidis biofilms were transferred to agar plates with antibiotics at several concentrations for one hour, and so Figure 3 displays colony viability data under these conditions. The data is expressed as percent viable cells relative to the control. The control colonies averaged 5.8 × 108 total viable cells (±7%) with an average areal density of 3.1 × 108 viable cells/cm2. For both the 4 and 14 mM concentrations of tetracycline, about 50% fewer viable cells were counted as compared to the control. Both of these concentrations were above the saturation concentration of tetracycline in aqueous solution, so the actual concentration of antibiotic was likely the same in both cases. Neither the 0.8 or 0.08 mM samples showed any significant inhibition of colony biofilm cell growth during one hour of treatment. By contrast, the minimal inhibitory concentration of tetracycline for planktonic S. epidermidis cells was 0.86 μM.
Figure 3.
Percentage of viable cells relative to control for S. epidermidis colony biofilms grown for one hour on tryptic soy agar with 0.08, 0.8, 4, and 14 mM (nominal) concentrations of tetracycline.
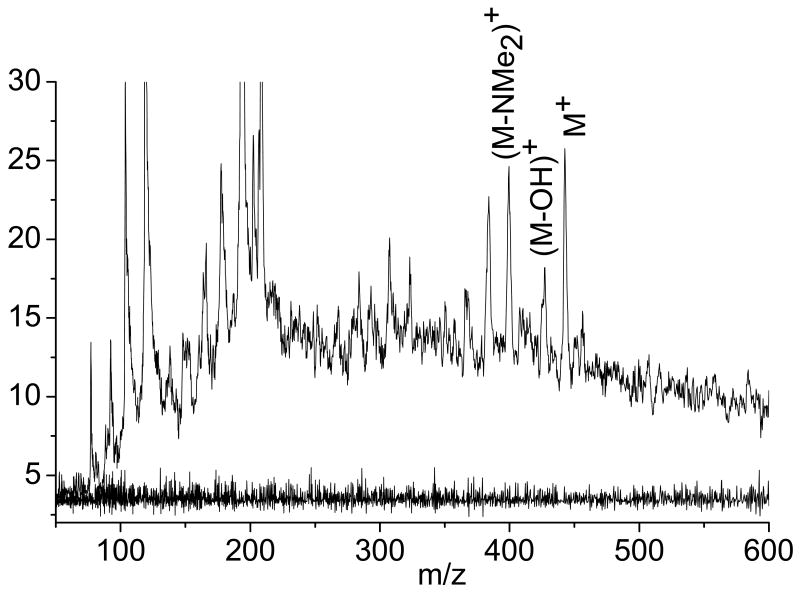
Figure 4 (top trace) displays LDPI-MS of a S. epidermidis colony biofilm transferred for one hour to an agar plate with 8 μM tetracycline with the laser desorption only spectrum displayed underneath (lower trace). LDPI-MS displayed the same characteristic ions for tetracycline above m/z 300, including the M+ parent ion at m/z 444, the [M-OH]+ fragment at m/z 427, the [M-NMe2]+ fragment at m/z 400, and the [M-OHNMe2]+ fragment at m/z 383. No significant ion signal was observed by laser desorption only from the tetracycline treated biofilm (bottom trace), as was the case for sulfadiazine. LDPI-MS with obvious peaks from tetracycline were also obtained from biofilms similarly prepared at 14, 4, 0.8 and 0.08 mM tetracycline concentrations (data not shown).The antibiotic concentration in Figure 4 were similar to the concentrations of several other drugs detected by MALDI-MS in tissue [5, 6], indicating that LDPI-MS can reach useful limits of detection at least for this antibiotic. No matrix was added to any of the biofilms analyzed by LDPI-MS.
Figure 4.
LDPI-MS (top trace) and laser desorption only (bottom trace, no postionization) of 8 μM tetracycline in S. epidermidis colony biofilms.
LDPI-MS of the 8 μM tetracycline in biofilm (Figure 4) showed much higher ion signal below m/z 300 than observed for 11 mM sulfadiazine in the biofilm (Figure 2). The more intense low mass signal for this low tetracycline concentration was due to the use of higher laser desorption powers of ∼11 MW/cm2 compared to ∼3 MW/cm2 for sulfadiazine. The higher laser power used at the lower concentration could have enhanced the low mass ion signal by ablation of a larger volume of biofilm per laser shot, generating more neutrals that could be postionized. Increased fragmentation of the antibiotic and biofilm components could have lead to the formation of small species that could be postionized. High power laser desorption of tetracycline may have formed fragments with ionization potentials below the 7.87 eV postionization photon energy while sulfadiazine either did not fragment (at the lower power densities) or the additional fragments it did form (beyond m/z 182/183) did not display ionization potentials below 7.87 eV.
4. Concluding Remarks
Prior work demonstrated that imaging MS of small molecules within intact biofilms is feasible by LDPI-MS [7]. LDPI-MS is experimentally just a MALDI-MS experiment with a postionization laser, so most of the same considerations that allow imaging with MALDI-MS also permit imaging with LDPI-MS. Furthermore, LDPI-MS may allow higher sensitivity given its ability to exploit the larger neutral yields from laser desorption [8]. However, perhaps the greater advantage of LDPI-MS with 7.87 eV radiation is its selectivity towards species with low ionization potentials. This selectivity can be used to target analytes directly or via their chemical derivatization in situ to reduce interferences from the multitude of other compounds present in complex biological media such as bacterial biofilms.
LDPI-MS achieved micromolar LOD in bacterial biofilms without addition of any matrix compound. Therefore, addition of matrix in future experiments should allow a dramatic increase in LOD by enhancing neutral desorption yields. Prior studies have shown that some matrix compounds and mixtures can be added in such a fashion as to enhance desorption without increasing ionization yields [8, 11].
Acknowledgments
This work was funded by the National Institutes of Health via grant EB006532. The authors thank Dr. Berdan Aydin for assistance in collecting scanning electron micrographs of the biofilms.
Abbreviations
- LDPI-MS
laser desorption postionization mass spectrometry
- VUV
vacuum ultraviolet
References
- 1.Götz F. Staphylococcus and biofilms. Mol Microbiol. 2002;43:1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- 2.Stewart PS, Mukherjee PK, Ghannoum M. Biofilm Antimicrobial Resistance. In: Ghannoum M, O'Toole GA, editors. Microbial Biofilms. ASM Press; Washington, D.C.: 2004. pp. 250–268. [Google Scholar]
- 3.Todd PJ, Schaaff TG, Chaurand P, Caprioli RM. Organic ion imaging of biological tissue with secondary ion mass spectrometry and matrix-assisted laser desorption/ionization. J Mass Spectrom. 2001;36:355–369. doi: 10.1002/jms.153. [DOI] [PubMed] [Google Scholar]
- 4.Wilkins CL, Lay JO. Identification of Microorganisms by Mass Spectrometry. Wiley; New York: 2006. [Google Scholar]
- 5.Reyzer ML, Caprioli RM. MALDI-MS-based imaging of small molecules and proteins in tissues. Curr Opin Chem Biol. 2007;11:29–35. doi: 10.1016/j.cbpa.2006.11.035. [DOI] [PubMed] [Google Scholar]
- 6.Stoeckli M, Staab D, Schweitzer A. Compound and metabolite distribution measured by MALDI mass spectrometric imaging in whole-body tissue sections. Inter J Mass Spectrom. 2007;260:195–202. [Google Scholar]
- 7.Edirisinghe PD, Moore JF, Skinner-Nemec KA, Lindberg C, Giometti CS, et al. Detection of in-situ derivatized peptides in microbial biofilms by laser desorption 7.87 eV postionizaton mass spectrometry. Anal Chem. 2007;79:508–514. doi: 10.1021/ac0615605. [DOI] [PubMed] [Google Scholar]
- 8.Dreisewerd K. The desorption process in MALDI. Chem Rev. 2003;103:395–425. doi: 10.1021/cr010375i. [DOI] [PubMed] [Google Scholar]
- 9.Finch JW, Toerne KA, Schram KH, Denton MB. Evaluation of a hydrogen laser vacuum ultraviolet source for photoionization mass spectrometry of pharmaceuticals. Rap Comm Mass Spectrom. 2005;19:15–22. doi: 10.1002/rcm.1742. [DOI] [PubMed] [Google Scholar]
- 10.Hanley L, Edirisinghe PD, Calaway WF, Veryovkin IV, Pellin MJ, et al. 7.87 eV postionization of peptides containing tryptophan or derivatized with fluorescein. Appl Surf Sci. 2006;252:6723–6726. [Google Scholar]
- 11.Edirisinghe PD, Moore JF, Calaway WF, Veryovkin IV, Pellin MJ, et al. Vacuum ultraviolet postionization of aromatic groups covalently bound to peptides. Anal Chem. 2006;78:5876–5883. doi: 10.1021/ac0605997. [DOI] [PubMed] [Google Scholar]
- 12.Zhou M, Wu C, Akhmetov A, Edirisinghe PD, Drummond JL, et al. 7.87 eV laser desorption postionization mass spectrometry of adsorbed and covalently bound bisphenol A diglycidyl methacrylate. J Amer Soc Mass Spectrom. 2007;18:1097–1108. doi: 10.1016/j.jasms.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu KD, Stewart PS, Xia F, Huang CT, McFeters GA. Spatial physiological heterogeneity in Pseudomonas aeruginosa biofilm Is determined by oxygen availability. Appl Environ Microbiol. 1998;64:4035–4039. doi: 10.1128/aem.64.10.4035-4039.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang CT, Xu KD, McFeters GA, Stewart PS. Spatial patterns of alkaline phosphatase expression within bacterial colonies and biofilms in response to phosphate starvation. Appl Environ Microbiol. 1998;64:1526–1531. doi: 10.1128/aem.64.4.1526-1531.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart PS, Rayner J, Roe F, Rees WM. Biofilm penetration and disinfection efficacy of alkaline hypochlorite and chlorosulfamates. J Appl Microbiol. 2001;91:525–532. doi: 10.1046/j.1365-2672.2001.01413.x. [DOI] [PubMed] [Google Scholar]
- 16.Fraud S, Maillard JY, Denyer SP, Kaminski MA, Hanlon GW. A simulated oral hygiene model to determine the efficacy of repeated exposure of amine oxide on the viability of Steptococcus mutas biofilms. Eur J Oral Sci. 2007;115:71–76. doi: 10.1111/j.1600-0722.2007.00419.x. [DOI] [PubMed] [Google Scholar]
- 17.Wei J, Buriak JM, Siuzdak G. Desorption/ionization mass spectrometry on porous silicon. Nature. 1999;399:243–246. doi: 10.1038/20400. [DOI] [PubMed] [Google Scholar]