Abstract
Optimal performance of LC-MS/MS platforms is critical to generating high quality proteomics data. Although individual laboratories have developed quality control samples, there is no widely available performance standard of biological complexity (and associated reference data sets) for benchmarking of platform performance for analysis of complex biological proteomes across different laboratories in the community. Individual preparations of the yeast Saccharomyces cerevisiae proteome have been used extensively by laboratories in the proteomics community to characterize LC-MS platform performance. The yeast proteome is uniquely attractive as a performance standard because it is the most extensively characterized complex biological proteome and the only one associated with several large scale studies estimating the abundance of all detectable proteins. In this study, we describe a standard operating protocol for large scale production of the yeast performance standard and offer aliquots to the community through the National Institute of Standards and Technology where the yeast proteome is under development as a certified reference material to meet the long term needs of the community. Using a series of metrics that characterize LC-MS performance, we provide a reference data set demonstrating typical performance of commonly used ion trap instrument platforms in expert laboratories; the results provide a basis for laboratories to benchmark their own performance, to improve upon current methods, and to evaluate new technologies. Additionally, we demonstrate how the yeast reference, spiked with human proteins, can be used to benchmark the power of proteomics platforms for detection of differentially expressed proteins at different levels of concentration in a complex matrix, thereby providing a metric to evaluate and minimize preanalytical and analytical variation in comparative proteomics experiments.
Access to proteomics performance standards is essential for several reasons. First, to generate the highest quality data possible, proteomics laboratories routinely benchmark and perform quality control (QC)1 monitoring of the performance of their instrumentation using standards. Second, appropriate standards greatly facilitate the development of improvements in technologies by providing a timeless standard with which to evaluate new protocols or instruments that claim to improve performance. For example, it is common practice for an individual laboratory considering purchase of a new instrument to require the vendor to run “demo” samples so that data from the new instrument can be compared head to head with existing instruments in the laboratory. Third, large scale proteomics studies designed to aggregate data across laboratories can be facilitated by the use of a performance standard to measure reproducibility across sites or to compare the performance of different LC-MS configurations or sample processing protocols used between laboratories to facilitate development of optimized standard operating procedures (SOPs).
Most individual laboratories have adopted their own QC standards, which range from mixtures of known synthetic peptides to digests of bovine serum albumin or more complex mixtures of several recombinant proteins (1). However, because each laboratory performs QC monitoring in isolation, it is difficult to compare the performance of LC-MS platforms throughout the community.
Several standards for proteomics are available for request or purchase (2, 3). RM8327 is a mixture of three peptides developed as a reference material in collaboration between the National Institute of Standards and Technology (NIST) and the Association of Biomolecular Resource Facilities. Mixtures of 15–48 purified human proteins are also available, such as the HUPO (Human Proteome Organisation) Gold MS Protein Standard (Invitrogen), the Universal Proteomics Standard (UPS1; Sigma), and CRM470 from the European Union Institute for Reference Materials and Measurements. Although defined mixtures of peptides or proteins can address some benchmarking and QC needs, there is an additional need for more complex reference materials to fully represent the challenges of LC-MS data acquisition in complex matrices encountered in biological samples (2, 3).
Although it has not been widely distributed as a reference material, the yeast Saccharomyces cerevisiae proteome has been extensively used by the proteomics community to characterize the capabilities of a variety of LC-MS-based approaches (4–15). Yeast provides a uniquely attractive complex performance standard for several reasons. Yeast encodes a complex proteome consisting of ∼4,500 proteins expressed during normal growth conditions (7, 16–18). The concentration range of yeast proteins is sufficient to challenge the dynamic range of conventional mass spectrometers; the abundance of proteins ranges from fewer than 50 to more than 106 molecules per cell (4, 15, 16). Additionally, it is the most extensively characterized complex biological proteome and the only one associated with several large scale studies estimating the abundance of all detectable proteins (5, 9, 16, 17, 19, 20) as well as LC-MS/MS data sets showing good correlation between LC-MS/MS detection efficiency and the protein abundance estimates (4, 11, 12, 15). Finally, it is inexpensive and easy to produce large quantities of yeast protein extract for distribution.
In this study, we describe large scale production of a yeast S. cerevisiae performance standard, which we offer to the community through NIST. Through a series of interlaboratory studies, we created a reference data set characterizing the yeast performance standard and defining reasonable performance of ion trap-based LC-MS platforms in expert laboratories using a series of performance metrics. This publicly available data set provides a basis for additional laboratories using the yeast standard to benchmark their own performance as well as to improve upon the current status by evolving protocols, improving instrumentation, or developing new technologies. Finally, we demonstrate how the yeast performance standard, spiked with human proteins, can be used to benchmark the power of proteomics platforms for detection of differentially expressed proteins at different levels of concentration in a complex matrix.
EXPERIMENTAL PROCEDURES
Generation of Yeast Protein Performance Standard
An SOP for preparation of the yeast performance standard was developed based on the approach of Piening et al. (12) with modifications to allow for scale-up. Production was outsourced to Boston Biochem (Cambridge, MA). The full protocol is given in supplemental Section A; initial characterization of the preparation is presented in supplemental Section B. In brief, S. cerevisiae strain BY4741 (MATa, leu2Δ0, met15Δ0, ura3Δ0, his3Δ1) was grown in a 10-liter batch of rich (yeast extract peptone dextrose) medium at 30 °C in a fermentor to an A600 of 0.93. The yeast were harvested by continuous flow centrifugation (yield, 5.4 g wet weight), and the cell pellet was washed three times with ice-cold water. The cells were lysed by incubation with ice-cold trichloroacetic acid (10% final concentration in 160-ml total volume) for 1 h at 4 °C. The protein precipitate was collected by centrifugation, washed twice with 160 ml of cold 90% acetone, and pelleted again. The resulting material was lyophilized and stored at −80 °C. The total yield of lyophilized yeast lysate was ∼0.75 g.
Preparation of Digested Yeast Lysate for Study Sample Preparation
Lyophilized yeast lysate (∼11 mg) was reconstituted in 50 mm ammonium bicarbonate containing 2 mg/ml RapiGest SF (Waters), heated at 60 °C for 45 min, and sonicated for 5 min on ice. Next, 50 mm DTT in 50 mm ammonium bicarbonate was added to yield a final DTT concentration of 5 mm, and the sample was incubated at 60 °C for 30 min. After cooling to room temperature, 200 mm iodoacetamide in water was added to yield a final concentration of 10 mm, and the alkylation reaction was left to proceed at room temperature in the dark for 30 min. To quench alkylation, 100 mm DTT in 50 mm ammonium bicarbonate was added to the sample to yield a final concentration of 10 mm. Prior to the addition of trypsin, an additional volume of 50 mm ammonium bicarbonate was added to the sample to reduce the RapiGest concentration to 0.1%. Trypsin (0.5 μg/μl in 20 mm aqueous HCl) was then added to the yeast lysate sample in a 1:50 ratio to the total protein amount. The sample was digested overnight (18 h) at 37 °C with gentle swirling. After digestion, to inactivate trypsin and cleave the RapiGest, concentrated trifluoroacetic acid was added to the sample to yield a concentration of 0.5%. The sample was then incubated again at 37 °C for 60 min followed by centrifugation at 10,000 rpm for 10 min. The supernatant was transferred to a new sample tube and lyophilized to dryness; after lyophilization, the dried digest was resuspended in 0.1% aqueous formic acid to yield a concentration that would correspond to ∼60 ng/μl total yeast protein prior to digestion. Where indicated, 48 human proteins (Sigma UPS1) were spiked into the reconstituted yeast performance standard (supplemental Section C).
LC-MS/MS Methods
Each laboratory was asked to follow an SOP for collection of all data in Study 6. A detailed description of the SOP is provided in supplemental Section C. Parameters and settings specified in the SOPs were derived by a combination of consensus among the participants and limited method optimization studies. The SOPs do not represent fully optimized methods and are not intended to be prescriptive for the field. The SOPs were used instead to minimize variation due to factors that could be anticipated and controlled. Each laboratory was allowed to use its own favorite protocol for Study 8, and the individual protocols are summarized in supplemental Section D. Four models of mass spectrometer were used: LTQ, LTQ-XL, LTQ-XL-Orbitrap, and LTQ-Orbitrap (see supplemental section J). In each case, MS/MS spectra were collected in the LTQ. For the LTQ-Orbitrap instruments, MS1 spectra used to determine the precursors selected for MS/MS were collected at 60,000 resolution in the Orbitrap. These high resolution scans enabled precursor selection to be limited to precursors that exhibited both a charge of 2+ or higher and an isotope cluster from which the monoisotopic peak could be discerned. The low resolution MS1 scans on LTQ instruments did not enable these precursor selection criteria. A complete description of the acquisition parameters and other instrument configuration parameters can be found in supplemental Sections C, D, and J.
Database Search Pipeline
For Studies 6 and 8, centroided tandem mass spectra were converted to peak lists in mzXML format by the msConvert tool of ProteoWizard 1.6.0 (21). The software was configured to centroid MS scans. Peptides were identified against the S. cerevisiae Genome Database orf_trans_all, downloaded April 6, 2007. These 6,718 sequences were augmented by 48 UPS1 sequences (Sigma) (supplemental Section E), 23 NCI20 sequences (supplemental Section F), and 74 contaminant protein sequences; the full database was then doubled in size by adding the reversed version of each sequence. The FASTA file is available at http://cptac.tranche.proteomecommons.org/. The MyriMatch database search algorithm version 1.6.0 (22) identified tandem mass spectra to peptide sequences. Semitryptic peptide candidates were included as possible matches. The configuration defined proteolytic cleavage sites after any Lys or Arg (whether or not Pro was the next residue) or after a Met at the N terminus of a protein, allowing for up to two missed cleavages. Potential modifications included oxidation of methionines, formation of N-terminal pyroglutamine, deamidation of Asn-Gly motifs, and carbamidomethylation of cysteines, all as variable modifications. For the LTQ, precursors were allowed to be up to 1.25 m/z from the average mass of the peptide. For the Orbitrap, precursor ions were required to fall within 10 ppm of the database peptide with ppm computed from m/z values. To retain identifications in which the peptide monoisotope had been miscalled by the instrument control software, MyriMatch also sought matches in which a neutron had been added to or subtracted from each database peptide. Fragment ions were uniformly required to fall within 0.5 m/z of the monoisotope. IDPicker version 2.5 (23, 24) applied a 2% false identification rate per raw file at the peptide-spectrum match level and applied parsimony to the protein lists, requiring all proteins to match at least two distinct peptide sequences and to match at least 13 spectra (one per instrument per study). The two-peptide rule was applied globally, not by instrument. Hence, proteins on the list might have a single peptide for a given instrument. In contrast, the two-peptide rule was applied per raw file in the statistics code that generated the outputs displayed in Tables I and III as well as Figs. 2 and 3. IDPicker reports can be downloaded from http://cptac.tranche.proteomecommons.org/.
Table I. Summary of LC-MS results, average and CV (in percentages), for unspiked yeast reference proteome.
pept., peptide(s); Avg, average.
| Total no. of yeast spectra |
Total no. of yeast pept. sequences |
Yeast proteins identified using |
CN50a |
Performance metricsb |
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 pept. |
2 pept. |
>2 pept. |
C-3A |
C-2A |
DS-2B |
IS-3B |
||||||||||||||
| Avg | CV | Avg | CV | Avg | CV | Avg | CV | Avg | CV | Avg | CV | Avg | CV | Avg | CV | Avg | CV | Avg | CV | |
| A. Study 8 (600 ng loaded on column) | ||||||||||||||||||||
| Instrument@Lab | ||||||||||||||||||||
| LTQ@73 | 7,600 | 2.0 | 5,401 | 1.3 | 343 | 1.1 | 181 | 5.3 | 574 | 1.9 | 18,039 | 2.7 | 10.1 | 1.5 | 32 | 1.0 | 5,374 | 1.2 | 0.4 | 0.4 |
| LTQ2@95 | 4,568 | 0.5 | 3,257 | 0.6 | 321 | 4.5 | 165 | 9.9 | 404 | 1.1 | 27,416 | 2.6 | 30.2 | 0.9 | 37 | 2.0 | 5,272 | 2.0 | 0.5 | 0.6 |
| LTQ-XLx@65 | 4,548 | 5.5 | 3,901 | 5.3 | 363 | 7.0 | 169 | 0.3 | 477 | 5.8 | 22,393 | 5.6 | 16.3 | 0.8 | 32 | 2.5 | 4,930 | 3.7 | 0.3 | 21.4 |
| Orbitrap@86 | 6,914 | 1.3 | 5,488 | 1.8 | 344 | 2.0 | 166 | 4.8 | 602 | 1.4 | 17,738 | 1.9 | 14.0 | 0.2 | 41 | 0.9 | 6,853 | 2.1 | 0.3 | 5.4 |
| OrbitrapO@65 | 5,998 | 0.9 | 5,023 | 1.8 | 363 | 4.0 | 190 | 8.7 | 572 | 3.6 | 17,959 | 1.6 | 19.4 | 1.0 | 32 | 1.9 | 4,569 | 2.2 | 0.3 | 4.7 |
| OrbitrapW@56 | 8,336 | 1.0 | 7,211 | 1.0 | 313 | 3.2 | 182 | 5.4 | 724 | 0.8 | 13,656 | 3.0 | 12.7 | 0.7 | 35 | 1.4 | 5,695 | 1.0 | 0.4 | 0.8 |
| Interlaboratory variation | ||||||||||||||||||||
| All LTQs | 5,572 | 31.5 | 4,186 | 26.3 | 343 | 6.1 | 171 | 5.0 | 485 | 17.6 | 22,616 | 20.7 | 18.9 | 47.3 | 34 | 7.4 | 5,192 | 4.4 | 0.4 | 17.0 |
| All Orbitraps | 7,083 | 16.6 | 5,907 | 19.5 | 340 | 7.4 | 179 | 7.0 | 633 | 12.7 | 16,451 | 14.7 | 15.4 | 20.1 | 36 | 10.5 | 5,706 | 17.4 | 0.3 | 5.6 |
| B. Study 8 (120 ng loaded on column) | ||||||||||||||||||||
| Instrument@Lab | ||||||||||||||||||||
| LTQ@73 | 4,583 | 6.5 | 3,563 | 4.9 | 278 | 5.5 | 118 | 12.5 | 429 | 3.2 | 26,282 | 4.9 | 10.1 | 1.5 | 27 | 1.3 | 4,038 | 1.3 | 0.3 | 5.5 |
| LTQ2@95 | 2,124 | 12.1 | 1,665 | 13.9 | 229 | 4.6 | 95 | 5.3 | 230 | 11.9 | 58,656 | 14.2 | 32.1 | 2.5 | 29 | 9.0 | 4,081 | 8.6 | 0.4 | 6.0 |
| LTQ-XLx@65 | 2,367 | 2.3 | 2,128 | 1.9 | 275 | 5.9 | 117 | 7.3 | 291 | 4.5 | 44,157 | 6.9 | 16.4 | 1.7 | 26 | 3.4 | 3,876 | 3.4 | 0.3 | 1.6 |
| Orbitrap@86 | 5,654 | 2.1 | 5,081 | 1.9 | 331 | 1.3 | 174 | 12.7 | 573 | 0.8 | 18,159 | 1.9 | 13.9 | 0.5 | 34 | 2.0 | 5,075 | 1.4 | 0.2 | 17.1 |
| OrbitrapO@65 | 4,383 | 1.1 | 3,868 | 1.1 | 347 | 2.5 | 178 | 1.1 | 474 | 3.2 | 22,615 | 1.2 | 18.2 | 1.4 | 25 | 2.5 | 3,365 | 0.5 | 0.2 | 1.7 |
| OrbitrapW@56 | 6,491 | 1.7 | 5,902 | 2.0 | 344 | 2.4 | 187 | 5.9 | 649 | 0.8 | 15,305 | 0.1 | 12.5 | 0.8 | 32 | 2.8 | 4,762 | 2.7 | 0.3 | 0.2 |
| Interlaboratory variation | ||||||||||||||||||||
| All LTQs | 3,025 | 44.8 | 2,452 | 40.4 | 261 | 10.5 | 110 | 11.8 | 317 | 32.1 | 43,032 | 37.6 | 19.5 | 50.5 | 27 | 7.3 | 3,998 | 5.3 | 0.3 | 23.8 |
| All Orbitraps | 5,509 | 19.3 | 4,951 | 20.7 | 341 | 2.5 | 180 | 3.5 | 565 | 15.6 | 18,693 | 19.7 | 14.9 | 17.3 | 30 | 13.4 | 4,401 | 18.0 | 0.2 | 19.6 |
| C. Study 6 SOP (unspiked yeast; 120 ng loaded on column) | ||||||||||||||||||||
| Instrument@Lab | ||||||||||||||||||||
| LTQ@73 | 2,941 | 3.8 | 2,434 | 3.4 | 326 | 2.3 | 146 | 9.8 | 320 | 0.8 | 35,474 | 3.4 | 20.8 | 0.3 | 29 | 4.1 | 4,238 | 4.5 | 0.3 | 16.0 |
| LTQ2@95 | 2,606 | 4.1 | 2,140 | 6.1 | 264 | 5.4 | 113 | 10.5 | 284 | 8.3 | 43,969 | 2.7 | 21.6 | 0.0 | 30 | 1.3 | 4,181 | 2.1 | 0.4 | 5.5 |
| LTQ-XLx@65 | 2,571 | 3.4 | 2,165 | 4.9 | 287 | 1.8 | 127 | 6.4 | 291 | 8.1 | 41,256 | 11.6 | 24.2 | 2.1 | 27 | 7.5 | 4,168 | 7.3 | 0.3 | 7.0 |
| Orbitrap@86 | 2,794 | 14.0 | 2,623 | 13.9 | 387 | 3.3 | 188 | 5.7 | 368 | 12.9 | 30,353 | 16.7 | 18.3 | 4.9 | 23 | 9.2 | 3,323 | 9.7 | 0.0 | 26.5 |
| OrbitrapO@65 | 5,022 | 3.7 | 4,627 | 4.4 | 391 | 4.9 | 212 | 6.6 | 565 | 4.0 | 18,218 | 5.2 | 19.9 | 1.5 | 29 | 5.8 | 4,378 | 6.0 | 0.3 | 1.6 |
| OrbitrapW@56 | 3,861 | 6.7 | 3,550 | 6.6 | 344 | 7.8 | 162 | 6.1 | 492 | 5.4 | 25,061 | 5.1 | 19.8 | 0.2 | 26 | 2.8 | 3,762 | 3.0 | 0.3 | 3.6 |
| OrbitrapP@65 | 4,453 | 7.9 | 4,116 | 7.3 | 341 | 4.3 | 165 | 3.0 | 439 | 5.3 | 21,777 | 8.7 | 15.5 | 2.1 | 30 | 1.4 | 4,489 | 3.9 | 0.4 | 2.1 |
| Interlaboratory variation | ||||||||||||||||||||
| All LTQs | 2,706 | 7.5 | 2,247 | 7.3 | 292 | 10.8 | 129 | 13.1 | 298 | 6.4 | 40,233 | 10.8 | 22.2 | 7.0 | 28 | 5.8 | 4,196 | 4.5 | 0.3 | 24.6 |
| All Orbitraps | 4,033 | 23.6 | 3,729 | 23.0 | 366 | 7.4 | 182 | 12.7 | 466 | 17.9 | 23,852 | 21.6 | 18.4 | 10.4 | 27 | 12.3 | 3,988 | 13.4 | 0.3 | 51.1 |
a CN50 values denote the copy number corresponding to 50% probability of detection for a randomly selected yeast protein. CN50 values are derived from logistic regression coefficients obtained by regressing yeast protein detection (yes/no) against log10 TAP copy number (12). CN50 is then converted to the copy number scale by taking 10 to the CN50 power. All mean and CV calculations are performed on the copy number scale. (CN50 values for all runs of all instruments are provided in supplemental Section H.)
b Four performance metrics (described in supplemental Section G and in Table II), designed to diagnose LC-MS issues, are provided for individual instruments. C-3A is median peak widths for unique peptides; C-2A is retention period over which 50% of the identified peptides eluted; DS-2B is the number of MS2 spectra produced over C-2A; IS-3B is the ratio of the number of 3+/2+ charge states for all peptide identifications.
Table III. Use of spiked yeast reference to estimate sensitivity and specificity of biomarker candidate discovery.
To demonstrate the utility of the reference proteome for benchmarking performance of biomarker discovery strategies, we used the Study 6 spiked yeast reference data to investigate the power of detecting potential biomarkers under typical case-control settings. The data were analyzed using the SASPECT method (25), which uses a probability model to make inferences about protein abundances based on peptide detection. Permutation testing is used to estimate the FDR.
| Case sample | No. identified biomarkersa | No. true positivesb | No. true negativesc | No. false positivesd | No. false negativese | Sensitivityf | Specificityg |
|---|---|---|---|---|---|---|---|
| Study 6 SOP (yeast + 0.25 fmol/μl UPS1) | 0 | 0 | 2,522 | 0 | 48 | 0.00 | 1.00 |
| Study 6 SOP (yeast + 0.74 fmol/μl UPS1) | 4 | 4 | 2,522 | 0 | 44 | 0.08 | 1.00 |
| Study 6 SOP (yeast + 2.2 fmol/μl UPS1) | 25 | 25 | 2,522 | 0 | 23 | 0.52 | 1.00 |
| Study 6 SOP (yeast + 6.7 fmol/μl UPS1) | 34 | 34 | 2,522 | 0 | 14 | 0.71 | 1.00 |
| Study 6 SOP (yeast + 20 fmol/μl UPS1) | 49 | 40 | 2,513 | 9 | 8 | 0.83 | >0.99 |
a Identified biomarkers are proteins with FDR ≤0.01.
b True positives (TP) are human proteins identified as “biomarkers” in the SASPECT analysis.
c True negatives (TN) are yeast proteins with FDR >0.01.
d False positives (FP) are yeast proteins with FDR ≤0.01.
e False negatives (FN) are human proteins with FDR >0.01.
f Sensitivity is calculated as TP/(TP + FN).
g Specificity is calculated as TN/(TN + FP).
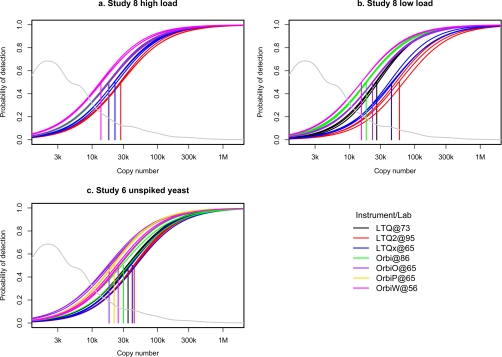
Fig. 2.
Detection efficiency of yeast proteins relative to their abundance. The gray curve shown in each panel denotes the TAP tag copy number distribution yeast proteins (derived from Ghaemmaghami et al. (16)); the abundance of proteins ranges from fewer than 50 to more than 106 molecules per cell, and only 9% of yeast proteins have copy numbers greater than 20,000. Logistic regression curves (in color) indicate the probability of protein detection as a function of copy number for each run. The vertical lines indicate the mean copy number (for each instrument) corresponding to 50% probability of detection (CN50). Smaller CN50 values indicate greater depth of proteome sampling. The graph indicates that, on average, only the most abundant yeast proteins have a high probability of being detected in this one-dimensional LC-MS analysis. a shows the results for high protein loading (600 ng on column) when each lab uses their typical (non-SOP) testing protocol (Study 8). b shows the results for low protein loading (120 ng on column) using the same (non-SOP) protocol. c shows the results obtained from the SOP (Study 6; 120 ng on column). As expected, CN50 is increased in the low protein loading group compared with the high loading group (p < 0.0001). For equivalent “detectability,” a randomly selected protein must be present at nearly 40,200 copies per cell in the low loading group versus 24,650 copies per cell at high loading (95% confidence interval for the difference 11,150 to 20,470).
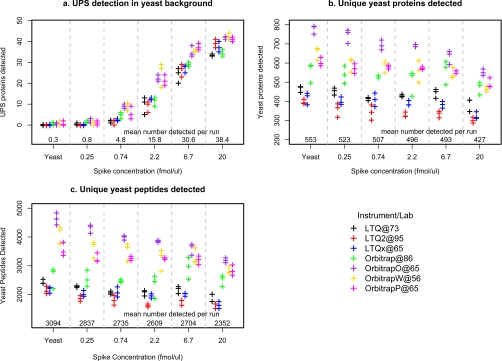
Fig. 3.
Detection efficiency of human proteins (UPS1) spiked into yeast background. This figure summarizes detection of UPS1 (supplemental Section E) and yeast proteins in the spiking experiments of Study 6. For all panels, the result of each RPLC run is indicated by a “+” plotting symbol; colors denote different instruments. Plotting symbols have been offset to avoid overlap of identical values. Protein detection is defined as observing two or more peptides mapping to the same protein (in a single RPLC run). On the x axis, “Spike concentration” refers to the concentration of the 48 equimolar human proteins (UPS1) spiked into the yeast matrix. Also on the x axis, “Yeast” refers to the unspiked matrix (i.e. 0 fmol/μl UPS1). a shows that the number of detected UPS1 proteins increases with increasing spike concentration. Note that at an equimolar spike concentration of 2.2 fmol/μl all instruments detect at least one UPS1 protein in each run. b and c show, respectively, the total number of yeast proteins and peptides detected per RPLC run. Different instruments show large variation in both the number of proteins/peptides detected and in the response to increasing spike concentration (p < 0.0001).
Statistical Methods
For Table I, we used a dimensionless measure, the coefficient of variation, to compare the within and between laboratory variation of the total number of spectra, sequences, and the total number of proteins. The coefficient of variation is the normalized measure of dispersion, computed as the ratio of the standard deviation to the mean of the data. The within lab CV% is computed as the ratio of the standard deviation of the response over all runs (times 100) for each lab to its mean. The between lab CV% is the ratio of the standard deviation of the means of each lab to its overall mean (times 100). For Tables I and II, a summary of the performance metrics is provided in supplemental Section G; a detailed description of these (and additional) metrics can be found in the accompanying study by Rudnick et al. (32). Furthermore, the software pipeline used to calculate the performance metrics is available for download from NIST.
Table II. Metrics for differential diagnosis of LC-MS performance issues.
ID'd, identified; peps, peptides; dev, deviation; ID, identification; S/N, signal to noise ratio.
| Metric codea | Description of metrica | Study 6 (unspiked yeast, 120 ng) |
Study 8 (120 ng) |
Study 8 (600 ng) |
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Mean | Mean intralab %dev | Interlab %dev | Mean | Mean intralab %dev | Interlab %dev | Mean | Mean intralab %dev | Interlab %dev | ||
| A. LTQs | ||||||||||
| Chromatography | ||||||||||
| C-2A | Retention period over which 50% of peps were ID'd (min) | 28.4 | 3.0 | 4.8 | 27.2 | 3.4 | 6.1 | 33.7 | 1.4 | 8.3 |
| C-2B | Peptide ID rate during C-2A | 46.4 | 4.4 | 5.5 | 51.5 | 3.1 | 30.0 | 61.7 | 1.8 | 26.3 |
| C-3A | Median peak width for unique peps (s) | 22.2 | 0.6 | 8.0 | 19.5 | 1.4 | 58.3 | 18.9 | 0.8 | 54.6 |
| Dynamic sampling | ||||||||||
| DS-1 | Median ratio of peps identified once/twice | 8.3 | 7.2 | 16.1 | 7.4 | 4.3 | 35.0 | 6.9 | 2.8 | 41.6 |
| DS-1B | Median ratio of peps identified twice/thrice | 5.1 | 12.2 | 16.6 | 6.6 | 10.2 | 24.6 | 5.5 | 7.0 | 22.4 |
| DS-2A | No. MS1 scans taken over C-2A | 541.0 | 2.2 | 2.6 | 715.7 | 3.8 | 21.8 | 959.6 | 1.6 | 22.0 |
| DS-2B | No. of MS2 spectra produced over C-2A | 4,195.7 | 3.2 | 0.9 | 3,998.2 | 3.3 | 2.7 | 5,192.2 | 1.7 | 4.5 |
| Ion source | ||||||||||
| IS-1A | Count where MS1 signal jumped >10× in adjacent full scans | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 3.7 | 66.7 | 0.0 |
| IS-1B | Count where MS1 signal fell >10× in adjacent full scans | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 4.0 | 66.7 | 0.0 |
| IS-2 | Median precursor m/z for IDs | 637.2 | 0.6 | 0.4 | 612.5 | 0.8 | 4.2 | 670.1 | 0.3 | 5.2 |
| IS-3B | Ratio of 3+/2+ for all IDs | 0.3 | 7.0 | 26.9 | 0.3 | 3.1 | 27.0 | 0.4 | 5.7 | 16.8 |
| MS1 | ||||||||||
| MS1-1 | MS1 ion injection time (ms) | 3.6 | 13.6 | 32.9 | 5.2 | 1.9 | 44.8 | 1.3 | 0.0 | 43.3 |
| MS1-2A | Median MS1 S/N value for spectra up to and including C-2A | 127.2 | 15.0 | 39.5 | 104.1 | 9.1 | 43.9 | 159.2 | 6.2 | 36.5 |
| MS2 | ||||||||||
| MS2-2 | Median S/N for ID'd MS2 spectra | 62.0 | 2.8 | 13.8 | 82.1 | 3.2 | 73.2 | 142.8 | 1.2 | 51.8 |
| Peptide identification | ||||||||||
| P-2C | No. of unique tryptic peps | 2,566.2 | 2.2 | 3.9 | 2,717.9 | 4.8 | 27.6 | 3,988.4 | 1.9 | 20.4 |
| Median | 3.0 | 5.5 | 3.2 | 27.0 | 1.8 | 22.0 | ||||
| B. Orbitraps | ||||||||||
| Chromatography | ||||||||||
| C-2A | Retention period over which 50% of peps were ID'd (min) | 27.1 | 3.3 | 12.6 | 30.1 | 1.9 | 15.3 | 36.3 | 1.1 | 12.1 |
| C-2B | Peptide ID rate during C-2A | 65.6 | 3.7 | 13.2 | 73.2 | 2.4 | 17.6 | 72.6 | 1.9 | 26.3 |
| C-3A | Median peak width for unique peps (s) | 18.4 | 1.6 | 11.2 | 14.9 | 0.7 | 20.0 | 15.4 | 0.5 | 23.2 |
| Dynamic sampling | ||||||||||
| DS-1 | Median ratio of peps identified once/twice | 17.8 | 3.6 | 20.5 | 21.1 | 5.5 | 28.7 | 19.1 | 5.3 | 18.7 |
| DS-1B | Median ratio of peps identified twice/thrice | 7.5 | 11.2 | 26.6 | 6.2 | 7.7 | 5.7 | 5.4 | 7.7 | 7.5 |
| DS-2A | No. of MS1 scans taken over C-2A | 529.0 | 3.3 | 11.3 | 656.9 | 2.2 | 11.0 | 902.2 | 0.7 | 14.0 |
| DS-2B | No. of MS2 spectra produced over C-2A | 3,988.0 | 4.0 | 13.7 | 4,400.8 | 1.2 | 20.7 | 5,705.6 | 1.3 | 20.0 |
| Ion source | ||||||||||
| IS-1A | Count where MS1 signal jumped >10× in adjacent full scans | 32.3 | 133.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| IS-1B | Count where MS1 signal fell >10× in adjacent full scans | 24.7 | 133.3 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 | 0.0 |
| IS-2 | Median precursor m/z for IDs | 608.6 | 0.9 | 3.9 | 593.1 | 0.6 | 2.1 | 639.6 | 0.3 | 3.3 |
| IS-3B | Ratio of 3+/2+ for all IDs | 0.3 | 6.3 | 56.4 | 0.2 | 4.8 | 21.2 | 0.3 | 2.7 | 5.0 |
| MS1 | ||||||||||
| MS1-1 | MS1 ion injection time (ms) | 85.7 | 5.0 | 39.3 | 109.3 | 6.0 | 28.0 | 29.8 | 4.4 | 33.7 |
| MS1-2A | Median MS1 S/N value for spectra up to and including C-2A | 481.9 | 7.5 | 37.7 | 574.4 | 3.4 | 27.6 | 652.0 | 2.4 | 16.0 |
| MS2 | ||||||||||
| MS2-2 | Median MS2 S/N for IDs | 66.6 | 5.0 | 34.1 | 76.4 | 5.0 | 33.4 | 143.6 | 2.4 | 25.3 |
| Peptide identification | ||||||||||
| P-2C | No. of unique tryptic peps | 3,463.8 | 4.2 | 22.8 | 4,329.3 | 1.8 | 26.8 | 5,069.1 | 0.8 | 22.4 |
| Median | 4.2 | 13.7 | 2.2 | 20.0 | 1.3 | 16.0 | ||||
a A series of metrics (see supplemental Section G and the accompanying study by Rudnick et al. (32)) was computed (from the historical benchmarking data sets described herein) to demonstrate average values obtained for both the LTQ instruments (part A) and the Orbitrap instruments (part B). Intra- and interlaboratory variation is also shown.
For Table III, we treated the spiked sample as a case sample and the pure yeast reference sample as a control sample. For each sample, we used the data resulting from all 21 independent LC-MS/MS replica runs (nine from LTQ instruments and 12 from Orbitrap instruments). The data were analyzed using the SASPECT (significant analysis of peptide counts) method described elsewhere (25). Briefly, SASPECT uses a probability model to make inferences about protein abundances based on peptide detection, and permutation testing is used to estimate the false discovery rate (FDR) of the analysis.
For Fig. 2, logistic regression was used to evaluate the association between TAP copy number and detection of yeast proteins in Studies 6 and 8. (“TAP copy number” refers to the estimated molecules per cell of a given protein, derived from the data of Ghaemmaghami et al. (16). The logarithm (base 10) of copy number was used as the regressor to improve model fit to the data. Although TAP copy number estimates include measurement error (16), this uncertainty was not reflected in the logistic regression analysis. We report a summary measure of performance for each RPLC run, the CN50. This statistic estimates the copy number corresponding to 50% probability of detection for a randomly selected yeast protein. Smaller CN50 values indicate greater depth of proteome sampling. CN50 is easily derived from the logistic regression coefficients. Mixed effects linear models were used for the subsequent analysis of CN50 values and for yeast peptide and protein counts in Studies 6 and 8. This procedure allows simultaneous estimation of potential effects of protein spikes as well as inter- and intrainstrument variability. (See supplemental Section H for the statistical analysis code for computing the CN50 values as well as a complete table of CN50 values.)
Public Access to Data
To manage the large number of data files generated for these studies, a password-protected web site was developed. This site, hosted at NIST, was designed as a portal used by the participating laboratories for initiating uploads and downloads of large data files. Information for each team including labs and instruments was preloaded into the system, stored in a MySQL database. At the beginning of each study, the participating instruments at each lab were added to the study, creating hyperlinks by which data could be uploaded. Importantly, all uploads were then unambiguously tied to the originating instrument and study along with a date stamp.
The data transfers were performed using Tranche, an open source, secure peer-to-peer file sharing tool. A custom user interface for use by participating laboratories was developed and added to the Tranche code base. This custom tool allowed the web site and database to communicate tracking information with Tranche via custom URLs. When uploads finished, the Tranche hash (a unique data identifier) and pass phrase were automatically recorded into the web site's database. These stored links allow for subsequent retrieval of the data files using the Tranche download tool. The Tranche hashes and pass phrases provide a simple and portable way to access data sets, including relatively large data sets, and can be easily associated with supporting annotation. Once published, the data will be made available to the community via Tranche at the data archival page, http://cptac.tranche.proteomecommons.org/.
Availability of Yeast Performance Standards
The certified yeast reference material for proteomics use is under development at the NIST to meet the long term needs of the community and will be available in 2010. In the interim, aliquots of the yeast performance standard described in this study are available through NIST.2
RESULTS
Production and Characterization of Yeast Performance Standard
A standard operating procedure for preparation of the yeast performance standard was developed based on the approach of Piening et al. (12) with modifications to allow for scale-up (supplemental Section A). Production was outsourced to a commercial vendor, and the protein preparation was transferred to NIST for initial characterization (supplemental Section B). In total, 0.75 g of protein was obtained from 10 liters of yeast culture. The certified yeast reference material for proteomics use is under development at NIST to meet the long term needs of the community. In the interim, aliquots of the yeast performance standard described in this study are available through NIST.
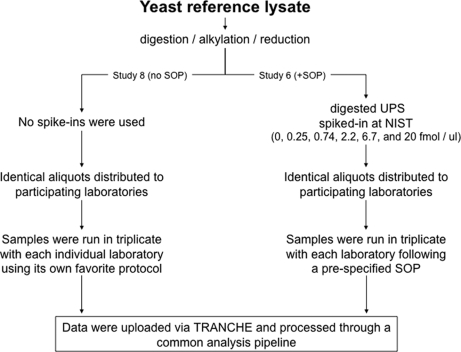
Identical aliquots of the trypsin-digested performance standard were distributed to multiple participating laboratories to generate a data set characterizing the performance standard and determining the degree of variation in the performance of LC-MS platforms across participating laboratories. Five independent laboratories, seven independent instruments, and four instrument models (LTQ, LTQ-XL, LTQ-XL-Orbitrap, and LTQ-Orbitrap) were included in the study. Two sets of analyses were performed (Fig. 1). For both sets, all sample processing (i.e. trypsin digestion, alkylation, and reduction) was done centrally at NIST, and all data were submitted and analyzed through a single analysis pipeline; hence, any experimental variation between laboratories was due to differences in LC-MS performance. In one set of analyses (“Study 6”), each sample was also run in triplicate (120 ng loaded on column); however, each laboratory was asked to follow a predefined SOP dictating HPLC and MS parameters (supplemental Section C). In addition, a series of samples was generated in which a mixture of 48 human proteins (Sigma UPS1) was spiked into the yeast performance standard at several concentrations, and each of these spiked samples was also analyzed in triplicate using the SOP. In a second set of analyses (“Study 8”), each participating laboratory was asked to perform six shotgun MS/MS runs of the performance standard (three runs of each of 120 and 600 ng loaded on column) with each laboratory using its own favorite LC-MS protocol.
Fig. 1.
Overview of analyses of yeast performance standard. Samples were processed centrally at NIST, and identical aliquots were distributed to the participating laboratories for LC-MS/MS analyses. Each sample was analyzed in triplicate, and the data were processed and analyzed centrally using a single analysis pipeline. For Study 6, each laboratory conformed to a prespecified SOP (supplemental Section C). For Study 8, no SOP was instituted, and the individual methods of each laboratory are described in supplemental Section D.
The results for the unspiked yeast performance standard are summarized in Table I. The unspiked samples in Study 6 produced 72,743 identified spectra that translated into 11,822 distinct peptides. (When spiked samples from Study 6 were also included, the search produced 407,836 high confidence identifications accounting for 17,904 distinct peptides.) Study 8 (all samples) produced 191,311 identified spectra for 17,333 distinct peptides. In Study 8, 120 ng on column yielded 76,808 identified spectra related to 11,622 peptides, and for the cases in which 600 ng were loaded, the search rendered 113,892 high confidence identifications accounting for 15,464 distinct peptides.
As expected, the Orbitrap instruments identified significantly more peptides than the LTQ instruments across all three data sets (Table I, parts A–C; p value = 0.013, t test). Detection sensitivity could also be monitored using the performance standard. The abundance of ∼3,800 yeast proteins has been estimated by quantitative Western blotting (16), and previous studies have shown strong correlation between these protein abundance measurements and detection efficiency by LC-MS (4, 12). This strong correlation is recapitulated in our analyses of the yeast performance standard (Fig. 2) with the Orbitrap instruments overall showing slightly higher detection efficiencies than the LTQ instruments (Table I, CN50 values; p value = 0.029, t test). Intra and interlaboratory variation was also calculated from the data set. Across the vast majority of parameters (Table I), intralaboratory variation was smaller than interlaboratory variation. For example, for depth of proteome sampling (Table I, CN50 values), intralaboratory variation was significantly lower than interlaboratory variation (p value ≤0.028 by one-sided paired two-sample t test).
Use of Performance Metrics for Diagnosing LC-MS Platform Problems
In work leading up to these interlaboratory studies, occasional runs were observed wherein the number of high confidence peptide identifications was significantly less than expected based on the bulk of data. In an effort to identify and monitor the factors contributing to this variability, a set of LC-MS performance metrics was developed. These metrics, described in detail in the accompanying study by Rudnick et al. (32), fall into six classes (chromatography, dynamic sampling, ion source, MS1 signal, MS2 signal, and peptide identifications), each designed to monitor the performance of a different aspect of the LC-MS platform (Table II). These metrics were computed for the yeast proteome reference data set. The 15 metrics showing the highest variation across the yeast reference data set are shown in Table II, and four of these (C-3A, C-2A, DS-2B, and IS-3B) are reported for individual instruments in Table I.
The performance metrics can be used to diagnose the underlying cause when suboptimal data are obtained. For example, early in the course of these experiments, it was noted that data from the LTQ@73 instrument (laboratory identities are coded numerically) consistently produced fewer peptide identifications than the other instruments in the study. Once the performance metrics were calculated, it became apparent that the retention period over which 50% of peptides were identified (performance metric C-2A) on LTQ@73 was significantly shorter than the average for the other LC-MS/MS platforms. In the affected data, this prime zone for peptide identification lasted only 21.93 min for LTQ@73 compared with an average of 31.03 min for the other platforms, indicating a contraction of the chromatography. Upon closer examination of the chromatography system, a degraded pump seal and contaminated check valve were identified despite no indication of performance degradation in measured flow rate or pressure. Following repair of the HPLC system, manual recalibration of the flow rates, and implementation of a lower flow rate for sample loading on the column, the retention time duration of the inner half of peptide identifications increased to 31.3 min (supplemental Section I).
In the transition from Study 6 (+SOP) to Study 8 (no SOP), the three instruments that had the most substantial changes to their procedures increased their numbers of matched spectra for the 120-ng sample by 56% (LTQ@73), 68% (Orbitrap@56), and 102% (Orbitrap@86). The other three instruments, all of which made more minor deviations from the SOP, increased or decreased their numbers of matched spectra by <20%. The number of peptides identified for LTQ@73, Orbitrap@56, and Orbitrap@86 correlated tightly with the median peak chromatographic width (metric C-3A; Table I, parts A and B), indicating the importance of this metric in peptide/protein identification yield. In both of these cases, smaller dimension packing material was used in these specific laboratories (supplemental Section D), accounting for the differences in peak widths observed. The sensitivity of Orbitrap@56 and Orbitrap@86 was also further improved by reducing the column diameters and flow rates from the Study 6 SOP configuration of 100-μm inner diameter at 600 nl/min. In Study 8, both instruments used 75-μm-inner diameter columns with flow rates of 200 (Orbitrap@56) and 400 nl/min (Orbitrap@86). Orbitrap@86 also had a further boost in Study 8 from improved ionization. In Table I, part C, for Study 6, the performance metrics flagged an ionization issue. Orbitrap@86 produced far fewer peptide identifications than the other Orbitraps in the study, and this instrument also showed a >3-fold reduction in the ratio of 3+/2+ peptide ions detected (Table I, part C, see IS-3B). The identification of triply charged peptides fell from an average of 30% of doubly charged peptides on comparable instruments to less than 10%, suggesting a problem during ionization.
Yeast Performance Standard Can Be Spiked with Exogenous Proteins to Assess Detection Efficiency
Western blotting is semiquantitative (16); hence, although we see good general correlation between protein copy number estimates and LC-MS detection efficiency (Fig. 2), we cannot determine the relationship between absolute protein concentrations and LC-MS detection efficiency using only the yeast proteins. To further quantify the detection sensitivity across the platforms, we spiked 48 human proteins (Sigma UPS1) into the yeast performance standard at a range of concentrations (0, 0.2, 0.74, 2.2, 6.7, and 20 fmol/μl) and determined the detection efficiency of these 48 equimolar proteins in the yeast matrix (Fig. 3). As expected, the number of human proteins detected increases with spike concentration (Fig. 3a). As the concentration of the spike proteins increases, the instruments showed a gradual decline in the number of yeast peptides/proteins identified, potentially due to ion suppression or competition with spiked peptides for MS/MS sequencing time (Fig. 3, b and c).
Yeast Standard, Spiked with Human Proteins, Can Be Used to Benchmark the Power of Proteomics Platforms for Detection of Differentially Expressed Proteins at Different Levels of Concentration in Complex Matrix
An increasingly common goal of proteomics is to discover proteins that are differentially expressed between two classes of samples. Frequently, biological samples are subjected to fractionation to improve depth of sampling. Even using standardized protocols, each step of sample processing introduces preanalytical variation that can lead to false positives and false negatives in comparative proteomics experiments, especially in label-free approaches where the two classes of samples are processed separately in parallel. Because of the high cost and effort to verify each potentially differentially expressed protein identified (26–28), it is imperative that the candidate discovery process is designed to minimize the FDR.
We used the Study 6 spiked yeast performance standard data for determining the FDR in a relatively simple scenario using a one-dimensional LC-MS/MS platform and a label-free approach to detect proteins differentially present in the Study 6 samples. This analysis is presented as a proof of concept for an approach that can also easily be applied to more complex analyses, such as those involving multidimensional separations. As described above, in Study 6, the standard human protein mixture was spiked into the yeast reference at five different levels. For each spike-in level, we compared the spiked sample with the unspiked yeast performance standard. Our goal was to detect the “differentially expressed” proteins between the two classes (spiked versus unspiked); because the matrix (yeast performance standard) was identical among all of the samples, only the spiked-in human proteins were differentially present. Hence, yeast proteins appearing as biomarkers are a measure of the false positive rate. Similarly, human proteins that the experimental work flow fails to identify as biomarkers provide a measure of the false negative rate.
For each sample, we used the data resulting from all 21 independent LC-MS/MS replica runs (nine from LTQ instruments and 12 from Orbitrap instruments). The data were analyzed using the SASPECT method (25), which uses a probability model to make inferences about protein abundances based on peptide detection. Permutation testing is used to estimate the FDR. The results for FDR ≤0.01 are shown in Table III. Not surprisingly, the more abundant the human proteins are, the easier they can be detected as differential. For the lowest spike-in level (0.25 fmol/μl), we could not detect any of the human proteins as differential, whereas for the highest spike-in level (20 fmol/μl), we are able to correctly identify 40 of 48 spiked-in proteins as differential.
DISCUSSION
The major purpose of this study was to provide a well characterized complex proteome performance standard (and an associated reference data set) for LC-MS benchmarking and to make this material available to the proteomics community for providing a means for comparing performance of LC/MS/MS platforms (i) over time (as a quality control), (ii) after the addition of new technologies (to evaluate their effectiveness compared with current technologies), and (iii) between laboratories (to inform optimization and troubleshooting). Although our studies were focused on commonly used ion trap instruments, the reference sample can be applied to any platform of interest to benchmark performance over time or between laboratories. For example, although our studies focused on CID for the MS/MS analysis, in view of the recent demonstrations of the complementary value of CID and electron transfer dissociation in proteome analyses (29, 30), it may be of interest to use the performance standard to further evaluate these two approaches.
Of note, the yeast performance standard is now the only true biological performance standard available as a well characterized preparation (obtainable from NIST; see “Experimental Procedures”). Individual laboratories using common ion trap instruments can now analyze the performance standard and determine how their platforms perform relative to the reference data summarized in this study, providing a measure of how their LC-MS platforms are performing relative to those in other laboratories in the community. For example, one can determine how many peptides and proteins their LC-MS system identifies from the yeast reference and how deeply (CN50) and reproducibly their platform samples the proteome; these results can be compared with the data described in this study. If the results differ significantly and the new data exceed the performance of the reference data, then a comparison of the methods used should reveal parameters that further optimize LC-MS performance in the community. If the results show subpar performance compared with the reference data set, this will facilitate the correction of potential underlying problems. Application of the performance metrics (Table II and the accompanying study by Rudnick et al. (32)) will facilitate the differential diagnosis of underlying problems as illustrated in this study.
The one-dimensional LC-MS/MS platforms we used are the simplest of shotgun analysis systems and represent the minimal depth of proteome coverage available with current methods. Multidimensional LC-MS/MS would substantially increase the numbers of identifications achieved. Because the focus of the CPTAC interlaboratory studies described here was on the performance of the reverse phase LC-MS/MS platform, we did not attempt to incorporate multidimensional peptide separations. (The combination of different peptide separation steps would have greatly complicated interpretation of sources of system variability.) Nevertheless, the yeast reference is well suited to benchmark more complex work flows that provide more in-depth proteome coverage (e.g. combining an upstream protein or peptide fractionation step with LC-MS/MS). For example, the CN50 value (Fig. 2 and Table I) associated with a given experimental work flow may serve as a useful performance metric for comparisons between alternative work flows and would also be a useful means of assessing the enhancement of proteome coverage by multidimensional protein and peptide fractionation steps.
As discussed elsewhere (2, 3), there is a need for multiple types of proteomics reference materials. For example, although yeast provides an invaluable complex performance standard, the “shape” (i.e. the abundance range of the constituent proteins) of the yeast proteome is not identical to all biological matrices (e.g. plasma), and optimization of some performance characteristics is likely to be context-specific. Additionally, the yeast performance standard is not useful for testing the performance of species-specific technologies, such as the immunodepletion columns designed to remove abundant proteins from human plasma (31) (unless human plasma is spiked into the yeast proteome). Hence, it would be of great value to the community to have several well characterized types of reference materials so that an appropriate material could be chosen to match the target application. Of note, because the abundances of the majority of the yeast proteins have been estimated (16), one can also imagine using the yeast performance standard as the spike. For example, if one wanted to benchmark the depth of coverage of their work flow in a plasma matrix, a small amount of the yeast reference could be spiked into the plasma, thus providing several thousand proteins whose relative abundances are known and whose detection efficiencies could be determined as a metric of performance.
An increasingly common goal of proteomics is to discover proteins that are differentially expressed between two classes of biological samples (e.g. treated versus untreated, case versus control, etc.). In such comparative proteomics experiments, it is desirable to minimize sources of variation to maximize the statistical power for detecting proteins that are differentially present. In such experiments, there are several types of variation, including biological, preanalytical, and analytical variation. Biological variation is inherent to the cell or organism being studied; it is a fact of nature. In contrast, preanalytical and analytical variations are introduced by our experimental protocols and instruments and thus to some extent can be manipulated. Preanalytical variation is introduced during sample collection, handling, and processing upstream of LC-MS, whereas analytical variation is specifically associated with changes in performance over time of the LC-MS system. A performance standard cannot be used to determine biological variation; however, a performance standard (such as yeast) is invaluable for measuring analytical and preanalytical variation to evaluate the effectiveness of interventions designed to minimize this variation. Where unavoidable variation remains, it is of use to characterize and measure variation during the course of an experiment so that it can be accounted for in the data analysis and interpretation as well as in planning experimental designs.
The results shown in Table III are presented as a proof of concept of the benchmarking that can be done using a spiked performance standard (yeast or other) where the proteome as been highly characterized. This analysis could be applied to more complex work flows typically used in comparative proteomics experiments where greater depth of coverage is desired; for example, the spiked samples could be subjected to off-line strong cation exchange chromatography prior to LC-MS, and the effect of this additional processing step on the sensitivity and specificity of detecting differential proteins could be determined and optimized.
Supplementary Material
Acknowledgments
The CPTAC Network includes: Broad Institute of MIT and Harvard: Steven A. Carr, Michael Gillette, Karl R. Clauser, Terri Addona, Susan Abbatiello, Ronald K. Blackman, Jacob D. Jaffe, Eric Kuhn, Hasmik Keshishian, and Michael Burgess; Buck Institute for Age Research: Bradford W. Gibson, Birgit Schilling, Jason M. Held, Bensheng Li, Christopher C. Benz, Gregg A. Czerwieniec, and Michael P. Cusack; California Pacific Medical Center: Dan Moore; Epitomics, Inc.: Xiuwen Liu and Guoliang Yu; Fred Hutchinson Cancer Research Center: Amanda G. Paulovich, Jeffrey R. Whiteaker, Lei Zhao, ChenWei Lin, Regine Schoenherr, Pei Wang, Peggy L. Porter, Constance D. Lehman, Diane Guay, JoAnn Lorenzo, Barbara R. Stein, Marit Featherstone, Lindi Farrell, Stephanie Stafford, and Julie Gralow; Hoosier Oncology Group: Linnette Lay and Kristina Kirkpatrick; Indiana University: Randy J. Arnold, Predrag Radivojac, and Haixu Tang; Indiana University-Purdue University Indianapolis: Jake Chen, Scott H. Harrison, Harikrishna Nakshatri, and Bryan Schneider; Lawrence Berkeley National Laboratory: Joe W. Gray, John Conboy, Anna Lapuk, Paul Spellman, Daojing Wang, and Nora Bayani; Massachusetts General Hospital: Steven J. Skates and Trenton C. Pulsipher; Memorial Sloan-Kettering Cancer Center: Paul Tempst, Hans Lilja, Mark Robson, James Eastham, Clifford Hudis, Brett Carver, Josep Villanueva, Kevin Lawlor, Arpi Nazarian, Lisa Balistreri, San San Yi, Alex Lash, John Philip, Yongbiao Li, Andrew Vickers, Adam Olshen, Irina Ostrovnaya, and Martin Fleisher; Monarch Life Sciences: Tony J. Tegeler and Mu Wang; National Cancer Institute: Henry Rodriguez, Tara R. Hiltke, Mehdi Mesri, and Christopher R. Kinsinger; National Cancer Institute-SAIC, Frederick, MD: Gordon R. Whiteley; National Institute of Standards and Technology, Gaithersburg, MD: Nikša Blonder, Bhaskar Godugu, Yuri Mirokhin, Pedi Neta, Jeri S. Roth, Paul A. Rudnick, Stephen E. Stein, Dmitrii Tchekhovskoi, Eric Yan, Xiaoyu (Sara) Yang, David M. Bunk, Karen W. Phinney, and Nathan G. Dodder; National Institute of Standards and Technology, Hollings Marine Lab, Charleston, SC: Lisa E. Kilpatrick; New York University School of Medicine: Thomas A. Neubert, Helene L. Cardasis, David Fenyo, Chongfeng Xu, Sofia Waldemarson, Steven Blais, and Åsa Wahlander; Plasma Proteome Institute: N. Leigh Anderson; Purdue University: Fred E. Regnier, Jiri Adamec, Charles Buck, Wonryeon Cho, Kwanyoung Jung, John Springer, and Xiang Zhang; Single Organism Software: Jayson A. Falkner; Texas A&M University: Cliff Spiegelman, Lorenzo Jorge Vega-Montoto, and Asokan Mulayath Variyath; University of Arizona: Dean D. Billheimer; University of British Columbia: Ronald C. Beavis and Dan Evans; University of California, San Francisco: Susan J. Fisher, Michael Alvarado, Mattheus Dahlberg, Penelope M. Drake, Matthew Gormley, Steven C. Hall, Keith D. Jones, Michael Lerch, Michael McMaster, Richard K. Niles, Akraporn Prakobphol, and H. Ewa Witkowska; University of Michigan: Philip C. Andrews, Mark Gjukich, Bryan Smith, and James Hill; University of North Carolina at Chapel Hill: David F. Ransohoff; University of Texas M. D. Anderson Cancer Center: Gordon B. Mills, Yiling Lu, Jonas Almeida, Katherine Hale, Doris Siwak, Mary Dyer, Webin Liu, Hassan Hall, and Natalie Wright; University of Victoria: Terry W. Pearson, Matt Pope, Martin Soste, and Morteza Razavi; University of Victoria-Genome British Columbia Proteomics Centre: Angela Jackson, Derek Smith, and Christoph Borchers; University of Washington: Michael J. MacCoss and Brendan MacLean; and Vanderbilt University School of Medicine: Daniel C. Liebler, Amy-Joan L. Ham, Lisa J. Zimmerman, Robbert J. C. Slebos, David L. Tabb, Carlos L. Arteaga, Ming Li, Melinda E. Sanders, Simona G. Codreanu, Misti A. Martinez, Corbin A. Whitwell, Constantin F. Aliferis, Douglas P. Hardin, Richard M. Caprioli, Dale S. Cornett, Eric I. Purser, Matthew C. Chambers, Scott M. Sobecki, Michael D. Litton, Darla Freehardt, De Lin, Sheryl L. Stamer, Anthony Frazier, Kimberly Johnson, Ze-Qiang Ma, Matthew V. Myers, Haixia Zhang, and Alexander Statnikov.
Footnotes
* This work was supported, in whole or in part, by an interagency agreement between the NCI, National Institutes of Health and the National Institute of Standards and Technology and by National Institutes of Health Grants U24CA126476-01, U24CA126485-01, U24CA126480-01, U24CA126477-01, and U24CA126479-01 as part of the NCI Clinical Proteomic Technologies for Cancer (http://proteomics.cancer.gov) initiative. A component of this initiative is the Clinical Proteomic Technology Assessment for Cancer (CPTAC) teams, which include the Broad Institute of MIT and Harvard (with the Fred Hutchinson Cancer Research Center, Massachusetts General Hospital, the University of North Carolina at Chapel Hill, the University of Victoria, and the Plasma Proteome Institute), Memorial Sloan-Kettering Cancer Center (with the Skirball Institute at New York University), Purdue University (with Monarch Life Sciences, Indiana University, Indiana University-Purdue University Indianapolis, and the Hoosier Oncology Group), University of California, San Francisco (with the Buck Institute for Age Research, Lawrence Berkeley National Laboratory, the University of British Columbia, and the University of Texas M. D. Anderson Cancer Center), and Vanderbilt University School of Medicine (with the University of Texas M. D. Anderson Cancer Center, the University of Washington, and the University of Arizona). Contracts from NCI, National Institutes of Health through SAIC to Texas A&M University funded statistical support for this work.
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental material.
2 Aliquots can be requested from NIST (proteome@nist.gov).
1 The abbreviations used are:
- QC
- quality control
- CPTAC
- Clinical Proteomic Technology Assessment for Cancer
- FDR
- false discovery rate
- NIST
- National Institute of Standards and Technology
- SOP
- standard operating procedure
- UPS1
- Universal Proteomics Standard
- LTQ
- linear trap quadrupole
- CV
- coefficient of variation
- SASPECT
- significant analysis of peptide counts
- TAP
- tandem affinity purification
- RPLC
- reverse phase LC.
REFERENCES
- 1.Klimek J., Eddes J. S., Hohmann L., Jackson J., Peterson A., Letarte S., Gafken P. R., Katz J. E., Mallick P., Lee H., Schmidt A., Ossola R., Eng J. K., Aebersold R., Martin D. B. ( 2008) The standard protein mix database: a diverse data set to assist in the production of improved Peptide and protein identification software tools. J. Proteome Res 7, 96– 103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barker P. E., Wagner P. D., Stein S. E., Bunk D. M., Srivastava S., Omenn G. S. ( 2006) Standards for plasma and serum proteomics in early cancer detection: a needs assessment report from the national institute of standards and technology—National Cancer Institute Standards, Methods, Assays, Reagents and Technologies Workshop, August 18–19, 2005. Clin. Chem 52, 1669– 1674 [DOI] [PubMed] [Google Scholar]
- 3.Vitzthum F., Siest G., Bunk D. M., Preckel T., Wenz C., Hoerth P., Schulz-Knappe P., Tammen H., Adamkiewicz J., Merlini G., Anderson N. L. ( 2007) Metrological sharp shooting for plasma proteins and peptides: the need for reference materials for accurate measurements in clinical proteomics and in vitro diagnostics to generate reliable results. Proteomics Clin. Appl 1, 1016– 1035 [DOI] [PubMed] [Google Scholar]
- 4.de Godoy L. M., Olsen J. V., de Souza G. A., Li G., Mortensen P., Mann M. ( 2006) Status of complete proteome analysis by mass spectrometry: SILAC labeled yeast as a model system. Genome Biol 7, R50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shevchenko A., Jensen O. N., Podtelejnikov A. V., Sagliocco F., Wilm M., Vorm O., Mortensen P., Shevchenko A., Boucherie H., Mann M. ( 1996) Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. U.S.A 93, 14440– 14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Washburn M. P., Wolters D., Yates J. R., 3rd ( 2001) Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol 19, 242– 247 [DOI] [PubMed] [Google Scholar]
- 7.Peng J., Elias J. E., Thoreen C. C., Licklider L. J., Gygi S. P. ( 2003) Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J. Proteome Res 2, 43– 50 [DOI] [PubMed] [Google Scholar]
- 8.Wei J., Sun J., Yu W., Jones A., Oeller P., Keller M., Woodnutt G., Short J. M. ( 2005) Global proteome discovery using an online three-dimensional LC-MS/MS. J. Proteome Res 4, 801– 808 [DOI] [PubMed] [Google Scholar]
- 9.Garrels J. I., McLaughlin C. S., Warner J. R., Futcher B., Latter G. I., Kobayashi R., Schwender B., Volpe T., Anderson D. S., Mesquita-Fuentes R., Payne W. E. ( 1997) Proteome studies of Saccharomyces cerevisiae: identification and characterization of abundant proteins. Electrophoresis 18, 1347– 1360 [DOI] [PubMed] [Google Scholar]
- 10.Perrot M., Sagliocco F., Mini T., Monribot C., Schneider U., Shevchenko A., Mann M., Jenö P., Boucherie H. ( 1999) Two-dimensional gel protein database of Saccharomyces cerevisiae (update 1999). Electrophoresis 20, 2280– 2298 [DOI] [PubMed] [Google Scholar]
- 11.de Godoy L. M., Olsen J. V., Cox J., Nielsen M. L., Hubner N. C., Fröhlich F., Walther T. C., Mann M. ( 2008) Comprehensive mass-spectrometry-based proteome quantification of haploid versus diploid yeast. Nature 455, 1251– 1254 [DOI] [PubMed] [Google Scholar]
- 12.Piening B. D., Wang P., Bangur C. S., Whiteaker J., Zhang H., Feng L. C., Keane J. F., Eng J. K., Tang H., Prakash A., McIntosh M. W., Paulovich A. ( 2006) Quality control metrics for LC-MS feature detection tools demonstrated on Saccharomyces cerevisiae proteomic profiles. J. Proteome Res 5, 1527– 1534 [DOI] [PubMed] [Google Scholar]
- 13.Nägele E., Vollmer M., Hörth P. ( 2004) Improved 2D nano-LC/MS for proteomics applications: a comparative analysis using yeast proteome. J. Biomol. Tech 15, 134– 143 [PMC free article] [PubMed] [Google Scholar]
- 14.Usaite R., Wohlschlegel J., Venable J. D., Park S. K., Nielsen J., Olsson L., Yates Iii J. R. ( 2008) Characterization of global yeast quantitative proteome data generated from the wild-type and glucose repression Saccharomyces cerevisiae strains: the comparison of two quantitative methods. J. Proteome Res 7, 266– 275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Picotti P., Bodenmiller B., Mueller L. N., Domon B., Aebersold R. ( 2009) Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell 138, 795– 806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. ( 2003) Global analysis of protein expression in yeast. Nature 425, 737– 741 [DOI] [PubMed] [Google Scholar]
- 17.Huh W. K., Falvo J. V., Gerke L. C., Carroll A. S., Howson R. W., Weissman J. S., O'Shea E. K. ( 2003) Global analysis of protein localization in budding yeast. Nature 425, 686– 691 [DOI] [PubMed] [Google Scholar]
- 18.Washburn M. P., Koller A., Oshiro G., Ulaszek R. R., Plouffe D., Deciu C., Winzeler E., Yates J. R., 3rd ( 2003) Protein pathway and complex clustering of correlated mRNA and protein expression analyses in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A 100, 3107– 3112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Futcher B., Latter G. I., Monardo P., McLaughlin C. S., Garrels J. I. ( 1999) A sampling of the yeast proteome. Mol. Cell. Biol 19, 7357– 7368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gygi S. P., Rochon Y., Franza B. R., Aebersold R. ( 1999) Correlation between protein and mRNA abundance in yeast. Mol. Cell. Biol 19, 1720– 1730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessner D., Chambers M., Burke R., Agus D., Mallick P. ( 2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24, 2534– 2536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabb D. L., Fernando C. G., Chambers M. C. ( 2007) MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res 6, 654– 661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Z. Q., Dasari S., Chambers M. C., Litton M. D., Sobecki S. M., Zimmerman L. J., Halvey P. J., Schilling B., Drake P. M., Gibson B. W., Tabb D. L. ( 2009) IDPicker 2.0: improved protein assembly with high discrimination peptide identification filtering. J. Proteome Res 8, 3872– 3881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang B., Chambers M. C., Tabb D. L. ( 2007) Proteomic parsimony through bipartite graph analysis improves accuracy and transparency. J. Proteome Res 6, 3549– 3557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Whiteaker J. R., Zhang H., Zhao L., Wang P., Kelly-Spratt K. S., Ivey R. G., Piening B. D., Feng L. C., Kasarda E., Gurley K. E., Eng J. K., Chodosh L. A., Kemp C. J., McIntosh M. W., Paulovich A. G. ( 2007) Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J. Proteome Res 6, 3962– 3975 [DOI] [PubMed] [Google Scholar]
- 26.Rifai N., Gillette M. A., Carr S. A. ( 2006) Protein biomarker discovery and validation: the long and uncertain path to clinical utility. Nat. Biotechnol 24, 971– 983 [DOI] [PubMed] [Google Scholar]
- 27.Anderson N. L., Anderson N. G., Haines L. R., Hardie D. B., Olafson R. W., Pearson T. W. ( 2004) Mass spectrometric quantitation of peptides and proteins using Stable Isotope Standards and Capture by Anti-Peptide Antibodies (SISCAPA). J. Proteome Res 3, 235– 244 [DOI] [PubMed] [Google Scholar]
- 28.Paulovich A. G., Whiteaker J. R., Hoofnagle A. N., Wang P. ( 2008) The interface between biomarker discovery and clinical validation: the tar pit of the protein biomarker pipeline. Proteomics Clin. Appl 2, 1386– 1402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Swaney D. L., McAlister G. C., Coon J. J. ( 2008) Decision tree-driven tandem mass spectrometry for shotgun proteomics. Nat. Methods 5, 959– 964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Coon J. J. ( 2009) Collisions or electrons? Protein sequence analysis in the 21st century. Anal. Chem 81, 3208– 3215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pieper R., Su Q., Gatlin C. L., Huang S. T., Anderson N. L., Steiner S. ( 2003) Multi-component immunoaffinity subtraction chromatography: an innovative step towards a comprehensive survey of the human plasma proteome. Proteomics 3, 422– 432 [DOI] [PubMed] [Google Scholar]
- 32.Rudnick P. A., Clauser K. R., Kilpatrick L. E., Tchekhovskoi D. V., Neta P., Blonder N., Billheimer D. D., Blackman R. K., Bunk D. M., Cardasis H. L., Ham A. J., Jaffe J. D., Kinsinger C. R., Mesri M., Neubert T. A., Schilling B., Tabb D. L., Tegeler T. J., Vega-Montoto L., Variyath A. M., Wang M., Wang P., Whiteaker J. R., Zimmerman L. J., Carr S. A., Fisher S. J., Gibson B. W., Paulovich A. G., Regnier F. E., Rodriguez H., Spiegelman C., Tempst P., Liebler D. C., Stein S. E. ( 2010) Performance metrics for liquid chromatography-tandem mass spectrometry systems in proteomics analyses. Mol. Cell. Proteomics 9, 225– 241 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.