Abstract
Deinococcaceae are a family of extremely radiation-tolerant bacteria that are currently subjected to numerous studies aimed at understanding the molecular mechanisms for such radiotolerance. To achieve a comprehensive and accurate annotation of the Deinococcus deserti genome, we performed an N terminus-oriented characterization of its proteome. For this, we used a labeling reagent, N-tris(2,4,6-trimethoxyphenyl)phosphonium acetyl succinimide, to selectively derivatize protein N termini. The large scale identification of N-tris(2,4,6-trimethoxyphenyl)phosphonium acetyl succinimide-modified N-terminal-most peptides by shotgun liquid chromatography-tandem mass spectrometry analysis led to the validation of 278 and the correction of 73 translation initiation codons in the D. deserti genome. In addition, four new genes were detected, three located on the main chromosome and one on plasmid P3. We also analyzed signal peptide cleavages on a genome-wide scale. Based on comparative proteogenomics analysis, we propose a set of 137 corrections to improve Deinococcus radiodurans and Deinococcus geothermalis gene annotations. Some of these corrections affect important genes involved in DNA repair mechanisms such as polA, ligA, and ddrB. Surprisingly, experimental evidences were obtained indicating that DnaA (the protein involved in the DNA replication initiation process) and RpsL (the S12 ribosomal conserved protein) translation is initiated in Deinococcaceae from non-canonical codons (ATC and CTG, respectively). Such use may be the basis of specific regulation mechanisms affecting replication and translation. We also report the use of non-conventional translation initiation codons for two other genes: Deide_03051 and infC. Whether such use of non-canonical translation initiation codons is much more frequent than for other previously reported bacterial phyla or restricted to Deinococcaceae remains to be investigated. Our results demonstrate that predicting translation initiation codons is still difficult for some bacteria and that proteomics-based refinement of genome annotations may be helpful in such cases.
A comprehensive parts list is required prior to accurate global modeling of the complex whole cellular unit. For this reason, the quality of genome annotation is a very important parameter for an efficient system biology study of the cell. Besides the combination of gene prediction tools, homology-based inference, and manual curation (1), the use of experimental data either from massive gene transcript sequencing (2) or massive protein sequence determination by mass spectrometry is currently considered with great interest (3–5). Proteogenomics consists in high throughput identification of proteins, their accurate partial sequencing by nano-LC-MS/MS shotgun approaches, and the integration of these data with the genome (6). Proteogenomics allows validation of predicted genes and, more importantly, correction of genome annotation errors such as discovery of unannotated genes; reversal of reading frames; identification of translational start sites, stop codon read-throughs, or programmed frameshifts; and detection of signal peptide processing and other maturation events at the protein level. Several studies dedicated to genome reannotation based on experimental proteomics evidences have been reported (7–15) and have paved the way for the proteogenomics approach. For example, the Mycobacterium smegmatis bacterium was subjected to an in-depth MS/MS analysis over the course of 25 different growth conditions (16). A set of 901 proteins was identified with most of them annotated as hypothetical. This study was further completed with mass spectrometry analysis and a resequencing procedure that showed that a significant proportion of interrupted coding sequences in this genome were sequencing errors (17). More recently, a reannotation of the genome was proposed after a novel mass spectrometry analysis in which 946 distinct proteins were uncovered (18).
Although proteogenomics has greatly improved over the last years, annotation of a significant number of N-terminal starts is still a challenging task whatever the genome under consideration. Large scale sequence determination of N-terminal peptides has been reported for the Aeropyrum pernix K1 crenarchaeon (19), the Halobacterium salinarum and Natronomonas pharaonis halophilic euryarchaeota (20), and the Shewanella oneidensis (21) and M. smegmatis bacteria (18). Among these reports, the most unexpected result consisted in the discovery that TTG was the most used translational initiation codon, far more common than ATG and GTG in A. pernix (19). In high throughput nano-LC-MS/MS studies, low sequence coverage is observed for most proteins. It results in a low coverage of N-terminal-most peptides. Over the last 7 years, a series of specific strategies have been devised to systematically catalogue N-terminal-most peptides (for a review, see Ref. 4). The most comprehensive studies reported up to now for a whole cellular proteome are based on methodologies consisting in a derivatization of N termini by a hydrophobic chemical reagent (18, 20). In the first method, named COFRADIC for combined fractional diagonal chromatography (22, 23), 2,4,6-trinitrobenzenesulfonic acid reacting with the N terminus of internal peptides was used to discard internal peptides by an appropriate reverse phase chromatography and focus the analysis on enriched N-terminal-most peptides. The second method consists in selective N terminus derivatization of intact proteins with N-tris(2,4,6-trimethoxyphenyl)phosphonium acetyl succinimide (TMPP),1 a labeling reagent that increases the hydrophobic character of the product, improves its ionization, and modifies the fragmentation pattern due to the positive charge introduced (18, 24).
We recently reported the proteogenomics analysis of Deinococcus deserti VCD115 bacterium, isolated from surface sand of the Sahara (25). This bacterium has an exceptional ability to withstand the lethal effects of DNA-damaging agents, including ionizing radiation, UV light, and desiccation. Accurate genome annotation of its 3455 genes was guided at the stage of primary annotation by an extensive proteome analysis. A set of 1348 proteins was uncovered after growth in standard conditions and proteome fractionation by phenyl-Sepharose chromatography. The alliance of proteomics and genomics high throughput techniques allowed identification of 15 genes that were not predicted by the two annotation softwares that were used. Surprisingly, we had to propose reversal of incorrectly predicted orientations of 11 genes. In this previous study, we checked the whole MS/MS data set for N-terminal peptides and found 212 distinct peptide signatures corresponding to N termini of 145 proteins. These data confirmed the starts of 112 proteins but also corrected the starts of 33 polypeptides that were incorrectly predicted even after manual inspection (25). Although several proteomics analyses have been carried out on Deinococcus radiodurans, the radioresistant model among bacteria (26), none has been specifically focused on genome reannotation (27–32). The genome annotation of D. radiodurans was among the first complete bacterial genome annotations ever reported (33). Those of Deinococcus geothermalis (34) and Thermus thermophilus HB27 (35) have been reported more recently, allowing a better genome coverage of the Deinococcus-Thermus phylum.
That one-fifth of the detected N termini were not correctly predicted in our previous D. deserti proteogenomics study (25) led us to develop a specific strategy for identifying N-terminal most-peptides on a very large scale for D. deserti VCD115 proteome. We labeled the proteome of cells harvested in exponential and prestationary growth phases with the TMPP reagent. The labeled products were digested with trypsin on one hand and chymotrypsin on the other. The resulting peptides were analyzed by nano-LC-MS/MS high resolution mass spectrometry. In this study, 664 N-terminal peptides from 341 proteins were characterized, leading to the validation of 278 and the correction of 63 translation initiation codons in the D. deserti VCD115 genome. Four new ORFs were also detected in its genome through the detection of peptidic signatures for the corresponding polypeptides. We found experimental evidences indicating that dnaA translation is initiated in Deinococcus spp. from a non-canonical ATC codon and report the use of non-canonical codons for three other genes. Furthermore, several corrections of D. radiodurans and D. geothermalis gene annotations are proposed based on comparative proteogenomics analysis, some affecting important genes involved in DNA repair mechanisms.
EXPERIMENTAL PROCEDURES
N-terminal Chemical Labeling of D. deserti Protein Extracts
D. deserti cells were grown and harvested during exponential growth and in the prestationary phase as described previously (25). Cells (1.5 g of wet material) were resuspended in 7.5 ml of cold 100 mm NaH2PO4/Na2HPO4 buffer (pH 8.20 at 20 °C) containing a protease inhibitor mixture (Complete Mini) from Roche Applied Science (one tablet/sample). Cells were disrupted by means of a Basic cell disrupter (Constant Systems Ltd.) operated at 1000 bars and centrifuged for 20 min at 20,000 × g at 4 °C to remove cellular debris. The buffer of a fraction of the resulting supernatant was further changed into 200 mm NaH2PO4/Na2HPO4 buffer (pH 8.20 at 20 °C) containing the protease inhibitor mixture (one tablet/7.5 ml) by means of chromatography onto a 5-ml HiTrap Desalting column (GE Healthcare). The sample (1 ml) was applied at a flow rate of 0.5 ml/min onto the column previously equilibrated with the final buffer. TMPP reagent (TMPP-AcOSu) was purchased from Sigma and dissolved at 0.2 m in 20% ACN, 80% H2O (v/v) just prior to use. To each protein extract (300 μg), 5 μl of TMPP-AcOSu solution was added. The mixtures were incubated at room temperature for 1 h under gentle agitation. They were then precipitated with trichloroacetic acid (10% (w/v) final). After centrifugation, proteins from the pellets were dissolved in lithium dodecyl sulfate loading sample buffer (Invitrogen) prior to 1D SDS-PAGE analysis.
1D SDS-PAGE Separation and In-gel Proteolysis
Proteins were resolved on 10% gradient NuPAGE (Invitrogen) gels. The gels were stained with Coomassie Blue Safe stain (Invitrogen). Each lane was excised into 2.5-mm-thick pieces from the top to the bottom. Each band was divided into two equal pieces for subsequent proteolysis with either trypsin or chymotrypsin. Protein bands were treated and in-gel proteolyzed with trypsin as described previously (25). Sequencing grade chymotrypsin (Roche Applied Science) was dissolved in 250 μl of 1 mm HCl solution to a final concentration of 0.1 μg/μl. Destained pieces of polyacrylamide gel were immersed in 50 μl of enzymatic solution consisting of 42 μl of 25 mm NH4HCO3, 2 μl of 1% CaCl2, and 6 μl of chymotrypsin at 0.1 μg/μl. They were incubated for 45 min on ice for complete rehydration. The excess proteolytic solution was then removed, and 75 μl of 25 mm NH4HCO3 was added. Samples were proteolyzed overnight at 25 °C. Peptides were then extracted as described previously (25).
Nano-LC-MS/MS Analysis
LC-MS/MS experiments were performed on an LTQ-Orbitrap XL hybrid mass spectrometer (ThermoFisher) coupled to an UltiMate 3000 LC system (Dionex-LC Packings). Peptide mixtures (0.3–2 pmol) were loaded and desalted on line in a reverse phase precolumn (Acclaim PepMap 100 C18, 5-μm bead size, 100-Å pore size, 5 mm × 300 μm) from LC Packings. They were resolved on a nanoscale Acclaim PepMap 100 C18 column (3-μm bead size, 100-Å pore size, 15 cm × 75 μm) from LC Packings at a flow rate of 0.3 μl/min. The LC gradient was optimized to detect the largest set of TMPP-labeled peptides. Peptides were separated using a 146-min gradient with aqueous solvent A (0.1% HCOOH) and solvent B (0.1% HCOOH, 80% CH3CN) developed as follows: 5–33% B in 45 min, 33–50% B in 90 min, 50–90% B in 1 min, 90% B for 10 min, 90–5% B in 1 min, and 5% B for 19 min. The full-scan mass spectra were measured from m/z 300 to 1800. The LTQ-Orbitrap XL mass spectrometer was operated in the data-dependent mode using the TOP5 strategy. In brief, a scan cycle was initiated with a full scan of high mass accuracy in the Orbitrap analyzer that was followed by MS/MS scans in the linear ion trap on the five most abundant precursor ions with dynamic exclusion of previously selected ions. This dynamic exclusion consisted of two acquisitions of MS/MS spectra of the most abundant ion during a period of 30 s and then excluding this ion for the followed fragmentations during the next 60 s. The activation type used was CID with standard normalized collision energy set at 30.
Whole Genome Database Mining
Peak lists were generated with the MASCOT DAEMON version 2.1.6 software (Matrix Science) from the LC-MS/MS raw data with default parameters. Using the MASCOT 2.1 search engine (Matrix Science), we searched all MS/MS spectra against a home-made polypeptide sequence database corresponding to a six-frame translation of the D. deserti genome sequence that was restricted to ORFs (defined from stop to stop) with at least 33 amino acids. The database comprises 65,801 polypeptide sequences, totaling 6,040,642 amino acids with an average of 92 amino acids per polypeptide. Searches for peptides were first performed with the following parameters: a mass tolerance of 5 ppm on the parent ion and 0.6 Da on the MS/MS, static modifications of carboxamidomethylated Cys (+57.0215), and dynamic modification of oxidized Met (+15.9949). To process data resulting from trypsin and chymotrypsin proteolysis, the maximum number of missed cleavage was set at 1 and 3, respectively. All peptide matches with a peptide score above its peptidic identity threshold set at p < 0.001 with the ORF database and rank 1 were filtered by the IRMa 1.16.0 software (36). Searches for trypsin and chymotrypsin full specificities were merged (supplemental Table S1). The criterion adopted for protein identification (supplemental Table S2) was very conservative with at least two peptides identified (with specific threshold set at p < 0.001). False-positive identification was quantified using a target-decoy database made by reversing ORF sequences. Although 14,305 non-redundant peptide assignments were found against the forward database, only 51 spectra assignments had scores above the threshold in the reversed database, giving a false discovery rate for peptides of less than 0.4% on the forward database. In the reversed database search, we did not observe any pair of peptides in the same ORF. At the protein level, the false-positive identification rate is considered low (0.0%). A second search round was performed with the same parameters but including dynamic TMPP-Ac (+572.1811) modification of N termini and Lys. All spectra assigned to TMPP-labeled peptides corresponding to a single signature of a given protein N terminus were manually verified, taking into account the chromatographic retention time, the reporter ions, and the a ion series.
Further data analysis was performed for semitrypsin and semichymotrypsin specificities for identification of TMPP-derivatized peptides. In this case, two types of dynamic modifications were sought: TMPP-Ac (+572.1811) on lateral chain of lysines and at the N terminus of polypeptides and TMPP-Ac-Met (+703.2216) at the N terminus of polypeptides (both values calculated after taking into account the additional positive charge introduced by TMPP and MASCOT requirements for taking into account this charge). We listed all TMPP-modified peptides resulting from both searches with MASCOT scores >10. This score threshold was determined after manual inspection of tens of spectra and corresponds to a threshold where a significant number of spectra are correctly assigned to TMPP-labeling peptides. The ORF library used as the search database for MASCOT comprises a large proportion of small ORFs resulting in a high degree of noise. For this reason, the 5606 TMPP-modified peptide identifications were filtered to remove false hits. First, peptides assigned to several loci were systematically removed. Second, redundant assignments to the same event due to the use of our two assignment strategies were reduced to a single event. Third, only peptide identifications corresponding to a possible initiation start (ATG, GTG, and TTG codons) were selected. This reduced our data set to one-fifth its original size (1081 positive queries instead of 5606). We then removed all the events with a score below 20 that correspond to a unique signature of N terminus as well as all multiple events with a maximum MASCOT score below 20. We further removed, after spectral manual inspection, 66 dubious items based on the absence of reporter ions, incomplete b and y series, or premature elution time. To reduce the search space for peptides not matching to current polypeptide annotations, the remaining spectra were further analyzed if the peptide sequence was longer than 6 amino acids and the ratio of protein length over total ORF length was greater than 0.5. The spectra of these N-terminal-most peptides were further analyzed by manual inspection when only one signature was detected (supplemental Table S4). For searching for the presence of TMPP-labeled peptide signatures demonstrating the use of non-canonical initiation codons, we came back to the first list of 5606 TMPP signatures. From this list, we removed all peptides assigned to several loci and selected only peptide identifications corresponding to a possible non-canonical initiation start (CTG, ATT, ATA, and ATC codons). The remaining 521 spectra were further analyzed if the MASCOT score was above 25, the peptide sequence was longer than 6 amino acids, and the ratio of protein length over total ORF length was above 0.5. A set of 49 putative TMPP-labeled peptide signatures remained after this in silico screening (supplemental Table S5).
N-terminal Maturation Predictions
The SignalP 3.0 server from the Technical University of Denmark was used for peptide signal predictions of the whole set of D. deserti protein sequences. Hidden Markov models and neural networks algorithms, both trained on Gram-negative bacteria, were used. The TatFind server version 1.4 and the LipoP 1.0 server were used as described (37–39).
Mass Spectrometry and Gene Reannotation Data Deposition
Mass spectrometry data were deposited in the PRIDE proteomics identifications database (40) under accession numbers 9863–9877. They are freely available at http://www.ebi.ac.uk/pride/init.do. The complete reannotation of the D. deserti genome can be accessed using GenBankTM accession numbers CP001114, CP001115, CP001116, and CP001117 for the main chromosome, plasmids P1, P2 and P3, respectively. We carefully checked the sequence similarities between the three Deinococcaceae sequenced genomes. Briefly, we generated the six-frame translation of each complete genome and compared homologous sequences on the full-length ORF (defined from stop to stop codons).
RESULTS
Evidence from Unlabeled Peptide Pool for Four Unannotated Genes
Total protein extracts were prepared from cells harvested in the exponential and stationary phases. Proteins were labeled with TMPP and resolved by 1D SDS-PAGE. Proteins were identified by nano-LC-MS/MS shotgun analysis after either trypsin or chymotrypsin proteolysis. Peptides were identified using the MASCOT search engine against a database consisting of a six-frame translation of the entire D. deserti genome. This database comprised 65,801 hypothetical protein sequences with a large fraction of short ORFs (68% of the ORFs have less than 80 residues). From the large body of MS/MS spectra (408,210) that were acquired from the 96 nano-LC-MS/MS runs, 123,800 spectra could be assigned to 14,305 unique peptides (p < 0.001) using the six-frame translation database. These peptides are listed in supplemental Table S1. They matched to 1246 proteins when using very stringent criteria (at least two peptides with p < 0.001). These polypeptides and the total number of tryptic and chymotryptic peptides that were identified per protein are listed in supplemental Table S2. We compared these results in terms of peptide identification to a search against the currently released D. deserti protein database. We found six confident peptide evidences for four unannotated ORFs: C_1731617_-1, P3_322634_-1, C_563543_-1, and C_1277035_1. The assignments for these peptide evidences (supplemental Table S3) were manually verified and no other assignment could be proposed. We checked that the four ORF coordinates did not overlap with those of previously annotated genes and found that potential promoter and Shine-Dalgarno sequences were present in the upstream region. On this basis, we propose the existence at these loci of four novel genes that we named Deide_14222, Deide_3p02315, Deide_04870, and Deide_10572. They all encode polypeptides with molecular masses in the 8–18-kDa range. The first ORF corresponds to a real orphan encoded by the 1731838–1731617 locus on the main chromosome. Its validation was made possible by the detection of two different peptides. Moreover, as described below, the translation initiation codon of this gene could be confirmed with the detection of a chemically derivatized peptide. The second ORF is located at locus 323056–323633 on plasmid P3 and flanked by genes encoding 5,10-methylenetetrahydromethanopterin reductase at its 5′ side and a LuxR family transcriptional regulator at its 3′ side. It codes for a protein exhibiting some similarities with an annotated protein from the Thiomicrospira crunogena XCL-2 γ-proteobacterium. The third specifies a polypeptide homologous to Dgeo_2021 and DR_0900 from D. geothermalis and D. radiodurans, respectively. These polypeptides are currently annotated as hypothetical proteins and are Deinococcus-Thermus phylum-specific. A multialignment with Dgeo_2021, DR_0900, and Deide_04870 points to a misannotation for Dgeo_2021, which has probably 20 conserved residues missing at its N terminus (supplemental Fig. S1). The fourth ORF encodes a polypeptide with similarities to Dgeo_1180 and DR_1256 and is exclusively found in Deinococcaceae. Although only one peptide was detected for Deide_04870 and Deide_10572, the high level of similarities with the homologues in other Deinococcaceae validated their existence.
Unlabeled Peptide Evidence for Extended N Termini
When listing all peptides identified with the ORF database that did not match to any hitherto released D. deserti protein sequences, we found 18 peptide signatures corresponding to extended N termini of 13 previously annotated ORFs. These peptide evidences are listed in supplemental Table S3. Two confidently assessed peptides were found to match upstream nucleic acid sequences of currently annotated Deide_07060, Deide_07960, Deide_10500, Deide_20150, and Deide_ 2p00190. These extensions were further confirmed by multisequence alignments of homologous protein sequences. From these alignments, translation initiation codons were identified for some genes only. Based on comparative proteogenomics, we noticed misannotation of D. radiodurans DR_0080 gene encoding the ornithine carbamoyltransferase homologous to Deide_07060, which has an unexpected extension of 45 amino acids at its N terminus. However, some initiation codons are difficult to assess as exemplified by Deide_07960/DR1870/Dgeo1678 where several GTG or TTG neighboring codons may be proposed as a start site. Deide_2p00190 is another example where a specific strategy to probe initiation codons would be necessary. Homology-based annotation of this ORF was not possible as this true orphan is found only in D. deserti. Fig. 1 shows that this gene was first annotated starting from the ATG codon located at 25483 on the P2 plasmid. Fig. 1 shows the two peptide signatures (LTQLGPEQGAEVQK and LIQAVQSSLAPTPR) that were identified matching to the P2_25138_-1 ORF. They indicate a probable extension of the gene at least 294 nucleotides upstream from the previous annotation start site. Only one peptide is found upstream from hitherto annotated sequences of Deide_05590, Deide_07640, Deide_13741, Deide_16590, Deide_19650, Deide_22950, Deide_2p01230, and Deide_2p01630. Visual inspection of multialignments of these polypeptides and close homologous sequences confirmed that some residues were missing in all of these cases. We thus propose the reannotation of these 13 genes.
Fig. 1.
The Deide_2p00190 polypeptide is covered by two tryptic peptides that do not match previous reported annotation. The previous annotated start codon is shown in blue. Two possible ATG translation initiation codons are shown in red. Tryptic peptides detected by LC-MS/MS are shown in bold red. The previous polypeptide sequence is shown in bold black. The stop codon is indicated with a * symbol.
Database Query Strategies for TMPP-labeled Peptide Assignment
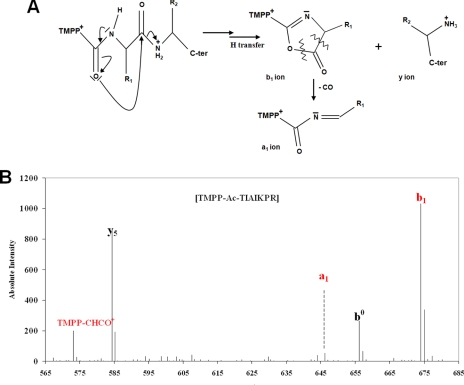
As recently reported by Gallien et al. (18), several criteria may be used to validate potential derivatized N-terminal-most peptides. TMPP labeling has a strong influence on (i) peptide fragmentation behavior because of its permanent positive charge and (ii) retention time on the reverse phase due to its hydrophobic character. We also observed here in the MS/MS spectra obtained with an LTQ-Orbitrap XL mass spectrometer that fragment ions from the a and b series are predominant in the fragmentation pattern. We specifically set MASCOT search parameters to take into account ions from the a series. Because the MASCOT search algorithm takes into account the two most preeminent peaks in 100 m/z windows when interpreting the spectra, this fragmentation behavior results in a low scoring of TMPP-modified peptide spectra by this search engine. Reporter ions may be observed at m/z 533.1940, 547.2097, and 573.1890 (monoisotopic theoretical values). They correspond to TMPP-H+ (C27H34O9P), TMPP-CH3+ (C28H36O9P), TMPP-CHCO+ (C29H34O10P), respectively. Moreover, the a1 and b1 ions are readily observed. Fig. 2A shows how peptide fragmentation can give these two specific ions by introduction of the carbonyl group by TMPP. Fig. 2B shows the spectrum in the 530–680 m/z range of an N-terminally TMPP-labeled peptide, TIAIKPR, corresponding to the N terminus of Deide_09040, which exemplifies the most intense of these reporter ions in most spectra with signals from m/z 573.16, 646.13, and 674.03 ions. We used mainly these three reporter ions to manually validate TMPP-labeled peptide assignments. For limiting the search space with the most appropriate rules, we devised a query strategy based on the six-frame translation database, two possible dynamic modifications, and a semienzyme digestion pattern. The first modification consists of TMPP labeling, resulting in the addition of C29H35O10P (574.1968 amu, monoisotopic theoretical value) and a positive charge at the N terminus of the peptide. When used with a semienzyme search mode, it allows the detection of translation initiation events at ATG (whether methionine maturation occurs or not), GTG, and TTG (if methionine is further removed) codons as well as intracellular proteolysis. The second mimics a theoretical “TMPP and methionine” labeling, resulting in the addition of C34H44N1O11P1S1 (705.2373 amu, monoisotopic theoretical value) and a positive charge at the N terminus of the peptide. When used with a semienzyme search mode, it allows the cataloguing of translation initiation at ATG, GTG, and TTG codons when methionine maturation does not occur. We listed all TMPP-modified peptides resulting from both database searches with specific criteria to avoid false positives. For example, only peptide identifications corresponding to a possible initiation start (ATG, GTG, and TTG codons) were selected. At this stage, 557 signatures matched the N termini of 278 different proteins previously annotated. They are reported in supplemental Table S4. Redundancy of signatures for some proteins arose from differences in terms of miscleavages, methionine oxidation status, and the use of two different proteases but also from incomplete initial methionine cleavage in some cases. It is worth noting that 31 signatures correspond to 23 proteins that were not listed in supplemental Table S2. The improved ionization efficiency of TMPP-labeled peptides and their hydrophobic shifted elution from the chromatography resulted in identification of these N-terminal-most peptides from proteins that were not identified with at least two peptides assigned with high confidence (p < 0.001) by means of the MASCOT search engine.
Fig. 2.
Fragmentation specificities of TMPP-modified peptides. A reaction scheme for formation of a1 and b1 ions from a TMPP-modified N-terminal most-peptide during CID is shown in A. These ions are not detected during fragmentation of unmodified peptides. The MS/MS spectrum of the TMPP-modified peptide TIAIKPR assigned to Deide_09040 is shown in B. The reporter ion TMPP-CHCO+ and the corresponding a1- and b1-specific ions are labeled in bold red.
Correction of Protein N Termini in D. deserti Genome
A total of 104 spectra were found not to correspond to hitherto known N termini. Three classes were defined among these signatures that are catalogued in supplemental Table S4.
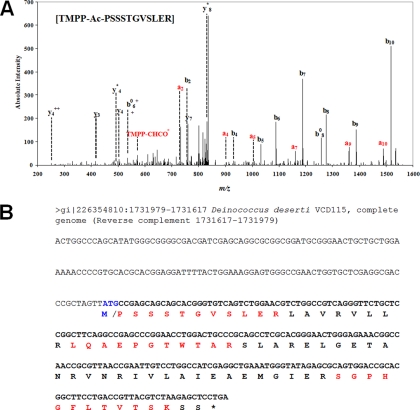
The first class (one item) corresponded to an identification matching a not previously annotated polypeptide sequence. This spectrum corresponds to the diagnostic peptide for C_1731617_-1 (Deide_14222) orphan N terminus identification. As shown in Fig. 3, manual interpretation of the corresponding spectrum validated this assignment. This PSSSTGVSLER peptide corresponds to the TMPP-labeled N terminus of Deide_14222 where the initial methionine has been removed. Such maturation is expected as the second residue is proline. The resulting 72-amino acid protein has a molecular mass of 7.7 kDa and a basic isoelectric point (10.5). We checked that no homologues could be encoded in any genomic DNA currently deposited in databases by Blastn with the D. deserti main chromosome 1731838–1731617 locus as query. No hint for a possible function could be found for this small orphan protein, but its small size and strong positive charge are two relevant characteristics similar to HU DNA-binding proteins (41).
Fig. 3.
Discovery of Deide_14222 gene. The MS/MS spectrum of the TMPP-modified peptide PSSSTGVSLER from Deide_14222 (C_1731617_-1 ORF) is shown in A. The ion reporter TMPP-CHCO+ and the series a ions are shown in red. Sequences of the Deide_14222 gene and the corresponding polypeptide are shown in B. The three detected peptides are shown in bold red. The translation initiation codon and excised methionine are shown in bold blue. The upstream nucleic acid region is shown in normal characters. The stop codon is indicated with a * symbol.
The second class (33 items) consisted of peptide signatures matching the upstream region of 20 previously annotated genes. Remarkably, four different peptides indicated a correction of the initiation codon for Deide_22800 as 17 residues are missing at the N terminus of the corresponding polypeptide. This gene encodes a peptidylarginine deiminase found in many bacteria, but wrong annotations were detected for the genes coding for D. radiodurans DR_2359 (3 additional residues) and D. geothermalis Dgeo_0034 (17 residues missing) homologues. The case of Deide_3p00250 is also worthy of mention. As this protein has no known homologue, annotation of the corresponding gene is far from straightforward. We detected its N-terminal-most peptide that revealed the absence of 63 residues in the previous annotation (a third of the new annotated sequence). The proposed new translation start for Deide_3p00250 is GTG. It has been validated after detection of the TMPP-modified peptide MNGYEPLEVVK, which exists only if GTG is used as coding the first methionine. The same applies for the cases of Deide_19360, Deide_10140, and Deide_02960. A new putative endonuclease (Deide_02842) sharing 30% identities with Bacillus subtilis BglI restriction enzyme was detected in the D. deserti genome, but not in the D. radiodurans or D. geothermalis genomes (25). We detected tryptic and chymotryptic TMPP-labeled peptides proving that 8 residues were missing in the previous annotation of Deide_02842. This enzyme is specifically overproduced after γ-irradiation in D. deserti.2 Accurate annotation of its gene is thus of importance to characterize the function of this endonuclease related to DNA repair.
The third class (70 items) consisted of N-terminal peptide signatures matching the internal sequence of 40 previously annotated genes, i.e. downstream of the previously annotated initiation codon. We checked whether these signatures could result from signal peptide maturation (see below). Five different TMPP-labeled peptide signatures were found for Deide_06640. They indicate the initiation codon located 18 bp downstream from the previously annotated start. This protein shares some similarities with the Spo0M protein from B. subtilis involved in the control of sporulation. No homologue has been yet reported in other bacteria from the Deinococcus-Thermus phylum. The results (supplemental Table S4) suggest that the 3-polyprenyl-4-hydroxybenzoate decarboxylase Deide_01420 start (18 additional residues) as well as that of its closest homologue, namely DR_0332 (22 extra residues), should be reannotated. Detection of the N-terminal-most peptide of Deide_1p01752 is of interest. This protein shares similarities only with another protein from D. deserti, Deide_3p00880, and both are real orphans. As shown in Fig. 4, information on the start of the former suggests reannotation of both genes. Three internal peptides are detected for the former, but none corresponded to the latter (Fig. 4), indicating that these homologous proteins may be produced in different growth conditions. The case of Deide_15130 is also of special interest. This gene encodes the DNA-directed polymerase I, a large protein of 923 residues that comprises two domains: 5′–3′ xeroderma pigmentosum G protein like exonuclease at its N terminus and the polymerase DNA PolA domain at its C terminus. Three and 35 extra residues were wrongly introduced in the course of Deide_15130 (gi:226356483) and DR_1707 (gi:7404361) annotations, respectively.
Fig. 4.
Reannotation of Deide_1p01752 and Deide_3p00880 homologues. A polypeptide sequence alignment of both proteins is shown with conserved residues. Senseless residues are shown in blue. Peptides detected by LC-MS/MS are shown in bold red. The specific N-terminal-most peptide that was labeled with TMPP is underlined.
Use of Non-canonical Translation Initiation Codons
Although considered inefficient, CTG, ATT, ATA, and ATC codons were reported to function as translation initiators in a very few cases (42–47). CTG is known to function as an initiator for RepA, a plasmid-encoded protein in Escherichia coli (47). The infC gene, which encodes translation initiation factor IF3, is known to be initiated at a rare ATT codon in a variety of bacteria (44) and at an ATA codon in some others (21). We checked whether D. deserti infC (Deide_06380) and its close Deinococcus and Thermus homologues start with non-canonical initiation codons or more conventional starts. Ten different peptides were assigned to a Deide_06380 polypeptide sequence with a threshold value below 0.001, but none corresponded to the most terminal peptide. Deide_06380 MS/MS sequence coverage is rather large (39%) but limited to the 21–181 region. When considering pfam05198/PRK00028 alignment, the translation initiation starts from D. deserti, D. geothermalis, D. radiodurans, Thermus thermophilus HB27, and Thermus aquaticus Y51MC23 orthologous infC genes are not properly assigned, and their hitherto N termini are heterogeneous. Inspection of a nucleic acid sequence multialignment indicates that for these five genes an ATA non-canonical translation initiation codon is probably the true start (supplemental Fig. S2). The specific mechanisms regulating infC expression, relying on a non-canonical initiation codon (48), appear to be widely conserved among bacteria. The gene specifying the tRNA nucleotidyltransferase/poly(A) polymerase PcnB is also known to start with an ATT codon in E. coli. The use of such non-canonical translation initiation codon is the basis of a specific regulation mechanism (42). We did not find any peptides corresponding to PcnB (Deide_05850) in our data set. We carefully analyzed the previous annotations of PcnB orthologues in the Deinococcus-Thermus phylum. On the basis of the nucleic acid sequence multialignment shown in supplemental Fig. S3, we propose that pcnB gene from D. deserti should be reannotated as starting with a standard ATG codon, whereas that of Meiothermus silvanus may start with an ATC codon.
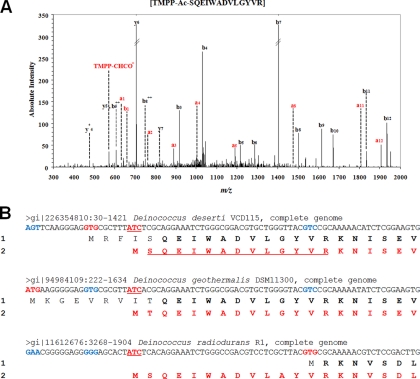
We checked for the presence of TMPP-labeled peptide signatures demonstrating the use of non-canonical initiation codons for other genes in D. deserti. We returned to the first list of TMPP signatures and selected only peptide identifications corresponding to a possible non-canonical initiation start (CTG, ATT, ATA, and ATC codons). A set of 49 putative TMPP-labeled peptide signatures remained after this in silico screening. They are reported in supplemental Table S5. The spectrum of the peptide with the highest MASCOT score (74.6) in this list is shown in Fig. 5. It is assigned to the SQEIWADVLGYVR peptide, which had been modified with TMPP. It corresponds to the N terminus of Deide_00010 if an ATC codon is the initiation codon and the initial methionine has been removed. This methionine maturation is expected because the second residue of the resulting polypeptide is Ser, a residue with a small lateral chain. This peptide assignment was manually confirmed with the presence of the TMPP-CHCO+ reporter ion, the a1 and b1 ions, and the a and b ion series quite complete. Remarkably, another TMPP-modified peptide SQEIWADVLGYVRK was found for the same N terminus because of a trypsin miscleavage. We discarded the possibility that this peptide resulted from a signal peptide maturation event because (i) this protein is cytoplasmic, and the resulting signal peptide is too short, and (ii) the number and the nature of the residues before this peptide are not conserved in close homologues (Fig. 5). The Deide_00010 gene encodes the chromosomal replication initiator protein DnaA. To our knowledge it has not been previously reported that synthesis of this conserved protein is initiated by a non-canonical codon in a bacterium. The genes specifying this crucial protein for DNA replication in bacteria also have to be reannotated in D. geothermalis and D. radiodurans: Dgeo0001 and DR0002, respectively. They start also with an ATC codon resulting in a 7-residue deletion and 12-residue extension, respectively, when compared with previous annotations reported in GenBank. As reported in Fig. 5, these reannotations confer a highly conserved N terminus for these proteins, whereas heterogeneity and poor sequence conservation are noted for previous annotated sequences. We did not confirm the assignment of the other putative TMPP-labeled peptides reported in supplemental Table S5 after manual inspection of the corresponding spectra. After the discovery of the Deide_00010 true translation start, we reanalyzed our previous set of unlabeled MS/MS data (25), searching for new non-canonical initiation starts. The ORF encoding RpsL, the S12 ribosomal conserved protein, was found to start with a CTG non-conventional initiation codon. The spectrum for the unique PTTQLLR peptide and an alignment of the corresponding nucleic acid loci in different Deinococcus-Thermus representatives are shown in supplemental Figs. S4 and S5, respectively. This CTG start is highly conserved among all these genomes.
Fig. 5.
Reannotation of DnaA in Deinococcus spp. The MS/MS spectrum of the TMPP-modified peptide SQEIWADVLGYVR from Deide_00010 is shown in A. The alignment of dnaA genomic loci from D. deserti, D. geothermalis, and D. radiodurans is shown in B. The translated polypeptide from the previous annotation and from the new corrected annotation are labeled 1 and 2, respectively. Previously annotated codons are shown in red, and their counterparts in other sequences are shown in blue. Correct initiation codons are underlined and in bold red. Conserved amino acid residues are indicated in bold. The peptide SQEIWADVLGYVR is underlined in the Deide_00010 sequence.
Another gene, Deide_03051, was previously found to be difficult to annotate (25). The encoded protein, 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase (HPPK), is a key enzyme in the folate biosynthetic pathway. It was not detected in our proteomics analysis. With the DnaA and RpsL results in hand, we carefully reanalyzed its possible translation initiation start. Supplemental Fig. S6 shows an alignment of Deide_03051 locus with those of its closest homologues (Dgeo_0398 and DR_0170) as well as a homologue (Y0787; Protein Data Bank code 2QX0) from Yersinia pestis whose structure was recently solved (49). We had to reannotate Deide_03051 as starting from a CTG non-canonical initiation codon as conserved residues crucial for the protein fold (a β-sheet and a short α-helix) were missing. It is worth mentioning that Y0787 and PcnB (see above) are encoded, perhaps not entirely coincidentally, in the same locus in Y. pestis, but this is not the case in D. deserti.
Signal Peptide Maturation
We further investigated for the presence of TMPP-labeled peptide signatures showing signal peptide maturation. For this, we first predicted whether each annotated D. deserti polypeptide encompasses a putative signal peptide and then analyzed whether an experimental TMPP-labeled peptide signature matches these predictions. SignalP 3.0 server (37) consists of two different predictors based on neural networks and hidden Markov model algorithms for prediction of classically secreted proteins. The choice of this bioinformatics tool was based on recent benchmark studies (50). We found that 1119 polypeptides from D. deserti were predicted to contain a signal peptide either by the neural networks or hidden Markov model approaches proposed by this server. Supplemental Table S6 reports 43 TMPP-labeled peptides matching exactly to the predicted matured polypeptides. Different peptides are detected for some cases because of (i) the use of two proteases to generate our MS/MS data set and (ii) trypsin or chymotrypsin miscleavages. Therefore, N-terminal maturation of 34 proteins as predicted by the SignalP 3.0 server was confirmed. As an alternative, we calculated the position of all signal peptide maturation events possible for all D. deserti polypeptides as well as their probabilities of occurrence by means of the LipoP 1.0 server (38), which predicts cleavage sites for signal peptidases I and II. This algorithm predicted that 702 proteins could contain a signal peptide (significantly less than SignalP prediction), and 5019 putative maturation events were listed as well as their computed probabilities occurrence (supplemental Table S6). This represents an average of seven maturation alternatives per polypeptide. Of the 5019 events, 23 cases were found to match TMPP-labeled N-terminal-most peptides. They correspond to 14 different polypeptides. Among these, one is remarkable because several maturation events were detected. As indicated in supplemental Table S6, Deide_15510 prepolypeptide is predicted to be proteolyzed at different positions with almost identical probabilities. We found TMPP-labeled N-terminal-most peptides for three different positions: 25, 26, and 27. The two most probable cleavage sites proposed by the LipoP 1.0 and SignalP 3.0 algorithms (positions 27 and 26) were thus confirmed with a TMPP-labeled signature. As our labeling strategy is applied on entire intact proteins, postdigestion trimming of proteolyzed peptides does not account at this stage for such diversity. This protein is specific for D. deserti and encompasses in its C-terminal region a copper binding domain usually found in the plastocyanin/azurin protein family. Proteolytic cleavage through cellular proteases is thought to be often specific and tightly regulated (21). According to our experimental results, signal peptide maturations may, at least in some cases, be heterogeneous. Our results could represent a precious training data set to further improve the prediction accuracy of a subcellular localization classifier tool. Finally, we predicted maturations via non-classical secretory pathways with the TatFind 1.4 server (39), which detected the presence of prokaryotic twin arginine translocation signal peptides in 24 D. deserti proteins (supplemental Table S6). None of these were found in our TMPP-labeled peptide data set, although eight were detected by internal peptides (supplemental Table S2). For example, the Deide_04460 protein is detected through 20 different peptides that cover 90% of the 87–353 sequence. This protein exhibits some similarities with zinc-dependent hydrolases (COG0491). The 87–353 region corresponds well to the mature form after removal of the predicted twin arginine translocation signal peptide. Two other proteins, Deide_07540 and Deide_08080, are homologous to desiccation-related proteins found in some bacteria and plants (51). Both sequences are well covered beyond their predicted twin arginine translocation signal peptide with 70% sequence coverage in the 80–352 and 88–330 regions, respectively.
Concluding Remarks
A total of 341 protein N termini were confidently identified in the D. deserti TMPP-labeled proteome. Among these, 63 were not correctly annotated in the first D. deserti genome annotation and should be modified accordingly. We carefully checked the sequence similarities between the three sequenced Deinococcus genomes. On the basis of our results, we propose that N termini of 37 and 100 additional proteins from D. geothermalis and D. radiodurans genomes, respectively, should be reannotated. These reannotations are listed in supplemental Table S4. When considering the manually validated TMPP-modified peptides, 664 unique signatures for N termini were identified with 398 tryptic and 266 chymotryptic sequences. These two digestions were thus found to be complementary. Our experimental N termini data set corresponds to 10% of the theoretical proteome. Additional experiments are required to further uncover poorly characterized proteins. As benchmarked here from this large experimental data set (i.e. almost 18% of N termini were wrong), we expect that a significant number of erroneous annotations have probably still to be corrected. For Mycobacterium, an overprediction of ATG as the translational start codon was recently reported (18). For Deinococcus spp. genome annotations, most of our start site corrections are not due to false estimations of GTG or TTG less frequent initiation codons. They are rather due to the lack of homologous sequences arising from the few complete Deinococcus-Thermus genomes that have been sequenced so far.
D. radiodurans and D. deserti were shown to be tolerant to massive doses of γ-irradiation. Their mechanisms of radiotolerance have been studied for many years (26, 52–55) but are not yet fully understood. To achieve a detailed view of these mechanisms, it is important to obtain a comprehensive and accurate annotation of their genomes. Our study has shown that some genes were still missing in the parts list of D. deserti, although this bacterium has been analyzed by a large shotgun proteomics study at the primary stage of genome annotation (25). Moreover, some of the proposed corrections affect important genes involved in DNA repair mechanisms: polA (Deide_15130 and DR_1707), ligA (Dgeo_0696 and DR_2069), ddrB (DR_0070), and genes encoding two different endonucleases (DR_2438 and Deide_02842). It is worth mentioning that our experimentally based reannotation of ddrB, shown to code for an alternative single-stranded DNA-binding protein induced by ionizing radiation, was proposed and verified in a recent independent study (56). Interestingly, the use of non-canonical translation initiation codons in Deinococcus is unusually important with at least four genes concerned: dnaA, rpsL, Deide_03051, and infC. In the case of infC, the ATT (AUU at the mRNA level) initiation codon was shown to be an essential cis-acting element in autogenous translational control of translation initiation factor IF3 expression in vivo (45). We expected that the rationale for a non-canonical codon for the dnaA and hppk (Deide_0351) translation initiation is also a translationally specific control of the expression of these two genes. Because the former is crucial for the initiation of DNA replication, we highlighted here a specific regulation mechanism linking DNA replication and translation. Such a link was recently proposed solely on the basis of gene neighborhoods for Archaea and Eukarya (57, 58). Further experiments are needed to characterize this important link between fundamental mechanisms for the bacterial cells. That the folate biosynthetic pathway is also controlled at the translation level through Deide_0351 is probably not fortuitous. Reduced folate derivatives participate in numerous reactions of bacterial intermediary metabolism, and a coordinated physiological response linking translation and intermediary metabolism appears logical. In the present study specific strategies to identify protein N termini at a genome-wide scale, such as chemical labeling of N termini, have proved to be useful to achieve a comprehensive and accurate genome annotation. The specific corrections to D. deserti genome are important.
Supplementary Material
Acknowledgments
We gratefully acknowledge Olivier Pible (iBEB, Service de Biochimie et Toxicologie Nucléaire (SBTN), Laboratoire Intéractions et Reconnaissance Moléculaires) for precious help concerning macroprogramming for the Excel software, Eric Quéméneur and Thierry Heulin for constant support, and Jérôme Garin and Christophe Bruley for the IRMa software.
Footnotes
* This work was supported by the Commissariat à l'Energie Atomique and Agence Nationale de la Recherche Grant ANR-07-BLAN-0106-02).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1–S6 and Tables S1–S6.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. S1–S6 and Tables S1–S6.
2 A. Dedieu, P. Guérin, A. de Groot, J. Armengaud, unpublished results.
1 The abbreviations used are:
- TMPP
- N-tris(2,4,6-trimethoxyphenyl)phosphonium acetyl succinimide
- HPPK
- 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase
- 1D
- one-dimensional
- LTQ
- linear trap quadrupole.
REFERENCES
- 1.Lima T., Auchincloss A. H., Coudert E., Keller G., Michoud K., Rivoire C., Bulliard V., de Castro E., Lachaize C., Baratin D., Phan I., Bougueleret L., Bairoch A. ( 2009) HAMAP: a database of completely sequenced microbial proteome sets and manually curated microbial protein families in UniProtKB/Swiss-Prot. Nucleic Acids Res 37, D471– D478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Denoeud F., Aury J. M., Da Silva C., Noel B., Rogier O., Delledonne M., Morgante M., Valle G., Wincker P., Scarpelli C., Jaillon O., Artiguenave F. ( 2008) Annotating genomes with massive-scale RNA sequencing. Genome Biol 9, R175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ansong C., Purvine S. O., Adkins J. N., Lipton M. S., Smith R. D. ( 2008) Proteogenomics: needs and roles to be filled by proteomics in genome annotation. Brief. Funct. Genomics Proteomics 7, 50– 62 [DOI] [PubMed] [Google Scholar]
- 4.Armengaud J. ( 2009) A perfect genome annotation is within reach with the proteomics and genomics alliance. Curr. Opin. Microbiol 12, 292– 300 [DOI] [PubMed] [Google Scholar]
- 5.Zivanovic Y., Armengaud J., Lagorce A., Leplat C., Guérin P., Dutertre M., Anthouard V., Forterre P., Wincker P., Confalonieri F. ( 2009) Genome analysis and genome-wide proteomics of Thermococcus gammatolerans, the most radioresistant organism known amongst the Archaea. Genome Biol 10, R70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jaffe J. D., Berg H. C., Church G. M. ( 2004) Proteogenomic mapping as a complementary method to perform genome annotation. Proteomics 4, 59– 77 [DOI] [PubMed] [Google Scholar]
- 7.Baerenfaller K., Grossmann J., Grobei M. A., Hull R., Hirsch-Hoffmann M., Yalovsky S., Zimmermann P., Grossniklaus U., Gruissem W., Baginsky S. ( 2008) Genome-scale proteomics reveals Arabidopsis thaliana gene models and proteome dynamics. Science 320, 938– 941 [DOI] [PubMed] [Google Scholar]
- 8.Bindschedler L. V., Burgis T. A., Mills D. J., Ho J. T., Cramer R., Spanu P. D. ( 2009) In planta proteomics and proteogenomics of the biotrophic barley fungal pathogen blumeria graminis f. sp. hordei. Mol. Cell. Proteomics 8, 2368– 2381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Castellana N. E., Payne S. H., Shen Z., Stanke M., Bafna V., Briggs S. P. ( 2008) Discovery and revision of Arabidopsis genes by proteogenomics. Proc. Natl. Acad. Sci. U.S.A 105, 21034– 21038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta N., Benhamida J., Bhargava V., Goodman D., Kain E., Kerman I., Nguyen N., Ollikainen N., Rodriguez J., Wang J., Lipton M. S., Romine M., Bafna V., Smith R. D., Pevzner P. A. ( 2008) Comparative proteogenomics: combining mass spectrometry and comparative genomics to analyze multiple genomes. Genome Res 18, 1133– 1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ishino Y., Okada H., Ikeuchi M., Taniguchi H. ( 2007) Mass spectrometry-based prokaryote gene annotation. Proteomics 7, 4053– 4065 [DOI] [PubMed] [Google Scholar]
- 12.Jungblut P. R., Müller E. C., Mattow J., Kaufmann S. H. ( 2001) Proteomics reveals open reading frames in Mycobacterium tuberculosis H37Rv not predicted by genomics. Infect. Immun 69, 5905– 5907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Romine M. F., Elias D. A., Monroe M. E., Auberry K., Fang R., Fredrickson J. K., Anderson G. A., Smith R. D., Lipton M. S. ( 2004) Validation of Shewanella oneidensis MR-1 small proteins by AMT tag-based proteome analysis. Omics 8, 239– 254 [DOI] [PubMed] [Google Scholar]
- 14.Shevchenko A., Jensen O. N., Podtelejnikov A. V., Sagliocco F., Wilm M., Vorm O., Mortensen P., Shevchenko A., Boucherie H., Mann M. ( 1996) Linking genome and proteome by mass spectrometry: large-scale identification of yeast proteins from two dimensional gels. Proc. Natl. Acad. Sci. U.S.A 93, 14440– 14445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wright J. C., Sugden D., Francis-McIntyre S., Riba-Garcia I., Gaskell S. J., Grigoriev I. V., Baker S. E., Beynon R. J., Hubbard S. J. ( 2009) Exploiting proteomic data for genome annotation and gene model validation in Aspergillus niger. BMC Genomics 10, 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang R., Prince J. T., Marcotte E. M. ( 2005) Mass spectrometry of the M. smegmatis proteome: protein expression levels correlate with function, operons, and codon bias. Genome Res 15, 1118– 1126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deshayes C., Perrodou E., Gallien S., Euphrasie D., Schaeffer C., Van-Dorsselaer A., Poch O., Lecompte O., Reyrat J. M. ( 2007) Interrupted coding sequences in Mycobacterium smegmatis: authentic mutations or sequencing errors? Genome Biol 8, R20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallien S., Perrodou E., Carapito C., Deshayes C., Reyrat J. M., Van Dorsselaer A., Poch O., Schaeffer C., Lecompte O. ( 2009) Ortho-proteogenomics: multiple proteomes investigation through orthology and a new MS-based protocol. Genome Res 19, 128– 135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamazaki S., Yamazaki J., Nishijima K., Otsuka R., Mise M., Ishikawa H., Sasaki K., Tago S., Isono K. ( 2006) Proteome analysis of an aerobic hyperthermophilic crenarchaeon, Aeropyrum pernix K1. Mol. Cell. Proteomics 5, 811– 823 [DOI] [PubMed] [Google Scholar]
- 20.Aivaliotis M., Gevaert K., Falb M., Tebbe A., Konstantinidis K., Bisle B., Klein C., Martens L., Staes A., Timmerman E., Van Damme J., Siedler F., Pfeiffer F., Vandekerckhove J., Oesterhelt D. ( 2007) Large-scale identification of N-terminal peptides in the halophilic archaea Halobacterium salinarum and Natronomonas pharaonis. J. Proteome Res 6, 2195– 2204 [DOI] [PubMed] [Google Scholar]
- 21.Gupta N., Tanner S., Jaitly N., Adkins J. N., Lipton M., Edwards R., Romine M., Osterman A., Bafna V., Smith R. D., Pevzner P. A. ( 2007) Whole proteome analysis of post-translational modifications: applications of mass-spectrometry for proteogenomic annotation. Genome Res 17, 1362– 1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gevaert K., Goethals M., Martens L., Van Damme J., Staes A., Thomas G. R., Vandekerckhove J. ( 2003) Exploring proteomes and analyzing protein processing by mass spectrometric identification of sorted N-terminal peptides. Nat. Biotechnol 21, 566– 569 [DOI] [PubMed] [Google Scholar]
- 23.Staes A., Van Damme P., Helsens K., Demol H., Vandekerckhove J., Gevaert K. ( 2008) Improved recovery of proteome-informative, protein N-terminal peptides by combined fractional diagonal chromatography (COFRADIC). Proteomics 8, 1362– 1370 [DOI] [PubMed] [Google Scholar]
- 24.Chen W., Lee P. J., Shion H., Ellor N., Gebler J. C. ( 2007) Improving de novo sequencing of peptides using a charged tag and C-terminal digestion. Anal. Chem 79, 1583– 1590 [DOI] [PubMed] [Google Scholar]
- 25.de Groot A., Dulermo R., Ortet P., Blanchard L., Guérin P., Fernandez B., Vacherie B., Dossat C., Jolivet E., Siguier P., Chandler M., Barakat M., Dedieu A., Barbe V., Heulin T., Sommer S., Achouak W., Armengaud J. ( 2009) Alliance of proteomics and genomics to unravel the specificities of Sahara bacterium Deinococcus deserti. PLoS Genet 5, e1000434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cox M. M., Battista J. R. ( 2005) Deinococcus radiodurans—the consummate survivor. Nat. Rev. Microbiol 3, 882– 892 [DOI] [PubMed] [Google Scholar]
- 27.Airo A., Chan S. L., Martinez Z., Platt M. O., Trent J. D. ( 2004) Heat shock and cold shock in Deinococcus radiodurans. Cell. Biochem. Biophys 40, 277– 288 [DOI] [PubMed] [Google Scholar]
- 28.Lipton M. S., Pasa-Tolic' L., Anderson G. A., Anderson D. J., Auberry D. L., Battista J. R., Daly M. J., Fredrickson J., Hixson K. K., Kostandarithes H., Masselon C., Markillie L. M., Moore R. J., Romine M. F., Shen Y., Stritmatter E., Tolic' N., Udseth H. R., Venkateswaran A., Wong K. K., Zhao R., Smith R. D. ( 2002) Global analysis of the Deinococcus radiodurans proteome by using accurate mass tags. Proc. Natl. Acad. Sci. U.S.A 99, 11049– 11054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lipton M. S., Romine M. F., Monroe M. E., Elias D. A., Pasa-Tolic L., Anderson G. A., Anderson D. J., Fredrickson J., Hixson K. K., Masselon C., Mottaz H., Tolic N., Smith R. D. ( 2006) AMT tag approach to proteomic characterization of Deinococcus radiodurans and Shewanella oneidensis. Methods Biochem. Anal 49, 113– 134 [PubMed] [Google Scholar]
- 30.Lu H., Gao G., Xu G., Fan L., Yin L., Shen B., Hua Y. ( 2009) Deinococcus radiodurans PprI switches on DNA damage response and cellular survival networks after radiation damage. Mol. Cell. Proteomics 8, 481– 494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmid A. K., Lipton M. S., Mottaz H., Monroe M. E., Smith R. D., Lidstrom M. E. ( 2005) Global whole-cell FTICR mass spectrometric proteomics analysis of the heat shock response in the radioresistant bacterium Deinococcus radiodurans. J. Proteome Res 4, 709– 718 [DOI] [PubMed] [Google Scholar]
- 32.Zhang C., Wei J., Zheng Z., Ying N., Sheng D., Hua Y. ( 2005) Proteomic analysis of Deinococcus radiodurans recovering from gamma-irradiation. Proteomics 5, 138– 143 [DOI] [PubMed] [Google Scholar]
- 33.White O., Eisen J. A., Heidelberg J. F., Hickey E. K., Peterson J. D., Dodson R. J., Haft D. H., Gwinn M. L., Nelson W. C., Richardson D. L., Moffat K. S., Qin H., Jiang L., Pamphile W., Crosby M., Shen M., Vamathevan J. J., Lam P., McDonald L., Utterback T., Zalewski C., Makarova K. S., Aravind L., Daly M. J., Minton K. W., Fleischmann R. D., Ketchum K. A., Nelson K. E., Salzberg S., Smith H. O., Venter J. C., Fraser C. M. ( 1999) Genome sequence of the radioresistant bacterium Deinococcus radiodurans R1. Science 286, 1571– 1577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Makarova K. S., Omelchenko M. V., Gaidamakova E. K., Matrosova V. Y., Vasilenko A., Zhai M., Lapidus A., Copeland A., Kim E., Land M., Mavrommatis K., Pitluck S., Richardson P. M., Detter C., Brettin T., Saunders E., Lai B., Ravel B., Kemner K. M., Wolf Y. I., Sorokin A., Gerasimova A. V., Gelfand M. S., Fredrickson J. K., Koonin E. V., Daly M. J. ( 2007) Deinococcus geothermalis: the pool of extreme radiation resistance genes shrinks. PLoS ONE 2, e955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Henne A., Brüggemann H., Raasch C., Wiezer A., Hartsch T., Liesegang H., Johann A., Lienard T., Gohl O., Martinez-Arias R., Jacobi C., Starkuviene V., Schlenczeck S., Dencker S., Huber R., Klenk H. P., Kramer W., Merkl R., Gottschalk G., Fritz H. J. ( 2004) The genome sequence of the extreme thermophile Thermus thermophilus. Nat. Biotechnol 22, 547– 553 [DOI] [PubMed] [Google Scholar]
- 36.Dupierris V., Masselon C., Court M., Kieffer-Jaquinod S., Bruley C. ( 2009) A toolbox for validation of mass spectrometry peptides identification and generation of database: IRMa. Bioinformatics 25, 1980– 1981 [DOI] [PubMed] [Google Scholar]
- 37.Emanuelsson O., Brunak S., von Heijne G., Nielsen H. ( 2007) Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc 2, 953– 971 [DOI] [PubMed] [Google Scholar]
- 38.Juncker A. S., Willenbrock H., Von Heijne G., Brunak S., Nielsen H., Krogh A. ( 2003) Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci 12, 1652– 1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rose R. W., Brüser T., Kissinger J. C., Pohlschröder M. ( 2002) Adaptation of protein secretion to extremely high-salt conditions by extensive use of the twin-arginine translocation pathway. Mol. Microbiol 45, 943– 950 [DOI] [PubMed] [Google Scholar]
- 40.Jones P., Côté R. G., Cho S. Y., Klie S., Martens L., Quinn A. F., Thorneycroft D., Hermjakob H. ( 2008) PRIDE: new developments and new datasets. Nucleic Acids Res 36, D878– D883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nguyen H. H., de la Tour C. B., Toueille M., Vannier F., Sommer S., Servant P. ( 2009) The essential histone-like protein HU plays a major role in Deinococcus radiodurans nucleoid compaction. Mol. Microbiol 73, 240– 252 [DOI] [PubMed] [Google Scholar]
- 42.Binns N., Masters M. ( 2002) Expression of the Escherichia coli pcnB gene is translationally limited using an inefficient start codon: a second chromosomal example of translation initiated at AUU. Mol. Microbiol 44, 1287– 1298 [DOI] [PubMed] [Google Scholar]
- 43.Hatfield D., Diamond A. ( 1993) UGA: a split personality in the universal genetic code. Trends Genet 9, 69– 70 [DOI] [PubMed] [Google Scholar]
- 44.Liveris D., Schwartz J. J., Geertman R., Schwartz I. ( 1993) Molecular cloning and sequencing of infC, the gene encoding translation initiation factor IF3, from four enterobacterial species. FEMS Microbiol. Lett 112, 211– 216 [DOI] [PubMed] [Google Scholar]
- 45.Polard P., Prère M. F., Chandler M., Fayet O. ( 1991) Programmed translational frameshifting and initiation at an AUU codon in gene expression of bacterial insertion sequence IS911. J. Mol. Biol 222, 465– 477 [DOI] [PubMed] [Google Scholar]
- 46.Sazuka T., Ohara O. ( 1996) Sequence features surrounding the translation initiation sites assigned on the genome sequence of Synechocystis sp. strain PCC6803 by amino-terminal protein sequencing. DNA Res 3, 225– 232 [DOI] [PubMed] [Google Scholar]
- 47.Spiers A. J., Bergquist P. L. ( 1992) Expression and regulation of the RepA protein of the RepFIB replicon from plasmid P307. J. Bacteriol 174, 7533– 7541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Butler J. S., Springer M., Grunberg-Manago M. ( 1987) AUU-to-AUG mutation in the initiator codon of the translation initiation factor IF3 abolishes translational autocontrol of its own gene (infC) in vivo. Proc. Natl. Acad. Sci. U.S.A 84, 4022– 4025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Blaszczyk J., Li Y., Cherry S., Alexandratos J., Wu Y., Shaw G., Tropea J. E., Waugh D. S., Yan H., Ji X. ( 2007) Structure and activity of Yersinia pestis 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase as a novel target for the development of antiplague therapeutics. Acta Crystallogr. Sect. D Biol. Crystallogr 63, 1169– 1177 [DOI] [PubMed] [Google Scholar]
- 50.Restrepo-Montoya D., Vizcaíno C., Niño L. F., Ocampo M., Patarroyo M. E., Patarroyo M. A. ( 2009) Validating subcellular localization prediction tools with mycobacterial proteins. BMC Bioinformatics 10, 134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Battista J. R., Park M. J., McLemore A. E. ( 2001) Inactivation of two homologues of proteins presumed to be involved in the desiccation tolerance of plants sensitizes Deinococcus radiodurans R1 to desiccation. Cryobiology 43, 133– 139 [DOI] [PubMed] [Google Scholar]
- 52.Blasius M., Sommer S., Hübscher U. ( 2008) Deinococcus radiodurans: what belongs to the survival kit? Crit. Rev. Biochem. Mol. Biol 43, 221– 238 [DOI] [PubMed] [Google Scholar]
- 53.Daly M. J. ( 2009) A new perspective on radiation resistance based on Deinococcus radiodurans. Nat. Rev. Microbiol 7, 237– 245 [DOI] [PubMed] [Google Scholar]
- 54.Daly M. J., Gaidamakova E. K., Matrosova V. Y., Vasilenko A., Zhai M., Venkateswaran A., Hess M., Omelchenko M. V., Kostandarithes H. M., Makarova K. S., Wackett L. P., Fredrickson J. K., Ghosal D. ( 2004) Accumulation of Mn(II) in Deinococcus radiodurans facilitates gamma-radiation resistance. Science 306, 1025– 1028 [DOI] [PubMed] [Google Scholar]
- 55.Zahradka K., Slade D., Bailone A., Sommer S., Averbeck D., Petranovic M., Lindner A. B., Radman M. ( 2006) Reassembly of shattered chromosomes in Deinococcus radiodurans. Nature 443, 569– 573 [DOI] [PubMed] [Google Scholar]
- 56.Norais C. A., Chitteni-Pattu S., Wood E. A., Inman R. B., Cox M. M. ( 2009) An alternative Deinococcus radiodurans SSB induced by ionizing radiation: the DdrB protein. J. Biol. Chem 284, 21402– 21411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Berthon J., Cortez D., Forterre P. ( 2008) Genomic context analysis in Archaea suggests previously unrecognized links between DNA replication and translation. Genome Biol 9, R71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Berthon J., Fujikane R., Forterre P. ( 2009) When DNA replication and protein synthesis come together. Trends Biochem. Sci 34, 429– 434 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.