Summary
A forward genetic screen in Arabidopsis led to the isolation of several arsenic tolerance mutants. ars5 is the strongest arsenate and arsenite resistant mutant identified in this genetic screen. Here, we report the characterization and cloning of the ars5 mutant gene. ars5 is shown to exhibit an increased accumulation of arsenic and thiol compounds during arsenic stress. Rough mapping together with microarray-based expression mapping identified the ars5 mutation in the alpha subunit F (PAF1) of the 26S proteasome complex. Characterization of an independent paf1 T-DNA insertion allele and complementation by PAF1 confirmed that paf1 mutation is responsible for the enhanced thiol accumulation and the arsenic tolerance phenotypes. Arsenic tolerance was not observed in a knockout mutant of the highly homologous PAF2 gene. However, genetic complementation of ars5 by over expression of PAF2 suggests that the PAF2 protein is functionally equivalent to PAF1 when expressed at high levels. No detectible difference was observed in total ubiquitinylated protein profiles between ars5 and wild type Arabidopsis, suggesting that the arsenic tolerance observed in ars5 is not derived from a general impairment in proteasome-mediated protein degradation. Quantitative RT-PCR showed that arsenic induces enhanced transcriptional activation of several key genes that function in glutathione and phytochelatin biosynthesis in wild type and this arsenic-induction of gene expression is more dramatic in ars5. The enhanced transcriptional response to arsenic and the increased accumulation of thiol compounds in ars5 compared to WT suggest the presence of a positive regulation pathway for thiol biosynthesis that is enhanced in the ars5 background.
Keywords: arsenic accumulation, glutathione, phytochelatins, micoarray-based cloning, qPCR
Introduction
Arsenic (As) is ubiquitous in the environment and found primarily in inorganic forms, with As5+ (arsenate) and As3+ (arsenite) as the most common As oxidation states (Brown et al., 2002). Inorganic arsenic has been identified as the main cause of cancers in humans, including urinary bladder, lung and non melanoma skin cancers (Lubin et al., 2007). Therefore, inorganic arsenic contamination in the environment poses a serious threat to human health. Arsenic is listed as the most toxic compound found at US Superfund sites (http://epa.gov/superfund/policy/cercla.htm). Irrigation of vegetables and crop plants with arsenic-contaminated water and accumulation of As by plants causes arsenic exposure to humans through daily diet (Alam et al., 2003). Thus, the identification of the processes driving arsenic accumulation in plants, as well as removal of arsenic from contaminated environments are important priorities to reduce As intake in humans. Arsenic removal methods using plant systems (phytoremediation) offer possible non-invasive and inexpensive alternatives to conventional excavation methods (Cobbett and Meagher, 2002; Pilon-Smits and Pilon, 2002; Tripathi et al., 2007).
Understanding the mechanisms that plants employ to respond to and detoxify arsenic is essential for optimal and efficient removal of arsenic from contaminated environments through the use of plants. Arsenic is present as either arsenate (AsO4−3) or arsenite (AsO3−3) depending on pH, redox potential, drainage conditions, and phosphorus content in soil (Marin et al., 1993). Arsenate, being similar to phosphate, mainly enters plants through phosphate transporters (Meharg and Macnair, 1992; Wang et al., 2002; Catarecha et al., 2007) and interferes with glycolysis and oxidative phosphorylation, inhibiting the synthesis of ATP (Carbonell et al., 1998; Hindmarsh, 2000).
Arsenic hyper-accumulation in the fern Pteris vittata decreased when the phosphate level was increased in the medium (Wang et al., 2002). Similarly, a mutation in a high affinity phosphate transporter (PHT1;1) in Arabidopsis resulted in a slower rate of arsenic uptake compared to wild type and conferred arsenic tolerance to the mutant (pht1;1–3) (Catarecha et al., 2007). Another major arsenic tolerance mechanism is thiolmediated chelation and sequestration of arsenic, in the form of arsenite. Arsenate is reduced to arsenite in plants by an arsenate reductase and this reduced form of arsenate is able to bind thiols with high affinity (Schmöger et al., 2000). Arsenite is also able to induce the synthesis of phytochelatins (PCs) and it is thought that PC-As complexes are transported and stored in vacuoles (Dhankher et al., 2002). Thiol compounds also function in the long distance transport of heavy metals between shoots and roots (Gong et al., 2003; Chen et al., 2006; Li et al., 2006; Mendoza-Cózatl et al., 2008). In addition, and in contrast to the results obtained with cadmium (Lee et al., 2003b; Li et al., 2004; Li et al., 2005), over expression of proteins that synthesize glutathione and phytochelatins resulted in an increased arsenic tolerance (Dhankher et al., 2002; Li et al., 2004, 2005, 2006; Gasic and Korban, 2007).
Only few genetic mutants are known in plants that cause arsenic resistance. We have developed a screen to isolate arsenic tolerant mutants (Lee et al., 2003a). The strongest arsenate and arsenite tolerant mutant isolated from this screen is ars5 (Sung et al., 2007). The ars5 mutant was identified as a double mutant in the ARS4 and ARS5 loci. The ARS4 locus encodes phytochrome A (Sung et al., 2007), however, the gene and mechanism underlying the ars5 phenotype have remained unknown even though the ars5 mutation causes a stronger degree of arsenic tolerance than ars4 (phyA) (Sung et al., 2007).
Here, we report the presence of a novel arsenic tolerance mechanism by the cloning and characterization of ars5, a null mutant defective in the gene encoding a subunit of the 20S proteasome core particle. ARS5 encodes the proteasome alpha subunit F1 protein (PAF1) of the 26S proteasome complex. 26S proteasome-mediated protein degradation has been implicated in regulation of signaling mechanisms important for plant responses to various environmental stresses (Smalle et al., 2003; Kurepa et al., 2008; Book et al., 2009). In addition, the present study shows that PAF1 is also a negative regulator of thiol-mediated arsenic tolerance.
Results
Arsenic tolerance in ars5
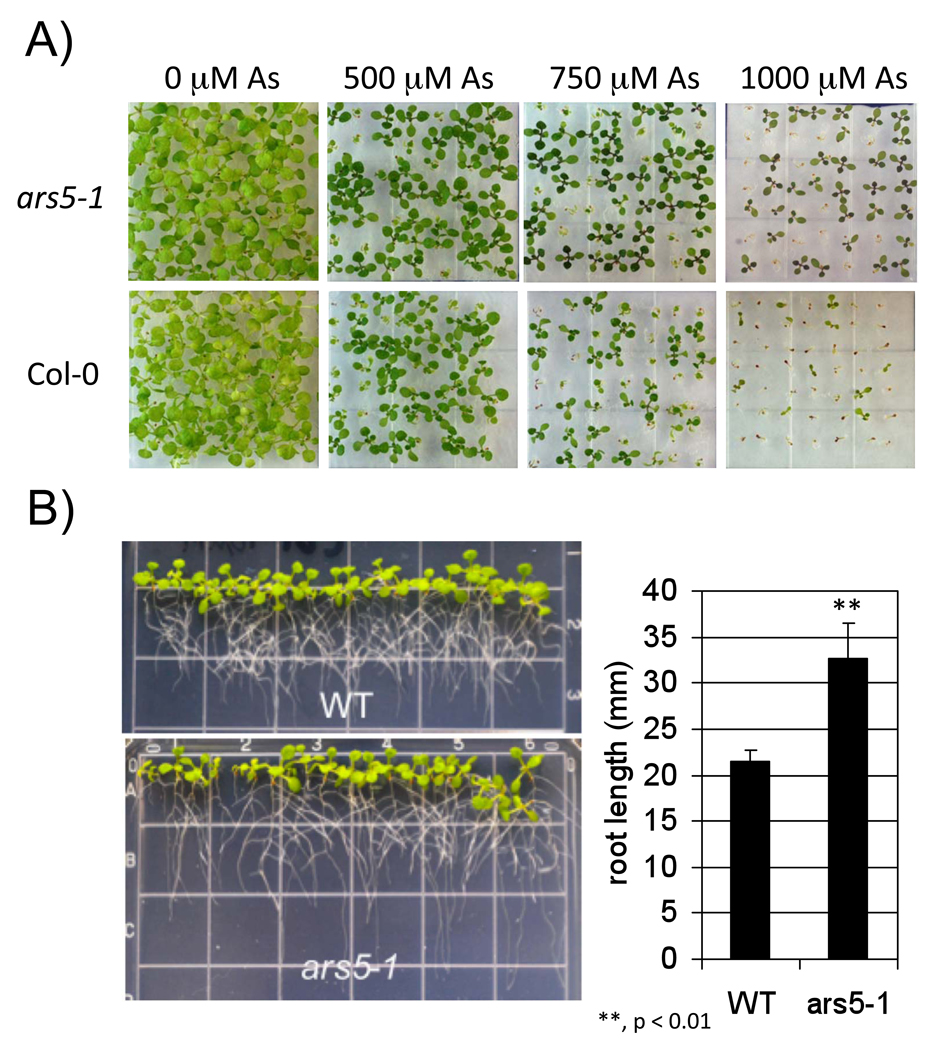
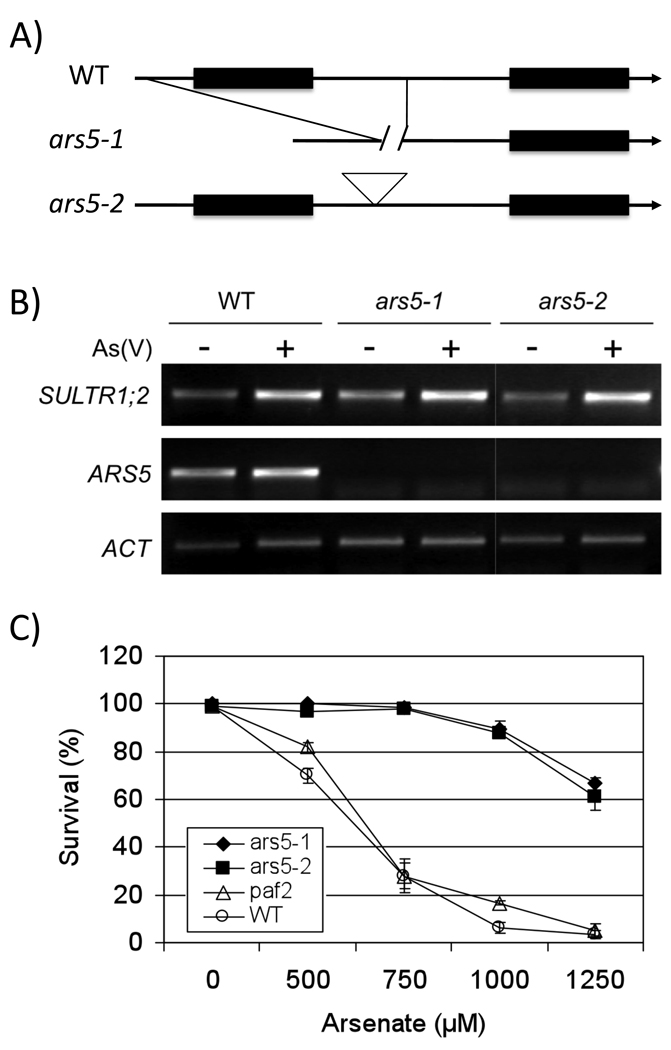
We have pursued screens for mutants with increased arsenic tolerance (Lee et al., 2003a; Sung et al 2007). One particular mutant, ars4ars5 contained two independent mutations and each mutation contributed to the overall arsenic tolerance. ars5 was isolated from the ars4ars5 double mutant by back-crossing the ars4ars5 double mutant with wild type Col-0 (Sung et al., 2007). The single ars5 mutant retained the major portion of arsenic tolerance from the ars4ars5 double mutant (Figure 1A). No major phenotypic differences were found between wild type and ars5-1 in the absence of arsenate under the imposed conditions. Wild type and ars5 (named ars5-1 from here on) seeds were germinated for one week on ¼ MS growth medium and exposed to a range of arsenate concentrations (Figure 1A), while germination and growth of wild type plants was inhibited by arsenate treatment, ars5-1 seedlings showed enhanced tolerance to arsenate at concentrations ranging from 500 µM to 1000 µM (Figure 1A). ars5-1 also showed an increased tolerance to arsenite in root growth assays in the presence of 10 µM AsIII (Figure 1B).
Figure 1. Arsenic tolerant seedling growth of ars5-1.
(A) WT and ars5-1 seeds were grown on minimal growth medium with 0, 500, 750, and 1000 µM arsenate (AsV). Images were photographed after one week of growth at 22 °C. (B) Seedlings (4 day-old) from WT and ars5-1 were grown on minimal medium and transferred to plates containing 10 µM arsenite (AsIII). Seedlings were grown for an additional week for root length measurements. ** denotes statistical differences (p < 0.01) between ars5-1 vs WT.
Arsenic accumulation is increased in ars5
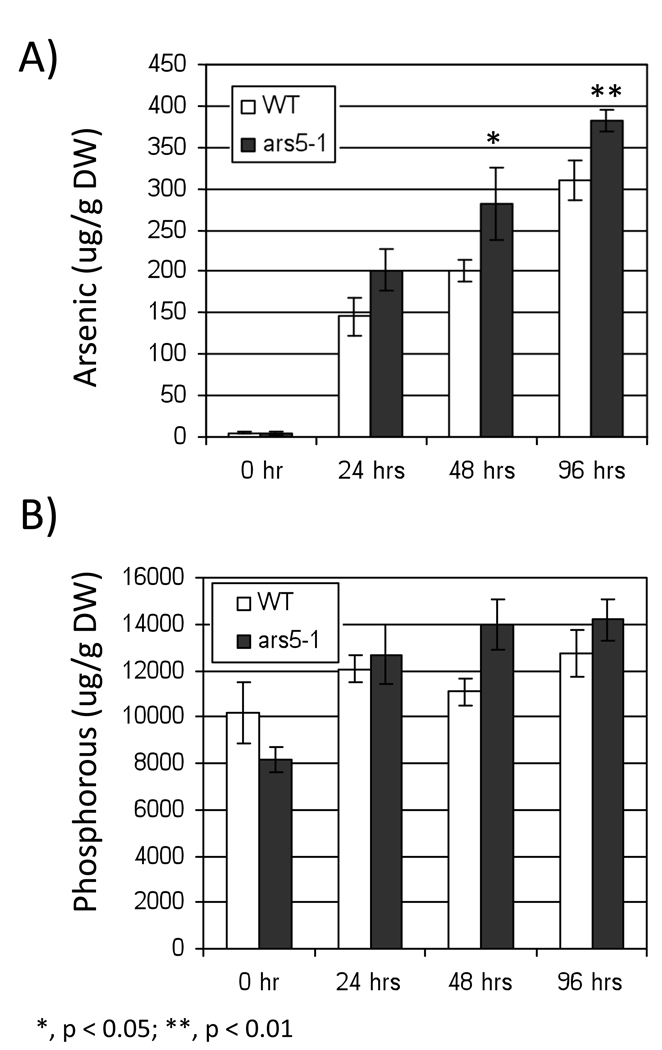
In some plants, arsenic tolerance has been related to a decreased rate of arsenic uptake. To test whether this is the case for arsenic tolerance in ars5-1, we analyzed the accumulation of arsenic and other elements in WT and ars5-1 seedlings exposed to 100 µM arsenate for 0, 24, 48, 96 hrs. ars5-1 plants exhibited a slight average increase in arsenic accumulation at 24, 48 and 96 hrs after arsenic exposure (Figure 2A, WT vs ars5-1 at 24 h P = 0.008), showing that the arsenic tolerance of ars5 did not result from a decreased arsenic accumulation. Phosphorus accumulation between ars5-1 and wild type did not show any statistically significant increase (Figure 2B), suggesting that phosphate metabolism is not related to the increased arsenic tolerance of ars5-1.
Figure 2. Arsenic and phosphorus accumulation in arsenate exposed wild type and ars5-1 seedlings.
One week-old seedlings were exposed to 100 µM potassium arsenate for 0, 24, 48, 96 hrs and the levels of arsenic (A) and phosphorus (B) were analyzed by ICP-OES. Y-axis shows the concentration of elements. Error bars represent standard error of the mean (n=4). The different levels of statistical significance between ars5-1 and wild type, at equal times of arsenic exposure, are denoted with asterisks (*, p < 0.05; **, p < 0.01).
Thiol synthesis is increased in ars5 mutants
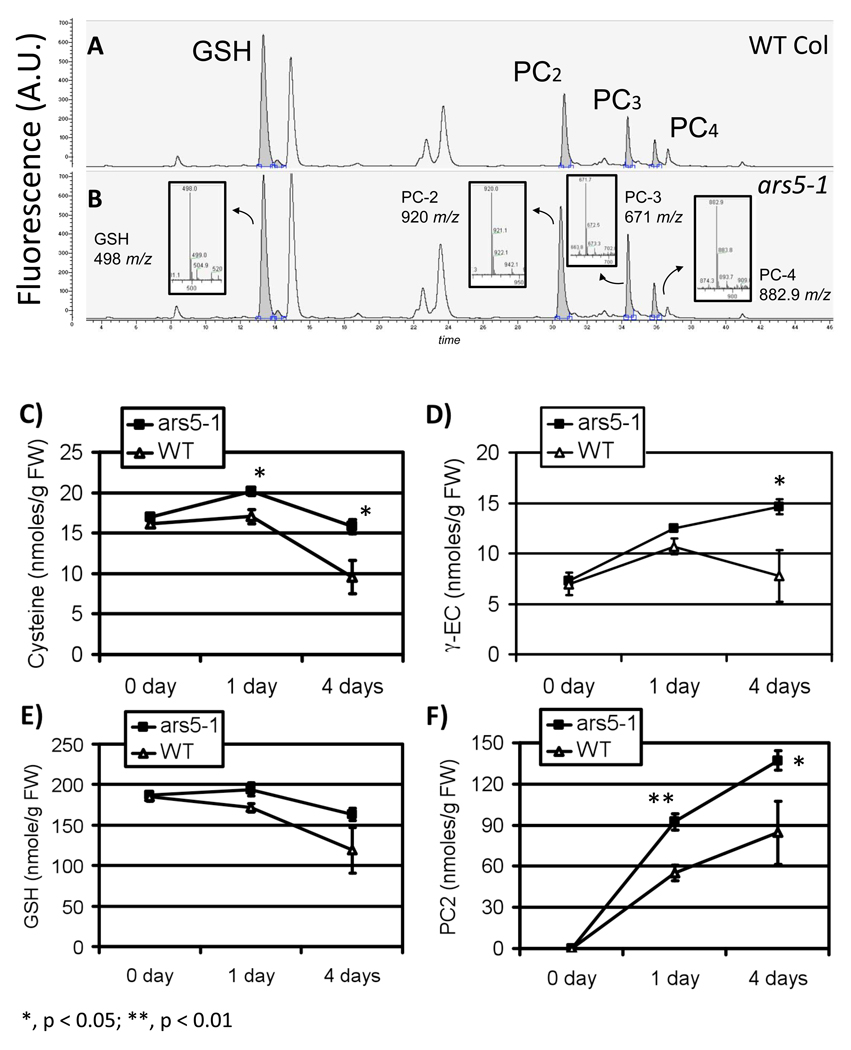
Fluorescence HPLC combined with tandem mass spectrometry analyses of thiols in WT and ars5-1 mutants (Figure 3A–B) showed that, during arsenic stress, ars5-1 contained more organic thiols (glutathione and phytochelatins) compared to WT seedlings (Figure 3C–F). These compounds are well-known arsenic chelating agents that have been described as key metabolites for arsenic tolerance in plants. Increased accumulation of these thiol-compounds could explain, at least in part, the increased arsenic tolerance phenotype of ars5-1 and also suggests that thiol biosynthesis is up-regulated in this mutant.
Figure 3. Thiol quantification in ars5-1 and WT.
HPLC-MS chromatograms of WT (A) or ars5-1 (B) exposed to 100 µM potassium arsenate for 96 hrs. Thiols were separated by HPLC, identified by tandem mass spectrometry and quantified by their respective bimane-derivative fluorescence. (C–F) Col-0 WT and ars5-1 seedlings (5 day-old) were treated with 100 µM arsenate for 0, 1, 4 days. The levels of (C) cysteine, (D) γ-EC, (E) GSH and (F) PC2 in whole seedlings were measured by fluorescence HPLC. Error bars represent SEM (n=3). The different levels of statistical significance between ars5-1 and wild type, at equal times of arsenic exposure, are denoted with asterisks (*, p < 0.05; **, p < 0.01).
ARS5 encodes an alpha subunit F (PAF1) of the 20S proteasome complex
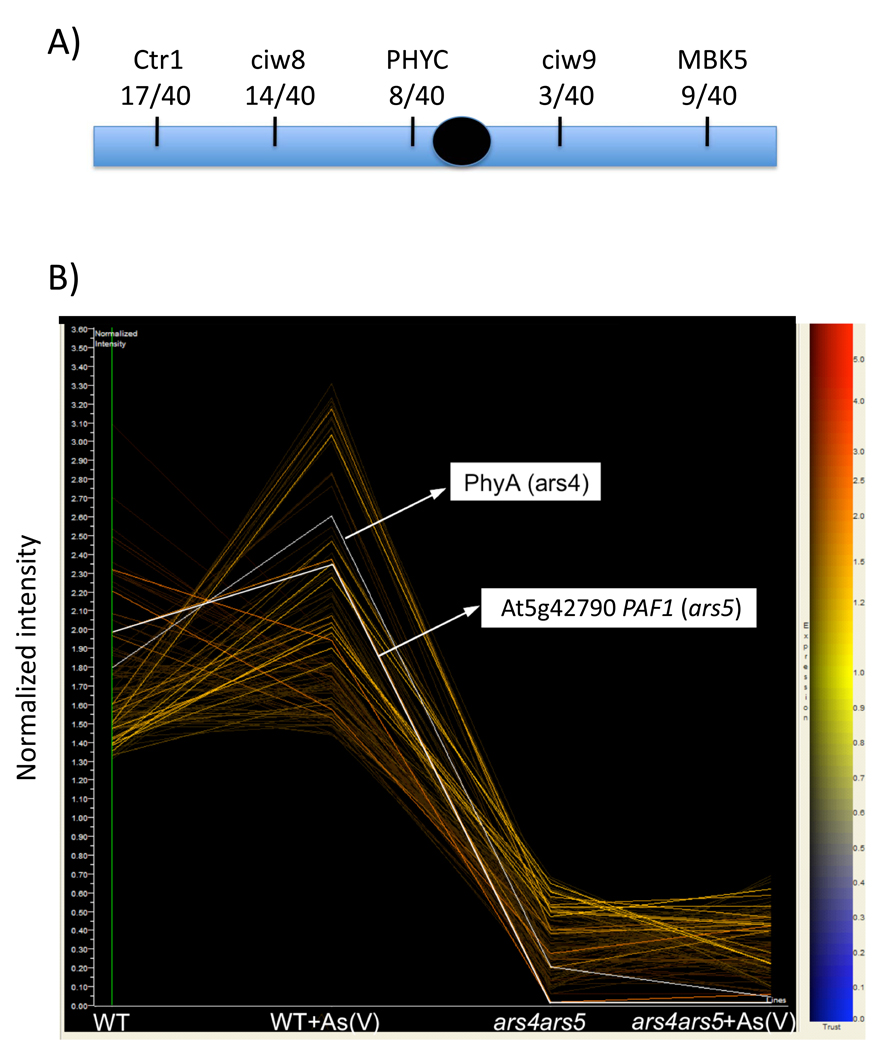
ars5-1 was isolated as an arsenic tolerant recessive mutant from the activation tagged ars4ars5 double mutant. To identify the ARS5 gene, a combined approach of genetic map-based cloning and microarray expression analysis was taken. Rough mapping of ars5-1 positioned the ARS5 locus in the region of the CIW9 marker on the lower arm of Chromosome 5 (Figure 4A). In order to identify genes with an altered expression in ars5-1, microarray expression data of ars4ars5 double mutant plants was investigated (Supplementary File 1). Including the ARS4/PHYA gene, we identified 110 genes whose expression was strongly reduced in the ars4ars5 double mutant compared to WT regardless of the presence or absence of any arsenic stress (Figure 4B; Supplementary File 1). Of the 110 genes, 15 genes are located on Chromosome 5 and only one candidate gene (At5g42790; Figure 4B), close to the CIW9 marker, contained a genetic mutation (deletion) within the gene (Figure 5A). PCR sequence analysis of the genomic region of At5g42790 revealed a deletion of a 1.4 kb genomic DNA fragment, encompassing part of the promoter and the entirety of the first exon of At5g42790 (Figure 5A).
Figure 4. Microarray-based mapping of ARS5.
(A) PCR-based markers on chromosome 5 are indicated at the top diagram. The number of recombination events per total number of meiotic events scored is given below each marker. (B) Microarray experiments of WT and ars4ars5 double mutant plants grown with or without arsenate identified 110 strongly repressed genes in ars4ars5 in the presence or absence of arsenic stress. Highlighted arrows show the expression values of ARS5 (At5g42790) and ARS4/PHYA (At1g09570). The vertical axis of the graph represents normalized gene expression levels.
Figure 5. Identification of the ars5 gene and analysis of arsenic tolerance of ars5-1, ars5-2 (paf1) and paf2.
(A) Two ars5 mutant alleles, ars5-1 and ars5-2 (paf1): ars5-1 was isolated from the ars4ars5 double mutant and has a 1.4Kb deletion spanning part of the promoter and the entire first exon of the ARS5 gene. ars5-2 is GABI T-DNA insertion allele (GABI_419_H03) and contains a T-DNA in intron 1. (B) RT-PCR analysis of WT, ars5-1, ars5-2 (SALK_098236), and paf2 (GABI_419_H03). Sultr1;2 (At1g78000) was used as a positive control of an arsenic-induced transcript. Transcript levels were analyzed by RT-PCR of ARS5 (At5g42790), PAF2 (At1g47250) and ACT7 (AT5G09810). (−): control, (+): arsenic stress (100 µM arsenate for 24 hours). (C) arsenic tolerant germination of ars5 and paf2 mutant alleles. Y-axis represents the percent of surviving seedlings under arsenic stress after 4 days at 22 °C. Seedlings with open green cotyledons were scored as surviving. X-axis represents the concentration of arsenate (µM). Error bars represent SEM (n=3).
The 26S proteasome complex is composed of two sub-particles, the 20S core particle and the19S regulatory particle. The 20S core particle consists of two rings formed by alpha subunits and two additional rings formed by beta subunits. Each ring is composed by seven subunits, named A to G. The At5g42790 gene encodes an alpha subunit F (PAF) of the 20S core particle and could be encoded by two paralogous genes: PAF1 (At5g42790) and PAF2 (At1g47250). We searched the public T-DNA insertional mutant populations and isolated a mutant allele of paf1 (GABI_419_H03) (Figure 5B). RT-PCR analysis showed no detectible PAF1 transcript in either ars5-1 or in the paf1 mutant (ars5-2, see below) (Figure 5B and Figure S1). The paf1 insertional mutant also showed an enhanced arsenic tolerance in germination assays (Figure 5C), an increased content of arsenic and elevated levels of GSH when compared to wild type (see below). Both ars5-1 and ars5-2 showed 80 % germination by the 4th day in growth medium containing 1 mM arsenate, compared to a 20 % germination rate of WT Col (Figure 5C). The wild type IC50 for arsenate was determined to be 742 ± 19 µM (n = 5, ± SD) while at an arsenate concentration of 1250 µM, more than 60% of ars5-1 and paf1 seeds were still able to germinate (Figure 5). These findings confirm that the arsenic resistance of ars5 is due to disruption of the PAF1 gene. Therefore, the paf1 mutant was named ars5-2.
To further explore the role of PAF proteins in arsenic tolerance in Arabidopsis, a paf2 T-DNA insertion mutant was isolated (SALK_098236). RT-PCR analysis showed no PAF2 transcript, indicating that paf2 is also a null mutant (data not shown). However, the paf2 mutant did not show arsenic tolerance under any of the conditions tested. To investigate the consequences of a paf1paf2 double knockout on arsenic tolerance in Arabidopsis, ars5-1 and ars5-2 were crossed with paf2 and a F2 population was screened for homozygous double mutants. Extensive PCR genotyping did not produce any homozygous double mutants indicating that the paf1paf2 double mutant is lethal. Furthermore, the absence of ars5-1ars5-1/paf2+ and ars5-1+/paf2paf2 alleles in the F2 populations indicates that gametophytes are not viable if both genes are missing in the gametophyte (Table 1).
Table 1.
Segregation in the F2 population from ars5-1/ars5-1 x paf2/paf2 crosses.
| ++ | +paf2 | ars5-1+ | ars5-1paf2 | |
| ++ | ++/++ | ++/paf2+ | ars5-1+/++ | X |
| +paf2 | ++/paf2+ | ++/paf2 paf2 | ars5-1+/paf2+ | X |
| ars5-1 | ars5-1+/++ | ars5-1+/paf2+ | ars5-1ars5-1/++ | X |
| ars5-1paf2 | X | X | X | X |
+: wild type copy of the gene
x: The offspring was lethal because gametophytes with the ars5-1paf2 double mutation did not survive.
PAF1 and PAF2 complement ars5
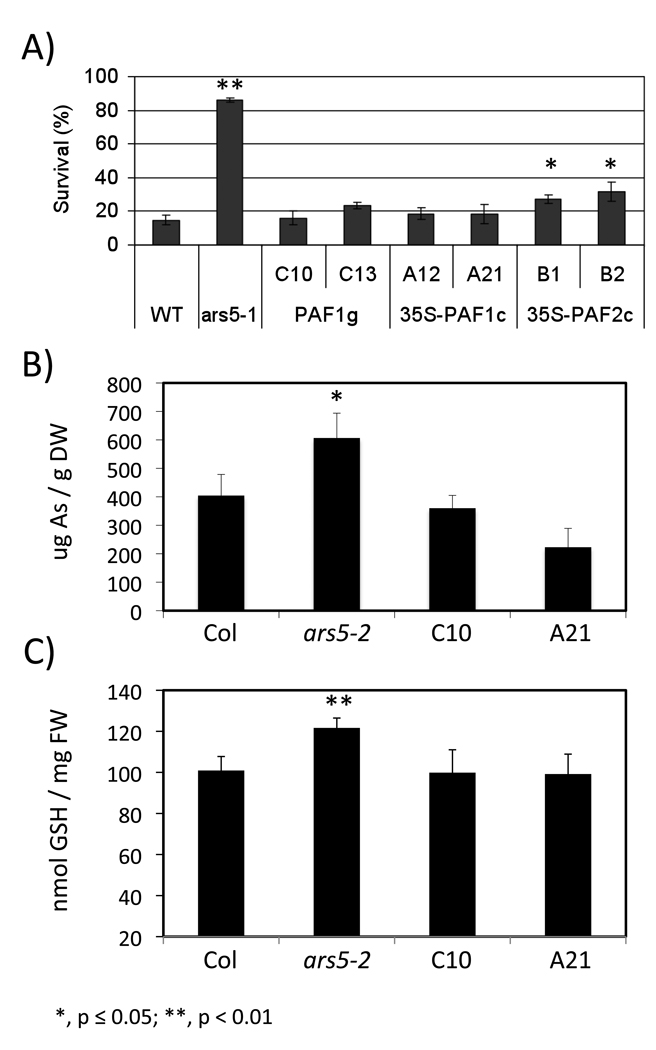
A 7 kb genomic DNA segment containing the native PAF1 promoter and the PAF1 coding region (PAF1g) complements the original ars5-1 mutant (Figure 6A). ars5-1 was also complemented by over-expressing the PAF1 or PAF2 cDNAs under the control of the 35S promoter (35S–PAF1c and 35S–PAF2c) (Figure 6A). Expression of the transgenes was confirmed by RT-PCR (Figure S2). The ars5-2 allele also showed increased levels of arsenic and glutathione content after 96 h of arsenic exposure (Figure 6B–C). ars5-1 complemented with either genomic or PAF1 cDNA showed restored levels of both arsenic and glutathione content compared to WT (Figure 6 B–C). These results confirmed that the arsenic tolerant phenotype of ars5-1 resulted from the null mutation of PAF1 and that both PAF1 and PAF2 are functionally capable of complementing ars5-1. However, the endogenous expression of PAF2 was not sufficient to replace the loss of PAF1 in the ars5-1 mutant.
Figure 6. Genetic complementation of ars5-1 and characterization of the ars5-2 allele.
(A) Seeds from Col-0 WT, ars5-1, two lines complemented with genomic DNA (C10 and C13; promoter and the coding region of PAF1, PAF1g), two lines complemented PAF1 cDNA (A12 and A21; 35S–PAF1c) and two lines complemented with PAF2 cDNA (B1 and B2; 35S–PAF2c) were germinated on minimal medium containing 1000 µM arsenate. Seedling survival was scored after 4 days at 22 °C. Error bars represent SEM (n=3). (B–C) Accumulation of arsenic (B) and GSH content (C), expressed in thiol (-SH) equivalents, in ars5-2 and two independent lines of ars5-1 complemented with a genomic fragment of PAF1 (C10) or with the PAF1 cDNA under the control of the 35S promoter (A21). Statistical differences between ars5-2 ars5-1 and ars5-1 complemented with PAF2 vs wild type are denoted with asterisks (*, p ≤ 0.05; **, p < 0.01). One week-old seedlings were exposed to 100 µM potassium arsenate for 96 hrs and the levels of arsenic were measured by ICP-OES and the glutathione content was measured by fluorescence HPLC. Error bars represent standard errors of the mean (n=3).
Increased expression of key metabolic genes in thiol biosynthesis is ars5
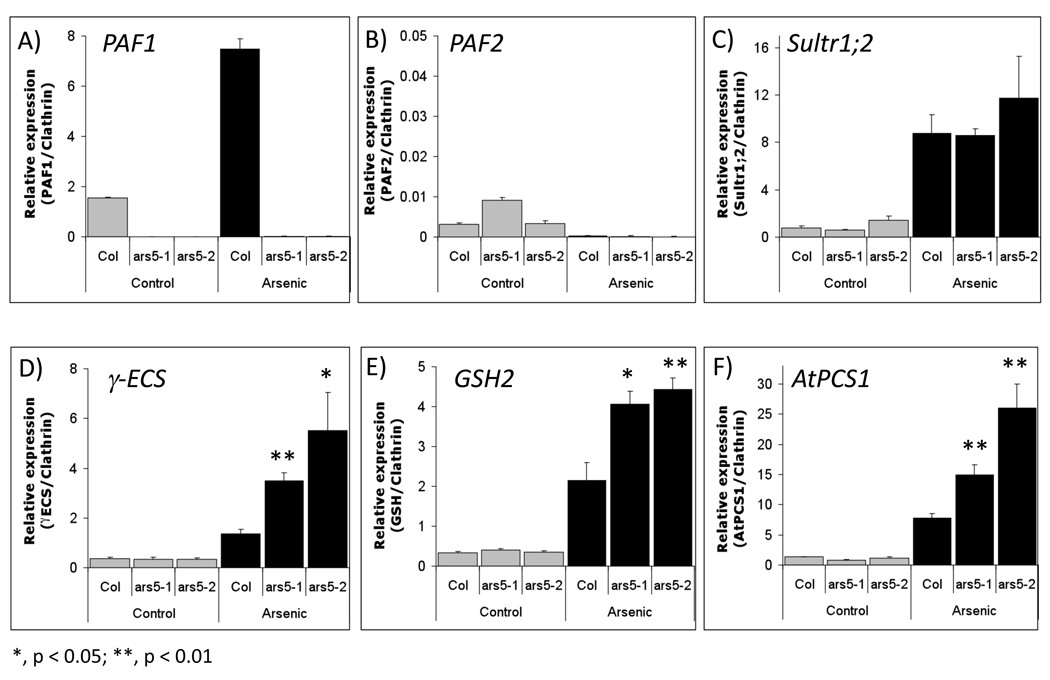
To test whether increased accumulation of thiol compounds in ars5-1 during arsenic exposure is the result of a transcriptional activation response, the expression of key metabolic genes involved in thiol biosythesis was analyzed. 5 day-old seedlings of WT, ars5-1 and ars5-2 were treated with 100 µM arsenate for 96 hrs and subjected to quantitative real-time qPCR analysis. PAF1, but not PAF2, was induced by arsenic treatment (Figure 7A–B), suggesting a role for PAF1 and the 26S proteasome during arsenic exposure. Sultr1;2, a sulfate transporter known to be induced by heavy metals (Herbette et al., 2006; Nocito et al., 2006) was also strongly-induced by arsenic (Figure 7C). qPCR also showed that three key metabolic genes that mediate the synthesis of thiol compounds, γ-glutamyl cysteine synthetase (γ-ECS), glutathione synthetase (GSH2) and interestingly also phytochelatin synthase 1 (PCS1), were induced under arsenic stress in wild type plants (Figure 7D–F). γ-ECS, GSH2, and in some instances PCS1, had been previously shown to be transcriptionally activated by cadmium (Clemens et al., 1999; Xiang and Oliver, 1998; Lee and Korban, 2002). Furthermore, the arsenic-mediated induction of these key metabolic genes was significantly enhanced in both ars5 mutants (Figure 7D–F).
Figure 7. Quantitative PCR analysis of PAF genes and genes involved sulfate assimilation and thiol biosynthesis.
Expression analysis (qPCR) of (A) PAF1, (B) PAF2, (C) Sultr1;2, (D) γ-ECS, (E) GSH synthetase and (F) AtPCS1 from Col-0 WT, ars5-1 and ars5-2 seedlings (5 day-old) treated or not with 100 µM arsenate for 96 hours. cDNA was synthesized from RNA obtained from whole seedlings and used for qPCR analysis. Expression was normalized to Clathrin. Error bars represent SEM (n=3). Statistical differences between ars5-1 and ars5-2 compared to wild type, both exposed to arsenic, are denoted with asterisks (*, p < 0.05; **, p < 0.01).
PAF1-mediated arsenic tolerance is not mediated by global protein stability
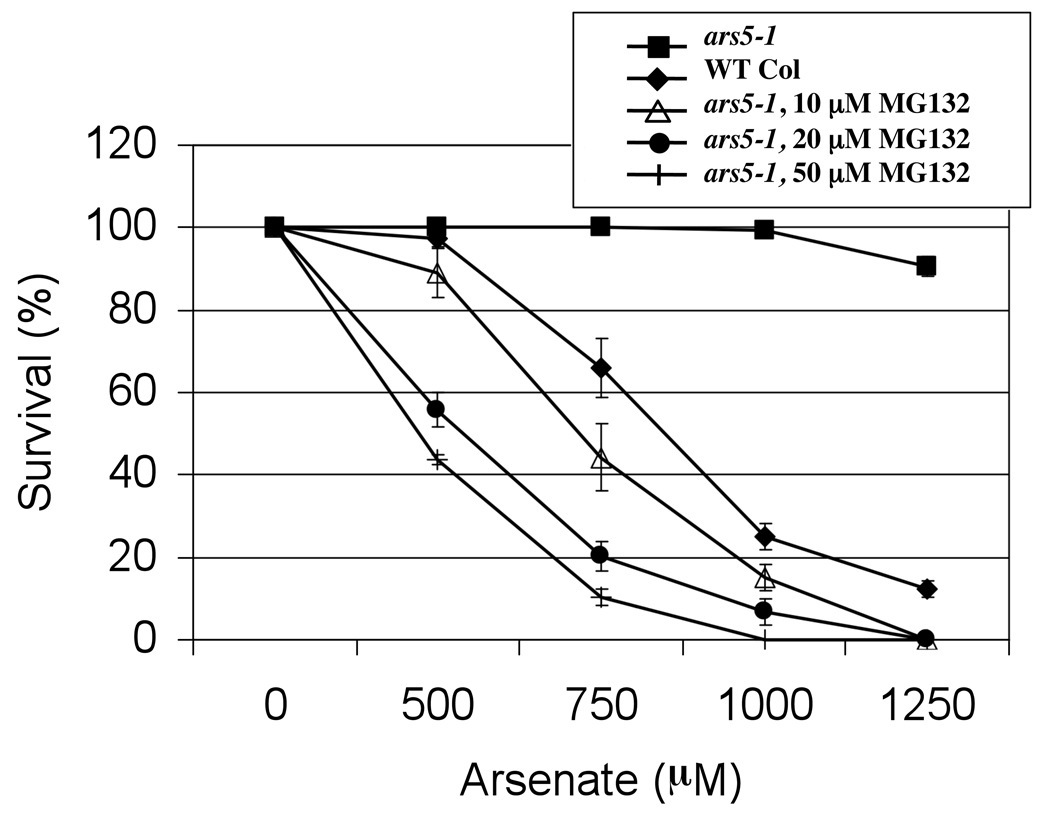
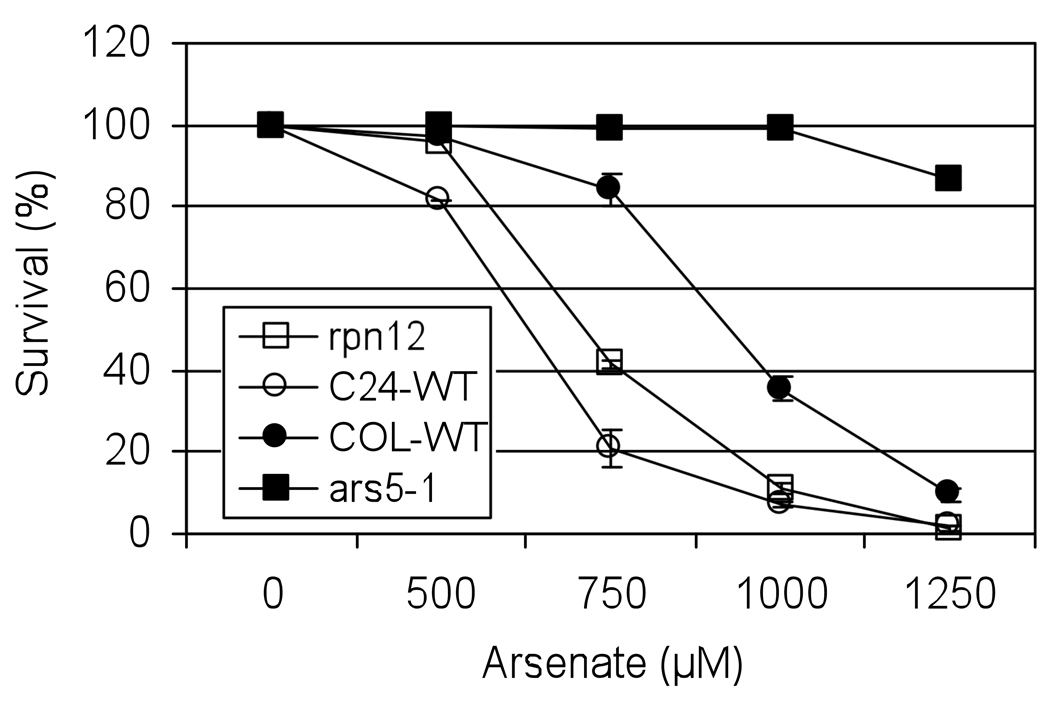
Multiple isoforms of the 26S proteasome complex exist in Arabidopsis (Yang et al., 2004; Kurepa et al., 2008). This raises the question whether the PAF1-containing isoform of the 26S proteasome is essential to global protein stability and how impairment in the proteasome-dependent protein degradation process may promote arsenic tolerance. To gain insight into these questions we followed two independent approaches. First, protein ubiquitination profiles between WT and ars5 were investigated and, second, the effect of inhibiting the 26S proteasome activity (using the proteasome inhibitor MG132) on arsenic tolerance in WT and ars5 mutants was evaluated. Western blot analyses, using an anti-ubiquitin antibody, did not detect any difference in the protein ubiquitination profiles between WT and ars5, both under normal conditions and during arsenic stress (Figure S3), which suggests that knocking out PAF1 did not compromise the global activity of the 26S proteasome. On the other hand, MG132 treatment failed to confer arsenic tolerance in wild type at concentrations ranging from 10 to 50 µM. Moreover, ars5-1 seeds treated with MG132 lost their arsenic tolerance (Figure 8), which suggest that inhibition of the proteasome complex in ars5-1 compromises seed viability. These results suggest that a specific function of PAF1 in the proteasome complex is responsible for the arsenic tolerance of the ars5 mutant, and that overall inhibition of the 26S proteasome cannot explain the arsenic resistance of ars5. To further evaluate this hypothesis, the arsenic tolerance of a different 26S proteasome mutant was analyzed. RPN12 encodes a regulatory subunit in the lid particle of the 26S proteasome (Smalle et al., 2002). rpn12 mutants, in the Arabidopsis C24 background, showed a slight increase in arsenic tolerance compared to WT (C24 background; Figure 9).
Figure 8. Proteasome inhibition reverts the arsenic tolerance phenotype of ars5.
Col-0 WT and ars5-1 seeds were germinated on minimal medium containing 0, 500, 750, 1000, 1250 µM arsenate. Where indicated, ars5-1 seeds were also germinated on plates containing 10, 20 and 50 µM MG132, a potent proteasome inhibitor. Germination of ars5 without MG132 treatment is shown as a positive control for arsenic tolerant germination. Seedling survival was scored on the 4th day based on expanded green cotyledons. Error bars represent SEM (n=3).
Figure 9. Arsenic tolerance in regulatory-particle mutants of the 26S proteasome (rpn12).
ars5-1 and rpn12 mutant seeds were germinated on minimal medium containing 0, 500, 750, 1000, 1250 µM arsenate together with their corresponding WT accession seeds (Col-0 or C24). Seedling survival was scored on the 4th day based on expanded cotyledons. Error bars represent SEM (n=3).
Discussion
ars5 reveals a novel mechanism for arsenic tolerance in Arabidopsis
Studies have demonstrated that arsenic tolerance can be achieved through independent mechanisms including active arsenic efflux (Rosen et al., 1988), mutations in transporters favoring the uptake of phosphate over arsenic (Catarecha et al., 2007) and through an increased synthesis of the arsenic-chelating thiol compounds, glutathione and phytochelatins (Dhankher et al., 2002; Li et al., 2004,2005, 2006; Gasic and Korban, 2007). However, it has remained unknown whether the glutathione-phytochelatin synthesis genes γ-ECS, GSH2 and AtPCS1 are induced by arsenic stress. Moreover, it is also not known how these arsenic tolerance mechanisms are regulated at the transcriptional level. In the present study, we cloned and characterized ARS5, a novel mechanism for arsenic tolerance in Arabidopsis. Enhanced arsenic tolerance in two independent PAF1 null mutants ars5-1 and ars5-2 (Figure 1A–B, Figure 5 A–C) and genetic complementation of ars5 with a PAF1 genomic DNA fragment and PAF1 cDNA (Figure 6 A–C) suggest that PAF1-containing 26S proteasome complex may negatively regulate the arsenic-induced transcription of the genes that mediate the synthesis of thiol compounds (Figure 3B–F, Figure 7 D–F).
Arsenic tolerance mechanism in ars5
Changes in phosphate uptake rates have been described as a mechanism able to confer enhanced arsenate tolerance (Lee et al., 2003a; Catarecha et al., 2007). However, ICP-OES analyses showed that arsenic tolerance in ars5 is not the result of active extrusion, decreased arsenic accumulation or an altered phosphate accumulation (Figure 2A–B). Moreover, ars5-1 also showed increased tolerance to arsenite (Figure 1B). The high reactivity of arsenite towards thiols (Schmöger et al., 2000) suggested that thiol metabolism could be contributing to the ars5-1 arsenic tolerance mechanism. Interestingly, ars5 contained an increased content of organic thiol compounds including phytochelatins compared to WT (Figure 3C–F). Thiol compounds represent a major mechanism for arsenic and heavy metal tolerance and accumulation in plants (Clemens et al., 1999; Ha et al., 1999; Vatamaniuk et al., 1999; Zhu et al., 1999a; Zhu et al., 1999b; Schmöger et al., 2000; Dhankher et al., 2002; Li et al., 2004,2005,2006; Gasic and Korban, 2007; Sung et al., 2007). Thiol compounds mediate complexation, translocation, and sequestration of heavy metals (Grill, 1987; Vatamaniuk et al., 1999; Gong et al., 2003; Chen et al., 2006; Mendoza-Cózatl et al., 2008). Thus, the increased accumulation of thiol compounds likely contributes to the enhanced arsenic tolerance and the increased arsenic accumulation in ars5 compared to wild type seedlings. Figure 7 shows that the increased thiol accumulation in ars5 correlates with an enhanced arsenic-induction of key genes that mediate thiol biosynthesis.
ARS5 is a negative regulator of arsenic tolerance
A plausible explanation for the inability of paf2 to produce an arsenic tolerant phenotype (Figure 5C), may be the difference in PAF2 expression compared to PAF1. Expression analyses, including wild type and ars4ars5 microarray analyses (Figure 4), as well as RT-PCR analysis indicate that PAF1 is highly expressed while PAF2 expression is very low. Based on the lethality of paf1paf2 double mutants, it is likely that the disruption of the highly expressed PAF1 resulted in the production of PAF2-containing 26S proteasomes. This isoform seems able to fulfill the 26S proteasome function in global protein stability (Figure S3) but is insufficient to repress the PAF1-dependent arsenic tolerance mechanism of ars5, which suggests that PAF1 might have additional functions other than protein degradation. This idea is further supported by the fact that expression of the PAF2 cDNA, under the control of the 35S promoter, did not fully complement the germination phenotype of ars5-1 compared to WT (Figure 6A). A similar relationship has been demonstrated for two RPN1 genes encoding different components of the 26S proteasome in Arabidopsis (Brukhin et al., 2005). When the highly expressed RPN1a is knocked out, the endogenous expression of RPN1b was not sufficient to counteract the embryo lethality resulting from the loss of RPN1a, but overexpression of RPN1b was able to complement a RPN1a null mutation (Brukhin et al., 2005). It has also been shown that Arabidopsis mutants defective in the regulatory particle (19S) of the 26S proteasome display an increased tolerance to oxidative stress and this tolerance is explained by an increased capacity to remove oxidized proteins, which is catalyzed by the 20S particle (Kurepa et al., 2008).
PAF1 negatively regulates the expression of genes involved in thiol biosynthesis
Regulation, at the transcriptional level, of arsenic tolerance mechanisms has been well documented for Synechocystis and yeast. De-repression/activation of the arsBCH operon (arsenic efflux mechanism) in Synechocystis is mediated by the transcription factor arsR (Lopez-Maury et al., 2003). In Saccharomyces cerevisiae, increased sulfate assimilation, directed towards an enhanced synthesis of glutathione, is mediated by Yap1p and Yap8p (AP1-like transcription factors), which also regulate the expression of other arsenic detoxification genes such as ACR2 (arsenate reductase) and ACR3 (arsenite transporter) (Wysocki et al., 2004; Di and Tamas, 2007; Thorsen et al., 2007). Yap8p degradation is mediated by the 26S proteasome and yeast mutants defective in 26S proteasome activity showed increased levels of Yap8p (Di and Tamas, 2007). In Schizosaccharomyces pombe, activation of genes related to sulfate assimilation and cysteine biosynthesis during cadmium exposure requires the ubiquitin-dependent degradation of the transcription factor Zip1 (Harrison et al., 2005).
Cadmium has been shown to activate transcription of genes that mediate GSH and PC biosynthesis in Arabidopsis (Xiang and Oliver, 1998; Lee and Korban, 2002). The results shown in Figure 7 suggest that arsenic induces the expression of γ-ECS, GSH2 and PCS1, and this arsenic-induction is even stronger in ars5 mutants. It is conceivable that, during arsenic stress, a positive regulator of sulfate assimilation and glutathione biosynthesis is rapidly degraded by the 26S proteasome, however, this positive regulator becomes stabilized in ars5, which in turn enhances the transcriptional arsenic-induced response. In embryonic stem cells, the 26S proteasome mediates repression of transcription of target genes by promoting both, the turnover of transcription factors and RNA polymerase II binding to DNA (Szutorisz et al., 2006). In the absence of PAF1, removal of repression of a positive regulatory component may function in the arsenic-induced enhancement of γ-ECS, GSH2, and AtPCS1 mRNA levels.
Experimental procedures
Arsenic tolerant germination
To analyze arsenic tolerant germination, seeds were plated on arsenate-containing minimal medium (Lee et al., 2003) with the indicated concentrations of potassium arsenate and stratified at 4°C for two days in the dark. After five days of growth at 22°C, 75% humidity, with a 16-h-light/8-h-dark photoperiod regime at approximately 75 µmol m−2 s−1 light intensity in a Conviron growth chamber (Controlled Environments Inc., Pembina, ND), germinating seeds were examined under a dissecting microscope and those seedlings for which green cotyledons had emerged from seed coats were scored as surviving.
Genetic segregation analysis of ars5-1 x paf2 F2 population
ars5-1 and paf2 mutant plants were reciprocally crossed. F1 seeds were self-pollinated to produce F2 seeds. 72 F2 plants were grown for 2 weeks and genotyped by genomic PCR. The primer pair of 5’-atcttcttgtatgagacaatatgaact-3’(F) and 5’-gcagatgtggataaaacagctaact-3’(R) distinguishes ars5-1 from WT by generating a 1.8 kb PCR product from the WT ARS5 gene and a 0.4 kb PCR product from the ars5-1 mutant gene. The primer pair 5’-ttgaagaacagaggaa-3’(F) and 5’-gatcttcttttgatgattcttgg-3’(R) generates a 1.5 kb PCR product from the WT PAF2 gene. The primer pair of 5’-ttgaagaacagaggaa-3’(F) and a T-DNA specific primer (5’-tggttcacgtagtgggccatcg-3’) distinguishes paf2 from WT by generating a 0.5 kb PCR product from the T-DNA inserted PAF2 gene in paf2 GABI_KAT mutant (GABI_419_H03). Segregation results were identical in both reciprocal crosses. Table 1 shows the segregation of F2 populations derived from ars5-1 x paf2.
ICP-OES measurements
To measure arsenic and phosphorus, plants were grown on 10 µm nylon mesh (Fisher Scientific, Pittsburgh, PA) on minimal medium with 1% agar for 5 days, then nylon meshes with seedlings were transferred to minimal medium plates containing 100 µM potassium arsenate. After exposure to arsenate for 24, 48, and 96 hours, seedlings were harvested and briefly rinsed twice with dH2O, then dried in a drying oven at 60 °C overnight, dry weight measured, and digested by boiling in concentrated nitric acid (Trace Metal grade, Sigma-Aldrich, St. Louis, MO, USA). The metal content of the digested samples was analyzed using Inductively Coupled Plasma Optical Emission Spectroscopy (ICP-OES; Varian Vista). Total concentrations of metals were normalized based on dry weight of plant samples as described previously (Gong et al., 2004).
Thiol measurements by fluorescence HPLC
Thiol compounds including cysteine, γ-glutamyl cysteine, glutathione (GSH) and phytochelatins (PCs) were analyzed using fluorescence detection HPLC essentially as described previously (Fahey and Newton, 1987; Mendoza-Cozatl et al., 2008). To analyze the levels of thiol compounds produced by plants in response to arsenic, plants were grown on 1/4MS media plates without sucrose for 5 days then transferred to fresh media plates containing 100 µM potassium arsenate. In order to minimize oxidation of thiol compounds during extractions, plant seedlings were flash-frozen in liquid nitrogen and ground and extracted with extraction buffer (6.3 mM Diethylenetriamine-pentaacetic acid (DTPA) in 0.1% trifluoroacetic acid (TFA)) bubbled with nitrogen gas. Thiols were then derivatized with mono-bromobimane (mBBr) as described (Sneller et al., 2000; Cazalé and Clemens, 2001) except that the peptides were separated with a Sunfire C18 column (5 µm, 4.6 × 250 mm; Waters) at 20°C and fluorescence was monitored on a Thermo Finnigan Fluoromonitor III fluorescence detector. The peaks of thiol compounds were identified by coupled parallel mass spectrometry measurements at the UCSD Superfund Biochemistry core as previously described (Chen et al., 2006; Mendoza-Cózatl et al., 2008) and quantified using Xcalibur software (Thermo Finnigan, Waltham, MA, USA). Thiol standards such as GSH, cysteine, γ-EC, and NAC were purchased from Sigma-Aldrich (St. Louis, MO, USA).
Western blot analyses
For Western analysis of phytochelatin synthase levels in Arabidopsis, one week-old seedlings of WT and ars5 were treated with 100 µM potassium arsenate for 0, 24, 48, 96 hours. Total proteins were extracted into an extraction buffer (50 mM Tris-HCl, pH 8.0). 30 µg of total proteins were loaded in each well in a 4–20% gradient SDS-PAGE and total proteins were transferred onto a PVDF membrane for 1 h at 100 V and then the membrane was blocked with 3% BSA in PBS overnight at 4°C. The protein bands on the PVDF membrane were probed against an anti-ubiquitin rabbit polyclonal antibody (1: 10,000 dilution) (PW9780-0025, Biomol, Plymouth Meeting, PA). Then the PVDF membrane was washed three times with 0.1% Triton X-100 in PBS. The membrane was then incubated with the secondary antibody conjugated with horseradish peroxidase (SuperSignal® West Dura kit, Pierce Biotechnology, Inc., Rockford, IL) for 1 hour at room temperature (1: 10,000 dilution). For chemiluminescence visualization, the reagents were added according to the manufacturer’s instructions and then incubated for 5 minutes in the dark (Pierce Biotechnology, Inc., Rockford, IL). Chemiluminescence signals were developed on X-ray film (Fisher Scientific, Pittsburgh, PA).
RT-PCR expression analyses
Total RNA was extracted from one week-old Arabidopsis seedlings using the TRIzol reagent (Life Technologies/Gibco-BRL). Isolated RNA samples were treated with DNA-Free kit (AMBION, Austin TX, USA) to remove any DNA contamination. cDNA was reverse transcribed from the DNA-free RNA using First Strand cDNA kit (GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA) and a Not I-d(T)18 primer. Actin 7 (ACT7, At5g09810) mRNA content was used as loading control. The transcript levels of ARS5 (At5g42750), PAF2 (At1g47290), SULTR1;2 (At1g7000) and ACT7 (At5g09810) were PCR amplified by corresponding primer sets. ARS5: 5’-tcacagccgacggtcgtgtattg-3’(F) and 5’-gatcttctctagaagaatcccca-3’(R), PAF2: 5’-tcactgctgatggtcgtgttctc-3’(F) and 5’-gatcttcttttgatgattcttgg-3’(R), SULTR1;2: 5’-tggggacgtaactacactttca-3’(F) and 5’-aaggcaaggcggagatattc-3’(R). PCR reactions were stopped at 15, 20, 25, 30, and 40 cycles and subjected to gel electrophoresis. After 30 PCR cycles DNA products showed unsaturated bands reflecting initial transcript levels as shown in Figure 5B.
Real time qPCR expression analyses
Total RNA was extracted from one week-old Arabidopsis seedlings as described above and 5 ug of each total RNA were DNase treated (DNA-free, Ambion) and used for cDNA synthesis (First strand cDNA synthesis kit, GE Healthcare Bio-Sciences Corp., Piscataway, NJ, USA). 65 ul of water were added to each 15 ul of total cDNA and 3 ul were used for qPCR reactions using a SYBR Green based real time qPCR kit (LightCycler FastStart DNA Master plus SYBR Green I, Roche). Quantitative expression analysis of qPCR reactions was carried out using Lightcycler 3.0 software (Roche). The genes analyzed and the primers used for each gene were as follows: CLATHRIN (loading control): 5’-atacgcgctgagttccc-3’(F)/ 5’-ctgactggccctgctt-3’(R), SULTR1;2(At1g78000): 5’-tggggacgtaactacactttca-3’(F)/ 5’-aaggcaaggcggagatattc-3’(R), PAF1(At5g42790): 5’-tcacagccgacggtcgtgtaatg-3’(F)/ 5’-gatcttctctagaagaatcccca-3’(R), PAF2(At1g47250): 5’-tcactgctgatggtcgtgttctc-3’(F)/ 5’-gatcttcttttgatgattcttgg-3’(R), γ-ECS(At4g23100): 5’-gatgtgaattgacttcagcacaa-3’(F)/ 5’-gaggagtaagagagggcatcaa-3’(R), GSH2(At5g27380): 5’-ataagcagcctagcattccat-3’(F)/ 5’-gcatagactctgaacgtgtaccta-3’(R), AtPCS1(At5g15630): 5’-cctcactgggttcctcttaaact-3’(F)/ 5’-tttcagcctagacttctcctctg-3’(R), and AtPCS2(At1g03980): 5’-gaaggaaagcaaatcttcaatgaa-3’(F)/ 5’-ttgctcctgaagaatgagcta-3’(R). Three independent biological samples were prepared and three independent qPCR reactions were carried out per sample.
Microarray experiments
Col-0 WT and ars4ars5 seeds were germinated on minimal media (Lee et al., 2003a), with or without 200 µM potassium arsenate. Total RNA was extracted from 10 day-old seedlings using the TRIzol reagent (Life Technologies/Gibco-BRL), labeled and hybridized using Affymetrix ATH1 chip arrays (Santa Clara, CA), containing approximately 22 500 Arabidopsis probes, at the University of California, San Diego Gene Chip Core facility. Raw data in Excel® format can be downloaded from Supplementary File 1.
Supplementary Material
Acknowledgments
We thank Dr. Felix Hauser for help provided during microarray data analysis. This research was supported by National Institute of Environmental Health Sciences grant number ES010337 (to JIS and EAK) and Department of Energy grant DOE-DE-FG02-03ER15449 and NSF grant IBN-0419695 (to JIS). DGMC is recipient of a PEW Latin American Fellowship 2006.
References
- Alam MGM, Snow ET, Tanaka A. Arsenic and heavy metal contamination of vegetables grown in Samta village, Bangladesh. Sci Total Environ. 2003;308:83–96. doi: 10.1016/S0048-9697(02)00651-4. [DOI] [PubMed] [Google Scholar]
- Book AJ, Smalle J, Lee KH, Yang P, Walker JM, Casper S, Holmes JH, Russo LA, Buzzinotti ZW, Jenik PD, Vierstra RD. The RPN5 Subunit of the 26s Proteasome Is Essential for Gametogenesis, Sporophyte Development, and Complex Assembly in Arabidopsis. Plant Cell. 2009;21:460–478. doi: 10.1105/tpc.108.064444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown K, Ross G American Council on Science and Health. Arsenic, drinking water, and health: a position paper of the American Council on Science and Health. Regul Toxicol Pharmacol. 2002;36:162–174. doi: 10.1006/rtph.2002.1573. [DOI] [PubMed] [Google Scholar]
- Brukhin V, Gheyselinck J, Gagliardini V, Genschik P, Grossniklaus U. The RPN1 Subunit of the 26S Proteasome in Arabidopsis Is Essential for Embryogenesis. Plant Cell. 2005;17:2723–2737. doi: 10.1105/tpc.105.034975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonell A, Aarabi M, Delaune R, Grambrell R, Patrick W. Arsenic in wetland vegetation: availability, phytotoxicity, uptake and effects on plants growth and nutrition. Sci Total Environ. 1998;217:189–199. [Google Scholar]
- Catarecha P, Segura MD, Franco-Zorrilla JM, Garcia-Ponce B, Lanza M, Solano R, Paz-Ares J, Leyva A. A Mutant of the Arabidopsis Phosphate Transporter PHT1;1 Displays Enhanced Arsenic Accumulation. Plant Cell. 2007;19:1123–1133. doi: 10.1105/tpc.106.041871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cazalé AC, Clemens S. Arabidopsis thaliana expresses a second functional phytochelatin synthase. FEBS Letters. 2001;507:215–219. doi: 10.1016/s0014-5793(01)02976-3. [DOI] [PubMed] [Google Scholar]
- Chen A, Komives EA, Schroeder JI. An Improved Grafting Technique for Mature Arabidopsis Plants Demonstrates Long-Distance Shoot-to-Root Transport of Phytochelatins in Arabidopsis. Plant Physiol. 2006;141:108–120. doi: 10.1104/pp.105.072637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemens S, Kim EJ, Neumann D, Schroeder JI. Tolerance to toxic metals by a gene family of phytochelatin synthases from plants and yeast. EMBO J. 1999;18:3325–3333. doi: 10.1093/emboj/18.12.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbett CS, Meagher RB. Arabidopsis and the Genetic Potential for the Phytoremediation of Toxic Elemental and Organic Pollutants. In: Somerville CR, Meyerowitz EM, editors. The Arabidopsis Book. Rockville, MD: American Society of Plant Biologists; 2002. doi/10.1199/tab.0032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhankher OP, Li Y, Rosen BP, Shi J, Salt D, Senecoff JF, Sashti NA, Meagher RB. Engineering tolerance and hyperaccumulation of arsenic in plants by combining arsenate reductase and gamma-glutamylcysteine synthetase expression. Nat Biotechnol. 2002;20:1140–1145. doi: 10.1038/nbt747. [DOI] [PubMed] [Google Scholar]
- Di Y, Tamas MJ. Regulation of the arsenic-responsive transcription factor Yap8p involves the ubiquitin-proteasome pathway. J Cell Sci. 2007;120:256–264. doi: 10.1242/jcs.03346. [DOI] [PubMed] [Google Scholar]
- Fahey RC, Newton GL. Determination of low-molecular weight thiols using monobromobimane fluorescent labeling and high performance liquid chromatography. Methods Enzymol. 1987;143:85–97. doi: 10.1016/0076-6879(87)43016-4. [DOI] [PubMed] [Google Scholar]
- Gasic K, Korban S. Transgenic Indian mustard ( Brassica juncea) plants expressing an Arabidopsis phytochelatin synthase (AtPCS1) exhibit enhanced As and Cd tolerance. Plant Mol Biol. 2007;64:361–369. doi: 10.1007/s11103-007-9158-7. [DOI] [PubMed] [Google Scholar]
- Gong JM, Lee DA, Schroeder JI. Long-distance root-to-shoot transport of phytochelatins and cadmium in Arabidopsis. Proc Natl Acad Sci U S A. 2003;100:10118–10123. doi: 10.1073/pnas.1734072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong JM, Waner DA, Horie T, Li SL, Horie R, Abid KB, Schroeder JI. Microarray-based rapid cloning of an ion accumulation deletion mutant in Arabidopsis thaliana. Proc Natl Acad Sci U S A. 2004;101:15404–15409. doi: 10.1073/pnas.0404780101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grill E. Phytochelatins, the heavy metal binding peptides of plants: characterization and sequence determination. Experientia Suppl. 1987;52:317–322. doi: 10.1007/978-3-0348-6784-9_28. [DOI] [PubMed] [Google Scholar]
- Ha SB, Smith AP, Howden R, Dietrich WM, Bugg S, O'Connell MJ, Goldsbrough PB, Cobbett CS. Phytochelatin synthase genes from Arabidopsis and the yeast Schizosaccharomyces pombe. Plant Cell. 1999;11:1153–1164. doi: 10.1105/tpc.11.6.1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison C, Katayama S, Dhut S, Chen D, Jones N, Bahler J, Toda T. SCFPof1-ubiquitin and its target Zip1 transcription factor mediate cadmium response in fission yeast. EMBO J. 2005;24:599–610. doi: 10.1038/sj.emboj.7600536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbette S, Taconnat L, Hugouvieux V, Piette L, Magniette M-LM, Cuine S, Auroy P, Richarud P, Forestier C, Bourguignon J, Renou J-P, Vavasseur A, Leonhardt N. Genome-wide transcriptome profiling of the early cadmium response of Arabidopsis roots and shoots. Biochimie. 2006;88:1751–1765. doi: 10.1016/j.biochi.2006.04.018. [DOI] [PubMed] [Google Scholar]
- Hindmarsh JT. Arsenic, its clinical and environmental significance. The Journal of Trace Elements in Experimental Medicine. 2000;13:165–172. [Google Scholar]
- Kurepa J, Toh-e A, Smalle JA. 26S proteasome regulatory particle mutants have increased oxidative stress tolerance. Plant J. 2008;53:102–114. doi: 10.1111/j.1365-313X.2007.03322.x. [DOI] [PubMed] [Google Scholar]
- Lee S, Korban SS. Transcriptional regulation of Arabidopsis thaliana phytochelatin synthase(AtPCS1) by cadmium during early stages of plant development. Planta. 2002;215:689–693. doi: 10.1007/s00425-002-0821-6. [DOI] [PubMed] [Google Scholar]
- Lee DA, Chen A, Schroeder JI. ars1, an Arabidopsis mutant exhibiting increased tolerance to arsenate and increased phosphate uptake. Plant J. 2003a;35:637–646. doi: 10.1046/j.1365-313x.2003.01835.x. [DOI] [PubMed] [Google Scholar]
- Lee S, Moon JS, Ko T-S, Petros D, Goldsbrough PB, Korban SS. Overexpression of Arabidopsis Phytochelatin Synthase Paradoxically Leads to Hypersensitivity to Cadmium Stress. Plant Physiol. 2003b;131:656–663. doi: 10.1104/pp.014118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dhankher OP, Carreira L, Balish RS, Meagher RB. Arsenic and mercury tolerance and cadmium sensitivity in Arabidopsis palnts expressing bacterial γ-glutamyl cysteine synthetase. Environ Tox Chem. 2005;24:1376–1386. doi: 10.1897/04-340r.1. [DOI] [PubMed] [Google Scholar]
- Li Y, Dankher OP, Carreira L, Smith AP, Meagher RB. The Shoot-Specific Expression of γ-Glutamylcysteine Synthetase Directs the Long-Distance Transport of Thiol-Peptides to Roots Conferring Tolerance to Mercury and Arsenic. Plant Physiol. 2006;141:288–298. doi: 10.1104/pp.105.074815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Dhankher OP, Carreira L, Lee D, Chen A, Schroeder JI, Balish RS, Meagher RB. Overexpression of Phytochelatin Synthase in Arabidopsis Leads to Enhanced Arsenic Tolerance and Cadmium Hypersensitivity. Plant Cell Physiol. 2004;45:1787–1797. doi: 10.1093/pcp/pch202. [DOI] [PubMed] [Google Scholar]
- Lopez-Maury L, Florencio FJ, Reyes JC. Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol. 2003;185:5363–5371. doi: 10.1128/JB.185.18.5363-5371.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin JH, Beane Freeman LE, Cantor KP. Inorganic Arsenic in Drinking Water: An Evolving Public Health Concern. J Natl Cancer Inst. 2007;99:906–907. doi: 10.1093/jnci/djm012. [DOI] [PubMed] [Google Scholar]
- Marin A, Pezeshki S, Masscheleyn P, Choi H. Effect of dimethylarsenic acid (DMAA) on growth, tissue arsenic and photosynthesis in rice plant. J Plant Nutr. 1993;16:865–880. [Google Scholar]
- Meharg AA, Macnair MR. Suppression of the High Affinity Phosphate Uptake System: A Mechanism of Arsenate Tolerance in Holcus lanatus L. J Exp Bot. 1992;43:519–524. [Google Scholar]
- Mendoza-Cózatl DG, Butko E, Springer F, Torpey JW, Komives EA, Kehr J, Schroeder JI. Identification of high levels of phytochelatins, glutathione and cadmium in the phloem sap of Brassica napus. A role for thiol-peptides in the long-distance transport of cadmium and the effect of cadmium on iron translocation. Plant J. 2008;54:249–259. doi: 10.1111/j.1365-313X.2008.03410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nocito FF, Lancilli C, Crema B, Fourcroy P, Davidian J-C, Sacchi GA. Heavy Metal Stress and Sulfate Uptake in Maize Roots. Plant Physiol. 2006;141:1138–1148. doi: 10.1104/pp.105.076240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilon-Smits E, Pilon M. Phytoremediation of metals using transgenic plants. Crit Rev Plant Sci. 2002;21:439–456. [Google Scholar]
- Rosen BP, Weigel U, Karkaria C, Gangola P. Molecular characterization of an anion pump. The arsA gene product is an arsenite(antimonate)-stimulated ATPase. J Biol Chem. 1988;263:3067–3070. [PubMed] [Google Scholar]
- Schmöger ME, Oven M, Gril IE. Detoxification of arsenic by phytochelatins in plants. Plant Physiol. 2000;122:793–801. doi: 10.1104/pp.122.3.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Babiychuk E, Kushnir S, Durski A, Vierstra RD. Cytokinin Growth Responses in Arabidopsis Involve the 26S Proteasome Subunit RPN12. Plant Cell. 2002;14:17–32. doi: 10.1105/tpc.010381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smalle J, Kurepa J, Yang P, Emborg TJ, Babiychuk E, Kushnir S, Vierstra RD. The Pleiotropic Role of the 26S Proteasome Subunit RPN10 in Arabidopsis Growth and Development Supports a Substrate-Specific Function in Abscisic Acid Signaling. Plant Cell. 2003;15:965–980. doi: 10.1105/tpc.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneller FEC, van Heerwaarden LM, Koevoets PLM, Vooijs R, Schat H, Verkleij JAC. Derivatization of phytochelatins from Silene vulgaris, induced upon exposure to arsenate and cadmium: Comparison of derivatization with Ellman's reagent and monobromobimane. J Ag Food Chem. 2000;48:4014–4019. doi: 10.1021/jf9903105. [DOI] [PubMed] [Google Scholar]
- Sung D-Y, Lee D, Harris H, Raab A, Feldmann J, Meharg A, Kumabe B, Komives EA, Schroeder JI. Identification of an arsenic tolerant double mutant with a thiol-mediated component and increased arsenic tolerance in phyA mutants. Plant J. 2007;49:1064–1075. doi: 10.1111/j.1365-313X.2006.03018.x. [DOI] [PubMed] [Google Scholar]
- Szutorisz H, Georgiou A, Tora L, Dillon N. The Proteasome Restricts Permissive Transcription at Tissue-Specific Gene Loci in Embryonic Stem Cells. Cell. 2006;127:1375–1388. doi: 10.1016/j.cell.2006.10.045. [DOI] [PubMed] [Google Scholar]
- Thorsen M, Lagniel G, Kristiansson E, Junot C, Nerman O, Labarre J, Tamás MJ. Quantitative transcriptome, proteome, and sulfur metabolite profiling of the Saccharomyces cerevisiae response to arsenite. Physiol Genomics. 2007;30:35–43. doi: 10.1152/physiolgenomics.00236.2006. [DOI] [PubMed] [Google Scholar]
- Tripathi RD, Srivastava S, Mishra S, Singh N, Tuli R, Gupta DK, Maathuis FJ. Arsenic hazards: strategies for tolerance and remediation by plants. Trends Biotechnol. 2007;25:158–165. doi: 10.1016/j.tibtech.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Vatamaniuk OK, Mari S, Lu YP, Rea PA. AtPCS1, a phytochelatin synthase from Arabidopsis: isolation and in vitro reconstitution. Proc Natl Acad Sci U S A. 1999;96:7110–7115. doi: 10.1073/pnas.96.12.7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Zhao F-J, Meharg AA, Raab A, Feldmann J, McGrath SP. Mechanisms of Arsenic Hyperaccumulation in Pteris vittata. Uptake Kinetics, Interactions with Phosphate, and Arsenic Speciation. Plant Physiol. 2002;130:1552–1561. doi: 10.1104/pp.008185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wysocki R, Fortier P-K, Maciaszczyk E, Thorsen M, Leduc A, Odhagen A, Owsianik G, Ulaszewski S, Ramotar D, Tamas MJ. Transcriptional Activation of Metalloid Tolerance Genes in Saccharomyces cerevisiae Requires the AP-1-like Proteins Yap1p and Yap8p. Mol Biol Cell. 2004;15:2049–2060. doi: 10.1091/mbc.E03-04-0236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang C, Oliver DJ. Glutathione metabolic genes coordinately respond to heavy metals and jasmonic acid in Arabidopsis. Plant Cell. 1998;10:1539–1550. doi: 10.1105/tpc.10.9.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang P, Fu H, Walker J, Papa CM, Smalle J, Ju Y-M, Vierstra RD. Purification of the Arabidopsis 26 S Proteasome: Biochemical and molecular analyses revealed the presence of multiple isoforms. J Biol Chem. 2004;279:6401–6413. doi: 10.1074/jbc.M311977200. [DOI] [PubMed] [Google Scholar]
- Zhu LY, Pilon-Smits EA, Jouanin L, Terry N. Overexpression of glutathione synthetase in indian mustard enhances cadmium accumulation and tolerance. Plant Physiol. 1999a;119:73–80. doi: 10.1104/pp.119.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu YL, Pilon-Smits EA, Tarun AS, Weber SU, Jouanin L, Terry N. Cadmium tolerance and accumulation in Indian mustard is enhanced by overexpressing gamma-glutamylcysteine synthetase. Plant Physiol. 1999b;121:1169–1178. doi: 10.1104/pp.121.4.1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.