Abstract
p38 kinases are members of the mitogen-activated protein kinase (MAPK) family that transduce signals from various environmental stresses, growth factors and steroid hormones. p38 is highly expressed in aggressive and invasive breast cancers. Increased levels of activated p38 are markers of poor prognosis. In this study we tested the hypothesis that blockade of p38 signaling would inhibit breast cancer cell proliferation. We studied breast cancer cell proliferation and cell cycle regulation upon p38 blockade by using three independent approaches: dominant-negative constructs, siRNA, and small molecule inhibitors. p38α and p38δ are the most abundant isoforms expressed by all examined human breast tumors and breast cancer cell lines. Expression of a dominant-negative p38 inhibited both anchorage-dependent and -independent proliferation of MDA-MB-468 cells. Silencing of p38α, but not p38δ, using siRNA suppressed MDA-MB-468 cell proliferation. Pharmacological inhibitors of p38 significantly inhibited the proliferation of p53 mutant and ER-negative breast cancer cells. While p38 has previously been considered as a mediator of stress-induced apoptosis, we propose that p38 may have dual activities regulating survival and proliferation depending on the expression of p53. Our data suggest that p38 mediates the proliferation signal in breast cancer cells expressing mutant but not wild-type p53. Since most of ER-negative breast tumors express mutant p53, our results provide the foundation for future development of p38 inhibitors to target p38 for the treatment of p53 mutant and ER-negative breast cancers.
Keywords: p38, breast cancer, estrogen receptor, p53
Introduction
It is predicted that in 2009, 192,370 women in the United States will develop breast cancer and 40,170 women will die from the disease (1). Selective estrogen receptor modulators (SERMs, such as tamoxifen and aromatase inhibitors) reduce ER-positive breast cancer recurrence by approximately 50% (2, 3). However, these agents are not effective in treating ER-negative breast cancer, which accounts for 30–40% of all breast cancers (4). The only effective targeted therapy for ER-negative breast cancer is trastuzumab (HerceptinR), a monoclonal antibody that targets the subset of breast cancer that overexpress HER-2 protein (5). Standard treatment for patients with ER-negative and HER-2-negative breast cancer is chemotherapy, which is non-specific and generally toxic. Thus, there is an urgent need to develop more effective agents that can treat ER-negative and HER-2-negative breast cancer.
Since several different growth factor pathways can stimulate breast cell proliferation, agents that target a single signal transduction pathway only have transient effects on the growth of breast cancer. It may be more effective to inhibit signal transduction at a more distal point in the cascade, where many mitogenic signals converge. One such point of convergence is the p38 mitogen-activated protein kinases (p38 MAPKs).
p38s are members of the MAPK family that also includes the extracellular signal-regulated kinases (ERKs) and the c-Jun N-terminal kinases (JNKs). p38s can be activated by multiple stimuli including various growth factors, inflammatory cytokines, and chemical/physical stresses (6). Activated p38 kinases can regulate cell growth, differentiation, apoptosis, and responses to inflammation or stress (6–8). Four mammalian p38 isoforms have been identified, p38α (9, 10), p38β (11), p38γ (12–14), and p38δ (15, 16). p38 isoforms are differentially expressed, produce responses in a cell type specific manner, and activate selective downstream substrates (15–18).
High levels of p38 have been correlated with highly invasive and poor prognostic breast cancers. Selective activation of p38 was found in Grade II or III intraductal tumors (19). p38 phosphorylation occurred in approximately 20% of primary breast carcinomas (20). Increased expression of phospho-p38 was correlated with HER-2 amplification and tamoxifen resistance (21). Davidson et al. observed significantly higher nuclear expression of phospho-p38 in breast carcinoma effusions, when compared with both primary tumors and lymph node metastases, making p38 a potential prognostic marker for patients with breast cancer effusions (22).
The role of p38 in regulating breast cancer cell proliferation has not been investigated. We hypothesized that blockade of p38 signaling would inhibit breast cancer cell proliferation. To test this hypothesis, we blocked p38 signaling in a panel of breast cancer cells using three independent approaches: dominant-negative constructs, siRNAs, and small molecule inhibitors. We found that blockade of p38 signaling significantly inhibited the proliferation of breast cancer cells with a p53 mutation (p53MUT). We propose that while p38 may function as a regulator of survival in the context of wild-type p53 (p53WT), it is a crucial regulator of proliferation when cells express p53MUT. These studies provide the foundation for future development of p38 inhibitors and clinical trials to target p38 signaling for the treatment of breast cancer, especially those with p53MUT and with a triple-negative (ER-negative, PR negative, and Her2 negative) molecular profile.
Material and Methods
Reagents, plasmids and cell lines
MCF-7 (ATCC, HTB-22, p10), T47D (ATCC, HTB-133, p16), BT474 (ATCC, HTB-20, p12), MDA-MB-361 (ATCC, HTB-27, p2), MDA-MB-231 (ATCC, HTB-26, p21), BT549 (ATCC, HTB-122, p4), MDA-MB-468 (ATCC, HTB-132, p8), HCC1937 (ATCC, CRL-2336, p5), SKBr3 (ATCC, HTB-30, p22), MDA-MB-453 (ATCC, HTB-131, p6), BT20 (ATCC, HTB-19, p5), MCF10A (ATCC, CRL-10317, p10), 184B5 (ATCC, CRL-8799, p5), HMEC (LONZA, CC-2551, p4) ZR75-30 (ATCC, CRL-1504, p8) and ZR75-1 (ATCC, CRL-1500, p10) cells were verified by morphology, growth curve analysis, and tested for mycoplasma. Phoenix A cells were a gift from Dr. Aubrey Thompson (Mayo Clinic, Jacksonville, FL). pcDNA3.1 vector expressing N-terminal Flag tagged dominant-negative (DN) human p38 (T180A/Y182P) cDNA was a gift from Dr. Rachel Schiff (Baylor College of Medicine, Houston, TX). MDA-MB-468 cells were transfected with pcDNA3.1/Flag-DNp38 or empty vector pcDNA3.1 using Fugene 6 (Roche, Indianapolis, IN) according to the manufacture’s recommendation. G418 resistant clones of MDA-MB-468 were screened for stable expression of Flag-DNp38. Alternatively, Flag-DNp38 cDNA was cloned into retroviral vector pBabe-puro3 (from Dr. Aubrey Thompson, Mayo Clinic). MDA-MB-468, MDA-MB-231 and MCF-7 cells were infected with retrovirus pBabe or pBabe-Flag-DNp38 produced using Phoenix A packaging cells, according to Dr. Garry Nolan’s protocol (Stanford University, Stanford, CA). Puromycin resistant pools of cells were screened for Flag-DNp38 expression. Two small molecule p38 inhibitors, SB203580 (Calbiochem, San Diego, CA) and AZ10164773 (obtained from AstraZeneca) were used in this study. Anisomycin and dimethyl sulphoxide (DMSO) were purchased from Sigma (St. Louis, MO). For anisomycin treatment, cells were cultured in serum free IMEM for 24 h and then treated with DMSO or 50ng/ml anisomycin for 15 min.
Western blot analysis
Cells lysates were prepared as described previously (23). 20μg of total protein extract was run on a 10% SDS-PAGE gel and transferred to a nitrocellulose membrane (Invitrogen). Primary antibodies specific for p38 (#9212), phospho-p38 (T180/Y182) (#9211), MAPKAPK-2 (#3042), JNK(#9252), phospho-JNK (#9251), ERK1/2 (#9102), phospho-ERK1/2 (#9101) and cyclin D1 (#2926) were purchased from Cell Signaling (Danvers, MA). Antibodies specific for Flag tag (#F-3165) and β-actin (#A-5441) were purchased from Sigma (St. Louis, MO). Anti- mouse (#NA931V) and anti-rabbit (#NA934V) secondary antibodies were obtained from Amersham (Piscataway, NJ).
Cell proliferation assays
Cells were plated in 6-well plates at 2 × 104 or 3 × 104 cells per well for slower growing cells. Cell proliferation was measured by counting cells using a hemocytometer. Each data point was performed in triplicate, and results are reported as average percentage ± standard deviation. Cell growth was also measured using the CellTiter 96TM Aqueous Non-Radioactive Cell Proliferation Assay (MTS assay, Promega, Madison, WI) according to the manufacturer’s protocol in the presence of SB203580 or AZ10164773. Each data point was performed in quadruplet, and results are reported as average absorption ± standard deviation.
Human Breast Tumors
37 human breast tumor samples were isolated from the Asterand tumor bank purchased by the Breast Center at Baylor College of Medicine. All studies were conducted with approval from the Institutional Review Boards at Baylor College of Medicine. Tumor tissue was flash frozen and stored in liquid nitrogen until RNA isolation was performed. Tumor tissue was disrupted by homogenization using a PRO Scientific rotor-stator homogenizer with Multi-Gen7 generators.
RNA preparation and quantitative RT-PCR
Total RNA was isolated using the RNeasy kit (QIAGEN, Valencia, CA) and probe based Quantitative RT-PCR (Q-RT-PCR) was performed as described previously (24). Primers are listed in supplemental Table 1. Data were reported as normalized copy numbers ± standard deviation.
siRNA transfection
Pools of siRNAs for p38α and p38δ were purchased from Dharmacon (Lafayette, CO). Additional siRNAs for p38α were purchased from Sigma (St. Louis, MO). Individual siRNAs for p53 and Wip1 were also purchased from Sigma. Catalog numbers or sequences are given in supplemental Table 2. siRNA transfection was performed at a final concentration of 10nM using DharmaFECT™ 1 (Dharmacon), according to the manufacture’s protocol.
Flow cytometry analysis
MDA-MB-468 cells were plated in 60mm tissue culture dishes at 1 × 105 cells per dish and maintained in serum free IMEM for 48h and then cells were harvested 0, 12, 24, 36, 48 and 72 h after serum stimulation and stained with propidium iodide (23). Cell cycle distribution for about 10000 cells was analyzed using a Coulter Epics-XL-MCL flow cytometer and XL System Software (Beckman Coulter, Fullerton, CA).
Anchorage independent cell growth
5 × 103 cells were suspended in 0.375% Seaplaque agar (FMC, Philadelphia, PA) in DMEM supplemented with 10% FBS, 1% Penicillin-Streptomycin-Glutamine, 10mM HEPES and 0.1% gentamycin. Suspended cells were layered over 0.75% agar base in the same medium. Microscopically visible colonies (average size 50 cells) were counted 4 weeks after plating by Giemsa staining. All experiments were performed in triplicates. Data reported represent average colony number per cm2 ± standard deviation.
Statistical Analyses
Analysis was performed using SAS software version 9.2 (SAS Institute Inc.). Friedman chi-square test and pair wise Wilcoxon signed-rank test with bonferroni adjustment were used to determine differences in the abundance of p38 isoforms in the human breast tumors. P < 0.05 was considered as statistically significant. We also employed a mixed linear model with random experiment effects to determine significance for all growth assays.
Results
p38 expression in human breast tumors and established breast cancer cell lines
To study p38 signaling in breast cancer, we measured p38 mRNA expression in 37 human breast tumors (see Material and Methods) using Q-RT-PCR. p38α is the most abundant of the four p38 isoforms in these human breast tumors (P < 0.0001) (Figure 1A). These tumors also express high levels of p38δ, but minimal expression p38β and p38γ. We then measured p38 mRNA levels in a panel of human breast cancer cell lines (Figure 1B). The expression pattern of p38 isoforms in these breast cancer cell lines is consistent with the human breast tumors, with p38α and p38δ isoforms being the most abundant. The RNA and protein levels do not exactly correlate with each other. This may relate to minor differences in RNA or protein stability in the cell lines, or alternatively reflects the semi-quantitative nature of western blot analysis. Finally, we measured the total p38 level in breast cancer cell lines using an antibody that recognizes all p38 isoforms (Figure 1C). The Western blot analysis showed that the p38 protein is expressed in all the examined breast cancer cell lines.
Figure 1. Expression of p38 in human breast tumors and cancer cell lines.
(A) mRNA levels of p38α, β, γ, and δ in 37 human breast tumors were measured by Q-RT-PCR. Genomic equivalent copies for each p38 isoform were normalized to cyclophilin. Statistical analysis was performed using Friedman chi-square test and pair wise Wilcoxon signed-rank test and found that p38α is the most abundant isoform (P < 0.0001).
(B) mRNA levels of p38α, β, γ, and δ in a panel of human breast cancer cells measured by Q-RT-PCR. Genomic equivalent copies for each p38 isoform were normalized to cyclophilin (n = 3).
(C) Total p38 protein levels in the cell lines were measured by Western blots. The level of total p38 protein was normalized to that of β-actin (n = 3).
Dominant-negative p38 inhibits proliferation of MDA-MB-468 and MDA-MB-231 cells, but not MCF-7 cells
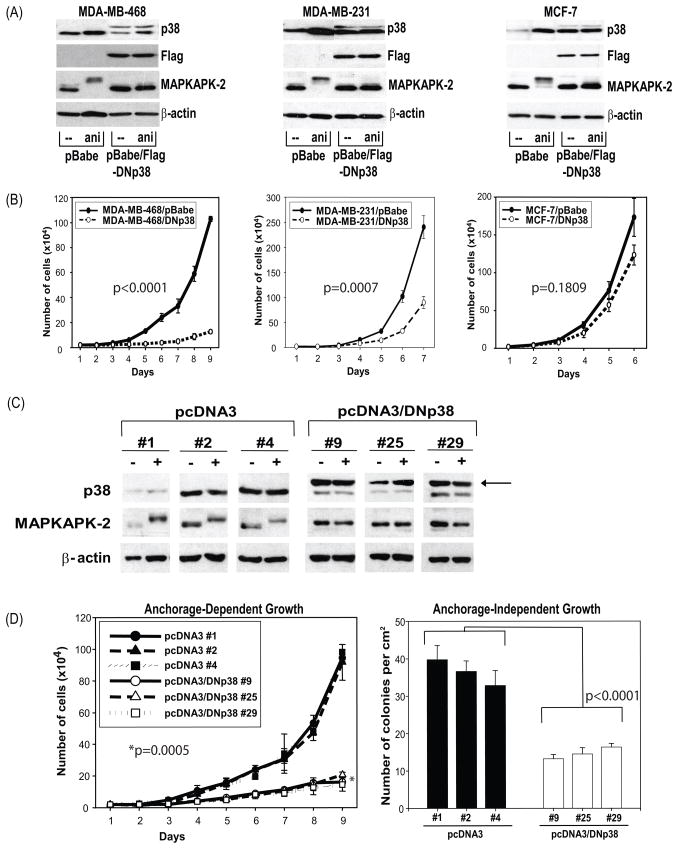
To further evaluate the role of p38 in regulating breast cancer cell proliferation, we expressed a dominant-negative form of p38 in several human breast cancer cell lines. This dominant-negative p38 was generated by mutating Thr180 to Ala and Tyr182 to Phe. The phosphorylation of Thr180 and Tyr182 is required for p38 kinase activity, but is not required for p38 to form complex with upstream kinases (i.e. MKK3 and MKK6) that phosphorylate p38 (25). Stable pools of MDA-MB-468, MDA-MB-231, and MCF-7 cells were created expressing this dominant-negative p38 (Figure 2A–B). MAPKAPK-2 has been previously shown to be regulated by p38 and anisomycin can phosphorylate p38 and subsequently phosphorylate MAPKAPK-2 (26). Expression of the dominant-negative p38 is visualized as a larger Flag-tagged p38 protein as well as by the anti-Flag antibody. This dominant-negative p38 protein inhibited phosphorylation MAPKAPK-2 as seen by the absence of a supershift in MAPKAPK-2 expression (Figure 2A), indicating that p38 signaling in the MDA-MB-468, MDA-MB-231, and MCF-7 cells was effectively blocked. Expression of the dominant-negative p38 also significantly inhibited the proliferation of MDA-MB-468 cells (P < 0.0001) and MDA-MB-231 cells (P = 0.0007) (Figure 2B). However, the growth of MCF-7 cells was not significantly suppressed in the presence of the dominant-negative p38 (P = 0.1809) (Figure 2B). MDA-MB-231 cells were not as effectively suppressed by the dominant-negative p38 as MDA-MB-468 cells even though they are both triple-negative cell lines and p53MUT. One possible reason for this difference in sensitivity to p38 blockade may be because MDA-MB-231 cells express an activated and mutant Ras protein which may provide an additional growth signal (thus making them slightly less sensitive to p38 blockade). Taken together these data provide evidence that p38 blockade can inhibit the proliferation of some breast cancer cells, particularly those that are p53MUT and triple-negative.
Figure 2. Expression of dominant-negative p38 inhibited MDA-MB-468 cell growth both in anchorage-dependent and independent manner.
(A) Stable pools were cultured in serum free IMEM for 24 h and treated with 50ng/ml anisomycin for 15 min. The levels of total p38, total MAPKAPK-2, Flag and β-actin were measured by Western blot.
(B) To measure growth, the stable pools of cells in A were plated in 6-well plates at 2 × 104 cells per well. Cell proliferation was measured by counting cells (n = 3) every 24 h. Analysis was performed using a mixed linear model and P values were as follows for MDA-MB-468 (P < 0.0001), MDA-MB-231 (P = 0.0007), and MCF-7 cells (P =0.1809).
(C) Clones of MDA-MB-468/pcDNA3 (#1, 2, and 4) and MDA-MB-468/pcDNA3-DNp38 (#9, 25, and 29) were plated maintained in serum free IMEM for 48 h. Medium was then replaced with full growth medium IMEM containing 10% heat inactivated FBS. Cells were harvested at either 0 (−) or 10 min (+) after serum stimulation. The levels of total p38, MAPKAPK-2, and β-actin were measured by Western blot. The arrow indicates the larger Flag-tagged p38 protein.
(D) The above pcDNA3 clones were plated in 6-well plates at 2 × 104 cells per well to measure anchorage-dependent growth. Cell proliferation was measured daily by counting cells (n = 3). Analysis was performed using a mixed linear model (P = 0.0005). The clones were also plated in soft agar in 60mm tissue culture dishes at 5 × 103cells per dish. Anchorage independent growth was measured by counting colonies formed in soft agar 4 weeks after plating (n = 3). Analysis was performed using a mixed linear model (P <0.0001).
Dominant-negative p38 inhibits anchorage-dependent and independent proliferation of MDA-MB-468 cells
Stable clones of MDA-MB-468 cells expressing the dominant-negative p38 were created to study the mechanisms by which p38 blockade induces growth suppression in breast cancer cells. In three independent clones of MDA-MB-468/Flag-DNp38 #9, #25 and #29, expression of the dominant-negative p38 blocked serum-induced phosphorylation of MAPKAPK-2 (Figure 2C), but not that of JNK or ERK1/2 (data not shown), suggesting that the blockade is specific for the p38 signaling pathway. These results were determined by comparing to control pcDNA3 clones #1, #2, and #4. The anchorage-dependent growth of MDA-MB-468/Flag-DNp38 cells was significantly repressed when compared to that of MDA-MB-468 transfected with only the empty vector (P = 0.0005) (Figure 2D). The expression of dominant-negative p38 also significantly suppressed anchorage-independent growth of MDA-MB-468 cells (P < 0.0001) (Figure 2D). These data indicate that expression of dominant-negative p38 inhibited the proliferation of MDA-MB-468 cells in an anchorage-dependent and -independent manner.
We then analyzed the cell cycle distribution of MDA-MB-468 cells expressing the dominant-negative p38. As shown in Figure S1A, 27% of the MDA-MB-468/pcDNA3 cells were in S phase 24 hours after serum stimulation. However, only 16% of MDA-MB-468/Flag-DNp38 cells were in S phase at that time point. There was a second G1/S progression 48 hours after serum stimulation. The percentage of cells in S phase was 15% and 10% for MDA-MB-468/pcDNA3s and MDA-MB-468/Flag-DNp38s, respectively. We did not observe apoptosis induced by expression of dominant-negative p38 (data not shown). These data suggest that the anti-proliferation effect caused by expression of dominant-negative p38 is due to inhibition of G1/S cell cycle progression. Since cyclin D1 is a major regulator of G1/S progression we measured RNA (Figure S1B) and protein levels (Figure S1C) of cyclin D1 and found the induction of cyclin D1 was blocked in MDA-MB-468 cells expressing dominant-negative p38 following serum stimulation.
Silencing of p38α, but not p38δ, by siRNA inhibits proliferation of MDA-MB-468 cells
To investigate the effect of individual p38 isoforms on breast cancer cell proliferation, we used siRNA to specifically silence p38α and p38δ. As shown in Figure 1, p38α and p38δ are the two most abundant isoforms in breast cancer cell lines and in human breast tumors. MDA-MB-468 cells were transfected with siRNAs to either p38α or p38δ. Cells transfected with siRNA dilution buffer (mock transfection) or non-specific (siLuciferase) siRNA were used as controls. After 24 hours, we re-plated the transfected cells for MTS growth assays, mRNA and protein analysis. As shown in Figure 3A, the p38α mRNA level was decreased by nearly 80% when p38α siRNA, but not p38δ siRNA was transfected. Likewise, p38δ expression is suppressed by p38δ siRNA, but not p38α siRNA. We did not detect p38 total protein by Western blot in cells transfected with p38α siRNA (Figure 3B). In addition, p38α siRNA by both pools of siRNA or by individual duplexes caused loss of MAPKAPK-2 protein expression. One possible reason might be that p38α is required for the stability of MAPKAPK-2. On the other hand, silencing of the p38δ gene with p38δ siRNA did not decrease p38 total protein or MAPKAPK-2 expression or phosphorylation (Figure 3B). These results suggest that p38δ does not phosphorylate MAPKAPK-2 in these cells, presumably because p38δ is not as abundant as p38α. With these data confirming the knockdown of p38 expression as well as suppression of p38 activity, we next tested the effect of the p38α siRNA or p38δ siRNA on MDA-MB-468 cell proliferation. As shown in Figure 3C, p38α siRNA alone inhibited cell proliferation by about 50% (P = 0.0463), while p38δ siRNA alone had no significant effect on cell proliferation (P = 0.3318). These data suggest that p38α, but not p38δ, is the major mediator of proliferation signaling in MDA-MB-468 cells. Blockade of p38 signaling by silencing p38α gene alone is sufficient to inhibit MDA-MB-468 cell proliferation.
Figure 3. Silencing p38α gene with siRNA inhibited MDA-MB-468 cell growth.
(A) Q-RT-PCR was used to measure p38α and p38δ mRNA levels 2 days after transfection. Relative p38α and p38δ mRNA abundance was normalized to that of cyclophilin (n = 3).
(B) 4 days after transfection, cells were treated with DMSO (-) or 50ng/ml anisomycin for 15 min. Levels of total p38, total MAPKAPK-2, and β-actin were measured by Western blot.
(C) 2 × 103 transfected cells were plated in 96-well plates. Proliferation was measured by MTS assay every 24 h (n = 4). p38α significantly inhibited the growth of MDA-MB-468 cells (P = 0.0463) while p38δ had no significant effect (P = 0.3318).
p38 inhibitors suppress breast cancer cell proliferation
To further confirm whether p38 is involved in regulating breast cancer cell proliferation, we blocked p38 signaling using two pharmacological p38 inhibitors: SB203580 and AZ10164773. Both of these compounds can bind the ATP-binding pocket of the p38 enzyme and inhibit p38 activity by blocking the binding of ATP. We measured the proliferation of a panel of ER-positive and ER-negative breast cancer cells using cell counts in the presence of increasing amounts of SB203580 or AZ10164773 (Figure S2A). AZ10164773 is a more potent inhibitor of breast cancer cell growth than SB203580. Of the cell lines analyzed, MDA-MB-468 cells are the most sensitive to p38 blockade. SB203580 blocked the proliferation of MDA-MB-468 cells by more than 50% at 9μM, while <0.1μM of AZ10164773 had the same effect. SB203580 and AZ10164773 inhibited the proliferation of other breast cancer cell lines tested (MDA-MB-361, BT474, MDA-MB-453, MDA-MB-231, and BT20) to various degrees (Table 1 and Supplemental Fig 2A). In contrast, the proliferation of MCF-7 and ZR75-1 cells is only moderately inhibited by either of the p38 inhibitors while normal breast cells (HMECs) and immortalized breast cells (MCF10A) were not inhibited. MCF-7 and ZR75-1 cells differ from the rest of the cell lines used in this study (27) in that they express p53WT (28, 29). These data suggest that p53 status may be associated with the cell’s sensitivity to p38 inhibitors rather than the levels of p38 protein. p38 protein levels, when normalized to beta-actin do not vary greatly between cell lines as displayed in Table 1. However, within the group of cells that are p53MUT, the triple-negative breast cancer cells are among the most sensitive to p38 inhibitors.
Table 1.
An overview of molecular features of breast cancer cell lines and their response to p38 inhibitors
| Molecular Subtype† | ER† | PR† | HER-2 Overexpression† | P53†† | SB203580 IC50 | AZ10164773 IC50 | p38 protein levels | |
|---|---|---|---|---|---|---|---|---|
| MCF-7 | Luminal | + | + | − | WT | 30μM | 20μM | 100% |
| ZR75-1 | Luminal | + | - | − | WT | 30μM | 8μM | 60% |
| BT474 | Luminal | + | + | + | MUT | 10μM | 4μM | 100% |
| MDA-MB-361 | Luminal | + | + | + | MUT | 20μM | 2μM | 100% |
| MDA-MB-468 | Basal A | − | − | − | MUT | 9μM | <0.1μM | 80% |
| MDA-MB-453 | Luminal | − | − | − | MUT | 6μM | <0.1μM | 100% |
| MDA-MB-231 | Basal B | − | − | − | MUT | 30μM | 2μM | 140% |
| BT20 | Basal A | − | − | − | MUT | 6μM | 0.1μM | 100% |
Biochemical effects of the small molecule inhibitors were tested in MBA-MB-468 cells (Figure S2B). The cells were incubated for 1 hour in the presence of DMSO or increasing amounts of SB203580 or AZ10164773. This was followed by stimulation with anisomycin for 15 minutes. Results indicate that the AZ10164773 compound is more effective at inhibiting p38 pathway activation compared to SB203580. AZ10164773 inhibited MAPKAPK-2 phosphorylation at the lowest dose tested and confirms that AZ10164773 inhibits p38 pathway activation more efficiently than SB203580.
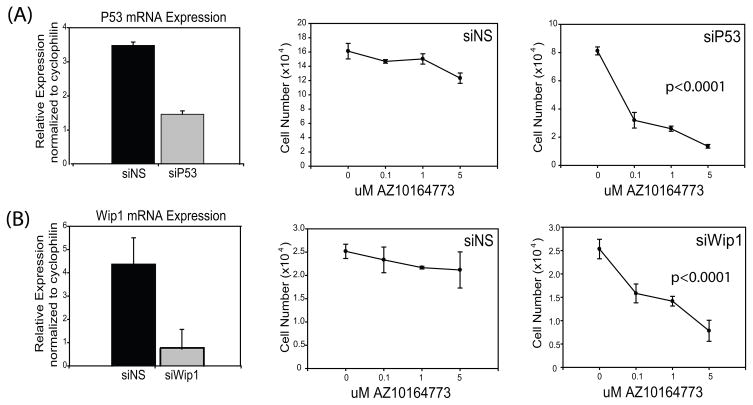
Knockdown of p53 or Wip1 in MCF7 and ZR75-30 cells sensitizes them to p38 inhibitors
We next tested whether knock down of p53WT causes cells resistant to p38 inhibitors to become sensitive to the growth suppressive effects of AZ10164773 (Figure 4). Using siRNA we effectively inhibited p53WT mRNA expression in MCF7 cells compared to non-specific siRNA (Figure 4A). As shown in Figure 4A, MCF7 cells transfected with non-specific siRNA remain resistant to AZ10164773, while MCF7 cells transfected with siRNA against p53WT became sensitive to this p38 inhibitor (P < 0.0001). Thus, loss of p53 expression causes breast cancer cells to become dependent on p38 for their growth.
Figure 4. siRNA against p53 and Wip1 causes addiction to p38 signaling.
(A) Q-RT-PCR was used to measure P53 mRNA levels 2 days after transfection of MCF-7 cells and normalized to cyclophilin (n=3). MCF-7 cells transfected with P53 siRNA and treated with increasing doses of AZ10164773 show sensitivity to the p38 inhibitor compared to non-specific siRNA knockdown when analyzed using a mixed linear model (P < 0.0001).
(B) Q-RT-PCR was also used to measure Wip1 mRNA as described above (n=3) and similar results were found when analyzed using a mixed linear model (P < 0.0001).
Wip1 is a protein phosphatase that is a p53 stimulated and can dephosphorylate p38. So we also wanted to test if knocking down Wip1 would have a similar effect on MCF7 cells as did knocking down p53WT. Wip1 was able to be effectively suppressed in the MCF7 cells treated with Wip1 siRNA (Figure 4B). MCF7 cells transfected with non-specific siRNA remain resistant to AZ10164773, while cells treated with Wip1 siRNA caused them to be sensitive to the p38 inhibitor (P < 0.0001) (Figure 4B). We also performed similar studies in ZR75-30 cells (which are p53WT and resistant to p38 inhibitors). These results showed that p53 or Wip1 knockdown in ZR75-30 cells also induced sensitivity to the p38 inhibitor (data not shown).
These siRNA studies demonstrate that just like p53MUT breast cancer cells, p53WT breast cancer cells in which p53 or Wip1 is reduced, become dependent on p38 signals for their growth, and become sensitive to p38 inhibitors. Loss of p53 alone might cause these cells to become resistant to other pathway inhibitors or the induction of stress, both of which were not tested in this study. However, these results demonstrate that the p38 pathway is an important transducer of proliferative signals in the setting of an impaired p53 pathway, such as occurs in ER-negative breast cancers with p53MUT.
Discussion
This study demonstrates that p38 is an important transducer of proliferative signals in human breast cancer cells. We show that blockade of p38 signaling can inhibit breast cancer cell proliferation, particularly those that are ER-negative and contain p53MUT. These studies show that sensitivity to p38 inhibitors is associated with p53MUT, and that in cells with p53WT, knockdown of p53 induces sensitivity to p38 blockade.
Depending on the nature of the stimuli and the cellular context, p38 can mediate a wide variety of cellular responses. p38 induces apoptosis in some cells (30–32), but prevents apoptosis in others (33–38). Likewise, p38 exhibits opposing effects on cell cycle regulation (39). p38 has been shown to be involved in cell proliferation and tumorigenesis (40, 41). Conversely a number of studies implicate p38 as a negative regulator of cell proliferation (42–44). The reasons for such discrepancy in p38’s role are still unclear.
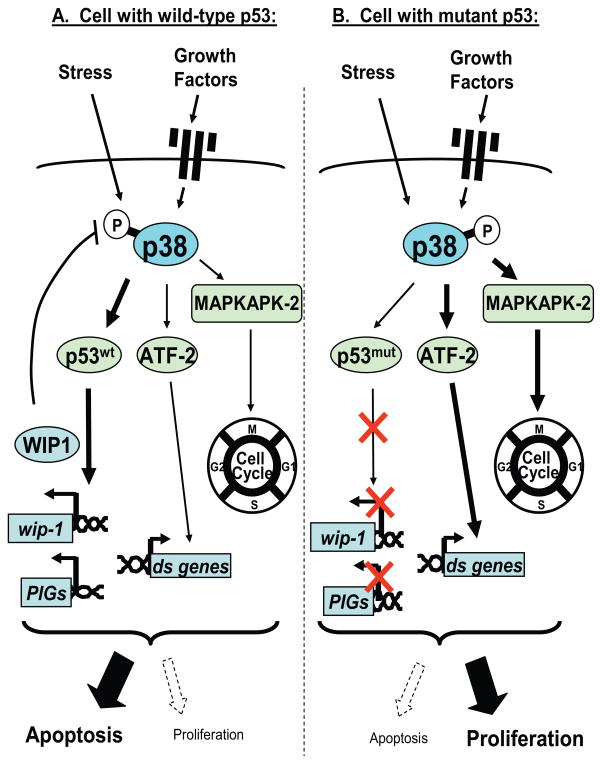
In breast cancer patients, high levels of p38 have been correlated with highly invasive and poor prognostic breast cancer (19–22). In this report we present direct evidence that p38 can transduce proliferative signals in breast cancer cells that have p53MUT (Figure 5) and that the proliferation of these cells is dependent upon intact p38 signaling. The p53 tumor suppressor gene is frequently mutated in breast cancer (45). Wip1 expression is regulated in a p53-dependent manner (46). Wip1 negatively regulates the activity of p38 by dephosphorylating Thr180, reducing p38-induced phosphorylation of p53 and suppressing p53-mediated transcription and apoptosis (47). Cells expressing p53WT maintain the negative feedback regulation of the p38-p53WT-Wip1 signaling. In the context of a cell expressing p53WT, signaling through p38 may predominantly lead to p53-mediated apoptosis rather than proliferation (Figure 5A). Conversely, cells that contain a p53MUT lose the ability to undergo p53-mediated apoptosis. Our data suggests that under this condition, the function of p38 in these cells is shifted to regulate proliferation (Figure 5B). p53MUT cells also lose the feedback regulation by Wip1. This may promote dependency upon p38 for their proliferation “p38-addiction”. In such p38-addicted cells, p38 blockade inhibits proliferation.
Figure 5. Proposed model for p38 signaling in p53 wild-type or mutant cells.
p38 is activated by various extracellular stimuli. In cells with p53WT (A), p38 phosphorylates and activates p53, leading to p53-dependent transcription and apoptosis. p53WT is required for the induction of Wip1, which can dephosphorylate and inactivate p38. Thus, there exists a negative feedback regulation among p38, p53WT, and Wip1. p38 can also regulate ATF-2, MAPKAPK-2 and other targets that are involved in regulating cell cycle and cell proliferation. However, in the context of p53WT, the predominant effect of p38 signaling may be regulating p53-dependent apoptosis rather than cell proliferation. On the other hand, when cells have p53MUT (B), they cannot undergo p53-dependent apoptosis and they lose the negative feedback regulation involving Wip1. Signaling through p38 continues to activate proliferative pathways and stimulates cell cycle progression and cell proliferation through mediators such as ATF-2 and MAPKAPK-2. In such p53MUT cells, blockade of p38 signaling suppresses cancer cell growth. (PIGs: p53-induced genes, ds genes: downstream genes).
MCF-7 and MDA-MB-361 cells have DNA amplification at 17q23, the region that contains the Wip1 gene (48), and thus overexpress the Wip1 protein. It is arguable that the high endogenous levels of Wip1 cause the cells to become independent of p38 for proliferation. However, MDA-MB-361 cells, despite having high endogenous Wip1 expression, have a p53 mutation and are relatively sensitive to p38 blockade. This suggests that it is the dynamic balance among p38-p53WT-Wip1, not the constitutive expression of Wip1 that is associated with the lack of growth suppression.
There is a significant negative correlation between levels of p53 and steroid hormone receptors (49). Most of the existing ER-negative breast cancer cell lines have p53 somatic mutations. Our data show that the ER-negative and p53MUT breast cancer cell lines are more sensitive to p38 blockade than are the ER-positive and p53WT breast cell lines. However, there may be a range of sensitivity depending on the acquisition of additional mutations that could confer a growth advantage. Our findings that p38 inhibitors significantly suppress the growth of ER-negative, p53MUT breast cancer cells suggest that targeting p38 signaling may be clinically useful for the treatment of the highly aggressive triple-negative breast cancers that are p53MUT. p38 inhibitors are now being tested for treatment of inflammatory diseases, including rheumatoid arthritis and cardiovascular diseases, and have been well tolerated with minimal side effects (50). Based on these studies, clinical trials of p38 inhibitors alone or in combination with standard chemotherapy should be considered for the treatment of patients with ER-negative, p53MUT breast cancer.
Supplementary Material
Acknowledgments
We thank Dr. Gordon Mills for his helpful discussions and critical reading of the manuscript, Dr. Aubrey Thompson for providing retroviral vector pBabe-puro3 and retrovirus Phoenix A packaging cells and Dr. Rachel Schiff for providing the dominant-negative p38 expression vector. This work was supported by Breast Cancer P50-SPORE 3 grant CA58183 (PB), Dan Duncan Cancer Center grant P30 CA125123 (SH), NIH NRSA T32 grant CA90221 (JM), a Komen Promise Grant KG081694 (PB) and a research grant from AstraZeneca Pharmaceuticals PLC.
References
- 1.Society AC. Breast Cancer Facts and Figures 2009. Atlanta: American Cancer Society; 2009. [Google Scholar]
- 2.Fisher B, Costantino JP, Wickerham DL, et al. Tamoxifen for prevention of breast cancer: report of the National Surgical Adjuvant Breast and Bowel Project P-1 Study. J Natl Cancer Inst. 1998;90:1371–88. doi: 10.1093/jnci/90.18.1371. [DOI] [PubMed] [Google Scholar]
- 3.Cummings SR, Eckert S, Krueger KA, et al. The effect of raloxifene on risk of breast cancer in postmenopausal women: results from the MORE randomized trial. Multiple Outcomes of Raloxifene Evaluation. Jama. 1999;281:2189–97. doi: 10.1001/jama.281.23.2189. [DOI] [PubMed] [Google Scholar]
- 4.American CS. Estimated new cancer cases and deaths by gender. Atlanta: American Cancer Society; 2002. [Google Scholar]
- 5.Tan AR, Swain SM. Adjuvant chemotherapy for breast cancer: an update. Semin Oncol. 2001;28:359–76. doi: 10.1016/s0093-7754(01)90130-7. [DOI] [PubMed] [Google Scholar]
- 6.Cano E, Mahadevan LC. Parallel signal processing among mammalian MAPKs. Trends Biochem Sci. 1995;20:117–22. doi: 10.1016/s0968-0004(00)88978-1. [DOI] [PubMed] [Google Scholar]
- 7.Ono K, Han J. The p38 signal transduction pathway: activation and function. Cell Signal. 2000;12:1–13. doi: 10.1016/s0898-6568(99)00071-6. [DOI] [PubMed] [Google Scholar]
- 8.Robinson MJ, Cobb MH. Mitogen-activated protein kinase pathways. Curr Opin Cell Biol. 1997;9:180–6. doi: 10.1016/s0955-0674(97)80061-0. [DOI] [PubMed] [Google Scholar]
- 9.Han J, Lee JD, Tobias PS, Ulevitch RJ. Endotoxin induces rapid protein tyrosine phosphorylation in 70Z/3 cells expressing CD14. J Biol Chem. 1993;268:25009–14. [PubMed] [Google Scholar]
- 10.Han J, Lee JD, Bibbs L, Ulevitch RJ. A MAP kinase targeted by endotoxin and hyperosmolarity in mammalian cells. Science. 1994;265:808–11. doi: 10.1126/science.7914033. [DOI] [PubMed] [Google Scholar]
- 11.Jiang Y, Chen C, Li Z, et al. Characterization of the Structure and Function of a New Mitogen-activated Protein Kinase (p38beta ) J Biol Chem. 1996;271:17920–6. doi: 10.1074/jbc.271.30.17920. [DOI] [PubMed] [Google Scholar]
- 12.Lechner C, Zahalka MA, Giot JF, Moller NP, Ullrich A. ERK6, a mitogen-activated protein kinase involved in C2C12 myoblast differentiation. Proc Natl Acad Sci U S A. 1996;93:4355–9. doi: 10.1073/pnas.93.9.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Z, Jiang Y, Ulevitch RJ, Han J. The primary structure of p38 gamma: a new member of p38 group of MAP kinases. Biochem Biophys Res Commun. 1996;228:334–40. doi: 10.1006/bbrc.1996.1662. [DOI] [PubMed] [Google Scholar]
- 14.Mertens S, Craxton M, Goedert M. SAP kinase-3, a new member of the family of mammalian stress-activated protein kinases. FEBS Lett. 1996;383:273–6. doi: 10.1016/0014-5793(96)00255-4. [DOI] [PubMed] [Google Scholar]
- 15.Kumar S, McDonnell PC, Gum RJ, Hand AT, Lee JC, Young PR. Novel homologues of CSBP/p38 MAP kinase: activation, substrate specificity and sensitivity to inhibition by pyridinyl imidazoles. Biochem Biophys Res Commun. 1997;235:533–8. doi: 10.1006/bbrc.1997.6849. [DOI] [PubMed] [Google Scholar]
- 16.Wang XS, Diener K, Manthey CL, et al. Molecular cloning and characterization of a novel p38 mitogen-activated protein kinase. J Biol Chem. 1997;272:23668–74. doi: 10.1074/jbc.272.38.23668. [DOI] [PubMed] [Google Scholar]
- 17.Enslen H, Raingeaud J, Davis RJ. Selective activation of p38 mitogen-activated protein (MAP) kinase isoforms by the MAP kinase kinases MKK3 and MKK6. J Biol Chem. 1998;273:1741–8. doi: 10.1074/jbc.273.3.1741. [DOI] [PubMed] [Google Scholar]
- 18.Knebel A, Morrice N, Cohen P. A novel method to identify protein kinase substrates: eEF2 kinase is phosphorylated and inhibited by SAPK4/p38delta. Embo J. 2001;20:4360–9. doi: 10.1093/emboj/20.16.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salh B, Marotta A, Wagey R, Sayed M, Pelech S. Dysregulation of phosphatidylinositol 3-kinase and downstream effectors in human breast cancer. Int J Cancer. 2002;98:148–54. doi: 10.1002/ijc.10147. [DOI] [PubMed] [Google Scholar]
- 20.Esteva FJ, Sahin AA, Smith TL, et al. Prognostic significance of phosphorylated P38 mitogen-activated protein kinase and HER-2 expression in lymph node-positive breast carcinoma. Cancer. 2004;100:499–506. doi: 10.1002/cncr.11940. [DOI] [PubMed] [Google Scholar]
- 21.Gutierrez MC, Detre S, Johnston S, et al. Molecular changes in tamoxifen-resistant breast cancer: relationship between estrogen receptor, HER-2, and p38 mitogen-activated protein kinase. J Clin Oncol. 2005;23:2469–76. doi: 10.1200/JCO.2005.01.172. [DOI] [PubMed] [Google Scholar]
- 22.Davidson B, Konstantinovsky S, Kleinberg L, et al. The mitogen-activated protein kinases (MAPK) p38 and JNK are markers of tumor progression in breast carcinoma. Gynecol Oncol. 2006 doi: 10.1016/j.ygyno.2006.01.034. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Lu C, Shen Q, Munoz-Medellin D, Kim H, Brown PH. AP-1 blockade in breast cancer cells causes cell cycle arrest by suppressing G1 cyclin expression and reducing cyclin-dependent kinase activity. Oncogene. 2004;23:8238–46. doi: 10.1038/sj.onc.1207889. [DOI] [PubMed] [Google Scholar]
- 24.Kim HT, Kong G, Denardo D, et al. Identification of biomarkers modulated by the rexinoid LGD1069 (bexarotene) in human breast cells using oligonucleotide arrays. Cancer Res. 2006;66:12009–18. doi: 10.1158/0008-5472.CAN-05-2515. [DOI] [PubMed] [Google Scholar]
- 25.Ashwell JD. The many paths to p38 mitogen-activated protein kinase activation in the immune system. Nat Rev Immunol. 2006;6:532–40. doi: 10.1038/nri1865. [DOI] [PubMed] [Google Scholar]
- 26.Sudo T, Kawai K, Matsuzaki H, Osada H. p38 mitogen-activated protein kinase plays a key role in regulating MAPKAPK2 expression. Biochemical and Biophysical Research Communications. 2005;337:415. doi: 10.1016/j.bbrc.2005.09.063. [DOI] [PubMed] [Google Scholar]
- 27.Neve RM, Chin K, Fridlyand J, et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell. 2006;10:515–27. doi: 10.1016/j.ccr.2006.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wasielewski M, Elstrodt F, Klijn JG, Berns EM, Schutte M. Thirteen new p53 gene mutants identified among 41 human breast cancer cell lines. Breast Cancer Res Treat. 2006;99:97–101. doi: 10.1007/s10549-006-9186-z. [DOI] [PubMed] [Google Scholar]
- 29.Concin N, Zeillinger C, Tong D, et al. Comparison of p53 mutational status with mRNA and protein expression in a panel of 24 human breast carcinoma cell lines. Breast Cancer Res Treat. 2003;79:37–46. doi: 10.1023/a:1023351717408. [DOI] [PubMed] [Google Scholar]
- 30.Sugden PH, Clerk A. Oxidative stress and growth-regulating intracellular signaling pathways in cardiac myocytes. Antioxidants & redox signaling. 2006;8:2111–24. doi: 10.1089/ars.2006.8.2111. [DOI] [PubMed] [Google Scholar]
- 31.Sumbayev VV, Yasinska IM. Role of MAP kinase-dependent apoptotic pathway in innate immune responses and viral infection. Scandinavian journal of immunology. 2006;63:391–400. doi: 10.1111/j.1365-3083.2006.001764.x. [DOI] [PubMed] [Google Scholar]
- 32.Baines CP, Molkentin JD. STRESS signaling pathways that modulate cardiac myocyte apoptosis. Journal of molecular and cellular cardiology. 2005;38:47–62. doi: 10.1016/j.yjmcc.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 33.Hendrickx N, Volanti C, Moens U, et al. Up-regulation of cyclooxygenase-2 and apoptosis resistance by p38 MAPK in hypericin-mediated photodynamic therapy of human cancer cells. The Journal of biological chemistry. 2003;278:52231–9. doi: 10.1074/jbc.M307591200. [DOI] [PubMed] [Google Scholar]
- 34.Kurosu T, Takahashi Y, Fukuda T, Koyama T, Miki T, Miura O. p38 MAP kinase plays a role in G2 checkpoint activation and inhibits apoptosis of human B cell lymphoma cells treated with etoposide. Apoptosis. 2005;10:1111–20. doi: 10.1007/s10495-005-3372-z. [DOI] [PubMed] [Google Scholar]
- 35.Sun Y, Sinicrope FA. Selective inhibitors of MEK1/ERK44/42 and p38 mitogen-activated protein kinases potentiate apoptosis induction by sulindac sulfide in human colon carcinoma cells. Molecular cancer therapeutics. 2005;4:51–9. [PubMed] [Google Scholar]
- 36.Navas TA, Nguyen AN, Hideshima T, et al. Inhibition of p38alpha MAPK enhances proteasome inhibitor-induced apoptosis of myeloma cells by modulating Hsp27, Bcl-X(L), Mcl-1 and p53 levels in vitro and inhibits tumor growth in vivo. Leukemia. 2006;20:1017–27. doi: 10.1038/sj.leu.2404200. [DOI] [PubMed] [Google Scholar]
- 37.Shi YY, Small GW, Orlowski RZ. Proteasome Inhibitors Induce a p38 Mitogen-activated Protein Kinase (MAPK)-dependent Anti-apoptotic Program Involving MAPK Phosphatase-1 and Akt in Models of Breast Cancer. Breast cancer research and treatment. 2006;100:33–47. doi: 10.1007/s10549-006-9232-x. [DOI] [PubMed] [Google Scholar]
- 38.Kocanova S, Buytaert E, Matroule JY, et al. Induction of heme-oxygenase 1 requires the p38(MAPK) and PI3K pathways and suppresses apoptotic cell death following hypericin-mediated photodynamic therapy. Apoptosis. 2007;12:731–41. doi: 10.1007/s10495-006-0016-x. [DOI] [PubMed] [Google Scholar]
- 39.Zarubin T, Han J. Activation and signaling of the p38 MAP kinase pathway. Cell research. 2005;15:11–8. doi: 10.1038/sj.cr.7290257. [DOI] [PubMed] [Google Scholar]
- 40.Maher P. p38 mitogen-activated protein kinase activation is required for fibroblast growth factor-2-stimulated cell proliferation but not differentiation. The Journal of biological chemistry. 1999;274:17491–8. doi: 10.1074/jbc.274.25.17491. [DOI] [PubMed] [Google Scholar]
- 41.Crawley JB, Rawlinson L, Lali FV, Page TH, Saklatvala J, Foxwell BM. T cell proliferation in response to interleukins 2 and 7 requires p38MAP kinase activation. The Journal of biological chemistry. 1997;272:15023–7. doi: 10.1074/jbc.272.23.15023. [DOI] [PubMed] [Google Scholar]
- 42.Lavoie JN, L’Allemain G, Brunet A, Muller R, Pouyssegur J. Cyclin D1 expression is regulated positively by the p42/p44MAPK and negatively by the p38/HOGMAPK pathway. J Biol Chem. 1996;271:20608–16. doi: 10.1074/jbc.271.34.20608. [DOI] [PubMed] [Google Scholar]
- 43.Casanovas O, Miro F, Estanyol JM, Itarte E, Agell N, Bachs O. Osmotic stress regulates the stability of cyclin D1 in a p38SAPK2-dependent manner. J Biol Chem. 2000;275:35091–7. doi: 10.1074/jbc.M006324200. [DOI] [PubMed] [Google Scholar]
- 44.Bulavin DV, Higashimoto Y, Popoff IJ, et al. Initiation of a G2/M checkpoint after ultraviolet radiation requires p38 kinase. Nature. 2001;411:102–7. doi: 10.1038/35075107. [DOI] [PubMed] [Google Scholar]
- 45.Davidoff AM, Kerns BJ, Pence JC, Marks JR, Iglehart JD. p53 alterations in all stages of breast cancer. J Surg Oncol. 1991;48:260–7. doi: 10.1002/jso.2930480409. [DOI] [PubMed] [Google Scholar]
- 46.Fiscella M, Zhang H, Fan S, et al. Wip1, a novel human protein phosphatase that is induced in response to ionizing radiation in a p53-dependent manner. Proc Natl Acad Sci U S A. 1997;94:6048–53. doi: 10.1073/pnas.94.12.6048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takekawa M, Adachi M, Nakahata A, et al. p53-inducible wip1 phosphatase mediates a negative feedback regulation of p38 MAPK-p53 signaling in response to UV radiation. Embo J. 2000;19:6517–26. doi: 10.1093/emboj/19.23.6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li J, Yang Y, Peng Y, et al. Oncogenic properties of PPM1D located within a breast cancer amplification epicenter at 17q23. Nature genetics. 2002;31:133–4. doi: 10.1038/ng888. [DOI] [PubMed] [Google Scholar]
- 49.Hassapoglidou S, Diamandis EP, Sutherland DJ. Quantification of p53 protein in tumor cell lines, breast tissue extracts and serum with time-resolved immunofluorometry. Oncogene. 1993;8:1501–9. [PubMed] [Google Scholar]
- 50.Lee MR, Dominguez C. MAP kinase p38 inhibitors: clinical results and an intimate look at their interactions with p38alpha protein. Curr Med Chem. 2005;12:2979–94. doi: 10.2174/092986705774462914. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.