Abstract
Type IIS restriction endonuclease BtsCI (GGATG 2/0) is a neoschizomer of FokI (GGATG 9/13) and cleaves closer to the recognition sequence. Although M.BtsCI shows 62% amino acid sequence identity to M.FokI, BtsCI and FokI restriction endonucleases do not share significant amino acid sequence similarity. BtsCI belongs to a group of Type IIS restriction endonucleases, BsmI, Mva1269I and BsrI, that carry two different catalytic sites in a single polypeptide. By inactivating one of the catalytic sites through mutagenesis, we have generated nicking variants of BtsCI that specifically nick the bottom-strand or the top-strand of the target site. By treating target DNA sequentially with the appropriate combinations of FokI and BtsCI nicking variants, we are able to generate long overhangs suitable for fluorescent labeling through end-filling or other techniques based on annealing of complementary DNA sequences.
INTRODUCTION
Most restriction endonuclease (REases) commonly used in molecular cloning are Type IIP enzymes which have the well-studied characteristic PD-Xn-E/DXK catalytic motif (X being any amino acid residue). These enzymes recognize palindromic double-stranded DNA (dsDNA) sequences ranging from 4 to 8 bp long and cleave both strands at symmetrical sites within the recognition sequences (1,2). For example, a BamHI dimer binds to the symmetric GGATCC sequence. The top- and bottom-strand cuts are probably introduced simultaneously following a serial of conformation changes (dimer interface changes, narrowing of the DNA binding cleft, C-terminal arm unfolding and binding to the minor groove, catalytic residues movement of more than 6 Å distance relative to scissile phosphate (3,4). In addition, recognition and catalysis of Type IIP REases are intimately coupled. Thus it has been difficult to alter recognition specificity while retaining respectable activity (5). For the less studied Type IIS REases, target sequence recognition and phosphodiester bond cleavage are carried out by separate domains (6–8). This separation of function allows the recognition of non-palindromes and cleavage outside the recognition sites. It also gives rise to a variety of DNA recognition and catalytic mechanisms. For example, in AlwI (GGATC 4/5), swapping the DNA cleavage domain with that of Nt.BstNBI (GAGTC 4/−) retains the AlwI recognition specificity but turns it into a top-strand nicking endonuclease (NEase), showing that the DNA recognition domain itself is sufficient to bind to the target DNA specifically (9). In terms of catalytic mechanism, BfiI (ACTGGC 5/4) has a phospholipase D-type catalytic site and does not require metal ions for catalysis (8). FokI (GGATG 9/13) has a DNA recognition domain that recognizes GGATG and a DNA cleavage domain that carries a canonical PD-Xn-DXK catalytic site. Structural and biochemical studies indicated that transient dimerization of FokI is required to make ds breaks (7,10). Recently, it has been reported that by mixing wild-type (wt) FokI with a mutant that carries mutations of crucial DNA-binding and catalytic residues, the heterodimeric enzyme nicks the bottom strand specifically (11).
Previously our group reported two Type IIS REases that contains two independent conserved PD-Xn-D/EXK catalytic motif variants. BtsI (GCAGTG 2/0) and BsrDI (GCAATG 2/0) are heterodimeric Type IIS REases that recognize similar 6-base sequences. Although each of their subunits carries a catalytic site, only the large subunit is active on its own and exhibits bottom-strand nicking activity. The small subunit by itself has no cleavage activity, but mixing the two subunits results in ds cleavage activity. When the catalytic site of the large subunits of BtsI and BsrDI are mutated, then combined with their wild-type small subunit partners, both enzymes exhibit only top-strand nicking activity (12). By comparing the amino acid sequences we found that BsrI (ACTGG 1/−1) and Mva1269I/BsmI (GAATGC 1/−1) have similar catalytic sites. Most interestingly, the two catalytic sites are located on the same polypeptide in these enzymes. The two catalytic sites also work on specific strands—mutation of residues in each of the two catalytic sites of Mva1269I has resulted in strand-specific DNA nicking mutants. In addition, Mva1269I cleavage follows a sequential order: the bottom strand is cleaved first and then the top strand (13).
Recently, NEases have been used to label dsDNA by nick translation for the detection of specific DNA sequences (14,15). Coupled with high-resolution imaging and DNA stretching, BbvCI sites have been mapped onto λ DNA with high speed and accuracy (16). Such mapping techniques likely provide rapid identification of pathogens and disease-related mutations, consequently speed up medical diagnostics. However, the limited number of NEases commercially available restricts the use of these applications.
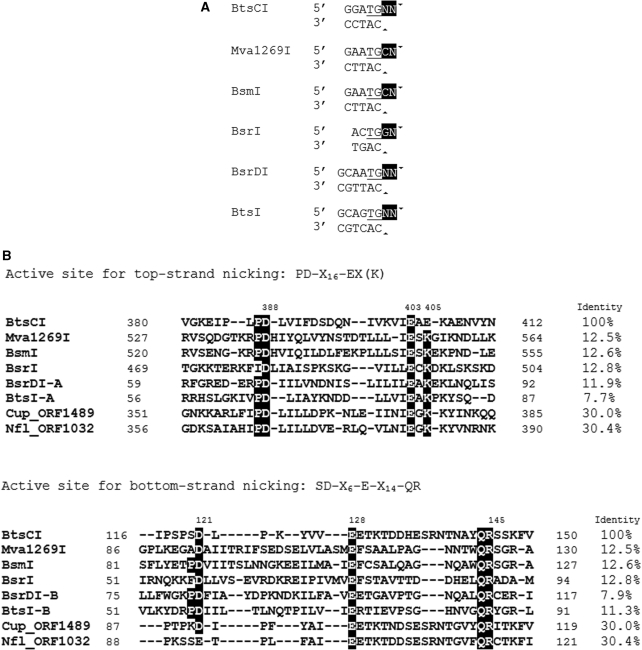
Several related Type IIS REases (Mva1269I, BsmI, BsrI, BsrDI and BtsI) have two catalytic sites, Cb and Ct. Cb is the catalytic site specific to the bottom-strand of the DNA while Ct is specific to the top-strand. Interestingly, BtsCI GGATG (2/0) and these Type IIS REases cleave their target sequences 2-bp downstream of the top strand dinucleotide TG, resulting in a 2-base 3′ overhangs (Figure 1A). In BtsI and BsrDI, the Cb is a variant of the canonical REase motif PD-Xn-E-Xn-QR whereas Ct is a canonical PD-Xn-EXK catalytic site. Inactivation of Cb or Ct by mutagenesis generates variants of BtsI or BsrDI that only nick the top or bottom strand (12). Interestingly, while the two catalytic sites are located in different subunits in heterodimeric BsrDI and BtsI, BsmI, BsrI and Mva1269I have both catalytic sites on a single polypeptide. It has been shown that in Mva1269I, mutation of residues in the catalytic sites resulted in nicking variants that bind DNA containing the target site as strongly as the wt Mva1269I (13).
Figure 1.
(A) The recognition sequences of BtsCI and related REases are aligned at the common TG bases (underlined). The common 2-base 3′ overhangs are highlighted. Arrowheads indicate the cleavage sites. (B) Amino acid sequence alignments of the two catalytic sites of BtsCI and related REases. Identical residues are shown as white on black. The identity indicates the percentage of amino acid sequence identity between BtsCI and related REases. The DNA sequence of BtsCI R-M system has been deposited in GenBank with accession no GQ449683.
In this work, we report the cloning of the BtsCI restriction-modification (R-M) system and engineering of strand-specific nicking variants. The presence of the conserved catalytic sites and creation of nicking variants by mutation of the putative catalytic residues confirm that BtsCI belongs to the Mva1269I/BsmI group of REases. We also demonstrate that sequential cleavage of BtsCI sites with FokI and BtsCI nicking variants can generate 11-base 3′ recessive ends suitable for labeling of DNA and ligation, or 9-base 5′ recessive ends for ligation. The long overhangs will be useful in hybridization-based DNA detection and capture.
MATERIALS AND METHODS
Restriction enzymes, DNA polymerases, DNA substrates, Escherichia coli strains (ER2683 and NEB Express), plasmid vectors pBR322, pUC19, pACYC184, DNA and protein ladders were obtained from New England Biolabs Inc. BstF5I was purchased from Sibenzyme.
Cloning of BtsCI R-M system
ApoI, NlaIII or Sau3AI partially digested genomic fragments from Bacillus thermosphaericus were cloned into EcoRI, SphI or BamHI digested, CIP-treated pUC19 with compatible cohesive ends, respectively. The plasmid DNA libraries were challenged by BstF5I digestion, an isoschizomer of BtsCI. Following BstF5I challenge, the survivors (resistant plasmids) were re-transformed into E. coli. Individual transformants were screened for resistance to BstF5I digestion and the inserts from resistant clones were sequenced and open reading frames (ORFs) were identified. The full-length BtsCI endonuclease gene (btsCIR) was obtained by inverse PCR walking and recloned into pUC19.
Strains for protein expression
The btsCIR and btsCIM genes were ligated to pUC19 and pACYC184, respectively. E. coli ER2683 or NEB Express was pre-modified by pACYC-btsCIM, followed by transformation with a second plasmid pUC19-btsCIR to generate the expression strain: ER2683 [pACYC-btsCIM, pUC19-btsCIR]. BtsCI expression was induced with addition of 0.25 mM IPTG to mid-log phase cell culture in LB medium for 3 h at 37°C. The culture was harvested and the cell pellet was kept frozen until lysis for protein purification.
Protein purification
Cell pellets from 1 L of IPTG-induced cells was resuspended in 40 ml of 20 mM Tris–HCl, pH 7.5, 1 mM EDTA and sonicated on ice. After centrifugation at 15 000 g at 4°C for 30 min, the supernatant was heated at 60°C for 30 min. The denatured E. coli protein was removed by centrifugation at 20 000 g for 30 min at 4°C. The supernatant was subjected to column chromatography through Heparin HyperD® M (Pall) and Q-Sepharose™ (GE Life Sciences) with linear gradients of NaCl (50 mM–1 M). Peak fractions were assayed for cleavage or nicking activity and pooled (detailed purification procedure available upon request).
Site-directed mutagenesis and screening for nicking variants
Mutations were introduced by a variation of the inverse PCR method using pUC19-btsCIR as template (17). The new constructs were sequenced to confirm the desired mutations.
Determination of oligomeric state
The molecular weight of BtsCI variants were determined by equilibrium gel filtration experiments (18) using an AKTA FPLC protein liquid chromatography system with a Superdex 200 10/300 GL column (GE Healthcare). The column was equilibrated with a buffer containing 20 mM Tris–HCl, pH 7.5, 200 mM NaCl. The calibration curve was generated by using the high molecular weight calibration kit (GE Healthcare).
Cleavage/nicking activity assays and identification of the cleaved strands and cleavage sites
Cleavage/nicking activity of BtsCI mutant cell extracts or purified enzymes were assayed on 0.5 µg of pUC19 DNA in NEBuffer 4 at 50°C for 1 h. The cleavage/nicked products were analyzed by gel electrophoresis using 1.5% agarose gels. There are five BtsCI sites on pUC19. Therefore, complete digestion results in fragment sizes of 181, 244, 287, 613 and 1361 bp as predicted by NEBcutter (19). As the closest neighboring sites are 181 bp apart, linearized DNA should not be observed under the agarose gel electrophoresis condition if the sites are predominantly nicked. For identification of the cleaved strands and cleavage sites, the nicked DNA was gel-purified and subjected to Sanger ‘run-off’ sequencing (BigDye® Terminator Cycle Sequencing Kit, Applied Biosystems).
Generation of specific overhangs and detection
Plasmid pUC19 (0.25 µg) was digested by FokI (5 units) in NEBuffer 4 at 37°C for 30 min. To generate 11-base 5′ overhang or 9-base 3′ overhang, five units of Nt.BtsCI or Nb.BtsCI was added to the FokI digested reactions and incubated at 50°C for 1 h. Digested product was ligated to a duplex oligonucleotide (∼100 bp long) containing the complementary overhang at room temperature for 2 h. Ligated product was detected by PCR where one of the primers anneals to the duplex oligonucleotide and the other on pUC19 near the FokI site. The PCR product was subjected to agarose gel electrophoresis.
DNA labeling by end filling using Cy3-dNTP
DNA sequences containing two BtsCI recognition sites were amplified from pUC19 using LongAmp Taq 2X master mix and digested by the indicated endonuclease(s). Digestion was terminated by adding EDTA to a final concentration of 20 mM. PCR product (0.1 µg) was labeled with Cy3-dNTP (PerkinElmer) by E. coli Pol I Klenow fragment at room temperature for 15 min and then analyzed by 6% PAGE in 1 × TBE. Labeled DNA was visualized using Typhoon 9400 laser scanner (GE Life Sciences; excitation—532 nm; emission—580 nm). Total DNA was visualized by UV illumination after staining with ethidium bromide (2 µg/ml).
RESULTS
Cloning of BtsCI R-M system
The methylase selection strategy (20) was used to clone the BtsCI modification (M) gene and part of the restriction (R) gene and inverse PCR walking was used to obtain the full-length R gene. Two genes, btsCIM (2064 bp) and btsCIR (1398 bp) were found in the BtsCI R-M system. We subcloned the btsCIM gene into pACYC184 to produce the pre-modified host (ER2683). The plasmid pACYC-btsCIM purified from E. coli was confirmed to be resistant to BtsCI digestion (modification of BtsCI sites by BtsCI methylase renders the plasmid resistance to BtsCI digestion). The btsCIR gene contained in a PCR fragment was then ligated into pUC19 and introduced into the pre-modified host. BtsCI REase activity was detected in IPTG-induced cell extracts (data not shown).
A BLASTP search indicated that M.BtsCI shares high sequence similarity to isoschizomers M.FokI (62% sequence identity/75% sequence similarity) and M.StsI (48% sequence identity/65% sequence similarity) (data not shown). The N-terminus of M.BtsCI is similar to M3.BstF5I and so is the C-terminus of M.BtsCI to M2.BstF5I, suggesting that M.BtsCI is a fusion of two functional methylases that modify each of the two strands. The amino acid sequences of BtsCI and FokI REases do not share significant similarity. However, two homologs of BtsCI endonucleases (putative endonucleases) were found from two sequenced microbial genomes (see below).
BtsCI catalytic motifs
Sequence alignment between BtsCI and similar enzymes reveals that BtsCI contains these two Mva1269I/BsmI-like catalytic sites, although the overall sequence identity is low (Figure 1B). A motif, SD-X6-E-X14-QR, was found near the N-terminus of BtsCI and a motif similar to the Ct of Mva1269I/BsmI-type REases was found near the C-terminus. Interestingly, although the Ct motif is the canonical PD-Xn-EXK for the Mva1269I/BsmI-like enzymes, the Ct motif in BtsCI is more BamHI-like and contains PD-Xn-EXE (E instead of K) (Figure 1B). This prompted us to verify the catalytic motifs and to engineer strand-specific nicking variants of BtsCI.
Generation of BtsCI nicking variants
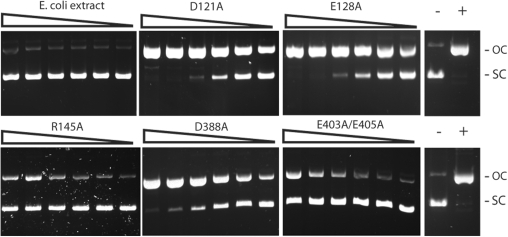
Single mutations D121A, E128A and R145A were introduced into the Cb motif in BtsCI based on analogous mutations D58A, E72A and R86A in the BtsI large subunit (Figure 1B) (12). Figure 2 shows that these mutants have lost most of their dsDNA cleavage activity and only nicked pUC19. Similarly, BtsCI mutants that carry the single mutation D388A or the double mutations E403A/E405A, the predicted catalytic residues in the Ct motif, have strong nicking activity. To confirm the cleavage sites of the nicking variants, purified mutants D121A/E128A, D388A and E403A/E405A were used to nick pUC19 or pBR322 DNA. The nicked DNA was then gel-purified and subjected to Sanger sequencing. Mutant D121A/E128A nicked the top strand at GGATGNN^, whereas mutants D388A and E403A/E405A nicked the bottom strand at ^CATCC (data not shown). This result supports the prediction that the N-terminal catalytic site is responsible for bottom-strand cleavage (Cb) and the C-terminal site is responsible for top-strand cleavage (Ct) for Mva1268I/BsmI-like REases.
Figure 2.
DNA nicking activity of BtsCI mutants. Two-fold serial dilutions of the clarified cell extracts of E. coli cultures that expressed the indicated BtsCI mutants were incubated with 0.5 µg of pUC19 as described in ‘Materials and Methods’ section. The cleavage products were analyzed on a 1% agarose gel. OC, open circle; SC, supercoiled; −, no cleavage; +, pUC19 nicked by Nt.BsmAI.
Improvement of the BtsCI nicking variants
Although mutants D388A and E403A/E405A show strong bottom-strand nicking activity, both of them still exhibit significant dsDNA cleavage at higher enzyme concentrations (data not shown). Similarly, the top-strand nicking mutants D121A, E128A, R145A and D121A/E128A also have detectable dsDNA cleavage activity (data not shown). To minimize the dsDNA cleavage activity, further mutagenesis was carried out.
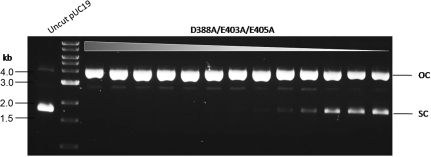
At first, D388 of Ct was mutated to the oppositely charged amino acid residue Lys (K). But D388K was inactive (data not shown). When amino acid substitutions D388A, E403A, and E405A were combined, the triple mutant D388A/E403A/E405A displayed high protein expression level, high bottom-strand nicking activity and minimal dsDNA cleavage activity (Figure 3), suggesting that the combination of the three amino acid substitutions completely inactivates the top-strand catalytic site.
Figure 3.
DNA nicking activity of the bottom-strand nicking BtsCI variant. Two-fold serial dilutions of the crude extract of BtsCI nicking variant (D388A/E403A/E405A) were incubated with pUC19 as described in ‘Materials and Methods’ section. The cleavage products were analyzed on a 1% agarose gel. OC, open circle; SC, supercoiled.
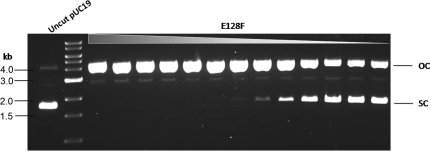
For top-strand nicking, combining the three mutations that gave rise to top strand-nicking activity (triple mutant D121A/E128A/R145A) did not eliminate the dsDNA cleavage activity (data not shown). Reversal of charge for the Cb catalytic residues (double mutant D121K/E128R) had a very low expression level and lower nicking activity compared to the double mutant D121A/E128A (data not shown). When E128, however, was substituted for Phe (F), the resulting mutant E128F has a high nicking activity and low dsDNA cleavage activity (Figure 4). It is possible that the bulky aromatic side chain of Phe provides a more effective interference to the cleavage reaction of Cb. We concluded that there are at least two strategies to improve nicking variants. The first one is by combination of multiple amino acid substitutions from imperfect nicking variants (construction of double or triple mutants). The second strategy is to carry out saturation mutagenesis on a particular amino acid residue and screen for improved enzyme properties.
Figure 4.
DNA nicking activity of top-strand nicking BtsCI variant. Two-fold serial dilutions of the cell extract of BtsCI nicking variant E128F were incubated with pUC19 as described in ‘Materials and Methods’ section. The cleavage products were analyzed on a 1% agarose gel. OC, open circle; SC, supercoiled.
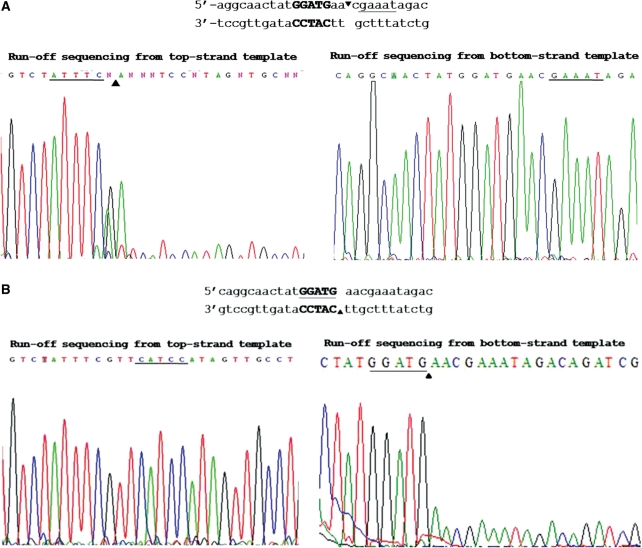
The target site and nicking strand specificity by BtsCI variants E128F and D388A/E403A/E405A were confirmed by Sanger ‘run-off’ sequencing (Figure 5). These two mutants were designated Nt.BtsCI and Nb.BtsCI, respectively, and were used in the following experiments.
Figure 5.
Run-off sequencing of nicked products by BtsI nicking variants. The cleavage product of mutants E128F (A) and D388A/E403A/E405A (B) were gel-purified and subjected to Sanger sequencing on both strands. The recognition sequence is bolded and the cleavage site was indicated by arrows. The extra A peak at the end of run-off sequence was added by the template independent terminal transferase activity of Taq DNA polymerase during sequencing. The extra A peak and drop off in peak signal indicate the nicking site (cleaved template).
BtsCI is a monomeric protein
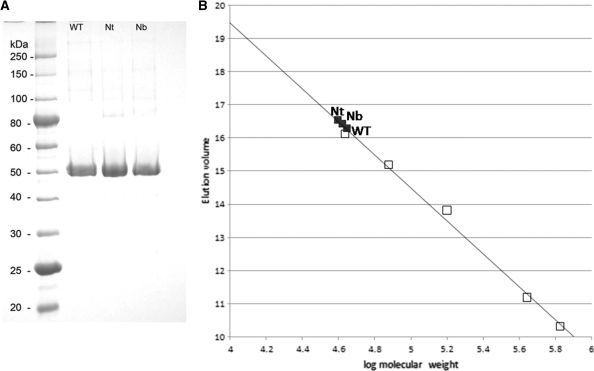
Wt-BtsCI, Nt.BtsCI and Nb.BtsCI were purified to over 95% purity (Figure 6A) and gel filtration was used to determine the oligomerization state of these proteins in solution. A single peak was obtained from each protein and was eluted with an apparent molecular mass of ∼40–45 kDa, consistent to being monomer protein (Figure 6B). Specific DNA cleavage (for the wt enzyme) or nicking activity (for the nicking variants) was present in the peak fractions (data not shown).
Figure 6.
(A) SDS–PAGE analysis of wt BtsCI and two nicking variants. (B) Determination of the oligomerization state of wt BtsCI, Nt and Nb nicking variants by gel filtration chromatography. The elution volume of the protein standard was plotted against the corresponding molecular weight (on the logarithmic scale). Standard proteins (open squares): Ovalbumin, 44 kDa; Conalbumin, 75 kDa; Aldolase, 158 kDa; Ferritin, 440 kDa; Thyroglobulin, 669 kDa. The R2 of the standard curve is 0.9937. The BtsCI proteins (black square) were located on the calibration curve and the corresponding molecular weights were calculated using calibration curve equation: elution volume = −4.9763 (log molecular weight) + 39.39.
Generation of long overhangs by combination of FokI and N.BtsCI digestion
BtsCI nicking variants can nick the top- or the bottom-strand of FokI-cleaved DNA and generate relatively long overhangs. DNA cut by FokI/Nt.BtsCI will have a 11-base 3′ recessive end (5′ overhang) whereas FokI/Nb.BtsCI digestions can generate a 9-base 5′ recessive end (3′ overhang) (Figure 7A). To generate the long overhangs, sequential digestion is necessary. It is partly because FokI and BtsCI have different optimum temperatures for cleavage activity. We also found that co-incubation of FokI and N.BtsCI at 37°C resulted in reduced FokI activity (data not shown), possibly due to competition between FokI and BtsCI for the same recognition sites.
Figure 7.
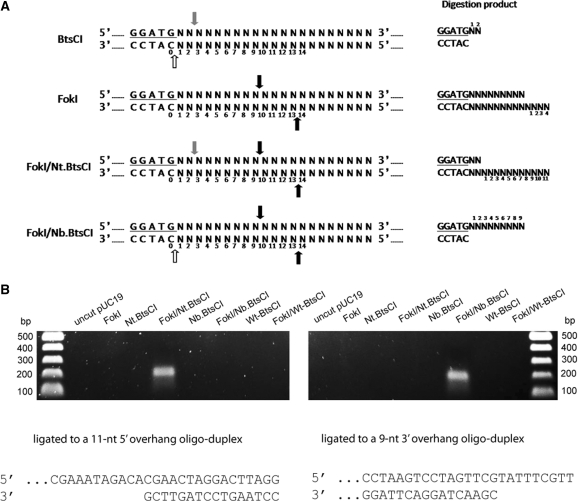
(A) Generation of overhangs. Cleavage sites for BtsCI, FokI and a combination of FokI/Nt.BtsCI and FokI/Nb.BtsCI are indicated. For BtsCI, a 2-nt 5′ recessive end is generated. For FokI, a 4-nt 3′ recessive end is generated. For FokI/Nt.BtsCI, an 11-nt 3′ recessive end is generated. For FokI/Nb.BtsCI, a 9-nt 5′ recessive end is generated. Grey arrows, BtsCI top-strand cleavage site; white arrows, BtsCI bottom-strand cleavage site; black arrows, FokI cleavage sites. (B) Annealing of oligonucleotides to the long overhangs for PCR. Plasmid pUC19 was cleaved by the indicated enzyme(s) and then ligated to a 100-bp long oligonucleotide as described in ‘Materials and Methods’ section. The ligation products were used as template for PCR that specifically detect the ligated DNA. Only the cleavage product of FokI/Nt.BtsCI can be annealed to the 11-nt 3′ recessive end oligonucleotide (left panel), whereas only the cleavage product of FokI/Nb.BtsCI can be annealed to the 9-nt 5′ recessive end oligonucleotide (right panel).
To confirm the generation of long overhangs, FokI/N.BtsCI digested pUC19 was ligated to a duplex oligonucleotide that contains the complementary overhang using T4 DNA ligase. The ligated product was then detected by PCR where one of the primers anneals to the oligonucleotide adapter and the other anneals to the pUC19 sequence. The specific PCR product (∼200 bp) could only be observed in the substrate digested by FokI/Nt.BtsCI or FokI/Nb.BtsCI, indicating the digestion is specific and the expected overhangs were generated (Figure 7B).
DNA labeling by end filling of the 11-nucleotide 3′ recessive end
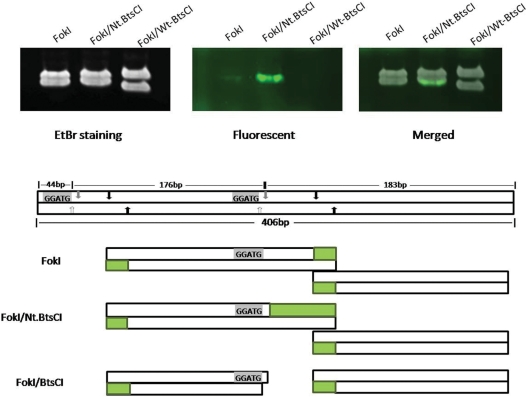
The 11-base 3′ recessive end generated by FokI/Nt.BtsCI can be used for labeling by the extension activity of DNA polymerases. A 406 bp DNA fragment containing two BtsCI sites was amplified by PCR and cleaved with FokI, FokI/Nt.BtsCI or FokI/Wt-BtsCI. The cleavage products were then extended using Klenow fragment in the presence of Cy3-dNTPs, followed by analysis by 6% PAGE. The lower panel of Figure 8 illustrates the expected cleavage products and the 3′ recessive ends that can be labeled by end filling. The FokI/Nt.BtsCI cleavage product has a strong fluorescent signal at the bottom band of the doublet (∼176 bp) after the extension reaction, indicating the overhang was labeled effectively. A weaker signal was detected in the FokI digested product as FokI generated a shorter 4-base overhang. On the other hand, the 2-base 5′overhang generated by BtsCI was not labeled.
Figure 8.
DNA labeling by end filling of the 11-nt 3′ recessive end. The 11-nt 3′ recessive end was generated by FokI/Nt.BtsCI. Strong fluorescent signal detected at the smaller DNA fragment (lower band) digested by FokI/Nt.BtsCI. The expected cleavage products and the 3′ recessive ends that can be labeled by end filling are illustrated at the lower panel. Green blocks, fluorescent labeled region; grey arrows, BtsCI top-strand cleavage site; white arrows, BtsCI bottom-strand cleavage site; black arrows, FokI cleavage sites.
DISCUSSION
Type IIS REases with two catalytic sites
Type IIS REases are an interesting group of REases in terms of the diversity of target sequence recognition mode and catalytic mechanism. Unlike the most common Type IIP REases that have one catalytic site per molecule that contains the DNA recognition residues, Type IIS REases invariably have separate DNA recognition and cleavage domains (12,21–23). This separation of function gives rise to a variety of DNA cleavage mechanisms. In this work we focus on BtsCI, which belongs to the Mva1269I/BsmI group of REases that carry two different catalytic sites on the same polypeptide. The two catalytic sites are similar to the canonical PD-Xn-D/EXK (X is any amino acid residue) but diverged. In BtsCI, the bottom-strand nicking catalytic site (Cb) conforms to the SD-X6-E-X14-QR motif of Mva1269I/BsmI-like REases, located near the N-terminus (Figure 1B). The top-strand catalytic site Ct, interestingly, is the canonical PD-Xn-EXK for the Mva1269I/BsmI-like enzymes, but becomes a BamHI-like PD-Xn-EXE in BtsCI (Figure 1B).
BtsI and BsrDI also contain these two catalytic sites but they are located in two separate subunits. Dimerization is required for BtsI and BsrDI to make dsDNA breaks. We have previously shown that mutations of residues of each of the catalytic sites resulted in strand-specific nicking variants for BtsI and BsrDI (12). Similar results have also been reported for Mva1269I (13). The cleavage/nicking of each strand by Mva1269I is sequential (i.e. bottom strand is nicked first, the bottom-strand nicked DNA is a suitable substrate for top-strand nicking. Top-strand nicking enzyme has very low activity).
In addition to Mva1269I/BsmI/BsrI/BtsCI family, a few Type IIS REases also contains separate catalytic sites each responsible for cleavage of a specific DNA strand. The BstNBI and BspD6I (GAGTC 4/6) systems reported by two independent groups are identical in amino acid sequences and were originally known as natural nicking enzymes (24,25). When the gene product of the neighboring ORF of BspD6I was purified and added to the BspD6I nicking enzyme, the enzyme cleaves on both strands (26). Similar to BtsI and BsrDI, the small subunit of BspD6I (the gene product of the neighboring ORF) has no cleavage activity on its own. More interestingly, the small subunit is highly homologous to the C-terminus of the BspD6I nicking enzyme. Mutagenesis work remains to be carried out to verify if mutation of the catalytic site of the small subunit results in nicking mutants that exclusively nicks the opposite strand.
BtsCI endonuclease shares very little amino acid sequence similarity to neoschizomers FokI and StsI. However, it shows 33% amino acid sequence identity to two conserved hypothetical proteins Cup_ORF1489 (463 aa, GI:57015558) and Nfl_ORF1032 (472 aa, GI:241320910), found in Campylobacter upsaliensis RM3195 and Neisseria flavescens SK114, respectively. Both proteins are accompanied by two DNA methylases (27,28). M1.Cup_ORF1489 and M2. Nfl_ORF1032 shows very high amino acid sequence homology to the N-terminal domain of M.BtsCI and the amino acid sequence of the M.BtsCI C-terminal domain is homologous M2.Cup_ORF1489 and M1. Nfl_ORF1032 (data not shown). Cup_ORF1489 and Nfl_ORF1032 are also predicted to carry two catalytic sites. The non-canonical motif, S(K)D(E)-X6-E-X14-QR, is more similar to BtsCI, while the canonical motif, PD-Xn-EXK, is more Mva1269I/BsmI-like (Figure 1B). It is very likely that the gene products of Cup_ORF1489 and Nfl_ORF1032 are BtsCI isoschizomers that recognize and cleave GGATG sequence.
Application of long overhangs
FokI and BtsCI are neoschizomers that cleave at a different distance downstream of GGATG. We took advantage of this property and generated 11-nt 3′ recessive ends or 9-nt 5′ recessive ends by sequential digestion with FokI and Nt.BtsCI or Nb.BtsCI, respectively. We showed that both ends could be ligated to complementary linkers (Figure 7B). The 11-nt 3′ recessive ends could be labeled by Klenow fragment using fluorescent-labeled dNTPs (Figure 8). The long overhangs can be used in a wide range of applications. Biotinylated oligonucleotides complementary to the overhangs can be used to pull out specific DNA fragments using streptavidin beads for detection and precipitation of protein–DNA complexes in high-throughput applications such as SAGE. The 11-base 3′ recessive ends contains more nt for high level labeling by fluorescent-labeled nucleotides than 4-base 3′ recessive ends generated by using a single REase (Figure 8). The relatively frequent 5-base recognition sequence of FokI/BtsCI can also allow rapid detection of DNA polymorphisms through direct and high level labeling through end filling.
To our knowledge, 9 nt is the longest overhang that a natural Type II REase can generate (TspRI, NNCASTGNN^, NEB catalog 2009/10, page 79). Terminases from some bacterial phages also produce long overhangs. However, phage terminases generally have low specific activity and requires ATP and concatemeric DNA for cleavage (29). The method described here, on the other hand, is highly efficient and specific to FokI/BtsCI sites.
Recently, Sanders and colleagues (11) reported the generation of strand-specific nicking activity for FokI by specifically inactivating the DNA-binding and/or the DNA cleavage activity. In their scheme, cleavage by a mixture of the wt FokI and a DNA binding-deficient and cleavage-deficient mutants give rise to bottom-strand cleavage activity whereas cleavage by using a combination of a DNA binding-deficient FokI variant and a cleavage-deficient FokI variant gives rise to top-strand nicking activity. Although only the top strand was exclusively cleaved in the latter case, the top strand is also cleaved to a significant extent in the former case because the presence of wt FokI results in homodimerization that cleaves both strands. In principle, cleavage by the bottom-nicking FokI heterodimer followed by Nt.BtsCI will generate DNA fragments each containing a 11-base 3′ recessive end capable of ligase-free DNA fragment assembly.
CONCLUSIONS
BtsCI and FokI display very little amino acid sequence homology despite their identical DNA recognition sequence. BtsCI joins BsrI, BsmI, Mva1269I, BbvCI, and heterodimeric BtsI, BsrDI and BstNBI/BspD6I as a group of Type IIS REases which contain two catalytic sites, each of which acts on a specific strand of the dsDNA substrate. BtsCI variants that nick a specific strand of the target site have been generated. Using FokI/N.BtsCI double digestion, one can generate 11-base 3′ recessive end or 9-base 5′ recessive end for hybridization-based DNA detection/capture and DNA end labeling.
FUNDING
Funding for open access charge: New England Biolabs.
Conflict of interest statement. None declared.
ACCESSION NUMBER
The DNA sequence of BtsCI restriction-modification system has been deposited in GenBank with accession No. GQ449683.
ACKNOWLEDGEMENTS
The authors thank Rich Roberts, Geoff Wilson and Bill Jack for critical comments and discussion; X. Pan for providing the native BtsCI-producing strain Bacillus thermosphaericus; Don Comb and Jim Ellard for financial support.
REFERENCES
- 1.Pingoud A, Jeltsch A. Structure and function of type II restriction endonucleases. Nucleic Acids Res. 2001;29:3705–3727. doi: 10.1093/nar/29.18.3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pingoud A, Fuxreiter M, Pingoud V, Wende W. Type II restriction endonucleases: structure and mechanism. Cell Mol. Life Sci. 2005;62:685–707. doi: 10.1007/s00018-004-4513-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newman M, Strzelecka T, Dorner LF, Schildkraut I, Aggarwal AK. Structure of Bam HI endonuclease bound to DNA: partial folding and unfolding on DNA binding. Science. 1995;269:656–663. doi: 10.1126/science.7624794. [DOI] [PubMed] [Google Scholar]
- 4.Newman M, Strzelecka T, Dorner LF, Schildkraut I, Aggarwal AK. Structure of restriction endonuclease BamHI and its relationship to EcoRI. Nature. 1994;368:660–664. doi: 10.1038/368660a0. [DOI] [PubMed] [Google Scholar]
- 5.Townson SA, Samuelson JC, Xu SY, Aggarwal AK. Implications for switching restriction enzyme specificities from the structure of BstYI bound to a BglII DNA sequence. Structure. 2005;13:791–801. doi: 10.1016/j.str.2005.02.018. [DOI] [PubMed] [Google Scholar]
- 6.Yonezawa A, Sugiura Y. DNA binding mode of class-IIS restriction endonuclease FokI revealed by DNA footprinting analysis. Biochim. Biophys. Acta. 1994;1219:369–379. doi: 10.1016/0167-4781(94)90061-2. [DOI] [PubMed] [Google Scholar]
- 7.Wah DA, Bitinaite J, Schildkraut I, Aggarwal AK. Structure of FokI has implications for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10564–10569. doi: 10.1073/pnas.95.18.10564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grazulis S, Manakova E, Roessle M, Bochtler M, Tamulaitiene G, Huber R, Siksnys V. Structure of the metal-independent restriction enzyme BfiI reveals fusion of a specific DNA-binding domain with a nonspecific nuclease. Proc. Natl Acad. Sci. USA. 2005;102:15797–15802. doi: 10.1073/pnas.0507949102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu Y, Lunnen KD, Kong H. Engineering a nicking endonuclease N.AlwI by domain swapping. Proc. Natl Acad. Sci. USA. 2001;98:12990–12995. doi: 10.1073/pnas.241215698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc. Natl Acad. Sci. USA. 1998;95:10570–10575. doi: 10.1073/pnas.95.18.10570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sanders KL, Catto LE, Bellamy SR, Halford SE. Targeting individual subunits of the FokI restriction endonuclease to specific DNA strands. Nucleic Acids Res. 2009;37:2105–2115. doi: 10.1093/nar/gkp046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu SY, Zhu Z, Zhang P, Chan SH, Samuelson JC, Xiao J, Ingalls D, Wilson GG. Discovery of natural nicking endonucleases Nb.BsrDI and Nb.BtsI and engineering of top-strand nicking variants from BsrDI and BtsI. Nucleic Acids Res. 2007;35:4608–4618. doi: 10.1093/nar/gkm481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Armalyte E, Bujnicki JM, Giedriene J, Gasiunas G, Kosinski J, Lubys A. Mva1269I: a monomeric type IIS restriction endonuclease from Micrococcus varians with two EcoRI- and FokI-like catalytic domains. J. Biol. Chem. 2005;280:41584–41594. doi: 10.1074/jbc.M506775200. [DOI] [PubMed] [Google Scholar]
- 14.Kuhn H, Frank-Kamenetskii MD. Labeling of unique sequences in double-stranded DNA at sites of vicinal nicks generated by nicking endonucleases. Nucleic Acids Res. 2008;36:e40. doi: 10.1093/nar/gkn107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kiesling T, Cox K, Davidson EA, Dretchen K, Grater G, Hibbard S, Lasken RS, Leshin J, Skowronski E, Danielsen M. Sequence specific detection of DNA using nicking endonuclease signal amplification (NESA) Nucleic Acids Res. 2007;35:e117. doi: 10.1093/nar/gkm654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao M, Phong A, Ha C, Chan TF, Cai D, Leung L, Wan E, Kistler AL, DeRisi JL, Selvin PR, et al. Rapid DNA mapping by fluorescent single molecule detection. Nucleic Acids Res. 2007;35:e16. doi: 10.1093/nar/gkl1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiu J, March PE, Lee R, Tillett D. Site-directed, Ligase-independent mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 2004;32:e174. doi: 10.1093/nar/gnh172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hummel JP, Dreyer WJ. Measurement of protein-binding phenomena by gel filtration. Biochim. Biophys. Acta. 1962;63:530–532. doi: 10.1016/0006-3002(62)90124-5. [DOI] [PubMed] [Google Scholar]
- 19.Vincze T, Posfai J, Roberts RJ. NEBcutter: a program to cleave DNA with restriction enzymes. Nucleic Acids Res. 2003;31:3688–3691. doi: 10.1093/nar/gkg526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Szomolanyi E, Kiss A, Venetianer P. Cloning the modification methylase gene of Bacillus sphaericus R in Escherichia coli. Gene. 1980;10:219–225. doi: 10.1016/0378-1119(80)90051-7. [DOI] [PubMed] [Google Scholar]
- 21.Zhu Z, Samuelson JC, Zhou J, Dore A, Xu SY. Engineering strand-specific DNA nicking enzymes from the type IIS restriction endonucleases BsaI, BsmBI, and BsmAI. J. Mol. Biol. 2004;337:573–583. doi: 10.1016/j.jmb.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 22.Samuelson JC, Zhu Z, Xu SY. The isolation of strand-specific nicking endonucleases from a randomized SapI expression library. Nucleic Acids Res. 2004;32:3661–3671. doi: 10.1093/nar/gkh674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heiter DF, Lunnen KD, Wilson GG. Site-specific DNA-nicking mutants of the heterodimeric restriction endonuclease R.BbvCI. J. Mol. Biol. 2005;348:631–640. doi: 10.1016/j.jmb.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 24.Higgins LS, Besnier C, Kong H. The nicking endonuclease N.BstNBI is closely related to type IIs restriction endonucleases MlyI and PleI. Nucleic Acids Res. 2001;29:2492–2501. doi: 10.1093/nar/29.12.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kachalova GS, Rogulin EA, Yunusova AK, Artyukh RI, Perevyazova TA, Matvienko NI, Zheleznaya LA, Bartunik HD. Structural analysis of the heterodimeric type IIS restriction endonuclease R.BspD6I acting as a complex between a monomeric site-specific nickase and a catalytic subunit. J. Mol. Biol. 2008;384:489–502. doi: 10.1016/j.jmb.2008.09.033. [DOI] [PubMed] [Google Scholar]
- 26.Yunusova AK, Rogulin EA, Artyukh RI, Zheleznaya LA, Matvienko NI. Nickase and a protein encoded by an open reading frame downstream from the nickase BspD6I gene form a restriction endonuclease complex. Biochemistry (Mosc) 2006;71:815–820. doi: 10.1134/s0006297906070157. [DOI] [PubMed] [Google Scholar]
- 27.Roberts RJ, Vincze T, Posfai J, Macelis D. REBASE—enzymes and genes for DNA restriction and modification. Nucleic Acids Res. 2007;35:D269–D270. doi: 10.1093/nar/gkl891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Rapp BA, Wheeler DL. GenBank. Nucleic Acids Res. 2000;28:15–18. doi: 10.1093/nar/28.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Catalano CE. The terminase enzyme from bacteriophage lambda: a DNA-packaging machine. Cell Mol. Life Sci. 2000;57:128–148. doi: 10.1007/s000180050503. [DOI] [PMC free article] [PubMed] [Google Scholar]