Abstract
T4 polynucleotide kinase/phosphatase (Pnkp) exemplifies a family of bifunctional enzymes with 5′-kinase and 3′-phosphatase activities that function in nucleic acid repair. The N-terminal kinase domain belongs to the P-loop phosphotransferase superfamily. The kinase is distinguished by a tunnel-like active site with separate entrances on opposite sides of the protein for the NTP phosphate donor and a 5′-OH single-stranded oligonucleotide phosphate acceptor. Here, we probed by mutagenesis the roles of individual amino acids that comprise the acceptor binding site. We thereby identified Glu57 as an important residue, by virtue of its participation in a salt bridge network with two catalytic residues identified previously: Arg38, which binds the 3′-phosphate of the terminal 5′-OH nucleotide, and the putative general base Asp35 that contacts the nucleophilic 5′-OH group. The 5′-OH nucleoside fits into a pocket lined by aliphatic amino acids (Val131, Pro132 and Val135) that make van der Waals contacts to the nucleobase. Whereas subtraction of these contacts by single alanine substitutions for Val131 or Val135 and glycine for Pro132 had modest effects on kinase activity, the introduction of bulkier phenylalanines for Val131 and Val135 were deleterious, especially V131F, which severely impeded both substrate binding (increasing Km by 15-fold) and catalysis (decreasing kcat by 300-fold).
INTRODUCTION
T4 polynucleotide kinase/phosphatase (Pnkp) exemplifies a family of repair enzymes that heal broken termini in RNA or DNA by converting 3′-PO4/5′-OH ends into 3′-OH/5′-PO4 ends, which are then sealed by RNA or DNA ligases. During T4 infection, Pnkp thwarts an RNA-based innate immune response in which the bacterium blocks viral protein synthesis by inducing site-specific breakage of host-cell tRNAs, to which the phage responds by repairing the broken tRNAs using Pnkp and a phage-encoded RNA ligase (1). T4 Pnkp catalyzes two reactions in this pathway: (i) the transfer of the γ phosphate from ATP to the 5′-OH terminus of RNA and (ii) the hydrolytic removal of a 3′-PO4 terminus from RNA (2–6).
T4 Pnkp is a homotetramer of a 301-aa polypeptide, which consists of an N-terminal kinase domain of the P-loop phosphotransferase superfamily and a C-terminal phosphatase domain of the DxD acylphosphatase superfamily. The homotetramer is formed via pairs of phosphatase–phosphatase and kinase–kinase homodimer interfaces (7–10). Essential constituents of the separate active sites for the 5′ kinase and 3′ phosphatase activities have been identified by alanine-scanning mutagenesis, guided initially by phylogenetic conservation of primary structure among Pnkp homologs (11,12) and subsequently by crystal structures of T4 Pnkp (10). Of the 20 amino acids within the kinase domain that were targeted for mutagenesis, six were found to be essential: Lys15, Ser16, Asp35, Arg38, Asp85 and Arg126.
The kinase domain consists of a central four-stranded parallel β-sheet flanked by three α helices on each side. The kinase active site is composed of (i) a classical P-loop motif (9GxxGxGKS16) that coordinates the β phosphate of the NTP donor via the main-chain amide nitrogens of the P-loop and the Lys15 side chain; (ii) essential side chain Arg126, which also coordinates the NTP β phosphate; (iii) essential side chain Arg38, which coordinates the 3′ phosphate of the 5′-OH acceptor nucleotide (Figure 1A), plus (iv) essential side chain Asp35, a putative general base that activates the 5′-OH for direct nucleophilic attack on the NTP γ phosphate. The active site is located within a tunnel through the heart of the enzyme. The tunnel entrance on the P-loop side admits the NTP phosphate donor and controls release of the NDP reaction product. The tunnel opening on the opposite face allows ingress of the 5′ end of a single-stranded polynucleotide to the kinase active site (7,8).
Figure 1.
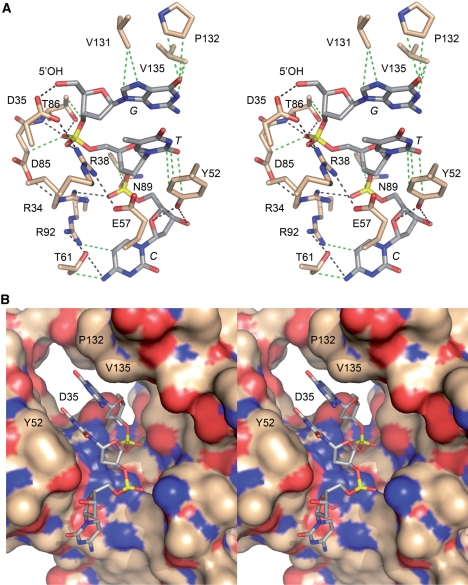
The 5′-OH polynucleotide acceptor site of T4 Pnkp. (A) A stereo view of the kinase active site is shown, highlighting enzymic contacts to a trinucleotide 5′-OH phosphate acceptor (PBD ID code 1RC8). The side chains that were subjected to mutational analysis here and previously are shown in stick representation with carbons colored beige; the residue numbers are specified. The 5′-GTC trinucleotide is depicted as a stick model with carbons colored gray and the nucleobases labeled in italics. Hydrogen-bonding and ionic interactions are depicted as black dashed lines. van der Waals contacts are depicted as green dashed lines. (B) Stereo view of the phosphate acceptor tunnel of the kinase domain with the protein rendered as a surface model and the 5′-GTC trinucleotide depicted as a stick model. The deep end of the tunnel is demarcated by Asp35, which coordinates the 5′-OH. Also labeled are Tyr52, which contacts the second and third nucleosides, and Val131 and Pro132, which contact the first nucleobase. The figure highlights the narrow tunnel aperture, which allows ingress of single-stranded nucleic acid, but would seem to exclude a nucleic acid duplex.
Further insights to the anatomy of the phosphate acceptor site were attained by Eastberg et al. (9), who soaked short 5′-OH DNA oligonucleotides (a 3mer and two different 5mers) into preformed Pnkp-ADP crystals and solved the structures of the substrate complexes. The observed electron density allowed modeling of up to 3 nt from the 5′-OH acceptor end. Figure 1 depicts the interactions of the kinase with HOGpTpC. The image in Figure 1B is a stereo view of the phosphate acceptor tunnel of the kinase domain with the protein rendered as a surface model and the 5′-GTC trinucleotide as a stick model. The deep end of the tunnel is demarcated by Asp35, which coordinates the 5′-OH. Figure 1B highlights the narrow tunnel aperture, which allows ingress of single-stranded nucleic acid, but would seem to exclude a nucleic acid duplex. When bound in the tunnel, the 5′-OH of the terminal HOGp nucleotide is oriented appropriately for attack on ATP modeled in the phosphate donor site. The image in Figure 1A shows that the enzyme-oligonucleotide interface consists of a network of hydrogen bonds and van der Waals contacts between amino acid side chains and the bases, sugars and phosphates of the HOGpTpC trinucleotide.
The structures of the acceptor complexes raised interesting questions about the functional architecture of the acceptor binding site, which, in principle, has to accomplish several tasks, including: accommodation of 5′-OH single-stranded nucleic acids with relatively little sequence bias; insistence on ingress of the 5′ ends of single-stranded nucleic acids while rejecting a 3′ oligonucleotide terminus; and exclusion of water as a futile nucleophile. The factors that aid these functions might include: shape complementarity between the tunnel surface and the 5′-OH oligonucleotide; and division of the tunnel into polar versus hydrophobic patches to orient the phosphates and bases, respectively. The enzyme might also rely on specific atomic contacts between the protein and nucleic acid atoms to bind and orient the phosphoacceptor. The aim of the present study was to examine by mutagenesis the contributions of the amino acids that line the tunnel to the 5′-kinase activity.
As it is well established that a mononucleoside 3′-phosphate (HONp) is the minimal phosphate acceptor for T4 Pnkp (2,4), we assume that the contacts required for phosphoryl transfer chemistry to nucleic acid (and for disfavoring hydrolysis) are those involving the terminal HONp nucleotide (referred to henceforth as the +1 nt). Indeed, prior mutational studies support this view, insofar as Arg38 and Asp35, which coordinate the 5′-OH and phosphate of the +1 nt, are essential for kinase activity (11,12). However, Thr86, which also donates a hydrogen bond to the +1 phosphate and makes van der Waals contact to the +1 ribose O3′ (Figure 1A), can be replaced by alanine without affecting kinase function (11). It is conceivable that the phosphate contact of the Thr86 Oγ is functionally redundant with the hydrogen bond donated to the same +1 phosphate oxygen from the Thr86 main chain amide (Figure 1A). Asp85, which is essential for kinase activity (11), contributes to the acceptor binding site via a van der Waals contact from Cβ to the +1 phosphate; it also interacts with Arg34, which donates a hydrogen bond to the +2 phosphate (HONpNpN) (Figure 1A). We suspect that neither of these contacts accounts for the complete loss of activity when Asp85 is replaced with alanine, insofar as Arg34 is dispensable for 5′-kinase activity with an oligonucleotide acceptor (11) and the substituted Ala85 Cβ should still contact the +1 nt. Rather, it is likely that the Asp85 is essential for the local structure of the kinase active site, via hydrogen bonding of its carboxylate oxygens with the Arg34 main chain amide nitrogen and the Gln64 side chain Nε (7,9).
Here, we interrogated by mutagenesis the functions of the other constituents of the phosphate acceptor site (Figure 1A). Our findings highlight the importance of Glu57 in positioning the catalytic Arg38 residue and the steric constraints on the hydrophobic pocket that accommodates the 5′ nucleoside base.
MATERIALS AND METHODS
Pnkp mutants
Missense mutations were introduced into the Pnkp ORF by using the two-stage polymerase chain reaction (PCR)-based overlap extension method as described previously (11,12). The PCR products were digested with NdeI and BamHI and then inserted into pET16b. The inserts were sequenced completely to confirm the desired mutations and exclude the acquisition of unwanted changes. The pET-Pnkp plasmids were introduced into Escherichia coli BL21(DE3). Recombinant protein production was induced by adjusting exponentially growing cultures (100 ml) to 0.3 mM IPTG and incubating them at 17°C for 15 h with continuous shaking. The wild-type and mutant His10-Pnkp proteins were purified from soluble bacterial lysates by Ni-agarose chromatography as described previously (11,12). Protein concentrations were determined by using the BioRad dye reagent with bovine serum albumin as the standard.
3′ Phosphatase assay
Reaction mixtures (25 µl) containing 100 mM imidazole (pH 6.0), 10 mM MgCl2, 5 mM DTT, 0.1 mg/ml bovine serum albumin (BSA), 1.6 mM 3′ dTMP (Sigma), and 0, 4.7, 9.4, 18.8, 37.5 or 75 ng of wild-type or mutant Pnkp as specified were incubated for 20 min at 37°C. The reactions were quenched by adding 75 µl of cold water and 1 ml of malachite green reagent (BIOMOL Research Laboratories). Phosphate release was determined by measuring A620 and interpolating the value to a phosphate standard curve. The phosphatase specific activity of each protein was determined from the slope of the titration curve. The results are shown in Figure 2B, where each datum is the average of three independent titration experiments ±SEM.
Figure 2.
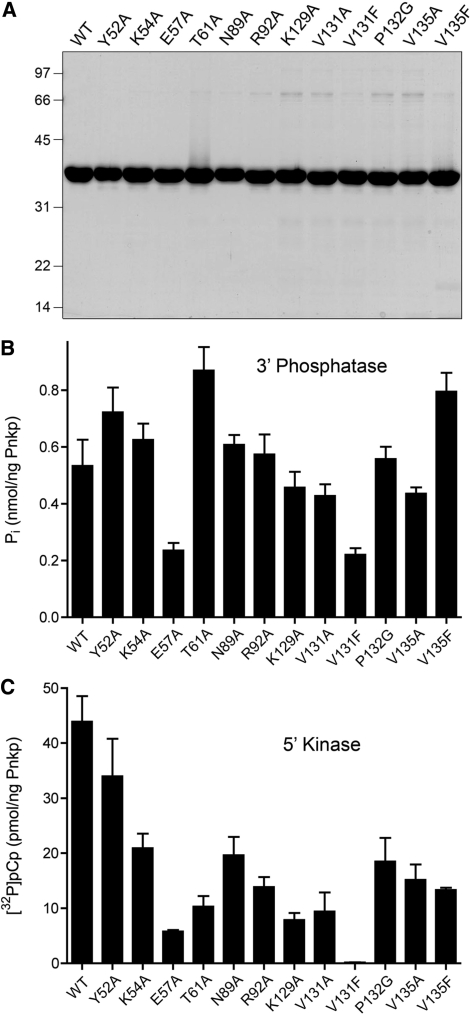
Pnkp mutants and their activities. (A) Aliquots (5 µg) of the nickel-agarose preparations of full-length wild-type (WT) Pnkp and the indicated mutants were analyzed by SDS-PAGE. Polypeptides were visualized by staining with Coomassie blue dye. The positions and sizes (in kDa) of marker proteins are indicated. 3′-phosphatase specific activities (B) and 5′ kinase-specific activities (C) were determined by enzyme titration as described under ‘Materials and methods’ section. Each datum in the bar graphs is the average of three titration experiments ±SEM.
5′ Kinase assays
3′-CMP substrate
Reaction mixtures (10 µl) containing 70 mM Tris–HCl (pH 7.6), 10 mM MgCl2, 5 mM DTT, 25 µM [γ32P]ATP, 1 mM 3′ CMP (Sigma) and increasing amounts of wild-type or mutant Pnkp were incubated for 20 min at 37°C. The reactions were quenched by adding 5 µl of 5 M formic acid. Aliquots of the mixtures were applied to a polyethyleneimine–cellulose TLC plate, which was developed with 1 M formic acid, 0.5 M LiCl. The [γ32P]ATP substrate and [α32P]pCp product were visualized and quantified by scanning the gel with a Fujix BAS2500 phosphorimager. The kinase specific activity of each protein was determined from the slope of the titration curve. The results are shown in Figure 2C, where each datum is the average of three independent titration experiments ±SEM.
Polynucleotide substrate
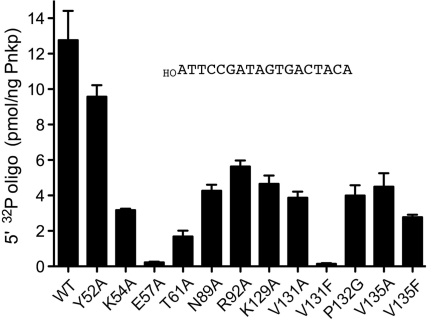
Reaction mixtures (10 µl) containing 70 mM Tris–HCl (pH 7.6), 10 mM MgCl2, 5 mM DTT, 25 µM [γ32P]ATP, 5 µM (50 pmol) of 5′-OH terminated 18-mer oligodeoxynucleotide d(ATTCCGATAGTGACTACA), and increasing amounts of wild-type or mutant Pnkp were incubated for 20 min at 37°C. The reactions were quenched by adding 6 µl of 95% formamide/20 mM EDTA. The products were analyzed by electrophoresis through a 15-cm 15% polyacrylamide gel containing 7 M urea in TBE (90 mM Tris–borate, 2.5 mM EDTA). The radiolabeled oligonucleotide products were visualized and quantified by scanning the gel with a Fujix BAS2500 phosphorimager. The kinase specific activity of each protein was determined from the slope of the titration curve. The results are shown in Figure 3, where each datum is the average of three independent titration experiments ±SEM.
Figure 3.
Mutational effects on 5′-kinase activity with an oligonucleotide phosphate acceptor. 5′ kinase specific activities with an 18-mer 5′-OH DNA oligonucleotide substrate (data as shown) were determined by enzyme titration as described under ‘Materials and methods’ section. Each datum in the bar graph is the average of three titration experiments ±SEM.
RESULTS AND DISCUSSION
Mutations in the polynucleotide kinase phosphate acceptor site
A goal of the present study was to extend the functional map of the kinase component of T4 Pnkp by mutating the amino acids that form the binding site for the 5′-OH terminated oligonucleotide phosphate acceptor. Alanines were introduced in lieu of the following residues highlighted in Figure 1: Tyr52, which makes stacking van der Waals interactions with the +2 base and hydrogen bonds to the +3 sugar; Glu57, which forms a salt bridge to the critical Arg38 side chain; Thr61, which makes a hydrogen bond and van der Waals contact to the +3 base; Asn89, which makes van der Waals contact to the +2 phosphate; Arg92, which makes a van der Waals contact to the +2 base and also a hydrogen bond to Thr61; and the Val131 and Val135 residues that make van der Waals contacts with the +1 base. Pro162, which contacts the +1 base, was changed to glycine. In order to gauge the steric constraints on the hydrophobic binding pocket for the +1 base, we also replaced Val131 and Val135 by bulkier phenylalanine side chains. In addition, we introduced alanines in lieu of two residues not depicted in Figure 1: Lys54, which is located near the entrance to the oligonucleotide acceptor site in the apoenzyme structure but is disordered in the oligonucleotide-bound enzyme; and Lys129, which donates a hydrogen bond from Nζ to the ribose O3′ of the ATP phosphate donor.
Wild-type Pnkp and the 12 mutants were produced in bacteria as His10-tagged fusions and purified from soluble bacterial extracts by Ni-agarose chromatography (Figure 2A). Pnkp’s 3′-phosphatase activity was measured by the release of inorganic phosphate from deoxythymidine 3′ monophosphate (5,6). Wild-type Pnkp released 0.53 nmol of Pi from 3′dTMP per ng of protein in the linear range of enzyme-dependence, which translates into a turnover number of ∼16 s−1. The specific activities of the Pnkp mutants were determined in parallel (Figure 2B). The mutants had activity similar to wild-type Pnkp (i.e. ±60%), as follows: Y52A (135%), K54A (117%), E57A (44%), T61A (163%), N89A (114%), R92A (108%), K129A (86%), V131A (80%), V131F (41%), P132G (105%), V135A (82%) and V135F (150%). Such prep-to-prep variations in phosphatase-specific activities among wild-type and ‘active’ Pnkp mutants were noted previously (10). Here, as is our previous studies (10,12), our operational criterion for a significant mutational effect is one that elicits at least a 4-fold reduction in specific activity compared to wild-type Pnkp. We construe the retention of 3′-phosphatase activity above this threshold to signify that none of the amino acid changes in the kinase domain grossly affected the global folding of the enzyme.
The wild-type and mutant Pnkp proteins were surveyed for 5′ kinase activity, which was measured as the transfer of 32P from 25 µM [γ-32P]ATP to 1 mM 3′-CMP (HOCp) to form [5′-32P]pCp. Wild-type Pnkp phosphorylated 44 pmol of HOCp per nanogram of protein in the linear range of enzyme-dependence, which translates into a turnover number of ∼81 min−1. The kinase-specific activities of the 12 Pnkp mutants were determined in parallel (Figure 2C). Most of the mutations elicited mild to moderate activity decrements, among which E57A, T61A, K129A and V131A met the 4-fold criterion of significance. By contrast, the V131F change was uniquely catastrophic, reducing kinase specific activity to 0.3% of the wild-type value.
Mutational effects on 3′-mononucleotide binding and catalysis
Mutational effects on the steady-state kinetic parameters of the kinase reaction were gauged by performing HOCp titration experiments with each enzyme preparation. (Exemplary titrations are shown in Supplementary Figure S1.) The results are compiled in Table 1. Wild-type Pnkp had a Km of 31 µM HOCp and a kcat of 98 min−1. Our observed Km value was in the same range as that reported by Novogrodsky et al. (4) for the monodeoxynucleotide HO(dC)p (Km 15 µM). Instructive findings emerged concerning the effects of mutations in the three hydrophobic residues (Val131, Pro132 and Val135) that pack against the same surface of the +1 purine base in the crystal structure depicted in Figure 1. Changing Pro132 to Gly had little effect on kcat (56 min−1), although it increased Km by 2-fold. The V135A change had little effect on Km but reduced kcat by 3-fold (to 33 s−1), whereas the V131A mutation increased Km and reduced kcat by ∼3-fold each, causing a net 11-fold decrement in catalytic efficiently (kcat/Km). This hierarchy of side-chain removal effects correlates with the distances from the respective side chain atoms to the +1 pyrimidine base observed in the crystal structure of Pnkp with a different trinucleotide (HOTpGpC) occupying the acceptor site (pdb ID 1RPZ) (9). In that structure, Val131 Cγ makes close van der Waals contacts to the pyrimidine C5 (3.3 Å) and C6 (3.4 Å) atoms, while Val135 Cγ is located farther away (3.9 Å) from the N3 and O4 (N4 in cytidine) atoms. More drastic effects on kinase activity were elicited by replacing the valines with phenylalanine. The V135F change resulted in 10-fold lower affinity for HOCp compared to wild-type Pnkp while maintaining a reduced kcat, so that the catalytic efficiency of V135F was 44-fold lower than wild type. The V131F mutant displayed a Km of 460 µM and a kcat of 0.33 s−1, corresponding to a 4400-fold decrement in catalytic efficiency (Table 1). We surmise that the Phe substitutions at Val135, and especially Val131, impede positioning the terminal 5′-OH nucleophile in the active site because of steric clash with the +1 base.
Table 1.
Mutational effects on 5′ kinase activity with HOCp acceptor
| Pnkp | Km HOCp (µM) | kcat (min−1) |
|---|---|---|
| WT | 31 ± 4 | 98 ± 16 |
| Y52A | 16 ± 3 | 69 ± 2 |
| K54A | 12 ± 1 | 80 ± 5 |
| E57A | 157 ± 22 | 3.3 ± 0.3 |
| T61A | 36 ± 4 | 21 ± 1.5 |
| N89A | 24 ± 6 | 50 ± 9 |
| R92A | 105 ± 7 | 74 ± 6 |
| K129A | 43 ± 9 | 33 ± 2 |
| V131A | 113 ± 13 | 32 ± 2 |
| V131F | 460 ± 46 | 0.33 ± 0.03 |
| P132G | 67 ± 7 | 56 ± 2 |
| V135A | 39 ± 6 | 33 ± 2 |
| V135F | 323 ± 28 | 23 ± 1.3 |
Reaction mixtures (10 µl) containing 70 mM Tris–HCl (pH 7.6), 10 mM MgCl2, 5 mM DTT, 25 µM [γ32P]ATP, wild-type or mutant Pnkp and varying concentrations of 3′-CMP were incubated for 20 min at 37°C. The extents of [α32P]pCp product formation were plotted as a function of 3′-CMP concentration. Km and kcat were obtained by nonlinear regression curve fitting of the data for each experiment to the Michaelis–Menten equation in Prism. The values shown are averages of three independent 3′-CMP titration experiments ±SEM. Representative 3′-CMP titration profiles are shown in Supplementary Figure S1.
The E57A change increased Km to 157 µM HOCp and lowered kcat to 3.3 s−1 (Table 1), resulting a 150-fold decrement in catalytic efficiency compared to wild-type Pnkp. Glu57 makes no direct contact with the kinase substrate; rather it makes a salt bridge to the catalytically essential Arg38 side chain, which coordinates the 3′-phosphate moiety of the 5′-OH nucleotide (Figure 1). We surmise that Glu57 is important to position Arg38 appropriately.
The Y52A and K54A mutations had little effect on kcat (69 s−1 and 80 s−1, respectively), but they had the distinctive effect of lowering Km for HOCp (to 16 and 12 µM, respectively) compared to wild-type Pnkp (Table 1). Neither Tyr52 nor Lys54 makes direct contact with the terminal 5′-OH nucleotide (Figure 1); rather, both are located toward the periphery of the oligonucleotide entry channel. Our results, showing higher affinity for the HOCp acceptor when either Tyr52 or Lys54 are absent, suggest that these side chains normally provide an obstacle to the ingress of a 3′-mononucleotide to the kinase active site.
Arg92 contacts the +3 nucleoside (Figure 1) and, at first glance, might not be expected to contribute to mononucleotide phosphorylation. Yet, the R92A change caused a 3-fold increase in Km, with little impact on kcat (Table 1). We suspect this result reflects the role of Arg92 in stabilizing the structure of the polynucleotide acceptor tunnel, by tethering two of the component α-helices via hydrogen bond donation from Arg92 to Thr61 Oγ (Figure 1) and Asn64 Oε. Mutating Thr61 to alanine had little impact on Km, but reduced kcat by a factor of five (Table 1). In addition to the hydrogen bond depicted in Figure 1, Thr61 makes van der Waals contacts via Cγ to Phe 65 and Arg92. Mutating Asn89 had little effect on the kinetic parameters for HOCp phosphorylation, in keeping with its lack of direct contact with the terminal nucleoside in the crystal structure (Figure 1). Finally, the effects of mutating Lys129 were consistent with its role as a constituent of the ATP-binding site of Pnkp; the K129A change had little effect on affinity for the HOCp acceptor, but reduced kcat by a factor of three (Table 1).
Mutational effects on kinase activity with an oligonucleotide substrate
The wild-type and mutant Pnkp proteins were tested for 5′ kinase activity with a 5′-OH terminated 18-mer DNA oligonucleotide phosphate acceptor. Transfer of 32P from 25 µM [γ-32P]ATP to 5 µM 5′-OH DNA was quantified as a function of input enzyme. Wild-type Pnkp phosphorylated ∼12 pmol of 18-mer per nanogram of protein in the linear range of enzyme dependence (Figure 3), which translates into a turnover number of ∼24 min−1. The kinase specific activities of the 12 Pnkp mutants were determined in parallel. The mutational effects on polynucleotide phosphorylation (Figure 3) were generally similar to the results obtained with HOCp (Table 1). The V131F and E57A changes were the most deleterious, reducing DNA kinase specific activity to 1% and 2% of the wild-type value, respectively (Figure 3).
Structure–activity relations at Glu57
To elucidate structure activity relations at Glu57, which we deemed important for polynucleotide kinase activity on the basis of the present alanine scan, we introduced conservative substitutions with glutamine and aspartate, purified the recombinant E57Q and E57D Pnkp proteins (in parallel with new preparations of wild-type Pnkp and the E57A mutant), and surveyed them for activity by enzyme titration. The E57Q and E57D proteins were both defective in 5′-kinase activity. Their respective specific activities in HOCp phosphorylation (at 1 mM 3′CMP substrate concentration) were 0.5% and 5.3% of wild-type, while E57A was 12% as active as wild type (normalized averages of three titration experiments; data not shown). The polynucleotide kinase specific activities of E57Q, E57D and E57A with the 18-mer DNA oligonucleotide phosphate acceptor were 0.1%, 4.6% and 2% of wild-type Pnkp, respectively (normalized averages of three titration experiments; data not shown). These results signify that an isosteric amide is unable to function in lieu of Glu57; indeed, the E57Q mutation was more deleterious to kinase activity than E57A or E57D.
CONCLUDING REMARKS
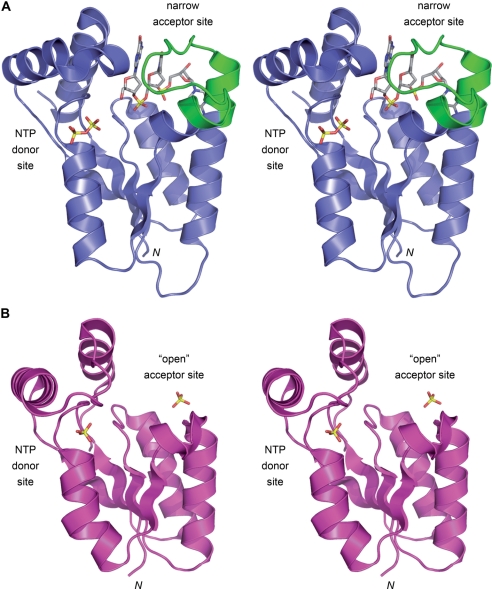
The relatively narrow tunnel of T4 Pnkp that comprises the 5′-OH polynucleotide phosphate acceptor site (Figures 1B and 4A) accounts for why the phage enzyme prefers to phosphorylate single-stranded nucleic acids or duplexes with 5′ single-stranded tails (13,14). Indeed, the in vivo substrates for Pnkp are broken tRNA stem–loops with a short 5′-OH single-strand segment (1,15). By contrast, the mammalian Pnkp enzyme, which is a DNA-specific kinase, has an ‘open’ nucleic acid binding site that can accommodate a duplex nucleic acid (16,17) (Figure 4B). A key difference when the T4 and mammalian kinase structures are superimposed is that mammalian Pnkp lacks the 22-aa segment of T4 Pnkp (colored green in Figure 4A), comprising the ends of two serial α-helices and the connecting loop (38RQSIMAHEERDEYKYTKKKEGI59), that forms one of the lateral walls and the roof of the tunnel. The open fold of the mammalian Pnkp allows it to phosphorylate its preferred substrates: 5′-OH DNA termini at duplex nicks, gaps and recessed 5′ ends within a 3′-tailed duplex (13,16). Unlike T4 Pnkp, which readily phosphorylates mononucleotides, mammalian Pnkp requires a polynucleotide phosphate acceptor with a minimum chain length of ∼8 deoxynucleotides (13). The inability of mammalian Pnkp to phosphorylate mononucleoside 3′-phosphates can be explained by the fact that it is missing a counterpart of T4 Pnkp Arg38 that coordinates the 3′-phosphate of the 5′-OH terminal nucleotide (18).
Figure 4.
Distinctive phosphate acceptor sites in T4 and mammalian polynucleotide kinases. The homologous tertiary structures of the kinase domains of T4 Pnkp (PDB 1RC8) and mammalian Pnkp (PDB IYJ5) were superimposed and then offset vertically. The T4 kinase fold is depicted in stereo in (A), with the elements homologous to mammalian Pnkp colored blue. The trinucleotide bound in the phosphate acceptor tunnel is shown as a sick model. The distinctive 22-aa segment of T4 Pnkp (38RQSIMAHEERDEYKYTKKKEGI59) that forms one of the lateral walls and the roof of the phosphate acceptor tunnel is colored green. The ADP phosphates (depicted as a stick model) demarcate the NTP phosphate donor site. The mammalian kinase fold is shown in stereo in (B), with a sulfate anion bound in the donor site (at the position corresponding to the ADP β phosphate) and a second sulfate sitting on the surface of the putative ‘open’ docking site for a duplex DNA phosphate acceptor.
Here, we relied on the structures of T4 Pnkp bound to short oligonucleotides (9) to guide a mutational analysis of the phosphate acceptor tunnel and thereby gained new insights to which constituents are important and why. In particular, we identified Glu57 as an important residue, by virtue of its participation in a salt-bridge network with two catalytic residues identified previously: the phosphate-binding Arg38 and the putative general base Asp35 (Figure 1). Mammalian Pnkp lacks a counterpart of Glu57.
The 5′-OH nucleoside fits into a pocket lined by several aliphatic amino acids that make van der Waals contacts with the nucleobase (Figure 1). Whereas subtraction of these contacts by single alanine substitutions for Val131 or Val135 and glycine for Pro132 had only modest effects on kinase activity, the introduction of bulkier phenylalanines for Val131 and Val135 were deleterious, especially V131F, which severely impeded both substrate binding and catalysis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (Grant GM42498). Funding for open access charge: National Institutes of Health grant GM42498.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Amitsur M, Levitz R, Kaufman G. Bacteriophage T4 anticodon nuclease, polynucleotide kinase, and RNA ligase reprocess the host lysine tRNA. EMBO J. 1987;6:2499–2503. doi: 10.1002/j.1460-2075.1987.tb02532.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Richardson CC. Phosphorylation of nucleic acid by an enzyme from T4 bacteriophage-infected Escherichia coli. Proc. Natl Acad. Sci. USA. 1965;54:158–165. doi: 10.1073/pnas.54.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Novogrodsky A, Hurwitz J. The enzymatic phosphorylation of ribonucleic acid and deoxyribonucleic acid: phosphorylation at 5′-hydroxyl termini. J. Biol. Chem. 1966;241:2923–2932. [PubMed] [Google Scholar]
- 4.Novogrodsky A, Tal M, Traub A, Hurwitz J. The enzymatic phosphorylation of ribonucleic acid and deoxyribonucleic acid: further properties of the 5′-hydroxyl polynucleotide kinase. J. Biol. Chem. 1966;241:2933–2943. [PubMed] [Google Scholar]
- 5.Becker A, Hurwitz J. The enzymatic cleavage of phosphate termini from polynucleotides. J. Biol. Chem. 1967;242:936–950. [PubMed] [Google Scholar]
- 6.Cameron V, Uhlenbeck OC. 3′-Phosphatase activity in T4 polynucleotide kinase. Biochemistry. 1977;16:5120–5126. doi: 10.1021/bi00642a027. [DOI] [PubMed] [Google Scholar]
- 7.Wang LK, Lima CD, Shuman S. Structure and mechanism of T4 polynucleotide kinase – an RNA repair enzyme. EMBO J. 2002;21:3873–3880. doi: 10.1093/emboj/cdf397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galburt EA, Pelletier J, Wilson G, Stoddard BL. Structure of a tRNA repair enzyme and molecular biology workhorse: T4 polynucleotide kinase. Structure. 2002;10:1249–1260. doi: 10.1016/s0969-2126(02)00835-3. [DOI] [PubMed] [Google Scholar]
- 9.Eastberg JH, Pelletier J, Stoddard BL. Recognition of DNA substrates by bacteriophage T4 polynucleotide kinase. Nucleic Acids Res. 2004;32:653–660. doi: 10.1093/nar/gkh212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhu H, Smith P, Wang LK, Shuman S. Structure-function analysis of the 3′-phosphatase component of T4 polynucleotide kinase/phosphatase. Virology. 2007;366:126–136. doi: 10.1016/j.virol.2007.03.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang LK, Shuman S. Domain structure and mutational analysis of T4 polynucleotide kinase. J. Biol. Chem. 2001;276:26868–26874. doi: 10.1074/jbc.M103663200. [DOI] [PubMed] [Google Scholar]
- 12.Wang LK, Shuman S. Mutational analysis defines the 5’-kinase and 3′-phosphatase active sites of T4 polynucleotide kinase. Nucleic Acids Res. 2002;30:1073–1080. doi: 10.1093/nar/30.4.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karimi-Busheri F, Weinfeld M. Purification and substrate specificity of polydeoxynucleotide kinases isolated from calf thymus and rat liver. J. Cell. Biochem. 1997;64:258–272. [PubMed] [Google Scholar]
- 14.Zhu H, Yin S, Shuman S. Characterization of polynucleotide kinase/phosphatase enzymes from mycobacteriophages Omega and Cjw1 and vibriophage KVP40. J. Biol. Chem. 2004;279:26358–26369. doi: 10.1074/jbc.M403200200. [DOI] [PubMed] [Google Scholar]
- 15.Schwer B, Sawaya R, Ho CK, Shuman S. Portability and fidelity of RNA-repair systems. Proc. Natl Acad. Sci. USA. 2004;101:2788–2793. doi: 10.1073/pnas.0305859101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bernstein NK, Williams RS, Rakovszky ML, Cui D, Green R, Galicia S, Koch CA, Cass CE, Durocher D, Weinfeld M, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol. Cell. 2005;17:657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 17.Bernstein NK, Hammel M, Mani RS, Weinfeld M, Pelikan M, Tainer JA, Glover JNM. Mechanism of DNA substrate recognition by the mammalian DNA repair enzyme, polynucleotide kinase. Nucleic Acids Res. 2005;37:6161–6173. doi: 10.1093/nar/gkp597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jilani A, Slack C, Matheos D, Zannis-Hadjopoulos M, Lasko DD. Purification of a polynucleotide kinase from calf thymus, comparison of its 3′-phosphatase domain with T4 polynucleotide kinase, and investigation of its effect on DNA replication in vitro. J. Cell. Biochem. 1999;73:188–203. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.