Abstract
Purpose
Age-related cataract is a multi-factorial disease with a poorly understood etiology. Numerous studies provide evidence that the human eye lens has evolved specific regulatory and protective systems to ameliorate lens damage associated with cataract. Other studies suggest that the presence of cataract is associated with the altered expression of specific genes including metallothionein IIa, osteonectin, transglutaminase 2, betaig-h3, multiple ribosomal proteins, ADAM9, and protein phosphatase 2A. Here, we sought to identify further gene expression changes that are associated with cataract and to cluster the identified genes into specific biological pathways.
Methods
Oligonucleotide microarray hybridization was used to analyze the full complement of gene expression differences between lens epithelia isolated from human age-related cataract relative to clear lenses. The expression levels of a subset of the identified genes were further evaluated by semi-quantitative RT-PCR. The identified genes were functionally clustered into specific categories and the probability of over-representation of each category was determined using the computer program EASE.
Results
412 transcripts were observed to be increased and 919 transcripts were observed to be decreased by 2 fold or more in lens epithelia isolated from age-related cataract relative to clear lenses. Of these, 74 were increased and 241 were decreased at the 5 fold level or greater. Seventeen genes selected for further confirmation exhibited similar trends in expression when examined by RT-PCR using both the original and separately prepared clear and cataract RNA populations. Functional clustering of the identified genes using the EASE bioinformatics software package revealed that, among others, transcripts increased in cataract are associated with transcriptional control, chromosomal organization, ionic and cytoplasmic transport, and extracellular matrix components while transcripts decreased in cataract are associated with protein synthesis, defense against oxidative stress, heat-shock/chaperone activity, structural components of the lens, and cell cycle control.
Conclusions
These data suggest that cataract is associated with multiple previously identified and novel changes in lens epithelial gene expression and they point to numerous pathways likely to play important roles in lens protection, maintenance, and age-related cataract.
The role of the eye lens is to focus incoming light onto the retina where visual information is then processed and transmitted to the brain. The lens is an excellent model for the study of age related diseases since it has no blood supply, contains some of the oldest cells in the body, grows throughout life, and is exposed to multiple environmental insults including toxic metals and UV-light which can result in oxidative stress [1]. Oxidative stress, combined with aging of the lens and consequential lens cell damage, is believed to contribute to age-related cataract formation, an opacity of the lens that results in blindness [1]. Cataract is a major health issue worldwide as it is the leading cause of world blindness. Surgical removal of the lens is the only known treatment. Cataract is an enormous economic burden, accounting for 12% of all Medicare expenses in the United States each year. With an aging American population cataract is, and will continue to be, a major economic and quality of life concern.
Despite the large number of studies documenting the biochemical and metabolic changes in the lens associated with age-related cataract, little is known about the changes in gene expression associated with this disease. To identify these changes we have focused on the lens epithelium since this monolayer of cells is essential for the growth, differentiation, and homeostasis of the entire organ [2,3]. The lens epithelium contains the highest levels of enzymes and transport systems in the lens [4–6] and is the first part of the lens exposed to environmental insults [5,6]. Multiple studies suggest that the lens epithelium is capable of communicating with the underlying fiber cells [7] and direct damage to the lens epithelium and its enzyme systems is known to result in cataract formation [1,8–10]. Importantly, the majority of transcription occurs in the epithelial cells of the lens, and therefore these cells make up the majority of lens cells capable of responding to environmental insults and/or the presence of cataract through altered gene expression. Since the lens epithelium is composed of a single cell-type it represents an ideal model for gene expression studies.
Although a multitude of lens culture studies have documented changes in the expression of numerous genes in response to H2O2, toxic metals, UV-light, and other stresses, and multiple studies have examined changes in gene expression in animal models of cataract, the full complement of gene expression differences that occur in lens epithelial cells of human age-related cataract is not known. Previous studies have used RT-PCR differential display and other techniques to identify differences in gene expression between human lens epithelial cells isolated from cataract relative to clear lenses. For instance, metallothionein IIa [11], osteonectin (also known as SPARC [12]), transglutaminase 2 [13], and betaig-h3 [14], are reported to be increased in cataract relative to clear lenses while multiple ribosomal proteins [15], ADAM9 [16], and protein phosphatase 2A [11] are reported to be decreased in cataract relative to clear lenses.
While these studies have provided important insight into the roles of specific gene expression changes in age-related cataract, information concerning individual gene expression changes is not adequate to reveal related clusters of genes whose identities are necessary to elucidate the biological pathways that are altered in age-related cataract. Although recent studies have examined the global changes in gene expression that occur in cultured human lens epithelial cells exposed to H2O2, a stress associated with cataract [17,18], to date no comprehensive study has documented the global gene expression changes occurring between human age-related cataract and clear lenses or reported the functional clustering of age-related cataract-specific genes. This information is necessary to identify those biological pathways altered in age-related cataract and is essential towards understanding the molecular basis for this disease. Despite the difficulty in obtaining sufficient numbers of human cataracts and clear lenses for this type of large-scale analysis, it is important that these studies be conducted with actual human lens epithelia since no tissue culture or animal model system can mimic the unique life history, physiology and genetic responses of the human lens.
We have used oligonucleotide microarrays to compare the global gene expression profiles between pooled age-matched human lens epithelia isolated from cataract and clear lenses. We demonstrate that more than 1,300 of the 22,215 genes surveyed have expression levels that differ by 2 fold or more in cataracts compared to clear lenses. Of these, 74 genes are increased and 241 genes are decreased in cataract relative to clear lenses at the level of 5 fold or greater. Functional clustering and over-representation analysis of the identified genes revealed that multiple biological pathways are significantly altered upon cataract formation including chaperones, oxidative stress, protein synthesis, and ion transport pathways. These data provide the basis for designing functional experiments to examine the roles of the identified genes in lens maintenance and protection and they provide insight into those mechanisms that may be important for the development of, and defense against, age-related cataract.
METHODS
Tissue collection and RNA preparation
Central lens epithelial tags (2–3 mm2) were obtained from patients undergoing cataract surgery at the Jules Stein Eye Institute, UCLA School of Medicine. The cataracts are representative of the entire population of patients undergoing cataract surgery and were obtained and classified by the same surgeon, according to a modified version of the Lens Opacities Classification Scale (LOCS)-III grading system. The cataracts used in this study were approximately 70% mixed, 20% nuclear, 5% cortical, and 2% posterior subcapsular. With the exception of cataract-type, age and sex, no further identifying information was available for individual lenses. Clear whole human lenses were obtained from organ donors within 24 h post-mortem by the Lions Eye Bank of Oregon and the West Virginia Eye Bank. Whole lenses were microscopically examined for opacities and those lenses exhibiting opacity were discarded from the present study. Clear lenses were micro-dissected for central epithelium (6–8 mm2) and contaminating fiber cells were removed. A total of 106 cataracts (average age 71.2 years) and 10 clear lens epithelia (average age 64.2 years) were used to obtain a sufficient amounts of RNA (2–5 μg) for the microarray study. An additional 50 cataracts (average age 70.8 years) and 10 clear lens epithelia (average age 63.3 years) were used for the secondary semi-quantitative RT-PCR confirmation studies. Another 50 cataracts (average age 68.7 years) and 10 clear lens epithelia (average age 57.0 years) were used for the control and tertiary semi-quantitative RT-PCR confirmation studies. Total RNA was isolated from these samples using the Trizol method.
Microarray procedure and analysis
The quality and quantity of RNA obtained from the cataract and clear lens epithelial tags was determined using a Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA) according to the manufacturers protocol. Briefly, a small amount of RNA from each sample was loaded on a microgel, electrophoresed, scanned and analyzed for the quantity and integrity of the 18s and 28s ribosomal RNA bands to ensure that the same amount of RNA was examined for both the cataract and clear lens samples.
First and second strand cDNAs were synthesized from 2–5 μg of total RNA using the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen, Gaithersburg, MD) and the oligo-dT24-T7 primer (5′-GGC CAG TGA ATT GTA ATA CGA CTC ACT AT-AGG GAG GCG G-3′) according to the manufacturer's instructions. cRNA was synthesized and labeled with biotinylated UTP and CTP by in vitro transcription using the T7 promoter coupled double-stranded cDNA as a template and the T7 RNA Transcript Labeling Kit (ENZO Diagnostics Inc., Farmingdale, NY). Briefly, double-stranded cDNAs synthesized from the previous steps were washed twice with 70% ethanol and resuspended in 22 μl of RNase-free H2O. The cDNA was incubated with 4 μl each of 10X Reaction Buffer, Biotin Labeled Ribonucleotides, DTT, RNase Inhibitor Mix, and 2 μl of 20X T7 RNA Polymerase for 5 h at 37 °C. The labeled cRNA was separated from unincorporated ribonucleotides by passing through a CHROMA SPIN-100 column (Clontech, Palo Alto, CA) and precipitated at −20 °C for 1 h to overnight.
The cRNA pellet was resuspended in 10 μl of RNase-free H2O and 10 μg was fragmented by heat and ion-mediated hydrolysis at 95 °C for 35 min in 200 μM Tris-acetate, pH 8.1, 500 mM KOAc, and 150 mM MgOAc. The fragmented cRNA was hybridized for 16 h at 45 °C to HG_U133A oligonucleotide arrays (Affymetrix, Santa Clara, CA) containing 22,283 probe sets representing 22,215 gene or extended sequence tag (EST) sequences. Arrays were washed at 25 °C with 6X SSPE (0.9 M NaCl, 60 mM NaH2PO4, 6 mM EDTA, and 0.01% Tween-20) followed by a stringent wash at 50 °C with 100 mM MES, 0.1 M (Na+), and 0.01% Tween-20. The arrays were then stained with phycoerythrein-conjugated streptavidin (Molecular Probes, Eugene, OR) and the fluorescence intensities were determined using a laser confocal scanner (Hewlett-Packard, Palo Alto, CA).
The scanned images were analyzed using Microarray Suite 5.0 software (Affymetrix), following user guidelines. Briefly, background signal intensities were calculated and used to determine if the signal intensity of an individual gene was statistically greater than the background intensity value. The signal intensity for each gene was calculated as the average intensity difference, represented by [Σ(PM-MM)/(number of probe pairs)], where PM and MM denote perfect-match and mismatch probes, respectively. Each reported gene value represents the average signal intensity of 10 separately hybridized gene signatures. Any gene whose MM value was saturated or fell within tau (τ) distance of the PM value was excluded from the analysis. τ is a parameter used in performing the One-Sided Wilcoxon's Signed Rank test for the detection call and represents a threshold that the discrimination score for a probe set must exceed in order for a gene to be regarded as being present in the sample. Each gene was then assigned a call of Present (P), meaning that its intensity value is statistically greater than that of the background level and/or falls outside of the calculated τ distance, or Absent (A) meaning that its intensity value is not statistically greater than that of the background level and/or falls within the calculated τ distance. All of the genes described in this study are rated as present in at least one, if not both, of the cataract and clear lens samples. Any gene that was determined to be absent in both the cataract and clear lens samples was excluded from this report.
The microarray data were normalized using the Microarray Suite 5.0 software (Affymetrix) by multiplying the output of the experimental array by a Normalization Factor so that its average intensity is the same as that of the baseline array. The Microarray Suite 5.0 software also requires scaling, in which the output of any array is multiplied by a scaling factor to make its average intensity equal to a defined target intensity. For these studies a standard target intensity of 250 was used.
Semi-quantitative RT-PCR confirmation
Seventeen genes were selected for use in semi-quantitative RT-PCR confirmations of the hybridization results. Gene-specific primers were designed using the BLAST program and GenBank database (National Center for Biotechnology Information, Bethesda, MD). All primers were designed to cross intron/exon boundaries. The primer sequences, GenBank accession numbers, annealing temperatures, product lengths, and PCR cycle numbers for all gene-specific primers used in this study are indicated in Table 1. Semi-quantitative RT-PCR was performed using 50 ng of RNA with a commercial RT-PCR system used in accordance with the manufacturer's protocol (One-Step; Invitrogen, Gaithersburg, MD). To provide further confidence in the data and to show that the PCR reactions are within the linear range of PCR cycles, 3 control genes, catalase, metal-responsive transcription factor 1 (MTF-1), and αB-crystallin, and two genes of interest, HSP27-1 and -2 were evaluated by RT-PCR using 50 ng of cataract RNA and 5 different amounts (5, 10, 30, 50, and 100 ng) of clear lens RNA. Products were separated by gel electrophoresis on 1.5% agarose gels and visualized by ethidium bromide staining. Product formation for indicated genes was linear over all of the PCR cycles used. All PCR products were sequenced to ensure product authenticity. All gels were scanned and the percent adjusted volume intensities of all of the RT-PCR products were determined using a Biorad gel documentation system (Biorad, Hercules, CA). These values were used to calculate the approximate fold changes of the selected genes between cataract and clear lens epithelia.
Table 1.
Primers used for RT-PCR
| Gene |
Abreviation |
Primer sequence |
Annealing temperature |
Product length |
Cycle number |
Accession number |
|---|---|---|---|---|---|---|
| Hsp27-1 | Hsp27-1 | CGCGCTCAGCCGGCAACTCAG | 64 | 419 | 27 | XM_055937 |
| Hsp27-1 | Hsp27-1 | AGGGGTGGGCATCCAGGCTAAGG | 64 | 419 | 27 | XM_055937 |
| Hsp27-2 | Hsp27-2 | TCCTGACCCCCACACTCTACCA | 61 | 421 | 27 | NM_001541 |
| Hsp27-2 | Hsp27-2 | GCTGCCTCCTCCTCTTCCTCTG | 61 | 421 | 27 | NM_001541 |
| aA-crystallin | aA | CCACCTCGGCTCCCTCGTCCTAAG | 64 | 492 | 25 | NM_000394 |
| aA-crystallin | aA | CCATGTCCCCAAGAGCGGCACTAC | 64 | 492 | 25 | NM_000394 |
| RPL13a | RPL13a | GTATGCTGCCCCACAAAACCA | 58 | 387 | 25 | XM_027885 |
| RPL13a | RPL13a | CAACGCATGAGGAATTAACAGTCTT | 58 | 387 | 25 | XM_027885 |
| Metallothionein IF | MTIF | GCTTCTCTCTTGGAAAGTCC | 55 | 226 | 30 | M10943 |
| Metallothionein IF | MTIF | GGCATCAGTCGCAGCAGCTG | 55 | 226 | 30 | M10943 |
| Metallothionein IH | MTIH | GAACTCCAGTCTCACCTCGG | 55 | 213 | 30 | X64834 |
| Metallothionein IH | MTIH | GACATCAGGCACAGCAGCTG | 55 | 213 | 30 | X64834 |
| Metallothionein IG | MTIG | GCCTCTTCCCTTCTCGCTTG | 55 | 234 | 30 | XM_048213 |
| Metallothionein IG | MTIG | GACATCAGGCGCAGCAGCTG | 55 | 234 | 30 | XM_048213 |
| Glutathione Peroxidase 1 | GPX-1 | GACCGACCCCAAGCTCATCACC | 60 | 333 | 30 | M21304 |
| Glutathione Peroxidase 1 | GPX-1 | ATCAACAGGACCAGCACCCATCTC | 60 | 333 | 30 | M21304 |
| Na+/H+ Exchanger II | Na+/H+ Ex | GCCATCTGTTTTGCGTTAGTGTTT | 56 | 530 | 23 | AF073299 |
| Na+/H+ Exchanger II | Na+/H+ Ex | GTTCGCTGACGGATTTGATAGAGA | 56 | 530 | 23 | AF073299 |
| Serine/ Threonine Protein Kinase | S/T PK | TGTTGGTGGGGATTTGCTTACTCT | 57 | 449 | 23 | NM_003607 |
| Serine/ Threonine Protein Kinase | S/T PK | CTTGGGCTGGAAACTGAAACCTCT | 57 | 449 | 23 | NM_003607 |
| Na+/K+ ATPase | Na+/K+ ATPase | AAAGTACAAAGATTCAGCCCAGAG | 52 | 419 | 23 | BC000006 |
| Na+/K+ ATPase | Na+/K+ ATPase | GGAGTTTGCCATAGTACGGATAAT | 52 | 419 | 23 | BC000006 |
| Secreted Apoptosis Related Protein | SARP | TTGTAATCCAGTCGGCTTGTTCTT | 56 | 478 | 23 | AF017987 |
| Secreted Apoptosis Related Protein | SARP | CTGGGCCTTTGCTGTCACTATTAC | 56 | 478 | 23 | AF017987 |
| Pleiotrophin | Pie. | GTTCCCCGCCTTCCAGTCCA | 60 | 430 | 23 | M57399 |
| Pleiotrophin | Pie. | TGCCCAGCCCACAGTCTCCA | 60 | 430 | 23 | M57399 |
| E3-Ubiquitin Ligase | UBE3-Lig | CAGGGAATGGTTGTATCTCTTGTC | 53 | 469 | 25 | AY014180 |
| E3-Ubiquitin Ligase | UBE3-Lig | AATGCCTCGTAAAAATCTCCAGTT | 53 | 469 | 25 | AY014180 |
| aB-crystallin | aB | AGCCGCCTCTTTGACCAGTTCTTC | 60 | 452 | 18 | NM_001885 |
| aB-crystallin | aB | GCGGTGACAGCAGGCTTCTCTTC | 60 | 452 | 18 | NM_001885 |
| Catalase | Cat | TACCCCTCCTGGACTTTTTACATC | 52 | 541 | 25 | NM_001752 |
| Catalase | Cat | CCTCATTCAGCACGTTCACATAGA | 52 | 541 | 25 | NM_001752 |
| Metal-responsive Transcription Factor 1 | MTF-1 | GGGCCAGGACCTCAGCACAAT | 59 | 445 | 25 | XM_001412 |
| Metal-responsive Transcription Factor 1 | MTF-1 | AGAAGCCCCAGCAACAACAGAAAG | 59 | 445 | 25 | XM_001412 |
The table lists the sequences, GenBank accession numbers, annealing temperatures, product lengths, and PCR cycle numbers for all gene-specific primers used in this study.
Functional clustering and over-representation analysis of differentially expressed genes
Genes identified to be differentially expressed by 2 fold or greater according to the microarray analysis were analyzed for significant functional clusters of genes using the EASE bioinformatics software package. This software package was used to rank functional clusters by statistical over-representation of individual genes in specific categories relative to all genes in the same category on the microarray. The functional clusters used by EASE were derived from the classification systems of the Gene Ontology, Proteome's “At A Glance,” SwissProt keywords, and Interpro protein domains.
RESULTS
Oligonucleotide microarray analysis
Analysis of gene expression differences between pooled age-matched cataract and clear lenses was conducted using Affymetrix HG_U133A microarrays as described in the methods section. In this analysis, only one hybridization was conducted for each RNA population due to the extremely large number of human lens epithelia required for this type of analysis and the limited availability of these tissues. Comparison of the gene expression data for 22,215 genes represented by 222,830 separate probe sets, each probe set containing 10 perfect match and 10 1 base pair mismatch probe sequences, between cataract and clear lens samples, identified 412 transcripts that were increased (Figure 1) and 919 transcripts that were decreased (Figure 2) by 2 fold or greater in cataract compared to clear lenses. Of the genes that exhibited increased expression in cataracts, 82% of them were increased by 2–5 fold, 13% by 5–9 fold, 3% by 9–15 fold, and 2% by greater than 15 fold (Figure 1). Of the genes that exhibited decreased expression in cataracts, 74% of them fell into the 2–5 fold range, 15% in the 5–9 fold range, 7% in the 9–15 fold range, and 4% in the 15 fold and greater range (Figure 2). Of the identified genes, 74 exhibited increased expression, of which 24 are ESTs or unknown gene products, and 241 exhibited decreased expression, of which 25 are ESTs or unknown gene products, at the 5 fold or greater level in cataract relative to clear lenses. These genes and their relative expression levels, intensity values and accession numbers are listed in Table 2. The raw microarray data, including intensity values and its statistical analysis, can be accessed in Appendix 1.
Figure 1.
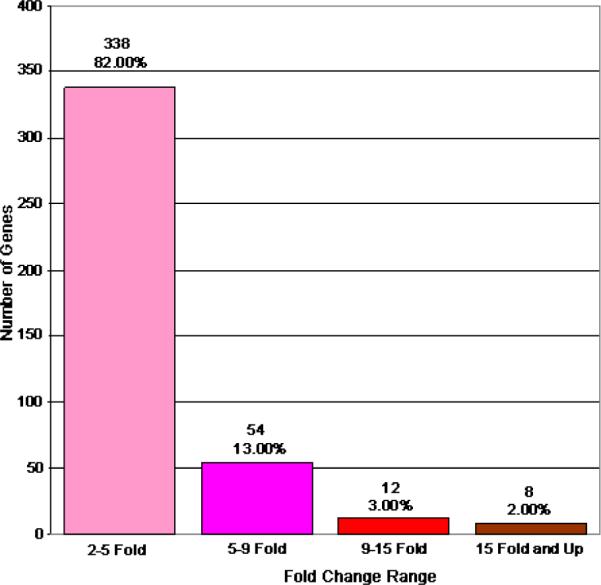
Genes increased 2 fold or greater between cataract and clear lenses. This figure graphically represents the genes whose expression levels are incresaed by 2 fold or greater in cataract relative to clear lenses. The total number of genes included in each fold change category are indicated. Percentages indicate the total number of genes in each category relative to the total number of increased genes (412) on the chip.
Figure 2.
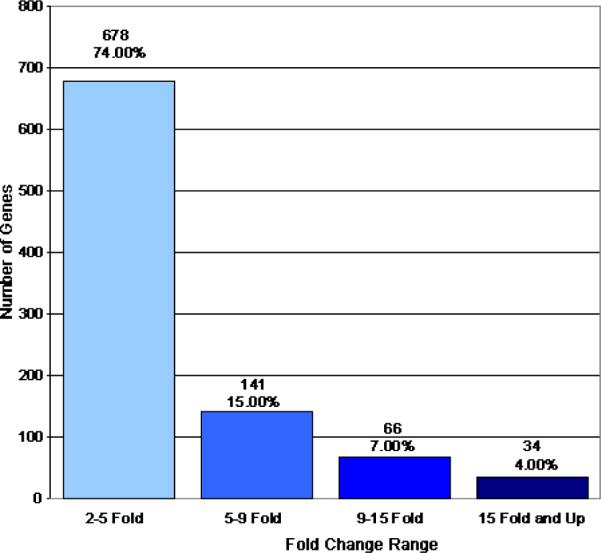
Genes decreased 2 fold or greater between cataract and clear lenses. This figure graphically represents the genes whose expression levels are decreased by 2 fold or greater in cataract relative to clear lenses. The total number of genes included in each fold change category are indicated. Percentages indicate the total number of genes in each category relative to the total number of decreased genes (919) on the chip.
Table 2.
Genes exhibiting differential expression in cataract relative to clear lenses
| Genes exhibiting increased expression in cataract relative to clear lenses | ||||
|---|---|---|---|---|
| Gene name |
Accesion number |
Normal signal intensity |
Cataract signal intensity |
Fold |
| nuclear phosphoprotein | BE796924 | 348.8 (P) | )1730.7 (P) | 5.28 |
| di-N-acetyl-chitobiase | NM_004388 | 117.2 (A) | )322.9 (P) | 5.28 |
| Hypothetical protein FLJ21551 | NM_024801 | 121 (P) | 524.5 (P) | 5.28 |
| Hypothetical protein PRO1048 | NM_018497 | 29.3 (A) | 261.1 (P) | 5.28 |
| EST | AA972711 | 354.4 (P) | 1919.5 (P) | 5.28 |
| Human erythroid-specific transcription factor EKLF | U65404 | 70.3 (P) | 408.9 (P) | 5.28 |
| Chromosome 14 clone | AC007956 | 154.4 (P) | 649.3 (P) | 5.66 |
| tetratricopeptide repeat domain 3 | AW510696 | 431.9 (P) | 1752.4 (P) | 5.66 |
| Hypothetical protein FLJ11827 | NM_025093 | 58.5 (P) | 338.9 (P) | 5.66 |
| ubinuclein 1 | T70262 | 397.9 (P) | 1981.9 (P) | 5.66 |
| alpha thalassemiamental retardation syndrome X-linked | AI650257 | 154.9 (P) | 852.1 (P) | 5.66 |
| Neuron-specific protein | NM_014392 | 54.5 (A) | 338.2 (P) | 5.66 |
| growth factor receptor-bound protein 10 | D86962 | 126.6 (P) | 544.2 (P) | 5.66 |
| Disabled homolog 2 (mitogen-responsive phosphoprotein) | NM_001343 | 237 (P) | 1096.6 (P) | 6.06 |
| Secreted apoptosis related protein 2 (SARP2) | AF017987 | 473.2 (P) | 3068.6 (P) | 6.06 |
| acid sphingomyelinase-like phosphodiesterase | AA873600 | 48.7 (A) | 264.1 (P) | 6.06 |
| EST | AI694562 | 2039.8 (P) | 14553.9 (P) | 6.06 |
| KIAA1641 protein | NM_025190 | 178.7 (P) | 878.1 (P) | 6.06 |
| Typtophan 2,3-dioxygenase | NM_005651 | 37.4 (A) | 324.8 (P) | 6.06 |
| adducin 3 (gamma) | AI763123 | 100.8 (A) | 379.3 (P) | 6.06 |
| Type II Golgi membrane protein | NM_014498 | 100 (A) | 618.9 (P) | 6.06 |
| EST | AA634446 | 13.3 (A) | 137.2 (P) | 6.5 |
| Na+H+ exchanger isoform 2 | AF073299 | 133.9 (A) | 1443.4 (P) | 6.5 |
| Ser-Thr protein kinase | NM_003607 | 1015.2 (P) | 3771 (P) | 6.5 |
| Sjogren syndrome antigen B | BG532929 | 47.8 (A) | 374.3 (P) | 6.5 |
| clone COL05464 | AK025143 | 68.1 (A) | 571.6 (P) | 6.5 |
| EST | BF592782 | 479.5 (P) | 3072.6 (P) | 6.5 |
| Bcl-2-associated transcription factor short form mRNA | AF249273 | 94.5 (P) | 518.1 (P) | 6.5 |
| eukaryotic translation initiation factor 4 gamma | BE966878 | 112.4 (P) | 612.6 (P) | 6.5 |
| Nijmegen breakage syndrome 1 (nibrin) | AI796269 | 83.6 (A) | 1188 (P) | 6.96 |
| DEADH (Asp-Glu-Ala-AspHis) box polypeptide 17 | AW188131 | 153.2 (A) | 1396 (P) | 6.96 |
| KIAA0876 protein | AW237172 | 128.9 (A) | 1181.2 (P) | 6.96 |
| Arginine methyltransferase | U79286 | 62.8 (A) | 366.1 (P) | 6.96 |
| Small nuclear RNA activating complex, polypeptide 1, 43 kD (SNAPC1) | NM_003082 | 145.6 (P) | 643.5 (P) | 6.96 |
| Zinc finger protein 161 (ZNF161) | NM_007146 | 81.9 (A) | 446.7 (P) | 6.96 |
| KIAA1641 protein | AB046861 | 32 (A) | 201.3 (P) | 6.96 |
| copine III | AA541758 | 69.6 (A) | 775.4 (P) | 6.96 |
| natural killer-tumor recognition sequence | AI361805 | 398.2 (P) | 2412.8 (P) | 6.96 |
| KIAA0480 gene product | AW299294 | 154 (P) | 997.5 (P) | 7.46 |
| Nerve growth factor (HBNF-1) | M57399 | 1448.1 (P) | 7425.6 (P) | 7.46 |
| natural killer-tumor recognition sequence | AI688640 | 95.4 (P) | 829 (P) | 7.46 |
| pleiorophin | BC005916 | 1187.8 (P) | 10502.3 (P) | 7.46 |
| nuclear receptor interacting protein 1 | AI824012 | 58.3 (A) | 383.9 (P) | 7.46 |
| EST | AW293343 | 84.3 (P) | 630.2 (P) | 7.46 |
| ATPase, Na+K+ transporting, beta 1 polypeptide | BC000006 | 1233.8 (P) | 14152 (P) | 8 |
| Glutathione peroxidase 2 | NM_002083 | 31.7 (A) | 257.1 (P) | 8 |
| transformer-2 alpha | AW978896 | 97 (A) | 618.3 (P) | 8 |
| Tubby like protein 1 | NM_003322 | 27.3 (A) | 211.3 (P) | 8 |
| EST | BF448315 | 197.7 (P) | 1500.5 (P) | 8 |
| DNA for HBV integration sites | X04014 | 80.7 (A) | 607.8 (P) | 8 |
| similar to widely-interspaced zinc finger motifs | AI828531 | 34.6 (A) | 273.6 (P) | 8 |
| cDNA DKFZp566M043 | AL050065 | 36.2 (A) | 322.8 (P) | 8.57 |
| secretory carrier membrane protein 1 | BF058944 | 177.3 (P) | 928.9 (P) | 8.57 |
| chondroitin sulfate proteoglycan 6 (bamacan) | AI373676 | 71.3 (P) | 1010.3 (P) | 8.57 |
| KIAA0594 protein | AW183677 | 39.1 (A) | 404.7 (P) | 9.19 |
| Claudin 1 (CLDN1) | NM_021101 | 41 (A) | 268.1 (P) | 9.85 |
| KIAA0256 gene product | N52532 | 71.8 (A) | 1709.6 (P) | 9.85 |
| HRIHFB2017 | AB015331 | 64.1 (A) | 368.4 (P) | 9.85 |
| KIAA0888 protein | AB020695 | 173.8 (A) | 2224.6 (P) | 10.56 |
| Osteomodulin | AI765819 | 26.4 (A) | 351.8 (P) | 11.31 |
| Bicaudal-D (BICD) | U90030 | 40.8 (A) | 888.1 (P) | 12.13 |
| EST | AI278204 | 46.2 (A) | 331.8 (P) | 12.13 |
| cDNA: FLJ21198 | AK024851 | 13.5 (A) | 217.6 (P) | 12.13 |
| KIAA0447 gene product | BE885244 | 45.2 (A) | 664 (P) | 13 |
| chloride channel 3 | AA902971 | 25.7 (A) | 221.7 (P) | 14.93 |
| Wiskott-Aldrich syndrome-like | BE504979 | 51.4 (A) | 686 (P) | 14.93 |
| Cofactor required for Sp1 transcriptional activation, subunit 2 | NM_004229 | 9.2 (A) | 196.8 (P) | 16 |
| KIAA0494 gene product | BC002525 | 15.5 (A) | 419.9 (P) | 17.15 |
| ring finger protein 15 | AU157590 | 62.5 (A) | 719.2 (P) | 19.7 |
| myeloidlymphoid or mixed-lineage leukemia | AA715041 | 39.2 (A) | 518.1 (P) | 19.7 |
| PRO2667 | AF119889 | 31.3 (A) | 717.7 (P) | 19.7 |
| cDNA DKFZp564M2422 | AL050388 | 4.2 (A) | 185.4 (P) | 19.7 |
| Similar to histamine N-methyltransferase | BC005907 | 10.4 (A) | 308 (P) | 27.86 |
| Testis-specific XK-related protein on Y | NM_004677 | 4.3 (A) | 124.2 (P) | 32 |
| Genes exhibiting decreased expression in cataract relative to clear lenses | ||||
|---|---|---|---|---|
| Gene name |
Accesion number |
Normal signal intensity |
Cataract signal intensity |
Fold |
| Jagged 1 | U73936 | 916.3 (P) | 69.9 (A) | 5.28 |
| Ribosomal protein, large, P0 | NM_001002 | 14191.2 (P) | 3138.4 (P) | 5.28 |
| Fibrillin 1 | NM_000138 | 404.2 (P) | 55.2 (A) | 5.28 |
| Similar to eukaryotic translation initiation factor 4A, isoform 1 | BC006210 | 2672.3 (P) | 494.9 (A) | 5.28 |
| EST | AI799802 | 228.4 (P) | 23.1 (A) | 5.28 |
| Zinc finger protein 219 | NM_016423 | 300.2 (P) | 53.3 (A) | 5.28 |
| Similar to eukaryotic translation initiation factor 3, subunit 8 | BC000533 | 2697.1 (P) | 471.5 (P) | 5.28 |
| heat shock cognate protein 54 | AB034951 | 1342.6 (P) | 152 (A) | 5.28 |
| Pyruvate kinase, muscle | NM_002654 | 1098.2 (P) | 221.9 (A) | 5.28 |
| IMP (inosine monophosphate) dehydrogenase 2 | NM_000884 | 1011.6 (P) | 124.8 (A) | 5.28 |
| EST | AI816291 | 458.9 (P) | 66.1 (A) | 5.28 |
| Translocase of inner mitochondrial membrane 23 homolog | NM_006327 | 435.6 (P) | 86.5 (A) | 5.28 |
| 4-hydroxyphenylpruvate dioxygenase | NM_002150 | 206.7 (P) | 36.9 (A) | 5.28 |
| Heat shock 27 kD protein 2 | NM_001541 | 1056.5 (P) | 172.3 (A) | 5.28 |
| Carbonyl reductase 1 | BC002511 | 589.2 (P) | 27.6 (A) | 5.28 |
| Proteasome (prosome, macropain) subunit, beta type, 4 | NM_002796 | 875.3 (P) | 143 (A) | 5.28 |
| Small membrane protein 1 | NM_014313 | 502 (P) | 78.7 (A) | 5.28 |
| Fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) | NM_004102 | 244 (P) | 48.2 (A) | 5.28 |
| Calpastatin | AF327443 | 300.3 (P) | 81.1 (A) | 5.28 |
| Myosin, light polypeptide, regulatory, non-sarcomeric | NM_006471 | 3899.8 (P) | 876.3 (P) | 5.28 |
| Proteolipid protein 2 (colonic epithelium-enriched) | NM_002668 | 437.4 (P) | 50.8 (A) | 5.28 |
| ribosomal protein L4 | AI953886 | 6333.2 (P) | 716.8 (P) | 5.28 |
| cDNA DKFZp586D1122 | AL050166 | 199.2 (P) | 29.6 (A) | 5.28 |
| poly(rC)-binding protein 2 | NM_005016 | 1855.1 (P) | 204.4 (A) | 5.28 |
| Metallothionein If gene | M10943 | 5381.9 (P) | 776.9 (A) | 5.66 |
| 3-hydroxy-3-methylglutaryl-Coenzyme A reductase | AL518627 | 159.3 (P) | 30.3 (A) | 5.66 |
| G8 protein | NM_016947 | 3539.8 (P) | 558.7 (P) | 5.66 |
| SMX5-like protein | AF196468 | 358.8 (P) | 39.1 (A) | 5.66 |
| Microtubule-associated proteins 1A1B light chain 3 | AF183417 | 423.7 (P) | 79.3 (A) | 5.66 |
| PRO2640 | AF116710 | 8064.3 (P) | 991.9 (P) | 5.66 |
| MYLE protein | NM_014015 | 471 (P) | 52.6 (A) | 5.66 |
| Cold shock domain protein A | NM_003651 | 1098.3 (P) | 144.3 (A) | 5.66 |
| kinesin 2 | AA284075 | 236.3 (P) | 40.9 (A) | 5.66 |
| Cell membrane glycoproein | NM_007002 | 368.8 (P) | 75.1 (A) | 5.66 |
| Biliverdin reductase | NM_000713 | 1583.4 (P) | 498.1 (P) | 5.66 |
| Nuclear localization signal deleted in velocardiofacial syndrome | NM_003776 | 970.5 (P) | 125.9 (A) | 5.66 |
| clone RP11-486O2 | AL356115 | 10470 (P) | 1310.9 (P) | 5.66 |
| proteasome (prosome, macropain) subunit, alpha type, 3 | NM_002788 | 452 (P) | 46 (A) | 5.66 |
| Cyclin D1 | BC000076 | 182.7 (P) | 21.7 (A) | 5.66 |
| Heat shock 70 kD protein 1B | NM_005346 | 1660.9 (P) | 397.7 (P) | 5.66 |
| CD24 signal transducer | L33930 | 736.9 (P) | 187.1 (A) | 5.66 |
| Zyxin related protein ZRP-1 | AF000974 | 792.4 (P) | 113.6 (A) | 5.66 |
| solute carrier family 2 (facilitated glucose transporter), member 3 | BE550486 | 210.6 (P) | 76.2 (A) | 5.66 |
| Tubulin, beta 5 | BC005838 | 3024.6 (P) | 533.4 (M) | 5.66 |
| weakly similar to LONGEVIY-ASSURANCE PROTEIN 1 | AK001105 | 1037.6 (P) | )178.7 (A) | 5.66 |
| clone 1033B10 | AL031228 | 565 (P) | 92 (A) | 6.06 |
| S-adenosylhomocysteine hydrolase (AHCY) | NM_000687 | 365.9 (P) | 59.1 (A) | 6.06 |
| ribosomal protein, large, P0 | AI953822 | 8792.1 (P) | 1133.5 (P) | 6.06 |
| Ovarian beta-A inhibin | M13436 | 4685.9 (P) | 898.3 (P) | 6.06 |
| MYG1 protein | NM_021640 | 583.2 (P) | 103.9 (A) | 6.06 |
| ribosomal protein L13 | AI186735 | 7108.6 (P) | 1468.8 (P) | 6.06 |
| Splicing factor arginineserine-rich 9 | NM_003769 | 1438.8 (P) | 422.6 (A) | 6.06 |
| HDCMB21P gene | AF072098 | 10344.7 (P) | 699.8 (P) | 6.06 |
| Goliath protein | NM_018434 | 340.6 (P) | 28.8 (A) | 6.06 |
| Eukaryotic translation initiation factor 2B, subunit, 1 (alpha, 26 kD) | NM_001414 | 281.1 (P) | 54.1 (A) | 6.06 |
| ribosomal protein L13 | AW574664 | 3994.8 (P) | 371.4 (A) | 6.06 |
| Proteasome (prosome, macropain) subunit, beta type, 7 | NM_002799 | 2050 (P) | 332.4 (M) | 6.06 |
| Tubulin, beta, 2 | BC004188 | 1048.5 (P) | 202.5 (A) | 6.06 |
| Phosphatidylethanolamine N-methyltransferase | NM_007169 | 670.3 (P) | 47.1 (A) | 6.06 |
| Adaptor-related protein complex 2, mu 1 subunit | NM_004068 | 863.9 (P) | 165.4 (A) | 6.06 |
| cDNA DKFZp564B076 | AL049313 | 470.2 (P) | 52.3 (A) | 6.06 |
| clone RP4-781L3 | AL121994 | 897.8 (P) | 150.3 (A) | 6.06 |
| Alpha-actinin-2 associated LIM protein mRNA, alternatively spliced product | AF002280 | 189.4 (P) | 28.7 (A) | 6.06 |
| Threonyl-tRNA synthetase | NM_003191 | 958 (P) | 97.8 (A) | 6.06 |
| MCP-1=monocyte chemotactic protein human, aortic endothelial cells | S69738 | 771 (P) | 73 (A) | 6.5 |
| eukaryotic translation elongation factor 1 gamma | BE963164 | 13185.4 (P) | 1579.7 (A) | 6.5 |
| Lectin, galactoside-binding, soluble, 1(galectin1) | NM_002305 | 2609.9 (P) | 94.5 (A) | 6.5 |
| CGI-44 protein; sulfide dehydrogenase like | NM_021199 | 1703.9 (P) | 151.3 (A) | 6.5 |
| DnaJ (Hsp40) homolog, subfamily B, member 1 | BG537255 | 532.8 (P) | 77.4 (A) | 6.5 |
| Fragile histidine triad gene | HN_002012 | 341.8 (P) | 59.8 (P) | 6.5 |
| Carboxypeptidase B1 | NM_001871 | 337.8 (P) | 35.4 (A) | 6.5 |
| Crystallin, beta B2 | NM_000496 | 20885.8 (P) | 3332.7 (P) | 6.5 |
| Meiotic recombination protein REC14 | AF309553 | 134.4 (P) | 34.9 (A) | 6.5 |
| Selenoprotein W, 1 | NM_003009 | 707.1 (P) | 39.3 (A) | 6.5 |
| mRNA for hMBF1alpha | AB002282 | 2012.1 (P) | 211.9 (A) | 6.5 |
| tudor repeat associator with PCTAIRE 2 | AW129593 | 2669.4 (P) | 387.8 (M) | 6.5 |
| EST | AV705559 | 593.1 (P) | 107.4 (A) | 6.5 |
| Clone: SMAP31-12 | AB059408 | 483.7 (P) | 68.1 (A) | 6.5 |
| Growth arrest and DNA damage inducible proteinbeta | AF087853 | 1895.6 (P) | 39.3 (A) | 6.5 |
| Crystallin, gamma B | NM_005210 | 721 (P) | 104.8 (A) | 6.5 |
| Eukaryotic translation elongation factor 1 delta (guanine nucleotide exchange protein) | NM_001960 | 3814.7 (P) | )480.1 (A) | 6.5 |
| FK506-binding protein 2 | NM_004470 | 503.6 (P) | 17.5 (A) | 6.5 |
| HLA class II region expressed gene KE2 | NM_014260 | 382.3 (P) | 30.3 (A) | 6.5 |
| Neuronal cell adhesion molecule | NM_005010 | 613.1 (P) | 94.8 (P) | 6.5 |
| polymerase (RNA) II (DNA directed) polypeptide J | BG335629 | 552.8 (P) | 31.8 (A) | 6.5 |
| Ribosomal protein L27a | NM 000990 | 12053.7 (P) | 1609.9 (P) | 6.5 |
| EST | L43577 | 354.2 (P) | 43 (A) | 6.96 |
| Tetraspan 3 | NM_005724 | 183 (P) | 21.3 (A) | 6.96 |
| phosphoserine aminotransferase | AI889380 | 4608.5 (P) | 970.4 (P) | 6.96 |
| Nuclear prelamin A recognition factor | NM_012336 | 482 (P) | 58.2 (A) | 6.96 |
| Zinc finger protein homologous to Zfp-36 in mouse | NM_003407 | 651.1 (P) | 63.6 (A) | 6.96 |
| cDNA DKFZp564J1516 | AL136601 | 192.2 (P) | 30.5 (A) | 6.96 |
| Antizyme inhibitor | NM_015878 | 232.8 (P) | 25.5 (P) | 6.96 |
| G protein-coupled receptor 39 | AL567376 | 257 (P) | 63.1 (A) | 6.96 |
| prostatic binding protein | BE969671 | 3392.7 (P) | 310.4 (P) | 6.96 |
| Tetratricopeptide repeat domain 2 | NM_003315 | 397.2 (P) | 43.4 (A) | 6.96 |
| Ribosomal protein S15 | NM_001018 | 15776.8 (P) | 2287.8 (P) | 6.96 |
| Hypothetical protein FLJ11730 | NM_022756 | 639.3 (P) | 105.1 (A) | 6.96 |
| kinesin 2 | AA284075 | 199.9 (P) | 21.7 (A) | 6.96 |
| Prefoldin 5 | NM_002624 | 2490.6 (P) | 261.6 (A) | 6.96 |
| Poly(A)-binding protein, cytoplasmic 4 (inducible form) | NM_003819 | 437.9 (P) | 32.9 (A) | 6.96 |
| Ribosomal protein L35 | NM_007209 | 6130.5 (P) | 732.7 (P) | 6.96 |
| Catenin (cadherin-associated protein), alpha 2 | NM_004389 | 350.7 (P) | 28.5 (A) | 6.96 |
| Hypothetical protein FLJ10493 | NM_018112 | 107.9 (P) | 17.8 (A) | 6.96 |
| Lysosomal-associated membrane protein 1 | NM_005561 | 888.3 (P) | 54.1 (A) | 6.96 |
| Human growth hormone-dependent insulin-like growth factor-binding protein | M31159 | 2003.6 (P) | 285.9 (P) | 6.96 |
| glutathione peroxidase 3 | AW149846 | 5548.5 (P) | 521 (P) | 6.96 |
| Prostatic binding protein | NM_002567 | 4056.2 (P) | 356.9 (A) | 7.46 |
| GMPR2 for guanosine monophosphate reductase isolog | NM_016576 | 584.3 (P) | 38.3 (A) | 7.46 |
| hemoglobin, alpha 1 | T50399 | 427.5 (P) | 75.7 (A) | 7.46 |
| Ribosomal protein L8 | NM_000973 | 5766.6 (P) | 462.4 (A) | 7.46 |
| F-box protein FLR1 | AF142481 | 771.2 (P) | 114.6 (A) | 7.46 |
| Homo sapiens, Similar to tubulin, beta, 4 | BC002654 | 1096.5 (P) | 127.3 (A) | 7.46 |
| Ribosomal protein L29 | NM_000992 | 1889.9 (P) | 228.7 (A) | 7.46 |
| KIAA0874 protein | AB020681 | 249.4 (P) | 45.6 (A) | 7.46 |
| CGI-91 protein | NM_016034 | 327.1 (P) | 49.7 (A) | 7.46 |
| Pre-mRNA splicing factor 2 p32 subunit | L04636 | 518.2 (P) | 46.5 (A) | 7.46 |
| Phosphoglycerate kinase 1 | NM_000291 | 2262.5 (P) | 332.5 (P) | 7.46 |
| Human 28S rRNA sequence | M11167 | 3708.3 (P) | 648.4 (P) | 7.46 |
| Similar to granulin | BC000324 | 480 (P) | 65.7 (A) | 8 |
| hypothetical protein FLJ10698 | AI951798 | 422.6 (P) | 49 (A) | 8 |
| solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 | AI961224 | 6069.8 (P) | 397.1 (A) | 8 |
| SKIP for skeletal muscle and kidney enriched inositol phosphatase | AI806031 | 30.6 (A) | 249.4 | 8 |
| Protein kinase | AF133207 | 2162.2 (P) | 316 (A) | 8 |
| Extracellular matrix protein 1 | U65932 | 1252.6 (P) | 150.3 (A) | 8 |
| Alpha II spectrin | U083867 | 843.9 (P) | 96.5 (A) | 8 |
| nucleophosminB23.2 | AB042278 | 655 (P) | 70.7 (A) | 8 |
| Ribosomal protein L4 | NM_000968 | 7153.3 (P) | 854.4 (P) | 8 |
| Phosphatidylcholine transfer protein | NM_021213 | 205 (P) | 23.8 (A) | 8 |
| SEC13 (S. cerevisiae)-like 1 | NM_030673 | 420.4 (P) | 37.6 (A) | 8 |
| Homo sapiens mRNA for puromycin sensitive aminopeptidase | AJ132583 | 303.3 (P) | 39.2 (A) | 8 |
| Eukaryotic translation initiation factor 3, subunit 4 (delta, 44 kD) | BC000733 | 1480 (P) | 131.3 (A) | 8 |
| SET translocation (myeloid leukemia-associated) | AI278616 | 459 (P) | 35.1 (A) | 8 |
| PRO1608 | AF119850 | 10333.9 (P) | 1251.4 (P) | 8 |
| Human bcl-1 mRNA | M73554 | 780.7 (P) | 139.7 (A) | 8.57 |
| ECSIT | NM_016581 | 238.5 (P) | 27.4 (A) | 8.57 |
| MCT-1 protein | NM_014060 | 328.5 (P) | 20.5 (A) | 8.57 |
| Human soluble protein Jagged mRNA | U77914 | 1063.3 (P) | 109.4 (A) | 8.57 |
| nidogen (enactin) | BF940043 | 608.8 (P) | 93.6 (A) | 8.57 |
| Mitochondrial robosomal protein S15 | NM_031280 | 132.4 (P) | 11.3 (A) | 8.57 |
| Proteasome (prosome, macropain) subunit, beta type, 1 | NM_002793 | 1915.3 (P) | 357.2 (A) | 8.57 |
| Translocase of inner mitochondrial membrane 17 (yeast) homolog A | BC004439 | 128.3 (P) | 7.6 (A) | 9.19 |
| Microfibrillar-associated protein 2, transcript variant 1 | NM_017459 | 233.4 (P) | 23.6 (A) | 9.19 |
| Ribosomal protein L4 | BC005817 | 7644.3 (P) | 816.1 (P) | 9.19 |
| Zinc finger protein 162 | NM_004630 | 734.7 (P) | 25.1 (A) | 9.19 |
| Tyrosine 3-monoxygenasetryptophan 5-monoxygenase activation protein, zeta polypeptide | BC003623 | 373.6 (P) | 32.5 (A) | 9.19 |
| Spinde pole body protein | NM_006322 | 215.4 (P) | 12.6 (A) | 9.19 |
| Glycogenin | NM_004130 | 407 (P) | 30.8 (A) | 9.85 |
| 6-pyruvoyl-tetrahydropterin synthasedimerization cofactor of hepatocyte nuclear factor alpha | NM_000281 | 217.7 (P) | 13.4 (A) | 9.85 |
| Moesin | NM_002444 | 974.5 (P) | 38.7 (A) | 9.85 |
| Nuclear autoantigenic sperm protein (histone-binding) | NM_002482 | 155.1 (P) | 16.4 (A) | 9.85 |
| Metalloprotease | NM_007038 | 220.2 (P) | 9.9 (A) | 9.85 |
| KIAA0116 protein | AL581473 | 822.7 (P) | 45.9 (P) | 9.85 |
| GAPDH | M33197 | 5091.6 (P) | 530.9 (A) | 9.85 |
| Brain acid-soluble protein 1 | NM_006317 | 7329.2 (P) | 674 (P) | 9.85 |
| HSPC177 | NM_016410 | 310.9 (P) | 30.8 (A) | 9.85 |
| glyceraldehyde-3-phosphate dehydrogenase | BF689355 | 9541.6 (P) | 1048.9 (P) | 9.85 |
| Latent transforming growh factor beta binding protein 3 | NM_021070 | 377.9 (P) | 39 (A) | 9.85 |
| U6 snRNA-associated Sm-like protein LSm7 | NM_016199 | 395.4 (P) | 37 (A) | 10.56 |
| GANP protein | AJ010089 | 462.1 (P) | 29.2 (A) | 10.56 |
| McKusick-Kaufman syndrome protein | NM_018848 | 533.2 (P) | 20.9 (A) | 10.56 |
| Clone image:3611719 | BC003542 | 167 (P) | 22.2 (A) | 10.56 |
| Cyclin G1 | BC000196 | 4919.8 (P) | 480.8 (A) | 10.56 |
| Microtubule associated protein | AI633566 | 402.3 (P) | 44.9 (P) | 10.56 |
| MM-1 beta | AB055804 | 1917.7 (P) | 106.5 (A) | 10.56 |
| transketolase | L12711 | 5334 (P) | 535.8 (P) | 10.56 |
| 78 kDa gastrin-binding protein | U04627 | 370.5 (P) | 24.3 (A) | 10.56 |
| SH3 domain binding glutamic acid-rich protein | NM_007341 | 682.4 (P) | 38.7 (A) | 10.56 |
| EEF1 gamma | NM_001404 | 9570.8 (P) | 935.6 (A) | 10.56 |
| phospholipase C, beta 3 | BE305165 | 419.7 (P) | 42.3 (A) | 10.56 |
| Glutathione peroxidase 3 | NM_002084 | 9749.5 (P) | 594.4 (P) | 11.31 |
| RD protein | L03411 | 506.3 (P) | 40.5 (A) | 11.31 |
| Adaptor-related protein complex 2 | NM_021575 | 225 (P) | 19.3 (A) | 11.31 |
| Phosphomannomutase 1 | NM_002676 | 238.4 (P) | 40.7 (A) | 11.31 |
| Quinone oxidoreductase homolog | BC000474 | 934 (P) | 50.2 (A) | 11.31 |
| HSPCO34 protein | NM_016126 | 217.9 (P) | 12.8 (A) | 11.31 |
| Ornithin decarboxylase antizyme 1 | AF090094 | 1153.7 (P) | 36.4(A) | 11.31 |
| JM5 protein | BC000464 | 327.3 (P) | 30.5 (A) | 12.13 |
| Retinitis pigmentosa 2 | NM_006915 | 33.1 (P) | 1.3 (A) | 12.13 |
| Guanine nucleotide binding protein (G protein), beta polypeptide 2-like 1 | NM_006098 | 3013.5 (P) | 315.4 (A) | 12.13 |
| Cytidine deaminase | NM_001785 | 324.7 (P) | 23.4 (A) | 12.13 |
| alpha-2-HS-glycoprotein | BG538564 | 40.6 (P) | )93.4 (A) | 12.13 |
| Ribosomal protein Ll1 | NM_000975 | 4539.6 (P) | 162.2 (A) | 12.13 |
| L-iditol-2 dehydrogenase | L29008 | 1540.6 (P) | 93.4 (A) | 12.13 |
| v-fos FBJ murine osteosarcoma viral oncogene somalRNAg | BC004490 | 394.2 (P) | 94.2 (P) | 12.13 |
| Crystallin beta B2 | NM_000496 | 18394.2 (P) | 1113.3 (P) | 12.13 |
| 28S ribosomal RNA gene | M27830 | 8810.6 (P) | 639.5 (P) | 12.13 |
| KIAA0230 gene | D86983 | 300.7 (P) | 19.7 (A) | 12.13 |
| Clone 24461 | AF070577 | 517.8 (P) | 16.7 (A) | 12.13 |
| MRJ gene for a member of the DNAJ protein family | BC002446 | 244.5 (A) | 19 (A) | 13 |
| HMG box mRNA, 3 end cds. | L07335 | 1053.5 (P) | 80.1 (A) | 13 |
| Beaded filament structural protein 2, phakinin | NM_003571 | 7986.8 (P) | 454.7 (A) | 13 |
| Adipose specific 2 | NM_006829 | 822.3 (P) | 103.3 (A) | 13 |
| NADH dehydrogenase (ubiquitone) 1 alpha subcomplex, 7 | NM_005001 | 537 (P) | 24 (A) | 13 |
| Histidyl-tRNA synthetase | NM_002109 | 1033.2 (P) | 33.8 (A) | 13 |
| Myristoylated alanine-rich protein kinase C substrate | NM_002356 | 195.8 (P) | 16.8 (A) | 13.93 |
| Id-2H complete cds. inhibitor of DNA binding 2, dominant negative helix-loop-helix protein | D13891 | 195.8 (P) | 16.8 (A) | 13.93 |
| Ancient ubiquitous protein 1 | NM_012103 | 258.7 (P) | 25.4 (A) | 13.93 |
| solute carrier family 1 (glutamateneutral amino acid transporter), member 4 | BF340083 | 5269.5 (P) | 333.4 (P) | 13.93 |
| signal peptidase complex | N99438 | 789.3 (P) | 51.9 (A) | 13.93 |
| ID4 helix-loop-helix DNA binding protein | AL022726 | 210.3 (P) | 16.1 (A) | 14.93 |
| proteasome (prosome, macropain) inhibitor subunit1 | BG029917 | 426.1 (P) | 28.4 (A) | 14.93 |
| Cysteine-rich protein 1 (intestinal) | NM_001311 | 583 (P) | 31 (A) | 14.93 |
| Epithelial membrane protein 1 | NM_001423 | 154.2 (P) | 10.6 (A) | 14.93 |
| EST | R06655 | 758.5 (P) | 32 (A) | 14.93 |
| Heterogeneous nuclear ribonucleoprotein AB | NM_004499 | 638.7 (P) | 33.7 (A) | 14.93 |
| HIV-1 TAR RNA binding protein | L22453 | 4646.4 (P) | 263 (A) | 14.93 |
| Calpain 4, small subunit (30 kDa) | NM_001749 | 866.5 (P) | 44.7 (A) | 16 |
| Archain 1 | NM_001655 | 635.5 (P) | 12.6 (A) | 16 |
| RuvB (E coli homolog)-like 2 | NM_006666 | 451.2 (P) | 31.5 (A) | 16 |
| peptidylprolyl isomerase B (cyclophilin B) | NM_000942 | 741.1 (P) | 24.4 (A) | 17.15 |
| Beaded filament structural protein 1, filensin | NM_001195 | 6597.1 (P) | 185.7 (A) | 17.15 |
| HSPC165 protein | NM_014185 | 393.4 (P) | 22.3 (A) | 17.15 |
| chimerin (chimaerin) 1 | BF339445 | 709.7 (P) | 44.2 (A) | 17.15 |
| Hypothetical protein FLJ11798 | NM_024907 | 510.9 (P) | 16.8 (A) | 18.38 |
| Saposin proteins A-D | M32221 | 554.2 (P) | 25.1 (A) | 18.38 |
| Uncharacterized hematopoietic stemprogenitor cells protein MDS032 | NM_018467 | 335.2 (P) | 25 (A) | 19.7 |
| Eukaryotic translation elongtion factor 2 | NM_001961 | 4841 (P) | 138.5 (A) | 21.11 |
| Lysyl oxidase-like 1 | NM_005576 | 824.8 (P) | 18.7 (A) | 21.11 |
| transketolase | BF696840 | 1557.5 (P) | 27 (A) | 22.63 |
| Alpha A crystallin | U66584 | 10945.5 (P) | 264.4 (P) | 22.63 |
| Crystallin beta B3 | NM_004076 | 2515.8 (P) | 64.9 (A) | 22.63 |
| glycoprotein M6A | BF939489 | 360.8 (P) | 19.6 (A) | 24.25 |
| Growth arrest and DNA-damage-inducible, alpha | NM_001924 | 1548.3 (P) | 31 (A) | 24.25 |
| pUb-R5 | AB033605 | 867 (P) | 30 (A) | 24.25 |
| solute carrier family 25 (mitochondrial carrier; adenine nucleotide translocator), member 6 | AA916851 | 1736.5 (P) | 20.8 (A) | 25.99 |
| ribosomal protein, large, P0 | AA555113 | 2885.2 (P) | 73.5 (A) | 27.86 |
| Lens intrinsic membrane protein 2 | NM_030657 | 7106.4 (P) | 144.3 (A) | 27.86 |
| Crystallin beta A3 | NM_005208 | 16082.8 (P) | 582.6 (M) | 27.86 |
| Microvascular endothelial differentiation gene 1 | NM_012328 | 110.3 (A) | 6.4 (A) | 27.86 |
| Crystallin gamma D | NM_006891 | 6226.2 (P) | 204.5 (A) | 29.86 |
| Ribosomal protein S9 | NM_001013 | 5469.9 (P) | 207.3 (A) | 32 |
| Phosphoglycerate kinase | S81916 | 477.3 (P) | 23.8 (A) | 42.22 |
| matrix Gla protein | AI653730 | 546.9 (P) | 14.5 (A) | 42.22 |
| Intersectin short isoform | AF114488 | 119.3 (P) | 4.3 (A) | 45.25 |
| Hypothetical protein PRO2577 | NM_018630 | 133.3 (P) | 6.8 (A) | 45.25 |
| Lengsin | NM_016571 | 2135 (P) | 17.1 (A) | 51.98 |
| Crystallin beta A2 | NM_005209 | 14598.1 (P) | 108.3 (A) | 55.72 |
| Crystallin beta B1 | NM_001887 | 7181.6 (P) | 68.5 (A) | 119.43 |
| Heat shock 27 kD protein 1 | NM_001540 | 3620.6 (P) | 28.2 (A) | 128 |
| Crystallin beta A4 | NM_001886 | 17523.3 (P) | 118.1 (A) | 168.9 |
In the third and fourth columns, the abbreviations P is present, statistically greater than background intensity values, A is absent, not statistically different than background intensity values, and M is marginal, possibly different than background intensity values.
Semi-quantitative RT-PCR confirmation
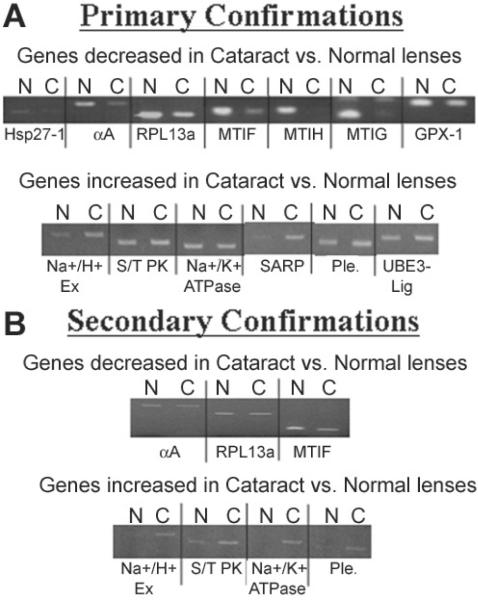
In order to confirm the accuracy of the microarray data, semi-quantitative RT-PCR was conducted with the original RNA samples used for the microarray experiments and 2 other sets of separately prepared cataract and clear lens RNA samples. Thirteen genes that were either increased or decreased by 2 fold or greater in cataracts were first examined using the same RNA samples that were used for the microarray studies. These included Na+/H+ exchanger isoform II (6.50 fold), serine/threonine protein kinase (3.73 fold), Na+/K+ ATPase (8.00 fold), secreted apoptosis related protein 2 (6.06 fold), pleiotrophin (7.46 fold), and E3-ubiquitin ligase (4.59 fold) which all exhibited increased expression in cataracts according to the microarray data. Heat shock protein 27-1 (128 fold), αA-crystallin (22.63 fold), ribosomal protein large subunit 13a (2.64 fold), metallothionein IF (5.66 fold), metallothionein IH (3.48 fold), metallothionein IG (3.73 fold), and glutathione peroxidase-1 (4.92 fold) all exhibited decreased expression in cataracts according to the microarray data.
Eleven of the 13 genes examined followed the same trends in gene expression as demonstrated by the microarray study (Figure 3A) using the original RNA samples including Na+/H+ exchanger isoform II, secreted apoptosis related protein 2, pleiotrophin, E3-ubiquitin ligase, heat shock protein 27-1, αA-crystallin, ribosomal protein large subunit 13a, metallothionein IF, metallothionein IH, metallothionein IG, and glutathione peroxidase-1. The two genes that did not follow the same trends in gene expression as demonstrated by the microarray data were serine/threonine protein kinase and Na+/K+ ATPase (Figure 3A).
Figure 3.
RT-PCR confirmation of gene expression differences. RTPCR confirmation of gene expression differences detected by microarray hybridization between cataract (C) and normal (N) lens epithelia. The expression levels of indicated genes were confirmed by RT-PCR. A: Genes examined using the same cataract and clear lens RNAs analyzed by microarray hybridization. B: Genes examined using separately prepared cataract and clear lens RNA samples.
A second sample of RNA was prepared from an additional 50 cataract and 10 age-matched clear lenses. Due to the limited amount of RNA recovered from the second population of cataracts, seven of the 13 aforementioned genes (including the two that did not confirm the microarray data using the first samples of RNA) were re-examined using the new samples of RNA. Of these, five of the seven genes exhibited similar trends as detected in the microarray analysis including Na+/H+ exchanger isoform II, pleiotrophin, metallothionein IF, serine/threonine protein kinase, and Na+/K+ ATPase (Figure 3B). The two genes that did not reconfirm in the second sample of RNA were αA-crystallin and ribosomal protein large subunit 13a.
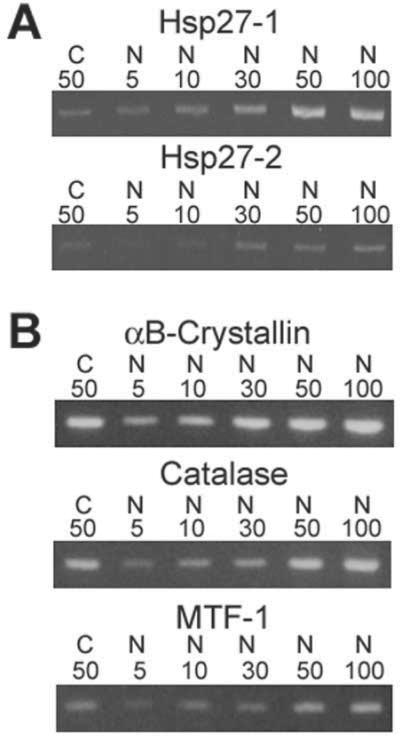
In order to further confirm the trends exhibited by the microarray study and to demonstrate that the PCR cycles used are within the linear range, we examined two particular genes of interest in a third sample of RNA prepared from another 50 cataract and 10 age-matched clear lenses. Consistent with the microarray data, Hsp 27 form 1 and 2 exhibited decreased expression in cataract relative to clear lenses using a fixed amount of cataract RNA (50 ng) and 5 different amounts of clear lens RNA (5, 10, 30, 50, and 100 ng, Figure 4). Heat shock protein 27 form 1 was decreased in cataract relative to clear lenses by approximately 10 fold while heat shock protein 27 form 2 was decreased in cataract by approximately 2 fold. Using this same sample of RNA we examined the expression levels of 3 genes (catalase, MTF-1, and αB-crystallin) that were unaltered between cataracts and clear lenses according to the microarray data as a further control. All 3 of these genes exhibited identical expression levels between cataract and normal lens epithlia, as predicted by the microarray analysis (Figure 4).
Figure 4.
Further RT-PCR confirmation of selected gene expression differences and control genes. RT-PCR confirmation of gene expression differences for HSP27-1 and -2 (A) and 3 control genes whose expression levels should be equal between cataract (C) and normal (N) lens epithelia (B). The total amount of RNA (ng) used in each reaction is indicated.
Densitometric gel scanning of all of the semi-quantitative RT-PCR products described in Figure 3 and Figure 4 was also conducted to further evaluate the data (Table 3). Although all of the calculated fold changes do not exactly match those detected by the microarray hybridization data, they importantly follow the same general trends in gene expression revealed by the microarray data. These combined confirmations suggest that the gene expression trends revealed by microarray analysis are approximately 84% accurate.
Table 3.
Fold changes and densitometry values of RT-PCR confirmations of the microarray data
| Gene |
Fold change in cataracts according to microarray data |
Percent adjusted volume for normal lens RNA |
Percent adjusted volume for cataractous lens RNA |
Calculated densitometry fold change in cataracts |
||
|---|---|---|---|---|---|---|
| Control genes shown in Figure 4: | ||||||
| aB-Crystallin | No Change | 21.62 | 17.52 | Decreased | 1.23 Fold | |
| Catalase | No Change | 26.55 | 18.38 | Decreased | 1.44 Fold | |
| MTF-1 | No Change | 22.29 | 24.46 | Increased | 1.10 Fold | |
| Primary confirmations shown in Figure 3A: | ||||||
| Hsp27-1 | Decreased | 128.00 Fold | 72.35 | 27.65 | Decreased | 2.62 Fold |
| aA-Crystallin | Decreased | 22.63 Fold | 78.27 | 21.73 | Decreased | 3.60 Fold |
| RPL13a | Decreased | 2.46 Fold | 60.37 | 39.63 | Decreased | 1.52 Fold |
| Metallothionein IF | Decreased | 5.66 Fold | 74.11 | 25.89 | Decreased | 2.86 Fold |
| Metallothionein IH | Decreased | 3.48 Fold | 86.92 | 13.08 | Decreased | 6.65 Fold |
| Metallothionein IG | Decreased | 3.73 Fold | 86.05 | 13.95 | Decreased | 6.17 Fold |
| Glutathione Peroxidase 1 | Decreased | 4.92 Fold | 60.98 | 39.02 | Decreased | 1.56 Fold |
| Na+/H+ Exchanger II | Increased | 6.5 Fold | 17.2 | 82.8 | Increased | 4.81 Fold |
| Serine/Threonine Protein Kinase | Increased | 6.5 Fold | 40.96 | 59.04 | Increased | 1.44 Fold |
| Na+/K+ ATPase | Increased | 8.00 Fold | 52.73 | 47.27 | Decreased | 1.12 Fold |
| Secreted Apoptosis Related Protein | Increased | 6.06 Fold | 17.78 | 82.22 | Increased | 4.62 Fold |
| Pleiotrophin | Increased | 7.46 Fold | 33.67 | 66.33 | Increased | 1.97 Fold |
| E3-Ubiquitin Ligase | Increased | 4.59 Fold | 34.84 | 65.16 | Increased | 1.87 Fold |
| Secondary confirmations shown in Figure 3B: | ||||||
| aA-Crystallin | Decreased | 22.63 Fold | 57.21 | 42.79 | Decreased | 1.34 Fold |
| RPL13a | Decreased | 2.46 Fold | 54.29 | 45.71 | Decreased | 1.19 Fold |
| Metallothionein IF | Decreased | 5.66 Fold | 67.16 | 32.84 | Decreased | 2.05 Fold |
| Na+/H+ Exchanger II | Increased | 6.5 Fold | 6.99 | 93.01 | Increased | 13.31 Fold |
| Serine/Threonine Protein Kinase | Increased | 6.5 Fold | 22.86 | 77.14 | Increased | 3.37 Fold |
| Na+/K+ ATPase | Increased | 8.00 Fold | 2.03 | 97.97 | Increased | 48.26 Fold |
| Pleiotrophin | Increased | 7.46 Fold | 14.78 | 85.22 | Increased | 5.77 Fold |
| Tertiary confirmations shown in Figure 4: | ||||||
| HSP27-1 | Decreased | 128 Fold | 34.74 | 4.23 | Decreased | 8.21 Fold |
| HSP27-2 | Decreased | 5.28 Fold | 20.49 | 15.69 | Decreased | 1.31 Fold |
Densitometric analysis of the RT-PCR confirmations shown in Figure 3A,B and Figure 4.
Functional clustering analysis of differentially expressed transcripts
The set of genes that exhibited either increased or decreased expression levels of 2 fold or greater was analyzed for significant enrichment with respect to various categories of gene function using the EASE bioinformatics package. Categories enriched within the mRNAs increased or decreased at the 2 fold or greater level with an EASE score of less than 0.05 are shown in Figure 5, Figure 6, Figure 7, Figure 8, and listed in Table 4. Because many genes have more than one function and are involved in various pathways, many of the identified genes appear in multiple categories.
Figure 5.
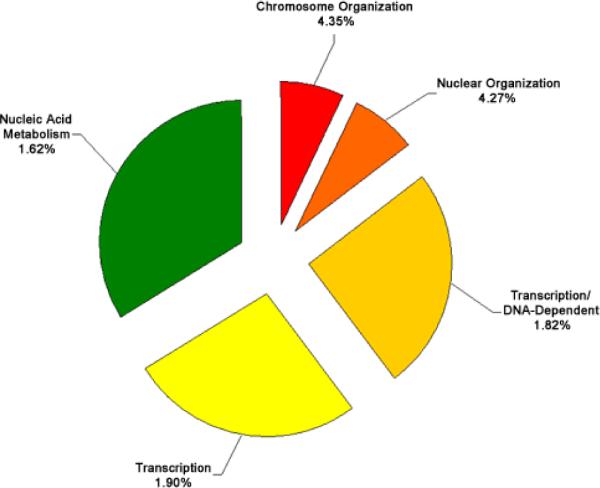
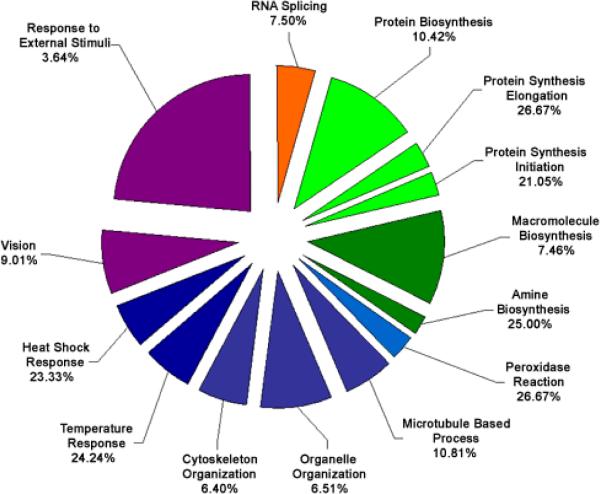
Functional cluster analysis of genes involved in biological processes which have increased expression levels in cataract versus clear lenses. Functional cluster analysis of genes involved in biological processes which have increased expression levels in cataract compared to clear lenses. The specific sub-categories of genes determined to be significantly altered using the statistical clustering program, EASE, are indicated. Percentages indicate the number of altered genes in each sub-category relative to their total representation on the microarray. Colors denote the approximate relative cellular location for which the genes in each sub-category function ranging from the nucleus to the plasma membrane (red to violet). Individual genes in each category are listed in Table 4. Pie piece size approximates the number of changed genes in each sub-category.
Figure 6.
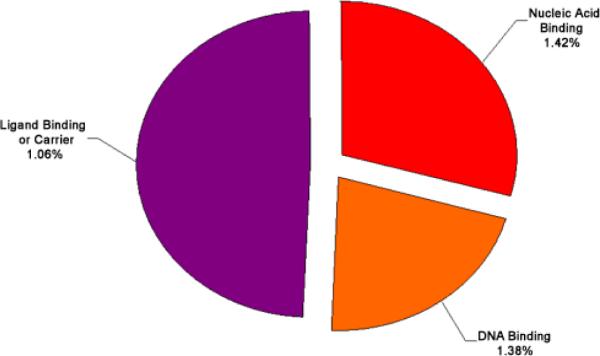
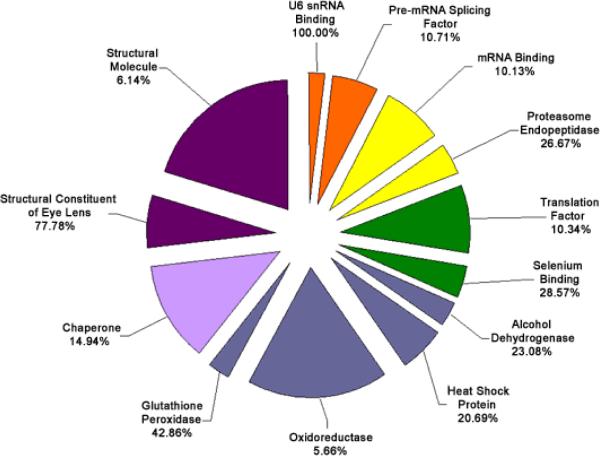
Functional cluster analysis of genes involved in molecular functions which have increased expression levels in cataract versus clear lenses. Functional cluster analysis of genes involved in molecular functions which have increased expression levels in cataract versus clear lenses. The specific sub-categories of genes determined to be significantly altered using the statistical clustering program, EASE, are indicated. Percentages indicate the number of altered genes in each sub-category relative to their total representation on the microarray. Colors denote the approximate relative cellular location for which the genes in each sub-category function ranging from the nucleus to the plasma membrane (red to violet). Individual genes in each category are listed in Table 4. Pie piece size approximates the number of changed genes in each sub-category.
Figure 7.
Functional cluster analysis of genes involved in biological processes which have decreased expression levels in cataract versus clear lenses. Functional cluster analysis of genes involved in biological processes which have decreased expression levels in cataract versus clear lenses. The specific sub-categories of genes determined to be significantly altered using the statistical clustering program, EASE, are indicated. Percentages indicate the number of altered genes in each sub-category relative to their total representation on the microarray. Colors denote the approximate relative cellular location for which the genes in each sub-category function ranging from the nucleus to the plasma membrane (red to violet). Individual genes in each category are listed in Table 4. Pie piece size approximates the number of changed genes in each sub-category.
Figure 8.
Functional cluster analysis of genes involved in molecular functions which have decreased expression levels in cataract versus clear lenses. Functional cluster analysis of genes involved in molecular functions which have decreased expression levels in cataract versus clear lenses. The specific sub-categories of genes determined to be significantly altered using the statistical clustering program, EASE, are indicated. Percentages indicate the number of altered genes in each sub-category relative to their total representation on the microarray. Colors denote the approximate relative cellular location for which the genes in each sub-category function ranging from the nucleus to the plasma membrane (red to violet). Individual genes in each category are listed in Table 4. Pie piece size approximates the number of changed genes in each sub-category.
Table 4.
Individual functionally clustered genes
| Increased In Cataract | |||
|---|---|---|---|
| Probe number |
Gene name |
Accession number |
Fold change |
| Biological Process | |||
| Chromosome organization | |||
| 200679 | high-mobility group (nonhistone chromosomal) protein 1 | NM_002128 | 4.5948 |
| 205062 | retinoblastoma-binding protein 1 (RBBP1) | NM_002892 | 4.9246 |
| 208859 | alpha thalassemiamental retardation syndrome X-linked | NM_000489 | 5.6569 |
| 209258 | chondroitin sulfate proteoglycan 6 (bamacan) | NM_005445 | 8.5742 |
| 209715 | heterochromatin protein homologue (HP1) | NM_012117 | 4.0000 |
| Nuclear organization | |||
| 200679 | high-mobility group (nonhistone chromosomal) protein 1 | NM_000489 | 4.5948 |
| 205062 | retinoblastoma-binding protein 1 (RBBP1) | NM_002892 | 4.9246 |
| 208859 | alpha thalassemiamental retardation syndrome X-linked | NM_000489 | 5.6569 |
| 209258 | chondroitin sulfate proteoglycan 6 (bamacan) | NM_005445 | 8.5742 |
| 209715 | heterochromatin protein homologue (HP1) | NM_012117 | 4.0000 |
| Transcription/DNA-dependent | |||
| 200679 | high-mobility group (nonhistone chromosomal) protein 1 | NM_002128 | 4.5948 |
| 201138 | Sjogren syndrome antigen B | NM_003142 | 6.4980 |
| 202173 | zinc finger protein 161 | NM_007146 | 6.9644 |
| 202600 | nuclear receptor interacting protein 1 | NM_003489 | 7.4643 |
| 202612 | cofactor required for Sp1 transcriptional activation, subunit 2 | NM_004229 | 16.0000 |
| 204771 | transcription termination factor, RNA polymerase I | NM_007344 | 4.0000 |
| 205062 | retinoblastoma-binding protein 1 (RBBP1) | NM_002892 | 4.9246 |
| 205070 | inhibitor of growth family, member 3 | NM_019071 | 4.2871 |
| 205443 | small nuclear RNA activating complex | NM_003082 | 6.9644 |
| 205596 | E3 ubiquitin ligase Smurf2 | NM_022739 | 4.5948 |
| 206848 | homeo box A7 | NM_006896 | 4.2871 |
| 208003 | nuclear factor of activated T-cells 5 | NM_006599 | 4.2871 |
| 208859 | alpha thalassemiamental retardation syndrome X-linked | NM_000489 | 5.6569 |
| 209088 | ubinuclein 1 | T70262 | 5.6569 |
| 210504 | erythroid-specific transcription factor | NM_006563 | 5.2780 |
| 212079 | myeloidlymphoid or mixed-lineage leukemia | NM_005933 | 19.6983 |
| 212492 | KIAA0876 protein | AW237172 | 6.9644 |
| Transcription | |||
| 200679 | high-mobility group (nonhistone chromosomal) protein 1 | NM_002128 | 4.5948 |
| 201138 | Sjogren syndrome antigen B | NM_003142 | 6.4980 |
| 201606 | nuclear phosphoprotein | NM_007062 | 5.2780 |
| 202173 | zinc finger protein 161 | NM_007146 | 6.9644 |
| 202600 | nuclear receptor interacting protein 1 | NM_003489 | 7.4643 |
| 202612 | cofactor required for Sp1 transcriptional activation, subunit 2 | NM_004229 | 16.0000 |
| 204771 | transcription termination factor, RNA polymerase I | NM_007344 | 4.0000 |
| 205062 | retinoblastoma-binding protein 1 (RBBP1) | NM_002892 | 4.9246 |
| 205070 | inhibitor of growth family, member 3 | NM_019071 | 4.2871 |
| 205443 | small nuclear RNA activating complex | NM_003082 | 6.9644 |
| 205596 | E3 ubiquitin ligase Smurf2 | NM_022739 | 4.5948 |
| 206848 | homeo box A7 | NM_006896 | 4.2871 |
| 208003 | nuclear factor of activated T-cells 5 | NM_006599 | 4.2871 |
| 208859 | alpha thalassemiamental retardation syndrome X-linked | NM_000489 | 5.6569 |
| 209088 | ubinuclein 1 | T70262 | 5.6569 |
| 210504 | erythroid-specific transcription factor | NM_006563 | 5.2780 |
| 212079 | myeloidlymphoid or mixed-lineage leukemia | NM_005933 | 19.6983 |
| 212492 | KIAA0876 protein | AW237172 | 6.9644 |
| Nucleic acid metabolism | |||
| 200679 | high-mobility group (nonhistone chromosomal) protein 1 | NM_002128 | 4.5948 |
| 201138 | Sjogren syndrome antigen B | NM_003142 | 6.4980 |
| 201606 | nuclear phosphoprotein | NM_007062 | 5.2780 |
| 202173 | zinc finger protein 161 | NM_007146 | 6.9644 |
| 202600 | nuclear receptor interacting protein 1 | NM_003489 | 7.4643 |
| 202612 | cofactor required for Sp1 transcriptional activation, subunit 2 | NM_004229 | 16.0000 |
| 202905 | Nijmegen breakage syndrome 1 (nibrin) | NM_002485 | 6.9644 |
| 204771 | transcription termination factor, RNA polymerase I | NM_007344 | 4.0000 |
| 205062 | retinoblastoma-binding protein 1 (RBBP1) | NM_002892 | 4.9246 |
| 205070 | inhibitor of growth family, member 3 | NM_019071 | 4.2871 |
| 205443 | small nuclear RNA activating complex | NM_003082 | 6.9644 |
| 205596 | E3 ubiquitin ligase Smurf2 | NM_022739 | 4.5948 |
| 206848 | homeo box A7 | NM_006896 | 4.2871 |
| 208003 | nuclear factor of activated T-cells 5 | NM_006599 | 4.2871 |
| 208835 | cisplatin resistance-associated overexpressed protein | AW089673 | 4.2871 |
| 208859 | alpha thalassemiamental retardation syndrome X-linked | NM_000489 | 5.6569 |
| 209024 | NS1-associated protein 1 | AF037448 | 4.9246 |
| 209088 | ubinuclein 1 | T70262 | 5.6569 |
| 209579 | methyl-CpG binding domain protein 4 | NM_003925 | 4.2871 |
| 209715 | heterochromatin protein homologue (HP1) | NM_012117 | 4.0000 |
| 210504 | erythroid-specific transcription factor | NM_006563 | 5.2780 |
| 212079 | myeloidlymphoid or mixed-lineage leukemia | NM_005933 | 19.6983 |
| 212492 | KIAA0876 protein | AW237172 | 6.9644 |
| Molecular Function | |||
| Nucleic acid binding | |||
| 200679 | high-mobility group (nonhistone chromosomal) protein 1 | NM_002128 | 4.5948 |
| 201138 | Sjogren syndrome antigen B | NM_003142 | 6.4980 |
| 201635 | fragile X mental retardation, autosomal homolog 1 | NM_005087 | 4.5948 |
| 202173 | zinc finger protein 161 | NM_007146 | 6.9644 |
| 202612 | cofactor required for Sp1 transcriptional activation, subunit 2 | NM_004229 | 16.0000 |
| 202905 | Nijmegen breakage syndrome 1 (nibrin) | NM_002485 | 6.9644 |
| 203567 | ring finger protein 15 | NM_006355 | 19.6983 |
| 204771 | transcription termination factor, RNA polymerase I | NM_007344 | 4.0000 |
| 205062 | retinoblastoma-binding protein 1 (RBBP1) | NM_002892 | 4.9246 |
| 205070 | inhibitor of growth family, member 3 | NM_019071 | 4.2871 |
| 206848 | homeo box A7 | NM_006896 | 4.2871 |
| 208003 | nuclear factor of activated T-cells 5 | NM_006599 | 4.2871 |
| 208325 | lymphoid blast crisis oncogene | NM_006738 | 4.2871 |
| 208624 | eukaryotic translation initiation factor 4 gamma | AF104913 | 6.4980 |
| 208859 | alpha thalassemiamental retardation syndrome X-linked | NM_000489 | 5.6569 |
| 209024 | NS1-associated protein 1 | AF037448 | 4.9246 |
| 209088 | ubinuclein 1 | T70262 | 5.6569 |
| 209579 | methyl-CpG binding domain protein 4 | NM_003925 | 4.2871 |
| 209715 | heterochromatin protein homologue (HP1) | NM_012117 | 4.0000 |
| 210504 | erythroid-specific transcription factor | NM_006563 | 5.2780 |
| 212079 | myeloidlymphoid or mixed-lineage leukemia | NM_005933 | 19.6983 |
| 212492 | KIAA0876 protein | AW237172 | 6.9644 |
| Ligand binding or carrier | |||
| 200679 | high-mobility group (nonhistone chromosomal) protein 1 | NM_002128 | 4.5948 |
| 201138 | Sjogren syndrome antigen B | NM_003142 | 6.4980 |
| 201242 | ATPase, Na+K+ transporting, beta 1 | BC000006 | 8.0000 |
| 201635 | fragile X mental retardation, autosomal homolog 1 | NM_005087 | 4.5948 |
| 201711 | RAN binding protein 2 | NM_006267 | 4.5948 |
| 201752 | adducin 3 (gamma) | NM_019903 | 6.0629 |
| 201777 | KIAA0494 gene product | NM_014774 | 17.1484 |
| 202082 | SEC14 | NM_003003 | 4.9246 |
| 202118 | copine III | NM_003909 | 6.9644 |
| 202173 | zinc finger protein 161 | NM_007146 | 6.9644 |
| 202600 | nuclear receptor interacting protein 1 | NM_003489 | 7.4643 |
| 202612 | cofactor required for Sp1 transcriptional activation, subunit 2 | NM_004229 | 16.0000 |
| 202831 | glutathione peroxidase 2 | NM_002083 | 8.0000 |
| 202905 | Nijmegen breakage syndrome 1 (nibrin) | NM_002485 | 6.9644 |
| 203567 | ring finger protein 15 | NM_006355 | 19.6983 |
| 204771 | transcription termination factor, RNA polymerase I | NM_007344 | 4.0000 |
| 205062 | retinoblastoma-binding protein 1 (RBBP1) | NM_002892 | 4.9246 |
| 205070 | inhibitor of growth family, member 3 | NM_019071 | 4.2871 |
| 205809 | Wiskott-Aldrich syndrome-like | NM_003941 | 14.9285 |
| 206848 | homeo box A7 | NM_006896 | 4.2871 |
| 207152 | neurotrophic tyrosine kinase, receptor, type 2 | NM_006180 | 4.0000 |
| 208003 | nuclear factor of activated T-cells 5 | NM_006599 | 4.2871 |
| 208325 | lymphoid blast crisis oncogene | NM_006738 | 4.2871 |
| 208624 | eukaryotic translation initiation factor 4 gamma | AF104913 | 6.4980 |
| 208859 | alpha thalassemiamental retardation syndrome X-linked | NM_000489 | 5.6569 |
| 209024 | NS1-associated protein 1 | AF037448 | 4.9246 |
| 209088 | ubinuclein 1 | T70262 | 5.6569 |
| 209258 | chondroitin sulfate proteoglycan 6 (bamacan) | NM_005445 | 8.5742 |
| 209466 | pleiotrophin | M57399 | 7.4643 |
| 209579 | methyl-CpG binding domain protein 4 | NM 003925 | 4.2871 |
| 209715 | heterochromatin protein homologue (HP1) | NM_012117 | 4.0000 |
| 210504 | erythroid-specific transcription factor | NM_006563 | 5.2780 |
| 212079 | myeloidlymphoid or mixed-lineage leukemia | NM_005933 | 19.6983 |
| 212492 | KIAA0876 protein | AW237172 | 6.9644 |
| 212926 | KIAA0594 protein | AW183677 | 9.1896 |
| 214464 | Ser-Thr protein kinase | NM_003607 | 6.4980 |
| 214933 | calcium channel, voltage-dependent, PQ type, alpha 1A | AA769818 | 4.9246 |
| DNA binding | |||
| 200679 | high-mobility group (nonhistone chromosomal) protein 1 | NM_002128 | 4.5948 |
| 202173 | zinc finger protein 161 | NM_007146 | 6.9644 |
| 202612 | cofactor required for Sp1 transcriptional activation, subunit 2 | NM_004229 | 16.0000 |
| 202905 | Nijmegen breakage syndrome 1 (nibrin) | NM_002485 | 6.9644 |
| 204771 | transcription termination factor, RNA polymerase I | NM_007344 | 4.0000 |
| 205062 | retinoblastoma-binding protein 1 (RBBP1) | NM_002892 | 4.9246 |
| 205070 | inhibitor of growth family, member 3 | NM_019071 | 4.2871 |
| 206848 | homeo box A7 | NM_006896 | 4.2871 |
| 208003 | nuclear factor of activated T-cells 5 | NM_006599 | 4.2871 |
| 208859 | alpha thalassemiamental retardation syndrome X-linked | NM_000489 | 5.6569 |
| 209088 | ubinuclein 1 | T70262 | 5.6569 |
| 209579 | methyl-CpG binding domain protein 4 | NM_003925 | 4.2871 |
| 209715 | heterochromatin protein homologue (HP1) | NM_012117 | 4.0000 |
| 210504 | erythroid-specific transcription factor | NM_006563 | 5.2780 |
| 212079 | myeloidlymphoid or mixed-lineage leukemia | NM_005933 | 19.6983 |
| 212492 | KIAA0876 protein | AW237172 | 6.9644 |
| Decreased In Cataract | |||
|---|---|---|---|
| Probe number |
Gene name |
Accession number |
Fold change |
| Biological Process | |||
| RNA splicing | |||
| 200826 | small nuclear ribonucleoprotein D2 polypeptide | NM_004597 | 4.5948 |
| 201698 | splicing factor, arginineserine-rich 9 | NM_003769 | 6.0629 |
| 202567 | small nuclear ribonucleoprotein D3 polypeptide | NM_004175 | 4.5948 |
| 204559 | U6 snRNA-associated Sm-like protein LSm7 | NM_016199 | 10.5561 |
| 208880 | putative mitochondrial outer membrane protein import receptor | AB019219 | 4.5948 |
| 209449 | SMX5-like protein | AF196468 | 5.6569 |
| Protein biosynthesis | |||
| 200005 | eukaryotic translation initiation factor 3, subunit 7 | NM_003753 | 4.9246 |
| 200689 | eukaryotic translation elongation factor 1 gamma | NM_001404 | 10.5561 |
| 201064 | poly(A)-binding protein, cytoplasmic 4 | NM_003819 | 6.9644 |
| 201263 | threonyl-tRNA synthetase | NM_003191 | 6.0629 |
| 201530 | eukaryotic translation initiation factor 4A, isoform 1 | NM_001416 | 4.2871 |
| 201632 | eukaryotic translation initiation factor 2B, subunit 1 | NM_001414 | 6.0629 |
| 201841 | heat shock 27 kDa protein 1 | NM_001540 | 128.0000 |
| 202021 | SUI1 isolog | AF083441 | 4.5948 |
| 202042 | histidyl-tRNA synthetase | NM_002109 | 12.9960 |
| 203113 | eukaryotic translation elongation factor 1 delta | NM_001960 | 6.4980 |
| 203725 | growth arrest and DNA-damage-inducible, alpha | NM_001924 | 24.2515 |
| 204102 | eukaryotic translation elongation factor 2 | NM_001961 | 21.1121 |
| 208856 | ribosomal protein, large, P0 | BC003655 | 4.5948 |
| 208887 | eukaryotic translation initiation factor 3, subunit 4 | BC000733 | 8.0000 |
| 210213 | translation initiation factor 6 | AF022229 | 4.0000 |
| Protein synthesis elongation | |||
| 200689 | eukaryotic translation elongation factor 1 gamma | NM_001404 | 10.5561 |
| 203113 | eukaryotic translation elongation factor 1 delta | NM_001960 | 6.4980 |
| 204102 | eukaryotic translation elongation factor 2 | NM_001961 | 21.1121 |
| 208856 | ribosomal protein, large, P0 | BC003655 | 4.5948 |
| Protein synthesis initiation | |||
| 201530 | eukaryotic translation initiation factor 4A, isoform 1 | NM_001416 | 4.2871 |
| 201632 | eukaryotic translation initiation factor 2B, subunit 1 | NM_001414 | 6.0629 |
| 202021 | SUI1 isolog | AF083441 | 4.5948 |
| 210213 | translation initiation factor 6 | AF022229 | 4.0000 |
| Macromolecule biosynthesis | |||
| 200005 | eukaryotic translation initiation factor 3, subunit 7 | NM_003753 | 4.9246 |
| 200689 | eukaryotic translation elongation factor 1 gamma | NM_001404 | 10.5561 |
| 201064 | poly(A)-binding protein, cytoplasmic 4 | NM_003819 | 6.9644 |
| 201263 | threonyl-tRNA synthetase | NM_003191 | 6.0629 |
| 201530 | eukaryotic translation initiation factor 4A, isoform 1 | NM_001416 | 4.2871 |
| 201632 | eukaryotic translation initiation factor 2B, subunit 1 | NM_001414 | 6.0629 |
| 201841 | heat shock 27 kDa protein 1 | NM_001540 | 128.0000 |
| 202021 | SUI1 isolog | AF083441 | 4.5948 |
| 202042 | histidyl-tRNA synthetase | NM_002109 | 12.9960 |
| 203113 | eukaryotic translation elongation factor 1 delta | NM_001960 | 6.4980 |
| 203725 | growth arrest and DNA-damage-inducible, alpha | NM_001924 | 24.2515 |
| 204102 | eukaryotic translation elongation factor 2 | NM_001961 | 21.1121 |
| 208856 | ribosomal protein, large, P0 | BC003655 | 4.5948 |
| 208887 | eukaryotic translation initiation factor 3, subunit 4 | BC000733 | 8.0000 |
| 210213 | translation initiation factor 6 | AF022229 | 4.0000 |
| Amine biosynthesis | |||
| 200790 | ornithine decarboxylase 1 | NM_002539 | 4.9246 |
| 201772 | antizyme inhibitor | NM_015878 | 6.9644 |
| 207621 | phosphatidylethanolamine N-methyltransferase | NM_007169 | 6.0629 |
| Peroxidase reaction | |||
| 200736 | glutathione peroxidae 1 (GPX1) | NM_000581 | 4.9246 |
| 201106 | glutathione peroxidase 4 (phospholipid hydroperoxidase) | NM_002085 | 4.2871 |
| 201348 | glutathione peroxidase 3 (GPX3) | NM_002084 | 11.3137 |
| 212013 | KIAA0230 gene | D86983 | 12.1257 |
| Microtubule-based process | |||
| 200712 | microtubule-associated protein, RPEB family, member 1 | AI633566 | 10.5561 |
| 200750 | GTP binding protein | AF054183 | 4.9246 |
| 203690 | spindle pole body protein (GCP3) | NM_006322 | 9.1896 |
| 204398 | microtubule-associated protein like echinoderm EMAP | NM_012155 | 4.9246 |
| 205191 | retinitis pigmentosa 2 | NM_006915 | 12.1257 |
| 208786 | microtubule-associated proteins 1A1B light chain 3 | AF183417 | 5.6569 |
| 208977 | tubulin, beta, 2 | BC004188 | 6.0629 |
| 209191 | Similar to tubulin, beta, 4 | BC002654 | 7.4643 |
| Organelle organization | |||
| 200712 | microtubule-associated protein, RPEB family, member 1 | AI633566 | 10.5561 |
| 200750 | GTP binding protein | AF054183 | 4.9246 |
| 200866 | saposin proteins A-D | M32221 | 18.3800 |
| 201707 | peroxisomal farnesylated protein | NM_002857 | 4.0000 |
| 201821 | translocase of inner mitochondrial membrane 17 | BC004439 | 9.1896 |
| 203690 | spindle pole body protein (GCP3) | NM_006322 | 9.1896 |
| 204398 | microtubule-associated protein like echinoderm EMAP | NM_012155 | 4.9246 |
| 205191 | retinitis pigmentosa 2 | NM_006915 | 12.1257 |
| 208786 | microtubule-associated proteins 1A1B light chain 3 | AF183417 | 5.6569 |
| 208977 | tubulin, beta, 2 | BC004188 | 6.0629 |
| 209191 | Similar to tubulin, beta, 4 | BC002654 | 7.4643 |
| Cytoskeleton organization | |||
| 200712 | microtubule-associated protein, RPEB family, member 1 | AI633566 | 10.5561 |
| 200750 | GTP binding protein | AF054183 | 4.9246 |
| 203690 | spindle pole body protein (GCP3) | NM_006322 | 9.1896 |
| 204398 | microtubule-associated protein like echinoderm EMAP | NM_012155 | 4.9246 |
| 205191 | retinitis pigmentosa 2 | NM_006915 | 12.1257 |
| 208786 | microtubule-associated proteins 1A1B light chain 3 | AF183417 | 5.6569 |
| 208977 | tubulin, beta, 2 | BC004188 | 6.0629 |
| 209191 | Similar to tubulin, beta, 4 | BC002654 | 7.4643 |
| Temperature response | |||
| 200064 | isolate Liv chaperone protein HSP90 beta | AF275719 | 4.9246 |
| 200664 | DnaJ (Hsp40) homolog, subfamily B, member 1 | BG537255 | 6.4980 |
| 200797 | myeloid cell leukemia sequence 1 (BCL2-related) | AI275690 | 4.0000 |
| 200800 | heat shock 70 kDa protein 1A | NM_005345 | 4.0000 |
| 201161 | cold shock domain protein A | NM_003651 | 5.6569 |
| 201841 | heat shock 27 kDa protein 1 | NM_001540 | 128.0000 |
| 202581 | heat shock 70 kDa protein 1B | NM_005346 | 5.6569 |
| 205824 | heat shock 27 kDa protein 2 | NM_001541 | 5.2780 |
| Heat shock response | |||
| 200064 | isolate Liv chaperone protein HSP90 beta | AF275719 | 4.9246 |
| 200664 | DnaJ (Hsp40) homolog, subfamily B, member 1 | BG537255 | 6.4980 |
| 200797 | myeloid cell leukemia sequence 1 (BCL2-related) | AI275690 | 4.0000 |
| 200800 | heat shock 70 kDa protein 1A | NM_005345 | 4.0000 |
| 201841 | heat shock 27 kDa protein 1 | NM_001540 | 128.0000 |
| 202581 | heat shock 70 kDa protein 1B | NM_005346 | 5.6569 |
| 205824 | heat shock 27 kDa protein 2 | NM_001541 | 5.2780 |
| Vision | |||
| 201563 | L-iditol-2 dehydrogenase | L29008 | 12.1257 |
| 201842 | EGF-containing fibulin-like extracellular matrix protein 1 | AI826799 | 4.9246 |
| 202766 | fibrillin 1 | NM_000138 | 5.2780 |
| 204398 | microtubule-associated protein like echinoderm EMAP | NM_012155 | 4.9246 |
| 205191 | retinitis pigmentosa 2 | NM_006915 | 12.1257 |
| 206777 | beta B2 crystallin | NM_000496 | 6.4980 |
| 206843 | beta A4 crystallin | NM_001886 | 168.8970 |
| 207399 | phakinin, beaded filament structural protein 2 | NM_003571 | 12.9960 |
| 207532 | gamma D crystallin | NM_006891 | 29.8571 |
| 207685 | beta B3 crystallin | NM_004076 | 22.6274 |
| Response to external stimulus | |||
| 200064 | isolate Liv chaperone protein HSP90 beta | AF275719 | 4.9246 |
| 200664 | DnaJ (Hsp40) homolog, subfamily B, member 1 | BG537255 | 6.4980 |
| 200797 | myeloid cell leukemia sequence 1 (BCL2-related) | AI275690 | 4.0000 |
| 200800 | heat shock 70 kDa protein 1A | NM_005345 | 4.0000 |
| 201064 | poly(A)-binding protein, cytoplasmic 4 | NM_003819 | 6.9644 |
| 201161 | cold shock domain protein A | NM_003651 | 5.6569 |
| 201315 | interferon induced transmembrane protein 2 | NM_006435 | 4.5948 |
| 201348 | glutathione peroxidase 3 (GPX3) | NM 002084 | 11.3137 |
| 201563 | L-iditol-2 dehydrogenase | L29008 | 12.1257 |
| 201841 | heat shock 27 kDa protein 1 | NM_001540 | 128.0000 |
| 201842 | EGF-containing fibulin-like extracellular matrix protein 1 | AI826799 | 4.9246 |
| 201891 | beta-2-microglobulin | NM_004048 | 4.2871 |
| 202581 | heat shock 70 kDa protein 1B | NM_005346 | 5.6569 |
| 202727 | interferon gamma receptor 1 | NM_000416 | 4.0000 |
| 202766 | fibrillin 1 | NM_000138 | 5.2780 |
| 203725 | growth arrest and DNA-damage-inducible, alpha | NM_001924 | 24.2515 |
| 203921 | carbohydrate (chondroitin 6keratan) sulfotransferase 2 | NM_004267 | 4.5948 |
| 204398 | microtubule-associated protein like echinoderm EMAP | NM_012155 | 4.9246 |
| 205081 | cysteine-rich protein 1 | NM_001311 | 14.9285 |
| 205191 | retinitis pigmentosa 2 | NM_006915 | 12.1257 |
| 205824 | heat shock 27 kDa protein 2 | NM_001541 | 5.2780 |
| 206777 | beta B2 crystallin | NM_000496 | 6.4980 |
| 206843 | beta A4 crystallin | NM_001886 | 168.8970 |
| 207399 | phakinin, beaded filament structural protein 2 | NM_003571 | 12.9960 |
| 207532 | gamma D crystallin | NM_006891 | 29.8571 |
| 207685 | beta B3 crystallin | NM_004076 | 22.6274 |
| 208650 | CD24 antigen | BG327863 | 4.9246 |
| 208791 | complement cytolysis inhibitor | M25915 | 4.2871 |
| 208910 | pre-mRNA splicing factor 2 p32 subunit | L04636 | 7.4643 |
| 209189 | v-fos FBJ murine osteosarcoma viral oncogene homolog | BC004490 | 12.1257 |
| 212013 | KIAA0230 gene | D86983 | 12.1257 |
| 216598 | monocyte chemotactic protein | S69738 | 6.4980 |
| Molecular Function | |||
| U6 snRNA binding | |||
| 204559 | U6 snRNA-associated Sm-like protein LSm7 | NM_016199 | 10.5561 |
| 209449 | SMX5-like protein | AF196468 | 5.6569 |
| Pre-mRNA splicing factor | |||
| 200826 | small nuclear ribonucleoprotein D2 polypeptide | NM_004597 | 4.5948 |
| 201698 | splicing factor, arginineserine-rich 9 | NM_003769 | 6.0629 |
| 202567 | small nuclear ribonucleoprotein D3 polypeptide | NM_004175 | 4.5948 |
| 204559 | U6 snRNA-associated Sm-like protein LSm7 | NM_016199 | 10.5561 |
| 208880 | putative mitochondrial outer membrane protein import receptor | AB019219 | 4.5948 |
| 209449 | U6 snRNA-associated Sm-like protein | AF196468 | 5.6569 |
| mRNA binding | |||
| 200826 | small nuclear ribonucleoprotein D2 polypeptide | NM_004597 | 4.5948 |
| 201064 | poly(A)-binding protein, cytoplasmic 4 | NM_003819 | 6.9644 |
| 201530 | eukaryotic translation initiation factor 4A, isoform 1 | NM_001416 | 4.2871 |
| 201698 | splicing factor, arginineserine-rich 9 | NM_003769 | 6.0629 |
| 202567 | small nuclear ribonucleoprotein D3 polypeptide | NM_004175 | 4.5948 |
| 204559 | U6 snRNA-associated Sm-like protein LSm7 | NM_016199 | 10.5561 |
| 208880 | putative mitochondrial outer membrane protein import receptor | AB019219 | 4.5948 |
| 209449 | U6 snRNA-associated Sm-like protein | AF196468 | 5.6569 |
| Proteasome endopeptidase | |||
| 200786 | proteasome (prosome,macropain)subunit,beta type, 1 | NM_002799 | 6.0629 |
| 200876 | proteasome (prosome,macropain)subunit,beta type, 1 | NM_002793 | 8.5742 |
| 201532 | proteasome (prosome,macropain)subunit,alpa type, 3 | NM_002788 | 5.6569 |
| 202243 | proteasome (prosome,macropain)subunit,beta type, 4 | NM_002796 | 5.2780 |
| Translation factor | |||
| 200005 | eukaryotic translation initiation factor 3, subunit 7 | NM_003753 | 4.9246 |
| 200689 | eukaryotic translation elongation factor 1 gamma | NM_001404 | 10.5561 |
| 201530 | eukaryotic translation initiation factor 4A, isoform 1 | NM_001416 | 4.2871 |
| 201632 | eukaryotic translation initiation factor 2B, subunit 1 | NM_001414 | 6.0629 |
| 202021 | SUI1 isolog | AF083441 | 4.5948 |
| 203113 | eukaryotic translation elongation factor 1 delta | NM_001960 | 6.4980 |
| 204102 | eukaryotic translation elongation factor 2 | NM_001961 | 21.1121 |
| 208887 | eukaryotic translation initiation factor 3, subunit 4 | BC000733 | 8.0000 |
| 210213 | translation initiation factor 6 | AF022229 | 4.0000 |
| Selenium binding | |||
| 200736 | glutathione peroxidase 1 | NM_000581 | 4.9246 |
| 201106 | glutathione peroxidase 4 (phospholipid hydroperoxidase) | NM_002085 | 4.2871 |
| 201194 | selenoprotein w, 1 (SEPW1) | NM_003009 | 6.4980 |
| 201348 | glutathione peroxidase 3 (GPX3) | NM_002084 | 11.3137 |
| Alcohol dehydrogenase | |||
| 201563 | L-iditol-2 dehydrogenase | L29008 | 12.1257 |
| 210609 | guinone oxidoreductase homolog | BC000474 | 11.3137 |
| 213540 | beta3-Galactosyltransferase | AL031228 | 6.0629 |
| Heat shock protein | |||
| 200064 | isolate Liv chaperone protein HSP90 beta | AF275719 | 4.9246 |
| 200664 | DnaJ (Hsp40) homolog, subfamily B, member 1 | BG537255 | 6.4980 |
| 200800 | heat shock 70 kDa protein 1A | NM_005345 | 4.0000 |
| 201841 | heat shock 27 kDa protein 1 | NM_001540 | 128.0000 |
| 202581 | heat shock 70 kDa protein 1B | NM_005346 | 5.6569 |
| 205824 | heat shock 27 kDa protein 2 | NM_001541 | 5.2780 |
| Oxidoreductase | |||
| 200736 | glutathione peoxidase 1 (GPX1) | NM_000581 | 4.9246 |
| 201106 | glutathione peroxidase 4 (phospholipid hydroperoxidase) | NM_002085 | 4.2871 |
| 201194 | selenoprotein w, 1 (SEPW1) | NM_003009 | 6.4980 |
| 201348 | glutathione peroxidase 3 (GPX3| | NM_002084 | 11.3137 |
| 201563 | L-iditol-2 dehydrogenase | L29008 | 12.1257 |
| 201892 | inosine monophosphate dehydrogenase 2 (IMPDH2) | NM_000884 | 5.2780 |
| 202201 | biliverdin reductase B(flavin reductase (NADPH)) | NM_000713 | 5.6569 |
| 202539 | 3-hydroxy-3-methylglutaryl-Coenzyme | M11058 | 12.9960 |
| 202785 | NADH dehydroxygenase (ubiquinone) 1 alpha subcomplex, 7 A reductase | NM_005001 | 5.6569 |
| 202839 | NADH dehydroxygenase (ubiquinone) 1 beta subcomplex, 7 | NM_004146 | 4.5948 |
| 203570 | lysyl oxidase-like 1 (LOXL1) | NM_005576 | 21.1121 |
| 206024 | 4-hydroxyphenylpyruvate dioxygenase | NM_002150 | 5.2780 |
| 208631 | 78 kDa gastrin-binding protein | U04627 | 10.5561 |
| 209213 | carbonyl reductase 1 | BC002511 | 5.2780 |
| 210609 | quinone oxidoreductase homolog | BC000474 | 11.3137 |
| 212013 | melenoma associated gene | D86983 | 12.1257 |
| 212224 | aldehyde dehydrogenase 1 (ALDH1) | AF003341 | 4.0000 |
| 213540 | contains BING5 gene, the gene for beta3-galactosyltransferase | AL031228 | 6.0629 |
| Glutathione peroxidase | |||
| 200736 | glutathione peroxidae 1 (GPX1) | NM_000581 | 4.9246 |
| 201106 | glutathione peroxidase 4 (phospholipid hydroperoxidase) | NM_002085 | 4.2871 |
| 201348 | glutathione peroxidase 3 (GPX3) | NM_002084 | 11.3137 |
| Chaperone | |||
| 200064 | isolate Liv chaperone protein HSP90 beta | AF275719 | 4.9246 |
| 200664 | DnaJ (Hsp40) homolog, subfamily B, member 1 | BG537255 | 6.4980 |
| 200800 | heat shock 70 kDa protein 1A | NM_005345 | 4.0000 |
| 200812 | chaperonin containing TCP1, subunit 7 | NM_006429 | 4.9246 |
| 200968 | peptidylprolyl isomerase B (cyclophilin B) | NM_000942 | 17.1484 |
| 201459 | RuvB | NM_006666 | 16.0000 |
| 201841 | heat shock 27 kDa protein 1 | NM_001540 | 128.0000 |
| 202416 | tetratricopeptide repeat domain 2 | NM_003315 | 6.9644 |
| 202581 | heat shock 70 kDa protein 1B | NM_005346 | 5.6569 |
| 202843 | microvascular endothelial differentiation gene 1 | NM_012328 | 27.8576 |
| 205191 | retinitis pigmentosa 2 | NM_006915 | 12.1257 |
| 205824 | heat shock 27 kDa protein 2 | NM_001541 | 5.2780 |
| 207132 | prefoldin 5 | NM_002624 | 6.9644 |
| Structural constituent of lens | |||
| 206746 | filensin, beaded filament structural protein 1 | NM_001195 | 17.1484 |
| 206777 | beta B2 crystallin | NM_000496 | 6.4980 |
| 206778 | beta B2 crystallin | NM_000496 | 12.1257 |
| 206843 | beta A4 crystallin | NM_001886 | 168.8970 |
| 207399 | phakinin, beaded filament structural protein 2 | NM_003571 | 12.9960 |
| 207532 | gamma D crystallin | NM_006891 | 29.8571 |
| 207685 | beta B3 crystallin | NM_004076 | 22.6274 |
| 207715 | crystallin, gamma B | NM_005210 | 6.4980 |
| Structural molecule | |||
| 200600 | moesin | NM_002444 | 9.8492 |
| 200696 | gelsolin | NM_000177 | 4.2871 |
| 201650 | keratin 19 | NM_002276 | 4.9246 |
| 202007 | nidogen (enactin) | BF940043 | 8.5742 |
| 202766 | fibrillin 1 | NM_000138 | 5.2780 |
| 203690 | spindle pole body protein | NM_006322 | 9.1896 |
| 203725 | growth arrest and DNA-damage-inducible, alpha | NM_001924 | 24.2515 |
| 205373 | catenin (cadherin-associated protein), alpha 2 | NM_004389 | 6.9644 |
| 206746 | filensin, beaded filament structural protein 1 | NM_001195 | 17.1484 |
| 206777 | beta B2 crystallin | NM_000496 | 6.4980 |
| 206778 | beta B2 crystallin | NM_000496 | 12.1257 |
| 206843 | beta A4 crystallin | NM_001886 | 168.8970 |
| 207399 | phakinin, beaded filament structural protein 2 | NM_003571 | 12.9960 |
| 207532 | gamma D crystallin | NM_006891 | 29.8571 |
| 207685 | beta B3 crystallin | NM_004076 | 22.6274 |
| 207715 | gamma B crystallin | NM_005210 | 6.4980 |
| 208611 | alpha II spectrin | U83867 | 8.0000 |
| 208856 | ribosomal protein, large, P0 | BC003655 | 4.5948 |
| 208977 | tubulin, beta, 2 | BC004188 | 6.0629 |
| 209191 | Similar to tubulin, beta, 4 | BC002654 | 7.4643 |
| 210987 | tropomyosin | M19267 | 4.0000 |
| 214953 | amyloid beta (A4) | X06989 | 4.0000 |
The table lists all genes comprising each of the sub-categories that were significantly altered between cataract and clear lenses.
The entire EASE data set can be accessed in Appendix 1. Statistically significant trends in biological processes (Figure 5) and molecular functions (Figure 6) with increased gene expression in cataract were chromosome organization, nuclear organization, transcription/DNA-dependent, transcription, nucleic acid metabolism, nucleic acid binding, ligand binding or carrier, and DNA binding. Statistically significant trends in biological processes (Figure 7) and molecular functions (Figure 8) with decreased gene expression in cataract were RNA splicing, protein biosynthesis, protein synthesis elongation, protein synthesis initiation, macromolecule biosynthesis, amine biosynthesis, peroxidase reaction, microtubule-based process, organelle organization, cytoskeleton organization, temperature response, heat shock response, vision, response to external stimulus, U6 snRNA binding, pre-mRNA splicing factor, mRNA binding, proteasome endopeptidase, translation factor, selenium binding, alcohol dehydrogenase, heat shock protein, oxidoreductase, glutathione peroxidase, chaperone, structural constituents of lens, and structural molecules. Specific examples of the genes included in each category are summarized in Table 4.
DISCUSSION
In the present study, we have compared the relative expression levels of more than half of the genes predicted to comprise the human genome between age-matched cataract and clear human lenses; we have confirmed the accuracy of the data set by semi-quantitative RT-PCR and clustered the differentially expressed genes into functional categories. This analysis has identified over 1,300 genes that are altered in cataract relative to clear lenses. Of these, 74 are increased and 241 are decreased at the 5 fold or greater level between cataract and clear lenses. Although limitations in obtaining sufficient numbers of cataract and clear lenses preclude the extensive analysis of individual genes at the mRNA and protein level, we estimate that the trends in gene expression detected in the microarray procedure are approximately 84% accurate based on semi-quantitative RT-PCR using separately isolated RNA populations. Although we cannot rule out the possibility that temporal and/or spatial differences between cataract and clear lenses may influence the results of the present study, we are confident that the differences in gene expression detected are truly cataract-specific since the lenses were approximately age-matched (cataract approximately 70.2 years and clear lenses approximately 61.5 years), controlled for the proportion of males and females between the two samples (approximately 45% male), obtained within 24 h post-mortem, and carefully dissected for central epithelium (2–3 mm cataract and 6–8 mm clear). The cataracts examined in this study were mostly mixed and nuclear (70% mixed, 20% nuclear, 5% cortical, and 2% posterior subcapsular) therefore, the effects in gene expression detected in the present survey most likely reflect general gene expression changes associated with age-related cataract and are unlikely to be related to specific types of cataracts, except for possibly nuclear. Large numbers of specific types of cataracts will need to be collected in order to analyze type-specific gene expression patterns. However, it is important to note that many of the same genes and their corresponding magnitude changes detected in the present study correlate almost exactly with the gene expression differences and magnitude changes detected between cataract epithelia and clear lens epithelia using an entirely different population of human subjects as well as a different type of hybridization screening [19,20]. This complementary study provides great confidence in the gene expression differences detected in the present survey.
The present study provides evidence for multiple novel differences in gene expression between cataract and clear human lenses. Although descriptions of all of the individual genes that exhibit altered expression are too cumbersome to report, and many of the detected gene expression differences involve ESTs with no known function, some observations can be made. The majority of genes whose expression levels are altered in cataract exhibit decreased expression. These genes function in diverse processes including protein synthesis, oxidative stress, membrane transport, structural proteins, chaperones, and cell cycle control proteins. Many of these processes represent metabolic systems designed to preserve lens homeostasis and their decreased expression may reflect the inability of the lens to maintain its internal environment in the presence of stress and/or cataract. Specific examples of individual genes that exhibit decreased expression in cataract include multiple ribosomal subunits involved in protein synthesis (including large subunits 21, 15, 13a, and 7a which were previously shown to be decreased in cataract relative to clear human lenses) [15], selenoprotein W1, a glutathione dependant antioxidant known to protect lung cells against H2O2 cytotoxicity [21] that could play a role in defending the lens against oxidative damage, Na/K ATPase, a membrane transporter likely to be critical for osmotic regulation of the lens (whose proteins levels have previously been shown to be decreased in lens epithelia isolated from human age-related cataract [22]), glutathione peroxidases 1, 3 and 4, important oxidative stress enzymes that are likely to play major roles in lens protection and maintenance [23], ferritin, which has been linked to hereditary hyperferritinemia-cataract syndrome [24], multiple crystallins and other lens structural components, Hsp70, a key ATPase activated chaperone [25], Hsp27-1, a small heat-shock protein likely to be important for lens protection [26], Hsp27-2, a small heat shock protein closely related to αB-crystallin [27] which may also be important for lens protection, and αA-crystallin that, in addition to its structural role in the lens, is also a small heat shock protein that can prevent protein aggregation in the lens [28].
The microarray data showing 21 large and small ribosomal subunit transcripts that have decreased expression levels of 2 fold or greater in cataracts is consistent with differential display results showing that 4 of the large ribosomal subunit transcripts are decreased in cataractous lenses [15]. This process reflects a generalized decrease in protein synthesis in cataractous lens epithelial cells.
We also found significant decreases in genes associated with oxidative stress such as glutathione peroxidase, the metallothionein I genes, quinone oxidoreductase, and transketolase. It has previously been demonstrated that glutathione peroxidase-1-deficient mice develop cataracts at an early age [23] and that the levels of glutathione peroxidase are significantly decreased in the plasma of patients with senile cataracts [29]. It is also known that oxidative stress occurs when the quinone oxidoreductase gene is damaged resulting in the production of oxygen radicals [30]. The down regulation of the quinone oxidoreductase gene would also result in the same outcome, an increase in the overall production of oxygen radicals. Others have shown that the loss of transketolase function, an enzyme that catalyzes two of three reactions for entry into the pentose-phosphate pathway, a major source of chemical reducing power, results in lens fiber cell degeneration [31].
Another major functional category exhibiting decreased gene expression in cataracts is the small heat shock proteins/chaperones. Small heat shock proteins (sHSPs) are a large family of proteins that, unlike the large HSPs that are mainly involved in protein folding, play an important role in protecting organisms against stress [26]. This study specifically found rather large decreases in many of the crystallin proteins as well as HSP27. Mice lacking the αA-crystallin gene develop cataracts at an early age [32] and a missense mutation in the gene has been genetically linked to one form of autosomal dominant congenital cataracts in mice [33] and humans [34,35].
Many of the genes encoding structural lens proteins also exhibited decreased expression in cataract. This includes many of the β- and γ-crystallins which are thought to be essential for lens clarity and refraction. Indeed, mutations in β-crystallin has also been related to cataract formation, including a nonsense mutation in βB1-crystallin [36] and a mutation in the βB2-crystallin gene [37]. Two other genes involved in lens structure are filensin and phakinin. These two genes together make up the lens-specific intermediate filament known as the Beaded Filament [38]. It has been shown that the filensin protein is absent in lenses that have posterior subcapsular cataracts [39].
One additional functional category exhibiting decreased expression in cataractous lenses is the cyclins. This includes cyclin D1, cyclin G1, and BCL-1. Although there are very few reports examining the roles of these genes in the lens or their effects, if any, on cataract formation, one group of researchers has demonstrated that overexpression of cyclin G1 in fetal human lens epithelial cells results in an increased incidence of apoptosis [40].
Fewer genes exhibited increased expression in cataract. These genes function in processes as diverse as transcriptional control, ion transport, cytoplasmic transport, ion regulation, Ca2+ homeostatsis, protein salvaging pathways, and extracellular matrix interactions. Many of the pathways that exhibit increased expression in cataract are also associated with transcriptional processes that may represent attempts by the lens to compensate for stresses related to cataract. Specific examples of individual genes include multiple zinc finger proteins (important for transcriptional regulation), Na/H exchangers (which play key roles in regulating intracellular pH levels [41]), multiple calcium transporters and chloride channels (important for the maintenance of cellular homeostasis), osteonectin (a calcium-binding protein that functions in the regulation of cell growth [42]), and adducin (a member of a gene family encoding cytoskeletal proteins [43]).
According to the EASE analysis, functionally related groups of genes that exhibit overall trends of increased expression in cataracts include peptidyl-prolyl cis-trans isomerases. Twenty five percent of cyclophilin-like peptidyl-prolyl cis-trans isomerases present on the microarray exhibited increased gene expression in cataract including RAN binding protein. The peptidyl-prolyl cis-trans isomerases catalyze the cis-trans isomerization of prolyl-peptide bonds [44–46]. Some peptidyl-prolyl cis-trans isomerases may also possess chaperone activity by binding to and inhibiting the formation of misfolded protein aggregates [47–49]. It is possible that these isomerases are increased in cataracts in an attempt to prevent the aggregation of proteins in the lens which occur during cataract formation. Splice variants of a new class of cyclophilin-related proteins, types I and II, have been isolated [50,51] and it was found that the type II isoform is identical to Ran-binding protein 2 (RanBP2) [52,53].
Ran-binding protein 2 is a component of the nuclear pore complex which mediates macromolecular transport between the nucleus and the cytoplasm of the cell and serves the cell's requirement for bi-directional, selective, diverse and high-volume transport between these two compartments [54]. Thirty to 40 different proteins, called nucleoporins, have been identified as components of the nuclear pore complex [55]. RanBP2, which exhibited increased expression in cataracts, is the largest nucleoporin and has been localized to the cytoplasmic filaments of the nuclear pore complex [56]. RanBP1, another cytosolic protein closely related to RanBP2, is also involved in nuclear transport [57] and exhibits increased expression in cataracts.
In addition to cytoplasmic transport, many genes associated with ionic transport also exhibit increased expression in cataracts. One gene in particular, cullin 5, which shares 96% homology with vasopressin-activated Ca2+-mobilizing receptor, is increased in cataract. Although its specific function is currently unknown, it is likely to be involved in the Ca2+ and cAMP dependent cell signaling pathways [58]. Organ culture studies of the bovine lens demonstrate that a marked decrease in protein synthesis and a net leakage of proteins is strongly associated with an increase in calcium concentration [59]. The activity of Ca2+-ATPase has also been shown to be reduced by 50% in the membranes of lens epithelia isolated from cataractous lenses compared to clear human lenses [60]. Oxidative stress has also been demonstrated to have an effect on the activity of Ca2+ transporters in the lens. For example, hydrogen peroxide decreases the activity of Ca2+ transporters in rabbit lenses [61]. These phenomenon are closely associated with our results demonstrating an increase in Ca2+ transporters, possibly in an attempt to overcome their decreased activity in cataractous lenses, as well as a decrease in genes associated with protein synthesis.
Another ion channel that demonstrated increased expression in cataracts is the Na+/H+ exchanger isoform 2. Electroneutral Na+-H+ exchange is present in virtually all cell types and mediates the exchange of extracellular Na+ for intracellular H+ and therefore plays an important role in regulating the intracellular pH level, cell volume, and transepithelial Na+ absorption [41]. Intracellular pH can affect many cell functions such as metabolic activity, protein synthesis, and cell growth rates [62]. Previous studies have demonstrated that the Na+/H+ exchangers play a significant role in regulating the intracellular pH of cultured bovine lens epithelial cells [63]. It is also known that the type I Na+/H+ exchanger is activated by hypertonicity in many cell types [64] and the epithelial cells of toad lenses exposed to hypertonic conditions become acidified, stimulating the Na+/H+ exchanger to return the pH of the epithelial cells back to normal levels [65].
Another major group of genes that exhibit increased expression in cataractous epithelia compared to normal clear epithelia encode extracellular matrix proteins. Specifically, adducin, a family member of genes encoding cytoskeletal proteins [43], was increased in cataract. A second gene, pleiotrophin, which is also an extracellular matrix protein that binds heparin [66] and is induced during wound repair [67], was also increased in cataracts. Claudin, a component of tight junction filaments capable of interacting adhesively with complementary molecules on adjacent epithelial cells [68], also exhibited increased expression in cataracts. Recent studies have found that overexpression of claudin-2 induces cation-selective channels in tight junctions of epithelial cells resulting in increased ion permeability [69]. Another extracellular matrix gene whose expression is increased in cataracts is supervillin, an F-actin bundling plasma membrane protein that contains functional nuclear localization signals [70]. Bamacan, a chondroitin sulfate proteoglycan that abounds in basement membranes and is thought to be involved in the control of cell growth and transformation [71], also exhibited increased expression in cataracts. One final extracellular matrix gene that was increased in cataracts is Osteonectin which has previously been demonstrated to be increased in human age-related cataracts [72].
In summary this report identifies the global gene expression changes associated with age-related cataract and provides evidence for specific biological pathways that are associated with this disease. It is not possible from this study to determine whether these gene expression differences are a cause of cataract formation or a response of the lens to the presence of the cataract. However, future confirmation at the protein level and functional analysis of the identified genes in tissue culture and animal model systems will eventually help define the individual roles that the identified genes play in lens maintenance, protection, and cataract. Analysis of the identified pathways will yield important information concerning the regulation of gene expression in age-related cataract and may aid in the development of therapeutic treatments to prevent or delay the onset of this disease.
ACKNOWLEDGEMENTS
The authors thank Divyen Patel of Genome Explorations Inc., for his technical advice and services, Tracy Cowell and Erik Peterson of the Kantorow laboratory for their suggestions and help throughout the course of this project and the Lions Eye Bank of Oregon and the West Virginia Eye Bank for providing the clear lenses used in this study. This research was supported by NIH grants EY13022 (MK) and EY03897 (JH).
Appendix 1 Raw affymetrix chip data with EASE analysis
These files are the raw affymetrix data and the entire EASE data analysis. The file entitled “raw-data.txt” is a list of all of the genes that are either increased or decreased by 2 fold or greater levels according to the affymetrix chip data. The list includes each gene's relative signal intensity, statistical probability, and description. The file entitled “increased-2-fold.txt” represents the EASE analysis data for all of the genes that are increased by 2 fold or greater levels according to the microarray data with the statistical analysis for each of the categories. This is the data that was used to create Figure 5 and Figure 6. The file entitled “decreased-2-fold.txt” represents the EASE analysis data for all of the genes that are decreased by 2 fold or greater levels according to the microarray data with the statistical analysis for each of the categories. This is the data that was used to create Figure 7 and Figure 8. The file entitled “scatter-plot.jpg” is a graphical representation of the expression differences between cataract and clear lenses. The Y-axis is the signal intensity value of the clear lens hybridization versus the cataract hybridization. The X-axis is the cataract signal intensity value. Each dot on this graph represents an individual gene. The blue dots are genes increased in cataract while the red dots represent genes decreased in cataract. The green lines indicate fold-chage values with the two most exterior lines representing 10 fold and the two most interior lines represent 2 fold changes. The middle lines represent 3 fold changes.
To access this data, click or select the words “data and analysis” in the online version of this article. This will initiate the download of a compressed (zip) archive. This file should be uncompressed with an appropriate program (the particular program will depend on your operating system). Once extracted, you will have a folder (or directory) containing eight files (one for each microarray). The files are tab delimited text. Most spreadsheet programs will import files in this format.
REFERENCES
- 1.Bron A, Brown NP. Lens Disorders: A Clinical Manual of Cataract Diagnosis. Butterworth-Heinemann; Oxford, UK: 1996. Biology of cataract; pp. 53–77. [Google Scholar]
- 2.Piatigorsky J. Lens differentiation in vertebrates. A review of cellular and molecular features. Differentiation. 1981;19:134–53. doi: 10.1111/j.1432-0436.1981.tb01141.x. [DOI] [PubMed] [Google Scholar]
- 3.Bloemendal H. Lens Proteins. In: Bloemendal H, editor. Molecular and cellular biology of the eye lens. Wiley; New York: 1981. pp. 1–47. [Google Scholar]
- 4.Reddy VN. Metabolism of glutathione in the lens. Exp Eye Res. 1971;11:310–28. doi: 10.1016/s0014-4835(71)80043-x. [DOI] [PubMed] [Google Scholar]
- 5.Spector A. Aging of the lens and cataract formation. In: Sekuler R, Kline D, Dismukes K, editors. Aging and human visual function. A.R. Liss; New York: 1982. pp. 27–43. [Google Scholar]
- 6.Reddan JR. Control of cell division in the ocular lens, retina and vitreous humour. In: McDevitt DS, editor. Cell biology of the eye. Academic Press; New York: 1982. pp. 299–375. [Google Scholar]
- 7.Rae JL, Bartling C, Rae J, Mathias RT. Dye transfer between cells of the lens. J Membr Biol. 1996;150:89–103. doi: 10.1007/s002329900033. [DOI] [PubMed] [Google Scholar]
- 8.Harding JJ, Crabbe MJC. The lens: development, proteins, metabolism and cataract. In: Davson H, editor. The eye. Vol 1B. Academic Press; Orlando (FL): 1984. pp. 207–492. [Google Scholar]
- 9.Spector A. Oxidative stress-induced cataract: mechanism of action. FASEB J. 1995;9:1173–82. [PubMed] [Google Scholar]
- 10.Hightower KR. The role of the lens epithelium in development of UV cataract. Curr Eye Res. 1995;14:71–8. doi: 10.3109/02713689508999916. [DOI] [PubMed] [Google Scholar]
- 11.Kantorow M, Kays T, Horwitz J, Huang Q, Sun J, Piatigorsky J, Carper D. Differential display detects altered gene expression between cataractous and normal human lenses. Invest Ophthalmol Vis Sci. 1998;39:2344–54. [PubMed] [Google Scholar]
- 12.Kantorow M, Horwitz J, Carper D. Up-regulation of osteonectin/SPARC in age-related cataractous human lens epithelia. Mol Vis. 1998;4:17. [PubMed] [Google Scholar]
- 13.Wan XH, Lee EH, Koh HJ, Song J, Kim EK, Kim CY, Lee JB, Kim SY, Yao K, Lee JH. Enhanced expression of transglutaminase 2 in anterior polar cataracts and its induction by TGF-beta in vitro. Br J Ophthalmol. 2002;86:1293–8. doi: 10.1136/bjo.86.11.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lee EH, Seomun Y, Hwang KH, Kim JE, Kim IS, Kim JH, Joo CK. Overexpression of the transforming growth factor-beta-inducible gene betaig-h3 in anterior polar cataracts. Invest Ophthalmol Vis Sci. 2000;41:1840–5. [PubMed] [Google Scholar]
- 15.Zhang W, Hawse J, Huang Q, Sheets N, Miller KM, Horwitz J, Kantorow M. Decreased expression of ribosomal proteins in human age-related cataract. Invest Ophthalmol Vis Sci. 2002;43:198–204. [PMC free article] [PubMed] [Google Scholar]
- 16.Lim JM, Lee JH, Wee WR, Joo CK. Downregulated expression of ADAM9 in anterior polar cataracts. J Cataract Refract Surg. 2002;28:697–702. doi: 10.1016/s0886-3350(01)01236-6. [DOI] [PubMed] [Google Scholar]
- 17.Goswami S, Sheets NL, Zavadil J, Chauhan BK, Bottinger EP, Reddy VN, Kantorow M, Cvekl A. Spectrum and range of oxidative stress responses of human lens epithelial cells to H2O2 insult. Invest Ophthalmol Vis Sci. 2003;44:2084–93. doi: 10.1167/iovs.02-0882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Spector A, Li D, Ma W, Sun F, Pavlidis P. Differential amplification of gene expression in lens cell lines conditioned to survive peroxide stress. Invest Ophthalmol Vis Sci. 2002;43:3251–64. [PubMed] [Google Scholar]
- 19.Maraini G, Ruotolo R, Rivetti C, Percudani R, Ottonello S. DNA array analysis of gene expression in the lens epithelium of pure nuclear cataract and transparent human lenses. ARVO Annual Meeting; Fort Lauderdale, FL. 2002 May 5–10. [Google Scholar]
- 20.Ruotolo R, Grassi F, Percudani R, Rivetti C, Martorana D, Maraini G, Ottonello S. Gene expression profiling in human age-related nuclear cataract. Mol Vis. 2003;9:538–48. [PubMed] [Google Scholar]
- 21.Jeong D, Kim TS, Chung YW, Lee BJ, Kim IY. Selenoprotein W is a glutathione-dependent antioxidant in vivo. FEBS Lett. 2002;517:225–8. doi: 10.1016/s0014-5793(02)02628-5. [DOI] [PubMed] [Google Scholar]
- 22.Tseng SH, Tang MJ. Na,K-ATPase in lens epithelia from patients with senile cataracts. J Formos Med Assoc. 1999;98:627–32. [PubMed] [Google Scholar]
- 23.Reddy VN, Giblin FJ, Lin LR, Dang L, Unakar NJ, Musch DC, Boyle DL, Takemoto LJ, Ho YS, Knoernschild T, Juenemann A, Lutjen-Drecoll E. Glutathione peroxidase-1 deficiency leads to increased nuclear light scattering, membrane damage, and cataract formation in gene-knockout mice. Invest Ophthalmol Vis Sci. 2001;42:3247–55. [PubMed] [Google Scholar]
- 24.Martin ME, Fargion S, Brissot P, Pellat B, Beaumont C. A point mutation in the bulge of the iron-responsive element of the L ferritin gene in two families with the hereditary hyperferritinemia-cataract syndrome. Blood. 1998;91:319–23. [PubMed] [Google Scholar]
- 25.Haslbeck M. sHsps and their role in the chaperone network. Cell Mol Life Sci. 2002;59:1649–57. doi: 10.1007/PL00012492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ganea E. Chaperone-like activity of alpha-crystallin and other small heat shock proteins. Curr Protein Pept Sci. 2001;2:205–25. doi: 10.2174/1389203013381107. [DOI] [PubMed] [Google Scholar]
- 27.Iwaki A, Nagano T, Nakagawa M, Iwaki T, Fukumaki Y. Identification and characterization of the gene encoding a new member of the alpha-crystallin/small hsp family, closely linked to the alphaB-crystallin gene in a head-to-head manner. Genomics. 1997;45:386–94. doi: 10.1006/geno.1997.4956. [DOI] [PubMed] [Google Scholar]
- 28.Horwitz J. Alpha-crystallin can function as a molecular chaperone. Proc Natl Acad Sci U S A. 1992;89:10449–53. doi: 10.1073/pnas.89.21.10449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xue AN, Cai QY, Wang SQ, Zhou AS, Li WX, Fu P, Chen XS. Antioxidant status in persons with and without senile lens changes. Biomed Environ Sci. 1996;9:144–8. [PubMed] [Google Scholar]
- 30.Pitkanen S, Robinson BH. Mitochondrial complex I deficiency leads to increased production of superoxide radicals and induction of superoxide dismutase. J Clin Invest. 1996;98:345–51. doi: 10.1172/JCI118798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Frederikse PH, Farnsworth P, Zigler JS., Jr. Thiamine deficiency in vivo produces fiber cell degeneration in mouse lenses. Biochem Biophys Res Commun. 1999;258:703–7. doi: 10.1006/bbrc.1999.0560. [DOI] [PubMed] [Google Scholar]
- 32.Brady JP, Garland D, Duglas-Tabor Y, Robison WG, Jr, Groome A, Wawrousek EF. Targeted disruption of the mouse alpha A-crystallin gene induces cataract and cytoplasmic inclusion bodies containing the small heat shock protein alpha B-crystallin. Proc Natl Acad Sci U S A. 1997;94:884–9. doi: 10.1073/pnas.94.3.884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cobb BA, Petrash JM. Structural and functional changes in the alpha A-crystallin R116C mutant in hereditary cataracts. Biochemistry. 2000;39:15791–8. doi: 10.1021/bi001453j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Litt M, Kramer P, LaMorticella DM, Murphey W, Lovrien EW, Weleber RG. Autosomal dominant congenital cataract associated with a missense mutation in the human alpha crystallin gene CRYAA. Hum Mol Genet. 1998;7:471–4. doi: 10.1093/hmg/7.3.471. [DOI] [PubMed] [Google Scholar]
- 35.Pras E, Frydman M, Levy-Nissenbaum E, Bakhan T, Raz J, Assia EI, Goldman B, Pras E. A nonsense mutation (W9X) in CRYAA causes autosomal recessive cataract in an inbred Jewish Persian family. Invest Ophthalmol Vis Sci. 2000;41:3511–5. [PubMed] [Google Scholar]
- 36.Mackay DS, Boskovska OB, Knopf HL, Lampi KJ, Shiels A. A nonsense mutation in CRYBB1 associated with autosomal dominant cataract linked to human chromosome 22q. Am J Hum Genet. 2002;71:1216–21. doi: 10.1086/344212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Graw J, Loster J, Soewarto D, Fuchs H, Reis A, Wolf E, Balling R, de Angelis MH. Aey2, a new mutation in the betaB2-crystallin-encoding gene of the mouse. Invest Ophthalmol Vis Sci. 2001;42:1574–80. [PubMed] [Google Scholar]
- 38.Gounari F, Karagianni N, Mincheva A, Lichter P, Georgatos SD, Schirrmacher V. The mouse filensin gene: structure and evolutionary relation to other intermediate filament genes. FEBS Lett. 1997;413:371–8. doi: 10.1016/s0014-5793(97)00937-x. [DOI] [PubMed] [Google Scholar]
- 39.Hess JF, Casselman JT, Kong AP, FitzGerald PG. Primary sequence, secondary structure, gene structure, and assembly properties suggests that the lens-specific cytoskeletal protein filensin represents a novel class of intermediate filament protein. Exp Eye Res. 1998;66:625–44. doi: 10.1006/exer.1998.0478. [DOI] [PubMed] [Google Scholar]
- 40.Kampmeier J, Behrens A, Wang Y, Yee A, Anderson WF, Hall FL, Gordon EM, McDonnell PJ. Inhibition of rabbit keratocyte and human fetal lens epithelial cell proliferation by retrovirus-mediated transfer of antisense cyclin G1 and antisense MAT1 constructs. Hum Gene Ther. 2000;11:1–8. doi: 10.1089/10430340050016102. [DOI] [PubMed] [Google Scholar]
- 41.Sangan P, Rajendran VM, Geibel JP, Binder HJ. Cloning and expression of a chloride-dependent Na+-H+ exchanger. J Biol Chem. 2002;277:9668–75. doi: 10.1074/jbc.M110852200. [DOI] [PubMed] [Google Scholar]
- 42.Sage EH, Bassuk JA, Yost JC, Folkman MJ, Lane TF. Inhibition of endothelial cell proliferation by SPARC is mediated through a Ca(2+)-binding EF-hand sequence. J Cell Biochem. 1995;57:127–40. doi: 10.1002/jcb.240570113. [DOI] [PubMed] [Google Scholar]
- 43.Gilligan DM, Sarid R, Weese J. Adducin in platelets: activation-induced phosphorylation by PKC and proteolysis by calpain. Blood. 2002;99:2418–26. doi: 10.1182/blood.v99.7.2418. [DOI] [PubMed] [Google Scholar]
- 44.Schmid FX. Prolyl isomerase: enzymatic catalysis of slow protein-folding reactions. Annu Rev Biophys Biomol Struct. 1993;22:123–42. doi: 10.1146/annurev.bb.22.060193.001011. [DOI] [PubMed] [Google Scholar]
- 45.Rudd KE, Sofia HJ, Koonin EV, Plunkett G, 3rd, Lazar S, Rouviere PE. A new family of peptidyl-prolyl isomerases. Trends Biochem Sci. 1995;20:12–4. doi: 10.1016/s0968-0004(00)88940-9. [DOI] [PubMed] [Google Scholar]
- 46.Rassow J, Pfanner N. Protein biogenesis: chaperones for nascent polypeptides. Curr Biol. 1996;6:115–8. doi: 10.1016/s0960-9822(02)00437-2. [DOI] [PubMed] [Google Scholar]
- 47.Freskgard PO, Bergenhem N, Jonsson BH, Svensson M, Carlsson U. Isomerase and chaperone activity of prolyl isomerase in the folding of carbonic anhydrase. Science. 1992;258:466–8. doi: 10.1126/science.1357751. [DOI] [PubMed] [Google Scholar]
- 48.Lilie H, Lang K, Rudolph R, Buchner J. Prolyl isomerases catalyze antibody folding in vitro. Protein Sci. 1993;2:1490–6. doi: 10.1002/pro.5560020913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rinfret A, Collins C, Menard R, Anderson SK. The N-terminal cyclophilin-homologous domain of a 150-kilodalton tumor recognition molecule exhibits both peptidylprolyl cis-transisomerase and chaperone activities. Biochemistry. 1994;33:1668–73. doi: 10.1021/bi00173a008. [DOI] [PubMed] [Google Scholar]
- 50.Ferreira PA, Pak WL. Characterization of vertebrate homologs of Drosophila photoreceptor proteins. In: Anderson RE, LaVail MM, Holyfield JG, editors. Degenerative diseases of the retina. Plenum Press; New York: 1995. pp. 263–274. [Google Scholar]
- 51.Ferreira PA, Hom JT, Pak WL. Retina-specifically expressed novel subtypes of bovine cyclophilin. J Biol Chem. 1995;270:23179–88. doi: 10.1074/jbc.270.39.23179. [DOI] [PubMed] [Google Scholar]
- 52.Yokoyama N, Hayashi N, Seki T, Pante N, Ohba T, Nishii K, Kuma K, Hayashida T, Miyata T, Aebi U, Fukui M, Nishimoto T. A giant nucleopore protein that binds Ran/TC4. Nature. 1995;376:184–8. doi: 10.1038/376184a0. [DOI] [PubMed] [Google Scholar]
- 53.Wu J, Matunis MJ, Kraemer D, Blobel G, Coutavas E. Nup358, a cytoplasmically exposed nucleoporin with peptide repeats, Ran-GTP binding sites, zinc fingers, a cyclophilin A homologous domain, and a leucine-rich region. J Biol Chem. 1995;270:14209–13. doi: 10.1074/jbc.270.23.14209. [DOI] [PubMed] [Google Scholar]
- 54.Walther TC, Pickersgill HS, Cordes VC, Goldberg MW, Allen TD, Mattaj IW, Fornerod M. The cytoplasmic filaments of the nuclear pore complex are dispensable for selective nuclear protein import. J Cell Biol. 2002;158:63–77. doi: 10.1083/jcb.200202088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rout MP, Aitchison JD. Pore relations: nuclear pore complexes and nucleocytoplasmic exchange. Essays Biochem. 2000;36:75–88. doi: 10.1042/bse0360075. [DOI] [PubMed] [Google Scholar]
- 56.Wilken N, Senecal JL, Scheer U, Dabauvalle MC. Localization of the Ran-GTP binding protein RanBP2 at the cytoplasmic side of the nuclear pore complex. Eur J Cell Biol. 1995;68:211–9. [PubMed] [Google Scholar]
- 57.Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–60. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- 58.Burnatowska-Hledin M, Zhao P, Capps B, Poel A, Parmelee K, Mungall C, Sharangpani A, Listenberger L. VACM-1, a cullin gene family member, regulates cellular signaling. Am J Physiol Cell Physiol. 2000;279:C266–73. doi: 10.1152/ajpcell.2000.279.1.C266. [DOI] [PubMed] [Google Scholar]
- 59.Duncan G, Jacob TJ. Calcium and the physiology of cataract. Ciba Found Symp. 1984;106:132–52. doi: 10.1002/9780470720875.ch8. [DOI] [PubMed] [Google Scholar]
- 60.Paterson CA, Zeng J, Husseini Z, Borchman D, Delamere NA, Garland D, Jimenez-Asensio J. Calcium ATPase activity and membrane structure in clear and cataractous human lenses. Curr Eye Res. 1997;16:333–8. doi: 10.1076/ceyr.16.4.333.10689. [DOI] [PubMed] [Google Scholar]
- 61.Borchman D, Paterson CA, Delamere NA. Oxidative inhibition of Ca2+-ATPase in the rabbit lens. Invest Ophthalmol Vis Sci. 1989;30:1633–7. [PubMed] [Google Scholar]
- 62.Bonanno JA. Regulation of corneal epithelial intracellular pH. Optom Vis Sci. 1991;68:682–6. doi: 10.1097/00006324-199109000-00002. [DOI] [PubMed] [Google Scholar]
- 63.Williams MR, Duncan G, Croghan PC, Riach R, Webb SF. pH regulation in tissue-cultured bovine lens epithelial cells. J Membr Biol. 1992;129:179–87. doi: 10.1007/BF00219513. [DOI] [PubMed] [Google Scholar]
- 64.Garnovskaya MN, Mukhin YV, Vlasova TM, Raymond JR. Hypertonicity activates Na+/H+ exchange through Janus kinase 2 and calmodulin. J Biol Chem. 2003;278:16908–15. doi: 10.1074/jbc.M209883200. [DOI] [PubMed] [Google Scholar]
- 65.Wolosin JM, Alvarez LJ, Candia OA. Stimulation of toad lens epithelial Na+/H+ exchange activity by hypertonicity. Exp Eye Res. 1989;48:855–62. doi: 10.1016/0014-4835(89)90068-7. [DOI] [PubMed] [Google Scholar]
- 66.Fath M, VanderNoot V, Kilpelainen I, Kinnunen T, Rauvala H, Linhardt RJ. Interaction of soluble and surface-bound heparin binding growth-associated molecule with heparin. FEBS Lett. 1999;454:105–8. doi: 10.1016/s0014-5793(99)00785-1. [DOI] [PubMed] [Google Scholar]
- 67.Deuel TF, Zhang N, Yeh HJ, Silos-Santiago I, Wang ZY. Pleiotrophin: a cytokine with diverse functions and a novel signaling pathway. Arch Biochem Biophys. 2002;397:162–71. doi: 10.1006/abbi.2001.2705. [DOI] [PubMed] [Google Scholar]
- 68.Gonzalez-Mariscal L, Betanzos A, Nava P, Jaramillo BE. Tight junction proteins. Prog Biophys Mol Biol. 2003;81:1–44. doi: 10.1016/s0079-6107(02)00037-8. [DOI] [PubMed] [Google Scholar]
- 69.Amasheh S, Meiri N, Gitter AH, Schoneberg T, Mankertz J, Schulzke JD, Fromm M. Claudin-2 expression induces cation-selective channels in tight junctions of epithelial cells. J Cell Sci. 2002;115:4969–76. doi: 10.1242/jcs.00165. [DOI] [PubMed] [Google Scholar]
- 70.Wulfkuhle JD, Donina IE, Stark NH, Pope RK, Pestonjamasp KN, Niswonger ML, Luna EJ. Domain analysis of supervillin, an F-actin bundling plasma membrane protein with functional nuclear localization signals. J Cell Sci. 1999;112(Pt 13):2125. doi: 10.1242/jcs.112.13.2125. [DOI] [PubMed] [Google Scholar]