Abstract
Study Objectives:
Narcolepsy with cataplexy is caused by a loss of orexin (hypocretin) signaling, but the physiologic mechanisms that result in poor maintenance of wakefulness and fragmented sleep remain unknown. Conventional scoring of sleep cannot reveal much about the process of transitioning between states or the variations within states. We developed an EEG spectral analysis technique to determine whether the state instability in a mouse model of narcolepsy reflects abnormal sleep or wake states, faster movements between states, or abnormal transitions between states.
Design:
We analyzed sleep recordings in orexin knockout (OXKO) mice and wild type (WT) littermates using a state space analysis technique. This non-categorical approach allows quantitative and unbiased examination of sleep/wake states and state transitions.
Measurements and Results:
OXKO mice spent less time in deep, delta-rich NREM sleep and in active, theta-rich wake and instead spent more time near the transition zones between states. In addition, while in the midst of what should be stable wake, OXKO mice initiated rapid changes into NREM sleep with high velocities normally seen only in transition regions. Consequently, state transitions were much more frequent and rapid even though the EEG progressions during state transitions were normal.
Conclusions:
State space analysis enables visualization of the boundaries between sleep and wake and shows that narcoleptic mice have less distinct and more labile states of sleep and wakefulness. These observations provide new perspectives on the abnormal state dynamics resulting from disrupted orexin signaling and highlight the usefulness of state space analysis in understanding narcolepsy and other sleep disorders.
Citation:
Diniz Behn CG; Klerman EB; Mochizuki T; Lin S; Scammell TE. Abnormal sleep/wake dynamics in orexin knockout mice. SLEEP 2010;33(3):297-306.
Keywords: EEG, orexin/hypocretin, narcolepsy, cataplexy, sleep
OVER 20 YEARS AGO, BROUGHTON AND COLLEAGUES HYPOTHESIZED THAT NARCOLEPSY IS BEST CONSIDERED A DISEASE OF STATE BOUNDARY CONTROL.1 They argued that sleepiness, cataplexy, hallucinations, and many other symptoms could be viewed as a breakdown of “whatever neurochemical ‘glues' or integrative neurophysiological mechanisms exist for sleep and wake state continuity.”1 This hypothesis is compelling, but it has been difficult to examine using conventional sleep scoring methods.
More recently, our understanding of narcolepsy has been greatly advanced by the discovery that narcolepsy with cataplexy is caused by a loss of functional signaling by the orexin (hypocretin) neuropeptides.2–5 The neurons producing orexins are active during wakefulness,6–8 and direct activation of these neurons can awaken mice from sleep.9 In addition, orexins probably stabilize wake and sleep; narcoleptic people, dogs, and mice lacking orexins have great difficulty remaining awake for long periods and also experience fragmented sleep.10–13
In earlier work, we found that the fragmented wakefulness of orexin deficiency is not a consequence of abnormal sleep homeostasis, poor circadian control, or defective fundamental arousal systems.10 However, conventional sleep scoring in 10- to 30-second epochs reveals little about the process of transitioning between states as cortical activity and behavior can change quite rapidly. Furthermore, conventional scoring simply identifies discrete states, so it can overlook important variations within states, such as the distinctions between light and deep NREM sleep or between drowsy wake and high levels of arousal. Therefore, to determine how orexin deficiency causes behavioral state instability we developed a state space analysis technique to examine the dynamics of sleep/wake behavior in orexin knockout (OXKO) mice, a model of narcolepsy.
The previous application of state space techniques to sleep recordings used local field potential data, but the variability in these signals prevented comparisons between animals.14–16 We adapted these techniques for analysis of EEG recordings in mice and developed metrics for inter-animal comparisons. State space techniques have high temporal resolution and analyze behavior as a continuum, rather than in discrete states, thus facilitating higher dimensional examination of state transitions. This approach enabled us to determine whether the state instability in this mouse model of narcolepsy reflects abnormal sleep/wake states, faster movements between states, or abnormal transition processes.
METHODS
Animals
Founder OXKO mice were on a C57BL/6J-129/SvEV background (T. Sakurai, Kanazawa University), and their offspring were backcrossed with C57BL/6J mice for 8 generations. We recorded sleep/wake behavior in 7 male OXKO mice and 6 wild type (WT) littermates, all 5-6 months old and weighing 30-35 g. All experiments were approved by the Institutional Animal Care and Use Committees of Beth Israel Deaconess Medical Center and Harvard Medical School.
Surgery and Electroencephalogram-Electromyogram Recordings
Mice were anesthetized with ketamine-xylazine (100 and 10 mg/kg, i.p.) and implanted with electroencephalogram (EEG) and electromyogram (EMG) electrodes as described previously.10 EEG signals were recorded using 2 ipsilateral stainless steel screws (1.5 mm to the right of the sagittal suture; 1 mm anterior to bregma and 1 mm anterior to lambda). EMG signals were acquired by a pair of multistranded stainless steel wires inserted into the neck extensor muscles. Nine days after surgery, mice were transferred to individual recording cages in a sound-attenuated chamber with a 12:12 h light/dark (LD) cycle (30 lux; lights on at 07:00 and off at 19:00) and a constant temperature of about 23°C. They had ad lib access to food and water and acclimated to the recording cables for another 5 days.
Two weeks after surgery, we recorded spontaneous sleep/wake behavior for 24 hours. EEG/EMG signals were amplified and analog filtered (low cut: 0.3 Hz; high cut: 1000 Hz; Model 12, Grass Technologies, West Warwick, RI) and then digitized at 512 Hz (Sleep Sign, Kissei Comtec, Matsumoto, Japan). Animals were video recorded during data collection.
Conventional Scoring of Behavioral States
For conventional scoring, we digitally filtered the signals (EEG: 0.3-30 Hz, EMG: 2-50 Hz) and then scored each 10-sec epoch as Wake, NREM sleep, or REM sleep with the aid of scoring software (Sleep Sign; Kissei Comtec, Matsumoto, Japan). We visually inspected and corrected this preliminary, semi-automatic scoring when appropriate. We scored epochs as Cataplexy using the recently published consensus definition: wake preceding cataplexy onset had to last ≥ 40 sec17,18; cataplexy onset was marked by an abrupt transition from wakefulness to periods of high EEG theta activity (4-9 Hz) and atonia in the nuchal muscles.10 Simultaneous video recordings showed that during cataplexy, the mouse was often prone or lying on its side in a posture atypical of sleep, and that the cataplexy often occurred outside of the usual nest. These episodes were always followed by a direct transition back to wakefulness.
Construction of the Two-Dimensional State Space and Definition of Clusters
Using an approach similar to that of Gervasoni and colleagues,14 we defined a 2-dimensional (2-D) state space using 2 spectral amplitude ratios calculated by dividing integrated spectral amplitudes at selected frequency bands. First, a sliding window Fourier transform was applied to each raw (0.3-256 Hz) EEG signal using a 2-sec window with a 1-sec step size. Then we calculated 3 spectral amplitude ratios by integrating the spectral energy over specific frequencies: 6.5-9/0.3-9 Hz for ratio 1 (plotted on the abscissa) and 0.3-20/0.3-55 Hz for ratio 2 (plotted on the ordinate).
These ratios were determined by a thorough search for parameters that optimized the separation between behavioral states. To distinguish between Wake and NREM sleep, we initially considered choices of ratio 2 that focused on the delta band (2-4 Hz), but the separation of clusters was optimal when we included all EEG activity between 0.3 and 20 Hz as shown in previous state space work.14,15 We defined ratio 1 as 6.5-9/0.3-9 Hz to emphasize high theta (6.5-9 Hz) frequencies because activity in this range dominates rodent REM sleep, and dysregulation of REM sleep is an important aspect of the narcolepsy phenotype. Note that the choice of frequency bands for ratio 1 (6.5-9 Hz) is slightly different from those proposed by Gervasoni et al. and empirically resulted in a better cluster separation for our data set. This difference may result from the different spectral properties of local field potentials versus EEG or from a difference in animal species used (rat versus mouse).
Next, we smoothed each second of data with a 20-sec wide Hann window. This technique substantially reduced within-state variability and minimized the effects of any EEG artifacts. Though this smoothing allowed us to include all data, it limited the precision of state space velocity calculations (see below). Finally, each second of EEG data was mapped into the 2-D state space based on its spectral content.
We validated the method by comparing the clustering of points in the 2-D state space against the conventionally-scored sleep/wake data and showed a general agreement between these 2 methods: distinct clusters of points correspond to distinct states of Wake, NREM sleep, and REM sleep (see Results). Therefore, for convenience, we refer to the clusters using the associated conventional scored data names (e.g., “NREM cluster” or “Wake cluster”). Note that the conventionally-scored data did not influence the creation of the clusters or the definition of the cluster core boundaries for Wake and NREM sleep (see below).
We also developed several new techniques for quantifying differences in sleep/wake behavior between WT and OXKO mice:
Peak-to-peak distances: To measure differences in the distributions of points within the Wake and NREM sleep clusters, we projected state space points into the ratio 2 axis. For each animal, the resulting projection had two distinct peaks corresponding to Wake (low ratio 2) and NREM sleep (high ratio 2). The point density in the region between clusters was reflected by the height of the lowest point between peaks (the trough). For each animal, we calculated the horizontal distance between peaks and the vertical trough height. To ensure that the inclusion of REM sleep did not affect our results, we then eliminated data scored as REM sleep or cataplexy from the analysis and reanalyzed the data. The results of that analysis were very similar (data not shown).
State space densities and cluster cores: To calculate state space densities, the state space was binned using a grid of 100 × 100 uniformly spaced boxes, and the density of points in each box was calculated. Therefore, the density of points reflects the relative abundance of the different behavioral states. Using state space density plots, cluster core boundaries were determined using a contour-based algorithm: after calculating 100 linearly spaced contours corresponding to point density over the entire space, sets of mutually excluding concentric contours were identified for each main cluster. Of these contours, the 95% most inclusive contour was used to delineate state-specific cluster core boundaries subject to a constraint that the boundaries did not overlap. Note that conventional sleep scoring information is not included in this procedure.
Over 24 h, mice spend relatively little time in REM sleep, so identification of the REM sleep cluster core boundaries required amplification of the data. To achieve this, we isolated the spectral ratio points corresponding to each 10-sec epoch of conventionally scored REM sleep. Each of the spectral points was smeared into a cluster of 9 points distributed on the nearest-neighbor 3 × 3 grid surrounding the initial point. We applied the contour-based algorithm to the smeared data and chose the 60% most inclusive contour to delineate the REM sleep core cluster boundary. The amplification also included data from the flanking 10-sec epochs to control for any variability in scoring the precise onset and offset of REM sleep. Although this choice led to the inclusion of some data points that were probably not REM sleep, the relative sparseness and heterogeneity of data associated with transitions in and out of REM sleep ensured that it did not strongly influence the densities used by the contour-based algorithm. We used the same approach to identify the boundaries of the Cataplexy cluster core as cataplexy also accounts for a relatively small percentage of behavior in OXKO mice.
We computed the areas enclosed by each cluster core boundary and the positive predictive value (the proportion of data within the boundary that was conventionally scored as the behavioral state corresponding to the boundary) and sensitivity (the proportion of data that was conventionally scored as a specific state that fell within the boundary corresponding to that state) of each boundary. In addition, to investigate the separation between states, we computed the centroids of points falling within cluster core boundaries and compared the distances between pairs of centroids of each cluster between genotypes.
State space velocities: For each animal, we created state space velocity maps to determine rates of change across the state space. The speed of spectral change was calculated as the distance between 2 consecutive data points (the grid velocity values represent velocities from the grid points), and in this sense, the temporal resolution was second by second. However, the smoothing of the data limits temporal resolution and probably reduces the differences in absolute velocities across the state space. After calculating these “second-by-second” velocities, we mapped them to a 100 × 100 grid with the values at each point representing the average velocities originating at that site. To examine velocities as mice moved in and out of REM sleep or between Wake and NREM sleep, we also calculated directional velocities in the horizontal (ratio 1) and vertical (ratio 2) directions, respectively.
We used a contour-based algorithm to delineate regions of fast velocities, and the areas enclosed by the fast velocity boundary were averaged within genotypes to determine group means and standard deviations. Finally, to investigate the types of transitions associated with the regions of high directional velocities, we split the directional velocities into their positive and negative parts to compare velocities in the up, down, right, and left directions.
Trajectory analysis: To better understand the patterns of EEG activity as mice move between stable states, we identified trajectories in the state space by tracking consecutive sequences of points. A trajectory was initiated at the first time step for which a point fell outside a cluster core boundary; the trajectory was terminated at the next time step for which the point fell within a cluster core boundary. We examined inter-state trajectories (e.g. moving from the Wake cluster core to the NREM sleep cluster core) as well as intra-state trajectories (e.g., leaving the Wake cluster core and returning to the Wake cluster core without crossing into another state). We analyzed trajectory paths, calculated transition probabilities, and determined the initiation and termination points for trajectories associated with each transition type.
Statistics: In the 2-dimensional state space, we used one-way ANOVA to compare centroid-to-centroid distances, core cluster areas, areas of high velocity, and mean trajectory durations between genotypes. In the 1-dimensional projection of the state space, we used one-way ANOVA to compare peak-to-peak distances and trough heights between genotypes. We report means ± standard deviation.
RESULTS
State Space Analysis Reliably Identifies Normal Behavioral States
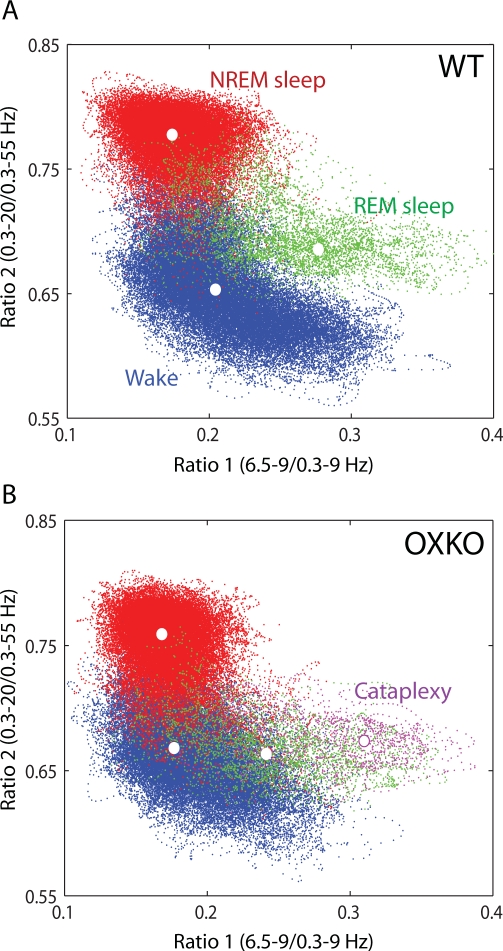
To investigate the dynamics of behavioral state instability in murine narcolepsy, we analyzed the second-by-second variations in spectral patterns of EEG activity (see Methods). In both WT and OXKO mice, states of Wake, NREM sleep, and REM sleep consistently mapped to distinct regions of the state space and gave rise to clusters corresponding to conventional scoring of these states (Figure 1). The dispersion of points within each cluster reflected variations in the depth of sleep or the intensity of wakefulness as measured by EEG power in the delta and theta bands, respectively. In OXKO mice, there appeared to be more overlap between the Wake and REM sleep clusters. Furthermore, cataplexy, brief episodes of atonia in the midst of wakefulness, mapped to a region overlapping REM sleep in these animals. Cataplexy did not occur in WT mice.
Figure 1.
Spectral ratios of EEG activity define a 2-dimensional state space with distinct clusters. Each plot shows 24 hours of EEG activity, and each point represents 1 second of EEG activity. In these graphs, the color of each state space point is determined from conventional scoring: Wake (blue), NREM sleep (red), REM sleep (green), and Cataplexy (magenta). Centroids of each cluster are denoted by white dots. A. State space analysis of a typical WT mouse. B. Same for a typical OXKO mouse. The relative locations of state clusters are conserved between WT and OXKO mice, but OXKO mice have cataplexy and show less cluster separation than WT mice.
Clusters in the State Space Differ Between Genotypes
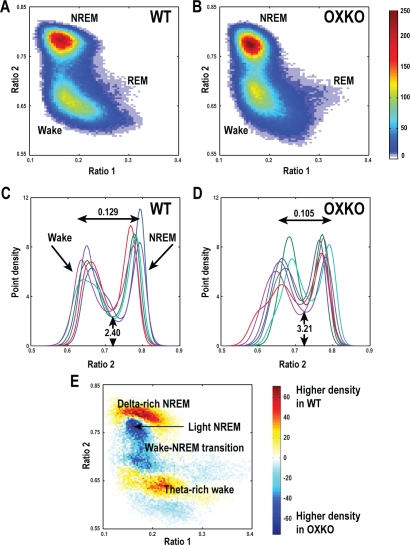
Within each genotype, state clusters were very similar, so we averaged state space plots across animals to examine the patterns typical of WT and OXKO mice (Figure 2). In both groups, most behavior was concentrated in clusters associated with Wake and NREM sleep. The general locations of the clusters were conserved between the two genotypes, but there were many important differences.
Figure 2.
OXKO mice have abnormal distributions of EEG activity during wake and sleep.
A, B—Point densities were averaged across mice of each genotype to create average WT and OXKO mouse density plots. WT mice (n = 6) have a Wake cluster that includes a region corresponding to active wakefulness with higher theta activity (high ratio 1), a highly concentrated NREM sleep cluster, and a relatively sparse REM sleep cluster, reflecting much less time spent in REM sleep compared to other states. In contrast, OXKO mice (n = 7) have a smaller Wake cluster concentrated closer to the transition region between Wake and NREM sleep. The color scale represents point densities with warm colors indicating higher densities.
C, D—State space densities for individual WT and OXKO mice projected into the ratio 2 dimension. Each color represents data for an individual mouse. The projection yields 2 peaks: the left peak is associated with Wake and the right peak is associated with NREM sleep. The horizontal peak-to-peak distances are shorter in OXKO mice (one-way ANOVA, P < 0.05). Trough height is higher in OXKO mice, reflecting more time spent in the transition region between Wake and NREM sleep (one-way ANOVA, P < 0.01).
E—Difference plot showing the average density pattern of OXKO mice subtracted from that of WT mice. Here the color scale highlights differences between genotypes: warm colors indicate regions where the average density is greater in WT mice, and cool colors indicate higher density in OXKO mice. WT mice spend more time in deep (delta-rich) NREM sleep and active (theta-rich) Wake, whereas OXKO mice spend more time in light NREM sleep and the Wake/NREM sleep transition region.
In OXKO mice, the Wake and NREM sleep clusters were closer together than in WT mice. To quantify this difference, we calculated the distances between cluster centroid pairs (see Figure 1 for examples of cluster centroids). The Wake and NREM sleep centroids were 16% closer in OXKO mice than in WT mice (0.12 ± 0.01 in WT vs. 0.10 ± 0.01 in OXKO, P < 0.05). In OXKO mice, the Cataplexy centroid was 60% further from the Wake centroid and 23% further from the NREM sleep centroid than the REM sleep centroid (0.13 ± 0.02 Cataplexy-Wake vs. 0.08 ± 0.03 REM-Wake; 0.17 ± 0.01 Cataplexy-NREM vs. 0.14 ± 0.02, P < 0 .01). This may reflect more power in theta frequencies during Cataplexy than in REM sleep. NREM-to-REM sleep and REM sleep-to-Wake centroid distances did not differ between the 2 genotypes.
To examine the separation between Wake and NREM sleep clusters in more detail, we projected the 2-D spectral density data into the ratio 2 dimension. This projection revealed 2 peaks corresponding to Wake and NREM sleep. In OXKO mice, the location of the Wake peak was more variable (0.669 ± 0.016 in OXKO vs. 0.65 ± 0.011 in WT) and horizontal peak-to-peak distances were significantly shorter (0.105 ± 0.012 in OXKO vs. 0.129 ± 0.014 in WT, P < 0.05), confirming the finding with cluster centroids. In addition, the height of the trough was higher in OXKO mice (3.21 ± 0.477 in OXKO vs. 2.40 ± 0.360 in WT, P < 0.01), showing that these animals spent more time in the transition region between Wake and NREM sleep.
To identify the regions in the 2-D state space that were associated with these differences in sleep/wake behavior, we subtracted the OXKO density plots from the WT density plots. This analysis revealed that OXKO mice spent less time in “deep” NREM sleep with high delta power and less time in “active” Wake rich in theta activity.19 Instead, OXKO mice spent more time in “light” NREM sleep and in the transition region between Wake and NREM sleep, consistent with the differences in peak-to-peak distances and trough heights in these animals. (The terms “deep” and “light“ reflect the vertical (ratio 2) location of these regions with respect to the peak of the NREM sleep cluster and indicate higher and lower than average delta power, respectively.) Therefore, although conventional scoring showed roughly normal amounts of Wake and NREM sleep in OXKO mice (Supplemental Table 1 available online at www.journalsleep.org), the greater amount of EEG activity in the transition region between Wake and NREM sleep may contribute to unstable sleep/wake behavior.
Large Regions of Fast State Space Velocities in OXKO Mice
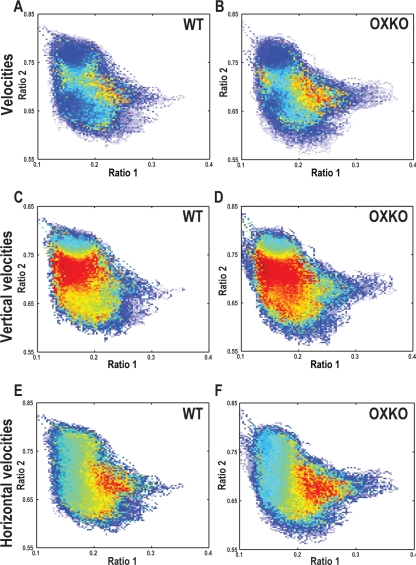
People and animals with narcolepsy often fall asleep very quickly (“sleep attacks”) or suddenly transition into cataplexy. To assess the dynamics of movement across the state space, we computed second-by-second velocities and averaged them across genotypes. In both WT and OXKO mice, state space velocities were slow within the Wake and NREM sleep clusters and fast between clusters (Figure 3), reflecting the stability of these states over time.
Figure 3.
OXKO mice have faster movements in certain regions of the state space. Plot colors represent the directionless (A, B), vertical (C, D), or horizontal (E, F) velocities of EEG activity originating in that region of the state space. Different color maps were used for each pair of panels to emphasize differences between genotypes.
A, B—Average directionless velocity plots in WT and OXKO mice distinguish between stable states where velocities are slow (cool colors) and transition regions where velocities are fast (warm colors).
C, D—The vertical (ratio 2) components of velocity vectors highlight the fast transitions between Wake and NREM sleep. In OXKO mice, this region of fast vertical velocities extends far into the middle of the Wake cluster.
E, F—The horizontal (ratio 1) components of the velocity vectors highlight the fast transitions between Wake and the REM sleep/Cataplexy clusters. For both horizontal and vertical velocities, the regions of fast transitions are larger in OXKO mice (one-way ANOVA, P < 0.01). This shows that regions of sleep/wake behavior that are normally stable are susceptible to rapid changes in OXKO mice, possibly contributing to the frequent state transitions in these animals.
Though WT and OXKO mice appeared to have similar velocity patterns, clear differences were apparent when velocities in the vertical (ratio 2) and horizontal (ratio 1) directions were analyzed separately. Movements around the state space are better described by vectors, with generally vertical trajectories as an animal moves between Wake and NREM sleep (changes in ratio 2), and horizontal trajectories as it transitions in and out of REM sleep/Cataplexy (changes in ratio 1)(Supplemental Movies 1 and 2 available online at www.journalsleep.org). Both WT and OXKO mice had very fast vertical velocities in the intermediate region between the Wake and NREM sleep clusters, but in OXKO mice, this region of fast velocities was 86% larger and extended well into the middle of the Wake cluster (0.022 ± 0.005 in OXKO vs. 0.012 ± 0.005 in WT, P < 0.05). Within this expanded region, upward movements in the Wake cluster were most common (data not shown) and are consistent with sudden transitions from Wake to NREM sleep. Both WT and OXKO mice had fast horizontal velocities as they transitioned from REM sleep to Wake, but in OXKO mice, the region of fast horizontal velocities was 72% larger and extended further into the REM sleep/Cataplexy region (0.018 ± 0.004 in OXKO vs. 0.010 ± 0.006 in WT, P < 0.05). In OXKO mice, fast horizontal rightward movements were more prominent than in WT mice, consistent with the presence of Wake to Cataplexy transitions (data not shown). In WT mice, horizontal movements were almost entirely leftward as these mice do not have direct transitions from Wake into the REM sleep region. These results demonstrate that OXKO mice can initiate rapid transitions towards NREM sleep and Cataplexy from regions of Wake that are much more stable (i.e., have many fewer transitions to other states) in WT mice.
Cluster Core Boundaries Reflect Differences in EEG Homogeneity
To examine the pathways underlying the transitions between states, we first defined cluster boundaries using a contour algorithm (Figure 4). These boundaries circumscribed “cluster cores,” stable regions in which the animal spent most of its time, and the relative areas of these cluster cores provided a measure of EEG homogeneity within the state. The area encompassed by the Wake core boundary was smaller in OXKO mice, suggesting a less diverse waking EEG. Most likely, this difference is a consequence of the reduction in theta-rich active waking we found in our density plot analysis (Figure 2). The area encompassed by the REM sleep core boundary was larger in OXKO mice, consistent with the more variable REM sleep-like behavior observed in these animals. Although cluster core boundaries for Wake and NREM sleep had high positive predictive value, 30% to 35% of the total EEG activity fell outside cluster core boundaries, indicating that the boundary technique was only moderately sensitive in encompassing these states.
Figure 4.
Using a contour algorithm, we delineated cluster core boundaries for each state.
A—Cluster core boundaries for Wake, NREM sleep, and REM sleep are drawn over a density plot for a representative WT mouse.
B—Cluster core areas provide a measure of EEG homogeneity with smaller clusters corresponding to less variable activity. The Wake cluster is smaller and the REM sleep cluster is larger in OXKO mice compared to WT mice (one-way ANOVA, P < 0.05).
C—Boundaries defined using the contour algorithm have high positive predictive value. In both genotypes, over 95% of data encompassed by the Wake boundary were independently scored as Wake using conventional scoring. The NREM sleep and REM sleep/Cataplexy cluster cores also have high positive predictive values.
D—The cluster core boundaries are moderately sensitive: 50% to 80% of all Wake and NREM sleep fall within these boundaries. The boundaries for REM sleep and Cataplexy are less sensitive, probably because these states are relatively uncommon, and data were sparse.
The cluster core boundaries of REM sleep and Cataplexy were more difficult to define because these states occur less frequently. About 70% of behavior within the REM sleep and Cataplexy boundaries was scored as such, but more than half of data scored as REM sleep or Cataplexy fell outside the cluster core boundaries, even in WT mice. Despite the reduced sensitivity of the cluster core boundaries for REM sleep and Cataplexy, comparisons with conventional scoring showed that, in general, at least part of each bout of REM sleep or Cataplexy fell within these boundaries. Therefore, transitions in and out of REM sleep and Cataplexy could be tracked using these cluster core boundaries.
Transitions in the State Space Reflect State Instability in OXKO Mice
By choosing cluster core boundaries with high positive predictive value, each cluster core was reliably associated with a single behavioral state. Each time a trajectory left a cluster core, it was classified as an inter-state or an intra-state transition depending on whether the next boundary it crossed corresponded to a different cluster core or a return to the original cluster core. Inter-state transitions followed distinct paths through the state space (Figure 5). For example, transitions from Wake to NREM sleep originated on the upper right region of the Wake cluster boundary, arced slightly through the transition region between the clusters, and then terminated on the lower right boundary of the NREM sleep cluster. In contrast, NREM sleep-to-Wake transitions began along the lower left boundary of the NREM cluster, and then arced downwards to the upper left boundary of the Wake cluster. Transition paths and mean transition trajectory durations were conserved between genotypes (one-way ANOVA, P > 0.05), suggesting that the process of transitioning between states was normal in OXKO mice. Despite the apparent normalcy of the transitions themselves, OXKO mice had higher probabilities of transitioning from Wake to NREM sleep or from Wake to REM sleep/Cataplexy (Table 1) consistent with previously described behavioral state instability in these animals.10,11
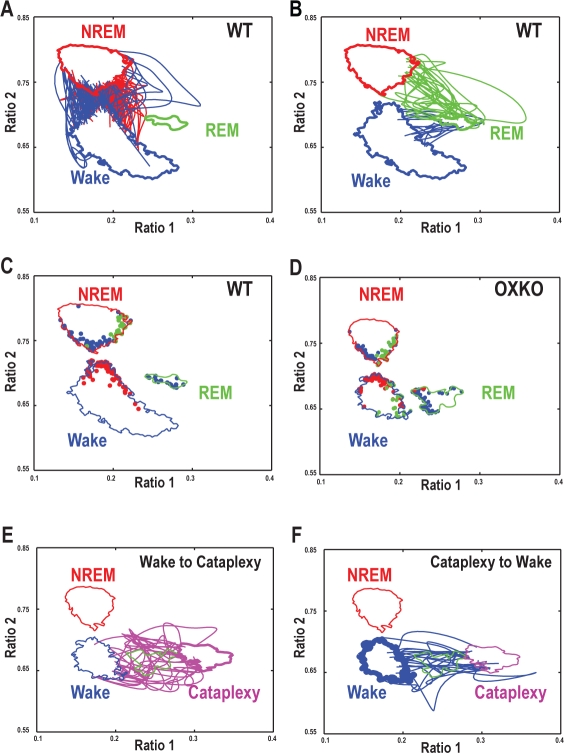
Figure 5.
The trajectories of individual transitions are normal in OXKO mice with the exception of transitions from Wake into Cataplexy. In all panels, the cluster core boundaries for each state for WT or for OXKO mice are shown (Wake - Blue; NREM - Red; REM-green; Cataplexy - magenta). Each panel also includes information about transition trajectories as described below.
A—Representative transition trajectories from Wake to NREM sleep (red lines) and from NREM sleep to Wake (blue lines) in an individual WT mouse. These transitions tend to arc right and left, respectively, suggesting that falling asleep and waking up are not simply inverse processes.
B—Representative transition trajectories from NREM sleep to REM sleep (green lines) and from REM sleep to Wake (blue lines) in an individual WT mouse.
C, D—Origins of transition trajectories in representative WT and OXKO mice are localized to specific regions of the state space. Origins are color-coded according to the terminating state: Wake (blue dots), NREM sleep (red dots), REM sleep (green dots). Origination regions are generally conserved between genotypes, although transitions from Wake to REM sleep/Cataplexy are common in OXKO mice (green dots on the right side of the Wake cluster core boundary). True Wake-to-REM sleep transitions almost never occur in WT mice, but due to the proximity of the Wake and REM sleep clusters, the boundary crossing approach occasionally identifies a few false positive Wake-to-REM sleep transitions.
E—Representative transition trajectories from Wake to Cataplexy (magenta lines) in an individual OXKO mouse. The direct movement from the Wake cluster to the Cataplexy/REM sleep region distinguishes these transitions from normal episodes of REM sleep that follow NREM sleep.
F—Representative transition trajectories from Cataplexy to Wake (blue lines) in an individual OXKO mouse. These transitions follow paths similar to transitions from REM sleep to Wake suggesting similar processes for terminating both Cataplexy and REM sleep. Furthermore, they constitute a reversal of the trajectories from Wake to Cataplexy indicating symmetry in EEG dynamics during transitions in and out of Cataplexy not observed with other transitions.
Table 1.
Inter- and intra-state transition probabilities show less stable Wake in OXKO mice.
| Termination state |
||||||
|---|---|---|---|---|---|---|
| Wake |
NREM sleep |
REM sleep/Cataplexy |
||||
| Origin state | WT | OXKO | WT | OXKO | WT | OXKO |
| Wake | 0.87 | *0.77 | 0.11 | *0.17 | 0.02 | *0.06 |
| NREM sleep | 0.10 | 0.14 | 0.86 | 0.82 | 0.03 | 0.04 |
| REM sleep/Cataplexy | 0.48 | 0.42 | 0.01 | 0.03 | 0.50 | 0.56 |
Transition probabilities show that Wake is less stable in OXKO mice compared to WT mice (*one-way ANOVA, P <0.01).
Analysis of intra-state transitions provided useful insights into the cohesion of behavioral states. Cluster cores were in the middle of the larger state clusters, and intra-state transition trajectories originated and terminated all around the core boundaries (data not shown), demonstrating cohesion of states even outside the core region. In OXKO mice, Wake-to-Wake transitions were less likely than in WT mice, demonstrating that if these animals left the core Wake region, they were more likely to transition to another state. This observation further supports the idea that waking behavior is less cohesive in OXKO mice. Interestingly, REM sleep-to-REM sleep transitions tended to be more likely in OXKO mice, though the challenge of defining REM sleep boundaries limits interpretation of this finding.
These results suggest that the patterns of EEG activity during state transitions are mostly normal in OXKO mice. However, OXKO mice have less stable cluster cores as reflected in the higher probability of inter-state transitions and reduced stability of Wake.
Cataplexy in OXKO Mice
OXKO mice had many episodes of Cataplexy, sudden transitions from wakefulness into a state with high EEG theta activity and atonia. Because Cataplexy occurs almost exclusively during the dark period and REM sleep occurs mainly during the light period, we analyzed transitions between Wake and Cataplexy in data from the 12-hour dark period. The Cataplexy cluster core was slightly above and to the right of the REM sleep cluster core, but the two regions had similar areas and often overlapped. Although WT mice appeared to have a few rare entries into the REM sleep region from the right border of the Wake cluster, these events are probably an artifact due to the proximity of the Wake and REM sleep cluster boundaries because true Cataplexy never occurred in WT mice.18 Despite the similarities in location for the Cataplexy and REM sleep clusters, the interstate transition trajectories clearly differentiate direct transitions from Wake to Cataplexy from the normal Wake to NREM sleep to REM sleep pattern (Figure 5).
DISCUSSION
To understand how orexin deficiency results in behavioral state instability, we used state space analysis of the EEG to examine sleep and wake states and the transitions between states, including velocities of these transitions, in OXKO mice. We found that state space clusters associated with Wake and NREM sleep were abnormally close in OXKO mice, suggesting that Wake and NREM sleep states are less distinct. In addition, OXKO mice spent more time in and near transition regions than WT mice. Finally, in OXKO mice, the fast velocities normally limited to state transition regions extended into cluster cores, indicating that behavioral states, especially Wake, are more labile. Each of these (possibly interrelated) factors may contribute to the unstable sleep/wake behavior of OXKO mice.
Causes of Behavioral State Instability in OXKO Mice
We found that the general locations of state space clusters associated with Wake, NREM sleep, and REM sleep are conserved in OXKO mice as suggested by prior studies.10,11 However, we identified two key differences between genotypes: first, the Wake and NREM sleep clusters are closer together in OXKO mice; second, OXKO mice spend less time in the extremes of the NREM sleep and Wake clusters (corresponding to large values of Ratio 2 and Ratio 1, respectively) and more time in the transition regions between the clusters. The reduced separation of clusters observed in OXKO mice suggests that Wake and NREM sleep are less distinct in these animals, while the differences in distributions within these states indicate that the quality of sleep and waking behavior is altered in OXKO mice.
The extreme regions of the NREM sleep and Wake clusters correspond to delta-rich (deep) NREM sleep and theta-rich (active) wakefulness. Transitions between Wake and NREM sleep do not originate in these regions, so, compared to WT mice, OXKO mice spend less time in typically “stable” areas of the Wake and NREM sleep clusters. This reduction in delta-rich NREM sleep in OXKO mice is consistent with the reduction in slow-wave activity described in people with narcolepsy.20,21 In fact, using a sleep deprivation protocol, Khatami and colleagues found that when slow-wave activity was increased in people with narcolepsy, individuals experienced less sleep fragmentation.21 It is possible that this improvement was associated with more time spent in stable regions of the NREM sleep cluster, as defined by this methodology.
In addition, OXKO mice spend more time in and near the Wake-NREM sleep transition region. Although this may contribute to the increased probability of state transitions observed in these animals, it is not the only factor increasing the likelihood of inter-state transitions in OXKO mice: dynamics also play an important role. In WT mice, high velocity vertical (ratio 2) and horizontal (ratio 1) movements occur only during transitions between states, but in OXKO mice, high velocities also occur within the Wake cluster core where velocities are normally slow. These inappropriately fast transitions out of what should be stable Wake are consistent with the rapid Wake to NREM sleep transitions described in narcoleptic dogs that skip through drowsiness and light sleep.13 In fact, these rapid transitions may underlie the “sleep attacks” and abrupt lapses in consciousness that are common in people with narcolepsy. Since the actual trajectories between states appear normal, orexin deficiency may not influence the general patterns of cortical activity during state transitions. Abnormal transition trajectories could be apparent in mice with state instability caused by abnormal thalamocortical signaling since changes in thalamocortical physiology would be directly reflected in the EEG.22–24
State Space Analysis of REM Sleep and Cataplexy
In OXKO mice, the Cataplexy and REM sleep clusters overlapped, though there were slight differences in centroid locations. The overlap in these patterns is consistent with previous work showing similarities in the spectral characteristics of these states.10,18,25 However, the state space approach clearly defined the abnormal transition trajectories from Wake to Cataplexy: paths originated in theta-rich regions of Wake and moved directly into the Cataplexy region without traveling through the NREM sleep region at all. Therefore, state space trajectories permit clear differentiation between abnormal transitions into Cataplexy and normal transitions from Wake to NREM sleep and then into REM sleep.
Advantages of the State Space Technique
As increasingly sophisticated genetic and physiologic techniques are applied to probe neuronal mechanisms involved in sleep/wake regulation, new measures of sleep/wake behavior are needed. Recently, several nonparametric and nonlinear methods to assess rest-activity rhythms have been introduced.26–29 These techniques have provided insights into altered rest-activity patterns associated with development, aging, and disease and have highlighted differences that are not detectable with standard linear power spectral analysis. Consistent with this trend, state space analysis provides a novel, non-categorical method for analyzing sleep/wake behavior.14–16 By enabling visualization of behavioral states as a continuum, state space analysis captures the richness of physiology better than conventional, categorical scoring of sleep/wake behavior. For example, the state space approach revealed clear differences in the depth of Wake and NREM sleep of OXKO mice that were not apparent with traditional spectral analysis, possibly because in these analyses transitional data is often excluded or diluted through averaging and because the state space approach employs the ratio of two frequency bands rather than a single frequency band.10,25
In addition, the fine temporal resolution of state space analysis allows investigation of state transitions and transition dynamics that is not possible with traditional methods. By tracking inter-state transition trajectories, we established that OXKO mice have normal patterns of EEG activity as they move between states. However, the high velocities identified in cluster cores for these animals showed that abrupt changes in state could originate in regions that are normally stable.
Although state space techniques were originally developed to analyze intracranial local field potentials, we have shown that they can be successfully applied to surface EEG recordings, thereby substantially broadening their applicability.14 In fact, the integrated neuronal information captured by surface EEG signals is more consistent across animals than intracranial field potentials that differ substantially depending on electrode placement. This consistency allows averaging within groups, thereby permitting detailed comparisons of animals with different sleep/wake phenotypes.
Limitations and Methodological Considerations
State space analysis of the EEG is especially useful for studying the dynamics of sleep/wake behavior, but it has some limitations. The separation between the Wake and REM sleep clusters was less well-defined using the surface EEG signal than it was with depth recordings, probably due to better detection of hippocampal theta activity with depth electrodes.14 Furthermore, because REM sleep and cataplexy together comprise less than 10% of all behavior, data corresponding to these states was sparse, and we had to amplify the data for our boundary analysis. The amplification partially relied upon conventional scoring which may have introduced some bias, and the resulting cluster core boundaries for REM sleep and Cataplexy still were less precise than the Wake and NREM sleep boundaries. Longer recordings and additional state space dimensions, possibly including an EMG dimension, should enable better resolution of the boundaries of these less common states.
More generally, state space analysis of the EEG reflects changes in cortical activity. Although these changes reliably reflect behavior, they do not directly address the role of orexin in stabilizing sleep/wake behavior through action on structures deeper in the brain.2,30 Furthermore, the use of state space techniques for comparison between genotypes requires an assumption that brain morphology is conserved. While this is a reasonable assumption for OXKO mice, it will be important to confirm these results in other rodent models of narcolepsy, such as ataxin-3 mice.31
Previous work has shown a high degree of coherent activity in cortex, hippocampus, striatum, and thalamus across states in normal animals,14 suggesting that a single channel of EEG is sufficient for state space analysis of sleep/wake behavior. However, reduced EEG coherence has been reported in people with narcolepsy32 and would support a hypothesized role for orexin across the cortex and in regions that coordinate cortical activity such as the basal forebrain and thalamus. The electrode locations in our study did not permit assessment of coherence, but future investigations with a more detailed EEG montage could use state space analysis to examine whether reduced cortical coherence contributes to sleep/wake instability.
Future Directions and Implications
This state space analysis provides strong support for Broughton's hypothesis that narcolepsy is caused by disruption of the boundaries between sleep and wake states.1 This approach enables visualization of these boundaries and reveals how abnormal dynamic processes contribute to state instability. Future work in which state space approaches are refined to incorporate additional details of the EEG and other physiologic measures should provide further insights into the mechanisms that disrupt state control in narcolepsy and other sleep disorders.
DISCLOSURE STATEMENT
This was not an industry supported study. Dr. Klerman has received investigator-initiated research support from Takeda (no salary support), Sepracor and Respironics and has participated in a speaking engagement for Sanofi-Aventis. Dr. Mochizuki has received research support from Jazz Pharmaceuticals. Dr. Scammell has received research support from Takeda and has participated in speaking engagements for Jazz Pharmaceuticals. The other authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
This study was supported by National Institutes of Health grants HL007901 (CDB), NS055367 (TES), HD045459 (EBK); NARSAD 2008 Young Investigator Award (SL); and AFOSR grant FA9550-08-1-0111 (CDB). OXKO mice were a kind gift from Takeshi Sakurai of Kanazawa University. The authors wish to thank Christian Baumann for helpful discussions of the work. We would also like to thank Erika Clark for technical support. Institution where work was performed: Beth Israel Deaconess Medical Center and Brigham and Women's Hospital.
Supplementary Table 1.
Using conventionally scored data we found that WT and OXKO mice spend similar percent time in states of wake, NREM sleep, and REM sleep for the 12 hour Light and Dark periods; in contrast to OXKO mice, WT mice did not spend any time in cataplexy. These findings are consistent with prior reports.10
| WT | OXKO | ||
|---|---|---|---|
| Wake | Light | 35 ± 3 | 36 ± 4 |
| Dark | 66 ± 4 | 63 ± 5 | |
| NREM | Light | 58 ± 4 | 58 ± 5 |
| Dark | 31 ± 3 | 31 ± 5 | |
| REM | Light | 8 ± 2 | 6 ± 2 |
| Dark | 2 ± 1 | 4 ± 2 | |
| Cataplexy | Light | 0 | 0.07 ± 0.1 |
| Dark | 0 | 2 ± 1 | |
REFERENCES
- 1.Broughton R, Valley V, Aguirre M, Roberts J, Suwalski W, Dunham W. Excessive daytime sleepiness and the pathophysiology of narcolepsy-cataplexy: a laboratory perspective. Sleep. 1986;9:205–15. doi: 10.1093/sleep/9.1.205. [DOI] [PubMed] [Google Scholar]
- 2.Peyron C, Tighe DK, van den Pol AN, et al. Neurons containing hypocretin (orexin) project to multiple neuronal systems. J Neurosci. 1998;18:9996–10015. doi: 10.1523/JNEUROSCI.18-23-09996.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thannickal TC, Moore RY, Nienhuis R, et al. Reduced number of hypocretin neurons in human narcolepsy. Neuron. 2000;27:469–74. doi: 10.1016/s0896-6273(00)00058-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lin L, Faraco J, Li R, et al. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–76. doi: 10.1016/s0092-8674(00)81965-0. [DOI] [PubMed] [Google Scholar]
- 5.Taheri S, Zeitzer JM, Mignot E. The role of hypocretins (orexins) in sleep regulation and narcolepsy. Annu Rev Neurosci. 2002;25:283–313. doi: 10.1146/annurev.neuro.25.112701.142826. [DOI] [PubMed] [Google Scholar]
- 6.Lee MG, Hassani OK, Jones BE. Discharge of identified orexin/hypocretin neurons across the sleep-waking cycle. J Neurosci. 2005;25:6716–20. doi: 10.1523/JNEUROSCI.1887-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mileykovskiy BY, Kiyashchenko LI, Siegel JM. Behavioral correlates of activity in identified hypocretin/orexin neurons. Neuron. 2005;46:787–98. doi: 10.1016/j.neuron.2005.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi K, Lin JS, Sakai K. Neuronal activity of orexin and non-orexin waking-active neurons during wake-sleep states in the mouse. Neuroscience. 2008;153:860–70. doi: 10.1016/j.neuroscience.2008.02.058. [DOI] [PubMed] [Google Scholar]
- 9.Adamantidis AR, Zhang F, Aravanis AM, Deisseroth K, de Lecea L. Neural substrates of awakening probed with optogenetic control of hypocretin neurons. Nature. 2007;450:420–4. doi: 10.1038/nature06310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mochizuki T, Crocker A, McCormack S, Yanagisawa M, Sakurai T, Scammell TE. Behavioral state instability in orexin knock-out mice. J Neurosci. 2004;24:6291–300. doi: 10.1523/JNEUROSCI.0586-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chemelli RM, Willie JT, Sinton CM, et al. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell. 1999;98:437–51. doi: 10.1016/s0092-8674(00)81973-x. [DOI] [PubMed] [Google Scholar]
- 12.Baker TL, Foutz AS, McNerney V, Mitler MM, Dement WC. Canine model of narcolepsy: genetic and developmental determinants. Exp Neurol. 1982;75:729–42. doi: 10.1016/0014-4886(82)90038-3. [DOI] [PubMed] [Google Scholar]
- 13.Nishino S, Riehl J, Hong J, Kwan M, Reid M, Mignot E. Is narcolepsy a REM sleep disorder? Analysis of sleep abnormalities in narcoleptic Dobermans. Neurosci Res. 2000;38:437–446. doi: 10.1016/s0168-0102(00)00195-4. [DOI] [PubMed] [Google Scholar]
- 14.Gervasoni D, Lin SC, Ribeiro S, Soares ES, Pantoja J, Nicolelis MA. Global forebrain dynamics predict rat behavioral states and their transitions. J Neurosci. 2004;24:11137–47. doi: 10.1523/JNEUROSCI.3524-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dzirasa K, Ribeiro S, Costa R, et al. Dopaminergic control of sleep-wake states. J Neurosci. 2006;26:10577–10589. doi: 10.1523/JNEUROSCI.1767-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin SC, Gervasoni D. Defining global brain states using multielectrode field potential recordings. In: Nicolelis MA, editor. Methods for neural ensemble recordings. 2nd ed. CRC Press; 2007. [PubMed] [Google Scholar]
- 17.Fujiki N, Cheng T, Yoshino F, Nishino S. Specificity of direct transition from wake to REM sleep in orexin/ataxin-3 transgenic narcoleptic mice. Exp Neurol. 2009;217:46–54. doi: 10.1016/j.expneurol.2009.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Scammell TE, Willie JT, Guilleminault C, Siegel JM. A consensus definition of cataplexy in mouse models of narcolepsy. Sleep. 2009;32:111–6. [PMC free article] [PubMed] [Google Scholar]
- 19.Czurko A, Hirase H, Csicsvari J, Buzsaki G. Sustained activation of hippocampal pyramidal cells by ‘space clamping’ in a running wheel. Eur J Neurosci. 1999;11:344–52. doi: 10.1046/j.1460-9568.1999.00446.x. [DOI] [PubMed] [Google Scholar]
- 20.Khatami R, Landolt HP, Achermann P, et al. Insufficient non-REM sleep intensity in narcolepsy-cataplexy. Sleep. 2007;30:980–9. doi: 10.1093/sleep/30.8.980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khatami R, Landolt HP, Achermann P, et al. Challenging sleep homeostasis in narcolepsy-cataplexy: implications for non-REM and REM sleep regulation. Sleep. 2008;31:859–67. doi: 10.1093/sleep/31.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Anderson MP, Mochizuki T, Xie J, et al. Thalamic Cav3.1 T-type Ca2+ channel plays a crucial role in stabilizing sleep. Proc Nat Acad Sci U S A. 2005;102:1743–8. doi: 10.1073/pnas.0409644102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joho RH, Ho CS, Marks GA. Increased gamma- and decreased delta-oscillations in a mouse deficient for a potassium channel expressed in fast-spiking interneurons. J Neurophysiol. 1999;82:1855–64. doi: 10.1152/jn.1999.82.4.1855. [DOI] [PubMed] [Google Scholar]
- 24.Wisor JP, Wurts SW, Hall FS, Lesch KP, Murphy DL, Uhl GR, Edgar DM. Altered rapid eye movement sleep timing in serotonin transporter knockout mice. Neuroreport. 2003;14:233–8. doi: 10.1097/00001756-200302100-00015. [DOI] [PubMed] [Google Scholar]
- 25.Willie JT, Chemelli RM, Sinton CM, et al. Distinct narcolepsy syndromes in orexin receptor-2 and orexin null mice: molecular genetic dissection of non-REM and REM sleep regulatory processes. Neuron. 2003;38:715–30. doi: 10.1016/s0896-6273(03)00330-1. [DOI] [PubMed] [Google Scholar]
- 26.Hu K, Van Someren EJ, Shea SA, Scheer FA. Reduction of scale invariance of activity fluctuations with aging and Alzheimer's disease: Involvement of the circadian pacemaker. Proc Nat Acad Sci U S A. 2009;106:2490–4. doi: 10.1073/pnas.0806087106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schwab K, Groh T, Schwab M, Witte H. Vol. 19. Woodbury, NY: Chaos; 2009. Nonlinear analysis and modeling of cortical activation and deactivation patterns in the immature fetal electrocorticogram; p. 015111. [DOI] [PubMed] [Google Scholar]
- 28.Carvalho-Bos SS, Riemersma-van der Lek RF, Waterhouse J, Reilly T, Van Someren EJ. Strong association of the rest-activity rhythm with well-being in demented elderly women. Am J Geriatr Psychiatry. 2007;15:92–100. doi: 10.1097/01.JGP.0000236584.03432.dc. [DOI] [PubMed] [Google Scholar]
- 29.Diniz Behn CG, Kopell N, Brown EN, Mochizuki T, Scammell TE. Delayed orexin signaling consolidates wakefulness and sleep: physiology and modeling. J Neurophysiol. 2008;99:3090–103. doi: 10.1152/jn.01243.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saper CB, Cano G, Scammell TE. Homeostatic, circadian, and emotional regulation of sleep. J Comp Neurol. 2005;493:92–8. doi: 10.1002/cne.20770. [DOI] [PubMed] [Google Scholar]
- 31.Hara J, Beuckmann CT, Nambu T, et al. Genetic ablation of orexin neurons in mice results in narcolepsy, hypophagia, and obesity. Neuron. 2001;30:345–54. doi: 10.1016/s0896-6273(01)00293-8. [DOI] [PubMed] [Google Scholar]
- 32.Park DH, Kwon JS, Jeong DU. Changes of EEG coherence in narcolepsy measured with computerized EEG mapping technique. Sleep Med Psychophysiol. 2001;8:121–8. [Google Scholar]