Abstract
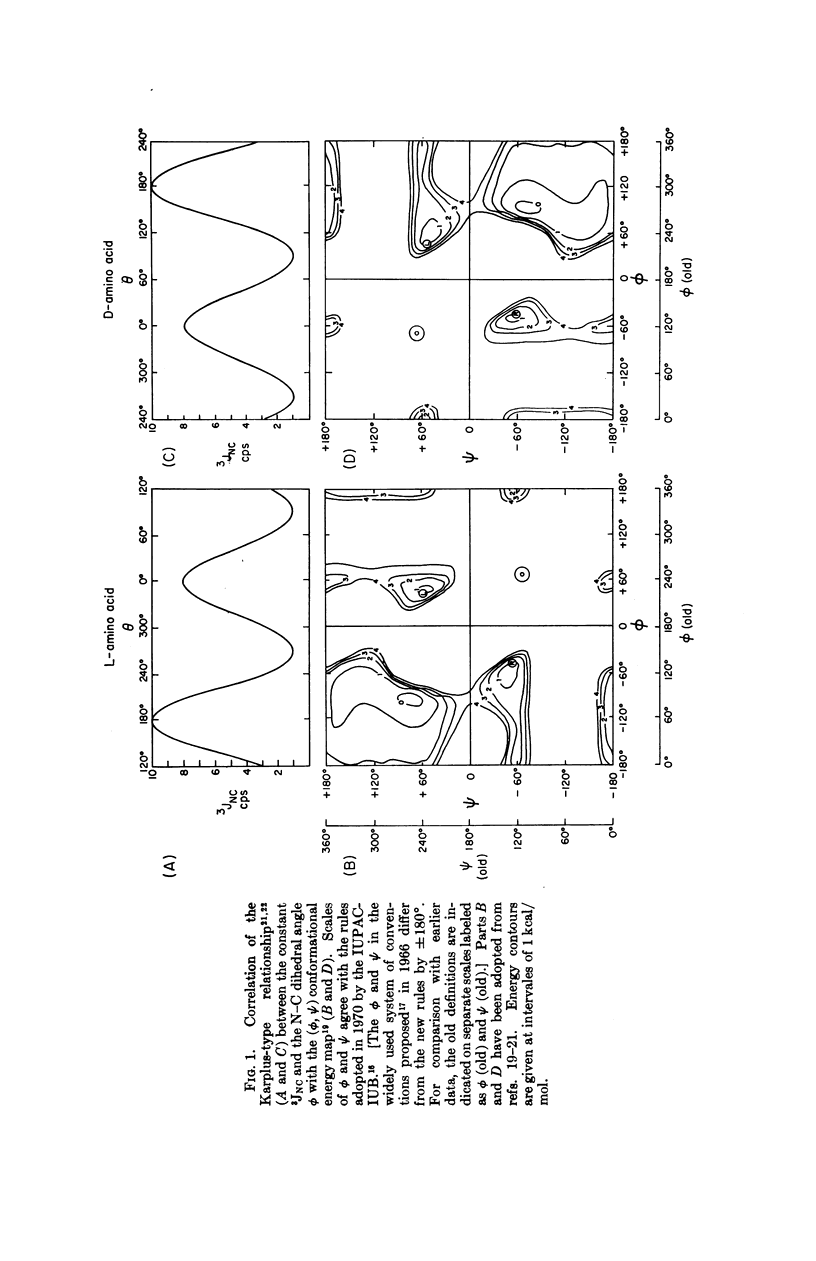
Simple criteria, based on the combined use of nmr spectral parameters and potential energy maps, are proposed for the conformational analysis of polypeptides and proteins. Experimentally determined coupling constants 3JNC for the N-Cα bond are consistent with the Karplus-Bystrov relationship. It is proposed therefore that 3JNC can be used to distinguish (a) between right-and left-handed α-helices, (b) between α-helical, β-pleated sheet, and randomly coiled forms of peptides. The average 3JNC for the random coil is predicted. The criteria proposed are valid for both L- and D-amino acids. Correlation between the Karplus-Bystrov relationship for 3JNC and the peptide conformational potential energy map limits the possible values of the N-Cα dihedral angle ϕ of each amino acid residue in a polypeptide and protein, and therefore presents a method of conformational analysis in solution superior to the use of either nmr or conformational maps alone. Nmr studies of hydrogen bonding or neighboring-group diamagnetic anisotropy reduce the number of possibilities consistent with the above criteria. A suggestion for evaluating the dihedral angle is presented. These criteria are useful provided the coupling constant is not obscured by line broadening.
Full text
PDF






Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bystrov V. F., Portnova S. L., Tsetlin V. I., Ivanov V. T., Ovchinnikov Y. A. Conformational studies of peptide systems. The rotational states of the NH--CH fragment of alanine dipeptides by nuclear magnetic resonance. Tetrahedron. 1969 Feb;25(3):493–515. doi: 10.1016/s0040-4020(01)83261-0. [DOI] [PubMed] [Google Scholar]
- Conti F. 220 Mc nuclear magnetic resonance spectra of gramicidin S in solution. Nature. 1969 Feb 22;221(5182):777–779. doi: 10.1038/221777b0. [DOI] [PubMed] [Google Scholar]
- Crippen G. M., Scheraga H. A. Minimization of polypeptide energy. 8. Application of the deflation technique to a dipeptide. Proc Natl Acad Sci U S A. 1969 Sep;64(1):42–49. doi: 10.1073/pnas.64.1.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edsall J. T., Flory P. J., Kendrew J. C., Liquori A. M., Nemethy G., Ramachandran G. N., Scheraga H. A. A proposal of standard conventions and nomenclature for the description of polypeptide conformation. J Biol Chem. 1966 Feb 25;241(4):1004–1008. [PubMed] [Google Scholar]
- Haynes D. H., Kowalsky A., Pressman B. C. Application of nuclear magnetic resonance to the conformational changes in valinomycin during complexation. J Biol Chem. 1969 Jan 25;244(2):502–505. [PubMed] [Google Scholar]
- Hvidt A., Nielsen S. O. Hydrogen exchange in proteins. Adv Protein Chem. 1966;21:287–386. doi: 10.1016/s0065-3233(08)60129-1. [DOI] [PubMed] [Google Scholar]
- Ivanov V. T., Laine I. A., Abdulaev N. D., Senyavina L. B., Popov E. M. The physicochemical basis of the functioning of biological membranes: the conformation of valinomycin and its K+ complex in solution. Biochem Biophys Res Commun. 1969 Mar 31;34(6):803–811. doi: 10.1016/0006-291x(69)90251-4. [DOI] [PubMed] [Google Scholar]
- Johnson L. F., Schwartz I. L., Walter R. Oxytocin and neurohypophyseal peptides: spectral assignment and conformational analysis by 220 MHz nuclear magnetic resonance. Proc Natl Acad Sci U S A. 1969 Dec;64(4):1269–1275. doi: 10.1073/pnas.64.4.1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopple K. D., Marr D. H. Conformation of cyclic peptides. The folding of cyclic dipeptides containing an aromatic side chain. J Am Chem Soc. 1967 Nov 22;89(24):6193–6200. doi: 10.1021/ja01000a035. [DOI] [PubMed] [Google Scholar]
- Kopple K. D., Ohnishi M. Conformations of cyclic peptides. II. Side-chain conformation and ring shape in cyclic dipeptides. J Am Chem Soc. 1969 Feb 12;91(4):962–975. doi: 10.1021/ja01032a029. [DOI] [PubMed] [Google Scholar]
- Laiken S. L., Printz M. P., Craig L. C. Tritium-hydrogen exchange studies of protein models. I. Gramicidin S-A. Biochemistry. 1969 Feb;8(2):519–526. doi: 10.1021/bi00830a010. [DOI] [PubMed] [Google Scholar]
- Markley J. L., Meadows D. H., Jardetzky O. Nuclear magnetic resonance studies of helix-coil transitions in polyamino acids. J Mol Biol. 1967 Jul 14;27(1):25–40. doi: 10.1016/0022-2836(67)90349-x. [DOI] [PubMed] [Google Scholar]
- Momany F. A., Vanderkooi G., Tuttle R. W., Scheraga H. A. Minimization of polypeptide energy. IV. Further studies of gramicidin S. Biochemistry. 1969 Feb;8(2):744–746. doi: 10.1021/bi00830a041. [DOI] [PubMed] [Google Scholar]
- Ohnishi M., Urry D. W. Temperature dependence of amide proton chemical shifts: the secondary structures of gramicidin S and valinomycin. Biochem Biophys Res Commun. 1969 Jul 23;36(2):194–202. doi: 10.1016/0006-291x(69)90314-3. [DOI] [PubMed] [Google Scholar]
- Pysh E. S. Conformations at local energy minimums for gramicidin S: optical calculations. Science. 1970 Jan 16;167(3916):290–292. doi: 10.1126/science.167.3916.290. [DOI] [PubMed] [Google Scholar]
- Ramachandran G. N., Venkatachalam C. M., Krimm S. Stereochemical criteria for polypeptide and protein chain conformations. 3. Helical and hydrogen-bonded polypeptide chains. Biophys J. 1966 Nov;6(6):849–872. doi: 10.1016/s0006-3495(66)86699-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts G. C., Jardetzky O. Nuclear magnetic resonance spectroscopy of amino acids, peptides, and proteins. Adv Protein Chem. 1970;24:447–545. doi: 10.1016/s0065-3233(08)60246-6. [DOI] [PubMed] [Google Scholar]
- Schwyzer R., Ludescher U. Conformational study of gramicidin S using the phthalimide group as nuclear magnetic resonance marker. Biochemistry. 1968 Jul;7(7):2519–2522. doi: 10.1021/bi00847a011. [DOI] [PubMed] [Google Scholar]
- Stern A., Gibbons W. A., Craig L. C. A conformational analysis of gramicidin S-A by nuclear magnetic resonance. Proc Natl Acad Sci U S A. 1968 Oct;61(2):734–741. doi: 10.1073/pnas.61.2.734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Victor T. A., Hruska F. E., Hikichi K., Danyluk S. S., Bell C. L. Nuclear magnetic resonance study of the structure and interactions of actinomycin D: temperature and solvent effects on the N--H and NH2 groups. Nature. 1969 Jul 19;223(5203):302–303. doi: 10.1038/223302a0. [DOI] [PubMed] [Google Scholar]


