Summary
Microfluidic reactors exhibit intrinsic advantages of reduced chemical consumption, safety, high surface-area-to-volume ratios, and improved control over mass and heat transfer superior to the macroscopic reaction setting. In contract to a continuous-flow microfluidic system composed of only a microchannel network, an integrated microfluidic system represents a scalable integration of a microchannel network with functional microfluidic modules, thus enabling the execution and automation of complicated chemical reactions in a single device. In this review, we summarize recent progresses on the development of integrated microfluidics-based chemical reactors for (i) parallel screening of in situ click chemistry libraries, (ii) multistep synthesis of radiolabeled imaging probes for positron emission tomography (PET), (iii) sequential preparation of individually addressable conducting polymer nanowire (CPNW), and (iv) solid-phase synthesis of DNA oligonucleotides. These proof-of-principle demonstrations validate the feasibility and set a solid foundation for exploring a broad application of the integrated microfluidic system.
Keywords: Integrated microfluidics; Chemical screening, In situ click chemistry; Sequential synthesis, Positron emission tomography probes; Oligonucleotide synthesis; Conducting polymer nanowires
Introduction
During the past century chemists studied reaction mechanisms and carried out chemical syntheses using reaction containers like beakers, round-bottomed flasks, test tubes and other specialized reaction apparatuses on their research benches. There have been enormous progresses made to achieve better understanding of reaction mechanisms,1 to establish a comprehensive collection of synthetic methodologies,2 as well as to accomplish syntheses of complicated nature products3 and artificial molecules.4 However, the conventional macroscopic reaction setting and their associated operation techniques have remained unchanged over the time. In the past two decades, significant efforts have been devoted to develop been devoted to develop microfluidic 5 reactors 6–16because these miniaturized devices offer advantages over the conventional reaction setting, such as reduced chemical consumption, safety, high surface-area-to-volume ratios, automation and improved control over mass and heat transfer. In addition, microfluidic reactors are amenable to interfacing with commonly used analytical instruments, e.g., UV–Vis spectroscopy,17 mass spectrometry (MS),18–20 gas chromatography (GC),21 liquid chromatography (LC) and LC/MS,22–24 offering a complete solution from reaction handling all the way to product analysis.
Fused glass25,26 and steel27,28 capillary tubes with inherent microscale internal dimensions, provide modular building blocks for assembly of microfluidic reactors on demand. These tube-based microfluidic networks can be applied for reactions with unique properties. Alternatively, photolithography and the associated microfabrication techniques cultivated by the contemporary semiconductor industry have set a solid foundation for computer-aided design (CAD) and scalable fabrication of chip-based microfluidic reactors with sizes smaller than a business card. Silicon and glass-based materials are compatible with these microfabrication processes and have been extensively used to produce a diversity of chip-based microreactors. 29–31 More recently, the advent of poly(dimethylsiloxane) (PDMS)-based softlithography32 technology offers a low cost and time-efficient approach for preparation of chip-based microreactors. It is important to note that from concept development to a working device, a PDMS-based microfluidic device can be designed and fabricated within three working days. However, the PDMS materials are susceptible33 to most of organic solvents or harsh chemicals/reagents, thus the PDMS-based microfluidic reactors are restricted to the reactions in aqueous solution or a few organic solvents (e.g., dimethyl sulfoxide and acetonitrile). Other plastic microfluidic devices have also been developed for conducting chemical reactions34–37 using different fabrication approaches. Based on reaction classification, we can simply divide the existing microfluidic reactors into two either continuous-flow or integrated microfluidic ones. We detail these two systems as the follows:
Continuous-flow microfluidics
A continuous-flow microfluidic system25–28 based on a internally connected microchannel network is the simplest microfluidic configuration that can be constructed by mechanical assembly of capillary tubes or lithographical fabrication of a on-chip microchannel network. Generally, reagents and solvents are introduced into a continuous-flow microfluidic reactor by syringes/syringe pumps or back pressures through tubing connections, and products are collected on the other end of the reactor. The characteristics of a continuous-flow microfluidic system confers great efficiency to different chemical reactions.10,38 For example, triphasic hydrogenation21 can be achieved with higher reaction efficiency, the inorganic synthesis of high-quality CdSe nanocrystals has been demonstrated,39,40 and chemical processes involving highly reactive intermediates can be executed with superior reaction selectivity. 41–44 However, challenges remain to explore the use of a continuous-flow microfluidic system for (i) parallel screening of chemical library in search of potential drug leads, and (ii) sequential synthesis of fine chemicals and pharmaceuticals, since cross-contamination between different reactions is difficult to avoid in a continuous flow setting.
Integrated microfluidics
In contrast to the continuous-flow microfluidic system, an integrated microfluidic system represents a scalable integration of a simple microchannel network with functional microfluidic modules, thus enabling the execution and automation of complicated chemical reactions and biological operations in a single device. Over the past 10 year, significant efforts have been devoted to the development of functional microfluidic modules that impart different utilities to microfluidic devices. For example, various valves 45–47 have been developed for isolation of distinct regions in a microreactor to avoid cross-contamination; different mixing modules have been utilized to overcome diffusion-limited mixing in the turbulence-free microfluidic environment; functioning pumps capable of delivering and metering fluidic components have been successfully integrated with microchannels. Among the existing integrated microfluidic systems, the PDMS-based integrated microfluidic system developed by Dr. Quake, is the most widely applied one, previously for biology,48 and now for chemistry.
Our research group at UCLA has pioneered in applying the PDMS-based integrated microfluidic system for complicated chemical reactions, including (i) parallel screening of in situ click chemistry libraries,49,50 (ii) multistep synthesis of radiolabeled imaging probes for positron emission tomography (PET),51 and (iii) sequential preparation of individually addressable conducting polymer nanowire (CPNW) electrode junctions. 52,53 In parallel, separate endeavors led by Dr. Quake came out with an integrated microfluidic reactor54 made of chemically resistant elastomer for sequential synthesis of DNA oligonucleotides. In this review, we would like to discuss how these PDMS-based integrated microfluidic reactors were developed to overcome a diversity of challenges encountered using macroscopic reaction setting. We hope these proof-of-principle demonstrations validate the feasibility and set a solid foundation for exploring a broad application of the integrated microfluidic reactors.
Screening microreactors for in situ click chemistry
In situ click chemistry
In situ click chemistry55,56 is a kinetically controlled target-guided synthesis (TGS)57–63 method for screening potential biligand inhibitors in a highly efficient manner. As one of the most widely used TGS approaches, in situ click chemistry enables selective assembly of a collection of complementary azide and acetylene building blocks inside the respective binding pockets of the target enzymes through a Huisgen cycloaddition reaction.64,65 Over the past eight years, such a TGS methodology has been successfully applied for the identification of inhibitors for a variety of biological targets, such as acetylcholine esterase (AchE),66 bovine carbonic anhydrase II (bCAII),56 HIV protease,67 and many other target proteins. Typically, an in situ click chemistry screening is conducted in 96-well plates.56 Since a stoichiometric amount of target protein is required for each in situ click reaction, the conventional experimental setting results in the significant consumption of the target proteins. The real challenge is that many interesting protein targets are notoriously difficult to obtain in large quantities, thus compromising the broad application of in situ click chemistry screening. Moreover, the conventional approach relies heavily upon manual operation, which limits screening throughput and fidelity of the outcomes. Therefore, it is imperative to develop a miniaturized and an automated platform capable of performing in situ click chemistry screening, on one hand to achieve an economical use of target proteins, and on the other hand to obtain an automated operation interface that rules out human operation error. We believe that the integrated microfluidics provides a great opportunity to overcome the challenges encountered by the conventional in situ click chemistry screening approach. Two generations of PDMS-based screening microreactors have been developed to enable highly efficient in situ click chemistry screening with significantly reduced sample consumption and gradually improved screening speed. During our proof-of-concept development, a known bovine carbonic anhydrase II (bCAII)56 in situ click chemistry system was employed as a model system.
1st-Generation screening microreactor
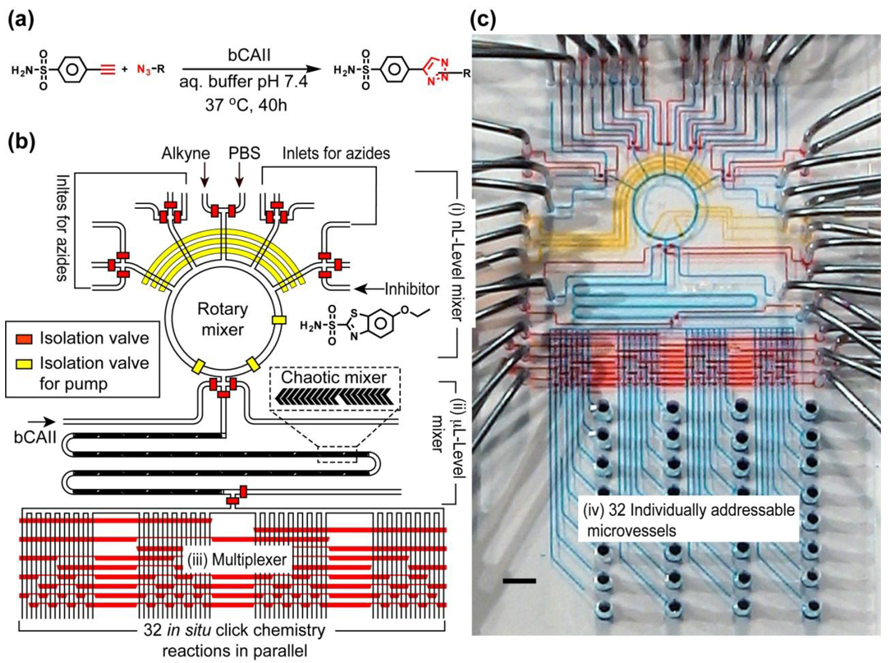
The 1st-generation screening microreactor 49 (Figure 1) was designed and fabricated to test the feasibility of a screening with 32 in situ click chemistry reactions (Figure 1a). In this pilot study, acetylenic benzenesulfonamide was used as the anchor molecule for screening a library of 20 complementary azides against the target enzyme bCAII. To achieve the on-chip screening, the reactor (Figure 1) incorporates four functional microfluidic components: (i) A nL-level rotary mixer responsible for selective sampling, precise metering, and rotary mixing of anchor molecule and complementary azides, (ii) a µL-level chaotic mixer68 for mixing µL-level bCAII solution with the reagent solutions generated in the rotary mixer, (iii) a multiplexer47 for directing each reaction mixture into one of the 32 individually addressable microvessels (component iv), which are millimeter-scale pin holes for storing the reaction mixtures. To fully utilize the automation performance of the integrated microfluidic system, a computer-controlled interface was employed to operate this screening microreactor for preparation of 32 reaction mixtures of the following types: (i) in situ click chemistry reactions in the presence of bCAII; (ii) control reactions with an inhibitor ethoxazolamide; (iii) thermal click reactions without bCAII; and (iv) a blank PBS solution containing only bCAII and a PBS solution utilized for the channel washing. After the library preparation, the microreactor with 32 different reaction mixtures (ca. 57 s reaction cycle−1) was incubated at 37 °C for 40 h, and the mixtures were collected for hit identification in a LC/MS. The 1st-generation screening microreactor validated the feasibility of performing a small-scale in situ click chemistry screening in an automated microfluidic setting, leading to a 5–12-fold improvement in sample/reagent consumption economy and enhanced experimental fidelity over the manually operated 96-well platform. The scalability of the integrated microfluidic allows further improvement of the next generation of devices, especially in increasing the number of screening reactions, reducing the consumption of samples and reagents, as well as accelerating operational speed.
Figure 1.
(a) In situ click chemistry reactions between an anchor molecule, acetylenic benzenesulfonamide and a library of 20 complementary azides in the presence of a target enzyme bCAII. (b) Schematic representation of the 1st-generation screening microreactor for testing the feasibility to perform a small scale screening of 32 in situ click chemistry reactions. The responsibilities of different hydraulic valves are illustrated by their colors: red for isolation valves and yellow for pump valves (for fluidic metering and circulation). (c) Optical image of the actual device. The various channels were loaded with food dyes to help visualize the different components of the microfluidic chip; the colors correspond to those in b), with blue indicating the fluidic channels.
2nd-Generation screening microreactor
The 2nd-generation screening microreactor50 (Figures 2) were subsequently developed for conducting up to 1024 in situ click chemistry reactions in conjunction with the use of an off-line hit identification approach. Similar to the 1st-generation device, the 2nd-generation screening microreactor (Figures 2) comprises also four functional microfluidic components: (i) a pair of microfluidic multiplexers47 for regulating the 2x16 individually addressed reagent inlets; (ii) a rotary mixer for mixing; (iii) a Page serpentine channel to accommodate each reaction mixture; and (iv) replaceable 20-cm long poly(tetrafluoroethylene) (PTFE) tubes for accommodating the 1024 reaction mixture slugs generated from the chip. Moreover, there were three vacuum suction membranes (green squares, Figure 2b) incorporated in the 2nd-generation device to reduce the time required for sample loading. In parallel, a miniaturized reverse phase clean-up step (ZipTip®) was developed to remove polar/charged reagents (i.e., DMSO and PBS salts) from the reaction mixtures that would otherwise interfere with direct electrospray ionization (ESI) mass spectrometry (MS) used for hit identification. This approach eliminated the need for a time-consuming liquid chromatographic step employed for the 1st-generation device. Again, a PC was employed to operate this 2nd-generation microreactor to handle an in situ click chemistry library composed of 16 acetylenes and 16 azides. Four different types of reaction conditions were tested in parallel to give 1024 individual reaction mixtures (ca. 17 s reaction cycle−1). After a sample clean-up process by ZipTip solid-phase extraction, the reaction mixtures were introduced into an ESI source of a triple quadrupole mass spectrometer for hit identification. At this moment our research team is restricted to analyzing the reaction results off-line, but in future developments, we intend to automate this aspect of the work as well.
Figure 2.
(a) Optical image of the 2nd-generation screening microreactor capable of carrying out 1024 in situ click chemistry reactions. The various channels were loaded with dyes to visualize the different components: red for isolation valves, yellow for isolation valves for pumping, green for vacuum and blue for fluidic channels. (b) Schematic representation of the device, illustaring the integration of different microfluidic modles for perfroming highly complicated sample prepaeration.
Comparison
A side-by-side comparison of sample/reagent consumption and operation times among the conventional 96-well approach and the two generations of screening microreactors are summarized in Table 1. In short, the 1st-generation screening microreactor enables a 5 to 12-fold sample economy over the conventional microliter platform. Further, a 20 to 50-fold of improvement in sample consumption and dramatically improved speed and throughput were achieved by the 2nd-generation screening microreactor. We demonstrated that the integrated microfluidic system can be applied for performing large-scale screening and promises to make lead discovery through in situ click chemistry more convenient and reliable, less expensive, and more diverse, through miniaturization of the library formation and development of a much sensitive hit identification approach. Although the systems have only been tested using a known bovine carbonic anhydrase II (bCAII) in situ click chemistry system. It is conceivable that such a miniaturized screening platform can be applied to screen other in situ click chemistry or TGS libraries in search of inhibitors for a variety of biological targets, including kinases which play critical roles in the malignant transformation of cancer.
Table 1.
Summary of the comparison among the conventional 96-well approach and the two generations of screening microreactors.
| Number of reactions |
Enzyme (bCAII) (µg) |
Alkyne (nmol) |
Azide (nmol) |
Total reaction volume (µL) |
Sample preparation time |
Hit identification time |
Detction methods |
|
|---|---|---|---|---|---|---|---|---|
| 96 well | 96 | 94 | 6 | 40 | 100 | few mins | few mins | LC-MS |
| 1st-Generation | 32 | 19 | 2.4 | 3.6 | 4 | 58 sec | 58 sec | LC-MS |
| 2nd-Generation | 1024 | 0.36 | 0.12 | 0.12 | 0.4 | 15 sec | 15 sec | MRM |
Multistep microreactors for PET imaging probe
FDG synthesis
Positron emission tomography (PET)69,70 is a noninvasive imaging technology for detailed mapping of biological processes in human and animal bodies. The development of sensitive and specific positron emitting radionuclide-labeled molecular probes is crucial for expanding the capability of targetspecific in vivo imaging for biological research and drug discovery. The commonly used positron emitting radionuclides have limited lifetimes, e.g., 18F, t1/2 = 109.7 min, 11C, t1/2 = 20.4 min, 13N, t1/2 = 9.96 min and 15O, t1/2 = 2.07 min. The short half-lifes of these radionuclides make rapid synthesis of doses essential. Among the commonly used PET imaging probes, the [18F]-labeled deoxyglucose analog, [18F]FDG,71 is the most commonly used PET probe for imaging normal and elevated metabolic states of disease processes of brain, heart and cancer. Presently, [18F]FDG is routinely produced in about 30–50 min using commercial synthesizers,72,73 which are expensive (ca. $140,000), have a physical size of approximately 80 × 60 × 40 cm, and produce about 10 to 100 doses in a single run. A unique aspect of [18F]FDG-PET imaging is that only nanogram mass (equivalent to 20 mCi radioactivity with 10 Ci/mmol specific activity) of the [18F]FDG is administered to a patient. Therefore, the preparation of [18F]FDG required expedited chemical kinetics and low-mass quantity of reaction platform, thus providing a unique opportunity74–77 for continuous microfluidics78–81 and integrated microfluidics.51,82 Moreover, the successful demonstration of chip-based [18F]FDG production give a conceptual model for the preparation of other molecules (including pharmaceuticals) because it includes common steps required in many chemical syntheses.
Multistep microreactor for synthesis of [18F]FDG
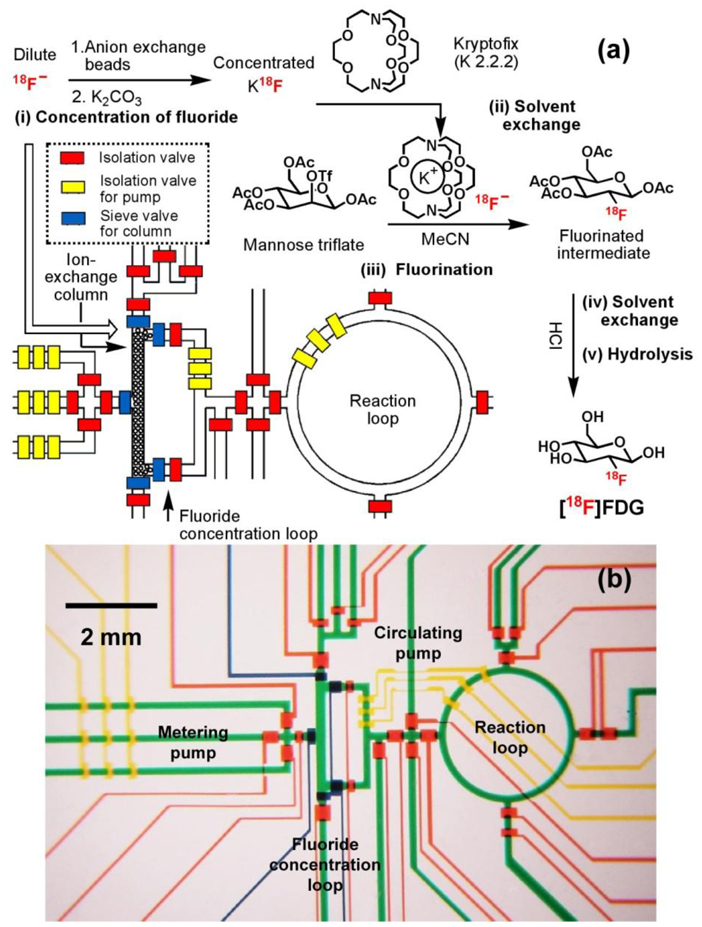
A multistep microreactor51 (Figure 3) capable of performing sequential chemical synthesis was developed for the synthesis of [18F]FDG. Similar to those applied in the commercial synthesizers, five sequential processes83 — (i) concentration of dilute [18F]fluoride using a miniaturized anion-exchange column located in the square-shaped fluoride concentration loop, (ii) solvent exchange from water to dry acetonitrile, (iii) fluorination of the D-mannose triflate precursor, (iv) solvent exchange back to water, and (v) acidic hydrolysis of the fluorinated intermediate in the ring-shaped reaction loop — produce nanogram (ng) levels of [18F]FDG. Like other integrated microfluidic devices, hydraulic valves with their delegated responsibilities, i.e., regular valves for isolation,45 pump valves for fluidic metering/circulation, and sieve valves84 for trapping anion exchange beads in the column module, were applied to control the automation of the multistep microreactor. In addition, the gas permeability of PDMS matrix allowed solvent exchange to occur within the microfluidic channel through direct evaporation, thereby enabling the sequential execution of anhydrous organic-solvent and aqueous-based chemical reactions. The automated device operation yielded the additional rewards of improved and accelerated chemical synthesis and high radiochemical purity (98%) of the resulting [18F]FDG. Besides [18F]FDG, other [18F]-labeled PET probes, for example, 3’-[18F]fluoro-3’-deoxythymidine ([18F]FLT)85 and 2-(1-[6-[(2-[18F]Fluoroethyl)methylamino]-2-naphthyl]ethylidene)malononitrile [18F]FDDNP86, whose synthetic procedures involve a nucleophilic [18F]-substitution reaction have also been successfully produced using similar microfluidic platforms.
Figure 3.
(a) Schematic representation of a PDMS-based microfluidic reactor used in the production of 2-deoxy-2-fluoro-D-glucose ([18F]FDG). Five sequential chemical processes produced nanogram (ng)-level of [18F]FDG. The operation of the device is controlled by pressure-driven valves, with their delegate responsibilities illustrated by their colors: red for isolation valves, yellow for isolation valves for pumping, and blue for sieve valves (for trapping anion exchange beads in the column module). (b) Optical micrograph of the central area of the microreactor. Various channels have been loaded with food dyes to help visualize different components of the microfluidic chip: colors as in (a), plus green for fluidic channels.
The results constitute a proof of principle for performing sequential chemical processes in this PDMS-based microfluidic reactor to produce nanogram-level [18F]FDG with enhanced operational efficiency and chemical economy. The advantages of an integrated microfluidic system with those of microfluidic environment provide a powerful tool to generalize, accelerate and diversify the preparation of radiolabeled imaging probes or any other compounds of limited supply or time sensitivity. In addition, this microfluidic reactor are capable of handling lower radioactivity on demand (amount enough for a single patient in each run). The devices have small dimensionality so that efficient shielding can be easily achieved. If the limitations associated with PDMS solvent/chemical resistance can be completely overcome,33,87 it is conceivable that the advantages of such automated chemical reaction circuits can help accelerate the drug discovery process by producing the small mass levels of drugs.
A solvent/chemical resistant integrated microreactor for solid-phase synthesis of oligonucleotide
Synthesis of oligonucleotide
Oligonucleotide synthesis plays a critical role in contemporary laboratory practice because it provides a rapid and inexpensive access to any desired oligonucleotide sequences.88 Nowadays, commercial solid-phase DNA synthesizers are capable of routinely producing single-stranded DNA or RNA molecules with around 15 to 25 bases for applications in gene sequencing, amplification, manipulation and detection. Automated solid-phase oligonucleotide synthesis is often carried out by a stepwise addition of A, C, G and T nucleoside building blocks, following a standard reaction cycle, including deblocking, coupling, capping and oxidation (Figure 4a). To miniaturize the automated solid-phase oligonucleotide synthesis in a PDMS-based integrated microfluidic chip is of great interest, reducing reagent consumption for chemical synthesis also offers the possibility of reducing waste proportionately and is thus environmentally friendly meanwhile it is essential to overcome the challenge associated with the poor solvent/chemical resistant performance33 of PDMS materials since the solid-phase synthesis requires the use of organic solvents and reagents. In a joint research endeavor led by Drs. DeSimone and Quake, a photocurable perfluoropolyether89,90 (PFPE) elastomer was developed91 to replace PDMS for the fabrication of solvent/chemical resistant integrated microfluidic devices. The fully cured PFPE matrix exhibits similar mechanical properties to those observed for PDMS. The multilayer softlithography fabrication approach can, therefore, be adopted for the fabrication of integrated microfluidic devices made of PFPE materials.
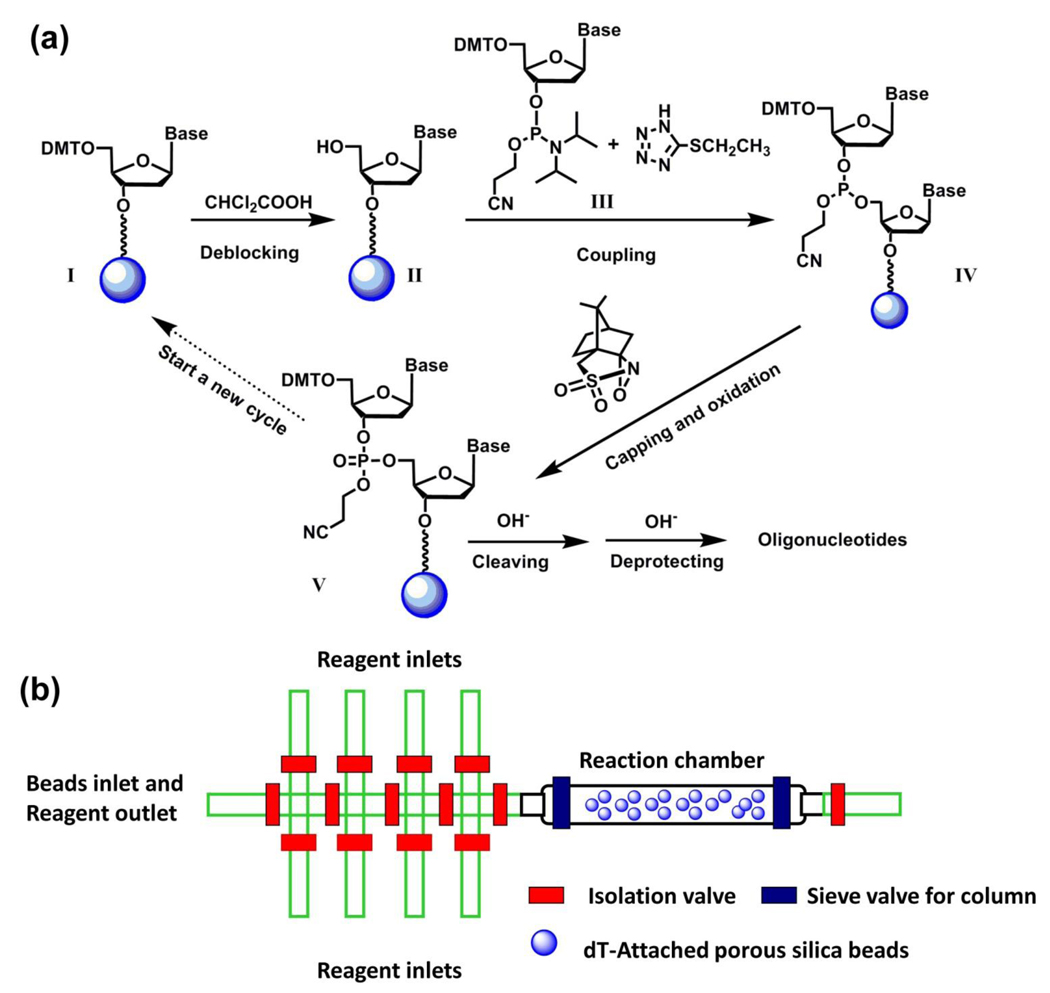
Figure 4.
(a) A solid-phase oligonucleotide synthesis by repeating a standard reaction cycle, including deblocking, coupling, capping and oxidation, by which A, C, G and T nucleoside building blocks were sequentially incorporated. (b) Schematic representation of PFPE-based integrated microreactor for solidphase synthesis of oligonucleotide. There were eight reagent inlets specifically assigned to different reagents/solvents, including acetonitrile, deblocking reagent, oxidizing reagent, activator, dT-CE phosphamidite, Pac-dA-CE phosphoramidite, iPr-Pac-dG-CE phophoramidite and Ac-dC-CE phosphoramidite. The ninth inlet at the left end of device serves two functions: (i) an inlet for beads loading during experimental setup and (ii) an outlet for reaction waste during the experiment.
PFPE-based integrated microreactor
In conjunction with a solid-phase oligonucleotide synthetic approach,92 an integrated microfluidic reactor54 made of PFPE elastomer (Figure 4b) has been developed for sequential synthesis of DNA oligonucleotides. In the device, there were nine individually addressed inlet channels for selective access to different solvents and reagents, including acetonitrile, deblocking/coupling/capping/ oxidation reagents, and phosphoramidite nucleotide precursors, as well as a reaction chamber in which porous silica beads were confined for the immobilization of the DNA oligonucleotides. As shown in Figure 4a, the synthesis of DNA oligonucleotides started from the precursor I which was pre-attached to the porous silica beads with a base-cleavable linker at the 3’-end. The 5’-position of this silica bead-attached precursor I was protected with dimethoxytrityl (DMT), which can be removed by the deblocking reagent to give free 5’-position in compound II for further coupling with phosphoramidite nucleotide precursor. A specific nucleotide precursor (III, for A, T, G and C nucleotides) can be introduced into the reaction chamber to couple with the surface-attached compound II, resulting in the product IV. (1S)-(+)-(10-camphorsulfonyl)oxaziridine (CSO) was utilized to oxidize93 phosphite triester IV to give phosphate triester V by the end of one reaction cycle. A single reaction cycle took about 9 min. The 20-mer DNA oligonucleotides with the sequence of 5’-CCG ACC TGG ATA CTG GCA TT-3’ were synthesized by performing multiple cycles of coupling reactions. The resulting DNA oligonucleotides were released from the beads by base treatments for sequential HPLC/MS characterization. This device is capable of producing 60 pmol DNA oligonucleotides, and only consuming less than 500 nL of 0.1 mol L−1 nucleotide precursor in each reaction cycle.
This work demonstrated that PFPE materials enable the use of organic solvents (i.e., acetonitrile and CH2Cl2) and harsh chemical reagents (e.g., phosphoramidite, dichloroacetic acid and CSO) in conjunction with an integrated microfluidic system. It is also a good example of integrating solid-phase synthetic protocol with an integrated microfluidic setting, which simplified the device operation processes, reduced chemical/solvent consumption and enabled an automated control with a PC interface. It is conceivable that a variety of integrated microfluidic reactors with similar design will be developed for the synthesis of other biopolymers, for example, oligopeptides.
Integrated microreactor for production of conducting polymer nanowires (CPNWs)
Conducting polymer nanowires
The production of one-dimensional nanostructured semiconducting materials for the ultrasensitive electronic sensing has received widespread attention. In the past two decades, significant progress has been achieved in the use of silicon nanowires94–96 and carbon nanotubes.97–100 Conducting polymer nanowires (CPNWs) are attractive alternatives to silicon nanowires and carbon nanotubes because of their tunable conductivity, flexibility, chemical diversity, and ease of processing. Our research group has previously demonstrated a template-free, site-specific, and scalable electrochemical method for the fabrication of individually addressable CPNW electrode junctions in a parallel-oriented array. These conducting electrochemically deposited CPNWs exhibited diameters ranging from 40 to 250 nm and have been applied as resistive sensors for detecting of several gas analytes at extremely low concentrations. 101–103
We foresaw that there are three profound advantages to carry out template-free electrochemical fabrication of CPNW electrode junctions in an integrated microfluidic device. First, the turbulence-free environment13 within the microchannel helps to improve the morphologies of CPNWs during their electrochemical growth. Second, the integrated microfluidic system assists the delivery of small amounts of monomer precursors and analytes, and enables the rapid exchange of nanoliter-level solutions in the same devices. Third, once the nanowires are grown, the entire device is ready for use, without the necessity of any post-fabrication processing.
Integrated microreactor for electropolymerization of conducting polymer nanowires (CPNWs)
The integrated microreactor56 (Figure 5a and b) capable of site-specific deposition of CPNWs, is composed two major components, including (i) a bottom silicon substrate with an array of Pt microelectrode junction pairs (each pair separated by a 2-µm-wide gap) for electropolymerization of CPNWs, and (ii) a 2-layer PDMS microfluidic component with individually addressable microchannel inlets for introduction of different monomer precursors, electrolyte and analysts. These two components were fabricated separately by standard photolithography and softlithography32 and then bonded together after oxygen plasma treatment. A galvanostatic current step method101,102 was applied for electropolymerization of CPNWs in the presence of precisely delivered aniline and pyrrole solutions, resulting in uniform polyaniline and polypyrrole nanowires with diameters ranged from 50 and 180 nm. (Figure 5c) We noted that the polyaniline and polypyrrole nanowires grown in the integrated microfluidic device were of better-defined morphologies, shorter growth times and experimental fidelity as a result of the turbulence-free environment in the device. After the electropolymerization of the CPNW electrode junctions, the entire device was applied for sensing of the NH3 and pH changes of buffered solutions after their fabrication within the microchannel.
Figure 5.
(a) Actual view of the microfabricted and assembled microreactor for electropolymerization of conducting polymer nanowires (CPNWs). (b) Micrograph of integrated microreactor, in which each microfluidic channel is 16 µm high and 100 µm wide and each of the five pairs of electrode junctions is separated by a 2-µm -wide gap. (c) SEM image of well-defined polyaniline nanowires grown in the microfluidic channels
Laminar flow guides the growth of CPNWs
As shown in Figure 5c, each electrochemical deposited CPNW electrode junction is composed of multiple randomly aligned CPNWs. Although the morphologies of on-chip fabricated CPNWs are dramatically improved compared to those observed in the macroscopic setting,101,102 the challenge remained to develop a CPNW sensor array, where only a single CPNW is grown across each pair of electrode junctions. To achieve this goal, we designed and fabricated a new type of microfluidic device, 57 in which hydrodynamically focused laminar stream104 was generated in situ for performing size-controllable, sitespecific electrochemical depositions of conducting polymer micropatterns and CPNWs across individually addressable electrode junction pairs. Similarly, this device (Figure 6a) is composed of (i) an array of 40 electrode junctions on a glass substrate for electrochemical deposition of CPNWs and (ii) an overlaying PDMS component for hydrodynamic focusing of a solution containing monomer precursors. The uniqueness of this approach is that the width of the monomer-containing focused stream can be altered by changing the flow rates of the surrounding sheath streams, and the position of the focused stream can be altered by the ratio of the sheath flow rates applied on both sides of the focused stream. Gathering the dynamic characteristics of flowing nature, as well as size and position-controllability, such a monomer-containing focused stream is defined as a dynamic template, which allows arbitrary spatial confinement of the redox-active monomer precursors for electrochemical deposition of CPNWs. More importantly, the deposition of CPNWs will not occur until an effective electrochemical potential is applied at the electrode junction located underneath the focused stream. Using this microfluidic setting, we have previously demonstrated the site-specific fabrication of an array of polypyrrole micropatterns with controllable widths ranging from 1 to 5 µm, and recently produced well aligned single CPNWs with uniform diameters of 300 nm. (Figure 6b) We also validated the fabrication fidelity and device scalability of this micro and nano-patterning approach by fabricating a sensor array composed of two different types of CP micropattern electrode junctions, i.e., seven Ppy and six carboxylic acid-substituted Ppy (COOH-Ppy) ones in a monolithic device. Two different sensing elements, i.e., a Ppy micropattern electrode junction and a COOH-Ppy one, selected from the sensor array were utilized to test the concept of electronic nose.105–107 We were able to distinguish (Figure 7) a library of saturated organic solvent vapors using this binary resistive sensor.
Figure 6.
(a) An Intedigitated microfluidic setup where a hydrodynamically focused laminar stream produced in can be employed as a dynamic template for site-specific electrochemical deposition of size controllable conducting polymer micropatterns across individually addressable electrode junction pairs. b) Scanning electron microscopy (SEM) image of 300-nm-wide Ppy nanopattern across a Pt electrode pair.
Figure 7.
(a) Real-time resistance responses (R) of a binary sensor composed of a Ppy- and a COOH-Ppy-based micropattern electrode junction upon periodic exposure to a library of saturated organic vapors (each 20 mL in volume). (b) Scatter plot summarizing the collective sensing responses to individual organic vapors. EA=ethyl acetate.
We have demonstrated that an integrated microfluidic platform capable of incorporating a variety of conducting polymer micropatterns and CPNWs (sensing elements) into a sensor array to achieve a sensing device with a broad diversity and specificity for recognition of a wide range of gas analytes. It is conceivable that this technology may be widely applied for micro and nanopatterning of other redox-active materials, including other conducting polymers, metals, inorganic semiconductors, and functional ceramic materials for broader application in other micro and nanoelectronic devices.
Conclusions and outlook
Overall, the review summarizes the proof-of-concept examples of a small collection of integrated microfluidic reactors for (i) screening chemical library, (i) producing imaging probes, (iii) preparing nanostructure materials, and (iv) performing solid-phase synthesis of DNA. From the stand point of chemistry, these results demonstrated a platform for performing complex synthetic processes on a single chip, in an automated fashion with precise digital control, faster reaction kinetics and improved yields, which revolutionizes the conventional way to perform chemical reactions. We foresee, in a boarder playing field, new-generation integrated microreactors could be integrated with other functioning platforms, e.g., a biological assay,108,109 for the broader exploration beyond a single functioning chemical platform on a chip. We envision new microfluidic platforms for a number of game systems: "Let's Play ____", where the blank is "drug discovery", "biomarker screening", "biochemical reaction kinetics", and "disease diagnosis" etc. where execution is rapid and learning curves are steep.
Acknowledgment
The authors appreciate research supports provided by National Institutes of Health, Department of Energy, Army Research Office and Defense Threat Reduction Agency.
Biographies

Wei-Yu Lin received his BS degree in Chemistry from Cheng-Kung University (1998) and his Ph.D. in Organic Chemistry from National Taiwan University (2006) in Taiwan. He is currently a postdoctoral fellow under the joint supervision between Prof. Clifton Shen and Prof. Hsian-Rong Tseng at UCLA. His has previously acquired extensive research experience on design, synthesis and characterization of a variety of electro-optical polymers. After joined UCLA, he has been working on the development of integrated microfluidic reactors for producing radioactive imaging probes for positron emission tomography as well as for performing large-scale screening of in situ click chemistry libraries in search of potent enzyme inhibitors.

Yanju Wang received her Ph.D. degree in Polymer Chemistry and Physics from Changchun Institute of Applied Chemistry, Chinese Academy of Sciences in 2002. Her Ph.D. thesis was focused on the production and characterization of polyaniline and its composite materials. From 2002 to 2004, she was a postdoctoral fellow at Max Planck Institute for Polymer Research, working on the development of conducting polymer-based DNA and protein sensors. She came to UCLA in 2005 with research interests to explore the applications of the integrated microfluidic system for producing of nanostructured materials, measuring kinase activities Leukemia specimens, performing large-scale screening of in situ click chemistry libraries.

Shutao Wang received his BS degree in Chemistry and MS degree in Inorganic Chemistry from Northeastern Normal University in 2000 and 2003, respectively. He then pursued his PhD study in Physical Chemistry under the supervision of Prof. Lei Jiang at Institute of Chemistry, Chinese Academy of Sciences, working on the development of bioinspired interfaces and responsive materials for application on controllable wettability. He joined UCLA as a postdoctoral fellow in 2007. Currently, his research has been focused on the development of novel micro/nano-platforms for synthesis of nanostructured materials, as well as early diagnosis of cancer metathesis.

Hsian-Rong Tseng is an Associate Professor in the in the Department of Molecular and Medical Pharmacology at University of California, Los Angles. He also holds membership in the Crump Institute for Molecular Imaging, Institute for Molecular Medicine and California NanoSystems Institute at UCLA. Prof. Tseng received his BS degree in Chemistry from Tunghai University (1993) and his PhD in Organic Chemistry from National Taiwan University (1998) in Taiwan. He conducted his postdoctoral studies with Prof. Sir Fraser Stoddart at UCLA, working on development of molecular electronic devices, before he joined the faculty of UCLA in 2003. Dr. Tseng’s current research focused on the development of microfluidic systems as enabling technologies for the fields of chemistry, in vitro molecular diagnostics and in vivo imaging.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Smith M, March J. March's advanced organic chemistry, reactions, mechanisms, and structure. 6th ed. Hoboken: Wiley-Interscience; 2007. [Google Scholar]
- 2.Li JJ. Name Reactions A Collection of Detailed Reaction Mechanisms. Berlin, Heidelberg: Springer-Verlag Berlin Heidelberg; 2006. [Google Scholar]
- 3.Nicolaou KC, Sorensen EJ. Classics in total synthesis : targets, strategies, methods. New York: VCH; 1996. [Google Scholar]
- 4.Balzani V, Credi A, Raymo FM, Stoddart JF. Angew. Chemie. Int. Ed. 2000;39:3349. doi: 10.1002/1521-3773(20001002)39:19<3348::aid-anie3348>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 5.Whitesides GM. Nature. 2006;442:368. doi: 10.1038/nature05058. [DOI] [PubMed] [Google Scholar]
- 6.deMello AJ. Nature. 2006;442:394. doi: 10.1038/nature05062. [DOI] [PubMed] [Google Scholar]
- 7.Ehrfeld W, Hessel V, Löwe H. Weinheim, New York: John Wiley & Sons., Wiley-VCH; 2000. [Google Scholar]
- 8.Fukuyama T, Rahman T, Sato M, Ryu I. Synlett. 2008:151. [Google Scholar]
- 9.Ahmed-Omer B, Brandt JC, Wirth T. Org. Bio. Chem. 2007;5:733. doi: 10.1039/b615072a. [DOI] [PubMed] [Google Scholar]
- 10.Fletcher PDI, Haswell SJ, Pombo-Villar E, Warrington BH, Watts P, Wong SYF, et al. Tetrahedron. 2002;58:4735. [Google Scholar]
- 11.Watts P, Haswell SJ. Chem. Soc. Rev. 2005;34:235. doi: 10.1039/b313866f. [DOI] [PubMed] [Google Scholar]
- 12.Watts P, Wiles C. Org. Bio. Chem. 2007;5:727. doi: 10.1039/b617327f. [DOI] [PubMed] [Google Scholar]
- 13.Watts P, Wiles C. Chem. Commun. 2007:443. doi: 10.1039/b609428g. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi J, Mori Y, Kobayashi S. Chem. Asian J. 2006;1:22. doi: 10.1002/asia.200600058. [DOI] [PubMed] [Google Scholar]
- 15.Jahnisch K, Hessel V, Lowe H, Baerns M. Angew. Chemie. Int. Ed. 2004;43:406. doi: 10.1002/anie.200300577. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Lin W-Y, Yu ZTF, Tseng H-R. Biol. Appl. Microfluid. 2008:183. [Google Scholar]
- 17.Chan EM, Alivisatos AP, Mathies RA. J. Am. Chem. Soc. 2005;127:13854. doi: 10.1021/ja051381p. [DOI] [PubMed] [Google Scholar]
- 18.Su J, Bringer MR, Ismagilov RF, Mrksich M. J. Am. Chem. Soc. 2005;127:7280. doi: 10.1021/ja051371o. [DOI] [PubMed] [Google Scholar]
- 19.Koster S, Verpoorte E. Lab Chip. 2007;7:1394. doi: 10.1039/b709706a. [DOI] [PubMed] [Google Scholar]
- 20.Schoenherr RM, Ye ML, Vannatta M, Dovichi NJ. Anal. Chem. 2007;79:2230. doi: 10.1021/ac061638h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kobayashi J, Mori Y, Okamoto K, Akiyama R, Ueno M, Kitamori T, et al. Science. 2004;304:1305. doi: 10.1126/science.1096956. [DOI] [PubMed] [Google Scholar]
- 22.Srbek J, Eickhoff J, Effelsberg U, Kraiczek K, van de Goor T, Coufal P. J. Sep. Sci. 2007;30:2046. doi: 10.1002/jssc.200700053. [DOI] [PubMed] [Google Scholar]
- 23.Van de goor TA. J. Chrom. Lib. 2007;72:165. [Google Scholar]
- 24.Brennen RA, Yin H, Killeen KP. Anal. Chem. 2007;79:9302. doi: 10.1021/ac0712805. [DOI] [PubMed] [Google Scholar]
- 25.Basheer C, Swaminathan S, Lee HK, Valiyaveettil S. Chem. Commun. 2005:409. doi: 10.1039/b414429e. [DOI] [PubMed] [Google Scholar]
- 26.Comer E, Organ MG. Chen. Eur. J. 2005;11:7223. doi: 10.1002/chem.200500820. [DOI] [PubMed] [Google Scholar]
- 27.Nielsen CA, Chrisman RW, LaPointe RE, Miller TE. Anal. Chem. 2002;74:3112. doi: 10.1021/ac020100i. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto S, Hanaoka T, Hamakawa S, Sato K, Mizukami F. Cata. Today. 2006;118:2. [Google Scholar]
- 29.Brivio M, Verboom W, Reinhoudt DN. Lab Chip. 2006;6:329. doi: 10.1039/b510856j. [DOI] [PubMed] [Google Scholar]
- 30.Kikutani Y, Horiuchi T, Uchiyama K, Hisamoto H, Tokeshi M, Kitamori T. Lab Chip. 2002;2:188. doi: 10.1039/b208382p. [DOI] [PubMed] [Google Scholar]
- 31.Jensen KF. MRS Bull. 2006;31:101. [Google Scholar]
- 32.Xia YN, Whitesides GM. Annl. Rev. Mater. Sci. 1998;28:153. [Google Scholar]
- 33.Lee JN, Park C, Whitesides GM. Anal. Chem. 2003;75:6544. doi: 10.1021/ac0346712. [DOI] [PubMed] [Google Scholar]
- 34.Eijkel JCT, Bomer J, Tas NR, van den Berg A. Lab Chip. 2004;4:161. doi: 10.1039/b315859d. [DOI] [PubMed] [Google Scholar]
- 35.Ryu KS, Shaikh K, Goluch E, Fan ZF, Liu C. Lab Chip. 2004;4:608. doi: 10.1039/b403305a. [DOI] [PubMed] [Google Scholar]
- 36.Fortier MH, Bonneil E, Goodley P, Thibault P. Anal. Chem. 2005;77:1631. doi: 10.1021/ac048506d. [DOI] [PubMed] [Google Scholar]
- 37.Yoon T-H, Park S-H, Min K-I, Zhang X, Haswell SJ, Kim D-P. Lab Chip. 2008;8:1454. doi: 10.1039/b804726j. [DOI] [PubMed] [Google Scholar]
- 38.Watts P, Haswell SJ. Curr. Opin. Chem. Biol. 2003;7:380. doi: 10.1016/s1367-5931(03)00050-4. [DOI] [PubMed] [Google Scholar]
- 39.Nakamura H, Yamaguchi Y, Miyazaki M, Uehara M, Maeda H, Mulvaney P. Chem. Lett. 2002:1072. doi: 10.1039/b208992k. [DOI] [PubMed] [Google Scholar]
- 40.Chan EM, Mathies RA, Alivisatos AP. Nano Letters. 2003;3:199. [Google Scholar]
- 41.Yoshida JI, Nagaki A, Yamada T. Chem. Eur. J. 2008;14:7450. doi: 10.1002/chem.200800582. [DOI] [PubMed] [Google Scholar]
- 42.Nagaki A, Iwasaki T, Kawamura K, Yamada D, Suga S, Ando T, et al. Chem-Asian J. 2008;3:1558. doi: 10.1002/asia.200800081. [DOI] [PubMed] [Google Scholar]
- 43.Kawaguchi T, Miyata H, Ataka K, Mae K, Yoshida J. Angew. Chemie. Int. Ed. 2005;44:2413. doi: 10.1002/anie.200462466. [DOI] [PubMed] [Google Scholar]
- 44.Nagaki A, Kim H, Yoshida J. Angew. Chemie. Int. Ed. 2008;47:7833. doi: 10.1002/anie.200803205. [DOI] [PubMed] [Google Scholar]
- 45.Unger MA, Chou HP, Thorsen T, Scherer A, Quake SR. Science. 2000;288:113. doi: 10.1126/science.288.5463.113. [DOI] [PubMed] [Google Scholar]
- 46.Grover WH, Ivester RHC, Jensen EC, Mathies RA. Lab Chip. 2006;6:623. doi: 10.1039/b518362f. [DOI] [PubMed] [Google Scholar]
- 47.Thorsen T, Maerkl SJ, Quake SR. Science. 2002;298:580. doi: 10.1126/science.1076996. [DOI] [PubMed] [Google Scholar]
- 48.Melin J, Quake SR. Annu. Rev. Biophys. Biomol. Struct. 2007;36:213. doi: 10.1146/annurev.biophys.36.040306.132646. [DOI] [PubMed] [Google Scholar]
- 49.Wang JY, Sui GD, Mocharla VP, Lin RJ, Phelps ME, Kolb HC, et al. Angew. Chemie. Int. Ed. 2006;45:5276. doi: 10.1002/anie.200601677. [DOI] [PubMed] [Google Scholar]
- 50.Wang Y, Lin W-Y, Liu K, Lin RJ, Selke M, Kolb HC, et al. Lab Chip. 2009;9:2281. doi: 10.1039/b907430a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee CC, Sui GD, Elizarov A, Shu CYJ, Shin YS, Dooley AN, et al. Science. 2005;310:1793. doi: 10.1126/science.1118919. [DOI] [PubMed] [Google Scholar]
- 52.Wang J, Bunimovich YL, Sui GD, Savvas S, Wang JY, Guo YY, et al. Chem. Commun. 2006:3075. doi: 10.1039/b604426c. [DOI] [PubMed] [Google Scholar]
- 53.Hou S, Wang S, Yu ZTF, Zhu NQM, Liu K, Sun J, et al. Angew. Chem. Int. Ed. 2008;47:1072. doi: 10.1002/anie.200704264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huang Y, Castrataro P, Lee CC, Quake SR. Lab Chip. 2007;7:24. doi: 10.1039/b613923j. [DOI] [PubMed] [Google Scholar]
- 55.Manetsch R, Krasinski A, Radic Z, Raushel J, Taylor P, Sharpless KB, et al. J. Am. Chem. Soc. 2004;126:12809. doi: 10.1021/ja046382g. [DOI] [PubMed] [Google Scholar]
- 56.Mocharla VP, Colasson B, Lee LV, Roper S, Sharpless KB, Wong CH, et al. Angew. Chemie. Int. Ed. 2005;44:116. doi: 10.1002/anie.200461580. [DOI] [PubMed] [Google Scholar]
- 57.Rideout D. Science. 1986;233:561. doi: 10.1126/science.3523757. [DOI] [PubMed] [Google Scholar]
- 58.Ramstrom O, Lehn JM. Nature Reviews Drug Discovery. 2002;1:26. doi: 10.1038/nrd704. [DOI] [PubMed] [Google Scholar]
- 59.Huc I, Lehn JM. Proc. Natl. Acad. Sci. USA. 1997;94:8272. doi: 10.1073/pnas.94.6.2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lehn JM, Eliseev AV. Science. 2001;291:2331. doi: 10.1126/science.1060066. [DOI] [PubMed] [Google Scholar]
- 61.Erlanson DA, Braisted AC, Raphael DR, Randal M, Stroud RM, Gordon EM, et al. Proc. Natl. Acad. Sci. USA. 2000;97:9367. doi: 10.1073/pnas.97.17.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Demko ZP, Sharpless KB. Angew. Chemie. Int. Ed. 2002;41:2110. [PubMed] [Google Scholar]
- 63.Lee LV, Mitchell ML, Huang SJ, Fokin VV, Sharpless KB, Wong CH. J. Am. Chem. Soc. 2003;125:9588. doi: 10.1021/ja0302836. [DOI] [PubMed] [Google Scholar]
- 64.Huisgen R. P Chem Soc London. 1961;357 [Google Scholar]
- 65.Kolb HC, Finn MG, Sharpless KB. Angew. Chemie. Int. Ed. 2001;40:2004. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 66.Lewis WG, Green LG, Grynszpan F, Radic Z, Carlier PR, Taylor P, et al. Angew. Chemie. Int. Ed. 2002;41:1053. doi: 10.1002/1521-3773(20020315)41:6<1053::aid-anie1053>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 67.Whiting M, Muldoon J, Lin YC, Silverman SM, Lindstrom W, Olson AJ, et al. Angew. Chemie. Int. Ed. 2006;45:1435. doi: 10.1002/anie.200502161. [DOI] [PubMed] [Google Scholar]
- 68.Stroock AD, Dertinger SKW, Ajdari A, Mezic I, Stone HA, Whitesides GM. Science. 2002;295:647. doi: 10.1126/science.1066238. [DOI] [PubMed] [Google Scholar]
- 69.Phelps ME. Proc. Natl. Acad. Sci. USA. 2000;97:9226. doi: 10.1073/pnas.97.16.9226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Phelps ME. PET : molecular imaging and its biological applications. New York: Springer; 2004. [Google Scholar]
- 71.Hamacher K, Coenen HH, Stocklin G. J. Nucl. Med. 1986;27:235. [PubMed] [Google Scholar]
- 72.Padgett HC, Schmidt DG, Luxen A, Bida GT, Satyamurthy N, Barrio JR. Appl. Radiat. Isot. 1989;40:433. doi: 10.1016/0883-2889(89)90213-x. [DOI] [PubMed] [Google Scholar]
- 73.Hamacher K, Blessing G, Nebeling B. Appl. Radiat. Isot. 1990;41:49. [Google Scholar]
- 74.Audrain H. Angew. Chemie. Int. Ed. 2007;46:1772. doi: 10.1002/anie.200603509. [DOI] [PubMed] [Google Scholar]
- 75.Elizarov AM. Lab Chip. 2009;9:1326. doi: 10.1039/b820299k. [DOI] [PubMed] [Google Scholar]
- 76.Miller PW. J. Chem. Technol. Biotechnol. 2009;84:309. [Google Scholar]
- 77.Lu SY, Pike VW. PET Chemistry. 2007;62:271. doi: 10.1007/978-3-540-49527-7_10. [DOI] [PubMed] [Google Scholar]
- 78.Gillies JM, Prenant C, Chimon GN, Smethurst GJ, Dekker BA, Zweit J. Appl. Radiat. Isot. 2006;64:333. doi: 10.1016/j.apradiso.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 79.Gillies JM, Prenant C, Chimon GN, Smethurst GJ, Perrie W, Hambletta I, et al. Appl. Radiat. Isot. 2006;64:325. doi: 10.1016/j.apradiso.2005.08.007. [DOI] [PubMed] [Google Scholar]
- 80.Lu SY, Watts P, Chin FT, Hong J, Musachio JL, Briard E, et al. Lab Chip. 2004;4:523. doi: 10.1039/b407938h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chun J, Lu S, Pike VW. J. Label. Compd. Radiopharm. 2009;52:S14. [Google Scholar]
- 82.Lin W-Y, Wang Y, Chen Y-C, Hou S, Satyamurthy N, Phelps ME, et al. Abstracts of Papers; 234th ACS National Meeting; August 19–23, 2007; Boston, MA, United States. ORGN; 2007. [Google Scholar]
- 83.Cai LS, Lu SY, Pike VW. Eur. J. Org. Chem. 2008:2853. [Google Scholar]
- 84.Hong JW, Studer V, Hang G, Anderson WF, Quake SR. Nat. Biotechnol. 2004;22:435. doi: 10.1038/nbt951. [DOI] [PubMed] [Google Scholar]
- 85.Shields AF, Grierson JR, Dohmen BM, Machulla HJ, Stayanoff JC, Lawhorn-Crews JM, et al. Nat. Med. 1998;4:1334. doi: 10.1038/3337. [DOI] [PubMed] [Google Scholar]
- 86.Liu J, Kepe V, Zabjek A, Petric A, Padgett HC, Satyamurthy N, et al. Mol. Imaging Biol. 2007;9:6. doi: 10.1007/s11307-006-0061-4. [DOI] [PubMed] [Google Scholar]
- 87.Toepke MW, Beebe DJ. Lab Chip. 2006;6:1484. doi: 10.1039/b612140c. [DOI] [PubMed] [Google Scholar]
- 88.Beaucage SL, Iyer RP. Tetrahedron. 1992;48:2223. [Google Scholar]
- 89.Willis PA, Greer F, Lee MC, Smith JA, White VE, Grunthaner FJ, et al. Lab Chip. 2008;8:1024. doi: 10.1039/b804265a. [DOI] [PubMed] [Google Scholar]
- 90.De Marco C, Girardo S, Mele E, Cingolani R, Pisignano D. Lab Chip. 2008;8:1394. doi: 10.1039/b803243b. [DOI] [PubMed] [Google Scholar]
- 91.Rolland JP, Van Dam RM, Schorzman DA, Quake SR, DeSimone JM. J. Am. Chem. Soc. 2004;126:2322. doi: 10.1021/ja031657y. [DOI] [PubMed] [Google Scholar]
- 92.Matteucci MD, Caruthers MH. J. Am. Chem. Soc. 1981;103:3185. [Google Scholar]
- 93.Ugi I, Jacob P, Landgraf B, Rupp C, Lemmen P, Verfurth U. Nucleos Nucleot. 1988;7:605. [Google Scholar]
- 94.Patolsky F, Lieber CM. Mater. Today (Oxford, U. K.) 2005;8:20. [Google Scholar]
- 95.Li Z, Chen Y, Li X, Kamins TI, Nauka K, Williams RS. Nano Lett. 2004;4:245. [Google Scholar]
- 96.Zheng G, Patolsky F, Cui Y, Wang WU, Lieber CM. Nat. Biotechnol. 2005;23:1294. doi: 10.1038/nbt1138. [DOI] [PubMed] [Google Scholar]
- 97.Kong J, Franklin NR, Zhou C, Chapline MG, Peng S, Cho K, et al. Science (Washington, D. C.) 2000;287:622. doi: 10.1126/science.287.5453.622. [DOI] [PubMed] [Google Scholar]
- 98.Dai H. Accounts Chem Res. 2002;35:1035. doi: 10.1021/ar0101640. [DOI] [PubMed] [Google Scholar]
- 99.Star A, Gabriel J-CP, Bradley K, Gruener G. Nano Lett. 2003;3:459. [Google Scholar]
- 100.Star A, Tu E, Niemann J, Gabriel J-CP, Joiner CS, Valcke C. Proc Natl Acad Sci U S A. 2006;103:921. doi: 10.1073/pnas.0504146103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang J, Chan S, Carlson RR, Luo Y, Ge G, Ries RS, et al. Nano Lett. 2004;4:1693. [Google Scholar]
- 102.Alam MM, Wang J, Guo YY, Lee SP, Tseng HR. J. Phys. Chem. B. 2005;109:12777. doi: 10.1021/jp050903k. [DOI] [PubMed] [Google Scholar]
- 103.Wang J, Coti KK, Wang J, Alam MM, Shyue J-J, Lu W, et al. Nanotechnology. 2007;18:424021/1. doi: 10.1088/0957-4484/18/42/424021. [DOI] [PubMed] [Google Scholar]
- 104.Knight JB, Vishwanath A, Brody JP, Austin RH. Phys Rev Lett. 1998;80:3863. [Google Scholar]
- 105.Albert KJ, Lewis NS, Schauer CL, Sotzing GA, Stitzel SE, Vaid TP, et al. Chem Rev. 2000;100:2595. doi: 10.1021/cr980102w. [DOI] [PubMed] [Google Scholar]
- 106.Janata J, Josowicz M. Nature Materials. 2003;2:19. doi: 10.1038/nmat768. [DOI] [PubMed] [Google Scholar]
- 107.McAlpine MC, Ahmad H, Wang DW, Heath JR. Nature Materials. 2007;6:379. doi: 10.1038/nmat1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- 109.Kamei K, Guo S, Yu ZT, Takahashi H, Gschweng E, Suh C, et al. Lab Chip. 2009;9:555. doi: 10.1039/b809105f. [DOI] [PubMed] [Google Scholar]