Abstract
Thylakoids are the chloroplast internal membrane systems that house light-harvesting and electron transport reactions. Despite the important functions and well-studied constituents of thylakoids, the molecular mechanism of their development remains largely elusive. A recent genetic study has demonstrated that plastidic type I signal peptidase 1 (Plsp1) is vital for proper thylakoid development in Arabidopsis (Arabidopsis thaliana) chloroplasts. Plsp1 was also shown to be necessary for processing of an envelope protein, Toc75, and a thylakoid lumenal protein, OE33; however, the relevance of the protein maturation in both of the two distinct subcompartments for proper chloroplast development remained unknown. Here, we conducted an extensive analysis of the plsp1-null mutant to address the significance of lumenal protein maturation in thylakoid development. Plastids that lack Plsp1 were found to accumulate vesicles of variable sizes in the stroma. Analyses of the mutant plastids revealed that the lack of Plsp1 causes a reduction in accumulation of thylakoid proteins and that Plsp1 is involved in maturation of two additional lumenal proteins, OE23 and plastocyanin. Further immunoblotting and electron microscopy immunolocalization studies showed that OE33 associates with the stromal vesicles of the mutant plastids. Finally, we used a genetic complementation system to demonstrate that accumulation of improperly processed forms of Toc75 in the plastid envelope does not disrupt normal plant development. These results suggest that proper maturation of lumenal proteins may be a key process for correct assembly of thylakoids.
Capture of light energy and subsequent electron transport reactions are key steps in photosynthesis. In eukaryotic cells, these reactions occur in thylakoids, the internal membrane systems of chloroplasts. Thylakoid constituents, i.e. membrane lipids, photosynthetic pigments, and various proteins, have been well defined (Schröder and Kieselbach, 2003; Peltier et al., 2004; Benning and Ohta, 2005; DellaPenna and Pogson, 2006; Masuda and Fujita, 2008), and genetic disruptions of their biosynthesis result in a severe reduction of thylakoid development (e.g. Dörmann et al., 1995; Jarvis et al., 1995; Hsieh and Goodman, 2005). Thylakoids depend for their lipid supply on the envelope membranes (Dörmann and Benning, 2002). Early cytological studies suggested that thylakoids derive from the inner envelope membrane by invagination followed by vesicle traffic and/or lateral transfer of membrane constituents (e.g. Mühlethaler and Frey-Wyssling, 1959; von Wettstein, 1959; Hoober et al., 1991). Several putative components for the vesicle trafficking have been identified (Hugueney et al., 1995; Kroll et al., 2001; Wang et al., 2004), although the mode of their functions remains elusive. Overall, molecular details of thylakoid development are not yet fully understood.
Proteins found in thylakoid membranes are encoded in either the chloroplast or nuclear genome, whereas lumenal proteins are encoded exclusively in the nucleus (Robinson and Mant, 2005). All of the nuclear-encoded thylakoid proteins identified so far are synthesized on cytosolic ribosomes with N-terminal extensions called transit peptides. These precursor proteins first traverse the double-membrane envelope via the general import machinery called the TOC and TIC complexes (for translocon at the outer- and inner-envelope-membranes of chloroplasts; Schnell et al., 1997), and their transit peptides are removed by a soluble metalloprotease, stromal processing peptidase (Richter et al., 2005). Extensive biochemical and genetic studies have identified four distinct pathways in the stroma to mediate sorting of these nuclear-encoded proteins to thylakoids (Robinson and Mant, 2005; Cline and Dabney-Smith, 2008). The cpSec (for chloroplast Sec) and cpTat (for chloroplast twin-arginine translocation) pathways catalyze protein targeting to the lumen. The cpSRP (for chloroplast signal recognition particle) pathway mediates sorting of the most abundant membrane proteins, LHCPs (for light-harvesting chlorophyll binding proteins), whereas a nonassisted spontaneous pathway targets several other integral membrane proteins in thylakoids. All known nuclear-encoded substrates of cpSec (e.g. OE33 [for oxygen evolving complex subunit 33] and plastocyanin), cpTat (e.g. OE23, OE17, and PsaN), and some spontaneous (e.g. CFoII, PsbW, and PsbX) pathways carry a cleavable thylakoid-targeting signal peptide that follows the stroma-targeting transit peptide in their N termini (Robinson and Mant, 2005). The signal peptides are necessary for the intraorganellar targeting and are removed in the lumen by a type I signal peptidase (SPase I) called thylakoidal processing peptidase (TPP).
SPases I are membrane-bound Ser proteases responsible for cleavage of intracellular and/or intraorganellar targeting sequences (signal peptides) from numerous proteins in both prokaryotic and eukaryotic cells (Paetzel et al., 2002). In bacteria, it was shown that signal peptide cleavage is required for some proteins to be released from the plasma membrane during or after their translocation from the cytoplasm (Dalbey and Wickner, 1985), and SPases I are essential for viability (Date, 1983; Cregg et al., 1996; Zhang et al., 1997; Zhbanko et al., 2005). In thylakoids, by contrast, removal of the signal peptide was shown in vivo to be dispensable for the function and assembly of a chloroplast-encoded substrate, cytochrome f, of the green alga Chlamydomonas reinhardtii (Kuras et al., 1995; Baymann et al., 1999). In addition, the precursor form of spinach (Spinacia oleracea) OE33 that contains both the stroma- and thylakoid-targeting sequences was able to reconstitute oxygen evolution activity in vitro (Popelkova et al., 2002). Nonetheless, the importance of signal peptide cleavage for thylakoid biogenesis remains largely unexplored. In 1998, the first plant TPP cDNA was identified from Arabidopsis (Arabidopsis thaliana) seedlings based on similarity of the encoded protein to a cyanobacterial SPase I in its sequence (Chaal et al., 1998). An immunoblot assay showed thylakoid localization of this protein (At2g30440), and an in vitro assay confirmed its processing activity against wheat (Triticum aestivum) OE23 (Chaal et al., 1998). More recent genetic and immunological studies showed that a second TPP isoform in Arabidopsis, named plastidic type I signal peptidase 1 (Plsp1), is involved in processing of an envelope protein, Toc75, and a lumenal protein, OE33 (Inoue et al., 2005; Shipman and Inoue, 2009). The genetic study also showed that Plsp1 is necessary for proper thylakoid development (Inoue et al., 2005), although the mode of its action remains elusive.
In this work, we conducted a more extensive characterization of plsp1-1 plastids to demonstrate the significance of protein maturation for thylakoid development. Disruption of the PLSP1 gene led to a reduction in the level of thylakoid proteins and to accumulation of improperly processed forms of multiple lumenal proteins, in addition to those of OE33 and Toc75. Plastids lacking Plsp1 accumulated stroma-localized vesicles of various sizes, at which OE33 was present. Finally, we used a genetic complementation system to show that complete maturation of the envelope protein Toc75 is dispensable for proper plant development. These results suggest that the maturation of lumenal proteins may be necessary for proper development of thylakoids.
RESULTS
Lack of Plsp1 Causes Morphological Defects of Arabidopsis Chloroplasts
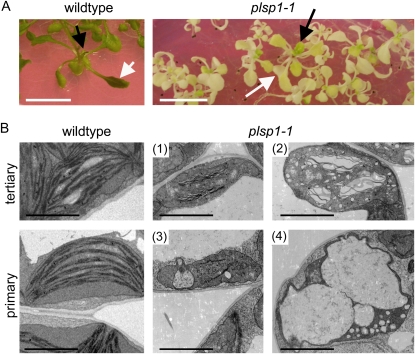
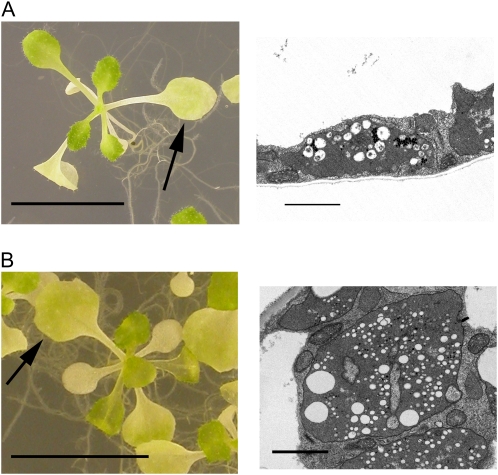
Disruption of PLSP1 expression by a T-DNA insertion caused a seedling lethal phenotype in which development of internal membranes was severely retarded in cotyledon plastids (Inoue et al., 2005). This phenotypic defect, which was observed when the plants were grown on media containing 1% Suc, clearly indicates the significance of Plsp1 for proper thylakoid development. However, further analysis of the mutant plants was difficult because of their poor growth. By contrast, when the mutant plants were grown on media supplemented with 3% Suc to support heterotrophic growth, they were able to develop multiple pale true leaves (Fig. 1A). Hence, seedlings grown under this condition were used for further studies. Interestingly, the severity of the pale phenotype appeared less in newly developed leaves than in more mature leaves. In electron micrographs, plastids from the youngest tertiary leaves of plsp1-1 plants were found to contain internal membrane structures (Fig. 1, B1), which appear to be poorly stacked lamellae, as well as multiple vesicular inclusions (Fig. 1, B2). By contrast, plastids in the oldest primary leaves of the mutant plants lacked detectable lamellae and granal membranes (Fig. 1, B3 and B4), similar to previously reported cotyledon plastids (Inoue et al., 2005), and many of them contained numerous vesicle-like structures in the stroma (Fig. 1, B4). The formation of these structures is unlikely to be a consequence of photooxidative damage because the plants examined were grown under 100 μmol m−2 s−1, which is generally considered a lower light condition (e.g. Havaux et al. 2005), and the vesicle-like structures were also observed in the mutant plants grown under even lower light conditions (30 and 60 μmol m−2 s−1; Fig. 2).
Figure 1.
Disruption of proper thylakoid formation in plsp1-1 plastids. A, Arabidopsis wild-type and plsp1-1 seedlings grown on solid media containing 3% Suc for 29 d. Bars = 1 cm. Newly developed tertiary and mature primary leaves examined in B are indicated with black and white arrows, respectively. B, Electron micrographs of plastids from newly developed tertiary leaves (top panels) and those from mature primary leaves (bottom panels) of wild-type and mutant plants. For the mutant leaves, representative images of four distinct plastid types (1–4) are shown. Bars = 2 μm.
Figure 2.
Light intensity does not affect the formation of stromal vesicles in plsp1-1 plastids. A and B, Shown at left are Arabidopsis plsp1-1 seedlings grown on solid media containing 3% Suc for 21 d in 30 μmol m−2 s−1 (A) or 60 μmol m−2 s−1 (B) light. Bars = 1 cm. Shown at right are electron micrographs of plastids from a mature primary leaf indicated with arrows. Bars = 2 μm. [See online article for color version of this figure.]
Proteomic Analysis Reveals the Presence of Photosynthetic Proteins in plsp1-1 Plastids
Previous immunoblotting of total protein extracts from etiolated seedlings revealed that plsp1-1 mutants contained near-wild-type levels of five chloroplast proteins, including the thylakoid lumenal protein OE33 (Inoue et al., 2005). We wished to test whether these and other chloroplast proteins were indeed in the mutant plastids. To this end, we prepared plastids from plsp1-1 seedlings and examined their proteome by tryptic digestion followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis and peptide identification according to the proteomics publishing guidelines (Bradshaw et al., 2006). Putative functions as well as subcellular and suborganellar locations of the identified proteins were assigned by cross-referencing the Plant Proteome Database (Sun et al., 2009) and the Arabidopsis Information Resource database (http://www.arabidopsis.org). Of the 113 proteins identified in the analysis, 94 were annotated as chloroplastic (Supplemental Table S1). This result indicated a successful plastid enrichment in the preparation from the mutant plants. Among proteins identified were those critical for proper chloroplast development, such as protein import, chloroplast gene expression, and various metabolic reactions, including a total of seven chloroplast-encoded proteins (Supplemental Table S1). Interestingly, despite the severe reduction of thylakoid development, plsp1-1 plastids accumulated various thylakoid photosynthetic proteins, including five independent TPP substrates (Table I); however, none of the identified peptides derived from the TPP substrates corresponded to a lumen-targeting sequence.
Table I. Photosynthetic thylakoid proteins identified in plsp1-1 plastids.
TPP substrates are indicated with asterisks.
| Group | Protein Name | Accession |
| PSII subunits | CP47 | ATCG00680 |
| PsbC | ATCG00280 | |
| PsbD/D2 | ATCG00270 | |
| PsbS | At1g44575 | |
| PsbO1/OE33* | At5g66570 | |
| PsbO2/OE33* | At3g50820 | |
| PsbP1/OE23* | At1g06680 | |
| PsbP2/OE23* | At2g30790 | |
| PSI subunits | PsaE | At2g20260 |
| PsaP | At2g46820 | |
| LHCPs | Lhcb1.1 | At1g29920 |
| Lhcb2.1 | At2g05100 | |
| Lhcb3 | At5g54270 | |
| Lhcb6 | At1g15820 | |
| CB26 | At4g10340 | |
| CB29 | At5g01530 | |
| ATP synthase subunits | α | ATCG00120 |
| β | ATCG00480 | |
| Plastocyanin | PETE2* | At1g20340 |
| Ferredoxin | FD2 | At1g60950 |
| Ferredoxin oxidoreductase | FNR1 | At5g66190 |
| FNR2 | At1g20020 |
Some But Not All Photosynthetic Proteins Accumulate at a Low Level, and a Subset of Thylakoid Lumenal Proteins Exist as Larger Forms in plsp1-1 Plastids
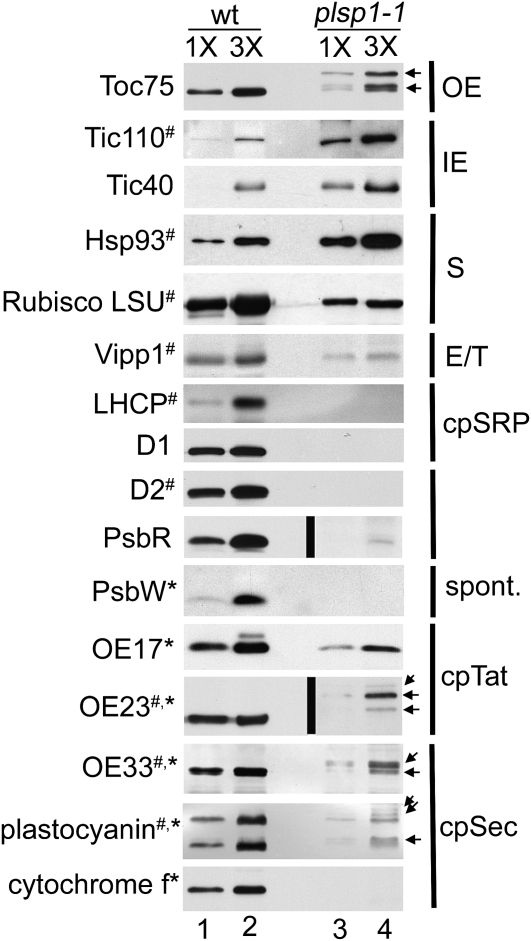
The proteomics results prompted us to ask two questions. The first was if the level of the identified proteins is comparable to that in wild type, as is the case of five chloroplast proteins examined previously (Inoue et al., 2005). The second question was if any of the TPP substrates exists as an incompletely matured form, similar to the case of OE33 previously demonstrated (Inoue et al., 2005), although no lumen-targeting sequence was identified by the proteomics study. To address these questions, we subjected the isolated mutant plastids to immunoblotting assays and compared the results with those of wild-type chloroplasts (Fig. 3). Analyzed proteins included those from various chloroplast subcompartments as well as substrates of all the four known thylakoid targeting pathways, including six TPP substrates. Due to the availability of antisera, we analyzed nine proteins identified in the plsp1-1 plastid proteome (Supplemental Table S1) and seven other proteins relevant to our study and/or known to be important for proper chloroplast development. Since the amount of proteins per plastid was comparable between wild-type and the mutant plants (approximately 0.6 pg), the sample loading was normalized based on the amount of proteins.
Figure 3.
The level of photosynthetic proteins in plsp1-1 plastids. Immunoblots of proteins (1.6 μg [1 × ] and 4.8 μg [3 × ]) from wild-type (wt) and plsp1-1 plastids using antisera against proteins indicated at left. Proteins identified by the LC-MS/MS analysis of plsp1-1 plastids (Supplemental Table S1) and TPP substrates are indicated with number signs and asterisks, respectively. Immunoreactive bands in the mutant plastids that appear to be larger than those in the wild type are shown with arrows at right. Suborganellar locations of nonthylakoid proteins are indicated at right: OE, outer envelope; IE, inner envelope; S, stroma. Vipp1 was found in the envelope and thylakoids (Li et al., 1994) and thus is indicated with E/T. For thylakoid proteins, targeting pathways, if elucidated previously, are indicated. Black lines indicate grouping of images from different exposures of the same blot (longer exposures for the blots containing proteins from plsp1-1 plastids).
As shown in Figure 3, proteins in the envelope membranes and stroma accumulated in plsp1-1 plastids to levels slightly less or significantly greater than the wild type, ranging from approximately 60% (large subunit of Rubisco) to > 300% (Tic110). By contrast, the amount of thylakoid proteins was much lower in the mutant plastids than that in the wild type. In particular, the level of thylakoid membrane proteins, i.e. LHCP, D1, D2, PsbW, and cytochrome f, was under the detection limit in plsp1-1 plastids.
Among proteins analyzed by the immunoblotting assays were five nuclear-encoded TPP substrates, OE33, OE23, OE17, plastocyanin, and PsbW, as well as one chloroplast-encoded substrate, cytochrome f (Fig. 3). Although multiple isoforms are present for the three OEC subunits and plastocyanin in Arabidopsis (Arabidopsis Genome Initiative, 2000), the methods used in this study allowed us to clearly distinguish among only those of plastocyanin (PETE1 and PETE2; Abdel-Ghany, 2009) in wild-type chloroplasts (Fig. 3). Among detectable proteins, three lumenal substrates, OE33, OE23, and plastocyanin, but not OE17, accumulate in the mutant plastids as larger forms than mature proteins in the wild type (indicated with arrows in Fig. 3). Thus, Plsp1 may be involved in maturation of multiple lumenal proteins.
Reduction in the Level of Thylakoid Membrane Proteins in plsp1-1 Appears to Depend on Posttranscriptional Regulation and Not on the Increased Level of Known Plastid Proteases
The immunoblotting assay revealed a reduction of some but not all photosynthetic proteins in plsp1-1 plastids. We wished to test whether this was caused transcriptionally, similar to the cases with various photosynthetic mutants (for review, see Rodermel, 2001). To this end, the amount of transcripts encoding Toc75, Tic110, OE33, OE23, PsbR, LHCP, and Plsp1 was examined by comparative reverse transcription-PCR (Supplemental Fig. S1). For LHCP, we analyzed accumulation of transcripts for two isoforms: Lhcb6, which was identified in the proteomics study (Supplemental Table S1), and Cab3, whose gene expression is known to be down-regulated by a chloroplast-nuclear retrograde signaling pathway (Susek et al., 1993). The level of cDNA derived from 18S ribosomal RNA (Kumar et al., 2003) was used as an internal control. Accuracy was ensured by monitoring amplification of products by multiple cycle numbers for each transcript in three biological replicates, as described before (Halford, 1999). As expected, we could not detect amplification of Plsp1 cDNA from plsp1-1 plants (Supplemental Fig. S1). By contrast, the signal intensities derived from transcripts encoding those that were at detectable but significantly lower levels in plsp1-1 plastids (OE23, OE33, and PsbR; see Fig. 3) were comparable between the two plants (Supplemental Fig. S1A). We also detected the presence of transcripts for both Lhcb6 and Cab3 in the mutant (Supplemental Fig. S1B). Their level appears only slightly lower than that in the wild type. Together, these data suggest the presence of posttranscriptional suppression of thylakoid protein accumulation in plsp1-1 plants.
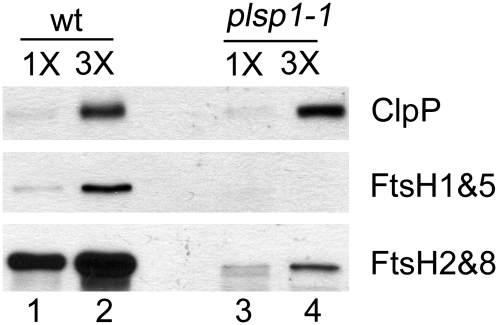
One potential mechanism for such a posttranscriptional regulation involves proteolysis. Several proteases have been identified in the chloroplast, although substrates of most of them are not known (Adam et al., 2006; Sakamoto, 2006). Nonetheless, up-regulation of several thylakoid proteases (FtsH2, FtsH5, and SppA) was suggested to play a role in accelerated turnover of thylakoid proteins in mutant Arabidopsis plants that had a reduced level of ClpR, a noncatalytic subunit of a stromal protease complex (Rudella et al., 2006). To test if this is also the case in plsp1-1 plastids, we compared the levels of known chloroplast proteases, FtsH and ClpP, in the mutant with those in the wild type. As shown in Figure 4, however, none of the proteases accumulated to a higher level in the mutant than in wild-type plastids. Similar to the other proteins in the same compartments of the mutant plastids, the levels of FtsH homologs (thylakoid membrane proteins) were significantly reduced in the mutant, whereas that of ClpP (a stromal protein) was near that of the wild type. Thus, the posttranscriptional reduction as seen for several photosynthetic proteins in plsp1-1 plastids (Fig. 3) is not due to a simple increase in the level of known proteases in chloroplasts. At this point, however, we cannot rule out the possibility that these proteases indeed play a role in degradation of thylakoid proteins in the mutant plastids because they may be activated by unknown regulatory mechanisms, as suggested (Adam et al., 2006; Sakamoto, 2006).
Figure 4.
The reduced level of some thylakoid proteins in plsp1-1 plastids does not depend on a simple increase in the level of known chloroplast proteases. Immunoblots of proteins (1.6 μg [1 × ] and 4.8 μg [3 × ]) from wild-type (wt) and plsp1-1 plastids using antisera against chloroplast proteases indicated at right.
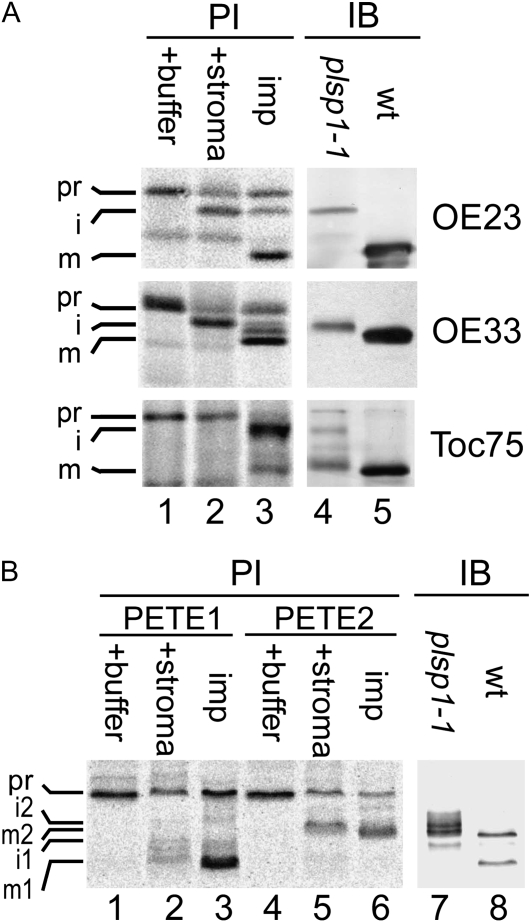
The Sizes of Multiple Unprocessed TPP Substrates in plsp1-Null Plastids Correspond to Those of the Stroma Intermediate Forms
The immunoblotting data indicate that plsp1-1 plastids accumulate one envelope protein (Toc75) and three lumenal proteins (OE33, OE23, and plastocyanin) as apparent immature forms (Fig. 3), suggesting that these proteins are Plsp1 substrates. Since plsp1-1 plastids appear to contain functional stromal processing peptidase (Inoue et al., 2005), nuclear-encoded Plsp1 substrates should lack the N-terminal stroma-targeting transit peptide but retain the thylakoid-targeting signal peptide. To test if this is the case, the mobility of immunoreactive proteins on SDS-PAGE was compared with that of radiolabeled proteins either treated with stromal extracts to remove the transit peptides only or imported into chloroplasts to mediate thylakoid targeting, thus removing both the transit peptides and signal peptides in vitro. As shown in Figure 5A, when the antisera against OE23 and OE33 were used, the mobility of the major immunoreactive proteins in the mutant plastids (lane 4) was comparable to that of the major isoforms lacking the stroma-targeting transit peptide (lane 2) but slower than that of the mature proteins found in the chloroplasts containing the imported proteins (lanes 3) and the immunoreactive proteins in wild-type chloroplasts (lane 5). For plastocyanin, the mutant plastids accumulated multiple immunoreactive proteins that appeared to correspond to the intermediate forms of the two isoforms (i1 and i2), as well as the mature form of PETE2 (m2), respectively (Fig. 5B, compare lanes 2, 5, and 7).
Figure 5.
Immature forms of Toc75 and thylakoid lumenal proteins in plsp1-1 plastids. Radiolabeled precursor proteins with various treatments (A, lanes 1–3; B, lanes 1–6) and 10 μg of proteins extracted from plsp1-1 and wild-type (wt) Arabidopsis seedlings (A, lanes 4 and 5; B, lanes 7 and 8) were separated by SDS-PAGE side by side, blotted on the same polyvinylidene fluoride membrane, and analyzed by phosphor imager (PI) or immunoblotting (IB) using antisera against proteins indicated at right (A) or plastocyanin (B). For treatments of radiolabeled precursors, they were incubated in buffer (+ buffer), stromal extracts (+ stroma), or intact chloroplasts from pea seedlings under the import condition (imp) for 5 min (OE23 and OE33), 20 min (Toc75), or 10 min (plastocyanin). Precursor (pr), intermediate (i), and mature (m) forms are indicated. For plastocyanin, two isoforms (PETE1 and PETE2) were analyzed (B).
Stromal Vesicles of plsp1-1 Plastids Contain a Thylakoid Lumenal Protein
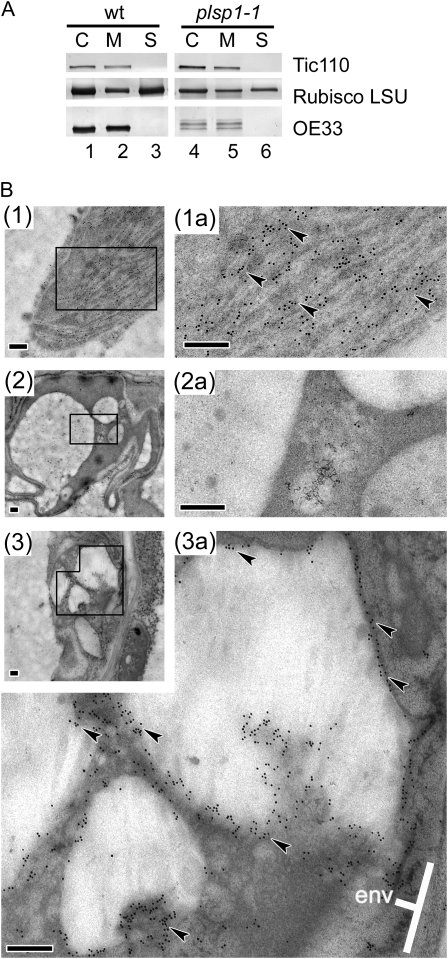
As shown in Figure 3, improperly processed forms of the lumenal protein OE33 accumulate in plsp1-1 at a significant level. It is unlikely that these proteins exist in the stroma because previous studies demonstrated that OE33 incorrectly sorted to the stroma is rapidly degraded by Clp protease (Halperin and Adam, 1996; Halperin et al., 2001), and plsp1-1 plastids accumulate a wild-type level of ClpP (Fig. 4). To further define the suborganellar location of the improperly processed forms of OE33, isolated wild-type and plsp1-1 chloroplasts were lysed hypotonically, separated into soluble and membrane fractions, and then examined by immunoblotting assays (Fig. 6A). Unprocessed forms of OE33 were recovered in the latter fraction, as was Tic110, while the stromal protein large subunit of Rubisco was found in both fractions (Fig. 6A, compare lanes 2 and 3 with 5 and 6). To determine the membrane at which OE33 is located, we probed sections of Arabidopsis leaf mesophyll cells with the antibody against OE33 and examined them under a transmission electron microscope (Fig. 6B). The specific signals were associated with the peripheral areas of stromal vesicles of plsp1-1 plastids (Fig. 6B3a), whereas they were nearly exclusively found in thylakoids in wild-type chloroplasts (Fig. 6B1a). These data suggest that the stromal vesicles of plsp1-1 plastids may be related to thylakoids.
Figure 6.
Localization of OE33 in stromal vesicles of plsp1-1 plastids. A, Immunoblots of proteins in total (C), membrane (M), and soluble (S) fractions of wild-type (wt) and plsp1-1 Arabidopsis plastids using antisera against proteins indicated at right. B, Immunogold localization of OE33 in wild-type (1) and plsp1-1 (2 and 3) Arabidopsis leaf mesophyll by electron microscopy. Tissue sections were probed with the anti-OE33 antibody (1 and 3) or preimmune sera (2). Images in 1a, 2a, and 3a correspond to the boxed regions in 1, 2, and 3, respectively, with a higher magnification. Some signals are indicated with black arrowheads, and the envelope membrane is indicated with “env.” Bars = 200 nm.
Disruption of Toc75 Maturation Does Not Result in a plsp1-1-Like Phenotype
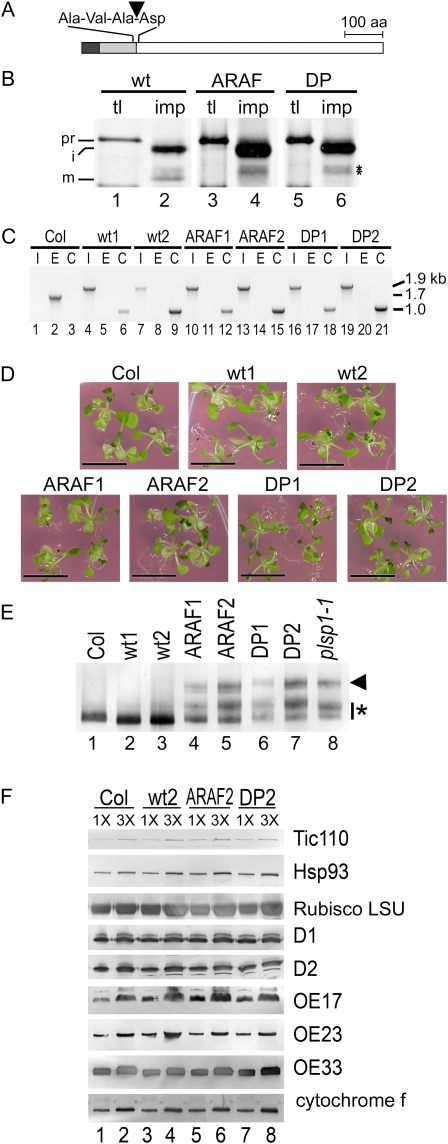
Previous genetic and cytological data (Inoue et al., 2005; Shipman and Inoue, 2009) as well as current results suggest that Plsp1 processes multiple proteins in the thylakoid lumen and is also responsible for complete maturation of the protein translocation channel Toc75 in the outer membrane of the chloroplast envelope. An important question is how thylakoid development is disrupted by the lack of Plsp1. One possible scenario is that the complete maturation of the protein-conducting channel Toc75 is required for import of sufficient amount of a single or multiple proteins that are necessary for proper thylakoid development. We tested this possibility by generating plants in which the endogenous Toc75 is replaced by an uncleavable form that cannot be completely matured even in the presence of Plsp1.
Proper recognition of substrates by SPase I usually requires small aliphatic residues, which in most cases are Ala, at positions −3 and −1 to the cleavage site. Furthermore, processing by SPase I is impaired if the first residue of the mature region of the substrate is Pro (Paetzel et al., 2002). Indeed, replacing the two conserved Ala residues with Gly and Pro dramatically reduced maturation but not import of pea (Pisum sativum) Toc75 into chloroplasts in vitro (Inoue and Keegstra, 2003). In this work, we generated constructs encoding two mutated forms of Arabidopsis Toc75: the first one (atToc75-ARAF) had substitutions of Arg and Phe for the two conserved Ala residues at 138 and 140, respectively, whereas the other protein (atToc75-DP) contained Pro instead of Asp at 141 immediately following the cleavage site (Fig. 7A). By import assay, we confirmed that both mutated forms are imported into chloroplasts but not properly matured in vitro (Fig. 7B, lanes 4 and 6).
Figure 7.
Accumulation of unprocessed forms of Toc75 does not cause plsp1-1 phenotypes. A, Schematic of atToc75 protein. The stroma-targeting sequence, envelope-targeting domain, and mature portion, which are predicted based on the sequence similarity with the pea homolog (Inoue and Keegstra, 2003), are indicated as black, gray, and white boxes, respectively. The amino acid (aa) residues at −3 to +1 to the second cleavage site (indicated with an arrowhead) are also shown. B, Results of import assay for wild-type (wt) and mutated (ARAF and DP) Toc75 proteins using chloroplasts from pea seedlings analyzed by fluorography. tl, Translation product (10% input); imp, proteins recovered after the assay. Precursor (pr), intermediate (i), and mature (m) forms are indicated. Improperly processed proteins derived from the mutated Toc75 precursors are indicated with asterisks. C, Genomic PCR of wild-type and mutant Arabidopsis seedlings. I, E, and C indicate reactions specific to amplify the inserted T-DNA into TOC75 (1.9 kb), part of the endogenous TOC75 (1.7 kb), and the transgene used to complement the mutation (1 kb), respectively. D, Seedlings of Arabidopsis wild-type (Col) and toc75 mutants complemented with constructs encoding wild-type (wt1 and wt2) or mutated forms of Toc75 (ARAF1, ARAF2, DP1, and DP2) grown on solid media containing 1% Suc for 4 week. Bars = 1 cm. E, Immunoblots of proteins (5.9 μg) in plastids isolated from wild-type and mutant Arabidopsis seedlings using antisera against Toc75. The apparent intermediate form that lacks the stroma-targeting sequence is indicated with an arrowhead, and other forms that are larger than the properly processed form with an asterisk. F, Immunoblots of proteins in wild-type and mutant Arabidopsis plastids containing 1 μg (1 ×) and 3 μg (3 ×) chlorophylls using antisera against various chloroplast proteins indicated at right. [See online article for color version of this figure.]
In order to introduce the uncleavable forms of Toc75 into plants, we needed to establish a genetic complementation system. In Arabidopsis, the functional Toc75 is encoded by a single gene, and its knockout causes an embryo-lethal phenotype (Baldwin et al., 2005; Hust and Gutensohn, 2006). Initial attempts to rescue the homozygous mutant by introducing an atToc75 open reading frame with either the constitutive cauliflower mosaic virus 35S promoter or a 2-kb genomic fragment preceding the coding sequence were not successful (A.J. Baldwin and R.L. Shipman-Roston, unpublished data). Consequently, we generated a construct containing a genomic sequence of atTOC75 that contains all seven exons and six introns, including 1 kb upstream and 500 bp downstream of the coding sequence, and transformed it into viable heterozygous toc75 plants. Among progenies, we were able to identify and confirm by genomic PCR multiple plants that were homozygous for the T-DNA insertion in the endogenous atTOC75 gene and were carrying the transgene (Fig. 7C, lanes 1–9). These plants were phenotypically indistinguishable from wild-type plants (Fig. 7D, compare Col, wt1, and wt2). Next, we transformed constructs encoding uncleavable forms of Toc75 into heterozygous toc75 mutants. Similar to the case with the nonmutated construct, we obtained multiple progenies homozygous for toc75 and carrying the transgene (Fig. 7C, lanes 10–21). The identity of the transgene for atToc75-ARAF and atToc75-DP was further confirmed by sequencing the PCR products that contained the mutated region, i.e. nucleotides 323 to 748, of atToc75 cDNA. All the lines were visually indistinguishable from wild-type plants (Fig. 7D) but distinct from homozygous plsp1-1 (Fig. 1A; Inoue et al., 2005).
Next, we isolated chloroplasts from these plants to examine the presence and size of Toc75 by immunoblotting (Fig. 7E). All the plants transformed with constructs encoding uncleavable forms of Toc75 accumulated multiple proteins recognized by the anti-Toc75 antibody (lanes 4–7). Some of the immunoreactive proteins, especially those in atToc75-ARAF mutants (lanes 4 and 5), appeared to migrate at a rate comparable to that of the mature Toc75 found in the wild type (lane 1). This suggests that processing at an alternative site may have allowed accumulation of a significant amount of almost properly processed Toc75 in vivo. Nonetheless, the accumulation pattern of these proteins in atToc75-ARAF and atToc75-DP mutants was comparable to that in the plsp1-1 plants (lane 8) but distinct from that in wild-type (lane 1) and the toc75 plants complemented with the wild-type constructs (lanes 2 and 3). We further confirmed that the level and size of a subset of chloroplast proteins were comparable to those in the wild type (Fig. 7F) and were distinct from those in plsp1-1 (Fig. 3). These data indicate that accumulation of improperly processed Toc75s alone does not cause the seedling-lethal phenotype of plsp1-1 and that maturation of lumenal proteins may be a key process for proper thylakoid development.
DISCUSSION
This work aims to help elucidate the molecular mechanism underlying the development of thylakoids, the chloroplast internal membrane systems essential for photosynthesis in higher plants. To this end, we analyzed Arabidopsis plastids that are devoid of stacked granal membranes due to the lack of the TPP homolog Plsp1. A previous study has shown that Plsp1 is necessary in vivo for maturation of a protein translocation channel in the outer envelope, Toc75, and a thylakoid lumenal protein, OE33 (Inoue et al., 2005). This study has extended the characterization of the mutant plastids, revealing that (1) the overall level of thylakoid proteins is reduced in plsp1-1 plastids; (2) Plsp1 is necessary for processing of at least two additional lumenal proteins, OE23 and plastocyanin, but not that of OE17 in vivo; (3) unprocessed forms of OE33 accumulate in stromal vesicles of the mutant plastids that lack obvious thylakoid membranes; and (4) incomplete maturation of Toc75 alone does not cause the phenotypic defects observed in plsp1-1 plants. Together, our data suggest that proper thylakoid formation may depend on complete maturation of lumenal proteins that may affect correct assembly of thylakoids.
The Potential Specificity of Chloroplast Type I Signal Peptidases
To date, signal peptidase activity of Plsp1 has not been demonstrated in vitro. Indeed, accumulation of intermediates in plsp1-1 plastids may arise as a secondary effect due to the loss of another activity or step affected by Plsp1. Nonetheless, genetic, cytological, and immunoblotting studies support the function of Plsp1 as a TPP in vivo (Inoue et al., 2005; Shipman and Inoue, 2009). Putative Plsp1 substrates in the thylakoid lumen identified by the previous and current studies include proteins that utilize both cpSec (OE33 and plastocyanin) and cpTat (OE23) pathways for their targeting. Another cpTat substrate, OE17, was found to exist as an apparent mature form in plsp1-1 plastids (Fig. 3B), which may indicate the presence of active TPP other than Plsp1. Previously, partially purified TPP from pea thylakoids was shown to efficiently process OE33 and OE23, but not OE17, in vitro (Halpin et al., 1989). Interestingly, in Arabidopsis, the residue at −3 to the TPP processing site of the two OE17 isoforms is Val, whereas that of OE23, OE33, and plastocyanin is Ala (Robinson and Mant, 2005; Zybailov et al., 2008). Thus, the activity of a TPP may depend on the recognition site within the substrate, instead of the pathway by which the substrate traverses across the membranes. Removal of the thylakoid-targeting signal from OE17 may be mediated by another TPP isoform, such as At2g30440 (Chaal et al., 1998), and/or an unidentified enzyme, which has a substrate preference distinct from that of Plsp1. Alternatively, in plsp1-1 plastids, the signal peptide of OE17 may not specifically be removed but eventually degraded by lumenal proteases. Genetic assays using Arabidopsis, such as disruption of genes encoding the other TPP isoforms or introducing a gene encoding OE33 with a substitution of Val for Ala at −3 to the TPP cleavage site into plsp1-1, may help address these possibilities.
The Nature of the plsp1-1 Plastid Stromal Vesicles
The presence of vesicular-like structures in the stroma, as seen in plsp1-1 plastids, often correlates with a severe or complete disruption of thylakoid formation in various mutant plants, such as those lacking 1-deoxyxylulose 5-phosphate synthase that catalyzes the initial step of plastid isoprenoid biosynthesis (Mandel et al., 1996), a membrane-bound cpSRP component, Alb3 (Sundberg et al., 1997), and an FtsH metalloprotease homolog, VAR2 (Takechi et al., 2000). However, properties of these vesicles remained undefined. The data presented in Figure 6 show that these vesicles may accumulate at least one protein, which usually exists in the thylakoid lumen. This protein, OE33, is sorted from the stroma to the lumen via the cpSec pathway in wild-type chloroplasts. Thus, the stromal vesicles of plsp1-1 plastids may also contain the functional cpSec translocon.
The data shown in Figure 6 also indicate that the unprocessed forms of OE33 are mainly present in the peripheral areas, instead of inside, of the stromal vesicles of plsp1-1 plastids. Whether these proteins are on the stromal or lumenal face of the vesicles remain unresolved. In the wild type, a significant fraction of properly processed OEC subunits exists in the soluble lumenal pool prior to assembly in PSII (Ettinger and Theg, 1991; Hashimoto et al., 1996, 1997). Previously, a population of unprocessed cpTat substrates, which were generated by introducing mutations around the TPP cleavage site, was found to associate with thylakoid membranes (Di Cola and Robinson, 2005; Frielingsdorf and Klösgen, 2007). These proteins were transiently trapped in large protein complexes, most probably containing the translocon, but resided primarily in the lipid bilayers (Frielingsdorf and Klösgen, 2007). Similarly, in the stromal vesicles of plsp1-1 plastids, OE33 may be stacked in the cpSec translocon or may attach to the lipid bilayers via its uncleaved signal peptide, which contains a hydrophobic domain.
Are these vesicles derived directly from the inner envelope and, thus, the precursors to thylakoids, or are they remnants of thylakoids? The presence of stromal lamellae in plastids of young but not old tissues of plsp1-1 plants (Fig. 1B) may support the second scenario. In this case, lumenal protein maturation would be prominent for the assembly and/or maintenance of thylakoids, rather than for the initiation of thylakoid development.
The Potential Mechanism Underlying Disruption of Proper Thylakoid Formation in plsp1-1 Plastids
The presence of OEC subunits is necessary to support the proper assembly and stability of PSII (Murakami et al., 2002, 2005; Ifuku et al., 2005; Yi et al., 2005, 2007; Ido et al., 2009). A previous in vitro study showed that a bacterially produced OE33 precursor protein folded properly, fully reconstituted oxygen evolution, but bound nonspecifically to the membrane fraction that contained PSII core components (Popelkova et al., 2002). Several genetic studies have demonstrated that reduction of thylakoid membrane protein accumulation by inhibition of the cpSRP pathway (Sundberg et al., 1997; Tzvetkova-Chevolleau et al., 2007; Asakura et al., 2008) or PSII assembly (Peng et al., 2006; Ma et al., 2007; Fu et al., 2007; Sirpiö et al., 2008) results in disruption of proper thylakoid biogenesis. Hence, in plsp1-1 plastids, a large amount of unprocessed OEC subunits may nonspecifically associate with lipid bilayers and prevent proper assembly of thylakoid membrane proteins, thus leading to the dissociation of the membrane system. Although not investigated in this study, other TPP substrates, including PSI and PSII components, such as PsaF, PsaN, PsbW, PsbX, and PsbY (Robinson and Mant, 2005), may also accumulate as unprocessed forms, causing disruption of thylakoid assembly in plsp1-null plastids. Alternatively, the unprocessed lumenal proteins may interfere with the translocon components for membrane protein integration. Replacement of the endogenous TPP substrates with an uncleavable form using a genetic complementation system (e.g. Liu et al., 2009), similar to the strategy used in this study with Toc75 (Fig. 7), may help address these possibilities.
MATERIALS AND METHODS
Plant Materials and Growth Condition
Arabidopsis (Arabidopsis thaliana; Columbia-0) plants were grown on Murashige and Skoog media (MP Biomedicals) supplemented with 3% Suc at 24°C, 19 h light (approximately 100 μmol m−2 s−1) d−1. To examine the effects of light intensity on the stromal vesicle formation, plants were grown on half-strength Murashige and Skoog media supplemented with 3% Suc at 24°C, 19 h light (approximately 60 μmol m−2 s−1) d−1. Neutral density filters (RoscoLux no. 97) were used for some plates to reduce the light intensity to approximately 30 μmol m−2 s−1. For isolation of plastids from plsp1-1 homozygous plants, 1 to 2 weeks after plsp1-1 heterozygous seeds were planted, green heterozygous and wild-type plants were manually removed. The remaining pale homozygous mutants were harvested after growing for another 2 to 3 weeks.
Immunoblotting
Immunoblotting assays were done as described by Inoue and Potter (2004). Amounts of proteins analyzed were quantified by Bradford's method using BSA as standard (Bradford, 1976). Immunoreaction was detected using secondary antisera conjugated with alkaline phosphatase and substrates, 5-bromo-4-chloro-3-indolyl phosphate/nitroblue tetrazolium (Bio-Rad Laboratories; for data shown in Figures 3 [plastocyanin], 5B, and 7, E and F) or those conjugated with horseradish peroxidase and ECL substrate (Pierce Biotechnology; for data shown in Figures 3 [all but plastocyanin], 4, and 5A). The antibody against Plsp1 was described by Shipman and Inoue (2009). Antibodies against OE33, OE23, OE17, and CF1γ were from S.M. Theg (University of California, Davis), those against Toc75, Hsp93, Tic40, Vipp1, LHCP, and Rubisco large subunit from K. Keegstra (Michigan State University), those against PsbR, D1, and D2 from E.-M. Aro (University of Turku), and those against FtsH1/FtsH5 and FtsH2/FtsH8 from W. Sakamoto (Okayama University). Antibodies against ClpP6, plastocyanin, and PsbW were from A. Clarke (Gothenburg University), M. Pilon (Colorado State University), and W. Schröder (Umeå Plant Science Center), respectively.
Chloroplast Preparations, Protein Import, and Stromal Processing Assay
Isolation of chloroplasts was done as described previously (Fitzpatrick and Keegstra, 2001), and the number of organelles was counted on a Hausser Scientific hemocytometer. Typically, mutant plants (20.9 g FW) from 27 plates (100 mm diameter × 25 mm height) yielded 1.54 × 107 chloroplasts containing 9.17 μg protein, which was estimated as described above for immunoblotting assay, while 2.44 × 109 chloroplasts containing 1.5 mg protein could be obtained from wild-type plants of 7.6 g FW from four plates. Fractionation of Arabidopsis chloroplasts and protein import assay with isolated pea (Pisum sativum) chloroplasts were done as described previously (Inoue et al., 2005). Radiolabeled precursor proteins were prepared as described (Inoue and Keegstra, 2003) with a plasmid encoding Arabidopsis Toc75 (At3g46740; Inoue and Keegstra, 2003) or those for two isoforms of plastocyanin (PETE1 [At1g76100] and PETE2 [At1g20340]) and one isoform each of OE33 (PsbO1 [At5g66570]) and OE23 (PsbP1 [At1g06680]), respectively, in pUNI51 (Yamada et al., 2003). For stromal processing assay, chloroplasts isolated from pea seedlings were resuspended in 5 mm HEPES KOH, pH 8.0, to 0.5 mg chlorophyll mL−1, incubated on ice under dark for 30 min, and then centrifuged at 15,500g for 30 min at 4°C. The resultant supernatant was designated as stromal extract, and its aliquot (60 μL) was incubated with 3 μL of radiolabeled precursor proteins for 90 min at room temperature. One-third of the reaction mixture was subjected to SDS-PAGE followed by fluorography analysis.
Proteomics Analysis of plsp1-1 Plastids
Proteins in the mutant plastids were precipitated by methanol/chloroform as described (Wessel and Flügge, 1984). An aliquot of the precipitated sample (approximately 4 μg protein) was resuspended in 50 μL of 50 mm ammonium bicarbonate supplemented with 10% acetonitrile, added tris(2-carboxyethyl)phosphine hydrochloride to 10 mm, and incubated at 95°C for 10 min, iodoacetamide to 15 mm and incubated in the dark at room temperature for 1 h, then dithiothreitol to 5 mm and incubated at room temperature for 10 min. Tryptic digestion was done with 30 ng of Sequencing Grade Modified Trypsin (Promega) in 50 mm ammonium bicarbonate at 4°C overnight. NanoLC-MS/MS analysis was performed on a LTQ-FT hybrid linear ion trap/7T Fourier transform ion cyclotron resonance mass spectrometer (Thermo-Scientific), equipped with a Nanospray ion source (Thermo-Scientific), a Surveyor MS pump (Thermo-Scientific), and a Surveyor micro-autosampler (Thermo-Scientific). The tryptic peptide mixture was separated on a 50 μm ID PicoFrit column packed in-house with Magic C18AQ (Michrom BioResources) to a length of 12 cm with a 100% methanol slurry of C18 reversed-phase material (100-Å pore size and 3-μm particle size) using a high-pressure cell pressurized with helium. The column was pre-equilibrated for 10 min at 2% solvent B (0.1% [v/v] formic acid in acetonitrile) and 98% solvent A (0.1% [v/v] formic acid in water) at a flow rate of 140 nL min−1. Separation was achieved by using a linear gradient from 2% to 50% solvent B in 110 min at a flow rate of 320 nL min−1. The LTQ-FT mass spectrometer was operated in the data-dependent acquisition mode using the TOP10 method: a full-scan MS acquired in the FTICR mass spectrometer was followed by 10 MS/MS experiments performed with the LTQ on the 10 most abundant ions detected in the full-scan MS.
For database searching, tandem mass spectra were extracted by BioWorks version 3.3. Charge state deconvolution and deisotoping were not performed. All MS/MS samples were analyzed using X! Tandem (www.thegpm.org; version TORNADO 2008.02.01.2). X! Tandem was set up to search International Protein Index Arabidopsis database v3.52 (March, 2009) assuming the digestion enzyme trypsin. X! Tandem was searched with a fragment ion mass tolerance of 0.40 D and a parent ion tolerance of 25 ppm. Iodoacetamide derivative of Cys was specified in X! Tandem as a fixed modification. Deamidation of Asn, oxidation of Met, sulfone of Met, Trp oxidation to formylkynurenin of Trp, and acetylation of the N terminus were specified in X! Tandem as variable modifications.
For protein identification, Scaffold (version Scaffold_2_03_01; Proteome Software) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at > 80.0% probability as specified by the Peptide Prophet algorithm (Keller et al., 2002). Protein identifications were accepted if they could be established at > 95.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (Nesvizhskii et al., 2003). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Reverse Transcription-PCR
RNA was extracted from 19- to 21-d-old Arabidopsis shoot tissue grown on plates using the RNeasy Plant Mini Kit (Qiagen) as per instructions. Total RNA isolated was quantified spectrophotometrically at 260 nm, and equivalent amounts (1.5 μg for one assay; 2 μg for two assays) were reverse transcribed using SuperScript III and random primers (Invitrogen). Primers used for PCR are listed in Supplemental Table S2, except those for the internal control (QuantumRNA 18S Internal Standards; Ambion). Detection was with ethidium bromide-stained agarose gel, and the signal quantification was done with ImageJ version 1.32j (http://rsbweb.nih.gov/ij/).
Electron Microscopy
Tissues were fixed in 2% paraformaldehyde and 2.5% glutaraldehyde as described previously (Shipman and Inoue, 2009). Antisera against OE33 used for the immunolocalization study was against residues 86 to 332 of PsbO1 (At5g66570) from Hsou-min Li (Academia Sinica). All grids were viewed on a Phillips CM120 Biotwin. Micrographs were taken with a Gatan MegaScan, model 794/20, digital camera.
Complementation of toc75-Null Plants
cDNA sequences encoding uncleavable forms of Arabidopsis Toc75 protein were prepared by modifying pGEMTEasy-atToc75, the plasmid encoding the wild-type sequence (Inoue and Keegstra, 2003), by using the QuikChange site-directed mutagenesis kit (Stratagene) and primers listed in Supplemental Table S2. Arabidopsis TOC75 genomic fragment of 4.7 kb, including 1 kb upstream and 0.5 kb downstream of the coding sequence, was amplified using BAC clone T6H20 (Arabidopsis Biological Resource Center; Alonso et al., 2003) as a template and primers listed in Supplemental Table S2 and then subcloned into pGEM-T Easy (Promega). Upon sequencing the whole insert, we identified one base change (from t to c) causing an amino acid change from Val-6 to Ala. The TOC75 genomic sequence was then subcloned into pDONR221 using the Gateway cloning system (Invitrogen) with primers listed in Supplemental Table S2, generating the entry clone pDONR-gTOC75wt. The entry clones for uncleavable forms of Toc75 were prepared by replacing the 569-bp ClaI-AvrII fragment of pDONR-gTOC75wt with that of the modified pGEMTEasy-atToc75, which contained the sequences for mutated residues. The expression clones were generated by performing an LR recombination reaction between these entry clones and the Gateway destination vector, pMDC100 (Curtis and Grossniklaus, 2003). Final constructs were confirmed by sequencing and transformed into viable heterozygous toc75-III-1 plants (Baldwin et al., 2005) by the Agrobacterium tumefaciens-mediated floral dip method (Clough and Bent, 1998). Resultant seeds were sowed on plates containing 20 μg mL−1 hygromycin and 40 μg mL−1 kanamycin to select for toc75-III-1 and the presence of the sequence derived from the pMDC100 expression vectors, respectively. Genomic PCRs were performed with primers listed in Supplemental Table S2 to confirm the presence of the endogenous TOC75 (E), T-DNA insertion into TOC75 (I), and transgene used to complement the mutation (C). Mutations for the uncleavable constructs were further confirmed by sequencing the genomic PCR products (C) with a primer, 5′-ACTGTACCATTAACCCAACG-3′.
Sequence data from this article can be found in the GenBank/EMBL data libraries under the accession numbers At1g06680, At1g20340, At1g76100, At2g30440, At3g24590, At3g46740, and At5g66570.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure S1. The reduced level of thylakoid proteins in plsp1-1 plastids appears to be due to a posttranscriptional regulation.
Supplemental Table S1. A list of proteins identified in plsp1-1 plastids.
Supplemental Table S2. Primer sets used in this study.
Acknowledgments
We thank G. Adamson and P. Kysar for help with the electron microscope studies, Drs. E.-M. Aro, A. Clarke, K. Ifuku, K. Keegstra, H.-m. Li, M. Pilon, W. Sakamoto, S.M. Theg, and W. Schröder for antibodies, and Dr. Stacey Harmer for help with the experiment using the neutral density filter. We also thank the current and past Inoue lab members and Dr. D. Potter for helpful comments on this work.
References
- Abdel-Ghany SE. (2009) Contribution of plastocyanin isoforms to photosynthesis and copper homeostasis in Arabidopsis thaliana grown at different copper regimes. Planta 229: 767–779 [DOI] [PubMed] [Google Scholar]
- Adam X, Rudella A, van Wijk KJ. (2006) Recent advances in the study of Clp, FtsH, and other proteases located in chloropalsts. Curr Opin Plant Biol 9: 234–240 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Asakura Y, Kikuchi S, Nakai M. (2008) Non-identical contributions of two membrane-bound cpSRP components, cpFtsY and Alb3, to thylakoid biogenesis. Plant J 56: 1007–1017 [DOI] [PubMed] [Google Scholar]
- Baldwin A, Wardle A, Patel R, Dudley P, Park SK, Twell D, Inoue K, Jarvis P. (2005) A molecular-genetic study of the Arabidopsis Toc75 gene family. Plant Physiol 138: 715–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baymann F, Zito F, Kuras R, Minai L, Nitschke W, Wollman FA. (1999) Functional characterization of Chlamydomonas mutants defective in cytochrome f maturation. J Biol Chem 274: 22957–22967 [DOI] [PubMed] [Google Scholar]
- Benning C, Ohta H. (2005) Three enzyme systems for galactoglycerolipid biosynthesis are coordinately regulated in plants. J Biol Chem 280: 2397–2400 [DOI] [PubMed] [Google Scholar]
- Bradford MM. (1976) Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bradshaw RA, Burlingame AL, Carr S, Aebersold R. (2006) Reporting protein identification data: the next generation of guidelines. Mol Cell Proteomics 5: 787–788 [DOI] [PubMed] [Google Scholar]
- Chaal BK, Mould RM, Barbrook AC, Gray JC, Howe CJ. (1998) Characterization of a cDNA encoding the thylakoidal processing peptidase from Arabidopsis thaliana. Implications for the origin and catalytic mechanism of the enzyme. J Biol Chem 273: 689–692 [DOI] [PubMed] [Google Scholar]
- Cline K, Dabney-Smith C. (2008) Plastid protein import and sorting: different paths to the same compartments. Curr Opin Plant Biol 11: 585–592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cregg KM, Wilding I, Black MT. (1996) Molecular cloning and expression of the spsB gene encoding an essential type I signal peptidase from Staphylococcus aureus. J Bacteriol 178: 5712–5718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis MD, Grossniklaus U. (2003) A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol 133: 462–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalbey RE, Wickner W. (1985) Leader peptidase catalyzes the release of exported proteins from the outer surface of the Escherichia coli plasma membrane. J Biol Chem 260: 15925–15931 [PubMed] [Google Scholar]
- Date T. (1983) Demonstration by a novel genetic technique that leader peptidase is an essential enzyme of Escherichia coli. J Bacteriol 154: 76–83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DellaPenna D, Pogson BJ. (2006) Vitamin synthesis in plants: tocopherols and carotenoids. Annu Rev Plant Biol 57: 711–738 [DOI] [PubMed] [Google Scholar]
- Di Cola A, Robinson C. (2005) Large-scale translocation reversal within the thylakoid Tat system in vivo. J Cell Biol 171: 281–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörmann P, Benning C. (2002) Galactolipids rule in seed plants. Trends Plant Sci 7: 112–118 [DOI] [PubMed] [Google Scholar]
- Dörmann P, Hoffmann-Benning S, Balbo I, Benning C. (1995) Isolation and characterization of an Arabidopsis mutant deficient in the thylakoid lipid digalactosyl diacylglycerol. Plant Cell 7: 1801–1810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ettinger WF, Theg SM. (1991) Physiologically active chloroplasts contain pools of unassembled extrinsic proteins of the photosynthetic oxygen-evolving enzyme complex in the thylakoid lumen. J Cell Biol 115: 321–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitzpatrick LM, Keegstra K. (2001) A method for isolating a high yield of Arabidopsis chloroplasts capable of efficient import of precursor proteins. Plant J 27: 59–65 [DOI] [PubMed] [Google Scholar]
- Frielingsdorf S, Klösgen RB. (2007) Prerequisites for terminal processing of thylakoidal Tat substrates. J Biol Chem 282: 24455–24462 [DOI] [PubMed] [Google Scholar]
- Fu A, He Z, Cho HS, Lima A, Buchanan BB, Luan S. (2007) A chloroplast cyclophilin functions in the assembly and maintenance of photosystem II in Arabidopsis thaliana. Proc Natl Acad Sci USA 104: 15947–15952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halford WP. (1999) The essential prerequisites for quantitative RT-PCR. Nat Biotechnol 17: 835. [DOI] [PubMed] [Google Scholar]
- Halperin T, Adam Z. (1996) Degradation of mistargeted OEE33 in the chloroplast stroma. Plant Mol Biol 30: 925–933 [DOI] [PubMed] [Google Scholar]
- Halperin T, Ostersetzer O, Adam Z. (2001) ATP-dependent association between subunits of Clp protease in pea chloroplasts. Planta 213: 614–619 [DOI] [PubMed] [Google Scholar]
- Halpin C, Elderfield PD, James HE, Zimmermann R, Dunbar B, Robinson C. (1989) The reaction specificities of the thylakoidal processing peptidase and Escherichia coli leader peptidase are identical. EMBO J 8: 3917–3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto A, Ettinger WF, Yamamoto Y, Theg SM. (1997) Assembly of newly imported oxygen-evolving complex subunits in isolated chloroplasts: sites of assembly and mechanism of binding. Plant Cell 9: 441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashimoto A, Yamamoto Y, Theg SM. (1996) Unassembled subunits of the photosynthetic oxygen-evolving complex present in the thylakoid lumen are long-lived and assembly-competent. FEBS Lett 391: 29–34 [DOI] [PubMed] [Google Scholar]
- Havaux M, Eymery F, Porfirova S, Rey P, Dörmann P. (2005) Vitamin E protects against photoinhibition and photooxidative stress in Arabidopsis thaliana. Plant Cell 17: 3451–3469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoober JK, Boyd CO, Paavola LG. (1991) Origin of thylakoid membranes in Chlamydomonas reinhardtii Y-1 at 38°C. Plant Physiol 96: 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh MH, Goodman HM. (2005) The Arabidopsis IspH homolog is involved in the plastid nonmevalonate pathway of isoprenoid biosynthesis. Plant Physiol 138: 641–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugueney P, Bouvier F, Badillo A, d'Harlingue A, Kuntz M, Camara B. (1995) Identification of a plastid protein involved in vesicle fusion and/or membrane protein translocation. Proc Natl Acad Sci USA 92: 5630–5634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hust B, Gutensohn M. (2006) Deletion of core components of the plastid protein import machinery causes differential arrest of embryo development in Arabidopsis thaliana. Plant Biol 8: 18–30 [DOI] [PubMed] [Google Scholar]
- Ido K, Ifuku K, Yamamoto Y, Ishihara S, Murakami A, Takabe K, Miyake C, Sato F. (2009) Knockdown of the PsbP protein does not prevent assembly of the dimeric PSII core complex but impairs accumulation of photosystem II supercomplexes in tobacco. Biochim Biophys Acta 1787: 873–881 [DOI] [PubMed] [Google Scholar]
- Ifuku K, Yamamoto Y, Ono T, Ishihara S, Sato F. (2005) PsbP protein, but not PsbQ protein, is essential for the regulation and stabilization of photosystem II in higher plants. Plant Physiol 139: 1175–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Baldwin AJ, Shipman RL, Matsui K, Theg SM, Ohme-Takagi M. (2005) Complete maturation of the plastid protein translocation channel requires a type I signal peptidase. J Cell Biol 171: 425–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Keegstra K. (2003) A polyglycine stretch is necessary for proper targeting of the protein translocation channel precursor to the outer envelope membrane of chloroplasts. Plant J 34: 661–669 [DOI] [PubMed] [Google Scholar]
- Inoue K, Potter D. (2004) The chloroplastic protein translocation channel Toc75 and its paralog OEP80 represent two distinct protein families and are targeted to the chloroplastic outer envelope by different mechanisms. Plant J 39: 354–365 [DOI] [PubMed] [Google Scholar]
- Jarvis P, Dörmann P, Peto CA, Lutes J, Benning C, Chory J. (1995) Galactolipid deficiency and abnormal chloroplast development in the Arabidopsis MGD synthase 1 mutant. Proc Natl Acad Sci USA 97: 8175–8179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Nesvizhskii AI, Kolker E, Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal Chem 74: 5383–5392 [DOI] [PubMed] [Google Scholar]
- Kroll D, Meierhoff K, Bechtold N, Kinoshita M, Westphal S, Vothknecht UC, Soll J, Westhoff P. (2001) VIPP1, a nuclear gene of Arabidopsis thaliana essential for thylakoid membrane formation. Proc Natl Acad Sci USA 98: 4238–4242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A, Silim SN, Okamoto M, Siddiqi MY, Glass ADM. (2003) Differential expression of three members of the AMT1 gene family encoding putative high-affinity NH4+ transporters in roots of Oryza sativa subspecies indica. Plant Cell Environ 26: 907–914 [DOI] [PubMed] [Google Scholar]
- Kuras R, Buschlen S, Wollman FA. (1995) Maturation of pre-apocytochrome f in vivo. A site-directed mutagenesis study in Chlamydomonas reinhardtii. J Biol Chem 270: 27797–27803 [DOI] [PubMed] [Google Scholar]
- Li HM, Kaneko Y, Keegstra K. (1994) Molecular cloning of a chloroplastic protein associated with both the envelope and thylakoid membranes. Plant Mol Biol 25: 619–632 [DOI] [PubMed] [Google Scholar]
- Liu H, Frankel LK, Bricker TM. (2009) Functional complementation of the Arabidopsis thaliana psbo1 mutant phenotype with an N-terminally His(6)-tagged PsbO-1 protein in photosystem II. Biochim Biophys Acta 1787: 1029–1038 [DOI] [PubMed] [Google Scholar]
- Ma J, Peng L, Guo J, Lu Q, Lu C, Zhang L. (2007) LPA2 is required for efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 19: 1980–1993 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Mandel MA, Feldmann KA, Herrera-Estrella L, Rocha-Sosa M, León P. (1996) CLA1, a novel gene required for chloroplast development, is highly conserved in evolution. Plant J 9: 649–658 [DOI] [PubMed] [Google Scholar]
- Masuda T, Fujita Y. (2008) Regulation and evolution of chlorophyll metabolism. Photochem Photobiol Sci 7: 1131–1149 [DOI] [PubMed] [Google Scholar]
- Mühlethaler K, Frey-Wyssling A. (1959) Entwicklung und struktur der proplastiden. J Biophys Biochem Cytol 6: 507–512 [PMC free article] [PubMed] [Google Scholar]
- Murakami R, Ifuku K, Takabayashi A, Shikanai T, Endo T, Sato F. (2002) Characterization of an Arabidopsis thaliana mutant with impaired psbO, one of two genes encoding extrinsic 33-kDa proteins in photosystem II. FEBS Lett 523: 138–142 [DOI] [PubMed] [Google Scholar]
- Murakami R, Ifuku K, Takabayashi A, Shikanai T, Endo T, Sato F. (2005) Functional dissection of two Arabidopsis PsbO proteins PsbO1 and PsbO2. FEBS J 272: 2165–2175 [DOI] [PubMed] [Google Scholar]
- Nesvizhskii AI, Keller A, Kolker E, Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal Chem 75: 4646–4658 [DOI] [PubMed] [Google Scholar]
- Paetzel M, Karla A, Strynadka NCJ, Dalbey RE. (2002) Signal peptidases. Chem Rev 102: 4549–4579 [DOI] [PubMed] [Google Scholar]
- Peltier JB, Ytterberg AJ, Sun Q, van Wijk KJ. (2004) New functions of the thylakoid membrane proteome of Arabidopsis thaliana revealed by a simple, fast, and versatile fractionation strategy. J Biol Chem 279: 49367–49383 [DOI] [PubMed] [Google Scholar]
- Peng L, Ma J, Chi W, Guo J, Zhu S, Lu Q, Lu C, Zhang L. (2006) LOW PSII ACCUMULATION1 is involved in efficient assembly of photosystem II in Arabidopsis thaliana. Plant Cell 18: 955–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popelkova H, Im MM, D'Auria J, Betts SD, Lydakis-Simantiris N, Yocum CF. (2002) N-terminus of the photosystem II manganese stabilizing protein: effects of sequence elongation and truncation. Biochemistry 41: 2702–2711 [DOI] [PubMed] [Google Scholar]
- Richter S, Zhong R, Lamppa G. (2005) Function of the stromal processing peptidase in the chloroplast import pathway. Physiol Plant 123: 362–368 [Google Scholar]
- Robinson C, Mant A. (2005) Biogenesis of the thylakoid membrane. Møller SG, , Plastids. Annual Plant Reviews, Vol 13 Blackwell, Oxford, pp 180–213 [Google Scholar]
- Rodermel S. (2001) Pathways of plastid-to-nucleus signaling. Trends Plant Sci 6: 471–478 [DOI] [PubMed] [Google Scholar]
- Rudella A, Friso G, Alonso JM, Ecker JR, van Wijk KJ. (2006) Downregulation of ClpR2 leads to reduced accumulation of the ClpPRS protease complex and defects in chloroplast biogenesis in Arabidopsis. Plant Cell 18: 1704–1721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W. (2006) Protein degradation machineries in plastids. Annu Rev Plant Biol 57: 599–621 [DOI] [PubMed] [Google Scholar]
- Schnell DJ, Blobel G, Keegstra K, Kessler F, Ko K, Soll J. (1997) A consensus nomenclature for the protein-import components of the chloroplast envelope. Trends Cell Biol 7: 303–304 [DOI] [PubMed] [Google Scholar]
- Schröder WP, Kieselbach T. (2003) Update on chloroplast proteomics. Photosynth Res 78: 181–193 [DOI] [PubMed] [Google Scholar]
- Shipman RL, Inoue K. (2009) Suborganellar localization of plastidic type I signal peptidase 1 depends on chloroplast development. FEBS Lett 583: 938–942 [DOI] [PubMed] [Google Scholar]
- Sirpiö S, Khrouchtchova A, Allahverdiyeva Y, Hansson M, Fristedt R, Vener AV, Scheller HV, Jensen PE, Haldrup A, Aro EM. (2008) AtCYP38 ensures early biogenesis, correct assembly and sustenance of photosystem II. Plant J 55: 639–651 [DOI] [PubMed] [Google Scholar]
- Sun Q, Zybailov B, Majeran W, Friso G, Olinares PD, van Wijk KJ. (2009) PPDB, the Plant Proteomics Database at Cornell. Nucleic Acids Res 37: D969–974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundberg E, Slagter JG, Fridborg I, Cleary SP, Robinson C, Coupland G. (1997) ALBINO3, an Arabidopsis nuclear gene essential for chloroplast differentiation, encodes a chloroplast protein that shows homology to proteins present in bacterial membranes and yeast mitochondria. Plant Cell 9: 717–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susek RE, Ausubel FM, Chory J. (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799 [DOI] [PubMed] [Google Scholar]
- Takechi K, Sodmergen, Murata M, Motoyoshi F, Sakamoto W. (2000) The YELLOW VARIEGATED (VAR2) locus encodes a homologue of FtsH, an ATP-dependent protease in Arabidopsis. Plant Cell Physiol 41: 1334–1346 [DOI] [PubMed] [Google Scholar]
- Tzvetkova-Chevolleau T, Hutin C, Noël LD, Goforth R, Carde JP, Caffarri S, Sinning I, Groves M, Teulon JM, Hoffman NE, et al. (2007) Canonical signal recognition particle components can be bypassed for posttranslational protein targeting in chloroplasts. Plant Cell 19: 1635–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein D. (1959) The effect of genetic factors on the submicroscopic structures of the chloroplast. J Ultrastruct Res 3: 235–237 [Google Scholar]
- Wang Q, Sullivan RW, Kight A, Henry RL, Huang JR, Jones AM, Korth KL. (2004) Deletion of the chloroplast-localized Thylakoid formation1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol 136: 3594–3604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessel D, Flügge UI. (1984) A method for the quantitative recovery of protein in dilute-solution in the presence of detergents and lipids. Anal Biochem 138: 141–143 [DOI] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al. (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Yi X, Hargett SR, Liu H, Frankel LK, Bricker TM. (2007) The PsbP protein is required for photosystem II complex assembly/stability and photoautotrophy in Arabidopsis thaliana. J Biol Chem 282: 24833–24841 [DOI] [PubMed] [Google Scholar]
- Yi X, McChargue M, Laborde S, Frankel LK, Bricker TM. (2005) The manganese-stabilizing protein is required for photosystem II assembly/stability and photoautotrophy in higher plants. J Biol Chem 280: 16170–16174 [DOI] [PubMed] [Google Scholar]
- Zhang YB, Greenberg B, Lacks SA. (1997) Analysis of a Streptococcus pneumoniae gene encoding signal peptidase I and overproduction of the enzyme. Gene 194: 249–255 [DOI] [PubMed] [Google Scholar]
- Zhbanko M, Zinchenko V, Gutensohn M, Schierhorn A, Klösgen RB. (2005) Inactivation of a predicted leader peptidase prevents photoautotrophic growth of Synechocystis sp. strain PCC 6803. J Bacteriol 187: 3071–3078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zybailov B, Rutschow H, Friso G, Rudella A, Emanuelsson O, Sun Q, van Wijk KJ. (2008) Sorting signals, N-terminal modifications and abundance of the chloroplast proteome. PLoS One 3: e1994. [DOI] [PMC free article] [PubMed] [Google Scholar]