Abstract
Summary: Caulobacter crescentus is an aquatic Gram-negative alphaproteobacterium that undergoes multiple changes in cell shape, organelle production, subcellular distribution of proteins, and intracellular signaling throughout its life cycle. Over 40 years of research has been dedicated to this organism and its developmental life cycles. Here we review a portion of many developmental processes, with particular emphasis on how multiple processes are integrated and coordinated both spatially and temporally. While much has been discovered about Caulobacter crescentus development, areas of potential future research are also highlighted.
INTRODUCTION
In the microbial world, resources are almost universally scant and are competed for in an evolutionary arms race. Different bacteria employ different strategies to survive. Some bacteria retreat to extreme niches and live on a metabolic fringe. Others live on the move, constantly altering gene expression in response to metabolic changes. Still others take a more long-term approach and construct complex strategies to survive, integrating multiple pathways, regulons and macromolecular assemblies. The bacterium Caulobacter crescentus is a potent example of the latter type, utilizing development to create a lifestyle that helps it survive in nutrient-limited environments.
Before tackling the vast knowledge about Caulobacter crescentus development, we must first define exactly what we mean by “development.” Microbial development is an elusive concept to pin down, likely due to the amazing diversity of model developmental systems. On the gross level, the production of Bacillus subtilis endospores does not resemble the multicellular organization of the Myxococcus xanthus fruiting body, which does not resemble the polar morphogenesis of C. crescentus. Yet all of these are considered model bacterial developmental systems, so where is the convergence? Development has been defined as “a series of stable or metastable changes in the form or function of a cell” (51) or as a series of “changes in form and function that play a prominent role in the life cycle of the organism” (25). Certainly something about development is intimately tied to changes in shape. For years microbiologists have been enthralled by pictures of B. subtilis sporulating or films of M. xanthus cells aggregating into a fruiting body, but the function of shape changes is the least well defined aspect of “development.” Changes in physiology often accompany development, but they are not necessarily an obligatory part of development. For example, while metabolic signals are integral to the developmental programs of B. subtilis and M. xanthus and certainly influence the pace of development in C. crescentus, the development of C. crescentus is part of its natural life cycle and therefore is not tied to a specific metabolic cue.
So what is the grand unifying factor that is common to all these systems? The key comes from the word “development” itself. To develop something, be it a housing project, a marketing brand, or even an idea, is to add complexity toward an elaborate purpose. In biological terms, development is the addition of complexity toward a selectable advantage. Therefore, the principle that unifies all the prokaryotic developmental systems is not that they share some fundamental mechanism but the facts that (i) they are all very complex and (ii) this complexity provides a selectable advantage by a change of form and/or function. The complexity of a given developmental system arises from the integration of multiple processes. Much of the research on development is spent not only teasing out the specifics of an individual process but also finding out how that process is integrated into the overall program of the organism. By metaphor, a given process is a thread, but multiple threads may be twined together to form a string, and the string itself is woven into a tapestry. It is the purpose of the prokaryotic development researcher to simultaneously see the thread individually and the tapestry as a whole.
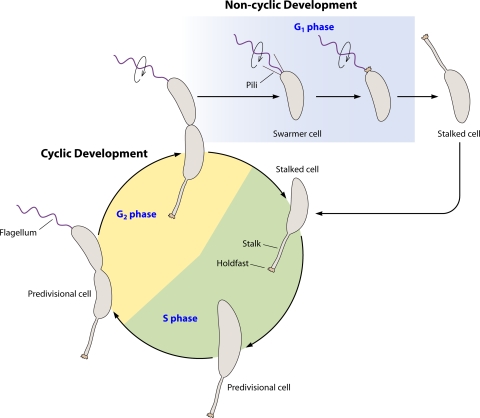
The tapestry of choice for this review is Caulobacter crescentus, a Gram-negative alphaproteobacterium. Though many Caulobacter species can be found in diverse environments, including ones with relatively high nutrient contents (142), highly toxic environments such as gold mines (95), or contaminated water or sediments (148, 175), C. crescentus is best known for living in oligotrophic aquatic environments. C. crescentus cells are found predominantly in two morphotypes. The first is the planktonic “swarmer cell,” which contains a single flagellum and multiple pili at one cell pole. The second is the sessile (and often surface-associated) “stalked cell,” where the polar flagellum has been replaced with a thin extension of the cell envelope known as a “prostheca” or “stalk.” The stalk is tipped with an adhesive organelle called the “holdfast.” The life cycle is depicted in Fig. 1. A stalked cell elongates the cell body, becoming a predivisional cell. A flagellum is produced at the pole opposite the stalk, and, once cell separation has occurred, pili are extruded. Thus, by segregating polar structures to different subcellular locations, a single predivisional cell can give rise to two different progeny cell types: a stalked cell (ostensibly the progenitor, or “mother cell”) and a swarmer cell (the progeny, or “daughter cell”). After a period of time, the swarmer cell differentiates into the stalked cell by extruding the holdfast, ejecting the flagellum, disassembling the pili, and extending the stalk from the same pole. The swarmer cell is unable to replicate its chromosome or perform cell division, whereas the stalked cell is the replication- and division-competent form (44). Therefore, the swarmer cell is in a presynthetic (G1) phase, while the stalked cell is in the synthesis (S) phase. In the late predivisional stage, the cell becomes incompetent for DNA replication and is in a postsynthetic (G2) phase. The stalked cell likely represents a terminally differentiated cell type, as a stalked cell reverting to a swarmer cell has never been observed.
FIG. 1.
Life cycle of Caulobacter crescentus. The cyclic developmental program begins with a stalked cell with an adhesive holdfast at the tip of the stalk. The stalked cell enters S phase, a cell state where it is competent for DNA replication. As the cell grows and replicates its DNA, it becomes a predivisional cell. During this time the cell becomes incompetent for DNA replication, entering the G2 phase. In the late predivisional stage, a flagellum is formed at the swarmer cell pole. After compartmentalization, flagellar rotation is activated (circular arrow) and pili are extruded. Cell separation leads to two different cell types. One cell is a stalked cell which reenters the cyclic developmental program and S phase, completing the circle. The other cell is a swarmer cell. The swarmer cell cannot replicate its chromosome yet is distinct from the predivisional cell and therefore is in a separate phase, referred to as G1. The holdfast is formed predominantly during the swarmer cell stage. Later the swarmer cell differentiates into a stalked cell. This differentiation comprises the noncyclic developmental program.
There exists a desire to understand a given process both temporally and spatially, not just for developmental biologists but for biologists from all fields. C. crescentus offers a remarkable experimental system for both aspects. There are multiple mechanisms by which swarmer cells can be isolated from a mixed cell population, thus allowing synchronization and the study of processes through time using large quantities of cells. Additionally, molecular biology techniques allow analysis of temporal events in individual cells. But it is the study of spatial processes that allows C. crescentus to truly shine. Not only does the cell have polarity (the definition of the cell poles from the bulk of the cell body), but it also has polar asymmetry (the differentiation of one pole from another). Different poles can be distinguished by morphological features, such as the stalk or the flagellum. The cell is bilaterally asymmetric as well, with one side concave and the other convex, leading to its namesake crescent shape. The importance of this bilateral asymmetry is currently unknown.
A concept that appears in older C. crescentus literature but appears to be missing in many recent articles is the idea that C. crescentus has, in fact, two distinct developmental programs. Though the life cycle is depicted as a circle, only one developmental program is cyclic. The stalked cell has a developmental program that involves the establishment of appendages at a defined pole at specific times and ultimately culminates with cell division and release of the swarmer cell. This program is cyclic in that the mother stalked cell returns to its predevelopmental stage. In fact, the stalked cell has often been likened to a stem cell in that it can give rise to different progeny but itself remains the same. On the other hand, the swarmer cell undergoes a noncyclic developmental program with holdfast synthesis, the shedding of the flagellum/disassembly of the pili followed by extension of the stalk. The swarmer cell does not return to the predevelopmental state, and thus its development is noncyclic. Swarmer cell development is not simply a morphological change that accompanies the early steps of cyclic stalked cell development. Certainly there are similar processes that occur in both circumstances. However, the signals that initiate these processes are largely unknown for both cell types, and there is no indication that signals are shared. Therefore, this review will consider swarmer cell development separate from stalked cell development, and each developmental event (cyclic versus noncyclic) will be analyzed separately.
CYCLIC DEVELOPMENT
A discussion of the cyclic developmental program of stalked cells will require discussion of the cell cycle. In fact, this developmental program is based largely on the way the cell cycle is elaborated. The bacterial cell cycle is the series of processes that lead to the duplication of the cell. These processes include DNA replication, chromosome segregation, establishment of the division plane, cytokinesis, and all the regulatory pathways that coordinate the processes. C. crescentus makes extensive use of feedback signaling such that one process is not allowed to proceed until a previous one has reached a satisfactory level of completion. Such stepwise progression through the cell cycle has caused some researches to refer to C. crescentus as hardwired and mechanical. Yet, C. crescentus is more than a membrane surrounding tiny, twirling gears. The signaling processes are dynamic, which in some cases proves essential for function.
Much of the internal circuitry that drives and coordinates different processes in C. crescentus makes use of bacterial two-component systems (for a review, see reference 242). As the name implies, two-component systems are composed of two protein partners: a histidine kinase and a response regulator. The histidine kinase is usually comprised of two portions, a signal-sensing portion and a kinase portion. In response to a signal, the kinase portion autophosphorylates on a conserved histidine residue using ATP. The phosphoryl group is then passed to a conserved aspartate on the receiver domain of the response regulator, which affects the action of an associated output domain. Occasionally a histidine kinase will have a receiver domain of its own, forming a hybrid histidine kinase, in which case the phosphoryl group is passed first to this receiver domain and then to a histidine phosphotransfer (Hpt) protein before finally reaching the response regulator. This extended pathway is termed a phosphorelay. It is thought that the function of the Hpt is to allow either the integration of another histidine kinase into the pathway leading to the response regulator or splitting of the pathway from a single histidine kinase to multiple response regulators through promiscuity of the Hpt. For traditional two-component systems, the histidine kinase is localized to the membrane, with the signal-sensing domain oriented outside the cell. An extracellular signal induces the phosphotransfer and subsequent activation of the response regulator, which often uses a DNA binding domain as an output. Thus, an extracellular signal is transformed to a change in gene expression. However, in C. crescentus, this simple paradigm is altered in new and inventive ways to accomplish the different developmental tasks.
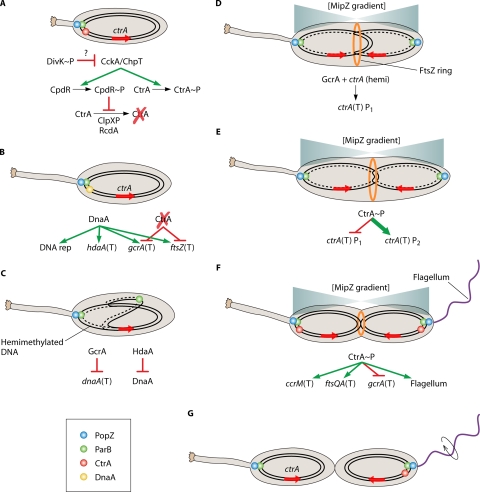
In the following sections, the cyclic developmental program will be described in a rough temporal order of events. We will discuss how C. crescentus regulates the initiation of chromosome replication, how chromosomes are segregated and how this leads to the establishment of the division plane, how biogenesis of polar organelles is coordinated with the cycle, and how both canonical and noncanonical two-component systems are used to integrate multiple processes and control cell fate. Aspects of cell cycle-coupled developmental processes are summarized in Fig. 2, while features of the two component systems that govern cell fate are summarized in Fig. 3. Given that the developmental program discussed here is cyclic, it technically has no beginning or end. However, we will begin with the initiation of chromosome replication.
FIG. 2.
Integral cell cycle components of the cyclic developmental program. (A) The initiation of the cyclic developmental program occurs during swarmer cell differentiation or in stalked cells that are the product of cell division, and it requires deactivation of CtrA. In the stalked cell that is the product of cell division, CtrA is deactivated after compartmentalization. In the stalked cell that is the product of swarmer cell differentiation (as indicated by a shortened stalk), activated CtrA (phosphorylated and not proteolytically degraded) is bound to and silences the origin of replication and must be deactivated (see “Modulation of CtrA Activity” in the text). DivK∼P leads to inhibition of CckA-mediated activation of CtrA by an unknown mechanism. Phosphoryl transfer from CckA to ChpT to CtrA ceases, preventing CtrA phosphorylation. Phosphoryl transfer from CckA to ChpT to CpdR also ceases, leading to decreased CpdR∼P levels and relieving inhibition of CtrA proteolysis. CtrA becomes deactivated (dephosphorylated and proteolytically degraded). In stalked cells originating from swarmer cell differentiation or cell division, the ParB-parS complex is bound to PopZ multimers at the stalked cell pole. (B) DnaA is synthesized and binds to the origin of replication vacated by CtrA, initiating DNA replication. DnaA also positively regulates transcription (T) of hdaA, gcrA, and ftsZ. The absence of activated CtrA allows transcription of gcrA as well as ftsZ. (C) One of the new ParB-parS complexes begins migration across the cell to the swarmer pole. DNA replication continues, leaving the replicated DNA in the hemimethylated state. GcrA represses transcription of dnaA, while HdaA inactivates existing DnaA, preventing additional initiation of DNA replication. (D) Segregated ParB-parS complexes allow establishment of MipZ gradients, with the lowest concentration at roughly midcell, determining the position of the FtsZ ring (orange ring). DNA replication past ctrA (red arrow) leaves the duplicated genes in the hemimethylated (hemi) state. Hemimethylation in combination with GcrA lead to activation of the weak ctrA P1 promoter. (E) The small amount of CtrA produced and activated represses the P1 promoter and activates the strong ctrA P2 promoter, leading a burst of CtrA synthesis (bold). (F) The burst of CtrA synthesis has multiple effects on the cell, including silencing the origin of replication, initiating CcrM synthesis which methylates the chromosomes, initiating FtsQA synthesis which allows cytokinesis to begin, blocking gcrA transcription, initiating flagellum biosynthesis, and other effects as well. (G) Once cytokinesis has completed, flagellum rotation is activated (circular arrow), and the two new cells are ready to separate. CtrA is deactivated in the stalked cell, allowing reinitiation of the cyclic developmental program. The mechanism for cell type-specific CtrA inactivation is described in Fig. 3.
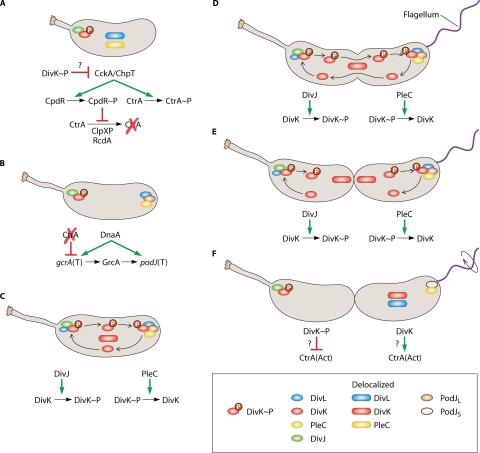
FIG. 3.
Cytokinesis-sensing mechanism. (A) As shown in Fig. 2, the developmental cycle represented begins with either a differentiating swarmer cell (as indicated by the shortened stalk) or a stalked cell immediately after cell division. The cyclic developmental cycle begins with DivJ and DivK∼P localized to the stalked pole. DivL and PleC are delocalized in the inner membrane. As described for Fig. 2, DivK∼P leads to deactivation of CtrA. (B) DnaA and the absence of activated CtrA lead to gcrA transcription (T). DnaA and GcrA lead to production of PodJ. PodJ localizes to the nascent swarmer pole and serves as a localization factor for PleC. DivL also becomes localized to this pole. (C) DivJ and PleC are located at opposite poles. DivK is phosphorylated by DivJ at the stalked pole and localizes there. DivL is found predominantly at the swarmer pole in the predivisional cell but is sometimes detected as a dimmer focus as the stalked pole, indicated by a smaller circle (see “DivL: a Wrench in the Works” in the text). This inconsistent DivL detection at the stalked pole could indicate a smaller protein pool at this location or a transient localization state. In either situation, DivL could serve as a DivK∼P localization factor at this pole. DivK∼P also diffuses to the swarmer pole (arrow trails) and forms a focus, again potentially through DivL. PleC dephosphorylates DivK∼P at the swarmer pole, causing it to delocalize. Delocalized DivK diffuses (arrow trails) and becomes rephosphorylated by DivJ at the stalked pole. (D) DivK phosphorylation/dephosphorylation cycling continues as the cell cycle progresses. (E) Immediately after cytokinesis completes, compartmentalization isolates DivJ and PleC enzymatic activities from each other. (F) As a consequence of compartmentalization, isolated DivJ activity leads to DivK phosphorylation in the stalked cell compartment, which then polarly localizes. Isolated PleC activity leads to DivK dephosphorylation and subsequently delocalization. The phosphorylation state of DivK affects activation/inactivation of CtrA in each cell compartment. CtrA activation/inactivation is accomplished by modulation of the CckA pathway, and though it is unknown exactly how DivK(∼P) interacts with this pathway, DivK∼P favors CtrA inactivation (see panel A). Conversely, DivK favors CtrA activation, perhaps simply by not favoring its inactivation, although other mechanisms have not been ruled out. As a result, CtrA is deactivated in the stalked cell compartment, while CtrA is activated in the swarmer cell compartment. PodJ is proteolytically processed to the short form, though PleC remains localized, and DivL and DivK become delocalized. Flagellar rotation is activated (circular arrow).
The Origin of Replication
Much study has gone into understanding the initiation of DNA replication in C. crescentus. This particular process is of interest because, unlike in Escherichia coli, it occurs only once every cell division cycle (150). Given the oligotrophic environment that C. crescentus usually inhabits, it seems unlikely that this organism would need to consistently employ multiple initiation events to increase the replication rate, like E. coli, and limiting initiation of replication to once per cell division has been hypothesized to help conserve energy. However, as will be discussed, there is evidence that the machinery controlling single replication per cell cycle is conserved among the alphaproteobacteria, some of which can inhabit nutrient-rich environments (see Evolutionary Role of Developmental Processes below).
The simplest mechanism for constraining DNA replication to once per cell cycle would be to control the DNA replication machinery in a cell cycle-dependent manner. Many genes encoding replication machinery are cell cycle regulated (74, 206). For example, DnaA (a protein essential for chromosome replication) is present throughout the cell cycle, though levels double just prior to replication initiation (292). DnaA synthesis is likely required, as DnaA synthesized in previous generations is deactivated (see “Regulation of DNA Replication and Methylation Machinery” below). Replicating plasmids are duplicated 20 times more efficiently in stalked cells than in replication-incompetent swarmer cells. Even so, detectable plasmid replication does occur in swarmer cells (151), indicating that replication machinery is present and competent throughout the cell cycle and that regulation of chromosome replication must use additional mechanisms. The C. crescentus origin of replication (Cori) was identified (19, 153), and plasmids bearing the Cori display cell cycle-dependent replication (153), indicating that at least a part of the mechanism that restricts replication initiation is found in the Cori itself.
The C. crescentus Cori represents a new archetype for bacterial origins of replication (152). Many of the essential elements are conserved with other bacterial oris, such as an AT-rich region, DnaA boxes, and an integration host factor (IHF) binding site (153). The exact number of DnaA boxes is still in question, but there is at least one essential box (153, 154). Unlike other oris, the Cori region is flanked by the hemE gene, encoding a protein involved in heme biosynthesis. The hemE gene is transcribed from both a weak promoter (Pw) and a strong promoter (Ps). The bulk of HemE protein synthesis is the result of Pw transcription, which is not cell cycle regulated (152). Mutations that abolish Ps activity also prevent chromosome replication initiation, suggesting a link between Ps promoter activity and initiation. Yet, this promoter lies partially in the AT-rich region, and alteration of sequences in this region could compromise initiation. Transcription from Ps may aid in melting this region. In DnaA-depleted cells Ps activity is increased but initiation is inhibited, indicating that while Ps activity is necessary for replication initiation, it is not sufficient (76). It is intriguing to speculate that hemE Ps activity may provide a mechanism for coupling DNA replication to metabolism, but secondary structure prediction indicates that the transcript from the Ps promoter folds such that the translational start codon is occluded (152). Coordinately, reporter activity displays only a minor increase in hemE translation rate when Ps activity is stimulated.
The cis-acting elements that provide cell cycle regulation for DNA replication initiation include five sets of inverted repeats that serve as binding sites for the essential response regulator CtrA. CtrA has been dubbed the “master regulator” for C. crescentus development. As will be demonstrated, much of the stalked cell developmental program involves the coordination of different stages of cell cycle progression with CtrA activity. As an indication of the importance of CtrA, chromosome immunoprecipitation experiments found 55 genes directly regulated by CtrA (132). Microarray experiments found that 144 genes (26% of all cell cycle regulated genes) have altered transcription due to direct or indirect CtrA activity (133). When phosphorylated on its conserved aspartate (D51), CtrA dimerizes, which increases its affinity for DNA (221, 222). Thus, the phosphorylated form of CtrA (CtrA∼P) represents its active form. In swarmer cells, CtrA is active and binds to the five sites (termed A to E) in the Cori region, thereby repressing replication initiation (10, 199). Specifically, the A and B sites work cooperatively to block Ps activity, site C overlaps the IHF binding site, and site E overlaps a DnaA box (199, 220, 221). Comparison of freshwater and marine Caulobacter oris demonstrates that while the number and position of CtrA binding sites are variable, the most conserved CtrA binding sites are always found in close proximity to DnaA binding sites, indicating that the most universally conserved function of CtrA in the ori is to modulate DnaA binding and subsequent replication initiation (216). This result underscores the importance of CtrA inhibiting replication. Therefore, in order to initiate DNA replication, CtrA must be deactivated.
Modulation of CtrA Activity
Phosphorylation is not the only mechanism that C. crescentus uses to regulate CtrA activity. A nonphosphorylatable but constitutively active allele, CtrAD51E, is not lethal as would be expected from a blockage in DNA replication (48). Interestingly CtrAD51E does not have an altered DNA binding affinity in vitro (222), indicating that activation by this allele occurs by a different mechanism, potentially by altering contacts with RNA polymerase. Constitutive activation of this CtrA allele is compensated for by cell cycle-regulated CtrA proteolysis. CtrA is degraded by the ClpXP ATP-dependent protease (107). At the time of replication initiation, ClpXP is localized to the stalked pole by CpdR, a response regulator lacking an output domain (96). It is not known how polar targeting regulates ClpXP function, but CtrA levels are stabilized in a cpdR mutant. ClpXP recognizes the terminal alanine-alanine dipeptide of CtrA; however, this is not sufficient for CtrA proteolysis, as cell cycle-regulated proteolysis also requires the N-terminal 56 amino acids (209). It was hypothesized that an adaptor protein would function in presenting CtrA to ClpXP, as has been seen for other proteins (290). This hypothesis was supported by the identification of RcdA, a protein required for targeting CtrA to the stalked pole in a ClpXP-dependent fashion; rcdA mutants have stabilized CtrA levels (160). However, recent in vitro evidence demonstrates that purified ClpXP efficiently degrades purified CtrA, and addition of purified RcdA has no effect on proteolysis, suggesting that RcdA is not an adaptor protein (33). The function of RcdA in CtrA proteolysis in vivo remains unknown.
Regulated phosphorylation and proteolysis are redundant in controlling CtrA activity. As stated above, a constitutively active CtrA allele is compensated for by regulated proteolysis. Conversely, a CtrA allele that is proteolysis resistant is not lethal due to regulated phosphorylation (48). Both activities are controlled by CckA, an essential hybrid histidine kinase (101). CckA is membrane bound, but it has no periplasmic sensing domain and has a receiver domain. CckA is present throughout the cell cycle but becomes localized predominantly to the swarmer cell pole in predivisional cells. Depletion of CckA leads to decreased CtrA phosphorylation and some CtrA destabilization (100, 101). The presence of a receiver domain suggests that CckA may utilize a phosphorelay, and indeed it was found that after autophosphorylation CckA transfers the phosphoryl group to an Hpt protein, ChpT (14). ChpT has two phosphoryl acceptors: CtrA and the aforementioned CpdR. CpdR is inactive for targeting ClpXP when phosphorylated (96). Therefore, when CckA is active, it activates CtrA by phosphorylation and prevents CtrA degradation by inactivating CpdR. Conversely, at the time of DNA replication initiation, CckA is deactivated, CtrA and CpdR are no longer phosphorylated, and this leads to CtrA deactivation by lack of phosphorylation combined with proteolysis. CckA is active during the mid- to late predivisional cell stage, coincident with CtrA activity (100). As stated, a proteolysis-resistant CtrA allele is not lethal due to regulated phosphorylation; however, it is not clear how cell cycle-regulated CtrA dephosphorylation is achieved. CckA is inactivated, yet it is unknown whether there is an active mechanism to dephosphorylate CtrA∼P or whether spontaneous dephosphorylation is sufficient to account for this deactivation. Similarly, the method of CpdR dephosphorylation is unknown. The signals and mechanism for repressing CckA kinase activity are unknown, though in the case of stalked cell development they involve the activity of DivJ/DivK (see “DivJ-DivK-PleC Outputs” below) (14).
As stated, deactivation of CtrA is critical to initiating cyclic development. This process not only occurs in differentiating swarmer cells but also must occur in stalked cells immediately after cell division. Stalked cells after cell division are often schematically represented as being devoid of CtrA, but this is not entirely accurate. While CtrA is necessarily deactivated at the start of cyclic development, it plays essential roles later in predivisional cells (see below). As such, activated CtrA is present and active throughout the predivisional cell during most of the cytokinesis process (48). Immediately after compartmentalization of the predivisional cell, CtrA is present in both compartments and deactivated specifically in the stalked compartment (see below).
Regulation of DNA Replication and Methylation Machinery
In addition to repressing the Cori, activated CtrA represses transcription of an unusual transcriptional regulator, gcrA (39). While GcrA lacks recognizable functional motifs, chromatin immunoprecipitation assays demonstrate that GcrA interacts with some cell cycle-regulated promoters, and microarray analysis shows that GcrA directly or indirectly affects transcription of 49 cell cycle-regulated genes, including DNA replication machinery components dnaQ, gyrA, and topoisomerase IV genes (87). Interestingly, GcrA negatively regulates expression of dnaA, but the gcrA promoter has a DnaA box which is necessary for transcription (39). This indicates not only that DnaA is a global transcriptional regulator but that GcrA provides feedback inhibition to prevent DnaA production at a time when the cell is competent for replication initiation. This may contribute to preventing multiple initiation events during a given cell cycle. It is not clear whether GcrA is the proposed negative regulator for conserved promoter elements found in other DNA replication machinery genes dnaN, dnaK, dnaX, and gyrB (119, 276).
DnaA functions in another initiation regulatory pathway as well. The Hda protein in E. coli associates with the β-clamp (DnaN) of the replisome, and once DNA is loaded into the clamp, Hda inactivates DnaA (118, 243). The C. crescentus homolog HdaA appears to function the same way (40). Expression of hdaA is positively regulated by DnaA in C. crescentus. This mechanism likely serves to balance the protein levels to ensure that enough HdaA is present in the cell to inactivate DnaA during replication. Therefore, once CtrA is inactivated and DnaA is active, replication is initiated and GcrA is produced. GcrA prevents further DnaA production while existing DnaA is inactivated by replisome-associated HdaA, thereby preventing reinitiation.
While the GcrA-DnaA and HdaA-DnaA feedback pathways help in preventing replication reinitiation, another aspect of this regulation seems to involve DNA methylation. Methylation control of reinitiation is not a new concept; in E. coli hemimethylated DNA is sequestered by the SeqA protein, preventing reinitiation until the chromosome is fully methylated by the Dam methylase (140, 258). However, despite the similarities in appearance between the two systems, the mechanism of methylation regulation in C. crescentus is markedly different from that in E. coli. To begin with, DNA methylation in C. crescentus is performed by the CcrM methylase, which is more closely related to the methylases of restriction modification systems than to the E. coli Dam methylase (291). Constitutive expression of CcrM leads to an increase in chromosome copy number in stalked cells (239, 291). Although evidence indicates that hemimethylated DNA is incompetent for replication initiation in both organisms (140, 291), CcrM is essential for viability in C. crescentus, while Dam in E. coli is not essential. CcrM monomers bind to hemimethylated DNA, catalyze the methylation of A bases in GANTC recognition sites using S-adenosylmethionine, and likely move processively down the DNA molecule due to a low release rate (12, 291). Though monomers are the catalytically active form, it was found that CcrM dimerizes in solution, perhaps as a mechanism to resist Lon proteolysis (219). Lon is known to target CcrM and is present throughout the cell cycle (280). CcrM is active only during a small time window in the predivisional cell, shortly before cytokinesis and compartmentalization, and the window of CcrM activity is determined by a spike in ccrM transcription that is able to overcome the constitutive rate of degradation by Lon. Therefore, the regulation of ccrM is key to controlling the methylation state of DNA, which in turn affects DNA replication initiation.
ccrM expression is coincident with and sensitive to DNA replication such that if DNA replication is inhibited, CcrM is not produced at high enough levels to overcome Lon degradation (241). Surprisingly, it was found that ccrM is positively regulated by CtrA (202). Since CtrA activity is cleared from the cell to allow DNA replication to initiate, how is CcrM expressed to remethylate the duplicated chromosomes? The results come from the clever way that CtrA is resynthesized.
The ctrA gene has two promoters, a weak P1 and a strong P2 (49). The weak P1 is activated first and requires two factors. First, it is positively regulated by GcrA (87). Second, it is active only in the hemimethylated form (203). Therefore, it is only once GcrA is active and the chromosome has been duplicated past the ctrA gene that ctrA expression increases. This result likely explains ccrM sensitivity to DNA replication inhibition. As CtrA levels steadily increase (and are presumably activated by the CckA pathway), CtrA∼P represses P1 and activates the strong P2 promoter, leading to a rapid and strong burst of CtrA (49). It is this burst of CtrA activity that is able to induce CcrM synthesis to a level necessary to overcome Lon degradation. Though Lon is present throughout the cell cycle, it was found to preferentially partition to the stalked cell (205), perhaps to ensure that residual CcrM activity in the mother stalked cell is removed prior to the next round of replication. The CtrA binding sites within the ctrA promoters are themselves unusual in that they have noncanonical spacing between the half-sites that severely reduces affinity for CtrA (233). However, physiological levels of CtrA rise high enough to overcome weak binding, and even constitutive occupancy of these binding sites throughout the cell cycle does not alter replication patterns due to the regulation of CtrA phosphorylation. This indicates that phosphorylation of CtrA does not regulate activity by altering affinity for binding sites but rather does so by another mechanism, possibly by altering binding contacts with other proteins (233), which is supported by the fact that CtrAD51E does not exhibit a change in DNA binding affinity in vitro compared to the wild-type nonphosphorylated allele (222).
Once the burst of CtrA activity leads to CcrM production high enough to overcome proteolysis, CcrM methylates the newly synthesized DNA strands, making them competent for replication initiation. However, because CcrM expression requires CtrA activity, the chromosomes become fully methylated only when CtrA is present and thus able to bind to the Coris and prevent reinitiation. Therefore, the simple mechanism of making the methylase dependent on CtrA ensures that the chromosome will be replicated only once per cell cycle. Yet, how exactly does methylation make the chromosome competent for replication? One possibility is that it is due to remethylation of the CcrM recognition sites in the Cori, similar to the situation in E. coli, where the SeqA protein recognizes hemimethylated sites in the ori and sequesters them from replication until they are remethylated (21, 227). However, comparison of multiple Caulobacter oris indicates that methylation sites within the ori are not conserved, with one marine Caulobacter strain not having any at all (216). It seems likely that methylation regulates replication competence in a different fashion. We have described above how methylation affects ctrA expression; methylation also affects expression of dnaA in C. crescentus. DnaA is actively degraded and protein levels decrease after the onset of replication (39, 77). Transcription of dnaA is cell cycle regulated, peaking prior to the initiation of replication (133, 292). The dnaA promoter is preferentially transcribed in the fully methylated state (38). Therefore, after replication initiates, GcrA prevents DnaA production and HdaA inactivates existing DnaA. Once replication has passed through the dnaA gene, it is further repressed by the hemimethylated state, whereas once replication passes through ctrA, the hemimethylation of P1 induces CtrA production, which in turn is able to produce CcrM, leading to methylation of the chromosome, including the dnaA promoter, making it competent for DnaA production. Therefore, methylation may make the chromosome competent for replication not by altering the Cori but by altering the ability of necessary replication proteins to be expressed. However, it is not clear what leads to a burst in DnaA synthesis prior to replication initiation, since the promoter region is fully methylated long before this time. Even more puzzling is that all the CtrA binding sites in the Cori region can be mutated together without leading to additional rounds of replication in a single cycle (10). Clearly there are other mechanisms, yet to be discovered, at work.
An unusual consequence of the C. crescentus methylation system is that different portions of the chromosome can be hemimethylated for a prolonged period of time. Whereas in E. coli newly duplicated chromosomes become fully methylated in less than 2 min (27, 244), portions of the C. crescentus chromosome can stay hemimethylated for up to 60% of the cell cycle (150, 291). It is known that the methylation state of certain promoters can have a significant impact on expression, with CtrA being a notable example. This means that where a gene is located on the overall chromosome with respect to the origin can have a significant effect on the timing of its methylation and thus cell cycle expression. It is interesting to think that not only where a gene is located with respect to other genes but also where it is found on the chromosomal map could have importance. An intriguing case study may be that of the C. crescentus chromosome terminus (ter) region. The ter region essentially remains fully methylated throughout the cell cycle; by the time it is duplicated, CcrM is already active (150). Whereas the ter regions in many organisms are locations with low conservation, weakly expressed genes, and a hot spot for integration of foreign DNA, the C. crescentus ter region is surrounded with essential and/or highly expressed genes (109), one of those being gcrA (39). Since this spot remains methylated for the longest time during the cell cycle, there may have been selection to reorganize the chromosome with important genes in this area so their expression would not be compromised by a lengthy period of hemimethylation. A few cases of chromosomal location regulating gene expression have been described before for B. subtilis. A portion of the chromosome becomes trapped in the forespore compartment during sporulation; later the rest of the chromosome is translocated into this compartment. Timely expression of some sporulation genes requires that they be located on the forespore-enclosed portion of the chromosome (124, 284); movement to locations on the chromosome excluded from the forespore compartment alters or abolishes expression.
Chromosome Segregation and Cytokinesis
Unlike some other bacterial systems, in which the ori is located to the midcell at replication initiation and sister chromosomes are segregated to opposite halves of the cell (72, 78, 270), the Cori in stalked cells is localized to the stalked pole, and when replication is initiated one Cori remains at the stalked pole while the other migrates across the length of the cell to the swarmer pole (110, 257). There is no preference for which Cori stays at which pole (151, 183). The mechanism by which sister chromosomes are segregated is still under investigation, but it employs a chromosomally carried parABS system. Par systems are used by some plasmids to ensure equal partitioning during cell duplication; however, chromosomal Par systems, though similar, are phylogenetically distinct from plasmid Par systems (67). The C. crescentus chromosomal Par system is different in that it is essential (165), unlike in many other bacteria such as B. subtilis (99).
The Par system is composed of two proteins, ParA and ParB, as well as a cis-acting element, parS, within the par operon located next to the Cori. Movement of the Cori region is indicative of parS movement and vice versa. When the parS sequence is moved elsewhere on the chromosome, chromosome segregation does not begin until DNA replication has reached the parS sequence (252), indicating that parS functions as a bacterial equivalent of a eukaryotic centromere. ParB binds to parS in a sequence-specific fashion and has three domains: an N-terminal ParA interaction domain, a middle DNA binding domain, and a C-terminal dimerization domain (60). While it has been known for some time that ParA has ATPase activity, only recently has it been found to form filaments in E. coli (54). Additionally, ParB is known to stimulate nucleotide exchange in ParA (52), leading to a hypothesis that the ParB-parS complex could bind to and cause depolymerization of ParA filaments, which could lead to the physical migration of ParB-parS across the cell.
The interaction between ParA and ParB is dynamic and multifaceted. This particular interplay may be important for maintaining proper stoichiometry between ParA and ParB in the cell. Both depletion of ParB and overexpression of ParA are lethal to C. crescentus (164). Overexpression of ParB can complement the lethality of ParA overexpression. In addition to forming a filament, ParA-ADP binds to single-stranded DNA, serving as a transcriptional repressor (52). How, or if, binding specificity of ParA-ADP for specific targets is conferred is not known, but ParA-ADP does seem to repress expression of the parAB operon and thus itself. As stated, ParA-ADP binds single-stranded DNA, but ParB-parS stimulates nucleotide exchange in ParA. ParA-ATP releases from single-stranded DNA, yet ParA-ATP disrupts the ParB-parS interaction (52). Therefore, ParB-parS levels serve to modulate the levels of ParA and ParB in the cell. An increased time of ParB-parS interaction would lead to increased ParA-ADP turnover, which would increase parAB transcription levels. This in turn would lead to an increase in ParA and subsequently ParA-ATP levels, which itself would lead to increased disruption of ParB-parS. While this action would also lead to an increase in ParB levels, the fact that there is only one parS region would quickly prove ParB-parS limiting.
The lethal nature of disruption of the Par system is puzzling. Disruption of Par systems in other bacteria leads to mild defects. In B. subtilis, disruption of the Par system causes production of anucleate cells but in only up to 3% of the total population, indicating that, while it is not optimal, B. subtilis can live without a Par system (99). Conversely, in C. crescentus disruption of the Par system causes cells to filament, suggesting that cytokinesis has been compromised. The interplay between chromosome segregation and cytokinesis has to do with the unique way that C. crescentus establishes the position of the division plane.
The position of the division plane in C. crescentus cells has long been a source of curiosity. Unlike in many other bacteria, the site of division is not exactly at the midcell. Instead, the division plane is shifted toward the swarmer pole such that the stalked cell that results from cell division is larger than the swarmer cell (245). It would be tempting to speculate that this asymmetry is the result of budding instead of normal cell division, but evidence indicates that cell growth occurs evenly on both sides of the division plane (1, 212, 228). The first step in division is polymerization of the tubulin homolog FtsZ into a ring-like structure associated with the inner membrane. There are two well-characterized systems for positioning the FtsZ ring and thus determining the cell division site. The first is the Min system in E. coli (for a review, see reference 141). In the Min system, Min proteins associate with the inner membrane and form a gradient, with the highest concentration at the cell pole and the lowest at the midcell. Min proteins inhibit FtsZ ring formation, and thus the Z-ring forms only where the Min protein concentration is lowest. However, C. crescentus does not have a Min system. Another system is nucleoid occlusion, in which a protein that inhibits cell division machinery binds nonspecifically to DNA (13, 285, 286). In this situation, the division plane cannot form where DNA is present. Once the sister chromosomes are properly partitioned, a DNA-free region is formed at the midcell, which allows the division plane to be formed. Initial evidence supported this model for C. crescentus because mutations that disrupted chromosome organization and packing were lethal (111, 112), and FtsZ rings did not form at midcell when DNA replication was inhibited and the chromosome was centrally located (197). However, cell invagination is observed before the ter regions have been resolved and separated from the midcell region, indicating that nucleoid occlusion is not utilized (110). The lethal nature of the disrupted chromosome organization may be due to alteration of some needed organization feature. The C. crescentus chromosome is highly ordered spatially in the cell, with particular genetic loci found consistently in the same position in the cell (257). The position of these loci is mirrored spatially when the chromosomes are segregated. It is not clear what function this organization serves, but disruption of the organization through disordering the chromosome could prove fatal.
Instead of the Min or nucleoid exclusion system, C. crescentus uses a system fundamentally similar to the Min system but mechanistically distinct. Division site formation is mitigated by the action of the essential MipZ protein, an ATPase related to ParA. MipZ disrupts FtsZ polymers by an unknown mechanism, though in vitro it was found to convert large, straight FtsZ filaments into short, curved filaments, a process which required the presence of ATP but not its hydrolysis (248). ATP hydrolysis is needed for the colocalization of MipZ with ParB. MipZ forms a gradient in the cell, with the highest concentration at the polar ParB-parS complexes. Given that the duplicated Coris with nearby parS sequences are located at opposite poles of the cell, the result is a concentration difference with MipZ levels lowest near the midcell, similar to the case for the Min system except that C. crescentus uses the Cori region as an orientation determinant. In fact, it has been shown that normal asymmetric division plane formation requires DNA replication initiation (197). In this manner, the Par system is essential because disruption of the system, either by depleting ParB or by altering chromosome segregation, would collapse the MipZ gradient and prevent proper FtsZ ring formation, leading to cell filamentation. How this system leads to slightly asymmetric FtsZ ring positioning is still under investigation.
In addition to spatial organization of cytokinesis, there is a temporal aspect as well. The first protein to localize to the division plane is FtsZ. FtsZ transcription is repressed by CtrA (122), and therefore degradation of CtrA, which allows initiation of DNA replication, also allows production of FtsZ. The FtsZ ring is stabilized by the N-terminal domain of FtsK (264). The C-terminal domain of FtsK is necessary to recruit ParC to the DNA replication machinery. ParC is part of the topoisomerase IV complex and is needed to decatenate and segregate sister chromosomes. Somehow this process is needed for Cori migration, as strains compromised in ParC and ParE (the other subunit in the topoisomerase IV complex, which is also needed to localize ParC to the replication machinery) often fail to fully migrate Cori regions to the opposite cell pole (263). However, the two domains of FtsK do not need to be on the same protein to maintain function, which precludes subcellular localization of the FtsK C-terminal domain in Cori migration.
While the FtsZ ring forms and is stabilized when CtrA is deactivated, cytokinesis cannot occur because other components of the machinery are missing. In particular, it has been found that the ftsQA operon is positively regulated by CtrA (277). Therefore, these critical cytokinesis components are synthesized only once DNA replication has proceeded to the point where CtrA expression is induced, coincident with CcrM expression. Thus, the timing of cytokinesis is measured against DNA replication and remethylation, with these processes coordinated in time through the action of the master regulator CtrA. Once cytokinesis has completed and cell separation has occurred, FtsZ is degraded in swarmer cells (122, 195) and stalked cells (198). FtsQ and FtsA are degraded in swarmer cells to a lesser extent than FtsZ (158). The mechanism of degradation for these proteins is unknown.
Given the importance of parS positioning to the cell cycle, it is not surprising that C. crescentus has a dedicated mechanism for maintaining its location at the cell poles. PopZ is a coiled-coil-rich protein that self assembles into large multimeric structures found at the cell poles (17, 53). One focus is found at the stalked pole, and a second focus forms concomitantly with parS migration at the new swarmer pole. PopZ directly interacts with ParB and likely serves as the ParB-parS polar anchor. In popZ mutants, the Coris are delocalized throughout the cell as well as seen to move about within a constrained area. It is tempting to speculate that PopZ may be involved in ParA polymerization; however, in time lapse experiments a third of observed cells displayed ParB-parS arriving at the cell pole prior to PopZ foci forming at that pole (17). Additionally it has been observed that Cori migration is unperturbed in a popZ mutant (53).
How PopZ localizes to a pole is a hotly debated topic. PopZ was found to form large complexes in zones free of chromosomal DNA, prompting a nucleoid exclusion hypothesis (53). However, given the polymeric structure of PopZ aggregates, it is unclear whether the aggregates formed specifically where DNA was absent or whether the aggregation of protein excluded DNA from that region. Alternatively, single-molecule imaging of PopZ suggests that oligomerization functions by a diffusion-capture mechanism, and therefore establishment of polar foci would require a polar PopZ-localizing factor that either nucleates PopZ or localizes a PopZ monomer that acts as a self-nucleator (17). It was found that depletion of the MreB protein, which forms an actin-like cytoskeleton in C. crescentus, prevents PopZ localization, suggesting that MreB may function in targeting a PopZ-localizing factor. Western blots detected some oligomerized PopZ in MreB-depleted cells (17). Cells treated with compound A22, which disrupts MreB filaments (71), were still able to form PopZ foci, suggesting that PopZ is able to oligomerize independent of MreB function, which would be expected for self-aggregation. PopZ was able to oligomerize in E. coli, which does not have a PopZ homolog and therefore would not be expected to contain a PopZ nucleator (53). The DNA exclusion hypothesis does not adequately explain how PopZ aggregates become polarly localized. While indeed the cell poles are relatively free of DNA, PopZ aggregates can form at multiple locations throughout filamentous cells, including linear stretches of the cell, even under native expression levels (53). These aggregate points do not appear to have DNA in them, but again, that could be a consequence of PopZ aggregation instead of a cause. Neither hypothesis, given the propensity for PopZ to self-aggregate, explains how a second PopZ focus is formed at the opposite pole during chromosome replication.
Cytoskeletons in prokaryotes function differently than those in eukaryotes. For example, depolymerization of actin and tubulin leads to instantaneous changes in eukaryotic cells, while depolymerization of MreB alters cell shape in bacteria only after growth occurs. It is not clear how the MreB cytoskeleton functions in cell polarity. The cytoskeleton is formed of multiple small filaments that display dynamic reorganization of monomers, though the filaments themselves are largely static (125). Throughout much of the cell cycle the MreB cytoskeleton exists as a helix that runs the length of the cell associated with the inner membrane (59, 70). When the division plane is formed, the MreB cytoskeleton condenses at the division plane. Once cell constriction begins, the cytoskeleton spirals back out into its helix formation. Depletion or overexpression of MreB leads to mislocalized Coris as well as other polar markers, and treatment with compound A22 prevents Cori migration (70, 71). Induction of MreB or removal of A22 restores Cori migration and polar marker localization, but in half of the cells the polarity of the markers is reversed. Yet, much of what is known about specific MreB function does not suggest a role in cell polarity. The major phenotype resulting from MreB depletion is a change in cell morphology. MreB is found only in rod-shaped cells (though not in all rod-shaped cells), and when MreB is depleted the cells round into spherical shapes (59, 114, 259). It is thought that this phenotype is the result of altered cell wall synthesis (59). This hypothesis is supported by the relocalization of MreB to the septal ring, as one of the functions of FtsZ is to localize cell wall synthesis machinery during cytokinesis (1). Additionally, MreB is important for cell shape but not chromosome segregation in Anabaena sp. strain PCC 7120 (90). Depletion and subsequent repletion of both MreB and the shape-determining protein RodA lead to budding and branching of both cells and ectopically positioned stalks in C. crescentus (261). Though the ectopic placement of stalks indicates misplaced poles, the production of branched stalks and cells is likely due to altered cell wall synthesis. Clearly, MreB disruption has large pleiotropic consequences for the cell, and it is not clear if the effects on polarity are direct or indirect.
Another potential PopZ-targeting factor could be the so-called “birth scar” protein TipN. TipN has two transmembrane domains and a coiled-coil-rich region (92, 130). In stalked cells TipN is located at the pole opposite the stalk until the division plane is formed, at which point it relocalizes to the division plane. After cell division, TipN is inherited by both cells and remains at the new pole formed by cell division until that cell matures and forms a new division plane. Therefore, TipN serves to demarcate the new pole from the old pole. tipN strains display mislocalized flagella, with approximately 50% of the cells forming a flagellum at the stalked pole, as well as somewhat mislocalized polar markers PleC and CpaE (see “Cytokinesis Sensing and Determination of Cell Fates: the DivJ-DivK-PleC System” and “Polar Localization of Histidine Kinases” below), though the degree of PleC mislocalization is a subject of debate. Additionally, tipN mutants display an altered bias in the location of the division plane such that swarmer cells are larger than stalked cells after division, in contrast to the wild-type bias (130). How TipN functions in overall cell polarity is not clear, as stalks in tipN cells still form at the old pole and DivJ localization is unaffected (see “Polar Localization of Histidine Kinases” below), and though flagella have seemingly randomized distributions to a given pole, it should be noted that the flagella still form at a cell pole instead of along the lateral portion of the cell.
A connection between TipN and PopZ comes from overexpression of TipN, which results in TipN foci ectopically located at random positions along the cell (130). Cells branch at the sites of these foci and form flagella at the end of the branches, as if each TipN focus establishes a new cell pole. These ectopic cell poles also localize PopZ foci (53). However, a direct interaction between PoPZ and TipN is not likely because PopZ foci are able to form at the swarmer cell pole in a normal cell cycle fashion in a tipN mutant (17). The MreB cytoskeleton shows a reduction in the ability to localize dynamically to the midcell in a tipN strain, instead preferentially staying in the helix formation (130). This result suggests that TipN can affect MreB function, which would be expected for the cell-branching phenotype of TipN overexpression, as altered cell wall growth would be needed to establish a new branch.
It is unknown how TipN itself localizes to the new pole. The coiled-coil-rich region would indicate protein-protein interactions; however, because TipN appears to serve as a marker for other polar functions to segregate to the new pole, this region may function more in directing other proteins such as TipF (see the next section) instead of the mechanism by which TipN gets directed. Clearly, how TipN, MreB, and PopZ function in cell polarity is a knot still to be untangled.
Flagellum Biosynthesis
The C. crescentus flagellum is a model of efficiency. Due to the small stall torque and working at below the knee rate (the rate above which energy from the proton motive force becomes utilized inefficiently and dissipated in the motor), C. crescentus swimming efficiency is an order of magnitude higher than that of E. coli or Vibrio alginolyticus (139). For many years the study of flagellum biosynthesis was the hallmark of C. crescentus research. It is an intricate and complex process that utilizes multiple different mechanisms of feedback, autoregulation, and checkpoint control (for reviews, see references 2, 23, 56, 196, and 281). It is also largely beyond the scope of this review. Instead, this review will focus on how flagellum biosynthesis is integrated and coordinated with the cell cycle. Accordingly, only a general knowledge of flagellum biosynthesis is required. Flagellum biosynthesis in C. crescentus is based largely on a hierarchy of genetic expression. Flagellum synthesis genes are categorized into four classes (I to IV), and the expression and subsequent assembly of proteins of one class of genes require signals from the successful action of proteins from the previous class.
Class I genes are those that lead to expression of class II genes based upon a cell cycle cue. The nature of class I genes was a mystery for some time. It was known that many class II genes had a conserved sequence element in their promoters that was thought to be the binding site of an alternative sigma factor, σR (240). However, it was later discovered that this sequence element was also conserved in the promoter element of ccrM, and it was later identified as the CtrA binding site (241). Thus, the product of the elusive class I regulator gene is the master regulator CtrA. From this, it is easy to understand how flagellum biosynthesis is cell cycle regulated (91, 218) and why it is susceptible to interruption in DNA replication (46, 184, 240, 277).
Among the class II genes are those encoding the MS ring, switch complex, and protein export apparatus, as well as genes encoding regulatory proteins sigma 54, FliX, and FlbD (167-171). FlbD is an NtrC-like transcriptional activator; it contains an N-terminal receiver domain, a central transcriptional activation domain, and a C-terminal DNA binding domain (201). Upon phosphorylation, NtrC-like activators bind to enhancer elements to help a given sigma factor activate transcription of targeted genes. In the case of FlbD, the enhancer elements are found in class III/IV genes and aid sigma 54 (rpoN)-based transcription (172, 173). FliX and FlbD form an important checkpoint in flagellum synthesis. FliX binds to FlbD and prevents activity (167). Upon completion of the MS ring, FliX repression of FlbD is relieved, though the sensing mechanism is not currently understood. FlbD is then activated by phosphorylation by an unknown kinase, though this activity is specific to the swarmer cell pole (274, 275), and FlbD/sigma 54 activity increases transcription of class III/IV genes, an act that also requires IHF (167). FliX is also needed for FlbD activation as well as repression. Mutant strains of fliX are nonmotile and fail to express class III and IV flagellar genes (166, 171). This phenotype can be bypassed by a gain-of-function mutation in flbD, indicating that FliX is needed for proper activation of FlbD (168, 170). While FlbD activates transcription of both class III (P ring, L ring, rod, and hook) and IV (flagellins) genes, translation of class IV genes is inhibited by the mRNA binding protein FlbT (7, 147). The mechanism by which class III gene product assembly completion is sensed and transduced, resulting in inactivation of FlbT, is unknown.
Mutations that prevent completion of class II gene product assembly also inhibit cytokinesis (73, 289). The inhibition of cytokinesis was shown to be the result of inactive FlbD, and it occurs temporally after the formation of the FtsZ ring (167). The mechanism of inhibition is not known, but a likely explanation would be a cytokinesis component that is under FlbD transcriptional control. Therefore, there are two checkpoints where flagellum biosynthesis is tied to the cell cycle, at CtrA activation of the flagellum hierarchy and at the inhibition of cytokinesis until class II gene products are assembled. The first checkpoint makes sense in that flagellum synthesis is needed only for the daughter swarmer cell and is inhibited until DNA replication occurs, signaling the production of a nascent swarmer cell. The logic of the second checkpoint is less evident. Here, cytokinesis is prevented until the base of the flagellum is formed. Why would the cell require this action to be completed prior to cytokinesis? One potential explanation may have to do with placing the flagellum on the cell.
TipN influences the site of flagellum assembly, since tipN mutants have randomly localized flagella with respect to swarmer versus stalked poles. TipN relocates from the swarmer pole to the division plane during cytokinesis in order to mark the new pole that is formed from cell division. If cytokinesis and TipN relocalization occur prior to establishment of the flagellum base, the flagellum may be formed at the wrong pole. Therefore, prevention of cytokinesis until the base is formed may be a mechanism of preserving proper flagellum localization. The influence of TipN on flagellum position may be mediated through its interaction with TipF. TipF has transmembrane, coiled-coil, and EAL domains; EAL domains have been shown to catalyze the degradation of cyclic di-GMP (c-di-GMP) (see “Swarmer → Stalked Cell Transition” below). Mutation of tipF as a whole or of the specific EAL domains results in strains that lack flagella (92). TipF localizes to the swarmer pole and the division plane in a TipN-specific manner. However, it is not clear that polar localization of TipF is required for its activity. Given that tipF mutants lack flagella instead of having mislocalized flagella, it seems unlikely that this protein specifically targets flagellum synthesis spatially. A better candidate is the protein PflI. pflI mutants have a 4- to 5-fold increase in mislocalized flagella, while other polar markers are unaffected (176). PflI localizes to the swarmer pole prior to establishment of the MS ring. It is unknown if TipN affects PflI localization.
Cytokinesis Sensing and Determination of Cell Fates: the DivJ-DivK-PleC System
The field of developmental regulation in C. crescentus was born from the study of the flagellum. Screens looking for nonmotile C. crescentus mutants yielded many strains that helped piece together the flagellum assembly hierarchy, but mixed in with these strains were mutants that had pleiotropic defects, mutants that not only were nonmotile but also failed to produce polar pili or stalks. Disregarded as useless for understanding flagellum assembly mechanics, these mutants would later prove instrumental in initiating the areas of research that dominate C. crescentus biology today. Principal among these mutants are pleC mutants.
pleC mutants produce flagella but are unable to rotate them, leaving them “paralyzed” and leading to a swarming defect on low-percentage-agar plates (55, 63, 232). These paralyzed flagella are also not shed during the swarmer → stalked cell transition. These mutants also do not produce pili (as measured by pilus-tropic phage sensitivity), holdfast, or stalks (229, 265). Intensive mapping techniques finally led to the identification of the pleC gene, encoding a histidine kinase (265). PleC is predicted to have at least four transmembrane domains and a cytoplasmic kinase region that demonstrates autokinase activity in vitro. It also has a periplasmic domain; however, the function of the periplasmic domain is unknown. A truncated allele of pleC that encodes only the cytoplasmic portion can partially complement a pleC mutant; this strain displays reduced swarming motility and phage sensitivity compared to the wild type.
PleC is produced throughout the cell cycle with its transcription under the control of the housekeeping sigma factor (145). While its expression level is constant throughout the cell cycle, the PleC pool itself displays dynamic changes in localization. Tracking the movement of PleC using fluorescent tags demonstrates that PleC is found as a focus at the flagellar pole in swarmer cells (45, 272). Upon swarmer cell differentiation, PleC becomes evenly distributed throughout the inner membrane until the cell begins replication, at which point PleC is relocalized at the swarmer cell pole in the predivisional cell. It remains at this pole during cell division, leading to the flagellar pole localization in swarmer cells. Initially it was not clear what function this dynamic localization served in signaling. Polar localization is not obligately required for PleC function, because the allele producing only the cytoplasmic portion and not likely to display a normal localization is able to partially complement a pleC deletion for polar morphogenesis. Nor was it clear why a histidine kinase, which was thought to simply transduce an extracellular signal to a change in gene expression, would need to be spatially restricted. It was only with the discovery of pleC suppressors, leading to mapping of an extensive signaling pathway, that the polar localization of PleC demonstrated its importance.
The polar development phenotype seen in pleC mutants, i.e., that of paralyzed flagella and no pilus, holdfast, or stalk synthesis, had actually been seen before. Cell division mutants blocked in cytokinesis displayed a paralyzed flagellum and no pili, nor did a stalk or holdfast form even as the cells became filamentous (91). Thinking that pleC might be involved in a pathway that connects polar development to the cell cycle led to the design of a screen where suppressors of a heat-sensitive allele of pleC that had a cold-sensitive cell division phenotype were isolated, leading to the discovery of DivJ and DivK (230). The histidine kinase DivJ lacks a periplasmic domain and likely has at least six transmembrane domains (177). DivK is a single-domain response regulator (it lacks an output domain) and is essential for viability (26, 80, 83). The isolation of pleC suppressor mutations in a histidine kinase and a response regulator is not necessarily surprising; cross talk between two-component systems has long been the bogeyman of these signaling systems. It was theoretically possible that altered signaling specificity of a DivJ-DivK system could complement a pleC defect. Yet, evidence suggested that this was not the case. Purified kinase portions of both PleC and DivJ can phosphorylate DivK in vitro (83), and yeast two-hybrid analysis demonstrated PleC-DivK and DivJ-DivK interactions in vivo with wild-type alleles (178), which is not surprising since the cytoplasmic portions of PleC and DivJ are quite similar (177). Phosphotransfer profiling in vitro demonstrated that DivJ and PleC display a clear kinetic preference for phosphotransfer to DivK over almost every other response regulator protein in C. crescentus (225). As a whole, these results suggested that DivK/DivJ suppressors were not altering a separate signaling network in a way that bypassed the pleC mutation but instead were modulating different aspects of the same regulatory network.
Given that PleC and DivJ are both histidine kinases and that DivK is a response regulator, an obvious question is: how do PleC and DivJ affect phosphorylation of DivK? The in vitro work suggested that both could act as kinases, yet a truly significant breakthrough occurred when it was found that PleC acts principally as a phosphatase of DivK in vivo, whereas DivJ had the expected kinase activity (159, 272). Therefore, PleC and DivJ have antagonistic activities on DivK, which is consistent with divJ and divK mutations acting as suppressors of pleC, particularly if considered in terms of phosphorylated DivK. pleC mutations would lead to hyperphosphorylation of DivK, whereas a mutation compromising the kinase activity of DivJ in a pleC mutant would swing the phosphorylation levels of DivK in the opposite directions (272). Similarly, an allele of DivK that limits phosphorylation would also compensate for hyperphosphorylation.
Another breakthrough came with analysis of the subcellular localization of DivJ and DivK. DivJ is absent in swarmer cells and produced during swarmer cell differentiation, at which point it is localized at the stalked pole (272), a process that requires the transmembrane domains (213). It remains at this position for the life of the stalked cell and does not change localization throughout the cell cycle. This means that DivJ is located at the pole opposite PleC during cell division, which serves to separate the antagonistic activities of these proteins spatially. Whereas DivJ has a simple localization pattern, the location of DivK throughout the cell cycle is dynamic. DivK is present and displays at least some detectable levels of phosphorylation throughout the cell cycle (102). It is delocalized in swarmer cells but becomes localized to the stalked pole in stalked cells. With the onset of replication, a second focus is formed at the swarmer pole. Following division, DivK remains localized in the stalked cell but becomes delocalized in the swarmer cell. Interestingly, divJ mutations cause DivK to always be delocalized, while pleC mutations cause DivK to always be strongly bipolar in predivisional cells, which suggests that the phosphorylation state of DivK affects its ability to form polar foci. This hypothesis was verified when a divK allele that could not be phosphorylated was shown to be delocalized just like a divJ mutant allele (129). This mutation was also lethal. This result means that separating DivJ and PleC across the length of the cell results in different amounts of DivK∼P in different parts of the cell. In fact, photobleaching and fluorescent resonance energy transfer (FRET) experiments demonstrated rapid shuttling of DivK between the cell poles, with a turnover rate of approximately 5 s (159), indicating that DivK is constantly moving between the poles with changing phosphorylation state. Thus, the DivJ-DivK-PleC system forms an elegant system for the cell to detect cytokinesis and produce different cell fates. DivJ, located at the stalked pole in the predivisional cell, phosphorylates DivK. DivK∼P then diffuses to the swarmer cell pole, where it becomes dephosphorylated by PleC. After cytokinesis, the different poles of the cell become compartmentalized (115), effectively separating DivJ and PleC activities. This results in a stalked cell compartment that has only DivJ, leading to predominantly phosphorylated DivK that then localizes as a focus. The swarmer cell compartment has only PleC activity, leading to predominantly dephosphorylated DivK that cannot localize to a polar focus. Therefore, the different cells inherit drastically different levels of phosphorylated DivK, which then affects cell fate (see “DivJ-DivK-PleC Outputs” below). The DivJ-DivK-PleC triumvirate constitutes the central mechanism of an elaborate system controlling development that involves many more proteins.
The DivJ-DivK-PleC system may appear to be well understood at this level, but there are still major questions that remain. While the levels of DivK∼P are reduced in a divJ mutant, they are not abolished, so what is providing residual DivK kinase activity? Similarly, it is odd that while DivK is essential for viability, DivJ and PleC are not (though loss of either does cause some growth defects) (83, 102). Does redundant kinase activity for DivK complement absence of DivJ? As PleC functions predominantly as a phosphatase, it is possible that the labile nature of protein phosphorylation causes DivK to spontaneously dephosphorylate fast enough to compensate for the loss of PleC activity, yet if this is true, why have PleC to begin with? Lastly, what exactly does DivK bind to at the poles? DivK forms a swarmer pole focus in pleC mutants (102), so it cannot bind to PleC at this pole. DivJ is delocalized in a pleC mutant (272), yet DivK clearly forms a stalked cell focus in this strain, suggesting that DivJ is not the stalked pole anchor. The polar anchoring of DivK may be explained by a single protein, DivL.
DivL: a Wrench in the Works
The same suppressor screen of pleC that identified divJ and divK also identified another gene, divL (230). DivL is unusual in that sequence similarity indicates that it is a histidine kinase (membrane bound but without a substantial periplasmic domain), but instead of the conserved histidine that becomes phosphorylated, it has a tyrosine residue (283). Though DivL was found in a suppressor screen for pleC and yeast two-hybrid analysis displayed a specific interaction between DivL and DivK (178), it is still not clear how DivL functions in the DivJ-DivK-PleC signaling pathway. For some time it was thought that DivL may be the kinase that phosphorylates CtrA (26, 283), but the discovery of the CckA-ChpT-CtrA pathway argues against this hypothesis (14, 100). Instead, we wish to present an alternative hypothesis.
DivL is present but dynamically localized throughout the cell cycle (214). It is dispersed in swarmer cells, but in stalked cells it is found predominantly at the stalk distal pole, though in a subpopulation of cells a dimmer second focus can be seen at the stalked pole. It then becomes dispersed in late predivisional cells. Though deletion of the discussed tyrosine residue causes lethality in the cell, alteration of the tyrosine to a histidine or phenylalanine and, surprisingly, deletion of the entire ATPase domain, preventing kinase activity, is not lethal (204, 214). This indicates that the essential function of DivL is independent of its kinase activity. Based upon these results, it has been proposed that DivL functions as the polar anchor for DivK∼P and presents the protein to PleC for phosphatase activity (204). A key to this hypothesis is that DivK is bipolarly localized in the pleC mutant. In this strain, DivK phosphorylation levels are elevated due to the lack of PleC phosphatase, which promotes polar focus formation in predivisional cells (102). DivL is found at the swarmer pole, and this localization is independent of PleC (214), so DivK∼P binding to DivL at this pole would explain that accumulation. DivJ is delocalized in the pleC mutant (272), but DivK∼P still forms a focus at the stalked pole in this strain (102). DivL is sometimes detected at the stalked pole (214) and could represent either a smaller protein population or a transient localization state at this pole, and this small amount of stalked pole DivL could be the anchor for DivK∼P at that pole. While this hypothesis is appealing, further verification is required.
DivJ-DivK-PleC Outputs
Though the DivJ-DivK-PleC system forms an intriguing cytokinesis-sensing mechanism and this mechanism has an impact on cell fate, as mutations in this system alter polar morphogenesis and cell division (55, 63, 230, 232), how the signals are transduced is still a mystery. What is the output for the DivJ-DivK-PleC system? Some evidence indicates that it works at the level of CtrA activation (see “Modulation of CtrA Activity” above). In a screen for divJ suppressors, mutations were found in DivL, as may be expected, but also in CckA (189). It was found that these mutations resulted in a decrease in CtrA∼P levels. Similarly, a screen for divK suppressors found mutations in DivL that, again, decrease CtrA∼P levels (204, 282). These results suggest a model where DivK∼P negatively regulates CtrA activation. In a divJ mutant, DivK∼P levels would be reduced, which would lead to overactivation of CtrA, and therefore suppressor mutations that result in reduced CtrA∼P levels would be expected (189). The temperature-sensitive allele of DivK used for the suppressor screen was likely a loss-of-function allele that would prevent inactivation of CckA and lead to overactive CtrA, suppressors of which would reduce CtrA activity (204, 282). Does DivK interact with/affect the CckA-ChpT signaling pathway directly? The answer is unknown. A direct interaction appears plausible given the evidence above, and another study found that DivK increased the relative proportion of nonphosphorylated CpdR, which would be predicted to increase ClpXP polar localization leading to increased CtrA degradation (97), indicating a direct interaction between DivK and the CckA-ChpT pathway. Yet recent findings show that CckA does not become displaced from the stalked pole by DivK during swarmer cell differentiation as had been originally proposed; in fact, CckA is not consistently found at this pole (8, 31). CckA is consistently localized to the swarmer cell pole during differentiation, and though localization does appear to be important for CckA activity, factors that influence localization have not been discovered (8). In a divL strain that suppresses the divK allele by leading to reduced CtrA∼P levels, it was found that CpdR∼P levels were unaltered (204). This result suggests that the CckA-ChpT signaling pathway is unaltered in this mutant and that the regulation of CtrA by DivK∼P occurs via a different mechanism than regulation of CckA localization and/or activity. DivK may regulate CtrA activity by multiple mechanisms, both direct and indirect, and such a model is consistent with redundant regulation of CtrA. One possible indirect mechanism is that DivK∼P may allosterically regulate the activity of DivL to make it principally dephosphorylate CtrA∼P. Such activity for a histidine kinase has been observed before (187). The in vitro phosphorylation data suggest a DivL-CtrA interaction (283), and the substitution of tyrosine for the conserved histidine may ensure that DivL displays only phosphatase activity. This hypothesis is attractive in that it would explain the regulated dephosphorylation of CtrA proteolysis-resistant alleles and it would not necessarily preclude the hypothesized function of DivL in DivK anchoring. The CckA-ChpT pathway was recently shown to be able to operate in reverse in vivo, becoming a phosphatase pathway on CtrA and CpdR prior to DNA replication, but it could not account for all of the in vivo dephosphorylation of these two targets (31). DivK regulation of DivL phosphatase activity could be the missing piece for CtrA dephosphorylation.
In the end, though, can the alteration of CtrA activity by the DivJ-DivK-PleC system explain all the effects that result from perturbation of the system? It seems unlikely, given that DivJ and PleC kinase/phosphatase activities function not only on DivK but also on the polar morphogenic regulator PleD (see “Swarmer → Stalked Cell Transition” below). Regardless, assuming that DivK∼P does negatively regulate CtrA activity, it is easy to see how this system leads to different cell fates. The outputs of the DivJ-DivK-PleC system remain a potent area of research.
Polar Localization of Histidine Kinases
One last aspect of this system for consideration is how the polarly localized histidine kinases become localized. While it is unknown how much polar localization contributes to their function (a thorough phenotypic analysis of delocalized histidine kinases has not been performed), the histidine kinases are clearly localized in wild-type cells, which raises the question of how this occurs. In the case of DivJ, its localization is dependent on the protein SpmX (200). The SpmX protein has a periplasmic muramidase domain and two transmembrane domains. SpmX has been shown to interact with DivJ in reciprocal coimmunoprecipitation experiments, with the interaction likely mediated by the transmembrane domains, as DivJ is predicted to have five transmembrane domains but very little protein sequence in the periplasm. The targeting of SpmX to the stalked pole depends on the muramidase domain, which is implicated in peptidoglycan binding. It has been hypothesized in the past that the peptidoglycan sacculus could serve as a polar targeting factor (134). Given that peptidoglycan synthesis occurs primarily at the midcell in C. crescentus, the polar peptidoglycan likely remains inert for most of the life cycle of the cell. If peptidoglycan is subject to modification, the oldest peptidoglycan could be chemically distinct from newly synthesized peptidoglycan, in which case a protein recognizing the modification could target to this area. In the case of the swarmer cell, the flagellar pole is the oldest pole. This pole later becomes the stalked pole, the same pole where DivJ localizes. However, there is a burst of peptidoglycan synthesis at this particular pole during the swarmer → stalked cell transition, leading to extension of the stalk itself (see “Stalk Biogenesis” below). This peptidoglycan would no longer be inert, and any chemical modification may be lost, which could potentially prevent SpmX recognition. Unfortunately, there is little evidence either supporting or contradicting inert peptidoglycan targeting. This hypothesis, for the time being, remains simply a hypothesis.
The regulation of spmX illuminates why DivJ is delocalized in a pleC mutant. spmX transcription requires sigma 54 as well as the sigma 54-activating protein TacA (200). TacA is necessary not only for spmX transcription but also for genes required in stalk biogenesis (see “Stalk Biogenesis” below). This finding is not surprising given that DivJ localization and stalk biogenesis occur at the same time. tacA itself is positively regulated by CtrA∼P. In a pleC mutant, DivK∼P levels are increased, which leads to a decrease in CtrA∼P levels (14), which then prevents TacA production. This in turn prevents SpmX production and prevents DivJ localization. Thus, the pleC disruption leads to DivJ delocalization. It should also be noted that pleC disruption leading to inhibition of TacA production also explains the stalkless phenotype of pleC mutants, since TacA is involved in expressing stalk biogenesis genes. Expression of tacA in a pleC mutant restores stalk biogenesis (200). Interestingly, DivJ is delocalized in a divL strain (204). It will be informative to see if spmX transcription is altered in this same strain. Even though the regulatory cascade outlined explains the delocalization of DivJ in a pleC mutant, it does not explain why spmX is not produced until swarmer cell differentiation even though CtrA is activated all throughout the swarmer cell phase. It has been found that spmX mRNA accumulates in early swarmer cells, but the protein is not produced until swarmer cell differentiation (200), indicating posttranscriptional regulation.
The localization of PleC is a complicated story. Single molecular imaging of fluorescently tagged PleC molecules indicates that the polar localization of PleC operates by a diffusion capture mechanism (45), which would require a PleC-targeting factor. That factor was found to be the protein PodJ. PodJ was identified in multiple developmental screens, and mutants have a phenotype similar to that of pleC mutants (43, 85, 229, 256). Both mutant strains lack pili and a holdfast and have difficulty ejecting the flagellum. However pleC mutants are stalkless and have a paralyzed flagellum, while podJ mutants produce stalks and the flagellum can rotate. podJ mutants do display a motility defect on swarm plates, but cells are seen to swim under the microscope.
The podJ gene is negatively regulated by CtrA and positively regulated by GcrA (43, 87). Therefore, PodJ is produced at around the time of DNA replication initiation and localizes to the swarmer pole (85, 256). PodJ is a large protein, containing 974 amino acids. The N-terminal two-thirds of the protein resides in the cytoplasm and has three predicted coiled-coil domains (85, 256). The protein then crosses the membrane, and the C-terminal third resides in the periplasm. This periplasmic domain has three tandem tetratricopeptide repeat domains (thought to be involved in protein-protein interaction), and the very C terminus is predicted to be a peptidoglycan binding domain. PleC is dispersed in a podJ mutant (85), indicating that PodJ is a polar targeting factor for PleC. How PodJ is targeted to swarmer pole is unknown. The presence of a periplasmic peptidoglycan binding domain would suggest a polar peptidoglycan recognition mechanism as postulated for SpmX, but mutants where the peptidoglycan binding domain has been truncated still demonstrate proper localization (135). However, these mutants do display altered proteolytic processing.
PodJ displays cell cycle-regulated proteolysis (85, 256). While the full-length form is found in predivisional cells, PodJ is proteolysed to a short form coincident with compartmentalization and flagellar rotation activation. The short form persists through the swarmer cell phase and is cleared from the cell during the swarmer → stalked cell transition. Proteolysis is performed by the coordinated action of two proteases. First, PodJ is cleaved in the periplasmic domain by the periplasmic protease PerP (29). perP mutations lead to stabilized long-form PodJ. The PerP cleavage event requires cell compartmentalization, DivK dephosphorylation, and, therefore, PleC. The perP gene has a CtrA binding site and was found to be positively regulated by CtrA (132). According to the model for the DivJ-DivK-PleC pathway, compartmentalization removes DivJ kinase activity from the swarmer cell compartment; DivK becomes predominantly dephosphorylated, preventing it from inhibiting CtrA activation; and CtrA becomes activated, thus leading to PerP production and PodJ cleavage. It is thought that degradation of the short form is regulated by the intramembrane protease MmpA, as mmpA mutants have stabilized short-form PodJ (30). However, recent results suggest that while MmpA is involved in PodJ proteolysis, degradation of the short form is regulated by a different mechanism (P. Curtis, unpublished results).
It has been difficult so far to attribute a function to the regulated proteolysis of PodJ, largely because there is still some residual processing in a perP mmpA mutant (29). However, a potential function is indicated by the podJ921 allele, which produces PodJ lacking the peptidoglycan binding domain. While PodJ921 still localizes to the swarmer pole, it undergoes immediate processing such that a long form is undetectable (135). This same strain also displays a reduction in phage sensitivity, suggesting that the periplasmic domain may be necessary for pilus biosynthesis. PodJ is necessary for localizing the pilus biogenesis protein CpaE (256). PodJ also is necessary to localize PleC, and though PleC is not necessary for CpaE localization, it is needed for CpaE release from the pole. It is also known that production of the pilus filament is inhibited when cell division is prevented (179, 231). These observations suggest a model where PodJ aids in pilus biogenesis by localizing CpaE in the predivisional cell but prevents extrusion of the pilus filament. Upon cell compartmentalization, PleC activity leads to PerP production, which removes the PodJ periplasmic domain, leading to pilus filament extrusion. Yet, overexpression of the structural subunit of the filament, pilin, causes an increase in the number of predivisional cells that have pili, suggesting that the pilus extrusion machinery is present and capable of functioning long before the pili are extruded (226). This result argues against PodJ serving as a brake for pilus extrusion, because it suggests that filament production is limited at the level of gene expression. Coordinately, the pilin gene is positively regulated by CtrA (226), and thus synthesis in the swarmer cell compartment would require PleC activity, as has been shown (256). Yet it is possible that overexpression of pilin is able to bypass a PodJ-dependent braking system; further experimentation is required. Why such regulation of pilus extrusion exists is not clear, but many of the isolated C. crescentus bacteriophages use the pilus as an infection mechanism. Making sure that the pili are extruded only after cell separation may be a mechanism to prevent infection of the parent cell, increasing its reproductive fitness. Additionally, it is known that pili aid in surface adhesion (16), and production of pili on predivisional cells could inhibit the efficiency of swarmer cell dispersal (see Evolutionary Role of Developmental Processes below). The flagellum also aids in adhesion (16), but it may also aid in separating daughter cells (Y. Brun, unpublished observation), which would necessitate earlier production of the flagellum.
An alternative hypothesis of how PodJ affects pilus synthesis is that PodJ could serve as a localization factor for the PleA protein. PleA is a lytic transglycosylase thought to be involved in reordering peptidoglycan at the pole so that the polar organelle machinery can be assembled through the sacculus (255). pleA mutants lack flagella and pili, and the machinery for each reaches a level of completion after assembly into the inner membrane but prior to passing through the sacculus. While PleA has the appropriate catalytic residues for transglycosylase activity, it is missing the domain needed for substrate targeting. It is possible that the peptidoglycan binding domain of PodJ could serve as the substrate-targeting domain, making sure that the flagellum and pili are assembled at the proper cell pole (135). However, while the podJ921 mutant has compromised pilus production, flagellum synthesis appears to be unaffected. Attempts to show PleA localization have been unsuccessful (255). These results suggest that the hypothesis of PodJ targeting PleA is incorrect. How PodJ influences pilus biosynthesis is still a mystery, but it obviously serves a greater purpose than just targeting PleC. The role in localizing PleC and CpaE suggests a larger scaffolding function in the cell. It is intriguing to consider other proteins that PodJ may localize. DivL is one potential target, since its localization pattern matches that of PodJ. Also, at least one chemotaxis sensory system is located at the same pole (6, 22, 123). The fact that the podJ mutant has functional flagella but displays swarming defects on soft agar plates suggests that the strain is compromised in chemotaxis. A potential interaction between PodJ and the chemotaxis machinery has yet to be investigated.
NONCYCLIC DEVELOPMENT
Swarmer cell differentiation represents perhaps the last great undiscovered continent in C. crescentus research. While some of the events that occur are known, they are known only on a gross level. A swarmer cell becomes a stalked cell; this entails loss of the flagellum and pili, production of a stalk, and derepression of DNA synthesis. We know some of the mechanisms that influence these processes, but the signaling behind them is largely unknown. Different proteins (FtsH) and systems (SsrA/SmpB) have been implicated in swarmer cell differentiation, as mutations in them alter the length of the swarmer phase, but there is no clear explanation as to their mechanism (61, 120, 121). As such, swarmer cell differentiation is a particularly intriguing area of study.
In the C. crescentus literature there is a subtle but important distinction in terminology for swarmer cell differentiation. Sometimes this event is referred to as the swarmer → stalked cell transition. Other times it is referred to as G1 → S. Occasionally these terms are interchanged, and while the meaning is understood, this is technically not correct. G1, S, and G2 refer to phases of the cell cycle analogous to the eukaryotic cell cycle, with S referring to the time of DNA synthesis and G1 and G2 referring to the presynthetic and postsynthetic gaps, respectively. Therefore, the G1 → S transition refers to a change in the cell state from replication incompetent to replication competent. However, swarmer → stalked cell transition describes a change in the morphology of the cell and does not necessarily refer to the competence of the cell to replicate its chromosome. Throughout this review, G1 → S refers to a change in chromosome replication competence, swarmer → stalked cell transition refers to morphological changes to cell, and swarmer cell differentiation refers to the combination of both events. While in wild-type cells these two events occur at the same time, they are genetically distinct and can be separated mechanistically. It is possible to get a swarmer cell to become a stalked cell but not replicate its chromosome by depleting DnaA or preventing CtrA inactivation (unsurprisingly, this is lethal) (89, 93). It is also possible to get a cell to replicate its chromosome without undergoing polar morphogenesis, as seen with the pleD mutant (3). This section not only reviews what is known about swarmer cell differentiation (aspects of which are shown in Fig. 4) but also highlights the areas of knowledge that are lacking.
FIG. 4.
Swarmer cell differentiation. (A) A mature swarmer cell is indicated by the presence of holdfast at the flagellar pole and the absence of pili. PodJS and PleC are localized to the flagellar pole. PleD is in the nonphosphorylated state and therefore delocalized, as is DivL. An unknown signal leads to DivJ synthesis. (B) DivJ is synthesized and localizes to the flagellar pole. For a brief period of time, PodJs, PleC, and DivJ all inhabit the same pole. DivJ and/or PleC phosphorylate PleD (see “Swarmer → Stalked Cell Transition” in the text), causing a subpopulation to localize to the developing pole. (C) PodJS is degraded by an unknown mechanism, and PleC becomes delocalized. PleD∼P catalyzes the formation of c-di-GMP. (D) Production of c-di-GMP leads to morphological changes in the cell through unknown mechanisms. The flagellum is ejected. (E) Stalk synthesis is initiated, which requires phosphorylation of the sigma 54 activator TacA through the ShkA/ShpA phosphorelay. The signal that leads to ShkA/ShpA/TacA activation is unknown. TacA activation leads to synthesis of the stalk length determinant StaR, as well as other, unidentified targets. Other factors are likely involved in stalk synthesis. It should be noted that the ShkA/ShpA/TacA pathway is present and active in the late predivisional and swarmer cell stages. The mechanism for controlling the timing of stalk synthesis is not understood. The stalk is extended with the holdfast at the tip. DivL localizes to the stalk-distal pole.
G1 → S Transition
The change from a replication-incompetent state to a replication-competent state involves the deactivation of CtrA with subsequent activation of GcrA; however, the signals that lead to these events are not known. In the case of a stalked cell that has just undergone division, an obvious possibility is that the compartmentalized DivJ leads to increased DivK∼P levels, which somehow inhibit CtrA activation. Thus, the cell immediately reinitiates the developmental cycle after division. While this action is observed under culture conditions, there is no evidence that cells in the wild immediately reinitiate DNA replication after division. In fact, it would seem unlikely to occur under severe nutrient-limiting conditions given that stalled replication forks often prove lethal to dividing cells, as shown for E. coli (79, 224). In the case of swarmer cell differentiation, DivJ is present at a lower level in swarmer cells than in stalked cells (272), which suggests that the signals to begin CtrA inactivation are different in swarmer cells and stalked cells. It is possible that the production and activation of DivJ are what governs the G1 → S transition, in which case the obvious question is: what governs DivJ production and activation? This is an intriguing, and still open, question. Given that SpmX production is regulated in a cell cycle fashion and in a similar manner as stalk biogenesis genes, it seems likely that the timing of DivJ production is controlled in a way that coordinates it with swarmer cell differentiation specifically. However, stalk formation and flagellum ejection are not needed for DivJ localization, suggesting that though DivJ production is timed with polar morphogenesis, its localization is not mechanistically tied (213). The specifics of DivJ production and how it affects the G1 → S transition are unknown.
Swarmer → Stalked Cell Transition
The developmental programs of C. crescentus have been referred to as “hardwired” in that various stages cannot be bypassed. For example, wild-type swarmer cells have never been observed to replicate and divide; instead, they must differentiate to stalked cells first. Even though stalkless phenotypes have been found, strains harboring these mutations must still enter a “stalked cell state,” displaying appropriate asymmetry and polar protein localization the same as a stalked cell despite the lack of an actual stalk (64, 213). However, one mutant comes close to approximating a swarmer cell division event.
Another screen for suppressors of pleC found mutations in pleD (232). pleD mutants produce few stalks, have pili, and are hypermotile (3, 4). Flagella are not shed and remain active throughout the cell cycle. A holdfast is formed, though later than in the wild type (137). Morphologically, then, pleD mutants resemble swarmer cells, but pleD is not essential and CtrA still displays appropriate cell cycle oscillation (3). Thus, pleD mutants closely resemble swarmer cells that are able to replicate and divide. As such, PleD does not appear to be involved in G1 → S but instead governs morphological differentiation. Despite the fact that both progeny cells resulting from cell division morphologically resemble swarmer cells, one cell is able to immediately reinitiate DNA replication while the other is replication incompetent. Therefore, one of the progeny cells is analogous to a stalked cell regardless of morphology.
PleD is an unusual response regulator protein, composed of an N-terminal receiver domain (D1), a central pseudoreceiver domain (the conserved phosphoryl-accepting aspartate is missing) (D2), and a C-terminal GGDEF domain (84). GGDEF domains, also known as DUF1, are widespread among bacteria and are often associated with sensing and/or receiving domains (65, 66), yet for many years the function of this particular domain was unknown. Eventually it was found that this domain catalyzes the production of the second messenger cyclic diguanylic acid (c-di-GMP) (188). c-di-GMP has been implicated in governing the switch between free-living and surface-associated lifestyles in many bacteria (106, 208, 223, 249). It is not surprising, then, that PleD diguanylate cyclase (DGC) activity is implicated in governing the swarmer (free-living)-to-stalked (surface-associated) cell transition.
PleD has been instrumental in elucidating the catalytic mechanism for DGC activity. The crystal structure of PleD shows that phosphorylated PleD forms a dimer that brings the GGDEF domains into the appropriate position to catalyze the condensation of two GTP molecules into c-di-GMP (28, 188, 269). The pseudoreceiver domain (D2) likely aids in phosphorylation-dependent dimerization, but the crystal structure also reveals a site between the D1/D2-GGDEF domain interface that binds two intercalated molecules of c-di-GMP (28, 269). Binding of c-di-GMP to this site (termed the I site) locks the GGDEF domains in place such that dimers cannot reorient and catalysis is inhibited. This provides a potent allosteric feedback inhibition mechanism that may have biological implications (see below).
The enzymatic activity of PleD is necessary for its role in governing morphological differentiation. It has been shown that PleD activity is needed to degrade the flagellar anchor protein FliF, which coincides with flagellum ejection (3). A surprising discovery was that pleD is carried in the same operon with divK (84). A pleD mutation suppresses all pleC mutations for motility, suggesting that pleD is a bypass suppressor and that PleC acts as a negative regulator of PleD (232). These observations suggest that PleD is part of the DivJ-DivK-PleC signaling system. In fact, the phosphotransfer profiling that demonstrated that DivJ and PleC have kinetic preferences for DivK over most response regulators also showed that they have the same kinetic preference for PleD (225). A divJ strain shows decreased PleD∼P, levels while a pleC strain shows increased PleD∼P, indicating that the established antagonistic kinase/phosphatase activities for DivK also act on PleD (4). Similar to DivK localization, PleD is diffuse in swarmer cells and only phosphorylated PleD localizes to the stalked pole during swarmer cell differentiation (186, 188). Based on epistasis experiments with pleC, divJ, and pleD mutants, it was found that conditions that favored decreased PleD∼P levels also favored motility, while conditions that favored increased PleD∼P also favored stalk formation (2). This is consistent with the compartmentalization of activities seen for DivK. The swarmer cell compartment formed after cytokinesis would have only PleC phosphatase activity, leading to decreased PleD∼P levels, conditions that favor motility. Conversely the stalked cell compartment would only have DivJ, favoring increased PleD∼P levels and holdfast and stalk formation. In this way, the DivJ-PleC system controls both G1/S phase balance (by altering phosphorylation of DivK) and swarmer/stalked phase balance (by altering PleD phosphorylation). Yet, recent in vitro evidence suggests that nonphosphorylated DivK acts as an allosteric regulator of PleC, causing it to reverse activities and principally phosphorylate PleD instead of dephosphorylate it (187). This contradicts the proposed isolated phosphatase activity of PleC in the swarmer cell in that compartmentalized PleC activity would rapidly lead to total DivK dephosphorylation (which prevents the G1 → S transition as per usual), but the nonphosphorylated DivK would lead to increased PleD phosphorylation, favoring the swarmer → stalked cell transition. These two activities therefore appear to be contradictory. The time spent as a swarmer cell would likely be dictated by the relative levels of DivK∼P/PleC, essentially placing a timer for swarmer cell differentiation in the cell. However, it seems likely the total length of such a timer based on enzymatic rates would be much shorter than the amount of time that swarmer cells can persist in culture under laboratory conditions and would not account for the various lengths of time that cells can persist in the swarmer stage, depending on nutrient availability. The in vivo analysis of this allosteric regulation effect looked only at phenotypic changes in cell surface attachment, which itself has multiple factors such as flagella, pili, and holdfast production (16, 57). Clearly, the observed in vitro effect of DivK on PleC activity needs to be modulated in vivo in order to account for the length and the variation of the swarmer phase.
Though there is much discussion of the relative coordination of PleD phosphorylation states, what must not be lost is the output of the signal, that of the second messenger c-di-GMP. Given how fond C. crescentus is of using two-component systems to coordinate development, why use a small-molecule messaging system? Is it simply a holdover from the evolutionary basis for the change in lifestyle? The gammaproteobacterium Pseudomonas fluorescens protein WspR can complement a pleD mutant, indicating how conserved some of these systems can be (146). Alternatively, the use of a second messenger may allow for a greater ability to integrate multiple inputs into a signal, as well as affect many more downstream targets than are afforded by a protein-based signal transduction mechanism. Potentially any protein that has a GGDEF domain or an EAL domain (the domain that catalyzes the breakdown of c-di-GMP to pGpG dinucleotide) can modulate the level of c-di-GMP. There are 13 predicted proteins in the C. crescentus genome that are thought to have GGDEF or EAL domains (50). There are also multiple mechanisms by which a c-di-GMP signal may cause an effect. DgrA and DgrB (PilZ-type proteins) have been shown to bind c-di-GMP (35). Binding of c-di-GMP by DgrA leads to destabilization of the flagellar motor protein FliL. Though the mechanism of action is not known, it does not act at the level of fliL transcription, suggesting that DgrA-mediated motility disruption functions on already-formed flagella. The PleD paralog PopA transduces c-di-GMP signaling by a different mechanism. PopA has a degenerate active site (and thus no catalytic activity) and a dispensable phosphorylation site; however, it does have the allosteric regulation (I) site, which is necessary for function. PopA targets RcdA to the stalked cell pole in stalked and predivisional cells (50). RcdA is a protein that mediates degradation of CtrA by ClpXP by an unknown mechanism (see “Modulation of CtrA Activity” above). PopA targets RcdA independently of ClpXP or CpdR. A popA mutant with an altered I site cannot target RcdA, leading to stabilized CtrA levels throughout the cell cycle. Decreasing the cellular level of c-di-GMP by ectopic expression of the P. aeruginosa EAL-containing protein PA5295 resulted in delocalized PopA. These results indicate that PopA binds to c-di-GMP at the I site, causing it to bind at the stalked pole. However, deletion of each of the GGDEF or EAL domain-encoding genes in C. crescentus had no effect on PopA localization, including deletion of PleD, which has been shown to be the major source for c-di-GMP production in C. crescentus (187). Since c-di-GMP levels are severely reduced in a pleD mutant, why is PopA targeting unaffected? The answer may lie in the I site itself.
Allosteric regulation of DGC activity by the I site is critical for function. One purpose for the I site may be to prevent runaway c-di-GMP synthesis, thereby depleting the cell of GTP. In support of this hypothesis, expression of DGCs without I site regulation is toxic to cells (34). The I site may have another function. Purified DgcA from C. crescentus had very little in vitro DGC activity despite having all the appropriate residues, whereas purified PleD had little difficulty displaying in vitro DGC activity (188). DgcA DGC activity could not be detected until an EAL protein was added to the reaction mixture (34). This result indicated that DgcA was particularly susceptible to I-site regulation, such that the nearly undetectable amounts of c-di-GMP produced by DgcA alone was enough to inhibit its further activity. It was not until the EAL protein was added to degrade the c-di-GMP produced that substantial enzymatic turnover of substrate was observed. Therefore, the degree to which a given I-site-containing protein binds to c-di-GMP could influence its activity. In the case of PopA, the I site may bind c-di-GMP with such affinity that the residual c-di-GMP in a pleD mutant is still enough to provide wild-type targeting. This I-site regulation mechanism potentially has larger implications for the cell. Based on activation (by phosphorylation, for example) of a given DGC with a given level of allosteric regulation, the cell may be able to modulate the level of c-di-GMP to various degrees, which may produce different phenotypic results. There is also potential modulation of c-di-GMP levels by expression/activity of EAL proteins. c-di-GMP signaling is a burgeoning field of research and should prove to be exciting, especially in how it affects C. crescentus development.
Removal of Swarmer Cell Polar Appendages
Part of the morphological changes that occur during swarmer cell differentiation is the loss of flagella and pili from the swarmer pole. In the case of the flagellum, the filament, hook, and distal rod portions are detected in the culture supernatant as a single structure, suggesting that flagella are ejected (117, 217, 236). The mechanism of ejection is not known, but it may involve proteolytic degradation of part of the basal body complex. FliF, a protein in the MS ring complex, is degraded at the swarmer → stalked cell transition, which may lead to flagellum release (108). Indeed, treatment of purified hook-basal body complexes with different proteases led to specific degradation of the MS ring and revealed intact rods (117). However, it should be noted that the rods revealed by proteolysis were full rods containing both the proximal and distal rod proteins, while rods attached to ejected flagella contain only the distal rod protein. The full extent of basal body proteolysis during swarmer cell differentiation is unexamined. Though the MS ring shows turnover during the cell cycle, FlgH, which forms the outer membrane pore, is stable (108). It is possible that the pore has a second unknown function; at the least it must be accounted for structurally as a very large outer membrane opening.
In addition to flagellum ejection, the pili are lost from the swarmer cell pole as well. The mechanism of pilus loss is an intriguing mystery. Though it shares similarity with the type IV pilus system, the C. crescentus pilus is an Flp-type pilus (251). This particular pilus system lacks the PilT protein, which is necessary for filament retraction (103). This is unsurprising in that Flp-type pili often form bundles, which would prevent retraction. However, there is anecdotal evidence that suggests that the C. crescentus pilus is retracted. First, unlike flagella, pili cannot be detected in culture supernatant, suggesting that they are not ejected (128). Second, electron microscopy of C. crescentus treated with pilus-tropic phage shows that at 15 s after infection the phage particles are bound along the length of the pilus at various distances from the cell body, whereas the same sample 15 min later shows nearly all the phage particles in direct contact with the cell surface, suggesting that the pilus has retracted (226). This observation does not preclude phage particles traveling along the length of the filament; however, an extended pilus filament is observed in the 15-s sample but not in the 15-min sample. If the C. crescentus pilus does indeed retract into the cell body, it does so by a novel mechanism and therefore is particularly intriguing for study.
Holdfast Biosynthesis
Less is known about the regulation of holdfast biosynthesis than about other polar morphogenetic events because most studies of development used the domesticated CB15 derivative strain NA1000 as the typical lab strain until recently. While this strain was, and continues to be, particularly useful because it can be synchronized using density gradients (58), it lacks a holdfast. Therefore, years of developmental research missed the holdfast simply because it was not there to observe in the various mutants. It is only in recent years that some laboratories have returned to the parental CB15 strain and included holdfast production in developmental analyses.
The holdfast is an adhesive organelle found at the tip of the stalk and has garnered interest in recent years due to the fact that it is an extremely strong biological adhesive able to withstand forces in the μN range (253). Though the holdfast is needed for strong permanent attachment of cells to surfaces, it has been found that flagella and pili are also needed for efficient initial attachment (16, 57, 137). Three lines of evidence indicate that the holdfast is composed principally of polysaccharide. First, the holdfast can be stained using fluorescein-conjugated wheat germ agglutinin, which specifically binds N-acetylglucosamine (162). Treatment with lysozyme, which is known to degrade N-acetylglucosamine polymers, increases the elasticity of the holdfast by 90% but it does not destroy the holdfast, suggesting that there are other components of the holdfast or that some of the glucosidic linkages are resistant to lysozyme (138). Second, many mutations that abolish holdfast production are found in genes that are predicted to encode polysaccharide biosynthesis machinery, including oligosaccharide synthesis (163, 250) and export (229). Third, the holdfast was observed to have physical properties of a polysaccharide gel by atomic force microscopy (138). It is unclear if protein components also function within the holdfast, though proteins that anchor the holdfast to the cell have been identified. Mutations in the holdfast attachment genes cause shedding of the holdfast from cells to various degrees (37, 127, 182, 229).
Surprisingly, evidence suggests that holdfast synthesis begins principally during the swarmer cell phase (137, 162, 182, 190, 191). This means that for a portion of the cell cycle the bulky polysaccharide gel, the whirling flagellum, and the extruded pili all inhabit the same pole. The confluence of organelles could prove problematic for function. The holdfast is ultimately placed at the tip of the extended stalk, but the vast majority of cytoplasmic proteins are excluded from the stalk interior (see “Stalk Biogenesis” below). Holdfast polysaccharide synthesis requires some cytoplasmic biosynthetic proteins (250). It is currently unknown if the machinery that synthesizes the oligosaccharide components can be separated from the export and attachment machinery. If it cannot, this would suggest that the bulk of holdfast synthesis must occur prior to stalk elongation. There is also the question of how the timing of holdfast production is regulated. It was found that de novo protein synthesis is not required for holdfast production in swarmer cells (137). This result suggests that the holdfast synthesis machinery is produced prior to swarmer cell differentiation and held in check. In congruence with this hypothesis, it was found that the holdfast attachment protein HfaA is produced maximally at the swarmer pole of predivisional cells (104). The signals that lead to holdfast production are not known.
Another intriguing aspect of the holdfast is its binding promiscuity. C. crescentus cells have been observed to attach via the holdfast to such diverse surfaces as plastic, Teflon, other bacterial cells, and even gold particles (182, 190; E. Quardokus, unpublished data). It seems that there is very little that the holdfast cannot attach to, except C. crescentus cells. In monocultures of C. crescentus, stalked cells are observed to gather into aggregates wherein the holdfasts of each cell bind to each other, forming the characteristic “rosette” (5, 182), but the holdfast never binds to the cell body of an adjacent cell (182). It would be tempting to speculate that the paracrystalline surface layer (S-layer) prevents holdfast attachment, but holdfast-body attachment is not observed in an S-layer-deficient strain. The basis for this discrimination in surface attachment is unknown.
Stalk Biogenesis
The stalk is a thin extension of the cell envelope, with inner and outer membranes and a peptidoglycan sacculus continuous with the cell wall of the main body. The stalk is extended from the flagellar pole during swarmer cell differentiation. Proteomic analysis of purified C. crescentus stalks showed remarkable similarity to the outer membrane proteome (98). The stalk is enriched in outer membrane transport proteins but lacks most cytoplasmic and inner membrane proteins (98, 262). This observation led to the hypothesis that the stalk aids in nutrient scavenging. While it is generally thought that the nutrient-scavenging ability of the stalk is the result of an increase in the surface area-to-volume ratio of the cell, this is not strictly true. In diffusion-limited environments, such as the boundary waters next to a surface as would be colonized by C. crescentus, doubling the number of receptors for a given molecule will increase the uptake rate by only ∼5% (260, 262). A much more effective strategy under these conditions is to space receptors out, particularly in a single linear dimension. Therefore, it is the lengthening of the cell by synthesizing a stalk that increases nutrient scavenging, not an increase in surface area. In fact, mathematical modeling predicts that a cell of normal shape with the same surface area of a stalked C. crescentus cell would still be 1.8 times less effective than the stalked cell in a diffusion-limited environment due to the difference in cell length. Accordingly, it is not surprising that C. crescentus lengthens its stalk in response to starvation of certain nutrients (75, 211). In environments that are not diffusion limited, the stalk still provides an advantage, but this time it is related to the increase in the surface area-to-volume ratio and not length per se. Therefore, the stalk provides nutrient uptake advantages in different ways depending on whether the environment is diffusion limited or not.
The regulation of stalk formation continues to confound researchers. There appear to be at least two pathways that regulate stalk biogenesis. It has been known for more than 40 years that stalks increase their length in response to phosphate starvation (211, 212). Phosphate starvation in E. coli causes the PhoR histidine kinase to autophosphorylate and then pass the phosphoryl group to the response regulator PhoB, increasing its affinity for the cis element pho box (143, 144, 266). PhoB∼P binding increases transcription of the Pho regulon, including the high-affinity phosphate transport system pstSCAB. In addition to phosphate transport, PstSCAB proteins are thought to form a complex with PhoR in the presence of excess phosphate and to repress the Pho regulon (268); mutations in pst genes cause constitutive activation of the Pho regulon (267). In C. crescentus, mutation of phoB or pst genes prevents the cell from modulating stalk length in response to phosphate starvation (75). phoB mutants have constitutively short stalks, while pst mutants have constitutively long stalks. Yet, neither set of mutants are stalkless, due to an alternative stalk biogenesis pathway.
In addition to phosphate starvation, stalk biogenesis occurs as part of the natural developmental cycle (81, 180, 246). Developmental regulation of stalk biogenesis requires sigma 54; sigma 54 mutants do not produce stalks in high-phosphate media (24). As mentioned above (see “Polar Localization of Histidine Kinases”), stalk biogenesis has been implicated in the function of the sigma 54-activating protein TacA (157). Phosphotransfer profiling experiments identified a phosphorelay that leads to the activation of TacA (15). A soluble cytoplasmic histidine kinase (ShkA) autophosphorylates and then passes the phosphate to an Hpt protein (ShpA), which then phosphorylates TacA (15, 287). Like tacA mutants, shkA and shpA mutants are stalkless under high-phosphate conditions. Microarray analysis identified potential downstream targets of this pathway, including a regulator of stalk length, StaR (15). Deletion of staR causes a decrease in stalk length, while overexpression causes an increase. Therefore, the ShkA-ShpA-TacA pathway would seem to form a single phosphorelay controlling cell cycle-dependent stalk formation. Yet the situation is not that simple.
As discussed before, one purpose for having a phosphorelay is to either integrate or branch a given signal, yet only a linear signaling pathway has been described in this situation. Reverse phosphotransfer profiling using ShpA demonstrated that it can pass a phosphoryl group to not only ShkA but to another hybrid histidine kinase encoded by CC0921 nearly as efficiently (15), suggesting that there may be another input into the system. There is also a matter of timing. tacA is under positive regulation by CtrA and is expressed in the predivisional cell (157). This in itself is not a problem in that TacA may not be activated by the ShkA-ShpA system until later, but expression analysis indicates that StaR, a target of this phosphorelay, is expressed in late predivisional cells (15), indicating that the phosphorelay is active long before stalk biogenesis begins. In addition to these observations is the fact that all the mutants that produce stalkless cells in high-phosphate media make stalks in low-phosphate media. Therefore, the integration point between phosphate level stalk regulation and cell cycle level stalk regulation is still unknown. It would be intriguing to think that the ShpA-TacA portion of the phosphorelay serves as the integration point, but the fact that mutants with mutations in the shared response regulator TacA still produce stalks under low-phosphate conditions argues against this. The integration may be at the point of controlling the stalk biogenesis machinery.
Unfortunately, because no completely stalkless mutants have been isolated, little is known about how the stalk is synthesized. Evidence indicates that it may be a special form of peptidoglycan synthesis and tied to cell division machinery. Treatment of C. crescentus cells with the β-lactam antibiotic amdinocillin prevents stalk elongation, though the stalk pole does seem to undergo a morphogenic event that resembles the start of stalk formation (215). This result brings up a question of how much of stalk biogenesis is the production of new peptidoglycan material and how much is remodeling of existing material. Depletion of MreB prevents stalk formation (261), once again providing a connection between MreB and peptidoglycan synthesis. Mutants that spontaneously shed stalks were found (192), indicating mislocalized cell division machinery to the base of the stalk. The peptidoglycan of isolated stalks shows an altered composition compared to the bulk cellular peptidoglycan, containing an enrichment of glucosamine (193). These results indicate that one of every five pairs of amino sugars in the stalk peptidoglycan is two glucosamine units, which cannot link to a peptide chain and thus reduce overall cross-linking. There is some evidence that glucosamine-enriched peptidoglycan is specific to the poles of the cell (210); perhaps the reduction of cross-linking is necessary to change the curvature of the sacculus. Not only would this result indicate that the stalk is an extension of the polar peptidoglycan, but it could provide a basis for chemical distinction of polar peptidoglycan for SpmX and/or PodJ. Alternatively, it has been shown that a fraction of C. crescentus peptidoglycan contains pentapeptide chains terminating in glycine residues; this also could be a mechanism of chemical discrimination (155). There is some evidence indicating that terminal glycine residues reduce cross-linking in C. crescentus peptidoglycan as well (156).
The interior of the stalk is also something of a mystery. The proteomics results suggest a lack of cytoplasmic proteins (98, 262), and it is known that ribosomes are excluded from the stalk interior (190). The stalk is known to contain plugs of peptidoglycan called “crossbands” that are not continuous with the sacculus and require FtsZ for formation (47, 113). These crossbands may provide a diffusion barrier. Yet, recently it was found that some proteins are targeted to the stalk interior (271). It appears that the cell has a mechanism of distinguishing and sorting proteins to the stalk.
METABOLIC INPUT INTO DEVELOPMENT
The metabolism of C. crescentus is an often overlooked but potentially interesting area of research outside of development. For example, genome analysis indicates that C. crescentus eschews the traditional OmpF-type outer membrane porin system used to take up nutrients in organisms such as E. coli and Vibrio cholerae and instead uses TonB-dependent outer membrane channels, with 65 members predicted in the genome (174). Microarray analysis demonstrates that growth on xylose, a component of the plant cell wall polymer xylan, also induces transcription of genes that are predicted to encode proteins used to metabolize other xylan components, indicating a robust regulon (88). Additionally, xylose metabolism is used as a genetic tool with ectopic expression of proteins controlled by the xylX promoter, but this tool is feasible only because C. crescentus does not display catabolite repression (161, 238, 247). C. crescentus also utilizes a PAS-kinase molecular oxygen sensing-signaling system (42). In Rhizobium species this system is used to modulate the activity of nitrogen fixation (62, 68, 69), but in C. crescentus it is used to modulate expression of terminal oxidase and other low-oxygen genes. C. crescentus is not a photosynthetic organism, but it has a light-sensing system which increases surface attachment in response to light (194). Yet, outside of these intriguing facets, there is also the question of how or even if metabolism affects developmental programs.
In laboratory culture, even though the growth rate of C. crescentus is dependent on the growth medium, the time spent in individual phases of development is approximately the same proportion of the cell cycle regardless of the total length of the cell cycle. This clockwork progression of the cell cycle has prompted researchers to speculate that metabolism has little input into modulating developmental progression in this organism; however, there is evidence to the contrary. The synthesis of membrane lipids may be used as a sensor of metabolic capability. The C. crescentus membrane is composed principally of phosphatidylglycerol (which is worthy of note in that it gives the membrane surface a net negative charge) (149), and phospholipid synthesis is confined to two specific periods of the cell cycle: (i) before initiation of stalk growth and DNA replication and (ii) between initiation of division site constriction and completion of DNA replication in predivisional cells (181). The timing of phospholipid synthesis itself is not surprising given that the developmental programs involve morphological changes that necessitate the addition of cell membrane; therefore, phospholipid synthesis would be required. Yet, phospholipid synthesis also uses precursors derived from the tricarboxylic acid (TCA) cycle, providing a potent link to central metabolism. Using fatty acid auxotrophs starved of exogenous fatty acids to inhibit phospholipid synthesis at different points in the cell cycle, it was found that inhibited cells stopped their developmental program closest to the phospholipid synthesis point (18, 86). Swarmer cells did not differentiate, and predivisional cells arrested after DNA replication and flagellum biosynthesis, not completing cell division. These results indicate that altering membrane synthesis was able to halt both swarmer cell differentiation and cytokinesis. Is membrane synthesis itself directly coupled to development? This seems unlikely. Late-stationary-phase C. crescentus cells grown in rich media become extremely elongated and helical (278), demonstrating that cells are able to synthesize phospholipids without an accompanying developmental program. It appears more likely that inhibiting membrane synthesis alters central metabolism and thus has a more indirect impact on development. These results strongly suggest that C. crescentus can alter progression through a developmental program based upon metabolic cues.
In addition to inhibition of phospholipid synthesis, swarmer cells can be prevented from differentiating by starvation of carbon or nitrogen sources (32, 77, 136). Part of this response is mediated by SpoT. The secondary metabolite (p)ppGpp accumulates in response to nitrogen starvation, though starvation of amino acid auxotrophs for their necessary amino acid does not elicit (p)ppGpp accumulation, indicating that C. crescentus has a noncanonical stringent response (32). In carbon-starved cells, inhibition of DNA replication is mediated by SpoT, as spoT mutants begin DNA replication even under carbon starvation conditions (136). Additionally, carbon or nitrogen starvation causes increased proteolytic turnover of DnaA and stabilization of CtrA, thus inhibiting DNA replication initiation (77). While SpoT mediates the G1 → S phase arrest for starved swarmer cells, it does not mediate morphological arrest. Starved spoT mutants initiate DNA replication but do not undergo morphogenesis to stalked cells (136). A potential target for morphogenesis regulation is c-di-GMP. Nitrogen starvation was found to reduce GTP levels (32). Since GTP is used to create c-di-GMP, reduced GTP levels may prevent c-di-GMP synthesis, thereby preventing the swarmer → stalked cell transition. Other regulatory mechanisms are also possible. For example, the protein PdeA has both GGDEF and EAL domains; however, the GGDEF domain is degenerate such that it can bind GTP but does not catalyze the formation of c-di-GMP (34, 36). Instead, binding of GTP modulates EAL c-di-GMP-degradation activity by reducing the Km to the physiological range. Thus, GTP serves to positively allosterically regulate c-di-GMP degradation in this enzyme. However, reduced GTP levels during nitrogen starvation would lead to reduced PdeA activity and increased c-di-GMP levels, and therefore it is unlikely that PdeA functions in this particular response. Yet, there are many predicted proteins in C. crescentus that are thought to contain GGDEF and EAL domains, and most have not been characterized. The principle of metabolic impact on c-di-GMP levels through GTP may still be sound.
Starvation leading to swarmer cell arrest may explain an unusual behavior observed in batch-grown C. crescentus cultures. It has been anecdotally observed that C. crescentus cultures in late exponential phase can contain an unusually large proportion of swarmer cells. The proportion of swarmer cells has been seen to rise from 35 to 60% between optical densities of 0.9 and 0.97 (94, 105). This phenomenon, affectionately called “swarmer burst,” may be the result of nutrient depletion. As the culture density increases and nutrients become depleted, the length of time that newly released swarmer cells stay in that phase may be prolonged, such that at the very end of exponential phase the majority of cells have accumulated in the swarmer phase. However, this burst does not seem to last. Cells entering stationary phase arrest at the predivisional stage (105, 278), suggesting that at some point of nutrient depletion the swarmer cells differentiate. This differentiation may be necessary for the cells to later enter the filamentous cell state with an accompanying increase in stress resistance (278). In this case, it seems that swarmer cell arrest can be attained metabolically only by an extreme nutrient downshift such that the cells become immediately depleted of enough nutrients to prevent morphological changes.
EVOLUTIONARY ROLE OF DEVELOPMENTAL PROCESSES
C. crescentus, like all organisms, must be viewed through the scope of evolution. Clearly this organism has evolved intricate and complex developmental systems, but what exactly is the fitness benefit resulting from them? The stalk provides an obvious advantage for scavenging nutrients, but what of the swarmer cell? One of the major purposes of the cyclic developmental program is to produce a swarmer cell offspring, and the noncyclic developmental program would not be necessary if there was no swarmer cell. So what is the fitness advantage provided by the swarmer cell phase?
From one perspective, the swarmer cell can be viewed as a prepubescent life stage. The phase is specific to the newly born, and the organism in question cannot reproduce. As such, it is very much like a child. Clearly childhood exists in other organisms, but what is the evolutionary cause? One of the prevailing theories among evolutionary biologists for the necessity of the prepubescent life stage is that it is the result of an energetic cost-benefit analysis (237). For organisms that take some time to reach reproductive maturity, it would simply cost the progenitor too much energy to carry the progeny to full maturity. For example, in humans a mother would have to carry a child for over a decade before it is ready to reproduce on its own. The cost to the mother would be too great. Instead, a child is born not fully mature and allowed to develop on its own. Does this theory carry over to C. crescentus swarmer cells? Given that the swarmer cell is not incapable of DNA replication but in fact has brakes specifically applied to prevent such an occurrence, the analogy to adolescence falls apart.
From a microbiological perspective, the swarmer cell phase is a conundrum. When it comes to bacteria, the one who divides fastest is supposed to win, particularly in the cutthroat world of multispecies environments. Therefore, it seems counterintuitive that this organism would have a stage of its life cycle that essentially makes it grow slower. So how does C. crescentus get by? It does not inhabit particularly extreme environments, nor is it overly metabolically robust, so it does not appear to inhabit uncompeted niches. Instead, the obligate swarmer stage may allow it to simply get to niches before the competition. The swarmer cell likely serves as a cell dispersal form, constantly forcing the organism to seek out new environments, which may be particularly useful in severely nutrient-limiting environments when the scant resources available can become depleted very quickly. Not every swarmer progeny will find a suitable reproductive environment (many probably will not), but the obligate dispersal stage increases the reproductive fitness of the species as a whole. Thus, C. crescentus may be more of a frontiersman, living where the going is roughest and constantly on the move from the main population of microbes following behind. Because it cannot compete with those organisms that grow faster on the same nutrients, it must stay ahead of the pack. Given this hypothesis, one might expect that Caulobacter species would be found preferentially in low-microbial-complexity environments. However, one study found Caulobacter species in multiple stages of a wastewater treatment plant, an environment of high microbial density and diversity (142). Yet, it should be noted that while Caulobacter species could be obtained from the system, there is no indication of how prevalent or how metabolically active they were in the community. It is also thought that organisms that store carbon in polyhydroxybutyrate granules and phosphate in polyphosphate have an advantage in the wastewater treatment system (41, 288); both are known characteristics of C. crescentus (190, 191).
Still, if the swarmer cell stage serves in dispersal, it is not clear why swarmer cell differentiation is necessary for reproduction. Genetically the two processes can be uncoupled, but in the wild type they are obligately connected. The swarmer cell state offers one type of advantage (mobility), and the stalked cell state offers other advantages (surface attachment and nutrient scavenging), but why has C. crescentus evolved a mechanism that hardwires reproduction to a certain cell stage instead of, for example, allowing a swarmer cell to produce another swarmer cells as it disperses? Does the replication machinery mechanistically require the stalk? The viability of tacA mutants under high-phosphate conditions suggests otherwise. The answer to the swarmer cell conundrum is not apparent, but one possibility involves metabolic efficiency. Duplication of a cell is clearly a metabolically demanding process; all cellular components have to be doubled. The stalk provides a nutrient-scavenging advantage, and thus the cell may be able to replicate faster as a stalked cell than as a swarmer cell. Even if nutrients are in excess, they may not always be so, in which case the stalk provides a long-term advantage. Therefore, the cost of (i) forcing the cell to remain in one place and (ii) undergoing morphogenesis, which itself is energetically costly, may be outweighed by the nutrient-scavenging benefit of the stalk. As pleD mutants approximate swarmer cell-only replication, their relative fitness compared to the wild type in an in situ environment may address this hypothesis.
The developmental programs of C. crescentus are intricate and complex, but are they specific to only this organism? What is the universal impact of this particular field of study? The principles described here are actually applicable to a wide array of organisms. Asticcacaulis biprosthecum is a closely related bacterium that produces a holdfast and swarmer cell life stage, but instead of a single polar stalk it has two laterally placed stalks, though the holdfast is still produced at the cell pole (185). Prosthecomicrobium and Ancalamicrobium produce many stalks over the surface of the cell body (234). Stalks are found on many budding bacteria, including Rhodomicrobium vannielii, Hyphomicrobium, and Hyphomonas (254, 273). Some bud off the cell body, whereas other bud through the stalk itself. It is very likely that some developmental mechanisms are conserved between C. crescentus and these organisms. Not only can understanding C. crescentus development illuminate development in these organisms, but studying these other prosthecate bacteria can show how developmental programs can be altered to lead to different outputs. Prosthecobacter fusiformis greatly resembles C. crescentus; both organisms produce a single polar stalk with a holdfast at the end, but unlike C. crescentus, P. fusiformis does not have a swarmer cell life stage (235). Instead, P. fusiformis produces two stalked progeny cells after division. Despite the similarities to C. crescentus, P. fusiformis does not belong to the alphaproteobacteria and likely has different mechanisms for polar development. Comparing the developmental programs of these two organisms may provide potent examples of convergent evolution.
Yet, the applicability of C. crescentus development does not stop at closely related bacteria or those having similar polar appendages. There is increasing evidence that many of the mechanisms elucidated for C. crescentus are conserved among the alphaproteobacteria. Many C. crescentus developmental genes are conserved in the Rhizobiales. The CcrM methylase is conserved and essential in Rhizobium meliloti (279). Overexpression of CcrM in this organism leads to cell division and DNA replication regulation defects similar to those in C. crescentus, and the R. meliloti and C. crescentus ccrM alleles are functionally interchangeable. CcrM is also conserved in Agrobacterium tumefaciens, and overexpression leads to cell filamentation and branching (116). Interestingly, flagellar proteins are cell cycle regulated in this organism and are expressed maximally when the cells are motile, indicating that A. tumefaciens may have a swarmer cell phase. Southern blotting techniques and more recent genomic analyses revealed that ctrA is found in many alphaproteobacteria, and it was found to be essential in Sinorhizobium meliloti and to contain promoter architecture similar to that in C. crescentus (9). C. crescentus ctrA could complement a ctrA S. meliloti mutant, but the opposite was not true. A divK homolog has also been found in S. meliloti and localizes to the old pole of the cell during division (129). Daughter cells with this pole are larger than the other daughter cells, similar to the case for C. crescentus. S. meliloti also has two CpdR homologs, one of which forms polar foci (126). Mutants of this homolog have gross morphology defects and reduced ClpXP polar localization and prevent bacteroid differentiation, thereby inhibiting effective symbiosis with alfalfa. These phenotypes could be partially complemented with the C. crescentus CpdR.
Gene conservation extends beyond the Rhizobiales. Not only is CtrA conserved in the obligate intracellular pathogen Rickettsia prowazekii, but the chromosomal origin of replication is similar in structure to the C. crescentus Cori (20). The R. prowazekii CtrA could partially complement a ctrA C. crescentus mutant. Multiple components of C. crescentus development are found in the mammalian pathogen Brucella abortis. DivK, PleC, and DivJ homologs are found in B. abortus, and an additional cytoplasmic histidine kinase, PdsH, may serve the same function as DivL (82). CcrM is conserved and essential in this organism, and like with the other organisms tested, overexpression leads to an increase in genomic copy number and altered cell division (207). Overexpression of CcrM also causes attenuation of cell replication inside murine macrophages, a phenotype that is not dependent on cell morphology defects, suggesting that gene regulation of essential pathogenesis factors may be altered. CtrA is essential in B. abortus, and overexpression leads to cell filamentation and branching (11). Some of the CtrA targets are conserved in B. abortus, such as ctrA and ccrM, but some, like divK, ftsZ, and the origin of replication, are not. However, B. abortus CtrA does regulate pleC, ftsE, and expression of Min system proteins, suggesting that while the exact targets regulated by CtrA are not the same as in C. crescentus, some of the processes are.
Yet, despite the conservation of many C. crescentus developmental genes, there are also indications that some of the functions may not be conserved. In B. abortus DivK and PdsH localize to the old pole, but PleC localizes to the division plane and DivJ is delocalized (82). CtrA is not essential in Rhodobacter capsulatus, and instead of regulating cell cycle functions, it appears to regulate structural genes for the gene transfer agent, a small phage-like particle that can transfer genes between R. capsulatus cells (131). No CtrA binding sites have been found in the S. meliloti or B. abortus oris (11, 82), indicating altered regulation of chromosome replication initiation. Therefore, the task facing researchers of the alphaproteobacteria is to determine how these developmental programs function in their systems, what different processes they regulate, and how the regulatory networks have evolved and have been rewired to produce different outcomes. Fortunately, the work with C. crescentus provides an excellent basis to begin questioning.
CONCLUDING REMARKS
In parsing through over 40 years of C. crescentus research, it is clear that while much has been learned about this fascinating organism, there is still much to discover. Yet, even for the discovered systems, many details have to be analyzed and reanalyzed. The two most common questions that usually pop into mind about any developmental protein are: is the timing of its synthesis developmentally regulated, and is it polarly localized? Certainly these are pertinent questions. The functions of many developmental proteins are absolutely dependent on these facets. CtrA must be cell cycle regulated to exert proper control over DNA replication initiation and polar development. PopZ must be polarly localized in order to properly anchor segregated chromosome origins of replication away from each other. But is this always the case? For many proteins it is unclear that cell cycle regulation and/or polar localization is necessary for their function. For example, different analyses indicate that divK transcription is cell cycle regulated (83, 133), but Western blotting shows that DivK protein levels are relatively unaltered throughout the cell cycle (102). In this case, transcription profiling is misleading, as the cell cycle regulation does not cause dramatic fluctuations in protein level and likely has little impact on DivK function. Instead, cell cycle regulation may simply be a means of increasing protein levels enough to compensate for dilution and loss of protein as the cell grows and divides. It was thought that polar targeting of CtrA by RcdA was necessary for efficient CtrA proteolysis since ClpXP was polarly localized, but recent evidence indicates that RcdA is not necessary for degradation of CtrA (see “Modulation of CtrA Activity” above). It is not clear, then, how much polar localization of these proteins factors into their developmental function. Certainly, if a protein is cell cycle regulated or polarly localized, it is for a reason, but the reason may simply be an increase in efficiency instead of an integral function of the system. These facets bear investigation not only for pathways yet to be discovered but for pathways that are already known as well.
Like a cell under a microscope, the image of C. crescentus is slowly coming into focus. Many of the developmental arcs are being closed into loops. Pathways are beginning to fold back on themselves, and the confusing phenotypes of earlier times are explained by the way that everything comes around again. With each new discovery and its integration into the previous knowledge, the scope of the intricacy of this organism grows larger and larger. It is often mind-boggling how such a small cell could house so much complexity. A major challenge for the future will be to understand how this developmental complexity provides selectable advantages to C. crescentus in its environment(s) and how this complexity can be modulated to adapt to different environmental conditions or modified by evolution to lead to different selectable phenotypes.
Acknowledgments
We thank members of the Brun laboratory for critical reading of the manuscript.
Research in our laboratory is supported by grants GM077648 and GM51986 from the National Institutes of Health and MCB0731950 from the National Science Foundation and by the Indiana METACyt Initiative of Indiana University, funded in part through a major grant from the Lilly Endowment, Inc. P.D.C. is supported by postdoctoral National Institutes of Health National Research Service Award F32GM084618 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Aaron, M., G. Charbon, H. Lam, H. Schwarz, W. Vollmer, and C. Jacobs-Wagner. 2007. The tubulin homologue FtsZ contributes to cell elongation by guiding cell wall precursor synthesis in Caulobacter crescentus. Mol. Microbiol. 64:938-952. [DOI] [PubMed] [Google Scholar]
- 2.Aldridge, P., and K. T. Hughes. 2002. Regulation of flagellar assembly. Curr. Opin. Microbiol. 5:160-165. [DOI] [PubMed] [Google Scholar]
- 3.Aldridge, P., and U. Jenal. 1999. Cell cycle-dependent degradation of a flagellar motor component requires a novel-type response regulator. Mol. Microbiol. 32:379-391. [DOI] [PubMed] [Google Scholar]
- 4.Aldridge, P., R. Paul, P. Goymer, P. Rainey, and U. Jenal. 2003. Role of the GGDEF regulator PleD in polar development of Caulobacter crescentus. Mol. Microbiol. 47:1695-1708. [DOI] [PubMed] [Google Scholar]
- 5.Alipour-Assiabi, E., G. Li, T. R. Powers, and J. X. Tang. 2006. Fluctuation analysis of Caulobacter crescentus adhesion. Biophys. J. 90:2206-2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alley, M. R., J. R. Maddock, and L. Shapiro. 1992. Polar localization of a bacterial chemoreceptor. Genes Dev. 6:825-836. [DOI] [PubMed] [Google Scholar]
- 7.Anderson, P. E., and J. W. Gober. 2000. FlbT, the post-transcriptional regulator of flagellin synthesis in Caulobacter crescentus, interacts with the 5′ untranslated region of flagellin mRNA. Mol. Microbiol. 38:41-52. [DOI] [PubMed] [Google Scholar]
- 8.Angelastro, P. S., O. Sliusarenko, and C. Jacobs-Wagner. 2010. Polar localization of the CckA histidine kinase and cell cycle periodicity of the essential master regulator CtrA in Caulobacter crescentus. J. Bacteriol. 192:539-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barnett, M. J., D. Y. Hung, A. Reisenauer, L. Shapiro, and S. R. Long. 2001. A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J. Bacteriol. 183:3204-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bastedo, D. P., and G. T. Marczynski. 2009. CtrA response regulator binding to the Caulobacter chromosome replication origin is required during nutrient and antibiotic stress as well as during cell cycle progression. Mol. Microbiol. 72:139-154. [DOI] [PubMed] [Google Scholar]
- 11.Bellefontaine, A. F., C. E. Pierreux, P. Mertens, J. Vandenhaute, J. J. Letesson, and X. De Bolle. 2002. Plasticity of a transcriptional regulation network among alpha-proteobacteria is supported by the identification of CtrA targets in Brucella abortus. Mol. Microbiol. 43:945-960. [DOI] [PubMed] [Google Scholar]
- 12.Berdis, A. J., I. Lee, J. K. Coward, C. Stephens, R. Wright, L. Shapiro, and S. J. Benkovic. 1998. A cell cycle-regulated adenine DNA methyltransferase from Caulobacter crescentus processively methylates GANTC sites on hemimethylated DNA. Proc. Natl. Acad. Sci. U. S. A. 95:2874-2879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bernhardt, T. G., and P. A. de Boer. 2005. SlmA, a nucleoid-associated, FtsZ binding protein required for blocking septal ring assembly over chromosomes in E. coli. Mol. Cell 18:555-564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Biondi, E. G., S. J. Reisinger, J. M. Skerker, M. Arif, B. S. Perchuk, K. R. Ryan, and M. T. Laub. 2006. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature 444:899-904. [DOI] [PubMed] [Google Scholar]
- 15.Biondi, E. G., J. M. Skerker, M. Arif, M. S. Prasol, B. S. Perchuk, and M. T. Laub. 2006. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol. Microbiol. 59:386-401. [DOI] [PubMed] [Google Scholar]
- 16.Bodenmiller, D., E. Toh, and Y. V. Brun. 2004. Development of surface adhesion in Caulobacter crescentus. J. Bacteriol. 186:1438-1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman, G. R., L. R. Comolli, J. Zhu, M. Eckart, M. Koenig, K. H. Downing, W. E. Moerner, T. Earnest, and L. Shapiro. 2008. A polymeric protein anchors the chromosomal origin/ParB complex at a bacterial cell pole. Cell 134:945-955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brassinga, A. K., B. Gorbatyuk, M. C. Ouimet, and G. T. Marczynski. 2000. Selective cell cycle transcription requires membrane synthesis in Caulobacter. EMBO J. 19:702-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brassinga, A. K., and G. T. Marczynski. 2001. Replication intermediate analysis confirms that chromosomal replication origin initiates from an unusual intergenic region in Caulobacter crescentus. Nucleic Acids Res. 29:4441-4451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brassinga, A. K., R. Siam, W. McSween, H. Winkler, D. Wood, and G. T. Marczynski. 2002. Conserved response regulator CtrA and IHF binding sites in the alpha-proteobacteria Caulobacter crescentus and Rickettsia prowazekii chromosomal replication origins. J. Bacteriol. 184:5789-5799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brendler, T., A. Abeles, and S. Austin. 1995. A protein that binds to the P1 origin core and the oriC 13mer region in a methylation-specific fashion is the product of the host seqA gene. EMBO J. 14:4083-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Briegel, A., H. J. Ding, Z. Li, J. Werner, Z. Gitai, D. P. Dias, R. B. Jensen, and G. J. Jensen. 2008. Location and architecture of the Caulobacter crescentus chemoreceptor array. Mol. Microbiol. 69:30-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown, J., A. Faulds-Pain, and P. Aldridge. 2009. The coordination of flagellar gene expression and the flagellar assembly pathway, p. 99-120. In K. F. Jarrell (ed.), Pili and flagella: current research and future trends. Caister Academic Press, Norfolk, United Kingdom.
- 24.Brun, Y. V., and L. Shapiro. 1992. A temporally controlled sigma-factor is required for polar morphogenesis and normal cell division in Caulobacter. Genes Dev. 6:2395-2408. [DOI] [PubMed] [Google Scholar]
- 25.Brun, Y. V., and L. J. Shimkets. 2000. Prokaryotic development: strategies to enhance surival, p.1-7. In Y. V. Brun and L. J. Shimkets (ed.), Prokaryotic development. American Society for Microbiology, Washington, DC.
- 26.Cabantous, S., V. Guillet, N. Ohta, A. Newton, and J. P. Samama. 2002. Characterization and crystallization of DivK, an essential response regulator for cell division and differentiation in Caulobacter crescentus. Acta Crystallogr. D Biol. Crystallogr. 58:1249-1251. [DOI] [PubMed] [Google Scholar]
- 27.Campbell, J. L., and N. Kleckner. 1990. E. coli oriC and the dnaA gene promoter are sequestered from dam methyltransferase following the passage of the chromosomal replication fork. Cell 62:967-979. [DOI] [PubMed] [Google Scholar]
- 28.Chan, C., R. Paul, D. Samoray, N. C. Amiot, B. Giese, U. Jenal, and T. Schirmer. 2004. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. U. S. A. 101:17084-17089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen, J. C., A. K. Hottes, H. H. McAdams, P. T. McGrath, P. H. Viollier, and L. Shapiro. 2006. Cytokinesis signals truncation of the PodJ polarity factor by a cell cycle-regulated protease. EMBO J. 25:377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen, J. C., P. H. Viollier, and L. Shapiro. 2005. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol. Microbiol. 55:1085-1103. [DOI] [PubMed] [Google Scholar]
- 31.Chen, Y. E., C. G. Tsokos, E. G. Biondi, B. S. Perchuk, and M. T. Laub. 2009. Dynamics of two phosphorelays controlling cell cycle progression in Caulobacter crescentus. J. Bacteriol. 191:7417-7429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chiaverotti, T. A., G. Parker, J. Gallant, and N. Agabian. 1981. Conditions that trigger guanosine tetraphosphate accumulation in Caulobacter crescentus. J. Bacteriol. 145:1463-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chien, P., B. S. Perchuk, M. T. Laub, R. T. Sauer, and T. A. Baker. 2007. Direct and adaptor-mediated substrate recognition by an essential AAA+ protease. Proc. Natl. Acad. Sci. U. S. A. 104:6590-6595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Christen, B., M. Christen, R. Paul, F. Schmid, M. Folcher, P. Jenoe, M. Meuwly, and U. Jenal. 2006. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 281:32015-32024. [DOI] [PubMed] [Google Scholar]
- 35.Christen, M., B. Christen, M. G. Allan, M. Folcher, P. Jeno, S. Grzesiek, and U. Jenal. 2007. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 104:4112-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Christen, M., B. Christen, M. Folcher, A. Schauerte, and U. Jenal. 2005. Identification and characterization of a cyclic di-GMP-specific phosphodiesterase and its allosteric control by GTP. J. Biol. Chem. 280:30829-30837. [DOI] [PubMed] [Google Scholar]
- 37.Cole, J. L., G. G. Hardy, D. Bodenmiller, E. Toh, A. Hinz, and Y. V. Brun. 2003. The HfaB and HfaD adhesion proteins of Caulobacter crescentus are localized in the stalk. Mol. Microbiol. 49:1671-1683. [DOI] [PubMed] [Google Scholar]
- 38.Collier, J., H. H. McAdams, and L. Shapiro. 2007. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc. Natl. Acad. Sci. U. S. A. 104:17111-17116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Collier, J., S. R. Murray, and L. Shapiro. 2006. DnaA couples DNA replication and the expression of two cell cycle master regulators. EMBO J. 25:346-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Collier, J., and L. Shapiro. 2009. Feedback control of DnaA-mediated replication initiation by replisome-associated HdaA protein in Caulobacter. J. Bacteriol. 191:5706-5716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Comeau, Y., J. Hall, R. E. W. Hancock, and W. K. Oldham. 1986. Biochemical model for enhanced biological phosphorus removal. Water Res. 20:1511-1521. [Google Scholar]
- 42.Crosson, S., P. T. McGrath, C. Stephens, H. H. McAdams, and L. Shapiro. 2005. Conserved modular design of an oxygen sensory/signaling network with species-specific output. Proc. Natl. Acad. Sci. U. S. A. 102:8018-8023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Crymes, W. B., Jr., D. Zhang, and B. Ely. 1999. Regulation of podJ expression during the Caulobacter crescentus cell cycle. J. Bacteriol. 181:3967-3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Degnen, S. T., and A. Newton. 1972. Chromosome replication during development in Caulobacter crescentus. J. Mol. Biol. 64:671-680. [DOI] [PubMed] [Google Scholar]
- 45.Deich, J., E. M. Judd, H. H. McAdams, and W. E. Moerner. 2004. Visualization of the movement of single histidine kinase molecules in live Caulobacter cells. Proc. Natl. Acad. Sci. U. S. A. 101:15921-15926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dingwall, A., W. Y. Zhuang, K. Quon, and L. Shapiro. 1992. Expression of an early gene in the flagellar regulatory hierarchy is sensitive to an interruption in DNA replication. J. Bacteriol. 174:1760-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Divakaruni, A. V., C. Baida, C. L. White, and J. W. Gober. 2007. The cell shape proteins MreB and MreC control cell morphogenesis by positioning cell wall synthetic complexes. Mol. Microbiol. 66:174-188. [DOI] [PubMed] [Google Scholar]
- 48.Domian, I. J., K. C. Quon, and L. Shapiro. 1997. Cell type-specific phosphorylation and proteolysis of a transcriptional regulator controls the G1-to-S transition in a bacterial cell cycle. Cell 90:415-424. [DOI] [PubMed] [Google Scholar]
- 49.Domian, I. J., A. Reisenauer, and L. Shapiro. 1999. Feedback control of a master bacterial cell-cycle regulator. Proc. Natl. Acad. Sci. U. S. A. 96:6648-6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Duerig, A., S. Abel, M. Folcher, M. Nicollier, T. Schwede, N. Amiot, B. Giese, and U. Jenal. 2009. Second messenger-mediated spatiotemporal control of protein degradation regulates bacterial cell cycle progression. Genes Dev. 23:93-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dworkin, M. 1985. Developmental biology of the bacteria. The Benjamin/Cummings Publishing Company, Inc., Menlo Park, CA.
- 52.Easter, J., Jr., and J. W. Gober. 2002. ParB-stimulated nucleotide exchange regulates a switch in functionally distinct ParA activities. Mol. Cell 10:427-434. [DOI] [PubMed] [Google Scholar]
- 53.Ebersbach, G., A. Briegel, G. J. Jensen, and C. Jacobs-Wagner. 2008. A self-associating protein critical for chromosome attachment, division, and polar organization in Caulobacter. Cell 134:956-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ebersbach, G., S. Ringgaard, J. Moller-Jensen, Q. Wang, D. J. Sherratt, and K. Gerdes. 2006. Regular cellular distribution of plasmids by oscillating and filament-forming ParA ATPase of plasmid pB171. Mol. Microbiol. 61:1428-1442. [DOI] [PubMed] [Google Scholar]
- 55.Ely, B., C. J. Gerardot, D. L. Fleming, S. L. Gomes, P. Frederikse, and L. Shapiro. 1986. General nonchemotactic mutants of Caulobacter crescentus. Genetics 114:717-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.England, J. C., and J. W. Gober. 2001. Cell cycle control of cell morphogenesis in Caulobacter. Curr. Opin. Microbiol. 4:674-680. [DOI] [PubMed] [Google Scholar]
- 57.Entcheva-Dimitrov, P., and A. M. Spormann. 2004. Dynamics and control of biofilms of the oligotrophic bacterium Caulobacter crescentus. J. Bacteriol. 186:8254-8266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Evinger, M., and N. Agabian. 1977. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J. Bacteriol. 132:294-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Figge, R. M., A. V. Divakaruni, and J. W. Gober. 2004. MreB, the cell shape-determining bacterial actin homologue, co-ordinates cell wall morphogenesis in Caulobacter crescentus. Mol. Microbiol. 51:1321-1332. [DOI] [PubMed] [Google Scholar]
- 60.Figge, R. M., J. Easter, and J. W. Gober. 2003. Productive interaction between the chromosome partitioning proteins, ParA and ParB, is required for the progression of the cell cycle in Caulobacter crescentus. Mol. Microbiol. 47:1225-1237. [DOI] [PubMed] [Google Scholar]
- 61.Fischer, B., G. Rummel, P. Aldridge, and U. Jenal. 2002. The FtsH protease is involved in development, stress response and heat shock control in Caulobacter crescentus. Mol. Microbiol. 44:461-478. [DOI] [PubMed] [Google Scholar]
- 62.Fischer, H. M. 1994. Genetic regulation of nitrogen fixation in rhizobia. Microbiol. Rev. 58:352-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fukuda, A., M. Asada, S. Koyasu, H. Yoshida, K. Yaginuma, and Y. Okada. 1981. Regulation of polar morphogenesis in Caulobacter crescentus. J. Bacteriol. 145:559-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fukuda, A., H. Iba, and Y. Okada. 1977. Stalkless mutants of Caulobacter crescentus. J. Bacteriol. 131:280-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Galperin, M. Y. 2004. Bacterial signal transduction network in a genomic perspective. Environ. Microbiol. 6:552-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Galperin, M. Y., A. N. Nikolskaya, and E. V. Koonin. 2001. Novel domains of the prokaryotic two-component signal transduction systems. FEMS Microbiol. Lett. 203:11-21. [DOI] [PubMed] [Google Scholar]
- 67.Gerdes, K., J. Moller-Jensen, and R. Bugge Jensen. 2000. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 37:455-466. [DOI] [PubMed] [Google Scholar]
- 68.Gilles-Gonzalez, M. A., G. S. Ditta, and D. R. Helinski. 1991. A haemoprotein with kinase activity encoded by the oxygen sensor of Rhizobium meliloti. Nature 350:170-172. [DOI] [PubMed] [Google Scholar]
- 69.Gilles-Gonzalez, M. A., G. Gonzalez, M. F. Perutz, L. Kiger, M. C. Marden, and C. Poyart. 1994. Heme-based sensors, exemplified by the kinase FixL, are a new class of heme protein with distinctive ligand binding and autoxidation. Biochemistry 33:8067-8073. [DOI] [PubMed] [Google Scholar]
- 70.Gitai, Z., N. Dye, and L. Shapiro. 2004. An actin-like gene can determine cell polarity in bacteria. Proc. Natl. Acad. Sci. U. S. A. 101:8643-8648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gitai, Z., N. A. Dye, A. Reisenauer, M. Wachi, and L. Shapiro. 2005. MreB actin-mediated segregation of a specific region of a bacterial chromosome. Cell 120:329-341. [DOI] [PubMed] [Google Scholar]
- 72.Glaser, P., M. E. Sharpe, B. Raether, M. Perego, K. Ohlsen, and J. Errington. 1997. Dynamic, mitotic-like behavior of a bacterial protein required for accurate chromosome partitioning. Genes Dev. 11:1160-1168. [DOI] [PubMed] [Google Scholar]
- 73.Gober, J. W., C. H. Boyd, M. Jarvis, E. K. Mangan, M. F. Rizzo, and J. A. Wingrove. 1995. Temporal and spatial regulation of fliP, an early flagellar gene of Caulobacter crescentus that is required for motility and normal cell division. J. Bacteriol. 177:3656-3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Gomes, S. L., J. W. Gober, and L. Shapiro. 1990. Expression of the Caulobacter heat shock gene dnaK is developmentally controlled during growth at normal temperatures. J. Bacteriol. 172:3051-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gonin, M., E. M. Quardokus, D. O'Donnol, J. Maddock, and Y. V. Brun. 2000. Regulation of stalk elongation by phosphate in Caulobacter crescentus. J. Bacteriol. 182:337-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Gorbatyuk, B., and G. T. Marczynski. 2001. Physiological consequences of blocked Caulobacter crescentus dnaA expression, an essential DNA replication gene. Mol. Microbiol. 40:485-497. [DOI] [PubMed] [Google Scholar]
- 77.Gorbatyuk, B., and G. T. Marczynski. 2005. Regulated degradation of chromosome replication proteins DnaA and CtrA in Caulobacter crescentus. Mol. Microbiol. 55:1233-1245. [DOI] [PubMed] [Google Scholar]
- 78.Gordon, S., J. Rech, D. Lane, and A. Wright. 2004. Kinetics of plasmid segregation in Escherichia coli. Mol. Microbiol. 51:461-469. [DOI] [PubMed] [Google Scholar]
- 79.Grigorian, A. V., R. B. Lustig, E. C. Guzman, J. M. Mahaffy, and J. W. Zyskind. 2003. Escherichia coli cells with increased levels of DnaA and deficient in recombinational repair have decreased viability. J. Bacteriol. 185:630-644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guillet, V., N. Ohta, S. Cabantous, A. Newton, and J. P. Samama. 2002. Crystallographic and biochemical studies of DivK reveal novel features of an essential response regulator in Caulobacter crescentus. J. Biol. Chem. 277:42003-42010. [DOI] [PubMed] [Google Scholar]
- 81.Haars, E. G., and J. M. Schmidt. 1974. Stalk formation and its inhibition in Caulobacter crescentus. J. Bacteriol. 120:1409-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hallez, R., J. Mignolet, V. Van Mullem, M. Wery, J. Vandenhaute, J. J. Letesson, C. Jacobs-Wagner, and X. De Bolle. 2007. The asymmetric distribution of the essential histidine kinase PdhS indicates a differentiation event in Brucella abortus. EMBO J. 26:1444-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hecht, G. B., T. Lane, N. Ohta, J. M. Sommer, and A. Newton. 1995. An essential single domain response regulator required for normal cell division and differentiation in Caulobacter crescentus. EMBO J. 14:3915-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hecht, G. B., and A. Newton. 1995. Identification of a novel response regulator required for the swarmer-to-stalked-cell transition in Caulobacter crescentus. J. Bacteriol. 177:6223-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Hinz, A. J., D. E. Larson, C. S. Smith, and Y. V. Brun. 2003. The Caulobacter crescentus polar organelle development protein PodJ is differentially localized and is required for polar targeting of the PleC development regulator. Mol. Microbiol. 47:929-941. [DOI] [PubMed] [Google Scholar]
- 86.Hodgson, D., P. Shaw, M. O'Connell, S. Henry, and L. Shapiro. 1984. Caulobacter crescentus fatty acid-dependent cell cycle mutant. J. Bacteriol. 158:156-162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Holtzendorff, J., D. Hung, P. Brende, A. Reisenauer, P. H. Viollier, H. H. McAdams, and L. Shapiro. 2004. Oscillating global regulators control the genetic circuit driving a bacterial cell cycle. Science 304:983-987. [DOI] [PubMed] [Google Scholar]
- 88.Hottes, A. K., M. Meewan, D. Yang, N. Arana, P. Romero, H. H. McAdams, and C. Stephens. 2004. Transcriptional profiling of Caulobacter crescentus during growth on complex and minimal media. J. Bacteriol. 186:1448-1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hottes, A. K., L. Shapiro, and H. H. McAdams. 2005. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol. Microbiol. 58:1340-1353. [DOI] [PubMed] [Google Scholar]
- 90.Hu, B., G. Yang, W. Zhao, Y. Zhang, and J. Zhao. 2007. MreB is important for cell shape but not for chromosome segregation of the filamentous cyanobacterium Anabaena sp. PCC 7120. Mol. Microbiol. 63:1640-1652. [DOI] [PubMed] [Google Scholar]
- 91.Huguenel, E. D., and A. Newton. 1982. Localization of surface structures during procaryotic differentiation: role of cell division in Caulobacter crescentus. Differentiation 21:71-78. [DOI] [PubMed] [Google Scholar]
- 92.Huitema, E., S. Pritchard, D. Matteson, S. K. Radhakrishnan, and P. H. Viollier. 2006. Bacterial birth scar proteins mark future flagellum assembly site. Cell 124:1025-1037. [DOI] [PubMed] [Google Scholar]
- 93.Hung, D. Y., and L. Shapiro. 2002. A signal transduction protein cues proteolytic events critical to Caulobacter cell cycle progression. Proc. Natl. Acad. Sci. U. S. A. 99:13160-13165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Iba, H., A. Fukuda, and Y. Okada. 1975. Synchronous cell differentiation in Caulobacter crescentus. Jpn. J. Microbiol. 19:441-446. [DOI] [PubMed] [Google Scholar]
- 95.Inagaki, F., K. Takai, H. Hirayama, Y. Yamato, K. H. Nealson, and K. Horikoshi. 2003. Distribution and phylogenetic diversity of the subsurface microbial community in a Japanese epithermal gold mine. Extremophiles 7:307-317. [DOI] [PubMed] [Google Scholar]
- 96.Iniesta, A. A., P. T. McGrath, A. Reisenauer, H. H. McAdams, and L. Shapiro. 2006. A phospho-signaling pathway controls the localization and activity of a protease complex critical for bacterial cell cycle progression. Proc. Natl. Acad. Sci. U. S. A. 103:10935-10940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Iniesta, A. A., and L. Shapiro. 2008. A bacterial control circuit integrates polar localization and proteolysis of key regulatory proteins with a phospho-signaling cascade. Proc. Natl. Acad. Sci. U. S. A. 105:16602-16607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Ireland, M. M., J. A. Karty, E. M. Quardokus, J. P. Reilly, and Y. V. Brun. 2002. Proteomic analysis of the Caulobacter crescentus stalk indicates competence for nutrient uptake. Mol. Microbiol. 45:1029-1041. [DOI] [PubMed] [Google Scholar]
- 99.Ireton, K., N. W. Gunther IV, and A. D. Grossman. 1994. spo0J is required for normal chromosome segregation as well as the initiation of sporulation in Bacillus subtilis. J. Bacteriol. 176:5320-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Jacobs, C., N. Ausmees, S. J. Cordwell, L. Shapiro, and M. T. Laub. 2003. Functions of the CckA histidine kinase in Caulobacter cell cycle control. Mol. Microbiol. 47:1279-1290. [DOI] [PubMed] [Google Scholar]
- 101.Jacobs, C., I. J. Domian, J. R. Maddock, and L. Shapiro. 1999. Cell cycle-dependent polar localization of an essential bacterial histidine kinase that controls DNA replication and cell division. Cell 97:111-120. [DOI] [PubMed] [Google Scholar]
- 102.Jacobs, C., D. Hung, and L. Shapiro. 2001. Dynamic localization of a cytoplasmic signal transduction response regulator controls morphogenesis during the Caulobacter cell cycle. Proc. Natl. Acad. Sci. U. S. A. 98:4095-4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jakovljevic, V., S. Leonardy, M. Hoppert, and L. Sogaard-Andersen. 2008. PilB and PilT are ATPases acting antagonistically in type IV pilus function in Myxococcus xanthus. J. Bacteriol. 190:2411-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Janakiraman, R. S., and Y. V. Brun. 1999. Cell cycle control of a holdfast attachment gene in Caulobacter crescentus. J. Bacteriol. 181:1118-1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Janakiraman, R. S., and Y. V. Brun. 1997. Transcriptional and mutational analyses of the rpoN operon in Caulobacter crescentus. J. Bacteriol. 179:5138-5147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Jenal, U. 2004. Cyclic di-guanosine-monophosphate comes of age: a novel secondary messenger involved in modulating cell surface structures in bacteria? Curr. Opin. Microbiol. 7:185-191. [DOI] [PubMed] [Google Scholar]
- 107.Jenal, U., and T. Fuchs. 1998. An essential protease involved in bacterial cell-cycle control. EMBO J. 17:5658-5669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Jenal, U., and L. Shapiro. 1996. Cell cycle-controlled proteolysis of a flagellar motor protein that is asymmetrically distributed in the Caulobacter predivisional cell. EMBO J. 15:2393-2406. [PMC free article] [PubMed] [Google Scholar]
- 109.Jensen, R. B. 2006. Analysis of the terminus region of the Caulobacter crescentus chromosome and identification of the dif site. J. Bacteriol. 188:6016-6019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Jensen, R. B. 2006. Coordination between chromosome replication, segregation, and cell division in Caulobacter crescentus. J. Bacteriol. 188:2244-2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Jensen, R. B., and L. Shapiro. 1999. The Caulobacter crescentus smc gene is required for cell cycle progression and chromosome segregation. Proc. Natl. Acad. Sci. U. S. A. 96:10661-10666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jensen, R. B., and L. Shapiro. 2003. Cell-cycle-regulated expression and subcellular localization of the Caulobacter crescentus SMC chromosome structural protein. J. Bacteriol. 185:3068-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Jones, H. C., and J. M. Schmidt. 1973. Ultrastructural study of crossbands occurring in the stalks of Caulobacter crescentus. J. Bacteriol. 116:466-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jones, L. J., R. Carballido-Lopez, and J. Errington. 2001. Control of cell shape in bacteria: helical, actin-like filaments in Bacillus subtilis. Cell 104:913-922. [DOI] [PubMed] [Google Scholar]
- 115.Judd, E. M., K. R. Ryan, W. E. Moerner, L. Shapiro, and H. H. McAdams. 2003. Fluorescence bleaching reveals asymmetric compartment formation prior to cell division in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 100:8235-8240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kahng, L. S., and L. Shapiro. 2001. The CcrM DNA methyltransferase of Agrobacterium tumefaciens is essential, and its activity is cell cycle regulated. J. Bacteriol. 183:3065-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kanbe, M., S. Shibata, Y. Umino, U. Jenal, and S. I. Aizawa. 2005. Protease susceptibility of the Caulobacter crescentus flagellar hook-basal body: a possible mechanism of flagellar ejection during cell differentiation. Microbiology 151:433-438. [DOI] [PubMed] [Google Scholar]
- 118.Kato, J., and T. Katayama. 2001. Hda, a novel DnaA-related protein, regulates the replication cycle in Escherichia coli. EMBO J. 20:4253-4262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Keiler, K. C., and L. Shapiro. 2001. Conserved promoter motif is required for cell cycle timing of dnaX transcription in Caulobacter. J. Bacteriol. 183:4860-4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Keiler, K. C., and L. Shapiro. 2003. tmRNA in Caulobacter crescentus is cell cycle regulated by temporally controlled transcription and RNA degradation. J. Bacteriol. 185:1825-1830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Keiler, K. C., and L. Shapiro. 2003. tmRNA is required for correct timing of DNA replication in Caulobacter crescentus. J. Bacteriol. 185:573-580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kelly, A. J., M. J. Sackett, N. Din, E. Quardokus, and Y. V. Brun. 1998. Cell cycle-dependent transcriptional and proteolytic regulation of FtsZ in Caulobacter. Genes Dev. 12:880-893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Khursigara, C. M., X. Wu, and S. Subramaniam. 2008. Chemoreceptors in Caulobacter crescentus: trimers of receptor dimers in a partially ordered hexagonally packed array. J. Bacteriol. 190:6805-6810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Khvorova, A., V. K. Chary, D. W. Hilbert, and P. J. Piggot. 2000. The chromosomal location of the Bacillus subtilis sporulation gene spoIIR is important for its function. J. Bacteriol. 182:4425-4429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kim, S. Y., Z. Gitai, A. Kinkhabwala, L. Shapiro, and W. E. Moerner. 2006. Single molecules of the bacterial actin MreB undergo directed treadmilling motion in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 103:10929-10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kobayashi, H., N. J. De Nisco, P. Chien, L. A. Simmons, and G. C. Walker. 2009. Sinorhizobium meliloti CpdR1 is critical for co-ordinating cell cycle progression and the symbiotic chronic infection. Mol. Microbiol. 73:586-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kurtz, H. D., Jr., and J. Smith. 1992. Analysis of a Caulobacter crescentus gene cluster involved in attachment of the holdfast to the cell. J. Bacteriol. 174:687-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Lagenaur, C., and N. Agabian. 1977. Caulobacter crescentus pili: structure and stage-specific expression. J. Bacteriol. 131:340-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Lam, H., J. Y. Matroule, and C. Jacobs-Wagner. 2003. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev. Cell 5:149-159. [DOI] [PubMed] [Google Scholar]
- 130.Lam, H., W. B. Schofield, and C. Jacobs-Wagner. 2006. A landmark protein essential for establishing and perpetuating the polarity of a bacterial cell. Cell 124:1011-1023. [DOI] [PubMed] [Google Scholar]
- 131.Lang, A. S., and J. T. Beatty. 2000. Genetic analysis of a bacterial genetic exchange element: the gene transfer agent of Rhodobacter capsulatus. Proc. Natl. Acad. Sci. U. S. A. 97:859-864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Laub, M. T., S. L. Chen, L. Shapiro, and H. H. McAdams. 2002. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc. Natl. Acad. Sci. U. S. A. 99:4632-4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Laub, M. T., H. H. McAdams, T. Feldblyum, C. M. Fraser, and L. Shapiro. 2000. Global analysis of the genetic network controlling a bacterial cell cycle. Science 290:2144-2148. [DOI] [PubMed] [Google Scholar]
- 134.Lawler, M. L., and Y. V. Brun. 2006. A molecular beacon defines bacterial cell asymmetry. Cell 124:891-893. [DOI] [PubMed] [Google Scholar]
- 135.Lawler, M. L., D. E. Larson, A. J. Hinz, D. Klein, and Y. V. Brun. 2006. Dissection of functional domains of the polar localization factor PodJ in Caulobacter crescentus. Mol. Microbiol. 59:301-316. [DOI] [PubMed] [Google Scholar]
- 136.Lesley, J. A., and L. Shapiro. 2008. SpoT regulates DnaA stability and initiation of DNA replication in carbon-starved Caulobacter crescentus. J. Bacteriol. 190:6867-6880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Levi, A., and U. Jenal. 2006. Holdfast formation in motile swarmer cells optimizes surface attachment during Caulobacter crescentus development. J. Bacteriol. 188:5315-5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Li, G., C. S. Smith, Y. V. Brun, and J. X. Tang. 2005. The elastic properties of the Caulobacter crescentus adhesive holdfast are dependent on oligomers of N-acetylglucosamine. J. Bacteriol. 187:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Li, G., and J. X. Tang. 2006. Low flagellar motor torque and high swimming efficiency of Caulobacter crescentus swarmer cells. Biophys. J. 91:2726-2734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lu, M., J. L. Campbell, E. Boye, and N. Kleckner. 1994. SeqA: a negative modulator of replication initiation in E. coli. Cell 77:413-426. [DOI] [PubMed] [Google Scholar]
- 141.Lutkenhaus, J. 2007. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76:539-562. [DOI] [PubMed] [Google Scholar]
- 142.MacRae, J. D., and J. Smit. 1991. Characterization of caulobacters isolated from wastewater treatment systems. Appl. Environ. Microbiol. 57:751-758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Makino, K., H. Shinagawa, M. Amemura, T. Kawamoto, M. Yamada, and A. Nakata. 1989. Signal transduction in the phosphate regulon of Escherichia coli involves phosphotransfer between PhoR and PhoB proteins. J. Mol. Biol. 210:551-559. [DOI] [PubMed] [Google Scholar]
- 144.Makino, K., H. Shinagawa, M. Amemura, S. Kimura, A. Nakata, and A. Ishihama. 1988. Regulation of the phosphate regulon of Escherichia coli. Activation of pstS transcription by PhoB protein in vitro. J. Mol. Biol. 203:85-95. [DOI] [PubMed] [Google Scholar]
- 145.Malakooti, J., S. P. Wang, and B. Ely. 1995. A consensus promoter sequence for Caulobacter crescentus genes involved in biosynthetic and housekeeping functions. J. Bacteriol. 177:4372-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Malone, J. G., R. Williams, M. Christen, U. Jenal, A. J. Spiers, and P. B. Rainey. 2007. The structure-function relationship of WspR, a Pseudomonas fluorescens response regulator with a GGDEF output domain. Microbiology 153:980-994. [DOI] [PubMed] [Google Scholar]
- 147.Mangan, E. K., J. Malakooti, A. Caballero, P. Anderson, B. Ely, and J. W. Gober. 1999. FlbT couples flagellum assembly to gene expression in Caulobacter crescentus. J. Bacteriol. 181:6160-6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mannisto, M. K., M. A. Tiirola, M. S. Salkinoja-Salonen, M. S. Kulomaa, and J. A. Puhakka. 1999. Diversity of chlorophenol-degrading bacteria isolated from contaminated boreal groundwater. Arch. Microbiol. 171:189-197. [DOI] [PubMed] [Google Scholar]
- 149.Mansour, J. D., S. Henry, and L. Shapiro. 1980. Differential membrane phospholipid synthesis during the cell cycle of Caulobacter crescentus. J. Bacteriol. 141:262-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Marczynski, G. T. 1999. Chromosome methylation and measurement of faithful, once and only once per cell cycle chromosome replication in Caulobacter crescentus. J. Bacteriol. 181:1984-1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Marczynski, G. T., A. Dingwall, and L. Shapiro. 1990. Plasmid and chromosomal DNA replication and partitioning during the Caulobacter crescentus cell cycle. J. Mol. Biol. 212:709-722. [DOI] [PubMed] [Google Scholar]
- 152.Marczynski, G. T., K. Lentine, and L. Shapiro. 1995. A developmentally regulated chromosomal origin of replication uses essential transcription elements. Genes Dev. 9:1543-1557. [DOI] [PubMed] [Google Scholar]
- 153.Marczynski, G. T., and L. Shapiro. 1992. Cell-cycle control of a cloned chromosomal origin of replication from Caulobacter crescentus. J. Mol. Biol. 226:959-977. [DOI] [PubMed] [Google Scholar]
- 154.Marczynski, G. T., and L. Shapiro. 2002. Control of chromosome replication in Caulobacter crescentus. Annu. Rev. Microbiol. 56:625-656. [DOI] [PubMed] [Google Scholar]
- 155.Markiewicz, Z., B. Glauner, and U. Schwarz. 1983. Murein structure and lack of dd- and ld-carboxypeptidase activities in Caulobacter crescentus. J. Bacteriol. 156:649-655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Markiewicz, Z., and Z. Kwiatkowski. 1985. Autolysis of Caulobacter crescentus grown in the presence of glycine. Acta Microbiol. Pol. 34:5-14. [PubMed] [Google Scholar]
- 157.Marques, M. V., S. L. Gomes, and J. W. Gober. 1997. A gene coding for a putative sigma 54 activator is developmentally regulated in Caulobacter crescentus. J. Bacteriol. 179:5502-5510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Martin, M. E., M. J. Trimble, and Y. V. Brun. 2004. Cell cycle-dependent abundance, stability and localization of FtsA and FtsQ in Caulobacter crescentus. Mol. Microbiol. 54:60-74. [DOI] [PubMed] [Google Scholar]
- 159.Matroule, J. Y., H. Lam, D. T. Burnette, and C. Jacobs-Wagner. 2004. Cytokinesis monitoring during development; rapid pole-to-pole shuttling of a signaling protein by localized kinase and phosphatase in Caulobacter. Cell 118:579-590. [DOI] [PubMed] [Google Scholar]
- 160.McGrath, P. T., A. A. Iniesta, K. R. Ryan, L. Shapiro, and H. H. McAdams. 2006. A dynamically localized protease complex and a polar specificity factor control a cell cycle master regulator. Cell 124:535-547. [DOI] [PubMed] [Google Scholar]
- 161.Meisenzahl, A. C., L. Shapiro, and U. Jenal. 1997. Isolation and characterization of a xylose-dependent promoter from Caulobacter crescentus. J. Bacteriol. 179:592-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Merker, R. I., and J. Smit. 1988. Characterization of the adhesive holdfast of marine and freshwater caulobacters. Appl. Environ. Microbiol. 54:2078-2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Mitchell, D., and J. Smit. 1990. Identification of genes affecting production of the adhesion organelle of Caulobacter crescentus CB2. J. Bacteriol. 172:5425-5431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Mohl, D. A., J. Easter, Jr., and J. W. Gober. 2001. The chromosome partitioning protein, ParB, is required for cytokinesis in Caulobacter crescentus. Mol. Microbiol. 42:741-755. [DOI] [PubMed] [Google Scholar]
- 165.Mohl, D. A., and J. W. Gober. 1997. Cell cycle-dependent polar localization of chromosome partitioning proteins in Caulobacter crescentus. Cell 88:675-684. [DOI] [PubMed] [Google Scholar]
- 166.Mohr, C. D., J. K. MacKichan, and L. Shapiro. 1998. A membrane-associated protein, FliX, is required for an early step in Caulobacter flagellar assembly. J. Bacteriol. 180:2175-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Muir, R. E., J. Easter, and J. W. Gober. 2005. The trans-acting flagellar regulatory proteins, FliX and FlbD, play a central role in linking flagellar biogenesis and cytokinesis in Caulobacter crescentus. Microbiology 151:3699-3711. [DOI] [PubMed] [Google Scholar]
- 168.Muir, R. E., and J. W. Gober. 2002. Mutations in FlbD that relieve the dependency on flagellum assembly alter the temporal and spatial pattern of developmental transcription in Caulobacter crescentus. Mol. Microbiol. 43:597-615. [DOI] [PubMed] [Google Scholar]
- 169.Muir, R. E., and J. W. Gober. 2004. Regulation of FlbD activity by flagellum assembly is accomplished through direct interaction with the trans-acting factor, FliX. Mol. Microbiol. 54:715-730. [DOI] [PubMed] [Google Scholar]
- 170.Muir, R. E., and J. W. Gober. 2001. Regulation of late flagellar gene transcription and cell division by flagellum assembly in Caulobacter crescentus. Mol. Microbiol. 41:117-130. [DOI] [PubMed] [Google Scholar]
- 171.Muir, R. E., T. M. O'Brien, and J. W. Gober. 2001. The Caulobacter crescentus flagellar gene, fliX, encodes a novel trans-acting factor that couples flagellar assembly to transcription. Mol. Microbiol. 39:1623-1637. [DOI] [PubMed] [Google Scholar]
- 172.Mullin, D., S. Minnich, L. S. Chen, and A. Newton. 1987. A set of positively regulated flagellar gene promoters in Caulobacter crescentus with sequence homology to the nif gene promoters of Klebsiella pneumoniae. J. Mol. Biol. 195:939-943. [DOI] [PubMed] [Google Scholar]
- 173.Mullin, D. A., and A. Newton. 1989. Ntr-like promoters and upstream regulatory sequence ftr are required for transcription of a developmentally regulated Caulobacter crescentus flagellar gene. J. Bacteriol. 171:3218-3227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Nierman, W. C., T. V. Feldblyum, M. T. Laub, I. T. Paulsen, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, M. R. Alley, N. Ohta, J. R. Maddock, I. Potocka, W. C. Nelson, A. Newton, C. Stephens, N. D. Phadke, B. Ely, R. T. DeBoy, R. J. Dodson, A. S. Durkin, M. L. Gwinn, D. H. Haft, J. F. Kolonay, J. Smit, M. B. Craven, H. Khouri, J. Shetty, K. Berry, T. Utterback, K. Tran, A. Wolf, J. Vamathevan, M. Ermolaeva, O. White, S. L. Salzberg, J. C. Venter, L. Shapiro, and C. M. Fraser. 2001. Complete genome sequence of Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 98:4136-4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.North, N. N., S. L. Dollhopf, L. Petrie, J. D. Istok, D. L. Balkwill, and J. E. Kostka. 2004. Change in bacterial community structure during in situ biostimulation of subsurface sediment cocontaminated with uranium and nitrate. Appl. Environ. Microbiol. 70:4911-4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Obuchowski, P. L., and C. Jacobs-Wagner. 2008. PflI, a protein involved in flagellar positioning in Caulobacter crescentus. J. Bacteriol. 190:1718-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Ohta, N., T. Lane, E. G. Ninfa, J. M. Sommer, and A. Newton. 1992. A histidine protein kinase homologue required for regulation of bacterial cell division and differentiation. Proc. Natl. Acad. Sci. U. S. A. 89:10297-10301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Ohta, N., and A. Newton. 2003. The core dimerization domains of histidine kinases contain recognition specificity for the cognate response regulator. J. Bacteriol. 185:4424-4431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Ohta, N., A. J. Ninfa, A. Allaire, L. Kulick, and A. Newton. 1997. Identification, characterization, and chromosomal organization of cell division cycle genes in Caulobacter crescentus. J. Bacteriol. 179:2169-2180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.O'Neill, E. A., and R. A. Bender. 1989. Cell-cycle-dependent polar morphogenesis in Caulobacter crescentus: roles of phospholipid, DNA, and protein syntheses. J. Bacteriol. 171:4814-4820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.O'Neill, E. A., and R. A. Bender. 1987. Periodic synthesis of phospholipids during the Caulobacter crescentus cell cycle. J. Bacteriol. 169:2618-2623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Ong, C. J., M. L. Wong, and J. Smit. 1990. Attachment of the adhesive holdfast organelle to the cellular stalk of Caulobacter crescentus. J. Bacteriol. 172:1448-1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Osley, M. A., and A. Newton. 1974. Chromosome segregration and development in Caulobacter crescentus. J. Mol. Biol. 90:359-370. [DOI] [PubMed] [Google Scholar]
- 184.Osley, M. A., M. Sheffery, and A. Newton. 1977. Regulation of flagellin synthesis in the cell cycle of Caulobacter: dependence on DNA replication. Cell 12:393-400. [DOI] [PubMed] [Google Scholar]
- 185.Pate, J. L., J. S. Porter, and T. L. Jordan. 1973. Asticcacaulis biprosthecum sp. nov. Life cycle, morphology and cultural characteristics. Antonie Van Leeuwenhoek 39:569-583. [DOI] [PubMed] [Google Scholar]
- 186.Paul, R., S. Abel, P. Wassmann, A. Beck, H. Heerklotz, and U. Jenal. 2007. Activation of the diguanylate cyclase PleD by phosphorylation-mediated dimerization. J. Biol. Chem. 282:29170-29177. [DOI] [PubMed] [Google Scholar]
- 187.Paul, R., T. Jaeger, S. Abel, I. Wiederkehr, M. Folcher, E. G. Biondi, M. T. Laub, and U. Jenal. 2008. Allosteric regulation of histidine kinases by their cognate response regulator determines cell fate. Cell 133:452-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Paul, R., S. Weiser, N. C. Amiot, C. Chan, T. Schirmer, B. Giese, and U. Jenal. 2004. Cell cycle-dependent dynamic localization of a bacterial response regulator with a novel di-guanylate cyclase output domain. Genes Dev. 18:715-727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Pierce, D. L., D. S. O'Donnol, R. C. Allen, J. W. Javens, E. M. Quardokus, and Y. V. Brun. 2006. Mutations in DivL and CckA rescue a divJ null mutant of Caulobacter crescentus by reducing the activity of CtrA. J. Bacteriol. 188:2473-2482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Poindexter, J. S. 1964. Biological properties and classification of the Caulobacter group. Bacteriol. Rev. 28:231-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Poindexter, J. S. 1981. The caulobacters: ubiquitous unusual bacteria. Microbiol. Rev. 45:123-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Poindexter, J. S. 1978. Selection for nonbuoyant morphological mutants of Caulobacter crescentus. J. Bacteriol. 135:1141-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Poindexter, J. S., and J. G. Hagenzieker. 1982. Novel peptidoglycans in Caulobacter and Asticcacaulis spp. J. Bacteriol. 150:332-347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Purcell, E. B., D. Siegal-Gaskins, D. C. Rawling, A. Fiebig, and S. Crosson. 2007. A photosensory two-component system regulates bacterial cell attachment. Proc. Natl. Acad. Sci. U. S. A. 104:18241-18246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Quardokus, E., N. Din, and Y. V. Brun. 1996. Cell cycle regulation and cell type-specific localization of the FtsZ division initiation protein in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 93:6314-6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Quardokus, E. M., and Y. V. Brun. 2003. Cell cycle timing and developmental checkpoints in Caulobacter crescentus. Curr. Opin. Microbiol. 6:541-549. [DOI] [PubMed] [Google Scholar]
- 197.Quardokus, E. M., and Y. V. Brun. 2002. DNA replication initiation is required for mid-cell positioning of FtsZ rings in Caulobacter crescentus. Mol. Microbiol. 45:605-616. [DOI] [PubMed] [Google Scholar]
- 198.Quardokus, E. M., N. Din, and Y. V. Brun. 2001. Cell cycle and positional constraints on FtsZ localization and the initiation of cell division in Caulobacter crescentus. Mol. Microbiol. 39:949-959. [DOI] [PubMed] [Google Scholar]
- 199.Quon, K. C., B. Yang, I. J. Domian, L. Shapiro, and G. T. Marczynski. 1998. Negative control of bacterial DNA replication by a cell cycle regulatory protein that binds at the chromosome origin. Proc. Natl. Acad. Sci. U. S. A. 95:120-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Radhakrishnan, S. K., M. Thanbichler, and P. H. Viollier. 2008. The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus. Genes Dev. 22:212-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Ramakrishnan, G., and A. Newton. 1990. FlbD of Caulobacter crescentus is a homologue of the NtrC (NRI) protein and activates sigma 54-dependent flagellar gene promoters. Proc. Natl. Acad. Sci. U. S. A. 87:2369-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Reisenauer, A., K. Quon, and L. Shapiro. 1999. The CtrA response regulator mediates temporal control of gene expression during the Caulobacter cell cycle. J. Bacteriol. 181:2430-2439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Reisenauer, A., and L. Shapiro. 2002. DNA methylation affects the cell cycle transcription of the CtrA global regulator in Caulobacter. EMBO J. 21:4969-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Reisinger, S. J., S. Huntwork, P. H. Viollier, and K. R. Ryan. 2007. DivL performs critical cell cycle functions in Caulobacter crescentus independent of kinase activity. J. Bacteriol. 189:8308-8320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Reuter, S. H., and L. Shapiro. 1987. Asymmetric segregation of heat-shock proteins upon cell division in Caulobacter crescentus. J. Mol. Biol. 194:653-662. [DOI] [PubMed] [Google Scholar]
- 206.Roberts, R. C., and L. Shapiro. 1997. Transcription of genes encoding DNA replication proteins is coincident with cell cycle control of DNA replication in Caulobacter crescentus. J. Bacteriol. 179:2319-2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Robertson, G. T., A. Reisenauer, R. Wright, R. B. Jensen, A. Jensen, L. Shapiro, and R. M. Roop II. 2000. The Brucella abortus CcrM DNA methyltransferase is essential for viability, and its overexpression attenuates intracellular replication in murine macrophages. J. Bacteriol. 182:3482-3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Ross, P., H. Weinhouse, Y. Aloni, D. Michaeli, P. Weinberger-Ohana, R. Mayer, S. Braun, E. de Vroom, G. A. van der Marel, J. H. van Boom, and M. Benziman. 1987. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 325:279-281. [DOI] [PubMed] [Google Scholar]
- 209.Ryan, K. R., E. M. Judd, and L. Shapiro. 2002. The CtrA response regulator essential for Caulobacter crescentus cell-cycle progression requires a bipartite degradation signal for temporally controlled proteolysis. J. Mol. Biol. 324:443-455. [DOI] [PubMed] [Google Scholar]
- 210.Satta, G., R. Fontana, P. Canepari, and G. Botta. 1979. Peptidoglycan synthesis in cocci and rods of a pH-dependent, morphologically conditional mutant of Klebsiella pneumoniae. J. Bacteriol. 137:727-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Schmidt, J. M. 1969. Caulobacter crescentus mutants with short stalks. J. Bacteriol. 98:816-817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Schmidt, J. M., and R. Y. Stanier. 1966. The development of cellular stalks in bacteria. J. Cell Biol. 28:423-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Sciochetti, S. A., T. Lane, N. Ohta, and A. Newton. 2002. Protein sequences and cellular factors required for polar localization of a histidine kinase in Caulobacter crescentus. J. Bacteriol. 184:6037-6049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Sciochetti, S. A., N. Ohta, and A. Newton. 2005. The role of polar localization in the function of an essential Caulobacter crescentus tyrosine kinase. Mol. Microbiol. 56:1467-1480. [DOI] [PubMed] [Google Scholar]
- 215.Seitz, L. C., and Y. V. Brun. 1998. Genetic analysis of mecillinam-resistant mutants of Caulobacter crescentus deficient in stalk biosynthesis. J. Bacteriol. 180:5235-5239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Shaheen, S. M., M. C. Ouimet, and G. marczynski. 2009. Comparative analysis of Caulobacter chromosome replication origins. Microbiology 155:1215-1225. [DOI] [PubMed] [Google Scholar]
- 217.Shapiro, L., and J. V. Maizel, Jr. 1973. Synthesis and structure of Caulobacter crescentus flagella. J. Bacteriol. 113:478-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Sheffery, M., and A. Newton. 1977. Reconstitution and purification of flagellar filaments from Caulobacter crescentus. J. Bacteriol. 132:1027-1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Shier, V. K., C. J. Hancey, and S. J. Benkovic. 2001. Identification of the active oligomeric state of an essential adenine DNA methyltransferase from Caulobacter crescentus. J. Biol. Chem. 276:14744-14751. [DOI] [PubMed] [Google Scholar]
- 220.Siam, R., A. K. Brassinga, and G. T. Marczynski. 2003. A dual binding site for integration host factor and the response regulator CtrA inside the Caulobacter crescentus replication origin. J. Bacteriol. 185:5563-5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Siam, R., and G. T. Marczynski. 2000. Cell cycle regulator phosphorylation stimulates two distinct modes of binding at a chromosome replication origin. EMBO J. 19:1138-1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Siam, R., and G. T. Marczynski. 2003. Glutamate at the phosphorylation site of response regulator CtrA provides essential activities without increasing DNA binding. Nucleic Acids Res. 31:1775-1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Simm, R., M. Morr, A. Kader, M. Nimtz, and U. Romling. 2004. GGDEF and EAL domains inversely regulate cyclic di-GMP levels and transition from sessility to motility. Mol. Microbiol. 53:1123-1134. [DOI] [PubMed] [Google Scholar]
- 224.Simmons, L. A., A. M. Breier, N. R. Cozzarelli, and J. M. Kaguni. 2004. Hyperinitiation of DNA replication in Escherichia coli leads to replication fork collapse and inviability. Mol. Microbiol. 51:349-358. [DOI] [PubMed] [Google Scholar]
- 225.Skerker, J. M., M. S. Prasol, B. S. Perchuk, E. G. Biondi, and M. T. Laub. 2005. Two-component signal transduction pathways regulating growth and cell cycle progression in a bacterium: a system-level analysis. PLoS Biol. 3:e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Skerker, J. M., and L. Shapiro. 2000. Identification and cell cycle control of a novel pilus system in Caulobacter crescentus. EMBO J. 19:3223-3234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Slater, S., S. Wold, M. Lu, E. Boye, K. Skarstad, and N. Kleckner. 1995. E. coli SeqA protein binds oriC in two different methyl-modulated reactions appropriate to its roles in DNA replication initiation and origin sequestration. Cell 82:927-936. [DOI] [PubMed] [Google Scholar]
- 228.Smit, J., and N. Agabian. 1982. Cell surface patterning and morphogenesis: biogenesis of a periodic surface array during Caulobacter development. J. Cell Biol. 95:41-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Smith, C. S., A. Hinz, D. Bodenmiller, D. E. Larson, and Y. V. Brun. 2003. Identification of genes required for synthesis of the adhesive holdfast in Caulobacter crescentus. J. Bacteriol. 185:1432-1442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Sommer, J. M., and A. Newton. 1991. Pseudoreversion analysis indicates a direct role of cell division genes in polar morphogenesis and differentiation in Caulobacter crescentus. Genetics 129:623-630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Sommer, J. M., and A. Newton. 1988. Sequential regulation of developmental events during polar morphogenesis in Caulobacter crescentus: assembly of pili on swarmer cells requires cell separation. J. Bacteriol. 170:409-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Sommer, J. M., and A. Newton. 1989. Turning off flagellum rotation requires the pleiotropic gene pleD: pleA, pleC, and pleD define two morphogenic pathways in Caulobacter crescentus. J. Bacteriol. 171:392-401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Spencer, W., R. Siam, M. C. Ouimet, D. P. Bastedo, and G. T. Marczynski. 2009. CtrA, a global response regulator, uses a distinct second category of weak DNA binding sites for cell cycle transcription control in Caulobacter crescentus. J. Bacteriol. 191:5458-5470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Staley, J. T. 1968. Prosthecomicrobium and Ancalomicrobium: new prosthecate freshwater bacteria. J. Bacteriol. 95:1921-1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Staley, J. T., J. A. Bont, and K. Jonge. 1976. Prosthecobacter fusiformis nov. gen. et sp., the fusiform caulobacter. Antonie Van Leeuwenhoek 42:333-342. [DOI] [PubMed] [Google Scholar]
- 236.Stallmeyer, M. J., K. M. Hahnenberger, G. E. Sosinsky, L. Shapiro, and D. J. DeRosier. 1989. Image reconstruction of the flagellar basal body of Caulobacter crescentus. J. Mol. Biol. 205:511-518. [DOI] [PubMed] [Google Scholar]
- 237.Stearns, S. C. 1992. The evolution of life histories. Oxford University Press, New York, NY.
- 238.Stephens, C., B. Christen, T. Fuchs, V. Sundaram, K. Watanabe, and U. Jenal. 2007. Genetic analysis of a novel pathway for d-xylose metabolism in Caulobacter crescentus. J. Bacteriol. 189:2181-2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Stephens, C., A. Reisenauer, R. Wright, and L. Shapiro. 1996. A cell cycle-regulated bacterial DNA methyltransferase is essential for viability. Proc. Natl. Acad. Sci. U. S. A. 93:1210-1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Stephens, C. M., and L. Shapiro. 1993. An unusual promoter controls cell-cycle regulation and dependence on DNA replication of the Caulobacter fliLM early flagellar operon. Mol. Microbiol. 9:1169-1179. [DOI] [PubMed] [Google Scholar]
- 241.Stephens, C. M., G. Zweiger, and L. Shapiro. 1995. Coordinate cell cycle control of a Caulobacter DNA methyltransferase and the flagellar genetic hierarchy. J. Bacteriol. 177:1662-1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 243.Su'etsugu, M., M. Takata, T. Kubota, Y. Matsuda, and T. Katayama. 2004. Molecular mechanism of DNA replication-coupled inactivation of the initiator protein in Escherichia coli: interaction of DnaA with the sliding clamp-loaded DNA and the sliding clamp-Hda complex. Genes Cells 9:509-522. [DOI] [PubMed] [Google Scholar]
- 244.Szyf, M., K. Avraham-Haetzni, A. Reifman, J. Shlomai, F. Kaplan, A. Oppenheim, and A. Razin. 1984. DNA methylation pattern is determined by the intracellular level of the methylase. Proc. Natl. Acad. Sci. U. S. A. 81:3278-3282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Terrana, B., and A. Newton. 1975. Pattern of unequal cell division and development in Caulobacter crescentus. Dev. Biol. 44:380-385. [DOI] [PubMed] [Google Scholar]
- 246.Terrana, B., and A. Newton. 1976. Requirement of a cell division step for stalk formation in Caulobacter crescentus. J. Bacteriol. 128:456-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Thanbichler, M., A. A. Iniesta, and L. Shapiro. 2007. A comprehensive set of plasmids for vanillate- and xylose-inducible gene expression in Caulobacter crescentus. Nucleic Acids Res. 35:e137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Thanbichler, M., and L. Shapiro. 2006. MipZ, a spatial regulator coordinating chromosome segregation with cell division in Caulobacter. Cell 126:147-162. [DOI] [PubMed] [Google Scholar]
- 249.Tischler, A. D., and A. Camilli. 2004. Cyclic diguanylate (c-di-GMP) regulates Vibrio cholerae biofilm formation. Mol. Microbiol. 53:857-869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Toh, E., H. D. Kurtz, Jr., and Y. V. Brun. 2008. Characterization of the Caulobacter crescentus holdfast polysaccharide biosynthesis pathway reveals significant redundancy in the initiating glycosyltransferase and polymerase steps. J. Bacteriol. 190:7219-7231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Tomich, M., P. J. Planet, and D. H. Figurski. 2007. The tad locus: postcards from the widespread colonization island. Nat. Rev. Microbiol. 5:363-375. [DOI] [PubMed] [Google Scholar]
- 252.Toro, E., S. H. Hong, H. H. McAdams, and L. Shapiro. 2008. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc. Natl. Acad. Sci. U. S. A. 105:15435-15440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Tsang, P. H., G. Li, Y. V. Brun, L. B. Freund, and J. X. Tang. 2006. Adhesion of single bacterial cells in the micronewton range. Proc. Natl. Acad. Sci. U. S. A. 103:5764-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Tyler, P. A., and K. C. Marshall. 1967. Pleomorphy in stalked, budding bacteria. J. Bacteriol. 93:1132-1136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Viollier, P. H., and L. Shapiro. 2003. A lytic transglycosylase homologue, PleA, is required for the assembly of pili and the flagellum at the Caulobacter crescentus cell pole. Mol. Microbiol. 49:331-345. [DOI] [PubMed] [Google Scholar]
- 256.Viollier, P. H., N. Sternheim, and L. Shapiro. 2002. Identification of a localization factor for the polar positioning of bacterial structural and regulatory proteins. Proc. Natl. Acad. Sci. U. S. A. 99:13831-13836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Viollier, P. H., M. Thanbichler, P. T. McGrath, L. West, M. Meewan, H. H. McAdams, and L. Shapiro. 2004. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. U. S. A. 101:9257-9262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.von Freiesleben, U., K. V. Rasmussen, and M. Schaechter. 1994. SeqA limits DnaA activity in replication from oriC in Escherichia coli. Mol. Microbiol. 14:763-772. [DOI] [PubMed] [Google Scholar]
- 259.Wachi, M., M. Doi, Y. Okada, and M. Matsuhashi. 1989. New mre genes mreC and mreD, responsible for formation of the rod shape of Escherichia coli cells. J. Bacteriol. 171:6511-6516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Wagner, J. K., and Y. V. Brun. 2007. Out on a limb: how the Caulobacter stalk can boost the study of bacterial cell shape. Mol. Microbiol. 64:28-33. [DOI] [PubMed] [Google Scholar]
- 261.Wagner, J. K., C. D. Galvani, and Y. V. Brun. 2005. Caulobacter crescentus requires RodA and MreB for stalk synthesis and prevention of ectopic pole formation. J. Bacteriol. 187:544-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Wagner, J. K., S. Setayeshgar, L. A. Sharon, J. P. Reilly, and Y. V. Brun. 2006. A nutrient uptake role for bacterial cell envelope extensions. Proc. Natl. Acad. Sci. U. S. A. 103:11772-11777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Wang, S. C., and L. Shapiro. 2004. The topoisomerase IV ParC subunit colocalizes with the Caulobacter replisome and is required for polar localization of replication origins. Proc. Natl. Acad. Sci. U. S. A. 101:9251-9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Wang, S. C., L. West, and L. Shapiro. 2006. The bifunctional FtsK protein mediates chromosome partitioning and cell division in Caulobacter. J. Bacteriol. 188:1497-1508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Wang, S. P., P. L. Sharma, P. V. Schoenlein, and B. Ely. 1993. A histidine protein kinase is involved in polar organelle development in Caulobacter crescentus. Proc. Natl. Acad. Sci. U. S. A. 90:630-634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Wanner, B. L. 1993. Gene regulation by phosphate in enteric bacteria. J. Cell Biochem. 51:47-54. [DOI] [PubMed] [Google Scholar]
- 267.Wanner, B. L. 1996. Signal transduction in the control of phosphate-regulated genes of Escherichia coli. Kidney Int. 49:964-967. [DOI] [PubMed] [Google Scholar]
- 268.Wanner, B. L., and J. A. Boline. 1990. Mapping and molecular cloning of the phn (psiD) locus for phosphonate utilization in Escherichia coli. J. Bacteriol. 172:1186-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269.Wassmann, P., C. Chan, R. Paul, A. Beck, H. Heerklotz, U. Jenal, and T. Schirmer. 2007. Structure of BeF3-modified response regulator PleD: implications for diguanylate cyclase activation, catalysis, and feedback inhibition. Structure 15:915-927. [DOI] [PubMed] [Google Scholar]
- 270.Webb, C. D., A. Teleman, S. Gordon, A. Straight, A. Belmont, D. C. Lin, A. D. Grossman, A. Wright, and R. Losick. 1997. Bipolar localization of the replication origin regions of chromosomes in vegetative and sporulating cells of B. subtilis. Cell 88:667-674. [DOI] [PubMed] [Google Scholar]
- 271.Werner, J. N., E. Y. Chen, J. M. Guberman, A. R. Zippilli, J. J. Irgon, and Z. Gitai. 2009. Quantitative genome-scale analysis of protein localization in an asymmetric bacterium. Proc. Natl. Acad. Sci. U. S. A. 106:7858-7863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Wheeler, R. T., and L. Shapiro. 1999. Differential localization of two histidine kinases controlling bacterial cell differentiation. Mol. Cell 4:683-694. [DOI] [PubMed] [Google Scholar]
- 273.Whittenbury, R., and C. S. Dow. 1977. Morphogenesis and differentiation in Rhodomicrobium vannielii and other budding and prosthecate bacteria. Bacteriol. Rev. 41:754-808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Wingrove, J. A., and J. W. Gober. 1994. A sigma 54 transcriptional activator also functions as a pole-specific repressor in Caulobacter. Genes Dev. 8:1839-1852. [DOI] [PubMed] [Google Scholar]
- 275.Wingrove, J. A., E. K. Mangan, and J. W. Gober. 1993. Spatial and temporal phosphorylation of a transcriptional activator regulates pole-specific gene expression in Caulobacter. Genes Dev. 7:1979-1992. [DOI] [PubMed] [Google Scholar]
- 276.Winzeler, E., and L. Shapiro. 1996. A novel promoter motif for Caulobacter cell cycle-controlled DNA replication genes. J. Mol. Biol. 264:412-425. [DOI] [PubMed] [Google Scholar]
- 277.Wortinger, M., M. J. Sackett, and Y. V. Brun. 2000. CtrA mediates a DNA replication checkpoint that prevents cell division in Caulobacter crescentus. EMBO J. 19:4503-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Wortinger, M. A., E. M. Quardokus, and Y. V. Brun. 1998. Morphological adaptation and inhibition of cell division during stationary phase in Caulobacter crescentus. Mol. Microbiol. 29:963-973. [DOI] [PubMed] [Google Scholar]
- 279.Wright, R., C. Stephens, and L. Shapiro. 1997. The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J. Bacteriol. 179:5869-5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Wright, R., C. Stephens, G. Zweiger, L. Shapiro, and M. R. Alley. 1996. Caulobacter Lon protease has a critical role in cell-cycle control of DNA methylation. Genes Dev. 10:1532-1542. [DOI] [PubMed] [Google Scholar]
- 281.Wu, J., and A. Newton. 1997. Regulation of the Caulobacter flagellar gene hierarchy; not just for motility. Mol. Microbiol. 24:233-239. [DOI] [PubMed] [Google Scholar]
- 282.Wu, J., N. Ohta, and A. Newton. 1998. An essential, multicomponent signal transduction pathway required for cell cycle regulation in Caulobacter. Proc. Natl. Acad. Sci. U. S. A. 95:1443-1448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 283.Wu, J., N. Ohta, J. L. Zhao, and A. Newton. 1999. A novel bacterial tyrosine kinase essential for cell division and differentiation. Proc. Natl. Acad. Sci. U. S. A. 96:13068-13073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284.Wu, L. J., and J. Errington. 1994. Bacillus subtilis SpoIIIE protein required for DNA segregation during asymmetric cell division. Science 264:572-575. [DOI] [PubMed] [Google Scholar]
- 285.Wu, L. J., and J. Errington. 2004. Coordination of cell division and chromosome segregation by a nucleoid occlusion protein in Bacillus subtilis. Cell 117:915-925. [DOI] [PubMed] [Google Scholar]
- 286.Wu, L. J., S. Ishikawa, Y. Kawai, T. Oshima, N. Ogasawara, and J. Errington. 2009. Noc protein binds to specific DNA sequences to coordinate cell division with chromosome segregation. EMBO J. 28:1940-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Xu, Q., D. Carlton, M. D. Miller, M. A. Elsliger, S. S. Krishna, P. Abdubek, T. Astakhova, P. Burra, H. J. Chiu, T. Clayton, M. C. Deller, L. Duan, Y. Elias, J. Feuerhelm, J. C. Grant, A. Grzechnik, S. K. Grzechnik, G. W. Han, L. Jaroszewski, K. K. Jin, H. E. Klock, M. W. Knuth, P. Kozbial, A. Kumar, D. Marciano, D. McMullan, A. T. Morse, E. Nigoghossian, L. Okach, S. Oommachen, J. Paulsen, R. Reyes, C. L. Rife, N. Sefcovic, C. Trame, C. V. Trout, H. van den Bedem, D. Weekes, K. O. Hodgson, J. Wooley, A. M. Deacon, A. Godzik, S. A. Lesley, and I. A. Wilson. 2009. Crystal structure of histidine phosphotransfer protein ShpA, an essential regulator of stalk biogenesis in Caulobacter crescentus. J. Mol. Biol. 390:686-698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 288.Yeoman, S., T. Stephenson, J. N. Lester, and R. Perry. 1986. Biotechnology for phosphorus removal during wastewater treatment. Biotechnol. Adv. 4:13-26. [DOI] [PubMed] [Google Scholar]
- 289.Yu, J., and L. Shapiro. 1992. Early Caulobacter crescentus genes fliL and fliM are required for flagellar gene expression and normal cell division. J. Bacteriol. 174:3327-3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Zhou, Y., S. Gottesman, J. R. Hoskins, M. R. Maurizi, and S. Wickner. 2001. The RssB response regulator directly targets sigma(S) for degradation by ClpXP. Genes Dev. 15:627-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.Zweiger, G., G. Marczynski, and L. Shapiro. 1994. A Caulobacter DNA methyltransferase that functions only in the predivisional cell. J. Mol. Biol. 235:472-485. [DOI] [PubMed] [Google Scholar]
- 292.Zweiger, G., and L. Shapiro. 1994. Expression of Caulobacter dnaA as a function of the cell cycle. J. Bacteriol. 176:401-408. [DOI] [PMC free article] [PubMed] [Google Scholar]