Abstract
Helicobacter pylori is known to be a major cause of gastric carcinoma and peptic ulceration. cagA positivity and vacA's signal regions and mid-regions are well-characterized markers of H. pylori's virulence. Recently, an intermediate region has been identified as another strong marker of H. pylori-associated disease, and its i1 allele has been linked with severe diseases in colonized hosts. The goal of this study was to determine the prevalence of the intermediate alleles in H. pylori isolates from China, Turkey, and Uruguay and from U.S. Africans and to compare their distribution with other well-characterized virulence factors. Originally, 123 H. pylori strains were studied, but 3 were excluded due to the failure to amplify the intermediate region in these samples. Therefore, a total of 120 strains were analyzed: 30 Chinese isolates, 35 Turkish isolates, 30 Uruguayan isolates, and 25 U.S. African isolates. The s type and the m type were determined by PCR amplification. The i type was identified by PCR amplification and DNA sequencing. CagA status was determined by PCR methodology. There was a strong correlation among CagA positivity, s1, and i1 in Chinese, U.S. African, and Uruguayan isolates, but less correlation among these markers in Turkish isolates. A new intermediate variant (i3) was identified in 25.7% of Turkish strains and 3.3% of the Chinese strains. In summary, the distribution of CagA positivity and s1 correlated with the i1 in the three populations, except in the Turkish population, which showed a disproportionate representation of the i3 allele. Phylogenetic mapping confirmed the i-typing method previously defined and adopted for this study. The phylogenetic tree showed country-specific correlation with the intermediate region. Our results showed that the i1 allele is strongly associated with CagA positivity and the vacA s1 allele, suggesting its role as a virulence marker and potential predictor for clinical outcome.
Colonization with Helicobacter pylori is the major etiological factor in gastritis, peptic ulcer disease (PUD) and gastric adenocarcinoma (GC). This microorganism affects more than half of the world's population and up to 70% of the adult population in developing countries (16). However, not all H. pylori infections lead to a clinical presentation. Disease outcome is a multifactorial phenomenon that includes host, bacterial, and environmental factors. Two virulence attributes of H. pylori have been shown to be strong predictors of clinical outcome in humans. The first is the cytotoxin-associated gene, cagA. Strains that are positive for the encoded protein, CagA, are strongly linked with PUD and GC (7, 21). CagA is translocated to epithelial cells, where it stimulates signaling pathways that enhance the interaction between bacteria and cell (27), activates transcription factors that leads to cellular proliferation and apoptosis (29), and disrupts apical junctions between epithelial cells, affecting the barrier function of the gastric mucosa (3). CagA+ strains are more prevalent in East Asian countries such as Japan and China than in Western countries. H. pylori infection leads to higher levels of inflammation and atrophy, as well as a higher occurrence of gastric cancer (5). In contrast, CagA− strains are rarely associated with disease.
VacA, the vacuolating cytotoxin encoded by the vacA gene, is another strong marker for H. pylori virulence. This toxin causes vacuolization of epithelial cells, disruption of the endosomal/lysosomal pathway, apoptosis through the mitochondrial release of cytochrome c, interference with cell signaling, and the inhibition of T-cell proliferation (9, 14). VacA is a mature 87-kDa monomer that is present in all H. pylori strains, but its cytotoxic activity has been shown to occur in only 50% of H. pylori strains (4). The secreted product assembles to form anion-selective membrane channels that are inserted into the lipid bilayer (13). This process is crucial for the vacuolating activity of H. pylori. However, the vacA gene is highly polymorphic, especially in two regions referred to as the signal region (s region) and the mid-region (m region), both of which have been well characterized as markers of determining virulence and progression to serious disease. The s region is classified as s1 or s2, and the m region is characterized as m1 or m2 (4). All s and m region combinations exist, although the occurrence of s2/m1 is very rare (17). The specific pairing of the alleles of these two regions are linked to differences in VacA toxicity and association with disease outcome. s1/m1 has been shown to be linked with duodenal and gastric ulceration (4), as well as with gastric cancer (19). Strains with s1 are associated with vacuolating activity, whereas strains with s2 have been shown to be nonvacuolating, causing less effective formation of membrane pores and apoptosis (18). The m region has been linked to the binding specificity of VacA to specific epithelial cells that are induced to undergo vacuolization (22). The m1 region is associated with vacuolization in a wider variety of cells than the m2 region. Therefore, the s1/m1 strains cause vacuolization in a wide variety of cells, whereas the s1/m2 strains are more restricted in the range of epithelial cells they can affect, leading to decreased toxicity in populations with increased VacA binding specificity to cell-surface receptors (22). The s2/m2 strains are neither linked with vacuolization of cells nor to the progression of gastritis to more serious disease.
Recently, a third area, the intermediate (i) region, of the vacA gene that regulates its protein's vacuolating activity has been identified between the s and m regions (25). Sequencing and amino acid analysis have characterized polymorphisms within the i region as clusters A, B, and C; however, only clusters B and C influenced the cytotoxin's activity. In particular, cluster C, with an additional three-residue insertion/deletion, has been shown to have a greater effect on determining the toxin's activity than cluster B. Based on the amino acid sequence differences, two phenotypes were described as i1 and i2 (25).
The i region is also associated with GC and PUD. In a previous study, it was demonstrated that the i type of VacA may be a better predictor of virulence than the m type. H. pylori isolates from Iranian and Western isolates, where both the i1 and the i2 alleles were present, all s1/m1 strains were i1, and all s2/m2 strains were i2. As expected, the s1/i1/m1 strains showed vacuolating activity, whereas the s2/i2/m2 strains did not. More importantly, the i types varied in the s1/m2 strains; s1/i1/m2 strains showed vacuolating activity, while s1/i2/m2 strains did not. Further, using statistical analysis, the i type was shown to be independent of the s and m types in its association with the increased risk of gastric carcinoma (25). The link between the i region and GC and PUD has been further confirmed by a recent study of an Italian population, where s1/i1/m1 strains were highly associated with gastric carcinoma (6). In addition, i1 was found to have a strong connection with the incidence of peptic ulceration. Therefore, based on this evidence in the Iranian and Italian populations, we believe that the determination of the i type may be important to identify H. pylori strains that are pathogenic and may cause progression to serious disease.
In Turkey and Uruguay, the prevalence of severe H. pylori-associated disease is known to be low (11, 12), whereas in Chinese and U.S. African populations, the prevalence of these diseases is high (1, 24). According to data from the International Agency for Research on Cancer, the incidence of gastric cancer varies among global populations. Between 1998 and 2002, the incidence of gastric cancer was 3,291 cases/100,000 population in Hong Kong, 2,025 cases/100,000 population among U.S. Africans, 1,584 cases/100,000 population in Uruguay, and 1,191 cases/100,000 population in Turkey (10). We predict that H. pylori isolates from Turkish and Uruguayan populations with the VacA s1/m2 genotype will be predominantly i2, whereas those from Chinese and U.S. Africans will be mostly i1, reflecting the greater incidence of PUD and/or GC in the population. Therefore, in the present study, we determined the prevalence of intermediate variants in VacA among H. pylori strains isolated from Turkish, Uruguayan, Chinese, and U.S. African populations using PCR amplification and DNA sequencing analysis.
MATERIALS AND METHODS
Source of bacteria.
We studied a total of 123 strains: 31 (25.2%) were isolated from an Uruguayan population, 37 (30.1%) were isolated from a Turkish population, 30 (24.4%) were isolated from a Chinese population, and 25 (20.3%) were isolated from U.S. Africans. H. pylori isolates were obtained from gastric biopsies of either the antrum or the corpus of endoscoped patients from each of these four countries.
PCR amplification of the vacA signal and mid-region.
DNA from the 123 strains had been previously extracted using the Promega Wizard (Promega, Madison, WI) and was used to analyze the vacA s and m types. vacA genotype was determined by PCR. vacA s and m type was determined using primers and conditions previously described to amplify the s1, s2, m1, and m2 regions (4). Both positive and negative controls were included. If the PCR amplification was not successful the first time, an additional amplification was performed before the samples were considered defective and thus excluded from analysis.
PCR amplification and DNA sequencing of the vacA intermediate region.
PCR was used to amplify the vacA intermediate region using primers (VacF1, forward; VacR9, reverse) under previously described conditions (25). The PCR products were purified by using a QIAquick DNA purification kit (Qiagen, Valencia, CA) and subjected to direct sequencing (SeqWright DNA Technology Services). The resulting nucleotide sequences from all of the strains were edited by using Sequencher (version 4.5). The same program was used to translate the nucleotide sequences into amino acid sequences. Both the nucleotide and the amino acid sequences were aligned by using CLUSTAL X (version 1.81) and Genedoc (version 2.6.0.2).
Typing and analysis of the vacA intermediate region.
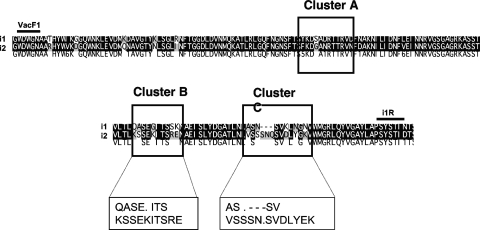
The criteria used for determining the i type (either i1 or i2) were based on the amino acid sequences at clusters B and C (25). Cluster A (amino acids [aa] 186 to 200) was not used for i typing because only 1 amino acid showed sequence divergence in the area and was not a reliable marker for different i-region alleles. Based on the amino acid sequence of strain 60190, i1 was defined by the presence of QASE.ITS (aa 232 to 239) within cluster B and AS.---SV (aa 256 to 263) within cluster C. The amino acid sequence of strain Tx30a was used to determine i2, which was defined by the presence of KSSEKITSRE (aa 232 to 241) at cluster B and VSSSN.SVDLYEK (aa 256 to 268) at cluster C. The position of clusters A, B, and C are based on the i-region amino acid sequence of Tx30a (Fig. 1). Some strains displayed discordant pairing of the clusters, in which the amino acid sequence at one cluster was i1-like and that of the other cluster was i2-like; such strains were defined as i3.
FIG. 1.
Comparison of the amino acid sequences of an i1 and i2 strain of H. pylori. The sequences of strain 60190 and Tx30a were used to define i1 and i2, respectively. Regions defined as clusters A, B, and C are indicated. VacF1 and i1R refer to the forward and reverse primers used to amplify the intermediate region.
Phylogenic analysis of H. pylori strains by the vacA intermediate region.
In addition to the determination of the i-type of each strain, a phylogenetic tree, based upon the 495-bp amplified sequence of the intermediate region PCR product, was created by the neighbor-joining method with MEGA (version 2.1). The stability of the tree was tested by performing 500 bootstrap replicates.
Analysis of the presence of cagA among H. pylori strains.
The forward primer ATCAAAAACCAATCGTTGATAAGAA and the reverse primer CATTTTTTTCTGCTTCTTGCCT were used to amplify a 500-bp fragment. The PCR was conducted with a 50-μl mixture containing 100 ng of DNA (1 μl), 5 μl of 10× buffer, 2.5 mM deoxynucleotide each (dATP, dCTP, dGTP, and dTTP), 1.25 U of Taq DNA polymerase and 0.1 to 0.5 μM each primer. Gene amplification was carried out through 30 cycles of denaturation (94°C) for 0.5 min, primer annealing (58°C) for 0.5 min, and extension (72°C) for 1.0 min. Gel electrophoresis on a 2% agarose gel was carried out for all samples to control which samples contained the empty site. Positive and negative controls were used.
Determination of the empty site by PCR.
Empty-site PCR was used to determine cag pathogenicity island (PAI) negative strains. When present, the cag PAI is located between the two protein coding H. pylori genes Hp0519 and Hp0549 (2). The empty-site forward primer binds to Hp0519, and the reverse primer binds to Hp0549. Since the cag PAI (40 kb in size) is too large to amplify with PCR, the amplification will yield a product when the cag PAI is absent. Thus, only cag PAI-negative strains amplified a product.
For the strains included in the present study, PCR was used to confirm the absence of cag PAI, which was a 550-bp segment containing the empty site. The forward primer CCAAATACATTTTGGTAAATAAAC and the reverse primer CTCTTTTTGTGCCTTTGATTGAA were used. The PCR was conducted with a 50-μl mixture containing 100 ng of DNA (1 μl), 5 μl of 10× buffer, 2.5 mM concentrations of each deoxynucleotide (dATP, dCTP, dGTP, and dTTP), 1.25 U of Taq DNA polymerase, and 0.1 to 0.5 μM each primer. Gene amplification was carried out through 30 cycles of denaturation (94°C) for 1 min, primer annealing (57°C) for 1 min, and extension (72°C) for 1.5 min. Gel electrophoresis on a 2% agarose gel was carried out for all samples to control which samples contained the empty site. Positive and negative controls were used.
Statistics.
The population-specific associations among the H. pylori genotypes were analyzed by using the χ2 test with Yates's continuity correction or the Fisher exact test. Differences were defined as being statistically significant if the P value was <0.05. The Epi-Info software (version 3.3.2) was used for all analyses.
RESULTS
Characterization of our strain population.
Bacterial isolates whose s, m, or i type failed to be clearly delineated were excluded from the present study. A total of 3 (2.4%) strains of the initial 123 strains were excluded from the analysis. None of the 30 Chinese and 25 U.S. African strains was excluded. Two of the initial 37 Turkey isolates and 1 of the initial 31 Uruguayan isolates were excluded because the intermediate region could not be amplified in these samples. Therefore, we are reporting the results from 120 (97.6%) strains.
The intermediate regions of all strains were classified according to the criteria previously mentioned in Materials and Methods. One strain classified as i1 had a single amino acid discrepancy at cluster B, encoding a K instead of an E at the fourth position. An i3 strain had one amino acid discrepancy at the second position of cluster C, encoding a Q instead of an S. One i2 strain had a shortened cluster C due to a failure to sequence the entire i region. The cluster B sequence for this strain only contained VSSSN.
cagA and vacA signal and mid-region typing of H. pylori isolates.
We determined the cagA status of each isolate from the four populations. A total of 68.3% of all isolates were CagA+, but the CagA status varied with the sampled population. U.S. African isolates had the greatest CagA positivity, followed closely by the Chinese isolates. The Turkish population was found to have the least CagA positivity (Table 1) . The Chinese strains were significantly more likely to be CagA+ than either of the Turkish or Uruguayan populations (Turkey, P < 0.003; Uruguay, P < 0.02). The difference between the Chinese and U.S. African isolates was not significant (P = 1).
TABLE 1.
CagA positivity and vacA signal region and mid-region types of 120 H. pylori strains isolated from patients in China, Turkey, and Uruguay and from U.S. Africans
| Strain population | No. of subjectsa | No. (%) that were CagA+ | No. of VacA s and m types |
||||
|---|---|---|---|---|---|---|---|
| s1 | s2 | m1 | m2 | Mixed | |||
| Chinese | 30 | 26 (86.7) | 30 | 0 | 2 | 28 | 0 |
| Turkish | 35* | 17 (48.6) | 20 | 15 | 8 | 31 | 4 |
| Uruguayan | 30* | 17 (56.7) | 17 | 14 | 14 | 17 | 1 |
| U.S. African | 25 | 22 (88) | 22 | 3 | 15 | 10 | 0 |
*, There are discrepancies between the total number of strains with those broken down into allele types due to the presence of one Turkish mixed strain and four Uruguayan mixed strains with both s1 and s2 and both m1 and m2 alleles in the signal and mixed regions.
The distribution of both the s and the m regions showed marked differences among the populations. s1 was the predominant signal region allele of all four countries, with a total prevalence of 74.2% for all isolates. The Chinese and U.S. African populations showed more a dramatic prevalence of s1 than the remaining two populations (100 and 88%, respectively). The Turkish and Uruguayan populations showed similar s1 prevalences of 57.1 and 56.7%. The occurrence of s1 in China was significantly greater than its occurrence in either Turkey or Uruguay (P < 0.0001). The difference in s1 prevalence among the Chinese and U.S. African populations were not statistically significant (P < 0.09).
The predominant mid-region allele in all four countries combined was the m2 allele, with a total prevalence of 71.7% for all isolates. The Chinese and Turkish populations showed a pronounced preference for this allele, with 93.3 and 88.6% of their strains classified as m2, respectively. The Uruguayan had a more modest m2 prevalence of 56.7%, and the U.S. African population had a prevalence of only 40%. The difference in m1 occurrence was significant between the Chinese and African-American populations (P < 0.001) and between the Chinese and Uruguayan populations (P < 0.002).
Upon examining the vacA s and m regions, the Chinese isolates showed a marked predominance of one genotype, whereas the U.S. African, Turkish, and Uruguayan populations showed a more balanced distribution among the different genotypes. Almost all of the Chinese isolates were s1/m2 (93.3%), with no observed s2/m2. The majority of isolates in the U.S. African population were s1/m1 (60%), and 28% were s1/m2. In the Turkish population, 50% of the isolates were s2/m2 (50%). A total of 36.7% were s1/m2, and 13.3% were s1/m1. Finally, the majority of the Uruguayan population was divided between s2/m2 and s1/m1 isolates (42.9 and 46.4%, respectively). We found that 10.7% were s1/m2 (see Table 3). In these analyses, mixed strains were not included.
TABLE 3.
Comparison of CagA positivity and vacA i1 type among H. pylori strains by country and VacA s and m types
| Strain population | No. of subjects | % CagA+/VacA i1a |
|
|---|---|---|---|
| s1/m1 | s1/m2 | ||
| Chinese | 30 | 6.7 | 80* |
| Turkish | 30b | 10.0 | 10.0 |
| Uruguayan | 28c | 46.4† | 7.1 |
| U.S. African | 25 | 60† | 20.0 |
*, P < 0.0001 compared to all other countries; †, P < 0.001 compared to isolates from China and Turkey.
Five strains with mixed results were excluded.
Two strains with mixed results were excluded.
Intermediate region type of vacA isolates.
The prevalences of i1 and i2 in all isolates was 58.3 and 36%, respectively. A total of 8.3% were characterized as i3, with all but one of them from the Turkish population. Another 3.3% were mixed strains: one strain was i1/i2, while the other three were i1/i3. Like the s and m regions, there were marked differences in the distribution of the i alleles in the various populations. The Chinese strains showed a marked predominance of the more virulent i1 allele (93.3%). i1 also made up the majority of the U.S. African strains (80%). The Turkish and Uruguayan stains had more modest i1 distributions (22.9 and 46.7%, respectively). The i2 allele was much more prevalent in Turkey and Uruguay, a finding that characterized nearly half of the isolates in each country. Statistical analysis confirmed these results. The i1 type was significantly more likely to occur in Chinese strains than in the strains from the Turkish or Uruguayan populations (Turkey, P < 0.001; Uruguay, P < 0.0001). The difference in i1 between the Chinese and U.S. African populations was not significant (Table 2).
TABLE 2.
vacA intermediate types of 120 H. pylori strains isolated from patients from China, Turkey, and Uruguay and from U.S. Africans
| Strain population | No. of subjects | No. of subjects (%) by intermediate region |
|||
|---|---|---|---|---|---|
| i1 | i2 | i3 | Mixeda | ||
| Chinese | 30 | 28 (93.3) | 1 (3.3) | 1 (3.3) | 0 (0) |
| Turkish | 35 | 8 (22.9) | 15 (42.9) | 9 (25.7) | 3 (8.6) |
| Uruguayan | 30 | 14 (46.7) | 15 (50) | 0 (0) | 1 (3.3) |
| U.S. African | 25 | 20 (80) | 5 (20) | 0 (0) | 0 (0) |
Mixed strains were those characterized as i1/i2 or i1/i3.
Intermediate region type and CagA status.
In all four populations, CagA positivity and i1 were similarly distributed among various s and m genotypes (Table 3). All s1/m1 isolates were CagA+ and i1, aside from two isolates. One s1/m1 Turkish isolate was CagA− but possessed the i1 allele. The other s1/m1 Uruguayan isolate was CagA+ but characterized as i2. In addition, all but one of the s2/m2 strains was CagA negative and i1 negative. This exception was an Uruguayan isolate that was i2 despite being CagA+. Therefore, the s1/m1 isolates were almost always associated with being CagA+ and i1. In contrast, s2/m2 isolates were rarely associated with these markers of poor disease outcome.
The proportions of CagA positivity and i1 in the s1/m2 isolates were more variable than those of the s1/m1 and s2/m2 groups across the four populations. In all populations, the majority of the s1/m2 isolates were CagA positive (China, 85.7%; Turkey, 90.9%; U.S. African subjects and Uruguay; 100%). However, the distribution of i1 among the s1/m2 isolates was less uniform: 92% of the Chinese s1/m2 isolates, 71.4% of the U.S. African s1/m2 isolates, 66.7% of the Uruguayan s1/m2 isolates, and 27.3% of Turkish s1/m2 isolates were i1. i1 was significantly more likely to occur in the Chinese population than in the Turkish s1/m2 population (P < 0.0001). The differences in i1 occurrence were not statistically significant in other populations. However, when all m2 strains are considered (including isolates with s1 or s2), the Chinese strains are significantly more likely to be i1 than any of the other three populations (U.S. African population, P < 0.008; Uruguay, P < 0.000001; Turkey, P < 0.00000).
Phylogenic mapping of vacA intermediate region.
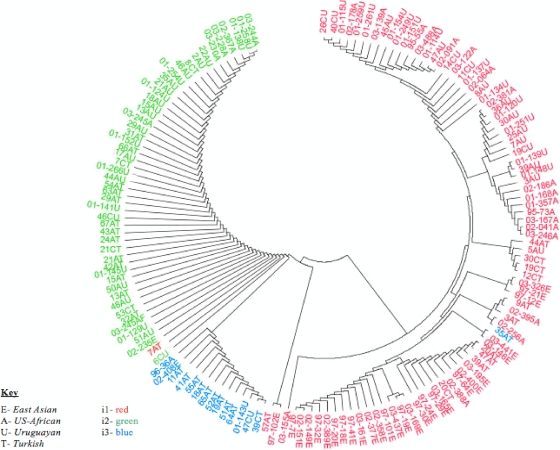
A phylogenetic tree was constructed to compare the vacA intermediate region of strains from the Chinese, Uruguayan, Turkish, and U.S. African populations, based on the amino acid sequence of the entire 495-bp region (including clusters A, B, and C) (Fig. 2). This tree reflects correlations between strains using the entire intermediate region rather than limiting sequence analysis to clusters B and C, which were the two regions by which the strains' i types were classified. As expected, the phylogenic mapping closely mirrored our classification of i1, i2, and i3 strains. The i1 strains were distinct from the other i2 and i3 strains. There was some country specific clustering among the i1 strains. All of the Chinese i1 strains originate from the same branch, with an association with some Turkish strains. Another point that requires further clarification is that there appears to be a clustering of H. pylori strains from the U.S. African and Uruguayan strains in the upper right corner of the tree. These observations suggest that all i1 strains are not identical. The clinical outcome of these i1-infected hosts should be assessed to determine whether the i1 strains from Uruguay and Turkey that are more closely related to the Chinese and U.S. African strains have a worse prognosis than those that are not.
FIG. 2.
Phylogenetic tree based on the amino acid sequences of the vacA intermediate region of strains from China, Uruguay, Turkey, and U.S. Africans. i types are separated by color: i1, red; i2, green; and i3, blue. Strains are labeled by region or country of origin: E, East Asia; U, Uruguay; T, Turkey; and A, United States. Preceding the country of origin, some strains are labeled by the area of the stomach from which they were isolated: A, antrum; C, corpus. Key: E, East Asian; A, U.S. African; U, Uruguayan; T, Turkish; i1, red; i2, green; i3, blue.
The i2 strains appear to be more homogenous. All strains, regardless of the countries from which they were obtained, have very similar amino acid sequences. This suggests that these strains are equally likely to show the absence of disease of the infected hosts.
All i3 variants were also clustered together on the phylogenetic map and originated from the same main branch as that of the i2 isolates, with the exception of one Turkish strain that appears to be more similar to the i1 strains. As shown in Table 2, almost all identified i3 strains were from Turkey, an area of low disease prevalence. The association of the i3 strains with i2 indicates that i3 variants may also be associated with a better clinical outcome than i1 strains. Further studies that assess the clinical outcome of the i3 hosts and test the cytotoxic activity of these strains must be done to confirm this hypothesis.
There was only one discrepancy between results of this tree and our genotyping results based on PCR and gene sequencing. One strain from Turkey, which we have typed as i1, based on clusters B and C, was shown to be more closely associated with the i2 strains than with the other i1 strains. This may be due to the inclusion of the entire intermediate region in phylogenetic mapping, which contrasted with our analysis, which only accounted for sequence differences in two clusters, shown to best predict H. pylori's cytotoxic activity. The i types of the rest of the strains were consistent, whether they were determined by sequencing at distinct clusters or by sequencing the entire intermediate region. This confirms the cluster B- and C-restricted definition defined previously (25) and justifies the use of a modified form of that definition in the present study as a reliable means for determining the i types of the isolates.
DISCUSSION
It is now well established that the prevalence of CagA in H. pylori strains vary among countries; those with high rates of severe disease have high rates of CagA+ strains, whereas those with mild diseases have lower rates of CagA+ strains. East Asian H. pylori strains have been shown to have CagA positivity approaching 100% (8, 30), and U.S. African H. pylori strains have been shown to have a CagA prevalence of ca. 80% (23, 28). Western European and Latin American H. pylori strains, represented by the Uruguayan strains in the present study, have been shown to possess a more moderate CagA prevalence of ca. 60 to 70% (15, 26). Finally, Middle Eastern H. pylori strains, represented by the Turkish strains, have been shown to possess a CagA positivity of <50% (20, 31). We selected H. pylori strains isolated from Chinese and U.S. African populations as representative groups from countries with high prevalence of GC and PUD, whereas strains from Turkish and Uruguayan populations were chosen for their low prevalence of such diseases. Our results confirmed that Chinese and U.S. African strains showed the highest prevalence of CagA+ strains, Uruguayan strains had the next highest prevalence, and followed by the Turkish strains with the lowest distribution of CagA.
The vacA signal regions and mid-regions are also H. pylori virulence markers that have been used to predict the clinical outcome of infected patients. The s1 allele is a well-established virulence marker, whose distribution matched our expectations. Notably, all of the Chinese isolates in our study were s1. In addition, neither the Chinese nor the U.S. African population possessed the s2 allele, which is associated with less virulent strains.
The vacA mid-region m1 allele, however, did not follow the expected distribution based on differences in disease severity among the four populations. Most of the Chinese, Turkish, and Uruguayan strains were m2, whereas most of the U.S. African strains were m1. Previous studies have shown that the m region of H. pylori strains isolated from East Asia showed country-specific differences. For example, most Chinese strains have been shown to be predominantly m2, whereas most Japanese strains were m1 (8, 30, 32). Strains from both countries have high CagA positivity, a high prevalence of s1, and a high incidence of severe disease; however, the strains show marked differences in the m region. These results suggest that the m region may play a minor, secondary role in predicting H. pylori's virulence compared to the cagA or vacA s region. The m region has been shown to determine the range of epithelial cells in which the infecting strain may cause vacuolation. m2 may be associated with lower virulence in previous studies because it narrows the range of cells that may be damaged, compared to m1 strains, that may cause vacuolization in a wider range of epithelial cell types. This distinction may not affect the clinical outcome of countries in which most of the population may have similar characteristics of epithelial cells of the gastric mucosa. Having the ability to cause vacuolization in a wide variety of epithelial cells may not confer an advantage over strains that possess a more narrow range of activity. Therefore, the low incidence of m1 in the Turkish and Chinese strains and the higher incidence in the U.S. African and Uruguayan strains may not reflect the virulence of different strains and their associated clinical outcomes.
Further, strains with the s1/m1 genotype have been associated with the most virulent strains, those with s1/m2 have been linked with less virulent strains, and those with s2/m2 are known to be the least virulent (4, 18, 19, 22). As expected from prior literature, all of our CagA+ strains were either s1/m1 or s1/m2, and none of our CagA+ strains (except for one Uruguayan isolate) were s2/m2. The s1/m2 population was more variable. They made up a significant proportion of Chinese isolates, and a very small portion of the Uruguayan isolates. Most of these s1/m2 strains were CagA+, with few country-specific differences.
Recently, the intermediate region of H. pylori's vacA has been shown to be a strong, independent predictor of the bacterium's pathogenicity. The presence of the i1 allele was found to be a more accurate virulence marker than either the s or the m region in Western and Iranian strains (25). It has also been studied in conjunction with the s and m regions to better characterize isolates that may lead to a poor clinical outcome. First, as expected, none of our s2/m2 strains were i1. Second, all s1/m1 isolates were i1 except for one Uruguayan strain, which was CagA+/s1/m1 but i2. The distribution of i1 among s1/m2 strains was more country specific. The majority of the Chinese s1/m2 strains were i1 (93.3%). The i1 distribution of U.S. African s1/m2 was the next highest (71.4%), followed by the Uruguayan s1/m2 strains (66.7%). In the Turkish s1/m2 population, only 27.3% were i1, although 90.9% of these had been CagA+. This distribution of the i1 allele reflects what has been predicted by previous studies and knowledge of disease distribution in these areas (8, 10, 15, 23, 26). From our results, it seems that CagA positivity is a less sensitive marker of strain virulence than i1 in the population of s1/m2.
In our study, there was a significant proportion of isolates that we characterized as i3, most of which were associated with the Turkish population. We have defined this allele as one that had the amino acid sequence of i1 at one cluster and the sequence of i2 at the other cluster that was examined. Of the 10 i3 strains that we identified, 9 were Turkish isolates. Furthermore, 8 of the 10 strains had an i1-like sequence at cluster B and an i2-like sequence at cluster C. This corresponded to the results of a previous study that identified a recombinant i1-i2 allele with this cluster B- and C-specific combination (25).
This i3 variant may reflect a genetic recombination between i1 and i2 strains, but the pathogenic and clinical significance of this allele is still unclear. However, building on results of the prior study that confirmed the low vacuolating activity of the i1-i2 recombinant allele (25), our study further suggests that this i3 allele may possess i2-like activity. First, most of our i3 isolates have an i2-like amino acid sequence in cluster C, which has been shown to be more predictive of vacuolating activity and have shown close correlation with disease outcome in colonized patients. Second, most of the i3 strains occur in the Turkish population found to have strains associated with better clinical outcome. Finally, our phylogenetic analysis further suggests that i3 may be associated with lower prevalence of disease since it has a closer association with i2 strains. It is necessary to confirm whether Turkey's disproportionate prevalence of i3 still exists with the assessment of a larger sample size or in H. pylori strains from another country in the Middle East.
Therefore, the prevalence of CagA positivity, s1, and i1 among Chinese, U.S. African, Turkish, and Uruguayan strains confirms findings of previous studies. The i region seems to possess an ability to predict the disease outcome of isolates from different areas of the world much like CagA positivity. We were unable to compare the i type with disease severity due to the lack of information from endoscoped patients, as well as the universality of the i1 allele in the East Asian population. The actual clinical outcome of infected hosts must be assessed to determine whether the i1 type was more representative of the group with a poor disease outcome than the hosts harboring the CagA+ or s1 strains. In addition, testing a greater number of isolates may show a more similar distribution of the three virulence factors among the different populations. A greater sample size isolated from more diverse populations may also better elucidate the nature of i3 and its link with disease progression of infected hosts.
Acknowledgments
This study was supported in part by the NYU Center for the Study of Asian American Health, a National Institutes of Health Research Center of Excellence (P60 MD000538-05).
We thank Can Gonen and Likay Simsek for their expertise and their collection of the Turkish biopsy samples and Selena Reynolds from Mount Holyoke College for her technical assistance. We especially thank John Atherton for his valuable input into this work.
Footnotes
Published ahead of print on 6 January 2010.
REFERENCES
- 1.Agha, A., and D. Y. Graham. 2005. Evidence-based examination of the African enigma in relation to Helicobacter pylori infection. Scand. J. Gastroenterol. 40:523-529. [DOI] [PubMed] [Google Scholar]
- 2.Akopyants, N. S., S. W. Clifton, D. Kersulyte, J. E. Crabtree, B. E. Youree, C. A. Reece, N. O. Bukanov, E. S. Drazek, B. A. Roe, and D. E. Berg. 1998. Analyses of the cag pathogenicity island of Helicobacter pylori. Mol. Microbiol. 28:37-53. [DOI] [PubMed] [Google Scholar]
- 3.Amieva, M. R., R. Vogelmann, A. Covacci, L. S. Tompkins, W. J. Nelson, and S. Falkow. 2003. Disruption of the epithelial apical-junctional complex by Helicobacter pylori CagA. Science 300:1430-1434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Atherton, J. C., P. Cao, R. M. Peek, M. K. R. Tummuru, M. J. Blaser, and T. L. Cover. 1995. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori: association of specific VacA types with cytotoxin production and peptic ulceration. J. Biol. Chem. 270:17771-17777. [DOI] [PubMed] [Google Scholar]
- 5.Azuma, T., S. Yamaaki, A. Yamakawa, M. Ohtani, A. Muramatsu, H. Suto, M. Y. Ito, M. Dojo, Y. Yamazaki, M. Kuriyama, Y. Keida, H. Higashi, and M. Hatakeyama. 2004. Association between diversity in the Src homology 2 domain-containing tyrosine phosphatase binding site of Helicobacter pylori CagA protein and gastric atrophy and cancer. J. Infect. Dis. 189:820-827. [DOI] [PubMed] [Google Scholar]
- 6.Basso, D., C. F. Zambon, D. P. Letley, A. Stranges, A. Marchet, J. L. Rhead, S. Schiavon, G. Guariso, M. Ceroti, D. Nitti, M. Rugge, M. Plebani, and J. C. Atherton. 2008. Clinical relevance of Helicobacter pylori cagA and vacA gene polymorphisms. Gastroenterology 135:91-99. [DOI] [PubMed] [Google Scholar]
- 7.Blaser, M. J., G. I. Perez-Perez, H. Kleanthous, T. L. Cover, R. M. Peek, P. H. Chyou, G. N. Stemmermann, and A. Nomura. 1995. Infection with Helicobacter pylori strains possessing cagA is associated with an increased risk of developing adenocarcinoma of the stomach. Cancer Res. 55:2111-2115. [PubMed] [Google Scholar]
- 8.Chen, X. J., J. Yan, and Y. F. Shen. 2005. Dominant cagA/vacA genotypes and coinfection frequency of Helicobacter pylori in peptic ulcer or chronic gastritis patients in Zhejiang Province and correlations among different genotypes, coinfection and severity of the diseases. Chin. Med. J. Engl. 118:460-467. [PubMed] [Google Scholar]
- 9.Cover, T. L., and S. R. Blanke. 2005. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Microbiol. 3:320-332. [DOI] [PubMed] [Google Scholar]
- 10.Curado, M. P., B. Edwards, H. R. Shin, H. Storm, J. Ferlay, M. Heanue, and P. Boyle (ed.). 2007. Cancer incidence in five continents, vol. IX. Publication no. 160. IARC Scientific Publications, Lyon, France.
- 11.DeStefani, E., L. Fierro, E. Barrios, and A. Ronco. 1994. Cancer mortality trends in Uruguay 1953-91. Int. J. Cancer 56:634-639. [DOI] [PubMed] [Google Scholar]
- 12.Erzin, Y., V. Koksal, S. Altun, A. Dobrucali, M. Asian, S. Erdamar, A. Dirican, and B. Kocazeybek. 2006. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA, and babA2 genotypes and correlation with clinical outcome in Turkish patients with dyspepsia. Helicobacter 11:574-580. [DOI] [PubMed] [Google Scholar]
- 13.Figueiredo, C., J. C. Machado, and Y. Yamaoka. 2005. Pathogenesis of Helicobacter pylori infection. Helicobacter 10(Suppl. 1):14-20. [DOI] [PubMed] [Google Scholar]
- 14.Gebert, B., W. Fischer, E. Weiss, R. Hoffmann, and R. Haas. 2003. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science 301:1099-1102. [DOI] [PubMed] [Google Scholar]
- 15.Ghose, C., G. I. Perez-Perez, L. J. van Doorn, M. G. Domínguez-Bello, and M. J. Blaser. 2005. High frequency of gastric colonization with multiple Helicobacter pylori strains in Venezuelan subjects. J. Clin. Microbiol. 43:2635-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Initiative for Vaccine Research. 2008. Helicobacter pylori. World Health Organization, Geneva, Switzerland. http://www.who.int/vaccine_research/diseases/soa_bacterial/en/index1.html.
- 17.Letley, D. P., A. Lastovica, J. A. Louw, C. J. Hawkey, and J. C. Atherton. 1999. Allelic diversity of the Helicobacter pylori vacuolating cytotoxin gene in South Africa: rarity of the vacA s1a genotype and natural occurrence of an s2/m1 allele. J. Clin. Microbiol. 37:1203-1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McClain, M. S., P. Cao, H. Iwamoto, A. D. Vinion-Dubiel, G. Szabo, Z. Shao, and T. L. Cover. 2001. The 12-amino-acid segment, present in type s2 but not s1 Helicobacter pylori VacA proteins, abolishes cytotoxin activity and alters membrane channel formation. J. Bacteriol. 183:6499-6508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moss, S. F., and P. Malfertheiner. 2007. Helicobacter and gastric malignancies. Helicobacter 12(Suppl. 1):23-30. [DOI] [PubMed] [Google Scholar]
- 20.Nimri, L. F., I. Matalka, K. Bani Hani, and M. Ibrahim. 2006. Helicobacter pylori genotypes identified in gastric biopsy specimens from Jordanian patients. BMC Gastroenterol. 6:27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nomura, A. M. Y., G. I. Perez-Perez, J. Lee, G. Stemmermann, and M. J. Blaser. 2002. Relation between Helicobacter pylori cagA status and risk of peptic ulcer disease. Am. J. Epidemiol. 155:1054-1059. [DOI] [PubMed] [Google Scholar]
- 22.Pagliaccia, C., M. de Bernard, P. Lupetti, X. Ji, D. Burroni, T. L. Cover, E. Papini, R. Rappuoli, J. L. Telford, and J. M. Reyrat. 1998. The m2 form of the Helicobacter pylori cytotoxin has cell type-specific vacuolating activity. Proc. Natl. Acad. Sci. U. S. A. 95:10212-10217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parsonnet, J., M. Replogle, S. Yang, and R. Hiatt. 1997. Seroprevalence of CagA-positive strains among Helicobacter pylori-infected, healthy young adults. J. Infect. Dis. 175:1240-1242. [DOI] [PubMed] [Google Scholar]
- 24.Prinz, C., S. Schwendy, and P. Voland. 2006. H. pylori and gastric cancer: shifting the global burden. World J. Gastroenterol. 12:5458-5464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rhead, J. L., D. P. Letley, M. Mohammadi, N. Hussein, M. A. Mohagheghi, M. E. Hosseini, and J. C. Atherton. 2007. A new Helicobacter pylori vacuolating cytotoxin determinant, the intermediate region, is associated with gastric cancer. Gastroenterology 133:926-936. [DOI] [PubMed] [Google Scholar]
- 26.Saribasak, H., B. A. Salih, Y. Yamaoka, and E. Sander. 2004. Analysis of Helicobacter pylori genotypes and correlation with clinical outcome in Turkey. J. Clin. Microbiol. 42:1648-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Segal, E. D., J. Cha, J. Lo, S. Falkow, and L. S. Tompkins. 1999. Altered states: involvement of phosphorylated CagA in the induction of host cellular growth changes by Helicobacter pylori. Proc. Natl. Acad. Sci. U. S. A. 96:14559-14564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Straus, E. W., H. Patel, J. Chang, R. M. Gupta, V. Sottile, J. Scirica, G. Tarabay, S. Iyer, S. Samuel, and R. D. Raffaniello. 2002. Helicobacter pylori infection and genotyping in patients undergoing upper endoscopy at inner city hospitals. Dig. Dis. Sci. 47:1575-1581. [DOI] [PubMed] [Google Scholar]
- 29.Tsutsumi, R., H. Higashi, M. Higuchi, M. Okada, and M. Hatakeyama. 2003. Attenuation of Helicobacter pylori CagA-SHP-2 signaling by interaction between CagA and C-terminal Src kinase. J. Biol. Chem. 278:3664-3670. [DOI] [PubMed] [Google Scholar]
- 30.Wong, B. C., Y. Yin, D. E. Berg, H. H. Xia, J. Z. Zhang, W. H. Wang, W. M. Wong, X. R. Huang, V. S. Tang, and S. K. Lam. 2001. Distribution of distinct vacA, cagA, and iceA alleles in Helicobacter pylori in Hong Kong. Helicobacter 6:317-324. [DOI] [PubMed] [Google Scholar]
- 31.Yilmaz, O., N. Sen, A. A. Küpelioğlu, and I. Simçsek. 2006. Detection of Helicobacter pylori infection by ELISA and Western blot techniques and evaluation of anti-CagA seropositivity in adult Turkish dyspeptic patients. World J. Gastroenterol. 12:5375-5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou, W., S. Yamazaki, A. Yamakawa, M. Ohtani, Y. Ito, Y. Keida, H. Higashi, M. Hatakeyama, J. Si, and T. Azuma. 2004. The diversity of vacA and cagA genes of Helicobacter pylori in East Asia. FEMS Immunol. Med. Microbiol. 40:81-87. [DOI] [PubMed] [Google Scholar]