Abstract
Cap hydrolysis is a critical control point in the life of eukaryotic mRNAs and is catalyzed by the evolutionarily conserved Dcp1-Dcp2 complex. In Saccharomyces cerevisiae, decapping is modulated by several factors, including the Lsm family protein Edc3, which directly binds to Dcp2. We show that Edc3 binding to Dcp2 is mediated by a short peptide sequence located C terminal to the catalytic domain of Dcp2. This sequence is required for Edc3 to stimulate decapping activity of Dcp2 in vitro, for Dcp2 to efficiently accumulate in P-bodies, and for efficient degradation of the RPS28B mRNA, whose decay is enhanced by Edc3. In contrast, degradation of YRA1 pre-mRNA, another Edc3-regulated transcript, occurs independently from this region, suggesting that the effect of Edc3 on YRA1 is independent of its interaction with Dcp2. Deletion of the sequence also results in a subtle but significant defect in turnover of the MFA2pG reporter transcript, which is not affected by deletion of EDC3, suggesting that the region affects some other aspect of Dcp2 function in addition to binding Edc3. These results raise a model for Dcp2 recruitment to specific mRNAs where regions outside the catalytic core promote the formation of different complexes involved in mRNA decapping.
mRNA degradation plays an important role in the control of gene expression. Removal of the 5′ N7-methylguanosine (m7G) cap structure is a critical step in several mRNA decay pathways and occurs following deadenylation in one of the major pathways of mRNA degradation (30). Decapping is catalyzed by a decapping holoenzyme consisting of the catalytic Dcp2 subunit and the coactivating Dcp1, which are conserved throughout eukaryotes (16). Dcp2 contains a Nudix/MutT domain, which binds to both the m7G cap and mRNA body and is critical for decapping catalysis (12, 28, 38, 39). Recent kinetic and structural studies using the Dcp proteins from Saccharomyces cerevisiae and Schizosaccharomyces pombe have indicated that Dcp1 strongly stimulates Dcp2 catalytic activity by binding the N-terminal region of Dcp2 (residues 1 to 101 in S. cerevisiae) and inducing a conformational change in Dcp2 so that the N-terminal region is brought to the proximity of the C-terminal catalytic domain (residues 102 to 245 in S. cerevisiae), which contains the Nudix/MutT motif (10, 33, 34).
In principle, three sequential events are required for mRNA engaged in translation to be eventually routed to decapping. First, the eukaryotic initiation factor eIF4E that protects the m7G cap should be displaced from the mRNA (32). Subsequently, the Dcp1-Dcp2 decapping complex needs to be recruited onto the mRNA. Finally, the decapping complex is activated to catalyze the removal of the m7G cap structure. Several factors are thought to function in these processes to promote decapping, including the DEAD-box helicase RCK/p54/Me31B/CGH-1/Dhh1, the RNA binding Lsm1-7 complex, the RNA binding protein Pat1, and the Lsm family protein Edc3, all of which are evolutionarily conserved (16). In yeast, Dhh1 and Pat1 appear to promote decapping at least in part by eliciting translational repression, which either directly or indirectly involves displacement of eIF4E (6). Other proteins appear to affect decapping by directly activating the decapping enzyme. For example, the Edc1 and Edc2 proteins can directly activate the decapping enzyme by unknown mechanisms (13, 31, 35). Similarly, the Edc3 protein does not appear to function in translation repression and instead directly binds and activates Dcp2 (P. Rajyaguru, T. Nissan, and R. Parker, submitted for publication).
In Saccharomyces cerevisiae, Edc3 is dispensable for efficient decapping of certain reporter transcripts (24) and also for maintenance of steady-state levels of most transcripts (1, 11). However, Edc3 becomes critical for efficient mRNA degradation of reporter transcripts when the decapping enzyme is compromised by mutations either in Dcp1 or in Dcp2 (24). Deletion of EDC3 also causes a synthetic growth defect when combined with another Lsm protein, Scd6 (9, 40), suggesting some redundancy in Edc3 function. Edc3 is also required for the specific degradation of some mRNAs. For example, the efficient degradation of the RPS28B mRNA and YRA1 pre-mRNAs requires Edc3 (1, 11). Moreover, in Drosophila S2 cells, Edc3 is required for the degradation of a subset of microRNA (miRNA) targets (14). Edc3 contains three functional domains: an N-terminal divergent Lsm (Sm-like) domain, which binds Dcp2 (8; Rajyaguru et al., submitted), a central FDF domain, which binds Dhh1 (8, 37), and a C-terminal YjeF-N domain, which self-interacts and promotes Edc3 dimerization (8, 26). Taken together, these results indicate that the Edc3 is a component of the decapping machinery that can both generally affect the activity of the decapping enzyme and be recruited for the regulation of specific mRNAs. An unresolved question is how Edc3 interacts with Dcp2 and the consequences of that interaction for general mRNA decapping and the control of specific mRNAs.
In this work, we demonstrate that a short peptide sequence just C terminal of the Nudix domain of Dcp2 interacts with Edc3. This peptide region is required for the binding of Dcp2 to the Lsm-FDF domain of Edc3, for the stimulation of Dcp1/Dcp2 decapping activity by Edc3 in vitro, and for efficient accumulation of Dcp2 into P-bodies (PBs) in vivo. Deletion of the peptide sequence, as well as mutations at residues within this region, causes a significant upregulation of a known substrate of Edc3-mediated decay, RPS28B mRNA. Surprisingly, degradation of the YRA1 pre-mRNA, whose decay is enhanced by Edc3, is not affected by removal of or mutations in this region, suggesting that the effect of Edc3 on YRA1 is independent of its interaction with Dcp2. Deletion of this region also results in a subtle but significant defect in decapping of the MFA2pG reporter mRNA, whereas the transcript is not noticeably affected by deletion of EDC3. This result implies that this region of Dcp2 has additional functions other than Edc3 binding and may represent an important site for the binding of additional decapping regulators.
MATERIALS AND METHODS
Yeast strains.
Yeast strains used in this study are listed in Table 1. All strains are in the yRP840/yRP841 background (19). Strains yRP2745, yRP2746, yRP2749, yRP2751, yRP2752, and yRP2753 were constructed from yRP841 by a PCR-based method using oligonucleotides oRP1450, oRP1449, oRP1461, oRP1463, oRP1464, oRP1462, and oRP1451 (27). Strains yRP2747 and yRP2748 were constructed from yRP1515 using oligonucleotides oRP1450, oRP1449, and oRP1451. Strains yRP2793 and yRP2794 were constructed from a cross between yRP1745 and yRP2792.
TABLE 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| yRP840 | MATaleu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG | 19 |
| yRP841 | MATαleu2-3,112 trp1 ura3-52 lys2-201 cup1::LEU2/PGK1pG/MFA2pG | 19 |
| yRP1346 | MATaleu2-3,112 trp1 ura3-52 his4-539 lys2-201 cup1::LEU2/PGK1pG/MFA2pG dcp2Δ::TRP1 | 12 |
| yRP1515 | MATαleu2-3,112 trp1 ura3-52 his4-539 lys2-201 cup1::LEU2/PGK1pG/MFA2pG dcp1-2::TRP1 | 12 |
| yRP1745 | MATaleu2-3,112 trp1 ura3-52 his4-539 cup1::LEU2/PGK1pG/MFA2pG edc3Δ::kanMX6 | 24 |
| yRP2745 | MATαleu2-3,112 trp1 ura3-52 lys2-201 cup1::LEU2/PGK1pG/MFA2pG dcp2(Δ301-970)::kanMX6 | This study |
| yRP2746 | MATαleu2-3,112 trp1 ura3-52 lys2-201 cup1::LEU2/PGK1pG/MFA2pG dcp2(Δ248-970)::kanMX6 | This study |
| yRP2747 | MATαleu2-3,112 trp1 ura3-52 his4-539 lys2-201 cup1::LEU2/PGK1pG/MFA2pG dcp1-2::TRP1 dcp2(Δ301-970)::kanMX6 | This study |
| yRP2748 | MATαleu2-3,112 trp1 ura3-52 his4-539 lys2-201 cup1::LEU2/PGK1pG/MFA2pG dcp1-2::TRP1 dcp2(Δ248-970)::kanMX6 | This study |
| yRP2749 | MATαleu2-3,112 trp1 ura3-52 lys2-201 cup1::LEU2/PGK1pG/MFA2pG DCP2::kanMX6 | This study |
| yRP2751 | MATαleu2-3,112 trp1 ura3-52 lys2-201 cup1::LEU2/PGK1pG/MFA2pG DCP2-3HA::kanMX6 | This study |
| yRP2752 | MATαleu2-3,112 trp1 ura3-52 lys2-201 cup1::LEU2/PGK1pG/MFA2pG dcp2(Δ301-970)-3HA::kanMX6 | This study |
| yRP2753 | MATαleu2-3,112 trp1 ura3-52 lys2-201 cup1::LEU2/PGK1pG/MFA2pG dcp2(Δ248-970)-3HA::kanMX6 | This study |
| yRP2792 | MATαleu2-3,112 trp1 ura3-52 lys2-201 cup1::LEU2/PGK1p2/MFA2pG dcp2Δ::TRP1 | This study |
| yRP2793 | MATaleu2-3,112 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp2Δ::TRP1 | This study |
| yRP2794 | MATaleu2-3,112 trp1 ura3-52 cup1::LEU2/PGK1pG/MFA2pG dcp2Δ::TRP1 edc3Δ::kanMX6 | This study |
Plasmids.
Plasmids used in this study are listed in Table 2. Plasmids pRP1891, pRP1892, and pRP1893 were constructed in multiple steps. First, pRP1667 (a gift from C. J. Decker) was digested with restriction enzymes SacI and HindIII to isolate the DNA fragment containing the GPD promoter, the NruI restriction site, a green fluorescent protein (GFP) open reading frame, and the ADH1 terminator. The fragment was cloned into corresponding sites in pRS416 to generate pRP1902. DNA fragments containing the 514-bp upstream region of DCP2 and a sequence encoding residues 1 to 970, 1 to 300, and 1 to 247 of Dcp2 were PCR amplified from the genome using a forward primer oRP1440 and reverse primers oRP1441, oRP1442, and oRP1443. The resultant fragments were digested with restriction enzymes SacI and BamHI and cloned into corresponding sites in pRP1902 to generate pRP1891, pRP1892, and pRP1893.
TABLE 2.
Plasmids used in this study
| Plasmid | Description | Source or reference |
|---|---|---|
| pRP250 | pRS416, centromeric vector with the URA3 marker | Lab stock |
| pRP1667 | pRS425 (lab stock) carrying a sequence encoding GFP and the ADH1 terminator | C. J. Decker |
| pRP1902 | pRS416 carrying a sequence encoding GFP and the ADH1 terminator | This study |
| pRP1903 | pRP416 carrying the DCP2 promoter, a sequence encoding GFP, and the ADH1 terminator | This study |
| pRP1891 | pRS416 carrying the DCP2 promoter, a sequence encoding Dcp2-GFP, and the ADH1 terminator | This study |
| pRP1892 | pRS416 carrying the DCP2 promoter, a sequence encoding Dcp2(1-300)-GFP, and the ADH1 terminator | This study |
| pRP1893 | pRS416 carrying the DCP2 promoter, a sequence encoding Dcp2(1-247)-GFP, and the ADH1 terminator | This study |
| pRP1894 | pRS416 carrying the DCP2 promoter, a sequence encoding Dcp2(243-970)-GFP, and the ADH1 terminator | This study |
| pRP1895 | pRS416 carrying the DCP2 promoter, a sequence encoding Dcp2(327-970)-GFP, and the ADH1 terminator | This study |
| pRP1278 | YCplac33 (18) carrying the DCP2 promoter, a sequence encoding Dcp2(Δ2-101)-GFP and the ADH1 terminator | C. J. Decker |
| pRP1904 | pRS416 carrying the DCP2 promoter, a sequence encoding Dcp2(102-300)-GFP and the ADH1 terminator | This study |
| pRP1905 | pRS416 carrying the DCP2 promoter, a sequencne encoding Dcp2(102-247)-GFP and the ADH1 terminator | This study |
| pRP1896 | pRS416 carrying the DCP2 promoter, a sequence encoding Dcp2(1-300) L255A K256A-GFP, and the ADH1 terminator | This study |
| pRP1906 | pRS416 carrying the DCP2 promoter, a sequence encoding Dcp2(1-300) R271A K273A-GFP and the ADH1 terminator | This study |
| pRP1907 | pRS416 carrying the DCP2 promoter, a sequence encoding Dcp2(1-300) L255A K256A R271A K273A-GFP, and the ADH1 terminator | This study |
| pRP1908 | pRS416 carrying the DCP2 promoter, a sequence encoding Dcp2(1-300) E252A-GFP, and the ADH1 terminator | This study |
| pRP1909 | pRS416 carrying the DCP2 promoter, a sequence encoding Dcp2(1-300) D268A-GFP, and the ADH1 terminator | This study |
| pRP1910 | pRS416 carrying the DCP2 promoter, a sequence encoding Dcp2(1-300) E252A D268A-GFP, and the ADH1 terminator | This study |
| pRP784 | pPROEX-1 | GibcoBRL |
| pRP1897 | pPROEX-1 carrying a sequence encoding Dcp2(1-300), to express N-terminal His-tagged protein in E. coli | This study |
| pRP1898 | pPROEX-1 carrying a sequence encoding Dcp2(1-247), to express N-terminal His-tagged protein in E. coli | This study |
| pRP1899 | pPROEX-1 carrying a sequence encoding Dcp2(1-300) L255A K256A, to express N-terminal His-tagged protein in E. coli | This study |
| pRP1900 | pGEX-6P-3, to express GST in E. coli | GE Healthcare |
| pRP1444 | pGEX-6P-3 carrying a sequence encoding the Lsm-FDF domain (aa 1-231) of Edc3, to express N-terminal GST-tagged protein in E. coli | C. J. Decker |
| pRP10 | Centromeric vector with the URA3 marker | 21 |
| pRP1207 | pRP10 carrying the DCP2 promoter, a sequence encoding full-length Dcp2, and the DCP2 terminator | 33 |
| pRP1453 | pRP10 carrying the DCP2 promoter, a sequence encoding Dcp2(1-300), and the DCP2 terminator | This study |
| pRP1450 | pRP10 carrying the DCP2 promoter, a sequence encoding Dcp2(1-247), and the DCP2 terminator | This study |
| pRP1449 | pRP10 carrying the DCP2 promoter, a sequence encoding Dcp2(1-227), and the DCP2 terminator | This study |
| pRP642 | pRS200, centromeric vector with the TRP1 marker | Lab stock |
| pRP1432 | pRS200 carrying the EDC3 promoter, a sequence encoding Edc3, and the EDC3 terminator | C. J. Decker |
To generate pRP1903, a DNA fragment containing the 514-bp upstream region of DCP2 was PCR amplified from the genome using primers oRP1440 and oRP1458. The resultant fragment was digested with restriction enzymes SacI and NruI and cloned into corresponding sites in pRP1902. Note that the plasmid contains a 7-bp random sequence downstream of the NruI site.
To generate pRP1894 and pRP1895, DNA sequences encoding residues 243 to 970 and 327 to 970 of Dcp2 were amplified from the genome with the initiation codon using forward primers oRP1459 and oRP1460 and reverse primer oRP1441. The resultant fragments were digested with restriction enzymes SacI and NruI and cloned into the corresponding site in pRP1903. Note that the plasmids contain a 6-bp random sequence upstream of the NruI site.
To generate pRP1904, a DNA fragment containing the 514-bp upstream region of DCP2, the initiation codon, and a sequence encoding residues 102 to 300 of Dcp2 were PCR amplified from the plasmid pRP1278 (a gift from C. J. Decker) using primers oRP1440 and oRP1442. The resultant DNA fragment was digested with restriction enzymes SacI and BamHI and cloned into corresponding sites in pRP1902. pRP1905 was constructed essentially in the same way using primers oRP1440 and oRP1443.
pRP1896 was constructed in multiple steps. First, a DNA fragment was PCR amplified from pRP1207 (33) using the primer pair oRP1447 and oRP1442. A second PCR was performed using the primer pair oRP1440 and oRP1448. The two PCR fragments were then mixed and fused by PCR amplification using primers oRP1440 and oRP1442. The resultant fragment was digested with restriction enzymes SacI and BamHI and cloned into corresponding sites in pRP1902. pRP1906, pRP1908, and pRP1909 were constructed essentially in the same way.
pRP1907 was constructed in multiple steps. First, a DNA fragment was PCR amplified from pRP1207 (33) using the primer pair oRP1467 and oRP1442. A second PCR was performed using the primer pair oRP1440 and oRP1468. The two PCR fragments were then mixed and fused by PCR amplification using primers oRP1440 and oRP1442. The resultant fragment was used as a template in PCR amplification using the primer pair oRP1465 and oRP1442. The resultant fragment was then mixed with another fragment amplified from pRP1207 (33) using oRP1466 and oRP1440, and the two fragments were fused by PCR using primers oRP1440 and oRP1442. The resultant fragment was digested with restriction enzymes SacI and BamHI and cloned into corresponding sites in pRP1902. pRP1910 was constructed essentially in the same way.
To generate pRP1897 and pRP1898, DNA fragments encoding residues 1 to 300 and 1 to 247 of Dcp2 were PCR amplified from pRP1207 (33) using forward primer oRP1444 and reverse primers oRP1445 and oRP1446, respectively; digested with restriction enzymes NdeI and BamHI; and cloned into corresponding sites in pRP784 (pPROEX-1; GibcoBRL).
Plasmids pRP1453, pRP1450, and pRP1449 were constructed in multiple steps. First, the DNA fragment containing the 3′ region of the DCP2 gene was PCR amplified from pRP1207 (33) using primers oRP1471 and oRP1472, digested with restriction enzymes SacI and BamHI, and cloned into corresponding sites in pRP10 (21), to generate pRP1454. Second, the DNA fragment containing the promoter region of DCP2 and N-terminal portions of the coding region (residues 1 to 300, 1 to 247, and 1 to 227, designated hereafter as regions 1-300, 1-247, and 1-227, respectively) was PCR amplified using forward primer oRP1473 and reverse primers oRP1474, oRP1475, and oRP1476; digested with restriction enzymes SacI and BglII; and cloned into corresponding sites in pRP1454.
To generate pRP1899, a DNA fragment was PCR amplified from pRP1896 using primers oRP1444 and oRP1445, digested with restriction enzymes NdeI and BamHI, and cloned into corresponding sites in pRP784.
Oligonucleotides.
Oligonucleotides used in this study are listed in Table S1 in the supplemental material.
Cell growth conditions.
For the microscopy and Western blot analysis shown here in Fig. 1B and C and 4B and C, as well as Fig. S1A and S1B in the supplemental material, cells were grown at 30°C in synthetic complete (SC) medium lacking uracil but containing 2% glucose as a carbon source, until an optical density at 600 nm (OD600) of 0.3 to 0.35 was reached. Cells were subsequently harvested by centrifugation, washed, resuspended in SC medium lacking uracil and a carbon source, and incubated in a shaking water bath at 30°C for 10 min. For the Northern blot analysis shown here in Fig. 5, 6 to D, and 7, as well as Fig. S4, S5B, and S6B in the supplemental material, cells were grown at 30°C in SC medium lacking uracil or tryptophan but containing 2% galactose until an OD600 of 0.3 to 0.35 was reached. For Western and Northern blot analyses shown in Fig. S3A, S3B, S3C, S3D, and S7 in the supplemental material, cells were grown at 24°C in YP medium (1% yeast extract and 2% peptone) containing 2% galactose until an OD600 of 0.3 to 0.35 was reached.
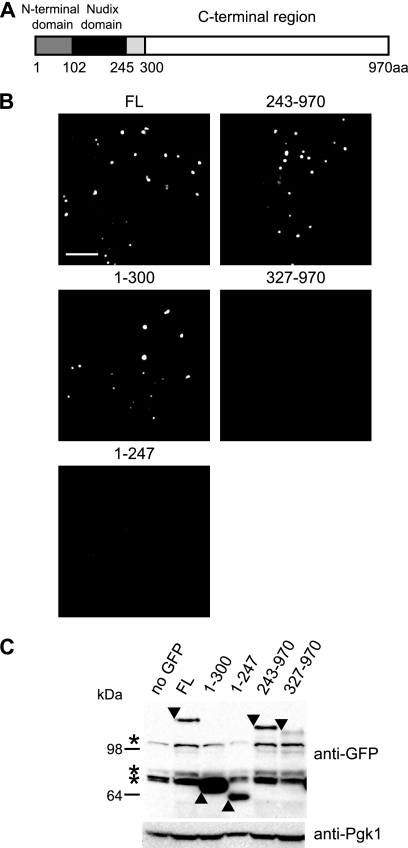
FIG. 1.
Region 248-300 of Dcp2 mediates its accumulation in P-bodies. (A) Domain architecture of S. cerevisiae Dcp2. The N-terminal globular domain is shown in gray. The Nudix domain is shown in black. Region 248-300 of Dcp2 is shown in light gray. (B) Localization of truncated Dcp2 proteins. Wild-type cells (yRP840) expressing the full-length (FL) or truncated Dcp2 protein (1-300, 1-247, 243-970, or 327-970) C-terminally tagged with GFP from plasmid pRP1891, pRP1892, pRP1893, pRP1894, or pRP1895 were used. Cells were cultured to mid-log phase and shifted to media lacking glucose for 10 min. A single focal plane image is shown. Bar, 5 μm. (C) Levels of the truncated Dcp2 proteins. The same strains as those described in the legend to panel B were cultured to mid-log phase and shifted to media lacking glucose for 10 min. pRP250 was used as a control vector (no GFP). Anti-GFP antibody was used in Western blot to detect GFP-tagged proteins. Anti-Pgk1 antibody was used to detect the endogenous Pgk1 protein as a loading control. Arrowheads indicate GFP-tagged proteins. Asterisks indicate nonspecific bands.
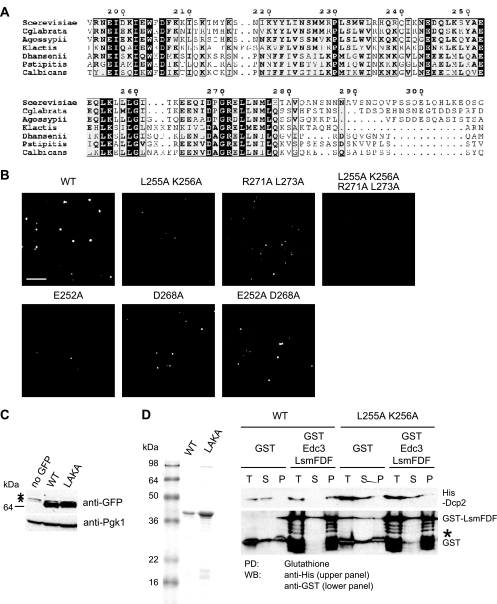
FIG. 4.
Mutations in region 248-300 of Dcp2 impair interaction between Dcp2 and Edc3. (A) Conservation of the region outside the catalytic core of Dcp2 across the seven Dcp2 fungal homologs (Saccharomyces cerevisiae, Candida glabrata, Ashbya gossypii, Kluveromyces waltii, Debaryomyces hansenii, Pichia stipitis, and Candida albicans). The numbers refer to residues of S. cerevisiae Dcp2. (B) Localization of Dcp2(1-300) with mutations. A wild-type (WT) strain (yRP840) expressing C-terminal GFP-tagged Dcp2(1-300) with or without the mutation (L255A K256A, R271A L273A, L255A K256A R271A L273A, E252A, D268A or E252A D268A) from plasmid pRP1892, pRP1896, pRP1906, pRP1907, pRP1908, pRP1909, or pRP1910 was cultured as described in the legend to Fig. 1B. A single focal image is shown. Bar, 5 μm. (C) Level of Dcp2 protein with the L255A K256A mutation. Wild-type cells (yRP840) expressing C-terminal GFP-tagged Dcp2(1-300) with or without the mutation from plasmid pRP1892 or pRP1896 were cultured to mid-log phase and shifted to media lacking glucose for 10 min. pRP250 was used as a control vector (no GFP). Anti-GFP antibody was used in the Western blot (WB) to detect GFP-tagged proteins. Anti-Pgk1 antibody was used to detect the endogenous Pgk1 protein as a loading control. Arrowheads indicate GFP-tagged proteins. Asterisks indicate nonspecific bands. (D) Coomassie blue-stained SDS-PAGE showing purified recombinant proteins used in the pulldown (PD) assay (left panel). An in vitro pulldown assay between purified GST-tagged Lsm-FDF domain of Edc3 and His-tagged Dcp2 (amino acids 1 to 300) or His-Dcp2(1-300) L255A K256A (right panel) is shown. The asterisk indicates partial degradation products of GST-Lsm-FDF.
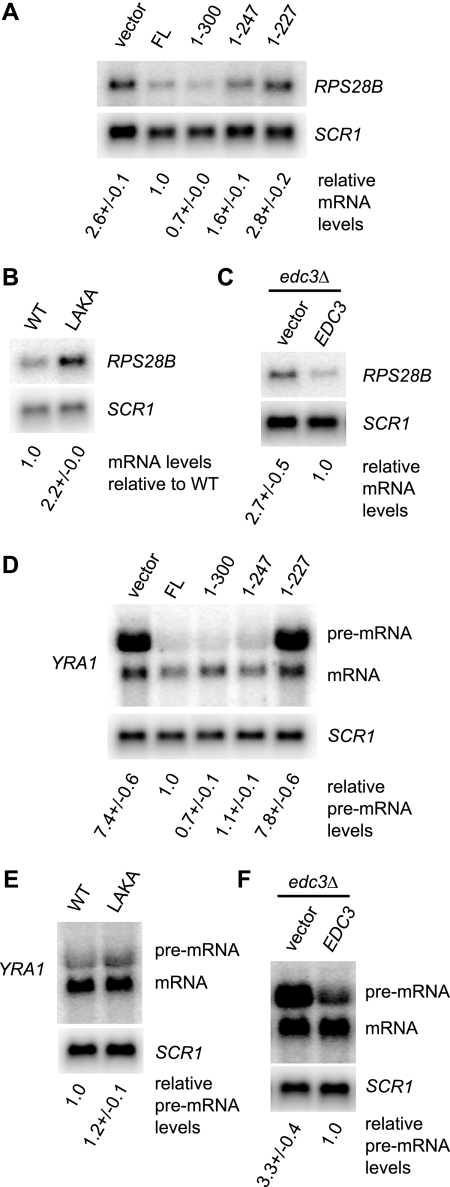
FIG. 5.
Region 248-300 of Dcp2 is required for efficient degradation of RPS28B mRNA but not for degradation of YRA1 pre-mRNA. (A) The RPS28B mRNA was analyzed in strain yRP1346 (dcp2Δ) expressing the full-length (FL) or truncated Dcp2 protein (1-300, 1-247, or 1-227) from plasmid pRP1207, pRP1453, pRP1450, or pRP1449. pRP10 was used as a control vector. Cells were grown in SC medium containing 2% galactose at 30°C. The RPS28B level normalized to the SCR1 RNA level in each strain relative to the normalized mRNA level in the strain expressing full-length Dcp2 is indicated below each lane. The average and standard deviation of values obtained with three independent transformants are shown. (B) RPS28B mRNA was analyzed in yRP1346 (dcp2Δ) expressing Dcp2(1-300)-GFP or Dcp2(1-300) L255A K256A-GFP from plasmid pRP1892 or pRP1896. Cells were grown in SC medium containing 2% galactose at 30°C. The mRNA level normalized to the SCR1 RNA level relative to the normalized mRNA level in the strain expressing wild-type Dcp2 is indicated. (C) Upregulation of the RPS28B mRNA by deletion of EDC3. Cells of strain yRP1745 (edc3Δ) complemented by wild-type Edc3 expressed from plasmid pRP1432 were grown in SC medium containing 2% galactose to mid-log phase at 30°C. pRP642 was used as a control vector. The mRNA level normalized to the SCR1 RNA level relative to the normalized mRNA level in the strain expressing wild-type Edc3 is indicated. (D) Analysis of the YRA1 pre-mRNA. The same RNA samples as described for panel A were analyzed. The YRA1 pre-mRNA level normalized to the SCR1 RNA level in each strain relative to the normalized pre-mRNA level in the strain expressing full-length Dcp2 is indicated below each lane. (E) The same RNA samples as described in the legend to panel B were analyzed. The YRA1 pre-mRNA level normalized to the SCR1 RNA level in each strain relative to the normalized pre-mRNA level in the strain expressing wild-type Dcp2 is indicated below each lane. (F) The same RNA samples as described for panel C were analyzed. The YRA1 pre-mRNA level normalized to the SCR1 RNA level in each strain relative to the normalized pre-mRNA level in the strain expressing wild-type Edc3 is indicated below each lane.
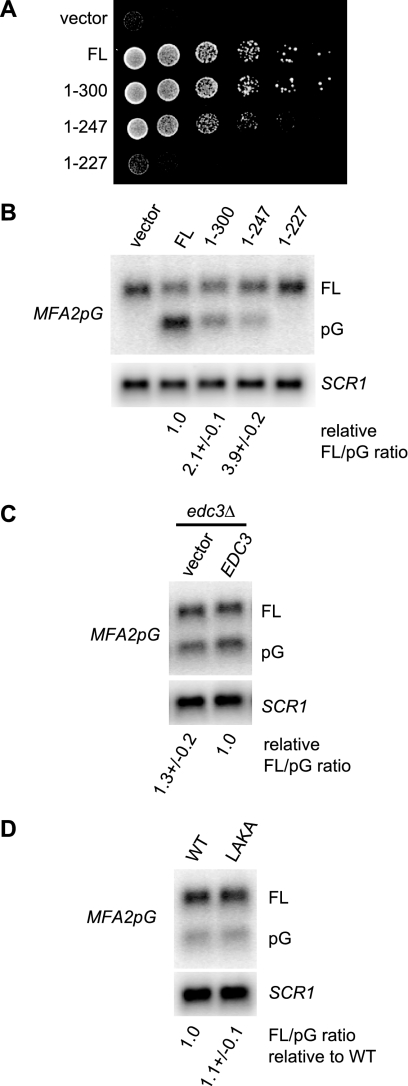
FIG. 6.
Region 248-300 of Dcp2 promotes decapping in vivo. (A) Deletion of region 248-300 causes a subtle but consistent growth defect. The growth phenotype of yRP1346 (dcp2Δ) expressing the full-length (FL) or truncated Dcp2 protein (1-300, 1-247, or 1-227) from plasmid pRP1207, pRP1453, pRP1450, or pRP1449 was analyzed by serial dilution spotting. pRP10 was used as a control vector. Cells were incubated on SC medium containing 2% glucose for 2 days at 30°C. (B) Region 248-300 of Dcp2 promotes decapping in vivo. Decay intermediates (pG) of the MFA2pG reporter mRNA were analyzed in the same strains described in the legend to Fig. 5A. Cells were grown in SC medium containing 2% galactose at 30°C to express the MFA2pG mRNA from the GAL promoter. “FL” and “pG” represent the full-length mRNA and the poly(G) decay intermediate, respectively. The ratio of the full-length mRNA to the poly(G) decay intermediate in each strain relative to the ratio in the strain expressing full-length Dcp2 is indicated below each lane. The average and standard deviation of values obtained with three independent transformants are shown. The SCR1 RNA was detected as a loading control. (C) Deletion of EDC3 does not affect the MFA2pG mRNA. The same RNA samples as those described in the legend to Fig. 5C were analyzed. The ratio of full-length mRNA to the poly(G) decay intermediate in each strain relative to the ratio in the strain expressing wild-type Edc3 is indicated below each lane. (D) The LAKA mutation does not affect the MFA2pG mRNA. The same RNA samples as those described in the legend to Fig. 5B were analyzed. The ratio of the full-length mRNA to the poly(G) decay intermediate in each strain relative to the ratio in the strain expressing wild-type Dcp2 is indicated below each lane.
FIG. 7.
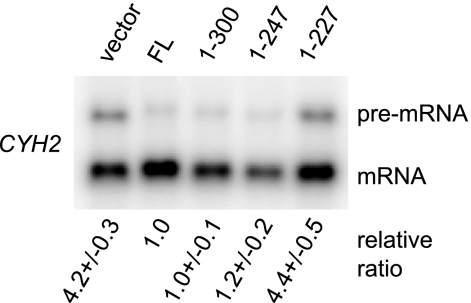
Region 248-300 of Dcp2 is dispensable for NMD. The same RNA samples as those described in the legend to Fig. 5A were analyzed. To assess relative accumulation, the CYH2 pre-mRNA/mRNA ratio was normalized to the ratio in the dcp2Δ strain expressing the full-length Dcp2 protein. Values represent the average and standard deviation of fold changes obtained with three independent transformants.
Microscopy.
Microscopy was performed essentially as described previously (2). All images were acquired using a Deltavision RT microscope system running softWoRx 3.5.1 software (Applied Precision, LLC), using an objective (UPlan sapo 100×, 1.4 NA; Olympus). They were collected using software (SoftWoRx) as 1,024- by 1,024-pixel files with a camera (CoolSNAP HQ, Photometrics) using 1-by-1 binning. A Z-series with 10 to 11 image planes and a step size of 0.8 μm was taken. All images were deconvolved using standard softWoRx deconvolution algorithms (enhanced ratio, medium noise filtering, 10 cycles). ImageJ (National Institute of Health) was used to adjust all images to equal contrast ranges.
Protein purification, in vitro protein-protein interaction assays, and Western blot analysis.
His-tagged Dcp2(1-300), Dcp2(1-247), and Dcp2(1-300) harboring the L255A K256A double mutation were expressed in Escherichia coli BL21 from plasmids pRP1897, pRP1898, and pRP1899. Purification of His-Dcp2(1-300) and His-Dcp2(1-247) was performed essentially as described previously (33). The glutathione S-transferase (GST)-tagged Lsm-FDF domain of Edc3 was expressed in E. coli BL21 from plasmid pRP1444 (a gift from C. J. Decker) and was purified using glutathione Sepharose 4B (GE Healthcare) after lysis in phosphate-buffered saline (PBS) buffer containing 1 mM dithiothreitol (DTT), 1% Triton X-100, 0.1 mM phenylmethylsulfonyl fluoride (PMSF), and complete protease inhibitor (Roche).
Binding experiments were performed at 4°C in 400 μl of binding buffer (50 mM HEPES, pH 7.0, 100 mM NaCl, 1 mM DTT, 2 mM MnCl2, 2 mM MgCl2, 1% Triton X-100, 10% glycerol, and 10 mg/ml bovine serum albumin [BSA]), essentially as described previously (8). Each protein was added to the binding reaction at a final concentration of approximately 0.5 μM. Glutathione Sepharose 4B was used to pull down GST-Lsm-FDF. Western analysis was performed by a standard method using monoclonal anti-His antibody (1:2,000; Novagen 70796) and polyclonal anti-GST antibody (1:2,000; Pharmacia 27-4577-01).
For Western analysis of GFP- and hemagglutinin (HA)-tagged Dcp2, total yeast extract was prepared as described previously (23). GFP- and HA-tagged versions of Dcp2 proteins were detected using monoclonal anti-GFP antibody (1:1,000; Covance MMS-118P) and monoclonal anti-HA 12CA5 antibody (1:2,500; Roche 1666606), respectively. For a loading control, the endogenous Pgk1 proteins were detected using monoclonal anti-Pgk1 antibody (1:2,000; Molecular Probes A-6457).
Northern blot analysis.
Total RNA was extracted by a hot phenol method. Northern blot analysis was conducted as previously described (4). The amount of MFA2pG, CYH2, YRA1, and RPS28B mRNA was quantified by oRP140, oRP1300, oRP1456, and oRP1439, respectively, using the Typhoon phosphoimager (Molecular Dynamics). As a loading control, the signal recognition particle 7S RNA (SCR1), a stable RNA polymerase III transcript, was detected by oRP100.
In vitro decapping assays.
Decapping reactions were carried out at 4°C under single-turnover conditions where enzyme is in at least 10-fold excess of substrate. The 343-nt 32P cap-radiolabeled MFA2 RNA substrate was prepared as described previously (25). Reaction buffer consisted of 50 mM Tris-Cl, 50 mM NH4Cl, 1 mM DTT, 0.1 mg/ml RNase-free BSA, and 0.01% NP-40 (pH 8.3) containing roughly 0.05 to 5 nM substrate. Assays were performed using 50 nM Dcp1-Dcp2(1-245) and Dcp1-Dcp2(1-315) in complex with 500 nM His-Edc3 (see Fig. S2 in the supplemental material). Decapping complexes in the presence or absence of Edc3 were incubated at 4°C for 30 min prior to initiation of the reactions. Time courses were monitored and analyzed as described previously (22). Dcp1-Dcp2(1-245) and Dcp1-Dcp2(1-315) were coexpressed in E. coli and purified as described previously (10). The constructs contain an N-terminal GB1 tag on Dcp1 and an N-terminal His tag on Dcp2.
RESULTS
Residues 248 to 300 of Dcp2 mediate its accumulation in P-bodies.
To identify the important functional domains of Dcp2, we first examined the ability of Dcp2 proteins harboring a series of deletions in the N terminus and/or C terminus to assemble into P-bodies (PBs). Although yeast Dcp2 consists of 970 residues, it has been shown that residues 1 to 300 are sufficient for decapping in vivo (12). Moreover, the N-terminal globular domain and the Nudix domain contained within residues 1 to 245 are known to be sufficient for decapping activity in vitro (Fig. 1A) (10). The truncated Dcp2 proteins were expressed from the DCP2 promoter with a C-terminal GFP tag on low-copy centromeric plasmids in wild-type cells, in which an intact genomic copy of DCP2 is present. We then observed the location of Dcp2 during glucose depletion, which is known to induce PB formation in yeast (36).
We observed that amino acids 1 to 300 of Dcp2 were sufficient for recruitment into P-bodies, which were similar to foci formed by full-length (FL) Dcp2 (Fig. 1B). In contrast, region 1-247 of Dcp2 was significantly impaired in accumulation in foci (Fig. 1B), suggesting that residues 248 to 300 of Dcp2 are important for its targeting and/or maintenance of Dcp2 in P-bodies. Surprisingly, region 243-970 of Dcp2, which lacks almost the entire domain sufficient for decapping in vitro, could accumulate in foci, whereas region 327-970 was unable to do so (Fig. 1B). We were not able to test if region 248-300 of Dcp2 is sufficient for its PB accumulation, because region 248-300 of Dcp2 fused to GFP was not stably expressed in yeast cells (data not shown). Taken together, these observations support the idea that region 248-300 of Dcp2 contains an element that mediates its accumulation in P-bodies.
The absence of a given Dcp2 fusion protein in P-bodies could be due either to a reduction in its cellular concentration or to a loss of specific protein-protein or protein-RNA interactions. To distinguish these possibilities, we examined the fusion proteins by Western blot analysis (Fig. 1C). We did observe that the level of Dcp2(1-247)-GFP was somewhat reduced compared to that of Dcp2(1-300)-GFP. Likewise, Dcp2(327-970)-GFP was reduced compared to Dcp2(243-970)-GFP. These observations raised the possibility that these constructs retain the intrinsic property to efficiently accumulate in P-bodies but fail to do so simply due to the reduction in their cellular concentration. However, we also observed that a Dcp2(1-300) construct with a pair of point mutations (LAKA) was impaired in its accumulation in P-bodies (see Fig. 4B below) but was expressed at a level comparable to that of Dcp2(1-300)-GFP (Fig. 4C). Moreover, both regions 102-300 and 102-247 of Dcp2 tagged with GFP were expressed well (see Fig. S1A in the supplemental material), and only the GFP fusion of region 102-300 accumulated in foci (Fig. S1B). Taken together, we interpret these observations to indicate that reduction of Dcp2(1-247)-GFP in focus formation is due, at least in part, to a loss of specific interactions that are dependent on region 248-300 of the protein.
Region 248-300 of Dcp2 is required for binding to Edc3 in vitro.
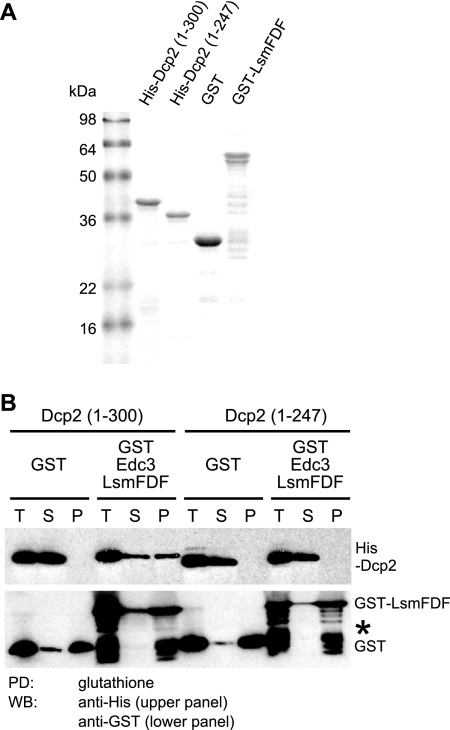
One possible explanation for why residues 248 to 300 are required for the accumulation of Dcp2 in P-bodies is that the region mediates interaction of Dcp2 with other P-body components. Because accumulation of Dcp2 in P-bodies is impaired by deletion of EDC3 and because region 1-300 of Dcp2 can interact with both the Lsm (amino acids 1 to 85) and FDF (amino acids 86 to 231) domains of Edc3 in vitro (8), we suspected that the lesion of residues 248 to 300 might affect the interaction between Dcp2 and Edc3. We tested this possibility by direct binding experiments with purified recombinant proteins (Fig. 2A).
FIG. 2.
Region 248-300 of Dcp2 is required for Edc3 binding in vitro. (A) Coomassie blue-stained SDS-PAGE showing purified recombinant proteins used in the pulldown assay. (B) In vitro pulldown (PD) assay between purified GST-tagged Lsm-FDF domain of Edc3 and His-tagged Dcp2 (amino acids 1 to 300) or His-Dcp2(1-247). Glutathione Sepharose was used to pull down either the GST-tagged Lsm-FDF of Edc3 or the GST portion alone as a control. Anti-His and anti-GST antibody were used in a Western blot (WB) to detect His-Dcp2 proteins and GST-tagged proteins, respectively. The amounts corresponding to the same volume in the binding reaction were loaded for all samples: T, total; S, supernatant; and P, pellet. The asterisk indicates partial degradation products of GST-Lsm-FDF.
We observed that a purified His-tagged Dcp2(1-247) could not be pulled down by GST-tagged Lsm-FDF of Edc3 (amino acids 1 to 231), whereas His-tagged Dcp2(1-300) could be efficiently pulled down, consistent with a previous result (Fig. 2B) (8). Together with the cytological data, the result suggests that residues 248 to 300 of Dcp2 are critical, and may be sufficient, for the interaction between Dcp2 and the decapping activator Edc3.
Region 248 to 300 of Dcp2 is required for stimulation of decapping activity by Edc3 in vitro.
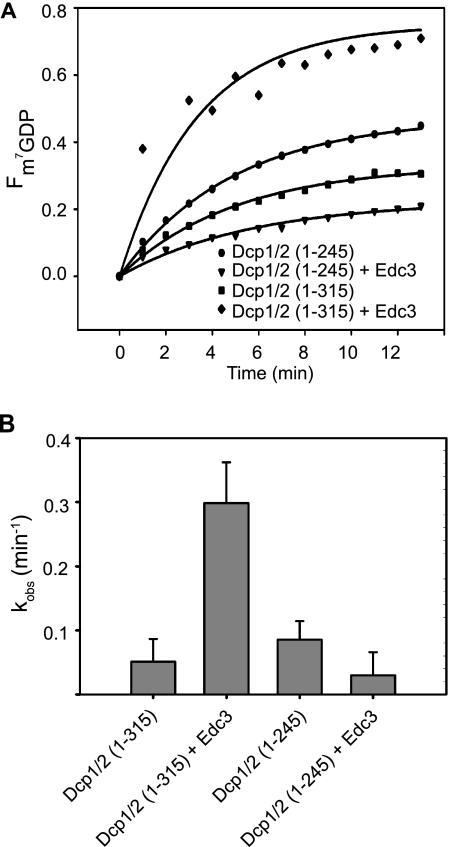
Purified Edc3 can stimulate decapping by the Dcp1-Dcp2(1-300) complex in vitro (Rajyaguru et al., submitted). If the interaction of residues 248 to 300 of Dcp2 with Edc3 is functionally important, we would predict that Dcp2 lacking this region would no longer be stimulated by Edc3 in vitro. To test this possibility, we examined the ability of recombinant Edc3 to stimulate decapping in vitro of both the Dcp1-Dcp2(1-245) complex and the Dcp1-Dcp2(1-315) complex. In a single-turnover assay (see Materials and Methods), we observed that Edc3 was unable to stimulate the activity of Dcp1-Dcp2(1-245), but did stimulate the activity of the Dcp1-Dcp2(1-315) complex (Fig. 3; see Fig. S2 in the supplemental material). We observed at least a 5-fold stimulation of activity upon addition of excess Edc3. These results are consistent with the idea that binding of Edc3 to Dcp2 through residues 248 to 300 of Dcp2 is required for Edc3 to stimulate the activity of Dcp2.
FIG. 3.
Region 246-315 of Dcp2 is required for decapping stimulation by Edc3 in vitro. (A) Time courses of the fraction of m7GDP released by decapping by recombinant purified GB1-Dcp1-His-Dcp2(1-245) and GB1-Dcp1-His-Dcp2(1-315) with and without recombinant purified His-Edc3 measured under single-turnover conditions using the 343-nt MFA2 RNA substrate. (B) Bar graphs of the observed first-order rate constants (kobs) fitted to the time courses depicted in panel A. Error bars represent the standard error.
Structural features and conservation of region 248-300.
Although yeast Dcp2 consists of 970 residues, the region outside the N-terminal globular domain and the Nudix domain (residues 1 to 245 of S. cerevisiae Dcp2) is not highly conserved among eukaryotes. To try to identify important functional residues in this region, we performed a BLAST search of DCP2 across several fungal species (Saccharomyces cerevisiae, Candida glabrata, Ashbya gossypii, Kluveromyces waltii, Debaryomyces hansenii, Pichia stipitis, and Candida albicans). This analysis revealed that the conservation extends to the 280th residue of S. cerevisiae Dcp2 and abruptly disappears in the region C terminal to the 280th residue (Fig. 4A). Interestingly, the conservation in this region appeared to be concentrated in two blocks: one from residues 245 to 261 and a second conserved region between residues 264 and 278. Consistent with the two possible significant regions of conservation, the putative homolog of Dcp2 from another fungal organism, Lodderomyces elongisporus, contains a spacer sequence that splits the homologous sequence into two parts at the 266th and 267th residues of S. cerevisiae (data not shown). Moreover, a secondary structure prediction identified two putative helical structures at residues 249 to 259 and 270 to 278 (5; data not shown).
To assess the significance of this conservation at the primary sequence level, we examined the effects of mutations at the conserved residues on accumulation of Dcp2 in P-bodies during glucose deprivation. We observed that amino acid substitutions at highly conserved residues (single mutations E252A and D268A; double mutations L255A K256A, E252A D268A, and R271A L273A; and quadruple mutation L255A K256A R271A L273A) reduced accumulation of Dcp2 in P-bodies to various extents (Fig. 4B). The expression level of the protein carrying the L255A K256A mutation, which appeared to have the greatest effect on PB accumulation, was comparable to that of the wild-type protein, suggesting that the reduction in PB accumulation was unlikely to be due to a decreased protein level (Fig. 4C). These results identify specific amino acids in this peptide region that modulate the function of this protein-protein interaction domain.
We then tested if the mutation affects the binding of Edc3 to Dcp2 in vitro. A purified His-tagged Dcp2(1-300) harboring the L255A K256A mutation was not pulled down by GST-tagged Lsm-FDF of Edc3 (Fig. 4D). This demonstrates that these residues are directly required for Edc3-Dcp2 interaction.
Dcp2-Edc3 interaction is required for control of RPS28B mRNA decay, but not for the decay of the YRA1 pre-mRNA.
The Edc3 protein has been specifically shown to be required for efficient decapping of the RPS28B mRNA and the YRA1 pre-mRNA (1, 11). In principle, those mRNA-specific roles of Edc3 could be due to its direct interaction with Dcp2 or could be due to other functions of Edc3. To determine the significance of the observed direct interaction between Edc3 and Dcp2 on mRNA metabolism and to determine how Edc3 modulates the RPS28B mRNA and the YRA1 pre-mRNA, we examined the control of these mRNAs in strains lacking region 248-300 of Dcp2 or in strains carrying the LAKA mutation in Dcp2, which is a more specific point mutation disrupting the Edc3-Dcp2 interaction, as shown above. If the effect of Edc3 on RPS28B mRNA or YRA1 pre-mRNA is modulated by its direct interaction with Dcp2, then we would expect increased levels of these mRNAs in the dcp2 mutant strains similar to those in an edc3Δ strain. In contrast, if Edc3 regulates the decay of these mRNAs independent of its direct interaction with Dcp2, then the levels of the mRNAs would be unchanged in the dcp2 mutant strains. It has been reported that deletion of DCP2 results in lethality in a certain yeast strain background (17) and causes a growth defect in other strain backgrounds (10, 12). In our analysis, we expressed either full-length or mutant Dcp2 proteins (1-300, LAKA, 1-247, and 1-227) in a viable dcp2Δ strain and examined the levels of the RPS28B or YRA1 pre-mRNA.
Consistent with previous results (1), we observed that the RPS28B mRNA was increased (2.7×) in the edc3Δ strain (Fig. 5C), demonstrating the regulation of the RPS28B mRNA levels by Edc3. Importantly, we observed an upregulation of the RPS28B mRNA in the dcp2Δ strain expressing either Dcp2(1-247) (1.6×) (Fig. 5A) or Dcp2 (LAKA) (2.2×) (Fig. 5B), whereas the mRNA level in the dcp2Δ strain expressing Dcp2(1-300) was comparable to that in the dcp2Δ strain expressing full-length Dcp2 (Fig. 5A). We also observed the same trends with strains in which C-terminal regions of the chromosomal DCP2 gene were deleted with or without C-terminal three-HA tags (see Fig. S3A in the supplemental material). The observed defect in turnover of RPS28B mRNA was unlikely to be due to a reduction in Dcp2 protein levels, since the level of the Dcp2(1-247)-3HA protein was similar to that of the Dcp2(1-300)-3HA protein (Fig. S3B). We interpret these observations to indicate that the regulation of RPS28B mRNA by Edc3 requires its interaction with Dcp2.
It is formally possible that the effects on RPS28B are due to functions of the region of Dcp2 independent of its interaction with Edc3 and that Edc3 and the region of Dcp2 act on the transcripts through parallel pathways. In principle, one can exclude this possibility by showing that deletion of EDC3 and mutations in DCP2 do not have additive effects on the levels of RPS28B mRNA. However, we were not able to address this issue, because deletion of EDC3 causes an upregulation of RPS28B mRNA to the same extent as the deletion of the DCP2 gene or expression of the functionally null Dcp2(1-227) mutant (Fig. 5A and C; see Fig. S4A and S4B in the supplemental material).
In contrast to RPS28B, the YRA1 pre-mRNA was at best only marginally upregulated by lesion of residues 248 to 300 or by the LAKA mutation (Fig. 5D and E; see Fig. S3C in the supplemental material), although its levels were increased significantly by deletion of EDC3 (Fig. 5F) (11). We interpret this observation to argue that the modulation of YRA1 pre-mRNA by Edc3 is largely independent of its direct interaction with Dcp2.
Region 248-300 of Dcp2 promotes the function of Dcp2 partly independent of its interaction with Edc3.
We noticed that strains carrying Dcp2(1-247) grow somewhat slower than strains expressing full-length Dcp2 or Dcp2(1-300), which was confirmed by serial dilution spotting of a viable dcp2Δ strain expressing either full-length or C-terminally-truncated Dcp2 proteins (1-300, 1-247, and 1-227) (Fig. 6A) and of strains containing chromosomal truncations of DCP2 either with or without three-HA tags (see Fig. S3E in the supplemental material). Since deletion of the EDC3 gene by itself does not cause a noticeable growth defect (see Fig. S5A in the supplemental material; compare EDC3 and edc3Δ expressing full-length Dcp2) (24), this observation is inconsistent with the idea that the binding to Edc3 is the sole function of region 248-300 of Dcp2. This implies that region 248-300 of Dcp2 may enhance the function of Dcp2 in part independently of Edc3.
To gain more insights into the Edc3-independent functions of region 248-300 of Dcp2, we analyzed mRNA decay of an additional decapping substrate in the presence and absence of residues 248 to 300 of Dcp2. Specifically, we used the MFA2pG reporter mRNA, which is an unstable transcript containing a highly structured poly(G) sequence that blocks 5′-to-3′ degradation (7) and whose decay rate is known to be unaffected by deletion of EDC3 alone (24). We expressed regions 1-300, 1-247, and 1-227 of Dcp2 in a dcp2Δ strain and examined the steady-state levels of the poly(G) (pG) fragment, which is a 5′-to-3′ degradation intermediate of the MFA2pG mRNA and indicative of efficient decapping (3, 19).
We observed substantial accumulation of the pG fragment in the cells carrying the region 1-300 or 1-247 of Dcp2 but not in the cells carrying region 1-227 of Dcp2 or an empty vector (Fig. 6B). Quantification of the signals revealed that deletion of residues 301 to 970 resulted in a decreased amount of the pG fragment (i.e., an increased ratio of the full-length mRNA to the fragment) and that additional deletion of residues 248 to 970 led to a further decrease in the amount of the pG fragment (Fig. 6B). We also observed the same trends in strains containing chromosomal truncations of DCP2 either with or without three-HA tags (see Fig. S3D in the supplemental material). In contrast, deletion of EDC3 did not affect the accumulation of the pG fragment (Fig. 6C; see Fig. S5B in the supplemental material), consistent with a previous observation that the half-life of the MFA2pG mRNA in edc3Δ is comparable to that in wild-type cells (24). These results suggest that both regions 248-300 and 301-970 of Dcp2 additively subtly promote decapping in vivo and are also consistent with the idea that region 248-300 of Dcp2 has additional functions beyond Edc3 binding. Consistent with this additional function being independent of Edc3, an edc3Δ Dcp2(1-247) strain showed the same effect on the MFA2pG mRNA as the Dcp2(1-247) strain by itself (see Fig. S5 in the supplemental material).
In contrast to deletion of region 248-300, the L255A K256A mutation did not noticeably affect either the growth rate or the MFA2pG mRNA, like deletion of EDC3 (Fig. 6D; see Fig. S6A and S6B in the supplemental material). Moreover, a strain with both the Dcp2-LAKA allele and the EDC3 deletion showed the same phenotype as either single mutant (Fig. S6). These results suggest that the LAKA mutation specifically impairs Edc3 binding but retains other functions of region 247-300 of Dcp2.
Region 248-300 of Dcp2 is dispensable for NMD.
The nonsense-mediated decay (NMD) pathway targets mRNAs with premature termination codons (PTCs) for rapid decay. In yeast, the decay process of NMD is initiated by decapping (29). To determine if region 248-300 of Dcp2 is required for this decay pathway, we analyzed the steady-state levels of a known NMD substrate, the CYH2 pre-mRNA, which is inefficiently spliced and contains a PTC (20). The level of the CYH2 pre-mRNA in the dcp2Δ strains expressing region 1-247 of Dcp2 was comparable to those in the strains expressing either full-length Dcp2 or region 1-300 of Dcp2 (Fig. 7), suggesting that region 248-300 of Dcp2 is dispensable for NMD. This result also supports the idea that the requirement of the region of Dcp2 is highly specific to each mRNP context.
DISCUSSION
Several observations indicate that residues 248 to 300 of Dcp2 are required for functional interaction between Dcp2 and the decapping activator Edc3 both in vitro and in vivo. First, the region is required for binding to Edc3 in vitro (Fig. 2 and 4). Second, the region is required for stimulation of decapping by Edc3 in vitro (Fig. 3). Third, the region, like Edc3, is required for efficient accumulation of Dcp2 in P-bodies (Fig. 1 and 4). Fourth, deletion of the region or mutations at residues preventing the Edc3-Dcp2 interaction (LAKA) cause upregulation of RPS28B mRNA, as does deletion of EDC3 (Fig. 5). Fifth, removal of region 248-300 of Dcp2 in strains with the hypomorphic dcp1-2 allele leads to a synergistic defect in decapping similar to that seen in edc3Δ dcp1-2 strains (see Fig. S7 in the supplemental material) (24). Taken together, these observations indicate that Edc3 interacts with this small peptide region, which allows Edc3 to activate the decapping activity of Dcp2.
An unresolved issue is exactly how binding of Edc3 to this region activates the activity of Dcp2. Region 248-300 of Dcp2 is located C terminal to the catalytic domain of Dcp2 and is not required for decapping activity (Fig. 3) (10). In the crystal structure of the Dcp1-Dcp2 complex from S. pombe, the corresponding region (residues 246 to 266) forms an alpha helix that extends along the back side of Dcp2 and connects the N- and C-terminal domains of Dcp2 (34). Although region 246-266 of S. pombe Dcp2 does not exhibit homology with S. cerevisiae Dcp2 at the primary sequence level, this structure raises several possibilities for how Edc3 binding at this site could affect activity. First, region 248-300 of Dcp2 might negatively regulate the catalytic activity normally, and binding of Edc3 might liberate Dcp2 from the inhibitory effect, although this possibility is inconsistent with the notion that the closed form of the Dcp1-Dcp2 complex is the more catalytically active form (10, 32). Moreover, this model is unlikely since it predicts that Dcp2(1-245) should be more active than Dcp2(1-315), which is not observed in comparable preparations of Dcp2 where the rates of decapping are similar (Fig. 3B). An alternative mechanism could be that Edc3 provides additional surfaces for RNA to bind to, thereby increasing the interaction of Dcp2 with the substrate. However, this appears unlikely since the Lsm domain of Edc3 is sufficient to activate Dcp2 and yet does not appear by itself to have RNA binding activity (26; Rajyaguru et al., submitted). One of the remaining possibilities is that binding of Edc3 to the region stabilizes the closed and active configuration of Dcp2. Future mechanistic studies will be required to test this possibility and to determine the details of this interaction and activation of decapping activity.
Several observations suggest that region 248-300 of Dcp2 might interact with other regulators of decapping. First, deletion of residues 248 to 300 of Dcp2 did not completely phenocopy the deletion of EDC3. Specifically, deletion of the region causes a subtle but consistent growth defect, whereas deletion of EDC3 does not result in a noticeable growth defect (Fig. 6A; see Fig. S5A in the supplemental material) (24). Second, the decapping defect of the MFA2pG mRNA caused by lesion of the region of Dcp2 is greater than that caused by deletion of EDC3 (Fig. 6B and C). It would be interesting to test if residues 248 to 300 of Dcp2 mediate binding to the other known decapping regulators.
Our results also suggest that Edc3 functions by two different mechanisms to modulate the decay of specific mRNAs. Specifically, because the deletion of residues 248 to 300 in Dcp2 phenocopies the deletion of EDC3 for the autoregulation of RPS28B, the interaction of Dcp2 and Edc3 appears to be required for Edc3 to promote the degradation of the RPS28B mRNA (Fig. 5A, B, and C). This is consistent with the earlier proposed model whereby binding of the Rps28B protein to the 3′ untranscribed region (UTR) of RPS28B mRNA would recruit Edc3 and the decapping enzyme to promote its degradation (1). In contrast, the interaction of Edc3 and Dcp2 does not appear to be required for the autoregulation of the YRA1 pre-mRNA (Fig. 5D, E, and F). This may suggest that specific features of this mRNA allow Edc3 to stimulate decapping independent of its direct interaction with Dcp2.
It is rather surprising that the Edc3-binding platform is located outside the conserved domain (residues 1 to 245) of Dcp2, given that both Dcp2 and Edc3 are evolutionarily conserved among eukaryotes. An important question is whether this type of sequence also exists in the Dcp2 proteins from other organisms. The Dcp2 homologs from various organisms, including humans, commonly contain divergent C-terminal extensions that are of very different lengths (38, 39). Although the conservation at the primary sequence level in the C-terminal regions appears to be restricted to certain fungal species, the use of regions outside the catalytic domain as docking platforms for regulators could be a universal feature of Dcp2. Alternatively, it is possible that other organisms have evolved a different strategy to maintain the integrity of the decapping complex. The existence of organism-specific decapping regulators might support this idea. For example, homologs of yeast Edc1 and Edc2, whose functional significance as decapping regulators has been demonstrated both in vivo and in vitro (13, 31), have not been recognized in higher eukaryotic genomes. On the other hand, Hedls/Ge-1/Edc4, which is essential for stable association of human Dcp1 and Dcp2, does not have an obvious homolog in yeast (15), and yeast Dcp1 and Dcp2 can form a stable complex by themselves (33). In any case, it is currently an open question if the C-terminal extensions of Dcp2 that exist in diverse organisms play an important role in the binding of decapping regulators.
In this work, we have identified a region of Dcp2 that mediates a functional interaction between Dcp2 and Edc3 both in vivo and in vitro. Although our results have extended the understanding of how Edc3 activates Dcp2, numerous questions remain for future study. As described above, it would be interesting to determine if other decapping regulators also bind to Dcp2 through this domain and, if so, to examine if these interactions can occur simultaneously or exclusively. To obtain deeper insights into functions of the decapping complex, it would be also important to determine the structure of this short peptide sequence in complex with Edc3 and possibly with other decapping regulators. Our results also support the notion that the decapping complex could function in multiple distinct manners, depending on each mRNP context. Therefore, it would be important to determine the specific modes of interactions among the decapping factors in each mRNP context and, eventually, in a specific local cellular context including macromolecular structures such as P-bodies.
Supplementary Material
Acknowledgments
We thank Carolyn J. Decker for helpful comments on the manuscript and for unpublished materials, Travis Dunckley for providing unpublished materials, Purusharth Rajyaguru and Tracy Nissan for allowing us to refer to their results before publication, and the Parker laboratory for support and discussion.
This work was financially supported by the Howard Hughes Medical Institute (R.P.) and NIH RO1 GM078360 (J.D.G.). Y.H. was a recipient of a postdoctoral fellowship from the Uehara Memorial Foundation. B.N.J. is a recipient of an NSF predoctoral fellowship.
Footnotes
Published ahead of print on 19 January 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Badis, G., C. Saveanu, M. Fromont-Racine, and A. Jacquier. 2004. Targeted mRNA degradation by deadenylation-independent decapping. Mol. Cell 15:5-15. [DOI] [PubMed] [Google Scholar]
- 2.Buchan, J. R., D. Muhlrad, and R. Parker. 2008. P bodies promote stress granule assembly in Saccharomyces cerevisiae. J. Cell Biol. 183:441-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao, D., and R. Parker. 2003. Computational modeling and experimental analysis of nonsense-mediated decay in yeast. Cell 113:533-545. [DOI] [PubMed] [Google Scholar]
- 4.Caponigro, G., D. Muhlrad, and R. Parker. 1993. A small segment of the MAT alpha 1 transcript promotes mRNA decay in Saccharomyces cerevisiae: a stimulatory role for rare codons. Mol. Cell. Biol. 13:5141-5148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cole, C., J. D. Barber, and G. J. Barton. 2008. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 36:W197-W201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coller, J., and R. Parker. 2005. General translational repression by activators of mRNA decapping. Cell 122:875-886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Decker, C. J., and R. Parker. 1993. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 7:1632-1643. [DOI] [PubMed] [Google Scholar]
- 8.Decker, C. J., D. Teixeira, and R. Parker. 2007. Edc3p and a glutamine/asparagine-rich domain of Lsm4p function in processing body assembly in Saccharomyces cerevisiae. J. Cell Biol. 179:437-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Decourty, L., C. Saveanu, K. Zemam, F. Hantraye, E. Frachon, J. C. Rousselle, M. Fromont-Racine, and A. Jacquier. 2008. Linking functionally related genes by sensitive and quantitative characterization of genetic interaction profiles. Proc. Natl. Acad. Sci. U. S. A. 105:5821-5826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Deshmukh, M. V., B. N. Jones, D. U. Quang-Dang, J. Flinders, S. N. Floor, C. Kim, J. Jemielity, M. Kalek, E. Darzynkiewicz, and J. D. Gross. 2008. mRNA decapping is promoted by an RNA-binding channel in Dcp2. Mol. Cell 29:324-336. [DOI] [PubMed] [Google Scholar]
- 11.Dong, S., C. Li, D. Zenklusen, R. H. Singer, A. Jacobson, and F. He. 2007. YRA1 autoregulation requires nuclear export and cytoplasmic Edc3p-mediated degradation of its pre-mRNA. Mol. Cell 25:559-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dunckley, T., and R. Parker. 1999. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 18:5411-5422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunckley, T., M. Tucker, and R. Parker. 2001. Two related proteins, Edc1p and Edc2p, stimulate mRNA decapping in Saccharomyces cerevisiae. Genetics 157:27-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eulalio, A., J. Rehwinkel, M. Stricker, E. Huntzinger, S. F. Yang, T. Doerks, S. Dorner, P. Bork, M. Boutros, and E. Izaurralde. 2007. Target-specific requirements for enhancers of decapping in miRNA-mediated gene silencing. Genes Dev. 21:2558-2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fenger-Gron, M., C. Fillman, B. Norrild, and J. Lykke-Andersen. 2005. Multiple processing body factors and the ARE binding protein TTP activate mRNA decapping. Mol. Cell 20:905-915. [DOI] [PubMed] [Google Scholar]
- 16.Franks, T. M., and J. Lykke-Andersen. 2008. The control of mRNA decapping and P-body formation. Mol. Cell 32:605-615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Veronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. Andre, A. P. Arkin, A. Astromoff, M. El-Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K. D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Guldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kotter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C. Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 18.Gietz, R. D., and A. Sugino. 1988. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene 74:527-534. [DOI] [PubMed] [Google Scholar]
- 19.Hatfield, L., C. A. Beelman, A. Stevens, and R. Parker. 1996. Mutations in trans-acting factors affecting mRNA decapping in Saccharomyces cerevisiae. Mol. Cell. Biol. 16:5830-5838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He, F., S. W. Peltz, J. L. Donahue, M. Rosbash, and A. Jacobson. 1993. Stabilization and ribosome association of unspliced pre-mRNAs in a yeast upf1- mutant. Proc. Natl. Acad. Sci. U. S. A. 90:7034-7038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heaton, B., C. Decker, D. Muhlrad, J. Donahue, A. Jacobson, and R. Parker. 1992. Analysis of chimeric mRNAs derived from the STE3 mRNA identifies multiple regions within yeast mRNAs that modulate mRNA decay. Nucleic Acids Res. 20:5365-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones, B. N., D. U. Quang-Dang, Y. Oku, and J. D. Gross. 2008. A kinetic assay to monitor RNA decapping under single-turnover conditions. Methods Enzymol. 448:23-40. [DOI] [PubMed] [Google Scholar]
- 23.Keogh, M. C., J. A. Kim, M. Downey, J. Fillingham, D. Chowdhury, J. C. Harrison, M. Onishi, N. Datta, S. Galicia, A. Emili, J. Lieberman, X. Shen, S. Buratowski, J. E. Haber, D. Durocher, J. F. Greenblatt, and N. J. Krogan. 2006. A phosphatase complex that dephosphorylates gammaH2AX regulates DNA damage checkpoint recovery. Nature 439:497-501. [DOI] [PubMed] [Google Scholar]
- 24.Kshirsagar, M., and R. Parker. 2004. Identification of Edc3p as an enhancer of mRNA decapping in Saccharomyces cerevisiae. Genetics 166:729-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.LaGrandeur, T. E., and R. Parker. 1998. Isolation and characterization of Dcp1p, the yeast mRNA decapping enzyme. EMBO J. 17:1487-1496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ling, S. H., C. J. Decker, M. A. Walsh, M. She, R. Parker, and H. Song. 2008. Crystal structure of human Edc3 and its functional implications. Mol. Cell. Biol. 28:5965-5976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 28.Lykke-Andersen, J. 2002. Identification of a human decapping complex associated with hUpf proteins in nonsense-mediated decay. Mol. Cell. Biol. 22:8114-8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Muhlrad, D., and R. Parker. 1994. Premature translational termination triggers mRNA decapping. Nature 370:578-581. [DOI] [PubMed] [Google Scholar]
- 30.Parker, R., and H. Song. 2004. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol. 11:121-127. [DOI] [PubMed] [Google Scholar]
- 31.Schwartz, D., C. J. Decker, and R. Parker. 2003. The enhancer of decapping proteins, Edc1p and Edc2p, bind RNA and stimulate the activity of the decapping enzyme. RNA 9:239-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwartz, D. C., and R. Parker. 2000. mRNA decapping in yeast requires dissociation of the cap binding protein, eukaryotic translation initiation factor 4E. Mol. Cell. Biol. 20:7933-7942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.She, M., C. J. Decker, N. Chen, S. Tumati, R. Parker, and H. Song. 2006. Crystal structure and functional analysis of Dcp2p from Schizosaccharomyces pombe. Nat. Struct. Mol. Biol. 13:63-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.She, M., C. J. Decker, D. I. Svergun, A. Round, N. Chen, D. Muhlrad, R. Parker, and H. Song. 2008. Structural basis of dcp2 recognition and activation by dcp1. Mol. Cell 29:337-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Steiger, M., A. Carr-Schmid, D. C. Schwartz, M. Kiledjian, and R. Parker. 2003. Analysis of recombinant yeast decapping enzyme. RNA 9:231-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Teixeira, D., U. Sheth, M. A. Valencia-Sanchez, M. Brengues, and R. Parker. 2005. Processing bodies require RNA for assembly and contain nontranslating mRNAs. RNA 11:371-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tritschler, F., J. E. Braun, A. Eulalio, V. Truffault, E. Izaurralde, and O. Weichenrieder. 2009. Structural basis for the mutually exclusive anchoring of P body components EDC3 and Tral to the DEAD box protein DDX6/Me31B. Mol. Cell 33:661-668. [DOI] [PubMed] [Google Scholar]
- 38.van Dijk, E., N. Cougot, S. Meyer, S. Babajko, E. Wahle, and B. Seraphin. 2002. Human Dcp2: a catalytically active mRNA decapping enzyme located in specific cytoplasmic structures. EMBO J. 21:6915-6924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang, Z., X. Jiao, A. Carr-Schmid, and M. Kiledjian. 2002. The hDcp2 protein is a mammalian mRNA decapping enzyme. Proc. Natl. Acad. Sci. U. S. A. 99:12663-12668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilmes, G. M., M. Bergkessel, S. Bandyopadhyay, M. Shales, H. Braberg, G. Cagney, S. R. Collins, G. B. Whitworth, T. L. Kress, J. S. Weissman, T. Ideker, C. Guthrie, and N. J. Krogan. 2008. A genetic interaction map of RNA-processing factors reveals links between Sem1/Dss1-containing complexes and mRNA export and splicing. Mol. Cell 32:735-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.