Abstract
Kinetoplast DNA in African trypanosomes contains a novel form of mitochondrial DNA consisting of thousands of minicircles and dozens of maxicircles topologically interlocked to form a two-dimensional sheet. The replication of this unusual form of mitochondrial DNA has been studied for more than 30 years, and although a large number of kinetoplast replication genes and proteins have been identified, in vitro replication of these DNAs has not been possible since a kinetoplast DNA primase has not been available. We describe here a Trypanosoma brucei DNA primase gene, PRI1, that encodes a 70-kDa protein that localizes to the kinetoplast and is essential for both cell growth and kinetoplast DNA replication. The expression of PRI1 mRNA is cyclic and reaches maximum levels at a time corresponding to duplication of the kinetoplast DNA. A 3′-hydroxyl-terminated oligoriboadenylate is synthesized on a poly(dT) template by a recombinant form of the PRI1 protein and is subsequently elongated by DNA polymerase and added dATP. Poly(dA) synthesis is dependent on both PRI1 protein and ATP and is inhibited by RNase H treatment of the product of PRI1 synthesis.
Trypanosoma brucei, the African trypanosome, is a protozoan parasite that causes sleeping sickness in humans and a similar disease in livestock (57). In addition to interest in T. brucei as a pathogen, this parasite is considered to be among the earliest-diverging eukaryotic organisms (54) and has many novel features, including antigenic variation of surface glycoproteins, trans splicing of precursors of nuclear mRNAs, and RNA editing of mitochondrial pre-mRNAs (31). In addition, trypanosomes have long been known for a characteristic form of mitochondrial DNA termed kinetoplast DNA, or kDNA, which can be seen as a brightly stained disc-shaped structure at the base of the flagellum in cells stained with fluorescent dyes (30).
Kinetoplast DNA consists of two types of circular DNAs, minicircles and maxicircles. The maxicircles encode mitochondrial ribosomal RNAs and genes involved in oxidative metabolism and are the equivalent of the mitochondrial genome in most other eukaryotic organisms. Minicircles encode small RNAs termed guide RNAs that serve as templates in the editing of maxicircle transcripts in a process involving the addition and/or deletion of U residues in primary transcripts (55). The minicircles and maxicircles are catenated together to form a single DNA network in each cell. An individual network contains thousands of minicircles and some 20 to 30 maxicircles. In T. brucei, the minicircles are approximately 1 kb in size and the maxicircles are 23 kb in size. This massive kDNA network is condensed into a tightly packed nucleoprotein disc that opens out into a two-dimensional oval-shaped sheet of interlocked circles upon removal of associated proteins (7). At no time in the cell cycle is the kDNA completely dissociated into its component minicircles and maxicircles.
A key breakthrough in dissecting the replication mechanism of kinetoplast minicircles was the finding that minicircles replicate free of the network (17). Individual minicircles are released one at a time from the network by a DNA topoisomerase, replicated, and then reattached to the network (35, 58). Replication of minicircles free of the network made possible the study of minicircle replication without the complication of network association. Studies with the insect parasite Crithidia fasciculata, as well as with T. brucei, have uncovered many features of minicircle replication (see references 27 and 51 for recent reviews).
In current models of kDNA replication, the minicircles are released from the network into the mitochondrial matrix between the kDNA disc and the mitochondrial membrane by a type II topoisomerase (35, 58). This space has been termed the kinetoflagellar zone, or KFZ. The universal minicircle sequence (UMS) binding protein (UMSBP) has been implicated in the initiation of minicircle replication and has been localized to two sites within the KFZ adjacent to the face of the kDNA disk (1). Minicircle replication intermediates have also been localized to the KFZ, further supporting the initiation of minicircle replication within that region, where unreplicated minicircles are released from the kDNA network (13). Replication of the free minicircles involves theta-type replication intermediates with coupled leading- and lagging-strand synthesis (4, 44, 45, 49). Initiation of replication occurs within a replication origin consisting of three domains (CSB-1, CSB-2, and CSB-3) that are conserved in minicircles of kinetoplastid parasites (43). Leading-strand synthesis initiates by an RNA priming mechanism at CSB-3, also known as the universal minicircle sequence. Lagging-strand synthesis is initiated at a hexamer sequence within CSB-1 and is also likely initiated by RNA priming (4, 38, 39, 44). Two proteins, UMSBP and P38, have been shown to bind to the minicircle replication origin sequences (1, 28, 40, 41, 50) and are essential for minicircle replication. Nascent minicircles are detected in the KFZ on the flagellar face of the kDNA disk (13) and are subsequently migrated by unknown mechanisms to antipodal sites flanking the edge of the kDNA disc where they undergo repair of discontinuities (19) and are reattached to the network. In contrast, maxicircles are never released from the kDNA network and undergo replication while topologically interlocked with minicircles and other maxicircles (5). Consequently, much less is known about maxicircle replication. Electron microscopy of maxicircles released from the kDNA networks by a type II DNA topoisomerase showed that maxicircles also replicate by a unidirectional theta-type mechanism. The maxicircle replication origin has not yet been defined at the DNA sequence level but does not appear to involve a UMS, since two UMS-related sequences present in the maxicircle are located several kilobases from where the origin has been mapped by electron microscopy (5). While RNA interference (RNAi) of several genes results in the loss of both minicircles and maxicircles, RNA interference of a mitochondrial DNA helicase (TbPIF2) results in preferential loss of maxicircles and overexpression of TbPIF2 results in a 3- to 6-fold increase in the abundance of maxicircles (29).
Many of the proteins expected to be involved in kDNA replication have been identified through enzyme purification and intracellular localization and, more recently, through genomic and proteomic approaches together with molecular genetic and RNA interference techniques. The list of known kinetoplast-specific replication proteins includes six DNA polymerases (23), six DNA helicases (29), two DNA ligases (12, 52, 53), DNA topoisomerases (3, 35, 48), histone-like DNA-binding proteins (20, 21, 32, 62), RNase H (15), a structure-specific DNA endonuclease (14, 16), minicircle origin-binding proteins (2, 28, 40, 50), mitochondrial RNA polymerase (18), and proteins of still-unknown function (26, 63). In spite of progress in identifying components of the replication machinery, it has not been possible to perform in vitro replication of minicircle or maxicircle DNA for the fundamental reason that DNA polymerases are unable to initiate DNA synthesis (24) and a kinetoplast-specific DNA primase has not been available. An enzyme with primase activity was purified from C. fasciculata over 10 years ago, but the gene encoding the primase was never cloned (25). We describe here a DNA primase gene encoding a kinetoplast-specific DNA primase that is essential for kinetoplast DNA replication in T. brucei and demonstrate the primase activity of the recombinant protein.
MATERIALS AND METHODS
Trypanosome growth.
T. brucei procyclic cell line 29-13 expressing T7 RNA polymerase and the tetracycline repressor was grown at 28°C in SM medium (11) containing 15% heat-inactivated fetal bovine serum (Invitrogen), 32 μg/ml G418, and 50 μg/ml hygromycin. YTaT.1 cells were grown in the same medium lacking drugs.
Synchronization of procyclic cells.
Synchronization of procyclic trypanosomes was performed by using a protocol developed independently in our laboratory which is essentially identical to one published recently (8). YTaT.1 cells (5.4 × 108 cells) were harvested with centrifugation for 10 min at 2,000 × g and resuspended in 100 ml of fresh medium with 250 μM hydroxyurea (HU). After 16 h, the cells were harvested as described above, washed two times with 20 ml of fresh medium without HU, and resuspended in 200 ml of fresh medium lacking HU. A zero-time sample was taken immediately, and additional aliquots were removed every 75 min for 15 h for RNA isolation and microscopic examination. For microscopic examination, the harvested cells were washed twice with phosphate-buffered saline (PBS), fixed for 10 min with 4% paraformaldehyde in PBS, washed twice with 0.1 M glycine in PBS, and stored at 4°C. The cells were subsequently stained with 4′,6′-diamidino-2-phenylindole (DAPI), and cell types counted by fluorescence microscopy.
RNA isolation and Northern analysis.
Total RNA was isolated from 2 × 107 cells by using a PureLink RNA isolation kit (Invitrogen), fractionated on a 1.2% agarose-0.22 M formaldehyde gel, and transferred to Hybond N membrane (Amersham). Specific mRNAs were detected by probing membranes with 32P-random-primed DNA probes, and signals were quantified using a phosphorimager.
Epitope tagging of PRI1.
A plasmid was constructed to express a C-terminal triple-hemagglutinin (HA) tag. The plasmid (pJH573) is derived from pC-PTP-NEO (47) by replacing a PspOMI-to-EcoRI fragment with an EagI-to-EcoRI fragment containing three consecutive HA tags followed by a stop codon. Primers Q38 and Q40 were used to amplify a region of the PRI1gene from T. brucei 29-13 genomic DNA with PspOMI sites at both ends. This PCR product was digested and cloned into the EagI site of pJH573 to create pJH580. This plasmid was digested with SmaI and transfected into YTaT.1 cells. Recombinants were selected with 20 μg/ml G418, and clones produced by limiting dilution.
Microscopy.
Cells harvested during the HU synchronization and uninduced and PRI1 RNAi-induced cells were harvested, resuspended in PBS, spotted onto poly-l-lysine-coated slides, and allowed to adhere for 5 min in a humid chamber. The cells were fixed in 4% paraformaldehyde in PBS for 10 min and washed twice in 0.1 M glycine in PBS, followed by 10 min in 0.025% Triton X-100 in PBS, and then stored in methanol at −20°C. The cells were stained with DAPI to monitor the loss of the kDNA. For protein immunolocalization, slides were removed from −20°C methanol, rehydrated by washing two times in PBS, and then blocked for 1 h at 37°C in 20% goat serum in PBS containing 0.05% Tween 20 (PBST). Next, the slides were incubated at 37°C for 2 h with anti-HA monoclonal antibody 12CA5 (Babco) at a 1:250 dilution and washed four times in PBST. The slides were then incubated for 1 h at 37°C in goat anti-mouse antibody conjugated to Alexa Fluor 594 at a 1:1,500 dilution, washed 4 times in PBST, and mounted in Prolong Gold antifade reagent with DAPI. Cells were imaged using a Zeiss upright light microscope (Zeiss Axio Imager Z1) with a 100× oil immersion objective. All images were rendered using Zeiss Axiovision software and a Zeiss digital charge-coupled-device camera (AxioCam MRm).
RNA interference.
A plasmid was constructed to express a stem-loop mRNA under tetracycline control. Plasmid pJH533 is derived from pLEW100 (61) by replacing a HindIII-to-BamHI fragment containing the luciferase gene with two multiple cloning sites flanking an XbaI-to-BsrG1 fragment of the luciferase gene. The 443-bp luciferase gene fragment acts as a “stuffer.” Primers Q11 and Q12 were used to amplify an 807-bp fragment of the PRI1 gene from T. brucei 29-13 genomic DNA. The PCR product was cloned into pCRII TOPO (Invitrogen). HindIII-to-EcoRV and BamHI-to-NotI fragments of this plasmid (pJH537) were cloned upstream and downstream, respectively, from the luciferase stuffer fragment to create pJH542. This plasmid was digested with NotI prior to transfecting T. brucei 29-13 cells. Cells with integrated plasmids were selected with 2.5 μg/ml phleomycin, and clonal lines were produced by limiting dilution. Three clones were evaluated for RNA loss and growth defect. One clone was selected for further analysis. Cell growth was monitored using a model Z2 Coulter counter (Beckman Coulter), and cultures were maintained between 2 × 105 and 1.0 × 107 cells/ml.
DNA isolation and Southern blot analysis.
Cloned T. brucei cells stably transfected with the RNAi vector were induced by the addition of 1 μg/ml tetracycline and harvested at various intervals. Total DNA was isolated as described previously (26). Analysis of free minicircle replication intermediates was performed by electrophoresis of total DNA samples on a 1.5% agarose gel and Southern blotting using a pool of five independently cloned T. brucei minicircles as the minicircle probe. A maxicircle probe was prepared by PCR amplification and cloning of a 2.6-kb fragment of the coding region of the maxicircle using primers MXF1 and MXR1. For analysis of total minicircle and maxicircle DNAs, the DNA was digested with HindIII and XbaI prior to electrophoresis and Southern blotting. The trypanosome hexose transporter probe (used as a loading control) was amplified from total DNA as described previously (26).
Fractionation of maxicircle DNAs.
The relative amounts of nicked/gapped (N/G) and covalently closed (CC) forms of maxicircle DNA in total DNA samples (2 × 106 cell equivalents) were determined by decatenation of the DNA by using human topoisomerase II (Affymetrix), followed by electrophoresis on a 0.6% agarose gel and Southern blotting as described above. Chromosomal DNA was detected with a probe for trypanosome hexose transporter as a loading control. Electrophoresis conditions were initially determined by gel electrophoresis of decatenated kDNA from 108 cells labeled for 60 min with 50 μM bromodeoxyuridine (BrdU) (29). The DNAs were blotted to nitrocellulose membranes, blocked with 2% bovine serum albumin, and incubated with anti-BrdU antibodies at 1:1,000 (Dako) prior to stripping and Southern blotting using the 2.6-kb maxicircle probe.
His10 tagging and purification of PRI1.
A plasmid with 10 histidine residues at the amino terminus was constructed for expressing PRI1 protein. The plasmid is derived from pGEX-2T (Pharmacia) by removing the glutathione S-transferase coding portion and creating an NdeI site at the start codon. Primers Q82 and Q84 were used to amplify pGEX-2T DNA. The resulting PCR product was digested with PspOMI and NdeI. Primers Q83 and Q78 were used to amplify T. brucei 29-13 genomic DNA to produce a PCR product representing amino acids 16 to 629 of PRI1 with Met-His10 at the amino terminus and an EcoRI site after the stop codon. This product was digested with NdeI and EcoRI and combined with the above-described fragment from pGEX-2T and a PspOMI-to-EcoRI fragment of pGEX-2T in a three-fragment ligation. The resulting plasmid (pJH598) was transferred into Escherichia coli BL21 cells. BL21(pJH598) cells were grown in L broth (MP Biomedicals) to an optical density of 0.6 and induced for 3 h at 37°C with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Harvested cells were lysed in metal chelate affinity chromatography buffer (Novagen) containing 0.1% Triton X-100 and sonicated. The lysate was clarified by centrifugation at 4°C and 16,000 × g for 15 min and purified on His-Bind resin (Novagen) according to the manufacturer's instructions. Peak protein fractions were pooled and passed over a 10-ml Sephadex G25 column equilibrated in 20 mM Tris, pH 8.0, 50 mM NaCl, 1 mM dithiothreitol (DTT). Glycerol was immediately added to the eluted fractions to a final concentration of 40% (vol/vol). Fractions were stored at −20°C.
DNA primase assay.
Coupled-reaction mixtures (25 μl) contained 250 ng poly(dT), 50 mM Tris-HCl, pH 8.0, 10 mM MgCl2, ATP as indicated below, and 8.3 μl PRI1 recombinant protein (125 ng/μl) in 50 mM Tris-HCl, pH 8.0, 30 mM NaCl, 1 mM DTT, and 40% glycerol. After 40 min at 30°C, E. coli Klenow DNA polymerase (1.5 units) and [α-32P]dATP (16 μM, 24,400 cpm/pmole) were included, and incubation was continued for an additional 40 min at 30°C. Where indicated, PRI1 was heat inactivated (20 min at 75°C) and then primers reannealed (75°C to 25°C for 20 min) after the first incubation. Prior to the second incubation, RNase H (2.5 units) was added or not for 30 min at 37°C. When RNase H was included, an additional heat inactivation (20 min at 65°C) was added prior to adding Klenow DNA polymerase and [α-32P]dATP (24,400 cpm/pmole). Reactions were terminated by spotting onto Whatman DE81 filter discs, washing three times in 500 ml of ammonium formate (pH 7.8) and once in 500 ml of 95% ethanol, and air drying prior to scintillation counting.
Western blotting.
Protein fractions were electrophoresed on SDS gels and transferred to membranes as described earlier (26). Rabbit anti-dnaG antibody was used at 1:1,000 dilution, and bands were visualized using peroxidase-conjugated anti-rabbit immunoglobulin G (Sigma) at 1:8,000 dilution as secondary antibody and the SuperSignal West Chemiluminescent system (Pierce). The anti-dnaG antibody recognizes PRI1 primase, as well as the dnaG protein.
Poly(rA) synthesis reactions.
A 175-μl reaction mixture contained the same components and concentrations as the coupled-reaction mixtures minus dATP and Klenow DNA polymerase and included [α-32P]ATP (100 μM, 18,000 cpm/pmole). After incubation at 30°C for the times indicated below, 50- or 75-μl aliquots were removed and extracted with phenol and then with chloroform, followed by ethanol precipitation. Pellets were resuspended in 20 mM Tris, pH 8.0, 10 mM MgCl2, 50 μM dATP and divided into either two or three equal portions. Klenow DNA polymerase (1.5 units) or RNase H (1.5 units) was added as indicated below, with further incubation at 30°C for 40 min. Reactions were stopped by adding an equal volume of sequencing gel loading buffer. Samples were electrophoresed on a 14- by 16- by 0.05-cm 20% polyacrylamide, 7 M urea gel at 12 W and exposed to film at −80°C.
Oligonucleotides used.
The oligonucleotides used were Q11, ATGCTCCGCATCACCTTACCATCACAGC; Q12, CCCTTGCGGAAGCCGGCGGGCGA; Q38, GGATATGGGCCCTGTTGGACGACGTGATGAATG; Q40, GGAATTGGGCCCTTGTGGAGGGATGCCGTTGGCACCACCG; Q78, TTCAAGGAATTCTTACGCTCCTTGTGGAGGGATGCCG; Q82, TGTTAGCGGGCCCATTAAGTTCTGTCTCGG; Q83,GAACCTCATATGCACCATCACCATCACCACCACCATCATCACGTTGCAAAGTTGG; Q84, CAATTAATCATCGGCTCGTATAATG; MXF1, CACAGCACCCGTTTCAGCACAG; and MXR1, TCTCCTCCCCCTAACCTTTCCTGC.
RESULTS
Identification of the PRI1 gene.
We initially identified a gene (Tb927.8.2550) in the GeneDB T. brucei genome database that contains two sequence elements (CATAGAGG and CATAGAAG) within approximately 500 nucleotides of the stop codon that are similar to the conserved octamer sequence (C/A)AUAGAA(G/A) which has been shown to be required for cyclic expression of several S-phase-expressed genes in the related parasite C. fasciculata (26, 33, 34, 36, 37). This sequence was also found to be predictive of S-phase gene expression in Leishmania major (26, 64). In addition, the encoded protein contains a C-terminal DNA primase PriCT-2 motif (22) and is predicted by MITOPROT (9) to have a probability of 0.98 of targeting to the mitochondrion with a 16-amino-acid cleavable leader sequence. The protein, which we term PRI1, has 629 residues and a predicted pI of 9.0 and is highly conserved in other kinetoplastids (see Fig. S1 in the supplemental material). PRI1 is a member of the archaeo-eukaryotic primase superfamily and belongs to the nucleocytoplasmic large-virus clade (22). A computational study of the protein sequences and structures of this superfamily revealed a unique active site shared by all members of the superfamily.
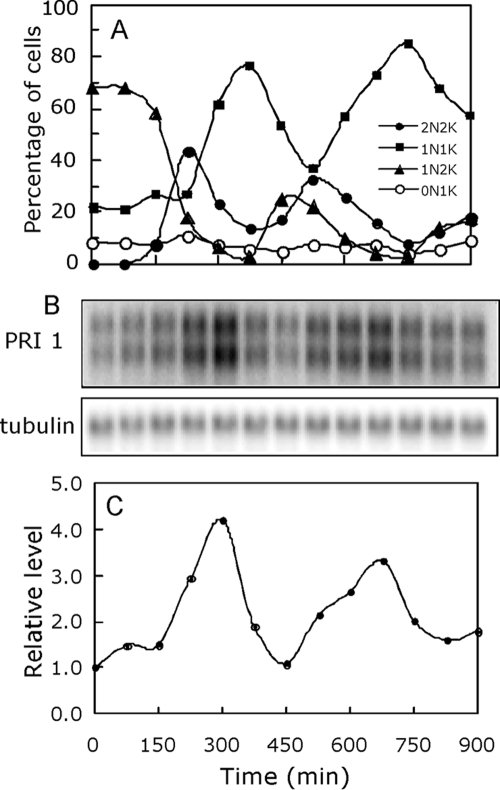
Examination of the PRI1 mRNA levels in a synchronized culture of T. brucei procyclic cells showed that the PRI1 mRNA does in fact exhibit cyclic expression. The PRI1 transcript level cycles in a synchronized culture and reaches a maximum level approximately 1.5 h prior to the increase in the frequency of cells containing two kinetoplasts (Fig. 1). The basis for the appearance of the PRI1 transcript as a doublet is unknown but might result from differences in the untranslated regions of the two alleles. Only coding sequence was used as the probe.
FIG. 1.
Cycling of the PRI1 mRNA levels during the cell cycle. (A) T. brucei procyclic cells synchronized by release from a hydroxyurea block were harvested at 75-min intervals, fixed in solution, stained with DAPI, and counted with a hemocytometer. The cells were scored for the number of nuclei and number of kinetoplasts in each cell. 2N2K, two nuclei and two kinetoplasts; 1N1K, one nucleus and one kinetoplast; 1N2K, one nucleus and two kinetoplasts; 0N1K, no nucleus and one kinetoplast. (B) RNA isolated from 2 × 107 cells at each indicated time interval was analyzed by Northern blotting using the PRI1 coding sequence and a PCR-amplified fragment of the tubulin gene (26) as probes. (C) Phosphorimager quantitation of the PRI1 mRNA levels from the results shown in panel B using the tubulin mRNA levels as loading controls.
Localization of epitope-tagged PRI1.
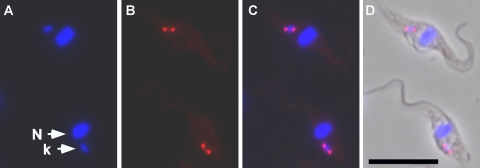
Since the PRI1 protein is predicted by MITOPROT to have an N-terminal mitochondrial targeting sequence, we examined its intracellular localization by expressing PRI1 protein in T. brucei with three copies of the hemagglutinin epitope tag at the C terminus. Fluorescence microscopy showed that the protein localizes to antipodal sites flanking the kDNA disc (Fig. 2). Similar localization has been observed for several other kinetoplast replication proteins and for minicircle replication intermediates (13, 27, 51).
FIG. 2.
Immunolocalization of PRI1. Cells stably expressing HA-tagged PRI1 were immunostained (red) with anti-HA monoclonal antibody and counterstained with DAPI (blue) for DNA. (A) DAPI; (B) anti-HA; (C) merged fluorescent images; (D) merged fluorescent images with phase-contrast image. Bar, 10 μM; N, nucleus; k, kinetoplast.
RNAi of PRI1.
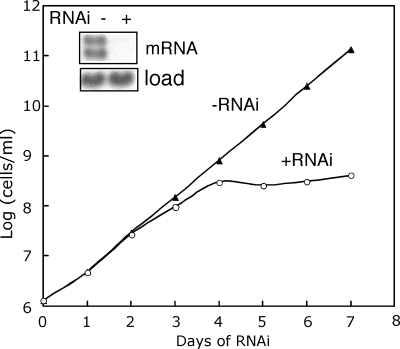
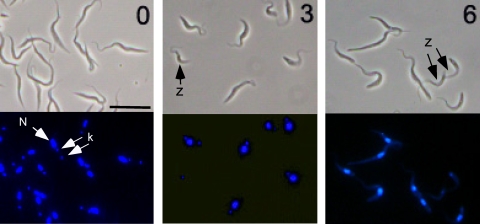
To address the function of PRI1 protein, we used the pJH533 vector that produces a stem-loop RNA upon induction with tetracycline (59). An 807-bp fragment of the PRI1 gene corresponding to residues 1 to 807 of the coding sequence was inserted into pJH533. This construct was transfected into T. brucei 29-13 cells to generate the PRI1 RNAi expression strain. A cloned cell line was obtained, and RNAi was induced by the addition of tetracycline. The cells were monitored for cell growth and PRI1 mRNA levels for 7 days. The PRI1 mRNA level was reduced more than 95% after 48 h of RNAi (Fig. 3). The cells ceased growing by 4 days after induction of RNAi, indicating that PRI1 is essential for cell growth. Similar to results of RNAi of other essential kDNA replication genes, we found that PRI1 RNAi results in loss of the kDNA as determined by DAPI staining of cells after 3 and 6 days of RNAi (Fig. 4). By day 3, there was a dramatic shrinkage of the kDNA, with approximately 32% of the cells having no visible kDNA. By day 6, the kDNA was so tiny that quantitation of the loss of the kinetoplast during RNAi was judged to be unreliable due to the difficulty in distinguishing cells with tiny kinetoplasts from those lacking kinetoplasts.
FIG. 3.
Effect of PRI1 depletion on the growth of procyclic T. brucei. Cloned procyclic T. brucei cells transfected with the PRI1 RNAi plasmid construct were incubated in culture medium containing 1 μg/ml tetracycline (+RNAi) or without tetracycline (−RNAi). The number of cells per milliliter on the y axis is the measured value times the dilution factor. The Northern blot (inset) shows the effect of RNAi (48 h) on the PRI1 mRNA level.
FIG. 4.
Shrinkage and loss of kDNA during RNAi of PRI1. Cells at 0, 3, and 6 days of RNAi were stained with DAPI and observed with a fluorescence microscope. Upper panels, phase contrast; lower panels, DAPI fluorescence. N, nucleus; k, kinetoplast; z, zoid (anucleate cell). Bar, 10 μm.
Loss of minicircles and maxicircles.
Total cell DNA was digested with restriction enzymes and analyzed by Southern blotting to determine the fate of minicircle and maxicircle DNAs during PRI1 RNAi (Fig. 5A). Maxicircles were lost at a greater rate than minicircles, resulting in a reduction of approximately 80% relative to the initial level within 3 to 4 days of RNAi (Fig. 5B), whereas total minicircle DNA was reduced by only approximately 20% of the initial level in the same time. Loss of minicircle DNA could possibly be a consequence of the loss of the maxicircles.
FIG. 5.
Effect of PRI1 RNAi on minicircle and maxicircle DNAs. (A) Kinetics of kDNA loss as determined by Southern blotting of total minicircles and maxicircles. After digestion with HindIII and XbaI, total cellular DNA (1 × 106 cell equivalents per lane) was fractionated on a 1.5% agarose gel containing 1 μg/ml ethidium bromide. Southern blots were probed for minicircle DNA, maxicircle fragment (Maxi), and a hexose transporter fragment as a loading control (Load). A 1.0-kb minicircle fragment (Mini) represents the full-length minicircle. (B) Phosphorimager quantitation of the relative levels of maxicircle and minicircle DNAs shown in panel A. (C) Effect of PRI1 RNAi on free minicircle replication intermediates. Total DNA was purified after various times of RNAi and then fractionated (5 × 105 cell equivalents per lane) on an agarose gel. Minicircle DNA was detected by Southern blotting using a minicircle probe. Intact kDNA does not enter the gel and remains in the well. The band near the top of the gel is due to chromosomal DNA. Covalently closed (CC), nicked/gapped (N/G), and catenated minicircles (cm) are indicated. (D) Phosphorimager quantitation of the relative levels of free minicircle species after various times of RNAi.
Free minicircles represent only a small percentage of the total minicircle content of the cell and are detected by Southern blotting of undigested whole-cell DNA (Fig. 5C). The free minicircles are separated on a one-dimensional agarose gel containing ethidium bromide. Covalently closed free minicircles represent minicircles that have been released from the kDNA network but have not been replicated. Nicked/gapped minicircles represent the products of minicircle replication that have not been reattached to the network, where they normally would undergo repair of most of the remaining discontinuities. The levels of both covalently closed and nicked/gapped free minicircles continued to increase for the first 3 days during PRI1 RNAi (Fig. 5D), implying a continued release of minicircles from the network. The essentially parallel accumulation of nicked/gapped free minicircles and covalently closed minicircles suggests that the released minicircles continued to replicate during the first 2 to 3 days of RNAi, perhaps as a result of residual PRI1 activity. We note the absence of the multiply gapped species migrating between the covalently closed and the nicked/gapped minicircles observed in earlier studies (45) and that accumulate during RNAi of a kinetoplast DNA polymerase (6). Since multiply gapped species result from multiple discontinuities in the discontinuously synthesized strand, the absence of these species during PRI1 RNAi suggests that PRI1 is not involved in the initiation of Okazaki fragments on minicircle DNA. The increased levels of the free minicircles by 3 days after induction of RNAi and the decrease in total minicircle DNA during that time indicates that the extent of minicircle replication and rejoining to the kDNA network was insufficient to maintain the network size.
The striking loss of maxicircle DNA during RNAi may also be the primary factor in the shrinkage and loss of the kDNA and suggests that PRI1 is required for maxicircle replication. To further examine the loss of maxicircle DNA during PRI1 RNAi, we first established gel conditions for fractionating CC and N/G maxicircles, according to the method of Liu et al. (29), as shown in Fig. 6A. Cells were pulse labeled for 60 min with BrdU to label replicating molecules. Purified kDNA networks were then decatenated with topoisomerase II and analyzed on a 0.6% agarose gel. The newly replicated maxicircles are identified by the presence of the BrdU label in N/G molecules but not in the CC molecules. We then induced PRI1 RNAi and decatenated the kDNA networks in total DNA (2 × 106 cell equivalents/lane) at various times during RNAi (Fig. 6B). Both CC and N/G maxicircles were rapidly lost during the first day of RNAi (Fig. 6B), consistent with the results shown in Fig. 5A and supporting an involvement of PRI1 in maxicircle replication. The increased levels of N/G maxicircles and corresponding decrease in the levels of CC maxicircles during the first 12 h of RNAi (Fig. 6C) possibly result from incomplete blockage of maxicircle replication early in the knockdown of PRI1 expression.
FIG. 6.
Loss of N/G and CC maxicircles during RNAi. (A) Incorporation of BrdU into maxicircle DNA. Wild-type cells (6 × 106 cells per ml) were labeled for 60 min with 50 μM BrdU. Isolated kDNA (from 108 cells) was decatenated with human topoisomerase II and fractionated on a 0.6% agarose gel containing ethidium bromide at 0.25 μg/ml. A Western blot of the gel was probed with anti-BrdU antibody (left panel), and after stripping, the blot was probed with a maxicircle DNA probe (right panel). (B) Southern blot of topoisomerase II-treated total DNA (2 × 106 cell equivalents/lane) during PRI1 RNAi. The gel was run, Southern blotted, and probed as described for panel A and then stripped and reprobed with a chromosomal probe as a loading control. (C) Phosphorimager quantitation of the levels of N/G and CC maxicircles relative to the levels at day 0.
DNA primase activity of recombinant PRI1.
The PRI1 gene was expressed in E. coli as a recombinant protein with 10 histidine residues at the N terminus, to facilitate purification. Extracts from cells induced for high-level expression of recombinant PRI1 were loaded onto a metal chelate column and eluted with increasing levels of imidazole. Peak fractions of the protein eluted from the metal chelate column were then passed over a G25 Sephadex column (Fig. 7A), and eluted fractions were assayed for primase activity in a coupled reaction (10) with Klenow DNA polymerase on a poly(dT) template (Fig. 7B). The His-tagged protein eluted as a single major band that migrated consistent with a 70-kDa polypeptide on an SDS polyacrylamide gel. The minor faster-migrating band on the gel in Fig. 7A has not been identified but possibly represents a breakdown product of the major species.
FIG. 7.
Coelution of His10-tagged PRI1 and DNA primase activity from a G25 Sephadex column after purification by metal chelate chromatography. (A) A His10-tagged PRI1 expression plasmid was expressed in E. coli BL21, purified from the cell lysate by chromatography on a His-Bind column (Novagen), and desalted by passage over a G25 Sephadex column. Fractions were analyzed by electrophoresis on a 10% SDS gel and stained with Coomassie blue. M, markers; L, load. (B) An 8.3-μl amount of each fraction was assayed for primase activity in a coupled reaction with Klenow DNA polymerase and [α-32P]dATP on a poly(dT) template. Each reaction mixture contained 1 mM ATP, and the incorporation of [α-32P]dAMP was assayed after 40 min at 30°C (closed circles). Cells containing the empty vector were induced and subjected to the same extraction and purification scheme and assayed for primase activity (filled squares). (C) Western blot analysis of pooled fractions. Lanes 1 to 3, fractions from total extract, His-Bind eluate, and G25 eluate from induced cells expressing PRI1; lanes 4 to 6, fractions from total extract, His-Bind eluate, and G25 eluate from induced cells containing the empty vector; lane 7, dnaG protein.
Cells containing the empty vector were also induced in parallel and subjected to the same purification protocol to investigate the possibility that a contaminating bacterial primase was responsible for the observed activity. No primase activity was detected in these fractions (Fig. 7B). To specifically address the possibility that the E. coli dnaG primase might contaminate the purified PRI1 protein, we performed Western blots of fractions from cells expressing PRI1 or from cells containing the empty vector (Fig. 7C). Polyclonal antisera against dnaG recognized not only dnaG but also PRI1 and its apparent breakdown product. While the dnaG protein was detected both in extracts of cells containing the PRI1 expression vector and in extracts of cells containing the empty vector, it was not detected in either case in the eluates from the metal chelate columns or the G25 columns. The recovery of primase activity in the purified fractions lacking the E. coli dnaG primase (Fig. 7B and C) and the absence of primase activity in purified fractions from cells expressing the empty vector (Fig. 7B) rule out dnaG primase as the source of primase activity associated with PRI1.
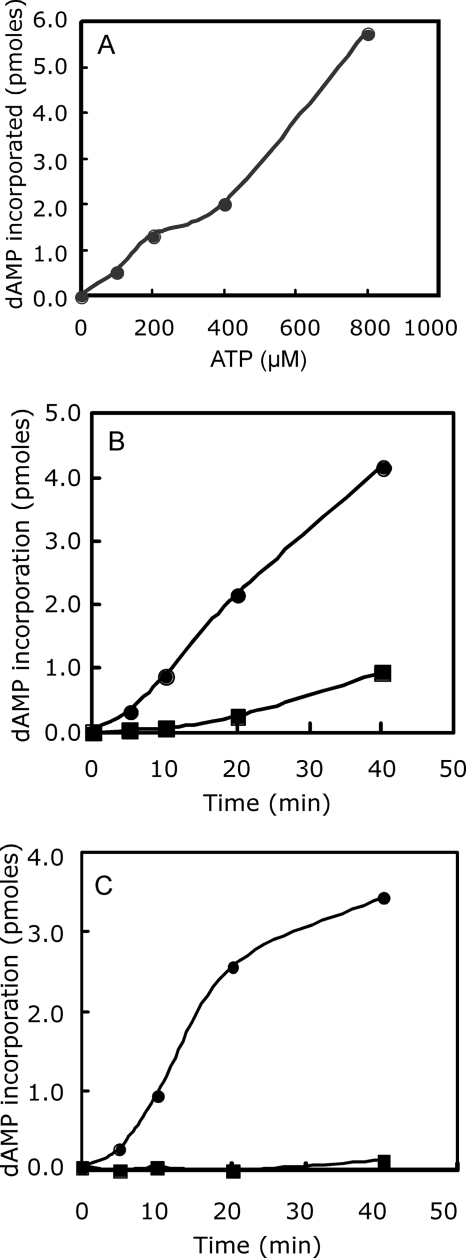
To further characterize the activity associated with PRI1, the peak fractions from the G25 column from cells expressing PRI1 were assayed in a coupled reaction with Klenow DNA polymerase in the presence of increasing concentrations of ATP (Fig. 8A). Poly(dA) synthesis required ATP and increased linearly with the ATP concentration up to 800 μM ATP. No poly(dA) synthesis was observed in the absence of PRI1. Synthesis of poly(dA) was also stimulated significantly by first preincubating the poly(dT) template with PRI1 and ATP prior to the addition of Klenow DNA polymerase and [α-32P]dATP (Fig. 8B). When the product of the PRI1 reaction was treated with RNase H prior to the addition of Klenow DNA polymerase and [α-32P]dATP, the synthesis of poly(dA) was abolished (Fig. 8C). These results indicate that the product of the PRI1 reaction is a 3′-hydroxyl-terminated oligoriboadenylate in a hybrid structure with poly(dT).
FIG. 8.
Activity of recombinant PRI1 protein. (A) ATP-dependent synthesis of poly(dA). Fraction 5 from the G25 Sephadex column was assayed for primase activity in the presence of various levels of ATP. Each reaction mixture contained 1 μg of recombinant PRI1, 1 unit of Klenow polymerase, and 16 μM [α-32P]dATP (24,400 cpm/pmole). Incorporated radioactivity was determined after 40 min at 30°C. (B) Effect of preincubation of the poly(dT) template with PRI1 and ATP. Closed circles, preincubation for 40 min prior to adding Klenow DNA polymerase and [α-32P]dATP; closed squares, coupled reaction without preincubation. (C) Effect of RNase H on priming of poly(dA) synthesis. Closed circles, poly(dA) synthesis in a reaction mixture with 40 min of incubation with PRI1, ATP, and poly(dT) at 30°C followed by 20 min at 75°C and then 40 min at 30°C with Klenow DNA polymerase and [α-32P]dATP. Closed squares, poly(dA) synthesis in a reaction mixture with 40 min of incubation with PRI1, ATP, and poly(dT) followed by 20 min at 75°C, 30 min with 2.5 units of RNase H at 37°C, 20 min at 65°C, and 40 min at 30°C with Klenow DNA polymerase and [α-32P]dATP.
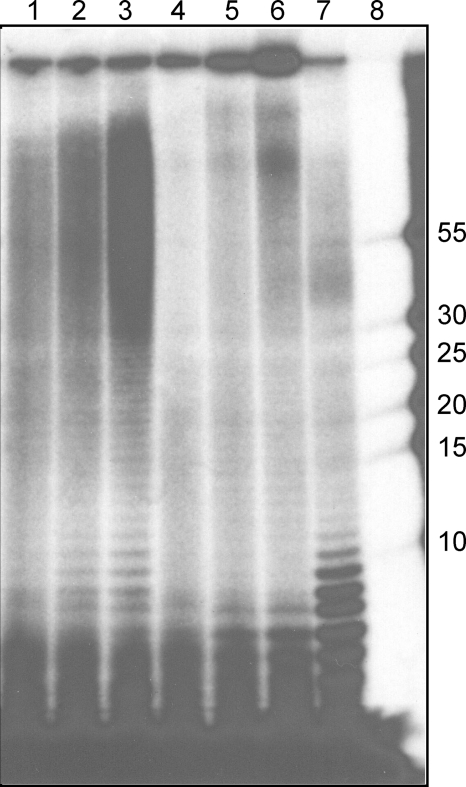
The oligoribonucleotide products of the PRI1 reaction were analyzed by acrylamide gel electrophoresis following incubation for 40, 80, or 120 min in reaction mixtures containing [α-32P]ATP (Fig. 9, lanes 1 to 3). PRI1 synthesized a ladder of poly(A) oligonucleotides that ranged up to 70 to 80 residues long. Longer incubation times increased the yield of products in this size range but did not result in a significant increase in the average length of the products. A subsequent chase in the absence of PRI1 and the presence of Klenow polymerase and dATP (lanes 4 to 6) resulted in the depletion of poly(A) oligonucleotides of 8 residues or more in length by elongation of the poly(A) with deoxynucleotides, and the products appeared in the well. To confirm that the poly(A) products of PRI1 are in a hybrid structure, we treated an equal aliquot of sample 3 with ribonuclease H (lane 7), which degraded the poly(A) to yield oligo(A) fragments of 10 nucleotides or shorter.
FIG. 9.
Elongation of oligo(A) primers by Klenow DNA polymerase. To analyze the PRI1 products, the reaction mixture (175 μl) contained poly(dT) template, PRI1 (7.3 μg), and 100 μM [α-32P]ATP (18,000 cpm/pmole) as substrate. After incubation at 30°C for the times indicated below, the reaction mixtures were processed as described in Materials and Methods. Lanes 1 to 3, incubation with PRI1 for 40, 80, and 120 min; lanes 4 to 6, equal volumes of each of samples 1 to 3 were incubated for an additional 40 min at 30°C with Klenow DNA polymerase (1.5 units) and dATP; lane 7, an equal volume of sample 3 incubated with RNase H (1.5 units) for 20 min at 37°C; lane 8, 32P-labeled DNA oligonucleotide markers of lengths indicated on the right (an adjacent overloaded marker lane has been cut off).
DISCUSSION
Six mitochondrial DNA polymerases and six mitochondrial DNA helicases have been identified in the genome of T. brucei (6, 23, 29, 46). Numerous other mitochondrial DNA replication genes and proteins in T. brucei and related kinetoplastid parasites have been identified earlier, based on protein purification and immunolocalization. These include minicircle origin-binding proteins, DNA topoisomerases, and other essential replication proteins. How these proteins interact with one another and how they participate in the replication of minicircles and maxicircles is still largely unknown and can best be revealed by the reconstitution of replication reactions in vitro with purified proteins. A significant impediment to the investigation of kDNA replication in vitro has been the lack of a mitochondrial DNA primase. Although a primase activity was purified from the insect trypanosomatid C. fasciculata over 10 years ago, the gene was never cloned, and the enzyme is no longer available (25). Since all known DNA polymerases are unable to initiate a DNA strand, it is vital to have a kinetoplastid mitochondrial DNA primase in order to allow the development of an in vitro mitochondrial DNA replication system. A search of the T. brucei genome revealed a putative mitochondrial DNA primase gene (PRI1) with multiple copies of a conserved octamer consensus sequence in the 3′ flanking sequence that has been shown to be required for the posttranscriptional regulation of S-phase expression of certain DNA replication genes in C. fasciculata (33, 34, 36, 37, 42). The presence of the consensus sequence in flanking sequences in Leishmania major was also shown to correlate with S-phase expression of the mRNA (26, 64). We show here that the PRI1 mRNA levels cycle and reach a maximum level shortly before doubling of the number of kinetoplasts (one nucleus and two kinetoplasts) and that PRI1 encodes a protein that localizes to the mitochondrion of T. brucei. In addition, the PRI1 protein is essential both for cell growth and for kDNA replication. Both minicircle and maxicircle DNAs are lost during PRI1 RNAi.
Intracellular enzyme localization has been valuable in implicating proteins in kDNA replication, with several distinct localizations having been observed. Replication proteins have been localized throughout the kDNA, at opposite faces of the kDNA disk, at the flagellar face of the disk, and at antipodal sites adjacent to the disk. The PRI1 protein appears to have the latter type of localization. Other proteins that localize to the antipodal sites include topoisomerase II; DNA polymerase β; structure-specific endonuclease I (SSE 1); two minicircle origin-binding proteins (UMSBP and P38); DNA ligase kβ; KAP4, a DNA binding protein; and P93, a protein of unknown function. It is unknown whether these proteins exist in multimeric complexes. The DNA ligase kβ and DNA topoisomerase II do not precisely colocalize (12), suggesting that there is not just a single complex containing all of these proteins.
Four lines of evidence suggest that an RNA primer is synthesized by PRI1 and is subsequently elongated by DNA polymerase action. (i) Synthesis of poly(dA) on a poly(dT) template by Klenow DNA polymerase depends on the prior or concomitant incubation of the template with PRI1 and ATP. In the absence of PRI1, no synthesis of poly(dA) was observed. (ii) In a coupled reaction with both PRI1 and Klenow polymerase, the extent of poly(dA) synthesis was strongly stimulated by increasing levels of ATP. In the absence of ATP, no synthesis of poly(dA) was observed. (iii) Primed synthesis of poly(dA) by Klenow DNA polymerase is strongly inhibited by incubation of the primed template with RNase H prior to the addition of the Klenow DNA polymerase. (iv) The products synthesized by PRI1 and ATP on a poly(dT) template become elongated following heat inactivation of PRI1 and subsequent incubation with [α-32P]dATP and Klenow DNA polymerase. These results show that PRI1 synthesizes a 3′-hydroxyl-terminated oligoriboadenylate on a poly(dT) template.
We note that the poly(A) synthesized by PRI1 ranges in length up to at least 70 to 80 nucleotides, a result similar to that observed for the Drosophila melanogaster DNA polymerase/primase α with poly(dT) template (10). Oligoribonucleotides of this length are also synthesized on poly(dT) templates in vitro by the mammalian mitochondrial RNA polymerase, the enzyme that primes mitochondrial leading-strand synthesis and, possibly, lagging-strand synthesis as well (56, 60). The T. brucei mitochondrial RNA polymerase has been implicated in maxicircle replication based on RNAi of mitochondrial RNA polymerase (18). The mitochondrial RNA polymerase is predicted to have a molecular mass of 144 kDa and is related to the bacteriophage T7 family of RNA polymerases. Since maxicircles replicate by a unidirectional theta-type mechanism, it is reasonable to suppose that different enzymes might prime synthesis of leading and lagging strands. It remains to be determined whether oligoribonucleotides of the length observed here are synthesized by PRI1 on mitochondrial templates in vivo. RNA primers of less than 10 nucleotides are observed in vivo on the minicircle leading strand of nascent minicircles. Primers of this length could result from processing of prepriming RNAs or from rapid utilization of short primers by a mitochondrial DNA polymerase. It is also likely that additional mitochondrial primases exist in trypanosomes (unpublished data). An activity purified from the insect parasite Crithidia fasciculata synthesized poly(A) primers of 10 nucleotides or less on poly(dT) templates (25). The associated polypeptide was estimated to have a molecular mass of 28 kDa, less than half the size of the PRI1 protein.
With the large number of kinetoplast-specific replication proteins that are known, it is striking how little is known regarding the enzymatic mechanisms and protein interactions involved in initiating minicircle and maxicircle replication. RNA priming of minicircle DNA replication has been known for over 20 years, and yet, direct studies of the enzymatic mechanism of initiation of minicircle replication have not been possible. RNAi of PRI1 resulted in the loss of maxicircle DNA, consistent with PRI1 functioning in priming of the replication of maxicircle DNA. The lower rate of loss of minicircles during PRI1 RNAi suggests that their loss may be a secondary effect of the loss of maxicircles, although a direct role of PRI1 in minicircle replication is also possible. Now that trypanosome mitochondrial minicircle origin-binding proteins, DNA polymerases, DNA helicases, and RNA polymerase have been identified and characterized, the addition of a mitochondrial DNA primase to the repertoire of available replication proteins should allow us to begin in vitro studies of kDNA replication.
Supplementary Material
Acknowledgments
This work was supported by NIH grant GM53254 to D.S.R.
We thank Charlie McHenry for dnaG protein and anti-dnaG antibodies, George Cross for T. brucei 29-13 cells, Kent Hill for plasmid DNA and YTaT.1 cells, Josh Beck for assistance with microscopy, and Peter Bradley for use of his microscope.
Footnotes
Published ahead of print on 11 January 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Abu-Elneel, K., D. R. Robinson, M. E. Drew, P. T. Englund, and J. Shlomai. 2001. Intramitochondrial localization of universal minicircle sequence-binding protein, a trypanosomatid protein that binds kinetoplast minicircle replication origins. J. Cell Biol. 153:725-734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Avrahami, D., Y. Tzfati, and J. Shlomai. 1995. A single-stranded DNA binding protein binds the origin of replication of the duplex kinetoplast DNA. Proc. Natl. Acad. Sci. U. S. A. 92:10511-10515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakshi, R. P., and T. A. Shapiro. 2004. RNA interference of Trypanosoma brucei topoisomerase IB: both subunits are essential. Mol. Biochem. Parasitol. 136:249-255. [DOI] [PubMed] [Google Scholar]
- 4.Birkenmeyer, L., H. Sugisaki, and D. S. Ray. 1987. Structural characterization of site-specific discontinuities associated with replication origins of minicircle DNA from Crithidia fasciculata. J. Biol. Chem. 262:2384-2392. [PubMed] [Google Scholar]
- 5.Carpenter, L. R., and P. T. Englund. 1995. Kinetoplast maxicircle DNA replication in Crithidia fasciculata and Trypanosoma brucei. Mol. Cell. Biol. 15:6794-6803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chandler, J., A. V. Vandoros, B. Mozeleski, and M. M. Klingbeil. 2008. Stem-loop silencing reveals that a third mitochondrial DNA polymerase, POLID, is required for kinetoplast DNA replication in trypanosomes. Eukaryot. Cell 7:2141-2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, J., C. A. Rauch, J. H. White, P. T. Englund, and N. R. Cozzarelli. 1995. The topology of the kinetoplast DNA network. Cell 80:61-69. [DOI] [PubMed] [Google Scholar]
- 8.Chowdhury, A. R., Z. Zhao, and P. T. Englund. 2008. Effect of hydroxyurea on procyclic Trypanosoma brucei: an unconventional mechanism for achieving synchronous growth. Eukaryot. Cell 7:425-428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claros, M. G., and P. Vincens. 1996. Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241:779-786. [DOI] [PubMed] [Google Scholar]
- 10.Conaway, R. C., and I. R. Lehman. 1982. A DNA primase activity associated with DNA polymerase alpha from Drosophila melanogaster embryos. Proc. Natl. Acad. Sci. U. S. A. 79:2523-2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunningham, I. 1977. New culture medium for maintenance of tsetse tissues and growth of trypanosomatids. J. Protozool. 24:325-329. [DOI] [PubMed] [Google Scholar]
- 12.Downey, N., J. C. Hines, K. M. Sinha, and D. S. Ray. 2005. Mitochondrial DNA ligases of Trypanosoma brucei. Eukaryot. Cell 4:765-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drew, M. E., and P. T. Englund. 2001. Intramitochondrial location and dynamics of Crithidia fasciculata kinetoplast minicircle replication intermediates. J. Cell Biol. 153:735-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engel, M., and D. S. Ray. 1998. A structure-specific DNA endonuclease is enriched in kinetoplasts from Crithidia fasciculata. Nucleic Acids Res. 26:4733-4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Engel, M. L., J. C. Hines, and D. S. Ray. 2001. The Crithidia fasciculata RNH1 gene encodes both nuclear and mitochondrial isoforms of RNase H. Nucleic Acids Res. 29:725-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Engel, M. L., and D. S. Ray. 1999. The kinetoplast structure-specific endonuclease I is related to the 5′ exo/endonuclease domain of bacterial DNA polymerase I and colocalizes with the kinetoplast topoisomerase II and DNA polymerase beta during replication. Proc. Natl. Acad. Sci. U. S. A. 96:8455-8460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Englund, P. T. 1979. Free minicircles of kinetoplast DNA networks in Crithidia fasciculata. J. Biol. Chem. 254:4895-4900. [PubMed] [Google Scholar]
- 18.Grams, J., J. C. Morris, M. E. Drew, Z. Wang, P. T. Englund, and S. L. Hajduk. 2002. A trypanosome mitochondrial RNA polymerase is required for transcription and replication. J. Biol. Chem. 277:16952-16959. [DOI] [PubMed] [Google Scholar]
- 19.Hines, J. C., M. L. Engel, H. Zhao, and D. S. Ray. 2001. RNA primer removal and gap filling on a model minicircle replication intermediate. Mol. Biochem. Parasitol. 115:63-67. [DOI] [PubMed] [Google Scholar]
- 20.Hines, J. C., and D. S. Ray. 1997. Tandem arrangement of two genes encoding kinetoplast-associated H1 histone-like proteins. Mol. Biochem. Parasitol. 89:41-49. [DOI] [PubMed] [Google Scholar]
- 21.Hines, J. C., and D. S. Ray. 1998. The Crithidia fasciculata KAP1 gene encodes a highly basic protein associated with kinetoplast DNA. Mol. Biochem. Parasitol. 94:41-52. [DOI] [PubMed] [Google Scholar]
- 22.Iyer, L. M., E. V. Koonin, and D. D. Leipe. 2005. Origin and evolution of the archaeo-eukaryotic primase superfamily and related palm-domain proteins: structural insights and new members. Nucleic Acids Res. 33:3875-3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klingbeil, M. M., S. A. Motyka, and P. T. Englund. 2002. Multiple mitochondrial DNA polymerases in Trypanosoma brucei. Mol. Cell 10:175-186. [DOI] [PubMed] [Google Scholar]
- 24.Kornberg, A. 1988. DNA replication. J. Biol. Chem. 263:1-4. [PubMed] [Google Scholar]
- 25.Li, C. J., and P. T. Englund. 1997. A mitochondrial DNA primase from the trypanosomatid Crithidia fasciculata. J. Biol. Chem. 272:20787-20792. [DOI] [PubMed] [Google Scholar]
- 26.Li, Y., Y. Sun, J. C. Hines, and D. S. Ray. 2007. Identification of new kinetoplast DNA replication proteins in trypanosomatids based on predicted S-phase expression and mitochondrial targeting. Eukaryot. Cell 6:2303-2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, B., Y. Liu, S. A. Motyka, E. E. Agbo, and P. T. Englund. 2005. Fellowship of the rings: the replication of kinetoplast DNA. Trends Parasitol. 21:363-369. [DOI] [PubMed] [Google Scholar]
- 28.Liu, B., H. Molina, D. Kalume, A. Pandey, J. D. Griffith, and P. T. Englund. 2006. The role of p38 in replication of Trypanosoma brucei kinetoplast DNA. Mol. Cell. Biol. 26:5382-5393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, B., J. Wang, N. Yaffe, M. E. Lindsay, Z. Zhao, A. Zick, J. Shlomai, and P. T. Englund. 2009. Trypanosomes have six mitochondrial DNA helicases with one controlling kinetoplast maxicircle DNA replication. Mol. Cell 35:490-501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lukes, J., D. L. Guilbride, J. Votypka, A. Zikova, R. Benne, and P. T. Englund. 2002. Kinetoplast DNA network: evolution of an improbable structure. Eukaryot. Cell 1:495-502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lukes, J., H. Hashimi, and A. Zikova. 2005. Unexplained complexity of the mitochondrial genome and transcriptome in kinetoplastid flagellates. Curr. Genet 48:277-299. [DOI] [PubMed] [Google Scholar]
- 32.Lukes, J., J. C. Hines, C. J. Evans, N. K. Avliyakulov, V. P. Prabhu, J. Chen, and D. S. Ray. 2001. Disruption of the Crithidia fasciculata KAP1 gene results in structural rearrangement of the kinetoplast disc. Mol. Biochem. Parasitol. 117:179-186. [DOI] [PubMed] [Google Scholar]
- 33.Mahmood, R., J. C. Hines, and D. S. Ray. 1999. Identification of cis and trans elements involved in the cell cycle regulation of multiple genes in Crithidia fasciculata. Mol. Cell. Biol. 19:6174-6182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahmood, R., B. Mittra, J. C. Hines, and D. S. Ray. 2001. Characterization of the Crithidia fasciculata mRNA cycling sequence binding proteins. Mol. Cell. Biol. 21:4453-4459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Melendy, T., C. Sheline, and D. S. Ray. 1988. Localization of a type II DNA topoisomerase to two sites at the periphery of the kinetoplast DNA of Crithidia fasciculata. Cell 55:1083-1088. [DOI] [PubMed] [Google Scholar]
- 36.Mittra, B., and D. S. Ray. 2004. Presence of a poly(A) binding protein and two proteins with cell cycle-dependent phosphorylation in Crithidia fasciculata mRNA cycling sequence binding protein II. Eukaryot. Cell 3:1185-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mittra, B., K. M. Sinha, J. C. Hines, and D. S. Ray. 2003. Presence of multiple mRNA cycling sequence element-binding proteins in Crithidia fasciculata. J. Biol. Chem. 278:26564-26571. [DOI] [PubMed] [Google Scholar]
- 38.Ntambi, J. M., and P. T. Englund. 1985. A gap at a unique location in newly replicated kinetoplast DNA minicircles from Trypanosoma equiperdum. J. Biol. Chem. 260:5574-5579. [PubMed] [Google Scholar]
- 39.Ntambi, J. M., T. A. Shapiro, K. A. Ryan, and P. T. Englund. 1986. Ribonucleotides associated with a gap in newly replicated kinetoplast DNA minicircles from Trypanosoma equiperdum. J. Biol. Chem. 261:11890-11895. [PubMed] [Google Scholar]
- 40.Onn, I., I. Kapeller, K. Abu-Elneel, and J. Shlomai. 2006. Binding of the universal minicircle sequence binding protein at the kinetoplast DNA replication origin. J. Biol. Chem. 281:37468-37476. [DOI] [PubMed] [Google Scholar]
- 41.Onn, I., N. Milman-Shtepel, and J. Shlomai. 2004. Redox potential regulates binding of universal minicircle sequence binding protein at the kinetoplast DNA replication origin. Eukaryot. Cell 3:277-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pasion, S. G., J. C. Hines, X. Ou, R. Mahmood, and D. S. Ray. 1996. Sequences within the 5′ untranslated region regulate the levels of a kinetoplast DNA topoisomerase mRNA during the cell cycle. Mol. Cell. Biol. 16:6724-6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ray, D. S. 1989. Conserved sequence blocks in kinetoplast minicircles from diverse species of trypanosomes. Mol. Cell. Biol. 9:1365-1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ryan, K. A., and P. T. Englund. 1989. Replication of kinetoplast DNA in Trypanosoma equiperdum. Minicircle H strand fragments which map at specific locations. J. Biol. Chem. 264:823-830. [PubMed] [Google Scholar]
- 45.Ryan, K. A., and P. T. Englund. 1989. Synthesis and processing of kinetoplast DNA minicircles in Trypanosoma equiperdum. Mol. Cell. Biol. 9:3212-3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saxowsky, T. T., G. Choudhary, M. M. Klingbeil, and P. T. Englund. 2003. Trypanosoma brucei has two distinct mitochondrial DNA polymerase beta enzymes. J. Biol. Chem. 278:49095-49101. [DOI] [PubMed] [Google Scholar]
- 47.Schimanski, B., T. N. Nguyen, and A. Gunzl. 2005. Highly efficient tandem affinity purification of trypanosome protein complexes based on a novel epitope combination. Eukaryot. Cell 4:1942-1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scocca, J. R., and T. A. Shapiro. 2008. A mitochondrial topoisomerase IA essential for late theta structure resolution in African trypanosomes. Mol. Microbiol. 67:820-829. [DOI] [PubMed] [Google Scholar]
- 49.Sheline, C., and D. S. Ray. 1989. Specific discontinuities in Leishmania tarentolae minicircles map within universally conserved sequence blocks. Mol. Biochem. Parasitol. 37:151-157. [DOI] [PubMed] [Google Scholar]
- 50.Shlomai, J. 2002. Specific recognition of the replication origins of the kinetoplast DNA. Acta Microbiol. Immunol. Hung. 49:455-467. [DOI] [PubMed] [Google Scholar]
- 51.Shlomai, J. 2004. The structure and replication of kinetoplast DNA. Curr. Mol. Med. 4:623-647. [DOI] [PubMed] [Google Scholar]
- 52.Sinha, K. M., J. C. Hines, N. Downey, and D. S. Ray. 2004. Mitochondrial DNA ligase in Crithidia fasciculata. Proc. Natl. Acad. Sci. U. S. A. 101:4361-4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sinha, K. M., J. C. Hines, and D. S. Ray. 2006. Cell cycle-dependent localization and properties of a second mitochondrial DNA ligase in Crithidia fasciculata. Eukaryot. Cell 5:54-61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sogin, M. L., and J. D. Silberman. 1998. Evolution of the protists and protistan parasites from the perspective of molecular systematics. Int. J. Parasitol. 28:11-20. [DOI] [PubMed] [Google Scholar]
- 55.Stuart, K. D., A. Schnaufer, N. L. Ernst, and A. K. Panigrahi. 2005. Complex management: RNA editing in trypanosomes. Trends Biochem. Sci. 30:97-105. [DOI] [PubMed] [Google Scholar]
- 56.Tsurumi, T., and I. R. Lehman. 1990. Release of RNA polymerase from Vero cell mitochondria after herpes simplex virus type 1 infection. J. Virol. 64:450-452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wang, C. C. 1995. Molecular mechanisms and therapeutic approaches to the treatment of African trypanosomiasis. Annu. Rev. Pharmacol. Toxicol. 35:93-127. [DOI] [PubMed] [Google Scholar]
- 58.Wang, Z., and P. T. Englund. 2001. RNA interference of a trypanosome topoisomerase II causes progressive loss of mitochondrial DNA. EMBO J. 20:4674-4683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, Z., J. C. Morris, M. E. Drew, and P. T. Englund. 2000. Inhibition of Trypanosoma brucei gene expression by RNA interference using an integratable vector with opposing T7 promoters. J. Biol. Chem. 275:40174-40179. [DOI] [PubMed] [Google Scholar]
- 60.Wanrooij, S., J. M. Fuste, G. Farge, Y. Shi, C. M. Gustafsson, and M. Falkenberg. 2008. Human mitochondrial RNA polymerase primes lagging-strand DNA synthesis in vitro. Proc. Natl. Acad. Sci. U. S. A. 105:11122-11127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wirtz, E., S. Leal, C. Ochatt, and G. A. Cross. 1999. A tightly regulated inducible expression system for conditional gene knock-outs and dominant-negative genetics in Trypanosoma brucei. Mol. Biochem. Parasitol. 99:89-101. [DOI] [PubMed] [Google Scholar]
- 62.Xu, C. W., J. C. Hines, M. L. Engel, D. G. Russell, and D. S. Ray. 1996. Nucleus-encoded histone H1-like proteins are associated with kinetoplast DNA in the trypanosomatid Crithidia fasciculata. Mol. Cell. Biol. 16:564-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao, Z., M. E. Lindsay, A. Roy Chowdhury, D. R. Robinson, and P. T. Englund. 2008. p166, a link between the trypanosome mitochondrial DNA and flagellum, mediates genome segregation. EMBO J. 27:143-154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zick, A., I. Onn, R. Bezalel, H. Margalit, and J. Shlomai. 2005. Assigning functions to genes: identification of S-phase expressed genes in Leishmania major based on post-transcriptional control elements. Nucleic Acids Res. 33:4235-4242. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.